Abstract
Background
Exosomal circular RNAs (circRNAs) are involved in the pathogenesis of prostate cancer (PCa) and chemotherapy resistance. This research aimed to explore the function and molecular mechanism of circRNA X-linked inhibitor of apoptosis (circ-XIAP) in docetaxel (DTX) resistance of PCa.
Methods
The expression of circ-XIAP, microRNA-1182 (miR-1182), tumor protein D52 (TPD52) was measured by quantitative real-time polymerase chain reaction (qRT-PCR). Exosomes were detected with transmission electron microscopy (TEM). Cluster of differentiation 63 (CD63), cluster of differentiation 9 (CD9) and TPD52 protein levels were detected by Western blot (WB). FIfty percent inhibitory concentration (IC50) of DTX and cell viability were determined using Cell Counting Kit-8 (CCK-8) assay. Colony formation assay was applied to assess colony-forming ability. Cell cycle distribution and apoptosis were analyzed by flow cytometry. Transwell assay was used for measuring cell migration and invasion. Dual-reporter luciferase assay was performed to confirm the interaction between miR-1182 and circ-XIAP or TPD52. The role of circ-XIAP in vivo was confirmed via the mice xenograft model.
Results
Circ-XIAP and TPD52 were upregulated and miR-1182 was downregulated in DTX-resistant PCa tissue specimens and cell lines. Circ-XIAP was also overexpressed in exosomes from DTX-resistant cells and could be transmitted via exosomes. Circ-XIAP knockdown enhanced DTX sensitivity by suppressing DTX-resistant cell proliferation, migration and invasion and inducing cell cycle arrest and apoptosis. Circ-XIAP directly targeted miR-1182, and the effects of circ-XIAP knockdown were reversed by downregulating miR-1182 in DTX-resistant cells. TPD52 was the target of miR-1182, and its upregulation weakened the promotive effect of miR-1182 on DTX sensitivity. Importantly, circ-XIAP depletion inhibited tumor growth and increased DTX sensitivity in vivo.
Conclusion
Exosomal circ-XIAP promoted DTX resistance of PCa by regulating miR-1182/TPD52 axis, providing a promising therapeutic target for PCa chemotherapy.
Keywords: prostate cancer, exosomes, circ-XIAP, miR-1182, TPD52
Introduction
Prostate cancer (PCa) is a common malignant tumor and is the fifth leading cause of cancer death in men (359,000 associated deaths in 2018).1 Although comprehensive treatments have been developed, the prognosis of PCa patients is still unsatisfactory because of the recurrence and metastasis.2 Docetaxel (DTX)-based chemotherapy is a first-line chemotherapy for the treatment of PCa that can prolong the survival of patients.3 However, many PCa patients gradually develop resistance to DTX treatment during chemotherapy, resulting in tumor recurrence.4 Hence, understanding the underlying mechanism of DTX resistance is of great significance to improve the prognosis of PCa patients.
Exosomes are a class of extracellular vesicles with a size of 40–100 nm, which originate from endosomal multivesicular bodies.5 Exosomes are secreted by most cells and contain complex and rich RNAs and proteins.6 Exosomes have been confirmed to be involved in regulating the tumor-normal communication in the tumor microenvironment.7 As key mediators of intercellular communication, exosomes can regulate the biological processes of cells and serve as crucial biomarkers for various diseases.8,9 Moreover, exosomes have been demonstrated to play an active role in tumor development (including PCa) and DTX resistance.10,11 Nevertheless, the components of exosomes secreted by different cells are complex and different, so more exosomal RNA molecules need to be discovered and explored.
CircRNAs, a special type of non-coding RNAs (ncRNAs), form continuous covalently closed-loops with neither 5ʹ-end cup nor 3ʹ-end ploy A tail.12 Moreover, circRNA have strong stability due to their covalently closed continuous loop structures.13 Increasing evidence indicates that circRNAs participate in regulating many physiological and pathological processes in human cancers.14,15 Moreover, circRNAs have been confirmed to be involved in participate in DTX resistance in many cancers, including PCa.16,17 Previous researches have indicated that circRNAs are stable and enriched in exosomes.18 Moreover, it has been reported that exosomal circRNAs can be served as potential biomarkers and new therapeutic targets for diseases.19 CircRNA X-linked inhibitor of apoptosis (circ-XIAP) is derived from mRNA back-splicing of XIAP gene and located at chrX: 123022468–123026623, and it acts as an oncogene in PCa.20 Nevertheless, the functions and molecular mechanism of exosomal circ-XIAP in DTX resistance of PCa have not been reported.
CircRNAs can function as microRNAs (miRNAs) sponges by competitively binding to miRNA response elements, thus regulating the biological activity of miRNA and the functional role of miRNA on target genes.21 MiR-1182 has been shown to inhibit proliferation, metastasis and tumorigenesis in many cancers.22,23 Recently, miR-1182 was found to function as a tumor suppressor in PCa.24 However, whether circ-XIAP can modulate the resistance of PCa to DTX through interacting with miR-1182 is still unclear. Tumor protein D52 (TPD52), a chromosome 8q21 amplification target, acts as an oncogene in many cancers, including PCa.25,26 Coincidently, by bioinformatics analysis, we found that both circ-XIAP and 3ʹUTR of TPD52 had the complementary binding sequence for miR-1182, which suggested the interactions among circ-XIAP, miR-1182 and TPD52 in DTX resistance of PCa.
In our research, we aimed to investigate the biological functions of exosome-transmitted circ-XIAP in DTX resistance of PCa, and search for the underlying mechanism mediated by circ-XIAP, hoping to find novel targets for the treatment of DTX-resistant PCa patients.
Materials and Methods
Patient Samples
PCa tissue specimens (N=52) were acquired from the PCa patients after surgical resection at The Central Hospital of Zhumadian. These PCa patients were divided into two groups based on their sensitivity to DTX: sensitive (N=23) and resistant (N=29). These tissues specimens were collected, timely frozen in liquid nitrogen and then kept in a refrigerator at −80°C after surgical resection. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki, These patients had provided informed consents. Our experiments were approved by the Research Ethics Committee of The Central Hospital of Zhumadian.
Cell Culture and Transfection
Four PCa cell lines (22Rv1, VCaP, DU145, and PC3) and human normal prostate epithelial cells (RWPE-1) were obtained from COBIOER (Nanjing, China). All cells were allowed to grow in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) that contained 10% FBS (Sigma-Aldrich, St. Louis, MO, USA) in a humidified air with 5% CO2 at 37°C. To establish DTX-resistant PCa cells (DU145/DTX and PC3/DTX), DU145 and PC3 cells were exposed to increasing doses of DTX (Sigma-Aldrich). Then, DTX-resistant cells were treated with DTX (5 nM) to maintain a resistant phenotype.
