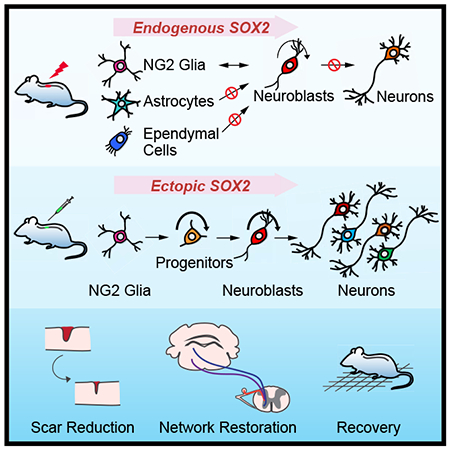
SUMMARY
Adult neurogenesis plays critical roles in maintaining brain homeostasis and in responding to neurogenic insults. However, the adult mammalian spinal cord lacks intrinsic capacity for neurogenesis. Here we show that spinal cord injury (SCI) unveils latent neurogenic potential of NG2+ glial cells, which can be exploited to produce new neurons and promote functional recovery after SCI. While endogenous Sox2 is required for SCI-induced transient reprogramming, ectopic Sox2 expression is both necessary and sufficient to unleash the full neurogenic potential of NG2 glia. Ectopic Sox2-induced neurogenesis proceeds through an expandable Ascl1+ progenitor stage and generates both excitatory and inhibitory propriospinal neurons, which make synaptic connections with ascending and descending spinal pathways. Importantly, Sox2-mediated reprogramming of NG2 glia reduces glial scarring and promotes functional recovery after SCI. These results reveal latent neurogenic potential of somatic glial cells, which can be leveraged for regenerative medicine.
Keywords: adult neurogenesis, spinal cord injury, NG2 glia, in vivo reprogramming, SOX2, astrocytes, ependymal cells, monosynaptic connections, rabies virus, lineage tracing
Graphical Abstract
In Brief
Tai et al. show that spinal cord injury (SCI) unveils latent neurogenic potential of NG2+ glial cells. SOX2 is both necessary and sufficient for neurogenic reprogramming of NG2 glia which promotes functional recovery after SCI, with new neurons forming synaptic connections with ascending and descending neural pathways.
INTRODUCTION
Neurogenesis persists in the adult mammalian brain and is modulated by many neurological conditions including injuries (Lie et al., 2004; Obernier et al., 2018). In contrast, new neurons are not normally detectable in the adult mammalian spinal cord (Horner et al., 2000). In response to spinal cord injury (SCI), several cell types including astrocytes and NG2 glia can proliferate; however, none of them are convincingly shown to generate mature neurons in vivo (Horky et al., 2006; Kang et al., 2010; Tripathi et al., 2010; Yamamoto et al., 2001). The central canal-lining ependymal cells, when isolated and cultured in vitro, can be induced to show stem cell-like properties giving rise to neurons, astrocytes, and oligodendrocytes (Meletis et al., 2008); nonetheless, they do not exhibit any stem cell function and fail to contribute to new cells in vivo after injury (Barnabe-Heider et al., 2010; Muthusamy et al., 2018; Ren et al., 2017; Shah et al., 2018).
In this study, we reexamined cellular responses to SCI by expression of the microtubule-associated protein doublecortin (DCX). DCX is normally expressed in neuroblasts and immature neurons and can serve as a reliable marker for adult neurogenesis (Couillard-Despres et al., 2005; Kempermann et al., 2004; Knoth et al., 2010). It is highly expressed in the developing spinal cord but completely turned off in the adult (Juhasova et al., 2015). Although DCX+ cells were previously observed surrounding the lesion site in the adult spinal cord (Ziv et al., 2006), their origin and final fate is not clear. Our genetic lineage mappings show that SCI induces transient DCX expression in NG2 glia but not astrocytes or ependymal cells. However, these injury-induced phenotypically reprogrammed NG2 glia fail to produce new neurons even in the presence of exogenous neurotrophic factors. We further show that SOX2 is both required and sufficient in a cell-autonomous manner for neurogenic reprogramming of NG2 glia. Ectopic SOX2 also enables these NG2 glia to generate propriospinal neurons that make synaptic connections with endogenous neurons located in the brain and spinal cord. Moreover, SOX2-mediated reprogramming of NG2 glia contributes to behavioral improvements after cervical SCI. Our data indicate that injury-induced transient neurogenic reprogramming of NG2 glia can be enhanced for neural regeneration.
RESULTS
SCI induces neurogenic reprogramming of NG2 glia
We first examined cellular responses to SCI by staining for DCX, a well-established molecular marker for immature neurons during adult neurogenesis (Couillard-Despres et al., 2005; Kempermann et al., 2004; Knoth et al., 2010). Consistent with previous reports (Juhasova et al., 2015; Su et al., 2014), DCX was not detectable in the spinal cord of sham-operated adult mice (Fig. 1A–C). The thoracic T7-9 level of the spinal cord was then injured through crush. When examined at one-week post-injury (wpi), approximately 5,000 DCX+ cells were clearly observed surrounding the GFAP+ lesion sites (Fig. 1A–C). We also detected about 740-3,200 DCX+ cells at 1 wpi in other SCI models induced by contusion, hemisection, or stab-wound (Fig. 1A–C). These cells often intermingled with GFAP+ reactive astrocytes surrounding but not far away from the lesion sites (Fig. 1C). A time-course analysis showed that the number of DCX+ cells drastically decreased at 2 wpi and subsequent later time points (Fig. 1D–F).
Figure 1. SCI-induced neurogenic reprogramming of NG2 glia.
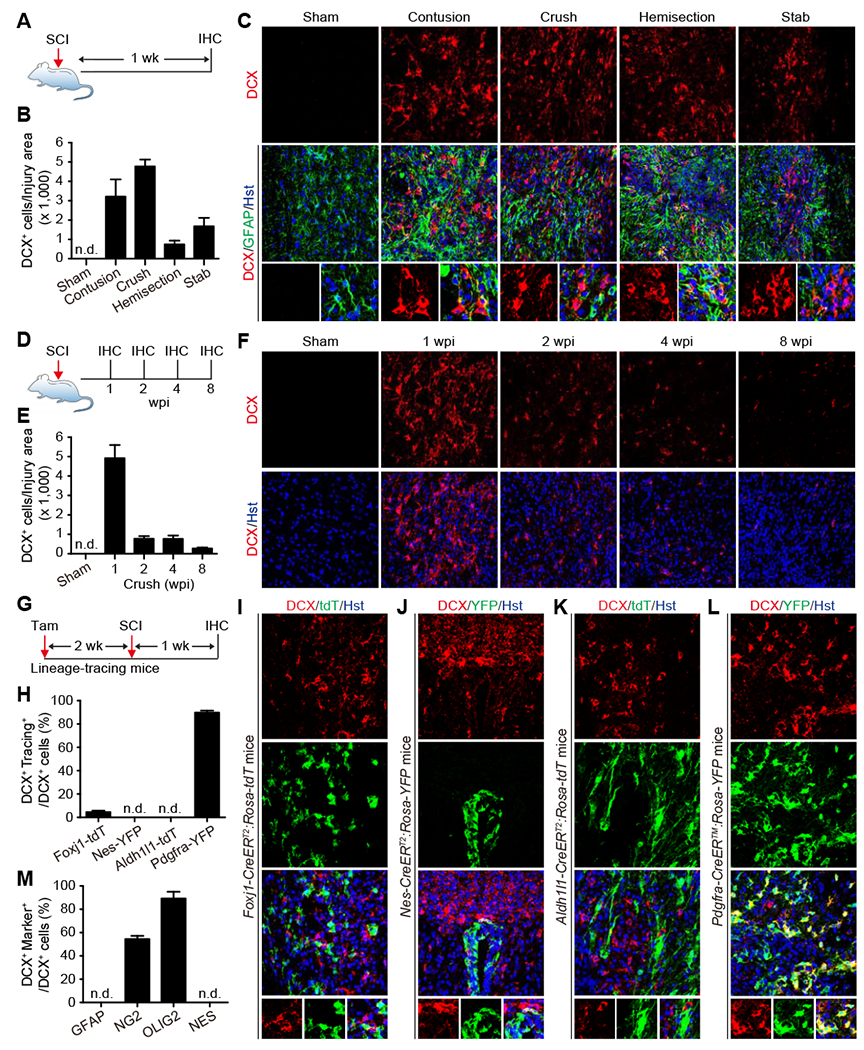
(A) Experimental design for SCI-induced DCX+ cells. IHC, immunohistochemistry; wk, week.
(B) Quantification of DCX+ cells induced by the indicated injury types (mean ± SEM; n=3 mice per condition). n.d., not detected.
(C) Confocal images of injury-induced DCX+ cells at one week post injury (wpi) in the adult mouse spinal cord. Enlarged views of the boxed regions are shown in the bottom panels. GFAP expression shows lesion areas. Nuclei are counterstained with Hoechst 33342 (Hst). Scale bars, 50 μm.
(D) Experimental design for time-course analysis of SCI-induced DCX+ cells.
(E) Quantification of crush-induced DCX+ cells at the indicated time-points (mean ± SEM; n=4-5 mice per time-point).
(F) Confocal images of crush-induced DCX+ cells through a time-course. Scale bars, 50 μm.
(G) Experimental design for genetic lineage tracings of SCI-induced DCX+ cells. Tam, tamoxifen.
(H) Quantification of genetically traced DCX+ cells at 1 wpi (mean ± SEM; n=3-7 mice per group).
(I) Lineage tracings showing SCI-induced DCX+ cells rarely originate from ependymal cells. An enlarged view of the boxed region is shown in the bottom panel. tdT is pseudocolored as green. Scale bars, 50 μm.
(J) Lineage tracings showing SCI-induced DCX+ cells do not come from NES+ cells located in the central canal. An enlarged view of the boxed region is shown in the bottom panel. Scale bars, 50 μm.
(K) lineage tracings showing SCI-induced DCX+ cells do not originate from resident astrocytes. An enlarged view of the boxed region is shown in the bottom panel. tdT is pseudocolored as green. Scale bars, 50 μm.
(L) lineage tracings showing SCI-induced DCX+ cells largely originate from NG2 glia. An enlarged view of the boxed region is shown in the bottom panel. Scale bars, 50 μm.
(M) Percentage of SCI-induced DCX+ cells expressing the indicated markers (mean ± SEM; n=3 mice per group).
See also Figure S1.
To determine the cell origin of these injury-induced DCX+ cells, we first traced the lineage of ependymal cells lining the central canal, since they were previously reported to show cellular plasticity and exhibit characteristics of neural stem cells following injury (Johansson et al., 1999). These cells were genetically labeled in a tamoxifen-dependent fashion in adult Foxj1-CreERT2;Rosa-tdTomato (tdT) mice (Fig. S1A) (Muthusamy et al., 2018; Ren et al., 2017). Two weeks post tamoxifen injection, we injured the T7-9 spinal cord with crush and conducted immunohistochemistry at 1 wpi (Fig. 1G). Although many DCX+ cells intermingled with tdT+ cells around the lesion site, only about 4.5% of DCX+ cells were traced with tdT (Fig. 1H, I). We also traced nestin-positive (NES+) ependymal cells lining the central canal in Nes-CreERT2;Rosa-YFP mice (Fig. S1B, C) (Lagace et al., 2007), since they were reported to show neural stem cell potential after injury (Meletis et al., 2008). However, none of the crush-induced DCX+ cells were YFP+ surrounding the central canal and lesion site (Fig. 1H, J). Together, these results indicate that ependymal cells are unlikely the cell origin for injury-induced DCX+ cells.
Astrocytes show plasticity following injury and they can be reprogrammed into neuroblasts and neurons in both the adult mouse brain and spinal cord (Gascon et al., 2016; Grande et al., 2013; Guo et al., 2014; Liu et al., 2015; Niu et al., 2015; Niu et al., 2013; Su et al., 2014; Torper et al., 2013; Wang et al., 2016). These cells and their derivatives were traced in adult Aldh1l1-CreERT2;Rosa-tdT mice after tamoxifen administration (Madisen et al., 2010; Srinivasan et al., 2016). Crush injury was performed 2 weeks later at the T7-9 spinal cord. When analyzed at 1 wpi, many DCX+ cells were consistently detected in the injury site, though none of them were traced with tdT (Fig. 1H, K). We finally traced NG2 glia and their progeny by using the Pdgfra-CreER™;Rosa-YFP mice (Kang et al., 2010; Srinivas et al., 2001). Unexpectedly, approximately 89.8% of crush-induced DCX+ cells were labeled with YFP (Fig. 1H, L). Consistent with this result, immunohistochemistry further revealed that about 89.3% and 54.5% of DCX+ cells expressed OLIG2 and NG2, respectively (Figs. 1M, S1D–E). In contrast, none of the injury-induced DCX+ cells were co-labeled with GFAP or NES (Figs. 1C, M, S1F). Together, these results indicate that SCI unveils a neurogenic potential of resident NG2 glia.
SOX2 is required cell-autonomously for SCI-induced reprogramming of NG2 glia
To understand how SCI induces cell reprogramming, we focused on SOX2, a stem cell factor essential for neurogenesis and neural development (Pevny and Nicolis, 2010). It is induced in glial cells of the adult brain and spinal cord following injury (Chen et al., 2019; Lee et al., 2013) and also required for injury-induced activation of adult cortical astrocytes (Chen et al., 2019). Consistent with these results, our immunohistochemistry showed a 3.6-fold increase of both the number of SOX2+ cells and the intensity of SOX2 expression in each cell surrounding the lesion site of the adult mouse spinal cord at 1 wpi (Fig. 2A–C). Cell type analysis further revealed that SOX2 was mainly detected in astrocytes and NG2 glia but not microglia and neurons (Fig. S2). Most intriguingly, nearly all (94%) of the SCI-induced DCX+ cells co-expressed SOX2 (Fig. 2D).
