Significance
The mechanisms of neurodegeneration are important targets for future treatments of the devastating motoneuron disease spinal muscular atrophy (SMA). Here, we show an altered signaling network in SMA models and patient cells with a reduced B-Raf expression in the network center. B-Raf is crucial for motoneuron survival, and we present data that B-Raf restoration ameliorates motoneuron loss and symptoms in SMA models.
Keywords: spinal muscular atrophy, SMA, neurotrophic signaling, Raf, 14-3-3
Abstract
Spinal muscular atrophy (SMA) is a motoneuron disease caused by deletions of the Survival of Motoneuron 1 gene (SMN1) and low SMN protein levels. SMN restoration is the concept behind a number of recently approved drugs which result in impressive yet limited effects. Since SMN has already been enhanced in treated patients, complementary SMN-independent approaches are needed. Previously, a number of altered signaling pathways which regulate motoneuron degeneration have been identified as candidate targets. However, signaling pathways form networks, and their connectivity is still unknown in SMA. Here, we used presymptomatic SMA mice to elucidate the network of altered signaling in SMA. The SMA network is structured in two clusters with AKT and 14-3-3 ζ/δ in their centers. Both clusters are connected by B-Raf as a major signaling hub. The direct interaction of B-Raf with 14-3-3 ζ/δ is important for an efficient neurotrophic activation of the MEK/ERK pathway and crucial for motoneuron survival. Further analyses in SMA mice revealed that both proteins were down-regulated in motoneurons and the spinal cord with B-Raf being reduced at presymptomatic stages. Primary fibroblasts and iPSC-derived motoneurons from SMA patients both showed the same pattern of down-regulation. This mechanism is conserved across species since a Caenorhabditis elegans SMA model showed less expression of the B-Raf homolog lin-45. Accordingly, motoneuron survival was rescued by a cell autonomous lin-45 expression in a C. elegans SMA model resulting in improved motor functions. This rescue was effective even after the onset of motoneuron degeneration and mediated by the MEK/ERK pathway.
Spinal muscular atrophy (SMA) is a neurodegenerative disease of newborns, infants, and young adults which preferentially affects lower motoneurons in the ventral horn of the spinal cord. As a consequence, patients suffer from muscle weakness and atrophy often resulting in respiratory insufficiency and early death. SMA is caused by homozygous deletions or mutations of the Survival of Motoneuron 1 (SMN1) gene (1). However, humans harbor the similar SMN2 gene which codes for the same protein but differs in a critical cytosine to thymine exchange within exon seven in an exonic splice enhancer region (2). Consequently, the SMN2 pre-mRNA is insufficiently spliced, resulting in low levels of functional full-length mRNA and protein (3). Thus, SMA is caused by low SMN protein levels. The number of SMN2 gene copies critically modifies the disease phenotype with a low number associated with severe forms and higher numbers with milder forms (4).
SMA is characterized by a neuromuscular phenotype starting in proximal muscles with hypotonia, fatigue, and paralysis. The disease is categorized in five different subtypes based on the clinical presentation (5). A small number of patients suffer from congenital SMA-Type 0 with a prenatal onset and a rapid disease progression. The majority of the patients are classified as severe Type 1. The onset is postnatal between 2 wk and 6 mo of age. Untreated patients are never able to roll or sit independently, and about two-thirds decease within the first 2 y of life. Type 2 patients develop first symptoms between 6 and 18 mo of age and are unable to walk independently. Type 3 patients have mild progressive muscle weaknesses with a normal life-expectancy, and Type 4 patients have some difficulties with gross motor functions only (5).
However, recently approved treatments dramatically changed the clinical situation. Nusinersen, an antisense oligonucleotide, and Risdiplam, a small molecule compound, both correct SMN2 pre-mRNA splicing (6–8). Onasemnogene Abeparvovec is a gene-therapy based on the delivery of a SMN cDNA by an adeno-associated virus (AAV) (9, 10). All treatments substantially enhance the survival and motor functions which will enhance the SMA prevalence. However, clinical and preclinical studies demonstrate that delayed interventions after disease onset led to limited clinical improvements. Moreover, there are a substantial number of nonresponders (7, 11, 12). Onasemnogene Abeparvovec, Nusinersen, and Risdiplam enhance the SMN-protein level thereby termed SMN-dependent treatments. Since SMN levels have already been restored by those drugs, other complementary therapies are needed which do not change the SMN levels: SMN-independent approaches (13). An intervention in the pathomechanisms downstream of SMN reduction is a promising strategy for the development of such SMN-independent approaches. Therefore, it is crucial to understand those mechanisms.
We and others previously identified dysregulated cellular signaling as potential mechanisms of motoneuron degeneration in SMA. Such pathways include the Rho kinase (ROCK) (14–16), the extracellular-regulated kinase (ERK) (17–19), the tumor protein p53 (20, 21), p38 mitogen-activated protein kinase (22), and the c-Jun N-terminal kinase 3 (JNK3) pathways (23), all of which were up-regulated in SMA. However, it is not clear whether and how those pathways interfere with each other. We previously showed a bidirectional crosstalk of the ERK and the ROCK pathways being important for the pathophysiology of SMA mice (18, 19). An isolated and reductionist view on single pathways may indeed obscure a global view on altered signaling in SMA. Network biology allows such a global view, which provides a basis for an informed decision for the most important master regulator of dysregulated signaling and motoneuron degeneration in SMA (13).
Here, we present a network biology approach for the identification of such critical signaling nodes within a network of altered signaling in a SMA mouse model. We identifed B-Raf and 14-3-3 ζ/δ as elements of a central and highly connected signaling hub in an SMA network. It has been previously shown that this is an important signaling hub which provides motoneuron function and survival in response to neurotrophic factors. B-Raf was reduced in SMA mice spinal cords already at presymptomatic stages. This reduction localizes to murine SMA motoneurons and was confirmed in SMA patient-derived motoneurons. Accordingly, we could rescue motoneuron loss and locomotion deficiency in vivo in a Caenorhabditis elegans SMA model by re-expression of the B-Raf homolog lin-45. Inhibition of MEK and ERK homologs, downstream targets of B-Raf, abrogated the neuroprotective function of B-Raf in C. elegans. Importantly, the rescue was obtained after onset of neurodegeneration and in a cell-autonomous, motoneuron-intrinsic manner.
Results
Dysregulated Signaling Pathways in SMA Spinal Cords Form a Network with B-Raf in Its Center.
