Summary
The initiation of DNA replication involves cell cycle-dependent assembly and disassembly of protein complexes, including the Origin Recognition Complex (ORC) and CDC6 AAA+ ATPases. We report that multiple short linear protein motifs (SLiMs) within intrinsically disordered regions (IDRs) in ORC1 and CDC6 mediate Cyclin-CDK dependent and independent protein-protein interactions, conditional on the cell cycle phase. A domain within the ORC1 IDR is required for interaction between the ORC1 and CDC6 AAA+ domains in G1, whereas the same domain prevents CDC6-ORC1 interaction during mitosis. Then, during late G1, this domain facilitates ORC1 destruction by a SKP2-Cyclin A-CDK2-dependent mechanism. During G1, the CDC6 Cy-motif cooperates with Cyclin E-CDK2 to promote ORC1-CDC6 interactions. The CDC6 IDR regulates self-interaction by ORC1, thereby controlling ORC1 protein levels. Protein Phosphatase 1 binds directly to a SLiM in the ORC1 IDR, causing ORC1 dephosphorylation upon mitotic exit, increasing ORC1 protein and promoting pre-RC assembly.
Keywords: DNA Replication, Cell Division Cycle, Cyclin-dependent protein kinases, Initiation, Short Linear Protein Motifs, PP1 phosphatase, Origin Recognition Complex, CDC6, protein degradation, liquid-liquid phase transition
Graphical Abstract
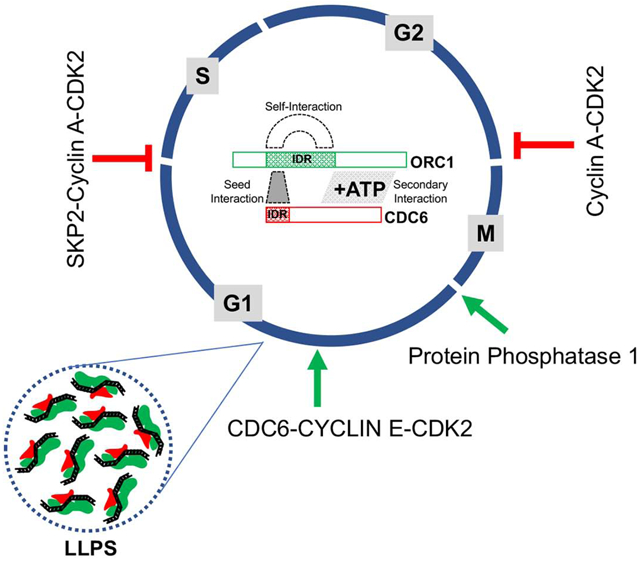
eTOC Blurb
Intrinsically disordered regions in the human DNA replication initiation proteins ORC1 and CDC6 contain short protein motifs that interact with multiple cyclin-dependent kinases, a phosphatase and with each other to control cell cycle regulated initiation of DNA replication, ORC1 intra-nuclear protein levels and the assembly of their AAA+ ATPase domains.
Introduction
In the budding yeast S. cerevisiae, DNA replication initiates at multiple origins following the assembly during G1 phase of a pre-Replicative Complex (pre-RC) at every potential origin that requires the six subunit Origin Recognition Complex (ORC), Cdc6, Cdt1 and the hetero-hexameric (Mcm2-7) complex (Bell and Labib, 2016; Parker et al., 2017). In contrast, many questions remain about pre-RC assembly in human cells. For example, it is not known when CDC6 binds to ORC owing to the fact that ORC1, the largest subunit of ORC, is ubiquitinated and targeted for degradation at the onset of S phase and then resynthesized in late G2, binding to chromosomes as cells progress into mitosis (DePamphilis, 2003; Kara et al., 2015; Kreitz et al., 2000; Méndez et al., 2002; Ohta et al., 2003; Tatsumi et al., 2003). Human CDC6, on the other hand, undergoes proteolytic degradation mediated by the APCCDH1 upon mitotic exit and during early G1 phase, to be re-synthesized and stabilized by Cyclin E-CDK2 dependent phosphorylation, a cycle completely opposite to that of S. cerevisiae Cdc6 (Cook et al., 2002; Coverley et al., 2002; Duursma and Agami, 2005; Mailand and Diffley, 2005). Thereafter, during S phase CDC6 is phosphorylated by Cyclin A-CDK2 and re-localizes to the cytoplasm (Delmolino et al., 2001; Jiang et al., 1999; Petersen et al., 2000).
In S. cerevisiae, ORC recognizes sequence specific origins and all of its subunits, including Orc1, are stable and consequently ORC remains bound to replication origin DNA for most of the cell cycle (Bell and Stillman, 1992; Diffley et al., 1994; Liang and Stillman, 1997; Tanaka et al., 1997). In contrast, yeast Cdc6 levels vary as a consequence of SCFCdc4 mediated degradation in S-phase, dependent on Cln-Cdc28 phosphorylation of Cdc6 (Figure 1A, bottom panel) (Perkins et al., 2001; Piatti et al., 1996). Yeast Cdc6 appears during late G2 phase and following mitotic exit it binds to ORC to assemble functional pre-RCs (Baum et al., 1998; Cocker et al., 1996; Drury et al., 2000; Liang et al., 1995; Piatti et al., 1995; Weinreich et al., 2001). Thus, the levels of yeast Cdc6 are reminiscent of human ORC1 (Figure 1A). Human ORC binds DNA in an apparent sequence-independent manner and with CDC6 and CDT1 can assemble MCM2-7 onto DNA in vitro (Vashee et al., 2003). Human ORC1 and CDC6 are also involved directly in regulation of CCNE1 gene expression in mid G1 phase to influence the decision of whether cells will proliferate or not (Hossain and Stillman, 2016).
Figure 1. Dynamic interaction between ORC1 and CDC6 proteins during the human cell cycle.
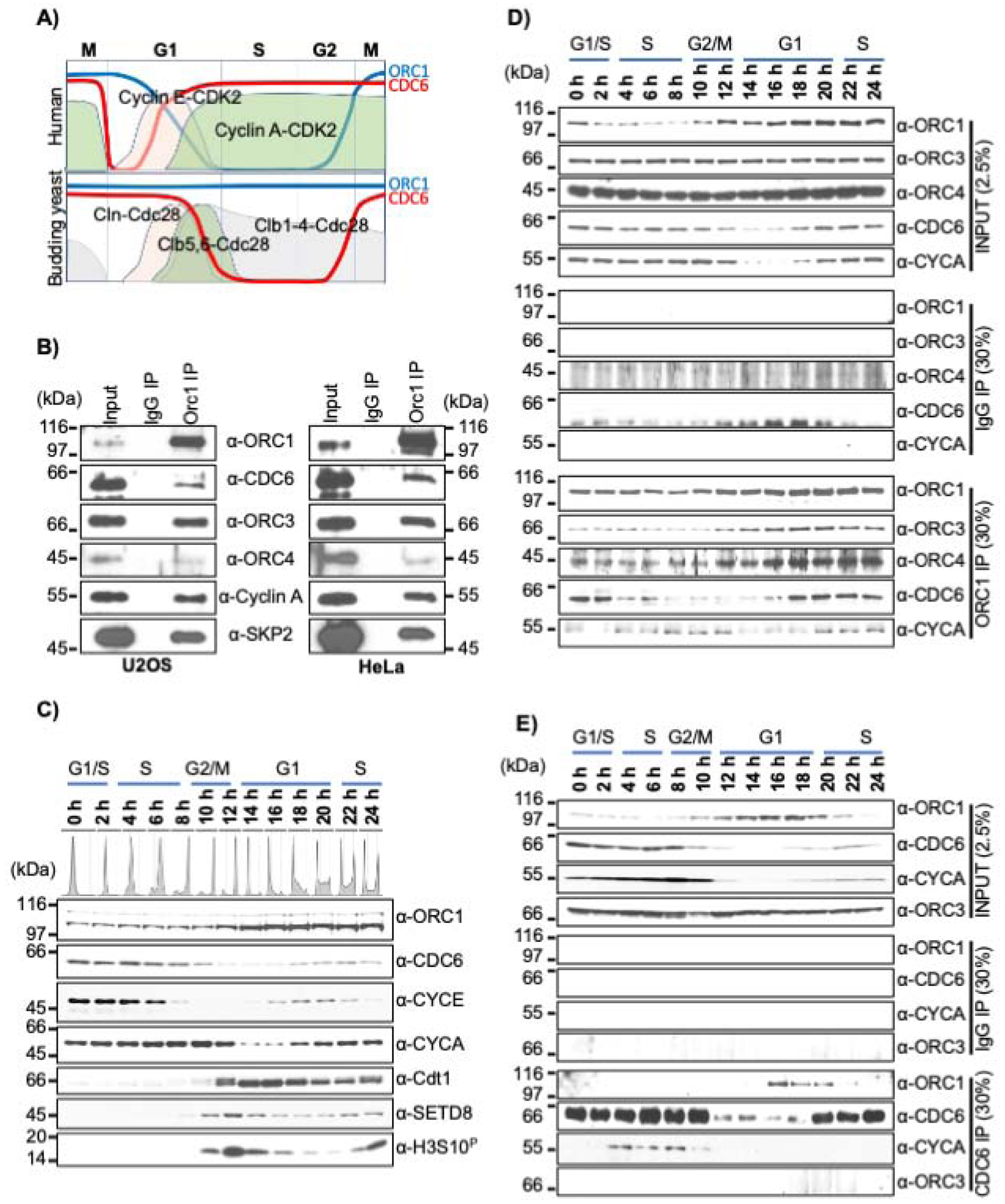
(A) Schematic of dynamic expression pattern of human and the yeast S. cerevisiae ORC1, CDC6 and Cyclin-CDK kinases across the cell division cycle. (B) Immunoprecipitation of ORC1 from asynchronous U2OS (left panel) and HeLa (right panel) cell lysates showing interactions with CDC6, ORC3, ORC4, Cyclin A and SKP2 proteins. Input and IP are 5% and 30%, respectively. Molecular weight markers, kDa. (C) Dynamic expression profile of pre-RC and cell cycle proteins detected by immunoblotting of extracts from double thymidine block and released synchronized HeLa cells. DNA content is indicated. CYCE and CYCA denotes Cyclin E and Cyclin A, respectively. (D-E) Double thymidine synchronized and released HeLa cell lysate prepared at different time points were immunoprecipitated either with an ORC1 antibody (D) or a CDC6 antibody (E) and immunoblotted as indicated. The input and IgG IP denote loading control and mock IP in the experiment, respectively.
It has long been known that Cyclin-CDKs regulate the timing of pre-RC assembly and function, but how they do this in human cells is unclear (Coverley et al., 2002; Li et al., 2004). The activity of Cyclin-CDKs requires their substrates to harbor a specific Cyclin-CDK recognition motif (Cy motif with consensus R/KxL) (Adams et al., 1996; Takeda et al., 2000; Wohlschlegel et al., 2001), although other Cyclin binding motifs have been reported (Örd et al., 2020). The N-terminal regions in both ORC1 and CDC6 harbor the R/KxL type Cy motif as well as multiple CDK phosphorylation sites, and both exist in predicted IDRs of each protein (Figure S1) (Hemerly et al., 2009; Schulman et al., 1998; Wood and Endicott, 2018). The N-terminal regions of yeast Orc1 and Cdc6 also contain predicted IDRs, with no apparent Cy motif in yeast Orc1 (Figure S1). In S. cerevisiae, Cdc6 and Orc6 contain a Cy motif specific for S phase Clb5-Cdc28 kinase (Örd et al., 2019; Weinreich et al., 2001; Wilmes et al., 2004) and Cdc6 also binds to the mitotic Clb2/Cdc28 kinase that inhibits pre-RC assembly (Mimura et al., 2004).
Apart from large structured domains in proteins with known functions, many short regulatory motifs have been identified in proteins, frequently within IDRs (Russell and Gibson, 2008). These Short Linear protein Motifs (SLiMs) can mediate many functions (Roey and Davey, 2015; Roey et al., 2014). In this report, we show that the human ORC1 IDR contains multiple SLiMs that perform varied regulatory functions. Cyclin A-CDK2 and protein phosphatase 1 (PP1) bind motifs in the ORC1 IDR, controlling ORC phosphorylation and CDC6 interaction at different stages of the cell cycle. CDC6 binds to a domain that overlaps with the Cy motif in the IDR of ORC1 and promotes ORC1-CDC6 AAA+ ATPase domain interactions, and also regulates ORC1 intra- and inter-molecular interactions that control the abundance of ORC1 in the cell. After pre-RC assembly in mid G1 phase, Cyclin A-CDK2 recruits the SCFSKP2 ubiquitin ligase to ORC1, causing destruction of ORC1 as cells enter S phase. We propose that differences in the ORC-CDC6 interaction cycle between budding yeast and human cells are mediated, in large part, by the acquisition of multiple SLiMs in the IDRs of ORC1 and CDC6 proteins that control the initiation of DNA replication. The same IDRs are known to mediate DNA-dependent liquid phase separation of ORC and CDC6 (Parker et al., 2019), we propose that DNA-regulated liquid phase separation involving ORC1 and CDC6 controls ORC1 protein levels in cells.
Results
Dynamic interaction between ORC1 and CDC6 proteins during the human cell cycle.
It is known that yeast Cdc6 and human CDC6 directly bind to yeast Orc1 and human ORC1, respectively (Hossain and Stillman, 2016; Liang et al., 1995; Saha et al., 1998; Weinreich et al., 2001). Because of the dynamic expression pattern of human ORC1 and CDC6 across the cell cycle (Figure 1A), we investigated the biochemical interactions between these proteins. ORC1 immunoprecipitation from asynchronous U2OS and HeLa cells showed co-precipitation of other ORC subunits, CDC6, Cyclin A and SKP2 (Figure 1B). To further explore ORC1 and CDC6 association across the human cell cycle, HeLa suspension cells were synchronized using a double thymidine block and following release, cell extracts were used for monitoring protein levels (Figure 1C). Cyclin E and Cyclin A varied as expected, undergoing proteasome-mediated degradation during late G1 or early S phase and during prometaphase of mitosis, respectively (Elzen and Pines, 2001; Geley et al., 2001; Nakayama et al., 2001; Ohtsubo et al., 1995). CDT1 and SETD8 were absent in S phase but were resynthesized after mitosis, while histone H3S10 phosphorylation marked the occurrence of mitotic cells at the 12-hour time point (Figure 1C).
The interaction between ORC1 and CDC6 with each other and other proteins was investigated by immunoprecipitation (Figure 1D and E). The level of ORC1 protein remained low until early G2-phase due to proteasome-mediated degradation during late G1 and early S phase. Upon exit from mitosis, ORC1 bound to ORC3 and ORC4 proteins across the cell cycle. Human ORC1 also interacted with Cyclin A protein in early S phase, late G2 and early mitotic cells when both the proteins were present. Interestingly, ORC1 bound to CDC6 only during mid-G1 phase and into early S phase, while interaction was not detected during G2-M phases, despite both proteins being present in mitosis (Figure 1D). In a reciprocal immunoprecipitation with CDC6 antibody, ORC1 and CDC6 proteins interacted only during mid-G1 phase (Figure 1E). Like Cyclin A, CDC6 protein was expressed across the cell cycle except in early G1-phase, but again, CDC6 was not able to co-precipitate ORC1 protein during the G2-M phase of the cell cycle. In contrast to ORC1, CDC6 immunoprecipitation did not show any interaction with ORC3 protein, but interacted with Cyclin A during S-phase, mediating export of CDC6 from the nucleus to the cytoplasm (Petersen et al., 1999). Thus, ORC1 and CDC6 proteins only interacted within a short window during mid to late G1-phase of the human cell cycle.
ATP dependent interaction between ORC1 and CDC6 requires a domain that overlaps with the CDC6 cyclin binding (Cy) motif.
The observation that CDC6 bound to ORC1 during G1 phase but not during mitosis prompted biochemical studies to investigate their interaction and its regulation. Recombinant GST-CDC6 protein (CDC6 tagged with glutathione S transferase) interacted directly with maltose binding protein (MBP) tagged ORC1 and ORC2 proteins, but not other ORC subunits, in MBP pull-down assays (Figure S2A), consistent with structural studies of human ORC in which, through in silico modelling, CDC6 was docked in between ORC1 and ORC2, consistent with the yeast and Drosophila ORC-Cdc6 structures (Bleichert et al., 2018; Jaremko et al., 2020; Schmidt and Bleichert, 2020; Tocilj et al., 2017; Yuan et al., 2017). The interaction between GST-CDC6 and MBP-ORC1 was enhanced by ATP (Figures 2A and S2B). Next, many fragments of CDC6 were constructed and binding assays showed that amino acids 1–110 within CDC6 were necessary and sufficient to bind to ORC1 protein (Figures 2B, S2C, S2D and S2E). This was surprising since the AAA+ domains of Orc1 and Cdc6 interact in the yeast and Drospohila ORC-Cdc6_Cdt1-Mcm2-7 (OCCM) complex bound to origin DNA and ORC stimulates the ATPase activity of Cdc6 (Randell et al., 2006; Schmidt and Bleichert, 2020; Speck and Stillman, 2007; Yuan et al., 2017). Nevertheless, the 1–110 region of GST-CDC6 bound to MBP-ORC1 protein while the AAA+ containing 110–560 fragment of Cdc6 did not bind (Figures 2B and S2E). Moreover, internal deletions of small N-terminal regions (Δ11–20, Δ21–30, Δ51–70 and Δ71–90) within full length GST-CDC6 protein still bound to MBP-ORC1 protein while deletion of 90–100 (Δ91–100) and 100–110 (Δ101–110) in CDC6 completely lost their interaction with ORC1 (Figure S2F). Thus, the 90–110 region of CDC6 protein was essential for its interaction with ORC1.
