Abstract
Background
Circular RNAs (circRNAs) are a class of non-coding RNAs that have been demonstrated to play important roles in tumorigenesis. However, how circRNAs regulate the progression of hepatocellular cancer (HCC) remains unclear.
Methods
In the present study, circRNA microarray analyses were performed with HCC tissues to identify circRNAs that are differentially expressed. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis was conducted on HCC cell lines and tissues, and circ0097009 was found to be significantly upregulated. The functions of circ0097009 in HCC were investigated by a series of experiments, including cell proliferation, invasion, and mouse xenograft assays. Additionally, luciferase assays and RNA immunoprecipitation (RIP) assays were used to explore the interactions of circ0097009, microRNA-1261 (miR-1261), and solute carrier family 7 member 11 (SLC7A11) in HCC.
Results
Microarray analysis and qRT-PCR verified that circRNA, circ0097009, was significantly upregulated in HCC tissues and cell lines. Knockdown of circ0097009 inhibited the proliferation and invasion of HCC cells. Luciferase reporter assays showed that circ0097009 and SLC7A11 directly bound to miR-1261. Subsequent experiments showed that circ0097009 and SLC7A11 reciprocally regulated their expression via miR-1261 sponging by circ0097009.
Conclusions
Circ0097009 acts as a competing endogenous RNA to regulate the expression of SLC7A11, a key regulator of cancer cell ferroptosis, by sponging miR-1261 in HCC. Circ0097009 may be used as a diagnostic biomarker for HCC and as a potential target for HCC therapy.
Keywords: Circular RNAs, miR-1261, SLC7A11, competitive endogenous RNAs, hepatocellular carcinoma
Introduction
Primary liver cancer is the 6th most commonly diagnosed cancer and the 4th leading cause of cancer-related death worldwide (1). There are approximately 841,000 new cases of liver cancer and 782,000 deaths from liver cancer annually (1,2). Hepatocellular carcinoma (HCC) accounts for approximately 75–85% of all liver cancer cases, and most HCC cases are diagnosed at an advanced stage, with the loss of opportunity for curative treatment (2-4). Although targeted therapies and immunotherapies have developed rapidly in recent years, the prognosis of HCC remains poor (5-7). Some studies have reported that many genes are involved in the tumorigenesis of HCC; however, the underlying mechanisms are still largely unknown (8-10). Therefore, the aim of the present study was to find novel molecular targets to develop more effective therapeutic strategies for HCC (11).
Circular RNAs (circRNAs), a class of non-coding RNAs, have been found to be widely expressed in mammals (12,13). Many circRNAs have been identified, but their potential functions are poorly understood (12,14). Few studies have explored the role of circRNAs in HCC (15-19). Yao et al. found that circZKSCAN1 suppresses HCC cell growth, invasion, and migration (16). Han et al. reported that circMTO1 acts as a sponge for microRNA (miR)-9 to inhibit HCC progression (17). Revealing the role that circRNAs play in HCC is critical to gaining an in-depth understanding of the molecular mechanisms underlying HCC progression and finding new biomarkers or therapeutic targets for HCC (20).
It has been reported that circRNAs can function as competitive endogenous RNAs (ceRNAs) to co-regulate other RNAs by competing for shared miRNAs (14). Kong et al. found that cirPLK1 inhibited triple-negative breast cancer progression by sponging miR-296-5p (21). circSMARCA5 is involved in the growth and metastasis of HCC by functioning as a ceRNA for miR-17-3p and miR-181b-5p (19). Collectively, these findings suggest that circRNAs can function as miRNA sponges to regulate cancer occurrence and development.
In the present study, we performed microarray analysis to investigate the expression profiles of circRNAs in HCC tissues. A significantly upregulated circRNA, hsa_circ_0097009 (circ0097009), was detected by quantitative real-time PCR in HCC tissues and cell lines. Circ0097009, derived from the solute carrier family 5 member 8 (SLC5A8), has not been reported in tumors. We further investigated the functions of circ0097009 in HCC and found that circ0097009 knockdown inhibited cell proliferation and invasion. Additionally, luciferase reporter and RNA immunoprecipitation (RIP) assays showed that circ0097009 bound to miR-1261. Furthermore, the catalytic subunit solute carrier family 7 member 11 (SLC7A11), a key regulator of cancer ferroptosis, was a direct target of miR-1261. Therefore, our findings indicated that circ0097009 might act as a ceRNA to regulate the expression of SLC7A11 by sponging miR-1261, and circ0097009 may be used as a potential therapeutic target in HCC. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-997).
