Abstract
The conserved endoplasmic reticulum (ER) membrane protein TRAPα (translocon-associated protein, also known as signal sequence receptor 1, SSR1) has been reported to play a critical but unclear role in insulin biosynthesis. TRAPα/SSR1 is one component of a four-protein complex including TRAPβ/SSR2, TRAPγ/SSR3, and TRAPδ/SSR4. The TRAP complex topologically has a small exposure on the cytosolic side of the ER via its TRAPγ/SSR3 subunit, whereas TRAPβ/SSR2 and TRAPδ/SSR4 function along with TRAPα/SSR1 largely on the luminal side of the ER membrane. Here, we have examined pancreatic β-cells with deficient expression of either TRAPβ/SSR2 or TRAPδ/SSR4, which does not perturb mRNA expression levels of other TRAP subunits, or insulin mRNA. However, deficient protein expression of TRAPβ/SSR2 and, to a lesser degree, TRAPδ/SSR4, diminishes the protein levels of other TRAP subunits, concomitant with deficient steady-state levels of proinsulin and insulin. Deficient TRAPβ/SSR2 or TRAPδ/SSR4 is not associated with any apparent defect of exocytotic mechanism but rather by a decreased abundance of the proinsulin and insulin that accompanies glucose-stimulated secretion. Amino acid pulse labeling directly establishes that much of the steady-state deficiency of intracellular proinsulin can be accounted for by diminished proinsulin biosynthesis, observed in a pulse-labeling as short as 5 minutes. The proinsulin and insulin levels in TRAPβ/SSR2 or TRAPδ/SSR4 null mutant β-cells are notably recovered upon re-expression of the missing TRAP subunit, accompanying a rebound of proinsulin biosynthesis. Remarkably, overexpression of TRAPα/SSR1 can also suppress defects in β-cells with diminished expression of TRAPβ/SSR2, strongly suggesting that TRAPβ/SSR2 is needed to support TRAPα/SSR1 function.
Keywords: diabetes, insulin secretion, pancreatic beta cell, preproinsulin, translocation
1 |. INTRODUCTION
The translation of (pre)proinsulin represents, by far, the single greatest commitment of the protein biosynthetic machinery of pancreatic β-cells.1 It is not possible to generate functional insulin unless the initial translation product successfully crosses the ER membrane from the cytosolic side where polyribosomes generate thousands of nascent preproinsulin molecules per second, to the ER lumen where the signal-cleaved proinsulin can fold in preparation for intracellular transport.2 The protein-conducting Sec61 channel has been shown to be essential for this translocation process.3 However, growing evidence suggests that translocation of various secretory proteins across the ER membrane may differ in their efficiency4,5 and that preproinsulin is one of the proteins that may not, on its own, efficiently engage the Sec61 mechanism.6,7 The translocation of such proteins may require the assistance of the Translocation-Associated Protein (TRAP) complex, also known as the Signal Sequence Receptor (SSR) complex.
The TRAP/SSR complex,8 composed of four subunits (TRAPα/SSR1, TRAPβ/SSR2, TRAPγ/SSR3, and TRAPδ/SSR4), is “an auxiliary protein complex” that assists in translocation of proteins whose primary structure bearing signal peptides are not optimal for efficient direct interaction with the Sec61 translocon.9 In pancreatic β-cells exposed ≥ 24 hours to high glucose, the mRNAs encoding TRAP subunits are among the most highly upregulated in the entire β-cell transcriptome.10 Expression of TRAP subunits is also likely to be important in the biosynthesis of some other peptide prohormones.11
The TRAP/SSR complex actually has peptide exposure on both sides of the ER membrane.12 Whereas TRAPγ/SSR3 is the subunit uniquely designed for its cytosolic disposition, the cytosolic side of the ER membrane exposes only ~60 residues of TRAPα/SSR1 and a seemingly negligible fraction of TRAPβ/SSR2 or TRAPδ/SSR4.13 Curiously, a recent report has proposed that in a signal sequence-specific manner, translocation of nascent preproinsulin critically involves selective interaction with TRAPβ/SSR2 on the cytosolic side of the ER membrane.14
In contrast to TRAPγ/SSR3, there is dramatically greater peptide exposure on the luminal side of the ER membrane for TRAPα/SSR1, TRAPβ/SSR2, and TRAPδ/SSR4.12,15 Recent published evidence has suggested that deficiency of the type 2 diabetes-associated gene, TRAPα/SSR1, impairs preproinsulin translocation and leads to notably diminished insulin storage in pancreatic β-cells.7 With this in mind, here, we have examined the behavior of pancreatic β-cells bearing genetic deficiency of TRAPβ/SSR2, or TRAPδ/SSR4, which are luminal partners of TRAPα/SSR1. We report that deficiency of either of these TRAP/SSR subunits recapitulates a defect in insulin storage in pancreatic β-cells, caused largely by deficient proinsulin biosynthesis, and deficiency of TRAPβ/SSR2 in particular causes dramatically diminished protein levels of TRAPα/SSR1. However, despite the recently proposed role of TRAPβ/SSR2 to engage the preproinsulin signal peptide,14 we find that overexpression of TRAPα/SSR1 helps to restore insulin storage in β-cells devoid of TRAPβ/SSR2, suggesting alternatively that the primary role of TRAPβ/SSR2 in preproinsulin translocation is to support TRAPα/SSR1 function.
2 |. MATERIALS AND METHODS
2.1 |. Reagents and antibodies
Lipofectamine 2000 for transfection, Lipofectamine RNAiMAX, 4–12% NuPage gels, LDS sample loading buffer, Met/Cys-deficient Dulbecco’s modified Eagle’s medium, and all other tissue culture reagents were from Invitrogen (Carlsbad, CA, USA). The siRNAs were from ThermoFisher (Waltham, MA, USA). The rodent (mouse/rat) insulin chemiluminescence ELISA kit was from ALPCO company (Salem, NH, USA). Protein A-Agarose was from Repligen (Waltham, MA, USA). 35S-amino acid mixture was from PerkinElmer (Waltham, MA). Rabbit anti-SSR1 antibody and mouse anti-rat proinsulin antibody CCI-17 were from Novus Biologicals (Littleton, CO, USA). Guinea pig anti-insulin was from Merck-Millipore (Billerica, MA, USA). Rabbit anti-SSR2 was from Proteintech (Rosemont, IL, USA). Rabbit polyclonal anti-SSR3 and anti-SSR4 were from Abmart (Shanghai, China). Mouse monoclonal anti-tubulin was from Sigma (St. Louis, MO, USA). Horseradish peroxidase-conjugated antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA, USA). Clarity Western ECL Substrate was from Bio-Rad (Hercules, CA, USA).
