Abstract
Background
Peroxisome proliferators-activated receptors γ (PPARγ) and secreted frizzled related protein 5 (SFRP5) are abnormally expressed in liver cells. But their role in the transformation of non-alcoholic fatty liver (NAFL) to non-alcoholic steatohepatitis (NASH) remains to be studied. We aimed to explore the role of S-nitrosylation (SNO) in the conversion of NAFL to NASH via the peroxisome PPARγ/SFRP5 pathway.
Methods
A normal diet and methionine-choline deficient diet were used to construct the NAFL and NASH mouse models, respectively. The differences between the SNO of PPARγ in both models were measured by irreversible biotinylation. Quantitative reverse transcription PCR (qRT-PCR) and Western blotting were used to detect the effect of SNO on the expression of PPARγ messageRNA (mRNA) and protein in L02 hepatocytes. Nubiscan software, luciferase reporter gene, and chromatin immunoprecipitation assay (CHIP) were used to verify the targeting relationship between PPAR and SFRP5. The expression of tumor necrosis factor α (TNFα), interleukin-1β (IL-1β), and interleukin-6 (IL-6), which are indicators for the activation of Kupffer cells, were determined by enzyme linked immunosorbent assay (ELISA) after co-cultivation of L02 hepatocytes and Kupffer macrophages, as well as the exogenous regulation of SNO, PPARγ, and SFRP5 in hepatic L02 cells.
Results
The NAFL and NASH mouse models were successfully constructed, and the level of PPARγ SNO in the NAFL model was significantly lower than the NASH model (P<0.05). The level of PPARγ was significantly downregulated after increasing the SNO of L02 cells, respectively (P<0.05). Nubiscan software and CHIP confirmed that PPARγ could bind to the promoter region of SFRP5 (P<0.05). Overexpression of PPARγ and SFRP5 could significantly downregulate the expression of TNFα, IL-1β, and IL-6 (P<0.05) correspondingly, while increasing the SNO level of L02 cells could restore the expression levels of TNFα, IL-1β, and IL-6.
Conclusions
SNO promoted the activation of macrophage Kupffer cells by inhibiting the PPARγ/SFRP5 pathway in L02 hepatocytes, thereby promoting the conversion of NAFL into NASH.
Keywords: Non-alcoholic fatty liver (NAFL), non-alcoholic steatohepatitis (NASH), S-nitrosylation (SNO), peroxisome proliferators-activated receptors γ (PPARγ), secreted frizzled related protein 5 (SFRP5)
Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most common cause of chronic liver disease in the world. It is expected to become the most common indication for liver transplantation by 2030 and is also considered to be a risk factor for cardiovascular disease (1,2). NAFLD can develop from benign non-alcoholic fatty liver (NAFL) to more severe non-alcoholic steatohepatitis (NASH). Nearly 25% of NASH patients could develop cirrhosis, and some of them could even progress to liver cancer (3,4). Clarifying the key links and molecular mechanisms of NAFL’s progression towards NASH will provide potential drug targets and treatment strategies for reversing NASH. It is currently believed that the abnormal activation of Kupffer cells, an intrinsic macrophage of the liver, is the core link leading to lobular inflammation (5), and the occurrence of hepatic lobular inflammation is the main pathological manifestation that differentiates NASH from NAFL (6). Therefore, exploring the molecular mechanism of abnormal activation of Kupffer cells has become the focus of current research to reverse NASH.
SFRP5 belongs to the SFRP family and contains a cysteine-rich domain homologous to the wingless/integrated (Wnt) binding site (7). A large number of studies have shown that the c-Jun N-terminal kinase (JNK) and nuclear transcription factor-κB (NF-κB) pathways are classic inflammatory pathways that can be activated by Kupffer cells during the conversion of NAFL to NASH (8). PPARγ is one of the members of the peroxisome Proliferator-Activated Receptor (PPAR) subfamily encoding nuclear receptors (9). A recent study showed that the expression level of PPARγ in liver cells under NASH was significantly lower than that in normal and NAFL states, which is consistent with the expression change of SFRP5, and the down-regulation of PPARγ expression mediated the occurrence of liver inflammation (10,11). It is suggested that PPARγ and SFRP5 may participate in the transformation of NAFL to NASH by mediating Kupffer cell activation. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1070).
Methods
Cell lines, laboratory animals, and reagents
Human normal L02 liver cells and macrophage Kupffer cells were purchased from American ATCC Company (USA). Ob/Ob mice were purchased from Nanjing Junke Bioengineering Co., Ltd. (China). Short hairpin RNA- peroxisome proliferators-activated receptors γ/pcDNA3.1- peroxisome proliferators-activated receptors γ (sh-PPARγ/pcDNA3.1-PPARγ) and Short hairpin RNA- secreted frizzled related protein 5/pcDNA3.1-secreted frizzled related protein 5 (sh-SFRP5/pcDNA3.1-SFRP5) were constructed in Guangzhou Huijun Biotechnology Co., Ltd. (China) Dulbecco’s Modified Eagle Medium (DMEM), fetal bovine serum, penicillin, and streptomycin were purchased from American HYCLONE company (USA). Transient transfection kits, Lipofectamine 2000, Trizol, and PrimeScriptTM reverse transcription PCR (RT-PCR) kits were purchased from Thermo Fisher (United States). The protein extraction kit, butyleyanoacrylate (BCA) protein concentration determination kit, and SDS-polyacrylamide gel electrophoresis (SDS-PAGE) rapid preparation kit were purchased from Beijing Solable Technology Co., Ltd. (China). Maleimide biotin was purchased from Wuhan Aimeijie Technology Co., Ltd. (China) PPARγ, SFRP5 primary and secondary antibodies and CHIP kits were purchased from Abcam (UK), and ELISA kits were purchased from R & D Systems (USA).
