Abstract
Inflammatory bowel disease (IBD) is a chronic inflammatory disease affecting various parts of the gastrointestinal tract. A majority of the current evidence points out the involvement of intestinal dysbiosis in the IBD pathogenesis. Recently, the association of intestinal fungal composition With IBD susceptibility and severity has been reported. These studies suggested gene polymorphisms in the front line of host defense against intestinal microorganisms are considered to play a role in IBD pathogenesis. The studies have also detected increased susceptibility to fungal infections in patients carrying IBD-related mutations. Therefore, a literature search was conducted in related databases to review articles addressing the mycobiota-genotype association in IBD.
Keywords: Inflammatory bowel disease, IBD, Fungal microbiota, Intestinal mycobiota, Single nucleotide polymorphisms, SNPs
Inflammatory bowel disease pathogenesis
Inflammatory bowel disease (IBD) is a chronic relapsing disease affecting various parts of the gastrointestinal tract and encompasses two common disorders: Crohn’s disease (CD) and Ulcerative Colitis (UC). IBD is a worldwide issue, especially in urban and westernized countries among young individuals [1], assumed to result from an improper and continuous inflammatory response to commensal microbes in a genetically susceptible host [2]. So far, the pathogenesis of the disease is considered to be a combination of genetic predisposition and environmental factors. The majority of current evidence emphasizes the involvement of intestinal dysbiosis in IBD pathogenesis [3]. While intestinal epithelial cells (IECs) are constantly exposed to microbial components; they are regarded not only as a structural but also a functional barrier in the front line of host defense against intestinal microorganisms. The functional alteration of these cells is hypothesized to be associated with IBD [4]. Bacteria as the predominant organisms of the gastrointestinal tract gained the greatest attention in IBD microbial studies [5–7]. Nonetheless, the association of intestinal fungal composition with mucosal inflammation in both CD and UC has recently become into consideration [8–11]. Of note, increased IBD flares were associated with increased global fungal load accompanied by alteration of certain fungal species in the microbiota [12–14].
To date, numerous gene polymorphisms are found to be connected to IBD susceptibility [15] and severity; for instance, an increased colitis severity was driven by activation of Leucine-rich repeat kinase 2 (LRRK2), an important enzyme that regulates innate immunity through nuclear factor kappa B (NF-κB) signaling pathway [16]. Some articles studied the association of specific intestinal bacterial microbiota with gene polymorphisms [17, 18]. However, few have focused on the role of fungal subsets in the intestine. The purpose of this study was to discuss the association of fungal flora with IBD and review the articles connecting the gene polymorphisms with intestinal mycobiota in IBD cases.
Anti-Saccharomyces cerevisiae antibody
The first sparks of fungi role in IBD pathogenesis flared by detecting elevated levels of anti-Saccharomyces cerevisiae mannan antibodies (ASCA) in the sera of IBD-affected patients since the early 90 s [19, 20]. A twin study in 2005 has detected ASCA in CD cases more frequently compared with healthy controls [21]. ASCA was also found commonly in CD patients with a positive family history of IBD [22] and even in unaffected relatives of CD patients [23]. ASCA was not only detected in answer to Saccharomyces antigens but also in response to Candida albicans or the presence of anti-β2 glycoprotein I antibodies in CD patients [24, 25]. Marrakchi et al. revealed a positive correlation of caspase recruitment domain-containing protein 15 (CARD15)/nucleotide-binding oligomerization domain-containing protein 2 (NOD2) gene mutation, an important intracellular pattern recognition receptor (PRR) that is expressed by dendritic cells (DCs), macrophages, and IECs [26], with ASCA expression in IBD-affected patients [27].
IBD affecting intestinal mycobiota
In addition to animal studies, some articles are conveying the alteration of intestinal mycobiome in human subjects with IBD. Ott et al. first described significantly higher fungal diversity in patients with CD in comparison with healthy controls, albeit no disease-specific fungal species were present in the CD and UC group [28]. Ever since, many studies have consistently shown an elevated abundance of Candida sp. in IBD fecal samples [29–31]. Lewis et al. have reported an increased amount of S. cerevisiae [29], whereas Hoarau et al. reported a reduction in intestinal S. cerevisiae abundance in IBD patients [31]. Another study in 2009 reported a significantly elevated C. albicans population obtained from fecal samples of CD patients (44%) and their healthy relatives (38%) compared to healthy controls [22]. Li et al. assessed 19 patients with active CD and 7 healthy individuals and discovered increased fecal fungal richness and diversity in C. albicans, Aspergillus clavatus, Cryptococcus neoformans, and a decrease in S. cerevisiae in CD patients. The diversity of the fecal fungal community was also positively correlated with serum C-reactive protein level and the CD activity index [13]. Another study in 2016, revealed a significant increase in global fungal load in both inflamed and non-inflamed mucosa compared with healthy subjects (HS). However, no significant differences in fungal diversity were observed between the groups [12].
Unlike most similar articles, Chehoud et al. demonstrated pediatric IBD to be associated with reduced fungal diversity in the host gut microbiota. Specific Candida taxa were also found to have increased abundance in the IBD samples [30]. An additional study with de-novo pediatric IBD cases revealed a shift from the Ascomycota-predominant mycobiota in HS to a different fungal spectrum with a predominance of Basidomycetes in patients with de-novo IBD without the conflicting impact of antibiotics or immunosuppression [32]. Later, another study investigated the possible fungal dysbiosis index in IBD; the fecal fungal composition of 235 patients with IBD and 38 HS showed an increased Basidiomycota-to-Ascomycota ratio that was dramatically higher in patients with IBD flares compared to patients in remission and HS [8]. There was also a negative correlation between the abundance of S. cerevisiae and C. albicans in fecal samples of IBD subjects, suggesting a competitive environment between these two species in the gut [8, 33]. The study also described a complex fungal-bacterial interaction in the fecal composition of subjects [8].
