Abstract
Biomonitoring of human exposure to environmental chemicals has gained momentum in recent years. Biomonitoring methods often include analysis of a single class of chemicals with similar chemical properties. In this study, we describe a method that involves solid-phase extraction (SPE) coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS) and capable of measuring 121 environmental chemicals comprising plasticizers (PMs; n=45), environmental phenols (EPs; n=45), and pesticides (n=31) through a single extraction of urine. Urine samples were incubated with 20 μL of β-glucuronidase/arylsulfatase (4000 units/mL urine) (from Helix pomatia) buffered at pH 5.5 for 2 h at 37°C for optimal deconjugation conditions. We compared two extraction methods, namely liquid-liquid extraction and SPE, and the latter with ABS Elut NEXUS® cartridges was optimized to yield best extraction efficiencies. For increased resolution and chromatographic separation, two methods involving Ultra AQ C18® and Betasil™ C18® columns were used. The MS/MS analyses were performed under both negative and positive ionization modes. The optimized method yielded excellent intra- and inter-day variabilities (relative standard deviation: 0.40–11%) and satisfactory recoveries (80–120%) for >95% of the analytes. The limits of detection were ≤ 0.1 for 101 analytes and between 0.1 and 1.0 ng/mL for 18 analytes. The optimized SPE LC-MS/MS method was validated through the analysis of standard reference materials and proficiency test urine samples and further applied in the analysis of 21 real urine samples to demonstrate simultaneous determination of 121 environmental chemicals in urine samples.
Keywords: Endocrine disruptors, Human biomonitoring, Urine, LC-MS/MS, Multi-class method
1. Introduction
Humans are continuously exposed to toxic environmental chemicals via multiple exposure routes (inhalation, ingestion, and dermal uptake) and sources (food, water, indoor and outdoor air) on a daily basis. Chemical exposure in humans has been a topic of public health concern for decades [1]. As increasing number of new chemicals is constantly introduced into the market, there is an ongoing interest in the assessment of these chemicals’ exposures and resulting health effects in humans [2]. For instance, ~150,000 chemical substances have been registered in the European Union’s regulatory program namely Registration, Evaluation, Authorization and Restriction of Chemicals (REACH) [3], which signifies the broad range of chemicals to which humans can be exposed. Among various methods available for the assessment of chemical exposures, biomonitoring has become the most health-relevant approach. Nevertheless, ultra-trace level (picogram to microgram quantities) determination of environmental chemicals in human specimens can be challenging due to the lack of quantitative analytical methods, analytical standards, and cost among several others. Furthermore, many environmental chemicals, following exposure, undergo biologic transformation in human bodies [4]. Oxidation, reduction, hydrolysis, hydroxylation, sulfation, and alkylation followed by phase II conjugation with glucuronide or other biomolecules can confound the identification of a chemical’s exposure biomarker [5]. Lack of complete understanding of toxico-kinetics of xenobiotics, especially that of emerging chemicals, poses challenges in biomonitoring.
Traditionally, biomonitoring methods were developed with a focus on measuring a defined pollutant class in a single analysis. For example, phthalate metabolites, environmental phenols (EPs), and current use pesticides were analyzed in urine using a separate extraction and sample preparation steps for each of the chemical classes. This is due to the fact that chemicals within a class that possess similar physico-chemical properties (such as solubility, polarity, ionizability, and chromatographic properties) are amenable for optimal extraction and quantification in a single method. Nevertheless, considering the fact that humans are exposed to hundreds, if not thousands, of chemicals application of specific analytical methods for each of the chemical classes can be costly and time-consuming. Furthermore, each of the analytical methods would consume an aliquot of human specimen used in extraction, which would necessitate requirement of large volumes for biomonitoring of multiple chemical classes. Therefore, a robust analytical method capable of measuring a wide range of chemical classes in a single extraction would alleviate the need for large sample volumes while reducing the time and cost of analysis [6–8].
One of the resource/labor-intensive steps in bioanalysis is sample preparation, which is required for the removal of bulk of interferences and endogenous compounds (i.e., proteins and phospholipids) from target chemicals prior to instrumental analysis. A robust sample preparation method that offers acceptable accuracy and precision in multi-class chemical analysis can be challenging, as polar functional groups in each chemical class can have different properties and therefore, are not amenable simultaneous extraction. Owing to the improvement in sensitivities and robustness of the latest generation of high-performance liquid chromatography coupled to tandem mass spectrometry (HPLC-MS/MS) instruments, a trend towards the development and application of multi-class analytical methods is envisaged in recent biomonitoring studies (Table 1) [9–19].
Table 1.
Published multiple-class analytical methods for endocrine disrupting chemicals in human urine
| Analyte | Urine volume | Enzyme Type | Sample preparation | Sample extraction | Instrumental method | LOD (ng/mL) | Reference |
|---|---|---|---|---|---|---|---|
| Total: 35 analytes 16 BRPs, 12 OH-PAHs, 5 OH-PBDEs, TCS, and TBBPA |
2.0 mL | β-glucuronidase/arylsulfatas e enzyme (Helix pomatia, β-glucuronidase activity ≈ 100,000 U/mL; sulfatase activity ≈ 47,500 U/mL) | enzyme amount: 10 μL (i.e., 50 U/100 μL urine); buffer: 1 mL of 1 M NaOAc (pH 5.5); incubation time: overnight at 37 °C | Oasis HLB SPE cartridge (500 mg, 6 mL) |
Single injection: Instrument: LC-MS/MS; LC column: Poroshell 120 EC-C18 (100 mm × 4.6 mm, 2.7 μm); Mobile phase: 5 mM NH4Ac in water and ACN; Run time: 27 min; Injection volume: 10 μL; |
0.008–0.161 | [10] |
| Total: 21 analytes 7 BPs, 7 parabens, 5 benzophenones, TCC, and TCS |
1.0 mL | β-glucuronidase enzyme (Type HP-2 from Helix pomatia, β-glucuronidase activity ≥ 100,000 U/mL; sulfatase activity ≈ 7500 U/mL) | enzyme amount: 2.0 μL (i.e., 20 U/100 μL urine); buffer: 0.1 mL of 1 M NH4Ac (≈ 6.0); incubation time: 24 h at 37 °C | vortex-assisted dispersive liquid-liquid microextraction |
Single injection: Instrument: LC-MS/MS; LC column: Brownlee Aq C18 column (100 mm × 4.6 mm, 5 μm); Mobile phase: water and MeOH; Run time: 12 min; Injection volume: 10 μL; |
0.01–0.20 | [9] |
| Total: 19 analytes 5 BADGEs, 5 benzophenones, 7 parabens, TCC, and TCS |
0.5 mL | β-glucuronidase/arylsulfatas e enzyme (Helix pomatia, β-glucuronidase activity ≈ 145,700 U/mL; sulfatase activity ~ 887 U/mL) | enzyme amount: 0.15 μL (i.e., 4.4 U/100 μL urine); buffer: 0.3 mL of 1 M NH4Ac (no pH adjustments); incubation time: 12 h at 37 °C; | liquid-liquid extraction with EtAc |
Two injections: Instrument: LC-MS/MS; LC column: Betasil C18 (2.1 mm × 100 mm, 5 μm); 1st injection for 5 BADGEs: Mobile phase: water and MeOH; Run time: 30 min; Injection volume: 10 μL; 2nd injection for remaining analytes: Mobile phase: 0.1% FA in water and MeOH; Run time: 30 min; Injection volume: 10 μL; |
0.06–0.6 | [11] |
| Total: 21 analytes 7 BPs, 7 parabens, 5 benzophenones, TCS, and TCS |
5.0 mL | β-glucuronidase enzyme (type HP-2 from Helix pomatia, β-glucuronidase activity ≈ 197,000 U/mL) | enzyme amount: 20 U/100 μL urine; buffer: not provided; incubation time: overnight at 37 °C; | air-assisted liquid-liquid microextraction |
Single injection: Instrument: LC-MS/MS; LC column: Atlantis T3 dC18 column (75 mm × 2.1 mm, 3.0 μm); Mobile phase: water and MeOH; Run time: 10 min; Injection volume: 10 μL; |
0.01–0.30 | [12] |
| Total: 12 analytes 7 PhMs, 4 parabens, and BP-3 |
3.0 mL | β-glucuronidase enzyme (Type HP-2 from Helix pomatia, β-glucuronidase activity ≥ 200,000 U/mL) | enzyme amount: 25 pL (i.e., 167 U/100 μL urine); buffer: 0.75 mL of 1 M NaOAc (pH 4.5); incubation time: overnight at 37 °C; | Bond Elut Certify SPE cartridge (130 mg, 10 mL) |
Single injection: Instrument: LC-MS/MS; LC column: Kinetex Phenyl-Hexyl column (2.1 mm × 100 mm, 1.7 μm); Mobile phase: 0.1% HAc in both water and ACN; Run time: 20 min; Injection volume: 5 μL; |
0.09–0.37 | [13] |
| Total: 23 analytes 8 BPs and 15 OH-PAHs |
0.5 mL | β-glucuronidase enzyme (from E. coli K12, β-glucuronidase activity ≈ 140 U/mL | enzyme amount: 20 μL (i.e., 0.6 U/100 μL urine); buffer: 1 mL of 0.5 M NH4Ac (pH 6.0); incubation time: 14 h at 37 °C; | liquid-liquid extraction with mixture of EtAc:pentane:toluene (5:4:1, v:v) |
Two injections: Instrument: LC-MS/MS; 1st injection for 8 BPs: LC column: Betasil C18 (2.1 mm × 100 mm, 5 μm); Mobile phase: 0.1% NH4OH in water and MeOH; Run time: 20 min; Injection volume: 10 μL; 2nd injection for 15 OH-PAHs: LC column: Agilent Eclipse Plus C18 column (100 mm × 4.6 mm, 3.5 μm) Mobile phase: water and MeOH; Run time: 20 min; Injection volume: 2 μL; |
0.003–0.36 | [14] |
| Total: 19 analytes 14 PhMs and 5 BPs |
0.05 mL | β-glucuronidase enzyme (from E. coli K12, β-glucuronidase activity ≈ 200 U/mL | enzyme amount: 0.5 μL (i.e., 0.2 U/100 μL urine); buffer: 0.025 mL of 1 M NH4Ac (pH 6.5); incubation time: 1.5 h at 37 °C; | Online Strata X SPE cartridge (20 mm × 2.0 mm, 25 μm) |
Single injection: Instrument: LC-MS/MS; LC column: Synergi MAX-RP column (150 mm × 3.0 mm; 4 μm); Mobile phase: 0.05% HAc in both water and ACN; Run time: 13 min; Injection volume: 50 μL; |
0.005–0.39 | [15] |
| Total: 6 analytes 4 PhMs, BPA, and 4-n-NP |
0.9 mL | β-glucuronidase enzyme (from abalone, β-glucuronidase activity > 100,000 U/mL, arylsulfatase activity < 20,000 U/mL) | enzyme amount: 0.5 U/100 μL urine; buffer: 0.25 mL of 1 mM NH4Ac (no pH adjustment); incubation time: 2 h at 37 °C; | ABS Elut NEXUS SPE cartridges (60 mg, 3 mL) |
Two injections: Instrument: LC-MS/MS; LC column: Acquity UPLC BEH Phenyl column (100 mm × 2.1 mm, 1.7 μm); 1st injection for PhMs: Mobile phase: 0.1% FA in water and MeOH; Run time: 22 min; Injection volume: 5 μL; 2nd injection for BPA and 4-n-NP: Mobile phase: 0.05% NH4OH in water and MeOH; Run time: 14 min; Injection volume: 5 μL; |
0.3–0.5 | [16] |
| Total: 260 analytes pesticides |
0.1 mL | no enzymatic deconjugation step | Quick, Easy, Cheap, Effective, Rugged, and Safe (QuEChERS) method |
Single injection: Instrument: LC-MS/MS; LC column: Kinetex C18 column (100 mm × 2.1 mm, 2.6 μm); Mobile phase: 5 mM AF and 0.1% FA in both water and MeOH; Run time: 15 min; Injection volume: 4 μL; |
10 | [17] | |
| Total: 13 analytes pesticides |
1.0 mL | acidic hydrolysis using 5 mL of HCl (50%) and heating in a 90 °C water bath for 45 min for deconjugation | Liquid-liquid extraction with hexane followed by derivatization with N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide |
Single injection: Instrument: GC-MS; GC column: Restek RTX-35 column (30 m × 0.25 mm, 0.25 μm); Run time: 30 min; Injection volume: 1 μL; |
0.025–0.10 | [18] | |
| Total: 10 analytes pesticides |
1.0 mL | β-glucuronidase enzyme (type H-1 from Helix pomatia with a specific activity of ~500 U/mg) | enzyme amount: 56 units/100 μL urine; buffer: 0.75 mL of 0.2 M NaOAc; incubation time: > 6 h at 37 °C; | Oasis HLB SPE cartridge (30 mg, 1 mL) |
Single injection: Instrument: LC-MS/MS; LC column: Betasil C18 column (100 mm × 2.1 mm; 3 μm); Mobile phase: 0.1% HAc in water and ACN; Run time: 20 min; Injection volume: 30 μL; |
0.1–0.6 | [19] |
| Total: 121 analytes 45 plasticizers and metabolites, 34 environmental phenols, 31 pesticides, 11 OH-PAHs; |
0.5 mL | β-glucuronidase/arylsulfatas e enzyme (Helix pomatia, β-glucuronidase activity ≈ 100,000 U/mL; sulfatase activity ≈ 47,500 U/mL) | enzyme amount: 20 μL (i.e., 400 units/100 μL urine); buffer: 0.5 mL of 1 M NH4Ac (pH 5.5); incubation time: overnight at 37 °C | ABS Elut NEXUS SPE cartridge (60 mg, 3 mL) |
Two injections: Instrument: LC-MS/MS; 1st injection for 43 analytes: LC column: Ultra AQ C18 column (2.1 mm × 100 mm, 3 μm); Mobile phase: 0.1% HAc in both water and MeOH; Run time: 12 min; Injection volume: 5 μL; 2nd injection for 78 analytes: LC column: Betasil C18 column (100 mm × 2.1 mm, 5 μm) Mobile phase: water and ACN; Run time: 9 min; Injection volume: 5 μL; |
≤ 0.1 for 101 analytes 0.1–1.0 for 18 analytes ≥ 1.0 for 2 analytes |
the present study |
OH-PAHs: monohydroxylated polycyclic aromatic hydrocarbons; BRPs: brominated phenols; OH-PBDEs: hydroxyl polybrominated diphenyl ethers; TCC: triclocarban; TCS: triclosan; TBBPA: tetrabromobisphenol A; BPs: bisphenols; BADGEs: bisphenol A diglycidyl ethers; PhMs: phthalate metabolites; BPA: 4,4’-(1-methylethylidene)bisphenol; NH4Ac: ammonium acetate; FA: formic acid; HAc: acetate acid; NH4OH: ammonium hydroxide; AF: ammonium formate; ACN: acetonitrile; MeOH: methanol; EtAc: ethyl acetate; NaOAc: sodium acetate; LOD: limit of detection.
A recent investigation of the extant data for chemicals not measured by the United States National Health and Nutrition Examination Survey (NHANES), identified those that are used in consumer products for which toxicity data are available and likelihood of human exposure was considerable [20]; this investigation highlighted the need for biomonitoring of chemical classes such as: alternative flame retardants, alternative plasticizers, environmental phenols (EPs) especially bisphenol A (BPA) substitutes, and current-use pesticides. Further details of the selection criteria of these chemical classes are described elsewhere [20]. Thus, we selected the target analytes from the list of emerging chemicals identified by Pellizzari et al [20] and enhanced the list with legacy chemicals that can be analyzed in urine using the same sample preparation method. The target analytes selected for the present study include plasticizers such as phthalates, terephthalates, and organophosphate ester metabolites (all three classes collectively referred here as PMs), EPs including bisphenol analogues, chlorophenols, hydroxy polycyclic aromatic hydrocarbons (PAHs) and benzophenones and current-use pesticides including neonicotinoids. Phthalate plasticizers are substituted with emerging chemicals like terephthalates and organophosphate esters [21]. EPs are used as antimicrobial preservatives (e.g., parabens, triclocarban, and triclosan), sunscreen agents (e.g., benzophenones), and industrial compounds (e.g., chlorophenols and bisphenols and their halogenated derivatives) [10]. Numerous studies suggested that exposures to these chemicals are associated with endocrine disruption, cytotoxicity, genotoxicity, reproductive toxicity, and neurotoxicity [22, 23]. To the best of our knowledge, no previous study reported the analysis of multi-class chemicals comprising plasticizers, phenolic compounds and pesticides in urine using LC-MS/MS methods. In this study, we describe a multi-class targeted LC-MS/MS method using isotopic dilution mass spectrometry for simultaneous identification and quantification of 121 biomarkers, including 45 PMs, 45 EPs, and 31 pesticides in human urine, which can be applied in the assessment of population exposure to environmental chemicals. The method is capable of measuring several classes of chemicals in a single extraction, and would save cost and time in biomonitoring programs.