Construction of NAFL and NASH animal models
Ob/Ob mice were randomly divided into NAFL and NASH groups. NAFL group ob/ob mice were fed a normal diet, while NASH mice were fed with methionine and choline deficent (MCD). Normal diet formulas included L-amino acid 175.7 g/kg, maize starch 150.0 g/kg, sucrose 441.9 g/kg, cellulose 30.0 g/kg, dextrose maltose 50.0 g/kg, sodium bicarbonate 7.4 g/kg, corn oil 100.0 g/kg, vitamin mixture 10.0 g/kg, salt mixture 35.0 g/kg, choline 2 g/kg, and methionine 3 g/kg. The MCD diet was identical to the normal diet, but with the choline and methionine removed. Mice serum was collected for alanine transaminase (ALT), aspartate transaminase (AST), high density lipoprotein cholesterol (HDL-C), low density lipoprotein cholesterol (LDL-C), triglyceride (TG) and total cholesterol (TC) test to verify the success of the mouse model. Experiments were performed under a project license (No.: SYXK2019-0003) granted by institutional ethics committee of Shanxi Provincial People’s Hospital, in compliance with institutional guidelines for the care and use of animals.
Cell culture
For the human normal L02 hepatocyte culture, L02 was cultured in DMEM containing 1% penicillin and 10% fetal bovine serum, and was placed in an incubator at 37 °C with 5% carbon dioxide (CO2). To co-culture the human normal L02 hepatocytes and macrophage Kupffer cells, human normal L02 hepatocytes in the logarithmic growth phase of each treatment group were first trypsinized and digested. Next, the cells were centrifuged, resuspended in DMEM medium, and counted before use. The supernatant was then discarded from the six-well plate that was covered with macrophage Kupffer cells. The cells were then washed twice with phosphate buffer saline (PBS), and 2 mL of DMEM medium was then added to the plate. The incubated transwell chamber was then placed in the six-well plate, and 1.5 mL (containing 1×106 cells) of L02 cell suspension was added to the upper chamber. DMEM medium without Kupffer cells in the lower chamber was used as the control.
Cell transfection
Human normal L02 liver cells in the logarithmic growth phase were inoculated in 96-well plates, and the transfection process was carried out according to the instructions of the Lipofectamine 2000 kit. Specifically, after the transfection concentration was adjusted to 50 nmol/L, sh-PPARγ/pcDNA3.1-PPARγ sh-SFRP5/pcDNA3.1-SFRP5, and a negative control were transfected into the L02 cells respectively, and were designated specifically as the NC group, sh-PPARγ group, pcDNA3.1-PPARγ group, sh-SFRP5 group, and the pcDNA3.1-SFRP5 group. 24 h after transfection, the transfection effect of the L02 cells was examined under a fluorescent microscope.
Expression level of PPARγ mRNA in human normal L02 liver cells was measured with qRT-PCR
The total RNA of human normal L02 liver cells was extracted according to the instructions of the Trizol kit. After measuring the total RNA concentration, 0.97 µg of total RNA was taken and reverse-transcribed according to the PrimeScriptTM RT reagent kit instructions to generate complementary DNA (cDNA). Real-time PCR was then carried out according to the SYBR® Green Real-time PCR Master Mix kit Manual. The qPCR reaction system (20 µL) included 2 µL cDNA, 0.8 µL of the forward primer, 0.8 µL of the reverse primer, 10 µL SYBR Green Mix, 0.4 µL ROX reference dye, and 6 µL H2O. The qPCR reaction (40 cycle total) conditions were as follows: 50 °C heating for 2 min, followed by 95 °C pre-denaturation for 10 min, followed by 95 °C denaturation for 15 s, and eventually 60 °C annealing for 1 min. The qPCR primer sequences used in this study were as follows: U6F: forward 5'-CTTCGGCAGCACATATACTAAAAT-3', reverse 5'-AATATGGAACGCTTCACGA-3'; PPARγ: forward 5'-TGGTGTACGATCACTGCGAC-3', reverse 5'-CACTTCTGGAAGCGGCAGTA-3'. U6 was used as an internal reference, the fluorescence signal and cycle threshold (Ct) value were detected with the icycler software of BIO-RAD (BIO-RAD, USA), and the expression level of PPARγ mRNA was calculated by the 2−ΔΔCt method.
Expression level of PPARγ protein in human normal L02 liver cells was measured by Western blotting
Radio Immunoprecipitation Assay (RIPA) was used to separately lyse cells and tissues to extract total protein. The BCA method was used to measure the protein concentration before determining the sample volume per well. The samples were separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane, and then blocked with 5% skimmed milk powder for 1 h. Subsequently, the primary antibodies, PPARγ (1:1,000) and SFRP5 (1:1,000), were added and incubated overnight at 4 °C. Next, the primary antibodies were removed and the corresponding secondary antibodies, PPARγ (1:2,000) and SFRP5 (1:2,000), were added and blocked at room temperature for 1 h. The DAB reagent was then added for image development. Using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal reference, the protein band was quantitatively analyzed with Image J software (U.S. State Health Center, USA).