Figure 2. Cell-autonomous requirement of SOX2 for SCI-induced reprogramming of NG2 glia.
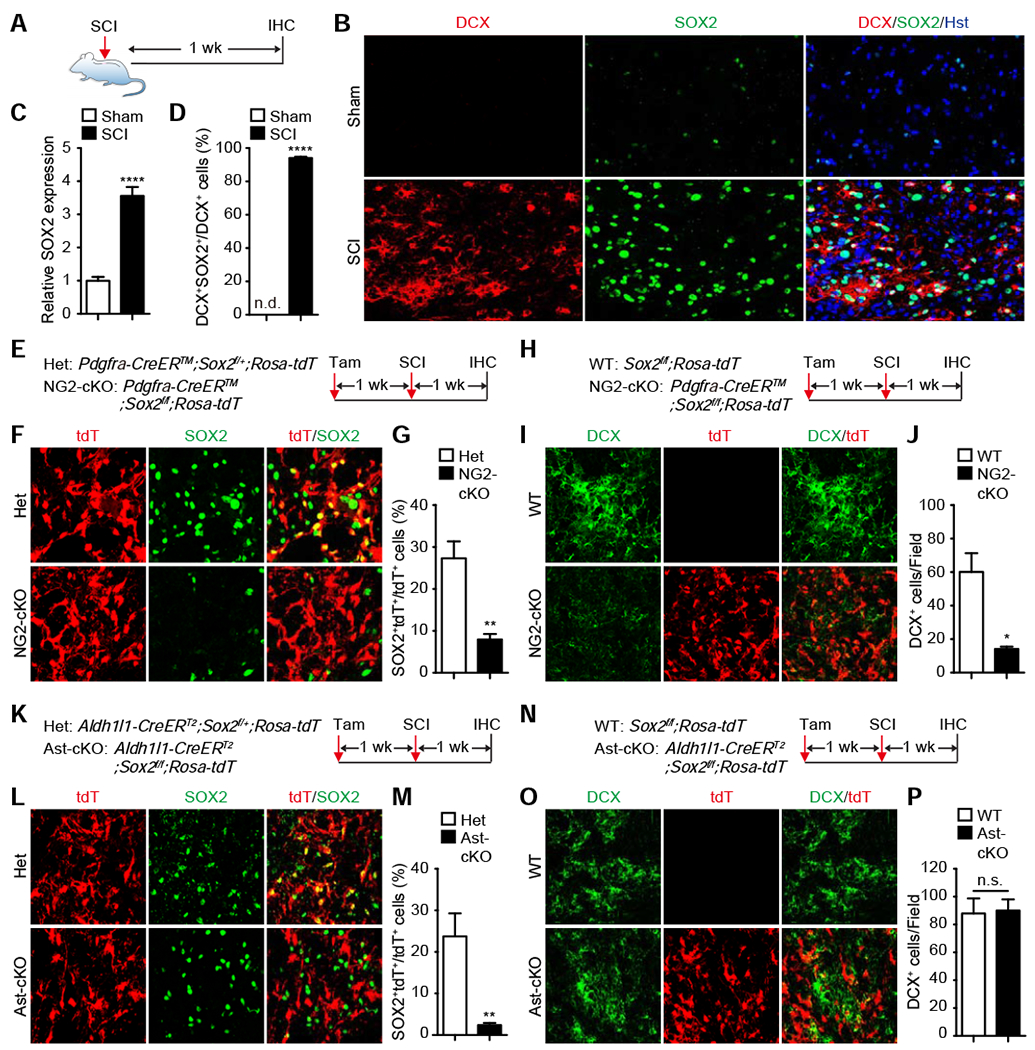
(A) Experimental design for analyzing SOX2 expression in SCI-induced DCX+ cells.
(B) Confocal images showing SOX2 expression in SCI-induced DCX+ cells. Scale bars, 50 μm.
(C) Quantification of relative SOX2 expression (mean ± SEM; n=6 mice per group; ****p<0.0001 by t-test).
(D) Percentage of DCX+ cells with SOX2 co-expression (mean ± SEM; n=6 mice per group; ****p<0.0001 by t-test).
(E) Experimental design for inducible deletion of Sox2 in NG2 glia (NG2-cKO). Het, heterozygotes.
(F) Confocal images showing inducible deletion of SOX2. Scale bars, 50 μm.
(g) Quantification of Sox2-deleted NG2 glia (mean ± SEM; n=4 mice per group; **p=0.0039 by t-test).
(H) Experimental design for inducible deletion of Sox2 in NG2 glia.
(I) Confocal images showing SCI-induced DCX+ cells in the indicated mouse spinal cord. Scale bars, 50 μm.
(J) Quantification of SCI-induced DCX+ cells in the indicated mouse spinal cords (mean ± SEM; n=3 mice per group; *p=0.0151 by t-test).
(K) Experimental design for inducible deletion of Sox2 in astrocytes (Ast-cKO).
(L) Confocal images showing inducible deletion of Sox2 in astrocytes. Scale bars, 50 μm.
(M) Quantification of Sox2-deleted astrocytes (mean ± SEM; n=6 mice per group; **p=0.0033 by t-test).
(N) Experimental design for inducible deletion of Sox2 in astrocytes.
(O) Confocal images showing SCI-induced DCX+ cells in the indicate mouse spinal cord. Scale bars, 50 μm.
(P) Quantification of SCI-induced DCX+ cells in the indicated mouse spinal cords (mean ± SEM; n=6 mice per group; n.s., not significant).
See also Figure S2.
To determine the role of SOX2 in SCI-induced DCX+ cells, we took a conditional knockout approach. Sox2 was deleted in NG2 glia after tamoxifen treatment of adult Pdgfra-CreER™;Sox2f/f;Rosa-tdT mice. One week later, contusion SCI was introduced and the mice were analyzed on the following week (Fig. 2E). Immunohistochemistry indicated a 3.4-fold reduction of the number of SOX2-expressing tdT+ cells in the lesion sites when compared to their controls (Fig. 2F, G). Correspondingly, the number of SCI-induced DCX+ cells reduced 76.5% in Sox2-deleted mice (Fig. 2H–J). By using the Aldh1l1-CreERT2;Sox2f/f;Rosa-tdT mice, we similarly examined whether SOX2 in adult astrocytes play any role for SCI-induced DCX+ cells. Tamoxifen treatment led to an about 10-fold reduction of SOX2-expressing astrocytes in the lesion sites (Fig. 2K–M). In contrast, the number of SCI-induced DCX+ cells remained unchanged after Sox2-deletion in these astrocytes (Fig. 2N–P). Together, these results show that SOX2 is required cell-autonomously for SCI-induced reprogramming of resident NG2 glia into DCX+ cells.
Elevated SOX2 is sufficient to drive neurogenic reprogramming of NG2 glia
These above results suggest that SCI-induced upregulation of SOX2 plays a critical role for generation of DCX+ cells from NG2 glia. To determine whether ectopic SOX2 alone was also sufficient to reprogram NG2 glia, we targeted these cells by using a human NG2 promoter (hNG2) in lentivirus. Cell type analysis of the hNG2-GFP reporter in adult mouse spinal cord showed that 54% of GFP+ cells expressed NG2, while the remaining cells were GFAP+ astrocytes (33.9%) and a few IBA1+ microglia (0.7%) and NeuN+ neurons (2.3%) (Fig. S3A–D). We then injected the hNG2-SOX2 virus into the adult spinal cord and analyzed at 4 weeks post virus-injection (wpv) (Fig. 3A), a time point at which very few injury-induced DCX+ cells were detectable (Fig. 1E). Excitingly, ectopic SOX2 induced approximately 15,000 DCX+ cells surrounding the injection area, whereas less than one thousand DCX+ cells were found in the control group (Fig. 3B, C).
Figure 3. Elevated SOX2 is sufficient to drive neurogenic reprogramming of NG2 glia.
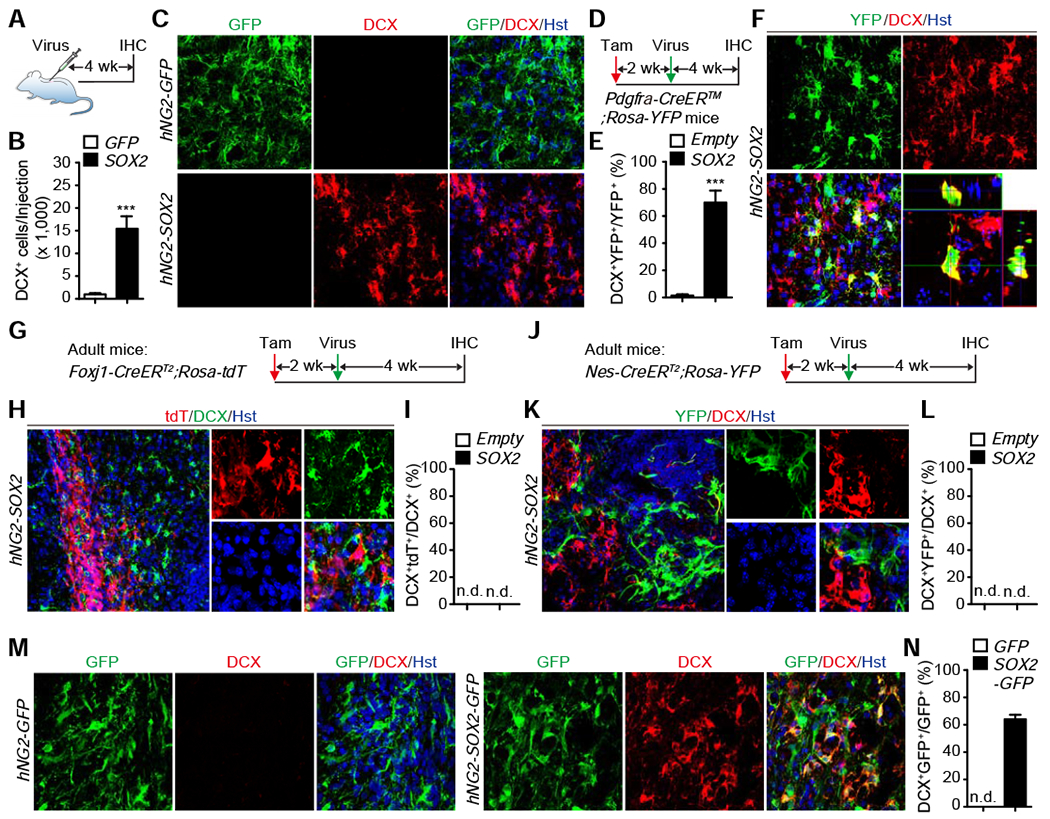
(A) Experimental design for analyzing SOX2-induced DCX+ cells in NG2 glia.
(B) Quantification of SOX2-induced DCX+ cells at 4 wpv (mean ± SEM; n=5 mice per group; ***p=0.0008 by t-test; wpv: weeks post virus-injections).
(C) Confocal images showing DCX+ cells induced by ectopic SOX2 in NG2 glia at 4 wpv. Scale bars, 50 μm.
(D) Experimental design to genetically trace NG2 glia-derived cells.
(E) Percentage of genetically traced cells converted into DCX+ cells at 4 wpv around the injection area (mean ± SEM; n=4 mice per group; ***p=0.0003 by t-test).
(F) Confocal images showing an origin of NG2 glia of SOX2-induced DCX+ cells. An orthogonal view of the boxed region is shown on the right. Scale bar, 50 μm.
(G) Experimental design to genetically trace derivatives of ependymal cells.
(H) Confocal images showing a non-ependymal cell origin of SOX2-induced DCX+ cells. An enlarged view of the boxed region is shown on the right. Scale bars, 50 μm.
(I) Percentage of SOX2-induced DCX+ cells from genetically traced ependymal cells at 4 wpv (n=3-7 mice per group).
(J) Experimental design to genetically trace derivatives of cells lining the central canal.
(K) Confocal images showing a non-central canal cell origin of SOX2-induced DCX+ cells. An enlarged view of the boxed region is shown on the right. Scale bars, 50 μm.
(L) Percentage of SOX2-induced DCX+ cells from genetically traced central canal cells at 4 wpv (n=3-4 mice per group).
(M) Confocal images showing reprogramming efficiency by SOX2 in NG2 glia. Scale bars, 50 μm.
(N) Quantification of reprogramming efficiency at 4 wpv (mean ± SEM; n=5 mice per group).
See also Figure S3.