Small interference RNA against circ-XIAP (si-circ-XIAP) and matched negative control (si-NC), circ-XIAP or TPD52 overexpression vector (circ-XIAP or TPD52) and matched negative control (vector), miR-1182 mimics (miR-1182) and match negative control (miR-NC), miR-1182 inhibitor (anti-miR-1182) and match negative control (anti-miR-NC) were synthesized by GeneCopoeia (Guangzhou, China). Lentivirus packaged plasmid sh-circ-XIAP and its negative control (sh-NC) were provided by RiboBio (Guangzhou, China). Cells were transfected by using Lipofectamine 3000 (Invitrogen).
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
TRIzol reagent (Invitrogen) was applied for isolation of total RNA. Total exosomal RNAs were acquired using the exoRNeasy Midi Kit (Qiagen, Valencia, CA, USA). The Prime Script RT reagent Kit (TaKaRa, Kusatsu, Japan) or miRNA cDNA synthesis kit (Ribobio) was applied for reverse transcription. Then, qRT-PCR was performed on a CFX Real-time PCR system (Bio-Rad, Hercules, CA, USA) using SYBR Green PCR Kit (Takara). Gene expression was detected according to the 2-ΔΔCt method, followed by normalization to GAPDH (for circ-XIAP, XIAP and TPD52) or U6 (for miR-1182). Primer sequences: circ-XIAP (F, 5ʹ-CATTCACTTGAGGAGTGTCTGG-3ʹ; R, 5ʹ-TTGTTCCCAAGGGTCTTCAC-3ʹ); XIAP (F, 5ʹ-TGGCAGATTATGAAGCACGGATC-3ʹ; R, 5ʹ-AGTTAGCCCTCCTCCACAGTGA-3ʹ); miR-1182 (F, 5ʹ-GCGGAGGGTCTTGGGAGG-3ʹ; R, 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ); TPD52 (F, 5ʹ-AACAGAACATTGCCAAAGGGTG-3ʹ; R, 5ʹ-TGACTGAGCCAACAGACGAAA-3ʹ). GAPDH (F, 5ʹ-GTCTCCTCTGACTTCAACAGCG-3ʹ; R, 5ʹ-ACCACCCTGTTGCTGTAGCCAA-3ʹ), U6 (F, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ; R, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ).
RNase R Treatment
To determine the circular characteristic of circ-XIAP, total RNA (2 µg) was treated with or without RNase R (6 units, Geneseed Biotech, Guangzhou, China) for 0.5 h at 37°C. Lastly, the enrichment of circ-XIAP and XIAP mRNA was tested by qRT-PCR analysis.
Subcellular Fractionation Location
The Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek, Thorold, Canada) was applied for isolating cytosolic and nuclear fractions. Afterwards, the expression was detected via qRT-PCR analysis in nuclear and cytosolic fractions. U6 or GAPD was used as a control for the nucleus and cytoplasm, respectively.
Cell Counting Kit-8 (CCK-8) Assay
CCK-8 purchased from Boster (Wuhan, China) was used for analyzing 50% inhibitory concentration (IC50) value of DTX and cell viability. In brief, DU145/DTX and PC3/DTX cell suspension (100 µL, 2×103 cells) was added into a 96-well plate. CCK-8 reagent (10 µL) was added to per well after respective treatment. After incubation for 2–3 h, a microplate reader (Bio-Rad) was applied to examine the absorbance of per well at 450 nm. IC50 value of DTX was calculated using GraphPad Prism.
Exosome Isolation and Identification
ExoQuick precipitation kit (System Biosciences, Mountain view, CA, USA) was utilized for isolating exosomes from cell culture medium following the manufacturer’s instructions. In short, cell culture medium was collected and centrifuged (3000 × g, 15 min), followed by addition of ExoQuick precipitation solution to supernatant. After centrifugation (1500 × g, 30 min), exosome pellet was re-suspended in phosphate-buffered saline (PBS; Beyotime, Shanghai, China). Exosomes were visualized by transmission electron microscopy (TEM; JEOL, Akishima, Japan) and exosome protein markers (CD63 and CD9) were identified by WB assay.
Western Blot (WB) Assay
RIPA lysis buffer (Beyotime) was applied for extracting the total protein. After measurement of protein concentration, protein (about 40 µg/lane) was subjected to SDS-PAGE (Beyotime). Subsequently, the proteins were transferred to nitrocellulose membranes (Invitrogen), which were then blocked with 5% non-fat milk (Yili, Beijing, China) and incubated with primary antibody overnight at 4°C. The primary antibodies including cluster of differentiation 63 (CD63; 1:1000, ab118307), cluster of differentiation 9 (CD9; 1:500, ab223052), TPD52 (1:5000, ab155296), and GAPDH (1:2000, ab37168) were bought from Abcam (Cambridge, UK). After that, the corresponding secondary antibody (1:4000, ab205718, Abcam) was used for the combination with the primary antibody. At last, the visualization of protein blots was achieved by an enhanced chemiluminescence reagent (Solarbio, Beijing, China).
Colony Formation Assay
Transfected cells (DU145/DTX and PC3/DTX) were plated into 12-well plates. The cells were then cultured for 14 days, and the medium was replaced as needed. After that, the colonies were fixed with ethanol (75%, Beyotime), followed by staining with violet (0.1%, Beyotime). At last, colonies (>50 cells/colony) were counted and photographed.
Flow Cytometry Analysis
For analysis of cell cycle progression, DU145/DTX and PC3/DTX cells were collected, followed by fixing with ice-cold ethanol (75%, Beyotime) at −20°C for 12 h. Next, the fixed cells were subsequently incubated with propidium iodide (PI; Sangon Biotech, Shanghai, China) and RNase A (Sangon Biotech) in the darkness for 0.5 h. The cells were analyzed by flow cytometry (BD Biosciences, Franklin, NJ, USA) for determination of cell cycle distribution. The apoptosis assay was conducted with the Annexin V-FITC/PI apoptosis detection kit (Sangon Biotech) via suspending the cells in binding buffer (400 μL) containing Annexin V-FITC and PI, incubating them in the darkness for 20 min. Finally, flow cytometry was utilized to quantify cell apoptosis.