Level of S-nitrosylation (SNO) of PPARγ in human normal liver cells was measured by irreversible biotinylation
Cells were lysed with RIPA solution, mixed, and centrifuged. The supernatant was then taken and s-methyl methanethiosulfonate (MMTS) was added. Protein from each treatment group was resuspended, reacted at 50 °C for 30 min, and shaken every 4 min. Subsequently, two volumes of ice-acetone were added, and the samples were reacted at −20 °C for 10 min, and after centrifugation, the excess MMTS was discarded. The obtained protein was then resuspended in Hens buffer, and 0.2 mmol/L maleimide biotin and 10 mmol/L ascorbate were added, which was then reacted at room temperature for 1 h. Excess biotin was removed after precipitation with acetone. The sample was then boiled in Hens buffer containing 200 mmol/L dithiothreitol (DTT) for 15 min to remove potential disulfide bonds between molecules. After that, the neutralization reaction was carried out, and the biotinylated protein was purified and eluted. Western blotting was then used to detect the SNO level of PPARγ.
Dual-luciferase reporter gene experiment
pGL-SFRP5 containing different truncated lengths of the SFRP5 promoter region and Renilla luciferase reporter plasmid were transfected into L02 cells according to the instructions of the Lipofectamine 2000 kit. 24 h after transfection of the pGL-3-2284 containing the SFRP5 promoter region and Renilla luciferase reporter plasmid, the cells were divided into dimethyl sulfoxide (DMSO) group, PPARγ antagonist (GW9662) group, Rosiglitazone (RSG) group, Adenovirus-PPARγ (AD-PPARγ) group, and RSG + AD-PPARγ group, and treated with the PPARγ agonist, rosiglitazone, as well as the PPARγ agonist rosiglitazone + adenovirus that overexpresses PPARγ. Following the treatments, the relative activity of luciferase was expressed by the ratio of firefly luciferase activity and renal luciferase activity according to the instructions of the dual-luciferase reporter gene kit.
The targeting relationship between PPARγ and SFRP5 was verified by Nubiscan software and CHIP
The Nubiscan online software was used to predict the binding site of the SFRP5 promoter region, and a similarity score greater than 0.8 was used as the inclusion criterion. Normal human L02 hepatocytes in the logarithmic growth phase were taken and 1% formaldehyde was used for cell fixation. After standing at 37 °C for 10 min, glycine with a final concentration of 0.125 M was then added. After mixing, the cells were then placed at room temperature for 5 min to end the fixation. After fixation, the medium was discarded, the cells were washed with cold PBS three times, and the cells were then scraped into a 15 mL eppondorf (EP) tube and centrifuged at 800 ×g for 5 min to collect the cells. After centrifugation, 0.5 ml of cell lysis buffer was added to resuspend the cells. The cells were then sonicated, the sonication solution was centrifuged at 2,000 ×g for 5 min, and the supernatant was then transferred to a new EP tube to precipitate the DNA fragment. PPARγ antibody and immunoglobulin G (IgG) antibody were then added to the supernatant, 10% of the final solution was taken, and Protein A magnetic beads were added and mixed with the solution. Protein/DNA complexes were collected after 12 h of incubation at 4 °C, and DNA was purified using a gel recovery kit. QRT-PCR was then used to determine the enrichment of PPARγ protein in the promoter region of the SFRP5 gene.
The expression levels of TNFα, IL-1β, and IL-6 secreted by macrophages detected with ELISA
The Kupffer cell supernatant of each treatment group was collected by centrifugation at 1,000 r/min. After centrifugation for 5 min the supernatant was collected. The concentration of TNFα, IL-1β, and IL-6 in the culture supernatant was then determined according to the instructions of the ELISA kit. Specifically, the concentrations of TNFα, IL-1β, and IL-6 were calculated based on a standard curve associating the standard concentration and absorbance (A) value.
Statistical analysis
The experimental data were analyzed with SPSS 20.0 statistical software (IBM, New York, USA), and the figures were drawn with Graphpad 8.0 software (GraphPad Software, USA). The t-test was used for comparison between groups, and single-factor analysis of variance was used for comparison among multiple groups. P<0.05 was used to indicate a statistically significant difference.
Results
Successfully constructed NAFL and NASH mouse models
As shown in Table 1, we first detected ALT, AST, HDL-C, LDL-C, TG and TC in the serum of the constructed NAFL and NASH mice models. Compared with normal mice, the serum levels of ALT, AST, LDL-C, TG and TC in NAFL mice were significantly increased, while HDL-C was significantly decreased (P<0.05). In addition, the levels of ALT, AST, LDL-C, TG and TC in the serum of NASH mice were significantly higher than those of NAFL mice, while the levels of HDL-C were the opposite (P<0.05). According to the above data, we have successfully constructed NAFL and NASH mouse models.
Table 1. Comparison of biochemical indexes among each groups of mice.
| NC | NAFL | NASH | |
|---|---|---|---|
| ALT (U/L) | 22.14±4.21 | 57.61±6.41* | 91.87±11.19# |
| AST (U/L) | 34.11±5.72 | 61.77±8.52* | 89.16±7.81# |
| HDL-C (mmol/L) | 0.89±0.26 | 0.46±0.28* | 0.27±0.09# |
| LDL-C (mmol/L) | 2.81±0.57 | 4.59±1.33* | 6.94±1.12# |
| TG (mmol/L) | 0.86±0.32 | 2.56±0.83* | 5.67±1.19# |
| TC (mmol/L) | 1.92±0.49 | 6.84±1.57* | 9.69±1.99# |
*P<0.05 vs. NC group; #P<0.05 vs. NAFL group. NC, negative control; NAFL, non-alcoholic fatty liver; NASH, non-alcoholic steatohepatitis; ALT, alanine transaminase; AST, aspartate transaminase; HDL-C, high density lipoprotein cholesterol; LDL-C, low density lipoprotein cholesterol; TG, triglyceride; TC, total cholesterol.