To confirm NG2 glia as the cell origin for SOX2-induced DCX+ cells, we employed a genetic lineage tracing approach by using the Pdgfra-CreER™;Rosa-YFP mice. Consistent with previous results (Kang et al., 2010), cell type analysis showed that about 57% and 100% of YFP+ cells expressed NG2 and OLIG2, respectively, while markers for GFAP+ astrocytes, IBA1+ microglia, and NeuN+ neurons were non-detectable (Fig. S3E–G). These mice were injected with hNG2-SOX2 virus 2 weeks post tamoxifen administration and analyzed after another 4 weeks (Fig. 3D). Approximately 70% of SOX2-induced DCX+ cells were labeled with YFP (Fig. 3E, F). To further confirm SOX2 was sufficient to induce DCX+ cells from NG2 glia, we used the Cre-dependent hNG2-FLEX-SOX2 virus so that SOX2 expression was restricted to NG2 glia after tamoxifen treatment of the adult Pdgfra-CreER™;Rosa-tdT mice (Fig. S3H). When examined at 4 weeks post tamoxifen administration, 94.8% of SOX2-induced DCX+ cells were traced by tdT (Fig. S3I–K). We also examined the contribution of ependymal cells lining the central canal and found that none of the SOX2-induced DCX+ cells were genetically traced in either Foxj1-CreERT2;Rosa-tdT (Fig. 3G–I) or Nes-CreERT2;Rosa-YFP mice (Fig. 3J–L). The reprogramming efficiency was then determined by using the co-expressed GFP in hNG2-SOX2-IRES-GFP virus. About 64% of the GFP+ cells expressed DcX+, indicating that SOX2 induced efficient reprogramming of NG2 glia (Fig. 3M, N). Together, these results reveal that ectopic SOX2 alone can robustly reprogram endogenous NG2 glia toward DCX+ cells in the adult mouse spinal cord.
SCI-reprogrammed NG2 glia fail to generate mature neurons
DCX+ neuroblasts proliferate during the early stage of adult neurogenesis (Kempermann et al., 2004). We analyzed SCI-induced DCX+ cells through BrdU incorporation, which was administered for 1 week in drinking water immediately following crush injury (S4A). About 45.2% of DCX+ cells incorporated BrdU when examined at 1 wpi (Fig. S4B, C). Immunostaining also showed 25.1% of SCI-induced DCX+ cells expressed Ki67, an endogenous marker for cells in active cell cycles (Fig. S4B, C). ASCL1, a master regulator expressed in neural progenitors and critical for neuronal differentiation and adult neurogenesis (Kim et al., 2011; Raposo et al., 2015), was detected in 28.6% of SCI-induced DCX+ cells (Fig. S4D, E). To investigate the fate of SCI-induced ASCL1+ cells, we traced them in adult Ascl1-CreERT2;Rosa-tdT knockin mice after 6 tamoxifen-treatments immediately pre- and post-SCI (Fig. S4F). Although only a few cells were traced when examined at 4 wpi, cell type analysis showed that tdT+ cells were predominantly co-labeled with NG2 (85.2%) or OLIG2 (86.9%), whereas other cell type-specific markers were rarely observed (Fig. S4G–L).
To more broadly examine whether SCI induced any newly mature neurons, we treated adult mice immediately following crush injury with BrdU in drinking water for 8 weeks (Fig. S4M). However, only 0-8 BrdU+NeuN+ cells were detected around the lesions (Fig. S4N, O). Such a low number of mature neurons might be due to the lack of neurotrophic factors in the adult injured spinal cord. We then injected lentivirus immediately following SCI to express BDNF and NOG, which were previously shown to promote long-term neuronal survival and maturation in the adult mouse brain and spinal cord (Cho et al., 2007; Niu et al., 2013; Wang et al., 2016). Neither virus injections nor the expression of BDNF-NOG influenced the number of SCI-induced DCX+ cells (Fig. S4P–R). When examined at 8 wpi/wpv, however, we still only observed 0-20 BrdU+NeuN+ neurons in the whole injured spinal cord area (Fig. S4S, T). Together, these results indicate that SCI only induces transient phenotypic switch of NG2 glia to DCX+ cells, which eventually fail to become mature neurons.
Elevated SOX2 is required for NG2 glia to produce mature neurons
Since ectopic SOX2 was more robust than SCI alone in inducing DCX+ cells from NG2 glia (Fig. 3), we next investigated in detail the underlying cellular process. When examined at 4 weeks post injection of hNG2-SOX2 virus, 96% and 15% of SOX2-induced DCX+ cells could be respectively labeled by BrdU and Ki67 (Fig. 4A–C), indicating that they passed through a proliferative progenitor/neuroblast state. Correspondingly, ectopic SOX2 induced nearly 10,000 ASCL1+ cells, many of which were also labeled with DCX (Fig. 4D–F). By using the adult Pdgfra-CreER™;Rosa-tdT mice, we confirmed that NG2 glia were the cell origin for these induced ASCL1+ cells (Fig. 4G–I). Similarly, we verified that SOX2-induced ASCL1+ cells gave rise to DCX+ cells by using the Ascl1-CreERT2;Rosa-tdT lineage-tracing mice (Fig. 4J–L). Together, these results clearly indicate that SOX2-induced DCX+ cells from NG2 glia pass through an ASCL1+ progenitor stage. Such a feature of induced DCX+ cells resembles endogenous neuroblasts during adult neurogenesis in the mammalian brain (Kempermann et al., 2004; Kim et al., 2011).
Figure 4. Ectopic SOX2 reprograms NG2 glia into mature neurons.
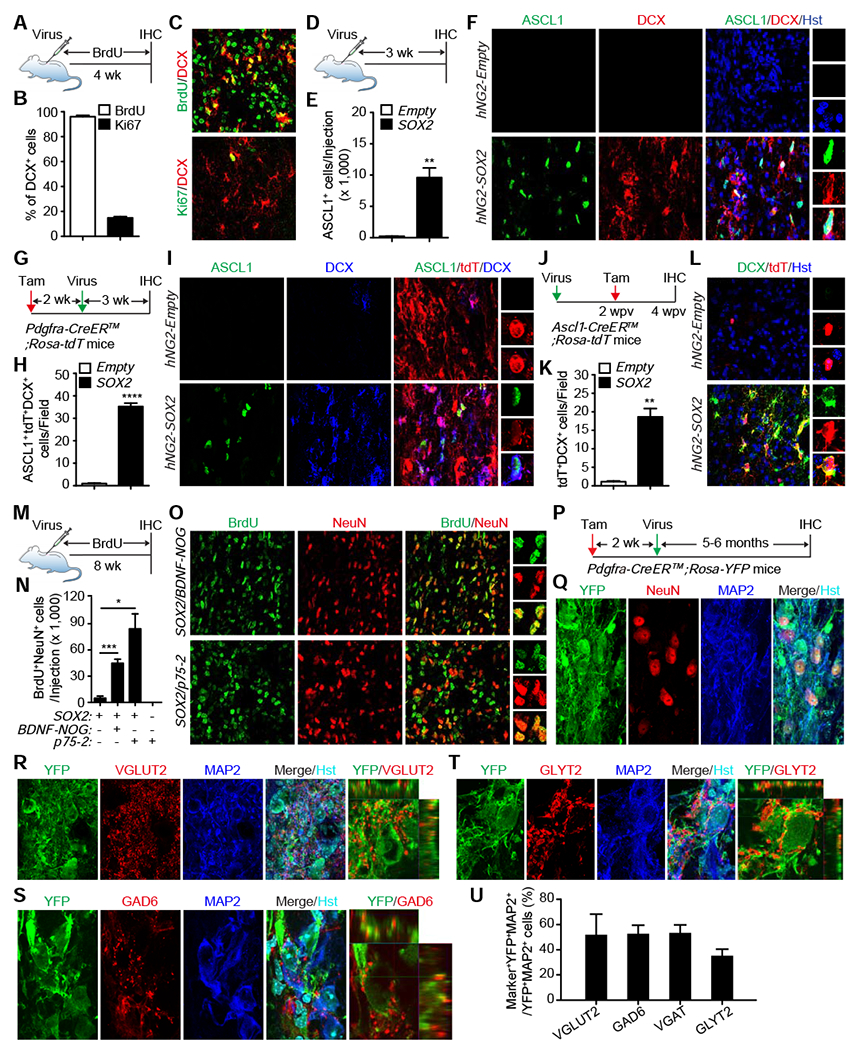
(A) Experimental design to determine proliferation of SOX2-induced DCX+ cells.
(B) Percentage of SOX2-induced DCX+ cells going through proliferation (mean ± SEM; n=5 mice per group).
(C) Confocal images showing proliferation of SOX2-induced DCX+ cells at 4 wpv. Scale bars, 50 μm.
(D) Experimental design for analyzing ASCL1+ progenitors.
(E) Quantification of virus-induced ASCL1+ progenitors at 3 wpv (mean ± SEM; n=3 mice per group; **p=0.0036 by t-test).
(F) Confocal images showing SOX2-induced ASCL1+progenitors at 3 wpv. Scale bars, 50 μm.
(G) Experimental design to determine the NG2 glia origin for SOX2-induced ASCL1+ cells.
(H) Quantification of SOX2-induced ASCL1+ progenitors from NG2 glia at 3 wpv (mean ± SEM; n=3 mice per group; ****p<0.0001 by t-test).
(I) Confocal images showing SOX2-induced ASCL1+ progenitors and DCX+ cells originating from NG2 glia at 3 wpv. Scale bars, 50 μm.
(J) Experimental design to determine the lineage relationship of SOX2-induced ASCL1+ progenitors and DCX+ cells.
(K) Quantification of DCX+ cells from SOX2-induced ASCL1+ progenitors at 4 wpv (mean ± SEM; n=3 mice per group; **p=0.0015 by t-test).
(L) Confocal images showing lineage-traced DCX+ cells from SOX2-induced ASCL1+progenitors at 4 wpv. Scale bars, 50 μm.
(M) Experimental design for analyzing SOX2-induced new neurons.
(N) Quantification of SOX2-induced new neurons from NG2 glia (mean ± SEM; n=3-6 mice per group; *p=0.0162 and ***p=0.0006 by t-test).
(O) Confocal images of SOX2-induced new neurons from NG2 glia. Enlarged views of the boxed regions are shown in the right panels. Scale bars, 50 μm.
(P) Experimental design for analyzing SOX2-induced neurons from NG2 glia.
(Q) Confocal images of NG2 glia-derived neurons expressing markers for mature neurons. Examples of these neurons are indicated by arrows. Some of the YFP−NeuN−MAP2− cells are outlined with dotted circles. Scale bars, 20 μm.
(R-T) Confocal images of NG2 glia-derived neurons with the indicated subtype-specific markers. Asterisks indicate examples of NG2 glia-derived neurons, whereas arrowheads point to signal co-localization in orthogonal views. Some of the YFP−GLYT2−MAP2− cells are outlined with dotted circles. Scale bars, 20 μm.
(U) Quantification of subtypes of NG2 glia-derived neurons (mean ± SEM; n=100-500 YFP+MAP2+ cells from 3 mice for each maker).
We next investigated whether SOX2-induced DCX+ cells could become mature neurons, which were traced by BrdU (Fig. 4M). Excitingly, SOX2 alone induced nearly 5,000 BrdU+NeuN+ neurons (Fig. 4N, O). The inclusion of BDNF-NOG increased the number of new neurons to more than 44,000. We also examined p75-2, a mutant form of neurotrophic factor NT3, which has a reduced affinity to p75NTR and can greatly enhance cell survival and axonal growth after SCI (Enomoto et al., 2013). Interestingly, p75-2 resulted in approximately 84,000 new BrdU+NeuN+ neurons per injection (Fig. 4N, O). On the other hand, DCX+ cells were rarely observed at this later time point (Fig. S5A, B), consistent with the notion that DCX+ neuroblasts are transient cell states during adult neurogenesis (Kempermann et al., 2004).
The subtypes of SOX2-induced neurons from NG2 glia were then determined through genetic lineage tracing in the spinal cord of adult Pdgfra-CreER™;Rosa-YFP mice (Fig. 4P). These neurons could be clearly identified by the co-expression of YFP and the mature neuronal marker NeuN and MAP2 in both the white and gray matter (Fig. 4Q). The presynaptic marker SYN1 was also robustly detectable, indicating potential synaptic connections with other neurons (Fig. S5C, D). A survey of markers for neuronal subtypes showed an approximately equal ratio of excitatory (VGLUT2+) and inhibitory (GAD6+ or VGAT+) neurons (Figs. 4R, S, U, S5E). Some of the inhibitory neurons also expressed GLYT2, a marker for glycinergic neurons (Fig. 4T, U). Together, these data indicate that NG2 glia can be coerced to produce diverse neuronal subtypes in the adult mouse spinal cord.
NG2 glia-derived neurons form trans-synaptic connections
Neuron-neuron connections are the cellular basis for function. We examined monosynaptic connections of NG2 glia-derived neurons by using a tracing method based on a recombinant rabies virus (Osakada and Callaway, 2013; Vivar et al., 2012; Wickersham et al., 2007). The EnvA-pseudotyped RVdG-eGFP rabies virus lacks the envelope glycoprotein (G-deleted) but expresses the reporter eGFP. The receptor TVA and rabies glycoprotein (RVg) are expressed in a Cre-dependent manner in the mouse line Rosa-TVAg (also known as RphiGT) (Takatoh et al., 2013). To specifically target the NG2 glia-derived neurons, we used the knockin allele of Ascl1-CreERT2 mice (Kim et al., 2011), since ASCL1 is uniquely induced during the neurogenic process (Fig. 4D–L). We created the Ascl1-CreERT2;Rosa-tdT;Rosa-TVAg triple knockin mouse line, such that TVAg is exclusively expressed in the induced tdT+ neurons after tamoxifen treatment during the early neurogenic process (Fig. 5A). TVA expression in the induced but not endogenous neurons allows RVdG-eGFP virus to enter the cells. The simultaneously expressed rabies glycoprotein (g) allows retrograde spreading of the viral particles to other neurons with direct presynaptic contacts with the induced neurons (Fig. 5A).
Figure 5. SOX2-induced neurons from NG2 glia form synaptic connections.