2. Materials and methods
2.1. Reagents and chemicals
HPLC-grade water, methanol (MeOH), ethyl acetate (EtAc), acetonitrile (ACN), methyl tert-butyl ether (MTBE), dichloromethane (DCM), acetone, hexane, ammonium acetate (NH4Ac), and ACS-grade formic acid (FA) were obtained from J.T. Baker (Phillipsburg, NJ, USA). ACS-grade acetic acid (HAc) was purchased from Macron Fine Chemicals (Phillipsburg, NJ, USA). HPLC-grade phosphoric acid (PA) was obtained from Burdick & Jackson (Muskegon, MI, USA). HPLC-grade ammonium formate (AF) and ACS-grade sodium phosphate monobasic monohydrate (SPMM) and ammonium hydroxide solution (NH4OH; 28.0–30.0% NH3 basis) were purchased from Sigma-Aldrich (St Louis, MO, USA). Three types of β-glucuronidase enzymes, namely, β-glucuronidase/arylsulfatase liquid enzyme from Helix pomatia (ALS; β-glucuronidase activity ≈ 100,000 units/mL; sulfatase activity ≈ 47,500 units/mL; Item #10127698001), and β-glucuronidase from E.coli-K12 (K12; β-glucuronidase activity ≈ 140 units/mg; Item #03707601001) and Helix pomatia (HP; β-glucuronidase activity 68,800 units/mL; sulfatase activity ≤ 5000 units/mL; Item #152284) were purchased from Roche Life Science through Sigma Aldrich (Mannheim, Germany) and MP Biomedicals (Santa Cruz, CA, USA), respectively. The enzyme K12 contained only glucuronidase activity whereas ALS and HP contained both glucuronidase and sulfatase activities with a greater sulfatase activity in the former.
Analytical standards (NSs) for 121 target chemicals and 92 isotopically labeled internal standards (ISs) of 95–99.9% purity were purchased from multiple suppliers including Cambridge Isotope Laboratories (Andover, MA, USA), Sigma-Aldrich (St Louis, MO, USA), AccuStandard (New Haven, CT, USA), Dr. Ehrenstorfer GmbH (Augsburg, Germany), Toronto Research Chemicals (North York, ON, Canada), TCI America (Portland, OR, USA), and C/D/N Isotopes (Pointe-Claire, Quebec, Canada) (Table S1 in the Supporting Information; SI). Chemical names, CAS numbers, and their abbreviations used in this study are given in Table 2. Majority of the analytical standards were purchased at a concentration of 1000 μg/mL or 100 μg/mL in an organic solvent. Some standards were purchased as neat compounds. Stock solutions of neat chemicals were prepared in ACN or MeOH and a small fraction of that was also prepared in acetone or hexane, at a concentration range of 100–1000 μg/mL. Known concentrations of individual stock solutions were transferred into 15 mL glass tubes for the preparation of a mixture of NSs or ISs at a concentration of 1 μg/mL. These solutions were diluted serially with water:ACN (8:2 v/v) for use in method development and analysis. All intermediate and working standard solutions were stored at −20 °C.
Table 2.
Compound name, abbreviation, CAS number, MRM (quantification) transition, collision energy (CE), method classification (MC), and retention time (RT) for 121 target compounds analyzed in the present study
| No. | Compound name | Synonym | CAS number | QT | CE | MC | RT |
|---|---|---|---|---|---|---|---|
| Plasticizers and metabolites | |||||||
| 1 | phthalic acid | PA | 88-99-3 | 165>77 | −25 | MSM1_NEG | 3.77 |
| 2 | mono-methyl phthalate | mMP | 4376-18-5 | 179>77 | −25 | MSM1_NEG | 4.37 |
| 3 | mono-ethyl phthalate | mEP | 2306-33-4 | 193>77 | −25 | MSM1_NEG | 5.06 |
| 4 | mono-iso-propyl phthalate | mIPrP | 4376-18-5 | 207>77 | −25 | MSM1_NEG | 5.59 |
| 5 | mono-n-propyl phthalate | mPrP | 4376-19-6 | 207>77 | −25 | MSM1_NEG | 5.77 |
| 6 | mono-iso-butyl phthalate | mIBP | 30833-53-5 | 221>77 | −25 | MSM1_NEG | 6.34 |
| 7 | mono-butyl phthalate | mBP | 131-70-4 | 221>77 | −25 | MSM1_NEG | 6.40 |
| 8 | mono-pentyl phthalate | mPeP | 24539-56-8 | 235>77 | −25 | MSM1_NEG | 6.95 |
| 9 | mono-hexyl phthalate | mHxP | 24539-57-9 | 249>77 | −25 | MSM1_NEG | 7.40 |
| 10 | mono-(3-carboxypropyl) phthalate | mCPP | 66851-46-5 | 251>103 | −25 | MSM1_NEG | 4.56 |
| 11 | mono-benzyl phthalate | mBzP | 2528-16-7 | 255>183 | −16 | MSM1_NEG | 6.47 |
| 12 | mono-2-heptyl phthalate | mHpP | 129171-03-5 | 263>77 | −25 | MSM1_NEG | 7.62 |
| 13 | mono-octyl phthalate | mOP | 5393-19-1 | 277>125 | −20 | MSM1_NEG | 8.03 |
| 14 | mono-(2-ethylhexyl) phthalate | mEHP | 4376-20-9 | 277>134 | −21 | MSM1_NEG | 7.89 |
| 15 | mono-(2-ethyl-5-oxohexyl) phthalate | mEOHP | 40321-98-0 | 291>121 | −25 | MSM1_NEG | 6.29 |
| 16 | mono-(2-ethyl-5-hydroxyhexyl) phthalate | mEHHP | 40321-99-1 | 293>121 | −26 | MSM1_NEG | 6.52 |
| 17 | mono-iso-decyl phthalate | mIDP | 31047-64-0 | 305>155 | −25 | MSM1_NEG | 8.39 |
| 18 | mono-(2-ethyl-5-carboxypentyl) phthalate | mECPP | 40809-41-4 | 307>159 | −25 | MSM1_NEG | 6.39 |
| 19 | mono-[2-(carboxymethyl)hexyl] phthalate | mCMHP | 82975-93-7 | 307>159 | −25 | MSM1_NEG | 7.00 |
| 20 | mono-(7-carboxyheptyl) phthalate | mCHpP | 856869-57-3 | 307>159 | −25 | MSM1_NEG | 6.49 |
| 21 | mono-carboxy-iso-octyl phthalate | mCIOP | 898544-09-7 | 321>173 | −20 | MSM1_NEG | 6.77 |
| 22 | mono-carboxy-iso-nonyl phthalate | mCINP | 1373125-93-9 | 335>187 | −21 | MSM1_NEG | 7.16 |
| 23 | 2-(((9-hydroxydecyl)oxy)carbonyl) benzoic acid | mHiDP | not available | 321>121 | −35 | MSM1_NEG | 7.30 |
| 24 | monohydroxy-iso-nonyl phthalate | mHiNP | 898544-10-0 | 307>121 | −25 | MSM1_NEG | 6.95 |
| 25 | cyclohexane-1,2-dicarboxylic acid-mono (hydroxy-isononyl) ester | mHNCH | 1637562-52-7 | 313.3>153 | −25 | MSM1_NEG | 7.40 |
| 26 | cyclohexane-1,2-dicarboxylic acid-mono (oxo-isononyl) ester | mONCH | 1588520-62-0 | 311.4>153 | −25 | MSM1_NEG | 7.20 |
| 27 | mono-2-(propyl-6-oxoheptyl)-phthalate | mPOHP | 1373125-92-8 | 319.3>121.1 | −22 | MSM1_NEG | 7.08 |
| 28 | mono-2-(propyl-6-hydroxy-heptyl)-phthalate | mPHHP | 1372605-11-2 | 321.2>121 | −35 | MSM1_NEG | 7.31 |
| 29 | cyclohexane-1,2-dicarboxylic acid-monocarboxy isooctyl ester | mCOCH | 1637562-51-6 | 327.4>173.1 | −22 | MSM1_NEG | 7.28 |
| 30 | mono-2-(propyl-6-carboxy-hexyl)-phthalate | mPCHP | 1412411-10-9 | 335.3>187.3 | −15 | MSM1_NEG | 7.16 |
| 31 | mono-ethyl terephthalate | mETP | 713-57-5 | 192.9>119.9 | −28 | MSM1_NEG | 6.29 |
| 32 | mono-tert-butyl terephthalate | mTBTP | 20576-82-3 | 221>119.8 | −30 | MSM1_NEG | 7.18 |
| 33 | mono-benzyl terephthalate | mBzTP | 18520-63-3 | 255.3>119.9 | −25 | MSM1_NEG | 7.38 |
| 34 | triethyl phosphate | TEP | 78-40-0 | 183>99.1 | 35 | MSM2_POS | 3.09 |
| 35-1 | tri-n-butyl phosphate | TNBP | 126-73-8 | 267.1>99 | 22 | MSM2_POS | 5.12 |
| 35-2 | tri-iso-butyl phosphate | TIBP | 126-71-6 | 267.1>99 | 22 | MSM2_POS | |
| 37 | tris(2-chloroethyl) phosphate | TCEP | 115-96-8 | 284.9>63.1 | 40 | MSM2_POS | 3.42 |
| 38 | tripropyl phosphate | TPP | 513-08-6 | 225.1>99 | 35 | MSM2_POS | 3.93 |
| 39 | triphenyl phosphate | TPhP | 115-86-6 | 327.1>77.1 | 46 | MSM2_POS | 4.98 |
| 40 | tris(2-butoxyethyl) phosphate | TBOEP | 78-51-3 | 399.1>199 | 20 | MSM2_POS | 5.51 |
| 41-1 | di-n-butyl phosphate | DNBP | 107-66-4 | 209>78.9 | −35 | MSM1_NEG | 6.92 |
| 41-2 | di-iso-butyl phosphate | DIBP | 6303-30-6 | 209>78.9 | −35 | MSM1_NEG | |
| 43 | diphenyl phosphate | DPhP | 838-85-7 | 248.9>93.1 | −40 | MSM1_NEG | 6.28 |
| 44 | bis(2-methylphenyl) phosphate | BMPP | 35787-74-7 | 277>107 | −40 | MSM2_NEG | 2.66 |
| 45 | bis(1,3-dichloro-2-propyl) phosphate | BDCIPP | 72236-72-7 | 316.9>35 | −30 | MSM2_NEG | 1.74 |
| Environmental phenols | |||||||
| 46 | methyl paraben | MeP | 99-76-3 | 151>92 | −32 | MSM2_NEG | 3.11 |
| 47 | ethyl paraben | EtP | 120-47-8 | 165>92 | −32 | MSM2_NEG | 3.34 |
| 48 | n-propyl paraben | PrP | 94-13-3 | 179>92 | −32 | MSM2_NEG | 3.61 |
| 49 | n-butyl paraben | BuP | 94-26-8 | 193>92 | −40 | MSM2_NEG | 3.93 |
| 50 | benzyl paraben | BzP | 94-18-8 | 227>92 | −32 | MSM2_NEG | 3.85 |
| 51 | heptyl paraben | HeP | 1085-12-7 | 235>92 | −40 | MSM2_NEG | 5.24 |
| 52 | benzophenone-1 | BP-1 | 131-56-6 | 213>91 | −35 | MSM2_NEG | 3.71 |
| 53 | benzophenone-2 | BP-2 | 131-55-5 | 245>91 | −40 | MSM2_NEG | 3.10 |
| 54 | benzophenone-3 | BP-3 | 131-57-7 | 227>211 | −35 | MSM2_NEG | 4.78 |
| 55 | benzophenone-6 | BP-6 | 131-54-4 | 273>123 | −24 | MSM2_NEG | 4.40 |
| 56 | benzophenone-8 | BP-8 | 131-53-3 | 243>93 | −38 | MSM2_NEG | 4.04 |
| 57 | 4-hydroxybenzophenone | 4-OH-BP | 1137-42-4 | 197>92 | −42 | MSM2_NEG | 3.47 |
| 58 | 4,4’-(1-methylethylidene)bisphenol | BPA | 80-05-7 | 227>212 | −26 | MSM2_NEG | 3.48 |
| 59 | 4,4’-di-hydroxydiphenylmethane | BPF | 620-92-8 | 199>77 | −34 | MSM2_NEG | 3.25 |
| 60 | 4,4’-sulfonyldiphenol | BPS | 80-09-1 | 249>108 | −35 | MSM2_NEG | 2.99 |
| 61 | 2,2-bis(4-hydroxyphenyl)butane | BPB | 77-40-7 | 241>211 | −39 | MSM2_NEG | 3.69 |
| 62 | 4,4’-cyclo-hexylidenebisphenol | BPZ | 843-55-0 | 267>173 | −35 | MSM2_NEG | 3.99 |
| 63 | 4,4’-(1-phenylethylidene)bisphenol | BPAP | 1571-75-1 | 289>273 | −43 | MSM2_NEG | 3.82 |
| 64 | 2,2-bis(4-hydroxyphenyl) hexafluoropropane | BPAF | 1478-61-1 | 335>265 | −31 | MSM2_NEG | 3.81 |
| 65 | 4,4’-(1,4-phenylenediisopropylidene) bisphenol | BPP | 2167-51-3 | 345>330 | −36 | MSM2_NEG | 4.68 |
| 66-1 | 2,4,5-trichlorophenol | 2,4,5-TCP | 95-95-4 | 195>35 | −35 | MSM2_NEG | 2.85 |
| 66-2 | 2,4,6-trichlorophenol | 2,4,6-TCP | 88-06-2 | 195>35 | −35 | MSM2_NEG | |
| 68 | 2,3,5,6-tetrachlorophenol | 2,3,5,6-TeCP | 935-95-5 | 231>35 | −60 | MSM2_NEG | 3.94 |
| 69 | 2,3,4,6-tetrachlorophenol | 2,3,4,6-TeCP | 58-90-2 | 231>35 | −60 | MSM2_NEG | 4.34 |
| 70 | 2,3,4,5-tetrachlorophenol | 2,3,4,5-TeCP | 4901-51-3 | 231>35 | −60 | MSM2_NEG | 4.77 |
| 71 | pentachlorophenol | PCP | 87-86-5 | 265>35 | −60 | MSM2_NEG | 3.65 |
| 72 | triclocarban | TCC | 101-20-2 | 313>160 | −25 | MSM2_NEG | 5.14 |
| 73 | triclosan | TCS | 3380-34-5 | 287>35 | −43 | MSM2_NEG | 5.24 |
| 74 | 3,3’,5-trichlorobisphenol A | TCBPA | 40346-55-2 | 329.1>278 | −35 | MSM2_NEG | 4.36 |
| 75 | 2,2’,6,6’-tetrachlorobisphenol A | TeCBPA | 79-95-8 | 365.1>314 | −37 | MSM2_NEG | 4.76 |
| 76 | 3,3’,5,5’-tetrabromobisphenol A | TBBPA | 79-94-7 | 542.8>420 | −57 | MSM2_NEG | 5.21 |
| 77 | bisphenol A diglycidyl ether | BADGE | 1675-54-3 | 358>191 | 16 | MSM2_POS | 4.70 |
| 78 | bisphenol A (2,3-dihydroxypropyl) glycidyl ether | BADGEH2O | 76002-91-0 | 376>209 | 20 | MSM2_POS | 3.56 |
| 79 | bisphenol A bis(2,3-dihydroxypropyl) glycidyl ether | BADGE2H2O | 5581-32-8 | 394>209 | 20 | MSM2_POS | 3.00 |
| 80 | 1-hydroxynaphthalene | 1-Nap | 90-15-3 | 143>115 | −38 | MSM2_NEG | 3.69 |
| 81 | 2-hydroxynaphthalene | 2-Nap | 135-19-3 | 143>115 | −38 | MSM2_NEG | 3.58 |
| 82-1 | 2-hydroxyfluorene | 2-Fluo | 2443-58-5 | 180.9>179.9 | −30 | MSM2_NEG | 3.94 |
| 82-2 | 3-hydroxyfluorene | 3-Fluo | 6344-67-8 | 180.9>179.9 | −30 | MSM2_NEG | |
| 82-3 | 9-hydroxyfluorene | 9-Fluo | 1689-64-1 | 180.9>179.9 | −30 | MSM2_NEG | |
| 85-1 | 2-hydroxyphenanthrene | 2-Phen | 605-55-0 | 193.1>164.9 | −45 | MSM2_NEG | 4.09 |
| 85-2 | 3-hydroxyphenanthrene | 3-Phen | 605-87-8 | 193.1>164.9 | −45 | MSM2_NEG | |
| 87-1 | 1-hydroxyphenanthrene | 1-Phen | 2433-56-9 | 193.1>164.9 | −45 | MSM2_NEG | 4.25 |
| 87-2 | 9-hydroxyphenanthrene | 9-Phen | 484-17-3 | 193.1>164.9 | −45 | MSM2_NEG | |
| 89 | 4-hydroxyphenanthrene | 4-Phen | 7651-86-7 | 193.1>164.9 | −45 | MSM2_NEG | 4.35 |
| 90 | 1-hydroxypyrene | 1-Pyr | 5315-79-7 | 217.1>188.9 | −45 | MSM2_NEG | 4.63 |
| Pesticides | |||||||
| 91 | nitenpyram | nitenpyram | 150824-47-8 | 271.1>126 | 50 | MSM2_POS | 2.88 |
| 92 | thiamethoxam | thiamethoxam | 153719-23-4 | 291.8>211.1 | 18 | MSM2_POS | 2.57 |
| 93 | imidacloprid | imidacloprid | 138261-41-3 | 256>209 | 32 | MSM2_POS | 2.95 |
| 94 | acetamiprid | acetamiprid | 135410-20-7 | 222.8>126 | 23 | MSM2_POS | 3.00 |
| 95 | thiacloprid | thiacloprid | 111988-49-9 | 253>126 | 27 | MSM2_POS | 3.15 |
| 96 | clothianidin | clothianidin | 210880-92-5 | 250>169.1 | 20 | MSM2_POS | 2.90 |
| 97 | flonicamid | flonicamid | 158062-67-0 | 230>203 | 20 | MSM2_POS | 2.83 |
| 98 | N-desmethyl thiamethoxam | N-Dmt | 171103-04-1 | 278>132 | 45 | MSM2_POS | 3.02 |
| 99 | N-desmethyl-acetamiprid | N-Dma | 190604-92-3 | 209>126 | 35 | MSM2_POS | 2.90 |
| 100 | thiacloprid-amide | Ta | 676228-91-4 | 271.1>126.1 | 36 | MSM2_POS | 2.88 |
| 101 | imidaclothiz | imidaclothiz | 105843-36-5 | 262>181 | 32 | MSM2_POS | 3.00 |
| 102 | 6-chloronicotinic acid | 6-Cn | 5326-23-8 | 155.9>111.9 | −15 | MSM1_NEG | 4.33 |
| 103 | sulfoxaflor | sulfoxaflor | 946578-00-3 | 275.9>213 | −21 | MSM2_NEG | 3.19 |
| 104 | 4-nitrophenol | 4-nitrophenol | 100-02-7 | 138>108 | −24 | MSM2_NEG | 3.17 |
| 105 | 2,4-dichlorophenoxyacetic acid | 2,4-D | 94-75-7 | 219>161 | −20 | MSM2_NEG | 1.31 |
| 106 | 3,5,6-trichloro-2-pyridinol | TCPY | 6515-38-4 | 196>35 | −40 | MSM1_NEG | 6.83 |
| 107 | trans-3-(2,2-di-chlorovinyl)-2,2-dimethyl-cyclopropane-1-carboxylic acid | trans-DCCA | 55701-07-0 | 207>35 | −40 | MSM1_NEG | 6.92 |
| 108 | cis-3-(2,2-di-chlorovinyl)-2,2-dimethyl-cyclopropane-1-carboxylic acid | cis-DCCA | 55667-40-8 | 207>35 | −40 | MSM1_NEG | 7.10 |
| 109 | 3-phenoxybenzoic acid | 3-PBA | 3739-38-6 | 213>93 | −30 | MSM1_NEG | 7.15 |
| 110 | 4-fluoro-3-phenoxybenzoic acid | 4F-3PBA | 77279-89-1 | 231>93 | −35 | MSM1_NEG | 7.17 |
| 111 | 2,4,5-trichlorophenoxyacetic acid | 2,4,5-T | 93-76-5 | 255>197 | −20 | MSM1_NEG | 7.30 |
| 112 | pyrimethanil | pyrimethanil | 53112-28-0 | 200.1>107 | 34 | MSM2_POS | 4.21 |
| 113 | dinotefuran | dinotefuran | 165252-70-0 | 203>129.1 | 20 | MSM2_POS | 1.30 |
| 114 | metribuzin | metribuzin | 21087-64-9 | 215.1>187 | 10 | MSM2_POS | 4.88 |
| 115 | atrazine | atrazine | 1912-24-9 | 216>174 | 25 | MSM2_POS | 3.70 |
| 116 | cyprodinil | cyprodinil | 121552-61-2 | 226>93 | 50 | MSM2_POS | 4.92 |
| 117 | metalaxyl | metalaxyl | 57837-19-1 | 280.2>220 | 20 | MSM2_POS | 3.67 |
| 118 | tebuconazole | tebuconazole | 107534-96-3 | 308.2>70 | 45 | MSM2_POS | 4.37 |
| 119 | propiconazole | propiconazole | 60207-90-1 | 342.1>159 | 42 | MSM2_POS | 4.72 |
| 120 | tetraconazole | tetraconazole | 112281-77-3 | 372>159 | 38 | MSM2_POS | 4.19 |
| 121 | azoxystrobin | azoxystrobin | 131860-33-8 | 404>372.1 | 20 | MSM2_POS | 4.07 |
QT: quantification ion transition; CE: collision energy (volt); MSM1_NEG: method 1 under negative mode; MSM2_NEG: method 2 under negative mode; MSM2_POS: method 2 under positive mode; MC: method classification; RT: retention time (min); There is no chromatographic resolution between compounds with same ID; therefore, sum concentrations are expressed for these isomers by using the present developed method.
