Significance
The MGA contains a large gene expression dataset from the developing brain of the common marmoset. However, the data represent the gene expression patterns across the entire brain, and methods to isolate region-specific gene expression patterns will provide important information regarding primate brain structure, function, and cellular and molecular organization. We have developed a system, GePS, that systematically identifies genes expressed in selected regions of the common marmoset brain. Comprehensive, comparative gene expression analysis with mice revealed many genes with species- and region-specific expression. Moreover, by combining GePS with the gene expression comparison and cortical area projection mapping systems, we were able to provide further insight into the molecular organization of these species- and region-specific gene expression patterns.
Keywords: gene expression, database, nonhuman primate, evolution, developmental disorder
Abstract
Precise spatiotemporal control of gene expression in the developing brain is critical for neural circuit formation, and comprehensive expression mapping in the developing primate brain is crucial to understand brain function in health and disease. Here, we developed an unbiased, automated, large-scale, cellular-resolution in situ hybridization (ISH)–based gene expression profiling system (GePS) and companion analysis to reveal gene expression patterns in the neonatal New World marmoset cortex, thalamus, and striatum that are distinct from those in mice. Gene-ontology analysis of marmoset-specific genes revealed associations with catalytic activity in the visual cortex and neuropsychiatric disorders in the thalamus. Cortically expressed genes with clear area boundaries were used in a three-dimensional cortical surface mapping algorithm to delineate higher-order cortical areas not evident in two-dimensional ISH data. GePS provides a powerful platform to elucidate the molecular mechanisms underlying primate neurobiology and developmental psychiatric and neurological disorders.
The mammalian brain contains many functionally distinct regions that are extensively interconnected, enabling rapid and accurate information processing. Each region contains diverse cell types that differ in their molecular, morphological, electrophysiological, and functional characteristics (1, 2). Advanced neuroscience technologies, which are generally optimized and extensively applied in mice, provide complicated approaches to analyze these characteristics and model the genetic basis of this regional- and cell-type diversity. However, clear cognitive differences arise from interspecies differences in these key characteristics, and most contemporary technologies are difficult to apply in species whose study involves extensive ethical, practical, and experimental impediments. The ability to compare gene expression patterns between primates and model species such as mice is crucial for understanding the human brain (3, 4).
Recent developments in technologies, such as microarray, single-cell RNA-sequencing (RNA-seq), and Drop-seq, have allowed for high-resolution genome-wide expression analysis in specific regions of the brain or in thousands of individual cells simultaneously (5–10). However, these methods provide limited anatomical information and no morphological observations. Slide-seq solves these problems by transferring RNA from tissue sections onto a surface covered in DNA-barcoded beads, allowing the preservation of positional information (11). However, the unbiased nature of next-generation sequencing technologies may hinder the molecular identification of specific cell types or their direct analysis. In situ hybridization (ISH) databases of humans and nonhuman primates (NHPs) have revealed brain gene expression profiles at cellular resolution, while preserving key morphological and anatomical characteristics, ultimately providing important information about the genetic conservation of analogous brain regions (12). However, these studies are highly resource intensive, requiring substantial time and cost. Consequently, such efforts have been limited, focusing on a few brain regions in limited species (13). Thus, interspecific comparison of brain-cell heterogeneity has not been possible.
Here, we describe the development and implementation of an unbiased gene expression profiling system (GePS) for use with the Marmoset Gene Atlas (MGA) (https://gene-atlas.brainminds.riken.jp/) (14), which is adaptable for use with other atlases. The system, developed as a part of “Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS),” a brain mapping program in Japan (15, 16), enables systematic and automatic analysis of gene expression across brain regions. Brain/MINDS aims to develop knowledge-based tools to explore primate-specific brain structure, function, and connectivity in health and disease. Specifically, we analyzed the expression patterns of identified risk genes for psychiatric disorders in the neonatal marmoset brain. There are >2,000 gene expression profiles for the neonatal common-marmoset brain in the MGA. Although each ISH section is complemented by an adjacent neuroanatomically stained section (visualizing the Nissl substance), enabling users to compare gene expression against a brain atlas, mapping gene expression patterns over the entire brain is both time and labor intensive. A large mouse ISH database, the Allen Brain Atlas (ABA; http://developingmouse.brain-map.org/), demonstrated that an anatomic gene expression atlas (AGEA) can characterize local gene expression in the mouse brain without prior knowledge of classical anatomy (17). To enhance the MGA in this manner, we developed GePS to systematically and automatically identify gene expression patterns in specific regions of the neonatal marmoset brain.
GePS, including the gene expression comparison system and cortical surface projection mapping function, is publicly available as an open resource on our website (https://gene-atlas.brainminds.riken.jp/). It provides primate-specific whole brain–gene expression patterns and will be a valuable platform in the field of primate neuroscience, helping reveal primate-specific brain functions and connections and providing insights into brain evolution and pathology.
Results
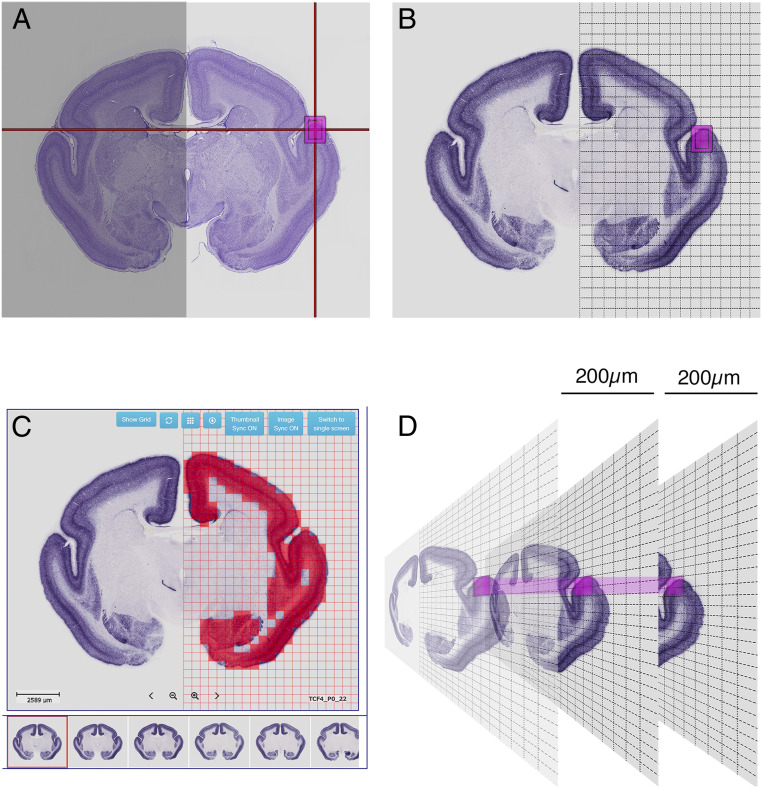
The MGA contains coronally sectioned ISH data from marmoset brains at different ages, including 2,000 gene expression profiles from neonatal common-marmoset brains. To enable automatic, systematic, and unbiased comparisons of gene expression patterns between brain regions and species, we developed GePS (http://www.progress-wave.co.jp/). GePS detects ISH signals using an automated image segmentation algorithm, which detects signals in individual grids and averages the signal intensity of the entire grid area to obtain positive and negative results. As the ISH color reaction varies between genes, brains, and sections, unbiased comparisons of signal-to-noise ratios in a given grid area were performed to determine whether the nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate reaction provided a positive signal. Fig. 1A shows a Nissl substance-stained section from the reference atlas, with a position indicator to search for genes expressed within the selected area. All ISH sections were overlaid with a grid (Fig. 1 A and B), and the gene expression detection software was used to sum the number and intensities of expressing pixels (Fig. 1C). Different grids were tested (from 20 × 20 pixel grids to 200 × 200 pixel grids), and we concluded that 200 × 200 pixel grids provide the most accurate result. When grid size becomes smaller, a chance to pick dust, scratch, and background becomes higher and provides false-positive results more often. Therefore, we decided to use 200 × 200 pixel grids for the automated signal identification system. Fig. 1C depicts the grid system for detecting the positive signal in each grid (red squares indicate 200 × 200 pixel grids). The anatomical location of the position indicator on the reference atlas was then transferred to a grid position on a selected plane of ISH data. When the sections had rostral–caudal misalignment, the software also searched for gene expression in one rostral section and one caudal section ∼200 µm from the selected section (Fig. 1D).
