Abstract
The sonic hedgehog (Shh) pathway is highly activated in a variety of malignancies and plays important roles in tumorigenesis, tumor growth, drug resistance, and metastasis. Our recent study showed that the inhibitors of the Shh pathway such as cyclopamine (CP), a Smothened (SMO) inhibitor, and GANT61, a Gli1 inhibitor, have modest inhibitory effects on thyroid tumor cell proliferation and tumor growth. The objective of this study was to determine whether autophagy was induced by inhibition of the Shh pathway and could negatively regulate GANT61-induced apoptosis. Here we report that inhibition of the Shh pathway by Gli1 siRNA or by cyclopamine and GANT61 induced autophagy in SW1736 and KAT-18 cells, two anaplastic thyroid cancer cell lines; whereas Gli1 overexpression suppressed autophagy. Mechanistic investigation revealed that inhibition of the Shh pathway activated TAK1 and its two downstream kinases, the c-Jun-terminal kinase (JNK) and AMP-activated protein kinase (AMPK). GANT61-induced autophagy was blocked by TAK1 siRNA and the inhibitors of TAK1 (5Z-7-oxozeaenol, 5Z), JNK (SP600125), and AMPK (Compound C, CC). Inhibition of autophagy by chloroquine and 5Z and by TAK1 and Beclin-1 siRNA enhanced GANT61-induced apoptosis and its antiproliferative activity. Our study has shown that inhibition of the Shh pathway induces autophagy by activating TAK1, whereas autophagy in turn suppresses GANT61-induced apoptosis. We have uncovered a previously unrecognized role of TAK1 in Shh pathway inhibition-induced autophagy and apoptosis.
Subject terms: Thyroid cancer, Macroautophagy, Apoptosis, Drug development
Introduction
Thyroid cancer is one of the most common endocrine malignancies1. Thyroid cancers are divided into several different tumor types based on the pathological characteristics and the origins of tumor cells2. Well differentiated papillary (PTCs) and follicular (FTCs) thyroid cancers are derived from thyroid follicular epithelial cells and may progress into poorly differentiated or anaplastic thyroid carcinomas (ATCs)1. Medullary thyroid cancers (MTCs) are derived from the C cells of the thyroid gland. Surgery, thyroid hormone therapy, and radioiodine can cure most differentiated PTCs and FTCs but are not effective for poorly differentiated thyroid cancer3. Approximately 15–20% of thyroid cancer patients develop recurrence in their lifetime1. Currently, there is no effective therapy to treat ATC. The mean survival of ATC patients is only 2–6 months4.
The Shh pathway has been implicated in a plethora of tumor promoting activities including cell proliferation, epithelial to mesenchymal transition (EMT), angiogenesis, drug resistance, and metastasis5. The Shh pathway is activated when one of the hedgehog ligands (Sonic, Indian, or Desert) engages with their shared Patched (Ptch) receptor, a 12-pass transmembrane protein. Smoothened (Smo), a G-protein-coupled seven-pass transmembrane protein, then dissociates from Ptch and is translocated into the primary cilia where it becomes activated6,7. Activated Smo disrupts the interaction between Sufu and Gli1, a latent zinc-finger transcription factor that regulates the expression of many genes involved in cell proliferation, apoptosis, stem cell self-renewal, and drug resistance. The PTCH and SMO genes are frequently mutated in basal cell carcinomas and neuroblastomas. The Shh pathway is also highly activated in various tumor types ranging from extremely malignant pancreatic and lung cancers to the relatively benign well differentiated thyroid cancer. The Shh pathway can be cross-activated by the phosphatidylinositol-3 (PI)-3 and mitogen-activated protein (MAP) kinase pathways, and through paracrine activation by Shh-secreting tumor stromal cells7,8. Targeting the Shh pathway is a potential novel therapeutic strategy to treat cancers6. Indeed, several Smo inhibitors have been approved for treating basal cell carcinomas (BCCs) and neuroblastomas but these inhibitors have little success in cancers that lack the PTCH or SMO gene mutation6. Our recent study showed that GANT61, a Gli1 inhibitor, is able to control the growth of thyroid cancer stem cell-derived tumor but not effective to control the growth of bulk cell-derived tumors in a xenograft mouse model9. Understanding the molecular mechanisms for the poor performance of Gli1 inhibitors is imperative to improve the anticancer activity of Shh pathway inhibitors.
Autophagy is an evolutionarily conserved self-digestion process used by eukaryotic cells for energy recycling and for eliminating misfolded proteins and damaged organelles. Autophagy starts with the expansion of the crescent endosomal membrane that then engulfs the neighboring cargos in the cytosol to form a double-membraned vesicle, the autophagosome10. This process is triggered when cells sense various forms of stress such as nutrient and energy starvation, protein aggregates, and intracellular microbes10. Biochemically, nutrient and energy deprivation leads to ULK1 activation by the mammalian target of rapamycin (mTOR) inaction and AMPK activation, respectively. mTOR phosphorylates UNC-51-like kinase 1 (ULK1)S757 and inhibits ULK1 activity, whereas AMPK phosphorylates ULK1S555/777/317 and activates ULK1 (ref. 10). ULK1 activates a cascade of signaling molecules involved in the formation of the pre-initiation complex and the autophagosomes10. Mature autophagosomes fuse with lysosomes to form autolysosomes in which engulfed contents are degraded by proteases.
Autophagy has been implicated in playing an important role in antagonizing the therapeutic effects of various anticancer drugs. Several studies have shown that inhibition of the Shh pathway can induce autophagy. For example, GANT61, a Gli1 inhibitor, induces autophagy, whereas Shh suppresses autophagy in hepatocellular carcinoma cells11. GANT61 induces autophagy in MCYN-amplified neuroblastoma cell lines12,13. Vismodegib, a Smo inhibitor, induces autophagy in human lung cancer cells14. However, the mechanisms of Shh pathway inhibition-induced autophagy remain largely unknown. Whether autophagy represses or enhances the Shh pathway inhibition-mediated anticancer activity remains controversial. Our present study focuses on the mechanisms of Shh pathway inhibition-induced autophagy in thyroid cancer cells and its role in apoptosis. Here we report that inhibition of the Shh pathway induced autophagy by activating TAK1, a MAP kinase kinase kinase (MAP3K) that then activated two downstream kinases, JNK and AMPK. Inhibition of autophagy enhanced GANT61-induced apoptosis in two thyroid tumor cell lines. Our study uncovers a previously unrecognized role of TAK1 in mediating Shh pathway inhibition-induced autophagy and apoptosis.