2.2. Urine samples and enzymatic treatment
Pooled urine samples collected from healthy adult volunteers, two standard reference materials (SRM 3672 and 3673) purchased from National Institute for Standards and Technology (NIST; Gaithersburg, MD, USA), and nine proficiency test (PT) samples obtained from three different providers were used in method development and validation. In addition, 21 de-identified individual adult urine samples collected from 11 males and 10 females in 2017 in Albany, New York, USA, were analyzed to demonstrate the feasibility of the developed method. All samples were stored frozen at −20 °C until analysis.
Majority of the target analytes were expected to be excreted as glucuronide or sulfate conjugates in urine. Enzymatic deconjugation steps were optimized to yield total concentrations of all target analytes [24, 25]. Further details of optimization of enzymatic deconjugation are given below. Among three different types of β-glucuronidase/arylsulfatase tested, ALS type from Sigma-Aldrich yielded higher recoveries within 2 h of incubation. The conditions for enzymatic deconjugation were optimized as: 500 μL of urine taken in a polypropylene (PP) tube, fortified with 2.5 ng each of ISs followed by the addition of 500 μL of 1 M NH4Ac buffer (pH 5.5) containing 20 μL of ALS enzyme (2000 units). After gentle mixing, samples were incubated for 2 h at 37°C with shaking at 100 rpm (Jeio Tech Co., Seoul, Korea). Quality control (QC) samples include analyses of reagent/procedural blanks (HPLC grade water used instead of urine), matrix blanks (pooled urine), matrix spike samples (analytes spiked in pooled urine at three different concentrations: 1, 10, and 20 ng/mL), two SRMs, and nine PT samples..
2.3. Sample extraction
After incubation of samples with β-glucuronidase/arylsulfatase, the reaction was quenched by the addition of 1.0 mL of 0.145 M SPMM buffer (pH 2.0; prepared by dissolving 20 g SPMM and 10 mL of PA in 990 mL of water). The sample was then subjected to ABS Elut NEXUS® solid phase extraction (SPE) (60 mg, 3 mL; Agilent Technologies, Santa Clara, CA, USA). Whereas several combinations of conditioning of cartridges followed by elution of analytes were followed, optimal recovery and reproducibility were found under the following conditions: preconditioning with 1.5 mL of ACN followed by 1.5 mL of 0.145 M SPMM buffer (pH 2.0), loading of samples, washing with 1.5 mL of 0.1 M FA in water and 1.5 mL of 5% MeOH in water (to remove matrix interferences) and elution of target compounds with 1.0 mL of ACN, 1.0 mL of EtAc, and 1.0 mL of MeOH:DCM (1:1, v/v). The eluate was collected into a 15 mL PP tube and concentrated to near-dryness under a gentle nitrogen stream. The residue was reconstituted in 250 μL of water:ACN (8:2, v/v) mixture, vortexed, and transferred into amber glass vials for LC-MS/MS analysis.
2.4. Instrumentation
The HPLC system comprised ExionLC solvent valve, ExionLC AD pump, ExionLC autosampler, ExionLC controller, and ExionLC AC column oven (SCIEX, Redwood City, CA, USA). Mass spectrometric analysis was performed with an AB SCIEX QTRAP 5500+ triple quadrupole mass spectrometer (Applied Biosystems, Foster City, CA, USA) equipped with an electrospray ionization source operated in both positive and negative modes. The optimal LC-MS/MS conditions, other method development and quality assurance protocols are described in detail below.
3. Results and discussion
A total of 121 analytes grouped into three categories as PMs, EPs and pesticides were targeted in the present study (Table 2). The method development and validation consisted of the following protocols in that order: (1) optimization of mass spectrometric parameters followed by liquid chromatographic separation, (2) optimization of sample preparation and extraction protocols, (3) method validation through the analysis of SRMs, PT and fortified samples, and (4) demonstration of feasibility through the analysis of real urine samples. Analytical methods for individual classes of the target chemicals studied here were developed, validated and successfully applied in several biomonitoring studies in our laboratory [11, 26, 27]. The current method combines salient features from several of those earlier methods into a single robust method for extraction and analysis.
3.1. Optimization of LC-MS/MS conditions
The choice of aqueous-organic mobile phase additives and LC columns plays vital roles in chromatographic separation. Various combinations of mobile phases [two organic mobile phase solvents (MeOH and ACN) and five additives (HAc, FA, NH4Ac, AF, and NH4OH)] and four reverse-phase columns [Ultra AQ C18® (3 μm, 100 mm × 2.1 mm, Restek, Bellefonte, PA, USA), Betasil™ C18® (5 μm, 100 mm × 2.1 mm, Thermo Scientific, West Palm Beach, FL, USA), Eclipse Plus C18® (3 μm, 100 mm × 4.6 mm, Agilent, Santa Clara, CA, USA), and Kinetex C18® (1.3 μm, 50 mm × 2.1 mm, Phenomenex, Torrance, CA, USA)] were tested in various combinations by analyzing the standard mixture (both NSs and ISs) at 10 ng/mL dissolved in the mobile phase. It was found that peaks of several PMs and EPs did not resolve adequately under a single LC condition. For instance, under the mobile phase compositions of 0.1% HAc in water and MeOH, PMs showed excellent chromatographic separation and peak shapes whereas signal intensities of several EPs were considerably suppressed. We tested MeOH, ACN and a combination of MeOH and ACN as mobile phases for the separation and analysis of EPs and pesticides. We found that ACN, as the organic mobile phase solvent, yielded narrower and well resolved chromatographic peaks with all four LC columns tested. Mobile phase additives such as NH4Ac or AF enhanced the signals for pesticides. However, it was found that the presence of a buffer in mobile phases affected the retention, signal intensity and resolution of some bisphenols (e.g. BPA and BPF), benzophenones (e.g. BP-3), and bisphenol A diglycidyl ethers (e.g., BADGE, BADGE·H2O, and BADGE·2H2O). Enhancement in signal intensities was achieved for some EPs such as BPA and BADGE, with NH4OH as the mobile phase additive; but analytes such as BPS, 2,4-D, and dinotefuran were not retained efficiently in the analytical column and eluted within 1 min, under those alkaline conditions. On the other hand, poor chromatographic separation and peak shapes of OH-PAHs resulted from acidic mobile phase or the presence of buffer. Among the four LC columns tested, Ultra AQ C18® column enabled excellent chromatographic separation of PMs, with symmetric peaks. Good separation and peak shapes were achieved for 2,3,4,6-TeCP, 2,3,5,6-TeCP, and 2,3,4,5-TeCP by Betasil™ C18® column with water and ACN as the mobile phase. Therefore, two separate injections were required with two different LC conditions to accomplish excellent sensitivity, peak shape and chromatographic separation of 121 target compounds.
The first LC method involved chromatographic separation of target analytes using an Ultra AQ C18® column. The HPLC mobile phase comprised 0.1% HAc in water (A) and 0.1% HAc in MeOH (B). The initial mobile phase composition was 5% B, held for 1 min, and then increased to 45% B within 0.2 min, and held for 1.3 min. Then, the composition was increased to 70% B within 2.2 min, then to 99% B in 2.0 min, and held for 3.0 min. Return to initial mobile phase conditions and column equilibration were accomplished in the last 2.3 min, with a total run time of 12 min. The MS/MS method was in the negative ionization mode (MSM1_NEG). Curtain gas (CUR), collision activated dissociation gas (CAD), source temperature (TEM), nebulizer gas (GS1), heater gas (GS2), and turbo ion spray voltage (IS) were set at 20 psi, 8 psi, 600°C, 60 psi, 60 psi, and 5500 V, respectively. For the second LC method, the chromatographic separation of target analytes was achieved with a Betasil™ C18® column. The HPLC mobile phase comprised water (A) and ACN (B) with the following gradient program; 20% B for 1 min, increased to 60% B within 0.2 min, then to 99% A within 4.3 min, held for 1.5 min, decreased to 20% B within 0.5 min, and equilibrated for 1.5 min, with a total run time of 9 min. The MS/MS analysis proceeded under both negative (MSM2_NEG) and positive (MSM2_POS) ionization modes. CUR, CAD, TEM, GS1, GS2, and IS were set at 20 psi, 8 psi, 600°C, 60 psi, 70 psi, and −4500/+4500 V, respectively. All target analytes were grouped based on their peak shape, separation, and sensitivity under the two separate methods. Accordingly, MSM1_NEG (mostly PMs), MSM2_NEG (mostly EPs), and MSM2_POS (mostly pesticides) yielded optimal analytical performance for 45, 45, and 31 of the 121 target compounds, respectively (see Table 2 for specific method classification). The compound specific m/z transitions of multiple reaction monitoring and optimized MS/MS parameters acquired by direct infusion of each analyte at 10 ng/mL using an in-built syringe pump are shown in Tables S2–S4. Typical chromatograms obtained from MSM1_NEG, MSM2_NEG, and MSM2_POS under the optimized mobile phase conditions are shown in Figures S1–S3 (1 ng /mL) and Figures 1–3 (10 ng/mL). Compound specific retention times are listed in Table 2.
Figure 1. Total ion chromatogram (TIC; A) and extracted ion chromatograms (B–F) of 43 target compounds injected (5 μL) at a concentration of 10 ng/mL measured using mass spectrometric method 1 under negative mode (M1_NEG).
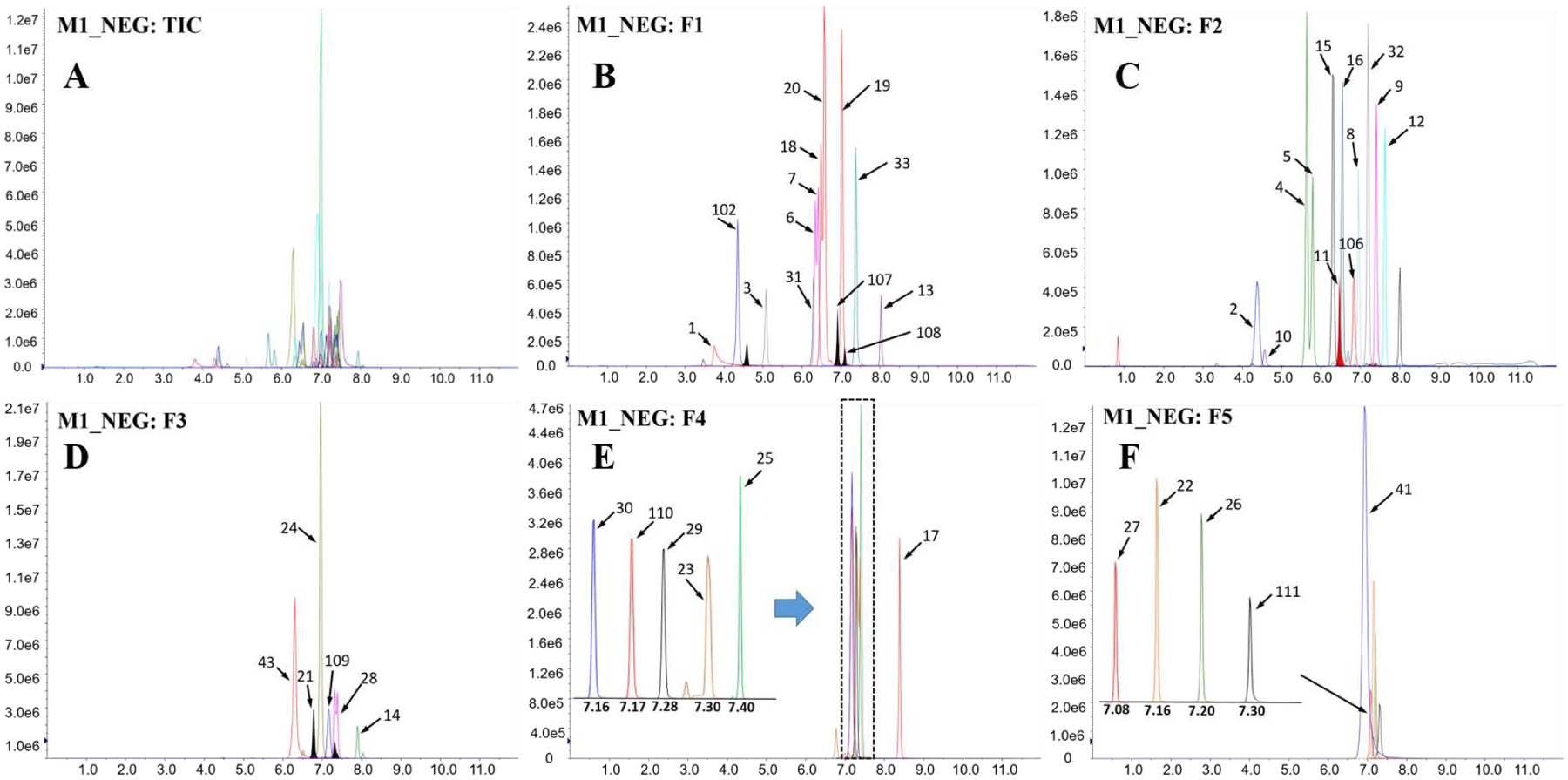
For the sake of distinction of individual compounds, ion chromatograms were extracted and divided into five fractions (i.e., F1–F5) depending on their retention times and signal intensities. Compound numbers assigned on the peaks are shown in Table 1. The x- and y-coordinates are retention time (min) and signal intensity (cps), respectively.