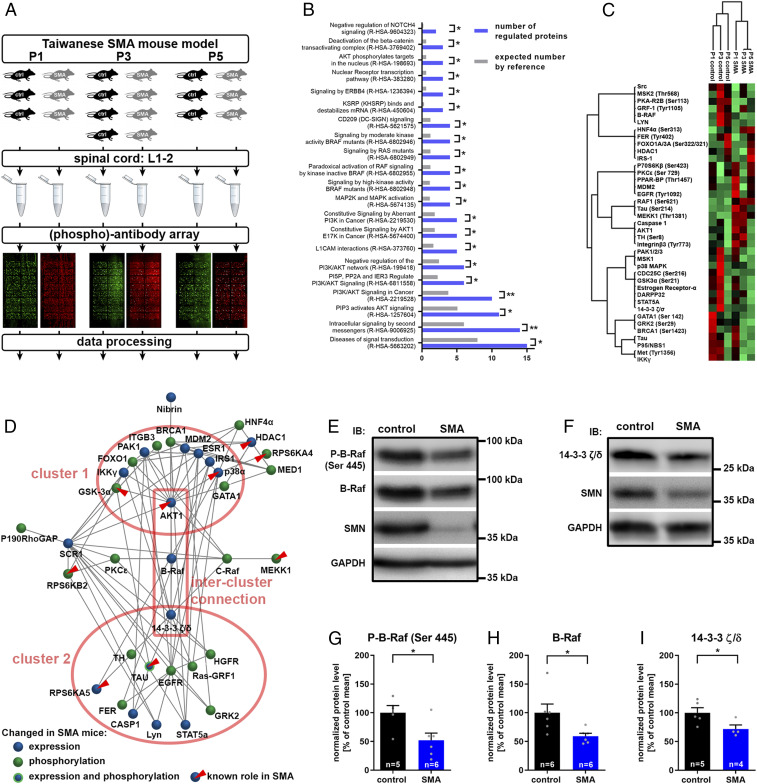
Several dysregulated signaling pathways are involved in motoneuron degeneration in SMA. However, it is not clear whether and how these pathways interact with each other. Therefore, we designed a rigorous experimental approach for a global and unbiased detection of altered signaling at presymptomatic stages. We employed severe Taiwanese SMA mice (24), which are well characterized with regard to the kinetics of the neuromuscular phenotype (19). Postnatal day 1 (P1) and 3 (P3) animals were included for the detection of presymptomatic changes and P5 animals to confirm that alterations persist at the onset of the disease (Fig. 1A). The lumbar spinal cords were screened with an array containing over 1,300 antibodies able to detect and quantify nonphosphoproteins and phosphoproteins involved in diverse signaling pathways. We identified 20 (phospho)proteins that became differentially expressed or phosphorylated between SMA and control conditions in postnatal development (SI Appendix, Fig. S1). An additional 19 (phospho)proteins were persistently dysregulated during the observational period (SI Appendix, Fig. S2). Next, we performed a gene ontology (GO) enrichment analysis for a global view on the signaling pathways altered in SMA mice. The set of dysregulated (phospho)proteins was compared to the total set of (phospho)proteins detectable on the array (Fig. 1B). We used the reactome database as the GO category, giving information about an involvement of distinct signaling pathways (25). Interestingly, 15 signaling proteins in the set of 39 proteins are designated members of the reactome pathway Diseases of signal transduction (R-HSA-5663202), reflecting a significant enrichment compared to the expected number in the reference set (Fig. 1B). GO analysis revealed two major pathways which were linked to dysregulated signaling in SMA: the PI3K/AKT signaling as well as MAPK signaling with an emphasis on Raf kinases (Fig. 1B). Signaling pathways often form cascades where an upstream kinase phosphorylates a downstream target, and a dysregulation of a signaling protein may induce a coregulatory pattern in its downstream targets. Therefore, the dysregulated (phospho)proteins were subjected to a hierarchical clustering analysis based on the normalized signal intensities (Fig. 1C). (Phospho)proteins which were regulated in a similar way, were grouped in ten subclusters. Some of those coregulatory subclusters represent known pathways. For example, Estrogen Receptor-α activates GSK3α via phosphorylation at Ser21 (26), and both the receptor and phospho-GSK3α (Ser21) group to the same cluster (Fig. 1C). However, since inhibitory interactions and compensatory mechanisms play a major role in signaling, the (phospho)proteins must not necessarily be coregulated. Indeed, we did not see a clear clustering by PI3K/AKT and MAPK pathways as suggested by the GO-enrichment analysis (Fig. 1 B and C). Therefore, we performed a network analysis on dysregulated signaling proteins, allowing identification of clusters irrespective of a common regulatory pattern. Moreover, critical hubs may be identified (Fig. 1D). Phosphoproteins and nonphosphoproteins form the nodes; the connections between the nodes represent known physical and functional interactions derived from the BioGrid database (27). The inclusion of functional interactions is important to reduce the impact of false-negatives. Those would induce network gaps if physical interactions were included only. The network forms two clusters with AKT as a hub and 14-3-3 ζ/δ as the second hub; 14-3-3 ζ/δ is a major regulator of Raf-MAPK signaling. Thus, the topology of the network well reflects the enriched GO terms in the set of dysregulated proteins (Fig. 1B). Moreover, we confirmed a number of (phospho)proteins and pathways which have been previously shown to play a role in SMA pathogenesis. Those are distributed over both clusters and include AKT1 (28), MEKK1 (18, 19), HDAC1 (29), GSK3 (30, 31), RPS6 kinases (31, 32), TAU (33), and p38α (22, 23) (Fig. 1D), the latter of which we validated by Western blot analysis showing an increase in total as well as activated p38 in SMA mice spinal cords (SI Appendix, Fig. S3). The two clusters are connected by B-Raf as the central intercluster node (Fig. 1D). B-Raf is a member of the Raf-family serine/threonine kinases and an integral part of the Ras-Raf-MEK-ERK signaling axis. It is essential for survival of motoneurons in response to neurotrophic factors while the other two Raf isoforms, A-Raf and C-Raf, are dispensable (34). The array data revealed a developmental regulation of B-Raf being down-regulated in SMA mice at presymptomatic P3 and at onset P5 (SI Appendix, Fig. S1E). B-Raf directly interacts with 14-3-3 ζ/δ, which critically regulates its activation (35); 14-3-3 ζ/δ regulation revealed a similar developmental expression pattern compared to B-Raf with a presymptomatic down-regulation (SI Appendix, Fig. S1D). We employed immunoblotting to validate their expression in another litter in the thoracic spinal cord (Fig. 1 E and F). B-Raf as well as its binding-partner 14-3-3 ζ/δ were both down-regulated in SMA mice (Fig. 1 H and I). A reduced expression may not necessarily result in a reduced B-Raf activation. B-Raf is activated by a direct interaction with Ras at the plasma membrane. This leads to a conformational change releasing the autoinhibitory domain and to phosphorylation at N-terminal residues (36). Phosphorylation on Serine-445 by the casein kinase 2 reduces the level of autoinhibition and is critical for B-Raf activation (37, 38). Thus, we evaluated total levels of B-Raf phosphorylated at Serine-445. In accordance with an expressional reduction, the housekeeping-normalized level of phosphorylated B-Raf was significantly reduced in the spinal cord of SMA mice compared with controls (Fig. 1 E and G). B-Raf is activated by 14-3-3 ζ/δ binding. Since 14-3-3 ζ/δ expression is reduced in SMA mice, B-Raf activity may be reduced by two different mechanisms—by an expressional reduction impacting phosphorylated B-Raf and/or by a reduced 14-3-3 ζ/δ binding with a subsequent B-Raf inactivation.
Fig. 1.
Systems biology of changed signaling in SMA and validation of critical signaling hubs. (A) Experimental setup for the screening of nonphosphoproteins and phosphoproteins in SMA. Taiwanese SMA mice (SMN−/−; hSMN2tg/0) and control littermates (SMN+/−; hSMN2tg/0) were dissected at the two presymptomatic time points P1 and P3 or at onset P5. Protein lysates of lumbar spinal cord segments 1 through 2 (L1-2) samples were prepared and analyzed on phospho explorer arrays for the quantification of phosphoprotein and total protein content of about 1,300 targets. (B) A list of the whole array targets served as a reference for the GO enrichment analysis in the set of dysregulated targets. The number of proteins with a reactome-pathway GO tag in the set of dysregulated proteins (blue bars) is compared to the expected number of proteins in the reference set (gray bars). The reactome-pathway identifier is given in parentheses. Fisher’s exact t test with *P ≤ 0.05 and **P ≤ 0.01. (C) A hierarchical cluster analysis was performed with normalized array intensities ranging from low intensities (red) to high intensities (green) constraint to a maximal number of 10 clusters. (D) Network analysis of altered (phospho)proteins in SMA mice which are connected by edges based on known interactions from the Biogrid database. The connectivity determined the topology of the network. Nodes which have been previously determined to be involved in SMA pathogenesis are highlighted (red triangles). (E and F) Representative Western blots of Th3-13 spinal cord segments from P5 control and SMA mice. (G–I) Western blots were densitometrically quantified, and the total amounts of (G) phospho-B-Raf (P-B-Raf), (H) B-Raf, and (I) 14-3-3 ζ/δ were calculated using glyceraldehyde 3-phosphate dehydrogenase for normalization. Bar graphs show mean + SEM with a Student’s t test P value given as *P ≤ 0.05.
B-Raf Is Presymptomatically Down-Regulated in SMA Mice Spinal Cords and in Motoneurons.
