Significance
Evolutionary innovations change how organisms interact with their environments. Trait modularization is thought to facilitate evolutionary innovation, while integration constrains diversification patterns. Cranial asymmetry in flatfishes was an innovation that allowed them to colonize, and dominate benthic aquatic habitats. In this study, we quantify rates of skull shape evolution, and integration across 102 species of flatfishes and their relatives and test for the effect of integration on rates of morphological diversification. We find that cranial asymmetry evolution was a rapid, integrated process. We additionally find that flatfishes are significantly more evolutionarily integrated than their relatives. These results suggest that trait integration plays a key role in the evolution of innovation by allowing traits to mount synchronous responses to selective pressures.
Keywords: geometric morphometrics, modularity, phylogeny
Abstract
Evolutionary innovations are scattered throughout the tree of life, and have allowed the organisms that possess them to occupy novel adaptive zones. While the impacts of these innovations are well documented, much less is known about how these innovations arise in the first place. Patterns of covariation among traits across macroevolutionary time can offer insights into the generation of innovation. However, to date, there is no consensus on the role that trait covariation plays in this process. The evolution of cranial asymmetry in flatfishes (Pleuronectiformes) from within Carangaria was a rapid evolutionary innovation that preceded the colonization of benthic aquatic habitats by this clade, and resulted in one of the most bizarre body plans observed among extant vertebrates. Here, we use three-dimensional geometric morphometrics and a phylogenetic comparative toolkit to reconstruct the evolution of skull shape in carangarians, and quantify patterns of integration and modularity across the skull. We find that the evolution of asymmetry in flatfishes was a rapid process, resulting in the colonization of novel trait space, that was aided by strong integration that coordinated shape changes across the skull. Our findings suggest that integration plays a major role in the evolution of innovation by synchronizing responses to selective pressures across the organism.
Evolutionary innovations are adaptations or exaptations that result in the colonization of novel regions of trait space, and shift the dynamic of ecosystem interactions in their respective environments. These innovations may involve a dramatic restructuring of an ancestral body plan, allow organisms novel access to ecological resources, and in (the case of key innovations) spur increases in lineage diversification (1–6).
Traditionally, much of the study of evolutionary innovation has focused on extrinsic drivers, such as ecological change or environmental stimuli, which have frequently been viewed as the principal triggers for evolutionary novelty (6–9). Recently, focus has shifted toward the effects of intrinsic (e.g., developmental) processes that may structure patterns of trait diversification (10–12). Among these processes, integration and modularity have emerged as important sources of insight as a growing consensus has found that patterns of trait covariation can constrain and facilitate responses to selection and strongly influence patterns of trait diversification at both contemporary and macroevolutionary timescales (13–20).
Modularity refers to the pattern whereby traits form complexes (i.e., modules) that exhibit a high degree of covariation within themselves, whereas the strength of covariation is far lower between these trait complexes (21, 22). This can result in the compartmentalization of trait complexes across an organism and has been hypothesized to allow modules to respond semiautonomously to different selective pressures (23–27). On macroevolutionary timescales, this modularization of different traits has been shown to result in mosaic patterns of evolution across organisms and is thought to facilitate morphological diversification as different traits are able to fine-tune responses to different selective pressures (27–29). Conversely, integration refers to a pattern whereby different traits exhibit a high degree of covariation (21, 30). Patterns of integration may be the result of pleiotropy or functional coupling (28, 30–33). There is less of a consensus on the macroevolutionary implications of phenotypic integration. Strong integration between traits has traditionally been hypothesized to constrain patterns of trait diversification along specific directions such that traits may exhibit more variation in some directions than others (30, 31). Furthermore, theoretical studies have found that evolutionary flexibility is negatively correlated with the magnitude of integration (27, 28, 34). However, more recent studies have found that the relationship between evolutionary integration and trait diversification is more complex than originally thought. A recent simulation study (28) found that trait integration can actually promote sizeable responses to selection, along lines that are parallel with the direction of trait covariation. This work also indicates that while a modular system has the freedom to explore a wider range of morphospace, this system is less likely to evolve maximally disparate phenotypes as compared to an integrated one that funnels variation along a more restricted trajectory. Indeed, several empirical studies have found rapid rates of shape evolution and high degrees of morphological disparity in clades that also exhibit tight evolutionary integration across the traits of interest (29, 35, 36). These studies suggest that evolutionary integration may play an important role in the evolution of innovation as it allows for clades to rapidly explore novel regions of morphospace.
The majority of our understanding of patterns of vertebrate trait diversification come from tetrapod systems, and primarily studies involving birds and mammals (14, 37–40). While these studies have been deeply informative, they help to explain less than half of the vertebrate story of diversification as these studies frequently exclude ray-finned fishes (Actinopterygii) (24). Ray-finned fishes comprise over half of the species diversity in vertebrates. They also exhibit a diverse array of evolutionary innovations that have allowed them to become arguably the most successful vertebrate radiation on the planet.
Flatfishes in particular represent a diverse clade of bottom-dwelling, teleost fishes that possess a striking evolutionary innovation: a degree of cranial asymmetry exceeding that of any other vertebrate lineage (41). Most flatfishes are completely blind on one side of their body, and instead feature both eyes on the same side of the head. This perplexing flatfish phenotype is achieved during the early developmental stages where one eye of a symmetrical larva gradually begins to migrate to the other side of its body, rendering one side “eyed” and the other “blind” (42). Developmental studies indicate that this orbital migration is driven by thyroid hormone expression and is paired with changes in swimming behavior (43), as well as asymmetrical visceral organ rearrangement (44). These developmental patterns recapitulate paleontological trends, with fossils indicating that flatfishes gradually became more asymmetrical from symmetrical ancestors (45). In addition to their cranial asymmetry, flatfishes possess a suite of adaptations that further allow them to exploit their benthic habitats; most nobly among these is their derived form of locomotion that combines strong, whole-body undulations (for fast escapes or burial) with finer-scale, dexterous undulations of their dorsal and anal fins (for slower cruising behaviors) (46–48). The evolution of this derived form of locomotion is coupled with additional changes in the roofing bones of the skull as they evolved projections to support the extended dorsal fin along the supraoccipital crest and the frontal bones (49, 50).
The flatfish body plan is thought to have rapidly arisen shortly after the Cretaceous–Paleogene boundary (66 Mya) (49). Comparative analyses show rapid bursts of body-shape diversification in flatfishes and their close relatives within the broader clade Carangaria, a diverse radiation including disparate lineages like billfishes and remoras in addition to flatfishes (49, 51, 52) (Fig. 1). This episode of phenotypic innovation is hypothesized to reflect the filling of newly available ecological roles in the early Cenozoic, matching patterns reported for other groups.