Fig. 1.
The gene expression detection software. (A) The Nissl reference atlas, with a position indicator to search for genes expressed in the selected area (magenta box). (B) All ISH sections were gridded (1 grid = 200 × 200 pixels or 500 × 500 μm). (C) The software indicates the gene expression in each grid. (D) The software also searches for gene expression in one rostral section and one caudal section 200 μm away from the selected section.
Identification of Gene Expression Enrichment in Specific Brain Regions by GePS.
To demonstrate GePS function in the neonatal common-marmoset brain, we first isolated genes expressed in sensory cortical areas, which are anatomically well characterized. The primary somatosensory cortex (S1) and primary visual cortex (V1) were selected (SI Appendix, Fig. S1), and 687 (S1) and 739 (V1) genes were expressed in each region (Datasets S1 and S2). Although some false positives were included, possibly due to ISH misalignment and strong background, GePS was able to identify genes expressed in selected brain regions (SI Appendix, Fig. S1).
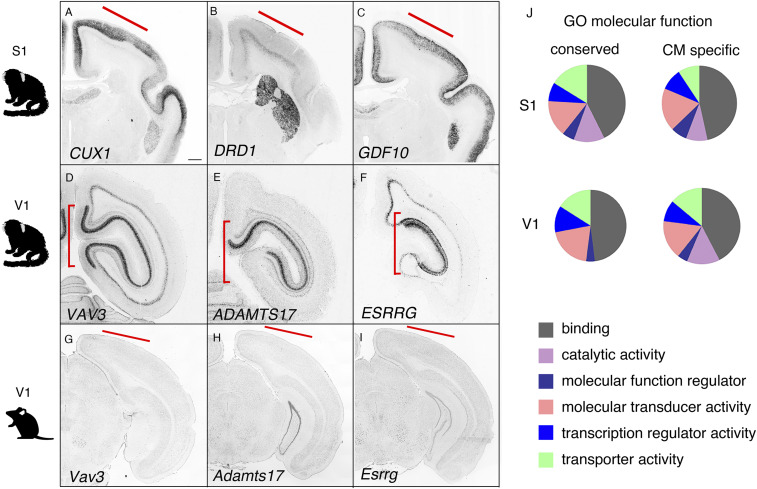
We next identified gene expression enrichment in selected marmoset brain regions for comparison with mouse gene expression patterns. To do this, we examined an equivalent time point in the developing mouse brain. Based on a previous report, the development in a postnatal day 0 marmoset brain is equivalent to that of an ∼2 to 3 wk old mouse (18, 19). For direct comparative gene expression analysis using the ABA-AGEA system, we compared genes expressed in the mouse and marmoset S1 and V1 regions, which are structurally conserved (Datasets S1 and S2). In the S1 region, 210 genes were expressed in both mice and marmosets, which we describe as conserved (ISH positive signal detected), 121 showed expression only in mice, and the rest were not determined due to lack of mouse ISH data. Conserved expression in mouse and marmoset was confirmed by ISH for genes such as cut-like homeobox 1 (CUX1; Fig. 2A). Fewer genes displayed marmoset-specific expression (20), including dopamine receptor 1 (DRD1) and growth differentiation factor 10 (GDF10; Fig. 2 B and C). Little to no Drd1 and Gdf10 expression was observed in P14 and P28 mouse cortexes (https://portal.brain-map.org/). Comparative gene expression analysis in V1 revealed more diverse gene expression patterns between mice and marmosets (conserved: 29; marmoset specific: 221; mouse specific: 92; n/d: 168; Dataset S2). We performed ISH on mouse brains to examine marker genes absent in the ABA, such as vav 3 oncogene (Vav3), a disintegrin like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 (Adamts17), and estrogen-related receptor gamma (Esrrg), and found that these marker genes were selectively expressed in the marmoset visual cortex (Fig. 2 D–I). We also performed gene ontology (GO) analysis on conserved, marmoset-specific and mouse-specific genes (Fig. 2J and Datasets S1 and S2). Interestingly, genes specifically expressed in the V1 showed significant enrichment for the GO term “catalytic activity,” and this effect was increased for marmoset-specific genes (Fig. 2J and Datasets S1 and S2). This indicates increased complexity in the enzymatic activity in the marmoset visual cortex, which supports the highly evolved visual circuitry in NHPs (21, 22). These results suggest that region-specific modulation in gene expression supports the specific functions of divergent cortical areas in the primate brain.
Fig. 2.
Gene expression in specific cortical areas. (A–C) Genes expressed in the marmoset S1 (CUX1, DRD1, and GDF10). (D–F) Genes expressed in the marmoset V1 (VAV3, ADAMTS17, and ESRRG). (G–I) Marmoset V1 marker genes not expressed in the P14 mouse visual cortex. (Scale bar in A: 1 mm for A–F, and 300 μm for G–I.) (J) GO analysis of the molecular functions of conserved and marmoset-specific genes.
Comparative Analysis of Thalamic Nuclei-Specific Markers.
The thalamus contains first- and higher-order nuclei, which either relay subcortical information to the cortex or from one cortical area to another (23). First-order nuclei process driving input from peripheral sensory receptors and transmit the information to specific cortical regions. For example, the lateral geniculate nucleus (LGN), ventrobasal thalamic nucleus (VB), and medial geniculate body (MGB) process input to the visual, sensory, and auditory cortexes, respectively (24). The thalamic nuclei are highly anatomically conserved, allowing direction comparison of gene expression.
To identify genes exclusively expressed in the thalamic nuclei, we developed a gene expression comparison system that enables the generation and comparison of gene expression lists from multiple regions (SI Appendix, Fig. S2). To demonstrate its function, we selected specific thalamic nuclei with clear boundaries from adjacent areas in terms of Nissl-substance staining (Fig. 3A).
Fig. 3.