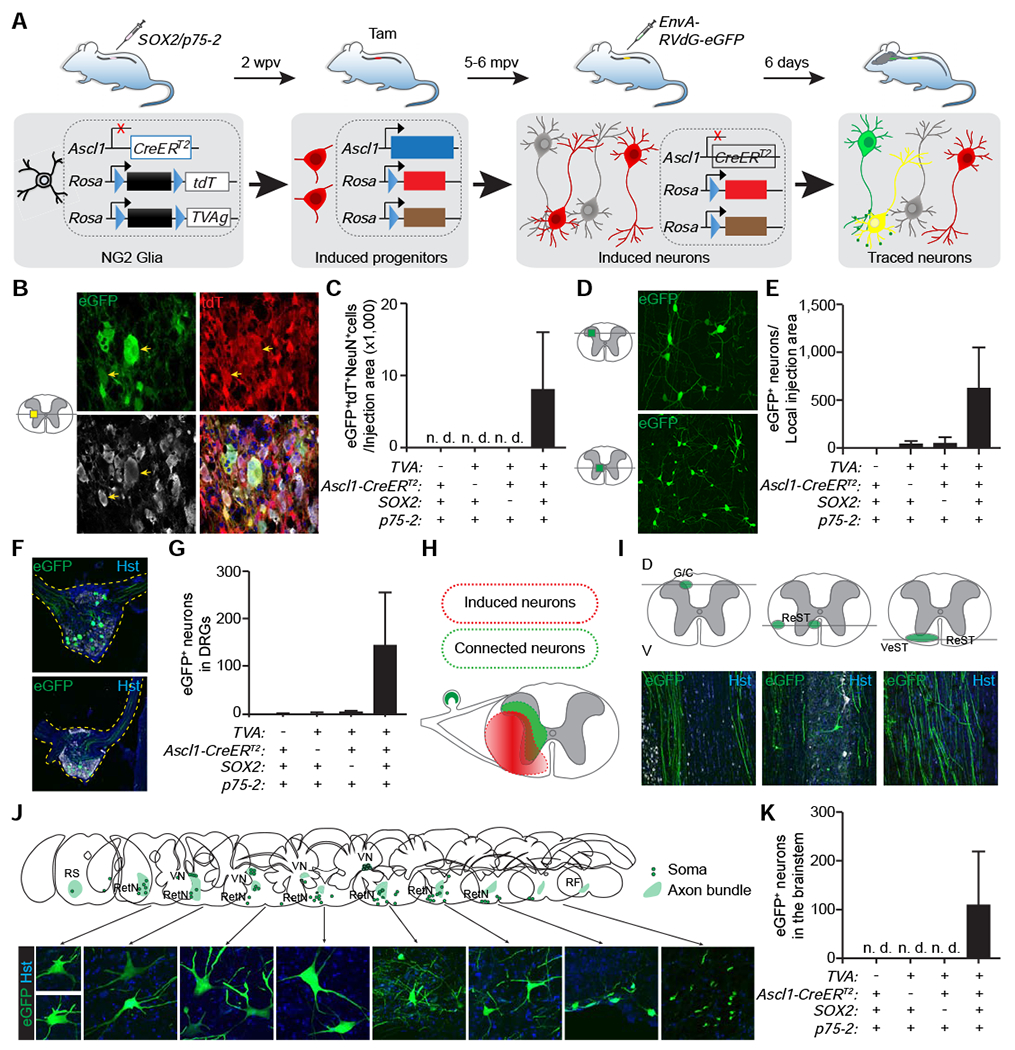
(A) Experimental design for analyzing monosynaptic connections of NG2 glia-derived neurons.
(B) Confocal images of cells surrounding the virus-injected spinal cord area. Induced neurons are traced with tdT, whereas cells harboring the engineered rabies virus are indicated by eGFP. Arrows show examples of “starter” cells (eGFP+tdT+NeuN+). Some of the eGFP−tdT−NeuN−> cells are outlined with dotted circles. Scale bar, 20 μm.
(C) Estimates of “starter” cells in the virus-injected spinal cord (mean ± SEM; n=3-6 mice per group).
(D) Confocal images of rabies-traced propriospinal neurons. Scale bars, 50 μm.
(E) Estimates of rabies-traced propriospinal neurons (mean ± SEM; n=3-6 mice per group).
(F) Confocal images of rabies-traced DRG neurons. Scale bars, 200 μm.
(G) Estimates of rabies-traced DRG neurons (mean ± SEM; n=3-6 mice per group).
(H) A schematic representation of spinal areas with induced neurons and rabies-traced neurons.
(I) Confocal images of rabies-traced axon bundles at the indicated spinal cord levels. Their approximate locations are marked in the top diagrams. G/C, gracile/cuneate tract; ReST, reticulospinal tract; VeST, vestibulospinal tract. Scale bars, 100 μm.
(J) Schematic representations and confocal images of rabies-traced neurons in the brainstem. Rabies-traced axon bundles and somas are indicated in the top diagrams. RS, rubrospinal tract; RetN, reticular nucleus; VN, vestibular nucleus; RF, reticular formation. Scale bars, 20 μm.
(K) Estimates of rabies-traced neurons in the brain/brainstem (mean ± SEM; n=3-6 mice per group).
See also Figure S6.
The specificity of the EnvA-pseudotyped RVdG-eGFP rabies virus was confirmed in Ascl1-CreERT2;Rosa-tdT mice, which lacked the TVA receptor expression. These mice were injected with lentiviruses expressing SOX2/p75-2, treated with tamoxifen at 2 wpv, injected with the recombinant rabies at 5-6 months post virus injection (mpv), and analyzed after 6 more days. Despite numerous SOX2-induced tdT+ cells surrounding the injected spinal cord, none of them were infected with the recombinant rabies, indicated by a lack of eGFP+ cells (Fig. S6A). We also examined the recombinant rabies in Ascl1-CreERT2;Rosa-tdT;Rosa-TVAg mice that were injected with the control lentivirus p75-2 alone. Only a few tdT+ cells with glia morphology were labeled by eGFP around the virus-injected spinal cord areas (Fig. S6B; SC). eGFP+ cells were not observed in other regions including the adjacent dorsal root ganglia (DRGs) of these control mice (Fig. S6B; DRG).
We therefore applied this recombinant rabies system to map the presynaptic connections of NG2 glia-derived neurons, which were induced by SOX2/p75-2 in the T7 spinal cords of adult Ascl1-CreERT2;Rosa-tdT;Rosa-TVAg mice (Fig. 5A). Horizontal sections of the spinal cords showed robust induction of new tdT+NeuN+ neurons that were only found in the injected spinal areas including the white and gray matters (Figs. 5B, S6C). Some of these cells were transduced with the recombinant rabies virus, indicated by coexpression of eGFP, tdT, and NeuN (Figs. 5B, C, S6D, E). Such neurons were not observed in control groups (Fig. 5C). The triple positive neurons could serve as “starter” cells, from which RVdG-eGFP virus could then transmit retrogradely to first order neurons. Many of these first order eGFP+ neurons were found throughout the gray matter especially surrounding the injected area in the ipsilateral side (Fig. 5D, E), although some of them were also in the rostral and caudal side. Interestingly, some neurons in the ipsilateral sides of the DRGs were also traced with eGFP, indicating monosynaptic connections with induced neurons in the spinal cord (Figs. 5F–H, S6C). Furthermore, some axon bundles in different horizontal levels of the white matter were labeled with strong eGFP signals (Fig. 5I). They mainly represent the ascending gracile/cuneate tract and the descending reticulospinal and vestibulospinal tracts, respectively. Examinations of serial cross sections through the brain and brainstem showed eGFP+ somas were mainly localized in the reticular nuclei and the vestibular nuclei (Fig. 5J–K). Together, these data indicate that NG2 glia-derived neurons can make monosynaptic connections with propriospinal neurons and those located in the brainstem and DRGs that form the ascending and descending pathways.
Reprogramming of NG2 glia reduces scar and promotes functional recovery following SCI
We next examined the biological function of reprogramming NG2 glia in a SCI model created by dorsal hemisection at the 5th cervical spina cord level (C5-DH; Fig. 6A) (Zhang et al., 2013). This SCI model was selected since it creates a clearly defined injury penumbra that allows precise virus injections. Furthermore, cervical injuries represent over half of the total SCIs in human patients especially young individuals (Norenberg et al., 2004).
Figure 6. SOX2-mediated reprogramming of NG2 glia promotes functional recovery following SCI.
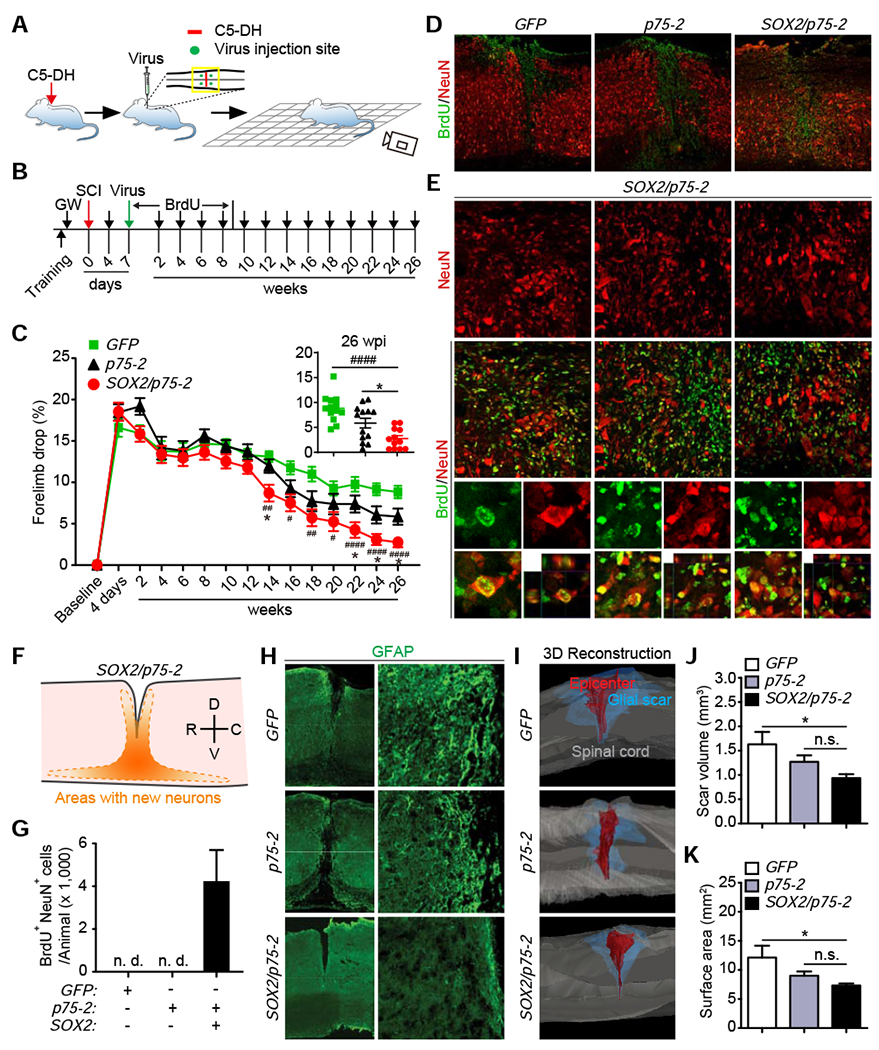
(A) A schematic drawing of the procedure. Adult mouse underwent dorsal hemisection at the 5th cervical level (C5-DH), followed by virus injections and behavioral tests.
(B) Experimental design for behavioral tests.
(C) Percentage of forelimb drops during grid-walking tests (mean ± SEM; n=12-13 mice per group; F(14,454)=176.5 and p<0.0001 for time-dependent effect; F(2,35)=4.299 and p=0.0214 for treatment-dependent effect; SOX2/p75-2 vs. GFP: ##p=0.0015 at 14 wks, #p= 0.0140 at 16 wks, ##p=0.0013 at 18 wks, #p=0.02223 at 20 wks, ####p<0.0001 at 22 wks, 24wks and 26 wks; SOX2/p75-2 vs. p75-2: *p=0.0261 at 14 wks, *p=0.0346 at 22 wks, *p=0.0488 at 24 wks, and *p=0.0383 at 26 wks). The inset graph shows performance of individual mouse at the endpoint 26 wks.
(D) Lower magnification views of spinal cord tissues with the indicated marker staining. Scale bars, 200 μm.
(E) Enlarged views of the boxed regions in Figure 6D. Arrowheads show examples of BrdU+NeuN+ neurons. Higher magnification views of the boxed regions are shown in the lower panels. Scale bars, 25 μm.
(F) A schematic of the distribution of SOX2-induced new neurons. D, dorsal; V, ventral; R, rostral; C, caudal.
(G) Quantification of new neurons in the injured spinal cord (mean ± SEM; n=6 mice per group).
(H) Low (stitched images) and high magnification views of GFAP-stained glial scar surrounding the lesion epicenter. Scale bars, 500 μm and 50 μm for the left and right panels, respectively.
(I) Representative 3D reconstructions of glial scar at the lesion site. Scale bars, 500 μm.
(J) Quantification of glial scar volume (mean ± SEM; n=8 mice per group; *p=0.0247).
(K) Quantification of glial scar surface area (mean ± SEM; n=8 mice per group; *p=0.0361).
See also Figure S7.