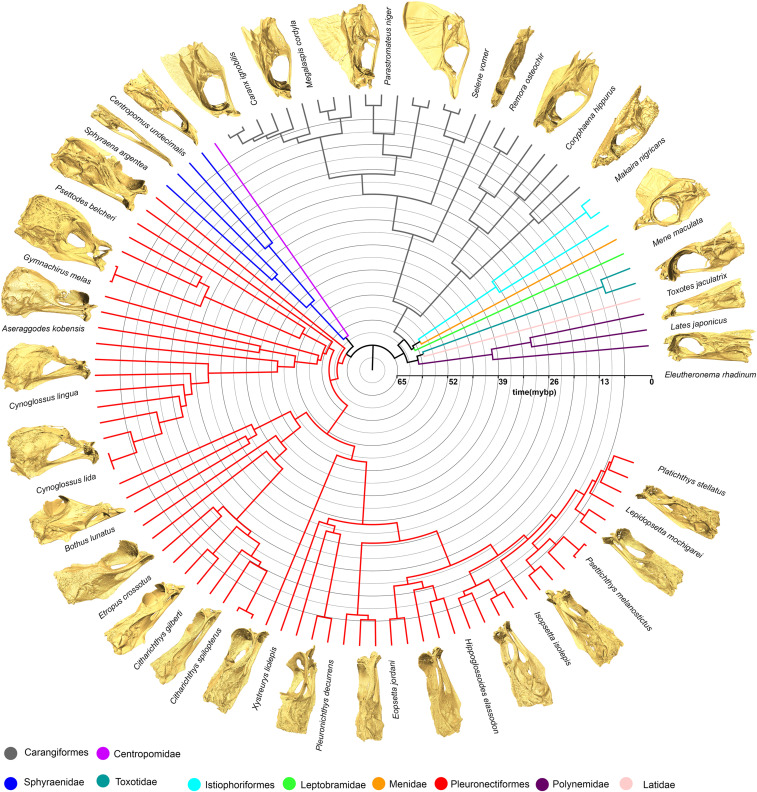
Fig. 1.
Phylogeny of 102 carangarian species included in the analyses of skull shape evolution. Insets depict representative skull shapes for each clade. Phylogeny based on Ribeiro et al. (52).
The striking and rapid evolutionary dynamics within Carangaria make this clade a tantalizing target for investigating the roles that integration and modularity play in trait diversification and, particularly, evolutionary innovation. Here we use three-dimensional geometric morphometrics and a cutting-edge phylogenetic comparative toolkit to study the evolution of the neurocranium and the evolution of cranial asymmetry across 102 carangarian species. We quantify shifts in the rate of skull shape evolution between species and across the neurocranium as a whole, while also quantifying patterns of integration and modularity between flatfishes and their relatives to test for the effect of integration and modularity on the evolution of innovation. We hypothesize that flatfishes underwent a rapid shift in their rates of skull shape evolution as a result of their orbital migration. We additionally hypothesize that flatfishes will exhibit higher levels of integration compared to their carangarian relatives as a result of their asymmetrical larval metamorphosis that involves coordinated changes across the body, and adaptations associated with their derived locomotory mode.
Results
Skull Shape Evolution in Carangaria.
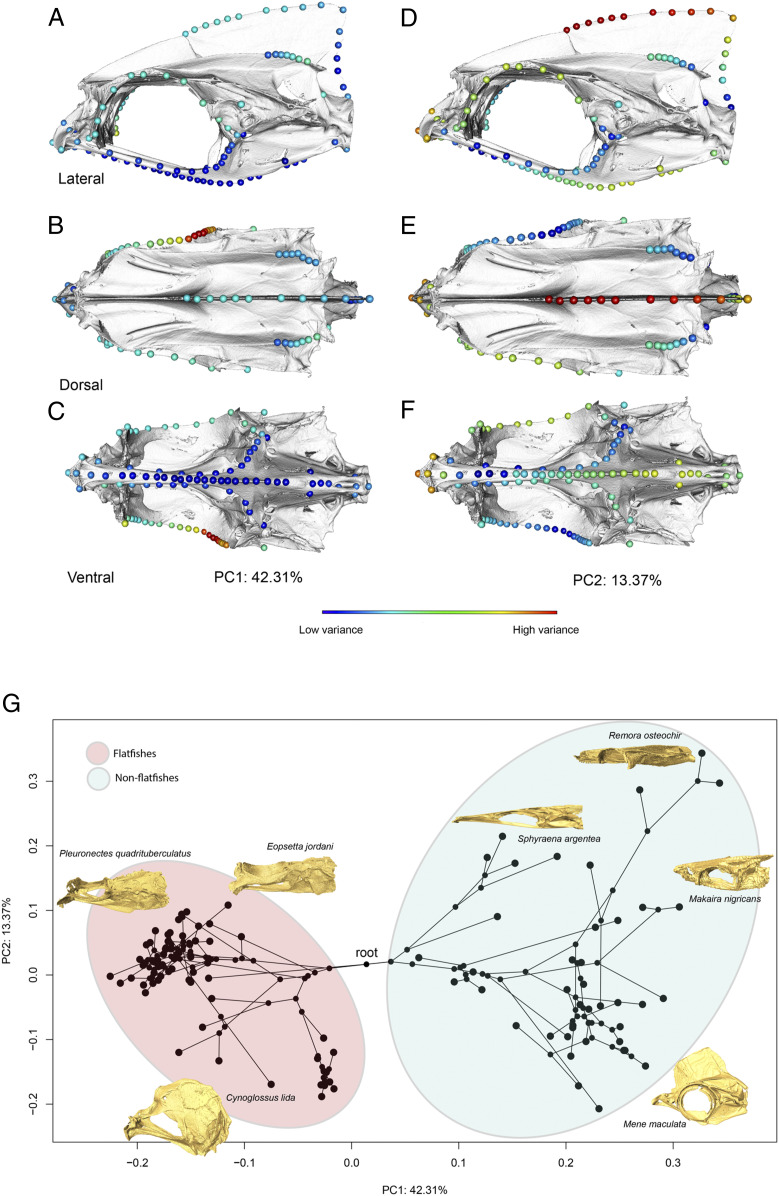
Carangarians exhibit a broad diversity of skull shapes (Fig. 2). An analysis of phylogenetic signal finds significant, but weak phylogenetic structure in neurocranium shape across this clade (P = 0.001; Blomberg’s K = 0.178). The phylogenetic signal is most apparent along the primary axis of shape variation (principal component 1, PC1), which corresponds overwhelmingly to variation in the shape and position of the right (migrating) orbit. Interestingly, aside from the shape divergence toward the root of the phylogeny, there appears to be rampant convergence among the flatfishes, particularly among species with elongated neurocrania (e.g., Pleuronectes quadrituberculatus). It’s possible that this convergence is driving the low phylogenetic signal observed in this clade. The results of the phylogenetically aligned component analysis reveal a similar pattern of dispersion (SI Appendix, Fig. S1). In both analyses, a strong asymmetrical pattern of shape variation can be observed across the neurocranium with the highest amount of variance concentrated at the right, frontal-sphenotic margin (Fig. 2). Using the phylogenetically aligned components analysis, we found that this variation in orbital position accounts for over 93% of the total shape variation for carangarians as a whole, suggesting that the early-diverging flatfish asymmetry is the primary contributor to phylogenetic signal in the shape data. In our phylomorphospace analysis, along the PC1 axis, flatfishes occupy a distinct region of morphospace with respect to their “nonflatfish” relatives, with the Alaska plaice (P. quadrituberculatus) exhibiting the lowest score and the common remora (Remora remora) exhibiting the highest score. The second PC axis (PC2) corresponds to variation in the skull depth and relative skull length (i.e., heterocephaly) with species like the roughscale tonguesole (Cynoglossus lida) and the moonfish (Mene maculata) exhibiting the lowest scores while more dorsoventrally compressed species, like the Petrale sole (Eopsetta jordani) and the Pacific barracuda (Sphyraena argentea), exhibit higher scores.