Results
Inhibition of the Shh pathway induces autophagy
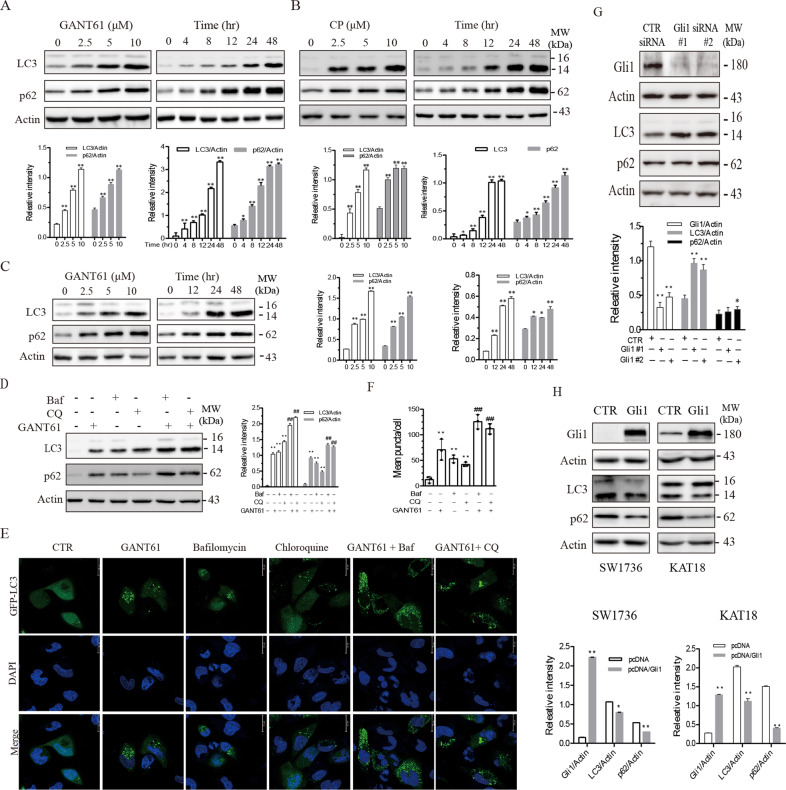
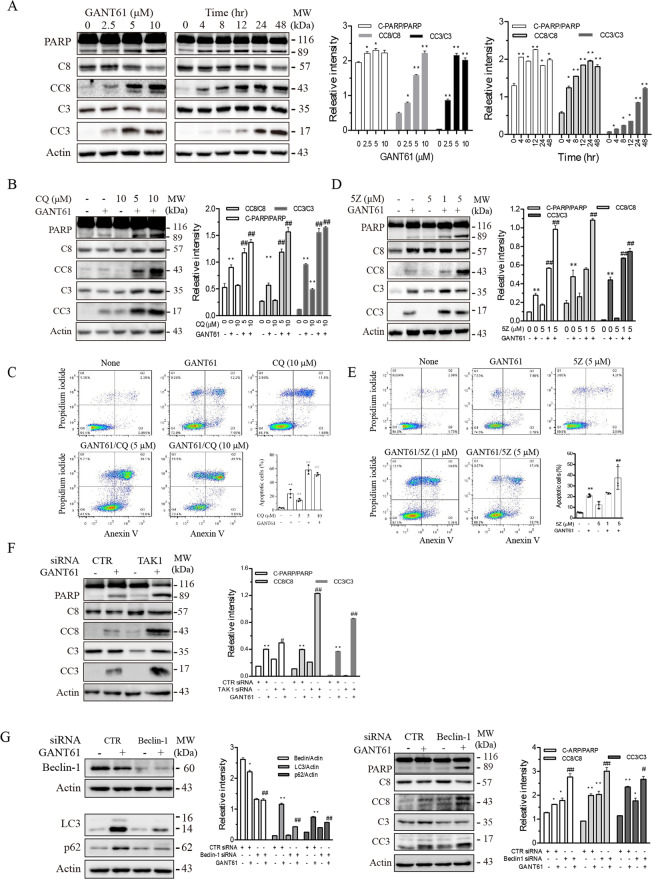
GANT61, a Gli1-specific inhibitor, induced LC3 lipidation in two anaplastic thyroid cancer cell lines, SW1736 (Fig. 1A) and KAT-18 cells (Fig. 1C), in a dose- and time-dependent manner. Consistently, cyclopamine (CP), an inhibitor of Smo, also increased microtubule-associated proteins 1A/1B light chain (LC3) lipidation in a dose- and time-dependent manner in SW1736 cells (Fig. 1B). p62 is a ubiquitin-binding protein that sequesters ubiquitinated proteins for lysosomal degradation through interacting with LC3-II in autophagosomes15. p62 levels are usually decreased in cells undergoing autophagy. Here we found that both GANT61 and CP increased p62 levels in a time- and dose-dependent manner in SW1736 and KAT-18 cells (Fig. 1A–C). To investigate if increased LC3 lipidation was due to the induction of autophagy or due to the stall of the autophagy flux, we investigated the effect of bafilomycin (Baf) and chloroquine (CQ) on GANT61-induced autophagy. As shown in Fig. 1D, GANT61 (10 μM), bafilomycin (10 nM), and CQ (10 μM) alone increased LC3 lipidation and p62 levels. The combination of GANT61 with bafilomycin or CQ further increased LC3 lipidation and p62 levels compared to bafilomycin or CQ alone. Confocal microscopy revealed that GANT61 (10 μM), bafilomycin (10 nM), and CQ (10 μM) all induced the formation of green fluoresce protein (GFP)-tagged LC3 puncta perinuclearly distributed in SW1736 cells (Fig. 1E). Enumeration of autophagosomes revealed that GANT61, bafilomycin, and CQ significantly increased the number of GFP-LC3 puncta (Fig. 1F). GANT61 in combination with bafilomycin or CQ further increased the number of GFP-LC3 puncta, compared to those treated with bafilomycin or CQ alone (Fig. 1E, F). These results collectively suggest that GANT61 induces autophagy, and that increased LC3-II levels is not due to the inhibition of the autophagy flux.
Fig. 1. Inhibition of the Shh pathway induces autophagy.
SW1736 (A) and KAT-18 (C) cells were treated with the indicated concentrations of GANT61 for 48 h or with 10 μM GANT61 for the indicated lengths of time. B SW1736 cells were treated with the indicated concentrations of cyclopamine (CP) for 48 h or with 10 μM CP for the indicated lengths of time. LC3, p62, and β-actin were analyzed by western blot. D The effect of bafilomycin and chloroquine on GANT61-induced autophagy. SW1736 cells were incubated in the absence or presence of GANT61 (10 μM) for 24 h. Bafilomycin (Baf) (10 nM) or chloroquine (CQ) (10 μM) was added and incubated for another 8 h. LC3 lipidation, p62, and actin levels were analyzed by western blot. Due to the low level of LC3 I, the ratio of LC3-II to β-actin was used for autophagy analysis in this and the remaining figures. Protein band density was analyzed by using NIH Image-J software. Data presented as the mean ± SD (n = 3) relative to control are shown in bar graphs. *p < 0.05, **p < 0.01, compared to untreated control; ##p < 0.01, compared to GANT61 (A–D). E, F GANT61 induces autolysosome formation. SW1736 cells transiently transfected with an expression vector encoding the GFP-LC3 gene were left untreated or treated with GANT61 (10 μM) for 24 h with or without bafilomycin (10 nM) or CQ (10 μM). After nuclear staining with DAPI, green puncta were visualized under a confocal microscope and statistically analyzed. Bar length: 20 μm. **p < 0.01, compared to untreated controls; ##p < 0.01, compared to GANT61. G SW1736 cells were transfected with control or Gli1 siRNA. After incubation for 48 hr, the cells were harvested and analyzed for Gli1, p62, LC3, and β-Actin by western blot. H SW1736 and KAT-18 cells were transfected with an empty vector or the vector encoding the GLI1 gene. After incubation for 48 h, the cells were harvested and analyzed for Gli1, p62, LC3, and β-Actin by western blot. Relative protein levels were analyzed by quantification of the density of the protein bands with NIH Image-J software and presented as bar graphs. Data are the mean ± SD of three experiments. *p < 0.05, **p < 0.01, compared to the transfection control (E, F).
Gli1 regulates autophagy
Gli1 is a transcription factor that plays a central role in executing many biological functions of the Shh pathway7,8. Here we investigated the role of Gli1 in mediating GANT61-induced autophagy. We first examined the effect of Gli1 knockdown on autophagy in SW1736 cells. As shown in Fig. 1G, Gli1 expression was very effectively suppressed in SW1736 cells transfected with two Gli1 siRNAs, compared to that transfected with the control siRNA. Gli1 knockdown led to increased LC3 lipidation and p62 levels. In contrast, Gli1 overexpression in SW1736 and KAT-18 cells significantly lowered LC3 lipidation and p62 levels (Fig. 1H).