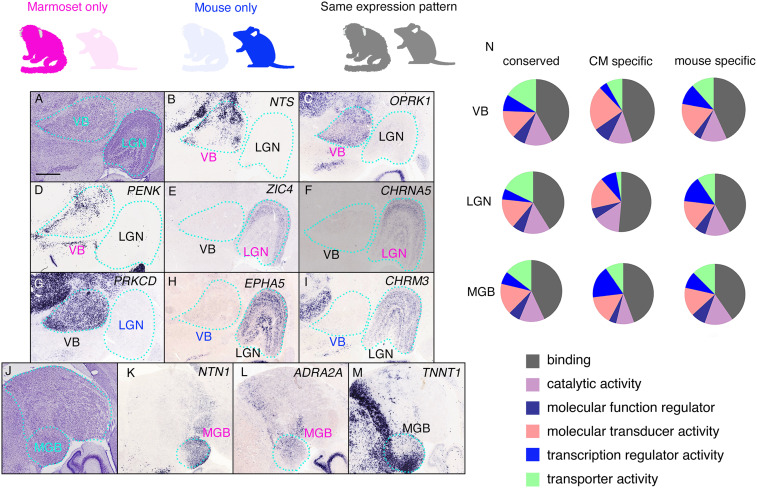
Gene-expression profiles in the thalamic nuclei. Three thalamic nuclei were selected, and their gene expression patterns were compared. Genes expressed in marmosets only, in mice only, and in both are indicated by magenta, blue, and black, respectively. (A) The Nissl reference atlas, showing the positions of the VB and LGN. (B–D) Marmoset- and VB-specific expression of NTS, OPRK1, and PENK. (E) Expression of PRKCD is specific to the marmoset VB. (E and F) ZIC4 and CHRNA5 display marmoset- and LGN-specific gene expression. (G) Expression of PRKCD is specific to the marmoset VB. (H and I) EPHA5 and CHRM3 are specifically expressed in the marmoset LGN. (J) The Nissl reference atlas showing the position of the MGB. (K and L) Marmoset- and MGB-specific expression of NTN1 and ADRA2A. (M) TNNT1 expression in the marmoset MGB with regional specification (medioventral strong). (N) GO analysis of the molecular functions of conserved and marmoset-specific genes. (Scale bar in A: 1 mm.)
First, 634 genes specific to the VB (i.e., not expressed in the LGN or MGB) were isolated and compared with genes expressed in the mouse VB (Dataset S3). Of these, 159 had conserved expression in the mouse and marmoset VB, 124 had expression only in mouse VB, 53 had marmoset-specific VB expression, and the rest were not determined due to the lack of mouse ISH data. Marmoset-specific VB expression included neurotensin (NTS), opioid receptor, kappa 1 (OPRK1), and preproenkephalin (PENK; Fig. 3 B–D). The marmoset specificity of these genes was confirmed using the ABA.
Next, we examined LGN-specific gene (659 genes) expression in mice and marmosets, and identified 183 genes with conserved expression, wherein 41 were marmoset-specific and 153 were mouse-specific (Dataset S4). Direct gene expression comparisons by ISH revealed marmoset LGN-specific expression for zinc finger protein of the cerebellum 4 (ZIC4) and cholinergic receptor, nicotinic, alpha polypeptide 5 (CHRNA5; Fig. 3 E and F), which are not expressed in the mouse dorsal (d)LGN (ABA site). This clearly demonstrates that several genes in anatomically conserved thalamic regions are expressed in a marmoset- and region-specific manner. We also identified genes expressed only in the mouse VB or LGN that displayed nuclei-specific expression in the marmoset brain. For example, protein kinase C delta (Prkcd), EPH receptor A5 (EphA5), and cholinergic receptor muscarinic 3 (Chrm3) were all expressed in the mouse VB and dLGN (SI Appendix, Fig. S3 A–F). However, in the marmoset thalamus, PRKCD was only expressed in the VB (Fig. 3G), and EPHA5 and CHRM3 were only expressed in the LGN (Fig. 3 L–I). These findings indicate the importance in examining both acquisitions and losses of gene expression in a comprehensive manner to reveal species-specific brain functions and circuit formations.
Finally, we identified genes including netrin 1 (NTN1) and adrenoreceptor alpha 2A (ADRA2A) as selectively expressed in the MGB (Fig. 3 J–L and SI Appendix, Fig. S3 G–J). Moreover, detailed analysis of conserved MGB marker genes revealed different expression patterns within the MGB. For example, troponin T1 (TNNT1) was expressed in the entire mouse MGB but displayed regional expression in the marmoset medioventral MGB (Fig. 3M and SI Appendix, Fig. S3 K and L). These results demonstrate the power of combining the GePS gene expression comparison system and the ABA AGEA to reveal species- and region-specific marker genes. Moreover, the ISH expression database is a powerful tool to reveal species-specific cell populations in conserved brain regions. GO analysis revealed enrichment of different molecular functions in various regions, suggesting that the gene expression profiles of the thalamic nuclei have evolved alongside connecting cortical areas (Fig. 3N).
GePS Detects Specific Subnuclear Gene Expression Patterns.
We next examined if GePS could detect genes expressed specifically in the dorsal (parvocellular) and ventral (magnocellular) regions of the LGN (SI Appendix, Fig. S4A). Several genes had relatively specific expression in the magnocellular layer, which receives input from the ipsilateral eye; these genes included gamma-aminobutyric acid receptor subunit delta (GABRD), ceramide kinase (CERK), neurensin 2 (NRSN2), and cytochrome B5 reductase 1 (CYB5R1; SI Appendix, Fig. S4 B–E). Genes specifically expressed in the parvocellular layer, which receives input from the contralateral eye, included neuropeptide Y receptor Y2 (NPY2R) and uncoupling protein 2 (UCP2; SI Appendix, Fig. S4 F and G). None of these genes displayed region-specific expression in the mouse dLGN. These results suggest that the connectivity, anatomy, and molecular profile of the marmoset LGN is highly evolved and distinct from the mouse LGN. We also identified marker genes with patchy expression in the marmoset LGN, including galectin 1 (LGALS1), secreted phosphoprotein 1 (SPP1), sodium channel subunit beta-3 (SCN3B), gamma-aminobutyric acid type A receptor subunit alpha 1 (GABRA1), and cerebellin 1 precursor (CBLN1; SI Appendix, Fig. S4 H–L). These results suggest relatively high cellular diversity and connectivity or reflect variation in gene expression within a single cell type rather than diversity in cell types in the marmoset LGN, similar to the cortex (25).
Marmoset-Specific Gene Expression in the Striatum.