SNO levels of PPARγ in NAFL and NASH mouse models
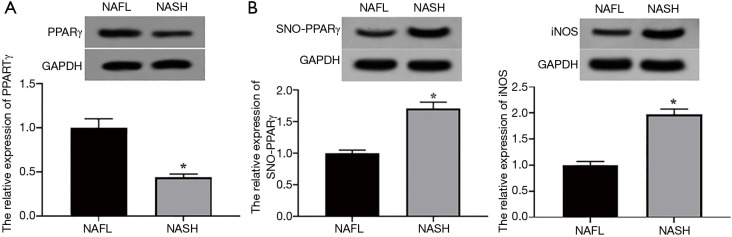
Western blotting results showed that the expression level of PPARγ in the NASH mouse model increased significantly compared with the NAFL group (P<0.05, Figure 1A). The expression level of SNO-PPARγ in the NAFL mouse model was significantly lower than that of the NASH mouse model (P<0.05, Figure 1B). Also, the expression level of inducible nitric oxide synthase (iNOS) in the NAFL mouse model was significantly lower than that of the NASH mouse model (P<0.05, Figure 1C). These results indicated that the SNO level of PPARγ in NASH mice was abnormally higher than that in NAFL mice.
Figure 1.
SNO levels of PPARγ in NAFL and NASH mouse models. (A) The expression levels of PPARγ in NAFL and NASH models. (B)The expression levels of SNO-PPARγ in NAFL and NASH models; (C) the expression levels of iNOS in NAFL and NASH models. *P<0.05 NAFL vs. NASH group. PPARγ, Peroxisome proliferators-activated receptors γ; SFRP5, secreted frizzled related protein 5; SNO, S-nitrosylation; iNOS, inducible nitric oxide synthase; NAFL, non-alcoholic fatty liver; NASH, non-alcoholic steatohepatitis.
The effect of SNO on the expression of PPARγ protein and mRNA in L02 hepatocytes
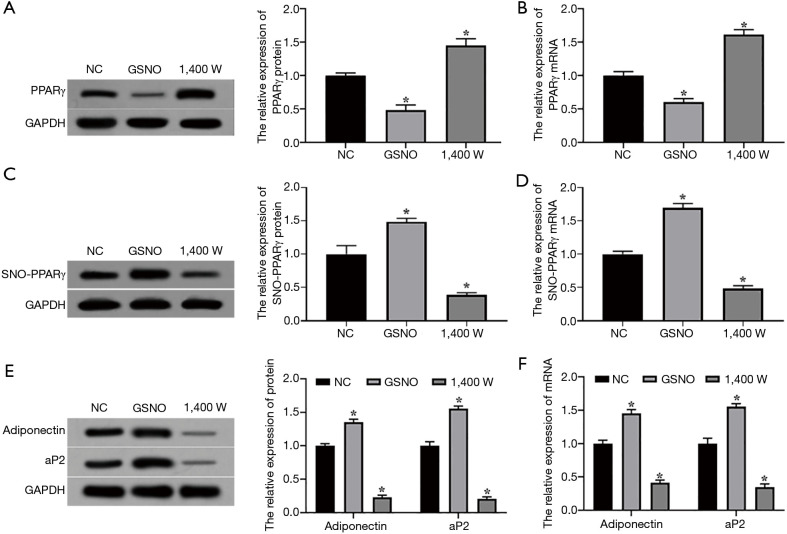
Western blotting results showed that after SNO enhancer S-Nitrosoglutathione (GSNO) and inhibitor 1,400 W treatment of L02 hepatocytes, the expression level of the PPARγ protein in the GSNO group was significantly lower than that in the NC group, while the expression level of the PPARγ protein in the 1,400 W group was significantly higher than that in the NC group (P<0.05, Figure 2A). Consistent with this, the qRT-PCR results showed that the expression level of PPARγ mRNA in the GSNO group was significantly lower than that in the NC group, and the expression level of PPARγ mRNA in the 1,400 W group was significantly higher than that in the NC group (P<0.05, Figure 2B). As expected, compared with the NC group, the expression level of SNO-PPARγ protein in the GSNO group was significantly increased, while the expression level of SNO-PPARγ protein in the 1,400 W group was significantly down-regulated (P<0.05), Figure 2C. The expression of SNO-PPARγ mRNA in the GSNO group was significantly higher than that in the NC group, while the 1,400 W group was significantly lower than that in the NC group (P<0.05), Figure 2D. Also, the protein expression levels of adiponectin and adapter protein2 (ap2), which are the target genes of PPARγ in the GSNO group, were significantly higher than those of the NC group. Not surprisingly, the protein expression levels of adiponectin and ap2 in the 1,400 W group were significantly lower than those of the NC group (P<0.05, Figure 2E). As expected, the mRNA expression levels of adiponectin and ap2 in the GSNO group were significantly higher than those in the NC group, while the mRNA expression levels of the adiponectin and ap2 proteins in the 1,400 W group were significantly lower than those in the NC group (P<0.05, Figure 2F). These results indicated that SNO modification can down-regulate the expression levels of the PPARγ protein and mRNA in L02 hepatocytes.