Figure 3. Total ion chromatogram (TIC; A) and extracted ion chromatograms (B–D) of 31 target compounds injected (5 μL) at a concentration of 10 ng/mL measured using mass spectrometric method 2 under positive mode (M2_POS).
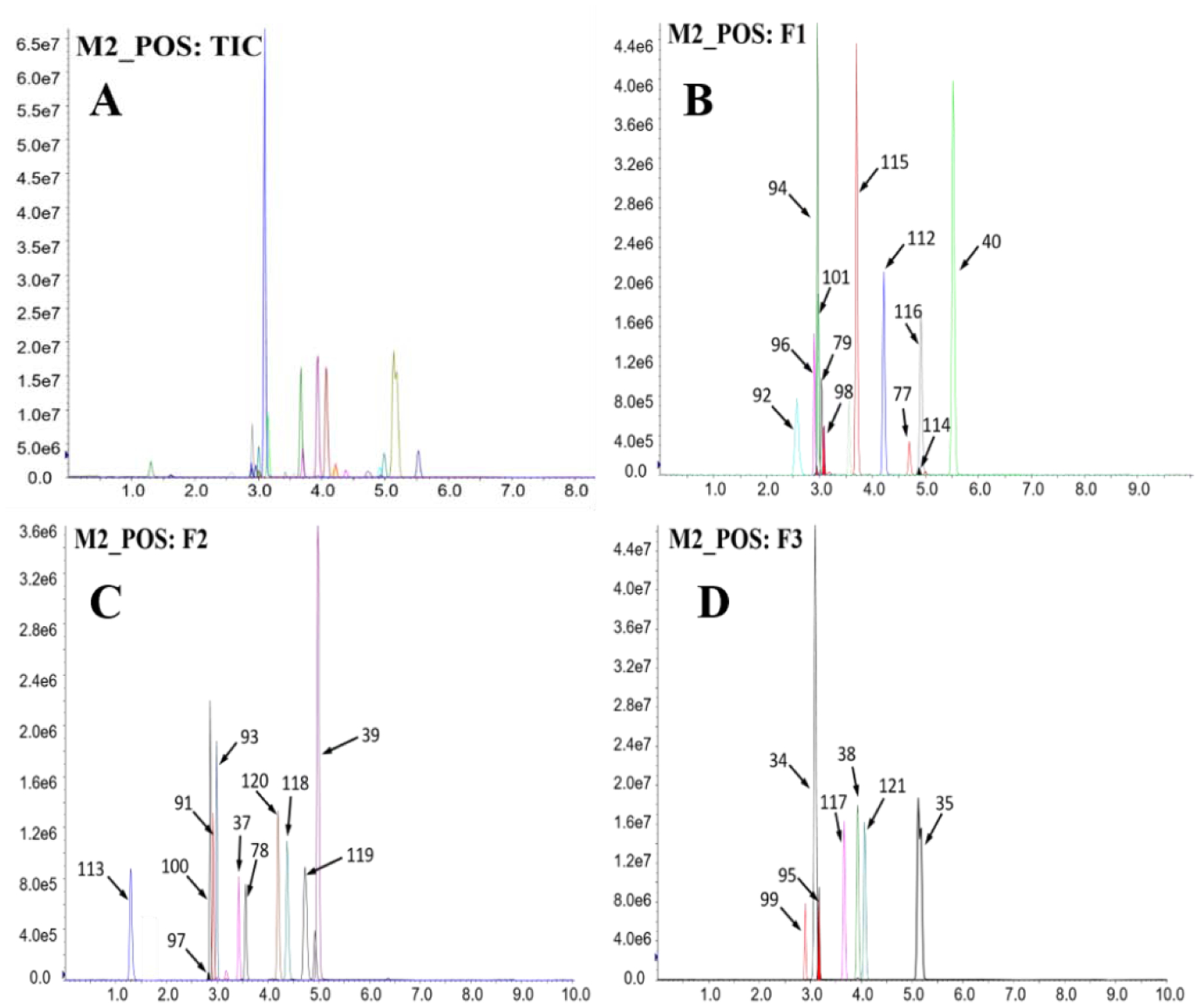
For the sake of distinction of individual compounds, ion chromatograms were extracted and divided into five fractions (i.e., F1–F3) depending on their retention times and signal intensities. Compound numbers assigned on the peaks are shown in Table 1. The x- and y-coordinates are retention time (min) and signal intensity (cps), respectively.
3.2. Sample preparation
3.2.1. Enzymatic hydrolysis
Xenobiotics undergo conjugation reactions to form various hydrophilic conjugates, such as O-glucuronides (i.e., PMs and EPs) [28, 29], N-glucuronides (i.e., TCC) [30], and sulfates (i.e., EPs) [31–33], prior to biliary or urinary excretion. Several factors such as age, gender, race and environmental factors determine the rate of glucuronidation and that can result in dramatic differences in the formation of glucuronidated metabolites. Thus, glucuronide/sulfate deconjugation is an important step in analysis of exposure to environmental chemicals. In this study, glucuronidase reaction was optimized through a full factorial design to examine the effect of enzyme type (ALS, HP, and K12), enzyme amount (10, 20, 30, and 40 μL/sample), buffer pH (5.0, 5.5, 6.0 and 6.5) and incubation time (1, 2, 4, 6, 8, 12, and 24 h), for a total of 54 experiments. The effectiveness of deconjugation reaction was also evaluated by analyzing SRM 3672, for which certified and reference values are available for 17 EPs and 11 PMs targeted in this study. The experiments on enzymatic hydrolysis yielded similar results under all test conditions for OH-PAHs and PMs (except at buffer pH 5.0 for PMs; Table S5), but the results varied considerably among EPs (Figure 4). Based on the recoveries of analytes, ALS enzyme was found superior over the other types (Figure 4A). Satisfactory recoveries were obtained for 7 certified EPs with ALS (80–103%) enzyme in comparison to those of HP (65–97%) and K12 (50–94%) enzymes. Incomplete hydrolysis of MeP (50%), EtP (62%), and PrP (63%) with K12 enzyme may be due to its lack of sulfatase activity. Furthermore, a complete hydrolysis of N-glucuronidated TCC was achieved using ALS enzyme than that of HP and K12 enzymes. Measured TCC concentrations in 21 real urine samples incubated with ALS enzyme were 26% and 45% higher than those obtained using HP and K12 enzymes, respectively. We also tested the hydrolysis efficiency of 7 EPs with varying amounts of ALS enzyme (Figure 4B). Incubation with both 10 μL (80–103%) and 20 μL (82–105%) volumes of enzyme yielded similar recoveries for 7 EPs but a volume of 20 μL yielded higher recoveries for TCS than did 10 μL (95% vs 89%). Incubation with 30 and 40 μL volumes of enzymes resulted in inconsistent recoveries of EtP (80%) and TCS (125%). This may be due to alkyl/aryl sulfatase mediated hydrolysis of alkyl-ester bond of EtP to p-hydroxybenzoic acid [34]. It was reported that the addition of MeOH before incubation would quench aryl sulfatase activity [35], which was not observed in our study. We then tested deconjugation efficiencies at different pH (5.0, 5.5, 6.0 and 6.5; Figure 4C) and incubation times (1, 2, 4, 6, 8, 12, and 24 h; Figure 4D). Although the manufacturers recommended an optimal pH of 4.5–5.0 for the ALS enzyme activity, we found that pH 5.5 was optimal for MeP and PMs. With regard to the incubation time, complete hydrolysis of 7 EPs was accomplished within 2 h at 37°C. Furthermore, the recoveries of MeP and TCS decreased with increasing incubation time. It has been reported that the sulfatase activity can convert parent phthalate diesters into their monoester metabolites [36]. This was found for mEHP, in that the recoveries significantly increased after incubation beyond 12 h (Table S5). Under the optimized conditions of enzymatic deconjugation, 28 certified chemicals in SRM 3672 yielded recoveries of between 80 and 111% following incubation at 37°C for 2 h with 20 μL ALS enzyme (i.e., 4 units/μL urine) at pH 5.5.
Figure 4.
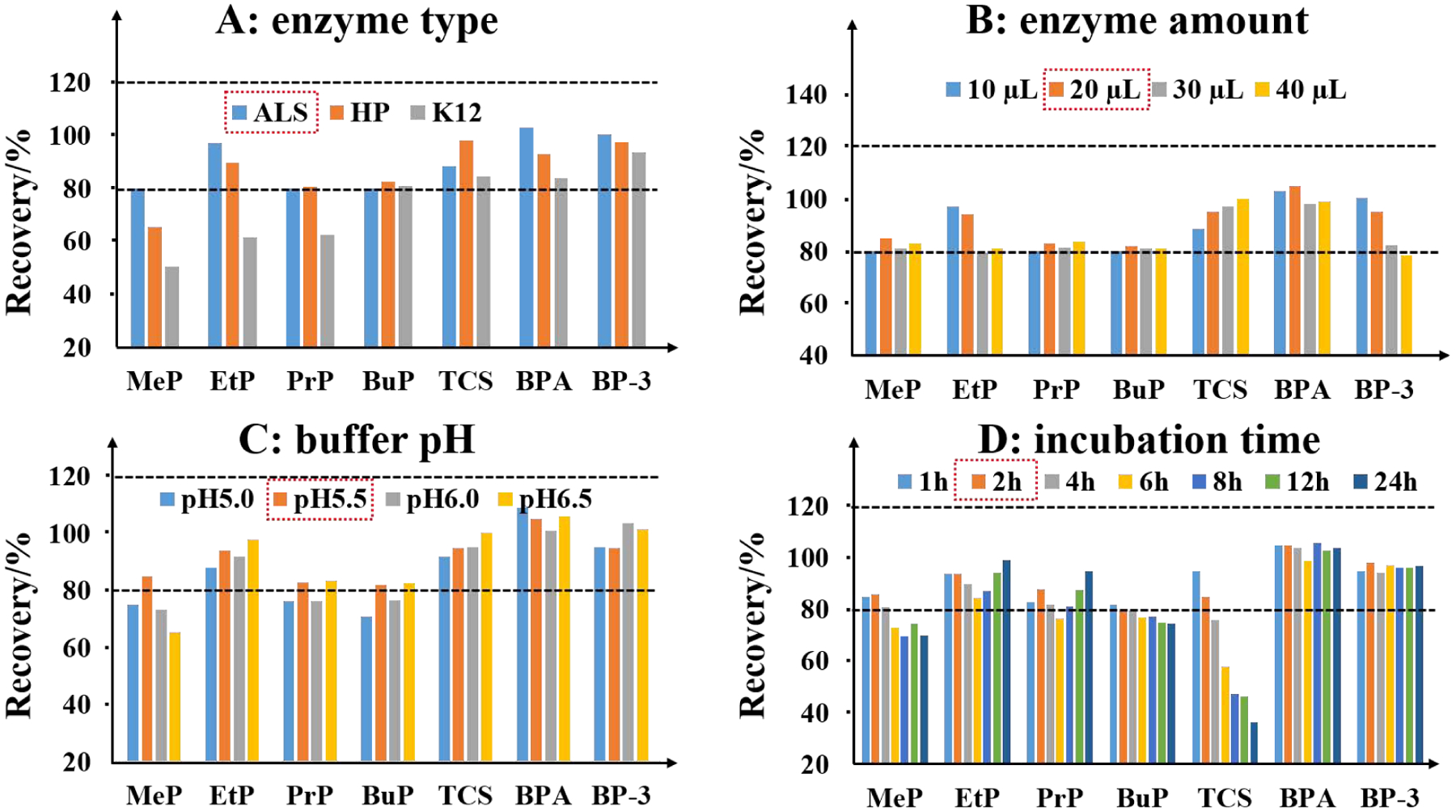
Recoveries of seven phenolic compounds certified in standard reference material 3672 (SRM 3672) with different enzymatic deconjugation conditions including enzyme type (A), enzyme amount (B), buffer pH (C), and incubation time (D).
3.2.2. Sample extraction
Conventional liquid-liquid extraction (LLE) with EtAc and solvent induced phase transition extraction (SPITE) with ACN, MTBE, and EtAc were examined. Fortified urine samples were extracted twice with 2 mL of EtAc or combinations of ACN, MTBE, and EtAc. The urine-solvent mixture was shaken in an orbital shaker (Eberbach Corp., Ann Arbor, MI, USA) at 180 strokes per min for 30 min and centrifuged at 4500 × g for 10 min. The supernatants were combined in a clean PP tube and purified by mixing with water or Q-sep™ QuEChERS dispersive-SPE (dSPE; containing 150 mg MgSO4 and 50 mg C18; Restek, Bellefonte, PA, USA). Some more polar compounds such as PA, mMP, mCPP, dinotefuran, and 2,4-D were lost in the extraction step. Furthermore, quantification of EPs such as BPA, BP-3, and BADGEs was affected by strong matrix effects even after purification by dSPE. dSPE step also increased the background level of contamination of some target compounds in blanks (e.g., mEHP and BPA). Our target analytes vary in polarity from moderate to very high. Therefore, LLE or SPITE method was not found suitable and we optimized the SPE method that has a broad range of retention for polar to nonpolar compounds at a wide pH range.
To maximize extraction efficiency and minimize matrix effects, three commonly used SPE cartridges in biomonitoring studies [10, 16, 37], namely ABS Elut NEXUS®, Oasis HLB® (60 mg, 3 mL; Waters, Milford, MA, USA) and Strata X-AW® (60 mg, 3 mL; Phenomenex, Torrance, CA, USA) were selected for optimization, by following manufacturers’ guidance, with some modifications. Specifically, cartridges were conditioned by passage of solvents - [1.5 mL of ACN followed by 1.5 mL of 0.145 M SPMM buffer (pH 2.0)] and [1.5 mL of MeOH followed by 1.5 mL of water] for ABS/HLB and X-AW cartridges, respectively. As charged molecules are not retained in ABS/HLB cartridges, enzymatically hydrolyzed sample was pH adjusted by the addition of 1 mL of 0.145 M SPMM buffer (pH 2; “pKa-rule”) prior to loading. For X-AW cartridge, 1.0 mL of 25 mM NH4Ac buffer (pH 6.5) was used. Then, the urine samples were loaded onto cartridges and washed with 2.0 mL of water and 2.0 mL of 25 mM NH4Ac buffer (pH 6.5) for ABS/HLB and X-AW cartridges, respectively. After dried under vacuum for 5 min, the target analytes were eluted by [1.5 mL of ACN and 1.5 mL of EtAc] and [1.5 mL of MeOH and 1.5 mL of 5% NH4OH in MeOH] for ABS/HLB and X-AW cartridges, respectively. The eluate was collected into a 15 mL PP tube and concentrated to near-dryness under a gentle nitrogen stream. The residue was reconstituted in 250 μL mixture of water:ACN (8:2, v/v), vortexed, and transferred into amber glass vials for LC-MS/MS analysis. Among the three SPE cartridges, ABS showed excellent recoveries for all target analytes except for TBBPA, TCBPA, TeCBPA, and OH-PAHs (<10%), which suggested that ACN and EtAc were not adequate to elute compounds with high log Kow values. Nevertheless, ABS cartridge was selected and the elution and washing steps were further optimized to recover chemicals with high log Kow. A mixture solvent of MeOH and DCM or acetone was used to test the elution and found that recoveries of the target compounds with high log Kow values increased (>90%) with 50% DCM in MeOH. Besides, we found that 1.5 mL of 0.1 M FA in water followed by 1.5 mL of 5% MeOH in water as washing solvents considerably reduced matrix interferences and ion suppression while improving sensitivity and robustness of the method.
3.3. Method validation
The method was validated by determining precision, accuracy, linearity (dynamic range), limit of detection (LOD), and matrix effect (Table 3). In this study, 92 of the 121 target compounds had corresponding IS available for isotopic dilution method of analysis. Alternative ISs, with properties similar to those of the target compounds, were used in the quantification of remaining 29 analytes. Instrument calibration range (linearity) was verified based on a 12-point matrix-matched calibration curve prepared in synthetic urine (Cerilliant, Round Rock, TX, USA), at concentrations ranging from 0.01 to 100 ng/mL (with 10 ng/mL ISs). More than 90% of the analytes showed excellent linearity with regression coefficient (r2) values >0.99 except for PA, mBzTP, TPhP, BP-6, BPB, BPP, Tetra-CBPA, BADGE·H2O, BADGE·2H2O, cis-DCCA, metribuzin, and propiconazole for which r2 values were >0.95 (Table 3).
