The environmental bacterium Pseudomonas aeruginosa, particularly multidrug-resistant clones, is often associated with nosocomial infections and outbreaks. Today, core genome multilocus sequence typing (cgMLST) is frequently applied to delineate sporadic cases from nosocomial transmissions.
KEYWORDS: whole-genome sequencing, Pseudomonas aeruginosa, cgMLST, health care-associated outbreak, typing
ABSTRACT
The environmental bacterium Pseudomonas aeruginosa, particularly multidrug-resistant clones, is often associated with nosocomial infections and outbreaks. Today, core genome multilocus sequence typing (cgMLST) is frequently applied to delineate sporadic cases from nosocomial transmissions. However, until recently, no cgMLST scheme for a standardized typing of P. aeruginosa was available. To establish a novel cgMLST scheme for P. aeruginosa, we initially determined the breadth of the P. aeruginosa population based on MLST data with a Bayesian approach (BAPS). Using genomic data of representative isolates for the whole population and all 12 serogroups, we extracted target genes and further refined them using a random data set of 1,000 P. aeruginosa genomes. Subsequently, we investigated reproducibility and discriminatory ability with repeatedly sequenced isolates and isolates from well-defined outbreak scenarios, respectively, and compared clustering applying two recently published cgMLST schemes. BAPS generated seven P. aeruginosa groups. To cover these and all serogroups, 15 reference strains were used to determine genes common in all strains. After refinement with the data set of 1,000 genomes, the cgMLST scheme consisted of 3,867 target genes, which are representative of the P. aeruginosa population and highly reproducible using biological replicates. We finally evaluated the scheme by reanalyzing two published outbreaks where the authors used single-nucleotide polymorphism (SNP) typing. In both cases, cgMLST was concordant with the previous SNP results and the results of the two other cgMLST schemes. In conclusion, the highly reproducible novel P. aeruginosa cgMLST scheme facilitates outbreak investigations due to the publicly available cgMLST nomenclature.
INTRODUCTION
The natural habitats of the Gram-negative bacteria Pseudomonas aeruginosa are water and soil, including sanitation and water installations in hospitals, and P. aeruginosa forms part of the normal flora in many healthy adults (1). P. aeruginosa is also an opportunistic human pathogen commonly associated with nosocomial infections (2). It can cause severe infections, especially in patients with underlying immunosuppressing conditions, and is well known in cystic fibrosis patients (3, 4). Besides sporadic infections, the environmental sources are frequently the source of nosocomial outbreaks (5–7).
The ubiquitous occurrence of this pathogen demands a high-resolution typing method to accurately identify the source of a possible outbreak and routes of transmission within a given setting. In the past, pulsed-field gel electrophoresis (PFGE) has been considered the standard method of bacterial typing, including P. aeruginosa. Driven by the technological advances of next-generation sequencing, however, whole-genome sequence (WGS)-based typing nowadays has become the gold standard for molecular subtyping. Besides the high interlaboratory reproducibility of WGS-based typing (8) and the higher discriminatory power (9, 10), PFGE is labor intensive and often challenging to implement (11). Whereas the technical challenges to generate WGS data were solved during recent years, data analysis is still a matter of debate, and two general principles are used to extract typing information from WGS data. Initially, extraction of single-nucleotide polymorphisms (SNPs) after mapping of read data on reference genomes was used to derive typing information. Whereas SNP typing is highly discriminatory, different sequencing platforms with different systematic sequencing biases and the use of different reference sequences for SNP detection complicate the establishment of a consistent nomenclature (12, 13). In analogy to the multilocus sequence typing (MLST) approach, which is based on the extraction of usually seven predefined housekeeping genes and subsequent gene-by-gene comparison with a central internet-based nomenclature database to determine an allelic profile (14), the core genome (cg)MLST was developed (15, 16). It relies on the comparison of hundreds to thousands of predefined target genes, the cgMLST scheme, thereby combining the ability to create a central nomenclature with the high discriminatory power of WGS-based typing.
Whereas WGS-based typing has already been successfully applied in investigating P. aeruginosa outbreaks using SNPs (17) and an ad hoc cgMLST scheme (18, 19), until recently (20, 21), there was no public cgMLST scheme for P. aeruginosa available. In this study, we therefore defined and evaluated a novel cgMLST scheme for WGS-based typing of P. aeruginosa that can serve as a basis for a central typing nomenclature.
MATERIALS AND METHODS
cgMLST target gene definition.
The first step in defining a stable cgMLST scheme for P. aeruginosa consists of defining a genome set representing the genetic diversity within the population of P. aeruginosa. We did this using the information available from the MLST database. However, choosing one representative for each MLST sequence type (ST) is not recommendable since many strains with different STs are closely linked to each other, resulting in an overrepresentation of some lineages. To overcome this issue, we applied the Bayesian analysis of population structure (BAPS) as previously described (22–24) with one minor modification, the maximum likelihood tree based on the concatenated sequences of all known STs (n = 3,309, as of 15 July 2019) that were downloaded from the MLST website (https://pubmlst.org) revealed that ST610 has a great phylogenetic distance from all other sequence types (data not shown). Since such outliers can interfere with BAPS analysis, this ST was excluded from the analysis, and partitioning was performed with data of the remaining 3,308 STs. Representative genomes of partitions far away from the center of the tree (assuming that these isolates were a different species) were checked by applying the fastANI algorithm (25) between them and the P. aeruginosa type strain DSM50071. If the identity was <95%, the partition was excluded (suggesting the ST of the respective partition represents a different species than P. aeruginosa) (26). Furthermore, to ensure that our data set represents the whole breadth of the P. aeruginosa population, we ran in silico genomic serotyping with the Pseudomonas aeruginosa serotyper (PAst) program (27) to check whether we had to add representative genomes for the total of 12 serogroups (27) not covered by the found BAPS partitions. Subsequently, we selected the representative genomes covering all BAPS partitions and serogroups by querying the NCBI database with the highest possible NCBI genome status (in the order “complete,” “chromosome,” “scaffold,” “contig”). In the case of alternative genomes within the same genome status, we chose the data set that showed the best percentage of found targets that passed the target scan and target quality control (i.e., sequence identity ≥90% and 100% overlap of the found targets to the corresponding genes of the reference genome) implemented in SeqSphere+ software (Ridom GmbH, Muenster, Germany).