2.2 |. Cell culture
INS832/13 rat insulinoma cells (ATCC, Manassas, VA, USA) were cultured in RPMI 1640 supplemented with 10% FBS, 1 mM sodium pyruvate, 10 mM Hepes, and 0.05 mM 2-mercaptoethanol (Sigma, St Louis, MO, USA).
2.3 |. Manipulating SSR2 and SSR4 expression in INS832/13 cells
For SSR2 or SSR4 gene suppression, INS832/13 cells were transfected with 20 nM SSR2 or SSR4 targeted siRNAs using Lipofectamine RNAiMAX. At 48 hours post transfection, immunoblotting was performed to evaluate cell levels of proinsulin, insulin, and SSR subunits.
SSR2/SSR4 null INS832/13 cells were generated using CRISPR-Cas9 mediated genome editing. The following guide design resources (https://zlab.bio/guide-design-resources) were used for designing single-guide RNAs. Nucleotide guide sequences (SSR2: 5′-CACCGCTGGTATGCTCAACGTCAAA-3′; SSR4: CACCGGATGGCATCTTTCGGCGCCC) were annealed and ligated into LentiCRISPRv2 vector and delivered to INS832/13 cells with Lipofectamine 2000. At 48 hours post-transfection, the culture medium was replaced by RPMI1640 containing puromycin (1 μg/mL). Puromycin-resistant pool and single clones were screened for SSR2 or SSR4 expression by immunoblotting.
For rescue experiments, plasmids encoding various flag-tagged SSR subunits were transfected into SSR2 or SSR4 deficient cells (SSR2/SSR4 null cells). At 48 hours after transfection, immunoblotting was performed to measure protein levels of proinsulin, insulin, and SSR subunits.
2.4 |. 35S-Met/Cys labeling and immunoprecipitation
INS832/13 cells were pulse labeled with 35S-amino acids and chased for the times indicated. The cells were then washed once with ice-cold PBS containing 20 mmol/L N-ethyl maleimide and then lysed in 500 μL RIPA buffer. Trichloroacetic acid (TCA)-precipitable counts were used to normalize total protein synthesis among samples. Cell lysates and medium were immunoprecipitated with anti-insulin at 4°C overnight. Anti-insulin IP products were washed twice with RIPA buffer and then boiled in SDS sample buffer with 100 mM DTT for 5 minutes and resolved in 4–12% NuPage gels, followed by phosphorimaging. Bands intensities were quantified using ImageJ software.
2.5 |. Immunofluorescence
Immunofluorescence was employed in SSR2/SSR4 null cells transfected with plasmid encoding either Flag-tagged SSR2 or SSR4 protein. Briefly, transfected INS832/13 cells monolayer grown in eight-well chamber slides were fixed with 4% paraformaldehyde for 30 minutes at room temperature, followed by permeabilization with 0.2% triton X-100 for an additional 12 minutes. After blocking, the samples were incubated with anti-Flag, anti-proinsulin, and anti-insulin antibodies. After incubated with appropriate secondary antibodies conjugated with different Alexafluor dyes as indicated, immunofluorescence images were acquired on a Nikon A1 confocal microscope (Melville, NY, USA).
2.6 |. Immunoblotting
Cell lysates were boiled in gel sample buffer with 100 mM DTT for 5 minutes, resolved by 4–12% gradient Nupage, electrotransferred to nitrocellulose, and incubated with diluted primary antibodies at 4°C overnight. Secondary antibodies were used at room temperature for 1 hours. Imaging was captured after incubation with the Clarity Western ECL Substrate according to the manufacturer’s instructions.
2.7 |. Glucose-stimulated insulin secretion assay
Glucose-stimulated insulin secretion assay was performed as previously described.16 Briefly, control or SSR2/SSR4 null cells were incubated with prewarmed Krebs-Ringer bicarbonate Hepes [KRBH; 0.5% bovine serum albumin, 129 mM NaCl, 5 mM NaHCO3, 4.8 mM KCl, 1.2 mM KH2PO2, 2.5 mM CaCl2, 1.2 mM MgSO4, and 10 mM Hepes (pH 7.4)] at 37°C for 2 hours. The preincubation medium was removed, and cells were then incubated with KRBH containing 2.5 mM glucose for additional 2 hours. The culture media were collected, and the cells were then stimulated with KRBH containing 25 mM glucose for another 2 hours. After collecting the stimulation media, cells were lysed with acid-ethanol. Rat insulin levels in both media and cell lysates were assayed using the STELLUX Insulin Rodent (Mouse/Rat) Chemiluminescence ELISA kit (ALPCO No. 80-INSMR-CH01).
2.8 |. Statistical analysis
All data were processed with GraphPad Prism 8 software and presented as means ± SD. Student’s t test and one-way ANOVA were used to determine significance between groups. A P value < .05 was considered as statistically significant.
3 |. RESULTS
3.1 |. TRAPβ/SSR2 and TRAPδ/SSR4 deficiency recapitulate proinsulin biosynthesis and insulin storage defects
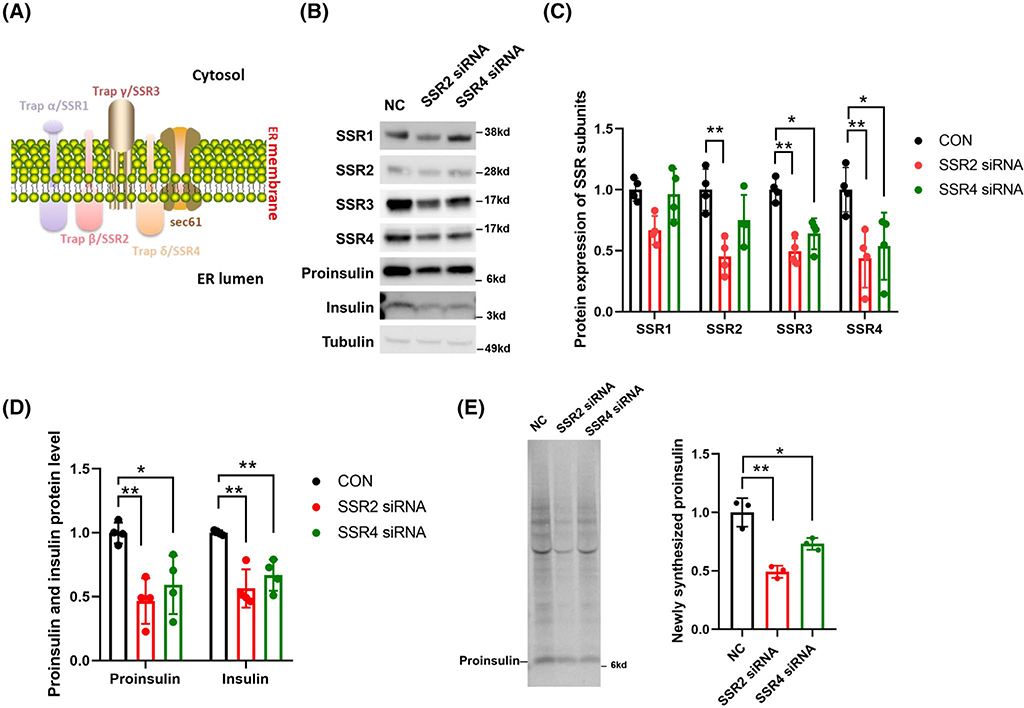
It is now established that deficiency of the type 2 diabetes gene, TRAPα/SSR1, leads to markedly diminished steady-state levels of proinsulin, as well as insulin storage, in pancreatic β-cells.7 As shown in Figure 1A, The TRAPα/SSR1 protein resides primarily on the luminal side of the ER membrane along with TRAPβ/SSR2 and TRAPδ/SSR4.13 To determine whether these two components of the complex are also required for proinsulin and insulin storage, we first attempted gene suppression in INS832/13 cells using siRNAs directed against TRAPβ/SSR2 or TRAPδ/SSR4—an approach that has previously been shown to be effective.17 Neither chosen siRNA was found to be highly efficient in achieving suppression at the protein level for TRAPβ/SSR2 and TRAPδ/SSR4in β-cells (Figure 1B,C), and neither changed the mRNA levels of subunits other than the one to which the siRNA was directed (discussed further in CRISPR/Cas9-mediated knockout cells, below). However, even a 50% knockdown of TRAPβ/SSR2 protein was accompanied by diminished levels of other subunits of the TRAP/SSR protein complex, whereas knockdown of TRAPδ/SSR4 was accompanied by partial depletion of TRAPγ/SSR3 protein without a major diminution of TRAPα/SSR1 or TRAPβ/SSR2 (Figure 1B,C). Remarkably, a decrease in steady-state proinsulin and insulin levels was associated with suppression of TRAPβ/SSR2 and, to a lesser extent, TRAPδ/SSR4 (Figure 1D).