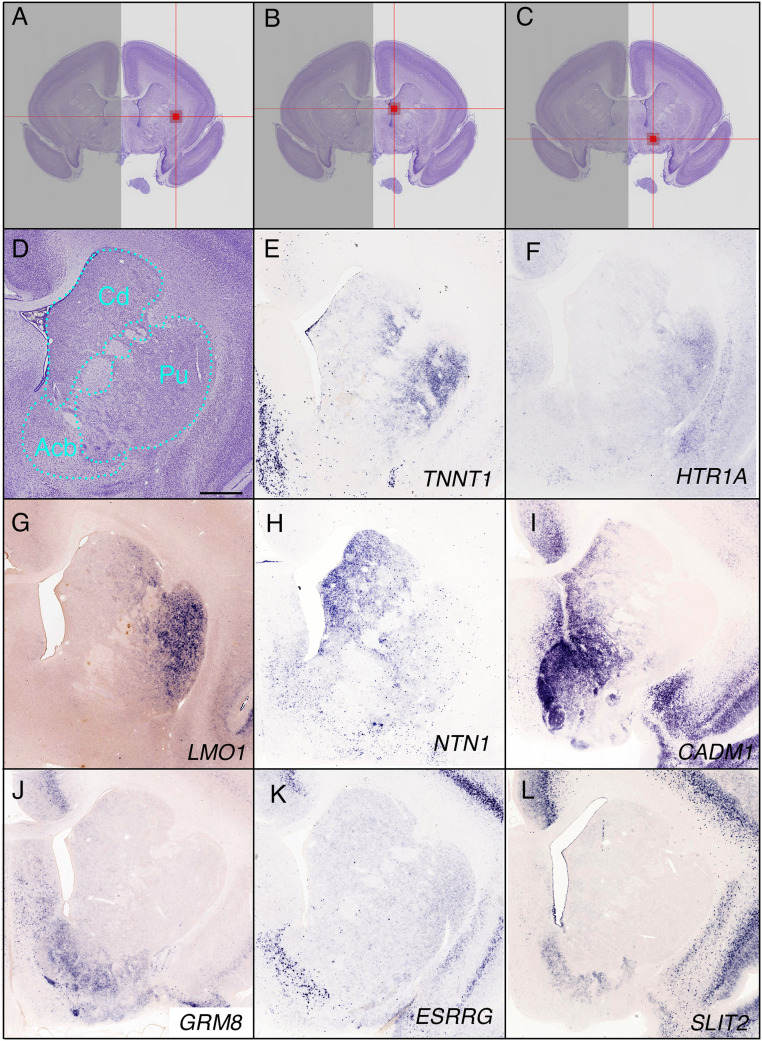
The primate striatum contains the nucleus accumbens (Acb), olfactory tubercle, caudate nucleus, and putamen. However, the borders of these regions are unclear, and many genes are expressed along a gradient in this region, as in mice (17). To identify genes with regional specificity in the striatum, we used the gene expression comparison system to identify region-specific markers of the putamen, caudate nucleus, and ventral regions of the striatum including the nucleus Acb (Fig. 4 A–D). We compared the expression of these genes in mice and marmosets to identify marmoset-specific putamen marker genes (Dataset S6), including TNNT1, 5-hydroxytryptamine (serotonin) receptor 1A (HTR1A), and LIM domain only 1 (LMO1; Fig. 4 E–G). In the caudate nucleus, 40 genes were marmoset specific (Dataset S7). ISH data confirmed the marmoset-specific expression of NTN1 and cell adhesion molecule 1 (CADM1) in the medial caudate region in a medial strong and lateral weak gradient manner (Fig. 4 H and I). Marmoset-specific ventral striatum markers (Dataset S8) included glutamate receptor, metabotropic 8 (GRM8), ESRRG, and slit guidance ligand 2 (SLIT2; Fig. 4 J–L). These results indicate increased diversity in cell types and connections or may reflect variation in gene expression within a single cell type.
Fig. 4.
Gene-expression profiles in the striatum. (A–C) The position of the indicator in the putamen (Pu; A), caudate nucleus (Cd; B), and nucleus accumbens (Acb; C) are indicated. (A) Grid position 15, 443, 272 (section number, column, raw), (B) grid position 15, 370, 252, and (C) grid position 15, 386, 317. (D) The Nissl reference atlas showing the position of the caudate nucleus. (E–G) Marmoset-specific gene expression in the putamen. TNNT1, HTR1A, and LMO1 are expressed along a lateral (strong) to medial (weak) gradient. (H and I) NTN1 and CADM1 are marmoset-specific caudate nucleus markers. CADM1 was also expressed in the Acb. (J–L) Marmoset-specific genes specifically expressed in the Acb (GRM8, ESRRG, and SLIT2). (Scale bar in D: 1 mm.)
Gene-Expression Profiles in Highly Derived Nuclei of the Primate Thalamus.
Primate pulvinar nuclei are greatly expanded in size and make connections with primate-specific cortical regions (26, 27). They can be divided anatomically into the medial (PM), lateral (PL), and inferior pulvinar (PI), which can be further divided into the medial (PIm), posterior (PIp), caudolateral, and caudomedial (PIcm) regions (Fig. 5A) (28–30). Mice also possess pulvinar nuclei, usually referred to as the lateral posterior nucleus of the thalamus (31). Conversely, the retinorecipient portion of the primate pulvinar is the PIm, which has no clear homolog in nonprimates (32). We identified genes expressed in three specific nuclei of the marmoset pulvinar (grid position 36, 420, 227 for PM, 564 genes; grid position 36, 468, 271 for PL, 412 genes; and grid position 36, 416, 275 for PI, 626 genes) and compared their expression in mice and marmosets (Dataset S9).
Fig. 5.
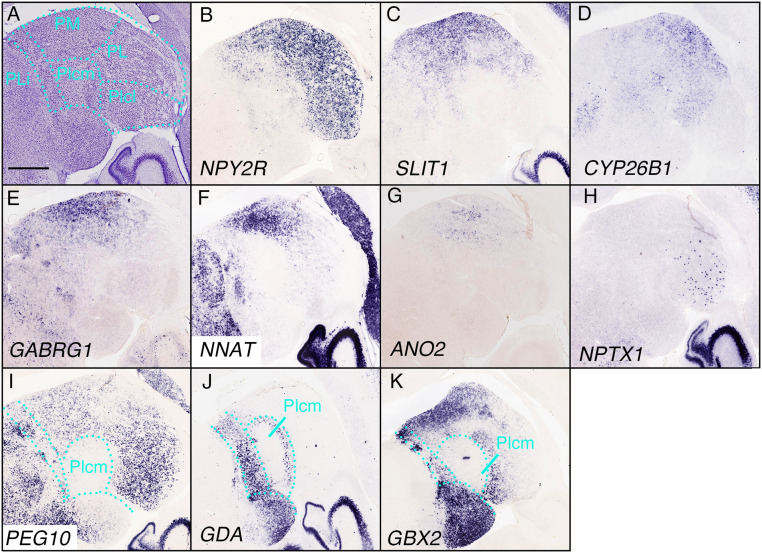
Gene-expression profiles in the pulvinar. (A) The Nissl reference atlas of the pulvinar. The medial (PM), lateral (PL), inferior (lateral and medial, Plcm and Plcl), and posterior limitans nucleus of the thalamus (PLi) are indicated. (B) NPY2R was expressed in the PM, PL, and Plcl. (C–E) Genes expressed in the PM and PL with less clear borders (SLIT1, CYP26B1, and GABRG1). (F and G) Specific expression of NNAT and ANO2 in the PM. (H) NPTX1 was expressed in a scattered manner in the PL. (I–K) Genes not expressed in the Plcm, but in the regions surrounding it (PEG10, GDA, and GBX2). (Scale bar in A: 1 mm.)
NPY2R was expressed in the PM, PL, and Plcl but not in the Plcm, whereas slit guidance ligand 1 (SLIT1), cytochrome P450, family 26, subfamily B, polypeptide 1 (CYP26B1), and gamma-aminobutyric acid type A receptor gamma 1 subunit (GABRG1) were expressed in the PM and PL but with less clear borders (Fig. 5 B–E). In comparison, neuronatin (NNAT) and anoctamin 2 (ANO2) were expressed more specifically in the PM, and ANO2 displayed a scattered pattern (Fig. 5 F and G). In the Pl, neuronal pentraxin 1 (NPTX1) was expressed in the Plcl in a stochastic manner (Fig. 5H). In addition, some genes were expressed widely in the pulvinar but not in the Plcm, such as paternally expressed 10 (PEG10; Fig. 5I). Other genes were expressed around the Plcm, including guanine deaminase (GDA) and GBX2 (Fig. 5 J and K).