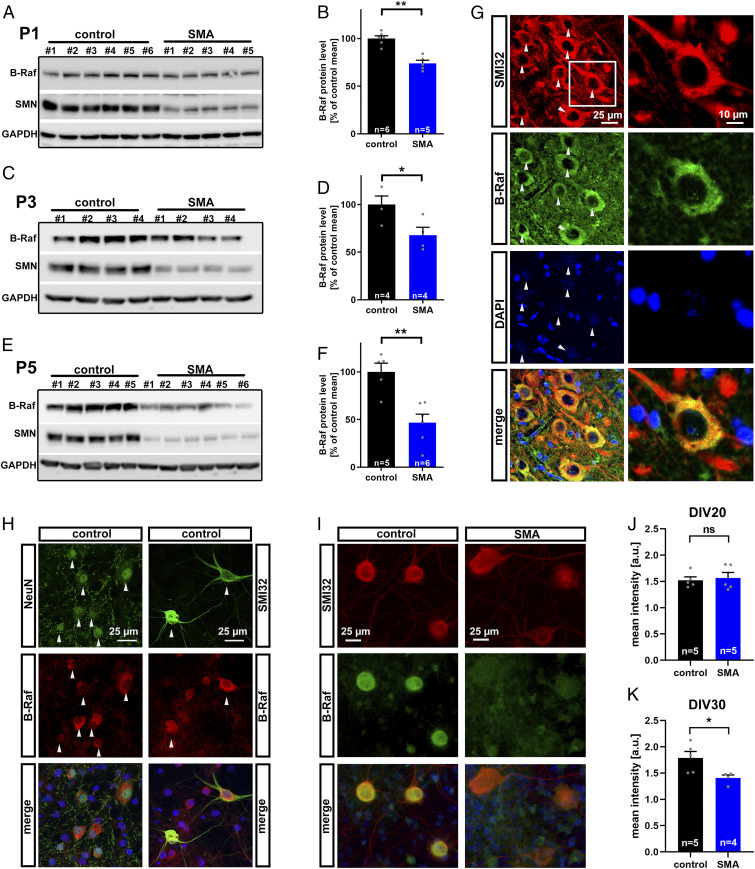
B-Raf is essential for motoneuron survival in response to neurotrophic factors and acts downstream of 14-3-3 ζ/δ being the critical output node in this complex (34). This renders B-Raf down-regulation a candidate mechanism for motoneuron degeneration in SMA. It is possible that B-Raf acts at earlier or at later stages of motoneuron degeneration. Therefore, we evaluated B-Raf expression over time. We used the lumbar spinal cord since those motoneurons are the earliest affected subpopulation in SMA mice (39). At P1, there was slight yet significant reduction in B-Raf expression between SMA and control mice (Fig. 2 A and B). At P3 this effect became more clear (Fig. 2 C and D), while there was a prominent B-Raf reduction in SMA animals below 50% at the disease onset at P5 (Fig. 2 E and F). Those SMA mice do not display gross motoneuron loss, indicating a functional rather than a morphological degeneration (40). This and the presymptomatic kinetics of the B-Raf reduction demonstrate that this is not a consequence of a possible reduction in motoneuron numbers. It has been previously reported that B-Raf is expressed in spinal cord neurons (34). Therefore, we evaluated B-Raf expression in motoneurons of the lumbar spinal cord (Fig. 2G). Importantly, motoneurons displayed a prominent staining for B-Raf compared with neighboring nonmotoneuronal cells which were devoid of a signal beyond background (Fig. 2G). Within the motoneurons, B-Raf localizes to the perikarya omitting SMI32-positive axons and dendrites (Fig. 2G). On the subcellular level, B-Raf was unevenly distributed forming punctae which may reflect activated signaling complexes at the membrane (Fig. 2G). This could be confirmed in primary embryonic spinal cord cultures in which B-Raf was preferentially expressed in neurons with a strong signal in motoneurons while other cells expressed comparatively lower B-Raf levels (Fig. 2H). The B-Raf expression was quantified in motoneurons from SMA mice compared with controls using an automated image analysis (Fig. 2I). While there was no decrease in early cultured motoneurons, there was a significant B-Raf reduction in mature motoneurons (Fig. 2 J and K).
Fig. 2.
Time course of B-Raf reduction and its localization. (A–F) Western blots of lumbar L1-5 spinal cord segments including densitometric quantification of B-Raf expression. (A and B) Litters were prepared at presymptomatic P1 and (C and D) P3 as well as at (E and F) P5 at onset of the first symptoms. (G) Lumbar spinal cord sections of P5 mice were analyzed by immunohistochemistry using the motoneuron marker SMI32 which reveals motoneurons in the ventral spinal cord positive for B-Raf (arrowheads) while SMI32-negative cells express background levels only. Individual motoneurons display a granule B-Raf distribution at the perikaryon (Insets). (H) Confocal images of primary E12.5 spinal cord cultures stained for B-Raf, SMI32, and the neuronal marker NeuN (arrowheads). (I) Primary E12.5 spinal cord cultures from control and SMA embryos stained at day of in vitro 30 (DIV30) for B-Raf and SMI32. (J and K) The B-Raf immunofluorescence intensity was quantified in SMI32-positive motoneurons at (J) DIV20 and (K) DIV30. All bar graphs show mean + SEM with a Student’s t test P value indicated by *P ≤ 0.05 and **P ≤ 0.01 and ns (not significant).
B-Raf Is Down-Regulated in SMA Patient Fibroblasts and iPSC-Derived Motoneurons.
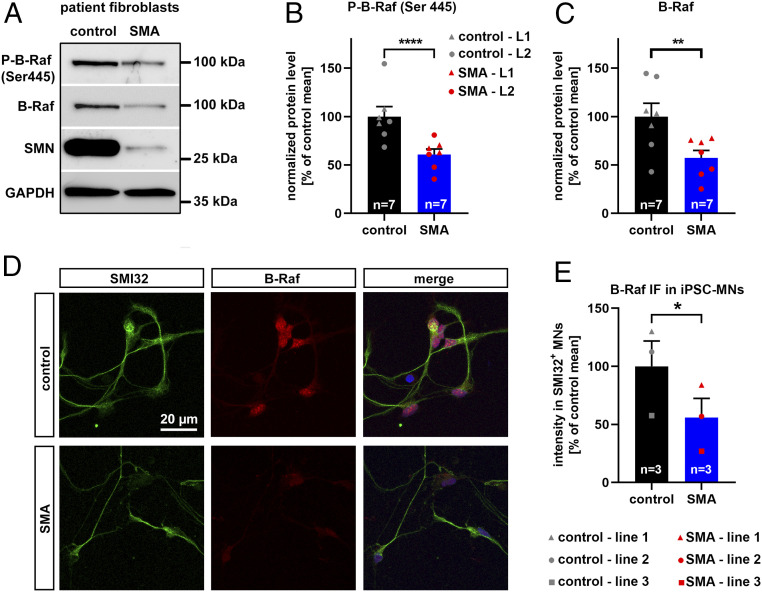
The SMN protein is ubiquitously expressed in all cell types and also ubiquitously reduced in SMA patients. Accordingly, there is growing evidence that SMA is a multisystem disorder (41). To test whether B-Raf is reduced in other cells than motoneurons, we cultured primary fibroblasts from severe SMA type I and SMA type 0 patients as well as from healthy control subjects and used Western blots for quantification of phosphoproteins and total B-Raf (Fig. 3 A–C). Interestingly, the levels of phosphorylated B-Raf as well as the amount of total B-Raf were both reduced in SMA patient fibroblast cultures (Fig. 3 B and C) similar to their reduction in SMA mice spinal cords. Therefore, we employed induced pluripotent stem cells (iPSCs) from SMA type I patients and healthy controls to generate motoneurons (Fig. 3D). Similar to primary spinal cord cultures, B-Raf preferentially localized to SMI32-positive motoneurons (Fig. 3D). Quantification of the signal in motoneurons revealed a clear reduction of B-Raf in patient-derived SMA motoneurons (Fig. 3E).
Fig. 3.
B-Raf expression in SMA patient cells. (A) Representative Western blot of primary fibroblasts from a SMA type I patient and a healthy control subject employing antibodies against phosphorylated B-Raf, total B-Raf, SMN, and GAPDH. (B) Densitometric quantification of the phospho-B-Raf and the (C) B-Raf signal both normalized by GAPDH. A SMA type I line (red dots) and a SMA type 0 line (red triangles) were used together with two different control lines. Biological independent passages were used in a paired experimental design. (D) Representative photomicrographs of iPSC-derived motoneurons identified by SMI32 immunopositivity. Motoneurons were generated from healthy control subjects and SMA type I patients and costained with B-Raf. (E) Three different control iPSC lines (gray data points) and three different SMA iPSC lines (red data points) were differentiated to motoneurons in a paired experimental setup with a control and SMA line treated in parallel each as a biological independent replicate (form of the data point). The B-Raf signal was quantified in SMI32-positive motoneurons. All bar graphs show mean + SEM with a paired-ratio Student’s t test P value indicated by *P ≤ 0.05, **P ≤ 0.01, and ****P ≤ 0.0001.
A B-Raf Homolog Partially Rescues Motoneuron Degeneration and Motor Functions in a C. elegans SMA Model.
