Abstract
COVID-19 is a newly emerged viral disease that is currently affecting the whole globe. A variety of therapeutic approaches are underway to block the SARS-CoV-2 virus. Among these methods, siRNAs could be a safe and specific option, as they have been tested against other viruses. siRNAs are a class of inhibitor RNAs that act promisingly as mRNA expression blockers and they can be designed to interfere with viral mRNA to block virus replication. In order to do this, we designed and evaluated the efficacy of six highly specific siRNAs, which target essential viral mRNAs with no predicted human genome off-targets. We observed a significant reduction in the copy number viral mRNAs after treatment with the siRNAs, and are expected to inhibit virus replication. We propose siRNAs as a potential co-therapy for acute SARS-CoV-2 infection.
Keywords: siRNAs, COVID-19, SARS-CoV-2, qRT-PCR
1. Introduction
Coronavirus disease 2019 (COVID-19) has been rapidly spread around the globe since December 2019. Due to the high transmission rate of this virus, it can affect vast number of people in a short time; and to this fact, there is an urgent need for a safe vaccine or therapeutic option for this viral infection. According to the pathological aspects and clinical stages of the disease, different classes of drugs have been used. They include antiviral agents, inflammation inhibitors, plasma, and hyperimmune immunoglobulins (Stasi et al., 2020; Bartoli et al., 2021). The antiviral agents mainly focus on virus replication inhibition either by protease inhibition -preventing the Gag/Pol polyprotein cleavage- or by interfering with the RNA-dependent RNA polymerase reaction; and they include Ritonavir-lopinavir, darunavir, Remdesivir, favipiravir, and ribavirin. Another possible approach is to block the virus entry path. The possible drugs that are capable of doing this are Chloroquine, hydroxychloroquine, arbidol. As the virus infection, in many cases, resulted in a severe inflammatory response, another option could be immunomodulatory agents. The other alternative is plasma-injected antibody therapy (Stasi et al., 2020; Bartoli et al., 2021; Wang and Guan, 2021). Besides, the nucleic acid-based treatments can be offered as a potential co-treatment for coronaviruses due to their high target specificity. In this regard, siRNA inhibitors can act as powerful therapeutic agents to combat the ongoing outbreak. Unique sequences that target the virus genome and do not interfere with the human genome can be used for the design of siRNAs to target the virus (de Fougerolles et al., 2007). These pre-designed siRNAs will target the viral RNA genome and its replication and terminate the proliferation of the virus (Dykxhoorn and Lieberman, 2006; Whitehead et al., 2009). Wu et al. and Li et al. used a similar siRNA approach with the SARS-CoV-1 virus RNA and inhibited the expression of viral proteins in infected cells (Li et al., 2005; Wu et al., 2005). They reported that upon the entry of the viral particles, direct interaction of viral RNA with introduced siRNAs within the upper airway epithelial cells resulted in degradation of viral RNA and blocked viral replication and the spread of SARS-CoV-1 particles (Li et al., 2005).
2. Materials and methods
For better therapeutic effects of potent siRNA- SARS-CoV-2 inhibitors and to minimize the effect of any probable mutation in the virus genome, we designed six selective siRNAs by the Oligo.7 software (Molecular Biology Insights, Colorado, USA) (Rychlik, 2007) for most of the virus mRNAs to ensure better efficiency (Table 1 ). The siRNAs were then used as a mixture consisting of an equal amount of each siRNA duplex. These siRNAs were designed to be highly specific for the virus mRNAs and to have the least interaction with human RNAs, especially mRNAs. This specificity was evaluated using the NCBI Basic Local Alignment Search Tool (BLAST). The RNA sequence of the virus (“Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome” (NCBI Reference Sequence: NC_045512.2) was aligned with human RNAs and similar sequences were excluded to ensure that no human mRNA would be affected. To minimize the off-target possibility of the siRNAs these sequences were re-checked with the human mRNAs by the BLAST to ensure there were no possible similarities between the designed siRNAs and the human-based mRNAs. Finally, each siRNA was checked by the BLAST to find which strains of the virus could be targeted by each siRNA.
Table 1.
