Blastomycosis due to Blastomyces dermatitidis and Blastomyces gilchristii is a significant cause of respiratory mycoses in North America with occasional reported outbreaks. We developed a highly sensitive, specific, and reproducible TaqMan duplex real-time PCR assay for the differentiation of B. dermatitidis and B. gilchristii.
KEYWORDS: Blastomyces, ecology, epidemiology, molecular assay, validation
ABSTRACT
Blastomycosis due to Blastomyces dermatitidis and Blastomyces gilchristii is a significant cause of respiratory mycoses in North America with occasional reported outbreaks. We developed a highly sensitive, specific, and reproducible TaqMan duplex real-time PCR assay for the differentiation of B. dermatitidis and B. gilchristii. The new assay permitted retrospective analysis of Blastomyces cultures (2005 to 2019) and primary clinical specimens from blastomycosis cases (2013 to 2019) from New York patients. We identified B. dermatitidis as the predominant pathogen in 38 cases of blastomycosis, while B. gilchristii was a minor pathogen involved in five cases; these findings expand understanding of blastomycosis in New York. The duplex real-time PCR assay could be implemented in reference and public health laboratories to further understand the ecology and epidemiology of blastomycosis due to B. dermatitidis and B. gilchristii.
INTRODUCTION
Until recently, Blastomyces dermatitidis was considered a single species within the genus Blastomyces and the sole etiologic agent of pulmonary and disseminated blastomycosis (1). This knowledge base was behind the development of several diagnostic tests (2–5). With the recent advances in fungal biology, several distinct genetic populations were identified as new species within the genus Blastomyces. Among the new species are Blastomyces gilchristii (6, 7), Blastomyces percursus, and Blastomyces silverae (8, 9). Blastomyces helicus was proposed as a new species responsible for rare blastomycosis cases in the western part of North America. Finally, B. percursus was proposed as a new species for blastomycosis cases from Africa (10, 11). In addition, taxonomic revisions assigned Emmonsia parva as Blastomyces parvus and Emmonsia helica as Blastomyces helicus within an expanded Blastomyces genus (9). Blastomyces dermatitidis is the predominant species in North America. Blastomyces gilchristii might be the leading cause of blastomycosis in northwestern Ontario and northern Wisconsin areas where the disease is hyperendemic (10). The ecological niche(s) of B. dermatitidis and B. gilchristii remains undefined. Similarly, the role of remaining rare Blastomyces species in the overall incidence and outcome of blastomycosis cases requires further investigations.
All Blastomyces spp. are thermally dimorphic fungi, existing as a mold form at ambient temperature in the environment. Blastomyces spp. convert to the pathogenic yeast form in vitro at 37°C or when a susceptible mammalian host inhales the conidia of the mold form. The yeast forms of these pathogens have distinguishable microscopic features: B. dermatitidis and B. gilchristii produce abundant, large (8- to 20-μm) broad-based budding yeasts, B. percursus produces large yeast-like cells from fragmented swollen hyphal cells, B. helicus produces variably shaped yeast cells (4 to 5 μm) in short chains, and B. parva and B. silverae produce thin-walled giant cells and a few broad-based budding yeast-like cells (12). A close examination of the yeast morphology might help in the species differentiation in the primary specimens. However, microscopic diagnosis requires expertise in mycology, which is not available in all laboratories. Culture and histopathology will help diagnose blastomycosis, but molecular methods are needed for accurate identification of newly described Blastomyces species. There are currently no molecular or serologic diagnostic tests for the rapid and accurate identification of species within the genus Blastomyces. An available commercial test (Gen-Probe, Inc., San Diego, CA) is somewhat limited, as it can be used only with pure cultures of B. dermatitidis. Also, culture-based identification is not feasible in all laboratories because of the occupational hazard associated with the fungus mold form. Earlier, we developed a rapid and specific real-time PCR assay to identify and detect the then-known four haplotypes of B. dermatitidis (5).
Outdoor exposure and proximity to water have been associated with blastomycosis (13). Blastomycosis is reportable in only five states in the United States, namely, Arkansas, Louisiana, Michigan, Minnesota, and Wisconsin (13). In New York State, blastomycosis is considered an emerging disease (http://www.nycasm.org/Alerts/Notification_101674.pdf). Several recent case series and case reports describe blastomycosis from New York. However, areas in New York where blastomycosis is endemic remain undefined, and it is not included in the list of reportable diseases (14–17). Likewise, in the absence of a skin test, it is difficult to assess the geographic location of likely exposure to the pathogen. Limited publications on canine blastomycosis suggest that the disease is prevalent in the Saint Lawrence River valleys on the New York-Canada border (14). Pneumonia is the most common manifestation of blastomycosis; approximately half of all cases are asymptomatic (18). However, Blastomyces infection can lead to a severe and fatal disease, often because of respiratory failure (19). Disseminated infection can involve any organ, often including cutaneous abscesses and osteomyelitis, and is frequently accompanied by fever, weight loss, and night sweats (2).
