This study examines the microbiological and epidemiological characteristics of toxigenic and nontoxigenic Corynebacterium isolates submitted to the national reference laboratory in Spain, between 2014 and 2019, in order to describe the current situation and improve our knowledge regarding these emerging pathogens. Epidemiological information was extracted from the Spanish Surveillance System.
KEYWORDS: Corynebacterium, Corynebacterium infections, diphtheria toxin, microbiology, diphtheria, epidemiology
ABSTRACT
This study examines the microbiological and epidemiological characteristics of toxigenic and nontoxigenic Corynebacterium isolates submitted to the national reference laboratory in Spain, between 2014 and 2019, in order to describe the current situation and improve our knowledge regarding these emerging pathogens. Epidemiological information was extracted from the Spanish Surveillance System. Microbiological and molecular characterization was carried out using phenotypic methods, multilocus sequence typing (MLST), whole-genome sequencing (WGS), and core genome MLST (cgMLST). Thirty-nine isolates were analyzed. Twenty-one isolates were identified as Corynebacterium diphtheriae (6 toxigenic), 14 as C. belfantii, 4 as C. ulcerans (3 toxigenic), and 1 as C. rouxii. One C. diphtheriae isolate was identified as nontoxigenic tox gene bearing (NTTB). Ages of patients ranged from 1 to 89 years, with 10% (3/30) of nontoxigenic and 22% (2/9) of toxigenic isolates collected from children less than 15 years. Twenty-five of the patients were males (17/30 in nontoxigenic; 8/9 in toxigenic). MLST identified 28 sequence types (STs), of which 7 were described for the first time in Spain. WGS analysis showed that 10 isolates, including 3 toxigenic isolates, harbored a variety of antibiotic resistance genes in addition to the high prevalence of penicillin resistance phenotypically demonstrated. Phylogenetic analysis revealed one cluster of isolates from family members. Risk information was available for toxigenic isolates (9/39); 3 patients reported recent travels to countries of endemicity and 3 had contact with cats/dogs. One unvaccinated child with respiratory diphtheria had a fatal outcome. Including nontoxigenic Corynebacterium infections in disease surveillance and using WGS could further improve current surveillance.
INTRODUCTION
Diphtheria is an acute infectious disease that affects the upper respiratory tract and occasionally the skin. Classical diphtheria is due to the production of a toxin during the infection by strains lysogenized by a bacteriophage (corynephage) harboring the toxin gene. Diphtheria toxin (DT) is produced by toxigenic strains of the human pathogen Corynebacterium diphtheriae as well as zoonotic C. ulcerans and C. pseudotuberculosis. C. diphtheriae isolates have traditionally been categorized into the four biovars Gravis, Mitis, Intermedius, and Belfanti (1). Recently, on the basis of genomic sequencing and biochemical and chemotaxonomic analyses, Dazas et al. proposed the name of C. belfantii for the strains previously considered C. diphtheriae bv. Belfanti (2) Additionally, a novel member of the diphtheriae species complex named C. rouxii has recently been described (3).
The incidence of diphtheria in Europe is very low, with a notification rate of less than 0.01 cases per 100,000 habitants (4, 5). The reported cases are mostly diagnosed among travelers, refugees, asylum seekers, or immigrants from countries of diphtheria endemicity (4–9). The incidence of diphtheria in Spain is similar to that in the rest of Europe (4). A highly effective toxoid-based vaccine for diphtheria exists that is considered to have saved millions of lives. Vaccination of diphtheria was introduced in Spain in the 1960s as a primary course of three doses in the first year of life. Boosters in the second year of life, childhood, adolescence, and adulthood (≥65 years) were gradually introduced in the national vaccination schedule (10). In 2015, there was a fatal case of respiratory diphtheria in an unvaccinated 6-year-old boy in Catalonia (11).
Spain had surveillance in place only for cases involving respiratory toxigenic C. diphtheriae prior to 2014, but since then, all toxigenic diphtheria cases must be reported following the European Union case definition. A confirmed case of diphtheria is defined as any person meeting the laboratory criteria (isolation of toxin-producing C. diphtheriae, C. ulcerans, or C. pseudotuberculosis from a clinical specimen) and at least one of the clinical forms (classic respiratory, mild respiratory, cutaneous, or diphtheria of other sites). As a result of the improving diagnostic capacity in hospitals (e.g., matrix-assisted laser desorption ionization–time of flight mass spectrometry [MALDI-TOF MS]), the reports of suspected cases of C. diphtheriae and C. ulcerans have increased over time. It has, however, highlighted the lack of knowledge regarding the prevalence and origins of both toxigenic and nontoxigenic corynebacteria circulating in the population. Nontoxigenic corynebacteria have been associated with severe diseases (12, 13). The aim of this study was to review the molecular and epidemiological characteristics of toxigenic and nontoxigenic isolates of corynebacteria, collected from 2014 to 2019 and to improve our knowledge regarding Corynebacterium sp. infections in Spain.
MATERIALS AND METHODS
Diphtheria surveillance.
The Epidemiology Surveillance Service of each autonomous region in Spain urgently notifies the Health Alert and Emergency Coordination Centre (CCAES) and National Centre for Epidemiology (CNE) for any suspected, probable, or confirmed case of respiratory diphtheria (14). The guidelines of the National Surveillance Network (RENAVE) are shown in Table 1.
TABLE 1.
Diphtheria surveillance as described in the guidelines of the National Surveillance Network
| Site of infection | Definition | Action requireda |
|---|---|---|
| Respiratory diphtheria | ||
| Suspected case | A person who meets the clinical criteria, i.e., upper respiratory tract disease with laryngitis, nasopharyngitis, or tonsillitis and a membrane or pseudomembrane. | Notify the CCAES and the CNE |
| Probable case | A person who meets the clinical criteria and has an epidemiologic link with a confirmed human or animal case. | |
| Confirmed case | A person who meets the clinical and laboratory criteria, i.e., isolation of toxigenic C. diphtheriae, C. ulcerans, or C. pseudotuberculosis, in a clinical specimen (Elek test positive). The test must be confirmed at the CNMb. | |
| Cutaneous diphtheria | ||
| Confirmed case | A person who meets the clinical criteria, i.e., chronic nonprogressive ulcerative lesion that may appear with a dirty gray membrane and the laboratory criteria, i.e., isolation of toxigenic C. diphtheriae, C. ulcerans, or C. pseudotuberculosis, in a clinical specimen (Elek test positive). The test must be confirmed at the CNM. | Notify CNE |
| Diphtheria in other localization | ||
| Confirmed case | A person who meets the clinical criteria, i.e., a conjunctiva or mucosal lesion and laboratory criteria, i.e., isolation of toxigenic C. diphtheriae, C. ulcerans, or C. pseudotuberculosis, in a clinical specimen (Elek test positive). The test must be confirmed at the CNM. | Notify CNE |
CCAES, Health Alert and Emergency Coordination Centre; CNE, National Centre for Epidemiology.
bCNM, National Centre for Microbiology.
