Significance
Increasing evidence suggests that evolutionary processes frequently shape ecological patterns; however, most microbiome studies thus far have focused on only the ecological responses of these communities. By using parallel field experiments and focusing in on a model soil bacterium, we showed that bacterial “species” are differentially adapted to local climates, leading to changes in their composition. Furthermore, we detected strain-level evolution, providing direct evidence that both ecological and evolutionary processes operate on annual timescales. The consideration of eco-evolutionary dynamics may therefore be important to understand the response of soil microbiomes to future environmental change.
Keywords: reciprocal transplant, Curtobacterium, adaptation, experimental evolution, ecotypes
Abstract
Microbial community responses to environmental change are largely associated with ecological processes; however, the potential for microbes to rapidly evolve and adapt remains relatively unexplored in natural environments. To assess how ecological and evolutionary processes simultaneously alter the genetic diversity of a microbiome, we conducted two concurrent experiments in the leaf litter layer of soil over 18 mo across a climate gradient in Southern California. In the first experiment, we reciprocally transplanted microbial communities from five sites to test whether ecological shifts in ecotypes of the abundant bacterium, Curtobacterium, corresponded to past adaptive differentiation. In the transplanted communities, ecotypes converged toward that of the native communities growing on a common litter substrate. Moreover, these shifts were correlated with community-weighted mean trait values of the Curtobacterium ecotypes, indicating that some of the trait variation among ecotypes could be explained by local adaptation to climate conditions. In the second experiment, we transplanted an isogenic Curtobacterium strain and tracked genomic mutations associated with the sites across the same climate gradient. Using a combination of genomic and metagenomic approaches, we identified a variety of nonrandom, parallel mutations associated with transplantation, including mutations in genes related to nutrient acquisition, stress response, and exopolysaccharide production. Together, the field experiments demonstrate how both demographic shifts of previously adapted ecotypes and contemporary evolution can alter the diversity of a soil microbiome on the same timescale.
Microbial communities respond quickly to environmental change (1, 2). These responses are typically associated with ecological processes; however, the potential for microbes to evolve and adapt to changes in the environment on ecological timescales remains largely unexplored in natural ecosystems. While evolutionary processes are typically considered over longer timescales, the short generation times, large populations, and high mutation rates indicative of microorganisms may allow for rapid adaptation. Laboratory studies have repeatedly demonstrated rapid evolution of bacterial populations (3) with consequences for organismal physiology (4), yet it remains unclear how these in vitro studies extend to in situ communities (5).
Both ecological and evolutionary processes likely contribute simultaneously (6, 7) to the response of a microbiome to changing environmental conditions (8). However, separating these processes for bacteria can be difficult as they occur along a continuum of temporal and genetic scales. In terms of ecological processes, microbiome composition can respond demographically, as selective forces promote the growth and survival of differentially adapted taxa within the bacterial community. Certainly, many studies have observed such shifts in taxonomic composition of 16S ribosomal RNA (rRNA)–defined taxa in response to simulated global changes (9), and these responses are considered an ecological process (e.g., species sorting). Few examples, however, link these responses to trait differences among bacterial taxa (10–13), precluding direct insights into whether these ecological shifts are due to adaptive differentiation among taxa as a result of past evolutionary divergence. Concurrently, the same selective forces can also shift the abundance of conspecific strains and alter the allele frequencies of preexisting genetic variation, which at this genetic scale is defined as an evolutionary process (14). Finally, evolution through de novo mutation can provide a new source of genetic variation that may allow for further adaptation to environmental change.
In this study, we aimed to capture this continuum of ecological and evolutionary processes that together produce the response of a microbiome’s diversity to environmental change (Fig. 1A). Studying evolution in microbial communities in situ, however, is challenging. For one, variation in highly conserved marker genes used in many microbiome studies (e.g., 16S rRNA) represents distant evolutionary divergences, and thus these regions are too conserved to detect locally adapted lineages (11, 12, 15), let alone recent evolutionary change within communities (16). To overcome this limitation, studies have leveraged shotgun metagenomic data (17, 18) and genome sequences of co-occurring, closely related strains (19, 20) to characterize evolutionary processes (e.g., recombination and gene flow) structuring the genetic diversity of bacterial lineages. However, these studies are also limited by an inherent challenge in microbiome research: delineating population boundaries, the fundamental unit of evolution. While progress has been made in defining microbial species (21–23), the high genetic heterogeneity within diverse microbial communities, such as soils, convolute the boundaries of the fine-scale patterns of genetic diversity within microbial taxonomic units (12). For instance, metagenome-assembled genomes are often composed of a composite of strains forming a large population of mosaic genomes (24) that may not fully capture the diversity of the local population (25). As such, it remains difficult to study evolutionary rates within microbial communities (however, see refs. 26, 27), and the extent and time scale at which evolutionary processes contribute to both standing and new genetic variation relative to ecological processes.
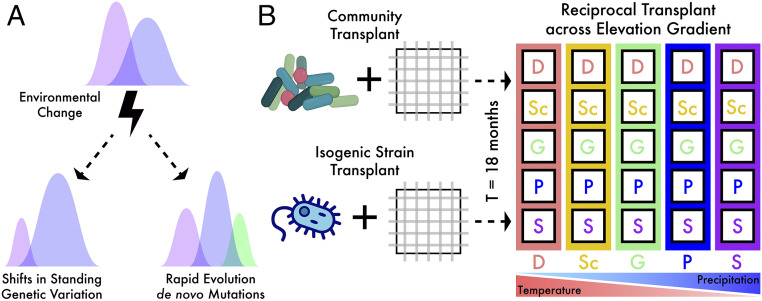
Fig. 1.
Microbial community transplant experiment. (A) Changes in microbial community composition can be due to a continuum of ecological and evolutionary processes. For instance, shifts in standing genetic variation can be attributed to both ecological and evolutionary processes depending on the level of biological resolution, while de novo mutations can be a result from evolutionary adaptation. (B) A schematic of the two parallel transplant experiments at the community and strain level. Inoculated litterbags were transplanted to all sites along an elevation gradient that covaried in temperature and precipitation. Site codes: D=Desert; Sc=Scrubland; G=Grassland; P=Pine-Oak; S=Subalpine.
Here, we asked the following question: can we characterize the ecological and evolutionary processes that are contributing concurrently to the response of a soil bacterial community to a changing environment? To answer this question, we utilized a field-based experimental approach to quantify the influence of both ecological and evolutionary processes on one focal soil bacterium in its natural environment, the genus Curtobacterium (28). Specifically, we transplanted the bacterium across an elevation gradient on a common resource (leaf litter) substrate (29) to assess its response to new climates in two parallel experiments over the same 18 mo time period (Fig. 1B). In both experiments, we used microbial cages [nylon mesh bags that allow for nutrient transport (30)] to manipulate microbial composition while restricting microbial migration to eliminate the introduction of new alleles and/or variants from dispersal (31). A reciprocal transplant design allowed for direct testing of microbial adaptation to abiotic conditions (i.e., moisture and temperature) in a natural setting.
In the first experiment, we conducted a reciprocal transplant of the entire microbial community (32) and tracked the ecological response of Curtobacterium ecotypes (33). A bacterial ecotype is defined as highly clustered genotypic and phenotypic strains occupying the same ecological niche, somewhat equivalent to a eukaryotic species (34). To test the hypothesis that Curtobacterium ecotypes are locally adapted to their climate conditions, we assessed the convergence of ecotype composition in the transplanted communities to that of control communities (those that remained in their native environment; Fig. 1B). We further hypothesized that the demographic shifts were due to differential adaptation to local climates as a result of trait variation among the ecotypes. Thus, we expected that the climate gradient would select for a strong trait–environment relationship (assessed by community-weighted mean (CWM) trait values) as typically observed in plant communities (35, 36).
In parallel, we conducted an in situ evolution experiment by transplanting an isogenic Curtobacterium strain across the same gradient to investigate the potential for rapid evolution on the same timescales. We hypothesized that a variety of genomic mutations would be associated with adaptation to local climate conditions. Therefore, we expected fewer genetic changes when the strain was transplanted to its original environment, the midelevation Grassland site, while the extreme sites of the gradient would impose stronger selective pressures resulting in greater genetic changes. We further expected to observe parallel mutations among replicates within a site, which would be indicative of adaptive events (37). Variation in such mutations across sites would suggest selection differences across the climate gradient. Together, the two experiments capture the simultaneous effects of both ecological and evolutionary processes on the response of a soil bacterium to new climates in the field.