BAD-1 is an important conserved adhesion-promoting protein and virulence factor of B. dermatitidis (20). Two large insertions were noted in the B. gilchristii BAD1 gene (21), while this gene was absent in African strains of Blastomyces spp. (22). Several conventional PCR assays were designed using the BAD1 gene (23) for the detection of B. dermatitidis DNA from clinical and soil samples (4, 24). Sidamonidze et al. (5) developed a real-time PCR assay targeting the BAD1 gene to identify B. dermatitidis in culture and primary specimens. In the present investigation, we focused on the sequence variations observed within BAD1 to develop a duplex real-time PCR assay for the differentiation of B. dermatitidis and B. gilchristii. We present data on the sensitivity, specificity, and reproducibility of the assay. The new assay was used for a retrospective analysis of culture and primary specimens from blastomycosis cases from 2005 to 2019 in New York. Our results highlight the idea that B. dermatitidis is the major pathogen in blastomycosis cases in New York, with B. gilchristii being involved in few cases.
MATERIALS AND METHODS
Strains, primary specimens, and DNA.
Seventy-nine isolates of Blastomyces spp. were evaluated in this study. Forty-eight clinical isolates came from the sporadic cases of blastomycosis between 2005 and 2019 from New York. Isolates from canine (6 isolates), feline (1 isolate), sea lion (2 isolates), bat (1 isolate), polar bear (1 isolate), soil (4 isolates), and human (2 isolates) sources were obtained from Gene M. Scalarone (Department of Biological Sciences, Idaho State University). American Type Culture Collection (ATCC) strains used were MYA2586 (soil), MYA2585 (dog), ATCC 56214 (human, Africa), and ATCC 56216 (human, Africa). Five well-characterized strains each of B. dermatitidis and B. gilchristii were obtained from the Public Health Laboratory, Public Health Ontario, Toronto, Ontario, Canada. The species-level identifications of Blastomyces cultures from Canada were confirmed by sequencing of the ITS2 and drk1 genes described in an earlier publication (7). Thirty-three primary clinical specimens from New York patients, including 12 paraffin-embedded tissues, six bronchial wash specimens, five lung tissue samples, three cerebrospinal fluid samples, two skin lesion tissue samples, two bone marrow samples, and one sample each of blood, brain lesion tissue, and sputum submitted for B. dermatitidis identification, were also part of this investigation. Additionally, DNA from closely and distantly related fungal pathogens was included as part of a specificity panel (5).
DNA extraction.
DNA samples from Blastomyces spp. were extracted using a Qiagen DNA minikit on the Qiacube automated extractor. In brief, fungal growth (approximately 5 by 5 mm) was removed from the agar surface using a sterile loop and added to a lysing reagent in a biosafety level 3 (BSL3) laboratory. The fungal suspension was incubated at 90°C for 1 h and then brought to a BSL2 laboratory for further processing. The heat-killed fungal suspension was homogenized three times in a Precellys homogenizer at 6,500 rpm for 15 s each time (program number 5; 6500-3x60-015). The homogenized suspension was transferred to a 2-ml screw-cap tube, leaving behind the beads. DNA from samples was extracted using the Qiagen DNA minikit in the QiaCube semiautomated DNA extractor, resulting in 50 µl of eluted DNA. All extracted DNAs were stored at −80°C. DNA of fungal species (yeasts and molds) other than Blastomyces spp. were procured from the Wadsworth Center Mycology Laboratory (WCML) DNA Collection Repository.
The Wadsworth Center Histopathology Core first sectioned DNA from paraffin-fixed tissues, and then the paraffin was dissolved with 1 ml of xylene, followed by two washes using 1 ml of 100% ethanol. The sample was then dried and extracted using the Qiagen DNA minikit as described above with an incubation temperature of 70°C instead of 90°C. For nonfixed tissues, an approximately 5- by 5-mm section of tissue was cut with a sterile blade, and extraction was done using the Qiagen DNA minikit as described for the isolates, with an incubation temperature of 70°C instead of 90°C.
Primers and probes.