The sequence of the siRNAs.
| siRNA sequence | GC content (%) | Target | |
|---|---|---|---|
| siRNA 1 | GTACTTTCTTTTGAACTTCTACA | 30% | surface glycoprotein |
| siRNA 2 | CAACAAAGATAGCACTTAA | 31% | orf1ab polyprotein |
| siRNA 3 | TCATACCACTTATGTACAA | 31% | orf1ab polyprotein |
| siRNA 4 | CCAAAATCATAACCCTCAAA | 35% | orf3a protein |
| siRNA 5 | AAACCTTCTTTTTACGTTTA | 25% | envelope protein |
| siRNA 6 | CGAACGCTTTCTTATTACAA | 35% | membrane glycoprotein |
Before testing the effect of the siRNAs on the proliferation of the virus, the effect of the siRNAs was tested on the normal unaffected Vero cells, obtained from the cell bank of the Pasteur Institute (Tehran, Iran), and their cell viability was compared with an untreated group. The cells were cultured in the proper culture medium, which was high glucose Dulbecco's Modified Eagle's Medium (DMEM) (Gibco, Grand Island, NY, USA) supplemented with 10% Fetal Bovine Serum (FBS) (Gibco, Grand Island, NY, USA). No sign of toxicity was observed in the uninfected siRNA-treated group according to the MTT assay (data was not shown). To test the efficacy of the siRNAs, the Vero cells were cultured in two main groups: an untreated SARS-CoV-2 infected group (I), and a siRNA-treated infected group (T). The I group was infected by a mixture of serum of the affected patients at a multiplicity of infection (MOI) of 0.05 (Kim, 2020; Sheahan et al., 2020). (All the individuals who donated serum signed consent forms.) Finally, the T group was infected in a similar condition; the only difference was that the siRNAs were added to the culture medium after infection, as a mixture with 10 mg/mL dosage (Kondratowicz et al., 2011). Forty-eight hours after the infection, the cells were harvested and entered the RNA extraction process for further investigations. Finally, the inhibition of viral RNA synthesis was measured by qRT-PCR, which is one of the most accurate methods for this infection according to Carter et al. (2020).
The equal number of cells was counted before RNA extraction. Total RNA of each group was extracted and purified by TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and RNeasy MinElute Cleanup Kit (QIAGEN, Hilden, Germany) respectively according to the manufacturer's instructions. DNase treatment was applied before the purification process to ensure the elimination of unwanted DNA contamination. An equal concentration of RNA (measured by Nano-Drop) was taken to the qRT-PCR reaction. As qRT-PCR is a promising method for detecting the quantity of this virus (Yan et al., 2020), a TaqMan-based PCR set was performed using commercial primers and probes for SARS-CoV-2 by StepOnePlus Real-Time PCR system. The sequence of the forward and reverse primers and the probe is as follows respectively. Forward 5′-AAATTTTGGGGACCAGGAAC-3′, reverse 5′-TGGCAGCTGTGTAGGTCAAC-3′, and probe 5′-FAM-ATGTCGCGCATTGGCATGGA-BHQ-3′. Since there is no acceptable reference gene yet to normalize the Ct of the qRT-PCR for this virus, we decided to report the Ct values themselves according to Shen et al. (2020).
3. Results
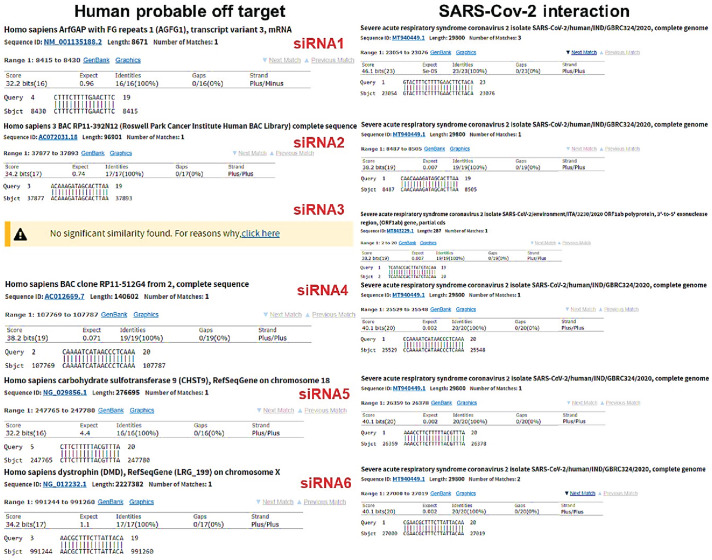
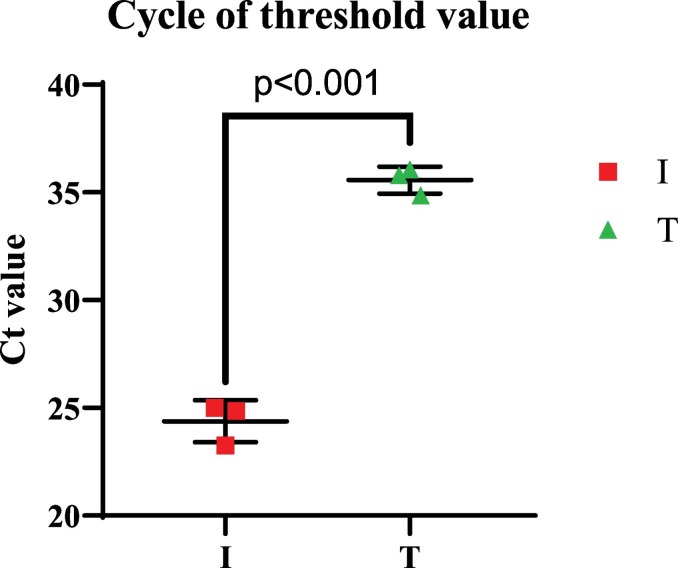
The BLAST results showed that all of the siRNAs could be matched with more than 5000 strains of the SARS-Cov 2 recognized from different parts of the world (more than 5000 accession numbers were matched in BLAST results) (data are not shown). Additionally, the BLAST results indicated that the chance of off-target effects in human cells was not significant for the siRNAs. The lowest E-value (the highest likelihood) of the BLAST results for human probable off-target and the actual SARS-Cov 2 target are shown in the Fig. 1 for each siRNA. In addition, for the qRT-PCR results, the group T showed significantly higher Ct value (Ct mean = 35.57 +/− 0.29) compared to the group I (Ct mean = 24.38 +/− 0.46) (Fig. 2 ).