During the second step of the cgMLST scheme definition, we extracted all genes that were present in all representative genomes found in the first step using the MLST+ target definer (version 1.5 [win]) function of SeqSphere+ software version 6.0.92 (Ridom GmbH) in default mode using the finished genome sequence of P. aeruginosa strain PAO1 (GenBank accession no. NC_002516.2 [24 January 2019]) as seed genome, i.e., starting point for target definition and naming. Available P. aeruginosa plasmid sequences were excluded (28 NCBI entries as of 30 July 2019). All genes of the reference genome that were common in all query genomes with a sequence identity ≥90% and 100% overlap formed the preliminary target gene set.
In the last step, we further optimized this preliminary target gene set by applying it to a randomly chosen set of P. aeruginosa genomes to determine whether these targets were actually found within most of the genomes. We therefore queried the NCBI SRA for P. aeruginosa data sets with the NCBI SRA filters “DNA,” “genome,” “paired,” and “Illumina” and removed all duplicates, which resulted in 6,124 data sets (as of 19 August 2019). Of these, 1,000 data sets were randomly chosen by generating a random number for each data set using Microsoft Excel and ordering the data sets according to the value of this random number. To ensure high-quality sequencing data, we then performed a fastANI quality control (QC) (25) (i.e., confirmation of the species P. aeruginosa) and Mash Screen (28) to detect potential contamination with other species and excluded the data sets not fulfilling the requirements. The remaining data sets (fastq file format) were subsequently de novo assembled using SKESA (29) followed by a read mapping onto the contigs using the software package BWA (30) (included in the SeqSphere+ software) with the option “mem” for mapping. Only records with an assembled coverage ≥70-fold were kept to ensure optimal assembly conditions (31). Using this final data set, we determined the presence of the preliminary cgMLST target gene set and moved all targets that were found in <95% of the SKESA-assembled data sets from the preliminary cgMLST scheme to the accessory gene set, which contains all genes from the seed genome PAO1 either not present in all scheme-defining sequences or present only in <95% of the randomly chosen set of genome sequences. A complete list of all used data sets can be found in Table S5 in the supplemental material.
Reproducibility and evaluation of the novel cgMLST scheme.
To investigate the reproducibility of the novel cgMLST scheme, we used 24 P. aeruginosa isolates from our routine surveillance efforts (19) that were detected at the University Hospital Muenster, Germany, during September and December 2019. We cultured these isolates twice and sequenced them independently. For repeated sequencing, we cultivated the 24 P. aeruginosa isolates that were frozen at −70°C and extracted the DNA for subsequent library preparation and sequencing either on an Illumina MiSeq or NextSeq platform (Illumina Inc., San Diego, CA, USA) as described previously (19). Isolates used for reproducibility testing are listed in Table S1.
To evaluate the novel scheme, we searched the PubMed database using the keywords “Pseudomonas aeruginosa,” “whole genome sequencing,” “molecular typing,” and “outbreak” with the search option “most recent” (as of 7 April 2020). We screened the results for suitable publications where an outbreak of P. aeruginosa in a hospital setting using genome data was analyzed and where the raw data and sufficient metadata were publicly available. We downloaded the fastq files, de novo assembled them using SKESA followed by using BWA for mapping as we did in the target gene definition, and finally analyzed the resulting contig sequences using the novel cgMLST scheme. The combination of the alleles of the found target genes in each strain formed an allelic profile that was used to generate minimum spanning trees (MST) by mutual comparison of each allele of the found target genes and summing up the number of different alleles between two isolates where possible (missing target genes were ignored by choosing the parameter “pairwise ignore missing values”). If possible (i.e., there are enough isolates left for a meaningful reanalysis), we aimed to only include isolates with an average sequencing coverage ≥50 to ensure sufficient sequence quality of the downloaded data sets (23).
The MST was compared to the phylogenetic tree given in the publications. To facilitate comparison with historical data, we also extracted the MLST ST from the genomic data.
Comparison of the novel cgMLST scheme with the two recently published cgMLST schemes.
Very recently, two other cgMLST schemes were published (20, 21). The schemes of Stanton et al. and de Sales et al. comprised 4,440 and 2,653 target genes, respectively. We imported the target genes of these two schemes into SeqSphere+ for comparison with our novel cgMLST scheme and determined clustering using the same methodology as for our novel scheme.
Software.
For MLST, cgMLST, and subsequent graphical representation of the results, we used SeqSphere+ software version 6.0 (Ridom GmbH).
Data availability.
All raw reads generated were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under accession no. PRJEB38241.
RESULTS
cgMLST target gene definition.
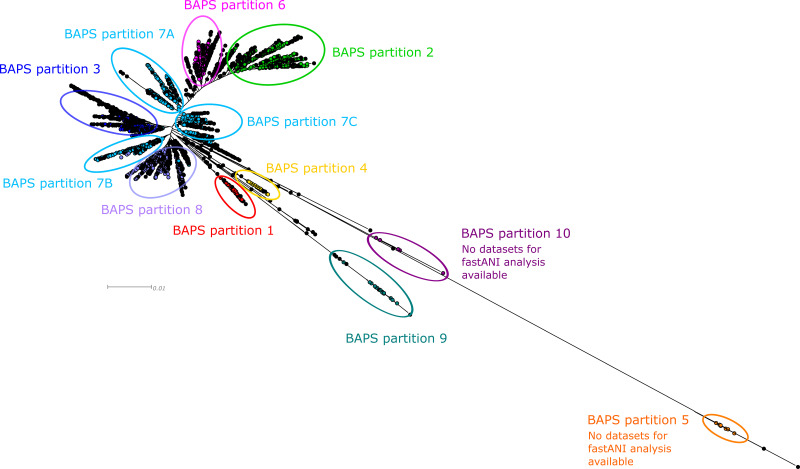
For BAPS analysis, we included, in total, 3,308 STs. Their phylogenetic relationship is shown in Fig. 1. Overall, 10 different partitions could be identified; whereas the center of the maximum likelihood tree contains the partitions 1 to 4 and 6 to 8, the partitions 5, 9, and 10 required further analysis since they were farther away from the center. To confirm or exclude their affiliation with the species P. aeruginosa, we performed a fastANI analysis of 27 available data sets of BAPS partition 9 (e.g., PA7 [GenBank accession no. NC_009656; ST1195] is assigned to BAPS partition 9). This analysis revealed <94.15% identity compared to the type strain genome DSM50071, suggesting that isolates from this partition belong to a different species than P. aeruginosa. No genome data were available for the STs of BAPS partitions 5 and 10, but since they were phylogenetically equal or even more distant from the center of the tree, fastANI values should be similar or even lower than those of the BAPS partition 9 representatives. As a consequence, we excluded representatives of the BAPS partitions 5, 9, and 10 from this study and subsequent scheme definition. The largest resulting partition 7 was further subdivided by visual inspection of the phylogram into the subpartitions 7A, 7B, and 7C, resulting in nine genomes representing the included BAPS (sub)partitions (1 to 4 and 6 to 8). Moreover, we added six representatives for the serogroups O1, O7, and O10 to O13 found by the in silico serotyping since they were neither assigned by a BAPS partition (serogroup O12) nor found within the same BAPS partition (serogroups O1, O7, O10, O11, and O13). Overall, we determined 15 genome data sets covering the fastANI-checked BAPS partitions and the 12 serogroups (Table 1), which were defined as query genomes to determine the preliminary cgMLST target genes.