Primers and probes for the real-time PCR assay were designed from the promoter region of the BAD1 gene using Geneious R9 9.1.6 software (Biomatters, Inc., San Diego, CA). The choice of BAD1 promoter was based upon our earlier successful utilization of this region for the singleplex real-time PCR assay for B. dermatitidis identification/detection (5). Multiple alignment of BAD1 promoter sequences revealed unique sequences for B. dermatitidis and B. gilchristii, which were used for primers and probes design (see Fig. S1 in the supplemental material). Oligonucleotide sequences of the primers and probes for B. gilchristii and B. dermatitidis are as follows: V2556 (B. gilchristii; forward), 5′-ATGGGTGCAAAATCCGCCTA-3′; V2558 (B. gilchristii; reverse), 5′-AATCTAGAAGCTGGAGCGCC-3′; V2557 (B. gilchristii; probe), 5′-FAM (6-carboxyfluorescein)-CCGTACTCC/ZEN/CTCCCCGGTACTCC-3′IABkFQ (Iowa Black fluorescent quencher); V2559 (B. dermatitidis; forward), 5′-GCAAAATCCGCCTACTACTA-3′; V2561 (B. dermatitidis; reverse), 5′-AGCTGAACCTGGAAGTATTG-3′; V2560 (B. dermatitidis; probe), 5′-Cy5-TCCCTACCC/TAO/CTGGCTACTTTTCT-3′IAbRQSp (Iowa Black red quencher spacer). The internal ZEN and TAO quencher was incorporated between bases 9 and 10 from the 5' end of the probes V2557 and V2560, respectively. This design decreased the distance between the dye and the quencher and was expected to reduce the background signal and achieve an improved dynamic range. The primers and probes were purchased from Integrated DNA Technologies, Inc. (Coralville, IA).
Real-time PCR assay.
Each DNA sample (isolate or primary specimen) was tested in duplicate in 20-μl reaction volumes using an optical 96-well plate. Each reaction mixture contained 1× PerfeCTa multiplex qPCR ToughMix (Quanta Biosciences), a 1,000 nM concentration of each B. gilchristii (V2556 and V2558) and B. dermatitidis (V2559 and V2561) primer, a 250 nM concentration of each B. gilchristii (V2557) and B. dermatitidis (V2560) probe, and 2 μl of DNA (approximately 1 to 10 ng) extracted from Blastomyces spp. or 5 μl of DNA extracted from primary clinical specimens. Each PCR run also included 2 μl (1 ng) of positive extraction control (M808; B. dermatitidis), 2 μl (1 ng) of positive amplification controls (M808 [B. dermatitidis] and MB97 [B. gilchristii]), and 2 μl of negative extraction (extraction reagents only) and negative amplification (sterilized nuclease-free water) controls. The unidirectional workflow kept the reagent preparation, specimen preparation, and amplification and detection areas separate to avoid cross-contamination. Cycling conditions on the ABI 7500 FAST system (Applied Biosystems, Thermo Fisher Scientific Inc., Waltham, MA) were initial denaturation at 95°C for 20 s, followed by 45 cycles of 95°C for 3 s and 60°C for 30 s. Based on limit of detection (LOD), a cycle threshold (CT) value of ≤38 was reported as positive and >38 was reported as negative. Specimens were reported as inconclusive if PCR inhibition was observed for the primary specimens.
Data availability.
GenBank accession numbers for BAD1 are MT822768 to MT822773; GenBank accession numbers for the internal transcribed spacer (ITS) genes are MT822762 to MT822767.
RESULTS
Assay sensitivity, specificity, and reproducibility.
The duplex real-time PCR assay was highly sensitive, with linearity over 5 logs, a correlation coefficient of >0.99, and amplification efficiency of >92%. The limit of detection (LOD) of the assay was 1 pg of genomic DNA (gDNA) per PCR within the linear range of the standard curve for B. dermatitidis and B. gilchristii (Fig. 1 and Table 1). The assay was highly specific, as it did not cross-react with other closely and distantly related fungal pathogens (see Table S2). The duplex real-time PCR assay correctly identified B. dermatitidis and B. gilchristii from 10 genomic-DNA samples submitted in a blind fashion from Ontario, Canada, confirming the assay validity (Table 2). Additionally, DNA from B. dermatitidis and B. gilchristii strains run on three different days and within the same day yielded consistent CT values with a cofficient of variance of <5.0, confirming assay reproducibility (Table S3A and B).
FIG 1.