Results
Overall Community Composition and Curtobacterium Abundances.
The five study sites span an elevation gradient that varies in climate (temperature and precipitation), plant vegetation and the leaf litter chemistry, and microbial community composition in the leaf litter layer of soil (SI Appendix, Fig. S1). Leaf litter microbial communities from each site were transplanted in a fully reciprocal manner on a common (Grassland) litter substrate to disentangle the effects of the abiotic factors (site) and the initial microbial community (inoculum). We first assessed microbial community composition with 120 shotgun metagenomic libraries (20 survey samples from nearby leaf litter outside of the litterbags + 100 litterbag samples 18 mo after transplantation) to compare to the overall community shifts assayed with 16S rRNA amplicons (32). Similar to prior observations, the bacterial communities shifted to reflect the surrounding abiotic environment, clustering largely by site after 18 mo (SI Appendix, Fig. S2A; permutational multivariate analysis of variance [PERMANOVA] P < 0.01) and explained 44% of variation in bacterial composition. Community composition also displayed significant legacy effects, with the initial inoculum accounting for 7.8% of the variation in bacterial composition 18 mo after transplantation (P < 0.01).
After confirming the demographic shifts at the community-level, we next investigated the response of Curtobacterium abundance. Outside of the litterbags (in the surrounding leaf litter), Curtobacterium was the third most abundant genus across all sites and the most abundant genus at the midelevation site, comprising 15.7 ± 1.7% of the Grassland community (green dashed lines in SI Appendix, Fig. S2B). Conversely, the relative abundance of Curtobacterium decreased at the ends of the climate gradient, comprising 1.3 and 1.5% of the total community in the Subalpine and Desert sites, respectively. Based on flow cytometry cell counts, we estimate that the absolute abundance of Curtobacterium was 1.4 × 108 cells per g of leaf litter in the Grassland site, an order of magnitude higher than all other sites (SI Appendix, Fig. S2B). Curtobacterium abundance also shifted in response to new environments within the transplanted litterbags. In general, Curtobacterium abundance increased in the transplant experiment, reaching an average abundance of 2.2 × 108 cells g−1 across all sites, likely reflecting an increase in performance on the common Grassland litter substrate in the transplanted bags. At the same time, Curtobacterium abundance responded to reflect the new climate conditions of the transplanted sites; abundance varied significantly in the litterbags across sites (ANOVA; F4,74 = 7.4, P < 0.001) but not by the initial inoculum or a site-by-inoculum interaction (P > 0.05).
Adaptive Differentiation of Curtobacterium Ecotypes to Local Climate.
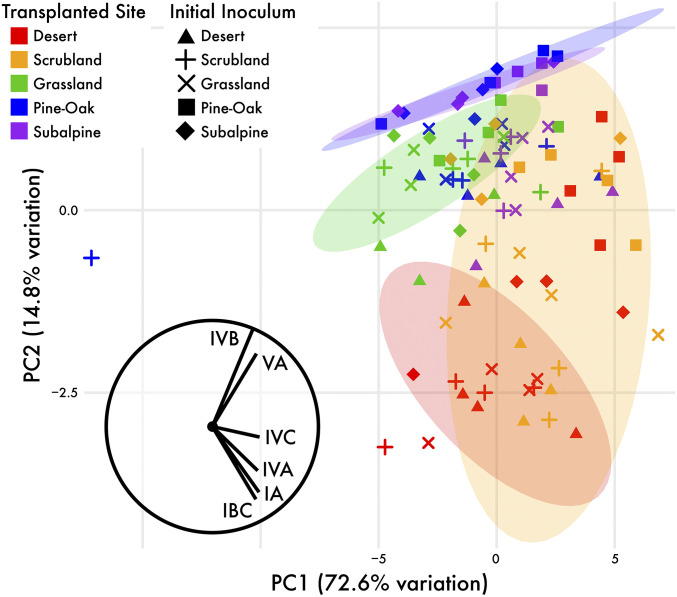
We next tracked shifts in Curtobacterium ecotypes when transplanted to new climates on a common leaf litter substrate. Given that the 16S rRNA gene is too conserved to delineate Curtobacterium ecotypes (12), we targeted 830 single-copy core genes within the metagenomic libraries to characterize their relative abundances. After 18 mo, Curtobacterium ecotypes shifted to reflect the native communities in the litterbags, demonstrating that the ecotypes are differentially adapted to the abiotic environment (i.e., temperature and precipitation). Specifically, when the different litter communities were placed in the same environment onto a common Grassland litter, the ecotype abundances converged toward the ecotype abundances of the native communities (Fig. 2). This trend was confirmed by the strong effect of site on ecotype composition, accounting for 24.5% of the variation (PERMANOVA; P < 0.01). The effect of the initial inoculum was still pronounced in driving ecotype composition after 18 mo, accounting for 13.4% of the total variation (P < 0.01). Indeed, the communities that were furthest from converging on the native ecotype composition tended to be those from the opposite ends of the gradient, resulting in a site-by-inoculum interaction explaining an additional 12.5% of the variation (P > 0.05). For instance, the Pine–Oak and Subalpine communities transplanted to the Desert remained quite different in ecotype composition from the Desert communities that stayed in their native environment (i.e., the red squares and diamonds outside the red ellipse in Fig. 2).
Fig. 2.
The composition of Curtobacterium diversity along the climate gradient after 18 mo of transplant. Principal Component Analysis (PCA) ordination plot depicting Curtobacterium composition in each metagenomic sample, colored by site with shapes representing inoculum. PC1 explains the largest amount of variation, which is driven by differences in absolute Curtobacterium abundances in the samples (SI Appendix, Fig. S2B). The ellipses are 75% CIs around the inoculum samples that remained in their native site (e.g., the red ellipse represents the spread of the red triangles, and the purple ellipse represents the spread of the purple diamonds). The vectors represent the contribution of the top six abundant ecotypes to compositional differences along the two axes.
The convergence of ecotype composition toward the native communities suggests fitness differences among the ecotypes with respect to local moisture and temperature conditions. To examine the traits that might underlie these fitness differences, we considered the relationship between CWM trait values of Curtobacterium ecotype composition across the climate gradient based on six traits previously assayed in vitro from representative isolates (33). These traits were originally chosen as they were hypothesized to relate to the ability of the strains to survive and reproduce on leaf litter across a range of temperatures and moisture stress. In the survey samples, CWM trait values exhibited a weak relationship to either average surface soil temperature or total precipitation (SI Appendix, Fig. S1) across the elevation gradient (Fig. 3A; mean r2 = 0.11 ± 0.10; all P > 0.05), suggesting factors other than climate are contributing to distributional patterns. However, in the transplanted litterbags with a controlled litter substrate, CWM trait values were strongly correlated with both temperature and precipitation (mean r2 = 0.35 ± 0.18; all P < 0.05).
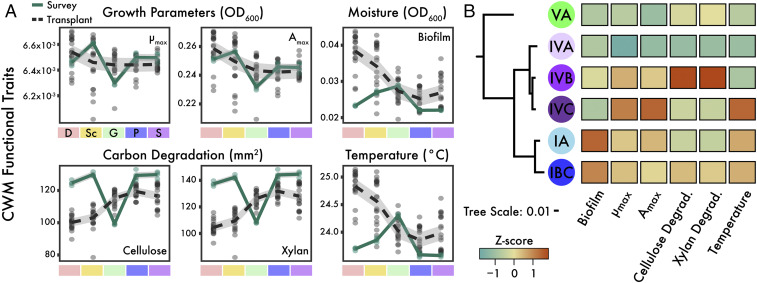
Fig. 3.
Correlation of Curtobacterium functional traits to field sites. (A) CWM values for Curtobacterium ecotypes’ traits along a climate gradient for survey (surrounding leaf litter outside the litterbags) and after 18 mo of transplant to the sites denoted on the x-axis. Smoothed averages (lines) were calculated from locally weighted smoothing with CIs (colored areas). (B) A heatmap showing normalized production of biofilms, growth measurements (μmax = maximum growth rate; Amax = maximum growth [OD600]), degradation for cellulose and xylan, and optimal temperature across all assays between the top six abundant Curtobacterium ecotypes. Phylogeny derived from multilocus alignment of Curtobacterium core genes.