Methods
Cell lines and culture
The human normal liver cell line LO2 and HCC cell lines HepG2, BEL-7402, and MHCC-97H were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA, Cat.# 11965092) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA, Cat.# 16140071) at 37 °C in 5% CO2. All cell lines were passaged in our laboratory for less than 6 months after resuscitation from frozen aliquots and were re-authenticated by DNA fingerprinting every 6 months. This study was approved by the Ethics Committee of Sun Yat-Sen University Cancer Center Health Authority, and was performed according to the ethical standards of the Declaration of Helsinki (as revised in 2013). Patient consent was obtained before the study commenced.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA from cells and tissues was isolated with TRIzol reagent (Life Technologies, Carlsbad, CA, USA, Cat.# 1559601). cDNA was synthesized using a PrimeScript RT reagent kit (Takara Bio, Dalian, China, Cat.# RR036A), and qRT-PCR was performed using SYBR Premix Ex Taq (Takara Bio, Dalian, China, Cat.# RR420A) in a CFX96 Real-time PCR system (Bio-Rad, Hercules, CA, USA). The abundances of target transcripts were evaluated using the housekeeping gene U6 or β-actin as the control. The relative fold change in expression with respect to the control sample was calculated by the 2−ΔΔCt method.
RNase R digestion experiment
RNA extracted from MHCC-97H cells was evenly divided into the following 2 groups: treatment with RNase R (Epicentre Technologies, Madison, WI, USA, Cat.# RNR07250) and treatment with buffer control. In the RNase R group, 2 µg of total RNA was incubated with RNase R (3 U/µg) for 20 min at 37 °C. β-Actin was used as the internal control.
Actinomycin D assay
Cells (1×105) were seeded into 6-well plates and treated with actinomycin D (2 mg/L; Sigma, St Louis, MO, USA, Cat.# A1410). All treated cells were collected at 8, 16, and 24 h for qRT-PCR analysis of circ0097009 and SLC7A11 mRNA.
Cell counting kit-8 (CCK-8) assay and Transwell assay
Cell proliferation was evaluated by a CCK-8 assay (Dojindo Laboratories, Kumamoto, Japan, Cat.# ck04). Cells (1×103) were seeded into 96-well plates and incubated at 37 °C for 24 h before transfection. CCK-8 solution (10 µL) was added to each well 48 h after transfection. After 2 h of incubation at 37 °C, the absorbance at 450 nM was measured using a microtiter plate reader (Epoch2; BioTek, Winooski, VT, USA). For the Transwell assay, cells (at a density of 1×104 cells/mL) were seeded into a migration chamber (BD Biosciences, Franklin Lakes NJ, USA), and medium (containing 10% FBS) was subsequently added into the lower chamber as an attractant. Cells were fixed with methanol after 24 h incubation, stained with 0.1% crystal violet, and then counted under a microscope. Three independent experiments were carried out.
Mouse xenograft model
All animal studies followed the guidelines for animal care and was approved by the Institutional Animal Experimental Ethics Committee of Sun Yat-Sen University Cancer Center. Four-week-old female BALB/c nude mice were subcutaneously inoculated with 2×106 cells (5 mice per group). Intratumoral injection (40 µL of si-circ0097009 or negative control siRNA) was administered every 4 days. Mice were sacrificed, and tumor weights were measured after 4 weeks. To establish lung metastases, HepG2 cells were treated with 20 µmol/L si-circ0097009. After 48 hours, cells (1×105) were intravenously injected into the tail veins of mice (6 mice per group). Eight weeks later, mice were sacrificed, and the lungs of the anaesthetized mice were removed, and the number of metastatic lung nodules was counted and verified by evaluation of hematoxylin-eosin (HE)-stained sections under a microscope.
Luciferase reporter assay
Luciferase reporter vectors with the full length of the SLC7A11 3’-untranslated region (UTR) or circ0097009 were constructed. We then generated mutant luciferase reporter vectors with a QuikChange XL Site-directed Mutagenesis Kit (Stratagene, LaJolla, CA, USA, Cat.# 200517). HepG2 cells were seeded into 96-well plates and co-transfected with a luciferase reporter vector and miR-1261 mimics or miR-1261 locked nucleic acids (LNA). After 48 h of incubation, luciferase activity was quantified with a dual-luciferase reporter assay (Promega, Madison, WI, USA). Cells (5×103) were plated, co-transfected with the constructed vectors and miR-1261 mimics, and incubated (48 h). Relative luciferase activity was then assessed with a dual-luciferase reporter assay system (Promega, USA).