Table 3.
Regression coefficient (linearity of calibration; r2), accuracy and precision from intra-day and inter-day measurements, recoveries, matrix effect results, and limit of detection (LOD) for 121 target compounds analyzed in this study.
| No. | Synonym | r2 | Accuracy and Precision (Intra-Day) | Accuracy and Precision (Inter-Day) | Recovery (%) | ME (%) 10 ng/mL | LOD (ng/mL) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 ng/mL | 10 ng/mL | 20 ng/mL | 1 ng/mL | 10 ng/mL | 20 ng/mL | 1 ng/mL | 10 ng/mL | 20 ng/mL | ||||||||||||||
| RE/% | RSD/% | RE/% | RSD/% | RE/% | RSD/% | RE/% | RSD/% | RE/% | RSD/% | RE/% | RSD/% | RSD/% | RSD/% | RSD/% | ||||||||
| 1 | PA | 0.9875 | −6.2 | 6.7 | 1.9 | 7.5 | −3.6 | 5.1 | −2.8 | 4.7 | 5.6 | 4.8 | −0.85 | 1.8 | 106 | 7.9 | 112 | 10 | 93 | 6.5 | −19 | 5.0 |
| 2 | mMP | 0.9923 | −3.7 | 6.2 | −11 | 9.5 | 0.55 | 2.5 | −0.90 | 5.7 | −7.2 | 4.0 | 1.7 | 1.5 | 94 | 1.0 | 92 | 3.8 | 95 | 4.4 | 31 | 0.052 |
| 3 | mEP | 0.9993 | −8.8 | 8.3 | −6.0 | 5.7 | −5.5 | 6.0 | −3.6 | 4.9 | −3.5 | 1.8 | −3.4 | 3.3 | 106 | 0.67 | 99 | 5.5 | 98 | 5.7 | 31 | 0.014 |
| 4 | mIPrP | 0.9991 | −3.7 | 3.3 | −12 | 9.6 | −1.4 | 4.8 | −2.3 | 1.7 | −8.7 | 3.4 | 0.73 | 2.5 | 99 | 2.1 | 93 | 0.92 | 97 | 9.0 | 29 | 0.012 |
| 5 | mPrP | 0.9991 | −5.9 | 6.2 | −10 | 8.2 | −1.8 | 5.2 | −4.4 | 5.2 | −7.2 | 4.3 | 0.27 | 3.8 | 98 | 4.1 | 94 | 4.3 | 94 | 3.7 | 32 | 0.106 |
| 6 | mIBP | 0.9985 | −4.3 | 7.8 | −13 | 9.1 | 9.6 | 8.4 | 0.91 | 4.4 | −10 | 7.0 | 14 | 4.2 | 96 | 3.5 | 98 | 4.8 | 90 | 0.39 | 28 | 0.238 |
| 7 | mBP | 0.9985 | −4.8 | 8.6 | −13 | 9.0 | −3.0 | 4.7 | 1.4 | 6.8 | −11 | 6.7 | −2.6 | 5.1 | 92 | 0.69 | 92 | 1.6 | 100 | 10 | 28 | 0.102 |
| 8 | mPeP | 0.9999 | −3.6 | 5.8 | −4.3 | 8.1 | −9.7 | 9.0 | −2.3 | 5.5 | −2.2 | 7.9 | −6.5 | 4.5 | 93 | 12 | 96 | 2.0 | 97 | 1.8 | 31 | 0.036 |
| 9 | mHxP | 0.9997 | −1.3 | 5.3 | −10 | 8.3 | −5.5 | 7.2 | −2.1 | 4.5 | −7.0 | 4.7 | −4.3 | 3.6 | 102 | 5.2 | 96 | 2.0 | 100 | 6.7 | 32 | 0.072 |
| 10 | mCPP | 0.9956 | −10 | 5.6 | −2.8 | 3.8 | −1.1 | 2.0 | −10 | 5.6 | −1.2 | 2.3 | −1.5 | 2.0 | 101 | 1.4 | 101 | 2.5 | 101 | 9.1 | 34 | 0.068 |
| 11 | mBzP | 0.9997 | −11 | 8.9 | −3.6 | 4.8 | −2.5 | 7.4 | −10 | 9.7 | −2.0 | 3.8 | −0.96 | 6.1 | 94 | 8.8 | 86 | 4.2 | 86 | 5.7 | 16 | 0.022 |
| 12 | mHpP | 0.9963 | −4.5 | 5.8 | −12 | 9.7 | −5.0 | 5.6 | −0.72 | 3.9 | −9.5 | 4.0 | −2.5 | 2.3 | 97 | 7.1 | 97 | 1.4 | 101 | 3.1 | 32 | 0.010 |
| 13 | mOP | 0.9983 | −1.0 | 4.8 | −6.5 | 5.6 | −7.4 | 8.1 | 1.2 | 3.3 | −4.4 | 3.9 | −5.0 | 4.6 | 101 | 3.7 | 94 | 1.2 | 97 | 10 | 26 | 0.042 |
| 14 | mEHP | 0.9912 | −4.8 | 5.1 | −14 | 6.9 | −4.6 | 7.9 | −1.9 | 2.5 | −10 | 3.9 | −0.62 | 3.2 | 114 | 8.9 | 106 | 5.9 | 94 | 4.5 | 29 | 2.0 |
| 15 | mEOHP | 0.9996 | −3.7 | 4.0 | −11 | 8.6 | −1.3 | 4.1 | −1.8 | 1.7 | −8.0 | 3.3 | 0.56 | 2.3 | 101 | 4.2 | 96 | 6.0 | 94 | 6.0 | 31 | 0.018 |
| 16 | mEHHP | 0.9995 | 0.98 | 5.6 | −9.6 | 8.4 | −0.74 | 4.0 | 1.6 | 6.0 | −6.2 | 5.1 | 0.99 | 2.8 | 101 | 2.2 | 98 | 5.2 | 100 | 4.5 | 30 | 0.012 |
| 17 | mIDP | 0.9998 | −2.0 | 6.2 | 3.1 | 5.0 | 0.73 | 3.8 | −0.62 | 5.3 | 2.2 | 4.0 | 2.0 | 3.1 | 94 | 2.2 | 84 | 2.1 | 85 | 1.6 | 14 | 0.020 |
| 18 | mECPP | 0.9972 | −9.6 | 9.4 | −5.4 | 3.7 | −12 | 8.0 | −3.5 | 3.6 | −3.9 | 3.1 | −12 | 7.3 | 107 | 0.66 | 86 | 5.9 | 98 | 6.1 | 32 | 0.060 |
| 19 | mCMHP | 0.9948 | 2.3 | 7.5 | 4.6 | 6.1 | −2.4 | 4.9 | 3.1 | 3.6 | −1.6 | 4.6 | 1.2 | 5.1 | 105 | 4.7 | 98 | 9.5 | 103 | 5.0 | 29 | 0.020 |
| 20 | mCHpP | 0.9992 | −9.4 | 6.3 | −4.4 | 7.0 | −0.46 | 6.6 | −7.2 | 4.9 | −2.0 | 5.8 | 3.2 | 3.1 | 103 | 7.5 | 102 | 0.70 | 98 | 7.6 | 49 | 0.080 |
| 21 | mCIOP | 0.9998 | −2.1 | 5.3 | −14 | 4.6 | −3.7 | 6.7 | 1.0 | 4.0 | −8.5 | 5.3 | −1.9 | 5.3 | 97 | 4.3 | 96 | 5.5 | 97 | 7.6 | 29 | 0.038 |
| 22 | mCINP | 0.9993 | −4.6 | 4.5 | −12 | 4.9 | −3.3 | 5.8 | −2.3 | 2.3 | −7.1 | 4.7 | −0.73 | 2.8 | 92 | 1.7 | 98 | 7.5 | 97 | 6.1 | 26 | 0.044 |
| 23 | mHiDP | 0.9982 | −1.8 | 3.2 | −11 | 7.4 | −1.2 | 4.0 | −0.17 | 1.8 | −7.7 | 3.9 | 0.38 | 2.9 | 101 | 1.9 | 98 | 9.4 | 99 | 5.0 | 20 | 0.020 |
| 24 | mHiNP | 0.9970 | −4.0 | 3.4 | −6.5 | 8.6 | −3.8 | 5.5 | −3.8 | 2.7 | −3.7 | 7.4 | −0.87 | 3.3 | 88 | 3.1 | 97 | 8.2 | 95 | 9.2 | 22 | 0.034 |
| 25 | mHNCH | 0.9997 | −6.4 | 5.4 | −11 | 8.8 | −7.6 | 8.3 | −3.5 | 3.1 | −8.5 | 4.1 | −4.5 | 4.6 | 102 | 0.70 | 98 | 3.4 | 96 | 8.8 | 21 | 0.018 |
| 26 | mONCH | 0.9994 | −4.3 | 6.2 | −9.8 | 8.9 | −10 | 7.4 | −1.9 | 4.1 | −5.4 | 3.6 | −8.7 | 2.9 | 101 | 6.7 | 94 | 3.5 | 92 | 2.6 | 19 | 0.046 |
| 27 | mPOHP | 0.9998 | −3.5 | 4.3 | −11 | 6.7 | −3.3 | 5.3 | −0.56 | 2.9 | −7.3 | 4.4 | −1.4 | 3.1 | 100 | 1.2 | 97 | 4.0 | 98 | 5.0 | 22 | 0.006 |
| 28 | mPHHP | 0.9989 | 0.05 | 3.3 | −9.1 | 8.3 | −3.2 | 4.9 | 2.2 | 2.1 | −5.6 | 3.0 | −1.0 | 2.2 | 102 | 1.3 | 96 | 4.5 | 94 | 2.6 | 22 | 0.014 |
| 29 | mCOCH | 0.9984 | −3.5 | 4.5 | −12 | 5.1 | 1.0 | 4.4 | −1.2 | 3.8 | −7.0 | 4.3 | 3.5 | 2.2 | 94 | 2.0 | 96 | 7.9 | 96 | 7.3 | 24 | 0.008 |
| 30 | mPCHP | 0.9990 | −1.9 | 4.2 | −13 | 4.7 | −3.4 | 5.4 | −0.19 | 3.7 | −8.4 | 5.3 | −0.91 | 3.1 | 95 | 3.8 | 97 | 2.4 | 98 | 3.9 | 25 | 0.012 |
| 31 | mETP | 0.9992 | −2.2 | 6.5 | −10 | 9.2 | 0.15 | 5.9 | 2.3 | 3.2 | −7.4 | 3.3 | 2.6 | 4.5 | 109 | 2.5 | 105 | 2.6 | 102 | 1.3 | 31 | 0.006 |
| 32 | mTBTP | 0.9982 | −1.6 | 7.0 | −12 | 9.2 | −0.09 | 3.1 | 1.0 | 7.0 | −9.0 | 5.1 | −1.1 | 3.0 | 90 | 9.0 | 96 | 2.2 | 98 | 0.73 | 30 | 0.026 |
| 33 | mBzTP | 0.9846 | −3.4 | 3.8 | −10 | 9.6 | −5.0 | 4.3 | −1.5 | 3.7 | −6.9 | 4.7 | −3.3 | 2.4 | 109 | 5.5 | 109 | 3.1 | 117 | 1.4 | 30 | 1.0 |
| 34 | TEP | 0.9973 | −3.3 | 5.0 | 5.2 | 3.2 | −2.5 | 4.4 | −4.5 | 4.8 | 4.9 | 3.6 | −2.3 | 3.4 | 106 | 5.3 | 109 | 2.6 | 102 | 1.0 | −88 | 0.028 |
| 35-1 | TNBP | 0.9953 | 2.0 | 2.8 | 3.3 | 2.5 | −0.63 | 3.0 | 0.89 | 2.2 | 3.1 | 1.6 | −2.2 | 2.0 | 96 | 0.44 | 94 | 2.2 | 88 | 1.7 | −56 | 0.016 |
| 35-2 | TIBP | |||||||||||||||||||||
| 37 | TCEP | 0.9995 | −4.8 | 6.1 | 2.1 | 4.5 | −6.2 | 5.1 | −4.1 | 6.0 | 0.82 | 3.6 | −5.4 | 4.1 | 97 | 8.3 | 104 | 3.4 | 102 | 2.0 | −18 | 0.028 |
| 38 | TPP | 0.9979 | 1.0 | 2.3 | −0.34 | 3.0 | −0.61 | 3.0 | −0.32 | 2.0 | −1.4 | 2.7 | −2.4 | 0.90 | 109 | 2.5 | 105 | 4.7 | 99 | 6.8 | −6 | 0.016 |
| 39 | TPhP | 0.9664 | −0.55 | 3.3 | 1.1 | 2.6 | −1.0 | 2.4 | 1.3 | 2.4 | 0.38 | 1.6 | −1.4 | 2.4 | 95 | 3.8 | 81 | 2.2 | 93 | 1.1 | −42 | 0.074 |
| 40 | TBOEP | 0.9978 | 4.3 | 3.5 | 2.1 | 2.5 | −0.12 | 3.0 | 3.1 | 3.3 | 2.0 | 1.8 | −2.0 | 0.90 | 96 | 3.7 | 92 | 2.0 | 97 | 9.4 | −63 | 0.036 |
| 41-1 | DNBP | 0.9998 | −4.6 | 3.0 | −6.30 | 5.6 | −3.2 | 4.3 | −3.5 | 1.1 | −3.9 | 2.4 | −1.4 | 2.4 | 101 | 1.8 | 99 | 6.5 | 97 | 3.7 | 23 | 0.030 |
| 41-2 | DIBP | |||||||||||||||||||||
| 43 | DPhP | 0.9980 | −3.2 | 2.8 | −7.7 | 6.9 | −2.1 | 3.8 | −1.8 | 0.90 | −4.7 | 2.4 | −0.52 | 2.0 | 98 | 3.9 | 101 | 0.70 | 102 | 9.7 | −2 | 0.028 |
| 44 | BMPP | 0.9990 | 3.7 | 4.4 | −2.0 | 3.7 | 5.6 | 5.3 | 0.93 | 3.5 | −3.7 | 1.8 | 3.9 | 4.8 | 103 | 2.7 | 94 | 8.3 | 94 | 6.3 | 11 | 0.050 |
| 45 | BDCIPP | 0.9997 | 8.9 | 8.6 | −3.0 | 2.3 | −0.29 | 2.6 | 4.4 | 2.8 | −2.3 | 1.8 | −0.38 | 2.7 | 97 | 5.2 | 103 | 0.69 | 103 | 5.5 | 20 | 0.200 |
| 46 | MeP | 0.9957 | 3.1 | 5.4 | −1.6 | 2.0 | −0.06 | 0.90 | 0.78 | 4.6 | −2.3 | 1.8 | 0.28 | 0.70 | 84 | 3.7 | 88 | 6.1 | 91 | 6.9 | 41 | 0.048 |
| 47 | EtP | 0.9964 | 0.78 | 7.2 | −0.86 | 1.6 | −0.44 | 2.3 | 0.41 | 7.4 | −1.5 | 0.70 | −0.94 | 2.1 | 95 | 1.2 | 96 | 3.6 | 96 | 9.1 | 37 | 0.040 |
| 48 | PrP | 0.9969 | −0.64 | 8.9 | −1.5 | 1.9 | −2.6 | 1.6 | 0.04 | 9.0 | −2.4 | 1.2 | −2.8 | 1.8 | 99 | 0.93 | 95 | 4.6 | 99 | 6.4 | 37 | 0.010 |
| 49 | BuP | 0.9920 | 0.97 | 3.4 | 1.4 | 1.3 | −0.94 | 1.3 | −0.64 | 3.3 | 1.1 | 0.60 | −0.86 | 1.3 | 80 | 2.6 | 95 | 3.2 | 100 | 6.0 | 30 | 0.016 |
| 50 | BzP | 0.9989 | 5.8 | 3.7 | −1.1 | 1.6 | −1.1 | 1.3 | 4.5 | 3.6 | −0.93 | 1.8 | −1.1 | 1.3 | 97 | 2.1 | 97 | 4.1 | 100 | 6.4 | 27 | 0.010 |
| 51 | HeP | 0.9983 | −6.3 | 8.6 | −0.96 | 3.0 | 0.19 | 1.1 | −2.5 | 4.6 | −1.2 | 3.0 | 0.43 | 0.80 | 72 | 3.6 | 84 | 2.0 | 89 | 4.7 | −31 | 0.010 |
| 52 | BP-1 | 0.9991 | 3.4 | 7.5 | 3.2 | 2.4 | −3.2 | 3.8 | 1.4 | 7.7 | 3.3 | 2.3 | −3.2 | 4.2 | 107 | 1.9 | 93 | 3.8 | 95 | 5.9 | 27 | 0.028 |
| 53 | BP-2 | 0.9987 | −4.7 | 4.7 | 1.6 | 1.5 | −0.75 | 1.8 | −2.6 | 4.4 | 1.4 | 1.6 | −0.71 | 2.0 | 107 | 3.4 | 86 | 5.7 | 85 | 8.3 | 40 | 0.012 |
| 54 | BP-3 | 0.9986 | −9.3 | 8.8 | −0.53 | 4.9 | −3.3 | 1.8 | −4.2 | 4.9 | 1.7 | 2.8 | −3.2 | 1.7 | 96 | 1.6 | 92 | 2.0 | 96 | 3.7 | −56 | 0.010 |
| 55 | BP-6 | 0.9857 | 0.10 | 2.9 | 1.1 | 2.5 | −1.3 | 1.8 | 1.6 | 2.0 | 2.4 | 1.8 | −1.4 | 1.8 | 88 | 1.5 | 100 | 5.9 | 104 | 4.7 | −53 | 0.028 |
| 56 | BP-8 | 0.9994 | 6.79 | 4.5 | −0.09 | 3.1 | −0.11 | 1.6 | 7.5 | 4.5 | 1.2 | 2.2 | −0.10 | 1.7 | 101 | 3.2 | 98 | 4.0 | 95 | 5.4 | −4 | 0.012 |
| 57 | 4-OH-BP | 0.9929 | −0.75 | 3.1 | −0.85 | 2.6 | −0.45 | 1.7 | −2.0 | 2.3 | −2.2 | 1.5 | −1.2 | 1.2 | 106 | 1.3 | 100 | 2.1 | 96 | 2.2 | 34 | 0.026 |
| 58 | BPA | 0.9953 | 0.79 | 5.8 | 1.4 | 4.8 | −0.45 | 2.0 | −1.3 | 2.6 | −0.89 | 3.4 | −0.34 | 2.2 | 85 | 6.4 | 94 | 9.1 | 94 | 7.3 | 48 | 0.124 |
| 59 | BPF | 0.9943 | 2.2 | 9.7 | 0.30 | 1.8 | 1.1 | 3.0 | 8.6 | 5.6 | −0.07 | 1.6 | −0.04 | 2.6 | 112 | 5.7 | 94 | 1.6 | 97 | 5.8 | 51 | 0.120 |