Fig. 1.
BLAST results for probable human off target and the main SARS-Cov 2 target. The BLAST results for the lowest expected value (E-value) (highest chance of targeting) are shown. The human probable off-target is on the left and the SARS-Cov 2 main target is on the right for each siRNA. All E-values for human probable off-target was greater than 0.05 (not statistically significant) and no E-values for SARS-Cov 2 interaction are greater than 0.01.
Fig. 2.
Ct value of the groups. The groups were tested in a triplicated manner and each test was repeated three times to minimize the operational errors. Student t-test was used to compare the statistical difference between the groups (p < 0.001). Data are presented as mean +/− SEM. I: untreated SARS-CoV-2 infected group, T: siRNA treated infected group.
4. Discussion
The mechanism of siRNA-COVID-19 inhibitors is to target and initiate the termination process for most critical mRNAs that encode the virus's essential proteins. These include the proteins that are translated first upon virus entry into the cells and encode the replicase (the very first process that is necessary for viral proliferation) and other structural proteins, which are used to form a complete virus (Du et al., 2009).
Because a higher Ct number is correlated with the lower copy number of the target gene, the copy number of the virus-based mRNA was lower in the T group. Therefore, it appears that siRNAs may in fact inhibit the prolifration of the virus.. Similar Ct values have recently been reported for patients with COVID. (Carter et al., 2020). Additionally, since the siRNAs did not affect the normal proliferation of the Vero cells, using these molecules in the airway system could be a sensible option as a co-treatment for the SARS-CoV-2. In other words, siRNAs appear to inhibit virus replication likely by degradation of siRNA-associated mRNA.
In comparison to proinflammatory cytokine treatments for the coronavirus family such as hemophagocytosis (Ware and Matthay, 2000; Yuen et al., 1998) or an interferon-gamma-related cytokine storm (Huang et al., 2005), intranasal delivery of siRNA into the upper respiratory tract provides an effective method for high-specificity inhibition by potent siRNA inhibitors of SARS-CoV-2. Furthermore, in this method, the exacerbation of symptoms and lung damage can be avoided as it provides a minimal induction of a proinflammatory antiviral cytokine response (Li et al., 2005). Besides, the current drugs for COVID-19 are phase-dependent and they should be carefully prescribed (Stasi et al., 2020; Bartoli et al., 2021); however, due to the nature of the disease and the unstoppable replication process of the virus, siRNAs can be administered in any phase of the disease.
Unfortunately, the lack of acceptable in vivo virus proliferation inhibition by siRNAs is the main limitation of this study.
One of the challenging barriers to the development of siRNA inhibitors clinically is their intracellular delivery process. However, the COVID-19 viral infection is a respiratory disease and the initial tissues affected by the virus are lung cells; thus, the pre-designed siRNAs can be delivered near the affected cells via an aerosolized method such as an inhaler (Dykxhoorn and Lieberman, 2006). This method has been previously tested for SARS-CoV-1 by Li et al. (2005) and this can be piloted in future studies for COVID-19.