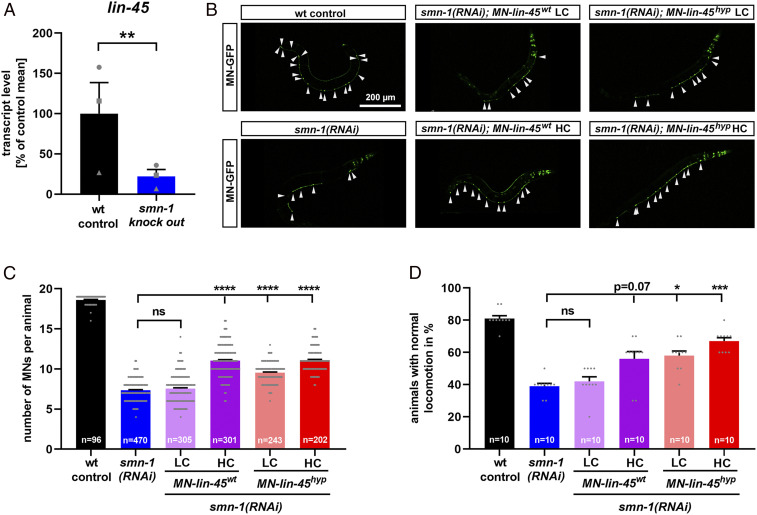
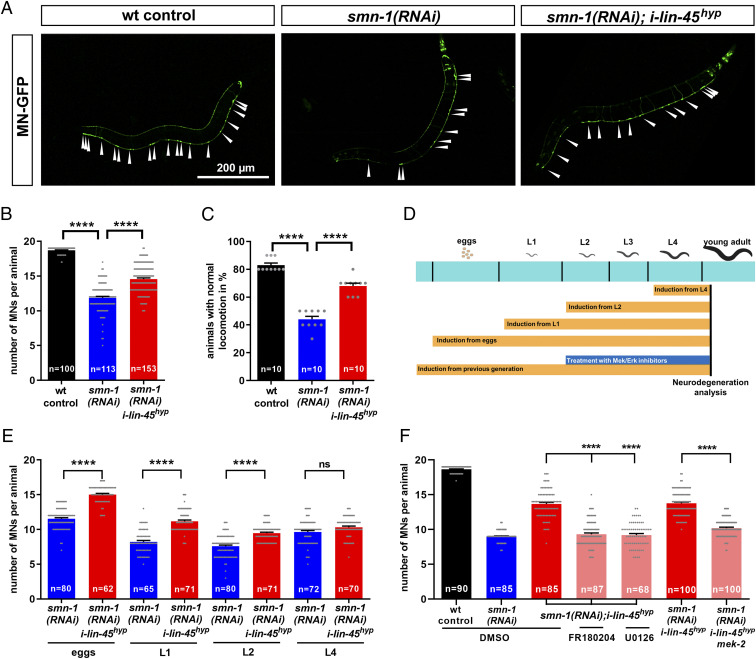
The motoneuron degeneration as a consequence of SMN loss is conserved among a broad range of organisms such as mice, zebrafish, Drosophila, and C. elegans. Similarly, the MAPK/ERK pathway is perfectly conserved, including C. elegans (42), with the B-Raf homolog lin-45 expressed in neurons as well as in motoneurons where it is involved in C. elegans locomotion (43–46). Therefore, we hypothesized that the reduction of B-Raf in response to SMN deficiency is conserved in C. elegans and quantified lin-45 transcript levels in a smn-1 knockout strain, which is the most severe SMA model in C. elegans with low maternal SMN levels only (47). In these presymptomatic animals, we observed a 78% reduction in lin-45 mRNA levels in smn-1 knockout compared to wild type (Fig. 4A), thus recapitulating the observations in SMA mice spinal cords and patient fibroblasts. To demonstrate that B-Raf down-regulation mediates smn-1 associated neurodegeneration, we performed a rescue approach in a SMA model with smn-1 silenced in the 19 motoneurons only, thus avoiding early lethality and pleiotropic effects in other cells (48). Motoneurons in those animals degenerate which can be monitored by the coexpression of green fluorescent protein (GFP) and by locomotion defects, thus underlining the validity of this in vivo model. We hypothesized a cell autonomous mechanism of lin-45–related motoneuron degeneration and re-established lin-45 levels in motoneurons only. Therefore, we created transgenic lines overexpressing the wild-type lin-45 under the motoneuron specific punc-47 promoter (MN-lin-45wt). The same promoter was used to express a hyperactive lin-45, allowing us to maximize possible rescue effects (MN-lin-45hyp). This S312A mutant is devoid of an inhibitory phosphorylation site, thus causing its hyperactivation (49). lin-45wt and lin-45hyp were expressed at different levels in a smn-1(RNAi) background. Confirming previous results, the silencing of smn-1 by motoneuron-specific RNAi induced motoneuron loss (Fig. 4 B and C). Coexpression of wild-type (MN-lin-45wt) or hyperactive lin-45 (MN-lin-45hyp) in motoneurons using a higher concentration (HC) partially rescued motoneuronal loss to similar levels, while only lin-45hyp at a lower concentration (LC) slightly but significantly rescued the neurodegeneration (Fig. 4 B and C). This result was confirmed at the functional level, since locomotion defects observed in smn-1(RNAi) were rescued only by HCs of both constructs (MN-lin-45wt HC and MN-lin-45hyp HC) or by LCs of the hyperactive variant (MN-lin-45hyp LC) (Fig. 4D). Together these results indicate a cell-autonomous conserved role of lin-45 in motoneurons and that lin-45 reduction is a major cause for motoneuron degeneration observed after smn-1 silencing.
Fig. 4.
A B-Raf homolog contributes to motoneuron degeneration in smn-1–deficient C. elegans. (A) Quantitative reverse transcription PCR analysis of the B-Raf homolog lin-45 expression in a C. elegans smn-1 KO mutant compared to wild type. Biological independent replicates (form of the data point) were performed in a paired experimental setup. (B) Representative images of C. elegans animals expressing GFP under the punc-25 promoter, which exclusively drives expression in motoneurons (MN-GFP): arrowheads show individual D-type motoneurons. smn-1 was silenced in D-type motoneurons only [smn-1(RNAi)]. Wild-type animals served as controls (wt control). The motoneuron promoter punc-47 drives the expression of lin-45 either as a wild type (MN-lin-45wt) or as a hyperactive mutant (MN-lin-45hyp) at LC or HC. (C) The motoneurons were identified as distinct GFP-expressing cells and quantified for each animal. (D) The backward locomotion was quantified as the percentage of animals with normal locomotion. All graphs show individual data points as scatterplots with bars and whiskers displaying mean + SEM. Kruskal–Wallis test was combined with Dunn’s multiple comparison test, with ****P ≤ 0.0001, ***P ≤ 0.001, *P ≤ 0.05 and ns (not significant) (C and D) or paired-ratio Student’s t test with **P ≤ 0.01 (A).
The B-Raf Homolog in C. elegans Is Able to Rescue Motoneuron Degeneration in Symptomatic Animals through the MAPK Pathway.
Progressive motoneuron degeneration is a hallmark of SMA which is recapitulated in C. elegans with a smn-1 knockdown in motoneurons only. The neurodegeneration starts from the first larval stage (L1) and increases with animal aging (48). We took advantage of this aspect to test if we could rescue the degeneration by expressing lin-45 at later stages, through heat-inducible transgenics (50). We employed the heat-inducible S312A mutated lines (i-lin-45hyp) since 1) unexpectedly there was no substantial expression of lin-45 from the well-established wild-type construct (i-lin-45wt) compared to the hyperactive variant (SI Appendix, Fig. S4 A–C), and 2) we wanted to maximize lin-45 activity to set up a robust rescue system. Similar to the cell autonomous lin-45 restoration, heat-induced i-lin-45hyp partially rescued motoneuron loss and motor functions in animals with a motoneuron specific smn-1 knockdown (Fig. 5 A–C), while there was no effect at low temperatures or in a wild-type background (SI Appendix, Fig. S4 D and E). While smn-1 silencing is obtained through a constitutive, noninducible, early promoter, we could express lin-45 from different stages using the heat-inducible promoter and characterize lin-45 kinetics in motoneuron degeneration (Fig. 5D). As expected, the most efficient rescue was obtained with a lin-45hyp expression started from the embryonic stage. However, an induction at L1 and even L2 larval stages was sufficient to substantially prevent motoneuron degeneration (Fig. 5E). This is extremely interesting since at L2 stage all 19 D-type motoneurons have already been generated (51), thus demonstrating that lin-45 protects from motoneuron loss rather than interfering with neurogenesis. Moreover, motoneuron degeneration starts at L1 in this model (48), and lin-45 is able to rescue the degeneration even after the onset of motoneuron loss. We further explored downstream mechanisms of this rescue effect using animals maintained under induction temperature from previous generation together with MAPK pathway inhibitors U0126 (MEK/mek-2 inhibitor) and FR180204 (ERK/mpk-1 inhibitor) (18, 52) (Fig. 5D). Inhibitors were applied at L2 stage to reduce general toxic effects and prevent interference with neurogenesis, and under these conditions we confirmed that treating wild-type or smn-1(RNAi) animals did not cause a reduction in motoneuron number (SI Appendix, Fig. S4F). Importantly, inhibition of the lin-45 downstream targets mek-2 or mpk-1 completely abrogated the i-lin-45hyp rescue effect on smn-1(RNAi) (Fig. 5E). We employed an additional genetic approach and generated a triple mutant with a mek-2 mutation together with smn-1(RNAi) and i-lin-45hyp. Since mek-2 knockout is lethal, we used the n1989 mutant, which is a viable weak allele in mek-2, consisting in a S217F substitution (53). Confirming results of pharmacological inhibition, the lin-45 rescue was abrogated by the n1989 mutation in mek-2 (Fig. 5F), which demonstrates that the rescue of motoneuron degeneration in smn-1(RNAi) animals is mediated by the mek-2/mpk-1 or MEK/ERK signaling cascade.