FIGURE 1.
SiRNA-mediated suppression of TRAPβ/SSR2 or TRAPδ/SSR4 in INS832/13 cells. A, Structure and subunit composition of the TRAP/SSR complex as predicted by bioinformatic analysis. B, Immunoblotting with the listed antibodies of INS832/13 cell lysates transfected with the indicated siRNAs. Proinsulin was measured with rodent-specific anti-proinsulin. Tubulin is a loading control. C, Quantitation (mean ± SD) of TRAP/SSR subunit protein levels from experiments like that in panel B (n = 4). *P < .05, **P < .01 comparing to WT. D, Quantitation (mean ± SD) of proinsulin and insulin protein levels from experiments like that in panel B (n = 4). *P < .05, **P < .01 compared to WT. E, Scrambled oligo (NC) or TRAPβ/SSR2 or TRAPδ/SSR4 siRNA-mediated gene suppression cells were pulse-labeled with 35S-Met/Cys for 10 minutes (left panel). Cell lysates (normalized to TCA-precipitable counts) were subjected to immunoprecipitation with anti-insulin, followed by SDS-PAGE and phosphorimaging. Quantitation of proinsulin (n = 3) is shown at right. *P < .05, **P < .01
To examine the rate of biosynthesis of newly synthesized proinsulin, INS832/13 cells with TRAPβ/SSR2 or TRAPδ/SSR4 gene suppression were pulse labeled for 10 minutes with 35S-amino acids, and the newly made proinsulin was immunoprecipitated with anti-insulin antibodies (normalized to TCA-precipitable counts). Proinsulin biosynthesis was significantly diminished in INS832/13 cells bearing suppression of either TRAPβ/SSR2 or TRAPδ/SSR4 (Figure 1E, quantitation at right).
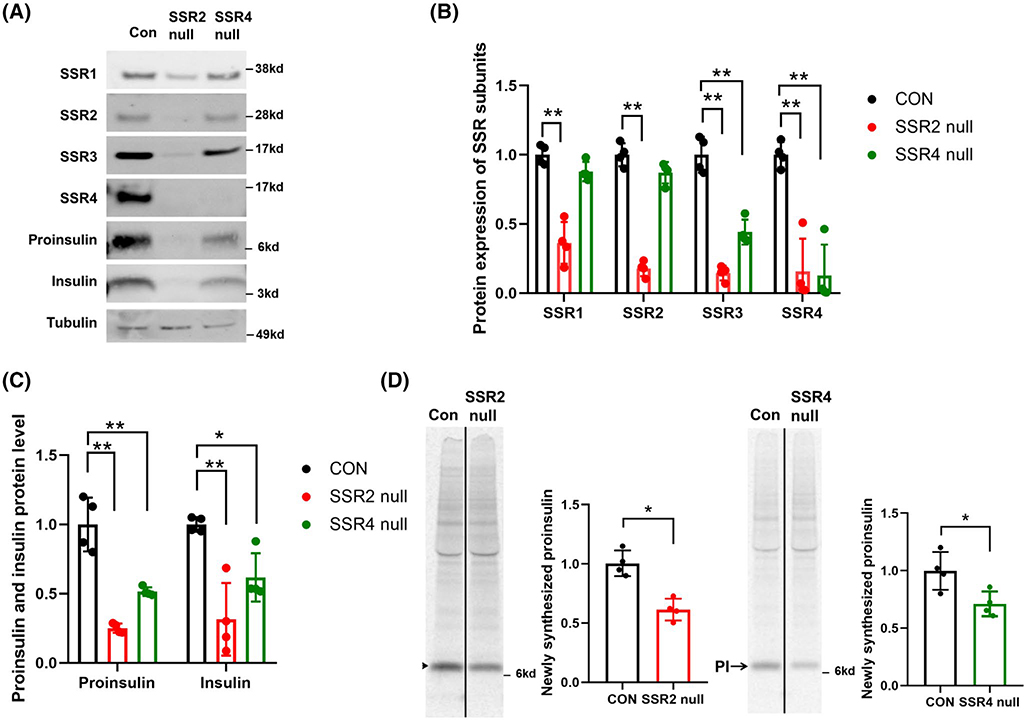
As these phenotypes were obtained with only a modest reduction of TRAPβ/SSR2 or TRAPδ/SSR4 protein levels, we proceeded to generate INS832/13 cells with CRISPR/Cas9-mediated null mutant of these gene products (see Section 2). By immunoblotting in TRAPδ/SSR4 null cells, TRAPδ/SSR4 protein was essentially undetectable, accompanied by a decrease of TRAPγ/SSR3, whereas intracellular protein levels of TRAPα/SSR1 and TRAPβ/SSR2 were nearly unchanged (Figure 2A,B). In contrast, deletion of TRAPβ/SSR2 led to a major decrease of all subunits of the TRAP complex (Figure 2A,B). In parallel with these changes, TRAPδ/SSR4 null cells exhibited a partial defect in the steady state levels of proinsulin (~50% decreased) and insulin (~30% decreased), while TRAPβ/SSR2 null cells exhibited a more severe defect in the levels of proinsulin (~75% decreased) and insulin (~50% decreased; Figure 2C). Diminished insulin content in TRAPδ/SSR4 or TRAPβ/SSR2 null cells was also independently suggested by immunofluorescence with anti-insulin antibodies (Figure S1). Moreover, after pulse labeling for 30 minutes with 35S-amino acids and normalizing to total protein synthesis (TCA-precipitable counts), TRAPδ/SSR4 null cells exhibited an ~30% decrease in the production of newly synthesized proinsulin, and TRAPβ/SSR2 null cells exhibited an ~50% decrease, as measured by anti-insulin immunoprecipitation, SDS-PAGE, and phosphorimaging (Figure 2D, quantitation at right). These data indicate that in TRAPβ/SSR2 or TRAPδ/SSR4 null cells, the predominant basis for the decrease in steady-state levels of proinsulin can be accounted for by inefficient proinsulin biosynthesis, which occurs without change of Ins mRNA levels (Figure S2), pointing to a post-transcriptional effect.