Microbiological characterization.
Microbiological procedures were carried out in accordance with the WHO manual for the laboratory diagnosis of diphtheria (15). Initially, the clinical sample is plated onto an enriched medium, Mueller-Hinton agar supplemented with 5% sheep blood and potassium tellurite, such as Hoyle’s tellurite medium. Potential corynebacterial colonies are small greyish colonies with a granular appearance on blood agar and have a gray to black pinpoint appearance on Hoyle’s medium. These colonies are then subcultured to Hoyle’s, Mueller-Hinton with 5% sheep blood and Tinsdale, where the colonies appear black with a brown halo (16). Subsequently, the Corynebacterium species and subspecies were identified using the API Coryne System adhering to the manufacturer’s instructions (bioMérieux, Durham, NC). PCR was used to distinguish between C. ulcerans, C. diphtheriae, and C. pseudotuberculosis as well as to detect the tox gene (17, 18). If the tox gene was detected, the phenotypic expression of the toxin was confirmed via the modified Elek test performed at the WHO Collaborating Centre for Diphtheria and Streptococcal Infections at Public Health England (15, 19).
Antimicrobial susceptibility.
Phenotypic antimicrobial susceptibility testing was performed using an Etest diffusion assay in accordance with EUCAST guidelines (20). To classify the sensitivity of the isolates to penicillin G, we used the breakpoints set by EUCAST. For erythromycin, the breakpoints are currently in preparation, so the corresponding CLSI breakpoints were used instead (21). Resistance genes in the WGS data were identified using ResFinder-3.2 (22).
Multilocus sequence typing.
The sequence type (ST) of the isolates received was assessed using the MLST scheme described by Bolt et al. (23). We included modified primers described by Both et al. to be able to amplify dnaE and dnaK of C. ulcerans (24). The assigned STs were obtained by uploading the allelic profiles to the PubMLST database (25).
Whole-genome sequencing.
For DNA extraction, we used isolates from blood agar plates. We extracted genomic DNA by using a modified protocol of the Qiamp DNA minikit (Qiagen, Germany).
Details of the WGS and its analysis can be found in the supplemental material.
The resulting core genome MLST (cgMLST) scheme consisted of 1,441 target loci for C. diphtheriae/C. belfantii and 1,209 for C. ulcerans. An accessory target scheme with 735 and 961 more loci was defined during the same process (for C. diphtheriae/C. belfantii and C. ulcerans, respectively).
We performed next-generation-based cgMLST with reference-based alignments after read trimming and assembling by using FastQC, Unicycler v.0.4.6, QUAST v.4.1, and Kmerfinder v.3.1, by the Bioinformatics Unit at the Instituto de Salud Carlos III. We performed in silico cgMLST by using the generated cgMLST or extended cgMLST scheme. After typing and assigning allele numbers, we calculated distances for tree building. Alleles with missing values in at least one sample and samples with missing values in more than 10% of distance columns were excluded from the comparison table. Subsequently, minimum spanning trees were generated. A cluster was defined as a group of closely related cgMLST-analyzed isolates differing by ≤5 alleles and subclusters with the same similarity threshold but after extended cgMLST.
We performed a comprehensive analysis of the prophages in all isolates using the PHASTER software (https://phaster.ca/). We also assessed the C. ulcerans isolates for the presence of a novel pathogenicity island harboring the DT as described by Dangel et al. (26). All isolates were assessed for the presence of the diphtheria toxin regulator gene (dtxR) via alignment using a dtxR reference sequence (GenBank accession number KU869770).
Data analysis.
The epidemiological data were described and presented based on their toxigenicity. Contact tracing was undertaken with every confirmed case. Secondary cases were identified only from the 2015 fatal case of respiratory toxigenic diphtheria. Ten asymptomatic cases were identified in 2 stages, as follows: 9 cases were identified from contacts of the confirmed case and the 10th case was identified from contacts of cases identified at the 1st stage (11). Contact tracing of a case of cutaneous diphtheria in 2019 did not identify any secondary cases, but one dog and two cats did test positive for Corynebacterium ulcerans. The isolates from contacts were excluded from the main analysis. Characterization of risk factors was possible only for patients with toxigenic diphtheria. The analysis was conducted using Stata 16 (StataCorp. 2019). The algorithm eBURST was used to conduct clonal analysis and visualize potential phylogenetic relationships between STs using Bionumerics (AppliedMaths) software (27).
Consent and ethical approval.
Samples and data used in this study were collected without patient identifiable data for diagnostic and surveillance purposes; therefore, a consent form was not required.
Data availability.
All sequence data have been deposited in the ENA database under accession numbers ERS5375606 to ERS5375642 (BioProject accession number PRJEB41500).
RESULTS
Epidemiological and microbiological characteristics.
From 2014 to 2019, C. diphtheriae, C. belfantii, and C. ulcerans were isolated from 39 specimens, and they were also isolated from 10 specimens obtained during contact tracing of the 2015 case. Specimens included throat swabs, aspirates, skin swabs, biopsy, sputum and blood.
The microbiological characteristics of the 39 isolates are shown in Table 2. Thirty isolates were nontoxigenic and nine were toxigenic, i.e., from confirmed cases. One patient was administered extensive antibiotic treatment before the sample was collected and the causative agent could therefore not be cultured for further analyses. However, the PCR analysis performed directly from the clinical sample demonstrated the presence of toxigenic C. diphtheriae. This finding, in combination with the clinical presentation, indicative of toxigenic respiratory diphtheria, lead to this patient being classified as a confirmed case. One nontoxigenic tox gene-bearing (NTTB) isolate was identified.
TABLE 2.
Microbiological and clinical profiles of nontoxigenic and toxigenic C. diphtheriae, C. belfantii, and C. ulcerans isolates in Spain, 2014 to 2019
| Characteristic | Total no. (%) of isolates (n = 39) | No. (%) of nontoxigenic isolates (n = 30) | No. (%) of toxigenic isolates (n = 9) |
|---|---|---|---|
| Type and subtype | |||
| C. diphtheriae | 21 (54) | 15 (50) | 6 (67) |
| Gravis | 7 (18) | 7 (23) | 0 (0) |
| Mitis | 13 (33) | 8 (27) | 5 (56) |
| Undetermineda | 1 (3) | 0 (0) | 1 (11) |
| C. belfantii | 14 (36) | 14 (47) | 0 (0) |
| C. ulcerans | 4 (10) | 1 (3) | 3 (33) |
| Antimicrobial resistancea | |||
| Penicillin G | |||
| Resistant (>0.125 mg/liter) | 29 (76) | 23 (77) | 6 (75) |
| Susceptible (≤0.125 mg/liter) | 9 (24) | 7 (23) | 2 (25) |
| Erythromycin | |||
| Resistant (>2 mg/liter) | 0 (0) | 0 (0) | 0 (0) |
| Intermediate (1 mg/liter) | 0 (0) | 0 (0) | 0 (0) |
| Susceptible (≤0.5 mg/liter) | 38 (100) | 30 (100) | 8 (100) |
| Sample type | |||
| Respiratory | 21 (54) | 18 (60) | 3 (33) |
| Cutaneous | 16 (41) | 10 (33) | 6 (67) |
| Other | 2 (5) | 2 (7) | 0 (0) |
One patient had undetermined subtype and antimicrobial resistance because of starting the antibiotic treatment prior to the specimen collection, therefore n = 38 in antimicrobial resistance.