RIP assay
HepG2 cells were transfected with MS2bs-circ0097009, MS2bs-circ0097009 mt, or blank control MS2bs-Rluc, along with MS2bp-GFP by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Forty-eight hours later, RIP was performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA, Cat.# 17-700). After purification of the RNA complexes, the miR-1261 level was measured. The anti-Argonaute 2 (Ago2)-RIP assay was performed with an anti-Ago2 antibody (Millipore, USA). After RNA purification, the levels of circ0097009, SLC7A11, and miR-1261 were quantified.
Western blot analysis
Total protein was extracted, separated by 10% SDS-PAGE and subsequently transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA). Membranes were then blocked with 5% skim milk for 1 h at room temperature, and then incubated with primary anti-SLC7A11 (1:1000; Abcam, Cambridge, MA, USA) or anti-GAPDH (1:1,000; Abcam, Cambridge, MA, USA, Cat.# ab8245) antibodies. A horseradish peroxidase (HRP)-labelled secondary antibody (1:5,000; Cell Signaling Technology, Danvers, MA, USA, Cat.# 7074) was used and detected by chemiluminescence.
Assessment of the glutathione (GSH)/oxidized GSH (GSSG) ratio
The GSH/GSSG Quantification Kit II (Dojindo, Shanghai, China, Cat.# G257-10) was used to measure the ratio of the GSH/GSSG levels. Sample preparation, standard solution preparation, and concentration detection were performed according to the manufacturer’s protocols. The relative levels were calculated with a microplate reader (Flexstation 3; Molecular Devices, San Jose, CA, USA).
Statistical analysis
Data are expressed as mean ± standard error of the mean (SEM). Comparisons among groups were analyzed by one-way ANOVA and the differences between groups were analyzed by two-tailed t tests by using SPSS 25.0(IBM, SPSS, Chicago, IL, USA) for Windows. P<0.05 was considered statistically significant.
Results
Circ0097009 is upregulated in HCC cell lines and tissues
CircRNA microarray analyses of 3 pairs of HCC tissues and matched para-carcinoma normal tissues showed that 6,024 circRNAs were downregulated with a fold change >2 and a P value of <0.05 in HCC tissue, while 10,720 circRNAs were upregulated, as determined by the same cut-off criteria, as we previously described (18). Of the top 5 upregulated circRNAs, we analyzed the expression level of circ0097009 in 3 HCC cell lines. It was found that circ0097009 was upregulated in HCC cells and tissues (Figure 1A,B). An RNase R digestion experiment was then performed to confirm the circular structure of circ0097009 (Figure 1C). In addition, an actinomycin D assay was carried out, and the results suggested that the circular transcript circ0097009 was more stable than the linear transcript solute carrier family 5 member 8 (SLC5A8) in 97H cells (Figure 1D).
Figure 1.
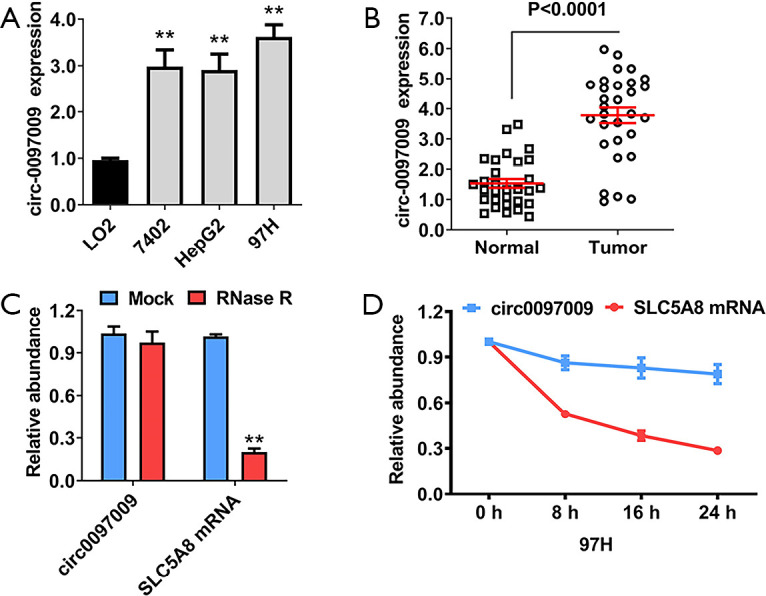
Circular RNA hsa_circ_0097009 (circ0097009) is upregulated in hepatocellular carcinoma (HCC) cell lines and tissues. (A) Expression level of circ0097009 in normal and HCC cell lines. (B) Expression level of circ0097009 in HCC tissues and matched normal tissues. (C) RNase R digestion experiment was performed to confirm the circular structure of circ0097009. (D) Stability of the circular transcript circ0097009 and linear transcript SLC5A8 in 97H cells, as assessed by actinomycin D assays. **P<0.01.