Inhibition of the Shh pathway leads to TAK1 activation
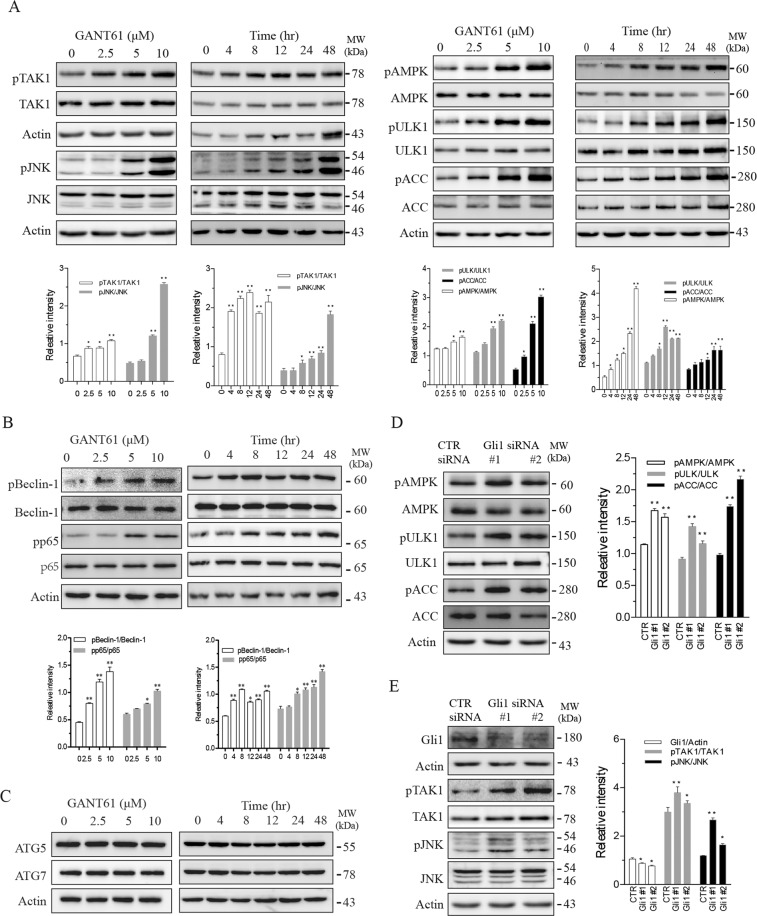
It has been increasingly recognized that TAK1, a serine/threonine kinase activated by various stimuli, plays an important role in autophagy and apoptosis16. TAK1 phosphorylates JNK and AMPK, both of which are involved in autophagy17. Here we tested if suppression of the Shh pathway led to TAK1 activation. As shown in Fig. 2A, GANT61 induced TAK1S412 phosphorylation in SW1736 cells in a time- and dose-dependent manner. GANT61 also increased phosphorylation of several downstream kinases, including JNKT183/185 and AMPKT172 as well as ULK1 (Unc-52-like kinase 1)S555 and ACC (acetyl-CoA carboxylase)S79, two substrates phosphorylated by AMPK. In addition to JNK, TAK1 phosphorylates and activates several other downstream molecules including NF-κB, p38, and ERK18. As shown in Fig. 2B, GANT61 increased p65S539 phosphorylation of NK-κB. Beclin-1 is an autophagy -related molecule downstream of AMPK10. Beclin-1 phosphorylation leads its dissociation from the Bcl-2-Beclin complex10. GANT61 dose- and time-dependently increased Beclin-1S93 phosphorylation (Fig. 2B). GANT61 did not affect the levels of Beclin-1 and other two autophagy-regulated proteins, ATG5 and ATG7 (Fig. 2B, C). Consistent with these observations, Gli1 suppression by two siRNAs also led to increased phosphorylation of JNKT183/185, AMPKT172, ULK1S555, and ACCS79 (Fig. 2D, E).
Fig. 2. Gli1 inhibition activates TAK1 and its downstream kinases.
A–C SW1736 cells were treated with the indicated concentrations of GANT61 for 48 h or with 10 μM GANT61 for the indicated lengths of time. D, E SW1736 cells were transfected with control or Gli1 siRNA and incubated for 48 h. Cell lysates were analyzed for protein phosphorylation and their total protein by western blot with the indicated antibodies. Protein band density was analyzed by using an NIH Image-J software. Data presented as the mean ± SD (n = 3) relative to control are shown in bar graphs. *p < 0.05, **p < 0.01, compared to the untreated control or the scrambled siRNA control.
TAK1 is required for GANT61-induced autophagy
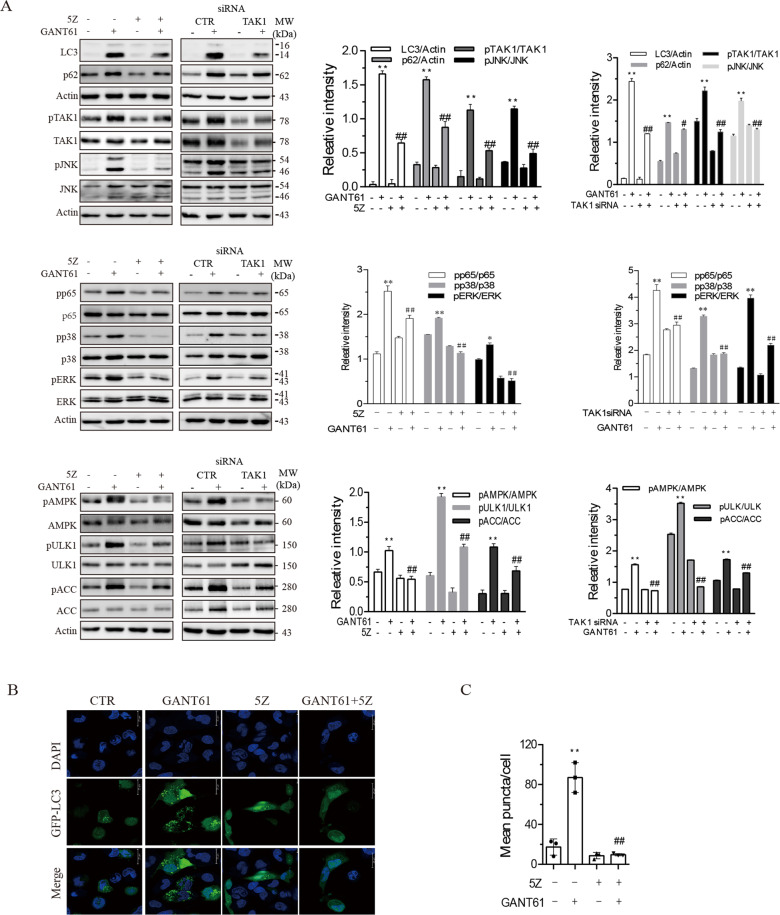
We next determined if inhibition of TAK1 by 5Z-7-oxozeaenol (5Z), a TAK1-specific inhibitor, could block GANT61-induced autophagy. As shown in Fig. 3A, 5Z blocked GANT61-induced LC3 lipidation and p62 expression. 5Z also blocked GANT61-induced phosphorylation of TAK1S412, JNKT183/185, p65S93, p38T180/182, ERKT202/204, AMPKT172, ULK1S555, and ACCS79. TAK1 siRNA very effectively suppressed TAK1 expression as well as TAK1S412 phosphorylation (Fig. 3A). TAK1 knockdown had similar effects on GANT61-induced protein phosphorylation and LC3 lipidation (Fig. 3A). Consistently, 5Z blocked GANT61-induced formation of autophagosomes in SW1736 cells (Fig. 3B, C).
Fig. 3. TAK1 mediates GANT61-induced autophagy.