Transwell Assay
Transwell assay was performed by transwell inserts (8 μm pore-size, (8 mm pores; Corning, Costar, NY, USA) either without Matrigel (for migration assay) or with Matrigel (for invasion assay) (BD Biosciences). Briefly, the serum-free media containing transfected cells was added to the top chamber, and the medium with 10% FBS was added to the lower part of the chamber. The cells on the bottom membrane surface were fixed in ethanol (Beyotime) and stained by crystal violet (Beyotime) after 24 h of incubation. Images were captured under a microscope (Leica, Wetzlar, Germany) at ×100 magnification.
Dual-Luciferase Reporter Assay
The putative binding sequence of miR-1182 and circ-XIAP or TPD52 was predicted by Circinteractome or TargetScan. Wide-type (WT) sequence of circ-XIAP or TPD52 3ʹUTR with binding sequence for miR-1182 was cloned into the pmirGLO vector (YouBia, Changsha, China) to construct WT-circ-XIAP or WT-TPD52 3ʹUTR. At the same time, mutant reporter plasmids (MUT-circ-XIAP or MUT-TPD52 3ʹUTR) without binding sequence for miR-1182 were generated in the same way. Then, the indicated vector and miR-NC/miR-1182 were co-transfected into DU145/DTX and PC3/DTX cells. After 48 h of co-transfection, dual-luciferase reporter assay system (Promega, Madison, WI, USA) was applied for examining the luciferase activity.
Immunohistochemistry Analysis
Human tumor tissues from sensitive and resistant tissues were fixed in formalin solution (Beyotime) and dehydrated in ethanol, embedded in paraffin, and cut. Next, consecutive thick sections (4 μm) were incubated with antibodies against TPD52 (1:5000, ab155296) for 12 h at 4°C. After that, the sections were incubated with secondary antibody (1:5000, ab205718, Abcam) for 1 h. The sections were stained by diaminobenzidine (DAB) complex, and the nuclei were counterstained with haematoxylin. At last, the sections were examined with a fluorescence microscope (Leica).
In vivo Tumor Model
BALB/c nude mice (N=32, male, 5 weeks, weighing 18 ± 2 g) used in this study were bought from Laboratory Animal, Inc., (Beijing, China). PC3/DTX cells (3×106/0.2 mL PBS, circ-XIAP silencing or control) were subcutaneously injected into nude mice. The circ-XIAP silencing or control group was randomly divided into two groups (N=8/group). After injection for 7 days, tumor volume was measured and one group was intraperitoneally injected with DTX (10 mg/kg) every 3 days. Tumors volume was calculated based on the formula: volume=length × width2 ×0.5. After 22 days, these mice were killed by cervical dislocation following exposure to CO2 (30% of the chamber volume was displace per minute by the flow of CO2), and tumor tissues were photographed, weighed and harvested for detecting the expression levels of circ-XIAP, miR-1182 and TPD52. The in vivo experiments obtained the approval from the Animal Care and Use Committee of The Central Hospital of Zhumadian. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Statistical Analysis
Experimental data from at least three independent experiments were displayed as mean ± standard deviation (SD) and analyzed by GraphPad Prism. Student’s t-test or a one-way ANOVA was used for assessing significance of differences between two groups or among multiple groups. Pearson correlation coefficient was applied for analyzing the correlation between miR-1182 and circ-XIAP or TPD52. P value <0.05 was considered to be statistically significant.
Results
Circ-XIAP Was Upregulated in PCa Cells and DTX-Resistant PCa Tissues and Cells
To explore whether circ-XIAP was dysregulated in PCa cells, its expression was detected by qRT-PCR. Compared to RWPE-1 cells, the expression of circ-XIAP was increased in PCa cells (22Rv1, VCaP, DU145, and PC3), especially in DU145 and PC3 cells (Figure 1A). Next, we chose DU145 and PC3 cells for further study. In contrast to the linear XIAP mRNA, circ-XIAP was resistant to RNase R (Figure 1B and C), indicating that circ-XIAP had a loop structure. Moreover, circ-XIAP was primarily located in the cytoplasm (Figure 1D and E). PCa tissues were divided into 2 groups: sensitive (N=23) group and resistant (N=29) group, according to the patient outcomes after DTX treatment. The data indicated that circ-XIAP level was enhanced in DTX-resistant tissue samples compared to DTX-sensitive tissue samples (Figure 1F). In addition, circ-XIAP was also upregulated in tumor tissues compared with the adjacent normal tissues (Supplementary Figure 1). Moreover, IC50 of DTX was significantly increased in DTX-resistant cells (DU145/DTX and PC3/DTX) cells relative to parental cells (Figure 1G), indicating that resistant cells respond less to DTX treatment. In addition, the level of circ-XIAP was higher in DTX-resistant cells than parental cells (Figure 1H). Collectively, circ-XIAP might play a vital role in DTX resistance.
Figure 1.
Relative expression of circ-XIAP in PCa cells and DTX-resistant PCa tissue samples and cells. (A) Circ-XIAP expression was detected via qRT-PCR in RWPE-1 and PCa cells (22Rv1, VCaP, DU145, and PC3). (B and C) After treatment with RNase R, circ-XIAP and linear XIAP mRNA expression were measured in DU145 and PC3 cells. (D and E) The subcellular location of circ-XIAP in DU145 and PC3 cells was analyzed by qRT-PCR. (F) Circ-XIAP expression was examined in DTX-sensitive (N=23) and DTX-resistant (N=29) tissues. (G) IC50 value of DTX was determined using CCK-8 analysis in DU145, DU145/DTX, PC3, and PC3/DTX cells. (H) The level of circ-XIAP was measured in RWPE-1, DU145, DU145/DTX, PC3, and PC3/DTX cells. **P<0.01, ***P<0.001, ****P<0.0001.
Circ-XIAP Could Be Delivered Through Exosomes
Exosomes act as crucial mediators of intercellular communication. Next, exosomes in DTX-resistant cell culture media were isolated. As shown in Figure 2A, exosomes of DU145/DTX and PC3/DTX cells were observed under a TEM. WB was conducted for detecting the levels of exosome protein markers (CD63 and CD9). Results showed that CD63 and CD9 were all highly enriched in cell exosomes, but not in supernatant (Figure 2B). Moreover, the expression of circ-XIAP in exosomes (exo-circ-XIAP) was measured. As presented in Figure 2C, exo-circ-XIAP expression in DU145 and PC3 cells was markedly higher than in RWPE-1 cells and strikingly lower than in DU145/DTX and PC3/DTX cells. Subsequently, we further explored whether circ-XIAP was secreted by incorporation into exosomes. Treatment of exosomes from DTX-resistant cells increased circ-XIAP expression in DU145 and PC3 cells (Figure 2D), suggesting that circ-XIAP-contained exosomes could be absorbed by recipient cells. GW4869 has been successfully used to block the secretion of exosomes from cells. As shown in Figure 2E, GW4869 treatment decreased circ-XIAP expression in DU145/DTX and PC3/DTX cells, indicating that reduction of exosomes would reduce the expression of circ-XIAP. These data illustrated that circ-XIAP was secreted via incorporation into exosomes in cells.