Knockdown of circ0097009 inhibits HCC tumor growth
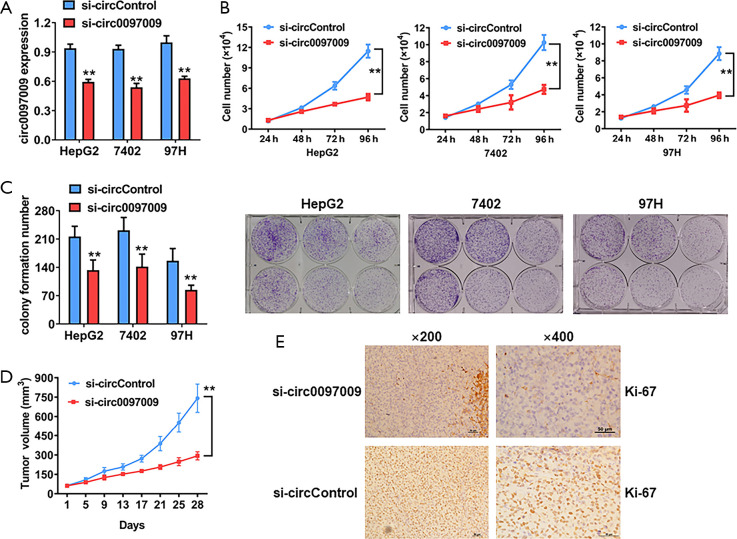
To evaluate the biological functions of circ0097009 in HCC, we used RNA interference to knock down the expression of circ0097009. qRT-PCR analysis showed that the inhibition was successful (Figure 2A). CCK-8 and colony formation assays revealed that the downregulation of circ0097009 significantly suppressed cell growth (Figure 2B,C). Xenograft experiments were then carried out to investigate the function of circ0097009 in tumor growth in vivo. We observed that the inhibition of circ0097009 significantly suppressed tumor growth (Figure 2D). Besides, immunohistochemistry sections of xenograft tumors supported that the inhibition of circ0097009 decreased the expression level of Ki-67 (Figure 2E).
Figure 2.
Knockdown of circ0097009 inhibits hepatocellular carcinoma tumor growth. (A) si-circ0097009 successfully knocked down circ0097009. (B) Cell proliferation was evaluated with a Cell Counting Kit-8 assay in HepG2, 7402, and 97H cells. (C) Number of colonies formed by HepG2, 7402, and 97H cells. (D) Tumor volume was measured every 4 days for 4 weeks. (E) Expression status of Ki67 in hematoxylin-eosin-stained sections of harvested xenograft tumors. **P<0.01.
Downregulation of circ0097009 inhibits the invasion and metastasis of HCC cells
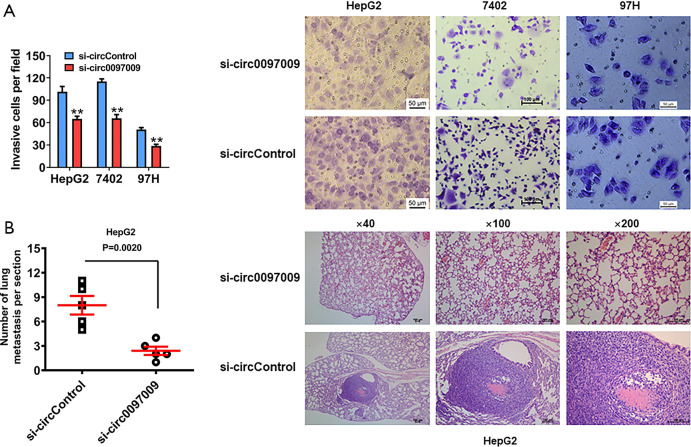
Transwell assay was used to assess the role of circ0097009 in the invasion of HCC cells. Silencing the expression of circ0097009 significantly impaired the invasive capacity of HepG2, 7402, and 97H cells (Figure 3A). To further investigate the function of circ0097009 in tumor invasion and metastasis in vivo, xenograft models were established, and the results showed that the inhibition of circ0097009 reduced the number of lung metastases (Figure 3B), indicating that circ0097009 plays an important role in the metastatic ability of HCC.
Figure 3.
Downregulation of circ0097009 inhibits the invasion and metastasis of hepatocellular carcinoma (HCC) cells. (A) Transwell assay was performed to assess the invasive ability of HCC cells. (B) Number of metastatic lung nodules was quantified, and hematoxylin-eosin-stained sections of metastatic lung nodules are shown. **P<0.01.