Behavior experiments were conducted in a randomized and blinded fashion. Three groups of viruses (GFP, p75-2, and SOX2/p75-2) were produced and individually coded with a randomized letter. Experimental wildtype mice were pretrained and tested for the grid-walking paradigm before SCI and 4 days following SCI (Fig. 6B). One week post C5-DH, mice were randomized into three groups and injected with the coded viruses (Fig. 6A, B). The identity of the injected viruses was blinded to the experimenters. Newly born neurons were traced through BrdU in drinking water for 8 weeks. Mice were then behaviorally examined at the indicated time points (Fig. 6B). Upon completion of the experiments, mouse groups were analyzed and then decoded.
The grid-walking paradigm, which requires mice to accurately place their paws on the bars while walking on the grid, examines both basic and skilled locomotion (Al-Ali et al., 2017; Sedy et al., 2008). Successfully walking on the grid depends more on the movement of shoulder and elbow. Both forelimbs were examined in this experiment. Statistical analyses through two-way ANOVA showed significant time- and treatment-dependent effects (Fig. 6C; F(14,454)=176.5 and p<0.0001 for time-dependent effect; F(2,35)=4.299 and p=0.0214 for treatment-dependent effect). Tukey’s post-hoc multiple comparisons test revealed that, when compared to either of the control groups, the SOX2/p75-2 group performed significantly better starting at 14 weeks post lesion (SOX2/p75-2 vs. GFP: ##p=0.0015 at 14 wks, #p= 0.0140 at 16 wks, ##p=0.0013 at 18 wks, #p=0.02223 at 20 wks, ####p<0.0001 at 22 wks, 24wks and 26 wks; SOX2/p75-2 vs. p75-2: *p=0.0261 at 14 wks, *p=0.0346 at 22 wks, *p=0.0488 at 24 wks, and *p=0.0383 at 26 wks).
Upon completion of behavioral tests, spinal cords were collected for histological analyses. We first confirmed generation of new neurons in this SCI model. These new neurons, indicated by BrdU+NeuN+, were mainly distributed surrounding the lesion/injection area of the SOX2/p75-2 group (Fig. 6D–F). Approximately 4,000 BrdU+NeuN+ neurons were detected in the group treated with SOX2/p75-2 virus but not in the other two control groups (Fig. 6G). We also examined glial scar by staining GFAP. Astrocytic boundaries were well defined surrounding the lesions (Fig. 6H). Interestingly, 3D reconstructions (Fig. S7) and quantifications showed that both scar volume and surface area were significantly reduced in SOX2/p75-2 group when compare to the GFP control (Fig. 6I–K; *p=0.0247 and *p=0.0361, respectively). The p75-2 group also showed moderate but not significant reduction of astrocytic scars (Fig. 6J, K). These results indicate that reprogramming of NG2 glia leads to generation of new neurons and reduction of glial scars, both of which may contribute to functional improvements post SCI.
SOX2-reprogrammed NG2 glia are bipotent and generate both neurons and oligodendrocytes
NG2 glia is the cell source for continued oligodendrogenesis in the adult central nervous system (Nishiyama et al., 2016; Tripathi et al., 2010). Reprogramming their fate for neurogenesis may lead to depletion of NG2 glia and disruption of oligodendrogenesis. We examined such a scenario by using the lineage tracing mouse line Ascl1-CreERT2;Rosa-tdT, since ASCL1 was uniquely induced by SOX2 from reprogrammed NG2 glia (Fig. 4D–L). Adult Ascl1-CreERT2;Rosa-tdT mice underwent C5-DH injury and were injected with viruses into the surrounding lesion area at 1 wpi (Fig. 7A, B). These mice were then administered with tamoxifen at 2 wpv and examined at 12 wpv (Fig. 7B). Numerous tdT+ cells were detected surrounding the lesion area of mice injected with the SOX2/p75-2 virus (Fig. 7C). In sharp contrast, only a few tdT+ cells were sparsely observed in the p75-2 control (Fig. 7C). Cell types were determined by using specific markers and showed that 45.8% of tdT+ cells were NeuN+ neurons in the SOX2/p75-2 group, whereas such NeuN+tdT+ cells were not detected in the p75-2 control (Fig. 7C, D, H). Interestingly, 34.6% and 43.5% of SOX2-induced tdT+ cells expressed the oligodendrocyte marker CC1 and OLIG2, respectively, while the number of GFAP+tdT+ cells was minimal (Fig. 7E–H). On the other hand, the few sparsely distributed tdT+ cells in the control p75-2 group all expressed CC1 or OLIG2 (Fig. 7D–H). Together, these results indicate that SOX2-reprogrammed NG2 glia are bipotent, generating both neurons and oligodendrocytes. As such, oligodendrogenesis is not abolished but maybe even enhanced by SOX2-mediated reprogramming of NG2 glia.
Figure 7. SOX2-reprogrammed NG2 glia are bipotent in the spinal cord with injury.
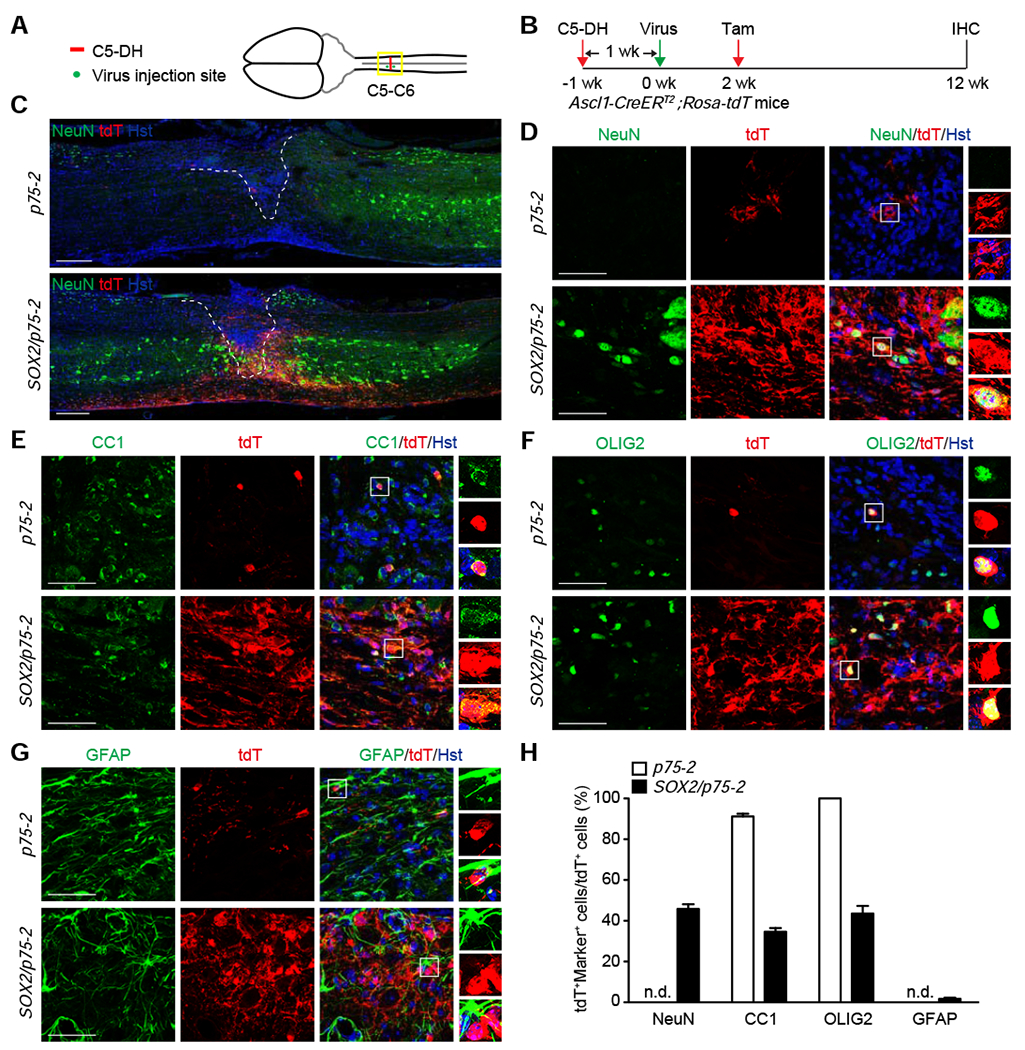
(A) A schematic drawing of C5-DH SCI and the locations of virus injections.
(B) Experimental design for analyzing the fates of SOX2-reprogrammed NG2 glia in adult Ascl1-CreERT2;Rosa-tdT mice.
(C) Stitched images of lower magnification views of spinal cords injected with the indicated viruses at 12 wpv. Scale bar, 250 μm.
(D-G) Confocal images of the indicated cell markers surrounding the injured spinal cord. Scale bars, 50 μm.
(H) Quantification of the fates of lineage traced cells (mean ± SEM; n=3-4 mice per group).
DISCUSSION
Multiple cell types can change their morphology and behavior in response to injury. This study shows that SCI elicits neurogenic potential of NG2 glia. Elevated SOX2 is both required and sufficient for neurogenic reprogramming of NG2 glia, which can eventually lead to functional neurogenesis in the normally non-neurogenic adult mouse spinal cord. Our results indicate that the expression level of SOX2 is critically important and a threshold must be overcome to enable NG2 glia to become robustly neurogenic.
NG2 glia, also known as oligodendrocyte precursor cells, are the major proliferative glial cells in the adult central nervous system (Buffo et al., 2008; Kang et al., 2010). They serve as a pool of progenitors to differentiate into oligodendrocytes and a few protoplasmic astrocytes (Kang et al., 2010; Nishiyama et al., 2016; Tripathi et al., 2010). In response to injury, NG2 glia increase their numbers and become a major component of the glial scar (Lytle and Wrathall, 2007; Tripathi and McTigue, 2007). Our finding that SCI-induces robust expression of DCX in these cells also indicate that they may undergo phenotypic reprogramming towards neurogenesis. However, such injury-induced phenotypic reprogramming is transient and incomplete for producing mature neurons. This is consistent with previous reports showing that NG2 glia are not neurogenic (Kang et al., 2010; Tripathi et al., 2010) but rather remain largely undifferentiated and contribute to neuroinflammation and axon regeneration failure after injury (Filous et al., 2014; Hackett and Lee, 2016; Levine, 2016). Reprogramming these cells by SOX2 not only provides new neurons but may also ameliorate the pathological microenvironment, indicated by glial scar reduction. Interestingly, SOX2-induced ASCL1+ progenitors from NG2 glia are bipotent, giving rise to both new neurons and oligodendrocytes.
We observed that SCI-induced DCX+ cells are not derived from ependymal cells that form the central canal. We traced central canal ependymal cells in both the Foxj1-CreERT2;Rosa-tdT and Nes-CreERT2;Rosa-YFP mice and failed to detect meaningful DCX expression in the lineage-mapped cells. These results are in agreement with multiple recent reports showing that ependymal cells lack properties of neural stem cells and rarely contribute new cells to the injury site (Muthusamy et al., 2018; Ren et al., 2017; Shah et al., 2018).
Similar to NG2 glia, adult astrocytes become reactive, proliferate, and contribute to glial scar formation following injury (Gotz et al., 2015; Sofroniew and Vinters, 2010). When isolated and cultured in vitro, reactive astrocytes can form neurospheres and gain multilineage differentiation potential (Gotz et al., 2015). Moreover, these cells can be in vivo reprogramed into neurons by multiple transcription factors including SOX2 (Gascon et al., 2016; Grande et al., 2013; Guo et al., 2014; Liu et al., 2015; Niu et al., 2015; Niu et al., 2013; Su et al., 2014; Torper et al., 2013). Notwithstanding, we fail to detect DCX expression in these cells or their derivatives following SCI. This is not because of lack of SOX2 expression, as the results from this study and others show that SOX2 is endogenously expressed in reactive astrocytes (Chen et al., 2019; Su et al., 2014). It could be that the SCI-induced endogenous SOX2 level fails to pass a threshold to enable neurogenic reprogramming of reactive astrocytes, suggesting that different cell types may require different levels of SOX2. Or, other factors in astrocytes maintain their fate in a more stable state. Furthermore, glial cells may also show regional differences in response to injury, as DCX expression is not detected in the injured adult mouse brain cortex that is enriched with reactive glial cells (Heinrich et al., 2014).
It is remarkable that endogenous SOX2 is absolutely required for SCI-induced neurogenic reprogramming of NG2 glia. SOX2 is normally expressed in NG2 glia and can be further up-regulated by SCI (Lee et al., 2013; Zhang et al., 2018). It remains to be determined how the injury signal is transmitted into the nucleus to control SOX2 expression. Although SOX2 can work as a pioneer transcription factor, the requirement of ectopic SOX2 for complete neurogenic reprogramming of NG2 glia suggests that some SOX2-regulated genomic sites may have low binding affinity and that transcription of such genomic sites is required for fate transition. Alternatively, transcriptional outputs of some SOX2 targets may well be determined by the expression levels of SOX2 and sustained high outputs of these targets are essential for fate transition. It should be interesting in the future to examine these scenarios through ATAC-seq, ChIP-seq, and RNA-seq under various conditions. Such investigations will provide significant insights into the molecular mechanisms underlying fate maintenance and reprogramming, which may be manipulated for neural regeneration.