Figure 2.
Circ-XIAP could be transferred via exosomes. (A) Exosomes secreted by DU145/DTX and PC3/DTX cells were observed by TEM. (B) WB assay was conducted for determination of CD63 and CD9 protein levels in the supernatant or exosomes of DU145/DTX and PC3/DTX cells. (C) Circ-XIAP expression in exosomes of RWPE-1, DU145, DU145/DTX, PC3, and PC3/DTX cells was examined. (D) Circ-XIAP expression in DU145 and PC3 cells treated with PBS or exosome was detected. (E) Circ-XIAP expression was measured in DU145/DTX and PC3/DTX cells treated with or without GW4869. **P<0.01, ***P<0.001, ****P<0.0001.
Knockdown of Circ-XIAP Enhanced DTX Sensitivity in DTX-Resistant PCa Cells
First, knockdown and overexpression efficiency of circ-XIAP were verified via qRT-PCR (Figure 3A). Since upregulation of circ-XIAP contributed to DTX resistance, we next examined whether knockdown of circ-XIAP would re-sensitize these cells to DTX treatment. Toward this purpose, we silenced circ-XIAP expression by transfection of si-circ-XIAP. We found that IC50 value of DTX was decreased in after knockdown of circ-XIAP (Figure 3B). Moreover, interference of circ-XIAP led to significant inhibition of cell viability and colony formation rate (Figure 3C and D), suggesting that circ-XIAP knockdown inhibited cell proliferation. In addition, flow cytometry results manifested that more cells were distributed in the G0/G1 phase, and fewer cells were distributed in S phase after downregulating circ-XIAP (Figure 3E), implying that cell cycle was arrested in G0/G1 phase. Furthermore, circ-XIAP downregulation exhibited significantly higher apoptosis rate (Figure 3F). Meanwhile, DU145/DTX and PC3/DTX cell migration and invasion were inhibited by circ-XIAP silence (Figure 3G and H). Next, we explored the role of circ-XIAP upregulation in parental PCa cells. Overexpression efficiency of circ-XIAP was shown in Supplementary Figure 2A. Overexpression of circ-XIAP increased IC50 value of DTX, cell viability, colony formation ability, migration, and invasion in DU145 and PC3 cells (Supplementary Figure 2B-2F). Together, these data implicated that circ-XIAP downregulation could re-sensitize DU145/DTX and PC3/DTX cells to DTX.
Figure 3.
Circ-XIAP knockdown increased DTX sensitivity in DTX-resistant PCa cells. (A) Circ-XIAP expression was determined in DU145/DTX and PC3/DTX cells after transfection with si-NC, si-circ-XIAP, vector, or circ-XIAP. (B–H) DU145/DTX and PC3/DTX cells were introduced with si-NC or si-circ-XIAP. (B and C) IC50 value of DTX and cell viability was examined via CCK-8 analysis. (D) The number of colonies was calculated using clone formation assay. (E and F) Flow cytometry was used to assess cell cycle distribution and cell apoptosis. (G and H) Transwell assay was utilized to evaluate cell migration and invasion (×100). **P<0.01, ***P<0.001, ****P<0.0001.
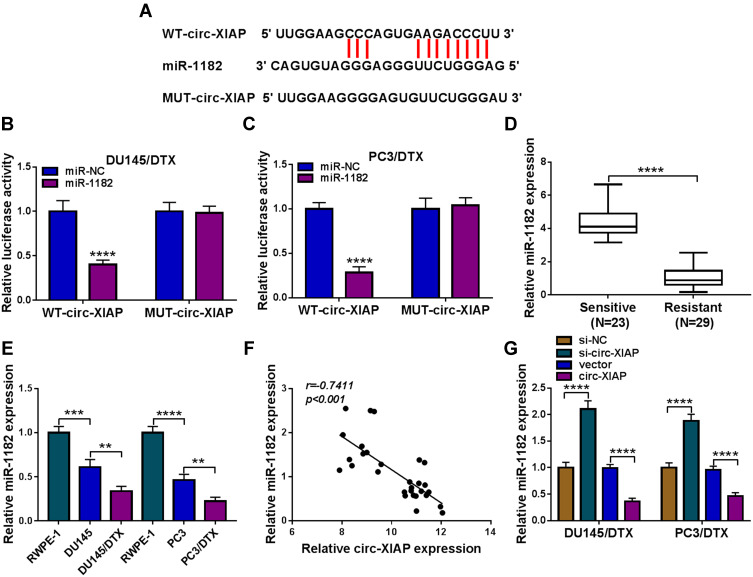
Circ-XIAP Was a Sponge of miR-1182
CircRNAs can exert their function as miRNA sponges.27 Next, we used online bioinformatics database (Circinteractome and circbank) to predict the potential targets of circ-XIAP. We found that two miRNAs (miR-507 and miR-1182) were overlapped in Circinteractome and Circbank (Supplementary Figure 3A). Moreover, we found that miR-507 and miR-1182 expression were increased by transfection of si-circ-XIAP, and the expression of miR-1182 was higher than that of miR-507 expression (Supplementary Figure 3B). Therefore, miR-1182 was selected for further study. As presented in Figure 4A, there existed potential binding sequence between circ-XIAP and miR-1182. To verify this prediction, dual-reporter luciferase assay was conducted. We observed that miR-1182 upregulation obviously decreased the luciferase activity in WT-circ-XIAP group but not in MUT-circ-XIAP group (Figure 4B and C). Moreover, miR-1182 was found to be lowly expressed in resistant tissues compared with sensitive tissues (Figure 4D). Similarly, miR-1182 expression in DU145 and PC3 cells was lower than RWPE-1 cells, and miR-1182 expression was the lowest in DU145/DTX and PC3/DTX cells (Figure 4E). A negative correlation between circ-XIAP and miR-1182 expression in resistant tissues was observed (Figure 4F). Furthermore, miR-1182 expression was increased when circ-XIAP was silenced in DTX-resistant cells, while miR-1182 expression was decreased after circ-XIAP overexpression (Figure 4G). These data concluded that circ-XIAP directly targeted miR-1182.
Figure 4.