Circ0097009 functions as a sponge for miR-1261
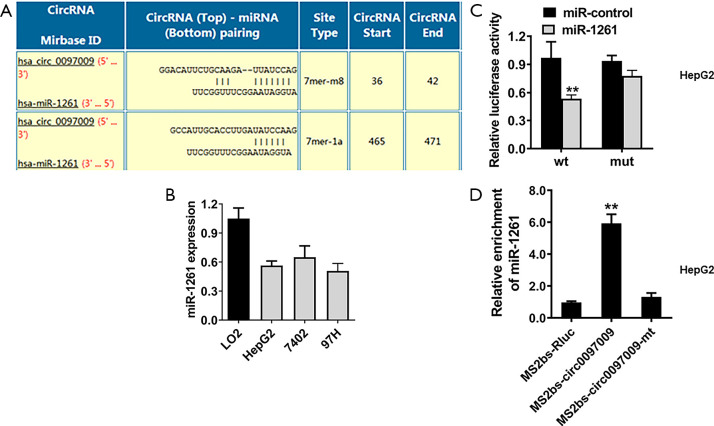
CircRNAs may function as ceRNAs to regulate miRNAs, leading to the release of control of corresponding miRNA-targeted genes. Therefore, we explored whether circ0097009 could act as a miRNA sponge and predicted the potential molecular target of circ0097009 based on the circRNA interactome (https://circinteractome.nia.nih.gov). We found that miR-1261 has a potential binding site within the circ0097009 sequence (Figure 4A). qRT-PCR showed that miR-1261 was downregulated in HCC cell lines (Figure 4B). Subsequently, we performed a luciferase reporter assay and found that after co-transfection with miR-1261 mimics, luciferase activity was inhibited; however, in cells transfected with the mutant luciferase reporter, there was no such effect (Figure 4C). To further validate the direct binding between circ0097009 and miR-1261, an MS2bp-MS2bs-based RIP assay was conducted, and the results suggested that miR-1261 was primarily enriched in the MS2bs-circ0097009 group (Figure 4D), indicating a direct interaction between circ0097009 and miR-1261. Therefore, our data confirmed that circ0097009 functionally interacts with miR-1261 and functions as a sponge for miR-346 in HCC.
Figure 4.
Circ0097009 functions as a sponge for microRNA-1261 (miR-1261). (A) Predicted direct binding sites of miR-1261 within the circ0097009 sequence. (B) Expression level of miR-1261 in the LO2 cell line and hepatocellular carcinoma cell lines. (C) Relative luciferase activity of HepG2 cells co-transfected with the miR-1261 mimics and the luciferase reporter vector containing the wild-type or mutant circ0097009 sequence. (D) MS2-based RNA immunoprecipitation assay in HepG2 cells transfected with MS2bs-circ0097009, MS2bs-circ0097009-mt, or MS2bs-Rluc. **P<0.01.
Knockdown of circ0097009 promotes ferroptosis in HCC
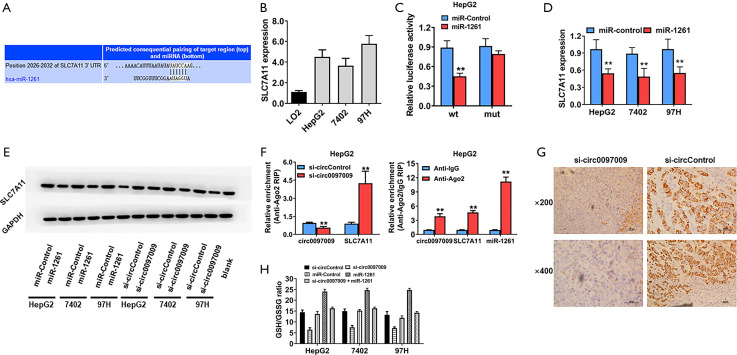
To further predict the potential downstream targets of miR-1261, we used the TargetScan algorithm, and SLC7A11 was identified as the candidate target oncogene (Figure 5A). To validate the predicted target interaction, the expression levels of SLC7A11 were evaluated in HCC cell lines and were found to be upregulated (Figure 5B). Subsequently, to confirm whether miR-1261 can directly bind to SLC7A11 mRNA, we performed a RIP assay and found that luciferase activity decreased after transfection with the luciferase reporter vector containing the wild-type SLC7A11 promoter and miR-1261 mimics, whereas no such effect was observed in cells transfected with the luciferase reporter vector containing the mutant SLC7A11 promoter (Figure 5C). Moreover, the miR-1261 mimics suppressed the expression of SLC7A11 in HepG2, 7402, and 97H cells, suggesting that SLC7A11 is regulated by miR-1261 (Figure 5D). Western blotting showed that, with the knockdown of circ0097009 and miR-1261, SLC7A11 expression was downregulated in HepG2, 7402, and 97H cells (Figure 5E). Additionally, Ago2-RIP assays were conducted, and the results revealed that circ0097009, SLC7A11, and miR-1261 were primarily enriched in Ago2 complexes. Moreover, the inhibition of circ0097009 significantly increased SLC7A11 enrichment in the Ago2 fraction and reduced circ0097009 enrichment in the Ago2 complexes (Figure 5F), indicating that circ0097009 competes with SLC7A11 for binding to miR-1261. In addition, HE-stained sections of harvested xenograft tumors confirmed that the knockdown of circ0097009 decreased the expression level of SLC7A11 (Figure 5G). Interestingly, SLC7A11, a component of system Xc-, has been identified as a key regulator of cancer cell ferroptosis (22). Therefore, it was concluded that the knockdown of circ0097009 promotes ferroptosis via the circ0097009/miR-1261/SLC7A11 axis. Moreover, we found that knockdown of circ0097009 led to a decrease in the GSH/GSSG ratio and abolished glutathione peroxidase 4 (GPX4) activation (Figure 5H). A reduction in the GSH/GSSG ratio causes deactivation of GPX4, which is another key regulator of ferroptosis and reduces lipid peroxides (23). Therefore, the knockdown of circ0097009 promotes ferroptosis in HCC cells via the following 2 independent pathways: system Xc- and GPX4.