| 60 | BPS | 0.9935 | −4.2 | 4.4 | 1.0 | 2.0 | −2.5 | 2.7 | −3.3 | 4.0 | 1.5 | 1.9 | −3.4 | 2.1 | 81 | 8.2 | 96 | 0.74 | 99 | 4.6 | 49 | 0.008 |
| 61 | BPB | 0.9533 | −8.5 | 6.5 | −2.3 | 2.6 | 0.59 | 2.6 | −1.6 | 6.8 | −1.5 | 2.2 | −0.37 | 2.0 | 119 | 4.3 | 88 | 2.6 | 89 | 2.9 | 42 | 0.112 |
| 62 | BPZ | 0.9947 | −4.2 | 8.2 | −0.14 | 4.0 | −2.5 | 2.0 | 0.87 | 4.7 | 2.3 | 1.3 | −1.8 | 0.80 | 103 | 6.2 | 95 | 6.3 | 97 | 6.2 | 38 | 0.102 |
| 63 | BPAP | 0.9945 | −8.2 | 9.8 | 2.2 | 2.3 | −1.5 | 2.0 | −2.4 | 7.0 | 3.1 | 1.7 | −1.1 | 1.3 | 98 | 3.1 | 97 | 5.6 | 91 | 7.7 | 35 | 0.080 |
| 64 | BPAF | 0.9996 | −2.6 | 4.9 | 0.33 | 1.8 | −1.2 | 1.6 | −0.05 | 3.0 | 0.36 | 2.0 | −1.8 | 1.2 | 105 | 3.3 | 93 | 2.6 | 90 | 5.9 | 33 | 0.054 |
| 65 | BPP | 0.9745 | −8.1 | 10 | −0.76 | 4.8 | −1.4 | 1.6 | −0.27 | 9.4 | 1.3 | 3.7 | −1.2 | 1.1 | 88 | 0.32 | 84 | 5.2 | 90 | 6.7 | −77 | 0.142 |
| 66-1 | 2,4,5-TCP | 0.9968 | 0.56 | 6.5 | −4.8 | 8.1 | 4.8 | 6.6 | 2.3 | 5.7 | −6.8 | 7.3 | 1.8 | 4.5 | 103 | 4.1 | 101 | 1.4 | 95 | 6.2 | 29 | 0.020 |
| 66-2 | 2,4,6-TCP | |||||||||||||||||||||
| 68 | 2,3,5,6-TeCP | 0.9961 | 1.6 | 4.4 | 2.6 | 2.8 | −0.69 | 0.50 | 1.5 | 4.1 | 3.0 | 2.2 | −0.53 | 0.40 | 96 | 2.5 | 106 | 3.1 | 94 | 5.6 | 22 | 0.046 |
| 69 | 2,3,4,6-TeCP | 0.9942 | −2.7 | 5.0 | 1.5 | 3.0 | 1.5 | 1.8 | −2.8 | 5.3 | 1.7 | 3.0 | 1.6 | 2.0 | 106 | 6.3 | 94 | 5.1 | 97 | 1.6 | −10 | 0.044 |
| 70 | 2,3,4,5-TeCP | 0.9937 | −0.43 | 4.2 | 2.2 | 2.7 | −1.3 | 1.9 | −1.0 | 4.5 | 2.5 | 2.6 | −1.4 | 2.0 | 104 | 4.0 | 91 | 1.7 | 104 | 8.8 | −21 | 0.014 |
| 71 | PCP | 0.9978 | 6.9 | 5.6 | −0.63 | 2.7 | −1.7 | 1.3 | 4.9 | 4.7 | −0.36 | 2.9 | −1.8 | 1.3 | 109 | 5.8 | 97 | 0.66 | 95 | 5.9 | 10 | 0.026 |
| 72 | TCC | 0.9966 | −6.4 | 6.2 | −2.4 | 4.7 | −2.2 | 2.2 | −3.9 | 4.7 | −2.5 | 3.6 | −2.0 | 2.2 | 85 | 0.25 | 84 | 1.4 | 87 | 3.2 | −98 | 0.034 |
| 73 | TCS | 0.9935 | −5.8 | 4.7 | −1.7 | 4.2 | −0.97 | 1.9 | −6.5 | 2.8 | −1.7 | 4.1 | −0.49 | 1.5 | 109 | 3.8 | 108 | 5.5 | 101 | 4.9 | −49 | 0.052 |
| 74 | TCBPA | 0.9912 | 1.1 | 4.9 | −3.3 | 3.5 | −4.4 | 2.1 | 2.9 | 4.2 | −4.4 | 3.5 | −4.9 | 1.4 | 109 | 6.7 | 104 | 2.6 | 103 | 7.2 | 14 | 0.080 |
| 75 | TeCBPA | 0.9875 | 3.9 | 9.8 | −5.0 | 5.7 | −1.3 | 3.9 | −1.4 | 4.3 | −4.4 | 3.2 | −3.2 | 2.7 | 112 | 5.7 | 105 | 6.3 | 98 | 5.9 | 23 | 0.072 |
| 76 | TBBPA | 0.9964 | −0.95 | 6.3 | 0.15 | 4.0 | 0.28 | 2.9 | −2.7 | 6.6 | 6.7 | 5.8 | −1.2 | 2.0 | 98 | 7.9 | 96 | 2.0 | 103 | 4.3 | 17 | 0.034 |
| 77 | BADGE | 0.9979 | −2.7 | 6.7 | 3.0 | 2.7 | −1.3 | 2.6 | −4.2 | 6.3 | 2.3 | 2.1 | −1.9 | 2.1 | 93 | 2.1 | 88 | 7.8 | 93 | 1.9 | −5 | 0.400 |
| 78 | BADGE·H2O | 0.9568 | −7.0 | 6.7 | 1.1 | 1.8 | −1.6 | 2.4 | −9.1 | 4.5 | 0.96 | 1.8 | −1.0 | 1.9 | 118 | 1.1 | 78 | 7.0 | 81 | 3.4 | 34 | 0.322 |
| 79 | BADGE·2H2O | 0.9690 | 1.4 | 6.1 | 1.2 | 3.3 | 2.6 | 3.5 | 0.29 | 6.8 | 1.0 | 2.4 | 1.8 | 2.8 | 128 | 9.4 | 62 | 0.72 | 68 | 6.8 | 42 | 0.146 |
| 80 | 1-Nap | 0.9909 | 3.8 | 8.0 | 2.6 | 2.2 | −1.2 | 3.0 | −1.5 | 5.4 | 2.0 | 1.8 | −1.5 | 2.4 | 86 | 4.6 | 94 | 2.4 | 97 | 2.5 | 60 | 0.080 |
| 81 | 2-Nap | 0.9967 | 6.2 | 7.5 | −0.92 | 1.7 | −2.0 | 1.2 | 6.9 | 7.7 | −0.17 | 1.4 | −2.1 | 1.2 | 75 | 5.0 | 92 | 2.3 | 97 | 4.0 | 34 | 0.050 |
| 82-1 | 2-Fluo | 0.9996 | 0.29 | 7.9 | 1.2 | 1.8 | 1.3 | 1.3 | −3.3 | 5.5 | 1.5 | 1.8 | 1.6 | 1.3 | 103 | 7.8 | 90 | 7.8 | 96 | 5.5 | 24 | 0.026 |
| 82-2 | 3-Fluo | |||||||||||||||||||||
| 82-3 | 9-Fluo | |||||||||||||||||||||
| 85-1 | 2-Phen | 0.9986 | −4.0 | 6.7 | 1.1 | 3.1 | 0.42 | 1.3 | −1.7 | 7.4 | 2.0 | 2.9 | −0.17 | 0.90 | 97 | 2.3 | 91 | 2.8 | 90 | 3.5 | 26 | 0.020 |
| 85–2 | 3-Phen | |||||||||||||||||||||
| 87–1 | 1-Phen | 0.9987 | 4.2 | 8.4 | 3.2 | 4.7 | −0.95 | 2.6 | −1.9 | 3.7 | 4.6 | 3.2 | −2.3 | 1.6 | 112 | 8.8 | 96 | 3.6 | 103 | 3.1 | 23 | 0.020 |
| 87–2 | 9-Phen | |||||||||||||||||||||
| 89 | 4-Phen | 0.9995 | 3.2 | 7.0 | 3.3 | 2.9 | 1.1 | 1.7 | 0.75 | 6.2 | 2.6 | 2.4 | 1.2 | 1.7 | 103 | 1.9 | 92 | 2.2 | 92 | 4.6 | 18 | 0.030 |
| 90 | 1-Pyr | 0.9991 | 0.56 | 4.7 | 9.7 | 10 | 6.1 | 10 | −0.50 | 4.9 | 11 | 7.4 | 1.8 | 7.6 | 106 | 6.3 | 103 | 2.5 | 97 | 4.7 | 17 | 0.020 |
| 91 | nitenpyram | 0.9993 | −2.3 | 5.9 | 6.8 | 3.8 | 1.1 | 6.0 | −2.8 | 6.2 | 5.3 | 2.4 | −0.74 | 4.6 | 111 | 0.64 | 101 | 6.3 | 95 | 6.3 | 60 | 0.108 |
| 92 | thiamethoxam | 0.9989 | 9.0 | 7.6 | 2.4 | 3.4 | 1.8 | 4.4 | 6.1 | 3.4 | 4.1 | 2.6 | 0.49 | 4.2 | 106 | 1.3 | 90 | 3.0 | 104 | 0.34 | 21 | 0.036 |
| 93 | imidacloprid | 0.9986 | 9.6 | 5.5 | 2.7 | 3.9 | 0.05 | 3.2 | 9.7 | 4.9 | 1.1 | 1.9 | −1.1 | 2.4 | 95 | 5.2 | 94 | 4.7 | 93 | 3.0 | 38 | 0.096 |
| 94 | acetamiprid | 0.9995 | 4.6 | 4.8 | 3.2 | 2.8 | 0.56 | 4.1 | 4.2 | 4.4 | 2.4 | 1.8 | −1.1 | 2.9 | 108 | 0.66 | 96 | 0.52 | 94 | 6.7 | 38 | 0.032 |
| 95 | thiacloprid | 0.9995 | 0.88 | 1.7 | 0.79 | 1.4 | 1.4 | 2.8 | 0.69 | 1.4 | 0.53 | 1.4 | −0.17 | 1.5 | 106 | 1.3 | 95 | 4.3 | 98 | 2.5 | 37 | 0.068 |
| 96 | clothianidin | 0.9989 | −6.0 | 4.1 | 6.3 | 3.2 | 0.19 | 5.7 | −7.0 | 4.2 | 5.3 | 2.1 | −3.2 | 1.8 | 105 | 2.0 | 91 | 2.0 | 105 | 2.3 | 61 | 0.026 |
| 97 | flonicamid | 0.9973 | 3.3 | 6.2 | −2.4 | 8.0 | −0.89 | 7.6 | 1.4 | 6.7 | −3.4 | 8.5 | 0.99 | 7.7 | 156 | 5.3 | 107 | 4.6 | 88 | 4.4 | 63 | 0.200 |
| 98 | N-Dmt | 0.9920 | 1.1 | 4.7 | 2.9 | 4.4 | 3.1 | 7.4 | −1.7 | 2.1 | 0.77 | 2.5 | 0.08 | 6.6 | 138 | 5.8 | 102 | 1.3 | 91 | 8.1 | 31 | 0.024 |
| 99 | N-Dma | 0.9996 | 0.82 | 3.7 | −0.12 | 3.3 | 2.8 | 4.1 | −0.85 | 3.0 | −1.2 | 2.7 | 1.0 | 1.7 | 114 | 8.6 | 100 | 9.2 | 99 | 3.9 | 63 | 0.060 |
| 100 | Ta | 0.9995 | −0.07 | 3.9 | 2.4 | 2.6 | 4.5 | 6.2 | −2.8 | 2.7 | 1.8 | 2.2 | 1.7 | 4.2 | 106 | 2.6 | 99 | 5.1 | 96 | 1.6 | 59 | 0.010 |
| 101 | imidaclothiz | 0.9976 | 7.8 | 4.4 | 6.0 | 3.4 | 2.7 | 3.8 | 5.3 | 2.7 | 7.3 | 2.9 | 1.9 | 1.6 | 95 | 2.1 | 104 | 4.7 | 91 | 11 | 37 | 0.048 |
| 102 | 6-Cn | 0.9991 | −6.8 | 5.9 | −12 | 6.4 | −1.9 | 4.8 | −4.1 | 3.4 | −8.3 | 4.4 | 0.29 | 2.1 | 103 | 6.9 | 95 | 1.6 | 100 | 3.1 | 31 | 0.046 |
| 103 | sulfoxaflor | 0.9998 | −0.09 | 6.6 | 2.1 | 2.5 | −0.40 | 2.0 | 0.05 | 5.0 | 2.1 | 2.6 | 0.12 | 1.4 | 106 | 1.3 | 95 | 8.7 | 98 | 4.3 | 42 | 0.008 |
| 104 | 4-nitrophenol | 0.9992 | 0.15 | 7.6 | 1.4 | 3.2 | 1.8 | 1.8 | 5.6 | 3.5 | 0.54 | 2.6 | 0.96 | 1.2 | 98 | 6.7 | 100 | 6.7 | 94 | 2.6 | 37 | 0.176 |
| 105 | 2,4-D | 0.9995 | 8.4 | 3.9 | 1.0 | 2.1 | −4.0 | 4.9 | 7.1 | 3.3 | 0.50 | 2.2 | −6.7 | 2.4 | 109 | 0.6 | 100 | 4.1 | 94 | 1.5 | 38 | 0.058 |
| 106 | TCPY | 0.9974 | −4.2 | 5.1 | −3.3 | 6.3 | −2.1 | 2.4 | −1.4 | 2.0 | 0.05 | 2.7 | −3.0 | 1.7 | 107 | 3.3 | 102 | 1.3 | 97 | 8.7 | 30 | 0.094 |
| 107 | trans-DCCA | 0.9990 | −2.3 | 5.3 | 1.3 | 4.3 | −0.69 | 2.9 | −1.9 | 3.6 | −2.0 | 4.2 | 0.98 | 1.9 | 102 | 1.3 | 103 | 2.0 | 100 | 0.71 | 23 | 0.166 |
| 108 | cis-DCCA | 0.9617 | −2.5 | 7.1 | −8.2 | 8.9 | −3.3 | 5.0 | 0.82 | 5.1 | −4.6 | 4.7 | −1.2 | 2.2 | 98 | 0.72 | 93 | 4.2 | 96 | 5.8 | 30 | 0.100 |
| 109 | 3-PBA | 0.9992 | −4.2 | 5.1 | −12 | 6.4 | −4.1 | 4.9 | −1.9 | 2.8 | −8.0 | 5.6 | −2.1 | 1.9 | 98 | 0.72 | 93 | 4.2 | 96 | 5.8 | 31 | 0.036 |
| 110 | 4F-3PBA | 0.9990 | −3.2 | 4.1 | −13 | 3.7 | −4.0 | 4.9 | −1.1 | 2.2 | −9.4 | 5.4 | −1.8 | 2.6 | 102 | 1.3 | 94 | 5.4 | 97 | 6.1 | 29 | 0.008 |
| 111 | 2,4,5-T | 0.9950 | −7.5 | 3.8 | −9.8 | 8.3 | −3.0 | 4.1 | −7.0 | 2.7 | −6.9 | 3.6 | −1.4 | 1.7 | 111 | 6.3 | 100 | 1.0 | 100 | 11 | 33 | 0.018 |
| 112 | pyrimethanil | 0.9992 | 1.7 | 3.2 | 2.4 | 1.9 | 1.3 | 3.5 | 2.4 | 3.1 | 1.8 | 1.6 | −0.73 | 1.4 | 107 | 4.6 | 94 | 3.2 | 101 | 6.3 | −4 | 0.058 |
| 113 | dinotefuran | 0.9998 | −12 | 11 | 3.9 | 3.8 | −0.45 | 5.8 | −7.4 | 10 | 4.0 | 2.9 | −1.7 | 4.8 | 100 | 3.0 | 100 | 0.92 | 90 | 9.4 | 34 | 0.010 |
| 114 | metribuzin | 0.9690 | −12 | 9.5 | 2.4 | 4.7 | 0.44 | 5.3 | −13 | 9.0 | 2.7 | 4.7 | −0.95 | 4.2 | 97 | 3.4 | 94 | 6.5 | 103 | 6.3 | 36 | 0.100 |
| 115 | atrazine | 0.9989 | 5.8 | 3.1 | 2.9 | 2.7 | 3.0 | 2.9 | 4.9 | 2.9 | 2.3 | 2.2 | 1.6 | 2.4 | 127 | 0.38 | 122 | 4.6 | 117 | 1.9 | 14 | 0.128 |
| 116 | cyprodinil | 0.9994 | 4.6 | 5.2 | 3.6 | 3.8 | −0.97 | 3.0 | 3.6 | 3.9 | 2.4 | 1.2 | −2.4 | 2.1 | 110 | 5.8 | 91 | 2.7 | 96 | 3.7 | −82 | 0.034 |
| 117 | metalaxyl | 0.9994 | 3.8 | 2.8 | 1.4 | 1.6 | 0.36 | 2.5 | 4.0 | 1.7 | 1.3 | 1.4 | −1.0 | 1.2 | 107 | 1.3 | 99 | 2.2 | 99 | 6.4 | 22 | 0.032 |
| 118 | tebuconazole | 0.9992 | −1.1 | 3.9 | 4.6 | 2.8 | −1.8 | 2.5 | −2.7 | 4.0 | 3.5 | 1.7 | −2.7 | 2.4 | 94 | 10 | 92 | 5.9 | 97 | 9.1 | 10 | 0.052 |
| 119 | propiconazole | 0.9743 | 2.8 | 2.1 | 1.0 | 3.7 | −2.3 | 3.2 | 1.9 | 1.8 | −0.40 | 2.1 | −4.0 | 1.7 | 107 | 2.7 | 77 | 4.0 | 83 | 2.1 | −12 | 0.012 |
| 120 | tetraconazole | 0.9991 | 3.9 | 6.0 | 1.2 | 2.7 | −1.2 | 3.2 | 5.5 | 5.1 | 0.27 | 2.2 | −2.8 | 2.2 | 112 | 7.1 | 106 | 6.0 | 102 | 9.0 | 8 | 0.072 |
| 121 | azoxystrobin | 0.9998 | 1.6 | 2.7 | 1.7 | 2.6 | 1.6 | 2.4 | 0.75 | 2.7 | 1.0 | 2.0 | 0.51 | 1.6 | 104 | 4.7 | 97 | 2.5 | 98 | 6.1 | 17 | 0.080 |
RE: relative error; RSD: relative standard deviation; ME: matrix effect; LOD: limit of detection; There is no chromatographic resolution between compounds with same ID; therefore, sum concentrations are expressed for these isomers by using the present developed method.