Fig. 2.
Skull shape diversity and evolution across 102 carangarian species. PC analysis showing the primary axes and regions of variation in neurocranium shape across Carangaria (A–F). (G) Phylomorphospace analysis showing the diversity of neurocranial shapes across carangarian fishes. Insets depict representative extremes for each PC axis.
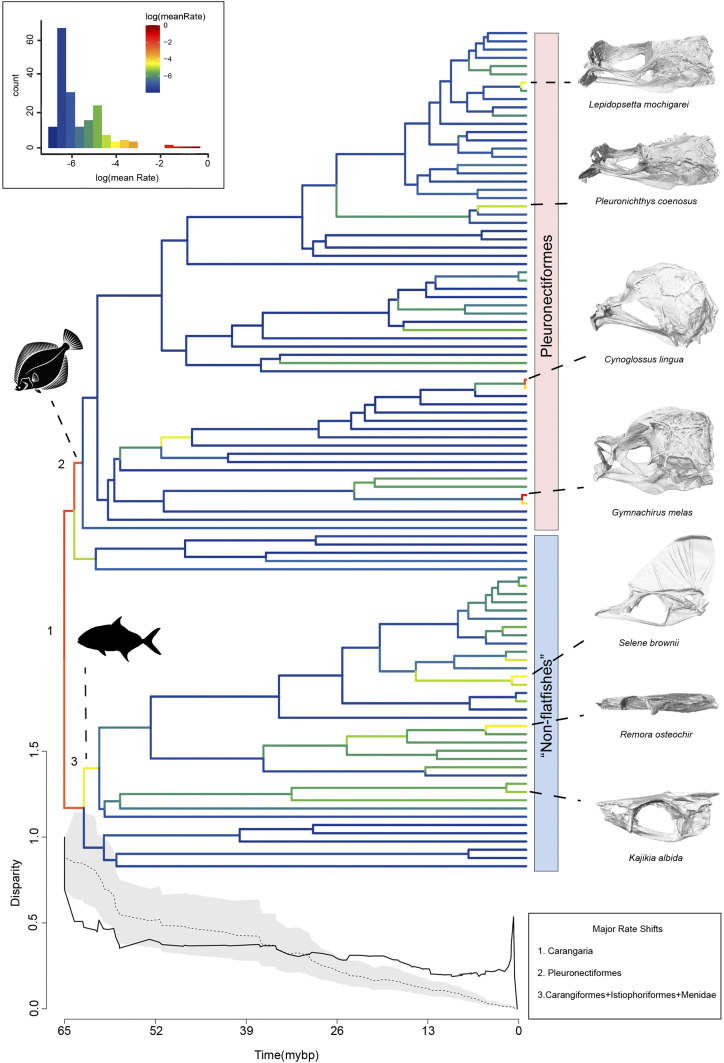
Tempo and Mode of Skull Shape Evolution.
Carangarians exhibit variable rates of skull shape evolution. We recover strong model support for a “variable-rate” model of trait evolution as opposed to a “single-rate” model (SI Appendix, Table S1). A disparity through time analysis indicates a rapid decline in subclade disparity that exceeds the Brownian motion expectation early in carangarian history (though on average, does not differ significantly across the entire history of the clade; mean disparity index = −0.012, P = 0.36). This rapid decline in subclade disparity is consistent with adaptively radiating lineages and suggests that skull shapes partitioned early in evolutionary history. After the initial rapid decline, subclade disparity remains fairly consistent over time, with a recent rapid spike less than 5 Mya coinciding with rapid rates of shape evolution occurring primarily within flatfishes specifically: Cynoglossus, Gymnachirus, and Lepidopsetta (Fig. 3). We found additional evidence for rapid shape divergences at the base of Carangaria using a BayesTraits analysis, which recovers a strong rate shift at the base of Carangaria. Additionally, we found that flatfishes underwent a rapid burst in the rate of skull shape diversification that coincided with the evolution of cranial asymmetry. Within flatfishes, rates of shape evolution generally decline with subsequent shifts in achirids and cynoglossids, coinciding with variation related to orbital position and relative skull length. Outside of flatfishes, we found the largest rate shifts among remoras (Echeneidae), bill-fishes (Istiophoridae), and lookdown jacks (Carangidae). These rate shifts coincide primarily with heterocephaly (variation between elongated and foreshortened skull shapes), dorsoventral compression, and the height of the supraoccipital crest. Comparisons of mean evolutionary rates between flatfishes and “nonflatfishes” found significantly higher rates of skull shape evolution among flatfishes (P < 0.001; rate ratio = 2.83). These results suggest that carangarians rapidly morphologically diversified early in their evolutionary history and that flatfishes continued to rapidly diversify in smaller subclades after initially evolving cranial asymmetry.
Fig. 3.
Tempo and mode of skull shape diversification in Carangaria. Results from BayesTraits and disparity though time analyses showing shifts in the rate of skull shape evolution across the carangarian phylogeny. Dashed line on DTT indicate Brownian motion expectation and shaded region indicates 95% confidence interval.
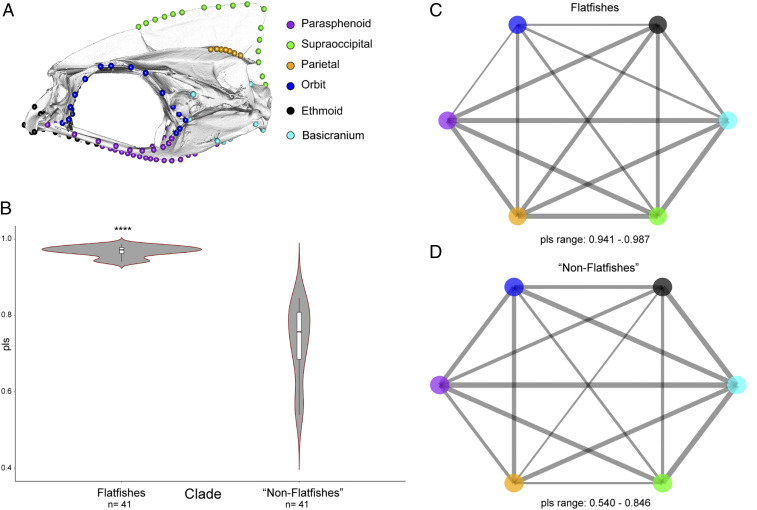
Evolutionary Modularity.
We found strong patterns of modularity across the carangarian neurocranium. We recovered strong model support for the six-module “bone” hypothesis encompassing the basicranium, ethmoid, orbit, parasphenoid, and supraoccipital regions (Table 1). Support for this model was retained after rerunning the analysis on a smaller subsample of landmarks (SI Appendix, Table S2). Support for the bone hypothesis was also recovered using the covariance ratio (CR) coefficient (CR = 0.943; P < 0.001). We subsequently divided the Carangaria into two subclades consisting of flatfishes (Pleuronectiformes) and nonflatfishes (Carangiformes, Istiophoriformes, Centropomidae, Leptobramidae, Menidae, Polynemidae, and Toxotidae) and reanalyzed patterns of phylogenetic integration and modularity. We found that flatfishes exhibit a strong signal of integration (partial-least squares [pls] = 0.968) while still exhibiting a weak but significant degree of modularity (CR = 0.96; P < 0.001). Meanwhile, we found that nonflatfishes exhibit significantly (P < 0.001) lower levels of integration (pls = 0.730) and significantly (P < 0.001) higher levels of modularity (CR = 0.782; P < 0.001) across the six regions of the neurocranium (Fig. 4 and SI Appendix, Fig. S2).