As opposed to Sokol and Mukhopadhya et al., Qiu and colleagues did not detect any significant difference in the abundance of Ascomycota, Basidiomycota, and the ratio of Ascomycota-to-Basidiomycota between the HS and UC patients. However, there was a prominent variation in the abundance of Aspergilli between the groups [11]. A recent report studied the cultivable intestinal mycobiota presented in feces obtained from 34 pediatric CD patients, 27 pediatric UC patients, and 32 healthy children. The authors observed increased load of S. cerevisiae and Candida sp. in IBD patients, which was in line with previous studies. Likewise, Di Paola et al. concluded that the presence of S. cerevisiae was associated with a favorable intestinal environment for beneficial bacterial genera, such as Faecalibacterium; whereas the absence of normal fungal flora or presence of unusual fungal species were conjugated with the presence of potential pathogenic bacteria that might lead to IBD [34]. The latest article by Nelson et al. reported an increased abundance of Candida sp. and a decreased Basidiomycota-to-Ascomycota ratio, in contrast to the previous literature, in CD cases [35]. Of note, the discrepancies between these studies might stem from different fungal extraction methods. In this regard, we provided additional information for these studies, including the fungal extraction method and the sample source, in Table 1.
Table 1.
IBD affecting intestinal mycobiota in IBD patients
| Number of patients | Sample/method | Result | References |
|---|---|---|---|
|
57 IBD patients 47 HS |
Intestinal mucosa 18S rDNA-based sequencing |
Significant higher fungal diversity in patients with CD in comparison with HS. No disease-specific fungal species were found in the CD and UC group | Ott et al. [28] |
|
41 CD families composed of: 129 patients and 113 healthy relatives 14 healthy controls families composed of 76 individuals |
Mouth swabs and Stool samples were processed using chromogenic medium. Mouth swabs were rubbed directly onto the medium. Stool samples were taken with an inoculation loop. Plates were incubated for 48 h at 37°℃C. The yeast species were differentiated using the specific color of the colonies. Presumptive identification of yeast species was confirmed by either Bichro-Latex-albicans for C. albicans, or the API 32C system for other species |
Top most prevalent mycobiome in CD patients: C. albicans mouth [26 (34.7%)] stool [13( 22%)] C. glabrate mouth [3 (4%)], stool [1 (1.7%)] C. tropicalis mouth [1 (1.3%)], stool 0 |
Standaert-Vitse et al. [22] |
|
19 patients with active CD 7 HS |
PCR targeting fecal fungal 18S rDNA gene | Decreased S. cerevisiae and overrepresented Aspergillus clavatus, C. albicans, and Cryptococcus neoformans proportions were present in CD patients | Li et al. [13] |
|
90 children with CD 26 HS children |
Sequence was acquired using the Illumina HiSeq method (Illumina) | Five yeasts including, S. cerevisiae, C. lusitaniae, Pichia jadinii (also known as and C. utiliz), C. albicans, and Kluyveromyces marxianus were positively associated with CD, particularly in the setting of greater bacterial dysbiosis | Lewis et al. [29] |
|
32 patients with IBD 90 HS |
PCR primers targeting fecal fungal the ITS rDNA gene | IBD samples had significantly lower fungal diversity The most commonly observed fungi were C. Pichia jadinii. C. parapsilosis, was also more common in the pediatric IBD samples. Cladosporium cladosporioides, was more common in HS | Chehoud et al. [30] |
|
9 multiplex families comprising 20 CD patients and their 28 cohabiting NCDR 4 unrelated healthy families 21 individuals with no history of CD (NCDU) living in the same geographic area |
PCR primers targeting fecal fungal ITS1 rDNA gene | Increased richness in the NCDU group compared to the CD or NCDR group but no difference in the mycobiome richness of CD patients and their healthy relatives. S. cerevisiae tended to increase in healthy (NCDR) individuals. C. tropicalis was significantly abundant in CD compared to NCDR group | Hoarau et al. [31] |
|
23 CD patients (16 in flare, 7 in remission) 10 HS |
Colonic mucosa ITS2, 16S, and 18S rDNA sequencing | Global fungi load was significantly increased in both inflamed and non-inflamed mucosa compared to HS. However, no significant differences in fungal diversity between the studied groups were observed | Liguori et al. [12] |
|
25 children with IBD 12 HS |
Colonic mucosa 18S rDNA sequencing |
A shift from the Ascomycota-predominant microbiota in HS to a different fungal spectrum with Basidomycetes predominance in patients with de-novo IBD | Mukhopadhya et al. [32] |
|
235 IBD patients 38 HS |