Figure 2. ATP-dependent ORC1 and CDC6 interaction region overlaps with their Cy motifs.
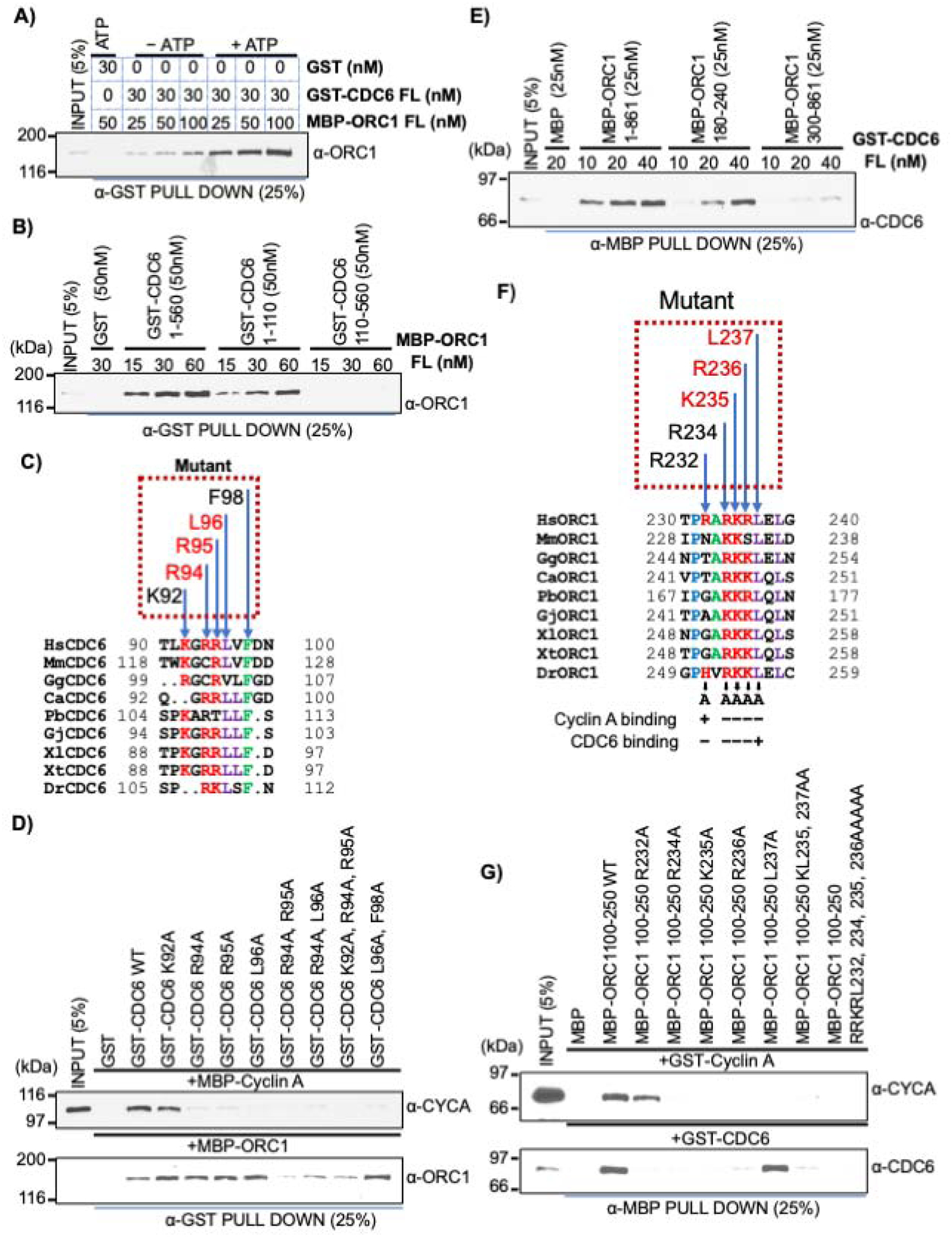
(A) Constant amounts of GST-CDC6 protein bound beads were incubated with varying concentrations of MBP-ORC1 protein in absence or presence of 1mM ATP and blotted with anti-ORC1 antibody. FL, full length protein. GST protein served as negative control. See also Figure S2B. (B) Equimolar amounts of GST-CDC6 wild type and N- and C-truncation fragments (1–110 and 110–560) were incubated with full length MBP-ORC1 protein, precipitated with anti-GST antibody and blotted with anti-ORC1 antibody. See also Figure S2E. (C) Multiple sequence alignment of the 90–100 amino acid region of CDC6 from vertebrate species (Homo sapiens, Hs; Mus musculus, Mm; Calypte anna, Ca; Gallus gallus, Gg; Python bivittatus, Pb; Gekko japonicus, Gj; Xenopus laevis, Xl; Xenopus tropicalis, Xt; Danio rerio, Dr). The dotted box above the alignment indicates the conserved residues of human CDC6 protein selected for making recombinant mutant proteins used in binding studies. The residues colored red in the dotted box represent Cy motif of human CDC6 protein. (D) Single (K92A, R94A, R95A, and L96A), double (R94A, R95A; R94A, L96A; L96A, F98A) and triple (K92A, R94A and R95A) mutants of GST-CDC6 full length protein, along with wild type protein, were incubated with either MBP-Cyclin A (top) or MBP-ORC1 (bottom) in a GST pull down assay. The proteins were detected by western blot using their respective antibodies as indicated. See also Figure S2G. (E) Increasing amounts of GST-CDC6 full-length protein binding to MBP-ORC1 wild (1–861) or fragments 180–240 aa or 300–861 aa in α-MBP antibody immunoprecipitate and blotted with anti-CDC6 antibody. See also Figure S2O. (F) Multiple alignment showing sequence conservation the 230–240 aa region of vertebrate ORC1 sequences containing the Cy motif (Homo sapiens, Hs; Mus musculus, Mm; Calypte anna, Ca; Gallus gallus, Gg; Python bivittatus, Pb; Gekko japonicus, Gj; Xenopus laevis, Xl; Xenopus tropicalis, Xt; Danio rerio, Dr). The dotted box above the alignment indicates the conserved residues of human ORC1 protein selected for making recombinant MBP-ORC1 mutants. The residues colored red in the dotted box represent the Cy motif of human ORC1 protein. A summary of binding to CDC6 and Cyclin A is shown below. (G) Human ORC1 single (R232A, R234A, K235A, R236A and L237A), double (K235A, L237A) or quadruple (R232A, R234A, K235A, R236A) mutants in the 100–250 aa region of recombinant MBP-ORC1 protein were incubated with either GST-Cyclin A (top) or GST-CDC6 (bottom) in an anti-MBP antibody pull down assay, then blotted with their respective antibodies as indicated. See also Figure S2P.
The CDC6 Cy motif, CDC694RRL96, that is required for its interaction with G1-S phase human cyclins (Cyclin E and Cyclin A) as well as the conserved LxF motif, CDC696LVF98, that mediates binding of yeast Cdc6 with mitotic cyclin were located in the 90–110 region of CDC6 protein (Figure 2C) (Hossain and Stillman, 2016; Örd et al., 2019; Pagliuca et al., 2011; Petersen et al., 1999; Takeda et al., 2000). Therefore, we tested whether the human CDC6 Cy and LxF motifs were involved in its interaction with ORC1 protein by making a series of point mutations within recombinant full-length GST-CDC6 protein and comparing the binding of these mutants to MBP-Cyclin A and MBP-ORC1. The results of multiple mutants showed that the ORC1 binding domain in CDC6 overlapped with the CDC6 Cy motif that binds Cyclin A, but they were clearly distinguishable (Figures 2D and S2G).
We next asked whether the Cy motif in human ORC1 protein, which binds Cyclin A (but not to Cyclin E) in an ORC1235KRL237-dependent manner, was required for ORC1 binding to CDC6 (Hemerly et al., 2009; Hossain and Stillman, 2012). The mutant MBP-ORC1235ARA237 (MBP-ORC1 A-A) bound to GST-CDC6 like the wild type protein, but as expected from the previous result, it did not bind to the GST-CDC6 double mutant (CDC6 AAL; R94A, R95A) protein (Figure S2H). Thus, the CDC6 domain that binds to ORC1 overlaps with the CDC6 Cy motif, whereas the ORC1 Cy motif in full length ORC1, defined by the ORC1235ARA237 mutation, was not required for binding CDC6.
The minimal region in ORC1 that binds CDC6 overlaps with the ORC1 Cy motif required for Cyclin A interaction
In a reciprocal binding test, MBP-ORC1 interacted with GST-CDC6 protein in an ATP-stimulated manner (Figure S2I). The minimal region within human ORC1 required for binding to CDC6 protein was mapped by systematically creating fragments of MBP-tagged ORC1 (Figures S2J, S2K, S2L, S2M and S2N). Fine mapping showed that the 180–240 region of MBP-ORC1 protein was necessary and sufficient to interact directly with GST-CDC6 while the 300–861 AAA+ domain-containing fragment of ORC1 protein showed only a weak interaction (Figures 2E and S2O). Again, it was surprising that the ORC1 AAA+ domain did not interact directly with CDC6.
Similar to CDC6, the minimal 180–240 fragment of human ORC1 that bound to CDC6 contained the well conserved Cy motif (Figure 2F). A series of single or double point mutations in the small 100–250 fragment of ORC1 that impact the Cy motif were examined for binding to Cyclin A and CDC6 (Figures 2F, 2G and S2P). The data showed that mutations within the core of the ORC1 Cy motif, ORC1235KRL237, affected binding of ORC1 to both Cyclin A and CDC6. But, point mutations on either end of the core Cy motif bound differentially to Cyclin A and CDC6. Mutation of L237, which normally binds to Cyclin A (Wang and Song, 2019) bound to CDC6 but not Cyclin A, whereas mutation of R232 disrupted binding of ORC1 to CDC6, but retained binding to Cyclin A (Figures 2F and 2G). Thus, the minimal domain of ORC1 that promotes binding to CDC6 contains sequences that highly overlap with the ORC1 Cy motif. When the full length ORC1 was used, the ORC1235ARA237 mutation did not disrupt CDC6 binding (Figure S2H). Thus, ORC1 binds in a cooperative manner to CDC6, involving both the 180–240 domain in the IDR and the 300–861 region containing the AAA+ domain, but the ORC1 AAA+ domain cannot stably bind CDC6 by itself.
ORC1-mediated liquid-liquid phase separation of ORC1-CDC6 protein complex.
The two domains in ORC1 and CDC6 that promote their interaction are located within the IDRs of both proteins. Based on a recent report showing that DNA replication initiation proteins in Drosophila melanogaster undergo liquid-liquid phase separation in presence of DNA (Parker et al., 2019), we investigated phase separation with purified human ORC1 and CDC6 proteins. The human MBP-ORC1 protein, but not the MBP-CDC6 protein bound to a 60 base-pair, biotin-tagged dsDNA in the presence of ATP (Figure 3A). Next, human ORC1 and CDC6 were expressed and purified from bacteria as MBP-PP-mEGFP and MBP-PP-mCherry protein fusions with a PreScission Protease (PP) site to investigate protease-mediated liquid droplet formation using a microscopy-based technique previously employed for phase separation studies (Burke et al., 2015; Schuster et al., 2018). Human MBP-PP-mEGFP-ORC1 remained soluble as no liquid droplets were observed under fluorescence microscopy in the absence of protease (Figure 3B, top panel). With the addition of protease, the MBP tag was removed and liquid droplets of mEGFP-ORC1 were observed (Figure 3B, middle panel), similar results were obtained in the presence of an equimolar amount of Cy5-DNA that colocalized with the mEGFP-ORC1 protein (Figure 3B, bottom panel). In contrast, droplet formation with protease cleavage of MBP-PP-mCherry-CDC6 WT in the absence (data not shown) or presence of Cy5-DNA (Figure S3A) was not observed. Similar results were obtained with MBP-PP-mCherry-CDC6 triple (K92A, R94A, R95A) mutant protein (Figure S3B). Since ORC1 and CDC6 form a tight complex, we investigated the appearance of CDC6 droplets upon mixing with ORC1 protein. Equimolar amounts of both MBP-PP-mEGFP-ORC1 and MBP-PP-mCherry-CDC6 WT resulted in colocalization of both the proteins in liquid droplets only upon protease addition, and when Cy5-DNA was added, it also co-localized with both ORC1 and CDC6 in the liquid droplets (Figure 3C, top and bottom panel; Figure S3C). Mixing the MBP-PP-mCherry-CDC6 (K92A, R94A, R95A) triple mutant protein with MBP-PP-mEGFP-ORC1 colocalized with or without the DNA only in the presence of protease, but with significantly reduced droplet size compared to the wild type mCherry-CDC6 protein (Figure 3D, top and bottom panel). No droplets were observed in the absence of the protease (Figure S3D). The results support the idea that the CDC6 protein can phase separate with ORC1 that is bound to dsDNA. The mutant CDC6 protein (K92A, R94A, R95A) that has reduced binding to ORC1 resulted in smaller liquid-phase droplets, even in the presence of DNA.
Figure 3. Phase separation of ORC1 and CDC6 proteins on DNA.
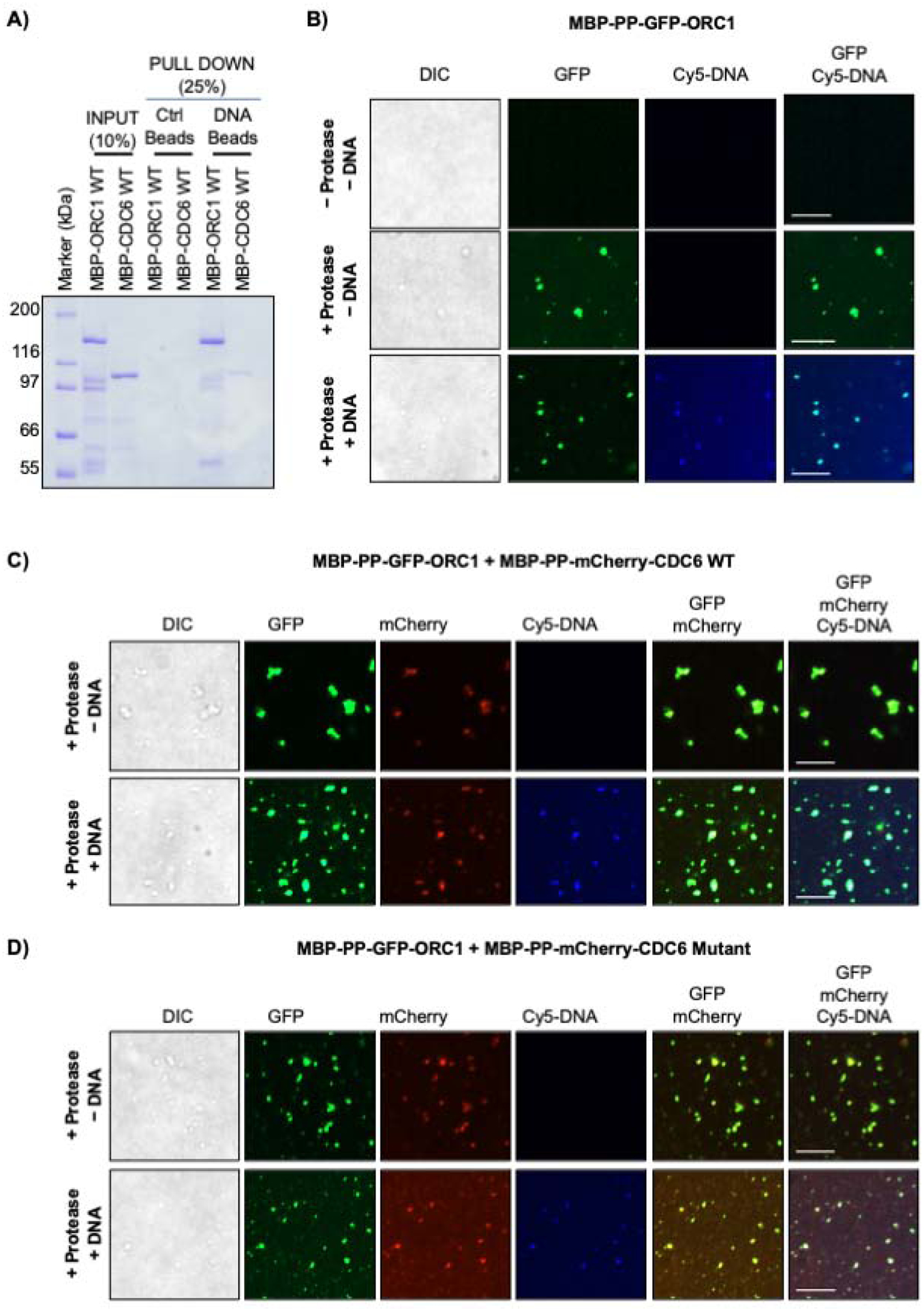
(A) MBP-ORC1 and MBP-CDC6 proteins were incubated with biotinylated dsDNA streptavidin beads in presence of 1mM ATP. Streptavidin beads alone served as control. (B) MBP-PP-GFP-ORC1 (4 μM) do not undergo a liquid-liquid phase separation without protease, but upon addition of protease in the absence or presence of 4 μM Cy5-dsDNA cleaved full length GFP-ORC1 forms liquid droplets. (C-D) MBP-PP-GFP-ORC1 (2 μM) protein was mixed with either 2 μM of MBP-PP-mCherry-CDC6 WT (C) or CDC6 Cy mutant protein (D) in the presence of protease without or with Cy5-dsDNA (4 μM). Merged images shows co-recruitment of cleaved GFP-ORC1 and mCherry-CDC6 (WT or mutant) with or without Cy5-dsDNA. See Figures S3C and S3D. In figures B-D, the images are representative of three independent experiments. The liquid droplets formed were captured after digestion with protease at 4°C for 16 hours. Scale bars 10 μm. PP; PreScission protease site.