Table 1.
Results for the evaluation of modularity hypotheses (using phylo.EMMLI) for 102 carangarian species
| Model | MaxL | K | n | AICc | dAICc | Model_L | Post_Pob |
| No.modules. default | −29182.5 | 2 | 17956 | 58369.09 | 22557.38 | 0 | 0 |
| Bone modules | |||||||
| Same within-module p + same between-module p | −19009.5 | 3 | 17956 | 38024.93 | 2213.216 | 0 | 0 |
| Separate within-module p + same between-module p | −18374.7 | 8 | 17956 | 36765.47 | 953.755 | 7.85E-208 | 7.85E-208 |
| Same within module p + separate between module p | −18518.6 | 17 | 17956 | 37071.16 | 1259.446 | 3.27E-274 | 3.27E-274 |
| Separate within module p + separate between module p | −17883.8 | 22 | 17956 | 35811.72 | 0 | 1 | 1 |
| Functional modules | |||||||
| Same within-module p + same between-module p | −25411.4 | 3 | 17956 | 50828.89 | 15017.18 | 0 | 0 |
| Separate within-module p + same between-module p | −22287.6 | 5 | 17956 | 44585.19 | 8773.472 | 0 | 0 |
| Same within module p + separate between module p | −25274.9 | 5 | 17956 | 50559.88 | 14748.17 | 0 | 0 |
| Separate within module p + separate between module p | −22151.1 | 7 | 17956 | 44316.18 | 8504.465 | 0 | 0 |
| Developmental modules | |||||||
| Same within-module p + same between-module p | −22261.3 | 3 | 17956 | 44528.56 | 8716.839 | 0 | 0 |
| Separate within-module p + same between-module p | −20675.5 | 6 | 17956 | 41363.03 | 5551.312 | 0 | 0 |
| Same within module p + separate between module p | −22059.1 | 8 | 17956 | 44134.13 | 8322.416 | 0 | 0 |
| Separate within module p + separate between module p | −20473.3 | 11 | 17956 | 40968.61 | 5156.892 | 0 | 0 |
Bold text indicates optimal model.
Fig. 4.
Phylogenetic integration across six modules of the carangarian skull (A). Violin plots showing results from a phylogenetic two-block partial least-squares for flatfishes and nonflatfishes showing significant differences in mean partial least squares values for each group (B). Network graphs showing the magnitude of integration between modules for flatfishes and nonflatfishes (C and D).
Mosaic Evolution.
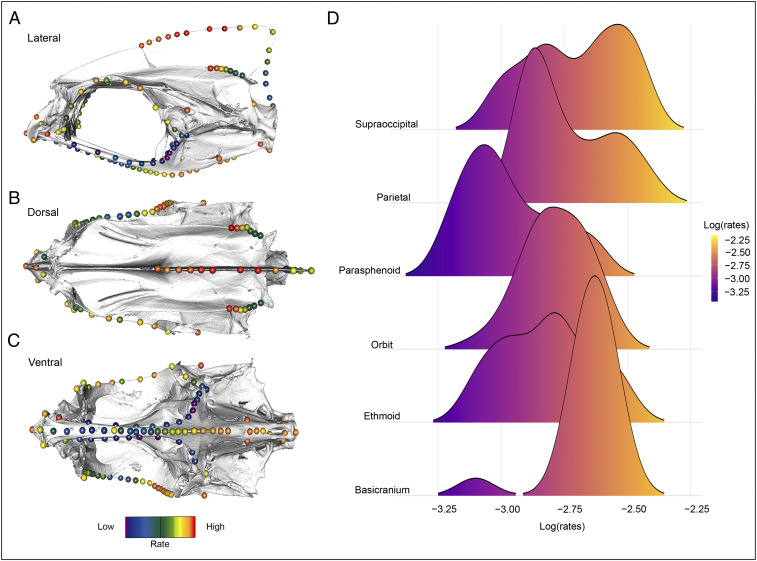
Among carangarians, we found that the fastest rates of shape evolution are concentrated in the ethmoid, orbital, and ventral basicranial regions of the neurocranium (all regions that undergo dramatic changes during flatfish metamorphosis), while the parasphenoid and parietal regions exhibit slower rates of shape evolution (Fig. 5 and SI Appendix, Table S3). Rate analyses (using σ2 mult) also recovered significant rate differences between modules (SI Appendix, Table S4). Mosaic patterns of evolution appear to be driven primarily by the slower-than-average rates of shape evolution in the anterior region of the parasphenoid, which differs significantly from every other region. Excluding the parasphenoid, no other significant rate differences were recovered between modules.
Fig. 5.
Mosaic evolution across the carangarian skull. Per landmark rates of shape evolution for 102 carangarian species (A–C). Ridgeline plot comparing per landmark rates across the six modules of the neurocranium (D).
Discussion
Evolutionary innovations allow the organisms that possess them to colonize novel regions of trait space, and grant them access to new and different adaptive zones. In this study, we examined a major evolutionary innovation in flatfish cranial asymmetry. We found that cranial asymmetry evolved within the Carangaria shortly after the K/Pg boundary and resulted in a rapid shift in rates of skull shape evolution at the base of Pleuronectiformes. This innovation allowed flatfishes to rapidly colonize a completely novel region of morphospace as they invaded benthic habitats. Among carangarians, we recovered asymmetrical patterns of variation across the skull driven by the orbital migration that flatfishes experienced at the macroevolutionary timescale. This degree of cranial asymmetry is not seen anywhere else in the vertebrate tree of life, even among other teleost fishes that inhabit the benthos (but see ref. 53). Typically, benthic fishes exhibit dorsoventrally flattened bodies to allow for partial burial in substrates (e.g., lizard fishes) (51, 54). Dorsoventrally compressed phenotypes are even present within carangarians (e.g., remoras), making the cranial asymmetry of flatfishes even more puzzling since some of their relatives were clearly able to evolve flattened skulls in a more typical fashion.
The role that integration and modularity play in the evolution of innovation is a hotly debated topic (23, 55, 56). Traditional hypotheses suggest that phenotypic diversification and novelty can be facilitated by the modularization of traits to allow them to behave semiautonomously over macroevolutionary timescales (23, 27, 56). In contrast to modularity, integration between traits across macroevolutionary timescales is thought to constrain patterns of trait diversification (29, 32, 57, 58; but see ref. 28). Here we found that the evolution of cranial asymmetry in flatfishes was an integrated process, with coordinated patterns of shape change across the neurocranium. This result may seem counterintuitive given the superficially modular nature of the orbital migration typified in flatfishes. However, the benthopelagic transition that flatfishes experience at the developmental level reveals a more integrated process across the skull and body that includes changes in bilateral pigmentation, hindbrain morphology, inner-ear morphology, swimming behavior, relative muscle masses between eyed and blind sides, and the relative placements and sizes of internal organs (59–61). Additional evidence for the integrated nature of the benthopelagic transition can be found in flatfish locomotion, which utilizes both the dorsal, and anal fins as paired appendages for benthic “walking” (46). This walking behavior is accomplished by the more anterior placement of the dorsal fin (relative to nonflatfishes), which interdigitates with the supraoccipital crest and other dermal-roofing bones in many species (e.g., Achirus). Similarly, widespread burial behavior exhibited across most flatfish species also involves coordinated sinusoidal undulations across the entire body and fins to cover the animal with sediment. These behavioral novelties almost certainly required integrated modifications to both axial and cranial morphologies.