Figure 2.
Effect of SNO on the expression of the PPARγ protein and mRNA in L02 hepatocytes. (A,B) The expression levels of the PPARγ protein and mRNA in L02 cells; (C,D) the expression levels of targeting gene of SNO-PPARγ protein and mRNA in L02 cells; (E,F) the expression levels of the target of the PPARγ protein and mRNA in L02 cells. Experimental vs. NC group, *P<0.05. NC, negative control; NAFL, non-alcoholic fatty liver; NASH, non-alcoholic steatohepatitis; GSNO, SNO enhancer S-Nitrosoglutathione; PPARγ, peroxisome proliferators-activated receptors γ; SNO, S-nitrosylation; ap2, adapter protein2.
Verification of the targeting relationship between PPARγ and SFRP5
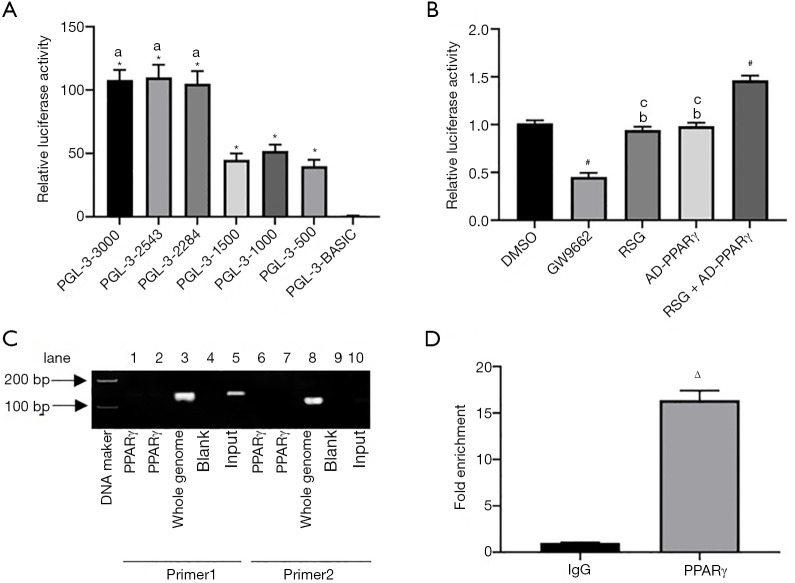
Nubiscan online software was used to predict the binding site of the SFRP5 promoter region (with a similarity score greater than 0.8 as the inclusion criterion). The SFRP5 promoter region contains three potential binding sites for PPARγ, as shown in Table 2. Luciferase reporter gene results showed that the relative fluorescence values of pGL-SFRP5 in different length groups were significantly higher than those in the pGL-3-Basic group (P<0.05, Figure 3A). Also, pGL-3-3000, pGL-3-2543, and pGL-3-2284 were significantly different from pGL-3-1500, pGL-3-1000, and pGL-3-500 (P<0.05, Figure 3A). On the other hand, there was no significant difference among pGL-3-3000, pGL-3-2543, pGL-3-2284, and pGL-3-1500, as well as between pGL-3-1000 and pGL-3-500, suggesting that there are important transcriptional activation cis response elements between −2,284 to −1,500 bp. Furthermore, the luciferase activity in the GW9662 group was lower than that of the DMSO group, and the luciferase activity of the RSG + AD-PPARγ group was higher than that of the DMSO group (P<0.05, Figure 3B). Meanwhile, the luciferase activity of the RSG and AD-PPARγ groups was the same as that of the DMSO group, suggesting that PPARγ can enhance the activity of the SFRP5 promoter. Based on these luciferase reporter gene experiment results, primer 1 and primer 2 of the DNA that bound to PPARγ as a template were designed, and further results through CHIP experiments showed that PPARγ can bind to the SFRP5 promoter region, as shown in Figure 3C. Consistent with this, qRT-PCR results showed that the Fold Enrichment was increased by 16.4 times with SFRP5 after using the PPARγ antibody (Figure 3D). The above experiments showed that PPARγ can bind to the SFRP5 promoter region and enhance the activity of the SFRP5 promoter.
Table 2. Possible binding regions of PPARγ to the SFRP5 promoter.
| Position | Score | P value | Site sequence |
|---|---|---|---|
| −2,543 | 0.811344 | 0.016892 | AGGGCAAGGGCTGACCA |
| −2,529 | 0.811344 | 0.016892 | TGACCACAGAAGGGCA |
| −2,284 | 0.803993 | 0.0139931 | TGCCAGCTCTGTCCAGGTCA |
PPARγ, peroxisome proliferators-activated receptors γ; SFRP5, secreted frizzled related protein 5.
Figure 3.
Verification of the targeting relationship between PPARγ and SFRP5. (A,B) Luciferase activity of each group; (C) the targeting relationship between PPARγ and SFRP5 was verified by CHIP; (D) fold enrichment of SFRP5. *P<0.05 vs. pGL-3-Basic group; aP<0.05 vs. pGL-3-1000 group; #P<0.05 vs. DMSO group; bP<0.05 vs. GW9662 group; cP<0.05 vs. RSG + AD-PPARγ group; ΔP<0.05 vs. IgG group. PPARγ, peroxisome proliferators-activated receptors γ; DMSO, dimethyl sulfoxide; GW9662, PPARγ antagonist; RSG, rosiglitazone; AD, adenovirus.
