Abstract
Inflammatory bowel disease (IBD) has become a global disease with accelerating incidence worldwide in the 21st century while its accurate etiology remains unclear. In the past decade, gut microbiota dysbiosis has consistently been associated with IBD. Although many IBD-associated dysbiosis have not been proven to be a cause or an effect of IBD, it is often hypothesized that at least some of alteration in microbiome is protective or causative. In this article, we selectively reviewed the hypothesis supported by both association studies in human and pathogenesis studies in biological models. Specifically, we reviewed the potential protective bacterial pathways and species against IBD, as well as the potential causative bacterial pathways and species of IBD. We also reviewed the potential roles of some members of mycobiome and virome in IBD. Lastly, we covered the current status of therapeutic approaches targeting microbiome, which is a promising strategy to alleviate and cure this inflammatory disease.
KEYWORDS: inflammatory bowel disease, pathogenesis, etiology, microbiome, dysbiosis, therapy
INTRODUCTION
Inflammatory bowel disease (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD), has become a global disease with accelerating incidence worldwide in the 21st century (Ng et al., 2018). IBD is characterized by chronic immune-mediated intestinal inflammation that is driven by genetic susceptibility, environmental and microbial factors (Ni et al., 2017; Imhann et al., 2018).
Microbial factors have been historically proven to be indispensable for the onset of IBD (Alhagamhmad et al., 2016) and advances in high-throughput sequencing has enabled us to elucidate the gut microbiome in IBD. Study of microbial etiology of IBD has been mainly focused on three directions: 1) the persistent pathogen theory 2) the excessive bacterial translocation theory and 3) the dysbiosis theory (De Hertogh et al., 2008; Kalischuk and Buret, 2010). The persistent pathogen theory hypothesizes IBD can be caused by persistent infection of an enteric pathogen like Mycobacterium avium subspecies paratuberculosis, Clostridium difficile, and adhesion-invasive Escherichia coli (AIEC). The excessive bacterial translocation theory suggests the excessive level of translocation of intestinal bacteria across the intestinal barrier is a cause of IBD. While, the dysbiosis theory hypothesizes that the shift of balance between “beneficial” vs. “detrimental” commensal bacteria can cause IBD.
The three theories are not mutually exclusive. For example, AIEC can be considered as both a persistent pathogen and detrimental commensal bacteria. The first two theories were comprehensively reviewed elsewhere (De Hertogh et al., 2008; Kalischuk and Buret, 2010); while in this review, we summarize the emerging evidences that imply the roles of dysbiosis in pathogenesis of IBD and the potential therapeutic options that target the gut microbiome to alleviate IBD.
POTENTIAL ROLES OF DYSBIOSIS IN PATHOGENESIS OF IBD
IBD is characterized by chronic immune-mediated intestinal inflammation that attacks the bowel. IBD has been consistently shown to be associated with gut dysbiosis (Kostic et al., 2014; Lynch and Pedersen, 2016). Although many IBD-associated dysbiosis have not been proven to be a cause or an effect of IBD, it is often hypothesized that at least some of alteration in microbiome is protective or causative.
Metagenomic studies have revealed microbial compositional changes in patients with IBD (Franzosa et al., 2019; Lloyd-Price et al., 2019) and metabolomic studies have revealed many defined microbial metabolites are depleted in individuals with IBD versus control individuals (Franzosa et al., 2019). Some of the depleted metabolites and related species are found to have anti-inflammatory effects and therefore are hypothesized to be protective; on the other hand, pro-inflammatory bacterial metabolites and species that are enriched in IBD patients are hypothesized to be causative, in terms of IBD.
In this review, we mainly focus on hypothesis that has both types of supporting evidences: 1) at least one association study in human IBD (rather than animal models); and 2) at least one pathogensis study in human, animal models, or cell models that explains the result in the association study.
Potentially protective bacterial pathways and species
The metabolic pathways encoded by the human gut microbiome produce numerous bioactive molecules that interact with the host. Typical bioactive molecules include short-chain fatty acids (SCFAs) and tryptophan derivatives that are produced by bacteria from dietary components, as well as secondary bile acids (BAs) that are bacteria-modified host products (Postler and Ghosh, 2017).
SCFAs (primarily acetate, propionate, and butyrate) produced by gut bacteria regulates protective immunity and reduces tissue inflammation (Furusawa et al., 2013; Kim et al., 2013). One study found that 12% of metabolic pathways were significantly different between IBD patients and healthy controls, and it confirmed a decrease in butanoate and propanoate metabolism genes in CD (Morgan et al., 2012). Another case-control analysis using shotgun metagenomic sequencing of stool samples from 1,792 individuals suggests the fermentation of pyruvate to butanoate, a butyrate precursor, was decreased in patients with IBD (Vich Vila et al., 2018). When looking at the bacterial species composition, a decreased amount of the commensal bacterium Faecalibacterium prausnitzii was reported in IBD patients compared with controls (Sokol et al., 2009; Hedin et al., 2016; Lloyd-Price et al., 2019). In vitro peripheral blood mononuclear cell stimulation by F. prausnitzii led to significantly lower IL-12 and IFN-γ production levels and higher secretion of the anti-inflammatory cytokine IL-10 (Sokol et al., 2008b). Additionally, various F. prausnitzii isolates have abilities to simulate IL-10 secretion by dendritic cells (DCs) (Rossi et al., 2016), which suggests the anti-inflammatory role of F. prausnitzii in colitis. A 15 kDa protein with anti-inflammatory properties, produced by F. prausnitzii, could alleviate colitis in mice by inhibiting the NF-κB pathway (Quevrain et al., 2016). Some F. prausnitzii strains are considered as candidates of next-generation probiotics (Martin et al., 2017).
Roseburia were also significantly reduced in IBD (Morgan et al., 2012), and IBD-genetic risk score was significantly associated with a decrease of Roseburia in healthy controls (Imhann et al., 2018). The depletion Roseburia hominis was observed in CD (Franzosa et al., 2019; Lloyd-Price et al., 2019) and UC (Machiels et al., 2014). In stool samples from patients with CD or UC, the strain abundance of Roseburia intestinalis decreased (Vich Vila et al., 2018). R. intestinalis are further reported as acetate-to-butyrate converters that reside in the intestinal mucus layer, where their anti-inflammatory effects may occur (Vich Vila et al., 2018).
In addition to reduced SCFA levels, decreased tryptophan metabolism levels were associated with a compromised epithelial barrier in IBD (Schirmer et al., 2019). Tryptophan can be converted by bacteria into bioactive indole-containing molecules that activate the aryl hydrocarbon receptor and down-regulates inflammation (Zelante et al., 2013). Indoleacrylic acid that promotes mucus production and suppresses inflammatory cytokine production was found reduced in patients with IBD (Wlodarska et al., 2017). Tryptophan-metabolizing pathways have been identified in some members of the human gut microbiota such as Clostridium sporogenes and E. coli (Williams et al., 2014; Dodd et al., 2017; Agus et al., 2018).
Primary BAs (PBAs) are produced by the host and then modified by bacteria into secondary BAs (SBAs) which mainly have anti-inflammatory activities. A normal bacterial BA metabolism plays an important role in modulating the host regulatory T (Treg) cell homeostasis (Song et al., 2020), as well as TH17 and Treg cell differentiation (Hang et al., 2019). Disrupted BAs metabolism has been observed in IBD patients, with fecal BAs pools skewed toward decreased SBAs and increased PBAs relative to healthy controls (Duboc et al., 2013; Franzosa et al., 2019). A more recent study also found that fecal BAs composition was altered (dominated by PBAs) in a sub-group of CD patients who did not sustain remission (Connors et al., 2019). PBAs cholate and its glycine and taurine conjugates were enriched in dysbiotic samples from participants with CD, and by contrast, the SBAs lithocholate and deoxycholate were reduced in dysbiosis (Lloyd-Price et al., 2019). Moreover, levels of lithocholic acid and deoxycholic acid (the most abundant gut SBAs), and genes required to convert PBAs to SBAs were reduced in stool from UC relative to familial adenomatous polyposis (FAP) (Sinha et al., 2020). Members of the Roseburia and unclassified Subdoligranulum species were associated with BAs metabolism, and they were both markedly reduced in IBD (Lloyd-Price et al., 2019). Ruminococcaceae was reduced in IBD compared to healthy people (Vich Vila et al., 2018; Lo Presti et al., 2019; Yilmaz et al., 2019) and is known to contain members (particularly Clostridium leptum) capable of SBAs generation, which ameliorate intestinal inflammation in a process reliant on the TGR5 bile acid receptor (Sinha et al., 2020). Supplementation of SBAs also reduces intestinal inflammation in three murine colitis models (Sinha et al., 2020).
Polysaccharide A (PSA) produced by Bacteroides fragilis directs the development of CD4+ T cells and induces the anti-inflammatory function of Tregs (Mazmanian et al., 2005; Round et al., 2011). PSA protects animals from experimental colitis depending on IL-10-producing CD41 T cells (Mazmanian et al., 2008). Individuals with IBD had a significantly lower percentage of the B. fragilis population with PSA promoter orientated “ON” (Blandford et al., 2019). Moreover, sphingolipids produced by B. fragilis regulate homeostasis of host intestinal natural killer T cells and confer protection against oxazolone-induced experimental colitis (An et al., 2014).