While our analyses recover strong patterns of evolutionary integration across extant flatfishes, the fossil record suggests that trait integration may have gradually increased over evolutionary time. Studies of stem flatfishes indicate that the evolution of asymmetry was a gradual, and stepwise process [contrary to the saltatory hypothesis proposed by Goldschmidt (45, 62, 63)]. While stem flatfishes exhibited some characteristic patterns of cranial asymmetry observed in extant flatfishes (primarily asymmetrical orbits), they retained a number of primitive and generalized morphologies as well [including a “generalized perciform caudal skeleton” (45), which suggests that cranial asymmetry may have predated the adaptations associated with flatfish locomotion (however, more complete fossils will be needed to confirm this). Many of these characters can still be observed in Psettodes, an early-branching, phenotypically conserved flatfish. Members of this genus exhibit incomplete orbital migration, pigmentation on both sides of the body, and left-handed and right-handed morphs that are present at equal frequencies in the population (considered a precursor to directional asymmetry) (42). This fluctuating asymmetry, may be indicative of a less-canalized developmental program, and could possibly suggest that the asymmetry in flatfishes arose originally from developmental instability in the cranial growth fields that may have subsequently been reorganized in later clades (64).
Taken together, our findings suggest that integration played a key role in coordinating changes across different traits during the benthopelagic transition in flatfishes. We further hypothesize that integration likely plays a key role in the evolution of innovation by distributing variation across multiple traits and allowing organisms to mount a coordinated response to selective pressures.
Materials and Methods
Morphological Sampling and General Shape Analyses.
Skull shape was quantified across 123 specimens representing 102 carangarian species (61 flatfishes, 41 nonflatfishes, ∼10% of total clade diversity, and 24 of the 27 carangarian families) (SI Appendix, Table S5). Specimens were micro-CT scanned at the University of Michigan and the University of Washington using a Nikon XT H 225 ST micro-CT scanner and a Bruker Skyscan 1173 micro-CT scanner, respectively, in conjunction with oVert and ScanAllFishes scanning initiatives. Additional scans were also downloaded from http://morphosource.org. Scans were segmented using Amira and the neurocranium was isolated from each specimen. Segmented neurocrania were converted to three-dimensional meshes and exported as .ply files.
Flatfishes are known to exhibit both sinistral and dextral forms. These drastic differences in asymmetry have the ability to confound geometric morphometric analyses. To account for these differences in asymmetry, we inverted the meshes for all sinistral (left-eyed) specimens in MeshLab turning them into dextral specimens. The inversion of the meshes effectively constrained the asymmetrical variation present in flatfish neurocrania to the right side for all specimens. While useful for standardizing our specimens, it bears noting that our approach may systematically underestimate shape variation and disparity in the flatfish skull.
After standardization, meshes were digitized with 134 (34 fixed; 100 sliding) three-dimensional landmarks using the Checkpoint software (SI Appendix, Fig. S3 and Table S6). Bone nomenclature follows Hilton et al. (65). Landmarks were not treated as bilaterally symmetrical due to the high degree of asymmetry present in flatfishes, which comprise over half of the shape data. Landmark coordinates were then imported into R and analyzed using the geomorph package (66). For specimens with more than one representative per species, landmark averages were calculated. Specimen coordinates were subjected to a generalized procrustes analysis to remove the effect of orientation and scaling between specimens (67). Semilandmarks were slid along their tangent directions while minimizing bending energy (68).
Allometric scaling can play an important role in structuring patterns of shape change (69, 70). To quantify the effect of allometric scaling on our skull shape data, we performed a phylogenetic generalized least-squares regression of shape vs. log(centroid-size) using the proc.pgls function in geomorph (71, 72). We found a small (r2 = 0.07) but significant (P = 0.0015) effect of size on shape (SI Appendix, Table S7). Due to its relatively small effect on shape variation and its general importance as relevant biological information, we elected not to remove the effect of allometric scaling from our shape data following the approach of Evans et al. (16) and Felice and Goswami (37).
Phylogenetic Reconstruction.
To study the evolution of shape change across Carangaria, we used the phylogeny of Ribeiro et al. (52). This time-calibrated phylogeny consists of 508 tips and comprises a phylogenetic backbone of Rabosky et al. (73), with additional carangarian taxa grafted onto it. We pruned the Ribeiro et al. (52) phylogeny in ape (74) to only include the taxa present in our study. The resulting pruned tree consisted of 102 carangarian species.
Phylogenetic Visualization.
To visualize major axes of shape variation and evolution, we utilized a phylomorphospace approach using our first two PCs of shape variation (accounting for 56% of total shape variance) and projected the Ribeiro et al. (52) phylogeny onto them (75). In order to compare phylogenetic trends in our data, we coupled our PC analysis with a phylogenetically aligned components analysis (76), which uses an ordinary least-squares approach to align the trait data to the axis of greatest phylogenetic signal and projected the first two components onto the aforementioned phylogeny. This approach allowed us to differentiate between stronger and weaker phylogenetic signals localized to a particular proportion of the shape axes. We additionally calculated phylogenetic signal in our shape data using the multivariate implementation of Blomberg’s K (kmult), to test for phylogenetic structure in our skull shape data (77).
Quantifying Clade Rates of Shape Evolution.
Rates of shape evolution across carangarian subclades were quantified using the variable-rates model implementation in the BayesTraitsV3 program (78). This Bayesian method uses a reversible-jump Markov chain Monte Carlo chain approach to estimate the probability of rate shifts in continuous trait data across a phylogeny. This method has the ability to detect clade and species-specific rate shifts in trait data. There are known difficulties with evolutionary model-fitting using data with high dimensionality (79). To reduce the dimensionality of our data, we used the first 20 PCs, which accounted for 95% of our total shape variance. While PC axes are mathematically orthogonal, and thus uncorrelated, trait variation can still be evolutionarily correlated. To account for this, we ran our analyses using the TestCorrel function, which constrains the correlation between trait axes to zero. We used uniform, uninformative priors and ran four independent chains each for 200 million generations, discarding the first 1 million as burn-in. The chain was sampled every 2 million generations after convergence using a stepping-stone sampler and convergence was evaluated before the first 1 million generations were removed as burn-in. Model convergence was evaluated for each model by running the analysis a second time and visually assessing the trace of the marginal likelihoods using Tracer (80) (SI Appendix, Fig. S4). We evaluated two models of trait evolution: a single-rate model that assumes a single Brownian motion rate of trait evolution across the phylogeny, and a variable-rates model that allows for rate heterogeneity and identifies regions of the tree where evolutionary rates differ across different branches and internal nodes (78). Model comparisons were performed by calculating Bayes factors from the marginal likelihoods of the single-rate and variable-rates models. The resulting output of the variable-rates analysis is a set of phylogenies where each branch is scaled by its Brownian motion rate of evolution. To summarize these rate shifts, we built a consensus tree out of the 100 trees produced using the BayesTrees program.