FIG 1.
Partitions as determined by BAPS mapped on a maximum likelihood (ML) tree generated by FastTree2. Coloring of the tree corresponds to the partitions determined by BAPS using aligned concatenated sequences of 3,308 P. aeruginosa MLST sequence types (ST). BAPS partition 7 was further subdivided manually into three subpartitions (7A to 7C) according to the branching of the tree. STs with significant admixture are given and colored in black.
TABLE 1.
P. aeruginosa reference strains used for cgMLST scheme definition
| BAPS partition no. | Strain | Serogroupa | FastANI similarity (%)b | MLST ST | NCBI genome status (no. of contigs) | GenBank accession no. |
|---|---|---|---|---|---|---|
| 1 | PA-VAP-2 | O3 | 99.28 | 2960 | Chromosome (1) | NZ_CP028331.1 |
| 2 | PAO1 | O5 | 99.34 | 549 | Complete (1) | NC_002516.2 |
| 3 | 97 | O4 | 99.27 | 234 | Complete (1) | NZ_CP031449.2 |
| 4 | ENV-567 | O9 | 97.50 | 1763 | Contig (68) | NZ_QZXH00000000.1 |
| 6 | PA1RG | O6 | 99.29 | 782 | Complete (1) | NZ_CP012679.1 |
| 7a | W45909 | O1 | 99.28 | 27 | Complete (1) | NZ_CP008871.2 |
| 7a | AR442 | O6 | 99.30 | 395 | Complete (1) | NZ_CP029090.1 |
| 7b | AR_0360 | O6 | 99.34 | 1712 | Complete (1) | NZ_CP027165.1 |
| 7c | LESB58 | O6 | 99.30 | 146 | Complete (1) | NC_011770.1 |
| 8 | PA8281 | O2 | 99.29 | 277 | Complete (1) | NZ_CP015002.1 |
| 2 | IOMTU 133 | O7 | 99.20 | 1047 | Complete (1) | NZ_AP017302.1 |
| 2 | PA14Or | O10 | 98.68 | 253 | Complete (1) | NZ_LT608330.1 |
| 2 | Ocean-1175 | O11 | 98.70 | 316 | Complete (1) | NZ_CP022525.1 |
| n.a.c | Carb01 63 | O12 | 99.20 | 111 | Complete (1) | NZ_CP011317.1 |
| 1 | ATCC 33360 | O13 | 98.09 | 3039 | Scaffold (318) | NZ_LJZG00000000.1 |
Determined in silico using the Pseudomonas aeruginosa serotyper (PAst) program (27).
Compared with genome sequence of P. aeruginosa type strain DSM50071 (GenBank accession no. NZ_CP012001).
n.a., not assigned.
Subsequent analysis of these genomes using the cgMLST target definer resulted in 4,378 target genes present in all query genomes with a sequence identity >90% and 100% overlap. These 4,378 genes cover 68.7% of the genome of P. aeruginosa strain PAO1. To further optimize the preliminary cgMLST scheme, we used 1,000 randomly chosen genome data sets to test for representativeness of the target gene set. Of the 1,000 data sets, 84 were excluded due to a failed Mash Screen contamination check, and, after SKESA assembly, another 426 isolates were excluded due to low coverage. Applying the preliminary cgMLST scheme on the remaining 490 quality-filtered genome data sets resulted in 511 targets that were found in <95%. These target genes were moved from the preliminary cgMLST scheme into the accessory gene set, resulting in 3,867 target genes as the final cgMLST scheme (Table S2 in the supplemental material).
Reproducibility of the novel cgMLST scheme.
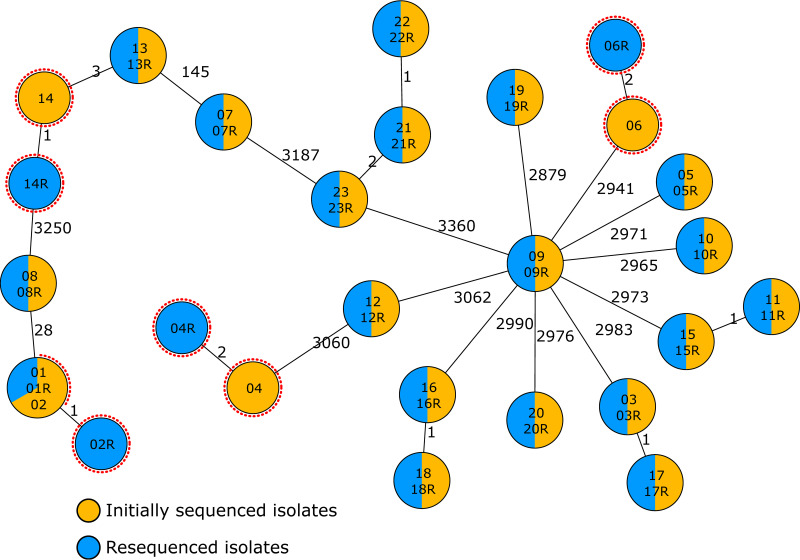
To test reproducibility of the novel cgMLST scheme, we compared typing results of independently sequenced 24 P. aeruginosa isolates using the novel cgMLST scheme. Here, the pairwise comparison resulted in 19 isolates exhibiting the identical allelic profile. In four pairs, the pairs differed in ≤2 alleles (Fig. 2). One isolate was excluded, as it exhibited contamination with another bacterial species. Overall, the analysis underlined the high reproducibility of WGS-based typing and, in particular, of the novel cgMLST scheme.