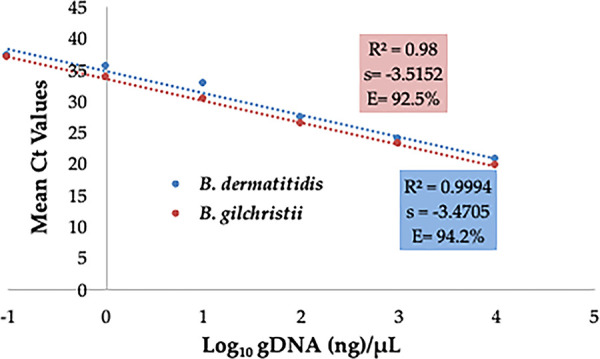
Sensitivity of duplex real-time PCR assay. Serial 10-fold dilution series of gDNA of B. dermatitidis and B. gilchristii were prepared, and PCR was run in duplicate. The slope (s), correlation coefficient (R2), and amplification efficiency (E) are shown.
TABLE 1.
Sensitivity study of B. dermatitidis and B. gilchristiia
| DNA concn (ng/µl) |
B. dermatitidis |
B. gilchristii |
||||
|---|---|---|---|---|---|---|
| CT 1 | CT 2 | Mean CT | CT 1 | CT 2 | Mean CT | |
| 10 | 20.73 | 20.70 | 20.72 | 19.85 | 19.76 | 19.80 |
| 1 | 23.88 | 23.87 | 23.88 | 23.10 | 23.08 | 23.09 |
| 0.1 | 27.25 | 27.51 | 27.38 | 26.54 | 26.44 | 26.49 |
| 0.01 | 32.96 | 32.60 | 32.78 | 30.59 | 30.15 | 30.37 |
| 0.001 | 35.96 | 35.27 | 35.62 | 33.68 | 33.64 | 33.66 |
| 0.0001 | 36.91 | 37.48 | 37.20 | 36.22 | 37.74 | 36.98 |
| 0.00001 | Undet | Undet | Undet | Undet | Undet | Undet |
Undet, undetermined.
TABLE 2.
Blind panel of DNA of B. dermatitidis and B. gilchristiia
| Sample no. | Mean CT in real-time PCR assayb |
Final ID | ||
|---|---|---|---|---|
| Singleplex | Duplex |
|||
| B. dermatitidis (Cy5) | B. gilchristii (FAM) | |||
| 1 | 23.02 | 23.48 | Undet | B. dermatitidis |
| 2 | 22.4 | Undet | 21.74 | B. gilchristii |
| 3 | 18.52 | 20.04 | Undet | B. dermatitidis |
| 4 | 22.31 | 22.64 | Undet | B. dermatitidis |
| 5 | 19.86 | Undet | 19.35 | B. gilchristii |
| 6 | 22.12 | Undet | 21.54 | B. gilchristii |
| 7 | 23.28 | Undet | 22.46 | B. gilchristii |
| 8 | 19.75 | 20.15 | Undet | B. dermatitidis |
| 9 | 22.23 | 22.39 | Undet | B. dermatitidis |
| 10 | 21.45 | Undet | 20.39 | B. gilchristii |
DNA was supplied by J.V.K. in a blind fashion.
Undet, undetermined.
Retrospective analysis of Blastomyces isolates and primary specimens.
The retrospective analysis of 79 isolates of Blastomyces spp. revealed 62 to be B. dermatitidis and 15 to be B. gilchristii (Table 3). One isolate (ATCC 56214), which was identified by singleplex real-time PCR assay as Blastomyces spp., was neither B. dermatitidis nor B. gilchristii by the duplex real-time PCR assay. It was later confirmed to be B. percursus by sequencing of the ribosomal genes and BLAST search. One strain of Blastomyces emzantsi (ATCC 21516), identified by sequencing, was not identified by the singleplex real-time PCR or the duplex real-time PCR assay, further confirming the specificity of the current duplex real-time PCR assay. Of 15 B. gilchristii isolates identified, 5 were well-characterized strains of B. gilchristii from Canada, 4 were from the Eagle River outbreak (Wisconsin) involving human, dog, and soil samples, and 6 isolates were recovered from five New York patients. All six New York isolates of B. gilchristii were also confirmed by sequencing of the BAD1 and ribosomal genes. Of 62 B. dermatitidis isolates identified, 5 were well-characterized strains of B. dermatitidis from Canada, 15 were from Illinois, Kentucky, Minnesota, Tennessee, and Wisconsin involving animals or soil and 42 isolates were from 38 patients from New York collected from 2005 to 2019. Of 33 primary specimens analyzed from 2013 to 2019, five specimens (one skin lesion, one bronchial wash, two tissue blocks of nasal masses, and one tissue block of unknown origin) from five patients were positive for B. dermatitidis DNA while none were positive for B. gilchristii DNA (Table 3).