Figure 5.
Knockdown of circ0097009 promotes ferroptosis in hepatocellular carcinoma (HCC) cells. (A) Predicted direct binding sites of microRNA-1261 (miR-1261) within the solute carrier family 7 member 11 (SLC7A11) sequence. (B) Expression level of SLC7A11 in the LO2 cell line and HCC cell lines. (C) Relative luciferase activity of HepG2 cells co-transfected with miR-1261 mimics and the luciferase reporter vector containing the wild-type or mutant SLC7A11 sequence. (D) Expression level of SLC7A11 after transfection with miR-1261 mimics. (E) Western blotting was performed to determine the expression levels of SLC7A11, glutathione peroxidase 4, and GAPDH. (F) RNA immunoprecipitation (RIP) assay detected the relative enrichment of circ0097009, SLC7A11, and miR-1261 in the anti-Argonaute 2 (Ago2) fraction (left); relative enrichment was detected by an Ago2-RIP assay (right). (G) Expression status of SLC7A11 in hematoxylin-eosin-stained sections of harvested xenograft tumors. (H) Glutathione (GSH)/oxidized GSH (GSSG) ratio was determined with a GSH/GSSG Quantification Kit. **P<0.01.
Discussion
A variety of miRNAs and long non-coding RNAs (lncRNAs) have been reported to regulate HCC progression. However, few studies have investigating the role of circRNAs, a family of non-coding RNAs, with covalently closed, single-stranded transcripts, in HCC (15). In the present study, we identified the expression profile of circRNAs in HCC and found that circ0097009 was upregulated in HCC tissues and cell lines. A series of in vitro and in vivo experiments showed that the knockdown of circ0097009 inhibited the growth and invasion of HCC cells. These results revealed that circ0097009 plays a vital role in HCC progression, indicating that it might be a diagnostic biomarker for, and a therapeutic target in, HCC.
Recently, numerous studies have revealed that pseudogenes, lncRNAs, and circRNAs can interact with or co-regulate each other by competing for miRNA binding (14,24), a phenomenon known as the ceRNA mechanism. CircRNAs have been shown to act as ceRNAs by sponging miRNAs to regulate gene expression in cancer (14,25). In the present study, we performed a bioinformatic analysis, and the results showed that circ0097009 contained a miRNA response element (MRE) for miR-1261. Functional assays were also conducted to confirm the direct relationship between circ0097009 and miR-1261. Previous studies found that miR-1261 plays a regulatory role in a variety of cancers. He et al. first reported that miR-1261 promotes the invasion and migration of prostate cancer cells by binding to the lncRNA prostate cancer antigen 3 (26); miR-1261 has been demonstrated to regulate the development of papillary thyroid cancer and glioma (27,28). Zhang et al. found that, in liver cancer, miR-1261 acts as a sponge for the lncRNA breast cancer anti-estrogen resistance 4 (BCAR4) to promote tumor progression by upregulating anaphase-promoting complex subunit 11 (ANAPC11) (29). In the present study, we found that miR-1261 can target both circ0097009 and SLC7A11, suggesting that circ0097009 might function as a miR-1261 sponge to regulate SLC7A11 expression through a ceRNA mechanism. There are several lines of evidence implying that circ0097009 functions as a ceRNA for SLC7A11 in HCC as a sponge for miR-1261. First, as mentioned, bioinformatic analyses showed that both the 3'-UTR of SLC7A11 and circ0097009 contain binding sites for miR-1261. Second, the luciferase reporter and MS2bp-MS2bs-based RIP assays verified this prediction. Third, the knockdown of circ0097009 reduced SLC7A11 expression. Finally, the inhibition of miR-1261 reversed the effect of circ0097009 knockdown. Collectively, these findings indicated that circ0097009 functions as a ceRNA to regulate SLC7A11 expression by sponging miR-1261.