PCR primers targeting fecal fungal ITS2 rDNA gene | S. cerevisiae reduction in patients with IBD (vs. healthy controls) and with flare (vs. remission). Higher Basidiomycota-to-Ascomycota abundance ratio in patients with IBD in flare (either UC or CD) but normal ratio in remission | Sokol et al. [8] |
| 14 UC patients15 HS | PCR primers targeting fecal fungal ITS1 and ITS2 rDNA gene | Wickerhamomyces, an unidentified genus of Saccharomycetales, Aspergillus, Sterigmatomyces, and Candida sp. showed an increasing trend in UC patients compared with HS. There was a marked difference in Aspergillus abundance between the groups. The proportions of Ascomycota and Basidiomycota were not significantly different between the groups | Qiu et al. [11] |
| 93 pediatric; 34 CD, 27 UC patients, 32 HS | PCR primers targeting fecal fungal ITS1-5.8-S-ITS2 regions of rDNA gene |
S. cerevisiae (n = 7 fecal samples) and other yeasts (Candida sp.; n = 5 samples) isolated from 19 CD patients S. cerevisiae is associated with a favorable gut environment for beneficial bacterial genera. Whilst, the absence of yeasts or the presence of other yeast species is connected with potential pathogenic bacteria |
Di Paola et al. [34] |
|
34 CD patients 47 HS without GI disease |
PCR primers targeting fecal fungal ITS1 rDNA gene | Candida sp. was most associated with CD and Cryptococcus sp. with non-CD. The Basidiomycota/Ascomycota abundance ratio was found to be significantly lower in CD patients | Nelson et al. [35] |
CD, Crohn disease; IBD, Inflammatory bowel disease; UC, Ulcerative colitis; ITS 1,2, Internal transcribed spacer 1,2; HS, Healthy subjects; C. albicans, Candida albicans; C. Tropicalis, Candida tropicalis; C. glabrate, Candida glabrate; sp., species; C. Pichia jadinii, Candida Pichia jadinii, C. parapsilosis, Candida parapsilosis; M. restricta, Malassezia restricta; M. sympodialis, Malassezia sympodialis; S. cerevisiae, Saccharomyces cerevisiae; NCDR, Non-CD relatives
Innate immunity against fungi
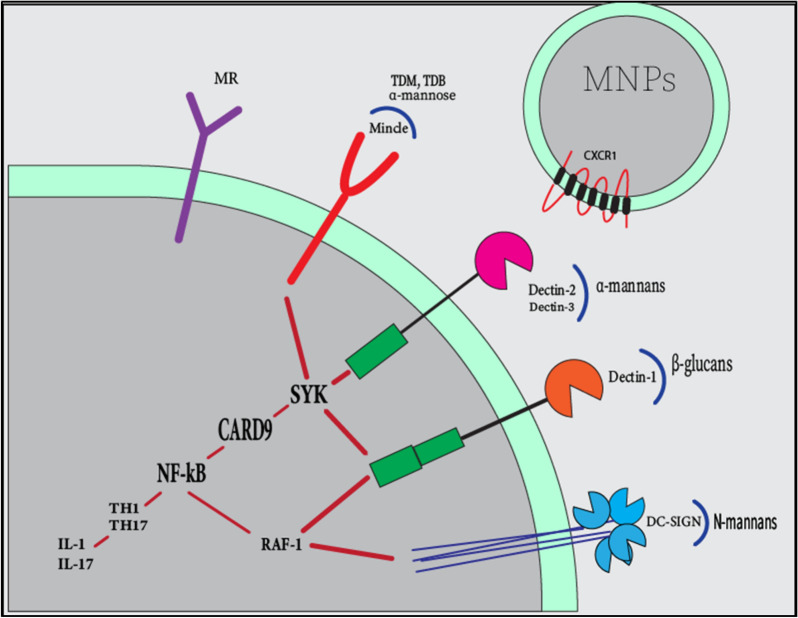
Several genetic polymorphisms have been detected in IBD over the years [15, 36]. The connection between various genetic polymorphisms with bacterial species in IBD patients has been widely studied [37–39]. Increased susceptibility to systemic fungal complications, such as candidemia was linked to polymorphisms of Interleukin 10 (IL-10) (rs1800896) [40], Toll-like receptors 1 (TLR-1) (rs5743611, rs4833095, rs5743618) [41], Toll-like receptors 2 (TLR-2) [42], caspase recruitment domain-containing protein 9 (CARD9) (G72S,R373P,Q295X) [43, 44], Toll-like receptors 4 (TLR-4) (rs4986790,rs4986791) [45], and Dectin-1 [46] since 2006. Chronic mucocutaneous candidiasis is also related to Toll-like receptors 3 (TLR3) (rs3775291) and Dectin-1 mutations [46–48]. Gene polymorphisms targeting innate immunity may play an important role in IBD. Although studies aiming at the role of intestinal fungi pathogenesis in IBD are scarce, studies focusing on innate immune pathways against intestinal bacteria and their inflammatory consequences have successfully revealed important roles for innate immunity in IBD. Similarly, fungal recognition in the gut may be also regulated by innate immunity [49]. Four main types of innate immune receptors that can recognize fungi through fungal Pathogen-associated molecular patterns (PAMPs) are TLRs, C-type lectin receptors (CLRs), NOD-like receptors (NLRs), and galectin 3 on antigen-presenting cells [50]. The most studied class are the CLRs which include Dectin-1, Dectin-2, Dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin receptor (DC-SIGN), Macrophage inducible Ca2+-dependent lectin receptor (MINCLE), and the Mannose Receptor (MR). Additionally, some CLRs can interact with TLRs to recognize fungi [51]. The β-glucan is the main PAMP that can be recognized by Dectin-1, although Dectin-1 can also recognize unidentified bacterial and endogenous ligands [52]. Dectin-2 has been recently shown to be the functional receptor for α-mannans and to be implicated in anti-bacterial immunity [53]. The α-mannose is also strongly suggested to be Mincle’s ligand, which has been implied in anti-mycobacterial immune activity [54]. The fractalkine receptor (CX3CR-1) expressed by intestinal-resident mononuclear phagocytes (MNPs), were also characterized to have a role in initiating immune responses against fungi [55]. Through fungal recognition, these pathways initiate the inflammatory cascade by predominantly driving the immune responses through spleen tyrosine kinase (SYK)-dependent, SYK-independent, and eventually NF-κB signaling pathway towards T helper 1 (TH1) and/or T helper 17 (TH17) immunophenotypes [56]. The brief signaling cascade leading to intestinal inflammation is available in Fig. 1.
Fig. 1.