Circ-XIAP directly interacted with miR-1182. (A) The binding sequence between circ-XIAP and miR-1182 was shown. (B and C) The luciferase activity in DU145/DTX and PC3/DTX cells co-transfected with WT-circ-XIAP/MUT-circ-XIAP and miR-1182/miR-NC was detected. (D) MiR-1182 level in DTX-resistant tissues and DTX-sensitive tissues was measured. (E) MiR-1182 expression was analyzed in RWPE-1, DU145, DU145/DTX, PC3, and PC3/DTX cells. (F) The association between the levels of miR-1182 and circ-XIAP in DTX-resistant tissues was analyzed. (G) The abundance of miR-1182 was detected in DU145/DTX and PC3/DTX cells after transfection with si-NC, si-circ-XIAP, vector, or circ-XIAP. **P<0.01, ***P<0.001, ****P<0.0001.
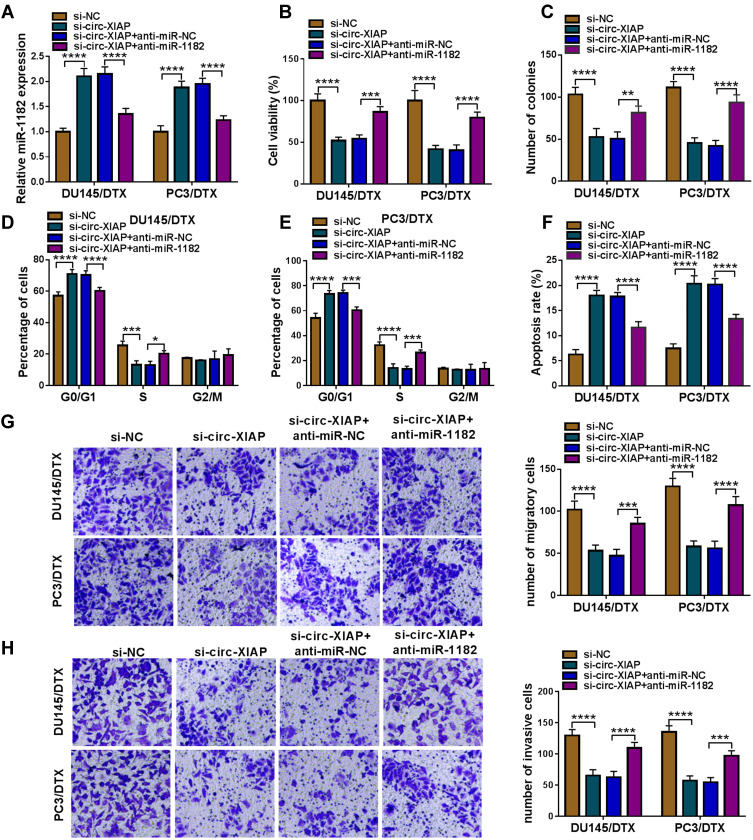
Silencing Circ-XIAP Enhanced DTX Sensitivity in DTX-Resistant PCa Cells by Upregulating miR-1182
To determine whether the function of si-circ-XIAP was mediated by miR-1182, rescue experiments were performed in DU145/DTX and PC3/DTX. The expression of miR-1182 was increased by downregulating circ-XIAP, which was attenuated by inhibiting miR-1182 (Figure 5A). Moreover, we found that miR-1182 inhibition reversed the suppressive effects of circ-XIAP silence on cell viability and colony-forming ability (Figure 5B and C). Additionally, miR-1182 silence weakened si-circ-XIAP-triggered cell cycle arrest and apoptosis (Figure 5D–F). Meanwhile, the inhibiting effects of circ-XIAP knockdown on migration and invasion were also counteracted by interference of miR-1182 (Figure 5G and H). Taken together, circ-XIAP regulated DTX sensitivity by targeting miR-1182.
Figure 5.
Downregulation of circ-XIAP improved sensitivity in DTX-resistant PCa cells by sponging miR-1182. DU145/DTX and PC3/DTX cells were transfected with si-NC, si-circ-XIAP, si-circ-XIAP + anti-miR-NC, or si-circ-XIAP + anti-miR-1182. (A) The expression of miR-1182 was tested. (B) CCK-8 assay was applied for analyzing cell viability. (C) Colony formation assay was used for evaluating colony-forming ability. (D–H) Cell cycle distribution, cell apoptosis, migration, and invasion were evaluated. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
TPD52 Was a Direct Target of miR-1182
To explore the mechanisms by which miR-1182 regulated DTX resistance, TargetScan was employed to search for the candidate targets. There are many mRNAs might be targeted by miR-1182. We selected six common mRNAs involved in the development of PCa. We found that AKT3, YY1 and TPD52 expression levels were decreased after overexpression of miR-1182, and TPD52 expression was the lowest (Supplementary Figure 4). Therefore, TPD52 was selected for further study. There were complementary sites between miR-1182 and TPD52 (Figure 6A), suggesting that miR-1182 might directly target TPD52. Moreover, miR-1182 overexpression evidently lowered the luciferase activity of WT-TPD52 3ʹUTR, while had no influence on the luciferase activity of MUT-TPD52 3ʹUTR (Figure 6B and C). We then investigated TPD52 expression in DTX-resistant tissues and cells. TPD52 mRNA and protein abundance were both higher in resistant tissues than sensitive tissues (Figure 6D and E). Immunohistochemistry analysis showed that TPD52 positive cells were increased in resistant tissues in comparison with sensitive tissues (Figure 6F). Likewise, TPD52 protein expression was found to be upregulated in DU145 and PC3 cells with respect to RWPE-1 cells, and TPD52 protein expression was further enhanced in DU145/DTX and PC3/DTX cells in contrast to parental cells (Figure 6G). Moreover, a distinct inverse correlation between miR-1182 expression and TPD52 mRNA in resistant tissues was revealed by Pearson correlation coefficient (Figure 6H). Inhibition and overexpression efficiency of miR-1182 were verified by qRT-PCR (Figure 6I). Additionally, we found that overexpression of miR-1182 inhibited the TPD52 protein level in DU145/DTX and PC3/DTX cells, and miR-1182 inhibition presented an opposite effect (Figure 6J). Therefore, we demonstrated that miR-1182 directly targeted TPD52 and negatively regulated its expression in DTX-resistant PCa cells.
Figure 6.