TABLE 3.
Validity of duplex real-time PCR assay for culture identification of Blastomyces spp. and direct detection from primary specimens
| Assay type and singleplex real-time PCR result (n) | No. of samples with duplex real-time PCR result |
|||
|---|---|---|---|---|
|
B. dermatitidis |
B. gilchristii |
|||
| Positive | Negative | Positive | Negative | |
| Culture identification | ||||
| Positive (78) | 62 | 16 | 15 | 63 |
| Negative (60) | 0 | 60 | 0 | 60 |
| Direct detection | ||||
| Positive (5) | 5 | 0 | 0 | 5 |
| Negative (28) | 0 | 28 | 0 | 28 |
Blastomycosis in New York.
Analysis of 43 cases of blastomycosis from New York (Table S1) revealed one to four cases per year from 2005 until 2016, but that number increased to 7 in 2017, and cases remained high, with 9 identified in 2018 and 8 identified in 2019 (Fig. 2). Men were more frequently infected than women with B. dermatitidis, while the number of cases was too small to derive this conclusion for B. gilchristii. Older people (over age 40) were found to be more prone to symptomatic infection irrespective of whether the agent was B. dermatitidis or B. gilchristii. B. dermatitidis was most commonly isolated from respiratory specimens, followed by skin/wounds/subcutaneous tissue and bone, while all six isolates of B. gilchristii were isolated from respiratory specimens (Table 4). Geographic distribution of blastomycosis cases revealed that the majority of patients presented with symptoms and lived in Mohawk Valley, Capital District, and Finger Lakes. Few cases were also reported from other regions in New York (Fig. S2).
FIG 2.
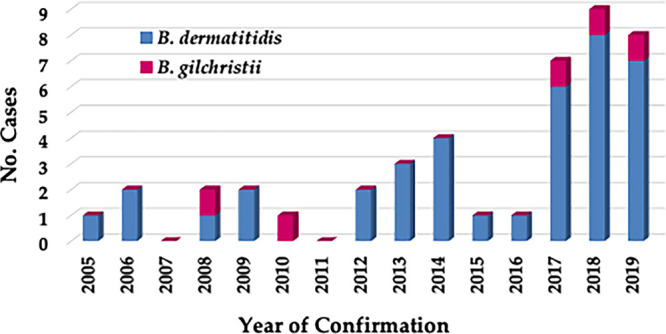
Blastomycosis cases in New York from 2005 to 2019. Blastomycosis cases confirmed earlier by singleplex real-time PCR assay for isolates and primary specimens were analyzed retrospectively with the newly developed duplex real-time PCR assay. Of 43 cases, 38 were confirmed as being caused by B. dermatitidis and 5 as B. gilchristii. There was a marked increase in blastomycosis cases from 2017 onward.
TABLE 4.
Characteristics of Mycology Laboratory (Wadsworth Center)-confirmed blastomycosis cases in New York from 2005 to 2019
| Characteristic | Value for group |
||
|---|---|---|---|
| Blastomycosis | B. dermatitidis | B. gilchristii | |
| No. of patients | 43 | 38 | 5 |
| Sex | |||
| M | 29 | 26 | 3 |
| F | 14 | 12 | 2 |
| Age (yr) | |||
| 20–29 | 4 | 4 | 0 |
| 30–39 | 4 | 4 | 0 |
| 40–49 | 8 | 7 | 1 |
| 50–59 | 11 | 10 | 1 |
| 60–69 | 13 | 10 | 3 |
| >69 | 3 | 3 | 0 |
| No. of specimens (n = 48) | |||
| Bronchial wash | 28 | 22 | 6 |
| Skin, wound, subcutaneous tissue | 15 | 15 | |
| Bone | 4 | 4 | |
| Brain | 1 | 1 | |
| No. of specimens positive for DNA (n = 5) | |||
| Bronchial wash | 1 | 1 | |
| Tissue block—nasal mass | 2 | 2 | |
| Tissue block—source not provided | 1 | 1 | |
| Skin wound fluid | 1 | 1 | |
DISCUSSION
In this study, we developed a duplex real-time PCR assay to differentiate B. dermatitidis from B. gilchristii. These two closely related species possibly overlap in their endemicity in North America. The duplex real-time PCR assay targeting the putative promoter region of the BAD1 gene was highly sensitive, specific, and reproducible. BAD1 and its promoter have been extensively used for the differentiation of Blastomyces species using restriction fragment length polymorphism (RFLP), PCR, and real-time PCR assays (5, 21, 24). The polymorphism in BAD1, its markedly different sizes (363 bp in B. dermatitidis and 663 bp in B. gilchristii), and the absence of this gene in African isolates of Blastomyces (5, 21, 22) confirmed the choice of this target as a highly specific one for the identification of B. dermatitidis and B. gilchristii in the present investigation.