FIG 2.
Reproducibility testing of P. aeruginosa cgMLST. Minimum spanning tree of 23 isolates that were sequenced twice as biological replicates. Each circle represents the genotype based on a unique allelic profile of up to 3,867 cgMLST genes (ignoring missing values in pairwise comparisons), and the numbers on connecting lines display the number of differing alleles. The circles are named according to the isolates and colored according to the status. Red dotted circles mark pairs of isolates that did not exhibit identical genotypes.
Evaluation of different outbreak scenarios and comparison with other cgMLST schemes.
The PubMed search yielded only four publications. Of these, only two studies fulfilled our search criteria and made their genomic data available (17, 32). No genomic data were provided by the other two studies (33, 34).
The first suitable publication described a P. aeruginosa outbreak where six patients isolates and six environmental isolates positive for P. aeruginosa collected from to the intensive care unit at Ninewells Hospital, Dundee, Scotland, between 2012 and May 2013, were analyzed using variable-number tandem repeats (VNTR), PFGE, and SNP typing (17). After assembling the isolates, we observed a generally low sequencing coverage. If we had followed the rule of excluding isolates with coverage below 50, we would have had to exclude 13 isolates out of 16. We therefore decided to initially include all isolates where at least 95% of the target genes were found, independent of the coverage. With the exception of one isolate (patient D), all isolates met this modified criterion. Indeed, this isolate exhibited the lowest coverage (12-fold) among all data sets, thereby explaining the low number of target genes found (see Table S3). We therefore decided to reanalyze the whole data set using the novel cgMLST scheme except for this one isolate (patient D).
In some cases, there were two P. aeruginosa isolates collected from the same patient or the same environmental site (e.g., “patient C,” “domestic service room,” and “ice machine”) (Table S3). Since these pairs from the same site did not differ in their allelic profile, we included only the isolate with the higher average coverage in the reanalysis (apparently, in the original paper, they did the same without explicitly mentioning it). Furthermore, one isolate was labeled to be collected from the “kitchen sink” (SRA accession no. ERR2022356; see Table S3). In the original publication, however, it was stated that “no P. aeruginosa was found there” (see Table 1 in reference 17), and no isolate labeled with “kitchen sink” appeared in the phylogenetic tree. We therefore decided to exclude this isolate as well, leaving, in total, 11 isolates for reanalysis.
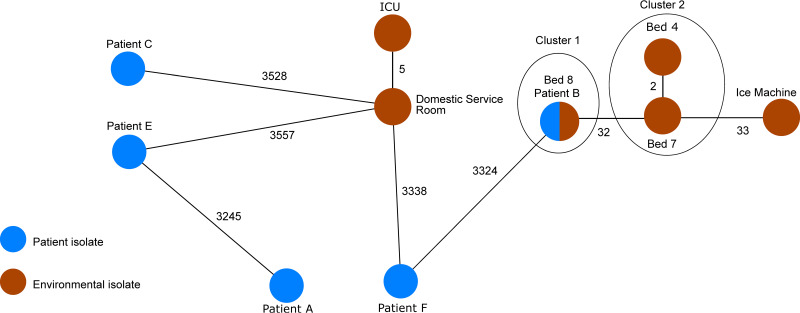
Five isolates with the same MLST ST and the same VNTR profile, according to the paper, required further analysis to determine whether they form a single outbreak cluster. The P. aeruginosa isolates from patient B and the handwash basin of bed 8 exhibited an identical allelic profile, indicating a nosocomial transmission. They were, however, only distantly related (difference of ≥32 alleles) to the isolates of the handwash basins of beds 4 and 7 and to the isolate of the ice machine (Fig. 3). Therefore, it is unlikely that these isolates belong to the same outbreak. These results are in agreement with the genomic analysis from the authors and achieved the same level of discrimination (17).
FIG 3.
Minimum spanning tree based on the allelic profiles of the novel P. aeruginosa cgMLST scheme of the genomic sequence data (n = 11 isolates) from Parcell et al. (17). Each circle represents the genotype based on a unique allelic profile of up to 3,867 cgMLST genes (ignoring missing values in pairwise comparisons), and the numbers on connecting lines display the number of differing alleles. The circles are named by the isolate labels and colored according to the status.
The second study analyzed in detail the P. aeruginosa epidemiology (environmental and patient isolates, follow-up isolates in case of long-term stay) at five different intensive care units (ICUs) of the University Hospital of Lausanne, Switzerland, between 2010 and 2014 with respect to possible outbreaks and epidemiological links between isolates using double-locus sequence typing and MLST for a broad overview and WGS for more detailed clustering, respectively (32). The whole WGS data set of this study consists of 153 different isolates in total. Almost all isolates exhibited sufficient coverage; only one isolate was excluded due to a coverage of 46. Two more isolates had to be excluded due to less than 95% of the target genes found. Another 7 isolated could not be included due to download issues, leaving 143 isolates for our reanalysis using the novel cgMLST scheme and the subsequent comparison with the previous findings (Table S3).
MLST separated the isolates into the following four STs: ST1076 (n = 67), ST17 (n = 44 isolates), ST253 (n = 31 isolate), and ST845 (n = 1 isolate), which corresponded to the DLST types used in the publication as follows: all DLST isolates 1 to 18 belonged to ST1076, DLST isolates 1 to 21 belonged to ST253, and DLST isolates 6 and 7 belonged to ST17 except for one isolate, which was found to be ST845.