We also noted an apparent trade-off in CWM values along the climate gradient. Traits associated with rapid growth and drought resistance (increased biofilm production and higher temperature preference) decreased when transplanted to wetter and colder (higher elevation) sites, while resource acquisition traits (cellulose and xylan degradation) showed an inverse relationship (Fig. 3A). In particular, after the 18 mo transplant, ecotypes IA and IBC were relatively abundant in the hotter, drier Desert, and Scrubland sites (Fig. 2 and SI Appendix, Fig. S3B). These ecotypes were the best biofilm producers, a trait associated with desiccation and water stress, and also preferred relatively warmer temperatures (Fig. 3B). Conversely, ecotypes IVB and VA were relatively more abundant in the colder, wetter Pine–Oak, and Subalpine sites (Fig. 2). While both ecotypes preferred lower temperatures, ecotype IVB was the best degrader of cellulose and xylan, two common polymeric carbohydrates in leaf litter (Fig. 3B). Collectively, these phenotypic patterns provide evidence that Curtobacterium ecotypes are differentially adapted to their local climates and, as a result, have the capacity to respond quickly to new climate conditions.
Rapid Evolution of a Single Curtobacterium Strain.
Variation among ecotypes represents distant diversification events over millions of years (38). As such, identification of adaptive changes arising from de novo genetic mutations would not be detected at the ecotype level. Therefore, in a second experiment, we tracked evolutionary changes of a single, isogenic strain across the same environmental gradient. The native Grassland strain (MMLR14002) belongs to the dominant Curtobacterium IBC ecotype (SI Appendix, Fig. S3A) and can degrade the polymeric carbohydrates found in leaf litter, cellulose and xylan, even at high temperatures (i.e., 37 °C) (12). We inoculated this strain onto sterile leaf litter and deployed the litterbags to all sites alongside the community transplant experiment.
We first wanted to validate the approach of the in situ evolution experiment and confirm that the strain established and proliferated in response to the transplant experiment. We sequenced the Curtobacterium population in the litterbags at the final time point (at 18 mo) across all sites and, in 20 out of 24 samples, reconstructed de novo assemblies of the ancestral strain (all >99.9% average nucleotide identity match). To track shifts in the strain’s abundance over the course of the experiment, we selected one bag from each site for additional sequencing across the course of the experiment. After normalizing the metagenomic data with flow cytometry cell counts, we found that Curtobacterium population sizes initially increased at the midelevation sites after the first 6 mo, while the extreme climate sites (i.e., Desert and Subalpine) saw a reduction in Curtobacterium abundance, suggesting more cell death than growth. After 6 mo, populations rapidly recovered and, by the end of the 18 mo transplant, all populations converged on a similar population size of 1 × 108 cells g−1 (SI Appendix, Fig. S4A), comparable to the total Curtobacterium abundances observed in the community transplant bags. Based on replication rates inferred from bidirectional sequencing coverage from the origin of replication (39), we estimated that 28.8 ± 16.6% of the population was actively replicating over the duration of the transplant experiment (SI Appendix, Fig. S4B).
We next evaluated whether the strain evolved over the course of the experiment. To do so, we deep sequenced the same litterbag over time from each site (1 bag × 5 sites × 3 time points = 15 total samples). This timeseries allowed for quantification of population-wide patterns of polymorphisms at each site. In total, we observed 8,435 unique variants across all sites and time points; however, many of these population polymorphisms were detected at extremely low frequencies (freqmean = 4.1 ± 0.4%; SI Appendix, Fig. S5A). After removing low-confidence variants, we detected a total of 268 mutations that occurred in 70 unique base pair (bp) positions in the ancestral genome. A majority of these mutations were introduced in the first 6 mo with an initial spike in mutation accumulation at the sites on the ends of the climate gradient (i.e., Desert and Subalpine). Notably, the “native” Grassland site, from which the strain was originally isolated and the common litter substrate was collected, experienced the lowest number of detectable mutations in this initial period. However, the total number of mutations detected at the Desert and Subalpine sites then decreased over time, and the number of mutations at the 18 mo time point was indistinguishable across sites (SI Appendix, Fig. S5B). The decreasing number of detectable mutations over time suggests that either mutations occurred at higher frequencies in the first 6 mo period, perhaps reflecting that selective pressures were stronger upon initial transplantation, or that drift was more pronounced during this period.
Of the 268 mutations observed in the timeseries litterbags, 52% occurred in intergenic regions, resulting in 107 short insertions and deletions (indels) and 33 base substitutions. The other 48% of mutations occurred in coding regions and, in particular, three coding regions that were consistently mutated by the integration of large indels (>40 bp) in nearly all samples (Table 1). While one of the three large indels was unannotated, the other two genes are thought to be related to nutrient acquisition, including a zinc transporter gene and a lactose transport system permease protein. Other mutations observed at all sites and time points included a nonsynonymous single-nucleotide polymorphism (SNP) in the relA gene, suggesting selection for this mutation being due to the transplant experiment. We also identified site-specific SNPs that were maintained over time. For example, a nonsynonymous mutation in the lutR gene, a transcriptional regulator, was detected at 12 mo in the Subalpine site and persisted in the population for the duration of the experiment (Table 1). While few mutations were observed across all time points within a site, a strong pattern of parallel mutations indicates that least some of the polymorphisms are adaptive.
Table 1.
Summary of mutations in coding regions found in the isogenic transplant litterbags
| Gene Name | Position | Specific mutation | Type of mutation | Occurrences in samples† | Mutation frequency‡ | Gene description |
| relA → | 732,949 | G → T | G196V | All samples | 62.0 ± 20.0% | Bifunctional (p)ppGpp |
| nonsynonymous | (GGC → GTC) | synthase/hydrolase | ||||
| marR → | 1,513,025 | A → C | A102A | Scrubland T1; | 32.1 ± 7.0% | Multiple antibiotic |
| synonymous | (GCA → GCC) | Pine–Oak T1-T3; | resistance protein | |||
| Subalpine T1-T3 | ||||||
| orf01789 ← | 1,895,438 | Δ54 bp | — | Desert T1-T3; | 12.8 ± 1.8% | Zinc transporter |
| deletion | Scrubland T1,T3; | |||||
| Grassland T1-T3; | ||||||
| Pine–Oak T1,T3; | ||||||
| Subalpine T1,T3 | ||||||
| lutR → | 2,623,478 | G → A | G77E | Subalpine T2-T3 | 13.4 ± 0.7% | HTH-type transcriptional |
| nonsynonymous | (GGG → GAG) | regulator | ||||
| wcaJ → | 3,114,825 | C → T | Q182* | Desert T1-T3; | 30.6 ± 6.2% | DP-glucose:undecaprenyl- |
| nonsense | (CAG → TAG) | Scrubland T2-T3; | phosphate glucose-1- | |||
| Grassland T1-T3 | phosphate transferase | |||||
| lacF ← | 3,355,556 | Δ46 bp | — | all samples | 14.7 ± 2.8% | Lactose transport system |
| deletion | permease protein |
T1 = population sample collected at 6 mo, T2 = 12 mo, and T3 = 18 mo.
Frequency calculated from samples where mutation was detected.
De Novo Mutations in Evolved Isolates.
To validate the results from the population polymorphisms, we also sampled the isogenic litterbags every 6 mo for reisolation of the ancestral strain and independently sequenced the isolates (reducing the chances that similar mutations were due to sequencing contamination). In total, we identified 783 de novo mutations across 112 evolved isolates that resulted in 121 unique bp mutations, including SNPs, deletions, and small insertions (Fig. 4A). Similar to the population-wide polymorphisms, 59.1% of the total mutations occurred in intergenic regions while mutations in coding regions were observed in 88 unique genes.