Recent research on Akkermansia muciniphila revealed another potential protective pathway against IBD. Initially, a study of 46 IBD and 20 control patients showed that the abundance of Akkermansia muciniphila reduced many fold in CD and in UC (Png et al., 2010). Although a contradictory research indicated A. muciniphila was sufficient for promoting intestinal inflammation in both specific-pathogen-free and germ-free IL10(− / −) mice model of IBD (Seregin et al., 2017), in a follow-up study, A. muciniphila strain ATCC BAA-835 was examined in gnotobiotic IL10(−/−) mice, and it did not promote short-term intestinal inflammation (Ring et al., 2019). A. muciniphila was shown to improve the gut barrier partially via its outer membrane protein Amuc_1100 that interacts with Toll-like receptor 2 (Plovier et al., 2017). The roles of A. muciniphila in modulating human immunological homeostatic was further demonstrated by the recent report that A. muciniphila induce homeostatic IgG production and antigen-specific T cell responses in mice (Ansaldo et al., 2019) and that A. muciniphila treatment ameliorated Dextran Sulfate Sodium (DSS)-induced UC in mice (Bian et al., 2019).
Potential causative bacterial pathways and species
The integrated human microbiome project has revealed a few metabolites, notably nicotinuric acid, taurine, and acylcarnitines are more abundant in IBD patients than controls (Lloyd-Price et al., 2019). Interestingly, taurine has been previously identified as a mucosal inflammasome activator (Levy et al., 2015). Therefore, these metabolites were suggested as potential causative metabolites and therapeutic targets. The acylcarnitines-related species are Roseburia hominis, Klebsiella pneumoniae, Haemophilus parainfluenzae, and Clostridium bolteae (Lloyd-Price et al., 2019). Bacterial genes with virulence-related functions were enriched in IBD patients (Erickson et al., 2012; Morgan et al., 2012), presumably due to overgrowth of functionally altered commensals termed pathobionts. Escherichia coli revealed an increased amount in patients with IBD (Lloyd-Price et al., 2019; Pittayanon et al., 2019) and the adherent invasive E. coli (AIEC) pathovar are associated specifically with ileal mucosa in IBD (Darfeuille-Michaud et al., 2004; Sepehri et al., 2011), suggesting AIEC may contribute to IBD pathogenesis (Mylonaki et al., 2005; Garrett et al., 2010).
The frequent recovery of E. coli adhering to intestinal mucosa of patients with CD (Darfeuille-Michaud, 2002; Martin et al., 2004; Prorok-Hamon et al., 2014) and UC (Kotlowski et al., 2007) has stimulated great interest. Interaction of AIEC with intestinal mucosa in the context of IBD include: (1) AIEC cross the mucous layer and resist antimicrobial peptides; (2) AIEC adhere to intestinal epithelial cells (IECs) via FimH and carcinoembryonic antigen related cell adhesion molecule 6 (CEACAM6), and lead to colonisation of the gut mucosa; (3) AIEC enter lamina propria and Peyer’s patches across M cells via long polar fimbriae (LPF) expression, and interact with immune cells (Palmela et al., 2018). AIEC can promote inflammation, survive and replicate, and escape autophagy when inside macrophages (Bringer et al., 2006). Besides, AIEC also have the ability to evade the host immune response by suppressing IFN-γ mediated signal transducer and activator of STAT1 in IECs, preventing an appropriate antimicrobial response (Ossa et al., 2013). AIEC strain NC101 harbors the pks pathogenicity island that encodes the biosynthetic machinery for synthesizing the genotoxin colibactin (Nougayrede et al., 2006). Monocolonization with the commensal NC101 promoted invasive carcinoma and tumorigenesis in azoxymethane-treated IL-10(−/−) mice (Arthur et al., 2012; Eaton et al., 2018).
In a recent study, an Enterococcus faecium strain that has adhesion gene was isolated from the feces of UC patients, promotes colitis and colonic cytokine expression (Seishima et al., 2019). A previous research showed colonic inflammation in IL10(−/−) mice inoculated with Enterococcus faecalis and faecium strains is associated with gene expression changes similar to those of human IBD (Barnett et al., 2010).
Another pathobiont that associated with IBD is enterotoxigenic Bacteroides fragilis (ETBF) (Prindiville et al., 2000; Zamani et al., 2017). ETBF induces focal colonic Stat3 activation and Th17 immune responses and then promotes mucosal permeability (Wick et al., 2014; Chung et al., 2018; Dejea et al., 2018). Genes for B. fragilis toxin (bft) encode secreted oncotoxins, and increase IL-17 in the colon (Dejea et al., 2018). Besides promoting IBD, ETBF are also possibly driving FAP and CRC (Thiele Orberg et al., 2017; Garrett, 2019).
Campylobacter concisus is another adherent, invasive proteobacterium that has been associated with IBD (Zhang et al., 2009; Man et al., 2010; Mahendran et al., 2011; Mukhopadhya et al., 2011; Kirk et al., 2016; Underwood et al., 2016). Although the natural colonization site of C. concisus is oral cavity, C. concisus can also colonize the intestinal tract. Intestinal colonization by bacteria from the oral cavity has been suggested to be extensively involved in inflammatory diseases (Cao, 2017; Dickson, 2018). Some C. concisus strains acquired zonula occludens toxin (zot) gene from a phage, and increased intestinal membrane permeability by affecting the tight junctions (Zhang et al., 2014). C. concisus Zot may have enteric pathogenic potential by damaging intestinal epithelial barrier, inducing intestinal epithelial and macrophage production of proinflammatory cytokines in particular TNF-α (Mahendran et al., 2016), thus triggering the relapse of IBD. C. concisus cause epithelial sodium channel dysfunction via IL-32-regulated ERK1/2, as well as claudin-8-dependent barrier dysfunction, both of which contribute to Na(+) malabsorption and enteritis (Nattramilarasu et al., 2020).
Another oral cavity and gastrointestinal bacterium, Fusobacterium varium, may be one of the pathogenic factors in UC. F. varium bacteria were present at a higher abundance in the colonic mucosa of patients with UC compared to healthy controls (Ohkusa et al., 2002). When administered by rectal enema in mice, F. varium was able to cause colonic mucosal inflammation (Ohkusa et al., 2003). F. varium invaded host intestinal epithelial cells, significantly increased the concentrations of IL-8 and TNF-α, and triggered host inflammatory reactions (Ohkusa et al., 2009). Genome analysis of a F. varium strain showed it possesses multiple virulence factors, including type V secretion system (T5SS) and Fusobacterium adhesion (FadA) paralogs, which involve in potential mucosal inflammation (Sekizuka et al., 2017).
Ruminococcus gnavus is part of the healthy gut microbiota in humans, but it is enriched in IBD (Png et al., 2010; Willing et al., 2010; Joossens et al., 2011; Nishino et al., 2018; Franzosa et al., 2019; Lloyd-Price et al., 2019). The increased level of R. gnavus has also been linked to spondyloarthritis (Breban et al., 2017), pouchitis in UC patients who have undergone a total colectomy (Machiels et al., 2017), and allergic diseases (Chua et al., 2018). 199 strain-specific genes involved in oxidative stress responses, adhesion, iron-acquisition, and mucus utilization were identified, potentially conferring an adaptive advantage for the R. gnavus clade in the IBD gut (Hall et al., 2017). R. gnavus produce and metabolize 2,7-anhydro-Neu5Ac to achieve nutritional competitive advantage in mucus against other bacteria (Tailford et al., 2015; Owen et al., 2017; Bell et al., 2019). In addition, R. gnavus synthesizes and secretes a pro-inflammatory complex polysaccharide, which potently induces TNF-α secretion by DCs via TLR4 (Henke et al., 2019).
Non-pylori Helicobacter also has numerous associations with IBD. In a cross-sectional study of 73 CD and 92 controls, CD is associated with the presence of enterohepatic Helicobacter spp. species DNA in intestinal biopsies (Laharie et al., 2009). Enterohepatic Helicobacter including H. hepaticus (Kullberg et al., 2001, 2006; Yang et al., 2013) and H. bilis (Jergens et al., 2007; Liu et al., 2011; Atherly et al., 2016) are often referred as pathobionts (Chai et al., 2017), because they have been shown to be capable of causing IBD-like disease in mice. H. hepaticus predominantly induces inflammatory TH17 cells in disease-susceptible IL-10-deficient animals and contributes to spontaneous colitis (Xu et al., 2018). Helicobacter pylori infection was reported to be negative associated with IBD (el-Omar et al., 1994; Sonnenberg and Genta, 2012; Rokkas et al., 2015), supporting a possible protective benefit of H. pylori infection against the development of IBD. Alternatively, IBD could be a protective factor against H. pylori infection. The presence of IBD-associated gastric mucosal lesions may create an inhospitable environment for H. pylori colonization (Castano-Rodriguez et al., 2017).
Dysbiosis in mycobiome
Besides bacterial dysbiosis, alterations in the eukaryotic fungal community (the “mycobiome”) are also important. Fungal composition in IBD is characterized with an increased Basidiomycota/Ascomycota ratio (Sokol et al., 2017), which was also skewed with higher values in CRC than control (Coker et al., 2019).
Candida spp. is significantly more abundant in patients with CD (Li et al., 2014; Lam et al., 2019) or IBD (Chehoud et al., 2015). In particular, Candida albicans were enriched in CD (Li et al., 2014), UC (Mar et al., 2016), as well as the general IBD patients (Sokol et al., 2017). Candida tropicalis are pathogenic fungus found in mouse intestine and when SPF mice were colonized with them, Clec7a(−/−) mice developed much severe colitis compared with uncolonized Clec7a(−/−) mice or colonized WT mice (Iliev et al., 2012; Tang et al., 2015). These findings suggest that fungal dysbiosis is associated with IBD and that Candida species are consistently associated with the inflamed gut (Li et al., 2019). In addition, a common skin resident fungus Malassezia restricta is specifically abundant in CD patients, and exacerbates colitis in mouse models through mechanisms requiring CARD9, a signaling protein involved in antifungal immunity (Limon et al., 2019).