A SW1736 cells were incubated in the absence or presence of GANT61 (10 μM) minus or plus 5Z (1 μM) for 48 h. Alternatively, SW1736 cells were transfected with scrambled or TAK1 siRNA (2.5 nmole each). After incubation for 4 h, the cells were left untreated or treated with GANT61 (10 μM) for 48 h. Cell lysates were prepared and analyzed for total and phosphorylated proteins by western blot. The expression levels were analyzed by quantification of the density of the protein bands with NIH Image-J software and presented as bar graphs. *p < 0.05; **p < 0.01, compared to the untreated control; #p < 0.05; ##p < 0.01, compared to GANT61-treated SW1736 cells. B, C Inhibition of GANT61-induced autolysosome formation by 5Z. SW1736 cells transiently transfected with the expression vector encoding the GFP-LC3 gene were incubated in the absence or presence of GANT61 (10 μM) minus or plus 5Z (1 μM) for 48 h. After nuclear staining with DAPI, the cells were examined under a confocal microscope for the presence of autophagosomes (B). The number of autophagosome puncta per cells was statistically analyzed (C). Bar length: 20 μm. **p < 0.01, compared to untreated control; ##p < 0.01, compared to GANT61.
AMPK and JNK are required for GANT61-induced autophagy
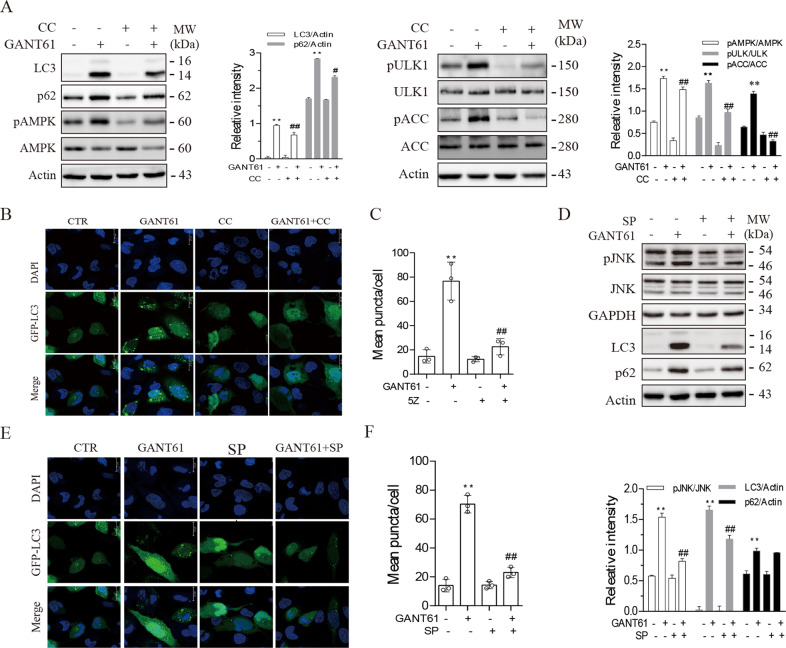
AMPK phosphorylates ULK1S555 and activates its enzymatic activity to trigger autophagy19,20. Here we tested if GANT61-induced autophagy was indeed mediated through TAK1-activated AMPK. As shown in Fig. 4A, compound C (CC), an inhibitor of AMPK, blocked GANT61-induced ULK1S555 phosphorylation and LC3 lipidation. CC inhibited AMPKT172 and ULK1S555 phosphorylation in GANT61-treated SW1736 cells. Consistently, CC blocked GANT61-induced formation of autophagosomes in SW1736 cells (Fig. 4B, C). JNK phosphorylates Bcl-2 and induces its dissociation with Beclin-1 (Klionsky et al.10). We then tested whether inhibition of JNK also blocked GANT61-induced autophagy. As shown in Fig. 4D, SP600125, a specific inhibitor of JNK, blocked GANT61-induced JNKT183/185 phosphorylation and LC3 lipidation. Consistently, SP600126 blocked GANT61-induced formation of autophagosomes in SW1736 cells (Fig. 4E, F).
Fig. 4. AMPK and JNK are required for GANT61-induced autophagy.
SW1736 cells were incubated in the absence or presence of GANT61 (10 μM) minus or plus compound C (5 μM) (A) or SP600125 (10 μM) (D) for 48 h. Cell lysates were prepared and analyzed for total and phosphorylated proteins by western blot. The expression levels were analyzed by quantification of the density of the protein bands with NIH Image-J software and presented as bar graphs. B, C, E, F SW1736 cells transiently transfected with the expression vector encoding the GFP-LC3 gene were incubated in the absence or presence of GANT61 (10 μM) minus or plus compound C (5 μM) (B, C) or SP600125 (10 μM) (E, F) for 48 h. After nuclear staining with DAPI, the cells were examined under a confocal microscope for the presence of autophagosomes (B, E). The number of autophagosome puncta per cells was statistically analyzed (C, F). Bar length: 20 μm. **p < 0.01, compared to untreated control; ##p < 0.01, compared to GANT61.
Autophagy inhibition enhances GANT61-induced apoptosis
It is well documented that autophagy antagonizes the antitumor effect of various anticancer drugs in part by suppressing apoptosis21. Here we investigated whether GANT61-induced autophagy also impacted GANT61-induced apoptosis. As shown in Fig. 5A, GANT61 induced caspase-3 (C3) and caspase-8 (C8) in a dose- and time-dependent manner but induced poly (ADP-ribose) polymerase (PARP) cleavage largely independent of the dose response. Notably, GANT61 activated caspase-8 and PARP slightly earlier than caspase-3 (Fig. 5A). Inhibition of autophagy by chloroquine (5 or 10 μM; Fig. 5B) and by inhibiting TAK1 activity with 5Z (1 or 5 μM; Fig. 5D) and by TAK1 siRNA (2.5 nmole; Fig. 5F) enhanced GANT61-induced caspase-8 and PARP cleavage more potently than caspase-3. To further confirm the effect of autophagy on GANT61-induced apoptosis, we conducted flow cytometry to analyze the expression of annexin V on the cell surface. As shown in Fig. 5C, E, chloroquine (5 or 10 μM) or 5Z (5 μM) in combination with GANT61 (10 μM) significantly increased the percentage of annexin V-positive cells, compared to those treated with GANT61, chloroquine, or 5Z alone. Finally, we investigated if inhibition of autophagy by Beclin-1 knockdown also potentiated GANT61-induced apoptosis. As shown in Fig. 5G, Beclin-1 siRNA significantly silenced Beclin-1 expression in SW1736 cells. Beclin-1 knockdown blocked GANT61-induced LC3 lipidation and p62 expression. Consistently, Beclin-1 silencing also enhanced GANT61-induced caspase-8, caspase-3, and PARP cleavage (Fig. 5G). These observations collectively suggest that inhibition of autophagy potentiates GANT61-induced apoptosis.
Fig. 5. Inhibition of autophagy enhances GANT61-induced apoptosis.
A SW1736 cells were treated with the indicated concentrations of GANT61 for 48 h or with 10 μM GANT61 for the indicated lengths of time. B, D SW1736 cells were incubated in the absence or presence of GANT61 (10 μM) minus or plus 5Z (1 or 5 μM) (B) or CQ (5 or 10 μM) (D) for 24 h. Cell lysates were prepared and analyzed for PARP, caspase-8 (C8), cleaved caspase-8 (CC8), caspase-3 (C3), and cleaved caspase-3 (CC3) by western blot. C-PARP cleaved PARP. C, E SW1736 cells were incubated in the absence or presence of GANT61 (10 μM) minus or plus 5Z (1 or 5 μM) (C) or CQ (5 or 10 μM) (E) for 24 h. Single-cell suspensions were stained for propidium iodide (PI) and annexin V followed by flow cytometry. Data in bar graphs are the mean ± SD of three independent experiments. F, G SW1736 cells were transfected with scrambled or TAK1 siRNA (F) or Beclin-1 siRNA (2.5 nmole each) (G). After incubation for 24 h, the cells were left untreated or treated with GANT61 for 24 h. Cell lysates were analyzed for p62, LC3, Beclin-1, TAK1, PARP, C8, CC8, C3, CC3, and β-Actin by western blot. The expression levels were analyzed by quantification of the density of the protein bands with NIH Image-J software and presented as bar graphs. *p < 0.05; **p < 0.01, compared to the untreated control; #p < 0.05; ##p < 0.01, compared to GANT61 (A–E).