Cyclin E and Cyclin A dependent CDK kinases differentially affect ORC1-CDC6 interactions
Cyclin E-CDK2 and Cyclin A-CDK2 kinases are known to phosphorylate both ORC1 and CDC6 and the initial ORC1-CDC6 protein-protein interaction involved residues that overlapped each of their Cy motifs in the IDR. Therefore, the interaction between ORC1 and CDC6 in the presence of purified GST-Cyclin E or GST-Cyclin A without their CDK2 kinase subunit partner was examined. Cyclin A bound to ORC1 better than Cyclin E in the absence of CDC6, but in the presence of CDC6, Cyclin A and Cyclin E both bound ORC1 with similar affinity (Figures 4A, 4B, S4A, S4B, S4C and S4D). Cyclins E and Cyclin A bind to different sites in ORC1, with Cyclin A requiring the canonical Cy motif, but Cyclin E binds to an N-terminal domain that is affected by mutations in Meier-Gorlin Syndrome (Hemerly et al., 2009; Hossain and Stillman, 2012). Importantly, using a sequential pull-down strategy, trimeric complexes of CDC6-ORC1-Cyclin A and CDC6-ORC1-Cyclin E were observed. In one approach, MBP-GFP-ORC1-HIS6 and GST-CDC6-HIS6 were incubated together in combination with either MBP-Cyclin E-HIS6 or MBP-Cyclin A-HIS6. In a two-step pull down (Figure 4C, left), both Cyclin E and Cyclin A associated the ORC1-CDC6 complex (Figure 4C, silver stain; Figure S4E, immunoblotted with HIS6 antibody). In an alternative approach, MBP-GFP-ORC1-HIS6 and MBP-Cyclin E-HIS6 or MBP-GFP-ORC1-HIS6 and MBP-Cyclin A-HIS6 were incubated with or without MBP-CDC6-HIS6 and sequential GST and anti-GFP precipitations confirmed that CDC6 remained associated with ORC1-Cyclin E or ORC1-Cyclin A complexes (Figure S4F, top and bottom panels). Thus, trimeric complexes of ORC1-CDC6-Cyclin A or ORC1-CDC6-Cyclin E exist.
Figure 4. G1-S Cyclin-CDK kinases have differential effects on ORC1-CDC6 protein interactions.
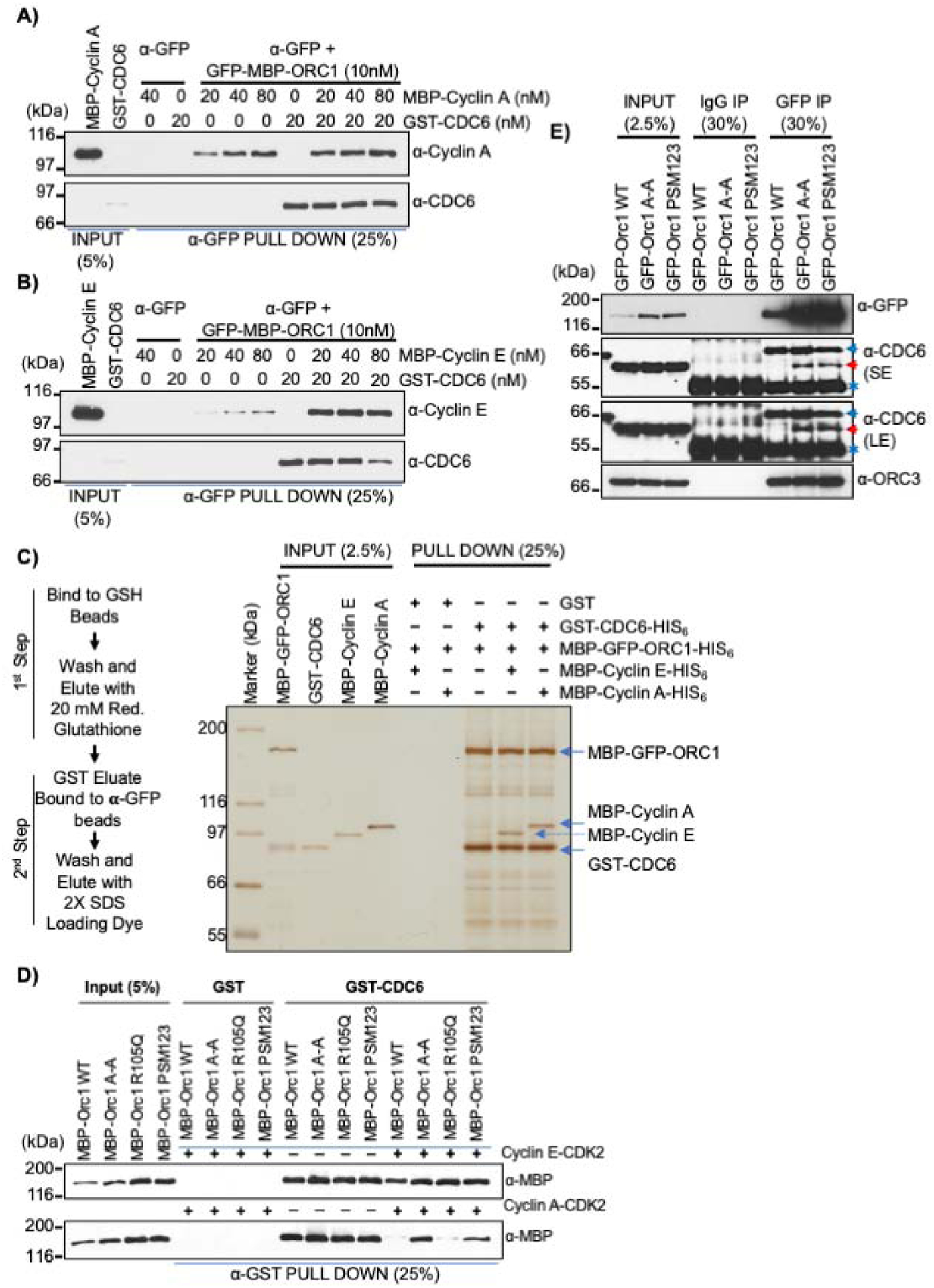
(A-B) In a α-GFP antibody pull-down assay, a constant amount of recombinant GFP-MBP-ORC1 protein was incubated with GST-CDC6 alone or with increasing molar amounts of MBP-Cyclin A (A) or MBP-Cyclin E (B) and the levels of Cyclin or CDC6 co-precipitated were detected by immuno-blotting. See Figures S4C and S4D. (C) Schematic outline of a two-step affinity purification of protein complexes using purified recombinant proteins (Left panel). MBP-GFP-ORC1-HIS6 (50nM) and GST-CDC6-HIS6 (50nM) were incubated together or in combination with MBP-Cyclin E-HIS6 (50nM) or MBP-Cyclin A-HIS6 (50nM) and control GST (50nM) protein were incubated with either MBP-Cyclin E-HIS6 or MBP-Cyclin A-HIS6 and MBP-GFP-ORC1-HIS6 as indicated. After two step purification, the protein complexes were detected with silver stain (right panel). The reaction contained 1mM ATP. See Figure S4E and S4F. (D) In a GST pull-down assay, GST-CDC6 protein was incubated with MBP-ORC1 WT or its mutants in the presence or absence of Cyclin E-CDK2 (top panel) or Cyclin A-CDK2 (bottom panel) with 1mM ATP. The western blot is probed with anti-MBP antibody and GST protein served as negative control. (E) Mitotic cells from stable GFP-ORC1 WT or ORC1 mutant cell lines were collected after nocodazole treatment and cell lysates were used for immunoprecipitation with anti-GFP antibody. The samples were further immunoblotted with CDC6 antibody and ORC3 antibody. A red arrow indicates specific CDC6 band, while a blue arrow and an asterisk symbol indicate non-specific and cross-reactive heavy IgG bands, respectively. SE, short exposure and LE, long exposure.
Next, the interaction between purified ORC1 and CDC6 proteins in the presence of active, purified G1-S kinases, Cyclin E-CDK2 and Cyclin A-CDK2 was examined. MBP-ORC1 protein was incubated with bead bound GST-CDC6 protein either in the presence or absence of purified Cyclin E-CDK2 or Cyclin A-CDK2 individually or together, and complexes bound to beads examined. Cyclin E-CDK2 did not alter the interaction between MBP-ORC1 and GST-CDC6 proteins, but Cyclin A-CDK2 alone or when both the kinases were present inhibited the interaction, a process requiring ATP (Figure S4G and Figure S4H). Although Cyclin E-CDK2 did not prevent the interaction between ORC1 and CDC6, it could remove the retinoblastoma (RB) tumor suppressor protein from the ORC1-CDC6 complex in an ATP-dependent manner (Figure S4I), confirming the previous observation that ORC1-RB interaction was prevented by Cyclin E-CDK2 kinase activity while having no effect on ORC1-CDC6 interaction, thereby relieving RB mediated repression of the CCNE1 gene (Hossain and Stillman, 2016).
Since Cyclin A-CDK2 kinase bound to the ORC1 Cy motif and inhibited the ORC1 interaction with CDC6 protein, we examined if this occurred by direct phosphorylation of ORC1. An ORC1 Cy mutant (ORC1 A-A; 235KRL237 to 235ARA237) - defective in its ability to bind Cyclin A-CDK2, an ORC1 R105Q mutant - defective in the ability of ORC1 to inhibit Cyclin E-CDK2 kinase activity, and an ORC1-PSM123 mutant (ORC1 Phosphorylation Site Mutant, S258A, S273A and T375A; - mutant in three Cyclin A-CDK2 kinase sites), were used in GST-CDC6 pull-down assays in the presence of Cyclin E-CDK2 or Cyclin A-CDK2. The interaction between either wild type or mutant MBP-ORC1 proteins with GST-CDC6 was unaffected in presence or absence of Cyclin E-CDK2 (Figure 4D, top panel). In sharp contrast, the binding of GST-CDC6 protein to the MBP-ORC1 A-A and PSM123 mutants was not blocked by Cyclin A-CDK2, while the kinase did inhibit the wild type and the ORC1 R105Q mutant binding to CDC6 (Figure 4D, bottom panel). Thus, phosphorylation of ORC1 by Cyclin A-CDK2 blocked its interaction with CDC6. Mutations in all three phosphorylation sites were required to block the inhibitory effect of Cyclin A-CDK2 on ORC1-CDC6 interaction (Figure S4J). These results reveal a previously unknown role for Cyclin A-CDK2 kinase in regulating the interaction between human ORC1 and CDC6.
The previous immunoprecipitation results demonstrated that ORC1 and CDC6 do not interact in mitosis, although both the proteins are present (Figures 1D and 1E). Human CDC6 is phosphorylated by Cyclin A-CDK2 (Petersen et al., 1999), as is ORC1 in Chinese Hamster cells (Li et al., 2004). To examine whether human ORC1 and CDC6 proteins were phosphorylated in mitosis, U2OS cells were arrested in G2/M phase by treatment with various concentrations of the microtubule polymerization inhibitor nocodazole for 16 hours, mitotic cells were collected following shake-off and cell extracts prepared at different time points following release from the mitotic block for immunoblotting. Both ORC1 and CDC6 proteins migrated faster in a gel within 2 hours after nocodazole release compared to mitotic arrested cells at 0 hour, indicating a change in phosphorylation upon mitotic exit (Figure S4K). Cyclin A, but not Cyclin E, is present in G2 to prometaphase of mitotic U2OS cells and is degraded after nocodazole release, while Cyclin E protein levels started rising at later time points in the next G1 phase (Figure S4K). Lambda phosphatase treatment of the mitotically arrested U2OS cell lysates confirmed that both human ORC1 and CDC6 were phosphorylated, moving faster in the gel like Cyclin F protein which is known to be phosphorylated in early mitosis (Choudhury et al., 2017; Mavrommati et al., 2018) (Figure S4L). A phosphor-specific RB antibody (S807/S811) was used as a control, showing the absence of the phospho-RB protein upon phosphatase treatment.
The interaction between CDC6 in mitosis with different ORC1 mutants was tested by tetracycline inducible GFP-ORC1 in U2OS cell lines, arresting them with nocodazole for 16 hours. GFP immunoprecipitation from mitotic cell lysates prepared from wild type GFP-ORC1 showed no interaction with endogenous CDC6 protein, but ORC1 did bind the ORC3 protein (Figure 4E). Interestingly, the two mutants of human ORC1 protein, GFP-ORC1 A-A and GFP-ORC1 PSM123 interacted with endogenous CDC6 protein along with ORC3 in GFP immunoprecipitation (Figure 4E). Thus, during late G2 phase Cyclin A-CDK2 binds to ORC1 in a Cy motif-dependent manner and phosphorylates ORC1, preventing CDC6 from binding to ORC1, thereby preventing pre-RC formation during mitosis. Despite Cyclin A being degraded during prometaphase (Elzen and Pines, 2001), ORC1 remains phosphorylated during mitosis when it binds to the mitotic chromosomes upon nuclear envelope breakdown (Kara et al., 2015; Okuno et al., 2001) and is dephosphorylated only after mitotic exit.
ORC1 Cy motif-dependent self-interaction and CDC6 control ORC1 protein levels
The Cyclin A-CDK2 kinase-dependent phosphorylation of ORC1 blocking its interaction with CDC6 created a paradox since the minimal region of ORC1 that bound to CDC6 was 180–240 aa, but the phosphorylation of ORC1 that occurred outside of this region (S258, S273 and T375) prevented CDC6 from binding to ORC1 during mitosis and in vitro (Figures 4D and 4E). Therefore, we tested whether these two regions of ORC1 cooperate with each other to regulate the ORC1-CDC6 interaction. In a GFP immunoprecipitation assay, GFP-MBP-ORC1 full length wild type protein co-precipitated MBP-ORC1 wild type or its Meier-Gorlin syndrome mutants (R105Q and/or R720Q), but not MBP-ORC4 protein, suggesting trans self-interaction between ORC1 molecules (Figure 5A). Intriguingly, the self-interaction increased with a Cy motif mutation in ORC1 that also affected CDC6 interaction (Figure 5B). Consistent with this observation, the ORC1-230-400-HIS6 fragment that contains the three CDK ORC1 phosphorylation sites interacted with the wild type MBP-ORC1-100-250 fragment, but not with the MBP-ORC1-100-250 Cy mutant fragment (Figure 5C, lanes 3 and 6), again suggesting an ORC1 intra- or inter-molecular interaction that involves the same domain (180–240) that also binds CDC6. Cyclin A-CDK2 kinase phosphorylation of ORC1 did not influence this self-binding (Figure 5C, lanes 4, 5, 7 and 8).
Figure 5. Self-interaction between the Cy motif containing domain and the BP5 basic patch of human ORC1 is blocked by CDC6 protein.