The retrospective analysis of Blastomyces cultures and primary specimens confirmed 43 cases of blastomycosis in New York from 2005 to 2019 based on the samples received in our facility. We noted an increase in number of cases in the recent years. The reason behind the observed increase in blastomycosis cases in New York are not apparent; our results are in agreement with other investigations describing the increase in case counts in New York (15, 17, 25, 26). Of interest was the identification of five of 43 blastomycosis cases due to B. gilchristii. Interestingly, patients infected with B. gilchristii resided in different parts of New York. Additional studies are needed to determine if these cases are due to patients traveling between regions or a focal niche of B. gilchristii in New York. We found that most blastomycosis cases were caused by B. dermatitidis, with concentrations of cases in Mohawk Valley and the Capital District, followed by the Finger Lakes. The regional aspect of geographic risk for blastomycosis is not well understood. It is challenging to track the progression from exposure to disease onset in patients. A skin test or another reliable marker of prior exposure is not available, and B. dermatitidis is rarely recovered from the environment (10). Furthermore, the recognition of clinical cases in New York is sporadic. Therefore, evidence regarding areas of endemicity in New York is equivocal. Our results are in agreement with other reports indicating that B. dermatitidis has a broader geographic distribution throughout North America while B. gilchristii has a limited distribution in the northern United States and certain Canadian provinces (27).
We know little about the geographic and phenotypic differences between B. dermatitidis and B. gilchristii. However, preliminary studies suggest that there may be a difference in the clinical manifestation of the diseases between the two species (28). There is also a report of an acute respiratory distress syndrome-related fatal case due to B. gilchristii (29). BAD1 has been implicated as the major virulence factor of B. dermatitidis (20). The significant difference in size of the BAD1 promoter due to two large insertions between B. dermatitidis and B. gilchristii noted above and the issue of whether these insertions have any influence on BAD1 expression and virulence need further investigation (21). To the best of our knowledge, no studies are available to demonstrate the comparative virulence of B. dermatitidis and B. gilchristii in vertebrate or invertebrate model systems. The highly sensitive and specific assay developed in the present study would allow more comprehensive surveillance of blastomycosis to monitor disease incidence. Systematic disease reporting and surveillance efforts will help diagnose new blastomycosis cases. Prompt diagnosis ensures prompt initiation of treatment to decrease illness and death. Finally, public awareness campaigns such as health advisories might be needed for blastomycosis in New York. In summary, the newly developed duplex real-time PCR assay would ensure accurate laboratory diagnosis of B. dermatitidis and B. gilchristii and help expand the understanding of the ecology and epidemiology of blastomycosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Wadsworth Center (WC) Tissue Culture & Media, Histopathology, and Applied Genomic Technologies Cores for providing media, fixed tissue sectioning, and Sanger sequencing, respectively.
This work was supported partly by the funds from the WC, New York State Department of Health (NYSDOH), and Centers for the Disease Control and Prevention (CDC) grant number NU50CK000516. In addition, Mitchell Kaplan was partially supported with National Science Foundation funding to the WC Research Experience for Undergraduates (REU) Program (grant DBI1757170).
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the Department of Health and Human Services or the CDC.
