Timely diagnosis of microorganisms in blood cultures is necessary to optimize therapy. Although blood culture media and systems have evolved for decades, the standard interval for incubation prior to being discarded as negative has remained 5 days. Here, we evaluated the optimal incubation time for the BacT/Alert Virtuo blood culture detection system (bioMérieux) using FA Plus (aerobic) and FN Plus (anaerobic) resin culture bottles in routine clinical use.
KEYWORDS: blood culture, Virtuo, time to positivity, BacT/Alert, FA Plus, FN Plus, Staphylococcus aureus, growth rate, Cutibacterium, Escherichia coli, Pseudomonas aeruginosa
ABSTRACT
Timely diagnosis of microorganisms in blood cultures is necessary to optimize therapy. Although blood culture media and systems have evolved for decades, the standard interval for incubation prior to being discarded as negative has remained 5 days. Here, we evaluated the optimal incubation time for the BacT/Alert Virtuo blood culture detection system (bioMérieux) using FA Plus (aerobic) and FN Plus (anaerobic) resin culture bottles in routine clinical use. Following institutional review board (IRB) approval, a retrospective review evaluated the outcomes of 158,710 bottles collected between November 2018 and October 2019. The number of positive blood bottles was 13,592 (8.6%); 99% of positive aerobic and anaerobic bottles flagged positive by 91.5 and 108 h, respectively. The mean (median) times to positivity were 18.4 h (15.6 h) for Staphylococcus aureus, 12.3 h (9.5 h) for Escherichia coli, 22.2 h (15.9 h) for Pseudomonas aeruginosa, and 48.9 h (42.9 h) for Candida spp. Only 175 bottles (0.1% of all bottles) flagged positive after 4 days of incubation; 89 (51%) of these bottles grew Cutibacterium (Propionibacterium) species. Chart review of blood cultures positive after 4 days (96 h) rarely had a clinical impact and sometimes had a negative impact on patient care. Finally, a seeded study of the HACEK group (i.e., Haemophilus, Aggregatibacter, Cardiobacterium, Eikenella, and Kingella), historically associated with delayed blood culture positivity, demonstrated no benefit to extended incubation beyond 4 days. Collectively, these findings demonstrated that a 4-day incubation time was sufficient for the Virtuo system and media. Implementation of the 4-day incubation time could enhance clinically relevant results by reducing recovery of contaminants and finalizing blood cultures 1 day earlier.
INTRODUCTION
Continuously monitored blood culture systems are used in clinical microbiology laboratories to test for life-threatening conditions like bacteremia, fungemia, and endocarditis. Historically, blood culture bottles were incubated on these systems for up to 7 days before being discarded and reported as “no growth.” Today, a 5-day minimum incubation is recommended for most commercial systems; however, there have been reports suggesting a 5-day incubation may not be necessary for modern blood culture systems (1–4). Optimization of the blood culture incubation period can support clinical care, with the goal of incubating long enough to recover clinically significant microorganisms while also minimizing the recovery of contaminants. The latter is important to minimize unnecessary antimicrobial therapy and can reduce waste in the clinical laboratory. In addition, a more rapid determination of a blood culture as negative has the potential to expedite patient discharges and more quickly deescalate broad-spectrum antibiotic usage to improve antimicrobial stewardship.
Our institution implemented the BacT/Alert Virtuo (Virtuo) blood culture detection system with FA Plus (aerobic) and FN Plus (anaerobic) resin culture bottles (bioMérieux) in mid-2018. This system has been shown to have a faster time to positivity compared to other systems using seeded cultures (5–7) and paired clinical cultures (8–10). These findings, along with anecdotal observations from the laboratory noting mostly growth of clinically insignificant microbes after 4 days of incubation, prompted a review of blood cultures from the Virtuo system. The goal of this work was to reassess optimal incubation time and microorganism recovery using a 1-year, retrospective analysis of Virtuo time-to-positivity data combined with medical chart review to determine clinical utility of a 5-day incubation. In addition, because they were encountered infrequently in the clinical specimens, a seeded study was included to evaluate HACEK microorganisms (Haemophilus, Aggregatibacter, Cardiobacterium, Eikenella, and Kingella).
MATERIALS AND METHODS
Clinical setting.
The central microbiology laboratory at the Barnes-Jewish Institute of Health in St. Louis, MO, serves Barnes-Jewish Hospital (a 1,250-bed tertiary care academic medical center) as well as a stand-alone academic children’s hospital and three community hospitals in the St. Louis metropolitan area.
Culture and identification.
Following approval from the Washington University in St. Louis Human Research Protection Office, all 158,710 blood culture bottles from November 2018 to October 2019 submitted to the laboratory were evaluated in the present study. The laboratory’s standard procedures recommend collecting blood cultures as a set consisting of two bottles: FA Plus (aerobic) and FN Plus (anaerobic) resin culture bottles. For adults, the recommended collection volume is 10 ml per bottle (i.e., 20 ml per set), and for children, approximately 1 ml of blood per year of patient age. No bottles were rejected by the laboratory or excluded from analyses based on blood volumes below the recommended volume. Bottles were continuously monitored until positive for up to 5 days at 37°C on the BacT/Alert Virtuo detection system (bioMérieux). While an extended incubation was available by physician request, there were no requests during the time frame of this study. The Virtuo system screens bottles almost continuously during the first 74.4 h of incubation then every 144 min thereafter. Positive bottles underwent Gram staining and were processed according to laboratory standard operating procedures. If no organisms were seen by Gram stain, acridine orange staining was performed and the bottles were subcultured to solid medium.
All positive bottles were subcultured to agar media: sheep’s blood agar, chocolate agar, MacConkey’s agar, or colistin-nalidixic acid agar if Gram stain reveals mixed microbes, including Gram-positive organisms; Campylobacter selective agar if Gram stain reveals curved Gram-negative bacilli; CandiSelect and Sabouraud dextrose agar if Gram stain reveals fungal elements or yeast; 7H11 agar if Gram stain reveals mycobacterium-like structures; and/or Brucella blood agar if an anaerobic bottle is flagged positive or if Gram stain is suggestive of anaerobic growth. Routine agars were incubated and imaged on the Kiestra Total Lab Automation system (Becton, Dickinson Microbiology Systems) (11). Anaerobic plates were incubated offline in conventional anaerobic chambers (either an Anaerobe Systems 580 or miniMACS anaerobic workstation). Microbial identification was routinely performed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) with a Bruker BioTyper (Bruker, Billerica, MA).
Data compilation.