A Cortical Surface Projection Mapping System to Delineate Cortical Areas Using Gene Expression.
The cerebral cortex is a continuous sheet of tissue with elaborate regional patterns. Within these broad regions, its complex functions are distributed among many anatomically distinct areas, which form maps that are similar between individuals and share topological features with other mammalian species. Using the expression patterns of multiple genes to demarcate distinct functional areas of the cerebral cortex is useful to directly compare mapping data from functional MRI (33, 34), adeno-associated virus-based injection (35), and diffusion tensor imaging (DTI) (36). This function provides comprehensive molecular cortical area maps of the primate cortex.
We first used GePS to identify genes specifically expressed in the prefrontal area (anosmin 1 [ANOS1]), S1 (semaphorin 3C [SEMA3C] and semaphorin 7A [John Milton Hagen blood group; SEMA7A]), temporal area (wingless-type MMTV integration site family, member 7b [WNT7B], decorin [DCN], and MET-proto oncogene [MET]), MT (visual area 5; CUX1), and V1 (ABI gene family, member 3 [NESH] binding protein [ABI3BP] and unc-5 netrin receptor D [UNC5D]; SI Appendix, Fig. S5).
ISH samples were collected every 196 µm (14). To project gene expression patterns onto the flat cortical map, we first registered them to a reference neonatal-marmoset MRI and generated segmentation expression masks of ISH sections using ImageJ. Even with great care, tissue sections are deformed by mechanical forces during processing, making MRI mapping challenging. We first removed the image background using image filters and morphological operators. Then, we scaled the images to 10% of their original size, converted them to grayscale, and inverted their intensities (SI Appendix, Fig. S6 A and B). Next, we aligned them spatially using a rigid body-stack alignment algorithm and improved the alignment using an elastic-stack alignment algorithm (37, 38). We improved the contrast by applying a histogram equalization and closed the gaps using a three-dimensional (3D) median filter. For final alignment with the MRI, we used the Advanced Normalization Tools (ANTS) image registration toolkit to align the 3D image stack with the 3D MRI image using a 3D deformable image transformation (39, 40). After mapping the gene expression data to the MRI reference space, we generated segmentation expression masks of ISH sections (SI Appendix, Fig. S6C). Finally, gene expression patterns in different lamina were projected onto the brain surface using a maximum intensity projection (SI Appendix, Fig. S6 D–F).
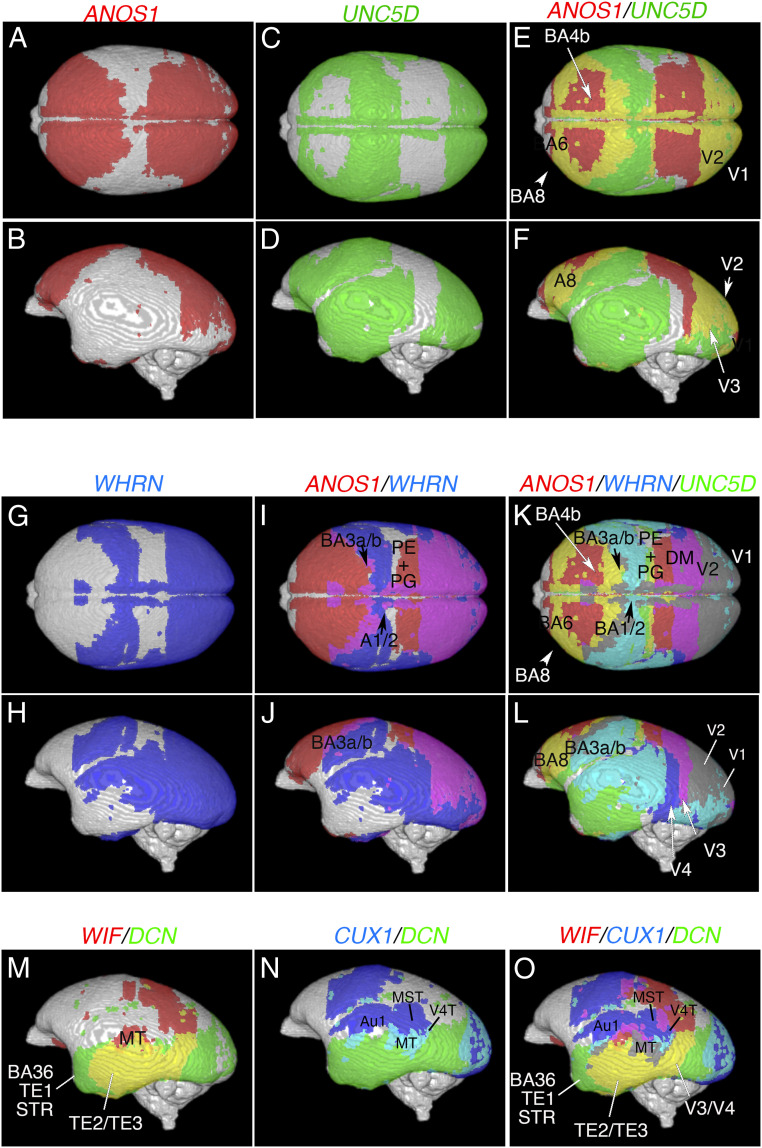
Cortical surface projection mapping provides the freedom to simultaneously compare gene expression patterns in different brains. For example, ANOS1 was expressed in the frontal, parietal (except for the somatosensory area), and occipital lobes but not in the temporal lobe (Fig. 6 A and B). In contrast, UNC5D was expressed in the parietal, occipital, and temporal lobes (Fig. 6 C and D). In an overlay image of the expression patterns, the primary motor cortex (BA4a/b), border of A6 and A8, and border of V1 and V2 (Fig. 6 E and F) were observed. Overlaying ANOS1 and WHRN revealed the borders of the BA1/2 and PE/PG (parietal area) regions in the parietal lobe (Fig. 6 G–J). Overlaying all three markers provided a clear border of the PE/PG and dorsomedial visual area in the dorsal view and the V1, V2, V3, and V4 regions in the lateral view (Fig. 6 K and L). Genes expressed in the temporal lobe, such as WNT inhibitory factor (WIF), DCN, and CUX1, allowed delineation of the AuA1, TE1 through 3 regions, MT (V5), medial superior temporal area, and visual area 4 transitional part (V4T; Fig. 6 M–O). This demonstrates that cortical surface projection mapping allows the comparison of gene expression in different brains and can reveal the borders of cortical areas when multiple genes are visualized.
Fig. 6.
Cortical surface projection mapping delineates cortical areas. Cortical area marker genes were projected onto the cortical surface and pseudocolored to compare the expression patterns of multiple genes. (A and B) Cortical surface projection mapping of ANOS1. (C and D) Cortical surface projection mapping of UNC5D. (E and F) Overlay image of ANOS1 and UNC5D expression. (G and H) Cortical surface projection mapping of WHRN. (I and J) Overlay image of ANOS1 and WHRN expression. (K and L) Overlay image of ANOS1, WHRN, and UNC5D expression. (M–O) Lateral views of WIF and DCN expression (M), CUX1 and DCN expression (N), and WIF, CUX1, and DCN expression (O). Cortical surface projection mapping delineates the A1/2, A3a/b, A4b, A6, A8, PE/PG (parietal area), DM (dorsomedial visual area), V1, V2, V3, V4, MT (V5), AuA1 (primary auditory cortex), A36/TE1/STR (temporal area 1, superior temporal area), TE2/TE3, MST (medial superior temporal area), and V4T (visual area 4, transitional part) areas.