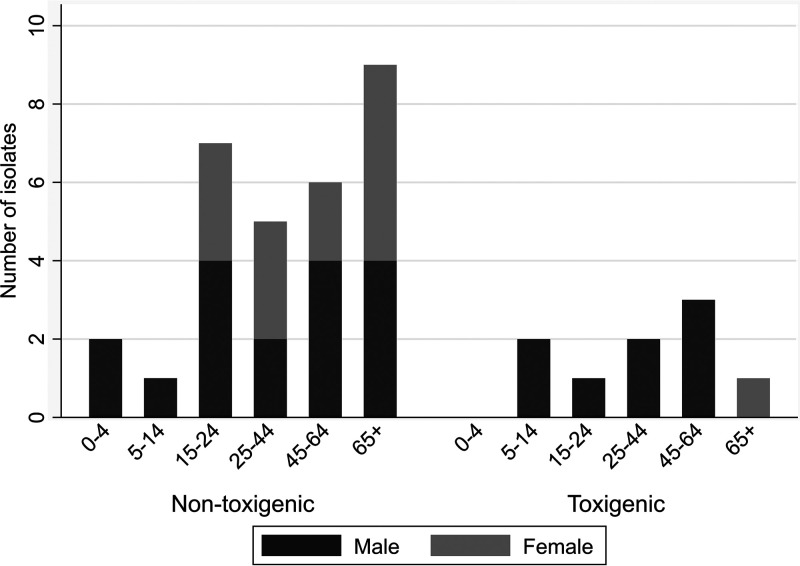
Overall, ages ranged from 1 to 89 years, and 64% (25/39) of the patients were males. Figure 1 shows the distribution of sex and age group per toxigenicity group. The ratio of male to female was 1.3:1 among the nontoxigenic isolates while in toxigenic isolates it was 8:1.
FIG 1.
Nontoxigenic and toxigenic C. diphtheriae, C. belfantii, and C. ulcerans isolates in Spain by age and sex groups, 2014 to 2019.
There were three cases with a respiratory presentation; one C. diphtheriae in 2015 and another in 2018, as well as one C. ulcerans in 2019. The 2015 case had a fatal outcome. The other four C. diphtheriae cases and the two C. ulcerans cases presented cutaneous manifestations. No fatalities were observed among C. ulcerans cases. Approximately half of the nontoxigenic patients (16/30) had available clinical information. Among them, five patients had respiratory symptoms, including rhinorrhoea, sinusitis, and pharyngitis. In eight patients, cutaneous symptoms, such as wounds and impetigo, were reported. Other recorded presentations included osteomyelitis and endocarditis (Table 3).
TABLE 3.
Corynebacterium isolates included in the study
| Species | Isolate | Isolation yr | Source | Disease | Gender | Age (yrs) | Risk factors | Biovara | Toxin PCR resulta,b | Elek test resulta,b | ST | Penicillin G MIC (mg/liter) | Erythromycin MIC (mg/liter) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Corynebacterium diphtheriae | 2014018 | 2014 | Cutaneous | Foot ulceration | M | 12 | Travel (Afghanistan), mixed infection S. pyogenes | Mitis | P | P | 389 | 0.38 | <0.016 |
| 2015002 | 2015 | Blood | Endocarditis | M | 62 | Cirrhosis, hepatitis C virus | Gravis | N | NA | 32 | 0.5 | <0.016 | |
| 2015004 | 2015 | Pharyngeal membrane | Diphtheria | M | 7 | Unvaccinated | Mitis | P | P | 377 | 0.38 | <0.016 | |
| 2015005 | 2015 | Pharyngeal swab | Asymptomatic | M | 8 | Contact | Mitis | P | P | 377 | 0.38 | <0.016 | |
| 2015006 | 2015 | Pharyngeal swab | Asymptomatic | M | 8 | Contact | Mitis | P | P | 377 | 0.38 | <0.016 | |
| 2015007 | 2015 | Pharyngeal swab | Asymptomatic | M | Contact | Mitis | P | P | 377 | 0.38 | <0.016 | ||
| 2015008 | 2015 | Pharyngeal swab | Asymptomatic | F | 0 | Contact | Mitis | P | P | 377 | 0.38 | <0.016 | |
| 2015009 | 2015 | Pharyngeal swab | Asymptomatic | M | 7 | Contact | Mitis | P | P | 377 | 0.38 | <0.016 | |
| 2015010 | 2015 | Pharyngeal swab | Asymptomatic | M | 35 | Contact | Mitis | P | P | 377 | 0.38 | <0.016 | |
| 2015011 | 2015 | Pharyngeal swab | Asymptomatic | M | 7 | Contact | Mitis | P | P | 377 | 0.38 | <0.016 | |
| 2015012 | 2015 | Pharyngeal swab | Asymptomatic | F | 8 | Contact | Mitis | P | P | 377 | 0.38 | <0.016 | |
| 2015013 | 2015 | Pharyngeal swab | Asymptomatic | F | 40 | Contact | Mitis | P | P | 377 | 0.38 | <0.016 | |
| 2015087 | 2015 | Pharyngeal swab | Asymptomatic | M | 8 | Contact | Mitis | P | P | 377 | 0.38 | <0.016 | |
| 2016005 | 2016 | Oropharyngeal swab | F | 19 | Unknown | Gravis | N | NA | 32 | 0.38 | <0.016 | ||
| 2016006 | 2016 | Cutaneous | M | 20 | Travel (Senegal) | Mitis | P | P | 484 | 0.25 | <0.016 | ||
| 2017001 | 2017 | Oropharyngeal swab | F | 22 | Unknown | Mitis | N | NA | 5 | 0.5 | <0.016 | ||
| 2017013 | 2017 | Cutaneous | Cutanea | F | 25 | Unknown | Gravis | N | NA | 96 | 0.38 | <0.016 | |
| 2017014 | 2017 | Pharyngeal swab | Pharyngitis | M | 26 | Unknown | Mitis | P | N | 212 | 0.75 | <0.016 | |
| 2017015 | 2017 | Cutaneous | Infected ulcer | M | 41 | Travel (Sri Lanka), mixed infection S. pyogenes | Mitis | N | NA | 422 | 0.38 | <0.016 | |
| 2017016 | 2017 | Cutaneous | Impetigo | M | 10 | Travel (Guinea), mixed infection S. pyogenes | Mitis | N | NA | 511 | 0.38 | <0.016 | |
| 2017017 | 2017 | Cutaneous | Impetigo | M | 4 | Travel (Guinea), mixed infection S. pyogenes | Mitis | N | NA | 297 | 0.75 | <0.016 | |
| 2017018 | 2017 | Cutaneous | Impetigo | M | 1 | Travel (Guinea) | Mitis | N | NA | 297 | 0.50 | <0.016 | |
| 2018042 | 2018 | Cutaneous | Wound infection | M | 18 | Immigrant, boat trip | Mitis | N | NA | 704 | 0.19 | <0.016 | |
| 2018043 | 2018 | Cutaneous | Wound infection | M | 16 | Immigrant, boat trip | Gravis | N | NA | 542 | 0.38 | <0.016 | |
| 2018046 | 2018 | Blood | Endocarditis | M | 21 | Unknown | Gravis | N | NA | 319 | 0.39 | <0.016 | |