The cascade of innate immune response against intestinal fungi. Several fungal cell wall polysaccharides initiates immune responses, Dectin-1 binds β-glucans, dectin-2 binds α-mannans, and Mincle attaches the glycolipid trehalose-6,6-dimycolate (TDM), trehalose-6,6-dibehenate (TDB), and α-mannose residues. DC-SIGN binds N-linked mannans. Dectin-1, dectin-2, and mincle begin intracellular signaling through the SYK activation. RAF-1 as an SYK-independent activator of NF-κB pathway actuated by DC-SIGN and dectin-1. NF-κB pathway leads to TH1 and TH17 activations and subsequent cytokine production. CX3CR-1 is expressed by intestinal-resident mononuclear phagocytes (MNPs) and participate in fungal recognition
Intestinal mycobiota-genotype association
As Table 2 represents, here, we concentrated on articles reporting the mutations of innate immunity components and resulted in the gut mycobiome alteration. In a recent article, Limon et al. expressed that colonization of the colonic mucosa with Malassezia restricta, a commensal fungus typically found on the skin, might increase IBD severity in patients with CARD9S12N risk allele. They found out that the CARD9S12N variant induces a potent pro-inflammatory cytokine response against M. restricta in IBD [57]. By examining the SYK-CARD9 signaling axis and gut fungi, Malik et al. also demonstrated the decreased occurrence of Ascomycetes along with elevation of Saccharomycetes in Card9−/− mice. They implied that a normal inflammasome assembly in an unperturbed SYK-CARD9 signaling axis led to protection against colitis and colon cancer and also promoted T cell-mediated anti-tumorigenic responses; thereby indicating that a healthy gut mycobiota could prevent the development of IBD [58]. According to Lamas et al., the fungal microbiota of wild type and Card9−/− mice with induced-colitis mainly were members of the Ascomycota, Basidiomycota, and Zygomycota phyla. However, there were different measurements at the days 0, 7, and 12, and both groups reached a peak at day 7 that was higher in Card9−/− mice. On day 7, Card9−/− mice showed decreased fecal Ascomycota, increased fecal Basidiomycota, and Zygomycota communities [59].
Table 2.
Intestinal mycobiota-genotype association in IBD
| Animal/ human sample | Fungal extraction | Mycobiota-genotype | References |
|---|---|---|---|
| CD patients Mucosal-tissue | The ITS1 rDNA sequencing | M. restricta (CARD9 S12N alleles) | Limon et al. [57] |
| Card9−/− mice feces | 18S ITS rDNA sequencing | Decreased Ascomycota, elevation of Saccharomycetes (CARD9) | Malik et al. [58] |
| Card9−/− mice feces | ITS2 rDNA sequencing | Ascomycota, Basidiomycota, and Zygomycota (CARD9) | Lamas et al. [59] |
| CX3CR1−/− mice, CD patients | enzyme-linked immunosorbent assay (ELISA) | Decreased antibody production against Candida sp. (CX3CR-1) [CX3CR-1 T280M (rs3732378)] | Leonardi et al. [62] |
| IBD patients Fecal samples | ITS2 rDNA sequencing |
Positive correlation: M. sympodialis [Dectin-1 (rs2078178, rs3901533)],[TLR1 (rs4833095, rs5743618)],[Mincle (rs10841845)] S. cerevisiae [CARD9 (rs10781499)], [TLR3 (rs3775291)] Ascomycota [DC-SIGN (rs2287886)], [TLR1 (rs5743611)] Basidiomycota [TLR1 (rs5743611)] Negative correlation: M. sympodialis [Dectin-1 (rs2078178, ‘T’allele 12)] S. cerevisiae [CARD9 (rs10781499, ‘A’ allele 21)] |
Sokol et al. [8] |
| Clec4d−/− mice feces | 18S rDNA sequencing | C. tropicalis (CLEC4D) | Wang et al. [64] |
| Clec7−/− mice feces | ITS1-2 rDNA sequencing | Increased Candida and Trichosporon sp. Decreased nonpathogenic Saccharomyces sp. | Iliev et al. [66] |
| CD patients Fecal sample | ITS1 rDNA sequencing | No differences were evident with NOD2 variances | Nelson et al. [35] |
ITS 1,2, Internal transcribed spacer 1, 2; CARD9; CARD9, Caspase recruitment domain-containing protein 9; TLR3, Toll-like receptors 3; TLR1, Toll-like receptors 1; CLEC4D, C-Type Lectin domain containing 4D; CLEC7A, C-Type Lectin domain containing 7A; DC-SIGN, Dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin receptor; MINCLE, Macrophage inducible Ca2+-dependent lectin receptor; NOD2, Oligomerization domain-containing protein 2; M. restricta, Malassezia restricta; M. sympodialis, Malassezia sympodialis; S. cerevisiae, Saccharomyces cerevisiae; C. tropicalis, Candida tropicalis; sp., species
CX3CR-1 T280M (rs3732378) is a common polymorphism that has been previously detected in extra-intestinal inflammatory diseases [60, 61]. In 2018, Leonardi et al. described that CX3CR1 + MNPs not only modifies adaptive immune responses to intestinal fungi and controls the mycobiota during experimental colitis in animal models (without changing bacterial communities), but is also connected with a decrease in antifungal antibody responses in CD patients. They concluded that intestinal mycobiota and CX3CR1-dependent immune responses might provoke extra-intestinal manifestations of inflammatory diseases [62]. Elevated antifungal antibodies detected in patients with alcoholic liver disease, Graves’ disease, spondyloarthritis, and systemic lupus erythematous corroborate this hypothesis [63]. Finally, the article provided evidence for CX3CR1 + MNPs as a mediator between gut mycobiome and both local and systemic immunity [55].
A previous study was conducted by Sokol et al. to examine the correlation between host genotype and fungal microbiota in IBD patients. The ten most significant connections between IBD-associated fungi taxa and single-nucleotide polymorphisms (SNPs) were as follows: Malassezia sympodialis association with Dectin-1 (rs2078178, rs3901533), TLR1 (rs4833095, rs5743618), and Mincle (rs10841845); S. cerevisiae with CARD9 (rs10781499) and TLR3 (rs3775291); Ascomycota with DC-SIGN (rs2287886) and TLR1 (rs5743611); and Basidiomycota with TLR1 (rs5743611). They also provided evidence supporting the negative correlation of M. sympodialis fecal abundance with Dectin-1 SNP (rs2078178, ‘T’allele 12) in medically refractory UC; M. sympodialis was also decreased during the IBD flares in patients. Moreover, the IBD-associated CARD9 variation (rs10781499, ‘A’ allele 21) was inversely correlated with the fecal abundance of S. cerevisiae. Lastly, they reported a decrease in fungal biodiversity only in UC and CD patients without ileal involvement [8].