S.C. conceived the study, supervised experiments, and wrote the manuscript. M.K. designed primers and probes under Y.Z.’s supervision, performed the majority of the experiments, and prepared graphs and tables. Y.Z. supervised M.K.’s work, performed a few key experiments, and prepared graphs and tables. V.C. contributed to the study design and edited the draft manuscript. J.V.K. and L.M. provided strains of Blastomyces dermatitidis and B. gilchristii and edited the draft manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Gilchrist TC, Stokes WR. 1898. A case of pseudo-lupus vulgaris caused by a Blastomyces. J Exp Med 3:53–78. doi: 10.1084/jem.3.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saccente M, Woods GL. 2010. Clinical and laboratory update on blastomycosis. Clin Microbiol Rev 23:367–381. doi: 10.1128/CMR.00056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein BS, Vergeront JM, Kaufman L, Bradsher RW, Kumar UN, Mathai G, Varkey B, Davis JP. 1987. Serological tests for blastomycosis: assessments during a large point-source outbreak in Wisconsin. J Infect Dis 155:262–268. doi: 10.1093/infdis/155.2.262. [DOI] [PubMed] [Google Scholar]
- 4.Bialek R, Cirera AC, Herrmann T, Aepinus C, Shearn-Bochsler VI, Legendre AM. 2003. Nested PCR assays for detection of Blastomyces dermatitidis DNA in paraffin-embedded canine tissue. J Clin Microbiol 41:205–208. doi: 10.1128/jcm.41.1.205-208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sidamonidze K, Peck MK, Perez M, Baumgardner D, Smith G, Chaturvedi V, Chaturvedi S. 2012. Real-time PCR assay for identification of Blastomyces dermatitidis in culture and in tissue. J Clin Microbiol 50:1783–1786. doi: 10.1128/JCM.00310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meece JK, Anderson JL, Fisher MC, Henk DA, Sloss BL, Reed KD. 2011. Population genetic structure of clinical and environmental isolates of Blastomyces dermatitidis, based on 27 polymorphic microsatellite markers. Appl Environ Microbiol 77:5123–5131. doi: 10.1128/AEM.00258-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown EM, McTaggart LR, Zhang SX, Low DE, Stevens DA, Richardson SE. 2013. Phylogenetic analysis reveals a cryptic species Blastomyces gilchristii, sp. nov. within the human pathogenic fungus Blastomyces dermatitidis. PLoS One 8:e59237. doi: 10.1371/journal.pone.0059237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dukik K, Muñoz JF, Jiang Y, Feng P, Sigler L, Stielow JB, Freeke J, Jamalian A, van den Ende BG, McEwen JG, Clay OK, Schwartz IS, Govender NP, Maphanga TG, Cuomo CA, Moreno LF, Kenyon C, Borman AM, de Hoog S. 2017. Novel taxa of thermally dimorphic systemic pathogens in the Ajellomycetaceae (Onygenales). Mycoses 60:296–309. doi: 10.1111/myc.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang Y, Dukik K, Muñoz JF, Sigler L, Schwartz IS, Govender NP, Kenyon C, Feng P, van den Ende BG, Stielow JB, Stchigel AM, Lu H, de Hoog S. 2018. Phylogeny, ecology and taxonomy of systemic pathogens and their relatives in Ajellomycetaceae (Onygenales): Blastomyces, Emergomyces, Emmonsia, Emmonsiellopsis. Fungal Divers 90:245–291. doi: 10.1007/s13225-018-0403-y. [DOI] [Google Scholar]
- 10.Schwartz IS, Kauffman CA. 2020. Blastomycosis. Semin Respir Crit Care Med 41:31–41. doi: 10.1055/s-0039-3400281. [DOI] [PubMed] [Google Scholar]
- 11.Chaturvedi V, de Hoog GS. 2020. Onygenalean fungi as major human and animal pathogens. Mycopathologia 185:1–8. [DOI] [PubMed] [Google Scholar]
- 12.Maphanga TG, Birkhead M, Muñoz JF, Allam M, Zulu TG, Cuomo CA, Schwartz IS, Ismail A, Naicker SD, Mpembe RS, Corcoran C, de Hoog S, Kenyon C, Borman AM, Frean JA, Govender NP. 2020. Human blastomycosis in South Africa caused by Blastomyces percursus and Blastomyces emzantsi sp. nov., 1967 to 2014. J Clin Microbiol 58:e01661-19. doi: 10.1128/JCM.01661-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein BS, Vergeront JM, DiSalvo AF, Kaufman L, Davis JP. 1987. Two outbreaks of blastomycosis along rivers in Wisconsin. Isolation of Blastomyces dermatitidis from riverbank soil and evidence of its transmission along waterways. Am Rev Respir Dis 136:1333–1338. doi: 10.1164/ajrccm/136.6.1333. [DOI] [PubMed] [Google Scholar]