Synaptic connections between neurons are the cellular basis for function. Studies show that propriospinal relay connections around injury sites can mediate pronounced spontaneous functional recovery without regeneration of direct projections from the brain past the lesion (Courtine et al., 2008). An agonist of the neuron-specific K+-Cl− co-transporter (KCC2) can restore consistent stepping ability in paralyzed SCI mice through reactivation of dormant relay pathways (Chen et al., 2018). These findings indicate that it is possible to restore function after SCI through relay connections around injury area. The SOX2-induced propriospinal neurons from NG2 glia may well form such relays surrounding the lesions. The transition through an ASCL1+ progenitor stage of SOX2-mediated neurogenesis enables us to precisely and specifically map relay connections of new neurons. This is accomplished by using a Cre-dependent recombinant rabies virus in mice harboring knockin alleles of Ascl1-CreERT2, Rosa-tdT, and Rosa-TVAg. Our results provide evidence that new spinal neurons can be directly targeted by endogenous neurons originating in the brainstem, spinal cord, and DRGs. The presynaptic neurons located in the brainstem mainly form the descending reticulospinal tracts (ReSTs) and vestibulospinal tracts (VeSTs). The ReSTs play a critical role in preparation of movements and postural control, whereas the VeSTs are responsible for initiation of limb and trunk extensor activity (Watson and Harvey, 2009). The DRGs contain cell bodies of sensory neurons that bring information from the periphery to interneurons in the spinal cord and some of those sensory neurons can also bifurcate to form the ascending gracile/cuneate pathways to relay sensory information to the brain. Our results indicate that new spinal neurons make presynaptic connections with neurons controlling sensorimotor function. Due to the lack of a robust, specific, and Cre-dependent monosynaptic anterograde tracing method, the immediate postsynaptic connections of the new spinal neurons await to be determined in the future.
Together, our results reveal a cellular and molecular mechanism underlying neural injury-induced cell plasticity, which can be further exploited for adult neurogenesis and relay formation in a region that has largely lost the ability to regenerate. Through expandable progenitors giving rise to both neurons and oligodendrocytes, reprogramming of NG2 glia not only ameliorates the pathological microenvironment but may also provide the much-needed new cells for network reconstruction and functional improvement following injury.
Limitations of Study
It is currently unknown whether glia-derived neurons have any deleterious effects in naive mice. Although no apparent behavioral abnormality was observed in home cages, future detailed behavioral tests are needed to address this issue. Postsynaptic connections of the glia-converted neurons are not assessed due to the lack of a robust Cre-dependent and monosynaptic anterograde tracing strategy. Since reprogramming of NG2 glia also reduces glial scar, the precise contribution of new neurons to functional recovery remains to be determined. Dissection of the neurocircuitry and expanded behavioral paradigms and injury types will further help understand the role of in vivo glia reprogramming. NG2 glia reprogramming not only generates neurons but also oligodendrocytes. More studies are needed to reveal the role of these new oligodendrocytes in SCI.
STAR ★ METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Chun-Li Zhang (chun-li.zhang@utsouthwestern.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All software was commercially or freely available and is listed in the STAR Methods description and Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Guinea Pig anti-ASCL1 | Jane E. Johnson lab (UT Southwestern Medical Center) | N/A |
| Rat monoclonal anti-BrdU | Bio-Rad | Cat# OBT0030; RID: AB_609568 |
| Mouse monoclonal anti-CC1 | Millipore | Cat# OP80; RRID: AB_2057371 |
| Goat polyclonal anti-DCX | Santa Cruz Biotechnology | Cat# sc-8066; RRID: AB_2088494 |
| Goat polyclonal anti-DsRed | Santa Cruz Biotechnology | Cat# sc-33354; RRID: AB_639922 |
| Rabbit polyclonal anti-DsRed | Takara Bio | Cat# 632496; RRID: AB_10013483 |
| Mouse monoclonal anti-GAD6 | Hybridoma Bank | N/A |
| Mouse monoclonal anti-GFAP | Sigma-Aldrich | Cat# G3893; RRID: AB_477010 |
| Chicken polyclonal anti-GFAP | Abcam | Cat# ab4674; RRID: AB_304558 |
| Chicken polyclonal anti-GFP | Aves Labs | Cat# GFP-1020; RRID: AB_10000240 |
| Rabbit polyclonal anti-GLYT2 | Synaptic Systems | Cat# 272003; RRID: AB_2619997 |
| Rabbit polyclonal anti-IBA1 | Wako | Cat# 019-19741; RRID: AB_839504 |
| Rabbit polyclonal anti-Ki67 | Abcam | Cat# ab15580; RRID: AB_443209 |
| Mouse monoclonal anti-MAP2 | Proteintech | Cat# 67015-1-Ig; RRID: AB_2882331 |
| Rabbit polyclonal anti-MAP2 | Proteintech | Cat# 17490-1-AP; RRID: AB_2137880 |
| Chicken polyclonal anti-Nestin | Aves Labs | Cat# NES; RRID: AB_2314882 |
| Rabbit monoclonal anti-NeuN | Abcam | Cat# ab177487; RRID: AB_2532109 |
| Rabbit polyclonal anti-NG2 | Millipore | Cat# AB5320; RRID: AB_11213678 |
| Rabbit polyclonal anti-OLIG2 | Millipore | Cat# AB9610; RRID: AB_570666 |
| Goat polyclonal anti-SOX2 | Santa Cruz Biotechnology | Cat# sc-17320; RRID: AB_2286684 |
| Rabbit polyclonal anti-SOX2 | Millipore | Cat# AB5603; RRID: AB_2286686 |
| Rabbit monoclonal anti-SYN1 | Cell Signaling Technology | Cat# 5297; RRID: AB_2616578 |
| Goat polyclonal anti-tdTomato | MyBioSource | Cat# MBS448092; RRID: AB_2827808 |
| Mouse monoclonal anti-VGAT | Synaptic Systems | Cat# 131011; RRID: AB_887872 |
| Mouse monoclonal anti-VGLUT2 | Synaptic Systems | Cat# 135421; RRID: AB_2619823 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202; RRID: AB_141607 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Guinea Pig IgG (H+L) | Jackson ImmunoResearch Laboratories | Cat# 706-545-148; RRID: AB_2340472 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson ImmunoResearch Laboratories | Cat# 703-545-155; RRID: AB_2340375 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Goat IgG (H+L) | Jackson ImmunoResearch Laboratories | Cat# 705-545-003; RRID: AB_2340428 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21206; RRID: AB_2535792 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11006; RRID: AB_2534074 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-21432; RRID: AB_2535853 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-31572; RRID: AB_162543 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-31570; RRID: AB_2536180 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31573; RRID: AB_2536183 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31571; RRID: AB_162542 |
| Alexa Fluor® 647 AffiniPure F(ab’)2 Fragment Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson ImmunoResearch Laboratories | Cat# 703-606-155; RRID: AB_2340380 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21247; RRID: AB_141778 |
| Alexa Fluor® 647 AffiniPure Donkey Anti-Goat IgG (H+L) | Jackson ImmunoResearch Laboratories | Cat# 705-605-147; RRID: AB_2340437 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| BrdU | Sigma-Aldrich | Cat# B5002 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664; RRID: IMSR_JAX:000664 |
| Mouse: Rosa-YFP: B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J | The Jackson Laboratory | JAX: 006148; RRID: IMSR_JAX:006148 |
| Mouse: Rosa-tdT: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory | JAX: 007914; RRID: IMSR_JAX:007914 |
| Mouse: Sox2f/f: STOCK Sox2tm1.1Lan/J | The Jackson Laboratory | JAX: 013093; RRID: IMSR_JAX:013093 |
| Mouse: Pdgfra-CreER™: B6N.Cg-Tg(Pdgfra-cre/ERT)467Dbe/J | The Jackson Laboratory | JAX: 018280; RRID: IMSR_JAX:018280 |
| Mouse: Ascl1-CreERT2: STOCK Ascl1tm1.1(Cre/ERT2)Jejo/J | The Jackson Laboratory | JAX: 012882; RRID: IMSR_JAX:012882 |
| Mouse: Nes-CreERT2: C57BL/6-Tg(Nes-cre/ERT2)KEisc/J | The Jackson Laboratory | JAX: 016261; RRID: IMSR_JAX:016261 |
| Mouse: Foxj1-CreERT2: STOCK Foxj1tm1.1(cre/ERT2/GFP)Htg/J | The Jackson Laboratory | JAX: 027012; RRID: IMSR_JAX:027012 |
| Mouse: Rosa-TVAg: B6;129P2-Gt(ROSA)26Sortm1(CAG_RABVgp4,-TVA)Arenk/J | The Jackson Laboratory | JAX: 024708; RRID: IMSR_JAX:024708 |
| Recombinant DNA | ||
| hNG2-GFP | This paper | N/A |
| hNG2-SOX2 | This paper | N/A |
| hNG2-Empty | This paper | N/A |
| hNG2-FLEX-Empty | This paper | N/A |
| hNG2-FLEX-SOX2 | This paper | N/A |
| hGFAP-Empty | This paper | N/A |
| hGFAP-mBDNF | This paper | N/A |
| hGFAP-mNoggin | This paper | N/A |
| pLV-p75-2 | Pantelis Tsoulfas Lab | RRID: Addgene_73036 |
| EnvA-pseudotyped RVdG-eGFP | The Vector Core at the Salk Institute | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | RRID:SCR_003070, https://imagej.net/Fiji |
| Graphpad Prism | Graphpad | RRID:SCR_002798), https://www.graphpad.com |
| ZEN | Zeiss | RRID:SCR_013672, https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| Neurolucida Imaging/tracing System (11.01 64-bit) | MBF Bioscience 185 Allen Brook Lane, Suite 101 Williston, VT 05495 USA | RRID:SCR_001775 https://www.mbfbioscience.com/neurolucida |
| Adobe Photoshop | Adobe | RRID:SCR_014199, https://www.adobe.com/products/photoshop.html |
| Adobe Illustrator | Adobe | RRID:SCR_010279, https://www.adobe.com/products/illustrator.html |
| Other | ||
| 33 gauge, 18-degree-beveled needle | Hamilton, Reno, NV | Cat# 22033 (Customized) |
| IH impactor | Precision Systems and Instrumentation, Lexington, KY | Model IH-0400 |
| Louisville Injury System Apparatus | Louisville Impactor System, Louisville, KY | N/A |
| stereotaxic apparatus | Stoelting Co., Wood Dale, IL | Cat# 51730 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Wild-type C57BL/6J mice (JAX: 000664; RRID: IMSR_JAX:000664) and the following transgenic mice were purchased from the Jackson Laboratory: Rosa-YFP (JAX: 006148; RRID: IMSR_JAX:006148) (Srinivas et al., 2001), Rosa-tdT (JAX: 007914; RRID: IMSR_JAX:007914) (Madisen et al., 2010), Sox2f/f (JAX: 013093; RRID: IMSR_JAX:013093) (Shaham et al., 2009), Pdgfra-CreER™ (JAX: 018280; RRID: IMSR_JAX:018280) (Kang et al., 2010), Ascl1-CreERT2 (JAX: 012882; RRID: IMSR_JAX:012882) (Kim et al., 2011) , Nes-CreERT2 (JAX: 016261; RRID: IMSR_JAX:016261) (Lagace et al., 2007), Foxj1-CreERT2 (JAX: 027012; RRID: IMSR_JAX:027012) (Muthusamy et al., 2018; Ren et al., 2017), Rosa-TVAg mouse line (RphiGT, JAX: 024708; RRID: IMSR_JAX:024708) (Takatoh et al., 2013). Both adult male and female mice at 2 months of age and older were used for all experiments unless otherwise stated. All mice were housed under a controlled temperature and a 12-h light/dark cycle with free access to water and food in the animal facility. Sample sizes were empirically determined. Animal procedures and protocols were approved by the Institutional Animal Care and Use Committee at UT Southwestern or Indian University School of Medicine.
METHOD DETAILS
Spinal Cord Injuries.
Adult mice were anesthetized with a cocktail of ketamine and xylazine. A laminectomy was performed at the indicated spinal segments. A crush injury was carried out by holding a pair of forceps (ZHTG Instrument, China) with a 0.1-mm spacer for 30 seconds, whereas a stab-wound injury was created by injecting a 33-gauge, 18-degree-beveled needle (Hamilton, Reno, NV; Cat# 22033, customized) into the spinal cord and holding in position for 5 min. A contusion injury was produced by using the IH impactor (Precision Systems and Instrumentation, Lexington, KY; Model IH-0400) with a 1-mm tip and a force of 50 kdyne. The C5 dorsal hemisection was created by using a VibraKnife attached to the Louisville Injury System Apparatus (Louisville Impactor System, Louisville, KY) (Al-Ali et al., 2017), which can achieve 0.01mm cutting accuracy (Zhang et al., 2013). The blade was 1.2-mm wide and was advanced 1.2 mm ventrally from the dorsal surface of the cord, extending beyond the central canal. Such a lesion completely transected the entire dorsal corticospinal tract and lateral corticospinal tract on both sides, which are critical for forelimb dexterous and general locomotor function. After surgery, animals were returned to their home cages and received manual bladder expression twice daily until the return of reflexive bladder controls.
Lentivirus Preparation and Intraspinal Injections.