Wang et al. described the role of Dectin-3 (a family member of CLRs) in recognizing Candida. tropicalis in experimental-colitis pathogenesis for the first time. They observed that C. tropical increased the disease burden in Clec4d−/− mice during the induced colitis. Since the C-Type Lectin domain containing 4D (CLEC4D) is the encoding gene for Dectin-3, Clec4d−/− mice were more susceptible to induced colitis due to the activation of the NF-κB signaling pathway64.
The impact of NOD2 variants on the intestinal bacterial community in CD patients has previously been described [65]. Thus, Nelson et al. investigated the presence of NOD2 polymorphisms in CD patients and its relation with fecal fungal diversity but did not find any significant correlation between NOD2 variants and specific intestinal fungi community [35].
Dectin-1 is the most important fungal PRR expressed by innate immune cells, such as macrophages, dendritic cells, and neutrophils. C-Type Lectin domain containing 7A (CLEC7A) is the gene that encodes Dectin-1. Clec7−/− mice with induced colitis had increased proportions of opportunistic pathogenic fungi including Candida sp. and Trichosporon sp. along with a decreased frequency of nonpathogenic Saccharomyces. Iliev et al. identified a significant association between CLEC7A SNP (rs2078178) and patients suffering from medically refractory UC and delineated the role of Dectin-1 as a fungal receptor during severe forms of colitis [66]. Other gene polymorphisms were also described to influence Dectin-1-associated immunity in IBD [16, 67]. Among these genes, LRRK2 has also been described as the familial Parkinson’s disease genetic risk factor. Multiple variations in LRRK2 comprising N2081D, rs11175593 LRRK2/MUC19, and rs11564258 LRRK2/MUC19 were associated with IBD as well [68]. Takagawa et al. suggested an increase in severity of colitis, mediated by increased Dectin-1–induced immunity, in (rs11564258) LRRK2/MUC19 polymorphism carriers [16]. Noteworthily, this variance (rs11564258) had the second-highest odds ratio in IBD patients of the European population [69]. Further studies are required to identify the intestinal mycobiota in the patients carrying this mutation.
Conclusion
In summary, the role of intestinal fungal mycobiota in IBD pathogenesis and severity index have been quite underrated. This review emphasizes that a majority of IBD-affected patients had increased diversity and richness of intestinal mycobiome, higher abundance of C. albicans and Basidiomycota-to-Ascomycota ratio, and a decreased proportion of S. cerevisiae despite a few contradictory results in other studies.
It is widely known that innate immunity takes part in intestinal fungal recognition and mutations in innate immunity mediators are linked to IBD pathogenesis. Even so, few articles aimed to examine the connection between gene polymorphisms and intestinal fungal dysbiosis in IBD.
Although DSS-induced colitis is a well-established experimental murine model with much resemblance to human IBD [70], we were able to find only three non-murine studies containing mycobiota-genotype data related to IBD patients. Additional evidence is needed to determine whether different gene polymorphisms can alter intestinal mycobiome or whether this information would be of use in providing novel insight into IBD pathogenesis. Therefore, our purpose was to highlight the importance of the matter and draw attention to this underappreciated aspect of IBD-associated research.
Acknowledgements
Not applicable.
Abbreviations
- IBD
Inflammatory bowel disease
- CD
Crohn’s disease
- UC
Ulcerative Colitis
- IECs
Intestinal epithelial cells
- LRRK2
Leucine-rich repeat kinase 2
- ASCA
Anti-Saccharomyces cerevisiae antibody
- CARD15
Caspase recruitment domain-containing protein 15
- NOD2
Nucleotide-binding oligomerization domain-containing protein 2
- PRR
Pattern recognition receptor
- DCs
Dendritic cells
- IL-10
Interleukin 10
- TLR-1
Toll-like receptors 1
- TLR-2
Toll-like receptors 2
- CARD9
Caspase recruitment domain-containing protein 9
- TLR-4
Toll-like receptors 4
- PAMPs
Pathogen-associated molecular patterns
- CLRs
C-type lectin receptors
- NLRs
NOD-like receptors
- DC-SIGN
Dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin receptor
- MINCLE
Macrophage inducible Ca2+-dependent lectin receptor
- MR
Mannose Receptor
- MNPs
Mononuclear phagocytes
- SYK
Spleen tyrosine kinase
- SNPs
Single-nucleotide polymorphisms
- TLR3
Toll-like receptors 3
- CLEC4D
C-Type Lectin domain containing 4D
- CLEC7A
C-Type Lectin domain containing 7A
- TH1
T helper 1
- TH17
T helper 17
- IL-1
Interleukin 1
- IL-17
Interleukin 17
Authors' contributions
All the authors read and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zuo T, Kamm MA, Colombel JF, Ng SC. Urbanization and the gut microbiota in health and inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2018;15:440–452. doi: 10.1038/s41575-018-0003-z. [DOI] [PubMed] [Google Scholar]
- 2.Endo K, Shiga H, Kinouchi Y, Shimosegawa T. Inflammatory bowel disease: IBD. Rinsho Byori. 2009;57:527–532. [PubMed] [Google Scholar]
- 3.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152:327–339.e324. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, Andoh A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018;11:1–10. doi: 10.1007/s12328-017-0813-5. [DOI] [PubMed] [Google Scholar]