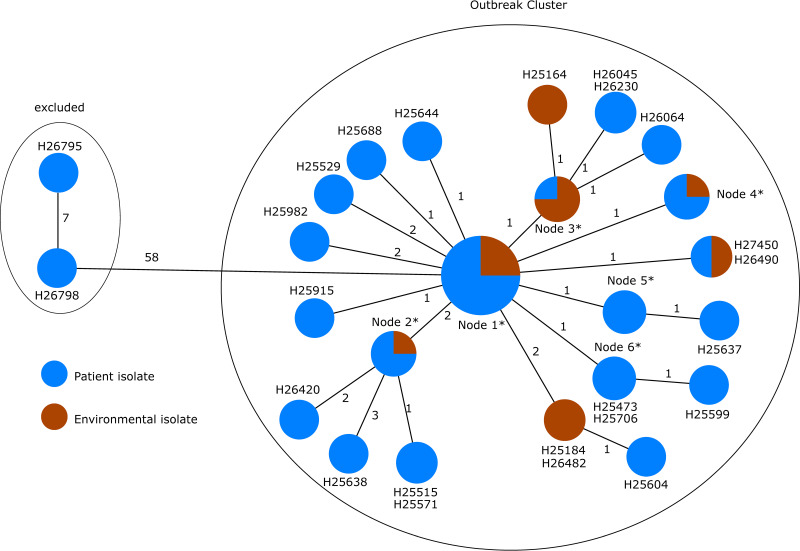
Among the 68 ST1076 isolates, cgMLST resulted in high genetic similarities (the distance matrix between all of these isolates yielded 9 alleles as maximum value [Table S4]) between almost all of these isolates (collected from patients and the environment) with the exception of isolates H26798 and H26795 (both isolates belong to patient 1; samples taken on 24 April 2010 and 31 March 2010), which clustered apart with an allelic distance of 58 alleles to the next closest isolate (Fig. 4). Based on these findings, respecting the epidemiological information, a clonal transmission of ST1076 excluding the isolates above is very likely, since the pairwise allelic differences of the other isolates are, at most, three. These results corroborate the findings of the original publication based on SNP typing (see Fig. 2 in reference 32).
FIG 4.
Minimum spanning tree based on the allelic profiles of the genome data of all isolates (n = 67) with the MLST ST1076 gathered from Magalhães et al. (32). Each circle represents the genotype based on a unique allelic profile of up to 3,867 cgMLST genes (ignoring missing values in pairwise comparisons), and the numbers on connecting lines display the number of differing alleles. The circles are named according to the isolates and colored according to the status. If more than two isolates belong to the same node, the node is marked with an asterisk, and they comprise the following isolates: H24445, H25328, H25469, H25525, H25624, H25689, H25692, H25716, H25727, H25776, H25841, H25905, H25913, H25954, H26060, H26069, H26071, H26073, H26076, H26078, H26410, H26927, H26928, H26929, H26932, H26934, and H26935 in node 1; H25179, H25509, H25524, and H25791 in node 2; H25162, H25163, H25784, and H25883 in node 3; H25305, H25471, H26166, and H26188 in node 4; and H25570, H25792, and H26413 in node 5.
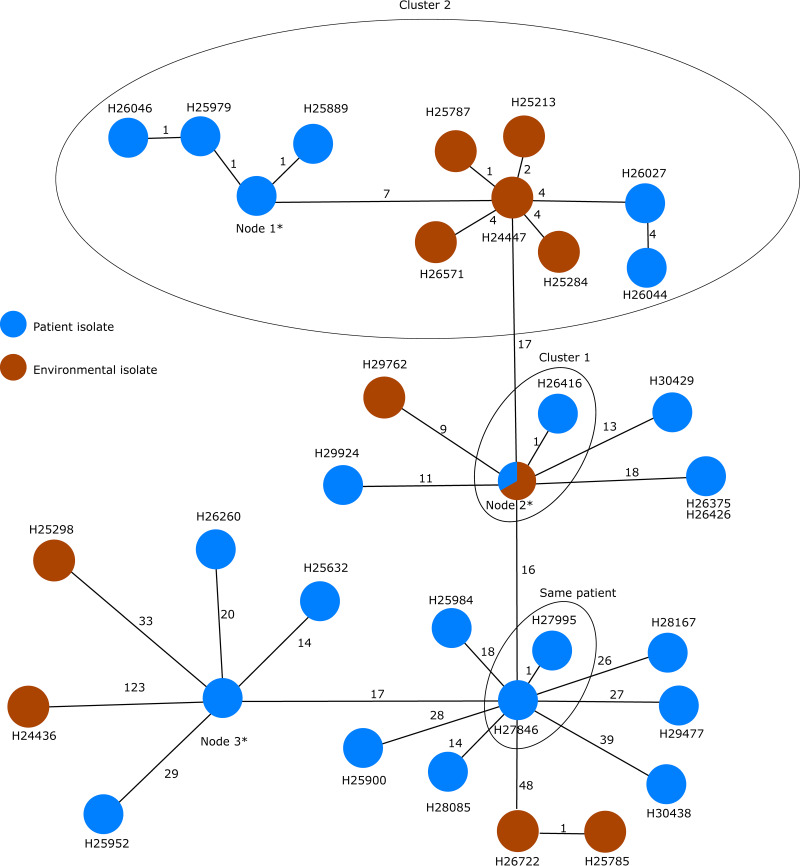
In contrast to the ST1076 isolates, cgMLST of the ST17 isolates revealed a much greater diversity between the isolates (Fig. 5). Since the allelic distance varied from 0 to 48 pairwise allelic differences, a single chain of transmission seems unlikely. Nonetheless, two clusters of similar isolates were found; cluster 1 includes isolates from a single patient (H26247, H26416, and H25508 from patient 11) and environmental isolates (H27791, H26524, H25961, and H25200) from the same ICU. It is worth noticing that the samples taken from the patient are dated November and December 2011, whereas the samples from the environment were taken in April, July, and November 2012 and March 2013. Cluster 2 was composed of 10 different isolates retrieved from 5 different patients between April 2010 and September 2010 with suspected epidemiological links and 5 different environmental isolates collected from ICU 3 and ICU 4, suggesting a local cluster. Similar to cluster 1, the environmental isolates were collected 2 years later in 2012. No further epidemiological information was given in the publication. Isolates recovered from the same patients are, as expected, highly similar (for example, H27846 and H27995 from patient 15 and H25718, H25723, and H25908 from patient 7). These results corroborate the findings of the original publication based on SNP typing (see Fig. 4 in reference 32).
FIG 5.