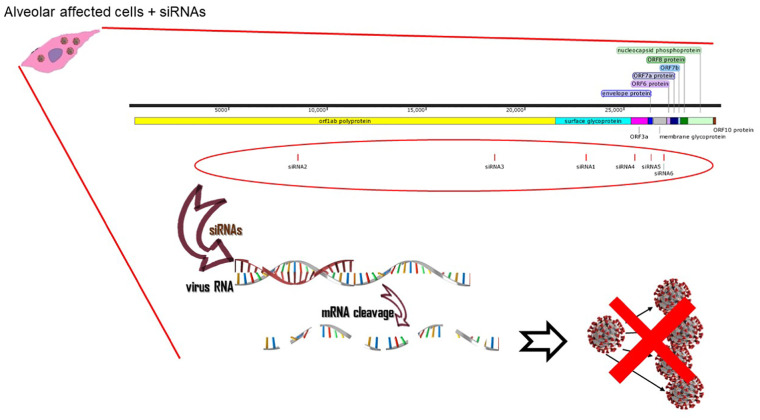
In conclusion, according to our initial results, we can state that this siRNA therapy can be used efficiently in vitro and there should be clinical practice in the next phase of this research (Fig. 3 ).
Fig. 3.
The schematic view of the virus replication inhibition by pre-designed siRNAs. The predesigned siRNAs interact with the viral mRNAs and this leads to viral mRNA cleavage. As a result, the replication of the virus is terminated due to the lack of its essential mRNAs.
Contributors
IN, MH and MB contributed equally in designing the research and drafted the original manuscript. TM, KG, and AK combined and edited the drafts, and partially supervised the manuscript. All authors subsequently revised the manuscript.
Declaration of interests
This work has been partially funded by the Natural Sciences and Engineering Research Council of Canada (NSERC). All other authors declare no competing interests.
Authors contribution
K. Ghaedi, T. L. Megraw, A.K. Kiani, I. Niktab Designed research; M. Haghparast Azad, I. Niktab, M-H Beigi, Performed research; K. Ghaedi, M. Haghparast Azad, I. Niktab and M-H Beigi Analyzed data; M. Haghparast Azad, I. Niktab M-H Beigi Wrote the first draft of paper. Final draft of paper was edited and approved by K. Ghaedi, T. L. Megraw, A.K. Kiani.
Acknowledgments
We appreciate all the efforts of Miss. Zahra Haghparast Azad for designing the graphs of this manuscript and we give our special thanks to Mrs. Jamile Mahdavinia for rationalizing the text of this manuscript.
References
- Bartoli A., et al. COVID-19 treatment options: a difficult journey between failed attempts and experimental drugs. Intern. Emerg. Med. 2021:1–28. doi: 10.1007/s11739-020-02569-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., et al. ACS Publications; 2020. Assay Techniques and Test Development for COVID-19 Diagnosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fougerolles A., et al. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6(6):443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., et al. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn D.M., Lieberman J. Knocking down disease with siRNAs. Cell. 2006;126(2):231–235. doi: 10.1016/j.cell.2006.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.J., et al. An interferon-γ-related cytokine storm in SARS patients. J. Med. Virol. 2005;75(2):185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., et al. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratowicz A.S., et al. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. 2011;108(20):8426–8431. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.-J., et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 2005;11(9):944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlik W. Springer; 2007. OLIGO 7 primer analysis software, in PCR primer design; pp. 35–59. [DOI] [PubMed] [Google Scholar]
- Sheahan T.P., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020:12(541). doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. Jama. 2020;323(16):1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasi C., et al. Treatment for COVID-19: An overview. Eur. J. Pharmacol. 2020:173644. doi: 10.1016/j.ejphar.2020.173644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Guan Y. COVID-19 drug repurposing: a review of computational screening methods, clinical trials, and protein interaction assays. Med. Res. Rev. 2021;41(1):5–28. doi: 10.1002/med.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware L.B., Matthay M.A. The acute respiratory distress syndrome. N. Engl. J. Med. 2000;342(18):1334–1349. doi: 10.1056/NEJM200005043421806. [DOI] [PubMed] [Google Scholar]
- Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-J., et al. Inhibition of SARS-CoV replication by siRNA. Antivir. Res. 2005;65(1):45–48. doi: 10.1016/j.antiviral.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Chang L., Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev. Med. Virol. 2020;30(3) doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K.-Y., et al. Clinical features and rapid viral diagnosis of human disease associated with avian influenza a H5N1 virus. Lancet. 1998;351(9101):467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]