The effect of SNO modification on the activation of macrophage Kupffer cells by regulating the PPARγ/SFRP5 pathway in L02 hepatocytes
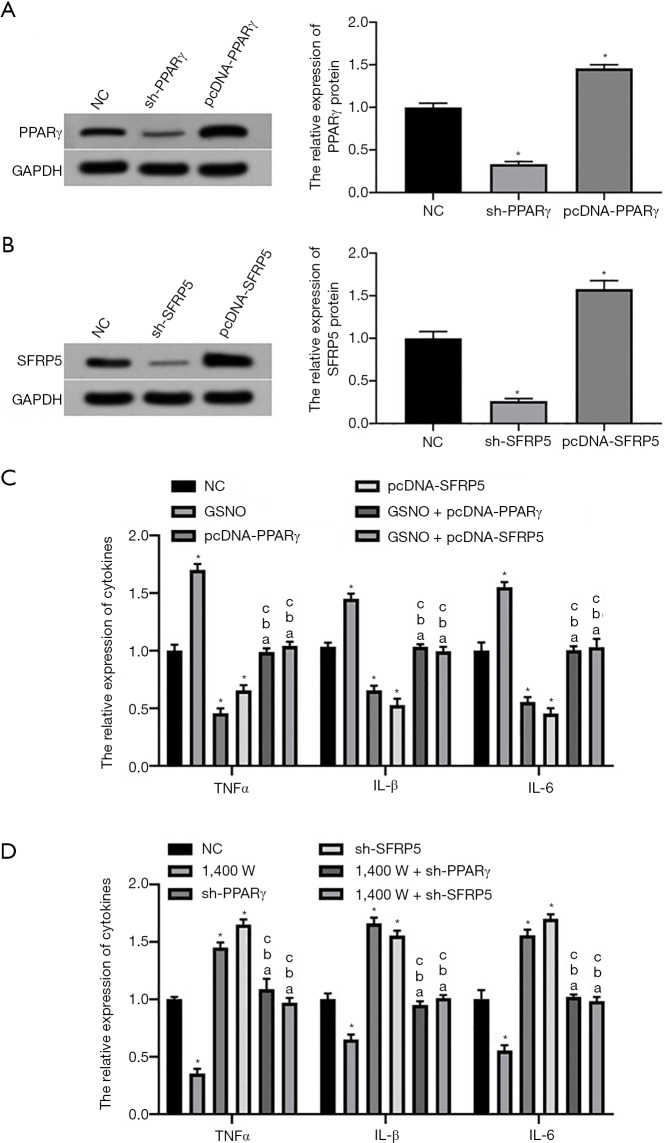
Western blotting results showed that the expression levels of PPARγ and SFRP5 proteins in L02 cells in the sh-PPARγ and sh-SFRP5 groups were significantly lower than those in the NC group, whereas the expression levels of PPARγ and SFRP5 proteins in L02 cells in the pcDNA-PPARγ and pcDNA-SFRP5 groups were significantly higher than those in the NC group (P<0.05, Figure 4A,B). ELISA results showed that after the co-culture of L02 hepatocytes with macrophage Kupffer cells, the expression levels of TNFα, IL-1β, and IL-6 secreted by Kupffer cells in the GSNO and 1,400 W groups were significantly higher and lower than those in NC group, respectively (P<0.05, Figure 4C,D). Moreover, the expression levels of TNFα, IL-1β, and IL-6 secreted by Kupffer cells in the pcDNA-PPARγ and pcDNA-SFRP5 groups were significantly lower than those in the NC group (P<0.05); however, the expression levels of TNFα, IL-1β, and IL-6 secreted by Kupffer cells in the sh-PPARγ and sh-SFRP5 groups were significantly higher than those in the NC group (P<0.05, Figure 4C,D). On the other hand, the expression levels of TNFα, IL-1β, and IL-6 secreted by Kupffer cells in the GSNO + pcDNA-PPARγ/SFRP5 and 1,400 W + sh-PPARγ/SFRP5 groups were not significantly different from the control group. The above results indicated that SNO modification activated macrophage Kupffer cells by inhibiting the PPARγ/SFRP5 pathway in L02 cells, thereby promoting the conversion of NAFL to NASH.
Figure 4.
Effects of SNO on macrophage Kupffer activation by regulating PPARγ/SFRP5 pathway in L02 hepatocytes. (A,B) The expression levels of PPARγ and SFRP5 protein in L02 cells; (C,D) the levels of TNFα, IL-1β, and IL-6 secreted by macrophage Kupffer cells. *P<0.05 vs. NC group; aP<0.05 vs. GSNO/1,400 W group; bP<0.05 vs. pcDNA/sh-PPARγ group; cP<0.05 vs. pcDNA/sh-SFRP5 group. NC, negative control; PPARγ, peroxisome proliferators-activated receptors γ; SFRP5, secreted frizzled related protein 5; SNO, S-nitrosylation; GSNO, SNO enhancer S-Nitrosoglutathione; TNFα, tumor necrosis factor α; IL-1β, interleukin-1β; IL-6, interleukin-6.
Discussion
As mentioned above, the abnormal activation of Kupffer cells, the intrinsic macrophages of the liver, plays an important role in the conversion of NAFL to NASH. The activation of Kupffer cells is a complex biological process that is regulated by multiple factors and is related to intestinal-derived endotoxin, the space-occupying effect of hepatocyte steatosis, and toxic lipid metabolites (12). Also, regulation of Kupffer cell activation by cytokines produced by the paracrine of hepatocytes is also an important factor that promotes NASH (13).