FIGURE 2.
Deletion of TRAPβ/SSR2 or TRAPδ/SSR4 in INS832/13 cells. A, TRAPβ/SSR2 or TRAPδ/SSR4 null cells and corresponding control cells (CON) were immunoblotted with the antibodies indicated. Tubulin is a loading control. B, Quantitation of TRAP/SSR subunits from experiments like that in panel A (n = 4). C, Quantitation of proinsulin and insulin from experiments like that in panel A (n = 4). *P < .05, **P < .01 compared to Control. D, TRAPβ/SSR2 or TRAPδ/SSR4 null cells were pulse-labeled with 35S-Met/Cys for 30 minutes (left panel) before immunoprecipitation with anti-insulin, followed by SDS-PAGE and phosphorimaging. Quantitation of newly-synthesized proinsulin (PI, mean ± SD) is shown *P < .05 compared to Control (n = 4)
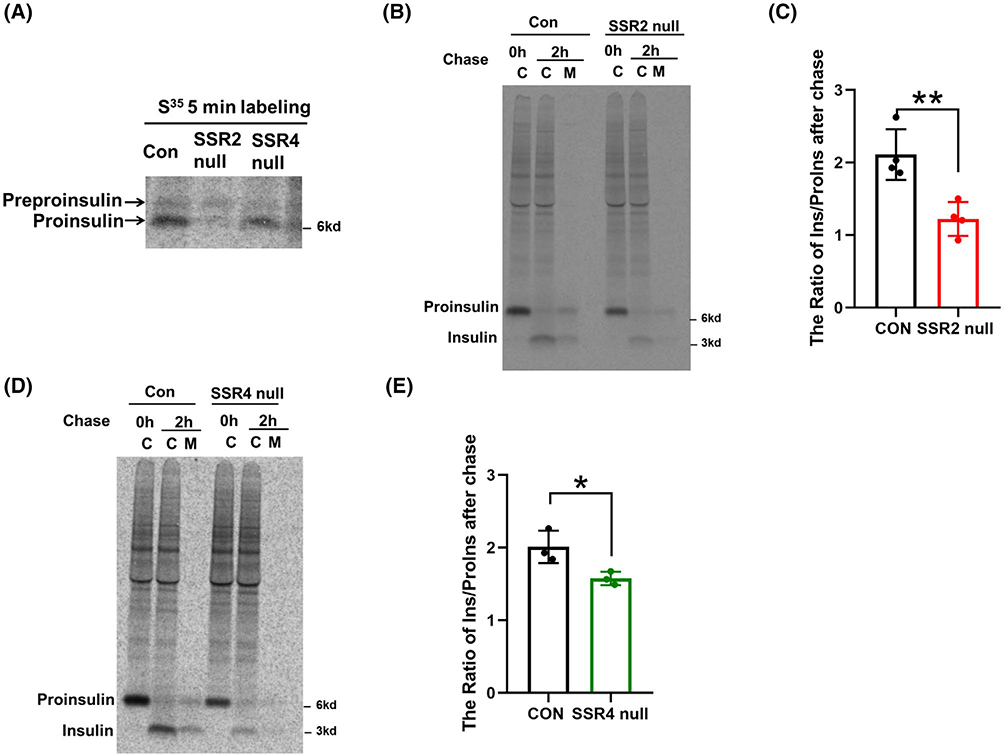
To further understand this effect, we repeated our experiments in TRAPβ/SSR2 or TRAPδ/SSR4 null cells using a pulse-labeling time of 5 minutes, in order to enrich for nascent polypeptide chains (with commensurately longer exposures for phosphorimaging). Reproducibly, TRAPδ/SSR4 null cells yielded a more modest decrease in newly synthesized proinsulin than that in TRAPβ/SSR2 null cells, and under these conditions, newly synthesized preproinsulin became more apparent (Figure 3A). Indeed, the data demonstrated a relative increase in the preproinsulin-to-proinsulin ratio in TRAPδ/SSR4 null cells and an even greater increase in TRAPβ/SSR2 null cells, consistent with the idea that these TRAP/SSR subunits contribute to the efficiency of preproinsulin translocation,7,14 which underlies the efficiency of proinsulin biosynthesis (Figures 2D and 3A).
FIGURE 3.