MiR-1182 directly targeted TPD52. (A) The binding sites between miR-1182 and TPD52 were displayed. (B and C) The luciferase activity of DU145/DTX and PC3/DTX cells co-transfected with WT-TPD52 3ʹUTR/MUT-TPD52 3ʹUTR and miR-1182/miR-NC was assessed. (D and E) TPD52 mRNA and protein expression in DTX-sensitive tissues and DTX-resistant tissues were measured. (F) Immunohistochemistry analysis was used to detect TPD52 positive cells in DTX-sensitive tissues and DTX-resistant tissues. (G) TPD52 protein expression was measured in RWPE-1, DU145, DU145/DTX, PC3, and PC3/DTX cells. (H) The correlation between miR-1182 expression and TPD52 mRNA expression in DTX-resistant tissues was analyzed using. (I and J) MiR-1182 expression and TPD52 protein level in DU145/DTX and PC3/DTX cells introduced with miR-NC, miR-1182, anti-miR-NC, or anti-miR-1182 were determined. **P<0.01, ***P<0.001, ****P<0.0001.
Overexpression of miR-1182 Improved DTX Sensitivity in DTX-Resistant PCa Cells by Targeting TPD52
To explore whether miR-1182 regulated DTX sensitivity by targeting TPD52, gain-of-function and rescue experiments were performed. WB data revealed that miR-1182 upregulation decreased the protein expression of TPD52, while TPD52 overexpression counteracted the inhibition effect of miR-1182 on TPD52 expression (Figure 7A). Moreover, we found that enforced expression of miR-1182 inhibited cell viability and colony-forming ability as well as induced cell cycle arrest and apoptosis in DU145/DTX and PC3/DTX cells, while these effects were reversed by addition of TPD52 (Figure 7B–F). Additionally, restoration of miR-1182 repressed migration and invasion of DU145/DTX and PC3/DTX cells, which could be abated by upregulating TPD52 (Figure 7G and H). These findings indicated that miR-1182 exerted its functions in DTX-resistant PCa cells by regulating TPD52.
Figure 7.
Upregulation of miR-1182 enhanced DTX sensitivity in DTX-resistant PCa cells by downregulating TPD52. DU145/DTX and PC3/DTX cells were transfected with miR-NC, miR-1182, miR-1182 + vector, or miR-1182 + TPD52. (A) The protein level of TPD52 was detected. (B and C) Cell viability and colony-forming ability were evaluated. (D–H) Cell cycle distribution, cell apoptosis, migration, and invasion were measured. **P<0.01, ***P<0.001, ****P<0.0001.
Circ-XIAP Positively Modulated TPD52 Expression by Acting as a Sponge for miR-1182
Next, we explored whether circ-XIAP could regulate TPD52 expression by sponging miR-1182. WB assay indicated that silence of circ-XIAP reduced TPD52 protein expression, whereas miR-1182 deficiency could neutralize this inhibition impact (Figure 8A and B), suggesting that circ-XIAP could regulate TPD52 expression by sponging miR-1182.
Figure 8.
Circ-XIAP regulated TPD52 expression by sponging miR-1182. (A and B) TPD52 protein expression was examined by WB in DU145/DTX and PC3/DTX cells after introduction with si-NC, si-circ-XIAP, si-circ-XIAP + anti-miR-NC, or si-circ-XIAP + anti-miR-1182. ***P<0.001, ****P<0.0001.
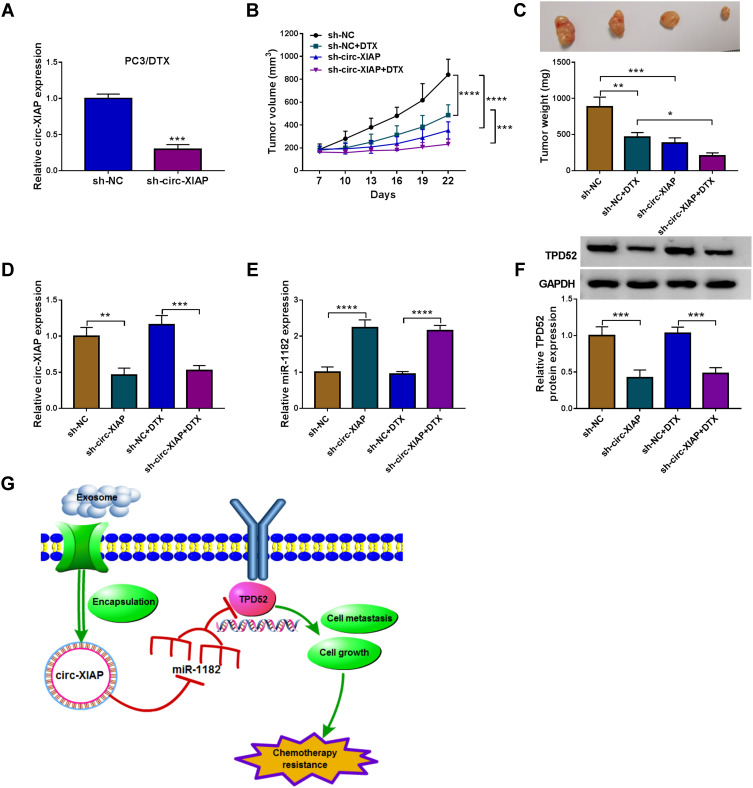
Downregulation of Circ-XIAP Hampered Tumor Growth and Enhanced DTX Sensitivity in vivo
Transfection efficiency of sh-circ-XIAP was detected by transfection of sh-circ-XIAP (Figure 9A). To assess the role of circ-XIAP in tumor growth and DTX sensitivity in vivo, the mice xenograft model was established. Consistent with in vitro experiments, circ-XIAP silence or DTX treatment group showed smaller tumor volume and weight versus than that in the sh-NC group, and combination of circ-XIAP knockdown and DTX treatment further decreased tumor volume and weight compared to only DTX treatment (Figure 9B and C). Moreover, circ-XIAP expression and TPD52 protein expression were significantly lower and miR-1182 expression was prominently higher in tumor tissues of sh-circ-XIAP group than that of sh-NC group (Figure 9D–F). The schematic diagram of the mechanism was presented in Figure 9G. Therefore, circ-XIAP silence suppressed tumor growth and promoted DTX sensitivity by upregulating miR-1182 and downregulating TPD52.
Figure 9.