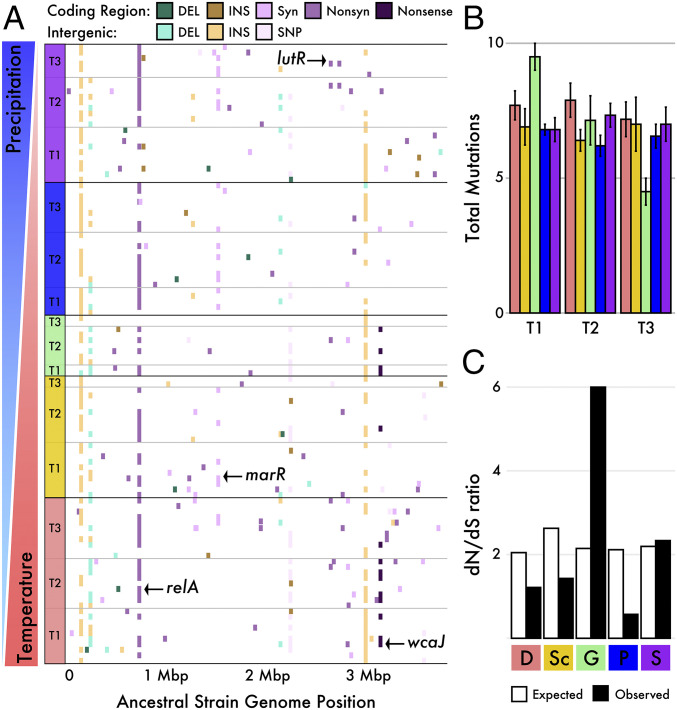
Fig. 4.
Mutation distribution across a Curtobacterium ancestral strain after transplant across the elevation gradient. (A) Mutations in 112 evolved clones isolated from five sites along the climate gradient at 6 (Time point 1), 12 (T2), and 18 mo (T3) intervals. Includes 30 strains from the Desert (D) site, 22 at Scrubland (Sc), 11 at Grassland (G), 24 at Pine–Oak (P), and 25 at Subalpine (S). Nonrandom mutations also observed in the population data are denoted for synonymous (syn), nonsynonymous (nonsyn), and nonsense mutations. (B) The total number of mutations per mutant strain by time point and site. (C) Lack of evidence for positive selection in the evolved populations in the base substitutions of expected (from base change spectrum) and observed dN/dS ratios.
Given the high number of total mutations, we sought to investigate whether these mutations were associated with site adaptation. While there were no differences in the total number of mutations by site or time point (Fig. 4B and SI Appendix, Fig. S6A; ANOVA; P > 0.05), there was a reduction in the average number of mutations in the “native” Grassland site after 18 mo, mirroring the results from the population sequencing of the litterbags (SI Appendix, Fig. S5B). Next, we investigated evidence for positive selection by assessing the ratio of nonsynonymous to synonymous mutations (dN/dS). At each site except for the Pine–Oak site, there was an excess of nonsynonymous relative to synonymous mutations (dN/dS > 1). However, once corrected for the sequence composition of genomic sites at risk for synonymous mutations in the ancestral genome, there was no deviation of the observed dN/dS ratios from those expected by chance (Fig. 4C; two-tailed binomial test, all P > 0.05).
Based on environmental variation across sites (SI Appendix, Fig. S1), certain functional genes might confer a selective advantage in a particular environment. To investigate this, we restricted our analysis to only mutations that resulted in amino acid changes in coding regions (i.e., indels and nonsynonymous or nonsense SNPs). There were minimal differences between the total number of mutations in amino acid sequences (average 1.5 mutations) and in the number of mutations by functional gene group (ANOVA; P > 0.05). For instance, a similar number of mutations were found across sites in genes related to signal transduction, transcription regulation, and cell membrane biosynthesis (SI Appendix, Fig. S6B). However, certain functional gene groups were unique to specific sites. Specifically, mutations in lipid metabolism, cell motility, and inorganic ion transport were only detected at the Desert site (SI Appendix, Fig. S6B), perhaps related to the drier and warmer conditions. Mutations in genes related to carbohydrate metabolism were also only observed at the ends of the elevation gradient (i.e., Desert and Subalpine).
Parallel Evolution Suggests Mutations Are Adaptive.
We next validated the presence of the population-wide mutations (Table 1) in our evolved isolates and examined their distribution across replicates within and across sites. Parallel mutations shared across sites may indicate adaptation to the transplant experiment (the litterbag conditions and common grass litter substrate), while mutations observed across replicates but unique to one or more sites provide candidate genes for environmental adaptation. For example, the nonsynonymous mutation in the relA gene, a stress-response protein that triggers amino acid biosynthesis rather than cell growth, appeared in all sites and time points within the population samples, and we observed 66/112 evolved strains with a mutation in this gene. Indeed, the probability of randomly observing the same mutation in the relA gene in either data set is extremely low (P < 2.0 × 10−35), suggesting that relA was under strong selection during the transplant experiment. In contrast, a parallel nonsense mutation in the wcaJ gene was only associated with the drier and warmer sites (i.e., Desert, Scrubland, and Grassland). The wcaJ variant was introduced within the first 6 mo of the transplant experiment at the Desert and Grassland sites (12 mo at the Scrubland) and remained at a 35.7 ± 6% frequency within the population at the final time point. Similarly, in the evolved isolates, we only detected this mutation in 14 strains isolated predominantly from the Desert and Grassland sites (Fig. 4A). The wcaJ gene provides another example of a nonrandom mutation (P < 6.1 × 10−20), indicating that this exopolysaccharide-producing enzyme may allow for adaptation to the extreme drought and high temperatures indicative of these sites (SI Appendix, Fig. S1C).
While both parallel evolution and selective sweeps are landmarks of adaptive evolution, most of the detected mutations remained at low frequencies (SI Appendix, Fig. S5A), and we did not observe enrichment in nonsynonymous mutations across sites (Fig. 4C). One possible reason for this result is that site-specific advantageous mutations did not proliferate and sweep through the population in the litterbag. For instance, based on in vitro growth rate measurements on Grassland litter leachate (SI Appendix, Fig. S7), we estimate that the ancestral population could replicate at a maximum rate of 5.45 generations per day, although we suspect that the growth rate in the field is likely much lower. Extrapolating from this estimate, the populations underwent, at most, 980 generations every 6 mo or <3,000 generations in total. We deduced that Curtobacterium population reached an estimated mutation rate of 5.28 × 10−7 mutations per base per year or 2.65 × 10−10 mutations per base per generation. These metrics also allowed for estimates of genomic diversity (θW) at each site, with the “native” Grassland site harboring the highest measure of diversity (θW = 2.6) while the higher elevation sites (i.e., Pine–Oak and Subalpine) were reduced (θW = 1.5 and 2.1, respectively). These values corresponded to differences in effective population sizes (Ne) ranging from 730 in the Pine–Oak to 1,320 individuals in the Grassland site, suggesting effective population sizes are orders of magnitude lower than the observed population sizes (SI Appendix, Fig. S5A). Although the small effective population sizes can allow for neutral genetic drift to occur, the observed parallel mutations suggest that Curtobacterium is adapting to the transplant experiment.
Discussion
While it is often assumed that ecological and evolutionary processes operate on different spatiotemporal scales, increasing evidence indicates that past evolutionary divergence (i.e., local adaptation) frequently influences contemporary ecological patterns and processes (40). In plants and animals, transplant experiments have been used to directly assess the extent of local adaptation within and across species (41, 42); however, similar experiments for microbes typically focus on the ecological response of the entire community (29, 32), limiting our understanding of how evolution shapes ecological patterns in microbiomes. Here, we sought to use this approach to characterize both the ecological and evolutionary processes contributing to genetic variation of a soil bacteria in response to changing environmental conditions. Within the community transplant experiment, we demonstrated that Curtobacterium ecotypes are locally adapted to the abiotic environment (moisture and temperature), where trait variation allowed for differential responses to new climate conditions. At the same time, the parallel isogenic transplant experiment demonstrated rapid evolution of a single Curtobacterium lineage in response to those same environmental conditions over the same timeframe. Together, the results demonstrate the potential for concurrent ecological and evolutionary processes to alter a microbiome’s diversity in response to climate change.