Fig. 5.
The B-Raf homolog lin-45 rescues motoneurons in smn-1–deficient C. elegans via MAPK pathway and after onset of motoneuron degeneration. (A) Representative images of C. elegans animals expressing GFP under the punc-25 promoter, which is exclusively expressed in motoneurons (MN-GFP): arrowheads show individual D-type motoneurons. smn-1 was silenced in D-type motoneurons only [smn-1(RNAi)]. Wild-type animals served as controls (wt control). The temperature-inducible promoter phsp-16 drives the expression of lin-45 hyperactive mutant (i-lin-45hyp). All genotypes were continuously incubated at 25 °C, which induces expression of the lin-45 hyperactive mutant. (B) Motoneurons were identified as distinct GFP-expressing cells and quantified for each animal. (C) The backward locomotion was quantified as the percentage of animals with normal locomotion. (D) Experimental setup of stage-specific temperature induction and lin-45 downstream partner pharmacological inhibition. Strains were grown at 15 °C, and the temperature was increased to 25 °C to induce i-lin-45 expression from different stages: eggs (0 h after eggs were laid), L1 larval stage (∼20 h post eggs laying), L2 larval stage (∼38 h post eggs laying), and L4 larval stage (∼62 h post eggs laying). Motoneuron degeneration was analyzed when animals reached the young adult stage. (E) Motoneurons were identified as distinct GFP expressing cells and quantified in smn-1 silenced animals combined with a temperature-mediated induction of i-lin-45hyp expression from different developmental stages, ranging from eggs to larval stages (eggs, L1, L2, and L4). (F) Motoneurons were identified as distinct GFP-expressing cells and quantified for each animal after treatments of the heat-inducible strain i-lin-45hyp with MEK (U0126, 20 µM) and ERK inhibitors (FR180204, 20 µM) or with a mek-2 mutation. All graphs show individual data points as scatterplots with bars and whiskers displaying mean + SEM. Kruskal–Wallis test was combined with Dunn’s multiple comparison test with ****P ≤ 0.0001 and ns (not significant).
Discussion
The exact mechanisms of motoneuron degeneration in SMA are still unknown. Here, we report the conserved down-regulation of the neurotrophic Raf-family kinase B-Raf signaling hub as a candidate mechanism which contributes to motoneuron degeneration in SMA. This hub localizes to the center of a network of altered signaling proteins, connects two clusters, and may thus be a master regulator. Moreover, expression of 14-3-3 ζ/δ was reduced as well, and both proteins are core components of the Raf/MEK/ERK pathway. A number of nodes around B-Raf have been previously identified as potential drivers of SMA pathology, indicating an integration of different pathomechanisms via Raf-dependent signaling. B-Raf and Akt may directly interact (54), and it is possible that this contributes to the reduced Akt activity observed in total spinal cord lysates of severe SMA mice (28). Increased tau phosphorylation contributes to loss of synapses at motoneurons, neuromuscular junction degeneration, and motoneuron degeneration in SMA mice (33). Interestingly, we found an increase in tau phosphorylation at Ser214, which mediates direct 14-3-3 ζ/δ binding (55). This could be a compensational regulation due to the loss of 14-3-3 ζ/δ. The interaction of tau and 14-3-3 proteins is important for microtubule homeostasis (56), which is altered in SMA (57). Recent reports showed that SMN directly associates with polyribosomes, including the ribosomal protein S6 (Rps6), a component of the small subunit (32). Several ribosomal S6 kinases (RPS6KA4, RPS6KA5, RPS6KB2) are nodes in the network around B-Raf, which indicates a connection of Rsp6 upstream regulators with altered signaling in SMA. Increased p38 levels have been shown in postmortem SMA patient spinal cord samples (23), and an inhibition of p38 prevented motoneuron degeneration in SMA mice via a p53-dependent mechanism (22). Interestingly, 14-3-3 proteins directly interact with p53 (58), and 14-3-3 ζ/δ regulates p38 activity (59), indicating a direct connection of B-Raf/14-3-3 with altered MAPK stress pathways in SMA. We have previously shown an increased activity of the MEK/ERK cascade in SMA mice (17–19). However, this did not localize to motoneurons but to non-neuronal cells (19). Those pleiotropic effects differ from the cell autonomous changes in motoneurons, which are caused by a loss of B-Raf and which depend on functional MEK/ERK downstream signaling.
In SMA mice, total B-Raf protein levels as well as the levels of its activated form were reduced at presymptomatic and onset stages (Fig. 6). This reduction localized to motoneurons in SMA mice and to patient-derived motoneurons where B-Raf is critical for motoneuron survival in response to neurotrophic factors: Primary embryonic motoneurons from B-Raf knockout mice were not able to survive in response to neurotrophic factors while knockouts of A-Raf and C-Raf did not change motoneuron survival (34). The B-Raf reduction was conserved in a C. elegans severe SMA model with reduced levels of the homolog lin-45. To avoid hindrances due to pleiotropic phenotypes observed in this severe model, we used a C. elegans conditional SMA model, through which we were already able to identify smn-1 interactors and factors playing a role in the prevention of neurodegeneration (60–62). In this model we could obtain a dose-dependent and cell-autonomous rescue of the neurodegeneration with a wild-type and a hyperactive version of lin-45. Moreover, motoneuron degeneration was rescued through the MEK/ERK cascade with a delayed lin-45 expression at a time point when motoneuron degeneration already started. This demonstrates that the degeneration of SMN-deficient motoneurons is partially caused by a lack of Raf expression. However, hyperactive lin-45 mutants caused a more robust rescue, which indicates the involvement of other Raf-related mechanisms beyond a reduced expression; 14-3-3 ζ/δ was less abundant in SMA mice as well, and it positively regulates the activity of B-Raf by binding to its activated Serine 445-phosphorylated form. This interaction is critical for the B-Raf kinase activity, and a reduced 14-3-3 ζ/δ expression may hinder full B-Raf activation irrespective of B-Raf restoration by expression of the lin-45 wild type (Fig. 6). The down-regulation of B-Raf and 14-3-3 protein levels could be a far down stream event in the pathomechanism or directly caused by molecular functions of the SMN protein. SMN is involved in spliceosome biogenesis, and it has been hypothesized that this results in impaired splicing and changed expression (63). Another mechanism which could influence expression relies on the interaction of the SMN protein with the zinc finger protein 1 (ZPR1), which prevents the formation of pathogenic DNA structures during transcription (64). Moreover, SMN regulates protein translation by a direct interaction with polyribosomes (32), and one or more of these mechanisms could in principle contribute to reduced B-Raf levels in SMA.
Fig. 6.
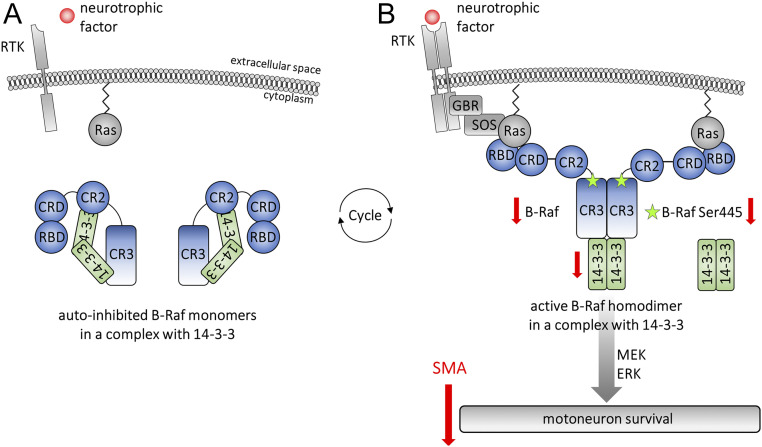
B-Raf and 14-3-3 in neurotrophic signaling. (A) B-Raf contains the catalytic conserved region 3 (CR3), the regulatory CR2, the Cys-rich domain 2 (CRD2), and the N-terminal RAS-binding domain (RBD). In its inactive form, B-Raf is stabilized by 14-3-3 proteins binding in the CR3 and CR2 region. (B) Neurotrophic factor binding to receptor tyrosine kinases (RTK) induces B-Raf recruitment to the membrane via RBD/CRD-Ras interaction and B-Raf phosphorylation at the critical activation site Serine-445 by kinases such as casein kinase 2 (CK2). The activated complex binds 14-3-3 via CR3 region only. Subsequent ATP-binding induces B-Raf kinase activity and the phosphorylation of its downstream target MEK. B-Raf, its activated and phosphorylated form as well as 14-3-3, are down-regulated in SMA motoneurons, which leads to motoneuron degeneration via the MEK/ERK signaling cascade.