Silencing circ-XIAP promoted DTX sensitivity in vivo. (A) The expression of circ-XIAP in PC3/DTX cells transfected with sh-NC (as the control) or sh-circ-XIAP was measured by qRT-PCR. (B–F) PC3/DTX cells stably knocked down circ-XIAP were inoculated into nude mice, and the nude mice were treated with or without DTX (10 mg/kg) after injection for 7 days. (B) Tumor volume was monitored every 3 days. (C) After 22 days of injection, the weight of the tumor tissues was measured. (D and E) Circ-XIAP and miR-1182 levels in tumor tissues of nude mice were determined. (F) TPD52 protein level was analyzed in tumor tissues. (G) Schematic diagram of the mechanism in which exosomal circ-XIAP modulated chemotherapy resistance by regulating miR-1182/TPD52 axis. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Discussion
PCa is frequent cancer that seriously threatens the health of men worldwide.28 Recently, the research on exosome function has attracted wide attention. It has been reported that exosome secretions, including circRNAs, not only regulate cancer progression but also affect chemosensitivity in human cancer.29,30 However, there are still few reports on the effect of exosomal circRNAs in DTX resistance of PCa. Therefore, the aim of our research was to probe the biological role of exosomal circ-XIAP in DTX resistance of PCa.
Recently, exosomes derived from tumor cells can stimulate tumor progression and serve as effective biomarkers for specific cancers.31,32 Moreover, the functions of exosomal circRNAs in cancer resistance have attracted increasing interest. For example, Han et al revealed that exosomal circ-HIPK3 facilitated temozolomide resistance and tumor progression via modulating miR-421/ZIC5 axis in glioma.33 Moreover, Hu et al found that exosome-derived circ_UBE2D2 promoted resistance of breast cancer to tamoxifen via regulating miR-200a-3p.34 Circ-XIAP has been found to be upregulated and promoted proliferation and migration in PCa.20 However, the roles of exosomal circ-XIAP in DTX resistance of PCa have not been reported. In our research, circ-XIAP expression was increased in DTX-resistant PCa cells and tissues. Next, cell exosomes were extracted from DTX-resistant PCa cells, and the vesicles were shown to be round shape with double membranes, and the diameter was from 40 nm to 100 nm under a TEM. Subsequently, we observed that circ-XIAP was significantly enriched in exosomes from DTX-resistant PCa cells and could be transferred to parental cells after treatment with exosomes. These data indicated that circ-XIAP could be transferred via exosomes. Moreover, circ-XIAP deletion mitigated DTX resistance by suppressing resistant cell growth, migration and invasion and inducing cell cycle arrest and cell apoptosis, indicating the involvement of circ-XIAP in the regulating DTX resistance.
To study the mechanism by which circ-XIAP functions in DTX resistance, we confirmed its intracellular localization, and found that circ-XIAP was predominantly located in the cytoplasm. Given that circRNAs have been widely studied as sponges for miRNAs and circ-XIAP had a high abundance in the cytoplasm, we next explored whether circ-XIAP acted as a sponge for miRNA. As we expected, miR-1182 was identified as a target of circ-XIAP. MiRNAs were reported to be involved in the pathological processes of diverse diseases through regulating cell growth, cell cycle, metastasis, apoptosis, and drug resistance.35,36 MiR-1182 has been demonstrated to suppress cell tumorigenesis in diverse cancers, such as ovarian cancer,37 gastric cancer,38 bladder cancer39 and so on. Moreover, Huang et al demonstrated that miR-1182 was lowly expressed in PCa tissue samples, and miR-1182 deficiency abolished the repressive impact of circABCC4 silence on PCa progression.24 Our study displayed that miR-1182 abundance was decreased in DTX-resistant PCa tissue samples and cells. Subsequent experiments showed that miR-1182 downregulation could abate the effects of si-circ-XIAP on DTX-resistant cell growth, cell cycle, apoptosis, migration, and invasion. In a word, our results implied that circ-XIAP knockdown enhanced DTX sensitivity by sponging miR-1182 in DTX-resistant PCa cells.
To explore how miR-1182 regulated the DTX resistance of PCa, possible targets of miR-1182 were predicted using the online tool (TargetScan). We demonstrated that TPD52 was targeted by miR-1182. TPD52, a tumor-promoting gene, is usually upregulated in many tumors, including PCa.40,41 More importantly, TPD52 also was reported to increase DTX resistance of PCa via suppressing LKB1/AMPK-mediated autophagy.42 Here, we uncovered that TPD52 level was elevated in DTX-resistant PCa tissue samples and cells. Moreover, TPD52 elevation could counteract the promotive effect of miR-1182 on DTX sensitivity. In addition, circ-XIAP inhibition could directly decrease TPD52 expression via sponging miR-1182. Meanwhile, circ-XIAP depletion restrained DTX-resistant tumor growth and improved DTX sensitivity via regulation of miR-1182 and TPD52 expression. These results suggested that circ-XIAP contributed to the development of DTX-resistant PCa cells by regulating miR-1182/TPD52 axis.
In conclusion, we revealed that exosome-mediated circ-XIAP could enhance DTX resistance of PCa by regulating miR-1182/TPD52 axis. Our study offered a novel insight into the roles of exosomes in PCa and offered a promising therapeutic target to improve chemotherapy efficiency for PCa patients.
Funding Statement
There is no funding to report.