A retrospective review evaluated the outcomes of 158,710 blood culture bottles, and, if positive, the microbial species/grouping. Bottles inoculated with non-blood specimens (e.g., pleural fluid) were excluded from analyses. Linking the recovered species to time to positivity required the merging of two large data sets. First, all time-to-positivity data were pulled from the Virtuo system’s MYLA middleware system using the culture accession number. Next, the reported species was pulled from the blood culture report logs using the Cerner Millennium laboratory information system. Using the accession number, cultures with a single positive bottle (with any number of reported species) or a single reported species (with any number of positive bottles) were linked. For cultures with multiple bottles with multiple species, these cultures required manual laboratory information system review to determine which species were recovered from which bottles.
Medical chart review.
A medical chart review in EPIC (Madison, WI) was performed on patients with a bottle time to positivity of greater than 4 days (96 h) of incubation. Patients’ electronic health records were reviewed for clinical actions prior to and after reporting of the blood culture. This included admission status, antimicrobial treatments, comorbidities, diagnoses, deceased status, and any mention of the culture in the medical chart, especially from infectious disease physicians. Examples of “No Impact” included (i) bottles with no microorganisms observed/recovered on a subculture, (ii) bottles that flagged positive after patient expired, (iii) bottles from cultures in a two-bottle set and where the companion bottle had a time to positivity of less than 4 days and reported the same microbe, (iv) bottles that were specifically called out in medical record as a “contaminant” by the physician, and (v) bottles that were from patients lost to follow-up.
Seeded study (HACEK).
Clinical isolates of HACEK microorganism recovered from frozen storage were first subcultured, and species were confirmed by MALDI-TOF MS. The species were Aggregatibacter aphrophilus, Cardiobacterium hominis, Eikenella corrodens, and Kingella denitrificans, identified using MALDI-TOF MS. For the seeded study, Escherichia coli strain ATCC 25922 was included as a positive control. After subculture, a few well-isolated colonies were used to make a 1.0 McFarland standard (∼3.0 × 108 CFU/ml). Subsequent dilutions were used to achieve a target inoculum of 30 CFU in 5 ml of human blood (packed red blood cell remnant units provided courtesy of the Barnes-Jewish Hospital transfusion service). This final concentration (∼6 CFU/ml) was determined by companion colony counts. If input colony counts were <10 or >80 CFU total on the agar plates using 0.1 ml of the prior ∼3 × 102-CFU/ml dilution, data were excluded from analysis and retested. A saline-only inoculum was used as a negative control. Aerobic and anaerobic bottles were inoculated in triplicate using standard practices and incubated on the Virtuo system per routine procedures for 5 days.
RESULTS
Cohort summary.
A total of 158,710 blood culture bottles were tested at the Barnes-Jewish Hospital microbiology laboratory between November 2018 and October 2019. Of these bottles, 13,592 (8.6%) flagged positive by the 5-day incubation cutoff (Fig. 1). The breakdowns of bottles from adults and children were 12,769 and 823, respectively.
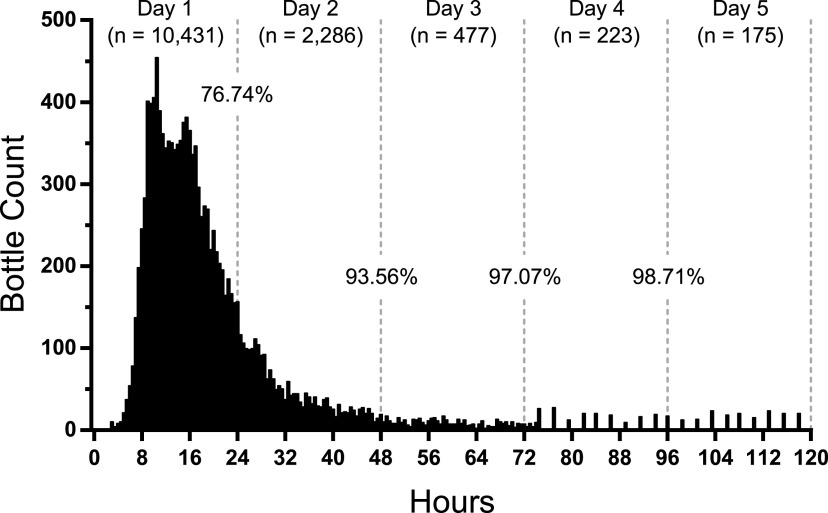
FIG 1.
Time to positivity for all positive blood culture bottles. Positive bottles (n = 13,592) were plotted in 30-min intervals. Of note, the BacT/Alert Virtuo system screens for positivity continuously for the first 74.4 h and every 144 min thereafter, resulting in greater column interspersion at later time points. After 4 days of incubation, 98.71% of all bottles flagged positive.
Time to positivity.
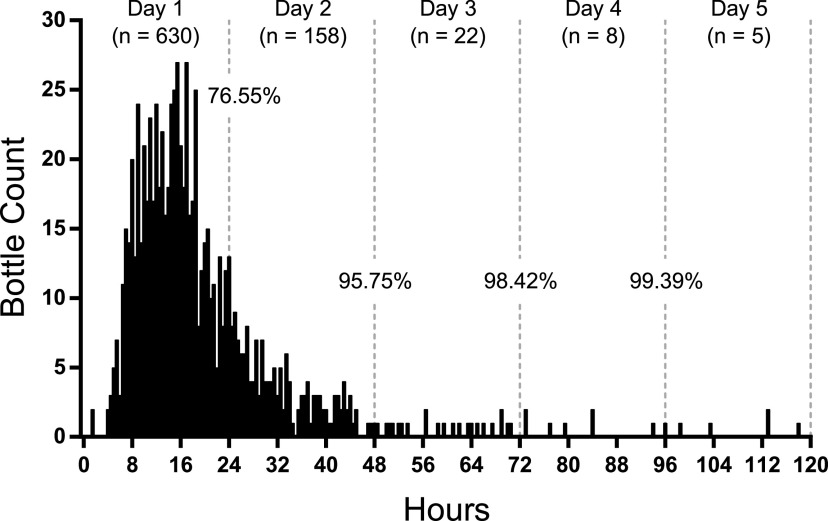
The times for 50% and 90% of bottles to signal positive were 16.5 and 38.5 h, respectively (Table 1). The mean time to positivity was 21.1 h. After 24 h of incubation, 10,431 bottles, or 76.74%, flagged positive. An additional 2,286 bottles flagged positive between 24 and 48 h, increasing the positivity rate to 93.56%. By the end of 4 days, 98.71% of bottles flagged positive—all but 175 bottles. Similar trends were observed from children (defined as ≤18 years of age [Fig. 2]). For the cultures collected from children, 630 bottles, or 76.55%, flagged positive after 24 h of incubation; by the end of 4 days, 99.39% flagged positive—all but five bottles.