We also quantified the rate of shape evolution between flatfishes and nonflatfishes using the compare.evol.rates function in geomorph (81). Significance was assessed using the phylogenetic simulation approach [see Adams and Collyer (79) for more information].
Disparity through Time.
A disparity through time (DTT) analysis was used to model changes in subclade shape disparity for carangarians under a Brownian motion model (using our entire shape dataset) (82). This method calculates changes in relative subclade disparity through time across every node in the phylogeny. We then compared the observed disparity of carangarians to a Brownian motion model that was simulated 1,000 times across the phylogeny. We used the observed and simulated disparities to calculate a morphological disparity index (MDI), which quantifies the difference in relative subclade disparity under a Brownian motion expectation. Negative MDI values are generally indicative of adaptively radiating clades (82, 83). The DTT analysis was performed in the R-package geiger (84).
Evolutionary Modularity.
Evolutionary modularity across different regions of the carangarian neurocranium was assessed using the phyloEMMLi approach; this method uses maximum-likelihood to compare different hypotheses of modularity across a shape dataset while accounting for phylogenetic nonindependence of the trait data (85). We fit three models of modularity (and a “no modularity” model) to our data (SI Appendix, Fig. S5). The first bone model consisted of six modules (vomer, parasphenoid, parietals, “basicranium,” supraoccipital, and “orbit”) roughly corresponding to individual bones; with the exception of the orbital and basicranial regions, which are composed of multiple smaller bones that were not densely sampled enough in the landmarking scheme to constitute separate module designation. The second model consisted of three “functional” modules (ethmoid, optic, and basicranium) corresponding to the location of the individual sensory capsules in the neurocranium (86). The otic region was not given an individual module designation apart from the basicranium due to the frequent lack of ossification in the area, which limited adequate landmark sampling. The third model consisted of four “developmental” modules (face, orbit, basicranium, and skull roof) following Evans et al. (16, 87, 88). The robustness of the EMMLI results were evaluated with a random subsampling down to 90% of the full landmark dataset, and EMMLi ran iteratively 100 times with an Akaike Information Criterion cutoff of 500, using the “subSampleEMMLi” function from the EMMLi v.0.0.3 R package. Mean results from the 100 subsamples were then compared to the analysis of the full dataset following the approach of Bon et al. (89).
Evolutionary modularity was also assessed using the CR coefficient (90), which quantifies the degree of modularity between shape variables while taking phylogeny into account. Significance is determined by comparing the observed CR coefficient with a randomly generated distribution.
Differences in the strength of modularity was assessed between flatfishes and nonflatfishes using the compare.CR function in geomorph (91). This function compares the strength of modularity between shape configurations using the CR effect sizes from each dataset.
Evolutionary Integration.
Evolutionary integration was assessed using the phylo.integration function in geomorph (92). This method uses a two-block partial least-squares analysis to quantify the degree of shape covariation between hypothesized modules while considering phylogenetic nonindependence. This analysis was run separately for flatfishes and nonflatfishes. Our best-supported modular configuration exceeded three modules, so we calculated an r coefficient value for each individual pairwise comparison using the phylo.integration function and tested for differences between the mean partial least-squares r coefficients for each group using a two-sample t test. The r coefficient is sensitive to differences in sample size, and differences in the number of variables between datasets (93). To account for this, we randomly removed 20 flatfish species prior to running the integration analyses to standardize the sample sizes between both groups.
Quantifying Mosaic Evolution.
Mosaic patterns of shape evolution across the neurocranium were assessed and quantified by calculating the per landmark rate of evolution for every landmark in the shape configuration using the Hotdots package in R (56). Landmark rates were then subdivided by module, and rate differences between modules were assessed using a Tukey’s honest significant difference to compare pairwise rate differences between modules while adjusting for multiple comparisons (94). Module rates of evolution were also quantified using the compare.multi.evol.rates function in geomorph which calculates rates of shape evolution from a priori defined modules [see Denton and Adams (81) for more information].
Supplementary Material
Acknowledgments
We thank Dr. Adam Summers and the oVert and ScanAllFishes initiatives for allowing us to micro-CT scan fishes for free; and Dr. Andrew Knapp for teaching K.M.E. how to invert specimen meshes. The National Science Foundation for Research Experiences for Undergraduates provided funding for S.-J.W. This study includes data produced in the Computed Tomography in Earth and Evironmental Sciences facility at University of Michigan, supported by the Department of Earth & Environmental Sciences and College of Literature, Science, and the Arts.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101330118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Alfaro M. E., Brock C. D., Banbury B. L., Wainwright P. C., Does evolutionary innovation in pharyngeal jaws lead to rapid lineage diversification in labrid fishes? BMC Evol. Biol. 9, 255 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burress E. D., Wainwright P. C., Adaptive radiation in labrid fishes: A central role for functional innovations during 65 My of relentless diversification. Evolution (2018). [DOI] [PubMed] [Google Scholar]
- 3.Dumont E. R., et al., Morphological innovation, diversification and invasion of a new adaptive zone. Proc. Biol. Sci. 279, 1797–1805 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goffredi S. K., et al., Evolutionary innovation: A bone-eating marine symbiosis. Environ. Microbiol. 7, 1369–1378 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Simpson G. G., Tempo and Mode in Evolution (Columbia University Press, 1944). [Google Scholar]
- 6.Price S. A., et al., Functional innovations and morphological diversification in parrotfish. Evolution 64, 3057–3068 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Heard S. B., Hauser D. L., Key evolutionary innovations and their ecological mechanisms. Hist. Biol. 10, 151–173 (1995). [Google Scholar]
- 8.Hunter J. P., Key innovations and the ecology of macroevolution. Trends Ecol. Evol. 13, 31–36 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Simpson G. G., The Baldwin effect. Evolution 7, 110–117 (1953). [Google Scholar]
- 10.Conith A. J., Hope S. A., Chhouk B. H., Craig Albertson R., Weak genetic signal for phenotypic integration implicates developmental processes as major regulators of trait covariation. Mol. Ecol. 30, 464–480 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons K. J., Trent Taylor A., Powder K. E., Albertson R. C., Wnt signalling underlies the evolution of new phenotypes and craniofacial variability in Lake Malawi cichlids. Nat. Commun. 5, 3629 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts R. B., Hu Y., Albertson R. C., Kocher T. D., Craniofacial divergence and ongoing adaptation via the hedgehog pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 13194–13199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y., Dokholyan N. V., The coordinated evolution of yeast proteins is constrained by functional modularity. Trends Genet. 22, 416–419 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Drake A. G., Klingenberg C. P., Large-scale diversification of skull shape in domestic dogs: Disparity and modularity. Am. Nat. 175, 289–301 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Du T. Y., Tissandier S. C., Larsson H. C., Integration and modularity of teleostean pectoral fin shape and its role in the diversification of acanthomorph fishes. Evolution 73, 401–411 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Evans K. M., Vidal-García M., Tagliacollo V. A., Taylor S. J., Fenolio D. B., Bony patchwork: Mosaic patterns of evolution in the skull of electric fishes (Apteronotidae: Gymnotiformes). Integr. Comp. Biol. 59, 420–431 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Gibson G., Wagner G., Canalization in evolutionary genetics: A stabilizing theory? BioEssays 22, 372–380 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Wagner G. P., Homologues, natural kinds and the evolution of modularity. Am. Zool. 36, 36–43 (1996). [Google Scholar]