Ferroptosis is a form of programmed cell death that is different from apoptosis, necroptosis, and autophagy (30). Iron accumulation and lipid peroxidation lead to the accumulation of reactive oxygen species (ROS), the key characteristic of ferroptosis (31). In recent years, ferroptosis induction has been regarded as a promising therapeutic approach for cancer (32,33). SLC7A11 and solute carrier family 3 member 2 (SLC3A2) are the 2 key members of system Xc-, which is a heterodimeric cystine/glutamate antiporter and the upstream molecule in the process of ferroptosis. As the key regulator of cancer cell ferroptosis, SLC7A11 can be regulated at the transcriptional level, and a reduction in the SLC7A11 level induces ferroptosis (34). Our findings indicated that the inhibition of circ0097009 decreased the expression level of SLC7A11, suggesting that circ0097009 is involved in the progression of HCC and may respond via the mechanism of ferroptosis by regulating SLC7A11.
Moreover, we also found that the knockdown of circ0097009 led to a decrease in the GSH/GSSG ratio and deactivation of GPX4. GPX4 is an antioxidant enzyme and another key regulator of ferroptosis (23). It has been reported that the overexpression of GPX4 causes resistance to ROS-induced tumor cell death and that silencing GPX4 sensitizes tumor cells to ROS-induced cell death (35). Therefore, on the basis of these findings, we concluded that circ0097009 promotes ferroptosis in HCC via the following 2 independent pathways: system Xc- and GPX4.
In conclusion, the findings of the present study indicated that circ0097009 is upregulated in HCC and that the knockdown of circ0097009 inhibits HCC cell growth and invasion. Moreover, circ0097009 functions as a ceRNA to regulate SLC7A11 expression by sponging miR-1261. The circ0097009/miR-1261/SLC7A11 axis mediates HCC progression by regulating ferroptosis. Therefore, circ0097009 may be a diagnostic biomarker for HCC and a potential target for HCC therapy.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: The study was funded by the National Natural Science Foundation of China (No. 81901850, Ning Lyu).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of Sun Yat-Sen University Cancer Center Health Authority, and was performed according to the ethical standards of the Declaration of Helsinki (as revised in 2013). Patient consent was obtained before the study commenced. All animal studies followed the guidelines for animal care and was approved by the Institutional Animal Experimental Ethics Committee of Sun Yat-Sen University Cancer Center.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-997
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-997
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-997). The authors have no conflicts of interest to declare.
(English Language Editor: R. Scott)
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Liu Z, Suo C, Mao X, et al. Global incidence trends in primary liver cancer by age at diagnosis, sex, region, and etiology, 1990-2017. Cancer 2020;126:2267-78. 10.1002/cncr.32789 [DOI] [PubMed] [Google Scholar]
- 3.Global Burden of Disease Liver Cancer C , Akinyemiju T, Abera S, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol 2017;3:1683-91. 10.1001/jamaoncol.2017.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi J, Han S, Kim N, et al. Increasing burden of liver cancer despite extensive use of antiviral agents in a hepatitis B virus-endemic population. Hepatology 2017;66:1454-63. 10.1002/hep.29321 [DOI] [PubMed] [Google Scholar]
- 5.Villanueva A. Hepatocellular Carcinoma. N Engl J Med 2019;380:1450-62. 10.1056/NEJMra1713263 [DOI] [PubMed] [Google Scholar]
- 6.Vitale A, Trevisani F, Farinati F, et al. Treatment of hepatocellular carcinoma in the Precision Medicine era: from treatment stage migration to therapeutic hierarchy. Hepatology 2020;72:2206-18. 10.1002/hep.31187 [DOI] [PubMed] [Google Scholar]
- 7.Hou J, Zhang H, Sun B, et al. The immunobiology of hepatocellular carcinoma in humans and mice: Basic concepts and therapeutic implications. J Hepatol 2020;72:167-82. 10.1016/j.jhep.2019.08.014 [DOI] [PubMed] [Google Scholar]
- 8.Craig AJ, von Felden J, Garcia-Lezana T, et al. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2020;17:139-52. 10.1038/s41575-019-0229-4 [DOI] [PubMed] [Google Scholar]