Additionally, there was a decreased proportion of Saccharomyces cerevisiae compared with healthy subjects in IBD (Sokol et al., 2017), and S. cerevisiae were depleted in CRC (Coker et al., 2019). S. cerevisiae UFMG A-905 showed protective potential in a murine model of acute UC (Tiago et al., 2015). S. cerevisiae CNCM I-3856 had been shown to reduce AIEC-induced ileal colitis in a mouse model, by inhibiting AIEC adhesion to enterocytes and restoring barrier function (Sivignon et al., 2015). In another study, however, S. cerevisiae colonization exacerbated intestinal disease and increased gut barrier permeability in a mouse model of colitis (Chiaro et al., 2017).
Dysbiosis in virome
Enteric virome is mainly consisted of bacteriophages. Among IBD subjects, the changes in virome composition reflected alterations in bacterial composition (Clooney et al., 2019). Caudovirales phage sequences were detected in intestinal washes and biopsy tissues of Australian pediatric CD patients (Wagner et al., 2013), and they were also observed in IBD patients from a UK cohort and two US validation cohorts (Norman et al., 2015). The abundance of intestinal Caudovirales phage families, including Siphoviridae, Myoviridae and Podoviridae, were elevated in a mouse model of colitis (Duerkop et al., 2018). Recent study showed phages from active UC patients induced more IFN-γ via a TLR9-dependent pathway, which is linked to aggravated intestinal inflammation and colitis (Gogokhia et al., 2019), suggesting that certain phages may trigger intestinal inflammation in the gut and contribute to IBD.
THERAPEUTIC APPROACHES TARGETING MICROBIOME
Probiotics, prebiotics and postbiotics
Probiotics are defined as live microorganisms which when administered in adequate amounts confer a health benefit on the host (Hill et al., 2014). Prebiotic is a substrate that is selectively utilized by probiotics conferring a health benefit (Gibson et al., 2017), while postbiotic is referring a bioactive molecule produced by a probiotic. American Gastroenterological Association Institute advised that probiotics may be considered for treatment of functional symptoms in IBD (Colombel et al., 2019). Probiotics could induce anti-inflammatory effects, improve (or restore) barrier function, and beneficially modulate the composition of the microbiome by inhibiting the growth of detrimental bacteria and promoting the growth of beneficial species (Abraham and Quigley, 2017).
As a prebiotic, inulin acts on IBD by retaining microbial populations, supporting epithelial barrier function, and defending against invasion and pathogens translocation (Akram et al., 2019).The probiotic cocktail VSL#3 (a mix of 4 lactobacilli, 3 bifidobacteria and 1 strain of Streptococcus) reduced recurrence and maintain remission in patients with CD (Fedorak et al., 2015) and UC (Bibiloni et al., 2005; Miele et al., 2009). A meta-analysis showed that probiotic cocktail VSL#3 was effective in inducing remission of active UC, and the probiotics may be as effective as 5-aminosalicylates (5-ASAs) in preventing relapse of quiescent UC (Derwa et al., 2017). Moreover, probiotic Lactobacillus reuteri ATCC 55730 (Oliva et al., 2012), and E. coli strain Nissle 1917 (Scaldaferri et al., 2016; Sonnenborn, 2016) also have shown efficacy in the treatment of UC; however, a percentage of adverse events such as diarrhea and abdominal pain were reported in patients treated with E. coli strain Nissle 1917 (Kruis et al., 2004; Sassone-Corsi et al., 2016). In contrast, Lactobacillus acidophilus La-5 and Bifidobacterium animalis subsp. lactis BB-12 (Probio-Tec AB-25) was demonstrated with no significant clinical benefit in comparison with placebo for maintaining remission in patients with UC (Wildt et al., 2011). Additionally, several randomized, double-blind trials indicated administration of Lactobacillus rhamnosus in children with gastroenteritis did not have better outcomes than those who received placebo (Freedman et al., 2018; LaMont, 2018; Schnadower et al., 2018). More recently, a multi-strain probiotic (Lactobacillus rhamnosus NCIMB 30174, Lactobacillus plantarum NCIMB 30173, Lactobacillus acidophilus NCIMB 30175 and Enterococcus faecium NCIMB 30176) is associated with decreased intestinal inflammation in patients with UC, but not with CD (Bjarnason et al., 2019).
In addition to the traditional probiotics mentioned above, next-generation probiotics (NGPs) including F. prausnitzii and A. muciniphila were proposed (O’Toole et al., 2017). Oral administration of either live F. prausnitzii or its supernatant (containing its postbiotics) markedly reduced the severity of TNBS colitis, partly due to secreted metabolites able to block NF-κB activation and IL-8 production (Sokol et al., 2008a). C57BL/6 male mice administered A. muciniphila once daily by oral gavage for 14 days improved DSS-induced colitis, which was evidenced by colon length shortening, histopathology scores and enhanced barrier function (Bian et al., 2019). A. muciniphila or a specific outer membrane protein Amuc_1100 (as a postbiotic) blunted colitis, with a reduction in infiltrating macrophages and CD8(+) cytotoxic T lymphocytes (CTLs) in the colon (Wang et al., 2020). In addition to looking for new probiotic species, synthetic biology techniques were used to improve existing probiotics. A recent research showed an E. coli Nissle 1917 strain, engineered to secrete the curli-fused trefoil factors, promotes intestinal barrier function and epithelial restitution, and enhance protective effects against colitis in mice (Praveschotinunt et al., 2019).
Phage therapy
Phages are highly specific and typically lyse a subgroup of strains within one bacterial species, indicating they have a limited impact on the overall composition of the subject’s microbiome and are likely to have a better safety profile than antibiotic therapy. A randomized trial of oral T4-like coliphages or a commercial Russian coliphage product therapy in 120 children with acute bacterial diarrhea in Bangladesh did not report any adverse events but failed to improve diarrhea outcome (Sarker et al., 2016). Another two clinical trials also showed oral bacteriophage are safe in healthy children and adults (McCallin et al., 2013; Sarker et al., 2017). Though they demonstrated the safety feature of phage therapy, more knowledge is needed on in vivo phage-bacterium interaction and assessing the efficacy in reducing severity of gastrointestinal diseases.
In recent years, phage therapy has re-gained attention as a therapeutic approach to combat infectious disease and non-communicable diseases. For instance, engineered phages was used for a human mycobacterial infection that are resistant to antibiotics (Dedrick et al., 2019), and phages that specifically targets cytolytic E. faecalis attenuated alcoholic liver disease in a recent study (Duan et al., 2019). AIEC are abnormally predominant on the ileal mucosa of IBD patients, and they bind to the CEACAM6 receptor expressed on the surface of epithelial cells (Barnich et al., 2007). Bacteriophages that targets AIEC reduced DSS-induced colitis symptoms on AIEC strain LF82-colonised CEABAC10 transgenic mice, expressing the human CEACAM6 receptor for AIEC, and significantly decreased the number of AIEC in faeces and in the adherent flora of intestinal sections (Galtier et al., 2017). Therefore, phages targeting AIEC strains are a promising new treatment for IBD.
Fecal microbiota transplantation (FMT)
FMT, where fecal microbiota from a healthy donor is transplanted into a patient’s GI tract, is already a successful therapy for recurrent Clostridium difficile infection (CDI) (Hamilton et al., 2012; van Nood et al., 2013; Hvas et al., 2019). The prevailing hypothesis is that FMT might correct the dysbiosis associated with IBD, leading to a restoration of the gut microbial homeostasis (Burrello et al., 2018). The restored colon microbial community could inhibit C. difficile by multiple mechanisms: competition for nutrients; direct suppression by antimicrobial peptides; bile-acid-mediated inhibition of spore germination and vegetative growth; and activation of immune-mediated colonization resistance (Khoruts and Sadowsky, 2016).
It also has received extended attention in the treatment of CD (Zhang et al., 2013; Cui et al., 2015) and UC (Moayyedi et al., 2015; Paramsothy et al., 2017a, 2019). Improved remission rates for patients treated with FMT, possibly dependent on donor fecal composition, the use of multiple FMTs, and early treatment (Moayyedi et al., 2015). FMT appears effective in UC remission induction, but long-term durability and safety remain unclear (Paramsothy et al., 2017b). A significant fraction of patients with recurrent CDI have IBD, and FMT is somewhat less effective in clearing CDI from patients with IBD compared with patients without IBD (Khoruts et al., 2016).
Some key issues should be followed: FMT indications; donor selection; preparation of faecal material; clinical management and faecal delivery; registries, monitoring of outcomes and ethical issues; basic requirements for implementing an FMT centre (Cammarota et al., 2017, 2019). Moreover, the gut fungal together with viral community in donor stool may affect the FMT outcome of treating IBD. It has been reported high abundance of Candida albicans in donor stool reduce FMT efficacy in CDI (Zuo et al., 2018). And when studying viral transfer following FMT, multiple recipients from a single donor displayed highly individualised virus colonisation patterns (Draper et al., 2018).