Autophagy inhibition enhances GANT61-induced antiproliferative effects
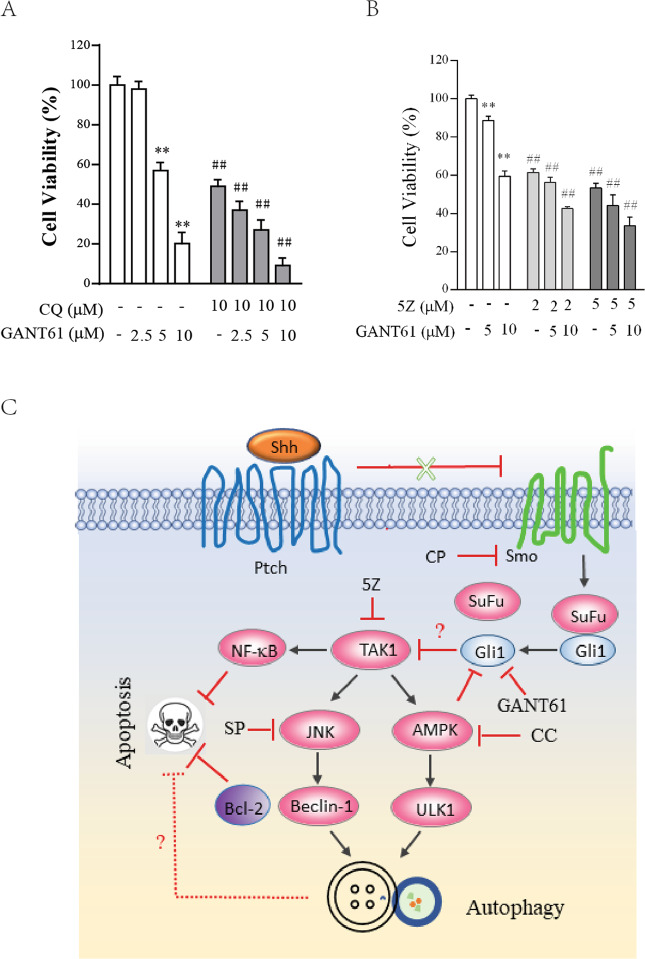
Finally, we investigated if inhibition of autophagy also enhanced GANT61-mediated antiproliferative activity. GANT61 incubated with SW1736 for 72 h (Fig. 6A) or 48 h (Fig. 6B) decreased SW1736 cell proliferation largely in a dose-dependent manner. CQ (10 μM) or 5Z (2 or 5 μM) alone inhibited SW1736 cell proliferation by ~50–60%. CQ (10 μM) or 5Z (2 or 5 μM) in combination with various concentrations of GANT61 had additive effects on cell proliferation, compared to those treated with CQ, 5Z, or GANT61 alone (Fig. 6A, B).
Fig. 6. Inhibition of autophagy potentiates the antiproliferative effect of GANT61.
SW1736 cells seeded in 96-well plates (2000 cells/well) were incubated with the indicated concentrations of GANT61 minus or plus CQ (10 μM) (A) for 72 h or 5Z (2 or 5 μM) (B) for 48 h. Cell viability was measured by using a CellTiter-Glo kit. Data are the mean ± SD of the triplicate from one representative of three independent experiments with similar results. **p < 0.01, compared to the untreated control; ##p < 0.01, compared to the counterparts in SW1736 left untreated or treated with the same concentrations of GANT61. C The mechanisms of Shh pathway inhibition-induced autophagy. Shh binding to Ptch releases the restriction of Smo activity. Activated Smo disrupts the interaction between Sufu and Gli1. Inhibition of the Shh pathway by the Smo inhibitor cyclopamine or by the Gli1 inhibitor GANT61 activates TAK1, which then activates JNK and AMPK. JNK promotes autophagy by phosphorylating Bcl-2 and dissociating it from Beclin-1. Free Beclin-1 becomes available for the initiation of autophagy. AMPK phosphorylates ULK1 and activates it. ULK1 is involved in the assembly of the pre-initiation complex and the early formation of autophagosomes. Activation of the autophagic pathway by inhibition of the Shh pathway attenuates apoptosis by multiple pathways including the activation of NF-kB and JNK by TAK1. Autophagy itself appears to also suppress apoptosis, albeit its underlying mechanism remains unknown. In addition, it is also not clear how Gli1 suppression leads to TAK1 activation.
Discussion
The Shh pathway is primarily recognized for its roles in tumor cell proliferation, drug resistance, metastasis, and stem cell self-renewal5. Several prior studies have shown that inhibition of this pathway induces autophagy in neuroblastomas and in breast, hepatocellular, and lung cancers11,12,14,22. However, the mechanisms of autophagy induction and its cytoprotective activity are poorly understood. Our present study provides evidence that inhibition of Gli1 and Smo activity by their specific inhibitors or by Gli1 siRNA induces autophagy through the activation of TAK1, a serine/threonine kinase that activates two downstream kinases, AMPK and JNK. AMPK activation leads to ULK1S555 phosphorylation and activation. Inhibition of JNK and AMPK activity by their specific inhibitors blocks GANT61-induced autophagy. We further show that blocking autophagy by chloroquine and 5Z and by TAK1 and Beclin-1 siRNA potentiates the apoptotic effect of GANT61 in SW1736 cells. Our study provides novel insights into the mechanisms by which inhibition of the Shh pathway induces autophagy.
TAK1 regulates a plethora of cellular functions including survival, apoptosis, differentiation, and the innate immune response18. TAK1 can be activated by various extracellular stimuli such as TGF-β, IL-1, TNF-α, LPS, DNA damage, microbial pathogens, and Toll-like receptors (TLR)23. TAK1 is a member of the MAP 3 kinase (MAP3K) family and activates multiple downstream MAP kinases including the JNK, ERK, and p38 pathways. TAK1 also activates IKKβ and its downstream NF-κB24. It has been increasingly appreciated that TAK1 can phosphorylate and activate AMPK to induce autophagy17. TAK1 is required for TGF-β−induced autophagy in murine mesangial cells through the TAK1-MKK3-p38 signaling pathway25 and is required for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced autophagy in human epithelial cells26. TAK1 activates AMPK to induce autophagy in response to S6K1 suppression27,28. TAK1 plays an important role in LPS and microbial pathogen-induced autophagy29,30. Our present study provides several lines of evidence showing that TAK1 activation plays an important role in Shh pathway inhibition-induced autophagy in thyroid tumor cell lines: (1) The inhibitors of Gli1 and Smo as well as Gli1 siRNA induced TAK1 phosphorylation, LC3 lipidation, and the formation of autophagosomes; (2) Gli1 overexpression decreased the basal levels of LC3 lipidation; (3) TAK1 inhibition by 5Z and by siRNA blocked GANT61-induced autophagy and phosphorylation of AMPK and JNK. These observations collectively suggest that TAK1 activation plays a crucial role in mediating Gli1 inhibition-induced autophagy (Fig. 6C).
AMPK can be activated by elevated intracellular AMP levels20 or by several protein kinases such as calcium/calmodulin-dependent kinase kinase (CaMKK), liver kinase B1 (LKB1), and TAK1 (Klionsky et al.10). Interestingly, AMPK activation by metformin phosphorylates Gli1 and downregulates its expression by destabilizing the Gli1 protein31–33. AMPK phosphorylates ULK1 at multiple sites, including S317, S468, S555, or S777. ULK1 phosphorylation and activation lead to autophagy induction10. We recently reported that TAK1 activation by inhibition of S6K1 activity or by LPS and microbial pathogens leads to AMPK activation and autophagy27–30,34. Consistent with these observations, our present study demonstrates that inhibition of the Shh pathway by GANT61 or by Gli1 knockdown led to increased AMPK and ULK1 phosphorylation and activation, and that AMPK activation contributed to GANT61-induced autophagy in thyroid cancer cell lines. It appears that the Shh pathway and AMPK regulate each other in a forward-feedback loop (Fig. 6C).