Evidence for adaptive differentiation in plants and animals is often inferred from the relationship between phenotypic traits of individuals within a species across different environments (43). In this way, variation in CWM trait values can allow for strong insights to the traits responsible for environmental distributions (35, 36). In microbes, however, identifying the traits associated with local adaptation in natural systems is particularly difficult. For example, we previously characterized co-occurring Curtobacterium ecotypes across the elevation gradient but could not explain biogeographic distributions based on in vitro trait measurements (33); this pattern was further supported here by the CWM trait distributions in the surrounding leaf litter (Fig. 3A). However, after controlling for a common litter substrate in the transplant experiment, we identified clear physiological trade-offs in the CWM trait values along the climate gradient. As such, ecotypes with trait values similar to the site-level CWM values occurred at relatively higher abundances after transplant to those particular sites, resulting in ecotype abundances converging on those of the native communities (Fig. 2). We conclude that trait variation among Curtobacterium ecotypes allows for adaptive differentiation and that the biogeographic distribution of ecotypes along this gradient is determined by both the climate and local litter substrate.
Our results also provide evidence that evolutionary processes, and specifically de novo mutations, can provide a novel source of genetic variation within just 18 mo in a soil bacterial community. We detected parallel mutations within and across sites, calculated mutation rates that are comparable to those observed in other natural systems (26, 44, 45), and estimated metrics of genomic diversity and effective population sizes. Nonetheless, our results largely indicated weak selective pressures by the environment over the 18 mo transplant experiment. Notably, the Curtobacterium strain was originally isolated from the Grassland site (1, 28) and may already be well adapted to the Grassland litter used in the transplant experiment. The Curtobacterium ecotype IBC is also present in leaf litter at all sites along the gradient, albeit at varying abundances (SI Appendix, Fig. S3A), suggesting that the average environmental conditions across the gradient may not impose a strong selective pressure in this lineage. Moreover, when we compared our observations to those made in laboratory experimental evolution studies (46), we did not observe typical signatures of genome evolution, such as selective sweeps or overrepresentation of nonsynonymous mutations (47). We suspect the highly heterogeneous matrix of soil habitats (48) may limit the spatial distribution of variants and depress opportunities for genetic exchange. Such spatial structural differences, combined with slower growth rates, may result in quite different evolutionary rates and dynamics in natural soil communities when compared to the well-mixed liquid cultures of laboratory experimental evolution studies.
Laboratory experimental evolution studies have provided insights into the adaptive responses of bacteria to temperature (3, 4), yet it remains unclear if these can be extrapolated to more complex trait responses, such as drought (49). Using a field-based experimental approach, we identified a set of functional genes that may be associated with adaptation to arid conditions (Table 1), including mutations in genes responsible for exopolysaccharide biosynthesis, nutrient acquisition, and stress response. While the detection of parallel mutations across sites does not preclude the possibility that mutations were present at low frequencies in the initial source population (i.e., inoculum), the repeated increase in frequency of these mutations similarly indicates that selection is a key process driving evolutionary adaptation to the transplant experiment. However, we recognize that further work is needed to confirm fitness effects. This would require comparing the growth of the evolved strains to the ancestor, which is not a trivial task under natural field conditions. Another caveat to our results is what we assessed the evolutionary response of the isogenic strain in isolation from the rest of the microbial community. The absence of biotic interactions with other community members is unrealistic and could alter metabolic evolution (50) and evolutionary dynamics (51) by excluding species interactions (52). Finally, many of the mutations may have been selected for by the common grass substrate or the litterbag environment rather than the climate conditions of the sites (i.e., relA; Table 1), although our previous work suggests that litterbag communities are similar to the surrounding natural community (29, 31, 32).
Our current study highlights a continuum of ecological and evolutionary processes, but future work should consider additional processes that may be equally important for soil microbiome diversity. In addition to de novo mutations, horizontal gene transfer promotes genetic innovation and can transform evolutionary landscapes over short timescales (53, 54). Population dynamics, including shifts in allele frequencies, can also provide fitness benefits that gradually impact evolutionary trajectories. Indeed, we previously identified genetically isolated populations within Curtobacterium ecotype IBC, where constrained recombination within populations promoted ecological specialization in localized microenvironments (20). Dispersal can also introduce novel diversity to a community (31, 55, 56) with the potential to impact environmental selection depending on dispersal rates (57). Field experiments that directly manipulate soil microbiomes afford the opportunity to identify how such processes contribute to the diversity and functioning of shifting microbiomes.
Materials and Methods
Field Sites.
We established four replicate plots (1 m2) at five sites across a climate gradient in Southern California (32). The five sites (from lowest to highest elevation) include the Sonoran Desert (33.648, −116.38; elevation = 275 m), coastal grassland (33.737, −117.70; 470 m), pinyon–juniper scrubland (33.610, −116.45; 1,280 m), pine–oak forest (33.683, −116.77; 1,710 m), and subalpine forest (33.823, −116.75; 2,250 m) as previously described (58). Sites are hereafter referred to as Desert, Grassland, Scrubland, Pine–Oak, and Subalpine, respectively. All sites experience Mediterranean climate patterns with a hot, dry summer and a cool, wet winter. The sites provide a natural environmental gradient as they range in mean annual air temperature from 10.3 to 24.6 °C and precipitation from 80 to 400 mm. Precipitation and soil temperature data were collected from nearby weather stations and iButton temperature sensors (32).
Community Transplant.
In October 2015, we inoculated survey leaf litter microbial communities from each site into microbial cages containing irradiated grassland leaf litter, as previously described (32). Briefly, grassland leaf litter was ground and homogenized, 5 g was transferred to nylon membrane bags with 0.22 µm pores (cat. no. SPEC17970; Tisch Scientific) and sterilized with at least 22 kGy gamma irradiation (Sterigenics). The pore size allows for movement of water and nutrients but prevents microbial migration (30). After sterilization, 50 mg of finely ground survey leaf litter communities were added to each nylon bag.
Metagenomic samples.
We performed a pairwise reciprocal transplant by placing inoculated litterbags in the four replicate plots at each site for 18 mo from October 2015 to April 2017 (5 sites × 5 inocula × 4 replicate plots = 100 litterbags). The four replicate 1 m2 plots were separated by >5 m. To assess the composition of the microbial communities outside the litterbags, litter adjacent to the plots (the mixture of decaying plant species present) was collected at initial deployment (5 sites × 4 replicate plots = 20 in situ survey samples). For all 120 litter samples, we extracted DNA from 0.05 g of ground leaf litter using the FastDNA SPIN Kit for Soil (Mo Bio) and cleaned the DNA with the Genomic DNA Clean and Concentrator kit (Zymo Research). Cleaned samples were diluted to 0.5 ng/μL and 1 ng of DNA was used for input for the Nextera XT library Prep kit for sequencing on the Illumina HiSeq4000 instrument with 150 bp paired-end reads (PE150). The raw data are deposited on the metagenomics analysis server (MG-RAST) (59) under the project ID mgp17355.
Metagenomic libraries were quality trimmed using the BBMap toolkit (60) (bbduk.sh qtrim = rl trimq = 10) and filtered to remove eukaryotic DNA. Specifically, we mapped all reads to a reference genome using Burrows–Wheeler Alignment (BWA) tool (61) from both an abundant grass (Lolium perenne; Accession: MEHO01000000) and fungus (Pyrenophora teres; Accession: NZ_AEEY00000000) found at the Grassland site. Filtered reads were merged with BBMap. If a read could not be merged with its counterpart, we included only the forward read in further analyses. Reads were then translated using Prodigal (62) with the metagenomic flag.
Bacterial community analysis.
To characterize the microbial communities, we utilized a computational pipeline to assign taxonomy using phylogenetic inference of 21 conserved single-copy marker genes against a reference marker gene database, as previously described (12). Briefly, the translated metagenomic paired-end reads were searched against the reference marker gene database with an initial BLASTp database with an E value of 1 × 10−5 and a secondary filter against the reference hidden Markov model (HMM) profiles with an E value ranging from 1 × 10−10 to 1 × 10−15 depending on the individual marker gene. Passed reads were aligned using ClustalO version 1.2.0 (63) to the corresponding reference package and placed onto the reference phylogenies using pplacer version 1.1.alpha17 (64). Relative abundances were calculated by generating single-branch abundance matrices and normalizing to the total number of marker genes present in each library. To test the effects of initial inoculum and transplanted sites on the transplanted microbial communities (N = 100), we generated a Bray–Curtis dissimilarity matrix as input for a permutational multivariate analysis of variance (PERMANOVA) for 999 permutations. The Bray–Curtis similarity matrix was also used for visualization with nonmetric multidimensional scaling in R.