- 14.Cote E, Barr SC, Allen C, Eaglefeather E. 1997. Blastomycosis in six dogs in New York State. J Am Vet Med Assoc 210:502–504. [PubMed] [Google Scholar]
- 15.Permpalung N, Kaewpoowat Q, Prasidthrathsint K, Chongnarungsin D, Hyman CL. 2013. Pulmonary blastomycosis: a new endemic area in New York state. Mycoses 56:592–595. doi: 10.1111/myc.12073. [DOI] [PubMed] [Google Scholar]
- 16.Spallone A, Tobin E, Chaturvedi S, Qian J. 2016. Central nervous system blastomycosis without travel history to unknown endemic area: a case report. Infect Dis Clin Pract 24:e30–e32. doi: 10.1097/IPC.0000000000000355. [DOI] [Google Scholar]
- 17.McDonald R, Dufort E, Jackson BR, Tobin EH, Newman A, Benedict K, Blog D. 2018. Notes from the Field: Blastomycosis cases occuring outside of region with known endmicity - New York, 2007–2017. MMWR Morb Mortal Wkly Rep 67:1077–1078. doi: 10.15585/mmwr.mm6738a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith JA, Kauffman CA. 2010. Blastomycosis. Proc Am Thorac Soc 7:173–180. doi: 10.1513/pats.200906-040AL. [DOI] [PubMed] [Google Scholar]
- 19.Mukkamala R, Mehta JB, Myers JW, Cole CP. 1997. Pulmonary blastomycosis with acute respiratory failure as predominant clinical feature. South Med J 90:847–850. doi: 10.1097/00007611-199708000-00017. [DOI] [PubMed] [Google Scholar]
- 20.Finkel-Jimenez B, Wüthrich M, Klein BS. 2002. BAD1, an essential virulence factor of Blastomyces dermatitidis, suppresses host TNF-alpha production through TGF-beta-dependent and -independent mechanisms. J Immunol 168:5746–5755. doi: 10.4049/jimmunol.168.11.5746. [DOI] [PubMed] [Google Scholar]
- 21.Meece JK, Anderson JL, Klein BS, Sullivan TD, Foley SL, Baumgardner DJ, Brummitt CF, Reed KD. 2010. Genetic diversity in Blastomyces dermatitidis: implications for PCR detection in clinical and environmental samples. Med Mycol 48:285–290. doi: 10.1080/13693780903103952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein BS, Aizenstein BD, Hogan LH. 1997. African strains of Blastomyces dermatitidis that do not express surface adhesin WI-1. Infect Immun 65:1505–1509. doi: 10.1128/IAI.65.4.1505-1509.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogan LH, Josvai S, Klein BS. 1995. Genomic cloning, characterization, and functional analysis of the major surface adhesin WI-1 on Blastomyces dermatitidis yeasts. J Biol Chem 270:30725–30732. doi: 10.1074/jbc.270.51.30725. [DOI] [PubMed] [Google Scholar]
- 24.Burgess JW, Schwan WR, Volk TJ. 2006. PCR-based detection of DNA from the human pathogen Blastomyces dermatitidis from natural soil samples. Med Mycol 44:741–748. doi: 10.1080/13693780600954749. [DOI] [PubMed] [Google Scholar]
- 25.Bethuel NW, Siddiqui N, Edmonds L. 2020. Pulmonary blastomycosis in rural upstate New York: a case series and review of literature. Ann Thorac Med 15:174–178. doi: 10.4103/atm.ATM_86_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Austin A, Tobin E, Judson MA, Hage CA, Hu K, Epelbaum O, Fantauzzi J, Jones DM, Gilroy S, Chopra A. 2020. Blastomycosis in the Capital District of New York State: a newly identified emerging endemic area. Am J Med doi: 10.1016/j.amjmed.2020.09.017. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 27.McTaggart LR, Brown EM, Richardson SE. 2016. Phylogeographic analysis of Blastomyces dermatitidis and Blastomyces gilchristii reveals an association with North American freshwater drainage basins. PLoS One 11:e0159396. doi: 10.1371/journal.pone.0159396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meece JK, Anderson JL, Gruszka S, Sloss BL, Sullivan B, Reed KD. 2013. Variation in clinical phenotype of human infection among genetic groups of Blastomyces dermatitidis. J Infect Dis 207:814–822. doi: 10.1093/infdis/jis756. [DOI] [PubMed] [Google Scholar]
- 29.Dalcin D, Rothstein A, Spinato J, Escott N, Kus JV. 2016. Blastomyces gilchristii as cause of fatal acute respiratory distress syndrome. Emerg Infect Dis 22:306–308. doi: 10.3201/eid2202.151183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GenBank accession numbers for BAD1 are MT822768 to MT822773; GenBank accession numbers for the internal transcribed spacer (ITS) genes are MT822762 to MT822767.