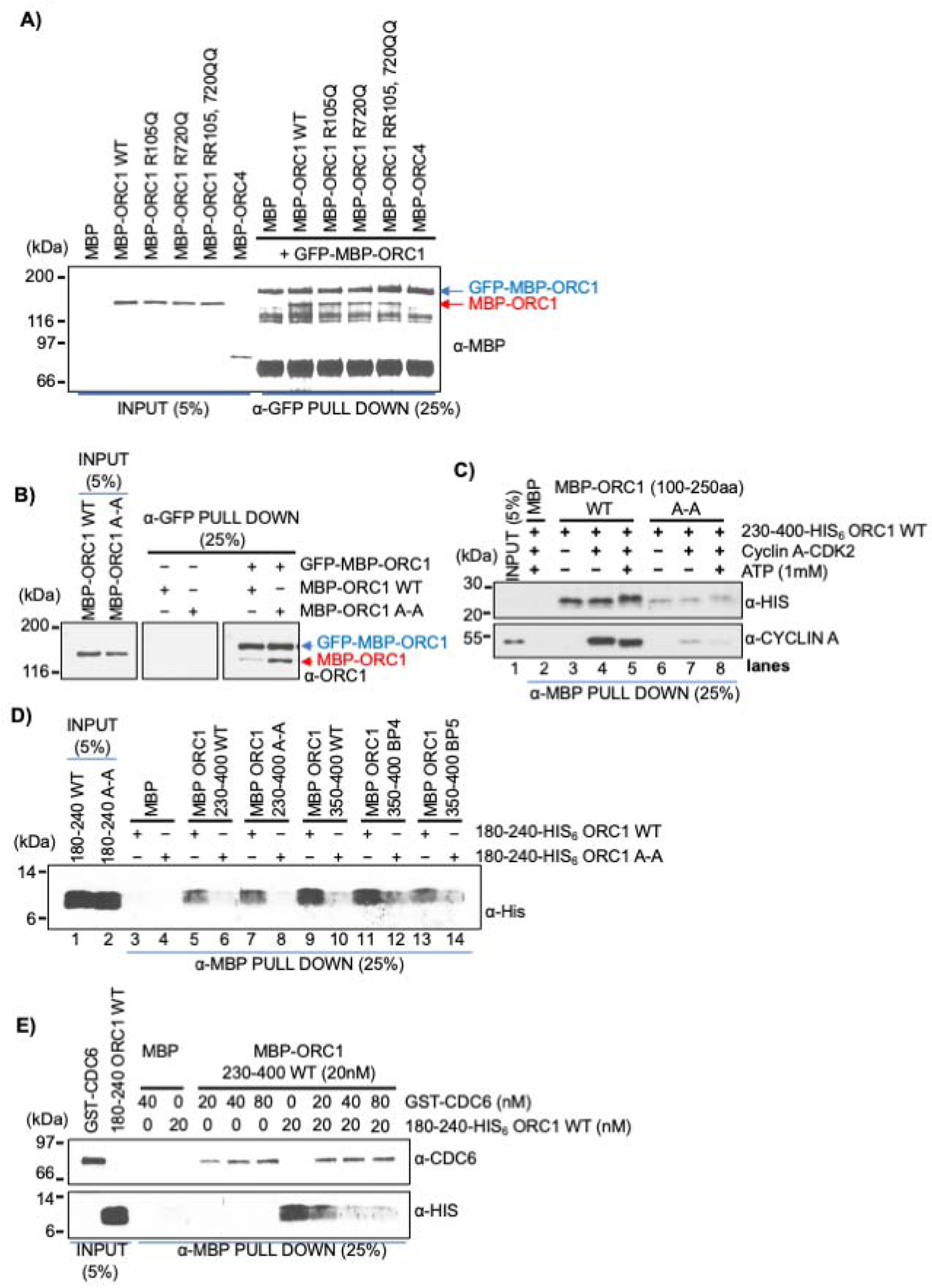
(A) MBP-GFP-ORC1 wild type protein bound to α-GFP antibody magnetic beads was incubated with either MBP-ORC1 WT or its mutants (R105Q; R720Q; R105Q, R720Q) or MBP-ORC4 and blotted with anti-GFP antibody in a GFP pull-down assay. (B) MBP-GFP-ORC1 protein was incubated with either MBP-ORC1 wild type (WT) or MBP-ORC1 Cy motif mutant (A-A) in the presence of 1mM ATP. MBP-GFP-ORC1 protein was immunoprecipitated with α-GFP antibody magnetic beads, then immunoblotted with the indicated antibodies (right panel). (C) ORC1 fragments, MBP-ORC1 100–250 aa WT or its A-A mutant proteins bound to α-MBP antibody magnetic beads were incubated with ORC1 230–400-His6 protein either alone or in presence of Cyclin A-CDK2 with or without 1mM ATP in MBP pull-down assays. Reactions were blotted with anti-HIS and anti-Cyclin A antibodies. (D) MBP-ORC1-230-400 WT or its Cy motif mutant along with MBP-ORC1-350-400 WT or its basic patch mutants (BP4 and BP5) bound to α-MBP antibody magnetic beads were incubated with His-tagged ORC1 protein fragments, 180–240 WT and 180–240 A-A and probed with anti-His antibody in MBP pull-down assays. See Figure S5B. (E) CDC6 competitively inhibits inter-molecular ORC1 interaction. The constant amount of recombinant MBP-ORC1-230-400 protein fragment bound to α-MBP antibody magnetic beads was incubated with either GST-CDC6 or ORC1-180-240-His6 individually or both together in MBP pull-down assays and immunoblotted with anti-CDC6 and anti-His antibodies. See Figure S5C.
The binding between the minimal domain of ORC1 that binds CDC6 (ORC1-180-240-HIS6; with and without the A-A mutation) and sub-fragments of the 230–400 domain of ORC1 were then examined, including a 230–400 fragment that harbors the Cy motif mutation (230–400 A-A) (Figures 5D and S5B). The wild type ORC1-180-240-HIS6 fragment bound to both wild type and Cy mutant versions of the MBP-ORC1-230-400 fragment (Figure 5D, lanes 5–8). The Cy (A-A) mutant ORC1-180-240-HIS6 did not bind to the MBP-ORC1-230-400 fragment. The ORC1-230-400 region is known to have a high abundance of positively charged residues that form basic patches that are involved in DNA binding in human ORC (Kawakami et al., 2015) and this fragment is related to a region in Drosophila and S. cerevisiae Orc1 that is required for ORC binding to DNA (Bleichert et al., 2018; Li et al., 2018; Schmidt and Bleichert, 2020). Comparative sequence analysis showed that human ORC1 contains two basic patches in this region, one previously reported as BP4 (Li et al., 2018) that is not well conserved, and another that we identified as a new basic patch and named BP5, which is highly conserved in the vertebrates (Figure S5A). MBP pull-down binding assays clearly show that mutation of BP5, but not BP4, in the ORC1-350-400 fragment disrupted its direct interaction with ORC1-180-240-HIS6 in Cy motif dependent manner (Figure 5D, lanes 9–14; Figure S5B). Since the ORC1-180-240 fragment does not overlap with the ORC1-350-400 fragment, this demonstrates that ORC1 molecules can interact in trans, but interaction between these two domains in cis is also possible.
Since the ORC1-180-240 fragment bound to CDC6 and the A-A mutation affects binding to CDC6, we investigated if CDC6 could disrupt the ORC1-ORC1 self-interaction between fragments 180–240 and 230–400. Increasing amounts of GST-CDC6 were added to binding assays between MBP-ORC1-230-400 and ORC1-180-240-HIS6 and it is clear that stoichiometric amounts of CDC6 inhibited the ORC1-ORC1 interaction (Figures 5E and S5C). Thus, CDC6 has the potential for controlling ORC1 self-interaction.
Dissociation of ORC1 from CDC6 by Cyclin A-CDK2-SKP2 promotes ORC1 degradation
As the levels of Cyclin A protein rise at the G1-S boundary and into S and G2 phases, the Cyclin A-CDK2 kinase disrupts ORC1 and CDC6 interaction, concomitantly leading to proteasome mediated degradation of human ORC1 protein, while phosphorylated CDC6 is exported to the cytoplasm (Méndez et al., 2002; Petersen et al., 1999). Human ORC1 is degraded by the SCFSKP2-activated proteasome complex using SKP2 as a F-box protein, but how ORC1 is targeted for degradation was unclear (Méndez et al., 2002). In U2OS cells, siRNA mediated depletion of effector molecules involved in various ubiquitin-mediated degradation pathways demonstrated that ORC1 degradation was solely dependent on SKP2 protein, as its loss stabilized ORC1 protein in a manner similar to Cyclin E protein degradation, another SCFSKP2 target (Figure S6A). In contrast, CDC6 protein was stabilized by loss of Cdh1, as expected (Petersen et al., 2000), but also by the loss of Cdt2 (Figure S6A) (Clijsters and Wolthuis, 2014). SETD8, which is known to be degraded by the CRL4Cdt2 pathway was used as a control (Centore et al., 2010; Petersen et al., 2000).
SCFSKP2 was depleted in double-thymidine synchronized U2OS cells and upon release, analyzed for protein levels at various times into S phase (Figure 6A). ORC1 remained degraded in control siRNA treated cells, while it was stabilized following SKP2 knockdown, as was Cyclin E, while the levels of Cyclin A, CDC6 and ORC3 proteins were not affected. SKP2 binds Cyclin A (Ji et al., 2006; Zhang et al., 1995) and ORC1 protein interacted with SKP2 only when Cyclin A was present across the synchronized HeLa cell cycle (Figure 6B). Cyclin A immunoprecipitation of a similar HeLa cell synchronization experiment confirmed that ORC1 and CDC6 interacted with Cyclin A when Cyclin A and either ORC1 or CDC6 were both present in the cells (Figure S6B). Since SKP2 associated with Cyclin A, it appears that Cyclin A levels during the G1-S transition and into S phase determine ORC1 protein levels.
Figure 6. Cyclin A-CDK2 recruits SPK2 to promote ORC1 degradation.
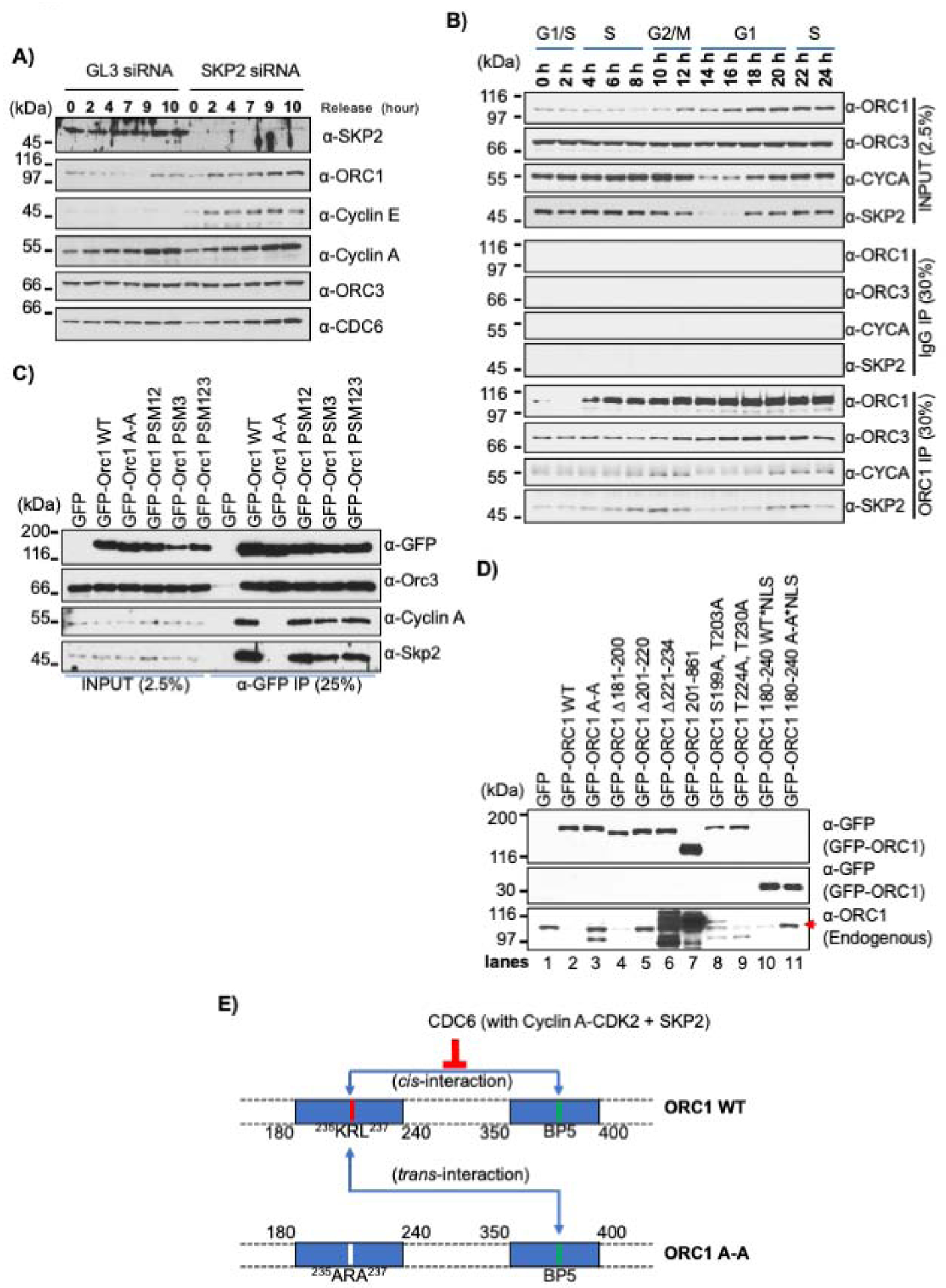
(A) U2OS cells transfected with either SKP2 or control GL3 siRNAs were synchronized by double thymidine block and harvested at indicated time points after release from the block. Cell lysates were immunoblotted as indicated. (B) Double thymidine synchronized HeLa cells collected at different time points were lysed and ORC1 immunoprecipitation analyzed by western blot. (C) HEK293 cells were transfected with GFP-ORC1 WT or its Cy motif and CDK phosphorylation site mutants as indicated for overexpression and cells were lysed after 36 hours. Cell lysate immunoprecipitated with anti-GFP antibody were analyzed by immunoblotting with indicated antibodies. (D) Cell lysate from HEK293 cells overexpressing GFP-tagged wild type ORC1 or its mutants [Cy mutant (A-A); internal deletion mutants from full length (Δ181–200, Δ201–220, and Δ221–234); N-terminal deletion (200–861); predicted phosphorylation site double mutants (S199A, T203A and T224A, T230A); NLS fused minimal region 180–240 WT or A-A mutants] were used either for immunoprecipitation in figure S6D or blotted with ORC1 antibody to detect endogenous level of ORC1 proteins. The input of anti-GFP blots from figure S6D is used again for comparison. Red arrow denotes endogenous ORC1 protein band. See also Figure S6D. (E) Schematic showing self-interaction of two distinct regions of ORC1: 180–240 that binds CDC6 as well as contains the Cy motif and 350–400 containing the BP5 basic patch. The cis-interaction is competitively inhibited by wild type CDC6 protein. Upon overexpression of the cis-interaction defective ORC1 Cy mutant, the mutant protein defective in binding to Cyclin A-SKP2 protein establishes a trans-interaction with its wild type counterpart, thus providing protection of wild type protein by sequestering it from the action of SCFSKP2 ubiquitylation and protein degradation.
Based on this observation, various ORC1 mutants were tested for interaction with Cyclin A and SKP2. Cells were transfected with either the GFP-ORC1 wild type plasmid, with the GFP-ORC1 A-A Cy motif mutant, or the GFP-ORC1 PMS123 phosphorylation mutant and then anti-GFP antibodies were used for co-immunoprecipitation of endogenous Cyclin A and SKP2 proteins. Neither Cyclin A nor SKP2 protein interacted with the GFP-ORC1 A-A Cy motif mutant protein, while mutation of the three ORC1 CDK phosphorylation sites had no effect on its binding to Cyclin A or the SKP2 protein (Figure 6C). It has been reported that the human ORC1 protein in Meier-Gorlin Syndrome derived patient cell lines had reduced protein stability in the chromatin fraction (Bicknell et al., 2011), but we found no loss of interaction between GFP-ORC1 MGS mutants (ORC1-F89S or ORC1-R105Q or ORC1 E127E) and SKP2 protein (Figure S6C).
These results suggested that the self-interaction between the Cy-motif-containing domain of ORC1 that also binds CDC6 (180–240) and the BP5 domain (350–400) might control ORC1 degradation. We therefore expressed N-terminally GFP-tagged mutants of either full length ORC1 or the ORC-180–240 domain that were tagged on the amino-terminus with GFP, and tested for self-interaction with endogenous ORC1 protein using anti-GFP immunoprecipitation (Figure 6D). Expression of GFP-ORC1 eliminated the expression of endogenous ORC1 protein, but the same protein harboring an A-A mutation that affects binding to Cyclin A-CDK2 and ORC1 self-interaction did not reduce endogenous ORC1 protein levels (Figure 6D, lanes 1–3). Smaller deletions in this domain of ORC1 did not reduce endogenous ORC1 levels, including Δ201–220, Δ221–234 and Δ1–200, but Δ181–200 reduced endogenous ORC1 levels like wild type ORC1 (Figure 6D, lanes 4–7). One deletion (Δ1–200) actually increased endogenous ORC1 levels because it was itself over-expressed (Figure 6D, lane 7). The phosphorylation mutants of ORC1 also reduced the endogenous level of ORC1 (Figure 6D, lanes 8 and 9). Interestingly, expression of the small domain (GFP-ORC-180–240) that binds to Cyclin A-CDK2, CDC6 and the ORC1 BP5 domain also reduced the levels of endogenous ORC1, but its A-A mutant did not (Figure 6D, lanes 10 and 11). The ability of ORC1 or its mutant derivatives to eliminate endogenous ORC1 levels when expressed as a GFP-fusion protein correlated precisely with binding to Cyclin A and SKP2 in anti-GFP immunoprecipitates (Figures 6D and S6D). This result strongly suggests that Cyclin A-CDK2 recruits SKP2 to ORC1 and causes ORC1 degradation, and of particular importance is the observation that over-expression of ORC1 or a fragment of ORC1 that can self-interact with the BP5 domain and bind Cyclin A-CDK2-SKP2 can degrade ORC1 in trans (i.e., degrade endogenous ORC1; Figure 6E).