Effect of TRAPβ/SSR2 or TRAPδ/SSR4 deficiency on insulin biogenesis. A, TRAPβ/SSR2 or TRAPδ/SSR4 deficient cells and control cells were pulse-labeled with 35S-Met/Cys for 5 minutes. The cells were lysed and immunoprecipitated with anti-insulin and analyzed by 4%−12% NuPage gel and phosphorimaging. B, TRAPβ/SSR2 or TRAPδ/SSR4 deficient cells were pulse-labeled for 30 minutes followed by 0 or 2 hours chase in complete culture medium. Lysates of cells (C) and 2 hours chase medium (M) were immunoprecipitated with anti-insulin and resolved by 4%−12% NuPage and phosphorimaging. The newly-synthesized insulin / proinsulin ratio after 2 hours chase from experiments like those in panel B or D was quantitated (mean ± SD; n = 4 for Figure 3B and n = 3 for Figure 3D) and is shown in panels C and E, respectively. *P < .05, **P < .01 compared to Control
3.2 |. TRAP-deficient β-cells exhibit an additional defect in proinsulin processing, but not stimulated secretion
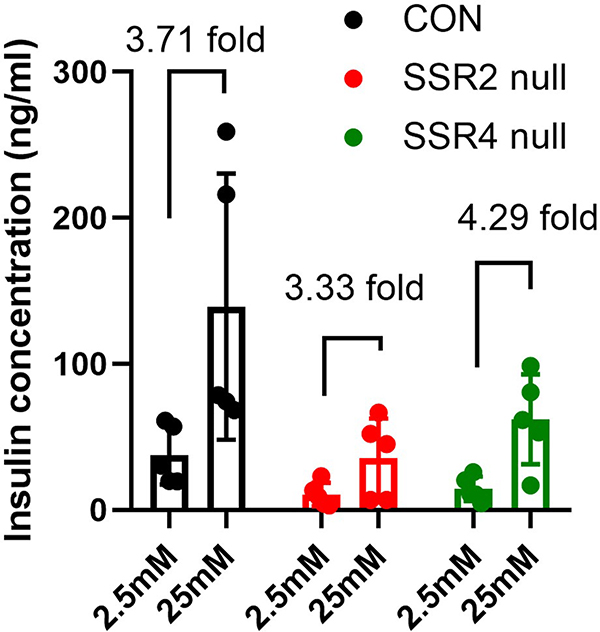
Although much of the decrease in proinsulin and insulin levels in TRAPβ/SSR2 or TRAPδ/SSR4 deficient β-cells could be accounted for diminished proinsulin biosynthesis, we extended our experiments to include a 2 hours chase, a time that is suitably designed to measure the efficiency of proinsulin-to-insulin conversion.18 At this chase time, by anti-insulin immunoprecipitation, SDS-PAGE, and phosphorimaging, TRAPβ/SSR2 null cells exhibited an ~40% decrease in proinsulin-to-insulin conversion (Figure 3B,C), and TRAPδ/SSR4 null cells exhibited an ~30% decrease in proinsulin-to-insulin conversion (Figure 3D,E). However, when TRAPβ/SSR2 null and TRAPδ/SSR4 null β-cells were acutely challenged, the glucose-stimulated insulin secretion in each case rose approximately fourfold (Figure 4). Together, these data indicate that a proinsulin processing defect coupled with diminished proinsulin biosynthesis account for the low steady state levels of proinsulin and insulin in TRAPβ/SSR2 null and TRAPδ/SSR4 null β-cells, although these cells are still able to proportionately release insulin from the (decreased) granule storage pool.
FIGURE 4.
Glucose-stimulated insulin secretion in TRAPβ/SSR2 or TRAPδ/SSR4 deficient cells. The cells (including control cells – CON) were incubated with prewarmed KRBH buffer containing 2.5 mM glucose for 2 hours followed by KRBH buffer containing 25 mM glucose for an additional 2 hours, as shown on the X-axis. Insulin released to the media was measured by ELISA (mean ± SD; n = 5)
3.3 |. Rescue of TRAP-deficient β-cell phenotypes upon re-expression of missing TRAP subunits
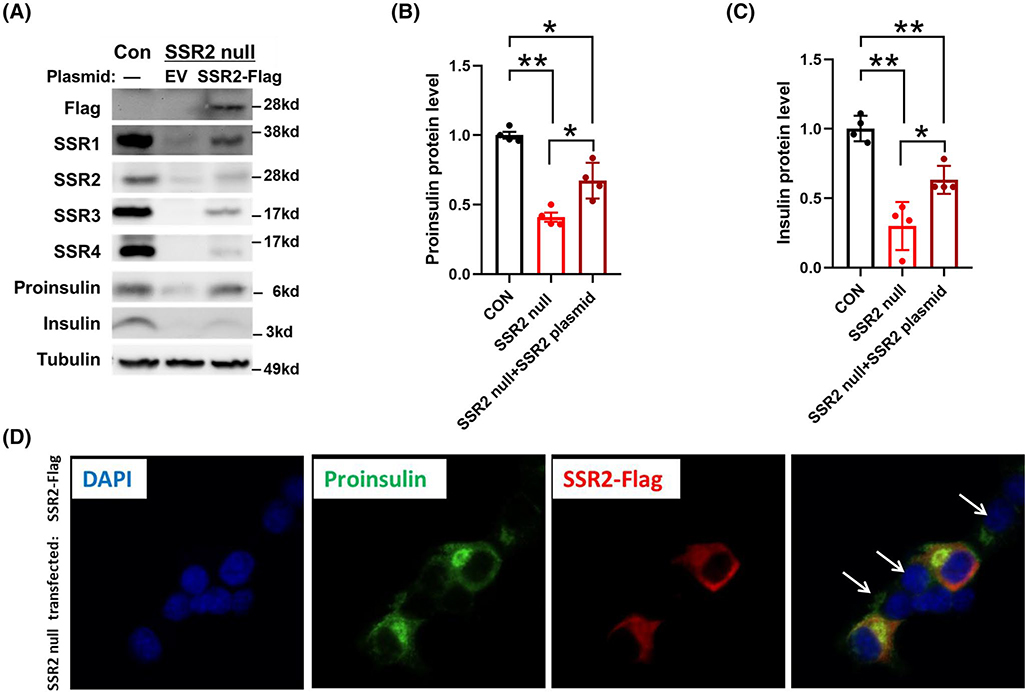
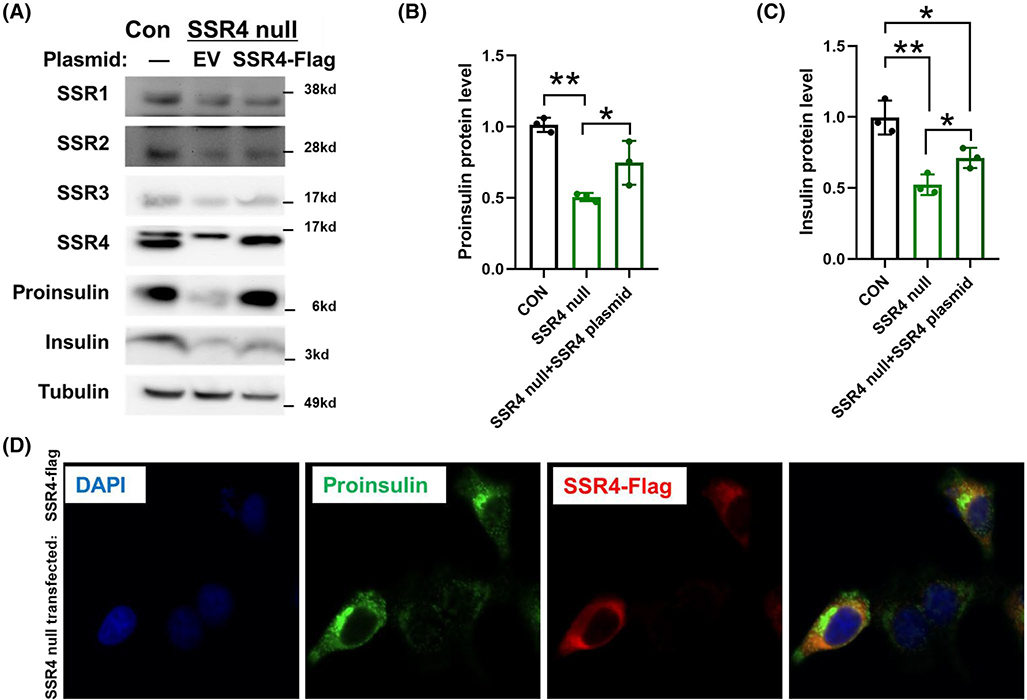
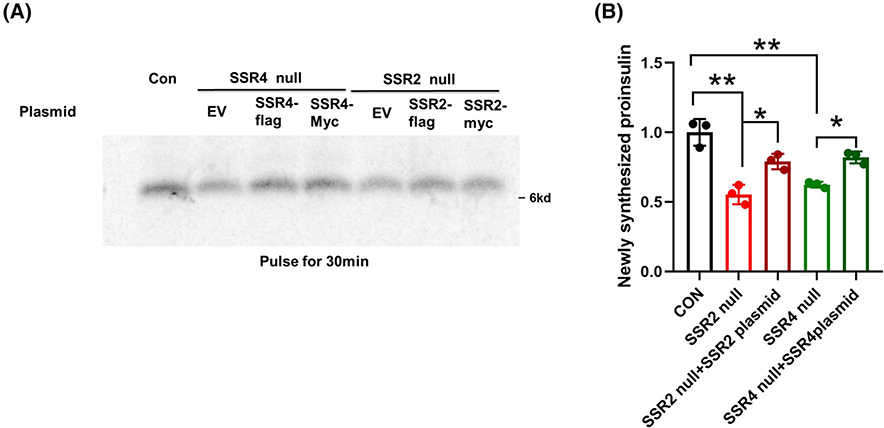