TABLE 1.
Comparison of blood culture times to positivity by indicated grouping
| Grouping | No. of bottlesa | Time to positivity (h)b |
||||
|---|---|---|---|---|---|---|
| Mean | T50 | T90 | T95 | T99 | ||
| All | 13,592 | 21.1 | 16.5 | 38.5 | 56.5 | 103.5 |
| Aerobic bottles | 6,940 | 20.0 | 15.5 | 36.5 | 54.0 | 91.5 |
| Anaerobic bottles | 6,652 | 22.3 | 17.5 | 39.5 | 59.5 | 108.0 |
| Bottles from children | 823 | 20.0 | 16.5 | 36.0 | 44.0 | 84.0 |
| S. aureus | 3,893 | 18.4 | 16.0 | 29.5 | 39.0 | 72.0 |
| Coagulase-negative Staphylococcus spp.c | 2,208 | 23.5 | 20.5 | 35.5 | 46.0 | 86.5 |
| E. coli | 1,382 | 12.3 | 10.0 | 17.5 | 27.5 | 66.0 |
| S. epidermidis | 1,248 | 20.9 | 19.5 | 29.5 | 34.5 | 54.5 |
| E. faecium/E. faecalis | 859 | 15.7 | 13.0 | 25.5 | 34.5 | 64.5 |
| Candida spp. | 314 | 48.9 | 43.0 | 82.0 | 108.0 | 115.5 |
| P. aeruginosa | 298 | 22.2 | 16.0 | 45.0 | 57.5 | 96.0 |
| S. pneumoniae | 192 | 10.6 | 10.5 | 14.5 | 17.5 | 30.5 |
| Cutibacterium spp. | 121 | 103.9 | 106.0 | 115.5 | 118.0 | 118.0 |
| B. fragilis | 76 | 31.8 | 28.0 | 52.0 | 57.5 | 60.0 |
| S. maltophilia | 25 | 25.9 | 21.5 | 56.0 | 57.5 | 79.5 |
| No growth | 46 | 33.0 | 14.5 | 94.0 | 106.0 | 118.0 |
The total numbers of aerobic and anaerobic bottles were 7,113 and 6,804, respectively.
Times to achieve 50, 90, 95, and 99% of bottles positive are indicated as T50, T90, T95, and T99, respectively.
The “Coagulase-negative Staphylococcus spp.” grouping includes isolates not otherwise specified by row in the table.
FIG 2.
Time to positivity for blood culture bottles collected from children. Positive bottles (n = 823) were plotted in 30-min intervals. After 4 days of incubation, 99.39% of bottles flagged positive.
Time to positivity under aerobic and anaerobic growth conditions.
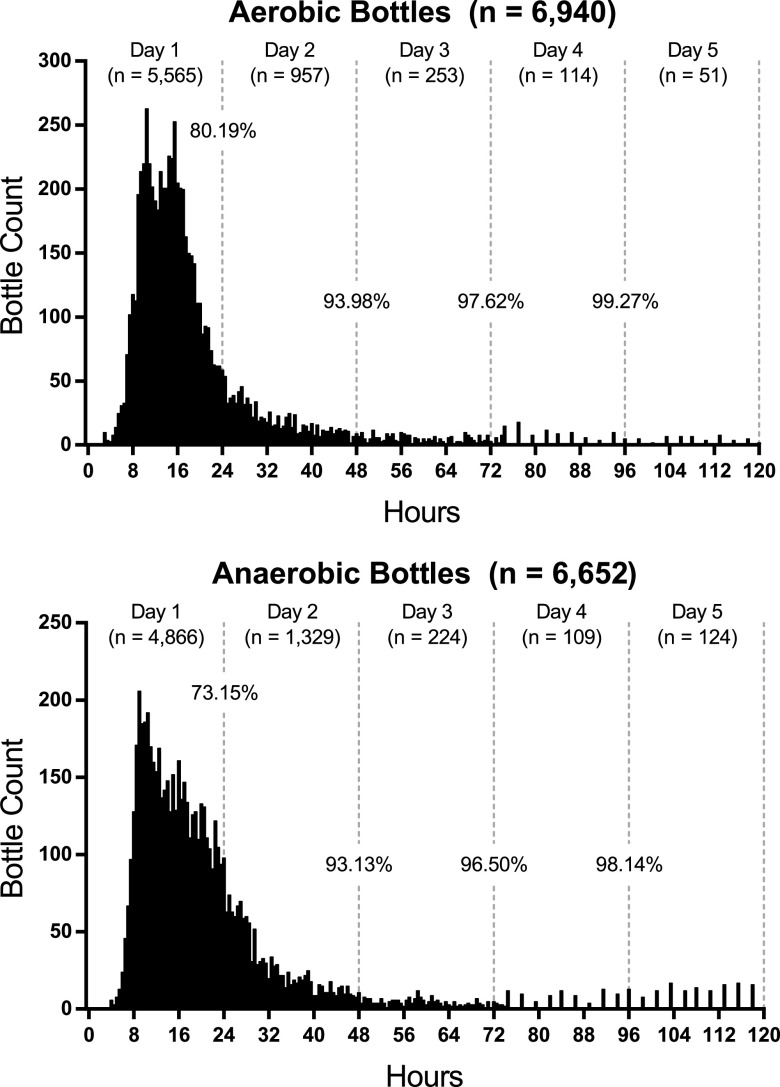
To compare differences between aerobic and anaerobic growth conditions, the aforementioned data were parsed into aerobic (FA) and anaerobic (FN) bottle types. For aerobic bottles, the times until 50% and 90% of the bottles signaled positive were 15.5 and 36.5 h, respectively (Table 1). The mean time to positivity was 20.0 h. After 24 h of incubation, 5,565 bottles, or 80.19%, flagged positive (Fig. 3, top panel). An additional 957 bottles flagged positive between 24 and 48 h, increasing the positivity rate to 93.98%. By the end of 4 days, 99.27% of bottles flagged positive—all but 51 bottles.
FIG 3.
Evaluation of time to positivity for aerobic and anaerobic blood culture bottles. Positive aerobic (n = 6,940) and anaerobic (n = 6,652) bottles were plotted in 30-min intervals. After 4 days of incubation, 99.27% of aerobic bottles and 98.14% of anaerobic bottles flagged positive.