Since human ORC1 A-A Cy motif mutant did not bind to Cyclin A and SKP2 protein, live cell microscopy was performed with U2OS cell lines stably expressing GFP-ORC1 wild type or mutants to investigate SCFSKP2 mediated protein degradation. The wild type GFP-ORC1 protein fluctuated during the cell cycle and was only present in mitosis and G1 phase, whereas the GFP-ORC1-A-A Cy motif mutant accumulated to high levels and was not degraded, indicating the loss of proteasome mediated control of ORC1 degradation (Figure S6E). In double thymidine synchronized and released cells that stably expressed GFP-ORC1 wild type or mutant proteins, the GFP-ORC1-A-A Cy motif mutant protein was more stable compared to wild type protein, while the levels of the GFP-ORC1 PMS123 phosphorylation mutant were only mildly affected (Figure S6F). These results describe a role for the Cy motif of human ORC1; controlling the dynamics of ORC1 protein levels by binding Cyclin A and SCFSKP2.
RVxF motif-dependent interaction of ORC1 with protein phosphatase PP1 controls ORC1 phosphorylation and protein stability
The observation that phosphorylated human ORC1 did not interact with CDC6 during mitosis, and the subsequent dephosphorylation of ORC1 upon mitotic exit led us to investigate the role of protein phosphatases in this dynamic behavior. Previous reports using yeast two-hybrid systems suggested that human ORC2 interacted with protein phosphatase PP1, which in turn controlled the association ORC with chromatin by dephosphorylating ORC2 protein (Lee et al., 2014a). We found that human ORC1 interacted with protein phosphatase PP1 along with its protein partner RIF1 in ORC1 immunoprecipitates from lysates of exponentially growing HeLa and U2OS cells (Figure S7A). Immunoprecipitation of all the three isoforms of GFP-PP1 (α-, β- and γ) from transiently transfected GFP-PP1 plasmids in HEK293 cells also detected an interaction between PP1 and endogenous ORC1 protein, along with RIF1 protein (Figure S7B) (Sreesankar et al., 2012). In MBP pull-down assays using purified proteins, GST-PP1α interacted with MBP-ORC1, MBP-ORC2 and MBP-CDC6, while no interaction was detected with other ORC subunits (Figure 7A). Mapping studies showed that the ORC1-200-300 IDR of ORC1 was necessary for its interaction with GST-PP1α (Figure S7C and S7D). The 200–300 amino acid region of human ORC1 harbors a conserved RVxF sequence (265KVAF268), a well-known PP1 binding motif, flanked by two CDK phosphorylation sites (Figure 7B) (Bollen et al., 2010; Hendrickx et al., 2009). MBP-ORC1 Δ260–280 mutant protein lacking the PP1 binding motif lost its interaction with GST-PP1α protein, while deletion of 20 amino acids from adjacent regions had no effect (Figure S7E). Mutation of the PP1 binding motif in MBP-ORC1 protein, from 265KVAF268 (MBP-ORC1 WT) to 265KAAA268 (MBP-ORC1-PP1mut) disrupted PP1 binding to ORC1 (Figure 7C).
Figure 7. ORC1 directly binds to PP1 protein phosphatase.
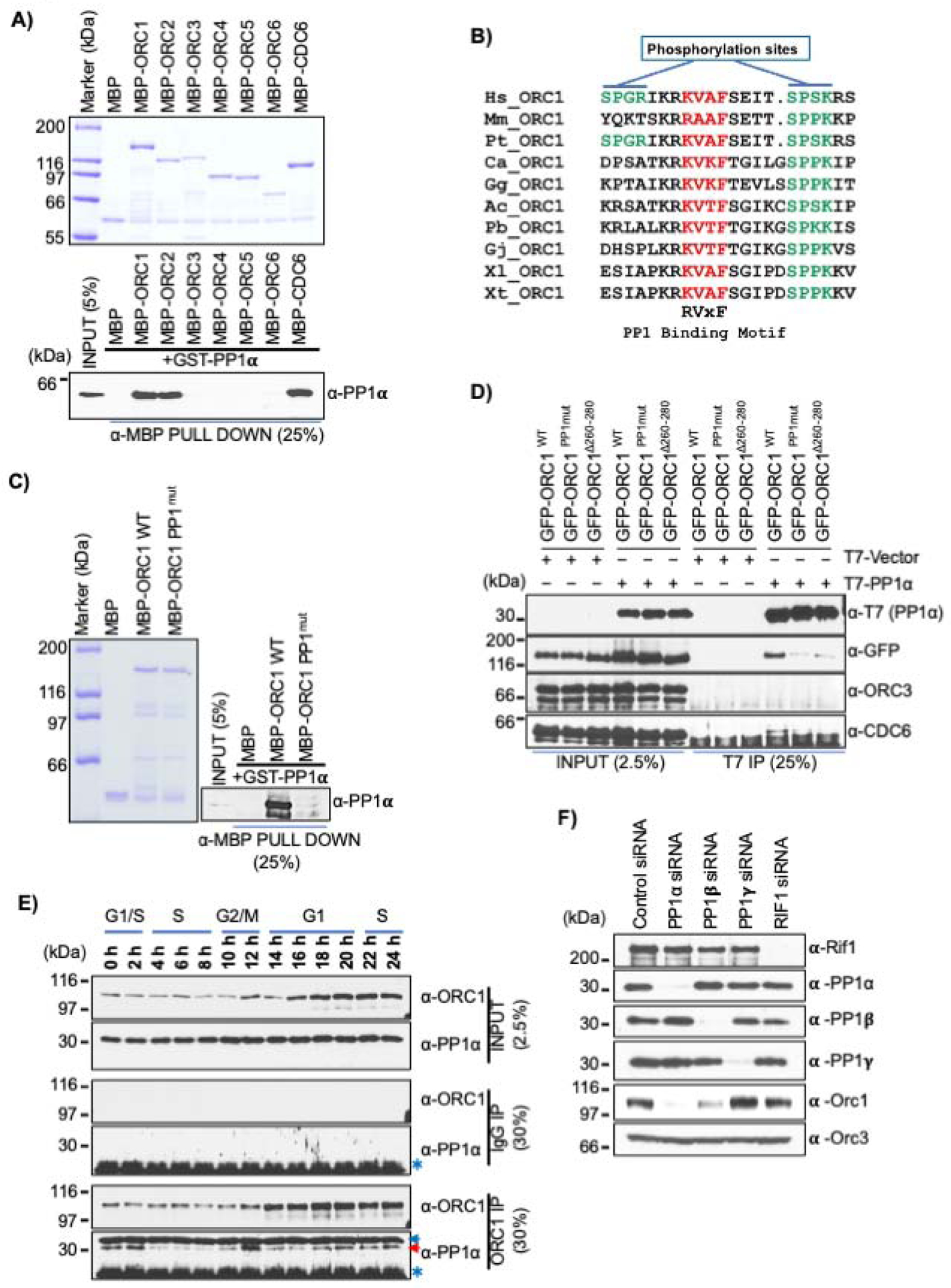
(A) Purified MBP tagged ORC and CDC6 proteins bound to α-MBP antibody magnetic beads were incubated with purified GST-PP1α protein in an MBP pull-down assay and blotted with anti-PP1α antibody (bottom panel); MBP-fused recombinant proteins shown in top panel. (B) Alignment of PP1 binding motif in ORC1 vertebrate species. The PP1 binding RVxF motif is highlighted in red, while the adjacent CDK phosphorylation sites are indicated in green. (Homo sapiens, Hs; Mus musculus, Mm; Pan troglodytes, Pt), Aves (Calypte anna, Ca; Gallus gallus, Gg), Reptiles (Anolis carolinensis, Ac; Python bivittatus, Pb; Gekko japonicus, Gj) and Amphibia (Xenopus laevis, Xl; Xenopus tropicalis, Xt). (C) MBP-ORC1 WT protein or corresponding PP1 binding mutant, MBP-ORC1PP1mut bound to α-MBP antibody magnetic beads were incubated with purified GST-PP1α protein in MBP pull-down assay and blotted with anti-PP1α antibody (right panel). MBP-fused recombinant proteins are shown in left panel. (D) HEK293 cells were co-transfected with GFP-ORC1 WT or its PP1 binding mutants (GFP-ORC1PP1mut and GFP-ORC1Δ260−280) with either T7-PP1α or empty T7 vector. Anti-T7 antibody precipitates were immunoblotted with the indicated antibodies. (E) HeLa cells were synchronized using double thymidine block and collected at different time points after release. Cell extracts were immunoprecipitated with anti-ORC1 or control IgG antibody and further immunoblotted with anti-ORC1 and anti-PP1α antibodies. The red arrow indicates specific PP1 band, while the blue arrow and an asterisk symbol indicates non-specific and cross-reactive light IgG bands, respectively. (F) siRNAs targeting PP1 isoforms and RIF1 were used to deplete endogenous proteins from asynchronous growing U2OS cells. Total cell lysate of siRNA treated U2OS cells were prepared after 36 hours of knockdown and immunoblotted with indicated antibodies.
HEK293 cells overexpressing T7-PP1α along with GFP-ORC1 WT or its mutants confirmed the in vitro data that the RVxF motif in human ORC1 was required for its interaction with PP1α, since T7-PP1α did not interact with the GFP-ORC1 Δ260–280 and GFP-ORC1 PP1mut mutants compared to GFP-ORC1 wild type (Figure 7D). Furthermore, T7-PP1α immunoprecipitation did not pull-down endogenous ORC3 protein, and while its interaction with endogenous CDC6 was very weak, it too, was dependent on the ORC1 RVxF motif.
The interaction between ORC1 and PP1α protein during the cell cycle was investigated by performing ORC1 immunoprecipitations from lysates of synchronized HeLa cells. ORC1 strongly interacted with PP1α during mitosis, while the interaction was slightly lowered at the G1-S transition (Figure 7E). Consistent with the observation that ORC1 protein was phosphorylated in mitosis, in vitro phosphorylation of MBP-ORC1 WT or its A-A Cy motif and CDK phosphorylation mutants by either Cyclin E-CDK2 or Cyclin A-CDK2 kinases did not impair its binding to GST-PP1α in MBP pull-down assays, while MBP-ORC1 PP1mut with GST-PP1α did not bind independent of phosphorylation (Figure S7F). Thus, human ORC1 phosphorylation does not affect its interaction with PP1α during mitosis.
Based on the results above, we treated U2OS cells with the protein synthesis inhibitor cycloheximide and compared the protein stability of ORC1 and CDC6 proteins over time. CDC6 protein was rapidly degraded, similar to Cyclin F, but the turn-over rate for ORC1 and Cyclin E proteins was slower (Figure S7G). Furthermore, to investigate the role of protein phosphorylation in human ORC1 protein degradation, cells were treated with roscovitine, a CDK inhibitor. Upon treatment, U2OS cells showed a rapid decrease in the stability of CDC6 protein with time, leading to the inference that the phosphorylation of endogenous CDC6 stabilizes the protein (Mailand and Diffley, 2005), also observed for Cyclin F protein. However, roscovitine treatment of U2OS cells showed an opposite effect on endogenous ORC1 protein, with an increase in the protein levels, indicating that phosphorylation of human ORC1 promoted its degradation, while no effect was seen on Cyclin E protein levels (Figure S7H).
Depletion of individual PP1 isoforms or RIF1 using siRNAs in U2OS cells showed that upon knockdown of PP1α the endogenous ORC1 protein level was reduced, while depletion of PP1β reduced ORC1 to a lesser extent and reducing PP1γ had no effect (Figure 7F). The results indicated that PP1α controls phosphorylation mediated degradation of ORC1. We did not observe any effect on ORC1 protein levels upon RIF1 depletion in contrast to a previous report indicating RIF1 mediated recruitment of RIF1-PP1 complex in controlling ORC1 protein stability (Hiraga et al., 2017).
Dominant negative phenotype of human ORC1 mutants defective in binding Cyclin A-CDK2 kinase and PP1 protein phosphatase
The phenotypes associated with ORC1 A-A, ORC1 PSM123 and the PP1 mutant were investigated. Cell proliferation and cell cycle profiles of stable U2OS cell lines harboring tetracycline inducible GFP-ORC1 wild type or mutants were monitored for 72 hours after depletion of endogenous ORC1 protein using a 3’-UTR specific ORC1 siRNA. Compared to control siRNA, the depletion of endogenous ORC1 from U2OS cells significantly retarded its proliferation, while the expression of GFP-ORC1 rescued the proliferation defect in siORC1 treated cells (Figures S8A & S8B). In contrast, expression of GFP-ORC1 mutants - GFP-ORC1 A-A, GFP-ORC1 PMS123 or GFP-ORC1PP1mut were not able to rescue the proliferation defect following endogenous ORC1 protein depletion (Figure S8A). Interestingly, when the GFP-ORC1 mutants were expressed, the cells proliferated slower in both target siORC1 treated and even control siRNA treated cells, demonstrating a dominant negative phenotype associated with these mutants. Flow cytometry data of EdU labelled cells shows that after tetracycline induction, the GFP-ORC1 A-A and GFP-ORC1 PMS123 cell lines had constant and high GFP expression of these mutant proteins in S phase, while the expression of GFP-ORC1 WT and GFP-ORC1PP1mut remained comparable and very low (Figure S8C and Figure S8E). During the G2/M phase, the expression of GFP-ORC1 A-A and GFP-ORC1 PSM123 mutants in the induced cell lines substantially increased compared to GFP-ORC1 WT, while the GFP-ORC1PP1mut was only marginally elevated (Figure S8D and Figure S8E).
Discussion
The ORC1 IDR contains multiple SLiMs, including a nuclear localization signal, a PP1 binding site, multiple CDK phosphorylation sites, the Cy motif that promotes binding of Cyclin A, but not Cyclin E, the BP5 motif that promotes ORC1 intramolecular interaction and a domain that binds to CDC6 (Figure S9, top panel). The ORC1 Cy motif overlaps extensively, but is not identical to the CDC6 binding domain. Likewise, the CDC6 IDR contains a Cy motif that binds both Cyclin A and Cyclin E, CDK phosphorylation sites and a degron. The amino acids within the CDC6 Cy motif contribute to ORC1 interaction (Figure S9, bottom panel).
The interaction between ORC1 and CDC6 was found to occur only during a short window during G1 phase, a period when Cyclin E-CDK2 was active. Even though ORC1 and CDC6 were present during mitosis, the two proteins did not bind due to Cyclin A-CDK2 phosphorylation of ORC1 that occurs during late G2 phase when ORC1 is re-stabilized. The cell cycle variation in protein levels and CDK phosphorylation of ORC1 in human cells resembles the levels and phosphorylation of S. cerevisiae Cdc6, where CDK phosphorylation prevents premature pre-RC assembly (Mimura et al., 2004). ORC1 remained phosphorylated during mitosis and was dephosphorylated by protein phosphatase PP1 upon mitotic exit, and the phosphorylated form of ORC1 could not bind CDC6, thereby contributing to prevention of premature pre-RC assembly until CDC6 is degraded by either CDH1- and CRL4Cdt2-dependent, ubiquitin-mediated proteolysis upon mitotic exit and entry into early G1 phase (Clijsters and Wolthuis, 2014; Petersen et al., 2000). A mutant ORC1 that could not bind PP1 was defective in cell proliferation and reduced the number of cells in S phase. Unlike a previous report, we did not observe increased levels of CDC6 upon removal of Cyclin F (Walter et al., 2016).
Multiple mechanisms involving Geminin inhibition of CDT1 function, CDT1 phosphorylation and CDK inhibition of pre-RC assembly all contribute to prevention of re-licensing of origins of DNA replication during S and G2 phases (Lygerou and Nurse, 2000; Wohlschlegel et al., 2000; Zhou et al., 2020). We suggest that as ORC1 binds mitotic chromosomes during mitosis (Kara et al., 2015; Okuno et al., 2001), the inhibition of CDC6 binding to ORC1 prevents assembly of a Mcm2–7 loading complex, but in this case rather than prevent re-licensing, it ensures new origin licensing is restricted to a post-mitotic period (Figure S10).
The mechanism of assembly of ORC proteins in human cells is different from that observed in Chinese hamster cells, since it has been reported that hamster CgOrc1 is stable throughout the cell cycle, is ubiquitylated upon G1-S phase transition, but is not degraded (Li et al., 2004; McNairn et al., 2005; Okuno et al., 2001). Moreover, the current understanding of chromatin binding of Orc1 in Chinese hamster CHO cells is confusing, with one report stating the Cyclin A-CDK2 blocks Orc1 from binding to chromatin during mitosis while another reporting that multiple CHO ORC subunits, including Orc1, bind to mitotic chromosomes (Li et al., 2004; Okuno et al., 2001). In human cells, ORC is assembled after mitotic exit (Figure S10).