- 6.Spalinger MR, Schmidt TS, Schwarzfischer M, Hering L, Atrott K, Lang S, Gottier C, Geirnaert A, Lacroix C, Dai X, et al. Protein tyrosine phosphatase non-receptor type 22 modulates colitis in a microbiota-dependent manner. J Clin Invest. 2019;129:2527–2541. doi: 10.1172/JCI123263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu LC. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis. J Biomed Sci. 2018;25:79. doi: 10.1186/s12929-018-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leonardi I, Paramsothy S, Doron I, Semon A, Kaakoush NO, Clemente JC, Faith JJ, Borody TJ, Mitchell HM, Colombel JF, et al. Fungal trans-kingdom dynamics linked to responsiveness to fecal microbiota transplantation (FMT) therapy in ulcerative Colitis. Cell Host Microbe. 2020;27:823–829.e823. doi: 10.1016/j.chom.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu F, Feng D, Ding C, Zhang T, Chen J, Yu Z, Zhao L, Xu Y, Zhu W, Gong J. Fungal dysbiosis aggravates pouchitis in a rat model of ileal pouch anal anastomosis. Inflamm Bowel Dis. 2020;26:1831. doi: 10.1093/ibd/izaa111. [DOI] [PubMed] [Google Scholar]
- 11.Qiu X, Ma J, Jiao C, Mao X, Zhao X, Lu M, Wang K, Zhang H. Alterations in the mucosa-associated fungal microbiota in patients with ulcerative colitis. Oncotarget. 2017;8:107577–107588. doi: 10.18632/oncotarget.22534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liguori G, Lamas B, Richard ML, Brandi G, da Costa G, Hoffmann TW, Di Simone MP, Calabrese C, Poggioli G, Langella P, et al. Fungal dysbiosis in mucosa-associated microbiota of Crohn's disease patients. J Crohns Colitis. 2016;10:296–305. doi: 10.1093/ecco-jcc/jjv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Wang C, Tang C, He Q, Li N, Li J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in crohn's disease. J Clin Gastroenterol. 2014;48:513–523. doi: 10.1097/MCG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiaro TR, Soto R, Stephens WZ, Kubinak JL, Petersen C, Gogokhia L, Bell R, Delgado JC, Cox J, Voth W, et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci Transl Med. 2017 doi: 10.1126/scitranslmed.aaf9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bank S, Skytt Andersen P, Burisch J, Pedersen N, Roug S, Galsgaard J, Turino YS, Brodersen JB, Rashid S, Rasmussen KB, et al. Polymorphisms in the inflammatory pathway genes TLR2, TLR4, TLR9, LY96, NFKBIA, NFKB1, TNFA, TNFRSF1A, IL6R, IL10, IL23R, PTPN22, and PPARG are associated with susceptibility of inflammatory bowel disease in a Danish cohort. PLoS ONE. 2014;9:e98815. doi: 10.1371/journal.pone.0098815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takagawa T, Kitani A, Fuss I, Levine B, Brant SR, Peter I, Tajima M, Nakamura S, Strober W. An increase in LRRK2 suppresses autophagy and enhances Dectin-1-induced immunity in a mouse model of colitis. Sci Transl Med. 2018;10:eaan8162. doi: 10.1126/scitranslmed.aan8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cousins DV, Whittington R, Marsh I, Masters A, Evans RJ, Kluver P. Mycobacteria distenct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable IS900 polymerase chain reaction: implications for diagnosis. Mol Cell Probes. 1999;13:431–442. doi: 10.1006/mcpr.1999.0275. [DOI] [PubMed] [Google Scholar]
- 18.Andoh A, Tsujikawa T, Sasaki M, Mitsuyama K, Suzuki Y, Matsui T, Matsumoto T, Benno Y, Fujiyama Y. Faecal microbiota profile of Crohn's disease determined by terminal restriction fragment length polymorphism analysis. Aliment Pharmacol Ther. 2009;29:75–82. doi: 10.1111/j.1365-2036.2008.03860.x. [DOI] [PubMed] [Google Scholar]
- 19.McKenzie H, Main J, Pennington CR, Parratt D. Antibody to selected strains of Saccharomyces cerevisiae (baker's and brewer's yeast) and Candida albicans in Crohn's disease. Gut. 1990;31:536–538. doi: 10.1136/gut.31.5.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vermeire S, Joossens S, Peeters M, Monsuur F, Marien G, Bossuyt X, Groenen P, Vlietinck R, Rutgeerts P. Comparative study of ASCA (Anti-Saccharomyces cerevisiae antibody) assays in inflammatory bowel disease. Gastroenterology. 2001;120:827–833. doi: 10.1053/gast.2001.22546. [DOI] [PubMed] [Google Scholar]
- 21.Halfvarson J, Standaert-Vitse A, Järnerot G, Sendid B, Jouault T, Bodin L, Duhamel A, Colombel JF, Tysk C, Poulain D. Anti-saccharomyces cerevisiae antibodies in twins with inflammatory bowel disease. Gut. 2005;54:1237–1243. doi: 10.1136/gut.2005.066860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Standaert-Vitse A, Sendid B, Joossens M, François N, Vandewalle-El Khoury P, Branche J, Van Kruiningen H, Jouault T, Rutgeerts P, Gower-Rousseau C, et al. Candida albicans colonization and ASCA in familial Crohn's disease. Am J Gastroenterol. 2009;104:1745–1753. doi: 10.1038/ajg.2009.225. [DOI] [PubMed] [Google Scholar]
- 23.Sutton CL, Yang H, Li Z, Rotter JI, Targan SR, Braun J. Familial expression of anti-Saccharomyces cerevisiae mannan antibodies in affected and unaffected relatives of patients with Crohn's disease. Gut. 2000;46:58–63. doi: 10.1136/gut.46.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Standaert-Vitse A, Jouault T, Vandewalle P, Mille C, Seddik M, Sendid B, Mallet JM, Colombel JF, Poulain D. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn's disease. Gastroenterology. 2006;130:1764–1775. doi: 10.1053/j.gastro.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Mankaï A, Layouni S, Ghedira I. Anti saccharomyces cerevisiae antibodies in patients with anti-β2 glycoprotein I antibodies. J Clin Lab Anal. 2016;30:818–822. doi: 10.1002/jcla.21942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramanan D, Tang MS, Bowcutt R, Loke P, Cadwell K. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity. 2014;41:311–324. doi: 10.1016/j.immuni.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marrakchi R, Bougatef K, Moussa A, Ouerhani S, Khodjet-el-Khil H, Messai Y, Mestiri O, Najar T, Benammar-Elgaaeid A. 3020insC insertion in NOD2/CARD15 gene, a prevalent variant associated with anti-Saccharomyces cerevisiae antibodies and ileal location of Crohn's disease in Tunisian population. Inflamm Res. 2009;58:218–223. doi: 10.1007/s00011-008-8139-x. [DOI] [PubMed] [Google Scholar]