A synthetic promoter from the human NG2 gene was subcloned into the lentiviral CS-CDF-CG-PRE vector to create the hNG2-GFP plasmid. Candidate genes were then sub-cloned by PCR into the hNG2-GFP vector at the AgeI and XhoI sites. Lentivirus generation and titer determination were prepared as previously described (Su et al., 2014; Wang et al., 2016). 2-3 μL of lentivirus (0.5-2 x 109 pfu/mL) was manually injected with a Hamilton syringe and a 33-gauge, 18-degree-beveled needle into the spinal parenchyma at the indicated spinal cord levels. Injections were performed at a rate of 0.3 μL/min. Following injections, the needle was held at the injection site for another 3 min and then slowly withdrawn within one min. For behavioral experiments with C5 dorsal hemisections, a total of 4 stereotaxic viral injections (1-2 x 109 pfu/ml) were made at 0.5 mm rostral and caudal to the incision to target the NG2 glia at the edge of the lesion. At each distance, bilateral injections (0.5 μl/site) were made according to the following coordinates: mediolateral (ML): 0.5 mm to the midline, and dorsoventral (DV): two injections at 0.4 mm and 0.8 mm each from the dorsal surface of the spinal cord.
Tamoxifen and BrdU Administration.
Tamoxifen (T5648, Sigma; Cat# T5648) was dissolved in a mixture of ethanol and sesame oil (1:9 by volume) at a concentration of 40 mg/mL and injected intraperitoneally at a daily dose of 1 mg/10 g body weight for 5-7 days. Proliferating cells were labelled in vivo through administration with BrdU (B5002, Sigma; Cat# B5002; 0.5g/L) in drinking water for the indicated durations.
Monosynaptic Tracing with Engineered Rabies Virus.
The EnvA-pseudotyped RVdG-eGFP rabies virus was obtained from the Vector Core at the Salk Institute. One μl diluted rabies virus (1.6 x 105 TU/ml) was stereotaxically injected into the previously operated spinal cord location 5-6 months post lentivirus injection. The brain, the brainstem, and the DRG-attached spinal cords were isolated 6 days following rabies virus injection. Series of 60-μm coronal brain/brainstem sections or horizontal spinal cord sections were then collected for confocal analysis.
Immunohistochemistry.
Mice were sacrificed with CO2 overdose and sequentially perfused with ice-cold phosphate-buffered saline (PBS) and 4% (w/v) paraformaldehyde (PFA) in PBS. Whole brains and spinal cords were carefully dissected out, post-fixed overnight with 4% PFA at 4°C, and cryoprotected with 30% sucrose in PBS for 48 h at 4 °C. Transverse sections or horizontal sections of a 1.5-cm segment of the spinal cord spanning the injection/injury sites were collected on a cryostat (Leica) set at 20-μm thickness. Coronal brain sections were collected at 60-μm thickness by using a sliding microtome (Leica). The immunostaining procedure was conducted as previously described (Wang et al., 2016). Images were collected on a Zeiss LSM 700 confocal microscope; image-stitching was performed in ZEN software (ZEISS) (RRID:SCR_013672). For cell number counts, every 10th serial 20-μm-thick spinal cord section and a total of 8-10 sections per animal were processed for staining. Confocal images were taken with a 20x objective and the ImageJ program (RRID:SCR_003070) was used for cell counts. The total cell counts from all the processed sections were then multiplied by 10 to arrive the estimated cell number per animal. The ImageJ program was also used for fluorescence intensity analysis.
Behavior Assessments.
All behavior experiments were conducted in a separate institution in a randomized and blinded fashion. Animals were randomized into groups and treated with letter-coded viruses. Experimenters were blinded to the identity of the treatments during the behavioral tests. Upon completion of the experiments, mouse groups were analyzed and then decoded. The grid-walking test was employed to assess sensorimotor coordination of the limbs. The test was performed according to a previously established protocol (Al-Ali et al., 2017; Wu et al., 2017). Briefly, 39 animals were pretrained by walking on a wire grid (10 mm x 10 mm) for 3 times with each lasting for 3 min each day for 3 days before the experiment. Then they were tested for 3 min at the predetermined experimental time points. The percentage of forepaw drops below the grid plane was calculated from the recorded videos.
Astroglial Scar Analysis.
Sagittal sections at 20 μm were cut using a cryostat. Astrocytes were identified by immunostaining for GFAP. 8-12 sagittal spinal cord sections, containing the reactive astroglia, spaced 200 μm were chosen for analysis using the Neurolucida system (MicroBrightfield, Inc.; RRID:SCR_001775). Images were all auto-stitched using the Neurolucida software. Briefly, the area of reactive astroglial scar was manually contoured in each section per every case, and all contours could be 3D reconstructed by cases using the Neurolucida software. Computational analysis of the astroglial scar, including the volume and surface area, were conducted using the Neurolucida software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean ± SEM. Statistical analysis for histological data was performed by homoscedastic two-tailed Student’s t-test. Two-way ANOVA and Tukey’s post hoc multiple comparisons were used for behavioral data. One-way ANOVA and Tukey’s post hoc multiple comparisons were applied for group analysis. A p value < 0.05 was considered significant. Significant differences are indicated by *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Supplementary Material
Highlights.
Spinal NG2 glia, but not astrocytes or ependymal cells, exhibit neurogenic potential
SOX2 is cell-autonomously required for SCI-induced reprogramming of NG2 glia
Elevated SOX2 is sufficient to induce functional neurogenesis from resident NG2 glia
Reprogramming NG2 glia rebuilds neural circuits and promotes injury recovery
ACKNOWLEDGEMENTS
We thank members of the Zhang laboratory for discussions and reagents, J. E. Johnson (UT Southwestern, USA) for ASCL1 antibody. C.L.Z. is a W. W. Caruth, Jr. Scholar in Biomedical Research. The work in Zhang laboratory was supported by the Welch Foundation (I-1724), the Decherd Foundation, the Texas Alzheimer’s Research and Care Consortium (TARCC2020), Kent Waldrep Foundation Center for Basic Research on Nerve Growth and Regeneration, and NIH Grants (NS099073, NS092616, NS111776, NS117065, and NS088095). The work in Xu laboratory was supported by NIH 1R01NS100531, 1R01NS103481, and 1R01NS111776.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Al-Ali H, Ding Y, Slepak T, Wu W, Sun Y, Martinez Y, Xu XM, Lemmon VP, and Bixby JL (2017). The mTOR Substrate S6 Kinase 1 (S6K1) Is a Negative Regulator of Axon Regeneration and a Potential Drug Target for Central Nervous System Injury. J Neurosci 37, 7079–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnabe-Heider F, Goritz C, Sabelstrom H, Takebayashi H, Pfrieger FW, Meletis K, and Frisen J (2010). Origin of new glial cells in intact and injured adult spinal cord. Cell stem cell 7, 470–482. [DOI] [PubMed] [Google Scholar]
- Buffo A, Rite I, Tripathi P, Lepier A, Colak D, Horn AP, Mori T, and Gotz M (2008). Origin and progeny of reactive gliosis: A source of multipotent cells in the injured brain. Proceedings of the National Academy of Sciences of the United States of America 105, 3581–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Li Y, Yu B, Zhang Z, Brommer B, Williams PR, Liu Y, Hegarty SV, Zhou S, Zhu J, et al. (2018). Reactivation of Dormant Relay Pathways in Injured Spinal Cord by KCC2 Manipulations. Cell 174, 1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhong X, Smith DK, Tai W, Yang J, Zou Y, Wang LL, Sun J, Qin S, and Zhang CL (2019). Astrocyte-Specific Deletion of Sox2 Promotes Functional Recovery After Traumatic Brain Injury. Cereb Cortex 29, 54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SR, Benraiss A, Chmielnicki E, Samdani A, Economides A, and Goldman SA (2007). Induction of neostriatal neurogenesis slows disease progression in a transgenic murine model of Huntington disease. J Clin Invest 117, 2889–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillard-Despres S, Winner B, Schaubeck S, Aigner R, Vroemen M, Weidner N, Bogdahn U, Winkler J, Kuhn HG, and Aigner L (2005). Doublecortin expression levels in adult brain reflect neurogenesis. The European journal of neuroscience 21, 1–14. [DOI] [PubMed] [Google Scholar]
- Courtine G, Song B, Roy RR, Zhong H, Herrmann JE, Ao Y, Qi J, Edgerton VR, and Sofroniew MV (2008). Recovery of supraspinal control of stepping via indirect propriospinal relay connections after spinal cord injury. Nature medicine 14, 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto M, Bunge MB, and Tsoulfas P (2013). A multifunctional neurotrophin with reduced affinity to p75NTR enhances transplanted Schwann cell survival and axon growth after spinal cord injury. Experimental neurology 248, 170–182. [DOI] [PubMed] [Google Scholar]
- Filous AR, Tran A, Howell CJ, Busch SA, Evans TA, Stallcup WB, Kang SH, Bergles DE, Lee SI, Levine JM, et al. (2014). Entrapment via synaptic-like connections between NG2 proteoglycan+ cells and dystrophic axons in the lesion plays a role in regeneration failure after spinal cord injury. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 16369–16384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon S, Murenu E, Masserdotti G, Ortega F, Russo GL, Petrik D, Deshpande A, Heinrich C, Karow M, Robertson SP, et al. (2016). Identification and Successful Negotiation of a Metabolic Checkpoint in Direct Neuronal Reprogramming. Cell stem cell 18, 396–409. [DOI] [PubMed] [Google Scholar]
- Gotz M, Sirko S, Beckers J, and Irmler M (2015). Reactive astrocytes as neural stem or progenitor cells: In vivo lineage, In vitro potential, and Genome-wide expression analysis. Glia 63, 1452–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande A, Sumiyoshi K, Lopez-Juarez A, Howard J, Sakthivel B, Aronow B, Campbell K, and Nakafuku M (2013). Environmental impact on direct neuronal reprogramming in vivo in the adult brain. Nature communications 4, 2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Zhang L, Wu Z, Chen Y, Wang F, and Chen G (2014). In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell stem cell 14, 188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett AR, and Lee JK (2016). Understanding the NG2 Glial Scar after Spinal Cord Injury. Frontiers in neurology 7, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C, Bergami M, Gascon S, Lepier A, Vigano F, Dimou L, Sutor B, Berninger B, and Gotz M (2014). Sox2-mediated conversion of NG2 glia into induced neurons in the injured adult cerebral cortex. Stem cell reports 3, 1000–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horky LL, Galimi F, Gage FH, and Horner PJ (2006). Fate of endogenous stem/progenitor cells following spinal cord injury. J Comp Neurol 498, 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner PJ, Power AE, Kempermann G, Kuhn HG, Palmer TD, Winkler J, Thal LJ, and Gage FH (2000). Proliferation and differentiation of progenitor cells throughout the intact adult rat spinal cord. The Journal of neuroscience : the official journal of the Society for Neuroscience 20, 2218–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson CB, Momma S, Clarke DL, Risling M, Lendahl U, and Frisen J (1999). Identification of a neural stem cell in the adult mammalian central nervous system. Cell 96, 25–34. [DOI] [PubMed] [Google Scholar]
- Juhasova J, Juhas S, Hruska-Plochan M, Dolezalova D, Holubova M, Strnadel J, Marsala S, Motlik J, and Marsala M (2015). Time course of spinal doublecortin expression in developing rat and porcine spinal cord: implication in in vivo neural precursor grafting studies. Cellular and molecular neurobiology 35, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Fukaya M, Yang JK, Rothstein JD, and Bergles DE (2010). NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron 68, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Jessberger S, Steiner B, and Kronenberg G (2004). Milestones of neuronal development in the adult hippocampus. Trends Neurosci 27, 447–452. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Ables JL, Dickel LK, Eisch AJ, and Johnson JE (2011). Ascll (Mashl) defines cells with long-term neurogenic potential in subgranular and subventricular zones in adult mouse brain. PloS one 6, e18472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoth R, Singec I, Ditter M, Pantazis G, Capetian P, Meyer RP, Horvat V, Volk B, and Kempermann G (2010). Murine features of neurogenesis in the human hippocampus across the lifespan from 0 to 100 years. PloS one 5, e8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace DC, Whitman MC, Noonan MA, Ables JL, DeCarolis NA, Arguello AA, Donovan MH, Fischer SJ, Farnbauch LA, Beech RD, et al. (2007). Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience 27, 12623–12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Wu J, Chung J, and Wrathall JR (2013). SOX2 expression is upregulated in adult spinal cord after contusion injury in both oligodendrocyte lineage and ependymal cells. Journal of neuroscience research 91, 196–210. [DOI] [PubMed] [Google Scholar]
- Levine J (2016). The reactions and role of NG2 glia in spinal cord injury. Brain research 1638, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie DC, Song H, Colamarino SA, Ming GL, and Gage FH (2004). Neurogenesis in the adult brain: new strategies for central nervous system diseases. Annu Rev Pharmacol Toxicol 44, 399–421. [DOI] [PubMed] [Google Scholar]