Minimum spanning tree based on the allelic profiles of the genome data of all isolates (n = 31) with the MLST ST253 gathered from Magalhães et al. (32). Each circle represents the genotype based on a unique allelic profile of up to 3,867 cgMLST genes (ignoring missing values in pairwise comparisons), and the numbers on connecting lines display the number of differing alleles. The circles are named with the isolates and colored according to the status. If more than two isolates belong to the same node, the node is marked with an asterisk, and they comprise the following isolates: H25209, H25532, and H25634 in node 1 and H25167, H26591, H26926, H26930, and H26933 in node 2.
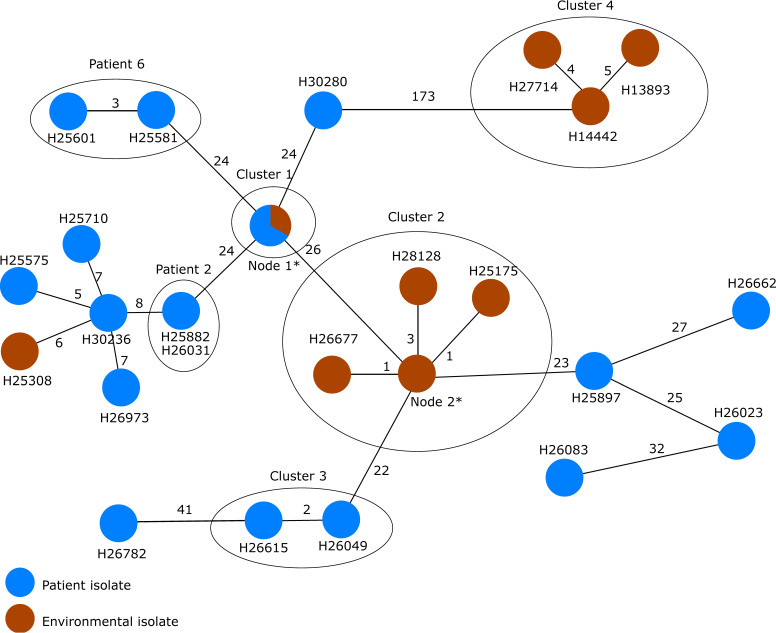
The isolates of ST253 were likewise diverse with clusters of highly similar isolates (Fig. 6). A single chain of transmission seems unlikely again since the allelic distance varied from 0 to 173 alleles (Table S4). Some isolates, however, were closely related or even identical in cgMLST, suggesting sporadic transmissions, e.g., isolates H25634 (isolated from patient 11), H25209 (isolated from patient 10), and H25532 (an environmental sample from ICU 2, where both patients were hospitalized at the same time) forming cluster 1. Moreover, as expected, isolates collected from the same patient at different days were highly similar, such as patient 2 with isolates H25882 (collection date, 29 April 2010) and H26031 (collection date, 3 May 2010) and patient 6 with isolates H25581 (collection date, 28 February 2011) and H25601 (collection date, 31 December 2011).
FIG 6.
Minimum spanning tree based on the allelic profiles of the genome data of all isolates (n = 45) with the MLST ST17 (including isolate H24436 with MLST ST845) gathered from Magalhães et al. (32). Each circle represents the genotype based on a unique allelic profile of up to 3,867 cgMLST genes (ignoring missing values in pairwise comparisons), and the numbers on connecting lines display the number of differing alleles. The circles are named according to the isolates and colored according to the status. If more than two isolates belong to the same node, the node is marked with an asterisk, and they comprise the following isolates: H26036, H26084, H26086, H26202, and H26203 in node 1; H25200, H25508, H25961, H26247, H26524, and H27791 in node 2; and H25718, H25723, and H25908 in node 3.
There were two further clusters of interest, cluster 2, consisting of eight environmental isolates that were retrieved between 23 March 2012 and 25 January 2013 in ICU 3, and cluster 3. In cluster 2, the isolates were all collected from the same environment, but during a relatively long period of time.
Interestingly, in cluster 3 (H26049 from patient 5 [27 October 2010], hospitalized in ICU 2, and H26615 from patient 12 [7 November 2012], hospitalized in ICU 4), the isolates originated from two patients who were admitted on two different ICUs 2 years apart, ruling out, as stated by the authors, an evident epidemiological link, e.g., by a common environmental source. Nevertheless, cgMLST and SNP typing grouped these two isolates together. These results corroborate the findings of the original publication based on SNP typing (see Fig. 3 in reference 32).
Using the schemes of Stanton et al. and de Sales et al., we created—similar to Fig. 3 to 6—MSTs to enable a visual comparison of the trees and the clustering of genotypes (Fig. S1 to S8). Although the numbers of different alleles and the number of genotypes varied among the different schemes, clustering of genotypes was concordant in all three cgMLST schemes.
DISCUSSION
In this study, we successfully defined a novel cgMLST scheme for P. aeruginosa comprising 3,867 targets. In a stepwise definition process, we were able to create a robust scheme that is highly representative of the P. aeruginosa population and highly reproducible using biological replicates. Moreover, we could demonstrate that cgMLST-based typing provides comparable results to SNP-based typing when applied to different outbreak scenarios and sporadic cases.
The definition and evaluation of the novel scheme warrant some additional comments. In contrast to previous studies, where we established cgMLST schemes for different pathogens, e.g., for Staphylococcus aureus (35), Listeria monocytogenes (22), or Clostridioides difficile (23), we have added an additional step during the development of the novel scheme: using 1,000 randomly chosen P. aeruginosa genome sequences, we were able to further improve the representativeness of the scheme in addition to the preceding BAPS analysis. Although this resulted in the removal of more than 500 target genes, which were only infrequently present in the query strains in comparison to the reference genomes, from the preliminary target gene set, the scheme still exhibited a discriminatory power similar to SNP-based typing approaches (Fig. 3 and 6).
In this study, we also tested the reproducibility of the novel scheme using a diverse set of P. aeruginosa isolates. This investigation was motivated by a recent study of Eyre and colleagues (36), where repeated typing of identical C. difficile DNA or isolates resulted in different typing results depending on the assembly algorithm used. We therefore decided to test our scheme with biological replicates, i.e., repeated cultivation from a frozen culture and subsequent DNA extraction, library preparation, and sequencing prior to the cgMLST analysis. Indeed, the whole process was highly reproducible with, at maximum, two alleles’ difference in a pairwise comparison.