CONCLUSION AND PERSPECTIVE
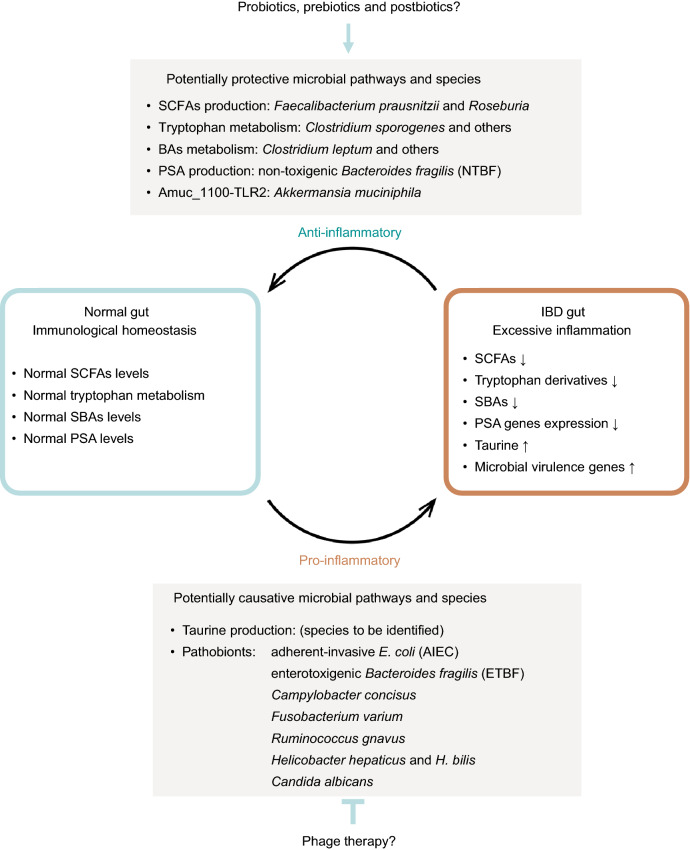
In the past decade there have been major advances in pathogenesis, pharmacological, and surgical interventions for both UC and CD. Current clinical applications for IBD diagnosis and treatment has been extensively reviewed by British Society of Gastroenterology consensus, and it highlights the importance of multidisciplinary research (Lamb et al., 2019). In this review we summarized the potential protective and causative microbial pathways and species in IBD (Fig. 1) as well as current status of therapeutic approaches targeting microbiome. Understanding of dysbiosis and the microbial pathways of specific microorganisms has suggested multiple strategies for modifying the intestinal microbiota to prevent or ameliorate IBD.
Figure 1.
Graphical summary of potentially protective and causative microbial bacterial pathways and species in IBD. SCFAs, tryptophan derivatives, secondary BAs and PSA gene expression are found depleted in human IBD gut. They are also proved to have anti-inflammatory effects in biological models and therefore are often proposed as protective factors. Prebiotics, probiotics or postbiotics targeting these factors are promising strategies to alleviate IBD. In contrast, taurine is found enriched in the metabolome and virulent genes are found enriched in microbiome of human IBD gut. Taurine and the virulence-gene-containing pathobionts also have pro-inflammatory effects in biological models and therefore are proposed as potentially causative factor for IBD. Phage therapies that target these factors are promising strategies to alleviate IBD. SCFAs, short-chain fatty acids; SBAs, secondary bile acids; PSA, polysaccharide A
Some symbiotic organisms such as AIEC, Campylobacter concisus, Fusobacterium varium, Ruminococcus gnavus, and Helicobacter species, are often referred to as pathobionts because they may cause disease under certain conditions. For AIEC, potential therapeutic strategies include targeting bacterial colonization of gut mucosa, such as the use of phage therapy, bacteriocins and anti-adhesion molecules. Bacteroides fragilis produce PSA and sphingolipids, which regulate homeostasis and protect animals from experimental colitis. In contrast, ETBF induce IL-17 in the colon and DNA damage in colonic epithelium, promoting IBD or even FAP and CRC.
Precise targeting of the metabolic pathways that are used by harmful bacteria may provide a new strategy to treat IBD. For example, tungstate-mediated microbiota editing reduced the severity of intestinal inflammation in mouse models of colitis (Zhu et al., 2018), and oral administration of sodium tungstate inhibited molybdoenzymes selectively decreased gut colonization with genotoxin-producing Enterobacteriaceae, thus reducing carcinogenesis in mouse models of colitis-associated CRC (Zhu et al., 2019).
Prebiotics are promising approaches to modify human microbiome. In the context of IBD, dietary fibers promote a selected group of SCFA-producing strains and regulate BAs profiles. SCFAs, particularly butyrate, promote the development of Treg cells and mucus production to down-regulate inflammatory signaling pathways and to strengthen the epithelial barrier. Restoration of SCFA producers by selected dietary fibers is a promising approach for managing IBD.
Traditional probiotics including Bacillus spp., Bifidobacterium spp., Lactobacillus spp., and S. cerevisiae have showed variably ameliorative effects on IBD; however, the number of patients in these trails are relatively small. Additionally, the major challenge of utilization traditional probiotics is that we do not understand the precise probiotic mechanisms of these bacteria in the context of IBD. In contrast, next-generation probiotics (NGPs or sometimes called live biotherapeutics) are based on the outcomes of mechanism studies. Some F. prausnitzii, Roseburia and A. muciniphila strains represent promising next-generation probiotic candidates. Notably, a normal bacterial BAs metabolism, especially SBAs, also play an important role modulating the host immunological homeostasis. The therapeutic potential of Clostridium sporogens and other SBA producing-species in IBD warrants additional investigations.
The safety of phages targeting intestinal pathogens is well documented for adults and children, based on data for several clinical trials in which no adverse events were reported. This is not surprising as phages are the most abundant viruses present in the human gastrointestinal tract. Phage-mediated targeting of E. faecalis ameliorated alcoholic liver disease, indicating precisely editing the intestinal microbiome is another promising direction. It would be interesting to test whether IBD could be treated by phages that target potentially causative bacteria reviewed in this article including AIEC. Besides bacteria, Candida species are consistently more abundant in IBD. Mycophage that targets Candida species may inhibit their colonization and contribute to the alleviation of IBD.
FMT can be used as a therapeutic option to treat CDI in the context of IBD when first line antibiotics are ineffective. The impact of phage on microbial dynamics is a factor that should be considered. Caudovirales phage are more significantly enriched in the intestine of individuals with IBD, which supports the notion that elevated Caudovirales phages might predict FMT failure and need for additional maintenance FMT delivery or escalation of treatment. Furthermore, Candida albicans, the fungal community that are more abundant in IBD patients compared to healthy individuals, compromises FMT efficacy in a mouse model of CDI. Therefore, further research is needed to explore whether pre-FMT eradication of C. albicans in some recipients might increase FMT success rates in some cases. US FDA recently issued a safety alert about the potential risk of transmission of pathogenic bacteria by FMT products and the resultant serious adverse reactions that may occur. It’s important to implement Shiga toxin-producing E. coli and enteropathogenic E. coli screening into the quality and safety protocols. Overall, FMT shows some evidence of benefit in IBD; however, it should only be used in the context of clinical trials until further high-quality evidence clarifies optimal administration protocol.
To conclude, we are excited to see the recent advances in microbiome research in IBD and anticipate studies in IBD pathogenesis provide more insights to facilitate therapeutic efforts to ameliorate this increasingly common disease.
ACKNOWLEDGEMENTS
We thank Drs. Xiaojian Wu, Zhen He and Jia Ke for helpful discussion. We thank Peijie Li for assisting preparation of this manuscript. This work has been supported by National Key Research and Development Program of China (No. 2017YFC1308800), the Fundamental Research Funds for the Central Universities (19ykzd33, Sun Yat-sen University) and the National Natural Science Foundation of China (Grant No. 31900056).
ABBREVIATIONS
AIEC, adherent-invasive E. coli; BAs, bile acids; BFT, B. fragilis toxin; CD, Crohn’s disease; CDI, Clostridium difficile infection; CEACAM6, Carcinoembryonic antigen related cell adhesion molecule 6; CRC, colorectal cancer; DCs, dendritic cells; DSS, dextran-sulfate sodium; ETBF, enterotoxigenic Bacteroides fragilis; FAP, familial adenomatous polyposis; FMT, fecal microbiota transplantation; IBD, inflammatory bowel diseases; IECs, intestinal epithelial cells; IL, interleukin; LPF, long polar fimbriae; NGPs, next-generation probiotics; NTBF, non-toxigenic Bacteroides fragilis; PBAs, primary BAs; PSA, polysaccharide A; SCFAs, short-chain fatty acids; SBAs, secondary BAs; TNF, anti-tumor necrosis factor; Treg cell, regulatory T cell; T5SS, type V secretion system; UC, ulcerative disease
COMPLIANCE WITH ETHICS GUIDELINES
Sheng Liu, Wenjing Zhao, Ping Lan, and Xiangyu Mou declare that they have no conflict of interest.
Contributor Information
Ping Lan, Email: lanping@mail.sysu.edu.cn.
Xiangyu Mou, Email: mouxy5@ms.sysu.edu.cn.