- 19.Badyaev A. V., Stress-induced variation in evolution: From behavioural plasticity to genetic assimilation. Proc. Biol. Sci. 272, 877–886 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frédérich B., Olivier D., Litsios G., Alfaro M. E., Parmentier E., Trait decoupling promotes evolutionary diversification of the trophic and acoustic system of damselfishes. Proc. Biol. Sci. 281, 20141047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson E. C., Miller R. L., Morphological Integration (University of Chicago Press, 1999). [Google Scholar]
- 22.Schlosser G., Wagner G. P., Modularity in Development and Evolution (University of Chicago Press, 2004). [Google Scholar]
- 23.Wagner G. P., Altenberg L., Perspective: Complex adaptations and the evolution of evolvability. Evolution 50, 967–976 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Larouche O., Zelditch M. L., Cloutier R., Modularity promotes morphological divergence in ray-finned fishes. Sci. Rep. 8, 7278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zelditch M. L., Carmichael A. C., Growth and intensity of integration through postnatal growth in the skull of Sigmodon fulviventer. J. Mammal. 70, 477–484 (1989). [Google Scholar]
- 26.Zelditch M. L., Wood A. R., Swiderski D. L., Building developmental integration into functional systems: Function-induced integration of mandibular shape. Evol. Biol. 36, 71–87 (2009). [Google Scholar]
- 27.Marroig G., Shirai L. T., Porto A., de Oliveira F. B., De Conto V., The evolution of modularity in the mammalian skull II: Evolutionary consequences. Evol. Biol. 36, 136–148 (2009). [Google Scholar]
- 28.Goswami A., Smaers J. B., Soligo C., Polly P. D., The macroevolutionary consequences of phenotypic integration: From development to deep time. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans K. M., Waltz B. T., Tagliacollo V. A., Sidlauskas B. L., Albert J. S., Fluctuations in evolutionary integration allow for big brains and disparate faces. Sci. Rep. 7, 40431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bookstein F. L., et al., Cranial integration in Homo: Singular warps analysis of the midsagittal plane in ontogeny and evolution. J. Hum. Evol. 44, 167–187 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Albertson R. C., Streelman J. T., Kocher T. D., Yelick P. C., Integration and evolution of the cichlid mandible: The molecular basis of alternate feeding strategies. Proc. Natl. Acad. Sci. U.S.A. 102, 16287–16292 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bookstein F. L., Integration, disintegration, and self-similarity: Characterizing the scales of shape variation in landmark data. Evol. Biol. 42, 395–426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper W. J., Wernle J., Mann K., Albertson R. C., Functional and genetic integration in the skulls of Lake Malawi cichlids. Evol. Biol. 38, 316–334 (2011). [Google Scholar]
- 34.Linde-Medina M., Boughner J. C., Santana S. E., Diogo R., Are more diverse parts of the mammalian skull more labile? Ecol. Evol. 6, 2318–2324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navalón G., Marugán-Lobón J., Bright J. A., Cooney C. R., Rayfield E. J., The consequences of craniofacial integration for the adaptive radiations of Darwin’s finches and Hawaiian honeycreepers. Nat. Ecol. Evol. 4, 270–278 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Segura V., Cassini G. H., Prevosti F. J., Machado F. A., Integration or modularity in the mandible of canids (Carnivora: Canidae): A geometric morphometric approach. J. Mamm. Evol. 28, 145–157 (2021). [Google Scholar]
- 37.Felice R. N., Goswami A., Developmental origins of mosaic evolution in the avian cranium. Proc. Natl. Acad. Sci. U.S.A. 115, 555–560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porto A., Shirai L. T., de Oliveira F. B., Marroig G., Size variation, growth strategies, and the evolution of modularity in the mammalian skull. Evolution 67, 3305–3322 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Santana S. E., Lofgren S. E., Does nasal echolocation influence the modularity of the mammal skull? J. Evol. Biol. 26, 2520–2526 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Sanger T. J., Mahler D. L., Abzhanov A., Losos J. B., Roles for modularity and constraint in the evolution of cranial diversity among Anolis lizards. Evolution 66, 1525–1542 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Black C. R., Berendzen P. B., Shared ecological traits influence shape of the skeleton in flatfishes (Pleuronectiformes). PeerJ 8, e8919 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer A. R., From symmetry to asymmetry: Phylogenetic patterns of asymmetry variation in animals and their evolutionary significance. Proc. Natl. Acad. Sci. U.S.A. 93, 14279–14286 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schreiber A. M., Asymmetric craniofacial remodeling and lateralized behavior in larval flatfish. J. Exp. Biol. 209, 610–621 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Youson J. H., “First metamorphosis” in Fish Physiology, Hoar W. S., Randall D. J., Eds. (Academic Press, San Diego, 1988), pp. 135–196. [Google Scholar]
- 45.Friedman M., The evolutionary origin of flatfish asymmetry. Nature 454, 209–212 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Fox C. H., Gibb A. C., Summers A. P., Bemis W. E., Benthic walking, bounding, and maneuvering in flatfishes (Pleuronectiformes: Pleuronectidae): New vertebrate gaits. Zoology (Jena) 130, 19–29 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Corn K. A., Farina S. C., Summers A. P., Gibb A. C., Effects of organism and substrate size on burial mechanics of English sole, Parophrys vetulus. J. Exp. Biol. 221, jeb176131 (2018). [DOI] [PubMed] [Google Scholar]
- 48.McKee A., MacDonald I., Farina S. C., Summers A. P., Undulation frequency affects burial performance in living and model flatfishes. Zoology (Jena) 119, 75–80 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Harrington R. C., et al., Phylogenomic analysis of carangimorph fishes reveals flatfish asymmetry arose in a blink of the evolutionary eye. BMC Evol. Biol. 16, 224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Futch C. R., Topp R. W., Houde E. D., Developmental osteology of the lined sole, Achirus lineatus (Pisces, Soleidae). Contrib. Mar. Sci. 16, 33–58 (1972). [Google Scholar]
- 51.Friedman S. T., et al., Body shape diversification along the benthic-pelagic axis in marine fishes. Proc. Biol. Sci. 287, 20201053 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ribeiro E., Davis A. M., Rivero-Vega R. A., Ortí G., Betancur-R R., Post-Cretaceous bursts of evolution along the benthic-pelagic axis in marine fishes. Proc. Biol. Sci. 285, 20182010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez C. M., Stiassny M. L. J., Can an eel be a flatfish? Observations on enigmatic asymmetrical heterenchelyids from the Guinea coast of West Africa. J. Fish Biol. 91, 673–678 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Maile A. J., May Z. A., DeArmon E. S., Martin R. P., Davis M. P., Marine habitat transitions and body-shape evolution in lizardfishes and their allies (Aulopiformes). Copeia 108, 820–832 (2020). [Google Scholar]
- 55.Wagner G. P., Zhang J., The pleiotropic structure of the genotype-phenotype map: The evolvability of complex organisms. Nat. Rev. Genet. 12, 204–213 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Felice R. N., Randau M., Goswami A., A fly in a tube: Macroevolutionary expectations for integrated phenotypes. Evolution 72, 2580–2594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Álvarez A., Perez S. I., Verzi D. H., The role of evolutionary integration in the morphological evolution of the skull of caviomorph rodents (Rodentia: Hystricomorpha). Evol. Biol. 42, 1–16 (2015). [Google Scholar]
- 58.Hallgrímsson B., et al., Deciphering the palimpsest: Studying the relationship between morphological integration and phenotypic covariation. Evol. Biol. 36, 355–376 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schreiber A. M., Flatfish: An asymmetric perspective on metamorphosis. Curr. Top. Dev. Biol. 103, 167–194 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Meyer D. L., von Seydlitz-Kurzbach U., Fiebig E., Bilaterally asymmetrical uptake of [14C]2-deoxyglucose by the octavo-lateralis complexes in flatfish. Cell Tissue Res. 214, 659–662 (1981). [DOI] [PubMed] [Google Scholar]
- 61.Helling K., Scherer H., Hausmann S., Clarke A. H., Otolith mass asymmetries in the utricle and saccule of flatfish. J. Vestib. Res. 15, 59–64 (2005). [PubMed] [Google Scholar]
- 62.Goldschmidt R., The Material Basis of Evolution (Yale University Press, 1982, reprint of 1940 edition).