The analysis of different outbreak scenarios and sporadic cases corroborated previous findings based on SNP typing. In-depth analysis of the outbreaks showed the need of a sophisticated and discriminatory method to accurately resolve complicated outbreak scenarios. As shown in Results, the analysis using the novel scheme delivers equal conclusions to the published literature, even for “high-risk clones” like ST253 that are frequently detected during nosocomial clusters (37).
At the first glance, some results may seem surprising. Within the isolates of ST253, cluster 3 was formed of isolates where no epidemiological link was found. However, this could be explained either by the fact that ST253 belongs to the group of high-risk clones that spread successfully under strong selection due to antibiotic resistance, thereby reducing genomic diversity, or by the fact that the epidemiological link is so hidden or complex that it cannot be discovered. Moreover, the environmental isolates of cluster 2 were very closely related, although they were collected within a period of 1 year. It is known that certain environmental conditions, such as during adaptation in cystic fibrosis patients (38) or under antibiotic pressure (39), could enhance the mutation rate in P. aeruginosa in comparison to wild-type strains under laboratory conditions (39); we therefore hypothesize that, in this case, the environmental conditions led to a reduced mutation rate resulting in very stable genotypes.
Most recently, two other cgMLST schemes were published (20, 21). Interestingly, this is the first time that, in parallel, different cgMLST schemes were available for the same pathogen. We have seen a similar situation for the classical MLST, for example, in Escherichia coli, where different schemes were used by the scientific community (40–42). We showed that all three schemes were, in principle, equally capable of resolving the analyzed outbreak scenarios. However, we believe that, in comparison to the two recently published schemes, our approach has two major advantages. First, we performed a statistical analysis, i.e., BAPS, to determine the population structure of the species to further substantiate the representativeness of our scheme and demonstrated the reproducibility of our scheme. Second, the allelic database is publicly available on the cgMLST server (https://www.cgMLST.org) (43). This enables not only users of the SeqSphere+ software but also any researcher worldwide to compare their allelic sequences to all known alleles of our scheme. This will ultimately facilitate interlaboratory comparisons of typing data. We therefore believe that there is room for more than one scheme and that the scientific community will decide which scheme is most suitable and convenient to use in the long run.
Our study is limited by the fact that isolates with highly similar cgMLST types do not necessarily mean that transmission took place. We saw examples of completely unrelated isolates with highly similar cgMLST types, e.g., isolates H26049 and H26615, as described above. Epidemiological information will always be needed to correctly interpret the detected clusters. This is, however, an intrinsic fact and valid for all typing results, irrespective of the applied typing method. Nevertheless, highly discriminatory methods like cgMLST enable a secure delineation of unrelated isolates and facilitate concentration on infection control measures (19).
In summary, we successfully established a novel cgMLST scheme for P. aeruginosa that can be used for detailed outbreak investigations, showed its reproducibility, and successfully evaluated it by reanalyzing published outbreaks.
Supplementary Material
ACKNOWLEDGMENT
We thank the team of the Institute of Hygiene for excellent laboratory work.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Walker J, Moore G. 2015. Pseudomonas aeruginosa in hospital water systems: biofilms, guidelines, and practicalities. J Hosp Infect 89:324–327. doi: 10.1016/j.jhin.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Moradali MF, Ghods S, Rehm BHA. 2017. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol 7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies JC. 2002. Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and persistence. Paediatr Respir Rev 3:128–134. doi: 10.1016/S1526-0550(02)00003-3. [DOI] [PubMed] [Google Scholar]
- 4.Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2:1051–1060. doi: 10.1016/S1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 5.Kayabas U, Bayraktar M, Otlu B, Ugras M, Ersoy Y, Bayindir Y, Durmaz R. 2008. An outbreak of Pseudomonas aeruginosa because of inadequate disinfection procedures in a urology unit: a pulsed-field gel electrophoresis-based epidemiologic study. Am J Infect Control 36:33–38. doi: 10.1016/j.ajic.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Jefferies JMC, Cooper T, Yam T, Clarke SC. 2012. Pseudomonas aeruginosa outbreaks in the neonatal intensive care unit–a systematic review of risk factors and environmental sources. J Med Microbiol 61:1052–1061. doi: 10.1099/jmm.0.044818-0. [DOI] [PubMed] [Google Scholar]
- 7.Costa D, Bousseau A, Thevenot S, Dufour X, Laland C, Burucoa C, Castel O. 2015. Nosocomial outbreak of Pseudomonas aeruginosa associated with a drinking water fountain. J Hosp Infect 91:271–274. doi: 10.1016/j.jhin.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Mellmann A, Andersen PS, Bletz S, Friedrich AW, Kohl TA, Lilje B, Niemann S, Prior K, Rossen JW, Harmsen D. 2017. High interlaboratory reproducibility and accuracy of next-generation-sequencing-based bacterial genotyping in a ring trial. J Clin Microbiol 55:908–913. doi: 10.1128/JCM.02242-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whaley MJ, Joseph SJ, Retchless AC, Kretz CB, Blain A, Hu F, Chang H-Y, Mbaeyi SA, MacNeil JR, Read TD, Wang X. 2018. Whole genome sequencing for investigations of meningococcal outbreaks in the United States: a retrospective analysis. Sci Rep 8:15803. doi: 10.1038/s41598-018-33622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. 2014. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One 9:e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salipante SJ, SenGupta DJ, Cummings LA, Land TA, Hoogestraat DR, Cookson BT. 2015. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J Clin Microbiol 53:1072–1079. doi: 10.1128/JCM.03385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. 2014. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearce ME, Alikhan N-F, Dallman TJ, Zhou Z, Grant K, Maiden MCJ. 2018. Comparative analysis of core genome MLST and SNP typing within a European Salmonella serovar Enteritidis outbreak. Int J Food Microbiol 274:1–11. doi: 10.1016/j.ijfoodmicro.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiden MCJ, van Jansen Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parcell BJ, Oravcova K, Pinheiro M, Holden MTG, Phillips G, Turton JF, Gillespie SH. 2018. Pseudomonas aeruginosa intensive care unit outbreak: winnowing of transmissions with molecular and genomic typing. J Hosp Infect 98:282–288. doi: 10.1016/j.jhin.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kossow A, Kampmeier S, Willems S, Berdel WE, Groll AH, Burckhardt B, Rossig C, Groth C, Idelevich EA, Kipp F, Mellmann A, Stelljes M. 2017. Control of multidrug-resistant Pseudomonas aeruginosa in allogeneic hematopoietic stem cell transplant recipients by a novel bundle including remodeling of sanitary and water supply systems. Clin Infect Dis 65:935–942. doi: 10.1093/cid/cix465. [DOI] [PubMed] [Google Scholar]