| 2018047 | 2018 | Cutaneous | Ankle laceration | F | 31 | Unknown | Mitis | N | NA | 134 | 0.094 | 0.032 | |
| 2018051 | 2018 | Cutaneous | Pharyngeal membrane, myocarditis | M | 53 | Unknown | NA | P | NA | 297 | NA | NA | |
| 2019026 | 2019 | Pharyngeal swab | M | 24 | Unknown | Gravis | N | NA | 32 | 0.38 | <0.016 | ||
| 2019027 | 2019 | Cutaneous | Leg necrotizing wound | M | 40 | Travel (Philippines) | Mitis | P | P | 458 | 0.5 | 0.016 | |
| 2019028 | 2019 | Oropharyngeal swab | F | 15 | Unknown | Gravis | N | NA | 32 | 0.19 | <0.016 | ||
| 2019030 | 2019 | Cutaneous | M | 34 | Travel (Pakistan) | Mitis | P | P | 377 | 0.25 | <0.016 | ||
| Corynebacterium belfantii | 2014013 | 2014 | Expectoration | M | 73 | Unknown | NA | N | NA | 81 | 0.5 | < 0.016 | |
| 2014017 | 2014 | Expectoration | F | 76 | Unknown | NA | N | NA | 23 | 0.5 | <0.016 | ||
| 2015001 | 2015 | Nasopharyngeal swab | Rhinorrhea, epistasis | F | 84 | Unknown | NA | N | NA | 482 | 0.25 | <0.016 | |
| 2015167 | 2015 | Cutaneous | Carcinoma in the atrial pavilion, costrosus area in scalp graft | M | 79 | Mixed infection S. aureus | NA | N | NA | 106 | 0.125 | <0.016 | |
| 2015202 | 2015 | Expectoration | Sinusitis | F | 79 | Unknown | NA | N | NA | 106 | 0.25 | <0.016 | |
| 2016001 | 2016 | Expectoration | Asymptomatic | F | 54 | Cystic fibrosis | NA | N | NA | 483 | 0.125 | <0.016 | |
| 2016007 | 2016 | Nasal biopsy specimen | F | 37 | Unknown | NA | N | NA | 42 | 0.25 | <0.016 | ||
| 2017002 | 2017 | Nasopharyngeal swab | Respiratory | M | 49 | Unknown | NA | N | NA | 92 | 0.19 | <0.016 | |
| 2017004 | 2017 | Bone biopsy specimen | Osteomyelitis | M | 79 | Unknown | NA | N | NA | 537 | 0.38 | <0.016 | |
| 2017008 | 2016 | Expectoration | Respiratory | M | 62 | Unknown | NA | N | NA | 163 | 0.047 | <0.016 | |
| 2018032 | 2018 | Expectoration | M | 85 | Unknown | NA | N | NA | 42 | 0.125 | <0.016 | ||
| 2018045 | 2018 | Expectoration | Respiratory | M | 89 | Unknown | NA | N | NA | 106 | 0.19 | <0.016 | |
| 2018048 | 2018 | Expectoration | F | 61 | Unknown | NA | N | NA | 163 | 0.094 | <0.016 | ||
| 2018076 | 2018 | Nasal swab | Sinusitis | F | 53 | Unknown | NA | N | NA | 670 | 0.38 | <0.016 | |
| Corynebacterium ulcerans | 2014016 | 2014 | Cutaneous | M | 62 | Unknown | NA | P | P | 325 | 0.064 | <0.016 | |
| 2016002 | 2016 | Bone biopsy specimen | F | 74 | Unknown | NA | N | NA | 325 | 0.125 | <0.016 | ||
| 2017009 | 2016 | Cutaneous | Chronic vascular ulcers | F | 85 | Animal contact (cat) | NA | P | P | 337 | 0.012 | 0.016 | |
| 2019012 | 2019 | Pharyngeal swab | Odynophagia, pharyngeal lesions | M | 60 | Animal contact (cat, dog) | P | P | 514 | 0.75 | 0.016 |
aNA, not applicable.
N, negative; P, positive.
The largest number of isolates was collected in 2017 and 2018, while the lowest number was in 2014 (Fig. 2). None of the 2017 isolates were identified as toxigenic.
FIG 2.
ST profiles of toxigenic and nontoxigenic C. diphtheriae, C. belfantii, and C. ulcerans isolates in Spain, 2014 to 2019. The asterisk (*) indicates the C. ulcerans isolates and underlined STs correspond to C. belfantii isolates. The STs that were identified once are gray, and those that have been identified in more than one specimen share a color.
A total of 28 sequence types (STs) were identified among the 39 C. diphtheriae, C. belfantii, and C. ulcerans analyzed, of which 21 (75%) appeared only once. The most common sequence type was ST-32 (10%). ST-325 was observed in a toxigenic C. ulcerans isolate in 2014 and then in 2016 in a nontoxigenic C. ulcerans. ST-297 was observed in 2017 as nontoxigenic C. diphtheriae and in 2018 appeared as toxigenic, resulting in respiratory diphtheria. Seven of them, namely, ST-377, ST-482, ST-483, ST-484, ST-511, ST-670, and ST-704, were newly allocated types.
Clonal analysis classified C. diphtheriae and C. belfantii isolates in 11 clonal complexes designated by eBurst groups and 1 singleton. The five toxigenic C. diphtheriae bv. Mitis pertained to five different groups (3, 9, 32, 50, and 51). Of the three toxigenic C. ulcerans, two were part of eBurst group 47, and the other was part of eBurst group 48. The nontoxigenic C. ulcerans was part of eBurst group 47 (see Fig. S1 in the supplemental material).
Genetic diversity.