For anaerobic bottles, a broader distribution was observed spanning the first 30 h. The times to reach 50% and 90% positivity were 17.5 and 39.5 h, respectively (Table 1). The mean time to flag positive was 22.3 h. After 24 h of incubation, 4,866 bottles, or 73.15%, flagged positive (Fig. 3, bottom panel). An additional 1,329 bottles flagged positive between 24 and 48 h, increasing the total positivity rate to 93.13%. By the end of 4 days, 98.14% of bottles flagged positive—all but 124 bottles.
Species-specific analyses.
A total of 217 different species or microbial “groupings” (e.g., Bacteroides fragilis group or coagulase-negative Staphylococcus species) were reported from positive bottles. Of these, 52 different species or microbial groupings were found in ≥20 bottles (see Table S1 in the supplemental material): these included 31 Gram-positive species or groupings, 17 Gram-negative bacteria, and 4 species of yeast. Forty-six bottles (0.3% of positive bottles) flagged positive, but no organism was ever recovered or detected by acridine orange staining.
Histograms of select medically relevant Gram-positive microbes [Staphylococcus aureus (n = 3,893), Staphylococcus epidermidis (n = 1,248), Streptococcus pneumoniae (n = 192), and Enterococcus faecium/Enterococcus faecalis (n = 859)] are shown in Fig. 4. All four species reached 95% and 99% times to positivity more quickly than the overall average (Table 1). S. pneumoniae had the shortest time to positivity of any microbe, with 98.44% of bottles with this microorganism flagging positive within 24 h and 100% within 48 h. The grouping coagulase-negative Staphylococcus was commonly found in the data set and exhibited a histogram similar to those of other Gram-positive microbes.
FIG 4.
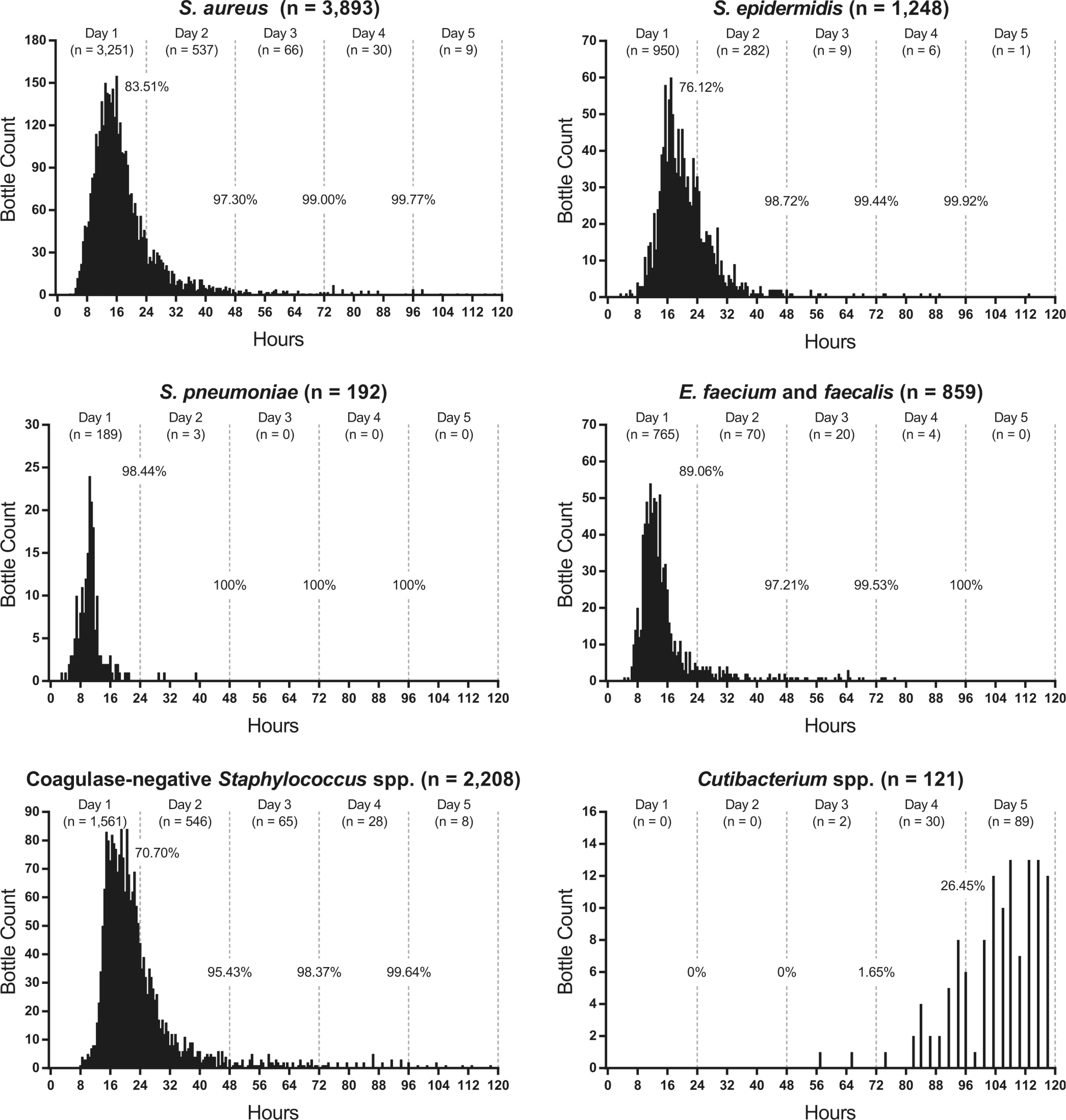
Evaluation of time to positivity for select Gram-positive bacteria. Positive bottles were plotted in 30-min intervals. For each grouping, greater than 99.6% of bottles flagged positive after 4 days, except for bottles with Cutibacterium spp., only 26.45% of which were positive.
Key Gram-negative bacteria recovered from positive bottles included Escherichia coli (n = 1,382), Pseudomonas aeruginosa (n = 298), Stenotrophomonas maltophilia (n = 25), and Bacteroides fragilis group (n = 76) (Fig. 5). E. coli reached 50% positivity faster than any other microbe, at 10 h (Table 1). While 94.43% of bottles flagged positive in 24 h, a long histogram tail was observed, with 25 bottles flagging positive over days 3 to 5. P. aeruginosa had 97.99% of bottles flag positive after 3 days and only two bottles flag positive after 4 days. The data sets for S. maltophilia and B. fragilis were noteworthy because the overwhelming majority of bottles (all but 1) flagged positive within 3 days.
FIG 5.