CDC6 is re-synthesized upon commitment to cell division by E2F1-dependent transcription (Yan et al., 1998). CDC6 and Cyclin E synthesis, and hence the majority of pre-RC formation in human cells, occurs after cells have committed to re-enter the cell division cycle, a decision regulated by activation of Cyclin D-CDK4/6 (Johnson and Skotheim, 2013; Narasimha et al., 2014) and also involving ORC1 binding to the Retinoblastoma protein and the histone methyltransferase SUV39H1 (Hossain and Stillman, 2016). The assembled Cyclin E-CDK2-CDC6 then promotes pre-RC assembly (Cook et al., 2002; Coverley et al., 2002), which involves ORC1 binding to CDC6 only during the mid-G1 phase. By mid G1 phase, the full ORC is assembled and CDC6 is recruited to form a MCM2–7 loading complex in cooperation with CDT1. It is known from studies in S. cerevisiae and Drosophila that CDC6 binding to ORC1 via its AAA+ domain enables the CDC6 ATPase activity to be stimulated by an intimate interaction between the AAA+ domains of CDC6 and ORC1 (Schmidt and Bleichert, 2020; Speck and Stillman, 2007; Yuan et al., 2017) [Feng, X., Barbon, M., Noguchi, Y., Stillman, B., Speck, C. and Li, H; submitted). Modeling of CDC6 into the structure of ORC confirms this interaction between human ORC1 and CDC6 (Tocilj et al., 2017). Thus, we were surprised to find that the initial mode of recruitment of CDC6 to ORC1 involves interactions between small domains in the IDRs of both proteins, not the AAA+ domains.
The IDR of CDC6 containing the Cy motif binds to the ORC1-180-240 domain, which also harbors a Cy motif. In ORC1, the amino acids that define its Cy motif are, for the most part, required for CDC6 and Cyclin A to bind ORC1. The absence of Cyclin A in mid G1 phase allows CDC6 to be recruited to ORC1 and the ORC1 Cy motif cannot bind to Cyclin E. This sets the stage for assembly of the previously observed ORC-CDC6-Cyclin E-CDK2 ternary complex on DNA that can assemble pre-RCs (Hossain and Stillman, 2016). Thus, the bringing together of the ORC1 and CDC6 AAA+ domains, which cannot by themselves interact, involves small domains within the IDR of both proteins, possibly assisted by formation of a liquid phase transition on DNA. We suggest that AAA+ ATPase assembly is mediated by these seed sequences in IDRs that regulate temporal assembly of ORC-CDC6 ATPase during the cell division cycle.
The use of small regulatory domains in ORC and CDC6 to assemble multimeric AAA+ domain protein complexes is reminiscent of the use of bacterial regulatory “receiver” R domains of Enhancer Binding Proteins (EPBs) to assemble functional AAA+ ATPases (Chen et al., 2008; Gao et al., 2020). These R domains respond to environmental conditions via two-component signal transduction to activate genes controlled by σ54 transcription factors. The R domains of EBPs lie outside of the AAA+ domains, but bring the AAA+ domains together so that they remodel the σ54–RNAP (RNA polymerase) holoenzyme at gene promoters. In an analogous way, we suggest that the CDC6 and ORC1 domains that mediate assembly of ORC-CDC6 complexes be called R (regulatory) domains because they control the assembly and disassembly of the ORC-CDC6 AAA+ ATPase complex and like the R domains of EBPs, they are controlled by protein phosphorylation (see Figure S9).
The IDRs of ORC1 and CDC6 undergo a liquid phase transition mediated by DNA (Parker et al., 2019). Purified ORC1 and CDC6 could also form a phase transition on DNA and ORC1 could self-interact between its CDC6-binding and Cy-motif-containing 180–240 domain and the BP5 domain, both of which exist within the ORC1 IDR. Moreover, the ORC1-ORC1 binding in trans controlled the level of ORC protein in cells and disruption of the Cy motif increased intracellular exo- and endogenous ORC1 protein levels. The ORC1 self-interaction was controlled by CDC6 binding. Therefore, we suggest that the self-interaction of ORC1 and its sequestration into a liquid phase with CDC6 provides a molecular mechanism for maintaining the appropriate levels of nuclear ORC1. This may be a general mechanism for controlling protein levels in cells (Jaremko et al., 2020; Parker et al., 2019; Siddiqui and Stillman, 2007; Tocilj et al., 2017).
Following pre-RC assembly at origins, Cyclin A-CDK2 is resynthesized at the G1-S phase transition to activate DNA replication and binds to ORC1 via its Cy motif, co-recruiting SKP2 to the complex. SKP2 depletion increased ORC1 levels, consistent with our previous studies (Méndez et al., 2002). The binding of SKP2 and Cyclin A-CDK2 to ORC1 is not affected by the three CDK sties that influence its interaction with CDC6 in mitosis, suggesting that the ORC1 Cy motif functions in a different manner at the G1-S phase transition than it does in mitosis. In mitosis, the phosphorylation of ORC1 prevents CDC6 binding, but in late G1 the binding of Cyclin A-CDK2 dissociates ORC1 from CDC6, but it also brings in a ubiquitin ligase that targets ORC1 for destruction. ORC1 is degraded in a spatially and temporarily regulated pattern in the nucleus that is similar to the temporal pattern of DNA replication in S phase (Kara et al., 2015). Furthermore, in early G1 phase, the APCCDH1 ubiquitin ligase antagonizes the SKP2 ubiquitin ligase (Bashir et al., 2004; Wei et al., 2004), thereby preventing premature ORC1 degradation. Thus, the ORC1 Cy motif at the G1-S phase transition functions to promote ORC1 destruction and is thus part of the ORC1 degron. In the ORC1 Cy mutant, ORC1 accumulated in S and G2 phases and greatly slowed cell proliferation, emphasizing the importance of maintaining the correct levels of ORC1 protein in the nucleus.
Multiple SLiMs are often found in IDRs, including IDRs that regulate protein kinases and many protein-protein assemblies such as transcription, splicing and protein location in cells (Gógl et al., 2019; Lee et al., 2014b) and this is also the case for the two ATPase subunits of the human replication initiator ORC1-CDC6. The diverse functions of the ORC1-CDC6 SLiMs, as protein-protein interaction motifs, enzyme accelerators for both kinases and a phosphatase, as a degron and as DNA binding motifs underscore the context dependency of these modules and show a remarkable complexity of function within the IDR. We suspect that the IDRs of ORC1 and CDC6 harbor other SLiM activities and need further investigation.
Limitations of the study
The demonstration of phase transition by ORC1 in vitro raises the issue of how ORC1 and CDC6 function in different physiological contexts in vivo. For example, fluorescence recovery after photobleaching (FRAP) demonstrated that ORC1 mobility in the nucleus is restricted compared to the mobility of ORC2, ORC3 and HP1 that all bind to ORC1 (Prasanth et al., 2010). One possibility that needs testing is whether ORC1 is tethered to nuclear structures, which need to be identified, and whether its various binding partners localize to different foci, such as sites of pre-RC assembly or heterochromatin. Furthermore, the nature of IDRs make investigation of the structure-function relationships between ORC and CDC6 challenging.
METHODS:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Bruce Stillman (stillman@cshl.edu).
Materials availability
The plasmids and cell line strains generated and used in this study are available through lead contact without restriction.
Data and code availability
The original images for figures in the paper are available at Mendeley database: http://dx.doi.org/10.17632/p8cn8srfvh.1. This study did not generate any unique datasets or code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture and cell synchronization, with siRNA treatment or transient transfection of expression plasmids.
U2OS, HeLa and HEK293 cells were obtained from the Cold Spring Harbor Laboratory cell culture collection and cultured in DMEM containing high glucose (Gibco) supplemented with 10% inactivated fetal calf serum and Penicillin/Streptomycin. HeLa suspension cells were grown in suspension in Joklik Modified Eagle’s Essential Minimal Medium (US Biologicals, M3867) supplemented with 5% calf serum (Hyclone, SH30087.04). All the cell lines tested negative for mycoplasma contamination. HeLa suspension cells were synchronized by double thymidine block and release as described previously (Siddiqui and Stillman, 2007). Briefly, cells were synchronized with 2.5mM Thymidine (Sigma, T1895–25G) for 16 hours and then released into fresh complete JMEM for 12 hours. Cells were again treated with 2.5mM Thymidine for 16 hours after which, cells released into fresh media were harvested as 0 hour (at G1/S boundary) and every two hours thereafter. To synchronize U2OS cells at G2/M boundary, 100 ng/ml of nocodazole was added to fresh medium for 16 hours. After 16 hours of block the cells were washed two times with 1x Phosphate Buffered Saline (PBS) and subsequently, released into the fresh media. Mitotic U2OS cells were collected by mitotic shake-off method. U2OS cells were synchronized at G1/S using double thymidine block and release protocol. Briefly, the U2OS cells were blocked for 16 hours with incubation of 2.5mM of thymidine, washed with PBS to release for 12 hours in drug free medium and again incubated with thymidine for 16 hours. The cells were washed and released after second thymidine block for different times. For depletion of proteins in G1/S synchronized U2OS cells, the U2OS cells transiently transfected with 100nM of siRNA’s (control GFP as well as SKP2 siRNA) using Lipofectamine RNAiMax after first thymidine block. Plasmids expressing exogenous genes were transfected using 5 μg of DNA using lipofectamine 2000 transfection reagents (Thermo Fisher Scientific). The U2OS cells were incubated 25 μg/ml of cycloheximide and 10 μg/ml of roscovitine drugs as indicated in the results. The sequences of the siRNAs used are listed in key resource table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse moclonal anti-ORC1 | CSHL Facility | Kara et. al., 2015 |
| Rabbit polyclonal anti-CDC6 | CSHL Facility | Hossain et al., 2016 |
| Rabbit polyclonal anti-ORC3 | CSHL Facility | Siddiqui et al., 2007 |
| Goat polyclonal anti-ORC4 | Abcam | Abcam Cat# ab9641, RRID:AB_296536 |
| Mouse monoclonal anti-Cyclin A | BD Biosciences | BD Biosciences Cat# 611269, RRID:AB_398797 |
| Rabbit polyclonal anti-Skp2 | Abcam | Abcam Cat# ab19877, RRID:AB_777950 |
| Mouse monoclonal anti-CDC6 | Millipore | Millipore Cat# 05550, RRID:AB_2276118 |
| Mouse monoclonal anti-Cyclin E | BD Biosciences | BD Biosciences Cat# 551159, RRID:AB_394079 |
| Mouse monoclonal anti-Cdt1 | CSHL Facility | PKS13 |
| Rabbit monoclonal anti-SETD8 | Cell Signaling Technology | Cell Signaling Technology Cat# 2996, RRID:AB_2254384 |
| Mouse monoclonal anti-Histone H3(Ser10) | Cell Signaling Technology | Cell Signaling Technology Cat# 9706, RRID:AB_331748 |
| Mouse monoclonal anti-MBP | New England Biolabs | New England Biolabs Cat# E8032, RRID:AB_1559730 |
| Goal polyclonal anti-GST | GE Healthcare | GE Healthcare Cat# 27-4577-01, RRID:AB_771432 |
| Mouse monoclonal anti-GST | CSHL Facility | N/A |
| Rabbit polyclonal anti-Cyclin F | Santa Cruz Biotechnology | Santa Cruz Biotechnology Cat# sc-952, RRID:AB_2071212 |
| Rabbit polyclonal anti-phospho RB (Ser807/811) | Cell Signaling Technology | Cell Signaling Technology Cat# 9308, RRID:AB_331472 |
| Rabbit polyclonal anti-GFP | Sigma-Aldrich | Sigma-Aldrich Cat# G1544, RRID:AB_439690 |
| Mouse monoclonal anti-GFP | Thermo Fisher Scientific | Thermo Fisher Scientific Cat# A-11120, RRID:AB_221568 |
| Rabbit polyclonal anti-6X His | Abcam | Abcam Cat# ab9108, RRID:AB_307016 |
| Mouse monoclonal anti-Cdh1 | Millipore | Millipore Cat# CC43, RRID:AB_2260255 |
| Rabbit polyclonal anti-Cdt2 | Novus | Novus Cat# NB100–40840, RRID:AB_789689 |
| Rabbit polyclonal anti-PPP1CA | Bethyl | Bethyl Cat# A300–904A, RRID:AB_2284190 |
| Mouse monoclonal anti-PP1 alpha | Santa Cruz Biotechnology | Santa Cruz Biotechnology Cat# sc-271762, RRID:AB_10708123 |
| Rabbit polyclonal anti-PP1 beta | Thermo Fisher Scientific | Thermo Fisher Scientific Cat# PA5-28225, RRID:AB_2545701 |
| Rabbit polclonal anti-PPP1CC | Bethyl | Bethyl Cat# A300-906A, RRID:AB_2168105 |
| Rabbit polyclonal anti-RIF1 | Bethyl | Bethyl Cat# A300–568A, RRID:AB_669806 |
| Bacterial and Virus Strains | ||
| BL21(DE3) Competent E. coli | New England Biolab | Cat# C2527I |
| XL10 Gold Ultracompetent Cells | Stratagene | Cat# 200314 |
| 5-alpha Competent E. coli | New England Biolab | Cat# C2987I |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lambda Protein Phosphatase | New England Biolab | Cat# P0753 |
| PreScission Protease | GE Healthcare | Cat# 27084301 |
| MG-132 | Sigma-Aldrich | Cat# 474787 |
| Cycloheximide | Sigma-Aldrich | Cat# C4859 |
| Doxycycline | Sigma-Aldrich | Cat# D9891 |
| Nocodazole | Sigma-Aldrich | Cat# M1404 |
| Hygromycin B | Thermo Fisher Scientific | Cat# 10687010 |
| Blasticidin S HCl | Thermo Fisher Scientific | Cat# A1113903 |
| Zeocin | Thermo Fisher Scientific | Cat# R25001 |
| Benzonase nuclease | Sigma-Aldrich | Cat# E1014 |
| Thymidine | Sigma-Aldrich | Cat# T1895 |
| Roscovitine | Sigma-Aldrich | Cat# 557364 |
| DMEM | Corning | Cat# 10–013-CV |
| JMEM medium | USBiological Life Sciences | Cat# M3867 |
| Fetal Bovine Serum | Sigma-Aldrich | Cat# F2442 |
| Tet System Approved FBS | Takara | Cat# 631101 |
| HyClone Calf Serum | Thermo Fisher Scientific | Cat# SH3008704 |
| Dynabeads ProteinG | Thermo Fisher Scientific | Cat# 10004D |
| Phusion high fidelity DNA polymerase | New England Biolab | Cat# M0530 |
| T7 gene 6 exonuclease | Thermo Fisher Scientific | Cat# 70025Z10KU |
| Amylose resin | New England Biolab | Cat# E8021 |
| Glutathione agarose | Thermo Fisher Scientific | Cat# 16100 |
| Ni-NTA resin | Qiagen | Cat# 30210 |
| Hemagglutinin peptide | Millipore Sigma | Cat# I2149 |
| Anti-HA Agarose | Thermo Fisher Scientific | Cat# 26181 |
| HRV-3C Protease, Biotin tagged | Millipore Sigma | Cat# SAE0110 |
| Critical Commercial Assays | ||
| Click-iT® Plus EdU Alexa Fluor® 647 Flow Cytometry Assay Kit | Thermo Fisher Scientific | Cat# C10635 |
| Experimental Models: Cell Lines | ||
| Human: U2OS | ATCC | Cat# HTB-96 |
| Human: HeLa | ATCC | Cat# CCL-2 |
| Human: HEK293T | ATCC | Cat# CRL-3216 |
| Human: U2OS TReX | Malecki et. al., 2006 | N/A |
| Deposited Data | ||
| Original Images | This Study | http://dx.doi.org/10.17632/p8cn8srfvh.1 |
| Oligonucleotides | ||
| siRNA targeting sequence: Control siRNA (GL3) # CUUACGCUGAGUACUUCGA |
Dharmacon | N/A |
| siRNA targeting sequence: Cdh1 siRNA# AAUGAGAAGUCUCCCAGUCAG |
Dharmacon | N/A |
| siRNA targeting sequence: Cdt2 siRNA# GAAUUAUACUGCUUAUCGA |
Dharmacon | N/A |
| siRNA targeting sequence: Cyclin F siRNA# UAGCCUACCUCUACAAUGA |
Dharmacon | N/A |
| siRNA targeting sequence: Skp2 siRNA# GAGACCAUCACCCAGCUGAAU |
Dharmacon | N/A |
| siRNA targeting sequence: PP1α siRNA# GAGACGCUACAACAUCAAA |
Dharmacon | N/A |
| siRNA targeting sequence: PP1β siRNA# AGAAGUUCGAGGCUUAUGU |
Dharmacon | N/A |
| siRNA targeting sequence: PP1γ siRNA# CUAUCCUACUAGAACUUGA |
Dharmacon | N/A |
| siRNA targeting sequence: RIF1 siRNA# AGAAUGAGCCCCUAGGGAA |
Dharmacon | N/A |
| siRNA targeting sequence: Orc1 UTR siRNA# GGAAAUGGCUCUCAUGUAU |
Dharmacon | N/A |
| Forward 5’[Biotin]- GAAGCTAGACTTAGGTGTCATATTGAACCTACTATGCCGAACTAGTTACGAGCTATAAAC-3’ |
SIGMA | N/A |
| Forward 5’[Cy5]- GAAGCTAGACTTAGGTGTCATATTGAACCTACTATGCCGAACTAGTTACGAGCTATAAAC-3’ |
SIGMA | N/A |
| Reverse 5’- GTTTATAGCTCGTAACTAGTTCGGCATAGTAGGTTCAATATGACACCTAAGTCTAGCTTC-3’ |
SIGMA | N/A |
| Recombinant DNA | ||
| pcDNAFRT5 | Thermo Fisher Scientific | Cat# V601020 |
| pOG44 | Thermo Fisher Scientific | Cat# V600520 |
| pMAL-c2E | Addgene | Cat# 75291 |
| pGEX-6P-1 | Sigma-Aldrich | Cat# GE28-9546-48 |
| pEGFP-C1 | BD Biosciences | Cat# 6084–1 |
| pECFP(C1)-PP1alpha | Addgene | Cat# 44233 |
| pEGFP(N3)-PP1beta | Addgene | Cat# 44223 |
| pEGFP(C1)-PP1gamma | Addgene | Cat# 44225 |
| mCherry2-C1 | Addgene | Cat# 54563 |
| mEGFP-C1 | Addgene | Cat# 54759 |
| Software and Algorithms | ||
| FlowJo Software | FlowJo, LLC | https://www.flowjo.com/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
Characterization of stable ORC1 mutants in U2OS cells
Wild type GFP-ORC1 as well as GFP-ORC1 A-A, GFP-ORC1 CDK and GFP-ORC1 PP1 mutants were cloned into the plasmid pcDNA ™5/FRT/TO (Life Technologies) under a tetracycline-regulated CMV-based promoter. The plasmids were integrated into a FRT-U2OS-TRex osteosarcoma cells (Malecki et al., 2006) via FRT-mediated recombination and the integrated transgenes were selected with hygromycin. The expression of ORC1 was assessed following treatment of cells with 1 μg/ml of Doxycycline.