Recent studies have found that SFRP5 can inhibit the binding of Wnt5a to its receptor, resulting in the inhibition of the Wnt pathway, which inhibits the activation of macrophages in fat, gastric mucosa, and other tissues (14,15). One study also found that SFRP5 was highly expressed in hepatocytes, and the expression level of SFRP5 in the liver cells of NASH patients was abnormally lower than that of normal people and NAFL patients (16), suggesting that the abnormal activation of Kupffer cells could be due to the decreased secretion of SFRP5 by liver cells, which could have important research significance in the progress of NASH.
At the same time, clinical trials have found that rosiglitazone (PPARγ agonist) can inhibit macrophage activation and improve liver inflammation in NASH patients (17). In addition, one study also found that during adipocyte differentiation, the expression changes of SFRP5 and PPARγ were completely congruent, and rosiglitazone could increase the expression of SFRP5 (18), suggesting that PPARγ may be an upstream regulatory gene of SFRP5. In this study, the luciferase reporter gene and CHIP experiments confirmed that PPARγ can bind to the SFRP5 promoter region and can enhance the SFRP5 promoter activity. Furthermore, overexpression or knockdown of PPARγ and SFRP5 can inhibit or promote the activation of Kupffer cells, thereby regulating the transformation of NAFL to NASH.
There are several ways to regulate PPARγ expression, including transcriptional regulation, epigenetic modification, ubiquitination modification, and S-nitrosation modification (19,20). SNO is a typical redox-dependent protein post-translational modification, which is closely related to the pathophysiological process of oxidative stress responses (21). In eukaryotic cells, NOS family proteins, especially inducible NOS (iNOS), are the main endogenous nitric oxide (NO) donors, which contribute to cellular SNO (22). Cumulative studies have shown that a large accumulation of iNOS and NO will promote protein SNO, which will participate in the occurrence and development of many diseases (23-25). In addition, studies have found that iNOS is the main NO synthase in hepatocytes, which has a high expression abundance in the cytoplasm (26), and PPARγ was initially expressed in the cytoplasm (27). Also, structural analysis found that there are 10 cysteine residues in the PPARγ protein, adipocytes, and bone marrow mesenchymal cells. Some of these cysteine residues can be modified by SNO, thereby reducing the expression of PPARγ and transcriptional activity (20,21). The above-mentioned evidence suggests that PPARγ and iNOS have the same subcellular localization, and the SNO modification of PPARγ also has a spatial and structural basis. This study found that the use of ordinary diet and MCD feeding could construct NAFL and NASH mouse models, and through irreversible biotin detection, it was found that the level of SNO modification of PPARγ in NASH was significantly higher than that of NAFL. Moreover, cell experiments showed that SNO modification can down-regulate the expression levels of the PPARγ protein and mRNA in L02 liver cells. At the same time, enhancing or inhibiting SNO modification in L02 cells can in turn promote or inhibit Kupffer cell activation.
In summary, this study explored the effect of SNO modification on Kupffer cell activation via the PPARγ/SFRP5 pathway in L02 hepatocytes. The results in this study showed that SNO modification inhibited the expression levels of PPARγ and SFRP5 in L02 cells, thereby abnormally activating Kupffer cells, and thus, promoting the conversion of NAFL to NASH.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No.: SYXK2019-0003) granted by institutional ethics committee of Shanxi Provincial People’s Hospital, in compliance with institutional guidelines for the care and use of animals.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1070
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1070
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1070). The authors have no conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.Byrne CD, Targher G. NAFLD: a multisystem disease. J Hepatol 2015;62:S47-64. 10.1016/j.jhep.2014.12.012 [DOI] [PubMed] [Google Scholar]
- 2.Marcuccilli M, Chonchol M. NAFLD, and Chronic Kidney Disease. Int J Mol Sci 2016;17:562. 10.3390/ijms17040562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eslam M, Valenti L, Romeo S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J Hepatol 2018;68:268-79. 10.1016/j.jhep.2017.09.003 [DOI] [PubMed] [Google Scholar]
- 4.Cobbina E, Akhlaghi F. Non-alcoholic fatty liver disease (NAFLD) - pathogenesis, classification, and effect on drug-metabolizing enzymes and transporters. Drug Metab Rev 2017;49:197-211. 10.1080/03602532.2017.1293683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robert O, Boujedidi H, Bigorgne A, et al. Decreased expression of the glucocorticoid receptor-GILZ pathway in Kupffer cells promotes liver inflammation in obese mice. J Hepatol 2016;64:916-24. 10.1016/j.jhep.2015.11.023 [DOI] [PubMed] [Google Scholar]