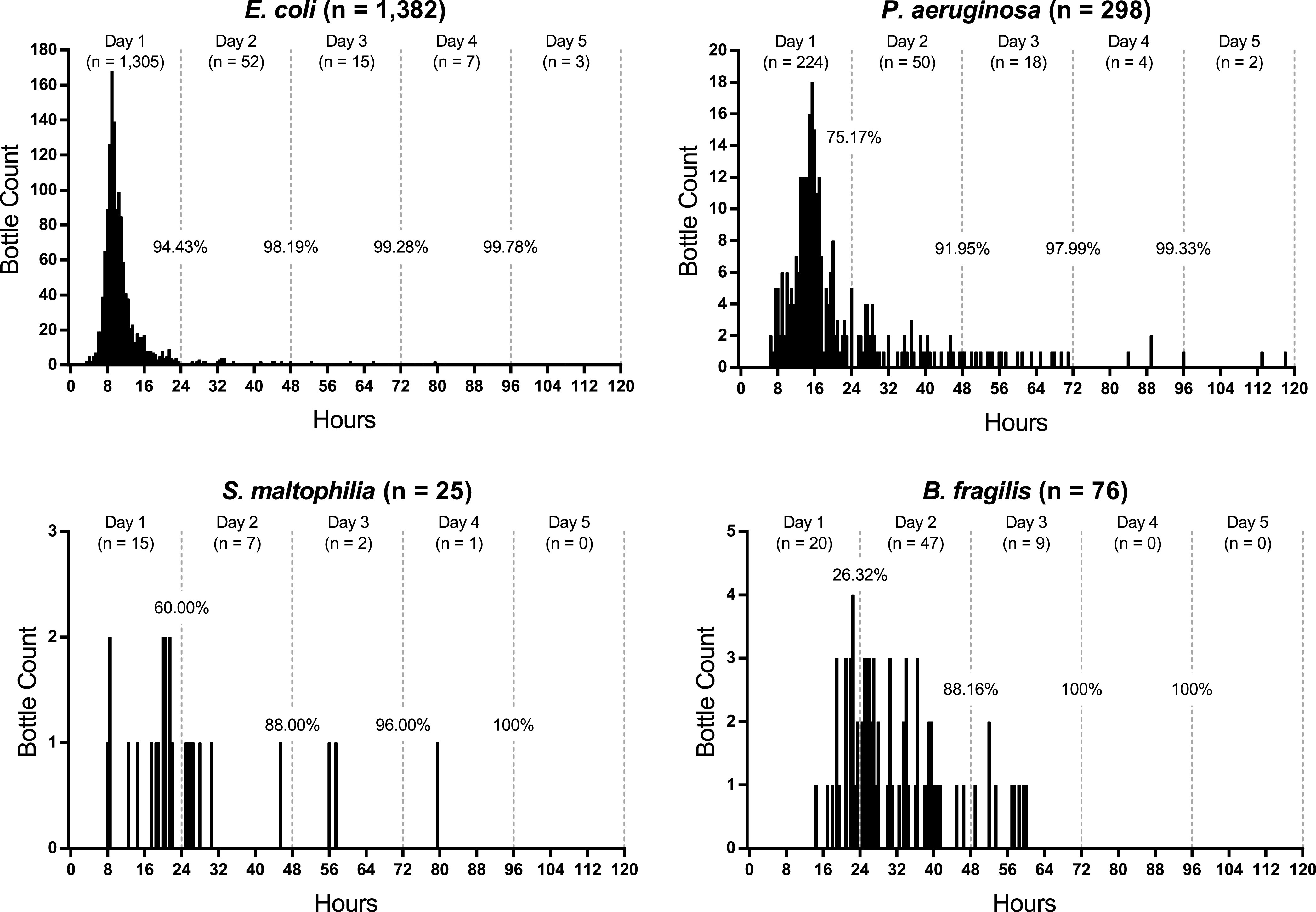
Evaluation of time to positivity for select Gram-negative bacteria. Positive bottles were plotted in 30-min intervals. For each grouping, greater than 99.3% of bottles flagged positive after 4 days.
To determine the growth patterns associated with fungemia, all Candida species were evaluated (Fig. 6). Time to positivity was more evenly distributed across the 5-day incubation time than the bacterial histograms. By the fourth day, 92.68% of bottles flagged positive, leaving 23 remaining. The delayed time to positivity was also evident in the times to achieve 50, 90, 95, and 99% positivity (Table 1).
FIG 6.
Evaluation of time to positivity for Candida species. Positive bottles (n = 314) were plotted in 30-min intervals. After 4 days of incubation, 92.68% of bottles flagged positive.
Cutibacterium (formerly Propionibacterium) species are often considered contaminants, although this microbe has been implicated in causing serious disease (12–14). The histogram of Cutibacterium species (Fig. 4) was strikingly different from those of other microbial histograms. Only two bottles flagged positive after 3 days (1.65%). An additional 30 bottles flagged positive by day 4 (26.45%), and 89 (73.55%) bottles flagged positive in the final 24 h. This delayed time to positivity was also notable in Table 1.
Clinical impact of positive bottles after 4 days.
Only 175 bottles (0.1% of all bottles and 1.3% of positive bottles) flagged positive after 4 days of incubation. There were 160 bottles that had no recorded impact in the patient’s medical record. Of these bottles, 38 were specifically mentioned by medical staff as being “contaminated.” Twenty-two bottles had a companion bottle in the culture flag positive with the same organism before 96 h. Three of the 175 bottles resulted in orders for additional tests that were never acted upon. Another bottle that grew Cutibacterium spp. delayed a patient’s discharge by 2 days. A different bottle that grew Cutibacterium spp. resulted in a visit to the Emergency Department, only to have repeat blood cultures show no growth. Another bottle grew Lactobacillus spp. that resulted in an unremarkable clinic visit. The remaining nine bottles caused a treatment adjustment. While it was difficult to determine true significance, there was documented reservation of true pathogenicity for at least three of the nine cultures, and three other cultures were from patients who expired in less than 96 h. Even by the most conservative of estimates, the last day of incubation was beneficial for <10 bottles (<0.006%).
Seeded experiment.