Mammals express three Raf isoforms with redundant as well as specific functions. B-Raf is the most potent activator of the MEK/ERK cascade followed by C-Raf and A-Raf (65). Consistently, A-Raf knockout mice survive during adulthood, and C-Raf knockouts die between embryonic day 10.5 (E10.5) and P0. B-Raf knockout mice are most severely affected dying between E10.5 and E12.5 (66–68). C-Raf may compensate for B-Raf loss during embryogenesis but not in postnatal development (69) in which B-Raf is prominently expressed in the mouse brain and spinal cord compared with other tissues (70). Both, B-Raf and C-Raf localize to spinal cord motoneurons and sensory neurons in the dorsal root ganglia. However, only B-Raf is critical for the survival of both neuronal cell types in response to the neurotrophic factors glial cell line-derived neurotrophic factor (GDNF), brain-derived neurotrophic factor (BDNF), and ciliary neurotrophic factor (CNTF) (motoneurons) and nerve growth factor (NGF) (sensory neurons) (34). About 38 germline mutations in the human B-Raf gene are associated with the LEOPARD, Noonan, and cardiofaciocutaneous syndromes. Next to a number of congenital phenotypes in the periphery, those patients exhibit neurological symptoms such as deafness or hypotonia. However, it is unclear whether there is a neuromuscular origin of these symptoms (71).
The reduced B-Raf expression in patient fibroblasts points toward a role of this mechanism also in peripheral SMA pathologies. The SMN protein is ubiquitously expressed and ubiquitously reduced in SMA patients. Not surprisingly, a number of peripheral phenotypes were reported, including liver, kidney, and vascularization defects (72–74). Recently, we reported a reduced longitudinal bone growth caused by an impaired chondrocyte proliferation (75). A reduction in B-Raf levels could be a potential underlying mechanism since B-Raf is a core component of MAPK signaling. In postmitotic neurons, B-Raf does not only provide neurotrophic survival but is also important for growth of sensory neuron projections to the ventral spinal cord, which is necessary for proper circuit formation (69). Impaired motoneuron survival and impaired growth of sensory neuron projections are central hallmarks of SMA pathogenesis. Thereby, a conditional knock-in of SMN in sensory neurons improved motoneuron function and motor functions of the surviving motoneurons, while there were minor effects on survival of motoneurons only (39, 76). Thus, not only motoneuron-intrinsic but also extrinsic, sensory neuron-mediated pathomechanisms contribute to the neuromuscular SMA phenotype. Interestingly, neuronal expression of a mutated gain-of-function B-Raf enhanced axonal regeneration of sensory neurons. More sensory fibers grow into the spinal cord in a dorsal root crush injury model expressing a constitutively active B-Raf (77). This demonstrates the regenerative capacity of such an approach which is a highly desirable property of an SMN-independent strategy for a combination with SMN-dependent treatments such as Nusinersen or Onasemnogene Abeparvovec. The translational potential of a B-Raf rescue is limited. Raf kinases are an essential part of the ERK signaling module acting downstream of a plethora of receptor tyrosine kinases. Dependent on the cellular context, Raf/ERK regulates growth, proliferation, differentiation, and survival. A prominent role of Raf/ERK signaling in mitotic cells is its positive regulation of proliferation. Not surprisingly, B-Raf is a proto-oncogene, and many gain-of-function mutations induce tumorigenesis. However, expression of such mutants alone is not sufficient to induce tumor growth in mice (78).
Network biology is a powerful tool to identify relevant signaling hubs in neurodegenerative diseases. Altered signaling proteins form a network at presymptomatic time points in SMA model mice with B-Raf in its center. The down-regulation of B-Raf is a putative SMA pathomechanism, which can be linked with motoneuron degeneration. The regenerative potential of the B-Raf signaling cluster comprising different regulatory proteins needs to be addressed in future studies.
Materials and Methods
SMA Mice.
Mouse breeding and tissue preparations were conducted according to the German Animal Welfare law approved by the Lower Saxony State Office for Consumer Protection and Food Safety (LAVES, reference numbers 15/1774 and 19/3309). Taiwanese SMA mice [FVB.Cg-Tg(SMN2)2Hung SMN1tm1Hung/J, Jackson Laboratory stock number: 005058] (24) were bred as described previously, resulting in 50% SMA and 50% heterozygous control littermates (29, 79). SMA animals are homozygous for the murine knockout allele and hemizygous for the transgenic allele with two human SMN2-copies. Heterozygous control littermates comprise the same human SMN2 allele but are heterozygous for the murine knockout allele. P1–5 mice were decapitated according to the German animal welfare law, and spinal cord segments were prepared immediately. Samples for array and Western blot analyses were snap-frozen in liquid nitrogen and stored at −80 °C. Samples for immunohistochemical analyses were fixed for 3 h in 4% (wt/vol) paraformaldehyde in PBS. Genotyping was performed as described previously (17).
Phospho Explorer Antibody Array.
The phospho explorer antibody arrays (Full Moon Biosystems, PEX100) were used together with the antibody array assay kit (Full Moon Biosystems, KAS02), which contains buffers for lysis, blocking, coupling, and detection. All procedures were performed according to the manufacturer’s guidelines. Briefly, lumbar segments 1 through 2 (L1-2) were pooled from different animals and lyzed in extraction buffer supplemented with phosphatase inhibitor mixture (PhosSTOP, Roche, 04906837001) and Complete EDTA-free protease inhibitor mixture (Roche, 11873580001). The lysate qualities were monitored via absorption spectra ranging between 200 and 300 nm. Proteins were biotinylated for 2 h with vortexing every 10 min. The proteins were detected after binding and washing using Cy3 conjugated streptavidin (Sigma-Aldrich, S6402). The arrays were scanned with an Agilent Microarray Scanner G2565CA. Each array contained 1,318 antibodies plotted in duplicates. After scanning, the background median was subtracted from the signal median, and the mean was calculated for each duplicate. The overall median of each array was used for normalization. The experimental setup was restricted to a single biological sample (pool) per time point with three different postnatal time points (P1, P3, P5). Given this screening character, statistical significance was detected comparing control and SMA samples irrespective of the postnatal time point, allowing a t test with three replicates using GraphPad Prism (version 8.3.0). Thereby we identified developmentally regulated targets (SI Appendix, Fig. S1). Additionally, we calculated the fold changes between SMA and control samples for each postnatal day by testing them against 1 (not regulated) by t test. This allowed us to identify persistently regulated targets (SI Appendix, Fig. S2).
Primary Spinal Cord Culture.
E12.5 embryos were dissected and prepared as described previously (19). Briefly, embryonic spinal cords were trypsinized and DNA-digested (0.5 g/L trypsin in PBS, trypsin: Worthington, Cat. No: 3707 and 0.5 g/L DNase I in PBS, DNase I: Worthington, Cat. No: 2139). After gentle mechanical tissue disruption, cells were plated on poly-ʟ-lysine–coated coverslips in DMEM (high glucose, Glutamax, Gibco), 5% (vol/vol) fetal calf serum (PAA, B15-001), Penicillin/Streptomycin (Pen/Strep, Gibco, 15140–122), 0.15% (wt/vol) glucose, and AmpB. Medium was changed at day of in vitro 1 (DIV1) to Neurobasal A (Invitrogen, 10888–022), Glutamax (Invitrogen, 35050–038), Pen/Strep (Invitrogen, 15140–122), B27 (Invitrogen, 17504–044), AmpB, 1 μM AraC, 50 ng/mL mGDNF (PeproTech, 450–44), 50 ng/mL hBDNF (PeproTech, 450–02), and 50 ng/mL rCNTF (PeproTech, 450–50) which was changed every 10th day until DIV11 at which cells were analyzed.
Patient and Control Subject Primary Fibroblast Cultures.
The control and the SMA type I patient lines have been established and characterized previously in compliance with the national and institutional ethical guidelines (80). The SMA type I patient line (ML39) was generated from a skin biopsy of an SMA type I patient with a homozygous SMN1 deletion and two SMN2 copies. The SMA type 0 patient cell line was generated in compliance with the Declaration of Helsinki, was approved by the institutional review board, and performed with informed consent of the participant. Fibroblasts were cultured as described previously (14) at 5% CO2 and 37 °C in DMEM (high glucose, Glutamax, Gibco) with 10% (vol/vol) fetal calf serum (PAA, B15-001) and Penicillin/Streptomycin (Pen/Strep, Gibco, 15140–122). Twenty-four hours after seeding, cells were lyzed in RIPA buffer (137 mM NaCl, 20 mM Tris⋅HCl [pH 7], 525 mM β-glycerophosphate, 2 mM EDTA, 1 mM sodium orthovanadate, 1% [wt/vol] sodium desoxycholate, 1% [vol/vol], Triton-X-100, protease inhibitor mixture by Roche, 11873580001) and phosphatase inhibitor mixture (Roche, 04906837001).
iPSC-Derived Motoneuron Cultures.
iPSCs from SMA type I patients and unaffected subjects have been generated as described previously, employing a viral nonintegrating protocol, demonstrating that these cells can differentiate into functional motoneurons (81–84). SMA line 1 harbored a heterozygous SMN1 deletion and the c.888+1G > C substitution on the other allele as well as one SMN2 copy. SMA line 2 is homozygous for the SMN1 deletion and has three SMN2 copies. SMA line 3 has a homozygous SMN1 deletion and two SMN2 copies. SMA and control iPSCs were tested for Mycoplasma spp. (MycoAlert kit, Lonza) and differentiated into motoneurons using a multistep protocol, based on embryoid body formation and addition of specific cytokines as described previously (85). To monitor the proper acquisition of a motoneuron phenotype, cells were fixed and stained utilizing established neuronal and motoneuronal markers. The studies involving human samples were conducted in compliance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and with national legislation and institutional guidelines.