References
- Abraham BP, Quigley EMM. Probiotics in inflammatory bowel disease. Gastroenterol Clin N Am. 2017;46:769–782. doi: 10.1016/j.gtc.2017.08.003. [DOI] [PubMed] [Google Scholar]
- Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Akram W, Garud N, Joshi R. Role of inulin as prebiotics on inflammatory bowel disease. Drug Discov Ther. 2019;13:1–8. doi: 10.5582/ddt.2019.01000. [DOI] [PubMed] [Google Scholar]
- Alhagamhmad MH, Day AS, Lemberg DA, Leach ST. An overview of the bacterial contribution to Crohn disease pathogenesis. J Med Microbiol. 2016;65:1049–1059. doi: 10.1099/jmm.0.000331. [DOI] [PubMed] [Google Scholar]
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR, Brown EM, Graham DB, Xavier RJ, Moon JJ, et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science. 2019;364:1179–1184. doi: 10.1126/science.aaw7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherly T, Mosher C, Wang C, Hostetter J, Proctor A, Brand MW, Phillips GJ, Wannemuehler M, Jergens AE. Helicobacter bilis infection alters mucosal bacteria and modulates colitis development in defined microbiota mice. Inflamm Bowel Dis. 2016;22:2571–2581. doi: 10.1097/MIB.0000000000000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett MP, McNabb WC, Cookson AL, Zhu S, Davy M, Knoch B, Nones K, Hodgkinson AJ, Roy NC. Changes in colon gene expression associated with increased colon inflammation in interleukin-10 gene-deficient mice inoculated with Enterococcus species. BMC Immunol. 2010;11:39. doi: 10.1186/1471-2172-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Colombel JF, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Investig. 2007;117:1566–1574. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A, Brunt J, Crost E, Vaux L, Nepravishta R, Owen CD, Latousakis D, Xiao A, Li W, Chen X, et al. Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut. Nat Microbiol. 2019;4:2393–2404. doi: 10.1038/s41564-019-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X, Wu W, Yang L, Lv L, Wang Q, Li Y, Ye J, Fang D, Wu J, Jiang X, et al. Administration of Akkermansia muciniphila ameliorates dextran sulfate sodium-induced ulcerative colitis in mice. Front Microbiol. 2019;10:2259. doi: 10.3389/fmicb.2019.02259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibiloni R, Fedorak RN, Tannock GW, Madsen KL, Gionchetti P, Campieri M, De Simone C, Sartor RB. VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am J Gastroenterol. 2005;100:1539–1546. doi: 10.1111/j.1572-0241.2005.41794.x. [DOI] [PubMed] [Google Scholar]
- Bjarnason I, Sission G, Hayee B. A randomised, double-blind, placebo-controlled trial of a multi-strain probiotic in patients with asymptomatic ulcerative colitis and Crohn’s disease. Inflammopharmacology. 2019;27:465–473. doi: 10.1007/s10787-019-00595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandford LE, Johnston EL, Sanderson JD, Wade WG, Lax AJ. Promoter orientation of the immunomodulatory Bacteroides fragilis capsular polysaccharide A (PSA) is off in individuals with inflammatory bowel disease (IBD) Gut Microbes. 2019;10:569–577. doi: 10.1080/19490976.2018.1560755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breban M, Tap J, Leboime A, Said-Nahal R, Langella P, Chiocchia G, Furet JP, Sokol H. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann Rheum Dis. 2017;76:1614–1622. doi: 10.1136/annrheumdis-2016-211064. [DOI] [PubMed] [Google Scholar]
- Bringer MA, Glasser AL, Tung CH, Meresse S, Darfeuille-Michaud A. The Crohn’s disease-associated adherent-invasive Escherichia coli strain LF82 replicates in mature phagolysosomes within J774 macrophages. Cell Microbiol. 2006;8:471–484. doi: 10.1111/j.1462-5822.2005.00639.x. [DOI] [PubMed] [Google Scholar]
- Burrello C, Garavaglia F, Cribiu FM, Ercoli G, Lopez G, Troisi J, Colucci A, Guglietta S, Carloni S, Guglielmetti S, et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun. 2018;9:5184. doi: 10.1038/s41467-018-07359-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarota G, Ianiro G, Tilg H, Rajilic-Stojanovic M, Kump P, Satokari R, Sokol H, Arkkila P, Pintus C, Hart A, et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut. 2017;66:569–580. doi: 10.1136/gutjnl-2016-313017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarota G, Ianiro G, Kelly CR, Mullish BH, Allegretti JR, Kassam Z, Putignani L, Fischer M, Keller JJ, Costello SP, et al. International consensus conference on stool banking for faecal microbiota transplantation in clinical practice. Gut. 2019;68:2111–2121. doi: 10.1136/gutjnl-2019-319548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. Intestinal inflammation induced by oral bacteria. Science. 2017;358:308–309. doi: 10.1126/science.aap9298. [DOI] [PubMed] [Google Scholar]
- Castano-Rodriguez N, Kaakoush NO, Lee WS, Mitchell HM. Dual role of Helicobacter and Campylobacter species in IBD: a systematic review and meta-analysis. Gut. 2017;66:235–249. doi: 10.1136/gutjnl-2015-310545. [DOI] [PubMed] [Google Scholar]
- Chai JN, Peng Y, Rengarajan S, Solomon BD, Ai TL, Shen Z, Perry JSA, Knoop KA, Tanoue T, Narushima S, et al. Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci Immunol. 2017;2(13):eaal5068. doi: 10.1126/sciimmunol.aal5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, Wu GD. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:1948–1956. doi: 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaro TR, Soto R, Zac Stephens W, Kubinak JL, Petersen C, Gogokhia L, Bell R, Delgado JC, Cox J, Voth W, et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci Transl Med. 2017;9(380):eaaf9044. doi: 10.1126/scitranslmed.aaf9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua HH, Chou HC, Tung YL, Chiang BL, Liao CC, Liu HH, Ni YH. Intestinal dysbiosis featuring abundance of Ruminococcus gnavus associates with allergic diseases in infants. Gastroenterology. 2018;154:154–167. doi: 10.1053/j.gastro.2017.09.006. [DOI] [PubMed] [Google Scholar]
- Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe. 2018;23(203–214):e205. doi: 10.1016/j.chom.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, O’Regan O, Ryan FJ, Draper LA, Plevy SE, Ross RP, et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe. 2019;26:764–778.e765. doi: 10.1016/j.chom.2019.10.009. [DOI] [PubMed] [Google Scholar]
- Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, Chan FKL, Sung JJY, Yu J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 2019;68:654–662. doi: 10.1136/gutjnl-2018-317178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombel JF, Shin A, Gibson PR. AGA clinical practice update on functional gastrointestinal symptoms in patients with inflammatory bowel disease: expert review. Clin Gastroenterol Hepatol. 2019;17:380–390.e381. doi: 10.1016/j.cgh.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors J, Dunn KA, Allott J, Bandsma R, Rashid M, Otley AR, Bielawski JP, Van Limbergen J. The relationship between fecal bile acids and microbiome community structure in pediatric Crohn’s disease. ISME J. 2019;14:702–713. doi: 10.1038/s41396-019-0560-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Feng Q, Wang H, Wang M, Peng Z, Li P, Huang G, Liu Z, Wu P, Fan Z, et al. Fecal microbiota transplantation through mid-gut for refractory Crohn’s disease: safety, feasibility, and efficacy trial results. J Gastroenterol Hepatol. 2015;30:51–58. doi: 10.1111/jgh.12727. [DOI] [PubMed] [Google Scholar]
- Darfeuille-Michaud A. Adherent-invasive Escherichia coli: a putative new E. coli pathotype associated with Crohn’s disease. Int J Med Microbiol. 2002;292:185–193. doi: 10.1078/1438-4221-00201. [DOI] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- De Hertogh G, Aerssens J, Geboes KP, Geboes K. Evidence for the involvement of infectious agents in the pathogenesis of Crohn’s disease. World J Gastroenterol. 2008;14:845–852. doi: 10.3748/wjg.14.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med. 2019;25:730–733. doi: 10.1038/s41591-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, DeStefano Shields CE, Hechenbleikner EM, Huso DL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592–597. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derwa Y, Gracie DJ, Hamlin PJ, Ford AC. Systematic review with meta-analysis: the efficacy of probiotics in inflammatory bowel disease. Aliment Pharmacol Ther. 2017;46:389–400. doi: 10.1111/apt.14203. [DOI] [PubMed] [Google Scholar]
- Dickson I. Gut microbiota: oral bacteria: a cause of IBD? Nat Rev Gastroenterol Hepatol. 2018;15:4–5. doi: 10.1038/nrgastro.2017.161. [DOI] [PubMed] [Google Scholar]
- Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551:648–652. doi: 10.1038/nature24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper LA, Ryan FJ, Smith MK, Jalanka J, Mattila E, Arkkila PA, Ross RP, Satokari R, Hill C. Long-term colonisation with donor bacteriophages following successful faecal microbial transplantation. Microbiome. 2018;6:220. doi: 10.1186/s40168-018-0598-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, White RC, Clarke TH, Nguyen K, Torralba M, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. 2019;575:505–511. doi: 10.1038/s41586-019-1742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, Thomas G, Barbu V, Humbert L, Despras G, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- Duerkop BA, Kleiner M, Paez-Espino D, Zhu W, Bushnell B, Hassell B, Winter SE, Kyrpides NC, Hooper LV. Murine colitis reveals a disease-associated bacteriophage community. Nat Microbiol. 2018;3:1023–1031. doi: 10.1038/s41564-018-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton K, Pirani A, Snitkin ES, Reproducibility Project: Cancer B, Iorns E, Tsui R, Denis A, Perfito N, Errington TM, Iorns E et al (2018). Replication Study: intestinal inflammation targets cancer-inducing activity of the microbiota. Elife 7:e34364. [DOI] [PMC free article] [PubMed]