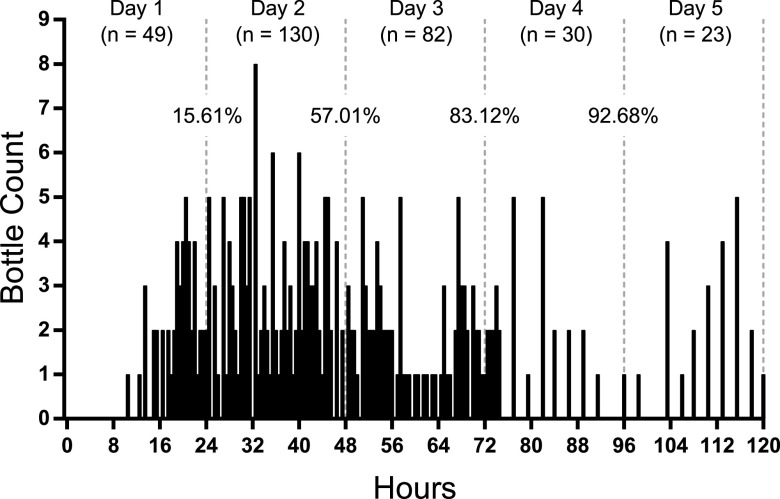
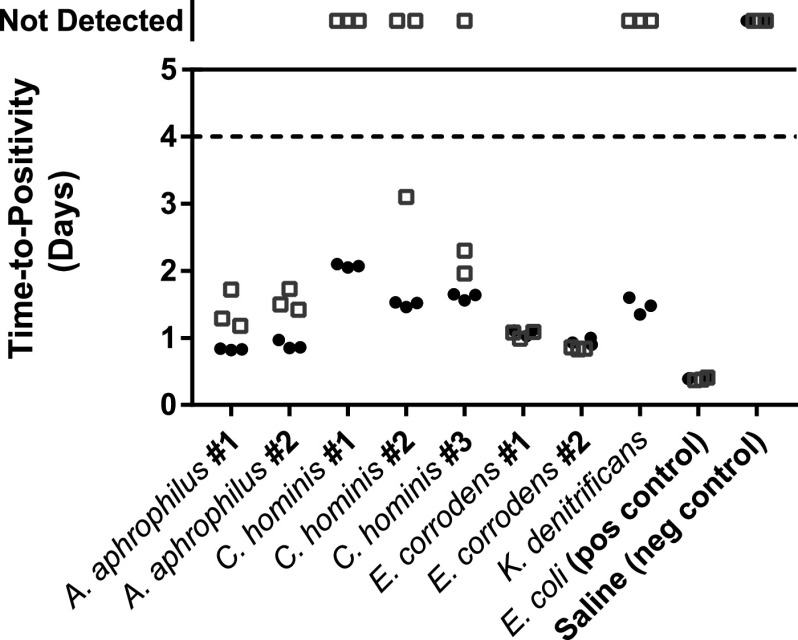
Historically, microbes in the HACEK group have been considered slow growing, and an extended incubation might be necessary for recovery. HACEK organisms were infrequently encountered in the retrospective review (e.g., Eikenella species, n = 3), with the exception of Haemophilus species (n = 31). To enhance the evaluable instances of the other HACEK organisms, a seeded study was performed in triplicate using representative species: Aggregatibacter aphrophilus, Cardiobacterium hominis, Eikenella corrodens, Kingella denitrificans, and Kingella kingae. The time to positivity of all isolates was less than 4 days for at least one bottle of the paired aerobic and anaerobic set (Fig. 7). Bottles that did not flag positive are shown at the top of the graph. Importantly, no bottles flagged positive between 4 and 5 days, suggesting no added benefit of incubation time past 4 days for the HACEK group.
FIG 7.
Seeded blood cultures for HACEK isolates. Aerobic and anaerobic bottles are shown as filled circles and open squares, respectively. The dashed line at 4 days denotes a theoretical 4-day cutoff. Bottles that did not flag positive for growth are shown above. No bottles flagged positive after 4 days of incubation.
S. aureus bacteremia.
Prolonged positivity and time to culture clearance are important blood culture metrics when assessing a patient’s bacteremia. As shown in Fig. 4, S. aureus was routinely recovered in ≤4 days, but nine bottles flagged positive after 4 days. None of these nine bottles resulted in therapeutic implications. For three bottles, the companion bottle in the blood culture set had already flagged positive in ≤4 days. Another three bottles had a different culture collected the same day that had already flagged positive with S. aureus. Two bottles were from patients with intermittent S. aureus-positive blood cultures that had flanking cultures collected before and after flagging positive. Finally, one bottle was the last S. aureus-positive blood culture in a series of cultures, with all subsequent blood cultures being negative.
Postimplementation.
Based on the data reported in this study, our institution implemented a 4-day incubation cutoff for blood cultures. Physicians still have the option to call the laboratory and request extended incubation. As a result, the laboratory is on track to report >60,000 blood culture tests 1 day earlier in just the first year of implementation. Another potential benefit has been a reduction in reporting of blood culture contaminants. While still preliminary, with only 2 months of data, we have seen a reduction in the recovery rate of Cutibacterium species relative to total positives (0.07%; predicted reduction based on shortened incubation, 0.23%; previous rate, 0.87%). Lastly, we were able to create additional instrument capacity without purchasing additional units in our Virtuo system by removing all the negative bottles 1 day earlier. This proved especially valuable in the era of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the corresponding massive increases in blood culture requests.
DISCUSSION
Bloodstream infections are a significant cause of morbidity and mortality in the United States, with an estimated burden of 72,000 to 85,000 deaths annually (15). Continuously monitored blood culture systems are a mainstay in clinical microbiology laboratories to test for these potentially life-threatening conditions. The goal of this study was to assess the optimal incubation time for blood cultures using the BacT/Alert Virtuo system paired with FA Plus (aerobic) and FN Plus (anaerobic) resin culture bottles. A retrospective review of 158,710 blood culture bottles collected over 1 year found only 175 (0.11%) flagged positive after 4 days of incubation. Chart review of these 175 bottles revealed that less than 10 bottles had a positive impact on clinical care, and some positives contributed to additional, unnecessary testing, delayed discharges, and unnecessary health care visits. These findings suggested that a routine incubation time of 4 days is sufficient for clinical laboratories using the Virtuo system. Optimizing culture incubation time to minimize the recovery of clinically insignificant cultures is appealing as blood culture contaminants can result in increased health care costs, prolonged length of stay, and laboratory waste (16–18).