C. elegans Methods.
Nematodes were grown and handled following standard procedures under uncrowded condition on nematode growth medium (NGM) agar plates seeded with Escherichia coli strain OP50 (86). All strains were kept at 20 °C unless otherwise noted. Genetic crosses were made to transfer transgenes and mutations in different backgrounds and were verified by phenotyping the clonal F2s using a dissecting microscope or by genotyping by polymerase chain reaction (PCR). For motoneuron specific expression of lin-45, we obtained transgenic lines. Two phenotypes caused by cell-specific smn-1(RNAi) were analyzed (48)—the death of motoneurons and a defect in locomotion—through microscopy analysis and a behavioral assay. The expression levels of lin-45 were detected by quantitative real-time PCR. To identify the downstream mechanism of lin-45–mediated rescue we used MAPK pathway inhibitors U0126 (MEK/mek-2 inhibitor) and FR180204 (ERK/mpk-1 inhibitor) (18, 52).
Further information is provided in the SI Appendix, Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Lisa Marie Walter and Nora Tula Detering (both Institute of Neuroanatomy and Cell Biology, Hannover Medical School) for their support in breeding the SMA mice; G. Zampi, F. La Rocca (Institute of Biosciences and BioResources), and D. Drongitis (Institute of Genetics and Biophysics) for technical support; the Caenorhabditis Genetic Center (funded by NIH Office of Research Infrastructure Programs P40OD010440) and Prof. M. Sundaram (University of Pennsylvania) for strains; Prof. J. McGhee (University of Calgary) for plasmids; and WormBase. Furthermore, we thank the Deutsche Muskelstiftung (DMS) for funding to P.C. and N. Hensel (Grant SMAPERIPHERAL), SMA Europe for a Post-Doctoral Fellowship to N. Hensel, and funding to P.C., N. Hensel, and E.D.S. (Grant SMATARGET) and the Italian Telethon Foundation (GGP16203) to E.D.S.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007785118/-/DCSupplemental.
Data Availability
Antibody Array Data and CellProfiler pipeline file data have been deposited in Figshare (https://doi.org/10.6084/m9.figshare.13668872) and Open Science Framework (https://doi.org/10.17605/OSF.IO/6PZYK).
References
- 1.Lefebvre S., et al., Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–165 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Lorson C. L., Hahnen E., Androphy E. J., Wirth B., A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 96, 6307–6311 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorson C. L., Androphy E. J., An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 9, 259–265 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Campbell L., Potter A., Ignatius J., Dubowitz V., Davies K., Genomic variation and gene conversion in spinal muscular atrophy: Implications for disease process and clinical phenotype. Am. J. Hum. Genet. 61, 40–50 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finkel R., Bertini E., Muntoni F., Mercuri E.; ENMC SMA Workshop Study Group , 209th ENMC international workshop: Outcome measures and clinical trial readiness in spinal muscular atrophy 7-9 November 2014, Heemskerk, The Netherlands. Neuromuscul. Disord. 25, 593–602 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Hua Y., et al., Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 478, 123–126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finkel R. S.et al.; ENDEAR Study Group , Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 377, 1723–1732 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Naryshkin N. A., et al., Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science 345, 688–693 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Meyer K., et al., Improving single injection CSF delivery of AAV9-mediated gene therapy for SMA: A dose-response study in mice and nonhuman primates. Mol. Ther. 23, 477–487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendell J. R., et al., Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 377, 1713–1722 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Mercuri E.et al.; CHERISH Study Group , Nusinersen versus sham control in later-onset spinal muscular atrophy. N. Engl. J. Med. 378, 625–635 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Al-Zaidy SA, et al., AVXS-101 (Onasemnogene Abeparvovec) for SMA1: Comparative study with a prospective natural history cohort. J. Neuromuscul. Dis. 6, 307–317 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Hensel N., Kubinski S., Claus P., The need for SMN-independent treatments of Spinal Muscular Atrophy (SMA) to complement SMN-enhancing drugs. Front. Neurol. 11, 45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nölle A., et al., The spinal muscular atrophy disease protein SMN is linked to the Rho-kinase pathway via profilin. Hum. Mol. Genet. 20, 4865–4878 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Bowerman M., Beauvais A., Anderson C. L., Kothary R., Rho-kinase inactivation prolongs survival of an intermediate SMA mouse model. Hum. Mol. Genet. 19, 1468–1478 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Hensel N., Rademacher S., Claus P., Chatting with the neighbors: Crosstalk between Rho-kinase (ROCK) and other signaling pathways for treatment of neurological disorders. Front. Neurosci. 9, 198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hensel N., et al., Analysis of the fibroblast growth factor system reveals alterations in a mouse model of spinal muscular atrophy. PLoS One 7, e31202 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hensel N., et al., Bilateral crosstalk of rho- and extracellular-signal-regulated-kinase (ERK) pathways is confined to an unidirectional mode in spinal muscular atrophy (SMA). Cell. Signal. 26, 540–548 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Hensel N., et al., ERK and ROCK functionally interact in a signaling network that is compensationally upregulated in Spinal Muscular Atrophy. Neurobiol. Dis. 108, 352–361 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Simon C. M., et al., Converging mechanisms of p53 activation drive motor neuron degeneration in spinal muscular atrophy. Cell Rep. 21, 3767–3780 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young P. J., et al., A direct interaction between the survival motor neuron protein and p53 and its relationship to spinal muscular atrophy. J. Biol. Chem. 277, 2852–2859 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Simon C. M., et al., Stasimon contributes to the loss of sensory synapses and motor neuron death in a mouse model of spinal muscular atrophy. Cell Rep. 29, 3885–3901.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genabai N. K., et al., Genetic inhibition of JNK3 ameliorates spinal muscular atrophy. Hum. Mol. Genet. 24, 6986–7004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh-Li H. M., et al., A mouse model for spinal muscular atrophy. Nat. Genet. 24, 66–70 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Fabregat A., et al., The reactome pathway knowledgebase. Nucleic Acids Res. 46, D649–D655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardona-Gomez P., Perez M., Avila J., Garcia-Segura L. M., Wandosell F., Estradiol inhibits GSK3 and regulates interaction of estrogen receptors, GSK3, and beta-catenin in the hippocampus. Mol. Cell. Neurosci. 25, 363–373 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Pathan M., et al., FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 15, 2597–2601 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Branchu J., et al., Shift from extracellular signal-regulated kinase to AKT/cAMP response element-binding protein pathway increases survival-motor-neuron expression in spinal-muscular-atrophy-like mice and patient cells. J. Neurosci. 33, 4280–4294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riessland M., et al., SAHA ameliorates the SMA phenotype in two mouse models for spinal muscular atrophy. Hum. Mol. Genet. 19, 1492–1506 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Butchbach M. E. R., et al., Protective effects of butyrate-based compounds on a mouse model for spinal muscular atrophy. Exp. Neurol. 279, 13–26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen P. C., et al., Identification of a maleimide-based glycogen synthase kinase-3 (GSK-3) inhibitor, BIP-135, that prolongs the median survival time of Δ7 SMA KO mouse model of spinal muscular atrophy. ACS Chem. Neurosci. 3, 5–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauria F., et al., SMN-primed ribosomes modulate the translation of transcripts related to spinal muscular atrophy. Nat. Cell Biol. 22, 1239–1251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller N., et al., Non-aggregating tau phosphorylation by cyclin-dependent kinase 5 contributes to motor neuron degeneration in spinal muscular atrophy. J. Neurosci. 35, 6038–6050 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiese S., et al., Specific function of B-Raf in mediating survival of embryonic motoneurons and sensory neurons. Nat. Neurosci. 