Curtobacterium ecotype analysis.
In order to track Curtobacterium ecotypes, we utilized a similar approach to leverage phylogenetic inference of Curtobacterium core genes that provided a robust phylogenetic signal (N = 830 orthologs), as previously described (33). Translated reads were searched against the Curtobacterium core gene database with a preliminary BLASTp filter (E value = 1 × 10−20) and a secondary HMMer filter (E value = 1 × 10−40). Similarly, reads were aligned and placed onto core gene phylogenies for classification to Curtobacterium clades and, when possible, ecotypes.
To incorporate differences in the absolute abundance of Curtobacterium among sites as well as biases from relative abundance data, total bacterial cell densities from transplanted bags were measured by flow cytometry and normalized by per gram of dry litter weight, as previously described (32). The relative abundances of 14 Curtobacterium lineages (ecotypes and other clades) in each sample were then multiplied by an estimate of total abundance of Curtobacterium from the community data. The resulting absolute abundances of Curtobacterium lineages were log transformed and then used to generate a Euclidean dissimilarity matrix (note that there are no zeros in this smaller dataset, mitigating the typical problem of counting double zeros when using Euclidean distance on community data). We tested the effects of treatment (site, inoculum, and interaction of the two) using a PERMANOVA for 999 permutations. Using the factoextra package in R, compositional differences were visualized using a principal component analysis ordination plot to determine which ecotypes were driving separation between sites.
To correlate ecotype distributions to trait variation, we assayed 24 representative strains from the six abundant ecotypes across the elevation gradient, as previously described (33). Briefly, functional traits included biofilm formation, growth parameters (μmax = maximum growth rate; Amax = maximum growth by absorbance [OD600]), and the degradation of cellulose and xylan. All traits were assayed in vitro across a temperature gradient ranging from 15 to 37 °C. We reanalyzed this trait data to correlate ecotype abundance to trait variation by calculating CWM trait values. Within each ecotype, we averaged trait performance across strains independent of the temperature of the assay. Temperature preference was then defined as the optimal temperature at which each strain performed best across all trait assays. Finally, the CWM trait values were calculated by weighting the measured ecotype–mean values by their absolute abundances in each metagenomic sample.
Isogenic Transplant.
Characterization of strain MMLR14002.
We transplanted an isogenic strain of Curtobacterium MMLR14002 from the most abundant ecotype across all sites, IBC. The strain was originally isolated at the grassland site in 2011, and this ecotype has been extensively characterized (12, 28, 33). To calculate growth rates of the ancestral strain, we grew (in triplicate) strain MMLR14002 shaking at 22 °C in both high-nutrient (Luria–Bertani [LB] media) and low-nutrient Loma Ridge (LR) litter leachate (the same substrate used in the litterbags collected from the Grassland site) and measured optical densities (OD600) for 48 h. The genome of MMLR14002 was previously reported (28); however, we amended the genome using a combination of PacBio long-read sequencing with the existing short-read data to assemble a complete reference genome (65). First, we performed a preliminary assembly with the long-read data using the PacBio Corrected Reads pipeline (length, 500; partitions, 200; genomeSize = 3,700,000) (66). Next, Illumina short-read data were quality trimmed using BBDuk (minlen = 25, qtrim = rl, trimq = r, k = 25) and mapped against the preliminary assembly with BWA. Finally, we hybridized the two datasets to generate a consensus assembly with pilon version 1.22 (67). We predicted coding regions of the complete assembly of the reference strain with prokka (68). The updated genome is available on National Center for Biotechnology Information (NCBI) under accession number PRJNA524191.
For the transplant, we grew a freezer stock of strain MMLR14002 on LR litter (from the Grassland site) leachate agar plates for 4 d at 22 °C. We transferred a single colony into 25 mL of liquid LR media for 3 d of growth, followed by an additional transfer to fresh LR media for 3 more d. We used 1 mL of the grown culture for frozen stocks of the ancestral strain prepared with glycerol for genome sequencing and long-term storage at −80 °C. The remainder of the culture was centrifuged to wash the cell pellet with 0.9% saline solution. After a third saline wash, we resuspended the cell pellet with 15 mL 0.9% saline solution and measured cell densities with optical density (OD) at 600 nm. Cells were diluted to cell densities resembling Curtobacterium abundances observed in the Grassland litter community (OD600 = 0.38). As in the community transplant, sterile grassland litter was prepared under identical protocols except for 2 g of ground leaf litter was added to each litterbag (instead of the 5 g used in the community transplant). We inoculated 25 sterile leaf litterbags (5 sites × 5 replicates) with 1 mL of diluted Curtobacterium cells. Bags were sealed with fabric glue, edges reinforced with duct tape, and deployed to all sites in chicken wire cages to prevent rodent interference and physical movement of bags.
At 6 mo intervals (until 18 mo), ∼1 g of each bag was transferred to a new litterbag containing 1 g of sterile Grassland litter, prepared under identical protocols. The five freshly inoculated bags were randomly distributed across the four replicate plots at each site to minimize plot effects. One bag from the Grassland site was unable to be located after initial deployment, reducing the number of replicates at this site from five to four litterbags. The remaining 1 g of litter was collected and stored for metagenomic sequencing, flow cytometry, and reisolation of the ancestral strain.
Metagenomic population samples.
To gain insights into the entire population within the isogenic litterbags, we constructed shotgun metagenomic libraries from a subset of samples from the experiment. For all 24 litterbags collected at the final time point (at 18 mo), we extracted DNA from 0.05 g of ground leaf litter using the FastDNA SPIN Kit for Soil (Mo Bio) and cleaned the DNA with the Genomic DNA Clean and Concentrator kit (Zymo Research). Cleaned samples were diluted to 0.5 ng/μL, and 1 ng of DNA was used for input for the Nextera XT library Prep kit for PE150 sequencing on the Illumina HiSeq4000 instrument. Since the gamma irradiation does not completely eliminate the presence of the original community or remove relic DNA, we first assessed the extent of possible contamination and recovery of Curtobacterium in the litterbags at the final time point. Briefly, we quality filtered the metagenomic reads with BBDuk and assembled all raw reads using the SPAdes genome assembler (69) with a “careful” iterative k-step ranging from k = 31 to 91. We then classified all contigs >1,000 bp using MegaBLAST against the NCBI nucleotide database (70) with an E value of 1 × 10−5. In total, 81 ± 8% of the assembled contigs classified as Curtobacterium or in its family, Microbacteriaceae, with minimal differences across sites. Reads were mapped to the reference genome with BBMap using a minid = 0.95, but we were unable to achieve sufficient depth across all samples for reliable variant calling (average depth = 28×). As such, we used this data to validate the presence of the ancestral strain in each litterbag by comparing the de novo assembly of the representative population genome to the ancestral strain.
Based on the population genome completeness and similarity to the ancestral strain, we selected one bag from each site to deeply sequence across all three time points (1 bag per site × 5 sites × 3 time points = 15 total litterbags). We isolated genomic DNA (gDNA) from 0.05 g leaf litter using the Zymobiomics DNA Mini kit (Zymo Research) following the protocol for soil. We quantified the DNA using the Qubit reagents and used the Nextera Flex kit (Illumina) to prepare the library for sequencing using a reduced volume protocol (71). The timeseries metagenomic samples were sequenced using PE150 reads on the NovaSeq6000 to produce 3.6 × 108 reads per sample for an average coverage of >4,000× to the reference genome that was evenly distributed across sites (SI Appendix, Fig. S8). For verification, we confirmed the recovery of the ancestral strain by performing an additional de novo assembly (all assemblies were 99.9 to 100% average nucleotide identity (ANI) match to reference strain) as described above. The timeseries metagenomic data were processed for variant calling by mapping quality filtered sequence data to the complete reference ancestral genome using both the Genome Analysis Toolkit (GATK) best practices pipeline (72) and breseq version 0.26.1 (73). To minimize the influence of sequencing error in the variant calling for calculating population polymorphisms, we first normalized the samples to kmer = 300 with BBNorm (target = 300, mindepth = 5) before running the breseq polymorphism mode pipeline. For the GATK pipeline, to account for the possibility of a mixed population, we increased the haploid count to 3, as the number of mutations called reached a saturation point with increasing haploid counts (SI Appendix, Fig. S9). Based on the recommendations from the GATK best practices pipeline, we filtered low-quality variants from further analyses (SNPs: quality by depth [QD] < 2.0 || Fisher strand Phred-scaled probability score [FS] > 60.0 || root mean square mapping quality score [MQ] < 40.0; indels: QD < 2.0 || FS > 200.0). High-confidence variants were further categorized by mutation type.