There is a precedent for reducing the standard incubation time of blood cultures on automated systems. For years, the standard was 7 days. Publications from the 1990s demonstrated that commonly used systems like the BacT/Alert could have a shortened incubation period (19–22). The optimal incubation length varied by blood culture system. Hardy et al. reported that ≥99% of cultures from pathogenic species were positive by 5 days and noted a 5-day incubation would significantly reduce recovered Cutibacterium (Propionibacterium) species by 77% (19), strikingly similar to the work presented here. The Difco ESP system was reported to have sufficient results, with a 4-day incubation detecting 97.8% of all positives (3). The AccuMed ESP-384 system with a 3-day incubation was considered sufficient, with 94.2% of positives detected (23). There are reports of the Virtuo system having an even faster time to positivity compared to other blood culture systems—BD Bactec FX (6, 24, 25) and the BacT/Alert 3D system (8–10, 24, 26–28)—although the vast majority of these investigations used seeded studies. It should be noted that the epidemiological prevalence of species throughout these studies was similar to that in the present study. The most commonly recovered microbes were coagulase-negative staphylococci, S. aureus, Enterococcus species, E. coli, and S. pneumoniae. As for the theoretically missed species with a reduced incubation, these were also similar: Cutibacterium (Propionibacterium) species, Candida glabrata, and microbes of questionable clinical significance. Taken together, the evidence within the present article supporting a 4-day incubation closely resembles that of the past that led to the now 5-day standard incubation time.
To our knowledge, this is the largest study of its kind evaluating the outcomes of blood cultures on the Virtuo system along with supplementary medical chart reviews. Even a study of this size will not include the rarest of slow-growing pathogens. To mitigate this limitation, a seeded experiment of the HACEK group was included and showed no difference between 4- and 5-day incubations. The HACEK group was historically often thought to require extended incubation for detection, but this has been refuted previously (29, 30). An additional limitation of this study is the findings may not be applicable to other blood culture systems or other laboratories with different local microbiological populations. Furthermore, a criticism of this work might be that the data come from a single microbiology laboratory, but cultures were received from five hospitals, including an academic hospital, community hospitals, and a children’s hospital. Lastly, there may be interest in establishing an even shorter incubation time in the future. If a 3-day incubation was proposed using our data set, the five most impacted species would be S. aureus, coagulase-negative Staphylococcus species, Cutibacterium species, Candida species, and Sphingomonas species. Future studies are needed to determine if shorter incubation periods are clinically feasible and could further optimize patient care and laboratory testing.
If an institution was to reduce the blood culture incubation time, national guidelines from pertinent organizations must be considered. The Clinical and Laboratory Standards Institute (CLSI) guidelines recommend a 5-day incubation time while also acknowledging 4 days, or even 3 days, might be adequate for some systems (31). The CLSI M47 document (published in 2007) states adequate data from a shorter incubation time may permit laboratories to adapt shorter protocols; this may be revised in updates to the M47 document. Other guidelines like those from the Infectious Diseases Society of America (IDSA) may be impacted by a shortened incubation period. For example, uncomplicated bacteremia is defined as patients with positive blood cultures and follow-up blood cultures performed on specimens obtained 2 to 4 days after the initial set that do not grow methicillin-resistant S. aureus (MRSA) (32). This 2- to 4-day time period may need to be adjusted in the setting of contemporary blood culture systems and media.
In conclusion, we found a 4-day incubation cutoff to detect the vast majority of microorganisms detected on the BacT/Alert Virtuo system. We project that a 4-day incubation on the Virtuo blood culture system not only will improve microbiological workflows but also will improve patient care.
Supplementary Material
ACKNOWLEDGMENTS
We thank the microbiology staff in the Central Laboratory at Barnes-Jewish Hospital for their ongoing efforts for the patients of the BJC Healthcare system. Thank you to colleagues in Laboratory and Genomic Medicine and Infectious Diseases for feedback on the study design and manuscript.
REFERENCES
- 1.Bourbeau PP, Pohlman JK. 2001. Three days of incubation may be sufficient for routine blood cultures with BacT/Alert FAN blood culture bottles. J Clin Microbiol 39:2079–2082. doi: 10.1128/JCM.39.6.2079-2082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourbeau PP, Foltzer M. 2005. Routine incubation of BacT/ALERT FA and FN blood culture bottles for more than 3 days may not be necessary. J Clin Microbiol 43:2506–2509. doi: 10.1128/JCM.43.5.2506-2509.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doern GV, Brueggemann AB, Dunne WM, Jenkins SG, Halstead DC, McLaughlin JC. 1997. Four-day incubation period for blood culture bottles processed with the Difco ESP blood culture system. J Clin Microbiol 35:1290–1292. doi: 10.1128/JCM.35.5.1290-1292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sepulveda J, Westblade LF, Whittier S, Satlin MJ, Greendyke WG, Aaron JG, Zucker J, Dietz D, Sobieszczyk M, Choi JJ, Liu D, Russell S, Connelly C, Green DA. 2020. Bacteremia and blood culture utilization during COVID-19 surge in New York City. J Clin Microbiol 58:e00875-20. doi: 10.1128/JCM.00875-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altun O, Almuhayawi M, Lüthje P, Taha R, Ullberg M, Özenci V. 2016. Controlled evaluation of the new BacT/Alert Virtuo blood culture system for detection and time to detection of bacteria and yeasts. J Clin Microbiol 54:1148–1151. doi: 10.1128/JCM.03362-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Somily AM, Habib HA, Torchyan AA, Sayyed SB, Absar M, Al-Aqeel R, Binkhamis AK. 2018. Time-to-detection of bacteria and yeast with the BACTEC FX versus BacT/Alert Virtuo blood culture systems. Ann Saudi Med 38:194–199. doi: 10.5144/0256-4947.2018.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menchinelli G, Liotti FM, Fiori B, De Angelis G, D'Inzeo T, Giordano L, Posteraro B, Sabbatucci M, Sanguinetti M, Spanu T. 2019. Corrigendum: in vitro evaluation of BACT/ALERT VIRTUO, BACT/ALERT 3D, and BACTEC FX automated blood culture systems for detection of microbial pathogens using simulated human blood samples. Front Microbiol 10:2688. doi: 10.3389/fmicb.2019.02688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs MR, Mazzulli T, Hazen KC, Good CE, Abdelhamed AM, Lo P, Shum B, Roman KP, Robinson DC. 2017. Multicenter clinical evaluation of BacT/Alert Virtuo blood culture system. J Clin Microbiol 55:2413–2421. doi: 10.1128/JCM.00307-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.She RC, Romney MG, Jang W, Walker T, Karichu JK, Richter SS. 2018. Performance of the BacT/Alert Virtuo Microbial Detection System for the culture of sterile body fluids: prospective multicentre study. Clin Microbiol Infect 24:992–996. doi: 10.1016/j.cmi.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Kim SC, Lee S, Kim S, Cho OH, Park H, Yu SM. 2019. Comparison of clinical performance between BacT/Alert Virtuo and BacT/Alert 3D blood culture systems. Ann Lab Med 39:278–283. doi: 10.3343/alm.2019.39.3.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey AL, Ledeboer N, Burnham CD. 2019. Clinical microbiology is growing up: the total laboratory automation revolution. Clin Chem 65:634–643. doi: 10.1373/clinchem.2017.274522. [DOI] [PubMed] [Google Scholar]