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Wu J, Cheng G, Zhang C, et al. Long noncoding RNA LINC01296 is associated with poor prognosis in prostate cancer and promotes cancer-cell proliferation and metastasis. Onco Targets Ther. 2017;10:1843–1852. doi: 10.2147/OTT.S129928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galsky MD, Vogelzang NJ. Docetaxel-based combination therapy for castration-resistant prostate cancer. Ann Oncol. 2010;21(11):2135–2144. doi: 10.1093/annonc/mdq050 [DOI] [PubMed] [Google Scholar]
- 4.Sekino Y, Oue N, Koike Y, et al. KIFC1 inhibitor CW069 induces apoptosis and reverses resistance to docetaxel in prostate cancer. J Clin Med. 2019;8(2):225. doi: 10.3390/jcm8020225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364–372. doi: 10.1016/j.tcb.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 6.Bei Y, Das S, Rodosthenous RS, et al. Extracellular vesicles in cardiovascular theranostics. Theranostics. 2017;7(17):4168–4182. doi: 10.7150/thno.21274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Yan IK, Kogure T, et al. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4(1):458–467. doi: 10.1016/j.fob.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vlassov AV, Magdaleno S, Setterquist R, et al. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940–948. doi: 10.1016/j.bbagen.2012.03.017 [DOI] [PubMed] [Google Scholar]
- 9.Ge R, Tan E, Sharghi-Namini S, et al. Exosomes in cancer microenvironment and beyond: have we overlooked these extracellular messengers? Cancer Microenviron. 2012;5(3):323–332. doi: 10.1007/s12307-012-0110-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J, Wang H, Zhang X, et al. Exosomal miR-200c suppresses chemoresistance of docetaxel in tongue squamous cell carcinoma by suppressing TUBB3 and PPP2R1B. Aging (Albany NY). 2020;12(8):6756–6773. doi: 10.18632/aging.103036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malla B, Zaugg K, Vassella E, et al. Exosomes and exosomal microRNAs in prostate cancer radiation therapy. Int J Radiat Oncol Biol Phys. 2017;98(5):982–995. doi: 10.1016/j.ijrobp.2017.03.031 [DOI] [PubMed] [Google Scholar]
- 12.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 13.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32 [DOI] [PubMed] [Google Scholar]
- 14.Arnaiz E, Sole C, Manterola L, et al. CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol. 2019;58:90–99. doi: 10.1016/j.semcancer.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 15.Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881.e813. doi: 10.1016/j.cell.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong X, Liu N, Liang Y, et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol Cancer. 2020;19(1):33. doi: 10.1186/s12943-020-01149-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y, Liu J, Huan J, et al. Downregulation of circular RNA hsa_circ_0000735 boosts prostate cancer sensitivity to docetaxel via sponging miR-7. Cancer Cell Int. 2020;20(1):334. doi: 10.1186/s12935-020-01421-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. doi: 10.1186/s12943-019-1041-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Y, Yang Y, Zhao X, et al. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis. 2019;10(11):792. doi: 10.1038/s41419-019-2028-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou XS, Han CQ, Zhang W. MiR-1182 inhibited metastasis and proliferation of ovarian cancer by targeting hTERT. Eur Rev Med Pharmacol Sci. 2018;22(6):1622–1628. doi: 10.26355/eurrev_201803_14569 [DOI] [PubMed] [Google Scholar]
- 23.Xiao J, Yu H, Ma Z. LINC00339 promotes growth and invasiveness of hepatocellular carcinoma by the miR-1182/SKA1 pathway. Onco Targets Ther. 2019;12:4481–4488. doi: 10.2147/OTT.S207397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C, Deng H, Wang Y, et al. Circular RNA circABCC4 as the ceRNA of miR-1182 facilitates prostate cancer progression by promoting FOXP4 expression. J Cell Mol Med. 2019;23(9):6112–6119. doi: 10.1111/jcmm.14477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byrne JA, Balleine RL, Schoenberg Fejzo M, et al. Tumor protein D52 (TPD52) is overexpressed and a gene amplification target in ovarian cancer. Int J Cancer. 2005;117(6):1049–1054. doi: 10.1002/ijc.21250 [DOI] [PubMed] [Google Scholar]
- 26.Dasari C, Yaghnam DP, Walther R, et al. Tumor protein D52 (isoform 3) contributes to prostate cancer cell growth via targeting nuclear factor-κB transactivation in LNCaP cells. Tumour Biol. 2017;39(5):1010428317698382. doi: 10.1177/1010428317698382 [DOI] [PubMed] [Google Scholar]
- 27.Su Q, Lv X. Revealing new landscape of cardiovascular disease through circular RNA-miRNA-mRNA axis. Genomics. 2020;112(2):1680–1685. doi: 10.1016/j.ygeno.2019.10.006 [DOI] [PubMed] [Google Scholar]
- 28.Schatten H. Brief overview of prostate cancer statistics, grading, diagnosis and treatment strategies. Adv Exp Med Biol. 2018;1095:1–14. [DOI] [PubMed] [Google Scholar]
- 29.Shi X, Wang B, Feng X, et al. circRNAs and exosomes: a mysterious frontier for human cancer. Mol Ther Nucleic Acids. 2020;19:384–392. doi: 10.1016/j.omtn.2019.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Zhang H, Yang H, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539–555. doi: 10.1002/1878-0261.12629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duijvesz D, Luider T, Bangma CH, et al. Exosomes as biomarker treasure chests for prostate cancer. Eur Urol. 2011;59(5):823–831. doi: 10.1016/j.eururo.2010.12.031 [DOI] [PubMed] [Google Scholar]
- 32.Li W, Li C, Zhou T, et al. Role of exosomal proteins in cancer diagnosis. Mol Cancer. 2017;16(1):145. doi: 10.1186/s12943-017-0706-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han C, Wang S, Wang H, et al. Exosomal circ-HIPK3 facilitates tumor progression and temozolomide resistance by regulating miR-421/ZIC5 axis in glioma. Cancer Biother Radiopharm. 2020. [DOI] [PubMed] [Google Scholar]
- 34.Hu K, Liu X, Li Y, et al. Exosomes mediated transfer of circ_UBE2D2 enhances the resistance of breast cancer to tamoxifen by binding to miR-200a-3p. Med Sci Monit. 2020;26:e922253. doi: 10.12659/MSM.922253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15(5):429. doi: 10.2174/138920101505140828161335 [DOI] [PubMed] [Google Scholar]
- 36.Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23. doi: 10.1016/j.critrevonc.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 37.Zong ZH, Du YP, Guan X, et al. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38(1):437. doi: 10.1186/s13046-019-1437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang D, Xiao YF, Zhang JW, et al. miR-1182 attenuates gastric cancer proliferation and metastasis by targeting the open reading frame of hTERT. Cancer Lett. 2015;360(2):151–159. doi: 10.1016/j.canlet.2015.01.044 [DOI] [PubMed] [Google Scholar]
- 39.Zhou J, Dai W, Song J. miR-1182 inhibits growth and mediates the chemosensitivity of bladder cancer by targeting hTERT. Biochem Biophys Res Commun. 2016;470(2):445–452. doi: 10.1016/j.bbrc.2016.01.014 [DOI] [PubMed] [Google Scholar]
- 40.Li J, Li Y, Liu H, et al. The four-transmembrane protein MAL2 and tumor protein D52 (TPD52) are highly expressed in colorectal cancer and correlated with poor prognosis. PLoS One. 2017;12(5):e0178515. doi: 10.1371/journal.pone.0178515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ummanni R, Teller S, Junker H, et al. Altered expression of tumor protein D52 regulates apoptosis and migration of prostate cancer cells. Febs j. 2008;275(22):5703–5713. doi: 10.1111/j.1742-4658.2008.06697.x [DOI] [PubMed] [Google Scholar]
- 42.Zeng J, Liu W, Fan YZ, et al. PrLZ increases prostate cancer docetaxel resistance by inhibiting LKB1/AMPK-mediated autophagy. Theranostics. 2018;8(1):109–123. doi: 10.7150/thno.20356 [DOI] [PMC free article] [PubMed] [Google Scholar]