To estimate consensus population sizes and replication rates, we utilized both the timeseries (N = 15) and final time point metagenomic data (N = 24). For the replication rate, we first mapped all reads to the reference genome with BBMap (minid = 0.95), sorted all reads, and reduced the resulting sam files with shrinksam (https://github.com/bcthomas/shrinksam). The mapped reads were then used as input for a modified replication rate estimator (39) as part of the iRep toolkit (74). Consensus population sizes were estimated using the percent of reads from each metagenomic library mapping back to the reference genome as a function of cell counts. Specifically, total bacterial cell densities were obtained by flow cytometry measurements of 0.2 g from each isogenic litterbag and normalized by per gram of dry litter weight.
Reisolation, variant calling, and mutation rate estimation.
Every 6 mo, we reisolated the ancestral strain from collected litter. We used 0.2 g of leaf litter from each litterbag (N = 24) that was suspended in 5 mL of 0.9% NaCl solution and vortexed for 5 min. Samples were serially diluted and plated on freshly prepared LR litter leachate agar plates. For each plate, 5 to 10 colonies were transferred three times on LB media agar plates to ensure clonal isolation and stored in glycerol solution at −80 °C until sequencing.
To verify that isolates belonged to Curtobacterium, we identified a subset of isolates from the final time point by PCR amplification of the 16S rRNA region, as previously described (33). Of these, we confidently identified 78% of the isolates as belonging to Curtobacterium, with an additional 10% as probable matches to the ancestral strain that were excluded based on low-quality base scores. Note that these percentages are similar to the percent of reads matching Curtobacterium in the metagenomic data obtained with the final time point litterbags (see above). Of the identified Curtobacterium isolates, we selected 124 isolates from across all sites and time points for whole-genome sequencing. Frozen stocks were grown in 10 mL liquid LB media for 48 h. We used 2 mL of grown cultures for gDNA extraction using the Wizard Genomic Purification Kit (Promega). Extracted DNA was diluted to 0.5 ng/μL for the Nextera XT Library Preparation Kit (Illumina Inc.) and pooled in equimolar concentrations for PE150 sequencing on the Illumina HiSeq4000.
Demulitplexed sequence data were quality filtered using BBDuk (qtrim = rl, trimq = 10, minlen = 25, ktrim = r, k = 25). Before calling variants against the ancestral strain, we performed de novo assemblies on the 124 sequenced strains using the SPAdes genome assembler with a “careful” iterative k-step ranging from k = 31 to 91. Assembled genomes were compared against the ancestral strain by calculating an ANI metric (75). We removed any assemblies with <99.9% ANI values to the ancestral strain and/or fragmented assemblies (>40 contigs from de novo assembly). The remaining 112 genomes were processed for variant calling. Specifically, we mapped the quality filtered sequence data to the complete reference ancestral genome using both the GATK best practices pipeline and breseq version 0.26.1. We observed no significant differences between pipelines and present only the results from the GATK pipeline. Passed variants from the GATK pipeline were further filtered to remove low-depth calls (overall depth of coverage [DP] < 20) and designated as insertions, deletions, and SNPs with further characterizations into whether the mutation was observed in coding regions or intergenic regions. Coding region SNPs were further characterized as synonymous, nonsynonymous, or nonsense mutations. Unique SNPs were used to calculate the expected dN/dS ratios with a 2:1 transition to transversion ratio (47) in R.
For mutation rate estimations, we identified the number of unique variants (SNPs and indels) across all reisolated clones. In total, we identified 139 unique polymorphic sites across the 112 mutant strains. By analyzing within each site, we can estimate Watterson (θw) with Eq. 1.
| [1] |
where S = number of unique polymorphic sites; an = number of genomes.
Assuming an average mutation rate of 0.001 mutations per genome per generation (45), we estimated the mutation rate for the ancestral strain (genome size = 3773875 bp). Finally, extrapolating from our growth rate measurements, we estimated the ancestral population replicates at a maximum rate of 5.45 generations per day. Accordingly, all values were used to calculate the effective population sizes (Ne) with Eq. 2.
| [2] |
where L = length of genome; μg = mutations per base per generation.
Supplementary Material
Acknowledgments
We thank Steven D. Allison, Mike Goulden, Adam C. Martiny, and Kathleen K. Treseder for their work in organizing the climate gradient project as well as Michaeline B. N. Albright, Nameer Baker, Sydney I. Glassman, Junhui Li, Chamee Moua, and Kendra Walters for assistance in field work. We would also like to thank Sergey Kryazhimskiy, Elsa Abs, Gabin Piton, and Brandon S. Gaut for advice on data analysis and insightful discussions on differentiating ecology and evolution in microbiomes. Finally, the authors thank Alejandra Rodriguez-Verdugo and Adam C. Martiny for their thoughts on previous versions of the manuscript, Andrew Oliver and Timothy M. Perez for statistical input, the University of California Natural Reserve System for access to the field sites, and the University of California Irvine High Performance Computing Cluster for computational resources. This work was supported by the Scripps Postdoctoral Scholar Fellowship to A.B.C., an NSF grant (DEB-1457160), and US Department of Energy Office of Science, Biological and Environmental Research award (DE-SC0020382) to J.B.H.M.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101254118/-/DCSupplemental.
Data Availability
The updated reference genome for Curtobacterium IBC strain MMLR14002 is available under the NCBI BioProject PRJNA524191. Paired-end sequence data were deposited in the NCBI Sequence Read Archive under the accession codes SAMN17506608-SAMN17506719 and SAMN17497431-SAMN17497469 for the evolved isolates and population litterbag metagenomics, respectively. Metagenomic sequences from the community transplant litterbags are available on MG-RAST under the project ID mgp17355. All other data and relevant code used can be found at GitHub, https://github.com/alex-b-chase/curtoEvol.