- el-Omar E, Penman I, Cruikshank G, Dover S, Banerjee S, Williams C, McColl KE. Low prevalence of Helicobacter pylori in inflammatory bowel disease: association with sulphasalazine. Gut. 1994;35:1385–1388. doi: 10.1136/gut.35.10.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson AR, Cantarel BL, Lamendella R, Darzi Y, Mongodin EF, Pan C, Shah M, Halfvarson J, Tysk C, Henrissat B, et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn’s disease. PLoS ONE. 2012;7:e49138. doi: 10.1371/journal.pone.0049138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorak RN, Feagan BG, Hotte N, Leddin D, Dieleman LA, Petrunia DM, Enns R, Bitton A, Chiba N, Pare P, et al. The probiotic VSL#3 has anti-inflammatory effects and could reduce endoscopic recurrence after surgery for Crohn’s disease. Clin Gastroenterol Hepatol. 2015;13:928–935.e922. doi: 10.1016/j.cgh.2014.10.031. [DOI] [PubMed] [Google Scholar]
- Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman SB, Williamson-Urquhart S, Farion KJ, Gouin S, Willan AR, Poonai N, Hurley K, Sherman PM, Finkelstein Y, Lee BE, et al. Multicenter trial of a combination probiotic for children with gastroenteritis. N Engl J Med. 2018;379:2015–2026. doi: 10.1056/NEJMoa1802597. [DOI] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Galtier M, De Sordi L, Sivignon A, de Vallee A, Maura D, Neut C, Rahmouni O, Wannerberger K, Darfeuille-Michaud A, Desreumaux P, et al. Bacteriophages targeting adherent invasive Escherichia coli strains as a promising new treatment for Crohn’s disease. J Crohns Colitis. 2017;11:840–847. doi: 10.1093/ecco-jcc/jjw224. [DOI] [PubMed] [Google Scholar]
- Garrett WS. The gut microbiota and colon cancer. Science. 2019;364:1133–1135. doi: 10.1126/science.aaw2367. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- Gogokhia L, Buhrke K, Bell R, Hoffman B, Brown DG, Hanke-Gogokhia C, Ajami NJ, Wong MC, Ghazaryan A, Valentine JF, et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe. 2019;25:285–299.e288. doi: 10.1016/j.chom.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9:103. doi: 10.1186/s13073-017-0490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MJ, Weingarden AR, Sadowsky MJ, Khoruts A. Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am J Gastroenterol. 2012;107:761–767. doi: 10.1038/ajg.2011.482. [DOI] [PubMed] [Google Scholar]
- Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, Ha S, Nelson BN, Kelly SP, Wu L, et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature. 2019;576:143–148. doi: 10.1038/s41586-019-1785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedin C, van der Gast CJ, Rogers GB, Cuthbertson L, McCartney S, Stagg AJ, Lindsay JO, Whelan K. Siblings of patients with Crohn’s disease exhibit a biologically relevant dysbiosis in mucosal microbial metacommunities. Gut. 2016;65:944–953. doi: 10.1136/gutjnl-2014-308896. [DOI] [PubMed] [Google Scholar]
- Henke MT, Kenny DJ, Cassilly CD, Vlamakis H, Xavier RJ, Clardy J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc Natl Acad Sci USA. 2019;116:12672–12677. doi: 10.1073/pnas.1904099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- Hvas CL, Dahl Jørgensen SM, Jørgensen SRP, Storgaard M, Lemming L, Hansen MM, Erikstrup C, Dahlerup JF. Fecal microbiota transplantation is superior to fidaxomicin for treatment of recurrent Clostridium difficile infection. Gastroenterology. 2019;156:1324–1332.e1323. doi: 10.1053/j.gastro.2018.12.019. [DOI] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van Dullemen HM, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67:108–119. doi: 10.1136/gutjnl-2016-312135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jergens AE, Wilson-Welder JH, Dorn A, Henderson A, Liu Z, Evans RB, Hostetter J, Wannemuehler MJ. Helicobacter bilis triggers persistent immune reactivity to antigens derived from the commensal bacteria in gnotobiotic C3H/HeN mice. Gut. 2007;56:934–940. doi: 10.1136/gut.2006.099242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- Kalischuk LD, Buret AG. A role for Campylobacter jejuni-induced enteritis in inflammatory bowel disease? Am J Physiol Gastrointest Liver Physiol. 2010;298:29. doi: 10.1152/ajpgi.00193.2009. [DOI] [PubMed] [Google Scholar]
- Khoruts A, Sadowsky MJ. Understanding the mechanisms of faecal microbiota transplantation. Nat Rev Gastroenterol Hepatol. 2016;13:508–516. doi: 10.1038/nrgastro.2016.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoruts A, Rank KM, Newman KM, Viskocil K, Vaughn BP, Hamilton MJ, Sadowsky MJ. Inflammatory bowel disease affects the outcome of fecal microbiota transplantation for recurrent Clostridium difficile infection. Clin Gastroenterol Hepatol. 2016;14:1433–1438. doi: 10.1016/j.cgh.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145(396–406):e391–e310. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- Kirk KF, Nielsen HL, Thorlacius-Ussing O, Nielsen H. Optimized cultivation of Campylobacter concisus from gut mucosal biopsies in inflammatory bowel disease. Gut Pathog. 2016;8:27. doi: 10.1186/s13099-016-0111-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. 2007;56:669–675. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruis W, Fric P, Pokrotnieks J, Lukas M, Fixa B, Kascak M, Kamm MA, Weismueller J, Beglinger C, Stolte M, et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut. 2004;53:1617–1623. doi: 10.1136/gut.2003.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg MC, Rothfuchs AG, Jankovic D, Caspar P, Wynn TA, Gorelick PL, Cheever AW, Sher A. Helicobacter hepaticus-induced colitis in interleukin-10-deficient mice: cytokine requirements for the induction and maintenance of intestinal inflammation. Infect Immun. 2001;69:4232–4241. doi: 10.1128/IAI.69.7.4232-4241.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg MC, Jankovic D, Feng CG, Hue S, Gorelick PL, McKenzie BS, Cua DJ, Powrie F, Cheever AW, Maloy KJ, et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J Exp Med. 2006;203:2485–2494. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laharie D, Asencio C, Asselineau J, Bulois P, Bourreille A, Moreau J, Bonjean P, Lamarque D, Pariente A, Soule JC, et al. Association between entero-hepatic Helicobacter species and Crohn’s disease: a prospective cross-sectional study. Aliment Pharmacol Ther. 2009;30:283–293. doi: 10.1111/j.1365-2036.2009.04034.x. [DOI] [PubMed] [Google Scholar]
- Lam S, Zuo T, Ho M, Chan FKL, Chan PKS, Ng SC. Review article: fungal alterations in inflammatory bowel diseases. Aliment Pharmacol Ther. 2019;50:1159–1171. doi: 10.1111/apt.15523. [DOI] [PubMed] [Google Scholar]
- Lamb CA, Kennedy NA, Raine T, Hendy PA, Smith PJ, Limdi JK, Hayee B, Lomer MCE, Parkes GC, Selinger C, et al. British Society of Gastroenterology consensus guidelines on the management of inflammatory bowel disease in adults. Gut. 2019;68:s1–s106. doi: 10.1136/gutjnl-2019-318484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMont JT. Probiotics for children with gastroenteritis. N Engl J Med. 2018;379:2076–2077. doi: 10.1056/NEJMe1814089. [DOI] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015;163:1428–1443. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang C, Tang C, He Q, Li N, Li J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J Clin Gastroenterol. 2014;48:513–523. doi: 10.1097/MCG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XV, Leonardi I, Iliev ID. Gut mycobiota in immunity and inflammatory disease. Immunity. 2019;50:1365–1379. doi: 10.1016/j.immuni.2019.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, et al. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe. 2019;25:377–388.e376. doi: 10.1016/j.chom.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Ramer-Tait AE, Henderson AL, Demirkale CY, Nettleton D, Wang C, Hostetter JM, Jergens AE, Wannemuehler MJ. Helicobacter bilis colonization enhances susceptibility to Typhlocolitis following an inflammatory trigger. Dig Dis Sci. 2011;56:2838–2848. doi: 10.1007/s10620-011-1701-3. [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti A, Zorzi F, Del Chierico F, Altomare A, Cocca S, Avola A, De Biasio F, Russo A, Cella E, Reddel S, et al. Fecal and mucosal microbiota profiling in irritable bowel syndrome and inflammatory bowel disease. Front Microbiol. 2019;10:1655. doi: 10.3389/fmicb.2019.01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- Machiels K, Sabino J, Vandermosten L, Joossens M, Arijs I, de Bruyn M, Eeckhaut V, Van Assche G, Ferrante M, Verhaegen J, et al. Specific members of the predominant gut microbiota predict pouchitis following colectomy and IPAA in UC. Gut. 2017;66:79–88. doi: 10.1136/gutjnl-2015-309398. [DOI] [PubMed] [Google Scholar]