JNK has been implicated in playing an important role in inducing autophagy under various settings. For example, nutrient deprivation activates JNK to phosphorylate Bcl-2 at multiple sites, including T69, S70, and S87 (Klionsky et al.10). Phosphorylated Bcl-2 dissociates from the Bcl-2-Beclin-1 complex and participates in the formation of the pre-initiation complex and induction of autophagy10. JNK activation is involved in autophagy induced by hypoxia and chemotherapy35. We reported earlier that JNK activation by TAK1 is also involved in S6K1 inhibition-induced autophagy27. RITA, a p53 activator, downregulates the Shh pathway in a p53-independent and reactive oxygen species-independent manner and activates JNK; JNK activation is required for RITA-induced autophagy36. Our present study shows that GANT61 induced JNK phosphorylation, and that inhibition of JNK activity by SP600125 partially blocked GANT61-induced autophagy. Mechanistic investigation revealed that JNK activation was mediated by its upstream TAK1 (Fig. 6C). Our study provides evidence suggesting that inhibition of the Shh pathway activates JNK1, which contributes to GANT61-induced autophagy.
In the past two decades, there have been extensive studies focusing on the role of autophagy in drug resistance to chemo- and target therapy21. For example, autophagy induced by the inhibitors of B-Raf kinase, EGFR, and other receptor tyrosine kinases renders tumor cells resistant to the induction of apoptosis and compromises their antitumor activity37,38. Autophagy is also involved in chemoresistance to many genotoxic anticancer drugs39. There has been great interest in combining an autophagy inhibitor plus an anticancer drug to achieve synergistic antitumor activity. Vismodegib, a Smo inhibitor, induces autophagy in two lung adenocarcinoma cell lines and has weak antiproliferative activity in vitro and antitumor activity in vivo14. Inhibition of autophagy by ATG5 or ATG7 siRNA or by chloroquine enhances the antitumor activity of vismodegib14. Inhibition of GANT61-induced autophagy enhances apoptosis in neuroblastoma12,13. Our present study shows that GANT61 induced autophagy and apoptosis in two thyroid tumor cell lines. Inhibition of autophagy by Beclin-1 or TAK1 knockdown or by chloroquine enhanced GANT61-induced apoptosis. These observations collectively suggest that autophagy compromises the antitumor activity of GANT61. In contrast, Wang et al.11 reported earlier that inhibition of autophagy by 3-methyadenine or Beclin-1 knockdown suppressed GANT61-induced apoptosis and cytotoxicity in hepatocellular carcinomas. It is not clear if these contradicting observations result from different types of cancer.
There are several unsolved issues in our current study. First, it is not clear how inhibition of the Shh pathway activated TAK1. TAK1 activation is complex and can be regulated by multiple mechanisms. TAK1 becomes activated when it interacts with TAK1 binding protein-1 (TAB1) plus TAB2 or TAB3, the latter is modified by K63-linked polyubiquitination following a variety of stimuli such as IL-1 and TNF-α18. O-linked GlcNAcylation of TAB1 by osmotic shock or IL-1 and phosphorylation by the calcium calmodulin-dependent kinase CaMKII also activate TAK1 (Mukhopadhyay et al.18) . TAK1 is also activated by TRAF6 through ataxia-telangiectasia mutated (ATM) in DNA-damaged tumor cells40–42. It is not clear if GANT61 activated TAK1 by activating ATM. Second, p62 is usually degraded in the autophagic process10. Our present study showed that GANT61 did not decrease but rather increased p62 levels in SW1726 and KAT-18 cells. Previous studies have shown that JNK activation induces p62 expression by autophagy inducers such as resveratrol or dehydroepiandrosterone10,43,44. We conducted RT-PCR and the p62 promoter-driven luciferase reporter assay and unexpectedly found that GANT61 did not increase p62 mRNA levels nor its promoter activity (data not shown). Kehl et al.45 recently reported that TAK1 can protect p62 from autophagy-mediated degradation by reducing p62 translocation into autophagosomes. Since TAK1 was readily activated by GANT61, we speculate that increased p62 levels in GANT61-treated cells are due to TAK1-mediated protection. Third, while inhibition of autophagy enhanced GANT61-induced apoptosis, chloroquine or 5Z only enhanced the antiproliferative activity of GANT61 modestly when they were used in combination with GANT61 (Fig. 6A, B). It is not clear if GANT61 in combination with chloroquine or 5Z could dramatically improve their antitumor activity in vivo in a mouse tumor xenograft model; Fourth, the mechanisms by which autophagy antagonizes GANT61-induced apoptosis remain to be investigated (Fig. 6C); Fifth, it is well known that NF-κB activation leads to Bcl-2 expression and confers resistance to apoptosis in tumor cells24,46. Since TAK1 activated by GANT61 also induced NF-κB activation (Fig. 2A), it is likely that 5Z potentiates GANT61-induced apoptosis in part by blocking NF-κB activation (Fig. 6C). Sixth, phosphorylation and activation of JNK leads to its dissociation with Bcl-2. Bcl-2 released from the JNK-Bcl-2 complex is translocated into the mitochondrial membrane to suppress apoptosis (Fig. 6C). Enhanced apoptosis by TAK1 inhibition may be contributed in part by blocking JNK phosphorylation (Fig. 6C). Thus, TAK1 activation may attenuate GANT61-induced apoptosis by multiple pathways (Fig. 6C).
In summary, our present study has shown that inhibition of the Shh pathway cross-activates the TAK1-AMPK pathway in two anaplastic thyroid cancer cell lines. TAK1 activation leads to the phosphorylation of its two downstream substrates, AMPK and JNK. Activation of AMPK and JNK contributes to GANT61-induced autophagy in thyroid tumor cell autophagy. Inhibition of autophagy enhances GANT61-induced apoptosis and antiproliferative activity. Our study suggests that inhibition of the Shh pathway induces autophagy by activating the TAK1-AMPK axis and provides new insights into the mechanisms of GANT61-induced autophagy (Fig. 6C).
Materials and methods
Reagents
Cyclopamine (CP) and Compound C (CC) were purchased from Selleck Chemicals LLC (Shanghai, China) and dissolved in dimethyl sulfoxide (DMSO). Bafilomycin, chloroquine, SP600125, and 5Z-7-oxozeaenol (5Z) were purchased from Sigma (St. Louis, MO). GANT61 was purchased from Medkoo Biosciences (Morrisville, NC) and dissolved in 100% ethanol. Antibodies against Gli1 (2643), ULK1 (8054S), ULK1S555 (5869S), AMPK (5831S), AMPKT172 (2535S), ACC (3662S), ACCS79 (3661S), Beclin-1S93 (14717S), Beclin-1(3495S), ATG5 (12994S), ATG7 (8558S), p65S539 (3033S), ERKS202/204 (3510S), ERK (4685S), p38T180/182 (9211S), p38 (8690S), TAK1 (5206S), TAK1S412 (9339S), JNK (9252S), JNKT183/185 (4668S), Beclin-1 (3495S), caspase-3 (9662S), cleaved caspase-3 (9664S), caspase-8 (4790S), cleaved caspase-8 (9694S), LC3 (3868S), p62 (5114S), and PARP (9532S) were purchased from Cell Signaling Technology, Inc. (Danvers, MA). Antibodies against p65 (sc-372), β-Actin (sc-47778), and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was obtained from Santa Cruz Biotechnology Inc. (San Diego, CA). Gli1 (SR301820C) siRNA ON-TARGETplus SMARTpools were synthesized by Dharmacon and purchased from Fisher Scientific (Pittsburg, PA). Beclin-1 siRNA (6246S) and TAK1 siRNA (6317S) were purchased from Cell Signaling Technology, Inc. (Danvers, MA). Fluorescein isothiocyanate (FITC)-labeled Annexin V Apoptosis Detection Kit (556547) were acquired from BD Biosciences, Inc. (Shanghai, China). Green fluorescence protein (GFP)-microtubule-associated protein 1 light chain 3 (GFP-LC3) plasmid was kindly provided by Dr. Quan Zhang (Yangzhou University, Yangzhou, China).