Gene Expression in Human, Marmoset, and Mouse Visual Cortical Layers.
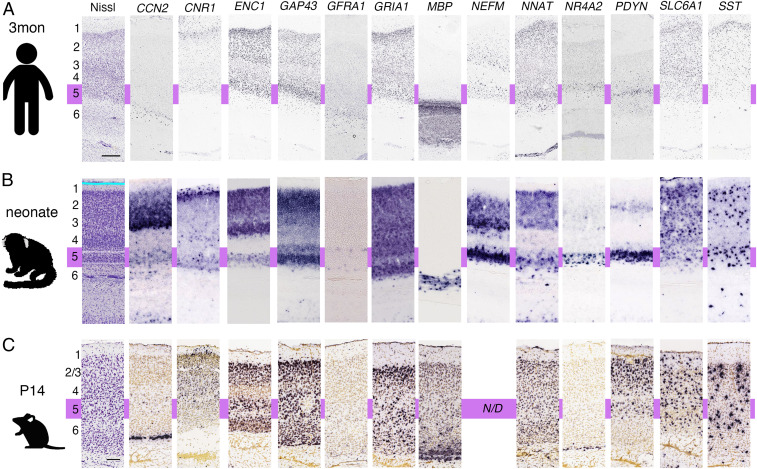
Our results demonstrate diverse gene expression patterns in the mouse and marmoset cortex, thalamus, and striatum, suggesting the possibility of different genotypes and phenotypes in genetically modified mouse models and marmosets. Predicting whether genetically modified marmoset models display more similar phenotypes to human patients than mouse models requires directly comparing gene expression patterns in marmoset and human V1. We compared ISH data from a 3 mo old human from the ABA (http://human.brain-map.org/ish/search) with neonatal marmoset and P14 mouse gene expression data, examining 13 genes with data in both the human gene expression database and the MGA (Fig. 7). Genes displaying the most dynamic differences between marmosets and humans included cellular communication network factor 2 (CCN2), a layer 6b marker gene in the mouse brain (41) (Fig. 7). There was no expression of CCN2 in marmoset layer 6; however, strong expression was observed in layer 5 and in upper layers. Prodynorphin (PDYN) expression in marmosets and humans was similar in layer 5, with additional expression in marmoset layer 2. In mice, Pdyn was expressed in layer 5, with additional widespread expression in upper layers. Similar gene expression and layer-distribution patterns in all three species were observed for cannabinoid receptor 1 (CNR1), ectodermal-neural cortex 1 (ENC1), glial cell line–derived neurotrophic factor family receptor alpha 1 (GFRA1), glutamate receptor, ionotropic, AMPA1 (alpha 1) (GRIA1), solute carrier family 6 (neurotransmitter transporter, GABA), member 1 (SLC6A1), and somatostatin (SST). The rest of the genes showed similar expression patterns between the marmoset and human visual cortexes but not in mouse cortexes, including growth-associated protein 43 (GAP43: expressed in layers 2, 3, and 5 in primates but distributed throughout the mouse cortical plate), myelin basic protein (MBA: restricted expression in deeper layers and white matter in primates but distributed throughout the mouse cortical plate), NNAT (strong layer 5 and upper layer expression in primates but distributed throughout the mouse cortical plate), and nuclear receptor subfamily 4 group A member 2 (NR4A2: strong layer 5 expression in primates but only observed in mouse layer 6). The results reveal more similar gene expression distribution patterns in marmosets and humans compared with mice. However, the number of genes tested and the human sample size were limited. To perform comparative gene expression analysis when more human samples become available, we developed the search engine “gene classification.” First, we used the Protein ANalysis THrough Evolutionary Relationships (PANTHER) pathway system (42), a freely available, comprehensive software system for relating protein sequence evolution to the evolution of specific protein functions and biological roles, to classify gene function. Next, we installed GePS in the “gene classification” search system, making it possible to search gene expression for a specific molecular function class or molecular pathway. We believe this makes it possible to search gene expression by cell-type–specific genes or genes that are important for developing neural circuits to compare their expressions in different species.
Fig. 7.
Gene expression in the human, marmoset, and mouse V1. A 3 mo old human (A), neonatal marmoset (B), and P14 mouse (C) gene expression patterns were compared to reveal the layer distributions of each gene. The approximate position of layer 5 is indicated by purple. (Scale bars in A (Nissl): 1 mm, 200 μm, and 100 μm for the human, marmoset, and mouse samples, respectively.)
Evaluation of the Marmoset as an Animal Model for Developmental Brain Disorders.
The above results suggest that the marmoset can be a good animal model for developmental brain disorders. Therefore, we compared genes associated with developmental brain disorders such as intellectual disability, autism spectrum disorder, epilepsy, attention deficit hyperactivity disorder, schizophrenia, and bipolar disorder in the cortex (S1, V1, and AuA1), thalamus (VB, LGN, and MGB), and striatum (caudate nucleus, putamen, and nucleus Acb). There were 602 relevant genes listed in the Developmental Brain Disorder Gene Database (https://dbd.geisingeradmi.org/), of which 71 had ISH data in the MGA and ABA (Dataset S10). Several genes showed species-specific expression in the thalamus and striatum, indicating that these regions may particularly contribute to species-specific behavior. It is not possible to test the functions of genes with marmoset-specific expression in the thalamus (lysine [K])-specific methyltransferase 2C (KMT2C), cardiotrophin-like cytokine factor 1 (CRLF), cholinergic receptor, nicotinic, alpha polypeptide 2 (CHRNA2), Meis homeobox2 (MEIS2), protocadherin 19 (PCDH19), and SLC6A in genetically modified mouse models; therefore, marmosets have the potential to reveal novel functions for developmental brain disorder genes and provide novel treatment targets for patients.
Discussion
The MGA contains a large gene expression dataset from the developing brain of the common marmoset. However, the data represent the gene expression patterns across the entire brain, and methods to isolate region-specific gene expression patterns will provide important information regarding primate brain structure, function, and cellular and molecular organization. We have developed a system, GePS, that systematically identifies genes expressed in selected regions of the common marmoset brain. Comprehensive, comparative gene expression analysis with mice revealed many genes with species- and region-specific expression. Moreover, by combining GePS with the gene expression comparison and cortical area projection mapping systems, we were able to provide further insight into the molecular organization of these species- and region-specific gene expression patterns.