4, 137–142 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Fischer A., et al., Regulation of RAF activity by 14-3-3 proteins: RAF kinases associate functionally with both homo- and heterodimeric forms of 14-3-3 proteins. J. Biol. Chem. 284, 3183–3194 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Lavoie H., Therrien M., Regulation of RAF protein kinases in ERK signalling. Nat. Rev. Mol. Cell Biol. 16, 281–298 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Tran N. H., Wu X., Frost J. A., B-Raf and Raf-1 are regulated by distinct autoregulatory mechanisms. J. Biol. Chem. 280, 16244–16253 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Ritt D. A., et al., CK2 Is a component of the KSR1 scaffold complex that contributes to Raf kinase activation. Curr. Biol. 17, 179–184 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Mentis G. Z., et al., Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron 69, 453–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powis R. A., Gillingwater T. H., Selective loss of alpha motor neurons with sparing of gamma motor neurons and spinal cord cholinergic neurons in a mouse model of spinal muscular atrophy. J. Anat. 228, 443–451 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamilton G., Gillingwater T. H., Spinal muscular atrophy: Going beyond the motor neuron. Trends Mol. Med. 19, 40–50 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Desideri E., Cavallo A. L., Baccarini M., Alike but different: RAF paralogs and their signaling outputs. Cell 161, 967–970 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Von Stetina S. E., et al., Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol. 8, R135 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaletsky R., et al., The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature 529, 92–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blazie S. M., et al., Alternative polyadenylation directs tissue-specific miRNA targeting in Caenorhabditis elegans somatic tissues. Genetics 206, 757–774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coleman B., Topalidou I., Ailion M., Modulation of gq-Rho signaling by the ERK MAPK pathway controls locomotion in Caenorhabditis elegans. Genetics 209, 523–535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Briese M., et al., Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum. Mol. Genet. 18, 97–104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallotta I., et al., Neuron-specific knock-down of SMN1 causes neuron degeneration and death through an apoptotic mechanism. Hum. Mol. Genet. 25, 2564–2577 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chong H., Lee J., Guan K. L., Positive and negative regulation of Raf kinase activity and function by phosphorylation. EMBO J. 20, 3716–3727 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kao G., Tuck S., Baillie D., Sundaram M. V., C. elegans SUR-6/PR55 cooperates with LET-92/protein phosphatase 2A and promotes Raf activity independently of inhibitory Akt phosphorylation sites. Development 131, 755–765 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Hobert O., Neurogenesis in the nematode Caenorhabditis elegans. WormBook 1–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Z., Wang B., He R., Zhao Y., Miao L., Calcium signaling and the MAPK cascade are required for sperm activation in Caenorhabditis elegans. Biochim. Biophys. Acta 1843, 299–308 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Kornfeld K., Guan K. L., Horvitz H. R., The Caenorhabditis elegans gene mek-2 is required for vulval induction and encodes a protein similar to the protein kinase MEK. Genes Dev. 9, 756–768 (1995). [DOI] [PubMed] [Google Scholar]
- 54.Jablonka S., Wiese S., Sendtner M., Axonal defects in mouse models of motoneuron disease. J. Neurobiol. 58, 272–286 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Sluchanko N. N., Seit-Nebi A. S., Gusev N. B., Phosphorylation of more than one site is required for tight interaction of human tau protein with 14-3-3zeta. FEBS Lett. 583, 2739–2742 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Chen Y., et al., 14-3-3/Tau interaction and tau amyloidogenesis. J. Mol. Neurosci. 68, 620–630 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Bora G., et al., Microtubule-associated protein 1B dysregulates microtubule dynamics and neuronal mitochondrial transport in spinal muscular atrophy. Hum. Mol. Genet. 29, 3935–3944 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Falcicchio M., Ward J. A., Macip S., Doveston R. G., Regulation of p53 by the 14-3-3 protein interaction network: New opportunities for drug discovery in cancer. Cell Death Discov. 6, 126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xing H., Zhang S., Weinheimer C., Kovacs A., Muslin A. J., 14-3-3 proteins block apoptosis and differentially regulate MAPK cascades. EMBO J. 19, 349–358 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mazzarella N., et al., Green kiwifruit extracts protect motor neurons from death in a spinal muscular atrophy model in Caenorhabditis elegans. Food Sci. Nutr. 7, 2327–2335 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Carlos Cáceres I., et al., Automated screening of C. elegans neurodegeneration mutants enabled by microfluidics and image analysis algorithms. Integr. Biol. 10, 539–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Giorgio M. L., et al., WDR79/TCAB1 plays a conserved role in the control of locomotion and ameliorates phenotypic defects in SMA models. Neurobiol. Dis. 105, 42–50 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Burghes A. H., Beattie C. E., Spinal muscular atrophy: Why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 10, 597–609 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kannan A., Jiang X., He L., Ahmad S., Gangwani L., ZPR1 prevents R-loop accumulation, upregulates SMN2 expression and rescues spinal muscular atrophy. Brain 143, 69–93 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pritchard C. A., Samuels M. L., Bosch E., McMahon M., Conditionally oncogenic forms of the A-Raf and B-Raf protein kinases display different biological and biochemical properties in NIH 3T3 cells. Mol. Cell. Biol. 15, 6430–6442 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pritchard C. A., Bolin L., Slattery R., Murray R., McMahon M., Post-natal lethality and neurological and gastrointestinal defects in mice with targeted disruption of the A-Raf protein kinase gene. Curr. Biol. 6, 614–617 (1996). [DOI] [PubMed] [Google Scholar]
- 67.Wojnowski L., et al., Craf-1 protein kinase is essential for mouse development. Mech. Dev. 76, 141–149 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Wojnowski L., et al., Endothelial apoptosis in Braf-deficient mice. Nat. Genet. 16, 293–297 (1997). [DOI] [PubMed] [Google Scholar]
- 69.Zhong J., et al., Raf kinase signaling functions in sensory neuron differentiation and axon growth in vivo. Nat. Neurosci. 10, 598–607 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Barnier J. V., Papin C., Eychène A., Lecoq O., Calothy G., The mouse B-raf gene encodes multiple protein isoforms with tissue-specific expression. J. Biol. Chem. 270, 23381–23389 (1995). [DOI] [PubMed] [Google Scholar]
- 71.Sarkozy A., et al., Germline BRAF mutations in Noonan, LEOPARD, and cardiofaciocutaneous syndromes: Molecular diversity and associated phenotypic spectrum. Hum. Mutat. 30, 695–702 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deguise M. O., et al., Abnormal fatty acid metabolism is a core component of spinal muscular atrophy. Ann. Clin. Transl. Neurol. 6, 1519–1532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allardyce H., et al., Renal pathology in a mouse model of severe spinal muscular atrophy is associated with downregulation of glial cell-line derived neurotrophic factor (GDNF). Hum. Mol. Genet. 29, 2365–2378 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Sintusek P., et al., Histopathological defects in intestine in severe spinal muscular atrophy mice are improved by systemic antisense oligonucleotide treatment. PLoS One 11, e0155032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hensel N., et al., Altered bone development with impaired cartilage formation precedes neuromuscular symptoms in spinal muscular atrophy. Hum. Mol. Genet. 29, 2662–2673 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Fletcher E. V., et al., Reduced sensory synaptic excitation impairs motor neuron function via Kv2.1 in spinal muscular atrophy. Nat. Neurosci. 20, 905–916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O’Donovan K. J., et al., B-RAF kinase drives developmental axon growth and promotes axon regeneration in the injured mature CNS. J. Exp. Med. 211, 801–814 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dankort D., et al., Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat. Genet. 41, 544–552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gogliotti R. G., Hammond S. M., Lutz C., Didonato C. J., Molecular and phenotypic reassessment of an infrequently used mouse model for spinal muscular atrophy. Biochem. Biophys. Res. Commun. 391, 517–522 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brichta L., et al., Valproic acid increases the SMN2 protein level: A well-known drug as a potential therapy for spinal muscular atrophy. Hum. Mol. Genet. 12, 2481–2489 (2003). [DOI] [PubMed] [Google Scholar]
- 81.Corti S., et al., Genetic correction of human induced pluripotent stem cells from patients with spinal muscular atrophy. Sci. Transl. Med. 4, 165ra162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nizzardo M., et al., Spinal muscular atrophy phenotype is ameliorated in human motor neurons by SMN increase via different novel RNA therapeutic approaches. Sci. Rep. 5, 11746 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rizzo F., et al., Key role of SMN/SYNCRIP and RNA-motif 7 in spinal muscular atrophy: RNA-seq and motif analysis of human motor neurons. Brain 142, 276–294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nizzardo M., et al., Synaptotagmin 13 is neuroprotective across motor neuron diseases. Acta Neuropathol. 139, 837–853 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maury Y., et al., Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 33, 89–96 (2015). [DOI] [PubMed] [Google Scholar]
- 86.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Antibody Array Data and CellProfiler pipeline file data have been deposited in Figshare (https://doi.org/10.6084/m9.figshare.13668872) and Open Science Framework (https://doi.org/10.17605/OSF.IO/6PZYK).