The LODs were calculated as 3 × Sb/a (Sb: intercept standard deviation; a: slope) from the regression equation obtained for low concentration range (ranging from 0.01 to 5 ng/mL) of spiked urine. The LODs ranged from ≤ 0.1 for 101 and from 0.1–1.0 ng/mL for 18 of the 121 target compounds. These values are comparable to bioanalytical methods reported previously (Table 1). The LODs for PA (5.0 ng/mL) and mEHP (2.0 ng/mL) were relatively high due to the fact that these two chemicals have high background values (many laboratory products and reagents contain these chemicals).
The accuracy is expressed as the relative error (RE, %) of an expected value, and its variation in repeatability (precision) is determined using the relative standard deviation (RSD). According to the guidance for bioanalytical method validation, acceptable accuracy and precision ranges are divided at two fortification levels [38]. First, from −20% to 20% with RSD ≤20% at the limit of quantitation (LOQ) level. Second, from −15% to 15% with RSD ≤15% at higher concentrations [44]. The accuracy and precision were determined at three fortification levels of 1, 10, and 20 ng/mL and the determination of intra- and inter-day variances of five measurements of three different levels injected daily for 6 days. At 1 ng/mL level, RE for the 121 analytes were −13% to 9.7% with RSDs ranging from 1.7% to 11.0% for intra-day measurements and −13% to 9.8% with RSDs from 0.90% to 10% for inter-day measurements (Table 3). At 10 and 20 ng/mL, RE ranges were −14% to 12% with RSDs of 0.6% to 10% for intra-day and −13% to 14% with RSDs of 0.4% to 10% for inter-day variances. Thus, the analytical method satisfied the criteria for accuracy and precision analyzed using fortified urine samples.
The extraction efficiency of the analytical method was considered excellent when the recovery rate of a target compound was near or equal to 100%. Analyte recovery and its variation (RSD) were also used in the determination of accuracy and precision in bioanalytical methods. Generally, a recovery rate of 80–120% with RSD < 20% is an acceptable range for those parameters [39]. The target analytes were fortified in QC samples (i.e., urine pool) at three different levels (i.e., 1, 10, and 20 ng/mL), to represent low, medium, and high concentrations. The QC samples were passed through the optimized sample preparation method described above (i.e., enzymatic incubation followed by SPE extraction). The recovery ranges of target analytes were 72–156% (RSD; 0.25–12%), 62–122% (RSD; 0.52–11%), and 68–117% (RSD; 0.34–12%) at the fortification levels of 1, 10, and 20 ng/mL, respectively (Table 3). Among the 121 analytes, 115, 117, and 120 analytes showed the recovery range of 80–120% with RSD values of <20% at fortification levels of 1, 10, and 20 ng/mL, respectively. Slightly lower recoveries for BADGE·2H2O (62% and 68% at medium and high levels) and higher recoveries for flonicamid (156% at low level) and N-Dmt (138% at low level) were found but the repeatability (RSD; 0.72–6.8%) was acceptable for these compounds.
The matrix effect (ME) was calculated using the following Equation (1):
| (1) |
where A is the peak area of an analyte in a pure solvent and B is the peak area of an analyte spiked in synthetic urine that has undergone sample preparation steps, at the same concentration as the standard solution. Matrix effect of each compound is expressed as the percentage enhancement (<0%) or suppression (>0%).
The results of matrix effect calculated for the 121 target analytes are shown in Table 3. The matrix effects ranged from soft (matrix effect within −20% to 0% or 0% to 20%), moderate (−50% to −20% or 20% and 50%), and strong (below −50% or above 50%) [40]. After optimization of SPE washing step, matrix effect for most compounds (83; 68.2% of total) fell within moderate range (Figure 5). Nevertheless, inclusion of IS as well as matrix-matched calibration curve enabled accurate analysis of target chemicals in urine.
Figure 5. Summary of matrix effects (ME) determined for 121 analytes analyzed in the study.
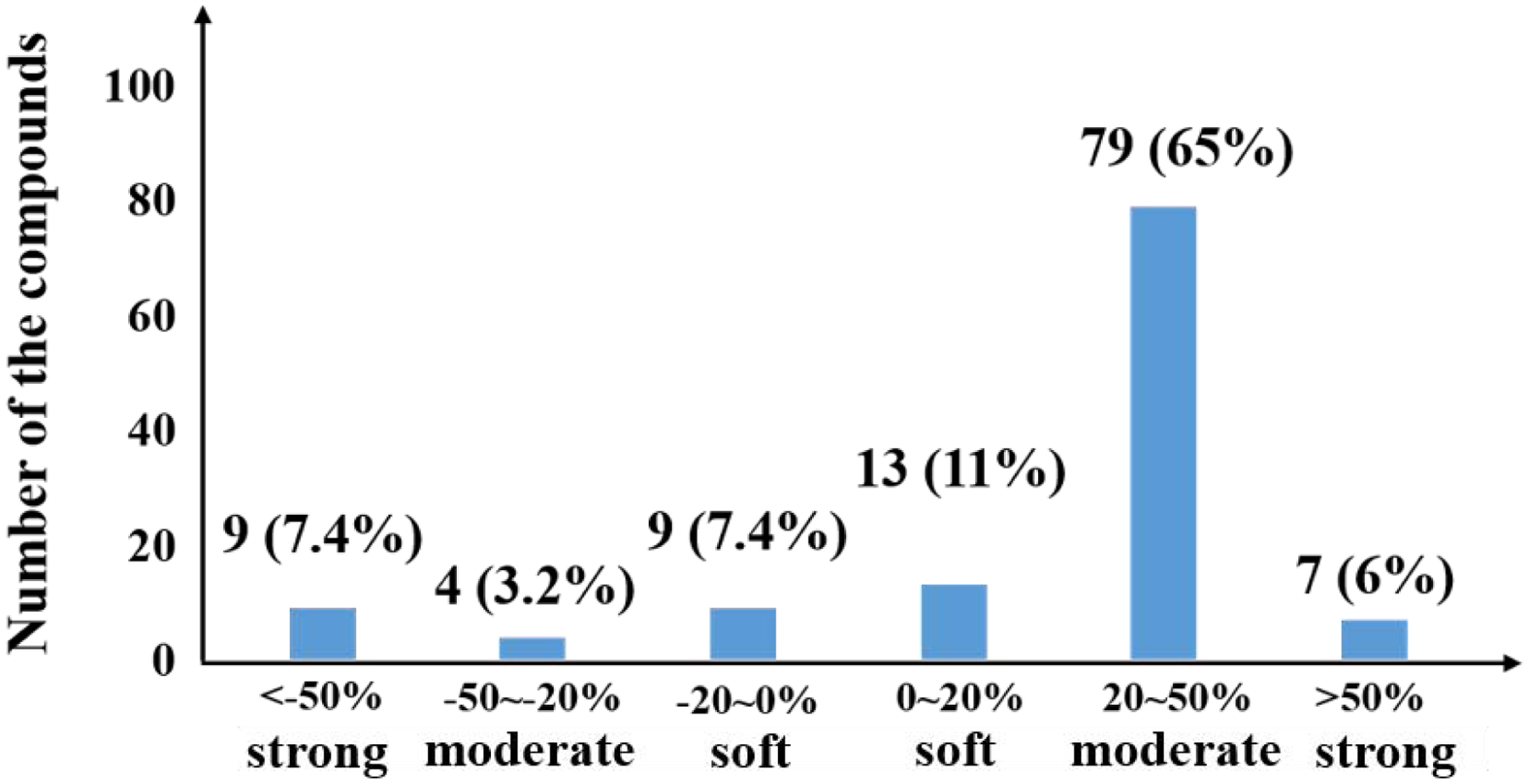
Matrix effects are classified into soft effect (−20% to 20%), moderate effect (−50% to −20% and 20% to 50%), and strong effect (<−50% and >50%). If ME ~0%, there is no matrix effect. ME> 0% indicates an ion-suppression and, ME <0% indicates an ion-enhancement.
The method was further validated through the analysis of NIST SRM 3672 and 3673 as well as nine different PT urine samples from the German-External Quality Assurance Scheme (G-EQUAS; n=4; 65/2020 9A/B and 14/15 A/B), External Quality Assessment Scheme for Organic Substances Program (OSEQAS; n=3; OS-U-E 2004, 2005, and 2006), and Centers for Disease Control and Prevention-Biomonitoring Quality Assurance for State Programs (CDC-BQASP; n=2; 201802001UPSU and 201802002UPSU). Our measurement of 121 target chemicals analyzed by the developed method is shown in Table S6. For those analytes that have been certified in the samples, our method yielded acceptable concentrations, further validating quantitative accuracy, precision and traceability.
3.4. Method application
The method was applied for the analysis of 21 urine samples collected from healthy adults from Albany area of New York state in 2017 (Table S7 and Figure 6). Forty target analytes including 20 PMs, 15 EPs, and 5 pesticides were found in >70% of the urine samples. The sum concentrations of the 121 compounds ranged from 82 to 629 ng/mL, with mean and median values of 215 and 192 ng/mL, respectively. The concentrations were found in the decreasing order of: ∑PMs (sum of 45 PMs; mean: 146 ng/mL) > ∑EPs (sum of 45 EPs; 42 ng/mL) > ∑pesticides (sum of 31 pesticides; 26 ng/mL). PA, mEP, mIBP, mBP, mBzP, mECPP, and mEHHP made up a vast majority of PMs detected in urine samples. BP-3 (mean: 13 ng/mL), 1-Nap (11 ng/mL), and MeP (9.7 ng/mL) were the major EPs found in urine. Among pesticides, TCPY (100%), thiamethoxam (95%), 4-nitrophenol (95%), and 2,4,5-T (90%) were frequently detected in samples. These results were similar to those reported in previous studies [41–43].
Figure 6. Concentrations (ng/mL; A) and profiles (%; B) of target chemicals found in 21 real urine analyzed.
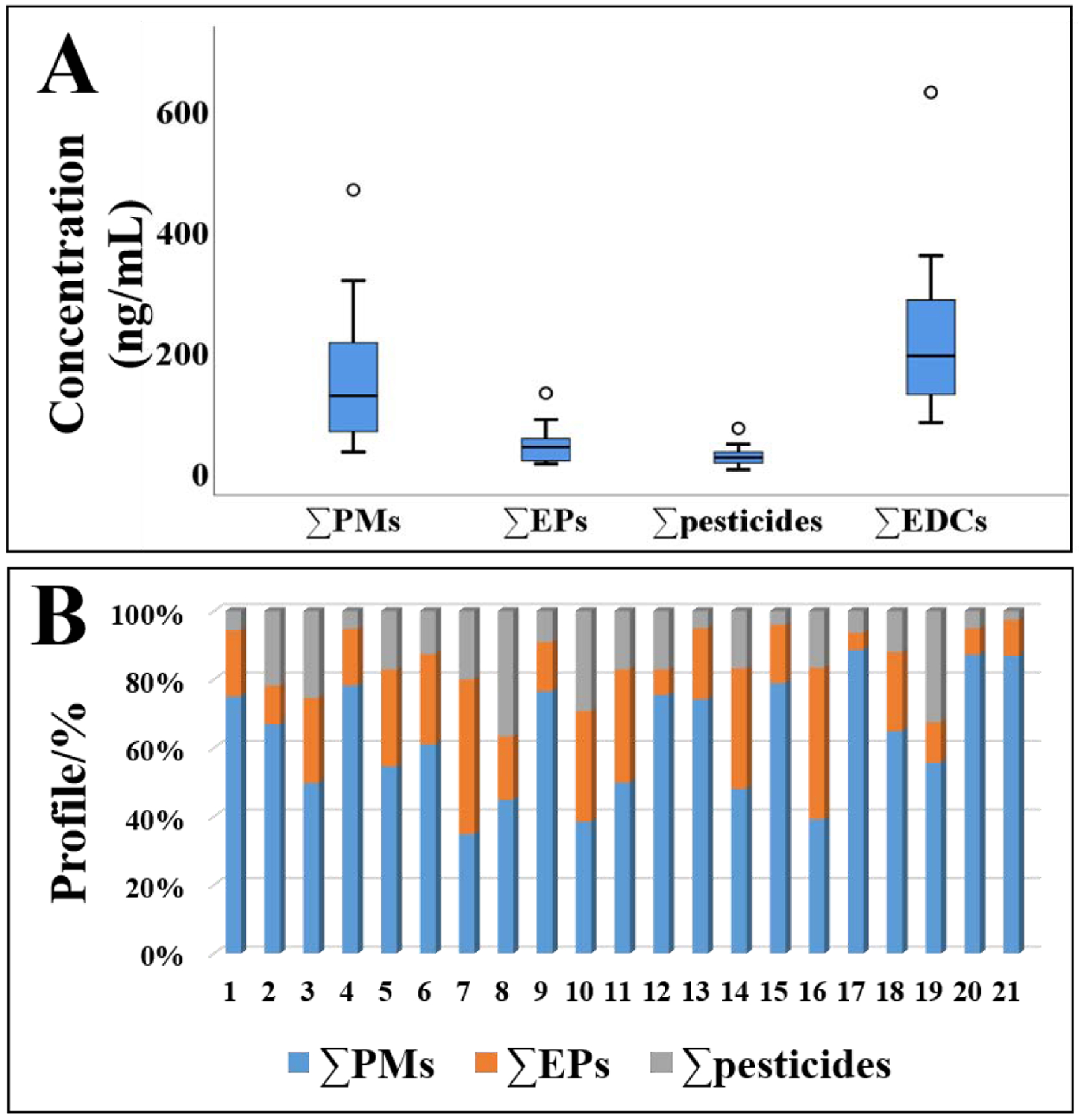
PMs: plasticizers and metabolites; EPs: environmental phenols; The black horizontal line inside each box represents median, the boxes represent 25th and 75th percentiles, whiskers represent a value of 1.5*SD and the dots represent outliers.