Cell lines and plasmid DNA
Two ATC cell lines, KAT-18 and SW1736, were kindly provided by Dr. Kenneth B. Ain, authenticated recently9. KAT-18 cells were used between 40 and 60 passages; SW1736 cells were used between 26 and 45 passages.
Western blot
SW1736 and KAT-18 cells seeded in six-well plates were treated with the indicated concentrations of cyclopamine or GANT61 for 48 h or transfected with a Gli1 expression vector according to the manufacturer’s instructions. Cell lysates were prepared as described47 and analyzed for the expression of the indicated proteins. For loading controls, β-actin was detected by a mouse monoclonal antibody. Relative protein levels were analyzed by quantifying the blots with NIH Image-J software and presented as bar graphs.
Gene knockdown
KAT-18 and SW1736 cells seeded in a six-well plate were transfected with Gli1 by using Lipofectamine RNAiMAX (Invitrogen Life Technologies, Grand Island, NY) according to the manufacturer’s instructions. A scrambled siRNA was included as a negative control. After incubation for 48 h, cell lysates were analyzed by Gli1, LC3, p62, and β-actin expression by western blot with their specific antibodies. To determine the effect of inhibition of autophagy on GANT61-induced apoptosis, SW1736 cells were transfected with TAK1 and Beclin-1 siRNA. A scrambled control siRNA was included as a negative control. After transfection for 48 h, the cells were left untreated or treated with GANT61 (10 μM) and then incubated for 24 h. Cell lysates were prepared and analyzed for TAK1, Beclin-1, caspase-8, caspase-3, PARP, and β-actin.
GFP-LC3 fluorescence analysis
SW1736 cells seeded on coverslips were transiently transfected with GFP-LC3 expression plasmid DNA using FuGENE6 following the manufacturer’s protocol. After incubation for 48 h, the cells were incubated in the absence or presence of GANT61 (10 μM) minus or plus bafilomycin (10 nM) or chloroquine (10 μM; Fig. 1D), 5Z (1 μM; Fig. 3B), CC (5 μM; Fig. 4B), SP600125 (10 μM; Fig. 4B). After incubation for 48 h, the cells were fixed in 100% methanol at −20 °C for 10 min. The coverslips were mounted with 50% glycerin in PBS containing 4,6-diamidino-2-phenylindole (DAPI; 0.5 μg/ml; Sigma Chemical Co.). Autophagosomes were examined under a Leica LP8 confocal microscope. The autophagosome puncta was examined under a Nikon fluorescence microscope. Autophagosome puncta in SW1736 cells treated with various drugs were counted in 10 randomly selected fields under a ×40 objective in a blinded fashion. Results represent the mean ± SD (standard error of deviation) of three independent experiments.
Apoptosis assay
SW1736 cells seeded in six-well plates were incubated in the absence or presence of GANT61 (10 μM) minus or plus 5Z (5 μM) (A) or CQ (10 μM) (B) for 24 h. Single-cell suspensions were prepared and stained for propidium iodide (PI) and annexin V by using a FITC Annexin V Apoptosis Detection kit following the manufacturer’s instructions. Single-cell suspensions were run in a Beckman Coulter flow cytometer (Model CyAn ADP). The fluorescence intensity was analyzed by using the FlowJo software. Annexin-positive cells were gated. The percentage of Annexin V-positive cells from three independent experiments were calculated and statistically analyzed by using the unpaired Student’s t test. Data in bar graphs are the mean ± SD of three independent experiments.
Cell viability assay
SW1736 cells seeded in 96-well plates (2000 cells/well) were incubated with the indicated concentrations of GANT61 minus or plus CQ (10 μM; Fig. 6A) for 72 h or 5Z (2 or 5 μM; Fig. 6B) for 48 h. Cell viability was measured by using a CellTiter-Glo kit (Promega, Madison, WI, USA). Data are the mean ± SD of the triplicate from one representative of three independent experiments with similar results.
Statistical analysis
The differences in the density of western blots, autophagosome puncta, cell viability, and apoptosis between different treatment groups were statistically analyzed by using an unpaired Student’s t test or Analysis of Variance (ANOVA) when the data set has three or more independent groups. The p value of <0.05 was considered statistically significant. All statistics was performed with SigmaPlot 11 software (Systat Software, Inc, San Jose, CA).
Acknowledgements
We thank Dr. Kenneth B. Ain (University of Kentucky Medical Center, Lexington, KY) for kindly providing KAT-18 and SW1736 cell lines, Dr. Tsutomu Kume (Northwestern University Feinberg School of Medicine) for pcDNA/Gli1 plasmid, and Dr. Quan Zhang (Yangzhou University, Yangzhou, China) for kindly providing GFP-LC3 plasmid.
Author contributions
S.L. conducted experiments and interpreted data; J.W. conducted experiments; Y.L. supervised experiments; Y.Z. conducted experiments; R.P. critically revised and edited the manuscript; X.X. conceived the idea, implemented execution of the project, and wrote the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (81672643) and the Priority Academic Program Development of Jiangsu Higher Education Institutions to Xiulong Xu, Yangzhou University Ph.D. student fellowship to Yurong Lu.