To confirm that the phenotype of CRISPR/Cas9 mediated TRAPβ/SSR2 deficient β-cells did not involve off-target genetic disruption, we transfected these cells to re-express Flag-tagged TRAPβ/SSR2. Successful transfection restored detectability of all the TRAP/SSR subunits (Figure 5A, upper panels), while partially rescuing the steady levels of proinsulin and, to a lesser extent, insulin (Figure 5A, lower panels; Figure 5B,C). As transfection of INS832/13 cells is inefficient, we could readily compare untransfected and transfected cells side by side upon immunofluorescence with anti-Flag antibodies. From this approach, it was apparent that for TRAPβ/SSR2 null cells in which TRAPβ/SSR2 protein expression was restored, proinsulin robustly re-appeared (Figure 5D). A generally similar outcome was observed in TRAPδ/SSR4 deficient β-cells transfected to re-express Flag-tagged TRAPδ/SSR4 (Figure 6A, upper panels), with increased proinsulin levels and partial restoration of insulin levels (Figure 6A, lower panels; Figure 6B,C). Additionally, transfected TRAPδ/SSR4 null cells also clearly differed from untransfected cells upon side-by-side comparison of proinsulin levels observed by immunofluorescence (Figure 6D). Moreover, re-expression of either Flag-tagged or Myc-tagged versions of the missing TRAP/SSR subunit increased the recovery of newly synthesized proinsulin in TRAPδ/SSR4 or TRAPβ/SSR2 deficient β-cells, respectively (Figure 7A,B), confirming that expression of these subunits are necessary for optimal proinsulin biosynthesis. Altogether, these data indicate that the decreased proinsulin level observed in TRAPβ/SSR2 and TRAPδ/SSR4 null mutated β-cells is a defect attributable to loss of specific TRAP/SSR subunits, rather than off-target effects.
FIGURE 5.
TRAPβ/SSR2 re-expression rescues proinsulin protein levels in TRAPβ/SSR2 null cells. A, TRAPβ/SSR2 null cells were transfected with empty vector (EV) or plasmid encoding Flag-tagged TRAPβ/SSR2. At 48 hours post transfection, these cells or INS832/13 wild type cells (Con) were lysed and immunoblotted with the indicated antibodies. Tubulin is a loading control. Quantitation of proinsulin and insulin from experiments like that in panel A was shown as panel B and C, respectively (n = 4). *P < .05, **P < .01compared to Control. D, Immunofluorescence of proinsulin (green) and Flag (red) in TRAPβ/SSR2 null cells transfected with Flag-tagged TRAPβ/SSR2 plasmid, containing both transfected and untransfected cells. Nuclei were counter-stained with DAPI (blue). For clarity in the merged image, white arrows point to the nuclei of some of the untransfected cells
FIGURE 6.
TRAPδ/SSR4 re-expression rescues proinsulin protein levels in TRAPδ/SSR4 deficient cells. A, TRAPδ/SSR4 null cells were transfected with empty vector (EV) or plasmid encoding Flag-tagged TRAPδ/SSR4. At 48 hours post transfection, these cells or INS832/13 wild type cells (Con) were lysed and immunoblotted with the indicated antibodies. Tubulin is a loading control. The proinsulin and insulin level presented in panel A was quantified as panel B and C (n = 3). *P < .05, **P < .01compared to Control. D, Immunofluorescence of proinsulin (green) and Flag (red) in TRAPδ/SSR4 null cells transfected with Flag-tagged TRAPδ/SSR4 plasmid, containing both transfected and untransfected cells. Nuclei were counter-stained with DAPI (blue)
FIGURE 7.