4. Conclusions
A rapid, cost-effective, and high-throughput method for simultaneous analysis of several classes environmental chemicals in human urine using SPE coupled with LC-MS/MS was developed and validated. The target analytes belonged to diverse chemical classes with a wide range of polarity which required a robust sample preparation condition (i.e., incubation at 37 °C for 1 h with 20 μL ALS enzyme at pH 5.5), extraction and cleanup protocol (i.e., Agilent ABS Elut NEXUS SPE cartridge), and instrumental analysis (i.e., two LC and MS methods). The optimized method was validated through multiple approaches for calibration range, accuracy/precision, detection limits, and matrix effects. The method was also validated through the analysis of standard reference materials and external quality assurance scheme proficiency test samples. The method enabled analysis of 121 chemicals with a total instrument run time of 21 min and requires a urine volume of 0.5 mL. The method was sensitive to detect trace levels of target chemicals found in urinary samples. This rapid method will support analysis of a wide range of pollutants from diverse chemical classes for large scale human biomonitoring programs.
Supplementary Material
Figure 2. Total ion chromatogram (TIC; A) and extracted ion chromatograms (B–D) of 47 target compounds injected (5 μL) at a concentration of 10 ng/mL measured using mass spectrometric method 2 under negative mode (M2_NEG).
For the sake of distinction of individual compounds, ion chromatograms were extracted and divided into three fractions (i.e., F1–F3) depending on their retention times and signal intensities. Compound numbers assigned on the peaks are shown in Table 1. The x- and y-coordinates are retention time (min) and signal intensity (cps), respectively.
Highlights.
A SPE-LC-MS/MS method to determine 121 chemicals in a single extraction was developed
The method was sensitive for the analysis of 45 plasticizers, 45 phenolics and 31 pesticides
Single extraction followed by two instrumental conditions were required for analysis
The method was validated through the analysis of reference materials and real urines
Acknowledgements
The research reported in this manuscript was supported by the National Institute of Environmental Health Sciences (NIEHS) under Award Number U2CES026542. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The use of trade names does not imply endorsement of the product by the authors or affiliated institution or the funding agency.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supporting Information (SI)
The SI contains details of analytical standards (Table S1), MS/MS parameters used in the analysis (Tables S2–S4), recoveries (%) of OH-PAHs and PMs under different enzymatic deconjugation conditions (Table S5), concentrations measured in several PT urine samples (Table S6), concentrations (ng/mL) in real urine samples (Table S7), and total ion chromatograms of target analytes injected at a concentration of 1 ng/mL each (Figures S1–S3).
References
- [1].Kahn LG, Philippat C, Nakayama SF, Slama R, Trasande L, Endocrine-disrupting chemicals: Implications for human health, Lancet Diabetes Endocrinol 8 (2020) 703–718. 10.1016/S2213-8587(20)30129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Escher BI, Stapleton HM, Schymanski EL, Tracking complex mixtures of chemicals in our changing environment, Science 367 (2020) 388–392. 10.1126/science.aay6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Oberg T, Iqbal MS, The chemical and environmental property space of REACH chemicals, Chemosphere 87 (2012) 975–981. 10.1016/j.chemosphere.2012.02.034. [DOI] [PubMed] [Google Scholar]
- [4].Janjua NR, Frederiksen H, Skakkebaek NE, Wulf HC, Andersson AM, Urinary excretion of phthalates and paraben after repeated whole-body topical application in humans, Int. J. Androl 31 (2008) 118–129. 10.1111/j.1365-2605.2007.00841.x. [DOI] [PubMed] [Google Scholar]
- [5].Wang Y, Zhu HK, Kannan K, A review of biomonitoring of phthalate exposures, Toxics 7 (2019) 1–28. 10.3390/toxics7020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Robles-Molina J, Lara-Ortega FJ, Gilbert-Lopez B, Garcia-Reyes JF, Molina-Diaz A, Multi-residue method for the determination of over 400 priority and emerging pollutants in water and wastewater by solid-phase extraction and liquid chromatography-time-of-flight mass spectrometry, J. Chromatogr. A 1350 (2014) 30–43. 10.1016/j.chroma.2014.05.003. [DOI] [PubMed] [Google Scholar]
- [7].Clark SB, Storey JM, Turnipseed SB, Optimization and validation of a multiclass screening and confirmation method for drug residues in milk using high-performance liquid chromatography/tandem mass spectrometry, J. AOAC Int 94 (2011) 383–393. 10.1093/jaoac/94.2.383. [DOI] [PubMed] [Google Scholar]
- [8].Schneider MJ, Lehotay SJ, Lightfield AR, Evaluation of a multi-class, multi-residue liquid chromatography-tandem mass spectrometry method for analysis of 120 veterinary drugs in bovine kidney, Drug Test. Anal. 4 Suppl 1 (2012) 91–102. 10.1002/dta.1359. [DOI] [PubMed] [Google Scholar]
- [9].Bocato MZ, Cesila CA, Lataro BF, de Oliveira ARM, Campiglia AD, Barbosa F, A fast-multiclass method for the determination of 21 endocrine disruptors in human urine by using vortex-assisted dispersive liquid-liquid microextraction (VADLLME) and LC-MS/MS, Environ. Res 189 (2020) 109883. 10.1016/j.envres.2020.109883. [DOI] [PubMed] [Google Scholar]
- [10].Lin MQ, Ma ST, Yu YX, Li GY, Mai BX, An TC, Simultaneous determination of multiple classes of phenolic compounds in human urine: Insight into metabolic biomarkers of occupational exposure to e-waste, Environ. Sci. Technol. Lett 7 (2020) 323–329. 10.1021/acs.estlett.0c00187. [DOI] [Google Scholar]
- [11].Asimakopoulos AG, Wang L, Thomaidis NS, Kannan K, A multi-class bioanalytical methodology for the determination of bisphenol A diglycidyl ethers, p-hydroxybenzoic acid esters, benzophenone-type ultraviolet filters, triclosan, and triclocarban in human urine by liquid chromatography-tandem mass spectrometry, J. Chromatogr. A 1324 (2014) 141–148. 10.1016/j.chroma.2013.11.031. [DOI] [PubMed] [Google Scholar]
- [12].Rocha BA, de Oliveira ARM, Barbosa F, A fast and simple air-assisted liquid-liquid microextraction procedure for the simultaneous determination of bisphenols, parabens, benzophenones, triclosan, and triclocarban in human urine by liquid chromatography-tandem mass spectrometry, Talanta 183 (2018) 94–101. 10.1016/j.talanta.2018.02.052. [DOI] [PubMed] [Google Scholar]
- [13].Dewalque L, Pirard C, Dubois N, Charlier C, Simultaneous determination of some phthalate metabolites, parabens and benzophenone-3 in urine by ultra high pressure liquid chromatography tandem mass spectrometry, J. Chromatogr. B 949 (2014) 37–47. 10.1016/j.jchromb.2014.01.002. [DOI] [PubMed] [Google Scholar]
- [14].Zhu HK, Wang L, Liu CG, Stryker Z, Loganathan BG, Kannan K, Phthalate metabolites, hydroxy-polycyclic aromatic hydrocarbons, and bisphenol analogues in bovine urine collected from China, India, and the United States, Environ. Sci. Technol 53 (2019) 11524–11531. 10.1021/acs.est.9b04178. [DOI] [PubMed] [Google Scholar]
- [15].Heffernan AL, Thompson K, Eaglesham G, Vijayasarathy S, Mueller JF, Sly PD, Gomez MJ, Rapid, automated online SPE-LC-QTRAP-MS/MS method for the simultaneous analysis of 14 phthalate metabolites and 5 bisphenol analogues in human urine, Talanta 151 (2016) 224–233. 10.1016/j.talanta.2016.01.037. [DOI] [PubMed] [Google Scholar]
- [16].Peng FL, Ji WL, Zhu F, Peng DH, Yang M, Liu R, Pu YP, Yin LH, A study on phthalate metabolites, bisphenol A and nonylphenol in the urine of Chinese women with unexplained recurrent spontaneous abortion, Environ. Res 150 (2016) 622–628. 10.1016/j.envres.2016.04.003. [DOI] [PubMed] [Google Scholar]
- [17].Shin Y, Lee J, Park E, Lee J, Lee HS, Kim JH, A quantitative tandem mass spectrometry and scaled-down QuEChERS approach for simultaneous analysis of pesticide multiresidues in human urine, Molecules 24 (2019) 1330. 10.3390/molecules24071330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hung CC, Simaremare SRS, Hsieh CJ, Yiin LM, Simultaneous determination of pyrethroid, organophosphate and carbamate metabolites in human urine by gas chromatography-mass spectrometry (GCMS), Appl. Sci 9 (2019) 879. 10.3390/app9050879. [DOI] [Google Scholar]
- [19].Centers For Disease Control And Prevention. Prevention, Specific Organophosphorous Pesticides, Synthetic Pyrethroids, and Select Herbicides (Universal Pesticides). Solid Phase Extraction-High-Performance Liquid Chromatography-Heated Electrospray Ionization Tandem Mass Spectrometry, (2013) https://www.cdc.gov/nchs/data/nhanes/nhanes_07_08/uphopm_e_met.pdf.
- [20].Pellizzari ED, Woodruff TJ, Boyles RR, Kannan K, Beamer PI, Buckley JP, Wang AL, Zhu YY, Bennett DH, Identifying and prioritizing chemicals with uncertain burden of exposure: Opportunities for biomonitoring and health-related research, Environ. Health. Perspect 127 (2019) 126001. 10.1289/EHP5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Young AS, Allen JG, Kim UJ, Seller S, Webster TF, Kannan K, Ceballos DM, Phthalate and organophosphate plasticizers in nail polish: Evaluation of labels and ingredients, Environ. Sci. Technol 52 (2018) 12841–12850. 10.1021/acs.est.8b04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Giulivo M, de Alda ML, Capri E, Barcelo D, Human exposure to endocrine disrupting compounds: Their role in reproductive systems, metabolic syndrome and breast cancer. A review, Environ. Res 151 (2016) 251–264. 10.1016/j.envres.2016.07.011. [DOI] [PubMed] [Google Scholar]
- [23].Sirohi D, Ramadhani RA, Knibbs LD, Environmental exposures to endocrine disrupting chemicals (EDCs) and their role in endometriosis: A systematic literature review, Rev. Environ. Health 36 (2021) 101–115. 10.1515/reveh-2020-0046. [DOI] [PubMed] [Google Scholar]
- [24].Zhang T, Song SM, Bai XY, He Y, Zhang B, Gui MW, Kannan K, Lu SY, Huang YY, Sun HW, A nationwide survey of urinary concentrations of neonicotinoid insecticides in China, Environ. Int 132 (2019) 105114. 10.1016/j.envint.2019.105114. [DOI] [PubMed] [Google Scholar]
- [25].Yao YM, Li MQ, Pan LY, Duan YS, Duan XY, Li YC, Sun HW, Exposure to organophosphate ester flame retardants and plasticizers during pregnancy: Thyroid endocrine disruption and mediation role of oxidative stress, Environ. Int 146 (2021) 106215. 10.1016/j.envint.2020.106215. [DOI] [PubMed] [Google Scholar]
- [26].Wang Y, Li W, Martinez-Moral MP, Sun H, Kannan K, Metabolites of organophosphate esters in urine from the United States: Concentrations, temporal variability, and exposure assessment, Environ. Int 122 (2019) 213–221. 10.1016/j.envint.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nayebare SR, Karthikraj R, Kannan K, Analysis of terephthalate metabolites in human urine by high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS), J. Chromatogr. B 1092 (2018) 473–479. 10.1016/j.jchromb.2018.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ye XY, Zhou XL, Furr J, Ahn KC, Hammock BD, Gray EL, Calafat AM, Biomarkers of exposure to triclocarban in urine and serum, Toxicology 286 (2011) 69–74. 10.1016/j.tox.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Albro PW, Corbett JT, Schroeder JL, Jordan S, Matthews HB, Pharmacokinetics, interactions with macromolecules and species differences in metabolism of DEHP, Environ. Health. Perspect 45 (1982) 19–25. 10.1289/ehp.824519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schebb NH, Franze B, Maul R, Ranganathan A, Hammock BD, In Vitro glucuronidation of the antibacterial triclocarban and its oxidative metabolites, Drug Metab. Dispos 40 (2012) 25–31. 10.1124/dmd.111.042283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Thayer KA, Doerge DR, Hunt D, Schurman SH, Twaddle NC, Churchwell MI, Garantziotis S, Kissling GE, Easterling MR, Bucher JR, Birnbaum LS, Pharmacokinetics of bisphenol A in humans following a single oral administration, Environ. Int 83 (2015) 107–115. 10.1016/j.envint.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Provencher G, Berube R, Dumas P, Bienvenu JF, Gaudreau E, Belanger P, Ayotte P, Determination of bisphenol A, triclosan and their metabolites in human urine using isotope-dilution liquid chromatography-tandem mass spectrometry, J. Chromatogr. A 1348 (2014) 97–104. 10.1016/j.chroma.2014.04.072. [DOI] [PubMed] [Google Scholar]
- [33].Ho KL, Yuen KK, Yau MS, Murphy MB, Wan Y, Fong BMW, Tam S, Giesy JP, Leung KSY, Lam MHW, Glucuronide and sulfate conjugates of tetrabromobisphenol A (TBBPA): Chemical synthesis and correlation between their urinary levels and plasma TBBPA content in voluntary human donors, Environ. Int 98 (2017) 46–53. 10.1016/j.envint.2016.09.018. [DOI] [PubMed] [Google Scholar]
- [34].Abbas S, Greige-Gerges H, Karam N, Piet MH, Netter P, Magdalou J, Metabolism of parabens (4-hydroxybenzoic acid esters) by Hepatic Esterases and UDP-glucuronosyltransferases in man, Drug Metab. Pharmacokinet 25 (2010) 568–577. 10.2133/dmpk.DMPK-10-RG-013. [DOI] [PubMed] [Google Scholar]
- [35].Dwivedi P, Zhou X, Powell TG, Calafat AM, Ye X, Impact of enzymatic hydrolysis on the quantification of total urinary concentrations of chemical biomarkers, Chemosphere 199 (2018) 256–262. 10.1016/j.chemosphere.2018.01.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Blount BC, Milgram KE, Silva MJ, Malek NA, Reidy JA, Needham LL, Brock JW, Quantitative detection of eight phthalate metabolites in human urine using HPLC-APCI-MS/MS, Anal. Chem 72 (2000) 4127–4134. [DOI] [PubMed] [Google Scholar]
- [37].Bastiaensen M, Van den Eede N, Su GY, Letcher RJ, Stapleton HM, Covaci A, Towards establishing indicative values for metabolites of organophosphate ester contaminants in human urine, Chemosphere 236 (2019) 124348. 10.1016/j.chemosphere.2019.124348. [DOI] [PubMed] [Google Scholar]
- [38].USFDA, Bioanalytical Method Validation Guidance for Industry, (2018) https://www.fda.gov/downloads/drugs/guidances/ucm070107.pdf.
- [39].European Commission, Guidance document on analytical quality control and method validation procedures for pesticide residues and analysis in food and feed, (2017) https://ec.europa.eu/food/sites/food/files/plant/docs/pesticides_mrl_guidelines_wrkdoc_2017-11813.pdf.
- [40].Ferrer C, Martinez-Bueno MJ, Lozano A, Fernandez-Alba AR, Pesticide residue analysis of fruit juices by LC-MS/MS direct injection. One year pilot survey, Talanta 83 (2011) 1552–1561. 10.1016/j.talanta.2010.11.061. [DOI] [PubMed] [Google Scholar]
- [41].Wen QP, Zhou YQ, Wang YJ, Li JF, Zhao HZ, Liao JQ, Liu HX, Li YY, Cai ZW, Xia W, Association between urinary paraben concentrations and gestational weight gain during pregnancy, J. Expo. Sci. Env. Epid 30 (2020) 845–855. 10.1038/s41370-020-0205-7. [DOI] [PubMed] [Google Scholar]
- [42].Zhou Y, Yao YA, Shao YJ, Qu WD, Chen Y, Jiang QW, Urinary bisphenol analogues concentrations and biomarkers of oxidative DNA and RNA damage in Chinese school children in East China: A repeated measures study, Environ. Pollut 254 (2019) 112921. 10.1016/j.envpol.2019.07.089. [DOI] [PubMed] [Google Scholar]
- [43].Zhang B, Zhang T, Duan Y, Zhao Z, Huang X, Bai X, Xie L, He Y, Ouyang J, Yang Y, Wu Y, Sun H, Human exposure to phthalate esters associated with e-waste dismantling: Exposure levels, sources, and risk assessment, Environ. Int 124 (2019) 1–9. 10.1016/j.envint.2018.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