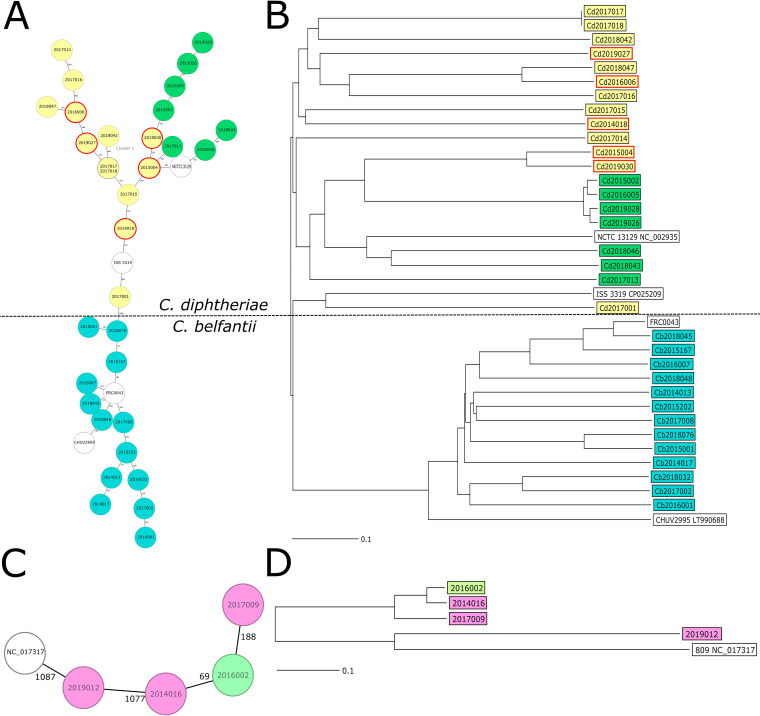
One of the C. belfantii isolates (Cb2017004) had missing values in more than 10% of the distance columns. Consequently, it was excluded from the cgMLST analysis. Finally, 1,091 (C. diphtheriae/C. belfantii scheme) and 1,174 alleles (C. ulcerans scheme) were used to generate the minimum spanning tree (MST) as 350 and 35 alleles with missing values in at least 1 sample were excluded, respectively. When mapping the results of the cgMLST into an MST, a high degree of diversity among the isolates collected was apparent (Fig. 3A). We identified a single cluster of two nontoxigenic C. diphtheriae isolates using a cluster threshold of five allelic differences. These two isolates originated from brothers that travelled from Guinea. As expected, the biovars of C. diphtheriae grouped, all toxigenic isolates were dispersed among the C. diphtheriae biovar Mitis (Fig. 3B), and C. belfantii clustered independently from C. diphtheriae. In the case of C. ulcerans, the toxigenic isolates did not group separately on the MST from the nontoxigenic isolates (Fig. 3C and D). We generated further MSTs based on the localization of the infection of the C. diphtheriae (see Fig. S2 in the supplemental material) that demonstrated no corresponding groupings. To further assess the phylogenetic relationships of the isolates obtained with regard to their epidemiological and clinical background, we generated a neighbor joining tree (Fig. 3B). The isolates exhibited no grouping based on time, origin, toxigenicity, or clinical manifestations.
FIG 3.
cgMLST analysis. (A) Minimum spanning tree of C. diphtheriae and C. belfantii isolates generated using a cluster distance threshold of ≤5 alleles. Species and biovars are color coded as follows: Corynebacterium belfantii (turquoise), Mitis (yellow), and Gravis (green); and all toxigenic isolates are marked with a solid red circle. The reference genomes used are white. (B) Neighbor joining tree generated from the C. diphtheriae cgMLST scheme. Reference genomes used were C. belfantii (FRC0043), C. diphtheriae bv. Gravis (NCTC 13129) and Mitis (ISS3319), and C. diphtheriae subsp. lausannense (CHUV2995). Isolates were color-coded according to their subtypes as in A, with red rectangles denoting the toxigenic isolates. (C) Minimum spanning tree of the C. ulcerans isolates generated using a cluster distance threshold of ≤5 alleles. Isolates are color-coded according to their toxigenicity; toxigenic isolates (purple), nontoxigenic isolates (green), and the reference genome used NCTC 017317 (white). (D) Neighbor joining tree of C. ulcerans isolates consisted of 1,209 target loci and 961 more loci for an accessory target scheme. Toxigenic isolates are denoted with a red rectangle.
Based on the molecular characterization of isolate 2017004 (registered as C. belfantii with a genome size of approximately 2.4 Mb), we compared our WGS assembly with the reference sequence of C. rouxii FRC0190 (GenBank accession number NZ_LR738855.1) (Fig. 4). Alleles with missing values in at least one sample were excluded from the comparison table, but we kept samples with missing values in more than 10% of distance columns. Finally, a 969 cgMLST scheme was used to generate the MST, and 472 alleles with missing values in at least 1 sample were excluded.
FIG 4.
cgMLST analysis of C. belfantii and C. rouxii. (A) Minimum spanning tree of C. belfantii and C. rouxii isolates generated using a cluster distance threshold of ≤5 alleles. Reference genomes for C. belfantii (FRC0043) and C. rouxii (FRC0190) are white. (B) Neighbor joining tree generated from the cgMLST scheme using the reference genomes for C. belfantii (FRC0043) and C. rouxii (FRC0190) (in white).
In silico antimicrobial resistance and virulence factors.
ResFinder analysis identified a total of 38 resistance genes with 2 or more resistance genes in 26% (10/39) of the isolates, of which all were identified in the C. diphtheriae isolates. A list of the resistance genes identified has been included in the supplemental material.
All strains were assessed for the presence of the dtxR gene required for toxin expression. This analysis demonstrated that of the C. diphtheriae isolates analyzed, 100% harbor the regulator gene.
PHASTER prophage analysis identified from one to four prophages in all isolates which were sequenced in this study (see Table S1 in the supplemental material). Further analysis of the genetic environment of the C. ulcerans tox gene did not identify pathogenicity islands.
Risk characterization.
Among the patients with toxigenic diphtheria, three had recent travels to African or Asian countries and one was an immigrant from a country where diphtheria is endemic. Two of the patients with toxigenic C. ulcerans had reported contact with companion animals. In the latter case, the individual had reported contact with a large number of animals, of which some were stray. Samples were taken from the animals, and one of the dogs and two of the cats were shown to harbor C. ulcerans. Interestingly, the cats harbored the same strain, while the dog harbored a different one. Two patients harboring a toxigenic isolate had documented vaccinations against diphtheria, while three reported vaccination but were unable to confirm it. The respiratory diphtheria case from 2015 was not vaccinated. Limited information was available for patients with nontoxigenic isolates, but among those with available data, four had a recent travel history and two were immigrants that had arrived in Spain by boat (Table 3).
DISCUSSION
In this report, we described the microbiological and epidemiological characteristics of C. diphtheriae, C. belfantii, C. ulcerans, and C. rouxii isolates submitted to the national reference laboratory (CNM) in Spain over the last 6 years (2014 to 2019). During this period, 39 isolates were submitted, of which 9 were toxigenic. The isolates exhibited a large degree of genetic diversity, belonging to 28 different sequence types, of which 7 were first described in this study. We also identified one NTTB isolate.