Ethical statement
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Edited by B. Zhivotovsky
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sumei Li, Jingxiang Wang
References
- 1.Fagin JA, Wells SA., Jr. Biologic and clinical perspectives on thyroid cancer. N. Engl. J. Med. 2016;375:1054–1067. doi: 10.1056/NEJMra1501993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibrahimpasic T, Ghossein R, Shah JP, Ganly I. Poorly differentiated carcinoma of the thyroid gland: current status and future prospects. Thyroid. 2019;29:311–321. doi: 10.1089/thy.2018.0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matrone A, et al. Differentiated thyroid cancer, from active surveillance to advanced therapy: toward a personalized medicine. Front. Endocrinol. 2019;10:884. doi: 10.3389/fendo.2019.00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabanillas ME, McFadden DG, Durante C. Thyroid cancer. Lancet. 2016;388:2783–2795. doi: 10.1016/S0140-6736(16)30172-6. [DOI] [PubMed] [Google Scholar]
- 5.Jeng KS, Chang CF, Lin SS. Sonic hedgehog signaling in organogenesis, tumors, and tumor microenvironments. Int. J. Mol. Sci. 2020;21:758. doi: 10.3390/ijms21030758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rimkus TK, Carpenter RL, Qasem S, Chan M, Lo HW. Targeting the sonic hedgehog signaling pathway: review of smoothened and GLI inhibitors. Cancers. 2016;8:22. doi: 10.3390/cancers8020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X, Lu Y, Li Y, Prinz RA. Sonic hedgehog signaling in thyroid. Cancer Front. Endocrinol. 2017;8:284. doi: 10.3389/fendo.2017.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsen LJ, Moller LB. Crosstalk of hedgehog and mTORC1 pathways. Cells. 2020;9:2316. doi: 10.3390/cells9102316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y, et al. Targeting the sonic hedgehog pathway to suppress the expression of the cancer stem cell (CSC)-related transcription factors and CSC-driven thyroid tumor growth. Cancers. 2021;13:418. doi: 10.3390/cancers13030418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition) (1) Autophagy. 2021;17:1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Han C, Lu L, Magliato S, Wu T. Hedgehog signaling pathway regulates autophagy in human hepatocellular carcinoma cells. Hepatology. 2013;58:995–1010. doi: 10.1002/hep.26394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carballo, G. B. et al. GANT-61 induces autophagy and apoptosis in glioblastoma cells despite their heterogeneity. Cell. Mol. Neurobiol. (2020). 10.1007/s10571-020-00891-6. [DOI] [PMC free article] [PubMed]
- 13.Wang J, et al. Inhibition of autophagy potentiates the efficacy of Gli inhibitor GANT-61 in MYCN-amplified neuroblastoma cells. BMC Cancer. 2014;14:768. doi: 10.1186/1471-2407-14-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan J, et al. Regulating autophagy facilitated therapeutic efficacy of the sonic Hedgehog pathway inhibition on lung adenocarcinoma through GLI2 suppression and ROS production. Cell Death Dis. 2019;10:626. doi: 10.1038/s41419-019-1840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moscat J, Diaz-Meco MT. Feedback on fat: p62-mTORC1-autophagy connections. Cell. 2011;147:724–727. doi: 10.1016/j.cell.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aashaq S, Batool A, Andrabi KI. TAK1 mediates convergence of cellular signals for death and survival. Apoptosis. 2019;24:3–20. doi: 10.1007/s10495-018-1490-7. [DOI] [PubMed] [Google Scholar]
- 17.Neumann D. Is TAK1 a direct upstream kinase of AMPK? Int. J. Mol. Sci. 2018;19:2412. doi: 10.3390/ijms19082412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukhopadhyay H, Lee NY. Multifaceted roles of TAK1 signaling in cancer. Oncogene. 2020;39:1402–1413. doi: 10.1038/s41388-019-1088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galluzzi L, Pietrocola F, Levine B, Kroemer G. Metabolic control of autophagy. Cell. 2014;159:1263–1276. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res. 2014;24:42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva VR, Neves SP, Santos L, Dias RB, Bezerra DP. Challlenges and therapeutic opportunities of autophagy in cancer therapy. Cancers. 2020;12:3461. doi: 10.3390/cancers12113461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, et al. Anti-proliferation of breast cancer cells with itraconazole: Hedgehog pathway inhibition induces apoptosis and autophagic cell death. Cancer Lett. 2017;385:128–136. doi: 10.1016/j.canlet.2016.10.034. [DOI] [PubMed] [Google Scholar]
- 23.Xu YR, Lei CQ. TAK1-TABs complex: a central signalosome in inflammatory responses. Front. Immunol. 2020;11:608976. doi: 10.3389/fimmu.2020.608976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mihaly SR, Ninomiya-Tsuji J, Morioka S. TAK1 control of cell death. Cell Death Differ. 2014;21:1667–1676. doi: 10.1038/cdd.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SI, et al. Autophagy promotes intracellular degradation of type I collagen induced by transforming growth factor (TGF)-beta1. J. Biol. Chem. 2012;287:11677–11688. doi: 10.1074/jbc.M111.308460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrero-Martin G, et al. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009;28:677–685. doi: 10.1038/emboj.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X, et al. Inhibition of p70 S6 kinase (S6K1) activity by A77 1726, the active metabolite of leflunomide, induces autophagy through TAK1-mediated AMPK and JNK activation. Oncotarget. 2017;8:30438–30454. doi: 10.18632/oncotarget.16737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J, et al. Inhibition of p70 S6 kinase activity by A77 1726 induces autophagy and enhances the degradation of superoxide dismutase 1 (SOD1) protein aggregates. Cell Death Dis. 2018;9:407. doi: 10.1038/s41419-018-0441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W, et al. Activation of TGF-beta-activated kinase 1 (TAK1) restricts Salmonella Typhimurium growth by inducing AMPK activation and autophagy. Cell Death Dis. 2018;9:570. doi: 10.1038/s41419-018-0612-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, et al. Toll-like receptor signalling cross-activates the autophagic pathway to restrict Salmonella Typhimurium growth in macrophages. Cell. Microbiol. 2019;21:e13095. doi: 10.1111/cmi.13095. [DOI] [PubMed] [Google Scholar]
- 31.Gonnissen A, Isebaert S, McKee CM, Muschel RJ, Haustermans K. The effect of metformin and GANT61 combinations on the radiosensitivity of prostate cancer cells. Int. J. Mol. Sci. 2017;18:399. doi: 10.3390/ijms18020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li YH, et al. AMP-activated protein kinase directly phosphorylates and destabilizes hedgehog pathway transcription factor GLI1 in medulloblastoma. Cell Rep. 2015;12:599–609. doi: 10.1016/j.celrep.2015.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan C, et al. Metformin exerts anticancer effects through the inhibition of the Sonic hedgehog signaling pathway in breast cancer. Int. J. Mol. Med. 2015;36:204–214. doi: 10.3892/ijmm.2015.2217. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Sheng T, et al. Role of TGF-beta-activated kinase 1 (TAK1) activation in H5N1 influenza A virus-induced c-Jun terminal kinase activation and virus replication. Virology. 2019;537:263–271. doi: 10.1016/j.virol.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Vasilevskaya IA, Selvakumaran M, Roberts D, O’Dwyer PJ. JNK1 inhibition attenuates hypoxia-induced autophagy and sensitizes to chemotherapy. Mol. Cancer Res. 2016;14:753–763. doi: 10.1158/1541-7786.MCR-16-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azatyan A, et al. RITA downregulates Hedgehog-GLI in medulloblastoma and rhabdomyosarcoma via JNK-dependent but p53-independent mechanism. Cancer Lett. 2019;442:341–350. doi: 10.1016/j.canlet.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Kwon Y, Kim M, Jung HS, Kim Y, Jeoung D. Targeting autophagy for overcoming resistance to anti-EGFR treatments. Cancers. 2019;11:1374. doi: 10.3390/cancers11091374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei W, Hardin H, Luo QY. Targeting autophagy in thyroid cancers. Endocr. Relat. Cancer. 2019;26:R181–R194. doi: 10.1530/ERC-18-0502. [DOI] [PubMed] [Google Scholar]
- 39.Perez-Hernandez M, et al. Targeting autophagy for cancer treatment and tumor chemosensitization. Cancers. 2019;11:1599. doi: 10.3390/cancers11101599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B, et al. The senescence-associated secretory phenotype is potentiated by feedforward regulatory mechanisms involving Zscan4 and TAK1. Nat. Commun. 2018;9:1723. doi: 10.1038/s41467-018-04010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hadian K, Krappmann D. Signals from the nucleus: activation of NF-kappaB by cytosolic ATM in the DNA damage response. Sci. Signal. 2011;4:pe2. doi: 10.1126/scisignal.2001712. [DOI] [PubMed] [Google Scholar]
- 42.Hinz M, et al. A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. Mol. Cell. 2010;40:63–74. doi: 10.1016/j.molcel.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 43.Puissant A, et al. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010;70:1042–1052. doi: 10.1158/0008-5472.CAN-09-3537. [DOI] [PubMed] [Google Scholar]
- 44.Vegliante R, Desideri E, Di Leo L, Ciriolo MR. Dehydroepiandrosterone triggers autophagic cell death in human hepatoma cell line HepG2 via JNK-mediated p62/SQSTM1 expression. Carcinogenesis. 2016;37:233–244. doi: 10.1093/carcin/bgw003. [DOI] [PubMed] [Google Scholar]
- 45.Kehl SR, et al. TAK1 converts Sequestosome 1/p62 from an autophagy receptor to a signaling platform. EMBO Rep. 2019;20:e46238. doi: 10.15252/embr.201846238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lork M, Verhelst K, Beyaert R. CYLD, A20 and OTULIN deubiquitinases in NF-kappaB signaling and cell death: so similar, yet so different. Cell Death Differ. 2017;24:1172–1183. doi: 10.1038/cdd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heiden KB, et al. The sonic hedgehog signaling pathway maintains the cancer stem cell self-renewal of anaplastic thyroid cancer by inducing snail expression. J. Clin. Endocrinol. Metab. 2014;99:2178–2187. doi: 10.1210/jc.2014-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]