In the present study, we developed a neonatal marmoset ISH-based coronal gene expression detection system and used it to demonstrate the strength of GePS. However, more comprehensive gene mapping at different time points will be important. In our future work, we plan to perform ISH at different marmoset developmental stages (1 mo, 3 mo, 6 mo, and 1 y). GePS is also applicable at these ages, which will provide an anatomical time course of gene expression in the marmoset brain. Moreover, the anatomical mapping resolution of GePS remains restricted to a voxel size of 200 µm on each side (1 grid = 200 × 200 pixels or 500 × 500 µm). As the current resolution may be too coarse to distinguish smaller nuclei or cell types in some regions, a new iteration of GePS with improved resolution is being developed. We believe that using this smaller grid system (20 × 20 pixels) will make it possible to show the quantitative analysis. The current grid size (200 × 200 pixels) makes quantitative assessment difficult, especially for sections which have weak signals and low signal-to-noise ratio. Moreover, we also tested the suitability of GePS for brain regions having more subdivisions, such as the hippocampus and amygdala. As we reported previously, detailed gene expression in neonatal marmosets revealed many different gene expression patterns in the specific region of the hippocampus (19). However, gene search in the hippocampus and amygdala using GePS did not provide detailed gene expression profiles due to grid size. Thus, developing a search function with a smaller grid is crucial for enabling the search of gene expression with cellular resolution.
GePS analysis of the MGA provides complementary and essential information allowing the discernment and characterization of discrete cell types and previously unrecognized molecular subdivisions. Moreover, as ISH is performed on preserved tissue with relatively low levels of RNA degradation, GePS also uniquely enables comparative analysis of different species, especially NHPs, from whom the high-quality tissue required for RNA-seq is not readily available. This will be critical to understanding the function and connectivity of highly evolved brain regions. Although the fundamental evolutionary mechanisms underlying brain structures are not understood, several lines of evidence have suggested that species-specific brain regions are either generated by duplicating existing areas (phylogenetic addition) or segregated from existing areas (phylogenetic segregation) and then further obtain their unique characters (20, 43, 44). It is possible that unique brain regions and duplicated or segregated regions share similar molecular properties, with similar functions and connectivity. For instance, the pulvinar, which is highly evolved in the primate brain and processes visual information, has a similar function to the LGN but a more complex connectivity. The primate pulvinar nuclei are greatly expanded in size and make connections with primate-specific cortical regions (26, 27). The PL and Pl have widespread connections with visual cortical association areas (31, 45, 46). The PIp and PIcm receive a high density of crudely topographic inputs from the superior colliculus (SC), whereas the PL receives punctate but topographically organized inputs from the SC, which seems to function as a secondary visual input route to the cortex (47, 48). A previous study used neuronal tracers to demonstrate that the Plm has widespread connections with layers 3 and 4 of the prefrontal cortex (45, 49). In the current study, detailed gene expression profiling within the LGN and pulvinar revealed region- and cell-type specific molecular markers. For example, NPY2R was specifically expressed in the PM, PL, and parvocellular layer of the LGN. Elsewhere, NPY2R expression was observed only in the anterior thalamic nuclei, specific cortical layers, and hippocampal CA2 region, with some scattered expression (https://gene-atlas.brainminds.riken.jp/gene-image/?gene=279-1). Shared NPY2R expression suggests that these brain regions also share similar cellular functions, connectivity, and evolutionary origins (32, 50–52).
Our ultimate goal is to understand how the emergence of complex human behavior, which was due to an expanded number of cortical areas (53), increases the complexity of cortical network organization. This can be revealed by observing the default mode network (DMN) in the human brain, which is quite different in health and disease (54). Utilizing the common marmoset as a model system required a platform that enabled direct comparison of human and NHP cortical area maps. In the current study, cortical surface projection mapping delineated several cortical areas that are not detectable through single gene expression analysis of coronally sliced ISH data. For instance, the borders for visual cortical areas (V1 through V4), labeled by coexpression of ANOS1, UNC5D, and WHRN, are visible only by cortical surface projection mapping, as these marker genes are expressed in different cortical layers that do not overlap in two-dimensional ISH images. Additional cortical surface projection maps and different gene combinations, which will be provided on the MGA site, have the potential to delineate further primate-specific cortical areas, providing insights into primate-specific cortical function and connectivity. Together with technologies such as microarrays, multiscale RNA-seq, and high-resolution human brain mapping by MRI, DTI, and DMN, the marmoset ISH database provides an invaluable reference tool that helps translate knowledge from rodents into primates and advance primate molecular neurobiology research (55).
Methods
Animals.
Animals were derived from a breeding colony at the RIKEN Center for Brain Science, Research Resource Division. Information of the weight, gender, and birth date of the animals is listed on the MGA website. The use of the marmosets followed the guidelines of and were approved by the RIKEN Institutional Animal Care Committee (W2020-2-022).
Image Collection.
Brains were sectioned to 28 µm on a freezing microtome. For ISH staining, 60 coronal sections at 196 μm intervals were used for each probe, allowing us to test eight different probes in each brain (14). Section images were converted and aligned with the originally developed reference atlas as previously described (14). All procedures (sampling, probe design, ISH, and image processing) have been previously described (14, 19).
Comparative Transcriptomics.
We used the ABA, a publicly available mouse ISH database, and its AGEA system to identify brain-region–specific gene expression patterns at P14 and P28 (56). The datasets were compared to marmoset region-specific gene lists to identify genes that were conserved, marmoset specific, mouse specific, and not determined. To identify overrepresented functional categories among different sets of expressed genes, we used the PANTHER database (http://go.pantherdb.org/) (42). GO terms in the molecular function, biological process, and cellular component domains were tested for enrichment in the conserved, marmoset-specific and mouse-specific gene sets. To examine the expression patterns of genes associated with developmental brain disorders, we sourced 71 genes from the Developmental Brain Disorder Gene Database (https://dbd.geisingeradmi.org/) and compared their expression.
Cortical Area Projection Mapping System.
The cortical area projection mapping system uses 3D reconstructions of gene expression patterns as input data and maps cortical patterns on the brain surface as a maximum intensity projection. In this system, a point on the surface is positive as long as at least one voxel in the projection path had a positive result. As an example, projection paths are illustrated by the yellow arrows in SI Appendix, Fig. S6. To determine projection trajectories, we calculated the normalized brain surface and used heat propagation to propagate directional information throughout the entire cortex.
Supplementary Material
Acknowledgments
We thank Akira Sato, Kanako Saga, Miyano Sakurai, Masae Sato, Sachie Suzuki, Keiko Yamashita, Tomoko Yoda, Mami Tanaka, Mao Yamaguchi, Makiko Someya, Masako Hama, and Karl Windak for technical assistance. We thank Dr. James Bourne (Monash University) and Dr. Seth Blackshaw (Johns Hopkins University) for critically reading the manuscript and providing comments to improve it. This work was supported by the RIKEN Center for Brain Science, the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST-program), and the Brain/MINDS project from the Japan Agency for Medical Research and Development (Grant JP20dm0207001) and, in part, by NIH Grant U01MH124619.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020125118/-/DCSupplemental.
Data Availability
Image data have been deposited in Tomomi Shimogori (https://gene-atlas.brainminds.riken.jp/) (57). All study data are included in the article and/or supporting information.