- 12.Corvec S. 2018. Clinical and biological features of Cutibacterium (formerly Propionibacterium) avidum, an underrecognized microorganism. Clin Microbiol Rev 31:e00064-17. doi: 10.1128/CMR.00064-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sohail MR, Gray AL, Baddour LM, Tleyjeh IM, Virk A. 2009. Infective endocarditis due to Propionibacterium species. Clin Microbiol Infect 15:387–394. doi: 10.1111/j.1469-0691.2009.02703.x. [DOI] [PubMed] [Google Scholar]
- 14.Dioguardi M, Alovisi M, Crincoli V, Aiuto R, Malagnino G, Quarta C, Laneve E, Sovereto D, Lo Russo L, Troiano G, Lo Muzio L. 2020. Prevalence of the genus Propionibacterium in primary and persistent endodontic lesions: a systematic review. J Clin Med 9:739. doi: 10.3390/jcm9030739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto M, Al-Hasan MN. 2013. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect 19:501–509. doi: 10.1111/1469-0691.12195. [DOI] [PubMed] [Google Scholar]
- 16.Gander RM, Byrd L, DeCrescenzo M, Hirany S, Bowen M, Baughman J. 2009. Impact of blood cultures drawn by phlebotomy on contamination rates and health care costs in a hospital emergency department. J Clin Microbiol 47:1021–1024. doi: 10.1128/JCM.02162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dempsey C, Skoglund E, Muldrew KL, Garey KW. 2019. Economic health care costs of blood culture contamination: a systematic review. Am J Infect Control 47:963–967. doi: 10.1016/j.ajic.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 18.Lalezari A, Cohen MJ, Svinik O, Tel-Zur O, Sinvani S, Al-Dayem YA, Block C, Moses AE, Oster Y, Salameh S, Strahilevitz J. 2020. A simplified blood culture sampling protocol for reducing contamination and costs: a randomized controlled trial. Clin Microbiol Infect 26:470–474. doi: 10.1016/j.cmi.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Hardy DJ, Hulbert BB, Migneault PC. 1992. Time to detection of positive BacT/Alert blood cultures and lack of need for routine subculture of 5- to 7-day negative cultures. J Clin Microbiol 30:2743–2745. doi: 10.1128/JCM.30.10.2743-2745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson ML, Mirrett S, Reller LB, Weinstein MP, Reimer LG. 1993. Recovery of clinically important microorganisms from the BacT/Alert blood culture system does not require testing for seven days. Diagn Microbiol Infect Dis 16:31–34. doi: 10.1016/0732-8893(93)90127-S. [DOI] [PubMed] [Google Scholar]
- 21.Cornish N, Kirkley BA, Easley KA, Washington JA. 1998. Reassessment of the incubation time in a controlled clinical comparison of the BacT/Alert aerobic FAN bottle and standard anaerobic bottle used aerobically for the detection of bloodstream infections. Diagn Microbiol Infect Dis 32:1–7. doi: 10.1016/S0732-8893(98)00057-1. [DOI] [PubMed] [Google Scholar]
- 22.Cornish N, Kirkley BA, Easley KA, Washington JA. 1999. Reassessment of the routine anaerobic culture and incubation time in the BacT/Alert FAN blood culture bottles. Diagn Microbiol Infect Dis 35:93–99. doi: 10.1016/S0732-8893(99)00064-4. [DOI] [PubMed] [Google Scholar]
- 23.Han XY, Truant AL. 1999. The detection of positive blood cultures by the AccuMed ESP-384 system: the clinical significance of three-day testing. Diagn Microbiol Infect Dis 33:1–6. doi: 10.1016/s0732-8893(98)00112-6. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Liu S, Chen H, Zhang X, Ling Y, Zhang N, Hou T. 2020. Comparative evaluation of BACTEC FX, BacT/ALERT 3D, and BacT/ALERT VIRTUO-automated blood culture systems using simulated blood cultures. Acta Clin Belg 29:1–8. doi: 10.1080/17843286.2020.1797343. [DOI] [PubMed] [Google Scholar]
- 25.Chung Y, Kim I-H, Han M, Kim HS, Kim H-S, Song W, Kim J-S. 2019. A comparative evaluation of BACT/ALERT FA PLUS and FN PLUS blood culture bottles and BD BACTEC Plus Aerobic and Anaerobic blood culture bottles for antimicrobial neutralization. Eur J Clin Microbiol Infect Dis 38:2229–2233. doi: 10.1007/s10096-019-03663-3. [DOI] [PubMed] [Google Scholar]
- 26.Congestrì F, Pedna MF, Fantini M, Samuelli M, Schiavone P, Torri A, Bertini S, Sambri V. 2017. Comparison of 'time to detection' values between BacT/ALERT VIRTUO and BacT/ALERT 3D instruments for clinical blood culture samples. Int J Infect Dis 62:1–5. doi: 10.1016/j.ijid.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Miller N, Brassinne L, Allemeersch D. 2018. Implementation of the new VIRTUO blood culture system: evaluation and comparison to the 3D system using simulated blood cultures. Acta Clin Belg 73:16–20. doi: 10.1080/17843286.2017.1331618. [DOI] [PubMed] [Google Scholar]
- 28.Totty H, Ullery M, Spontak J, Viray J, Adamik M, Katzin B, Dunne WM, Jr, Deol P. 2017. A controlled comparison of the BacT/ALERT 3D and VIRTUO microbial detection systems. Eur J Clin Microbiol Infect Dis 36:1795–1800. doi: 10.1007/s10096-017-2994-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petti CA, Bhally HS, Weinstein MP, Joho K, Wakefield T, Reller LB, Carroll KC. 2006. Utility of extended blood culture incubation for isolation of Haemophilus, Actinobacillus, Cardiobacterium, Eikenella, and Kingella organisms: a retrospective multicenter evaluation. J Clin Microbiol 44:257–259. doi: 10.1128/JCM.44.1.257-259.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baron EJ, Scott JD, Tompkins LS. 2005. Prolonged incubation and extensive subculturing do not increase recovery of clinically significant microorganisms from standard automated blood cultures. Clin Infect Dis 41:1677–1680. doi: 10.1086/497595. [DOI] [PubMed] [Google Scholar]
- 31.CLSI. 2007. Principles and procedures for blood cultures; approved guideline. CLSI document M47-A. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 32.Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, Rybak MJ, Talan DA, Chambers HF. 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin Infect Dis 52:e18–e55. doi: 10.1093/cid/ciq146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.