In well-vaccinated populations, diphtheria occurs sporadically. The surveillance systems must be sensitive and specific with protocols able to capture any diphtheria circulation in the population. Advances in laboratory techniques, including molecular studies, improve the specificity of surveillance and can help to better describe transmission routes and potential sources of infection. According to the guidelines of the RENAVE, only nine cases were classified as confirmed cases and therefore reported to the surveillance network. The updated WHO guidance on surveillance, clinical care, and outbreak response suggests a case-based surveillance with laboratory confirmation of all suspected cases, including a set of definitions and classification of cases (laboratory-confirmed classic respiratory diphtheria, laboratory-confirmed mild respiratory/asymptomatic diphtheria, nonrespiratory confirmed diphtheria, epidemiologically linked, clinically compatible, and discarded case) that more accurately depicts the situations occurring in surveillance practice (28). By adopting the definitions and classification of cases proposed by the WHO, information and knowledge of the clinical, epidemiological, microbiological, and molecular aspects of diphtheria in Spain would be significantly improved.
Prior to 2015, Spain had been diphtheria free for more than 30 years, and the laboratory capacity for the diagnosis of diphtheria in Spain relies on reference laboratories at regional and national levels. As there is no national microbiological surveillance program, it is difficult to estimate how extensive screening is for diphtheria in our country. Nevertheless, the increasing awareness of clinicians due to the fatal case in 2015 and the progressive introduction of MALDI-TOF analysis to routine hospital laboratories in Spain could be related to the increased number of Corynebacterium sp. isolates identified in 2017 and 2018. Those isolates are sent to the National Center for Microbiology for confirmation and characterization on a voluntary basis. However, similar to reports from other countries, the majority of isolates were identified as nontoxigenic Corynebacterium spp. and are therefore unlikely to be the causative agent (29–31).
Interestingly, the gender distribution of the toxigenic isolates was strongly skewed toward males (8:1), which is not reflected in the literature. Based on the small number of toxigenic isolates, we are hesitant to draw firm conclusions as to why this may be the case in Spain. Perhaps, as the immigrants arriving from countries where diphtheria is endemic are predominantly males, the observed tendency may be a result of that bias.
Less than a quarter (24%) of the isolates showed susceptibility to penicillin G. Resistance to this antibiotic was also observed in Algeria and Brazil, as well as in an outbreak among refugees from Northeast Africa and Syria in Switzerland (4, 26, 27). The interpretation of the EUCAST clinical breakpoints for corynebacteria should, however, be interpreted carefully, as they were developed for species other than C. diphtheriae. Moreover, in a recent study, a tentative epidemiological cutoff (TECOFF) of 0.5 mg/liter for penicillin G was suggested, as the wild-type mode for the tested C. diphtheriae and C. ulcerans isolates was above the EUCAST breakpoints (32). Currently, there are no EUCAST erythromycin breakpoints for Corynebacterium spp. According to CLSI breakpoints and the TECOFF proposed by Marosevi et al., all isolates were susceptible to erythromycin, which is in agreement with other authors, although isolates resistant to erythromycin have been reported in other settings (7, 33). As such, in severe cases, it may improve the patient outcome to administer erythromycin over penicillin as a first-line treatment.
The WGS data analysis using ResFinder identified several other resistance genes in silico. Three out of the nine toxigenic isolates as well as seven of the nontoxigenic isolates harbored a variety of resistance genes, but none of them conferred resistance to β-lactams or macrolides.
Similar to results from other authors (9, 26), we identified common prophage insertions in C. diphtheriae, C. belfantii, and C. ulcerans genomes, demonstrating that phage infections commonly occur in Spanish Corynebacterium spp. To draw further conclusions regarding the DT-mediated pathogenicity of our isolates, a more exhaustive study of the genetic environment and regulatory components is necessary.
The MLST results showed that there was a large variety in the sequence types of the isolates. According to the PubMLST database, ST-377, ST-482, ST-483, ST-484, ST-511, ST-670, and ST-704 have been first described in Spain. A few STs reoccurred over different years, such as ST-32, which was also the most common sequence type. ST-32 is a nontoxigenic C. diphtheriae clone that has been circulating in Europe, with the earliest isolate collected in France in 1984, as documented in PubMLST (25). Since then, it has been described in Germany, Algeria, Romania, and Belgium. A recent genomic study suggested that ST-32 was also endemic in Australia and was suspected to have enhanced virulence, with higher adhesion rates than toxigenic strains of C. diphtheriae (34). As the number of isolates pertaining to the other sequence types identified was very small, it is difficult to ascertain their origin in Spain. Noteworthy, ST-377 that was first described in the 2015 respiratory toxigenic diphtheria case reappeared at the end of 2019. However, analyzing the specimens using WGS instead of traditional MLST revealed that the 2019 ST-377 actually differed from the 2015 isolate by 284 different alleles.
With regard to the STs identified in C. ulcerans, ST-325 has been previously described in cats, dogs and humans in Germany, France and Belgium (25). Our study identified two isolates pertaining to this ST without epidemiological data available. ST-337 was first described in France in 1998, where it was isolated from a synovial fluid sample. In our study, this ST was identified from a wound exudate in a patient presenting with chronic vascular ulcers who also had a cat. As contact with companion animals is considered a primary risk factor for C. ulcerans infection (35), this could explain the origin of this infection; however, to confirm this infection, a sample would have needed to be analyzed. One of the C. ulcerans isolates we identified did have corresponding samples taken from contact animals which harbored the same ST-514 found in the human sample, suggesting this as a likely origin of infection. This finding demonstrates the urgent need to collect epidemiological data containing information on companion animals and appropriate samples when toxigenic C. ulcerans is identified.
As previously published, our findings support the conclusion that modern, low-cost WGS could replace traditional MLST methods in the surveillance of diphtheria to describe transmission events and sources of infection with the highest possible resolution (36).
The WGS genetic diversity studies demonstrated a profile typical of that expected in a country where the cases are imported. The only cluster we identified was found in closely linked individuals traveling together from a country where diphtheria is endemic. The grouping of Corynebacterium species, biovars, toxigenicity, and colonization sites appeared to have no epidemiological, geographical, or chronological link. With more complete clinical, epidemiological, and vaccination information of the nontoxigenic isolates circulating in Spain, it is highly probable that clusters would be identified.
Ongoing mass movements of travelers, refugees, asylum seekers, or immigrants from countries of diphtheria endemicity to areas of no endemicity and rising numbers of unvaccinated individuals have resulted in an increase in the global incidence of diphtheria (26). Unfortunately, it was not possible to understand the contribution of these factors to corynebacterial infections in Spain, as risk characterization was limited to the patients with confirmed toxigenic diphtheria. Important variables such as country of origin, travel history, contact with animals, and vaccination status were not collected for most of the nontoxigenic isolates, as it is currently not mandatory based on national surveillance guidelines. Of the confirmed cases, traveling to countries of diphtheria endemicity or having an origin from an country with endemicity seemed to be a risk factor. At least two patients with C. ulcerans reported contact with animals. In one case, the microbiological investigation identified one dog and two cats positive for C. ulcerans. Nontoxigenic Corynebacterium spp. are classified as an emerging pathogen, as they are increasingly associated with severe clinical outcomes, such as endocarditis and bacteremia (12, 13). Cases of invasive bacteremia due to nontoxigenic C. diphtheriae have been reported in certain at-risk populations (37). Endocarditis was observed in one of the patients with a nontoxigenic isolate in this study; however, it is not clear if this condition was attributed to infection by C. diphtheriae. Currently, the potential of these nontoxigenic Corynebacterium spp. to cause disease is likely to be underestimated by clinicians. Analysis of the WGS data demonstrated that all of the C. diphtheriae harbor the diphtheria toxin regulatory gene, meaning that these isolates are capable of becoming fully toxigenic if they acquire the toxin-bearing β-phage. Disease due to nontoxigenic variants is not vaccine preventable using the toxoid-based vaccine (1).