METHOD DETAILS
Plasmid Construction and Mutagenesis
Recombinant human MBP tagged ORC1 protein or its mutants, ORC2, ORC3, ORC4, ORC5, ORC6 and CDC6 were expressed and purified from bacteria by cloning into pMALp-c2E vector. MBP-ORC1 180–240 WT and A-A as well as MBP-ORC1 230–400 WT and A-A were also cloned in pMALp-c2E vector with an addition of PreScission protease site at N-terminus and 6XHis tag at C-terminus. The human ORC1 WT or its mutants were also cloned in pEGFP-C1 vector for transient over-expression in mammalian cells. Human CDC6 or its mutants and PP1α were also cloned in pGEX-6P1 vector to produce GST fusion proteins in bacterial cells. PP1α plasmids were also generated with a N-terminal T7 tag in the pLPC vector (McCurrach et al., 1997) (gift from Scott Lowe, Memorial Sloan Kettering Cancer Center) for mammalian expression. The mutant plasmids were generated following site directed mutagenesis protocol using Phusion high fidelity DNA polymerase (NEB). To generate internal deletions of MBP-ORC1 and GST-CDC6 plasmids, we employed phosphorothioate-modified PCR primers and T7 gene 6 exonuclease following a protocol previously described (Stoynova et al., 2004). Briefly, the 0.25 mM of modified forward and reverse primers at the two junctions of region of interest to be deleted were used in 50 μl PCR reaction containing 20 ng of template DNA, 5U Herculase Enhanced DNA polymerase, 0.5 mM dNTP and 1x polymerase buffer. After PCR amplification, the amplified DNA is treated with 10U/μl of DpnI enzyme at 37°C for 1 hour. After DpnI treatment, the PCR products were digested with 2U/μl of T7 Gene6 Exonuclease for 10 min at 37°C and enzyme was deactivated by heating at 80°C for 10 min, followed by cooling at 4°C. After final treatment, the plasmids were purified by Qiagen PCR purification kit and transformed in XL10 gold ultracompetent bacterial cells. The colonies were screened for positive deletion mutants by plasmid DNA sequencing. Oligonucleotide sequences used to generate the plasmids are available upon request.
Expression and purification of recombinant proteins
The MBP- and GST-fusion recombinant proteins were expressed and purified using amylose and Glutathione beads, respectively following a previously described protocol (Hossain and Stillman, 2012). Briefly, the MBP and GST fusion were transformed into E. coli BL21 cells with their respective plasmids, transformed bacterial cells were grown in LB media at 37°C till the O.D. of the cells reaches 0.7–0.9 and were induced for 12 hours with 0.3mM of IPTG at 16°C. The induced cells were pelleted, washed, and further lysed with sonication in a lysis buffer A containing 25mM Tris-HCl at pH 7.5, 150mM NaCl, 0.02% NP-40, 5mM benzamidine-HCl, 1mM phenylmethylsulfonylfluoride, Protease cocktail inhibitor tablets [Roche], 10% glycerol) plus 100μg/ml lysozyme. The lysed bacterial cells were centrifuged and the clarified supernatant is incubated with respective pre-washed Amylose or Glutathione agarose beads for 3 hours at 4°C. The bead bound proteins were washed with ten column volumes of buffer A plus 0.05% NP-40 + 500mM NaCl and further with ten column volumes of buffer A alone. Fusion protein was eluted in a stepwise manner with buffer A containing 20mM Maltose for MBP fusion proteins or 20 mM reduced glutathione, pH7.5 for GST fusion proteins. Fractions containing purified proteins were pooled, concentrated and dialyzed, and protein concentration was estimated using a standard Bradford protein assay. His-tagged ORC1 180–240 WT and A-A mutant were made after cleaving MBP protein from the N-terminus using PreScission protease and further purified using Ni-NTA resin (Qiagen). Cyclin A-CDK2 and Cyclin E-CDK2 kinases were expressed and purified from baculoviral infected insect cells as described previously (Hossain and Stillman, 2012). The insect Hi5 cells were transfected with baculovirus containing either His6-Cyclin A or His6-Cyclin E with CDK2-HA at a multiplicity of infection of 4–6 in T-175 flasks. After incubation for 68–72 hours at 30°C, cells were harvested, washed with PBS and then insect cells were in lysis buffer containing 50mM Tris-HCl (pH 7.5), 150mM NaCl, 2mM MgCl2, 0.02% NP-40, 5mM benzamidine-HCl, 1mM phenylmethylsulfonylfluoride, Protease cocktail inhibitor tablets (Roche), 10% glycerol. The cells were lysed by Dounce homogenization for 25–30 strokes followed by brief sonication for 2 minutes at 30% amplitude for 10 seconds ON or OFF pulse. The lysates were clarified by centrifugation at 14,000 rpm for 30 minutes, and the supernatant was incubated with prewashed Ni-NTA resin (Qiagen) at 4°C. The resin bound protein complex were washed three times with lysis buffer and stepwise eluted with 250 mM Imidazole in lysis buffer. Fraction containing Cyclin (E or A)-CDK2 were pooled together and further purified by Hemagglutinin (HA) affinity purification. For HA purification, pooled proteins from Ni-NTA purification were bound to anti-HA agarose (Pierce) at 4°C for 4 hours, beads were washed and stepwise eluted with lysis buffer containing 2mg/ml HA peptide. Fraction containing stochiometric amounts of Cyclin (E or A)-CDK2 were pooled, dialyzed, and protein concentration were estimated. The proteins were stored at −70°C after snap freezing in liquid nitrogen.
Binding assay
For MBP, GFP and GST pull-down assay, anti-MBP or anti-GFP or anti-GST antibodies were initially bound to gamma bind protein G magnetic beads, washed and further incubated with respective purified recombinant MBP- and GST-fusion proteins in binding buffer with following composition; 25mM Tris-Cl at pH 7.5, 150mM KCl, 0.15% Nonidet P-40, 0.1mM EDTA, 5 mM magnesium acetate, 1mM DTT. For MBP-ORC1 pull down, MBP-ORC1 protein bound beads were either incubated with GST-CDC6, GST-PP1α, Cyclin E-CDK2, Cyclin A-CDK2 or ORC1 180–240-His depending on context of experiments for 4–5 hours at 4°C. For GST-CDC6 pull down, the GST-CDC6 protein bound magnetic beads were either incubated MBP-ORC1 alone or in combination with Cyclin E-CDK2 or/and Cyclin A-CDK2 in the presence or absence of 1mM ATP. The beads were washed with binding buffer and was further immunoblotted with respective antibodies as indicated in the results.
DNA binding assay for MBP-ORC1 and MBP-CDC6 were performed by following the protocols described (Parker et al., 2019) as well as used the same dsDNA oligo sequence and single stranded oligos were further annealed to form dsDNA oligo. The biotinylated dsDNA oligo was bound to streptavidin magnetic beads in binding buffer containing 25mM Tris-Cl at pH 7.5, 150mM KCl, 0.15% Nonidet P-40, 0.1mM EDTA, 5 mM magnesium acetate, 1mM DTT at 4°C for 3–4 hours. The beads were further washed three times with binding buffer to remove the unbound DNA and were further incubated with purified MBP-ORC1 and MBP-CDC6 proteins or no protein with 1mM ATP at 4°C for 2 hours. The beads were washed with binding buffer, eluted with 2X SDS dye. The 8% SDS-PAGE gel was run and further stained with Coomassie Brilliant Blue R-250.
For sequential or two-step binding assay, the recombinant proteins (as indicated in figure legends) were mixed and incubated with glutathione beads in binding buffer with the following composition; 25mM Tris-Cl at pH 7.5, 50mM KCl, 0.05% Nonidet P-40, 0.1mM EDTA, 5 mM magnesium acetate, 1mM DTT. The bound proteins were washed and further eluted with 10mM reduced glutathione in binding buffer. The eluted protein complexes were subsequently incubated with anti-GFP antibodies bound to gamma bind protein G magnetic beads at 4°C for 4–5 hours. The protein complexes bound to anti-GFP beads were washed with binding buffer and was further silver stained and immunoblotted with anti-HIS6 antibody. All the protein incubation steps with beads contained 1mM ATP.
Liquid-liquid phase separation (LLPS) assay
Human ORC1 and CDC6 were cloned in mCherry2-C1 and mEGFP-C1 vectors (Addgene), respectively. GFP-ORC1 and mCherry-CDC6 were PCR amplified, cloned in E. coli expression pMAL-c2E vector and positive clones were transformed in BL21-DE3 cells for expression in bacteria. Full length MBP-PP-GFP-ORC1 wild type, MBP-PP-mCherry-CDC6 wild type and MBP-PP-mCherry-CDC6 mutant (K92A, R94A and R95A) proteins were purified from bacteria as described in above section. After the final purification, the purified recombinant proteins were buffer exchanged with LLPS buffer (50mM Tris pH 7.5, 100mM NaCl, 1mM MgCl2 and 1mM DTT) using 30K cutoff Amicon Ultra-0.5 centrifugal filter devices (Millipore Sigma). Biotinylated HRV-3C protease from Millipore Sigma is also buffer exchanged with LLPS buffer using 3K cutoff filtration devices. The concentrations of proteins were estimated using Bradford assay and stored at −70°C after flash frozen in liquid nitrogen. Phase separation assay were performed in LLPS buffer with the indicated concentrations of recombinant proteins in the presence or absence of Cy5-labelled DNA and/or 0.5 μg of HRV-3C in 20 μl total reaction volume. The reactions were incubated at 4°C for 16 hours to remove soluble MBP tag and were further spotted on chambered glass slides with coverslips for DIC and fluorescent imaging on Perkin Elmer spinning disk using 63x oil immersion objective lens.
Immunoprecipitation
For expression of proteins, HEK293 cells were transiently transfected with the indicated plasmids with lipofectamine 2000 transfection reagent (ThermoFisher Scientific). For immunoprecipitation, the cells were harvested and washed in PBS and lysed in a buffer containing 20mM Tris-HCl pH7.5, 300mM NaCl, 0.3% NP-40, 5mM MgCl2, 0.1 mM EDTA, 10% Glycerol, 1mM DTT, 1mM CaCl2, 20uM MG132 and protease as well as phosphatase inhibitor tablets (Roche). Benzonase (Sigma-Aldrich, St. Louis, MO) was added to the buffer and the suspension incubated for 30 min on ice with intermittent mixing. The concentration of NaCl and NP-40 was reduced to 150mM and 0.15%, respectively with dilution buffer after 30 minutes incubation on ice. The extract was centrifuged at 14, 000 rpm for 15 min at 4°C. The GFP-ORC1 and GFP-PP1 proteins were precipitated with anti-GFP specific antibodies as indicated in figure legends. The whole cell extract was first incubated with antibodies for 4 hours and subsequently, 2 hours with pre-washed gamma bind G sepharose beads with end-to end shaking at 4°C. The beads were washed 3 times with washing buffer containing 20mM Tris-HCl pH7.5, 150mM NaCl, 0.15% NP-40, 5mM MgCl2, 0.1mM EDTA, 10% Glycerol, 1mM DTT and protease as well as phosphatase inhibitor tablets from Roche. Finally, the washed beads were suspended in Laemmli sample buffer and 8% SDS-PAGE gels were run and immunoblotted.
For co-immunoprecipitation of ORC1, CDC6 and Cyclin A interacting proteins, extracts were prepared by resuspending the synchronized and asynchronous cells in Lysis Buffer A (20mM Tris-HCl pH 7.5, 250mM NaCl, 5mM MgCl2, 0.1mM EDTA, 0.3% NP40, 1mM DTT, 1mM CaCl2, 5% Glycerol supplemented with Benzonase, Roscovitine, MG132, protease inhibitor cocktail and phosphatase inhibitors) for 30 minutes on rotation at 4°C. Following lysis, the extract was diluted with equal volume of Dilution Buffer B (Buffer A without NaCl and NP-40) to adjust the final salt and detergent concentration to 125mM NaCl and 0.15% NP-40 respectively, and centrifuged to obtain clarified extract. Half of each extract was used to perform immunoprecipitations using either CDC6 (CS1881) or ORC1 antibody and corresponding isotype control bound to protein G dynabeads. The beads were washed and were then suspended in 2X Laemmli sample loading buffer, and analyzed by western blot.
For immunoprecipitations, the following antibodies were used: mouse monoclonal anti-GFP antibody (A11120; Invitrogen), rabbit polyclonal anti-CDC6 antibody (CS1881, CSHL facility), rabbit polyclonal anti-Cyclin A antibody (A305–254A; Bethyl), mouse monoclonal ORC1 78-1-172 (Kara et al., 2015) and mouse monoclonal anti-T7 antibody (CSHL Facility). For immunoblotting, the antibodies are indicated in the key resource table under antibodies section.
EdU incorporation and Cell cycle analyses of mutant ORC1
U2OS cells were cultured in DMEM containing high glucose (Gibco) supplemented with Penicillin/Streptomycin and 10% fetal bovine serum, or in case of tetracycline-inducible U2OS cells stably expressing GFP-ORC1 WT, GFP-ORC1 A-A, GFP-ORC1 CDK123 and GFP-ORC1 PP1mut in 10% Tet-free fetal bovine serum (Takara, 631101) and 150 μg/ml Hygromycin. On day 0 all cells were induced with 1 μg/ml doxycycline (Sigma, D9891–10G) and thereafter maintained in it for the remainder of the experiment. Cell lines were treated on day 0 with (a) 200nM of either control or 3’UTR ORC1 siRNA to deplete endogenous ORC1 to be collected 24 hours later, and (b) 100nM of either control or 3’UTR ORC1 siRNA on day 0 and again on day 1 (24 h), for samples to be collected on day 2 (48 h) and day 3 (72 h). All samples were treated with 10μM EdU for 2 hours prior to harvesting. Samples were either counted for growth curves, or prepared for cell cycle analyses using the Click-iT Plus EdU Alexa Fluor 647 flow cytometry assay kit (Invitrogen, C10635) as per the manufacturer’s protocol. Three-color flow cytometric analysis was performed using a BD LSRFortessa Dual SORP (BD Biosciences, San Jose, CA). The GFP emission was collected off the 488nm laser using a 505LP dicroic mirror and a 530/30 filter, Alexafluor 647 emission was collected off the 638nm laser using a 670/30 filter and the FxViolet emission was collected off the 407nm laser using the 450/50 filter.
Live cell microscopy
Tetracycline-inducible GFP-Orc1 WT and GFP-ORC1 A-A stable U2OS cell lines were used for live-cell imaging after addition of 1 μg/ml doxycycline. The cells were transferred to 6-well plates mounted onto the stage of spinning disk confocal microscope (Perkin Elmer) and kept at 37°C in DMEM medium with 10% FBS. Time-lapse images acquired with a 40X objective lens were captured with a CCD camera or the Perkin Elmer spinning disk acquisition setup.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data points in figures represent mean values, with error bars in all figures represent mean ± Standard Deviation. Number of replicates are indicated in the figure legends.
Supplementary Material
Highlights.
ORC1 and CDC6 IDRs have short linear motifs that bind many regulatory proteins.
ORC1 and CDC6 have short domains that promote assembly of their AAA+ ATPase domains
ORC1 levels in the cell are controlled by ORC1-ORC1 interactions and by CDC6
ORC1 and CDC6 form a liquid-liquid phase transition stimulated by DNA
Acknowledgements
We thank Patricia Wendel and Ariana Amendolara for technical help and Leemor Joshua-Tor for discussions and comments on the manuscript. The antibody and the microscopy shared resources of the Cold Spring Harbor Laboratory Cancer Center (P30CA045508). This research was supported by grants from the National Cancer Institute (P01CA13106) and the National Institute of General Medical Sciences (R01GM45436).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
Bruce Stillman is a member of the science advisory board of Circle Pharma and is an advisor to EnGeneIC and Pfizer. The authors declare no competing interests.