- 9.Topper MJ, Vaz M, Marrone KA, et al. The emerging role of epigenetic therapeutics in immuno-oncology. Nat Rev Clin Oncol 2020;17:75-90. 10.1038/s41571-019-0266-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kudo M. A Paradigm Change in the Treatment Strategy for Hepatocellular Carcinoma. Liver Cancer 2020;9:367-77. 10.1159/000507934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kloeckner R, Galle PR, Bruix J. Local and regional therapies for hepatocellular carcinoma. Hepatology 2021;73 Suppl 1:137-49. 10.1002/hep.31424 [DOI] [PubMed] [Google Scholar]
- 12.Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell 2019;176:869-81.e13. 10.1016/j.cell.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 14.Wu S, Turner KM, Nguyen N, et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 2019;575:699-703. 10.1038/s41586-019-1763-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dragomir MP, Kopetz S, Ajani JA, et al. Non-coding RNAs in GI cancers: from cancer hallmarks to clinical utility. Gut 2020;69:748-63. 10.1136/gutjnl-2019-318279 [DOI] [PubMed] [Google Scholar]
- 16.Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol 2017;11:422-37. 10.1002/1878-0261.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017;66:1151-64. 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 18.Bai N, Peng E, Qiu X, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res 2018;37:172. 10.1186/s13046-018-0838-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol 2018;68:1214-27. 10.1016/j.jhep.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 20.Zheng Q, Zhao J, Yu H, et al. Tumor-specific transcripts are frequently expressed in hepatocellular carcinoma with clinical implication and potential function. Hepatology 2020;71:259-74. 10.1002/hep.30805 [DOI] [PubMed] [Google Scholar]
- 21.Kong Y, Yang L, Wei W, et al. CircPLK1 sponges miR-296-5p to facilitate triple-negative breast cancer progression. Epigenomics 2019;11:1163-76. 10.2217/epi-2019-0093 [DOI] [PubMed] [Google Scholar]
- 22.Shi ZZ, Fan ZW, Chen YX, et al. Ferroptosis in carcinoma: regulatory mechanisms and new method for cancer therapy. Onco Targets Ther 2019;12:11291-304. 10.2147/OTT.S232852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seibt TM, Proneth B, Conrad M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic Biol Med 2019;133:144-52. 10.1016/j.freeradbiomed.2018.09.014 [DOI] [PubMed] [Google Scholar]
- 24.Stojic L, Lun ATL, Mascalchi P, et al. A high-content RNAi screen reveals multiple roles for long noncoding RNAs in cell division. Nat Commun 2020;11:1851. 10.1038/s41467-020-14978-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei Y, Chen X, Liang C, et al. A noncoding regulatory RNAs network driven by circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology 2020;71:130-47. 10.1002/hep.30795 [DOI] [PubMed] [Google Scholar]
- 26.He JH, Li BX, Han ZP, et al. Snail-activated long non-coding RNA PCA3 up-regulates PRKD3 expression by miR-1261 sponging, thereby promotes invasion and migration of prostate cancer cells. Tumour Biol 2016. [Epub ahead of print]. 10.1007/s13277-016-5450-y [DOI] [PubMed] [Google Scholar]
- 27.Wei H, Pan L, Tao D, et al. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem Biophys Res Commun 2018;503:56-61. 10.1016/j.bbrc.2018.05.174 [DOI] [PubMed] [Google Scholar]
- 28.Zhang F, Mai SR, Cao FP, et al. MiR-1261/circ-PTPRZ1/PAK1 pathway regulates glioma cell growth and invasion. Hum Cell 2019;32:540-7. 10.1007/s13577-019-00271-3 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Zhou H. LncRNA BCAR4 promotes liver cancer progression by upregulating ANAPC11 expression through sponging miR1261. Int J Mol Med 2020;46:159-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060-72. 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie Y, Hou W, Song X, et al. Ferroptosis: process and function. Cell Death Differ 2016;23:369-79. 10.1038/cdd.2015.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Z, Song J, Yung BC, et al. Emerging strategies of cancer therapy based on ferroptosis. Adv Mater 2018;30:e1704007. 10.1002/adma.201704007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stockwell BR, Friedmann Angeli JP, Bayir H, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 2017;171:273-85. 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D, Tavana O, Chu B, et al. NRF2 is a major target of ARF in p53-independent tumor suppression. Mol Cell 2017;68:224-32.e4. 10.1016/j.molcel.2017.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinowaki Y, Kurata M, Ishibashi S, et al. Glutathione peroxidase 4 overexpression inhibits ROS-induced cell death in diffuse large B-cell lymphoma. La [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as