- 63.Goldschmidt R., Some aspects of evolution. Science 78, 539–547 (1933). [DOI] [PubMed] [Google Scholar]
- 64.Palmer A. R., Strobeck C., Fluctuating asymmetry as a measure of developmental stability: Implications of non-normal distributions and power of statistical tests. Acta Zool. Fenn. 191, 13 (1992). [Google Scholar]
- 65.Hilton E. J., Johnson G. D., Smith-Vaniz W. F., Osteology and systematics of Parastromateus Niger (Perciformes: Carangidae), with comments on the carangid dorsal gill-arch skeleton. Copeia 2010, 312–333 (2010). [Google Scholar]
- 66.Adams D. C., Otárola‐Castillo E., geomorph: An R package for the collection and analysis of geometric morphometric shape data. Methods Ecol. Evol. 4, 393–399 (2013). [Google Scholar]
- 67.Rohlf F. J., Slice D., Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst. Biol. 39, 40–59 (1990). [Google Scholar]
- 68.Gunz P., Mitteroecker P., Bookstein F. L., “Semilandmarks in three dimensions” in Modern Morphometrics in Physical Anthropology. Developments in Primatology: Progress and Prospects, Slice D. E., Ed. (Springer, 2005), pp. 73–98. [Google Scholar]
- 69.Cheverud J. M., Relationships among ontogenetic, static, and evolutionary allometry. Am. J. Phys. Anthropol. 59, 139–149 (1982). [DOI] [PubMed] [Google Scholar]
- 70.Gould S. J., Allometry and size in ontogeny and phylogeny. Biol. Rev. Camb. Philos. Soc. 41, 587–640 (1966). [DOI] [PubMed] [Google Scholar]
- 71.Loy A., Mariani L., Bertelletti M., Tunesi L., Visualizing allometry: Geometric morphometrics in the study of shape changes in the early stages of the two-banded sea bream, Diplodus vulgaris (Perciformes, Sparidae). J. Morphol. 237, 137–146 (1998). [DOI] [PubMed] [Google Scholar]
- 72.Adams D. C., A method for assessing phylogenetic least squares models for shape and other high-dimensional multivariate data. Evolution 68, 2675–2688 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Rabosky D. L., et al., An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559, 392–395 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Paradis E., Claude J., Strimmer K., APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Sidlauskas B., Continuous and arrested morphological diversification in sister clades of characiform fishes: A phylomorphospace approach. Evolution 62, 3135–3156 (2008). [DOI] [PubMed] [Google Scholar]
- 76.Collyer M. L., Adams D. C., Phylogenetically aligned component analysis. Methods Ecol. Evol. 12, 359–372 (2020). [Google Scholar]
- 77.Adams D. C., A generalized K statistic for estimating phylogenetic signal from shape and other high-dimensional multivariate data. Syst. Biol. 63, 685–697 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Venditti C., Meade A., Pagel M., Multiple routes to mammalian diversity. Nature 479, 393–396 (2011). [DOI] [PubMed] [Google Scholar]
- 79.Adams D. C., Collyer M. L., Multivariate phylogenetic comparative methods: Evaluations, comparisons, and recommendations. Syst. Biol. 67, 14–31 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A., Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Denton J. S., Adams D. C., A new phylogenetic test for comparing multiple high-dimensional evolutionary rates suggests interplay of evolutionary rates and modularity in lanternfishes (Myctophiformes; Myctophidae). Evolution 69, 2425–2440 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Harmon L. J., Schulte J. A. 2nd, Larson A., Losos J. B., Tempo and mode of evolutionary radiation in iguanian lizards. Science 301, 961–964 (2003). [DOI] [PubMed] [Google Scholar]
- 83.Slater G. J., Price S. A., Santini F., Alfaro M. E., Diversity versus disparity and the radiation of modern cetaceans. Proc. Biol. Sci. 277, 3097–3104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harmon L. J., Weir J. T., Brock C. D., Glor R. E., Challenger W., GEIGER: Investigating evolutionary radiations. Bioinformatics 24, 129–131 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Goswami A., Finarelli J. A., EMMLi: A maximum likelihood approach to the analysis of modularity. Evolution 70, 1622–1637 (2016). [DOI] [PubMed] [Google Scholar]
- 86.Helfman G., Collette B. B., Facey D. E., Bowen B. W., The Diversity of Fishes: Biology, Evolution, and Ecology (John Wiley & Sons, 2009). [Google Scholar]
- 87.Langille R. M., Hall B. K., Role of the neural crest in development of the cartilaginous cranial and visceral skeleton of the medaka, Oryzias latipes (Teleostei). Anat. Embryol. (Berl.) 177, 297–305 (1988). [DOI] [PubMed] [Google Scholar]
- 88.Evans K. M., Waltz B., Tagliacollo V., Chakrabarty P., Albert J. S., Why the short face? Developmental disintegration of the neurocranium drives convergent evolution in neotropical electric fishes. Ecol. Evol. 7, 1783–1801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bon M., Bardua C., Goswami A., Fabre A.-C., Cranial integration in the fire salamander, Salamandra salamandra (Caudata: Salamandridae). Biol. J. Linn. Soc. Lond. 130, 178–194 (2020). [Google Scholar]
- 90.Adams D. C., Evaluating modularity in morphometric data: Challenges with the RV coefficient and a new test measure. Methods Ecol. Evol. 7, 565–572 (2016). [Google Scholar]
- 91.Adams D. C., Collyer M. L., Comparing the strength of modular signal, and evaluating alternative modular hypotheses, using covariance ratio effect sizes with morphometric data. Evolution 73, 2352–2367 (2019). [DOI] [PubMed] [Google Scholar]
- 92.Adams D. C., Felice R. N., Assessing trait covariation and morphological integration on phylogenies using evolutionary covariance matrices. PLoS One 9, e94335 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Adams D. C., Collyer M. L., On the comparison of the strength of morphological integration across morphometric datasets. Evolution 70, 2623–2631 (2016). [DOI] [PubMed] [Google Scholar]
- 94.Yandell B. S., Practical Data Analysis for Designed Experiments (CRC Press, 1997), vol. 39. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.