- 28.Ott SJ, Kühbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 29.Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, Bittinger K, Bailey A, Friedman ES, Hoffmann C, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's Disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, Wu GD. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:1948–1956. doi: 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. MBio. 2016 doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukhopadhya I, Hansen R, Meharg C, Thomson JM, Russell RK, Berry SH, El-Omar EM, Hold GL. The fungal microbiota of de-novo paediatric inflammatory bowel disease. Microbes Infect. 2015;17:304–310. doi: 10.1016/j.micinf.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam S, Zuo T, Ho M, Chan F, Chan P, Ng S. Fungal alterations in inflammatory bowel diseases. Alimentary Pharmacol Therapeutics. 2019;50:1159. doi: 10.1111/apt.15523. [DOI] [PubMed] [Google Scholar]
- 34.Di Paola M, Rizzetto L, Stefanini I, Vitali F, Massi-Benedetti C, Tocci N, Romani L, Ramazzotti M, Lionetti P, De Filippo C, Cavalieri D. Comparative immunophenotyping of Saccharomyces cerevisiae and Candida spp. strains from Crohn's disease patients and their interactions with the gut microbiome. J Transl Autoimmunity. 2020;3:10036. doi: 10.1016/j.jtauto.2020.100036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson A, Stewart CJ, Kennedy NA, Lodge JK, Tremelling M, Probert CS, Parkes M, Mansfield JC, Smith DL, Hold GL, et al. The impact of NOD2 Genetic variants on the gut mycobiota in Crohn's disease patients in remission and individuals without gastrointestinal inflammation. J Crohns Colitis. 2020 doi: 10.1093/ecco-jcc/jjaa220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choteau L, Vasseur F, Lepretre F, Figeac M, Gower-Rousseau C, Dubuquoy L, Poulain D, Colombel JF, Sendid B, Jawhara S. Polymorphisms in the mannose-binding lectin gene are associated with defective mannose-binding lectin functional activity in Crohn's disease patients. Sci Rep. 2016;6:29636. doi: 10.1038/srep29636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turpin W, Bedrani L, Espin-Garcia O, Xu W, Silverberg MS, Smith MI, Guttman DS, Griffiths A, Moayyedi P, Panaccione R, et al. FUT2 genotype and secretory status are not associated with fecal microbial composition and inferred function in healthy subjects. Gut Microbes. 2018;9:357–368. doi: 10.1080/19490976.2018.1445956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zakrzewski M, Simms LA, Brown A, Appleyard M, Irwin J, Waddell N, Radford-Smith GL. IL23R-protective coding variant promotes beneficial bacteria and diversity in the ileal microbiome in healthy individuals without inflammatory bowel disease. J Crohns Colitis. 2019;13:451–461. doi: 10.1093/ecco-jcc/jjy188. [DOI] [PubMed] [Google Scholar]
- 39.Yilmaz B, Spalinger MR, Biedermann L, Franc Y, Fournier N, Rossel J-B, Juillerat P, Rogler G, Macpherson AJ, Scharl M. The presence of genetic risk variants within PTPN2 and PTPN22 is associated with intestinal microbiota alterations in Swiss IBD cohort patients. PLoS ONE. 2018;13:e0199664–e0199664. doi: 10.1371/journal.pone.0199664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson MD, Plantinga TS, van de Vosse E, Velez Edwards DR, Smith PB, Alexander BD, Yang JC, Kremer D, Laird GM, Oosting M, et al. Cytokine gene polymorphisms and the outcome of invasive candidiasis: a prospective cohort study. Clin Infect Dis. 2012;54:502–510. doi: 10.1093/cid/cir827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plantinga TS, Johnson MD, Scott WK, van de Vosse E, Velez Edwards DR, Smith PB, Alexander BD, Yang JC, Kremer D, Laird GM, et al. Toll-like receptor 1 polymorphisms increase susceptibility to candidemia. J Infect Dis. 2012;205:934–943. doi: 10.1093/infdis/jir867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Netea MG, Van Der Graaf CA, Vonk AG, Verschueren I, Van Der Meer JW, Kullberg BJ. The role of toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J Infect Dis. 2002;185:1483–1489. doi: 10.1086/340511. [DOI] [PubMed] [Google Scholar]
- 43.Glocker EO, Hennigs A, Nabavi M, Schäffer AA, Woellner C, Salzer U, Pfeifer D, Veelken H, Warnatz K, Tahami F, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drewniak A, Gazendam RP, Tool AT, van Houdt M, Jansen MH, van Hamme JL, van Leeuwen EM, Roos D, Scalais E, de Beaufort C, et al. Invasive fungal infection and impaired neutrophil killing in human CARD9 deficiency. Blood. 2013;121:2385–2392. doi: 10.1182/blood-2012-08-450551. [DOI] [PubMed] [Google Scholar]
- 45.Van der Graaf CA, Netea MG, Morré SA, Den Heijer M, Verweij PE, Van der Meer JW, Kullberg BJ. Toll-like receptor 4 Asp299Gly/Thr399Ile polymorphisms are a risk factor for Candida bloodstream infection. Eur Cytokine Netw. 2006;17:29–34. [PubMed] [Google Scholar]