Re-expression of missing TRAP/SSR subunits increases newly-synthesized proinsulin in TRAPδ/SSR4 or TRAPβ/SSR2 deficient β-cells. A, TRAPδ/SSR4 or TRAPβ/SSR2 deficient β-cells were transfected with plasmids encoding Myc-tagged or Flag-tagged TRAPδ/SSR4 or TRAPβ/SSR2-KO, respectively. Both tags worked similarly. At 48 hours post transfection, the cells were pulse-labeled with 35S-Met/Cys for 30 minutes, followed by immunoprecipitation with anti-insulin, and analysis by 4%−12% NuPage gel and phosphorimaging. B, Quantitation of newly-synthesized proinsulin (mean ± SD) from experiments like those in panel A is shown. *P < .05, **P < .01compared to Control (n = 3)
3.4 |. TRAPα/SSR1 is an extragenic suppressor of TRAPβ/SSR2 or TRAPδ/SSR4 null mutant
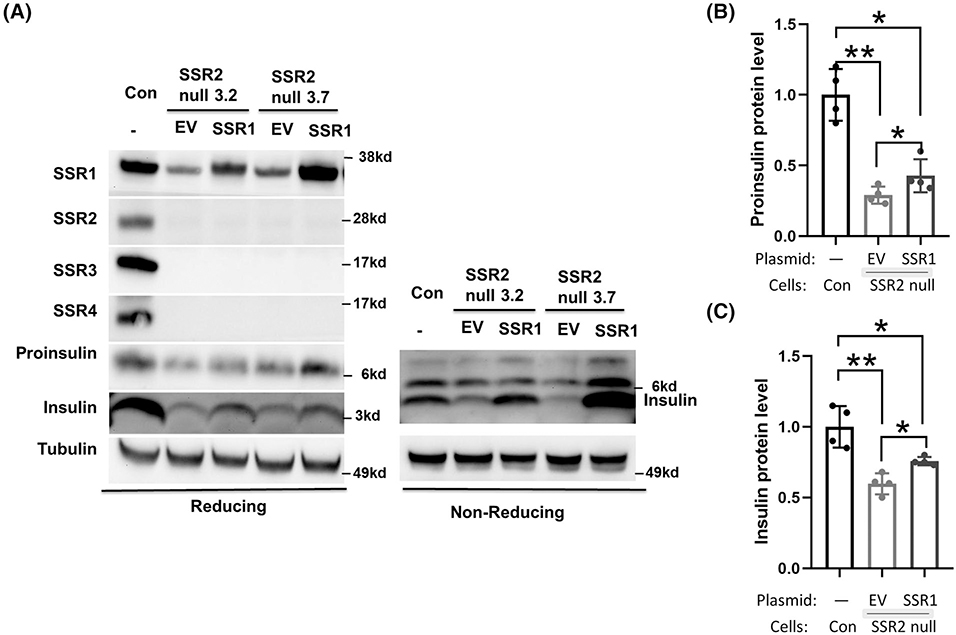
TRAPα/SSR1, TRAPβ/SSR2, and TRAPδ/SSR4 all have considerable exposure on the luminal side of the ER membrane,13 and of these three subunits, deficiency of TRAPα/SSR1 may have the most dramatic impact on preproinsulin translocation,7 whereas deficiency of TRAPβ/SSR2 or TRAPδ/SSR4 appears to have progressively weaker effects (this report). Recently, it has been proposed that TRAPβ/SSR2 offers a critical and selective interaction with nascent preproinsulin on the cytosolic side of the ER membrane.14 If so, we reasoned that the diminished levels of proinsulin and insulin observed in TRAPβ/SSR2 deficient β-cells would not be rescued by increased expression of TRAPα/SSR1. However, to our surprise, in two different pools of TRAPβ/SSR2 null cells, overexpression of TRAPα/SSR1 partially rescued proinsulin and insulin levels despite persistent absence of other TRAP/SSR subunits (Figure 8). These data suggest the possibility that instead of or in addition to acting as a preproinsulin interactor on the cytosolic side of the ER membrane, a crucial role of TRAPβ/SSR2 is to support the stability, and thus the function, of TRAPα/SSR1 in insulin biogenesis. TRAPβ/SSR2 strongly interacts on the ER luminal side with TRAPα/SSR1,13 which works in concert with TRAPδ/SSR4 not only to communicate with other resident protein complexes in the ER membrane12 but also to promote proinsulin and insulin levels in pancreatic β-cells (Figure S3).
FIGURE 8.
Overexpression of TRAPα/SSR1 partially rescues proinsulin and insulin levels in TRAPβ/SSR2 deficient β-cells. A, Two independent TRAPβ/SSR2-deficient clones were transfected with plasmid encoding Myc-tagged TRAPα/SSR1. At 48 hours post-transfection, cell lysates were subjected to immunoblotting under reducing conditions with the antibodies shown, or non-reducing conditions (for insulin). Tubulin is a loading control. Quantitation of proinsulin and insulin bands (mean ± SD) from experiments like those in panel A is shown in B and C, respectively (n = 4). *P < .05, **P < .01
4 |. DISCUSSION/CONCLUSIONS
A Genome-wide trans-ancestry meta-analysis identified TRAPα/SSR1 as a risk allele of T2D, and its SNPs are associated with an increased fasting blood glucose and reduced HOMA-β, suggesting that SSR1 may have a primary role in susceptibility of type 2 diabetes through β-cell dysfunction. Indeed, our recent report has indicated that TRAPα/SSR1 deletion in pancreatic β-cells impairs both preproinsulin translocation and additional steps that impact quantitatively on insulin secretion.7 The ER luminal domain of TRAPα/SSR1 heterodimerizes with TRAPβ/SSR2 and is also associated with the luminal domain of TRAPδ/SSR4 aligned alongside.13 Evidence from fibroblasts of patients with congenital disorder of glycosylation has suggested that depletion of TRAPδ/SSR4 is associated with a milder loss of other TRAP/SSR subunits than is seen upon depletion of TRAPγ/SSR3,13 which itself produces only a partial defect19,20 compared to an even more severe loss of TRAP/SSR subunits upon depletion of TRAPβ/SSR2 (this report). Together, the TRAP/SSR complex participates in gating the Sec61 translocon15 and associating with other protein complexes at the ER membrane.21 TRAP engagement in secretory polypeptide translocation appears rather closely timed to follow early events including signal peptide transfer to Sec61 and displacement of the signal recognition particle (SRP).12 Here we have analyzed in pancreatic β-cells the impact of depletion of the TRAPα/SSR1 luminal partners, TRAPβ/SSR2 and TRAPδ/SSR4. We find that even mild depletion of these subunits by siRNA-mediated knockdown leads to decreased proinsulin and insulin levels (Figure 1), and the phenotype is exacerbated in CRISPR/Cas9-mediated deletion (Figure S1) with TRAPβ/SSR2 more severe than TRAPδ/SSR4-deficient β-cells (Figure 2). Most of the diminished proinsulin and insulin levels can be accounted for by diminished proinsulin biosynthesis without change in INS mRNA levels (Figure S2)—compatible with defective co-translational translocation of preproinsulin into the ER lumen—but there is an additional decrease in the efficiency of insulin biogenesis from proinsulin (Figure 3). The net result is diminished insulin storage, although the actual fold-increase in glucose-stimulated insulin secretion appears equally robust TRAP/SSR-deficient cells (Figure 4). The proinsulin biosynthesis defect, as well as steady-state proinsulin and insulin levels, are at least partially reversed upon re-expression of the missing TRAP/SSR subunit (Figures 5–7 and S3).
Interestingly, deficiency/deletion of TRAPβ/SSR2 or TRAPδ/SSR4 decreases protein levels of other partner subunits (Figures 1B and 2A) with no effect on the mRNAs of corresponding subunits (Figure S4), suggesting that deficiency of SSR2/4 does not impair transcription of other subunits but affects their stability. Indeed, Nagasawa et al reported that SSR1 and SSR4 protein levels were reduced in the cells transfected with shRNAs of for SSR2 and depletion of SSR2 disrupted formation of the TRAP complex.22,23 In addition, a disease causing SSR4 mutant decreased protein levels of other SSR subunits.24 Importantly, steady-state proinsulin and insulin levels are also at least partially reversed upon overexpression from a plasmid encoding TRAPα/SSR1 even in cells devoid of TRAPβ/SSR2 (Figure 8). These data suggest that TRAPβ/SSR2 might not play an essential independent role in proinsulin and insulin biosynthesis but rather might play its essential role through heterodimerization and stabilization of TRAPα/SSR1, which is known to be critical for proinsulin and insulin production7—with the suggestion of a similar finding in cells deficient for TRAPδ/SSR4 (Figure S3).