References
- 1.Matulich K. L., et al., Temporal variation overshadows the response of leaf litter microbial communities to simulated global change. ISME J. 9, 2477–2489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amend A. S., et al., Microbial response to simulated global change is phylogenetically conserved and linked with functional potential. ISME J. 10, 109–118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tenaillon O., et al., The molecular diversity of adaptive convergence. Science 335, 457–461 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Kent A. G., Garcia C. A., Martiny A. C., Increased biofilm formation due to high-temperature adaptation in marine Roseobacter. Nat. Microbiol. 3, 989–995 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koskella B., Vos M., Adaptation in natural microbial populations. Annu. Rev. Ecol. Evol. Syst. 46, 503–522 (2015). [Google Scholar]
- 6.Hairston N. G. Jr, Ellner S. P., Geber M. A., Yoshida T., Fox J. A., Rapid evolution and the convergence of ecological and evolutionary time. Ecol. Lett. 8, 1114–1127 (2005). [Google Scholar]
- 7.Rudman S. M., et al., What genomic data can reveal about eco-evolutionary dynamics. Nat. Ecol. Evol. 2, 9–15 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Garud N. R., Pollard K. S., Population genetics in the human microbiome. Trends Genet. 36, 53–67 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Shade A., et al., Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3, 417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson Z. I., et al., Partitioning among Prochlorococcus ecotypes along environmental gradients. Science 311, 1737–1740 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Moore L. R., Rocap G., Chisholm S. W., Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393, 464–467 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Chase A. B., et al., Microdiversity of an abundant terrestrial bacterium encompasses extensive variation in ecologically relevant traits. mBio 8, e01809-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larkin A. A., Martiny A. C., Microdiversity shapes the traits, niche space, and biogeography of microbial taxa. Environ. Microbiol. Rep. 9, 55–70 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Barrett R. D. H., Schluter D., Adaptation from standing genetic variation. Trends Ecol. Evol. 23, 38–44 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Hunt D. E., et al., Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320, 1081–1085 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Chase A. B., Martiny J. B. H., The importance of resolving biogeographic patterns of microbial microdiversity. Microbiol. Aust. 39, 5–8 (2018). [Google Scholar]
- 17.Crits-Christoph A., Olm M. R., Diamond S., Bouma-Gregson K., Banfield J. F., Soil bacterial populations are shaped by recombination and gene-specific selection across a grassland meadow. ISME J. 14, 1834–1846 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bendall M. L., et al., Genome-wide selective sweeps and gene-specific sweeps in natural bacterial populations. ISME J. 10, 1589–1601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arevalo P., VanInsberghe D., Elsherbini J., Gore J., Polz M. F., A reverse ecology approach based on a biological definition of microbial populations. Cell 178, 820–834.e14 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Chase A. B., et al., Maintenance of sympatric and allopatric populations in free-living terrestrial bacteria. mBio 10, e02361-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain C., Rodriguez-R L. M., Phillippy A. M., Konstantinidis K. T., Aluru S., High-throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro B. J., Polz M. F., Ordering microbial diversity into ecologically and genetically cohesive units. Trends Microbiol. 22, 235–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordero O. X., Polz M. F., Explaining microbial genomic diversity in light of evolutionary ecology. Nat. Rev. Microbiol. 12, 263–273 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Tyson G. W., et al., Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428, 37–43 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Meziti A., et al., The reliability of metagenome-assembled genomes (MAGs) in representing natural populations: Insights from comparing MAGs against isolate genomes derived from the same fecal sample. Appl. Environ. Microbiol. 87, e02593-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denef V. J., Banfield J. F., In situ evolutionary rate measurements show ecological success of recently emerged bacterial hybrids. Science 336, 462–466 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Garud N. R., Good B. H., Hallatschek O., Pollard K. S., Evolutionary dynamics of bacteria in the gut microbiome within and across hosts. PLoS Biol. 17, e3000102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chase A. B., Arevalo P., Polz M. F., Berlemont R., Martiny J. B. H., Evidence for ecological flexibility in the cosmopolitan genus Curtobacterium. Front. Microbiol. 7, 1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker N. R., Khalili B., Martiny J. B. H., Allison S. D., Microbial decomposers not constrained by climate history along a Mediterranean climate gradient in southern California. Ecology 99, 1441–1452 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Allison S. D., et al., Microbial abundance and composition influence litter decomposition response to environmental change. Ecology 94, 714–725 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Albright M. B. N., Chase A. B., Martiny J. B. H., Experimental evidence that stochasticity contributes to bacterial composition and functioning in a decomposer community. mBio 10, e00568-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glassman S. I., et al., Decomposition responses to climate depend on microbial community composition. Proc. Natl. Acad. Sci. U.S.A. 115, 11994–11999 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chase A. B., et al., Emergence of soil bacterial ecotypes along a climate gradient. Environ. Microbiol. 20, 4112–4126 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Cohan F. M., What are bacterial species? Annu. Rev. Microbiol. 56, 457–487 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Muscarella R., Uriarte M., Do community-weighted mean functional traits reflect optimal strategies? Proc. Biol. Sci. 283, 20152434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonnier G., Shipley B., Navas M., Quantifying relationships between traits and explicitly measured gradients of stress and disturbance in early successional plant communities. J. Veg. Sci. 21, 1014–1024 (2010). [Google Scholar]
- 37.Christin P. A., Weinreich D. M., Besnard G., Causes and evolutionary significance of genetic convergence. Trends Genet. 26, 400–405 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Ochman H., Elwyn S., Moran N. A., Calibrating bacterial evolution. Proc. Natl. Acad. Sci. U.S.A. 96, 12638–12643 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korem T., et al., Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349, 1101–1106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urban M. C., et al., Evolutionary origins for ecological patterns in space. Proc. Natl. Acad. Sci. U.S.A. 117, 17482–17490 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ågren J., Schemske D. W., Reciprocal transplants demonstrate strong adaptive differentiation of the model organism Arabidopsis thaliana in its native range. New Phytol. 194, 1112–1122 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Clausen J., Keck D. D., Hiesey W. M., “Experimental studies on the nature of species. III. Environresponses of climatic races of Achillea” in Experimental Studies on the Nature of Species. III. Environresponses of Climatic Races of Achillea (Carnegie Institution of Washington, Publication 581, Washington, DC, 1948). [Google Scholar]
- 43.Ackerly D. D., Community assembly, Niche conservatism, and adaptive evolution in changing environments. Int. J. Plant Sci. 164, S165–S184 (2003). [Google Scholar]
- 44.Ghalayini M., et al., Evolution of a dominant natural isolate of Escherichia coli in the human gut over the course of a year suggests a neutral evolution with reduced effective population size. Appl. Environ. Microbiol. 84, e02377-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee H., Popodi E., Tang H., Foster P. L., Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, E2774–E2783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tenaillon O., et al., Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536, 165–170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wielgoss S., et al., Mutation rate inferred from synonymous substitutions in a long-term evolution experiment with Escherichia coli. G3 (Bethesda) 1, 183–186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nannipieri P., et al., Microbial diversity and soil functions. Eur. J. Soil Sci. 54, 655–670 (2003). [Google Scholar]
- 49.Martiny J. B. H., Jones S. E., Lennon J. T., Martiny A. C., Microbiomes in light of traits: A phylogenetic perspective. Science 350, aac9323 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Evans R., et al., Eco-evolutionary dynamics set the tempo and trajectory of metabolic evolution in multispecies communities. Curr. Biol. 30, 4984–4988.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheuerl T., et al., Bacterial adaptation is constrained in complex communities. Nat. Commun. 11, 754 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodríguez-Verdugo A., Ackermann M., Rapid evolution destabilizes species interactions in a fluctuating environment. ISME J. 15, 450–460 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smillie C. S., et al., Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Quistad S. D., Doulcier G., Rainey P. B., Experimental manipulation of selfish genetic elements links genes to microbial community function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190681 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langenheder S., Székely A. J., Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J. 5, 1086–1094 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Albright M. B. N., Martiny J. B. H., Dispersal alters bacterial diversity and composition in a natural community. ISME J. 12, 296–299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evans S., Martiny J. B. H., Allison S. D., Effects of dispersal and selection on stochastic assembly in microbial communities. ISME J. 11, 176–185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker N. R., Allison S. D., Extracellular enzyme kinetics and thermodynamics along a climate gradient in southern California. Soil Biol. Biochem. 114, 82–92 (2017). [Google Scholar]
- 59.Meyer F., et al., The metagenomics RAST server–A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bushnell B., BBMap Short Read Aligner (University of California; , Berkeley, CA, 2016), http//sourceforge.net/projects/bbmap. [Google Scholar]
- 61.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hyatt D., et al., Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sievers F., et al., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsen F. A., Kodner R. B., Armbrust E. V., pplacer: Linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11, 538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chakraborty M., Baldwin-Brown J. G., Long A. D., Emerson J. J., Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 44, e147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rhoads A., Au K. F., PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13, 278–289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walker B. J., et al., Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Bankevich A., et al., SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Federhen S., The NCBI Taxonomy database. Nucleic Acids Res. 40, D136–D143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bruinsma S., et al., Bead-linked transposomes enable a normalization-free workflow for NGS library preparation. BMC Genomics 19, 722 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van der Auwera G. A., et al., From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–11.10.33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deatherage D. E., Barrick J. E., Engineering and Analyzing Multicellular Systems (Springer, 2014), pp. 165–188. [Google Scholar]
- 74.Brown C. T., Olm M. R., Thomas B. C., Banfield J. F., Measurement of bacterial replication rates in microbial communities. Nat. Biotechnol. 34, 1256–1263 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rodriguez-R L. M., Konstantinidis K. T., The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4, e1900v1 (2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The updated reference genome for Curtobacterium IBC strain MMLR14002 is available under the NCBI BioProject PRJNA524191. Paired-end sequence data were deposited in the NCBI Sequence Read Archive under the accession codes SAMN17506608-SAMN17506719 and SAMN17497431-SAMN17497469 for the evolved isolates and population litterbag metagenomics, respectively. Metagenomic sequences from the community transplant litterbags are available on MG-RAST under the project ID mgp17355. All other data and relevant code used can be found at GitHub, https://github.com/alex-b-chase/curtoEvol.