- 46.Tam JM, Reedy JL, Lukason DP, Kuna SG, Acharya M, Khan NS, Negoro PE, Xu S, Ward RA, Feldman MB, et al. Tetraspanin CD82 organizes Dectin-1 into signaling domains to mediate cellular responses to Candida albicans. J Immunol. 2019;202:3256–3266. doi: 10.4049/jimmunol.1801384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nahum A, Dadi H, Bates A, Roifman CM. The biological significance of TLR3 variant, L412F, in conferring susceptibility to cutaneous candidiasis, CMV and autoimmunity. Autoimmun Rev. 2012;11:341–347. doi: 10.1016/j.autrev.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 48.Plantinga TS, van der Velden WJ, Ferwerda B, van Spriel AB, Adema G, Feuth T, Donnelly JP, Brown GD, Kullberg BJ, Blijlevens NM, Netea MG. Early stop polymorphism in human DECTIN-1 is associated with increased candida colonization in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2009;49:724–732. doi: 10.1086/604714. [DOI] [PubMed] [Google Scholar]
- 49.Underhill D, Braun J. Current understanding of fungal microflora in inflammatory bowel disease pathogenesis. Inflamm Bowel Dis. 2008;14:1147–1153. doi: 10.1002/ibd.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bourgeois C, Kuchler K. Fungal pathogens—a sweet and sour treat for toll-like receptors. Front Cell Infect Microbiol. 2012;2:142. doi: 10.3389/fcimb.2012.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vautier S, MacCallum DM, Brown GD. C-type lectin receptors and cytokines in fungal immunity. Cytokine. 2012;58:89–99. doi: 10.1016/j.cyto.2011.08.031. [DOI] [PubMed] [Google Scholar]
- 52.Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, Brown GD. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol. 2007;8:31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, Fujikado N, Kusaka T, Kubo S, Chung SH, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 54.Yamasaki S, Matsumoto M, Takeuchi O, Matsuzawa T, Ishikawa E, Sakuma M, Tateno H, Uno J, Hirabayashi J, Mikami Y, et al. C-type lectin Mincle is an activating receptor for pathogenic fungus, Malassezia. Proc Natl Acad Sci U S A. 2009;106:1897–1902. doi: 10.1073/pnas.0805177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern DPB, et al. CX3CR1(+) mononuclear phagocytes control immunity to intestinal fungi. Science. 2018;359:232–236. doi: 10.1126/science.aao1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richard ML, Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. 2019;16:331–345. doi: 10.1038/s41575-019-0121-2. [DOI] [PubMed] [Google Scholar]
- 57.Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, et al. Malassezia is associated with Crohn's disease and exacerbates colitis in mouse models. Cell Host Microbe. 2019;25:377–388.e376. doi: 10.1016/j.chom.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malik A, Sharma D, Malireddi RKS, Guy CS, Chang TC, Olsen SR, Neale G, Vogel P, Kanneganti TD. SYK-CARD9 signaling axis promotes gut fungi-mediated inflammasome activation to restrict colitis and colon cancer. Immunity. 2018;49:515–530.e515. doi: 10.1016/j.immuni.2018.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moatti D, Faure S, Fumeron F, Amara Mel W, Seknadji P, McDermott DH, Debré P, Aumont MC, Murphy PM, de Prost D, Combadière C. Polymorphism in the fractalkine receptor CX3CR1 as a genetic risk factor for coronary artery disease. Blood. 2001;97:1925–1928. doi: 10.1182/blood.V97.7.1925. [DOI] [PubMed] [Google Scholar]
- 61.Wan W, Murphy PM. Regulation of atherogenesis by chemokines and chemokine receptors. Arch Immunol Ther Exp. 2013;61:1–14. doi: 10.1007/s00005-012-0202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern DPB, et al. CX3CR1+, mononuclear phagocytes control immunity to intestinal fungi. Science. 2018;359:232–236. doi: 10.1126/science.aao1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li XV, Leonardi I, Iliev ID. Gut mycobiota in immunity and inflammatory disease. Immunity. 2019;50:1365–1379. doi: 10.1016/j.immuni.2019.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang T, Pan D, Zhou Z, You Y, Jiang C, Zhao X, Lin X. Dectin-3 deficiency promotes colitis development due to impaired antifungal innate immune responses in the gut. PLoS Pathog. 2016;12:e1005662. doi: 10.1371/journal.ppat.1005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kennedy NA, Lamb CA, Berry SH, Walker AW, Mansfield J, Parkes M, Simpkins R, Tremelling M, Nutland S, Parkhill J, et al. The impact of NOD2 variants on fecal microbiota in Crohn's disease and controls without gastrointestinal disease. Inflamm Bowel Dis. 2018;24:583–592. doi: 10.1093/ibd/izx061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Vries HS, Plantinga TS, van Krieken JH, Stienstra R, van Bodegraven AA, Festen EA, Weersma RK, Crusius JB, Linskens RK, Joosten LA, et al. Genetic association analysis of the functional c.714T>G polymorphism and mucosal expression of dectin-1 in inflammatory bowel disease. PLoS ONE. 2009;4:e7818. doi: 10.1371/journal.pone.0007818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rastegar AD, Dzamko N. Leucine rich repeat kinase 2 and innate immunity. Front Neurosci. 2020 doi: 10.3389/fnins.2020.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Franke A, McGovern DPB, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pakravan N, Kermanian F, Mahmoudi E. Filtered Kombucha tea ameliorates the leaky gut syndrome in young and old mice model of colitis. Iran J Basic Med Sci. 2020;22:1158. doi: 10.22038/ijbms.2019.36189.8622. [DOI] [PMC free article] [PubMed] [Google Scholar]