- Liu Y, Miao Q, Yuan J, Han S, Zhang P, Li S, Rao Z, Zhao W, Ye Q, Geng J, et al. (2015). Ascl1 Converts Dorsal Midbrain Astrocytes into Functional Neurons In Vivo. The Journal of neuroscience : the official journal of the Society for Neuroscience 35, 9336–9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JM, and Wrathall JR (2007). Glial cell loss, proliferation and replacement in the contused murine spinal cord. The European journal of neuroscience 25, 1711–1724. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletis K, Barnabe-Heider F, Carlen M, Evergren E, Tomilin N, Shupliakov O, and Frisen J (2008). Spinal cord injury reveals multilineage differentiation of ependymal cells. PLoS Biol 6, e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy N, Brumm A, Zhang X, Carmichael ST, and Ghashghaei HT (2018). Foxj1 expressing ependymal cells do not contribute new cells to sites of injury or stroke in the mouse forebrain. Scientific reports 8, 1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Boshans L, Goncalves CM, Wegrzyn J, and Patel KD (2016). Lineage, fate, and fate potential of NG2-glia. Brain research 1638, 116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Zang T, Smith DK, Vue TY, Zou Y, Bachoo R, Johnson JE, and Zhang CL (2015). SOX2 reprograms resident astrocytes into neural progenitors in the adult brain. Stem cell reports 4, 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Zang T, Zou Y, Fang S, Smith DK, Bachoo R, and Zhang CL (2013). In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat Cell Biol 15, 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norenberg MD, Smith J, and Marcillo A (2004). The pathology of human spinal cord injury: defining the problems. Journal of neurotrauma 21, 429–440. [DOI] [PubMed] [Google Scholar]
- Obernier K, Cebrian-Silla A, Thomson M, Parraguez JI, Anderson R, Guinto C, Rodas Rodriguez J, Garcia-Verdugo JM, and Alvarez-Buylla A (2018). Adult Neurogenesis Is Sustained by Symmetric Self-Renewal and Differentiation. Cell stem cell 22, 221–234 e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakada F, and Callaway EM (2013). Design and generation of recombinant rabies virus vectors. Nat Protoc 8, 1583–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny LH, and Nicolis SK (2010). Sox2 roles in neural stem cells. Int J Biochem Cell Biol 42, 421–424. [DOI] [PubMed] [Google Scholar]
- Raposo A, Vasconcelos FF, Drechsel D, Marie C, Johnston C, Dolle D, Bithell A, Gillotin S, van den Berg DLC, Ettwiller L, et al. (2015). Ascl1 Coordinately Regulates Gene Expression and the Chromatin Landscape during Neurogenesis. Cell reports 10, 1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Ao Y, O'Shea TM, Burda JE, Bernstein AM, Brumm AJ, Muthusamy N, Ghashghaei HT, Carmichael ST, Cheng L, et al. (2017). Ependymal cell contribution to scar formation after spinal cord injury is minimal, local and dependent on direct ependymal injury. Scientific reports 7, 41122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedy J, Urdzikova L, Jendelova P, and Sykova E (2008). Methods for behavioral testing of spinal cord injured rats. Neuroscience and biobehavioral reviews 32, 550–580. [DOI] [PubMed] [Google Scholar]
- Shah PT, Stratton JA, Stykel MG, Abbasi S, Sharma S, Mayr KA, Koblinger K, Whelan PJ, and Biernaskie J (2018). Single-Cell Transcriptomics and Fate Mapping of Ependymal Cells Reveals an Absence of Neural Stem Cell Function. Cell 173, 1045–1057 e1049. [DOI] [PubMed] [Google Scholar]
- Shaham O, Smith AN, Robinson ML, Taketo MM, Lang RA, and Ashery-Padan R (2009). Pax6 is essential for lens fiber cell differentiation. Development 136, 2567–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV, and Vinters HV (2010). Astrocytes: biology and pathology. Acta Neuropathol 119, 7–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Lu TY, Chai H, Xu J, Huang BS, Golshani P, Coppola G, and Khakh BS (2016). New Transgenic Mouse Lines for Selectively Targeting Astrocytes and Studying Calcium Signals in Astrocyte Processes In Situ and In Vivo. Neuron 92, 1181–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Niu W, Liu ML, Zou Y, and Zhang CL (2014). In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nature communications 5, 3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatoh J, Nelson A, Zhou X, Bolton MM, Ehlers MD, Arenkiel BR, Mooney R, and Wang F (2013). New modules are added to vibrissal premotor circuitry with the emergence of exploratory whisking. Neuron 77, 346–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torper O, Pfisterer U, Wolf DA, Pereira M, Lau S, Jakobsson J, Bjorklund A, Grealish S, and Parmar M (2013). Generation of induced neurons via direct conversion in vivo. Proceedings of the National Academy of Sciences of the United States of America 110, 7038–7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi R, and McTigue DM (2007). Prominent oligodendrocyte genesis along the border of spinal contusion lesions. Glia 55, 698–711. [DOI] [PubMed] [Google Scholar]
- Tripathi RB, Rivers LE, Young KM, Jamen F, and Richardson WD (2010). NG2 glia generate new oligodendrocytes but few astrocytes in a murine experimental autoimmune encephalomyelitis model of demyelinating disease. The Journal of neuroscience : the official journal of the Society for Neuroscience 30, 16383–16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivar C, Potter MC, Choi J, Lee JY, Stringer TP, Callaway EM, Gage FH, Suh H, and van Praag H (2012). Monosynaptic inputs to new neurons in the dentate gyrus. Nature communications 3, 1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Su Z, Tai W, Zou Y, Xu XM, and Zhang CL (2016). The p53 Pathway Controls SOX2-Mediated Reprogramming in the Adult Mouse Spinal Cord. Cell reports 17, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C, and Harvey AR (2009). Projections from the Brain to the Spinal Cord. In The Spinal Cord (Academic Press; ), pp. 168–179. [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, and Callaway EM (2007). Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53, 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Xiong W, Zhang P, Chen L, Fang J, Shields C, Xu XM, and Jin X (2017). Increased threshold of short-latency motor evoked potentials in transgenic mice expressing Channelrhodopsin-2. PLoS One 12, e0178803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Yamamoto N, Kitamura T, Nakamura K, and Nakafuku M (2001). Proliferation of parenchymal neural progenitors in response to injury in the adult rat spinal cord. Experimental neurology 172, 115–127. [DOI] [PubMed] [Google Scholar]
- Zhang S, Rasai A, Wang Y, Xu J, Bannerman P, Erol D, Tsegaye D, Wang A, Soulika A, Zhan X, et al. (2018). The Stem Cell Factor Sox2 Is a Positive Timer of Oligodendrocyte Development in the Postnatal Murine Spinal Cord. Mol Neurobiol 55, 9001–9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YP, Walker MJ, Shields LB, Wang X, Walker CL, Xu XM, and Shields CB (2013). Controlled cervical laceration injury in mice. J Vis Exp, e50030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Avidan H, Pluchino S, Martino G, and Schwartz M (2006). Synergy between immune cells and adult neural stem/progenitor cells promotes functional recovery from spinal cord injury. Proceedings of the National Academy of Sciences of the United States of America 103, 13174–13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All software was commercially or freely available and is listed in the STAR Methods description and Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Guinea Pig anti-ASCL1 | Jane E. Johnson lab (UT Southwestern Medical Center) | N/A |
| Rat monoclonal anti-BrdU | Bio-Rad | Cat# OBT0030; RID: AB_609568 |
| Mouse monoclonal anti-CC1 | Millipore | Cat# OP80; RRID: AB_2057371 |
| Goat polyclonal anti-DCX | Santa Cruz Biotechnology | Cat# sc-8066; RRID: AB_2088494 |
| Goat polyclonal anti-DsRed | Santa Cruz Biotechnology | Cat# sc-33354; RRID: AB_639922 |
| Rabbit polyclonal anti-DsRed | Takara Bio | Cat# 632496; RRID: AB_10013483 |
| Mouse monoclonal anti-GAD6 | Hybridoma Bank | N/A |
| Mouse monoclonal anti-GFAP | Sigma-Aldrich | Cat# G3893; RRID: AB_477010 |
| Chicken polyclonal anti-GFAP | Abcam | Cat# ab4674; RRID: AB_304558 |
| Chicken polyclonal anti-GFP | Aves Labs | Cat# GFP-1020; RRID: AB_10000240 |
| Rabbit polyclonal anti-GLYT2 | Synaptic Systems | Cat# 272003; RRID: AB_2619997 |
| Rabbit polyclonal anti-IBA1 | Wako | Cat# 019-19741; RRID: AB_839504 |
| Rabbit polyclonal anti-Ki67 | Abcam | Cat# ab15580; RRID: AB_443209 |
| Mouse monoclonal anti-MAP2 | Proteintech | Cat# 67015-1-Ig; RRID: AB_2882331 |
| Rabbit polyclonal anti-MAP2 | Proteintech | Cat# 17490-1-AP; RRID: AB_2137880 |
| Chicken polyclonal anti-Nestin | Aves Labs | Cat# NES; RRID: AB_2314882 |
| Rabbit monoclonal anti-NeuN | Abcam | Cat# ab177487; RRID: AB_2532109 |
| Rabbit polyclonal anti-NG2 | Millipore | Cat# AB5320; RRID: AB_11213678 |
| Rabbit polyclonal anti-OLIG2 | Millipore | Cat# AB9610; RRID: AB_570666 |
| Goat polyclonal anti-SOX2 | Santa Cruz Biotechnology | Cat# sc-17320; RRID: AB_2286684 |
| Rabbit polyclonal anti-SOX2 | Millipore | Cat# AB5603; RRID: AB_2286686 |
| Rabbit monoclonal anti-SYN1 | Cell Signaling Technology | Cat# 5297; RRID: AB_2616578 |
| Goat polyclonal anti-tdTomato | MyBioSource | Cat# MBS448092; RRID: AB_2827808 |
| Mouse monoclonal anti-VGAT | Synaptic Systems | Cat# 131011; RRID: AB_887872 |
| Mouse monoclonal anti-VGLUT2 | Synaptic Systems | Cat# 135421; RRID: AB_2619823 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202; RRID: AB_141607 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Guinea Pig IgG (H+L) | Jackson ImmunoResearch Laboratories | Cat# 706-545-148; RRID: AB_2340472 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson ImmunoResearch Laboratories | Cat# 703-545-155; RRID: AB_2340375 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Goat IgG (H+L) | Jackson ImmunoResearch Laboratories | Cat# 705-545-003; RRID: AB_2340428 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21206; RRID: AB_2535792 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11006; RRID: AB_2534074 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-21432; RRID: AB_2535853 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-31572; RRID: AB_162543 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-31570; RRID: AB_2536180 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31573; RRID: AB_2536183 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31571; RRID: AB_162542 |
| Alexa Fluor® 647 AffiniPure F(ab’)2 Fragment Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson ImmunoResearch Laboratories | Cat# 703-606-155; RRID: AB_2340380 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21247; RRID: AB_141778 |
| Alexa Fluor® 647 AffiniPure Donkey Anti-Goat IgG (H+L) | Jackson ImmunoResearch Laboratories | Cat# 705-605-147; RRID: AB_2340437 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| BrdU | Sigma-Aldrich | Cat# B5002 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664; RRID: IMSR_JAX:000664 |
| Mouse: Rosa-YFP: B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J | The Jackson Laboratory | JAX: 006148; RRID: IMSR_JAX:006148 |
| Mouse: Rosa-tdT: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory | JAX: 007914; RRID: IMSR_JAX:007914 |
| Mouse: Sox2f/f: STOCK Sox2tm1.1Lan/J | The Jackson Laboratory | JAX: 013093; RRID: IMSR_JAX:013093 |
| Mouse: Pdgfra-CreER™: B6N.Cg-Tg(Pdgfra-cre/ERT)467Dbe/J | The Jackson Laboratory | JAX: 018280; RRID: IMSR_JAX:018280 |
| Mouse: Ascl1-CreERT2: STOCK Ascl1tm1.1(Cre/ERT2)Jejo/J | The Jackson Laboratory | JAX: 012882; RRID: IMSR_JAX:012882 |
| Mouse: Nes-CreERT2: C57BL/6-Tg(Nes-cre/ERT2)KEisc/J | The Jackson Laboratory | JAX: 016261; RRID: IMSR_JAX:016261 |
| Mouse: Foxj1-CreERT2: STOCK Foxj1tm1.1(cre/ERT2/GFP)Htg/J | The Jackson Laboratory | JAX: 027012; RRID: IMSR_JAX:027012 |
| Mouse: Rosa-TVAg: B6;129P2-Gt(ROSA)26Sortm1(CAG_RABVgp4,-TVA)Arenk/J | The Jackson Laboratory | JAX: 024708; RRID: IMSR_JAX:024708 |
| Recombinant DNA | ||
| hNG2-GFP | This paper | N/A |
| hNG2-SOX2 | This paper | N/A |
| hNG2-Empty | This paper | N/A |
| hNG2-FLEX-Empty | This paper | N/A |
| hNG2-FLEX-SOX2 | This paper | N/A |
| hGFAP-Empty | This paper | N/A |
| hGFAP-mBDNF | This paper | N/A |
| hGFAP-mNoggin | This paper | N/A |
| pLV-p75-2 | Pantelis Tsoulfas Lab | RRID: Addgene_73036 |
| EnvA-pseudotyped RVdG-eGFP | The Vector Core at the Salk Institute | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | RRID:SCR_003070, https://imagej.net/Fiji |
| Graphpad Prism | Graphpad | RRID:SCR_002798), https://www.graphpad.com |
| ZEN | Zeiss | RRID:SCR_013672, https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| Neurolucida Imaging/tracing System (11.01 64-bit) | MBF Bioscience 185 Allen Brook Lane, Suite 101 Williston, VT 05495 USA | RRID:SCR_001775 https://www.mbfbioscience.com/neurolucida |
| Adobe Photoshop | Adobe | RRID:SCR_014199, https://www.adobe.com/products/photoshop.html |
| Adobe Illustrator | Adobe | RRID:SCR_010279, https://www.adobe.com/products/illustrator.html |
| Other | ||
| 33 gauge, 18-degree-beveled needle | Hamilton, Reno, NV | Cat# 22033 (Customized) |
| IH impactor | Precision Systems and Instrumentation, Lexington, KY | Model IH-0400 |
| Louisville Injury System Apparatus | Louisville Impactor System, Louisville, KY | N/A |
| stereotaxic apparatus | Stoelting Co., Wood Dale, IL | Cat# 51730 |