- Mahendran V, Riordan SM, Grimm MC, Tran TA, Major J, Kaakoush NO, Mitchell H, Zhang L. Prevalence of Campylobacter species in adult Crohn’s disease and the preferential colonization sites of Campylobacter species in the human intestine. PLoS ONE. 2011;6:e25417. doi: 10.1371/journal.pone.0025417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahendran V, Liu F, Riordan SM, Grimm MC, Tanaka MM, Zhang L. Examination of the effects of Campylobacter concisus zonula occludens toxin on intestinal epithelial cells and macrophages. Gut Pathog. 2016;8:18. doi: 10.1186/s13099-016-0101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Kaakoush NO, Leach ST, Nahidi L, Lu HK, Norman J, Day AS, Zhang L, Mitchell HM. Host attachment, invasion, and stimulation of proinflammatory cytokines by Campylobacter concisus and other non-Campylobacter jejuni Campylobacter species. J Infect Dis. 2010;202:1855–1865. doi: 10.1086/657316. [DOI] [PubMed] [Google Scholar]
- Mar JS, LaMere BJ, Lin DL, Levan S, Nazareth M, Mahadevan U, Lynch SV. Disease severity and immune activity relate to distinct interkingdom gut microbiome states in ethnically distinct ulcerative colitis patients. mBio. 2016 doi: 10.1128/mBio.01072-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- Martin R, Miquel S, Benevides L, Bridonneau C, Robert V, Hudault S, Chain F, Berteau O, Azevedo V, Chatel JM, et al. Functional characterization of novel Faecalibacterium prausnitzii strains isolated from healthy volunteers: a step forward in the use of F. prausnitzii as a next-generation probiotic. Front Microbiol. 2017;8:1226. doi: 10.3389/fmicb.2017.01226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McCallin S, Alam Sarker S, Barretto C, Sultana S, Berger B, Huq S, Krause L, Bibiloni R, Schmitt B, Reuteler G, et al. Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects. Virology. 2013;443:187–196. doi: 10.1016/j.virol.2013.05.022. [DOI] [PubMed] [Google Scholar]
- Miele E, Pascarella F, Giannetti E, Quaglietta L, Baldassano RN, Staiano A. Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis. Am J Gastroenterol. 2009;104:437–443. doi: 10.1038/ajg.2008.118. [DOI] [PubMed] [Google Scholar]
- Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Armstrong D, Marshall JK, Kassam Z, Reinisch W, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149:102–109.e106. doi: 10.1053/j.gastro.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhya I, Thomson JM, Hansen R, Berry SH, El-Omar EM, Hold GL. Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS ONE. 2011;6:e21490. doi: 10.1371/journal.pone.0021490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonaki M, Rayment NB, Rampton DS, Hudspith BN, Brostoff J. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:481–487. doi: 10.1097/01.mib.0000159663.62651.4f. [DOI] [PubMed] [Google Scholar]
- Nattramilarasu PK, Bucker R, Lobo de Sa FD, Fromm A, Nagel O, Lee IM, Butkevych E, Mousavi S, Genger C, Klove S, et al. Campylobacter concisus impairs sodium absorption in colonic epithelium via ENaC dysfunction and claudin-8 disruption. Int J Mol Sci. 2020;21(2):373. doi: 10.3390/ijms21020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2018;390:2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14:573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K, Nishida A, Inoue R, Kawada Y, Ohno M, Sakai S, Inatomi O, Bamba S, Sugimoto M, Kawahara M, et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol. 2018;53:95–106. doi: 10.1007/s00535-017-1384-4. [DOI] [PubMed] [Google Scholar]
- Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- Ohkusa T, Sato N, Ogihara T, Morita K, Ogawa M, Okayasu I. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol. 2002;17:849–853. doi: 10.1046/j.1440-1746.2002.02834.x. [DOI] [PubMed] [Google Scholar]
- Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Sato N. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 2003;52:79–83. doi: 10.1136/gut.52.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkusa T, Yoshida T, Sato N, Watanabe S, Tajiri H, Okayasu I. Commensal bacteria can enter colonic epithelial cells and induce proinflammatory cytokine secretion: a possible pathogenic mechanism of ulcerative colitis. J Med Microbiol. 2009;58:535–545. doi: 10.1099/jmm.0.005801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva S, Di Nardo G, Ferrari F, Mallardo S, Rossi P, Patrizi G, Cucchiara S, Stronati L. Randomised clinical trial: the effectiveness of Lactobacillus reuteri ATCC 55730 rectal enema in children with active distal ulcerative colitis. Aliment Pharmacol Ther. 2012;35:327–334. doi: 10.1111/j.1365-2036.2011.04939.x. [DOI] [PubMed] [Google Scholar]
- Ossa JC, Ho NK, Wine E, Leung N, Gray-Owen SD, Sherman PM. Adherent-invasive Escherichia coli blocks interferon-gamma-induced signal transducer and activator of transcription (STAT)-1 in human intestinal epithelial cells. Cell Microbiol. 2013;15:446–457. doi: 10.1111/cmi.12048. [DOI] [PubMed] [Google Scholar]
- O'Toole PW, Marchesi JR, Hill C. Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat Microbiol. 2017;2:17057. doi: 10.1038/nmicrobiol.2017.57. [DOI] [PubMed] [Google Scholar]
- Owen CD, Tailford LE, Monaco S, Suligoj T, Vaux L, Lallement R, Khedri Z, Yu H, Lecointe K, Walshaw J, et al. Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus. Nat Commun. 2017;8:2196. doi: 10.1038/s41467-017-02109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmela C, Chevarin C, Xu Z, Torres J, Sevrin G, Hirten R, Barnich N, Ng SC, Colombel JF. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut. 2018;67:574–587. doi: 10.1136/gutjnl-2017-314903. [DOI] [PubMed] [Google Scholar]
- Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, Leong RWL, Connor S, Ng W, Paramsothy R, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218–1228. doi: 10.1016/S0140-6736(17)30182-4. [DOI] [PubMed] [Google Scholar]
- Paramsothy S, Paramsothy R, Rubin DT, Kamm MA, Kaakoush NO, Mitchell HM, Castano-Rodriguez N. Faecal microbiota transplantation for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis. 2017;11:1180–1199. doi: 10.1093/ecco-jcc/jjx063. [DOI] [PubMed] [Google Scholar]
- Paramsothy S, Nielsen S, Kamm MA, Deshpande NP, Faith JJ, Clemente JC, Paramsothy R, Walsh AJ, van den Bogaerde J, Samuel D, et al. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology. 2019;156:1440–1454.e1442. doi: 10.1053/j.gastro.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Pittayanon R, Lau JT, Leontiadis GI, Tse F, Yuan Y, Surette M, Moayyedi P. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology. 2019;158(4):930–946.e1. doi: 10.1053/j.gastro.2019.11.294. [DOI] [PubMed] [Google Scholar]
- Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- Postler TS, Ghosh S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab. 2017;26:110–130. doi: 10.1016/j.cmet.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveschotinunt P, Duraj-Thatte AM, Gelfat I, Bahl F, Chou DB, Joshi NS. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat Commun. 2019;10:5580. doi: 10.1038/s41467-019-13336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prindiville TP, Sheikh RA, Cohen SH, Tang YJ, Cantrell MC, Silva J., Jr Bacteroides fragilis enterotoxin gene sequences in patients with inflammatory bowel disease. Emerg Infect Dis. 2000;6:171–174. doi: 10.3201/eid0602.000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prorok-Hamon M, Friswell MK, Alswied A, Roberts CL, Song F, Flanagan PK, Knight P, Codling C, Marchesi JR, Winstanley C, et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut. 2014;63:761–770. doi: 10.1136/gutjnl-2013-304739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermudez-Humaran LG, Pigneur B, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut. 2016;65:415–425. doi: 10.1136/gutjnl-2014-307649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring C, Klopfleisch R, Dahlke K, Basic M, Bleich A, Blaut M. Akkermansia muciniphila strain ATCC BAA-835 does not promote short-term intestinal inflammation in gnotobiotic interleukin-10-deficient mice. Gut Microbes. 2019;10:188–203. doi: 10.1080/19490976.2018.1511663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokkas T, Gisbert JP, Niv Y, O’Morain C. The association between Helicobacter pylori infection and inflammatory bowel disease based on meta-analysis. United Eur Gastroenterol J. 2015;3:539–550. doi: 10.1177/2050640615580889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi O, van Berkel LA, Chain F, Tanweer Khan M, Taverne N, Sokol H, Duncan SH, Flint HJ, Harmsen HJ, Langella P, et al. Faecalibacterium prausnitzii A2–165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci Rep. 2016;6:18507. doi: 10.1038/srep18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker SA, Sultana S, Reuteler G, Moine D, Descombes P, Charton F, Bourdin G, McCallin S, Ngom-Bru C, Neville T, et al. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh. EBioMedicine. 2016;4:124–137. doi: 10.1016/j.ebiom.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker SA, Berger B, Deng Y, Kieser S, Foata F, Moine D, Descombes P, Sultana S, Huq S, Bardhan PK, et al. Oral application of Escherichia coli bacteriophage: safety tests in healthy and diarrheal children from Bangladesh. Environ Microbiol. 2017;19:237–250. doi: 10.1111/1462-2920.13574. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540:280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaldaferri F, Gerardi V, Mangiola F, Lopetuso LR, Pizzoferrato M, Petito V, Papa A, Stojanovic J, Poscia A, Cammarota G, et al. Role and mechanisms of action of Escherichia coli Nissle 1917 in the maintenance of remission in ulcerative colitis patients: an update. World J Gastroenterol. 