REFERENCES
- Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, and Kaelin WG (1996). Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol 16, 6623–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir T, Dorrello NV, Amador V, Guardavaccaro D, and Pagano M (2004). Control of the SCFSkp2–Cks1 ubiquitin ligase by the APC/CCdh1 ubiquitin ligase. Nature 428, 190–193. [DOI] [PubMed] [Google Scholar]
- Baum B, Nishitani H, Yanow S, and Nurse P (1998). Cdc18 transcription and proteolysis couple S phase to passage through mitosis. Embo J 17, 5689–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, and Labib K (2016). Chromosome Duplication in Saccharomyces cerevisiae. Genetics 203, 1027–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, and Stillman B (1992). ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357, 128–134. [DOI] [PubMed] [Google Scholar]
- Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C, Martin C-A, Yeyati P, Sanna NA, Bober M, et al. (2011). Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet 43, 350–355. [DOI] [PubMed] [Google Scholar]
- Bleichert F, Leitner A, Aebersold R, Botchan MR, and Berger JM (2018). Conformational control and DNA-binding mechanism of the metazoan origin recognition complex. Proc Natl Acad Sci USA 115, E5906–E5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen M, Peti W, Ragusa MJ, and Beullens M (2010). The extended PP1 toolkit: designed to create specificity. Trends Biochem Sci 35, 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, and Fawzi NL (2015). Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell 60, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore RC, Havens CG, Manning AL, Li J-M, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, and Zou L (2010). CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell 40, 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Sysoeva TA, Chowdhury S, and Nixon BT (2008). Regulation and action of the bacterial enhancer-binding protein AAA+ domains. Biochem Soc T 36, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury R, Bonacci T, Wang X, Truong A, Arceci A, Zhang Y, Mills CA, Kernan JL, Liu P, and Emanuele MJ (2017). The E3 Ubiquitin Ligase SCF(Cyclin F) Transmits AKT Signaling to the Cell-Cycle Machinery. Cell Reports 20, 3212–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clijsters L, and Wolthuis R (2014). PIP-box-mediated degradation prohibits re-accumulation of Cdc6 during S phase. J Cell Sci 127, 1336–1345. [DOI] [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, and Diffley JFX (1996). An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 379, 180–182. [DOI] [PubMed] [Google Scholar]
- Cook JG, Park C-H, Burke TW, Leone G, DeGregori J, Engel A, and Nevins JR (2002). Analysis of Cdc6 function in the assembly of mammalian prereplication complexes. Proc Natl Acad Sci USA 99, 1347–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D, Laman H, and Laskey RA (2002). Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat Cell Biol 4, 523–528. [DOI] [PubMed] [Google Scholar]
- Delmolino LM, Saha P, and Dutta A (2001). Multiple Mechanisms Regulate Subcellular Localization of Human CDC6. J Biol Chem 276, 26947–26954. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML (2003). The ‘ORC cycle’: a novel pathway for regulating eukaryotic DNA replication. Gene 310, 1–15. [DOI] [PubMed] [Google Scholar]
- Diffley JFX, Cocker JH, Dowell SJ, and Rowley A (1994). Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78, 303–316. [DOI] [PubMed] [Google Scholar]
- Drury LS, Perkins G, and Diffley JFX (2000). The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr Biol 10, 231–240. [DOI] [PubMed] [Google Scholar]
- Duursma A, and Agami R (2005). p53-Dependent Regulation of Cdc6 Protein Stability Controls Cellular Proliferation. Mol Cell Biol 25, 6937–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzen N. den, and Pines J (2001). Cyclin a Is Destroyed in Prometaphase and Can Delay Chromosome Alignment and Anaphase. J Cell Biol 153, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Danson AE, Ye F, Jovanovic M, Buck M, and Zhang X (2020). Bacterial Enhancer Binding Proteins—AAA+ Proteins in Transcription Activation. Biomol 10, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters J-M, and Hunt T (2001). Anaphase-Promoting Complex/Cyclosome–Dependent Proteolysis of Human Cyclin a Starts at the Beginning of Mitosis and Is Not Subject to the Spindle Assembly Checkpoint. J Cell Biol 153, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gógl G, Kornev AP, Reményi A, and Taylor SS (2019). Disordered Protein Kinase Regions in Regulation of Kinase Domain Cores. Trends Biochem Sci 44, 300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly AS, Prasanth SG, Siddiqui K, and Stillman B (2009). Orc1 controls centriole and centrosome copy number in human cells. Science 323, 789–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx A, Beullens M, Ceulemans H, Abt TD, Eynde AV, Nicolaescu E, Lesage B, and Bollen M (2009). Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol 16, 365–371. [DOI] [PubMed] [Google Scholar]
- Hiraga S-I, Ly T, Garzón J, Hořejší Z, Ohkubo Y-N, Endo A, Obuse C, Boulton SJ, Lamond AI, and Donaldson AD (2017). Human RIF1 and protein phosphatase 1 stimulate DNA replication origin licensing but suppress origin activation. Embo Rep 18, 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M, and Stillman B (2012). Meier-Gorlin syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome reduplication. Gene Dev 26, 1797–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M, and Stillman B (2016). Opposing roles for DNA replication initiator proteins ORC1 and CDC6 in control of Cyclin E gene transcription. Elife 5, e12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaremko MJ, On KF, Thomas DR, Stillman B, and Joshua-Tor L (2020). The dynamic nature of the human Origin Recognition Complex revealed through five cryoEM structures. Elife 9, e58622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Goldin L, Ren H, Sun D, Guardavaccaro D, Pagano M, and Zhu L (2006). Skp2 Contains a Novel Cyclin A Binding Domain That Directly Protects Cyclin A from Inhibition by p27Kip1. J Biol Chem 281, 24058–24069. [DOI] [PubMed] [Google Scholar]
- Jiang W, Wells NJ, and Hunter T (1999). Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc Natl Acad Sci USA 96, 6193–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, and Skotheim JM (2013). Start and the restriction point. Curr Opin Cell Biol 25, 717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara N, Hossain M, Prasanth SG, and Stillman B (2015). Orc1 Binding to Mitotic Chromosomes Precedes Spatial Patterning during G1 Phase and Assembly of the Origin Recognition Complex in Human Cells. J. Biol Chem 290, 12355–12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H, Ohashi E, Kanamoto S, Tsurimoto T, and Katayama T (2015). Specific binding of eukaryotic ORC to DNA replication origins depends on highly conserved basic residues. Sci Rep 5, 14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitz S, Ritzi M, Baack M, and Knippers R (2000). The Human Origin Recognition Complex Protein 1 Dissociates from Chromatin during S Phase in HeLa Cells. J Biol Chem 276, 6337–6342. [DOI] [PubMed] [Google Scholar]
- Lee KY, Bae JS, Yoon S, and Hwang DS (2014a). Dephosphorylation of Orc2 by protein phosphatase 1 promotes the binding of the origin recognition complex to chromatin. Biochem Bioph Res Co 448, 385–389. [DOI] [PubMed] [Google Scholar]
- Lee R. van der, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, et al. (2014b). Classification of intrinsically disordered regions and proteins. Chem Rev 114, 6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vassilev A, and DePamphilis ML (2004). Role for Cdk1 (Cdc2)/Cyclin A in Preventing the Mammalian Origin Recognition Complex’s Largest Subunit (Orc1) from Binding to Chromatin during Mitosis. Mol Cell Biol 24, 5875–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Lam WH, Zhai Y, Cheng J, Cheng E, Zhao Y, Gao N, and Tye B-K (2018). Structure of the origin recognition complex bound to DNA replication origin. Nature 559, 217–222. [DOI] [PubMed] [Google Scholar]
- Liang C, and Stillman B (1997). Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Gene Dev 11, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Weinreich M, and Stillman B (1995). ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 81, 667–676. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, and Nurse P (2000). Cell cycle. License withheld--geminin blocks DNA replication. Science 290, 2271–2273. [DOI] [PubMed] [Google Scholar]
- Mailand N, and Diffley JFX (2005). CDKs Promote DNA Replication Origin Licensing in Human Cells by Protecting Cdc6 from APC/C-Dependent Proteolysis. Cell 122, 915–926. [DOI] [PubMed] [Google Scholar]
- Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, and Blacklow SC (2006). Leukemia-Associated Mutations within the NOTCH1 Heterodimerization Domain Fall into at Least Two Distinct Mechanistic Classes. Mol Cell Biol 26, 4642–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrommati I, Faedda R, Galasso G, Li J, Burdova K, Fischer R, Kessler BM, Carrero ZI, Guardavaccaro D, Pagano M, et al. (2018). β-TrCP- and Casein Kinase II-Mediated Degradation of Cyclin F Controls Timely Mitotic Progression. Cell Reports 24, 3404–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurrach ME, Connor TMF, Knudson CM, Korsmeyer SJ, and Lowe SW (1997). bax-deficiency promotes drug resistance and oncogenic transformation by attenuating p53-dependent apoptosis. Proc National Acad Sci 94, 2345–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNairn AJ, Okuno Y, Misteli T, and Gilbert DM (2005). Chinese hamster ORC subunits dynamically associate with chromatin throughout the cell-cycle. Exp Cell Res 308, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez J, Zou-Yang XH, Kim S-Y, Hidaka M, Tansey WP, and Stillman B (2002). Human Origin Recognition Complex Large Subunit Is Degraded by Ubiquitin-Mediated Proteolysis after Initiation of DNA Replication. Mol Cell 9, 481–491. [DOI] [PubMed] [Google Scholar]
- Mimura S, Seki T, Tanaka S, and Diffley JFX (2004). Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature 431, 1118–1123. [DOI] [PubMed] [Google Scholar]
- Nakayama K-I, Hatakeyama S, and Nakayama K (2001). Regulation of the Cell Cycle at the G1–S Transition by Proteolysis of Cyclin E and p27Kip1. Biochem Bioph Res Co 282, 853–860. [DOI] [PubMed] [Google Scholar]
- Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, and Dowdy SF (2014). Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife 3, e02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S, Tatsumi Y, Fujita M, Tsurimoto T, and Obuse C (2003). The ORC1 Cycle in Human Cells: II. Dynamic Changes in the Human ORC Complex During the Cell Cycle. J Biol Chem 278, 41535–41540. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, and Pagano M (1995). Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol 15, 2612–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, McNairn AJ, Elzen N. den, Pines J, and Gilbert DM (2001). Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. Embo J 20, 4263–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Örd M, Venta R, Möll K, Valk E, and Loog M (2019). Cyclin-Specific Docking Mechanisms Reveal the Complexity of M-CDK Function in the Cell Cycle. Mol Cell 75, 76–89.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Örd M, Puss KK, Kivi R, Möll K, Ojala T, Borovko I, Faustova I, Venta R, Valk E, Kõivomägi M, et al. (2020). Proline-Rich Motifs Control G2-CDK Target Phosphorylation and Priming an Anchoring Protein for Polo Kinase Localization. Cell Reports 31, 107757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MW, Botchan MR, and Berger JM (2017). Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit Rev Biochem Mol 52, 1–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MW, Bell M, Mir M, Kao JA, Darzacq X, Botchan MR, and Berger JM (2019). A new class of disordered elements controls DNA replication through initiator self-assembly. Elife 8, e48562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G, Drury LS, and Diffley JFX (2001). Separate SCFCDC4 recognition elements target Cdc6 for proteolysis in S phase and mitosis. Embo J 20, 4836–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen BO, Lukas J, Sørensen CS, Bartek J, and Helin K (1999). Phosphorylation of mammalian CDC6 by Cyclin A/CDK2 regulates its subcellular localization. Embo J 18, 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Denchi EL, Gieffers C, Matteucci C, Peters J-M, and Helin K (2000). Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Gene Dev 14, 2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, and Nasmyth K (1995). Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. Embo J 14, 3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Bohm T, Cocker JH, Diffley JF, and Nasmyth K (1996). Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Gene Dev 10, 1516–1531. [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Shen Z, Prasanth KV, and Stillman B (2010). Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc National Acad Sci 107, 15093–15098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JCW, Bowers JL, Rodríguez HK, and Bell SP (2006). Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Molecular Cell 21, 29–39. [DOI] [PubMed] [Google Scholar]
- Roey KV, and Davey NE (2015). Motif co-regulation and co-operativity are common mechanisms in transcriptional, post-transcriptional and post-translational regulation. Cell Commun Signal Ccs 13, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roey KV, Uyar B, Weatheritt RJ, Dinkel H, Seiler M, Budd A, Gibson TJ, and Davey NE (2014). Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem Rev 114, 6733–6778. [DOI] [PubMed] [Google Scholar]
- Russell RB, and Gibson TJ (2008). A careful disorderliness in the proteome: Sites for interaction and targets for future therapies. Febs Lett 582, 1271–1275. [DOI] [PubMed] [Google Scholar]
- Saha P, Chen J, Thome KC, Lawlis SJ, Hou Z, Hendricks M, Parvin JD, and Dutta A (1998). Human CDC6/Cdc18 Associates with Orc1 and Cyclin-cdk and Is Selectively Eliminated from the Nucleus at the Onset of S Phase. Mol Cell Biol 18, 2758–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, and Bleichert F (2020). Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6. Nat Commun 11, 4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Lindstrom DL, and Harlow E (1998). Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA 95, 10453–10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster BS, Reed EH, Parthasarathy R, Jahnke CN, Caldwell RM, Bermudez JG, Ramage H, Good MC, and Hammer DA (2018). Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat Commun 9, 2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui K, and Stillman B (2007). ATP-dependent Assembly of the Human Origin Recognition Complex. J Biol Chem 282, 32370–32383. [DOI] [PubMed] [Google Scholar]
- Speck C, and Stillman B (2007). Cdc6 ATPase activity regulates ORC × Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. The Journal of Biological Chemistry 282, 11705–11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreesankar E, Senthilkumar R, Bharathi V, Mishra RK, and Mishra K (2012). Functional diversification of yeast telomere associated protein, Rif1, in higher eukaryotes. BMC Genomics 13, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoynova L, Solórzano R, and Collins ED (2004). Generation of large deletion mutants from plasmid DNA. Biotechniques 36, 402–406. [DOI] [PubMed] [Google Scholar]
- Takeda DY, Wohlschlegel JA, and Dutta A (2000). A Bipartite Substrate Recognition Motif for Cyclin-dependent Kinases. J Biol Chem 276, 1993–1997. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, and Nasmyth K (1997). Loading of an Mcm Protein onto DNA Replication Origins Is Regulated by Cdc6p and CDKs. Cell 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, and Obuse C (2003). The ORC1 Cycle in Human Cells: I. Cell Cycle-Regulated Oscillation of Human ORC1. J Biol Chem 278, 41528–41534. [DOI] [PubMed] [Google Scholar]
- Tocilj A, On KF, Yuan Z, Sun J, Elkayam E, Li H, Stillman B, and Joshua-Tor L (2017). Structure of the active form of human origin recognition complex and its ATPase motor module. Elife 6, e20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, and Walter JC (2003). Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Gene Dev 17, 1894–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter D, Hoffmann S, Komseli E-S, Rappsilber J, Gorgoulis V, and Sørensen CS (2016). SCFCyclin F-dependent degradation of CDC6 suppresses DNA re-replication. Nat Commun 7, 10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, and Song J (2019). Structural basis for the ORC1-Cyclin A association. Protein Sci 28, 1727–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ayad NG, Wan Y, Zhang G-J, Kirschner MW, and Kaelin WG (2004). Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature 428, 194–198. [DOI] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Chen HH, and Stillman B (2001). Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proceedings of the National Academy of Sciences of the United States of America 98, 11211–11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, Archambault V, Austin RJ, Jacobson MD, Bell SP, and Cross FR (2004). Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Gene Dev 18, 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, and Dutta A (2000). Inhibition of Eukaryotic DNA Replication by Geminin Binding to Cdt1. Science 290, 2309–2312. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Takeda DY, and Dutta A (2001). Mutational Analysis of the Cy Motif from p21 Reveals Sequence Degeneracy and Specificity for Different Cyclin-Dependent Kinases. Mol Cell Biol 21, 4868–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DJ, and Endicott JA (2018). Structural insights into the functional diversity of the CDK–cyclin family. Royal Soc Open Biology 8, 180112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, DeGregori J, Shohet R, Leone G, Stillman B, Nevins JR, and Williams RS (1998). Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America 95, 3603–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Riera A, Bai L, Sun J, Nandi S, Spanos C, Chen ZA, Barbon M, Rappsilber J, Stillman B, et al. (2017). Structural basis of Mcm2–7 replicative helicase loading by ORC–Cdc6 and Cdt1. Nat Struct Mol Biol 24, 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kobayashi R, Galaktionov K, and Beach D (1995). pl9skp1 and p45skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell 82, 915–925. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Pozo PN, Oh S, Stone HM, and Cook JG (2020). Distinct and sequential re-replication barriers ensure precise genome duplication. Plos Genet 16, e1008988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original images for figures in the paper are available at Mendeley database: http://dx.doi.org/10.17632/p8cn8srfvh.1. This study did not generate any unique datasets or code.