Although biochemically, isolate 2017004 is almost indistinguishable from C. belfantii, our cgWGS analysis suggests that it is likely a member of the C. rouxii species, making this the first description of this species in Spain.
This was the first study to describe the microbiological and epidemiological characteristics of Corynebacterium isolates identified in Spain. Using modern techniques like WGS and cgMLST, we provided a detailed molecular characterization of the isolates. Introducing WGS in the routine diagnostics for corynebacteria will be an invaluable addition to the surveillance of diphtheria in Spain and will be useful for the harmonization of diagnostics within the European Union and with other countries. To gain insights into the nontoxigenic Corynebacterium circulating in Spain, it would be of interest to collect comprehensive epidemiological data from all patients with corynebacterial infections as per the new WHO guidelines.
In conclusion, this study was the first to describe the diphtheria isolates collected in Spain, after the implementation of the new surveillance guidelines in 2014. Nontoxigenic corynebacteria are a concern for public health, as they can cause severe outcomes and have the capacity to become fully toxigenic. Collecting relevant disease and exposure information from patients with nontoxigenic corynebacteria and encouraging their notification could strengthen the current surveillance and improve the epidemiological knowledge regarding these emerging pathogens. Furthermore, the introduction of WGS in describing corynebacteria would be an invaluable addition to the surveillance of diphtheria in Spain, due to its greater discriminative power over traditional techniques.
Supplementary Material
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We thank the clinicians and microbiologists at the National Health System who submitted isolates at the National Reference Laboratory and completed enhanced surveillance forms and the regional Epidemiological Surveillance Services. We are also grateful to the microbiologists at the WHO Collaborating Centre Laboratory for Diphtheria and Streptococcal Infections at Public Health England for performing the Elek tests. Moreover, we thank the head scientific coordinator of EUPHEM Aftab Jasir and the scientific coordinator of the EPIET fellowship program Frantiska Hruba for their guidance and feedback.
L.H.-L. and S.H.-L. proposed the concept of the analyses. L.H.-L., S.H.-L., S.P., and A.H. conducted the laboratory work and the WGS analyses. S.V. conducted the WGS assembly and quality control. D.P. guided by J.M.-C. and N.L.-P. performed the epidemiological analysis. A.H. and D.P. drafted the first version of the manuscript and provided the graphs and figures. N.L.-P., S.H.-L., J.M.-C., and L.H.-L. supervised the work and revised and approved the final manuscript. We alone are responsible for the views presented in the manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Sharma NC, Efstratiou A, Mokrousov I, Mutreja A, Das B, Ramamurthy T. 2019. Diphtheria. Nat Rev Dis Prim 5:81. doi: 10.1038/s41572-019-0131-y. [DOI] [PubMed] [Google Scholar]
- 2.Dazas M, Badell E, Carmi-Leroy A, Criscuolo A, Brisse S. 2018. Taxonomic status of Corynebacterium diphtheriae biovar Belfanti and proposal of Corynebacterium belfantii sp. nov. Int J Syst Evol Microbiol 68:3826–3831. doi: 10.1099/ijsem.0.003069. [DOI] [PubMed] [Google Scholar]
- 3.Badell E, Hennart M, Rodrigues C, Passet V, Dazas M, Panunzi L, Bouchez V, Carmi–Leroy A, Toubiana J, Brisse S. 2020. Corynebacterium rouxii sp. nov., a novel member of the diphtheriae species complex. Res Microbiol 171:122–127. doi: 10.1016/j.resmic.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 4.European Centre for Disease Prevention and Control. 2017. ECDC annual epidemiological report for 2017. European Centre for Disease Prevention and Control, Stockholm, Sweden. [Google Scholar]
- 5.Wagner KS, White JM, Lucenko I, Mercer D, Crowcroft NS, Neal S, Efstratiou A, Diptheria Suveillance Network. 2012. Diphtheria in the postepidemic period, Europe, 2000–2009. Emerg Infect Dis 18:217–225. doi: 10.3201/eid1802.110987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakovljev A, Steinbakk M, Mengshoel AT, Sagvik E, Brügger-Synnes P, Sakshaug T, Rønning K, Blystad H, Bergh K. 2014. Imported toxigenic cutaneous diphtheria in a young male returning from Mozambique to Norway, March 2014. Eurosurveillance 19:20835. doi: 10.2807/1560-7917.ES2014.19.24.20835. [DOI] [PubMed] [Google Scholar]
- 7.Nelson TG, Mitchell CD, Sega-Hall GM, Porter RJ. 2016. Cutaneous ulcers in a returning traveller: a rare case of imported diphtheria in the UK. Clin Exp Dermatol 41:57–59. doi: 10.1111/ced.12763. [DOI] [PubMed] [Google Scholar]
- 8.Rahman MR, Islam K. 2019. Massive diphtheria outbreak among Rohingya refugees: lessons learnt. J Travel Med 26. doi: 10.1093/jtm/tay122. [DOI] [PubMed] [Google Scholar]
- 9.Meinel DM, Kuehl R, Zbinden R, Boskova V, Garzoni C, Fadini D, Dolina M, Blümel B, Weibel T, Tschudin-Sutter S, Widmer AF, Bielicki JA, Dierig A, Heininger U, Konrad R, Berger A, Hinic V, Goldenberger D, Blaich A, Stadler T, Battegay M, Sing A, Egli A. 2016. Outbreak investigation for toxigenic Corynebacterium diphtheriae wound infections in refugees from Northeast Africa and Syria in Switzerland and Germany by whole genome sequencing. Clin Microbiol Infect 22:1003.e1–1003.e8. doi: 10.1016/j.cmi.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Ministerio de Sanidad CYBS. 2019. Consejo Interterritorial Sistema Nacional de Salud. Calendario común de vacunación a lo largo de toda la vida. Ministerio de Sanidad, Madrid, Spain. [Google Scholar]
- 11.Jané M, Vidal MJ, Camps N, Campins M, Martínez A, Balcells J, Martin-Gomez MT, Bassets G, Herrera-León S, Foguet A, Maresma M, Follia N, Uriona S, Pumarola T. 2018. A case of respiratory toxigenic diphtheria: contact tracing results and considerations following a 30-year disease-free interval, Catalonia, Spain, 2015. Euro Surveill 23:17-00183. doi: 10.2807/1560-7917.ES.2018.23.13.17-00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belko J, Wessel DL, Malley R. 2000. Endocarditis caused by Corynebacterium diphtheriae: case report and review of the literature. Pediatr Infect Dis J 19:159–163. doi: 10.1097/00006454-200002000-00015. [DOI] [PubMed] [Google Scholar]