References
- 1.Cadwell C. R., et al., Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol. 34, 199–203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bomkamp C., et al., Transcriptomic correlates of electrophysiological and morphological diversity within and across excitatory and inhibitory neuron classes. PLoS Comput. Biol. 15, e1007113 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlyle B. C., et al., A multiregional proteomic survey of the postnatal human brain. Nat. Neurosci. 20, 1787–1795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molnár Z., et al., New insights into the development of the human cerebral cortex. J. Anat. 235, 432–451 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sousa A. M. M., et al., Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Y., et al., Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macosko E. Z., et al., Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer S., et al., Multimodal single-cell analysis reveals physiological maturation in the developing human neocortex. Neuron 102, 143–158.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodge R. D., et al., Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tosches M. A., et al., Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 360, 881–888 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Rodriques S. G., et al., Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen E. H., Overly C. C., Jones A. R., The Allen Human Brain Atlas: Comprehensive gene expression mapping of the human brain. Trends Neurosci. 35, 711–714 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Zeng H., et al., Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell 149, 483–496 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimogori T., et al., Digital gene atlas of neonate common marmoset brain. Neurosci. Res. 128, 1–13 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Okano H., Mitra P., Brain-mapping projects using the common marmoset. Neurosci. Res. 93, 3–7 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Okano H., et al., Brain/MINDS: A Japanese national brain project for marmoset neuroscience. Neuron 92, 582–590 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Ng L., et al., An anatomic gene expression atlas of the adult mouse brain. Nat. Neurosci. 12, 356–362 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Romero-Grimaldi C., Moreno-López B., Estrada C., Age-dependent effect of nitric oxide on subventricular zone and olfactory bulb neural precursor proliferation. J. Comp. Neurol. 506, 339–346 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Mashiko H., et al., Comparative anatomy of marmoset and mouse cortex from genomic expression. J. Neurosci. 32, 5039–5053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krubitzer L., The magnificent compromise: Cortical field evolution in mammals. Neuron 56, 201–208 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Mundinano I. C., et al., Transient visual pathway critical for normal development of primate grasping behavior. Proc. Natl. Acad. Sci. U.S.A. 115, 1364–1369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahata T., et al., Activity-dependent expression of occ1 in excitatory neurons is a characteristic feature of the primate visual cortex. Cereb. Cortex 16, 929–940 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa Y., Shimogori T., Diversity of thalamic progenitor cells and postmitotic neurons. Eur. J. Neurosci. 35, 1554–1562 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Macchi G., Bentivoglio M., Minciacchi D., Molinari M., Trends in the anatomical organization and functional significance of the mammalian thalamus. Ital. J. Neurol. Sci. 17, 105–129 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Krienen F. M.et al., Innovations in primate interneuron repertoire. Nature 586, 262–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fosse V. M., Fonnum F., Biochemical evidence for glutamate and/or aspartate as neurotransmitters in fibers from the visual cortex to the lateral posterior thalamic nucleus (pulvinar) in rats. Brain Res. 400, 219–224 (1987). [DOI] [PubMed] [Google Scholar]
- 27.Bystron I., Blakemore C., Rakic P., Development of the human cerebral cortex: Boulder Committee revisited. Nat. Rev. Neurosci. 9, 110–122 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Tamietto M., Morrone M. C., Visual plasticity: Blindsight bridges anatomy and function in the visual system. Curr. Biol. 26, R70–R73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warner C. E., et al., Preservation of vision by the pulvinar following early-life primary visual cortex lesions. Curr. Biol. 25, 424–434 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Homman-Ludiye J., Kwan W. C., de Souza M. J., Bourne J. A., Full: Ontogenesis and development of the nonhuman primate pulvinar. J. Comp. Neurol. 526, 2870–2883 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Baldwin M. K. L., Wong P., Reed J. L., Kaas J. H., Superior colliculus connections with visual thalamus in gray squirrels (Sciurus carolinensis): Evidence for four subdivisions within the pulvinar complex. J. Comp. Neurol. 519, 1071–1094 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baldwin M. K. L., Balaram P., Kaas J. H., The evolution and functions of nuclei of the visual pulvinar in primates. J. Comp. Neurol. 525, 3207–3226 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Liu C., et al., A resource for the detailed 3D mapping of white matter pathways in the marmoset brain. Nat. Neurosci. 23, 271–280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hikishima K., et al., Atlas of the developing brain of the marmoset monkey constructed using magnetic resonance histology. Neuroscience 230, 102–113 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Watakabe A., et al., Application of viral vectors to the study of neural connectivities and neural circuits in the marmoset brain. Dev. Neurobiol. 77, 354–372 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mimura K., et al., Abnormal axon guidance signals and reduced interhemispheric connection via anterior commissure in neonates of marmoset ASD model. Neuroimage 195, 243–251 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Thévenaz P., Ruttimann U. E., Unser M., A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27–41 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Saalfeld S., Fetter R., Cardona A., Tomancak P., Elastic volume reconstruction from series of ultra-thin microscopy sections. Nat. Methods 9, 717–720 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Avants B. B., et al., A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Avants B. B., Epstein C. L., Grossman M., Gee J. C., Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoerder-Suabedissen A., Molnár Z., Molecular diversity of early-born subplate neurons. Cereb. Cortex 23, 1473–1483 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Mi H., Muruganujan A., Casagrande J. T., Thomas P. D., Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krubitzer L., Kaas J., The evolution of the neocortex in mammals: How is phenotypic diversity generated? Curr. Opin. Neurobiol. 15, 444–453 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Krubitzer L., Dooley J. C., Cortical plasticity within and across lifetimes: How can development inform us about phenotypic transformations? Front. Hum. Neurosci. 7, 620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romanski L. M., Giguere M., Bates J. F., Goldman-Rakic P. S., Topographic organization of medial pulvinar connections with the prefrontal cortex in the rhesus monkey. J. Comp. Neurol. 379, 313–332 (1997). [PubMed] [Google Scholar]
- 46.Kwan W. C., et al., Unravelling the subcortical and retinal circuitry of the primate inferior pulvinar. J. Comp. Neurol. 527, 558–576 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Stepniewska I., Kaas J. H., Architectonic subdivisions of the inferior pulvinar in New World and Old World monkeys. Vis. Neurosci. 14, 1043–1060 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Weller R. E., Steele G. E., Kaas J. H., Pulvinar and other subcortical connections of dorsolateral visual cortex in monkeys. J. Comp. Neurol. 450, 215–240 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Homman-Ludiye J., Mundinano I. C., Kwan W. C., Bourne J. A., Extensive connectivity between the medial pulvinar and the cortex revealed in the marmoset monkey. Cereb. Cortex 30, 1797–1812 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Turk-Browne N. B., The hippocampus as a visual area organized by space and time: A spatiotemporal similarity hypothesis. Vision Res. 165, 123–130 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Groen T., Kadish I., Michael Wyss J., Role of the anterodorsal and anteroventral nuclei of the thalamus in spatial memory in the rat. Behav. Brain Res. 132, 19–28 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Jankowski M. M., et al., The anterior thalamus provides a subcortical circuit supporting memory and spatial navigation. Front. Syst. Neurosci. 7, 45 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geschwind D. H., Rakic P., Cortical evolution: Judge the brain by its cover. Neuron 80, 633–647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Padmanabhan A., Lynch C. J., Schaer M., Menon V., Sciences B., The default mode network in autism. Biol. Psychiatry Cognit. Neurosci. Neuroimaging 2, 476–486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okano H., Current status of and perspectives on the application of marmosets in neurobiology. Annu. Rev. Neurosci. 44, 27–48 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Thompson C. L., et al., A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron 83, 309–323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimogori T., Marmoset Gene Atlas https://gene-atlas.brainminds.riken.jp/. Deposited 14 January 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Image data have been deposited in Tomomi Shimogori (https://gene-atlas.brainminds.riken.jp/) (57). All study data are included in the article and/or supporting information.