2016;22:5505–5511. doi: 10.3748/wjg.v22.i24.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Garner A, Vlamakis H, Xavier RJ. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol. 2019;17:497–511. doi: 10.1038/s41579-019-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnadower D, Tarr PI, Casper TC, Gorelick MH, Dean JM, O'Connell KJ, Mahajan P, Levine AC, Bhatt SR, Roskind CG, et al. Lactobacillus rhamnosus GG versus placebo for acute gastroenteritis in children. N Engl J Med. 2018;379:2002–2014. doi: 10.1056/NEJMoa1802598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seishima J, Iida N, Kitamura K, Yutani M, Wang Z, Seki A, Yamashita T, Sakai Y, Honda M, Yamashita T, et al. Gut-derived Enterococcus faecium from ulcerative colitis patients promotes colitis in a genetically susceptible mouse host. Genome Biol. 2019;20:252. doi: 10.1186/s13059-019-1879-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekizuka T, Ogasawara Y, Ohkusa T, Kuroda M. Characterization of Fusobacterium varium Fv113-g1 isolated from a patient with ulcerative colitis based on complete genome sequence and transcriptome analysis. PLoS ONE. 2017;12:e0189319. doi: 10.1371/journal.pone.0189319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepehri S, Khafipour E, Bernstein CN, Coombes BK, Pilar AV, Karmali M, Ziebell K, Krause DO. Characterization of Escherichia coli isolated from gut biopsies of newly diagnosed patients with inflammatory bowel disease. Inflamm Bowel Dis. 2011;17:1451–1463. doi: 10.1002/ibd.21509. [DOI] [PubMed] [Google Scholar]
- Seregin SS, Golovchenko N, Schaf B, Chen J, Pudlo NA, Mitchell J, Baxter NT, Zhao L, Schloss PD, Martens EC, et al. NLRP6 protects Il10(−/−) mice from colitis by limiting colonization of Akkermansia muciniphila. Cell Rep. 2017;19:733–745. doi: 10.1016/j.celrep.2017.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha SR, Haileselassie Y, Nguyen LP, Tropini C, Wang M, Becker LS, Sim D, Jarr K, Spear ET, Singh G, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. 2020;27(4):659–670.e5. doi: 10.1016/j.chom.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivignon A, de Vallee A, Barnich N, Denizot J, Darcha C, Pignede G, Vandekerckove P, Darfeuille-Michaud A. Saccharomyces cerevisiae CNCM I-3856 prevents colitis induced by AIEC bacteria in the transgenic mouse model mimicking Crohn’s disease. Inflamm Bowel Dis. 2015;21:276–286. doi: 10.1097/MIB.0000000000000280. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Dore J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, Geva-Zatorsky N, Jupp R, Mathis D, Benoist C, et al. Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature. 2020;577:410–415. doi: 10.1038/s41586-019-1865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg A, Genta RM. Low prevalence of Helicobacter pylori infection among patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2012;35:469–476. doi: 10.1111/j.1365-2036.2011.04969.x. [DOI] [PubMed] [Google Scholar]
- Sonnenborn U. Escherichia coli strain Nissle 1917-from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties. FEMS Microbiol Lett. 2016;363(19):fnw212. doi: 10.1093/femsle/fnw212. [DOI] [PubMed] [Google Scholar]
- Tailford LE, Owen CD, Walshaw J, Crost EH, Hardy-Goddard J, Le Gall G, de Vos WM, Taylor GL, Juge N. Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat Commun. 2015;6:7624. doi: 10.1038/ncomms8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Kamiya T, Liu Y, Kadoki M, Kakuta S, Oshima K, Hattori M, Takeshita K, Kanai T, Saijo S, et al. Inhibition of Dectin-1 signaling ameliorates colitis by inducing Lactobacillus-mediated regulatory T cell expansion in the intestine. Cell Host Microbe. 2015;18:183–197. doi: 10.1016/j.chom.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Thiele Orberg E, Fan H, Tam AJ, Dejea CM, Destefano Shields CE, Wu S, Chung L, Finard BB, Wu X, Fathi P, et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2017;10:421–433. doi: 10.1038/mi.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiago FC, Porto BA, Ribeiro NS, Moreira LM, Arantes RM, Vieira AT, Teixeira MM, Generoso SV, Nascimento VN, Martins FS, et al. Effect of Saccharomyces cerevisiae strain UFMG A-905 in experimental model of inflammatory bowel disease. Benef Microbes. 2015;6:807–815. doi: 10.3920/BM2015.0018. [DOI] [PubMed] [Google Scholar]
- Underwood AP, Kaakoush NO, Sodhi N, Merif J, Seah Lee W, Riordan SM, Rawlinson WD, Mitchell HM. Campylobacter concisus pathotypes are present at significant levels in patients with gastroenteritis. J Med Microbiol. 2016;65:219–226. doi: 10.1099/jmm.0.000216. [DOI] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, Kurilshikov A, Bonder MJ, Jiang X, Tigchelaar EF, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018;10(472):eaap8914. doi: 10.1126/scitranslmed.aap8914. [DOI] [PubMed] [Google Scholar]
- Wagner J, Maksimovic J, Farries G, Sim WH, Bishop RF, Cameron DJ, Catto-Smith AG, Kirkwood CD. Bacteriophages in gut samples from pediatric Crohn’s disease patients: metagenomic analysis using 454 pyrosequencing. Inflamm Bowel Dis. 2013;19:1598–1608. doi: 10.1097/MIB.0b013e318292477c. [DOI] [PubMed] [Google Scholar]
- Wang L, Tang L, Feng Y, Zhao S, Han M, Zhang C, Yuan G, Zhu J, Cao S, Wu Q, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8(+) T cells in mice. Gut. 2020 doi: 10.1136/gutjnl-2019-320105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick EC, Rabizadeh S, Albesiano E, Wu X, Wu S, Chan J, Rhee KJ, Ortega G, Huso DL, Pardoll D, et al. Stat3 activation in murine colitis induced by enterotoxigenic Bacteroides fragilis. Inflamm Bowel Dis. 2014;20:821–834. doi: 10.1097/MIB.0000000000000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildt S, Nordgaard I, Hansen U, Brockmann E, Rumessen JJ. A randomised double-blind placebo-controlled trial with Lactobacillus acidophilus La-5 and Bifidobacterium animalis subsp. lactis BB-12 for maintenance of remission in ulcerative colitis. J Crohns Colitis. 2011;5:115–121. doi: 10.1016/j.crohns.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Williams BB, Van Benschoten AH, Cimermancic P, Donia MS, Zimmermann M, Taketani M, Ishihara A, Kashyap PC, Fraser JS, Fischbach MA. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe. 2014;16:495–503. doi: 10.1016/j.chom.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, Jarnerot G, Tysk C, Jansson JK, Engstrand L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854.e1841. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- Wlodarska M, Luo C, Kolde R, d’Hennezel E, Annand JW, Heim CE, Krastel P, Schmitt EK, Omar AS, Creasey EA, et al. Indoleacrylic acid produced by commensal Peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017;22:25–37.e26. doi: 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, Littman DR. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature. 2018;554:373–377. doi: 10.1038/nature25500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I, Eibach D, Kops F, Brenneke B, Woltemate S, Schulze J, Bleich A, Gruber AD, Muthupalani S, Fox JG, et al. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS ONE. 2013;8:e70783. doi: 10.1371/journal.pone.0070783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz B, Juillerat P, Oyas O, Ramon C, Bravo FD, Franc Y, Fournier N, Michetti P, Mueller C, Geuking M, et al. Microbial network disturbances in relapsing refractory Crohn’s disease. Nat Med. 2019;25:323–336. doi: 10.1038/s41591-018-0308-z. [DOI] [PubMed] [Google Scholar]
- Zamani S, Hesam Shariati S, Zali MR, Asadzadeh Aghdaei H, Sarabi Asiabar A, Bokaie S, Nomanpour B, Sechi LA, Feizabadi MM. Detection of enterotoxigenic Bacteroides fragilis in patients with ulcerative colitis. Gut Pathog. 2017;9:53. doi: 10.1186/s13099-017-0202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang L, Man SM, Day AS, Leach ST, Lemberg DA, Dutt S, Stormon M, Otley A, O’Loughlin EV, Magoffin A, et al. Detection and isolation of Campylobacter species other than C. jejuni from children with Crohn’s disease. J Clin Microbiol. 2009;47:453–455. doi: 10.1128/JCM.01949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FM, Wang HG, Wang M, Cui BT, Fan ZN, Ji GZ. Fecal microbiota transplantation for severe enterocolonic fistulizing Crohn’s disease. World J Gastroenterol. 2013;19:7213–7216. doi: 10.3748/wjg.v19.i41.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lee H, Grimm MC, Riordan SM, Day AS, Lemberg DA. Campylobacter concisus and inflammatory bowel disease. World J Gastroenterol. 2014;20:1259–1267. doi: 10.3748/wjg.v20.i5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Buttner L, de Lima Romao E, Behrendt CL, Lopez CA, et al. Precision editing of the gut microbiota ameliorates colitis. Nature. 2018;553:208–211. doi: 10.1038/nature25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Miyata N, Winter MG, Arenales A, Hughes ER, Spiga L, Kim J, Sifuentes-Dominguez L, Starokadomskyy P, Gopal P, et al. Editing of the gut microbiota reduces carcinogenesis in mouse models of colitis-associated colorectal cancer. J Exp Med. 2019;216:2378–2393. doi: 10.1084/jem.20181939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T, Wong SH, Cheung CP, Lam K, Lui R, Cheung K, Zhang F, Tang W, Ching JYL, Wu JCY, et al. Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nat Commun. 2018;9:3663. doi: 10.1038/s41467-018-06103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]