- 6.Xu C, Wan X, Xu L, et al. Xanthine oxidase in non-alcoholic fatty liver disease and hyperuricemia: One stone hits two birds. J Hepatol 2015;62:1412-9. 10.1016/j.jhep.2015.01.019 [DOI] [PubMed] [Google Scholar]
- 7.Carstensen-Kirberg M, Hatziagelaki E, Tsiavou A, et al. Sfrp5 associates with beta-cell function in humans. Eur J Clin Invest 2016;46:535-43. 10.1111/eci.12629 [DOI] [PubMed] [Google Scholar]
- 8.Farrell GC, Van Rooyen D, Gan L, et al. NASH is an Inflammatory Disorder: Pathogenic, Prognostic and Therapeutic Implications. Gut Liver 2012;6:149-71. 10.5009/gnl.2012.6.2.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaur S, Nag A, Singh AK, et al. PPARγ-targeting Potential for Radioprotection. Curr Drug Targets 2018;19:1818-30. 10.2174/1389450119666180131105158 [DOI] [PubMed] [Google Scholar]
- 10.Batatinha HA, Lima EA, Teixeira AA, et al. Association Between Aerobic Exercise and Rosiglitazone Avoided the NAFLD and Liver Inflammation Exacerbated in PPAR-α Knockout Mice. J Cell Physiol 2017;232:1008-19. 10.1002/jcp.25440 [DOI] [PubMed] [Google Scholar]
- 11.Wu CW, Chu ES, Lam CN, et al. PPARgamma is essential for protection against nonalcoholic steatohepatitis. Gene Ther 2010;17:790-8. 10.1038/gt.2010.41 [DOI] [PubMed] [Google Scholar]
- 12.Sharma M, Mitnala S, Vishnubhotla RK, et al. The Riddle of Nonalcoholic Fatty Liver Disease: Progression From Nonalcoholic Fatty Liver to Nonalcoholic Steatohepatitis. J Clin Exp Hepatol 2015;5:147-58. 10.1016/j.jceh.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machado MV, Diehl AM. Pathogenesis of Nonalcoholic Steatohepatitis. Gastroenterology 2016;150:1769-77. 10.1053/j.gastro.2016.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao C, Bu X, Wang W, et al. GEC-derived SFRP5 inhibits Wnt5a-induced macrophage chemotaxis and activation. PLoS One 2014;9:e85058. 10.1371/journal.pone.0085058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouchi N, Higuchi A, Ohashi K, et al. Sfrp5 is an anti-inflammatory adipokine that modulates metabolic dysfunction in obesity. Science 2010;329:454-7. 10.1126/science.1188280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutiérrez-Vidal R, Vega-Badillo J, Reyes-Fermín LM, et al. SFRP5 hepatic expression is associated with non-alcoholic liver disease in morbidly obese women. Ann Hepatol 2015;14:666-74. 10.1016/S1665-2681(19)30761-6 [DOI] [PubMed] [Google Scholar]
- 17.He L, Liu X, Wang L, et al. Thiazolidinediones for nonalcoholic steatohepatitis: A meta-analysis of randomized clinical trials. Medicine (Baltimore) 2016;95:e4947. 10.1097/MD.0000000000004947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv C, Jiang Y, Wang H, et al. Sfrp5 expression and secretion in adipocytes are up-regulated during differentiation and are negatively correlated with insulin resistance. Cell Biol Int 2012;36:851-5. 10.1042/CBI20120054 [DOI] [PubMed] [Google Scholar]
- 19.Wei J, Bhattacharyya S, Jain M, et al. Regulation of Matrix Remodeling by Peroxisome Proliferator-Activated Receptor-γ: A Novel Link Between Metabolism and Fibrogenesis. Open Rheumatol J 2012;6:103-15. 10.2174/1874312901206010103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y, Gomes S A, Rangel E B, et al. S-nitrosoglutathione reductase-dependent PPARγ denitrosylation participates in MSC-derived adipogenesis and osteogenesis. J Clin Invest 2015;125:1679-91. 10.1172/JCI73780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin R, Fang L, Li Y, et al. Pro-inflammatory Macrophages suppress PPARγ activity in Adipocytes via S-nitrosylation. Free Radic Biol Med 2015;89:895-905. 10.1016/j.freeradbiomed.2015.10.406 [DOI] [PubMed] [Google Scholar]
- 22.Salim T, Sershen CL, May EE. Investigating the Role of TNF-α and IFN-γ Activation on the Dynamics of iNOS Gene Expression in LPS Stimulated Macrophages. PLoS One 2016;11:e0153289. 10.1371/journal.pone.0153289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen LN, Sun J, Yang XD, et al. The Brain NO Levels and NOS Activities Ascended in the Early and Middle Stages and Descended in the Terminal Stage in Scrapie-Infected Animal Models. Mol Neurobiol 2017;54:1786-96. 10.1007/s12035-016-9755-z [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Li G, Guo Y, et al. Regulation of ezrin tension by S-nitrosylation mediates non-small cell lung cancer invasion and metastasis. Theranostics 2019;9:2555-71. 10.7150/thno.32479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan L, Lin Z, Tang X, et al. S-Nitrosylation of Plastin-3 Exacerbates Thoracic Aortic Dissection Formation via Endothelial Barrier Dysfunction. Arterioscler Thromb Vasc Biol 2020;40:175-88. 10.1161/ATVBAHA.119.313440 [DOI] [PubMed] [Google Scholar]
- 26.Kaibori M, Okumura T, Sato K, et al. Inducible Nitric Oxide Synthase Expression in Liver Injury: Liver Protective Effects on Primary Rat Hepatocytes. Inflamm Allergy Drug Targets 2015;14:77-83. 10.2174/1871528114666160330113227 [DOI] [PubMed] [Google Scholar]
- 27.Liman N, Alan E. Region-specific localization of NOS isoforms and NADPH-diaphorase activity in the intratesticular and excurrent duct systems of adult domestic cats (Felis catus). Microsc Res Tech 2016;79:192-208. 10.1002/jemt.22619 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as