Co-translational translocation is linked to the translational de-repression that occurs when SRP dissociates from nascent preproinsulin at the ER membrane.25 There is currently no published evidence to indicate that TRAP association with the translocon participates in SRP-mediated translational de-repression of prepoinsulin biosynthesis. However, in a 5-min pulse labeling of pancreatic β-cells lacking a functional TRAP/SSR complex, we do not detect an actual accumulation newly synthesized preproinsulin but rather a diminution of newly synthesized proinsulin, suggesting immediate feedback of the translocation defect to the protein biosynthetic machinery (Figure 3A). Although they do not clarify whether the molecular mechanism specifically involves SRP dissociation, these data suggest a regulated coupling between preproinsulin translocation and the proinsulin biosynthesis that is required to maintain sufficient insulin storage for glycemic control in vertebrate organisms.
Supplementary Material
ACKNOWLEDGMENTS
The Liu lab is supported by research grants from National Key R&D Program of China 2019YFA0802502; the National Natural Science Foundation of China 81620108004, 81830025, and 81700699; and support from the Tianjin Municipal Science and Technology Commission (17ZXMFSY00150 and 18JCQNJC82100). The Arvan lab is supported by NIH RO1-DK48280 and R01-DK111174. We also acknowledge support of the Protein Folding Diseases Initiative of the University of Michigan. We thank members of the Peter Arvan, Ming Liu, Ling Qi, and Billy Tsai laboratories for helpful interactions and discussion.
Abbreviations:
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- SRP
signal recognition particle
- SSR
signal sequence receptor
- TRAP
translocon-associated protein
- TCA
trichloroacetic acid
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the Supporting Information section.
REFERENCES
- 1.Schuit FC, In’t Veld PA, Pipeleers DG. Glucose stimulates proinsulin biosynthesis by a dose-dependent recruitment of pancreatic beta cells. Proc Natl Acad Sci U S A. 1988;85:3865–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo H, Xiong Y, Witkowski P, et al. Inefficient translocation of preproinsulin contributes to pancreatic beta cell failure and late-onset diabetes. J Biol Chem 2014;289:16290–16302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu M, Weiss MA, Arunagiri A, et al. Biosynthesis, structure, and folding of the insulin precursor protein. Diabetes Obes Metab 2018;20(Suppl 2):28–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaguchi J, Conlon DM, Liang JJ, Fisher EA, Ginsberg HN. Translocation efficiency of apolipoprotein B is determined by the presence of beta-sheet domains, not pause transfer sequences. J Biol Chem 2006;281:27063–27071. [DOI] [PubMed] [Google Scholar]
- 5.Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science (New York, N Y ). 2014;346:1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo H, Sun J, Li X, et al. Positive charge in the n-region of the signal peptide contributes to efficient post-translational translocation of small secretory preproteins. J Biol Chem 2018;293: 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Itani OA, Haataja L, et al. Requirement for translocon-associated protein (TRAP)alpha in insulin biogenesis. Sci Adv 2019;5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann E, Gorlich D, Kostka S, et al. A tetrameric complex of membrane proteins in the endoplasmic reticulum. Eur J Biochem 1993;214:375–381. [DOI] [PubMed] [Google Scholar]
- 9.Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J Cell Biol 2003;160:529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webb GC, Akbar MS, Zhao C, Steiner DF. Expression profiling of pancreatic beta cells: glucose regulation of secretory and metabolic pathway genes. Proc Natl Acad Sci U S A. 2000;97:5773–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holthuis JC, van Riel MC, Martens GJ. Translocon-associated protein TRAP delta and a novel TRAP-like protein are coordinately expressed with pro-opiomelanocortin in Xenopus intermediate pituitary. Biochem J 1995;312(Pt 1):205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gemmer M, Forster F. A clearer picture of the ER translocon complex. J Cell Sci 2020;133. 10.1242/jcs.231340 [DOI] [PubMed] [Google Scholar]
- 13.Pfeffer S, Dudek J, Schaffer M, et al. Dissecting the molecular organization of the translocon-associated protein complex. Nat Commun 2017;8:14516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kriegler T, Kiburg G, Hessa T. Translocon-associated protein complex (TRAP)is crucial for co-translational translocation of preproinsulin. J Mol Biol 2020;432:166694. [DOI] [PubMed] [Google Scholar]
- 15.Lang S, Nguyen D, Pfeffer S, Forster F, Helms V, Zimmermann R. Functions and mechanisms of the human ribosome-translocon complex. Subcell Biochem 2019;93:83–141. [DOI] [PubMed] [Google Scholar]
- 16.Liu M, Hodish I, Rhodes CJ, Arvan P. Proinsulin maturation, misfolding, and proteotoxicity. Proc Natl Acad Sci U S A. 2007;104:15841–15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen D, Stutz R, Schorr S, et al. Proteomics reveals signal peptide features determining the client specificity in human TRAP-dependent ER protein import. Nat Commun 2018;9:3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steiner DF, Park SY, Stoy J, Philipson LH, Bell GI. A brief perspective on insulin production. Diabetes Obes Metab 2009;11 (Suppl 4):189–196. [DOI] [PubMed] [Google Scholar]
- 19.Ng BG, Lourenco CM, Losfeld M-E, et al. Mutations in the translocon-associated protein complex subunit SSR3 cause a novel congenital disorder of glycosylation. J Inherit Metab Dis 2019;42:993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dittner-Moormann S, Lourenco CM, Reunert J, et al. TRAPgamma-CDG shows asymmetric glycosylation and an effect on processing of proteins required in higher organisms. J Med Genet 2020;jmedgenet-2019–106279. [DOI] [PubMed] [Google Scholar]
- 21.Braunger K, Pfeffer S, Shrimal S, et al. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science (New York, N Y ). 2018;360:215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Replication DIG, Meta-analysis C, Asian Genetic Epidemiology Network Type 2 Diabetes C, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 2014;46:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagasawa K, Higashi T, Hosokawa N, Kaufman RJ, Nagata K. Simultaneous induction of the four subunits of the TRAP complex by ER stress accelerates ER degradation. EMBO Rep 2007;8:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Losfeld ME, Ng BG, Kircher M, et al. A new congenital disorder of glycosylation caused by a mutation in SSR4, the signal sequence receptor 4 protein of the TRAP complex. Hum Mol Genet 2014;23:1602–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolin SL, Walter P. Discrete nascent chain lengths are required for the insertion of presecretory proteins into microsomal membranes. J Cell Biol 1993;121:1211–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.