- 19.Mellmann A, Bletz S, Böking T, Kipp F, Becker K, Schultes A, Prior K, Harmsen D. 2016. Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol 54:2874–2881. doi: 10.1128/JCM.00790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sales R.d, Migliorini LB, Puga R, Kocsis B, Severino P. 2020. A core genome multilocus sequence typing scheme for Pseudomonas aeruginosa. Front Microbiol 11:49. doi: 10.3389/fmicb.2020.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanton RA, McAllister G, Daniels JB, Breaker E, Vlachos N, Gable P, Moulton-Meissner H, Halpin AL. 2020. Development and application of a core genome multilocus sequence typing scheme for the healthcare-associated pathogen Pseudomonas aeruginosa. J Clin Microbiol 58:e00214-20. doi: 10.1128/JCM.00214-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, Harmsen D, Mellmann A. 2015. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J Clin Microbiol 53:2869–2876. doi: 10.1128/JCM.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bletz S, Janezic S, Harmsen D, Rupnik M, Mellmann A. 2018. Defining and evaluating a core genome multilocus sequence typing scheme for genome-wide typing of Clostridium difficile. J Clin Microbiol 56:e01987-17. doi: 10.1128/JCM.01987-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Been M.d, Pinholt M, Top J, Bletz S, Mellmann A, van Schaik W, Brouwer E, Rogers M, Kraat Y, Bonten M, Corander J, Westh H, Harmsen D, Willems RJL. 2015. Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol 53:3788–3797. doi: 10.1128/JCM.01946-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wingett SW, Andrews S. 2018. FastQ screen: a tool for multi-genome mapping and quality control. F1000Res 7:1338. doi: 10.12688/f1000research.15931.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee I, Ouk Kim Y, Park S-C, Chun J. 2016. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 27.Thrane SW, Taylor VL, Lund O, Lam JS, Jelsbak L. 2016. Application of whole-genome sequencing data for O-specific antigen analysis and in silico serotyping of Pseudomonas aeruginosa isolates. J Clin Microbiol 54:1782–1788. doi: 10.1128/JCM.00349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ondov BD, Starrett GJ, Sappington A, Kostic A, Koren S, Buck CB, Phillippy AM. 2019. Mash Screen: high-throughput sequence containment estimation for genome discovery. Genome Biol 20:232. doi: 10.1186/s13059-019-1841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Souvorov A, Agarwala R, Lipman DJ. 2018. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol 19:153. doi: 10.1186/s13059-018-1540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv 1303.3997v2. http://arxiv.org/pdf/1303.3997v2.
- 31.Jünemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, Mellmann A, Goesmann A, Haeseler A.v, Stoye J, Harmsen D. 2013. Updating benchtop sequencing performance comparison. Nat Biotechnol 31:294–296. doi: 10.1038/nbt.2522. [DOI] [PubMed] [Google Scholar]
- 32.Magalhães B, Valot B, Abdelbary MMH, Prod'hom G, Greub G, Senn L, Blanc DS. 2020. Combining standard molecular typing and whole genome sequencing to investigate Pseudomonas aeruginosa epidemiology in intensive care units. Front Public Health 8:3. doi: 10.3389/fpubh.2020.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galdys AL, Marsh JW, Delgado E, Pasculle AW, Pacey M, Ayres AM, Metzger A, Harrison LH, Muto CA. 2019. Bronchoscope-associated clusters of multidrug-resistant Pseudomonas aeruginosa and carbapenem-resistant Klebsiella pneumoniae. Infect Control Hosp Epidemiol 40:40–46. doi: 10.1017/ice.2018.263. [DOI] [PubMed] [Google Scholar]
- 34.Willmann M, Bezdan D, Zapata L, Susak H, Vogel W, Schröppel K, Liese J, Weidenmaier C, Autenrieth IB, Ossowski S, Peter S. 2015. Analysis of a long-term outbreak of XDR Pseudomonas aeruginosa: a molecular epidemiological study. J Antimicrob Chemother 70:1322–1330. doi: 10.1093/jac/dku546. [DOI] [PubMed] [Google Scholar]
- 35.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eyre DW, Peto TEA, Crook DW, Walker AS, Wilcox MH. 2019. Hash-based core genome multilocus sequence typing for Clostridium difficile. J Clin Microbiol 58:e01037-19. doi: 10.1128/JCM.01037-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cabrolier N, Sauget M, Bertrand X, Hocquet D. 2015. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identifies Pseudomonas aeruginosa high-risk clones. J Clin Microbiol 53:1395–1398. doi: 10.1128/JCM.00210-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, Perez JL, Oliver A. 2008. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol 190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabot G, Zamorano L, Moyà B, Juan C, Navas A, Blázquez J, Oliver A. 2016. Evolution of Pseudomonas aeruginosa antimicrobial resistance and fitness under low and high mutation rates. Antimicrob Agents Chemother 60:1767–1778. doi: 10.1128/AAC.02676-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaureguy F, Landraud L, Passet V, Diancourt L, Frapy E, Guigon G, Carbonnelle E, Lortholary O, Clermont O, Denamur E, Picard B, Nassif X, Brisse S. 2008. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics 9:560. doi: 10.1186/1471-2164-9-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lacher DW, Steinsland H, Blank TE, Donnenberg MS, Whittam TS. 2007. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J Bacteriol 189:342–350. doi: 10.1128/JB.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohl TA, Harmsen D, Rothgänger J, Walker T, Diel R, Niemann S. 2018. Harmonized genome wide typing of tubercle bacilli using a web-based gene-by-gene nomenclature system. EBioMedicine 34:131–138. doi: 10.1016/j.ebiom.2018.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw reads generated were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under accession no. PRJEB38241.