- 13.Muttaiyah S, Best EJ, Freeman JT, Taylor SL, Morris AJ, Roberts SA. 2011. Corynebacterium diphtheriae endocarditis: a case series and review of the treatment approach. Int J Infect Dis 15:e584-8. doi: 10.1016/j.ijid.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Centro Nacional de Epidemiología. 2013. Protocolos de la Red Nacional de Vigilancia Epidemiológica, 2015 ed. Centro Nacional de Epidemiología, Madrid, Spain. https://www.isciii.es/QueHacemos/Servicios/VigilanciaSaludPublicaRENAVE/EnfermedadesTransmisibles/Documents/PROTOCOLOS/PROTOCOLOS%20EN%20BLOQUE/PROTOCOLOS_RENAVE-ciber.pdf. [Google Scholar]
- 15.Begg N, WHO Regional Office for Europe. 1994. Manual for the management and control of diphtheria in the European region/by Norman Begg. WHO Regional Office for Europe, Copenhagen, Denmark. [Google Scholar]
- 16.Public Health England. 2014. UK standards for microbiology investigations. Identification of Corynebacterium species. Public Health England, London, UK. [Google Scholar]
- 17.Pacheco LGC, Pena RR, Castro TLP, Dorella FA, Bahia RC, Carminati R, Frota MNL, Oliveira SC, Meyer R, Alves FSF, Miyoshi A, Azevedo V. 2007. Multiplex PCR assay for identification of Corynebacterium pseudotuberculosis from pure cultures and for rapid detection of this pathogen in clinical samples. J Med Microbiol 56:480–486. doi: 10.1099/jmm.0.46997-0. [DOI] [PubMed] [Google Scholar]
- 18.Mancini F, Monaco M, Pataracchia M, von Hunolstein C, Pantosti A, Ciervo A. 2012. Identification and molecular discrimination of toxigenic and nontoxigenic diphtheria Corynebacterium strains by combined real-time polymerase chain reaction assays. Diagn Microbiol Infect Dis 73:111–120. doi: 10.1016/j.diagmicrobio.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 19.Engler KH, Glushkevich T, Mazurova IK, George RC, Efstratiou A. 1997. A modified Elek test for detection of toxigenic corynebacteria in the diagnostic laboratory. J Clin Microbiol 35:495–498. doi: 10.1128/JCM.35.2.495-498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.European Committee on Antimicrobial Susceptibility Testing. 2020. Breakpoint tables for interpretation of MICs and zone diameters. https://www.eucast.org/clinical_breakpoints/.
- 21.CLSI. 2015. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. CLSI, Wayne, PA. [Google Scholar]
- 22.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolt F, Cassiday P, Tondella ML, Dezoysa A, Efstratiou A, Sing A, Zasada A, Bernard K, Guiso N, Badell E, Rosso M-L, Baldwin A, Dowson C. 2010. Multilocus sequence typing identifies evidence for recombination and two distinct lineages of Corynebacterium diphtheriae. J Clin Microbiol 48:4177–4185. doi: 10.1128/JCM.00274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Both L, Collins S, de Zoysa A, White J, Mandal S, Efstratiou A. 2015. Molecular and epidemiological review of toxigenic diphtheria infections in England between 2007 and 2013. J Clin Microbiol 53:567–572. doi: 10.1128/JCM.03398-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dangel A, Berger A, Konrad R, Sing A. 2019. NGS-based phylogeny of diphtheria-related pathogenicity factors in different Corynebacterium spp. implies species-specific virulence transmission. BMC Microbiol 19:28. doi: 10.1186/s12866-019-1402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization (WHO). 2018. Surveillance standards for vaccine-preventable diseases, second edition. WHO, Geneva, Switzerland. [Google Scholar]
- 29.Czajka U, Wiatrzyk A, Mosiej E, Formińska K, Zasada AA. 2018. Changes in MLST profiles and biotypes of Corynebacterium diphtheriae isolates from the diphtheria outbreak period to the period of invasive infections caused by nontoxigenic strains in Poland (1950–2016). BMC Infect Dis 18:121. doi: 10.1186/s12879-018-3020-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gower CM, Scobie A, Fry NK, Litt DJ, Cameron JC, Chand MA, Brown CS, Collins S, White JM, Ramsay ME, Amirthalingam G. 2020. The changing epidemiology of diphtheria in the United Kingdom, 2009 to 2017. Euro Surveill 25:1900462. doi: 10.2807/1560-7917.ES.2020.25.11.1900462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martini H, Soetens O, Litt D, Fry NK, Detemmerman L, Wybo I, Desombere I, Efstratiou A, Piérard D. 2019. Diphtheria in Belgium: 2010–2017. J Med Microbiol 68:1517–1525. doi: 10.1099/jmm.0.001039. [DOI] [PubMed] [Google Scholar]
- 32.Marosevic DV, Berger A, Kahlmeter G, Payer SK, Hörmansdorfer S, Sing A. 2020. Antimicrobial susceptibility of Corynebacterium diphtheriae and Corynebacterium ulcerans in Germany 2011–17. J Antimicrob Chemother 75:2885–2893. doi: 10.1093/jac/dkaa280. [DOI] [PubMed] [Google Scholar]
- 33.Barraud O, Badell E, Denis F, Guiso N, Ploy M-C. 2011. Antimicrobial drug resistance in Corynebacterium diphtheriae mitis. Emerg Infect Dis 17:2078–2080. doi: 10.3201/eid1711.110282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timms VJ, Nguyen T, Crighton T, Yuen M, Sintchenko V. 2018. Genome-wide comparison of Corynebacterium diphtheriae isolates from Australia identifies differences in the Pan-genomes between respiratory and cutaneous strains. BMC Genomics 19:869. doi: 10.1186/s12864-018-5147-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meinel DM, Margos G, Konrad R, Krebs S, Blum H, Sing A. 2014. Next generation sequencing analysis of nine Corynebacterium ulcerans isolates reveals zoonotic transmission and a novel putative diphtheria toxin-encoding pathogenicity island. Genome Med 6:113. doi: 10.1186/s13073-014-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seth-Smith HMB, Egli A. 2019. Whole genome sequencing for surveillance of diphtheria in low incidence settings. Front Public Heal 7:235. doi: 10.3389/fpubh.2019.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romney MG, Roscoe DL, Bernard K, Lai S, Efstratiou A, Clarke AM. 2006. Emergence of an invasive clone of nontoxigenic Corynebacterium diphtheriae in the urban poor population of Vancouver, Canada. J Clin Microbiol 44:1625–1629. doi: 10.1128/JCM.44.5.1625-1629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data have been deposited in the ENA database under accession numbers ERS5375606 to ERS5375642 (BioProject accession number PRJEB41500).