Abstract
SARS-CoV-2, the virus that causes COVID-19, has killed over 3 million people worldwide. Despite the urgency of the current pandemic, most available diagnostic methods for COVID-19 use RT-PCR to detect nucleic acid sequences specific to SARS-CoV-2. These tests are limited by their requirement of a large laboratory space, high reagent costs, multistep sample preparation, and the potential for cross-contamination. Moreover, results usually take hours to days to become available. Therefore, fast, reliable, inexpensive, and scalable point-of-care diagnostics are urgently needed. Here, we describe RAPID 1.0, a simple, handheld, and highly sensitive miniaturized biosensor modified with human receptor angiotensin-converting enzyme-2. RAPID 1.0 can detect SARS-CoV-2 using 10 μL of sample within 4 min through its increased resistance to charge transfer of a redox probe measured by electrochemical impedance spectroscopy. The sensitivity and specificity of RAPID for nasopharyngeal/oropharyngeal swab and saliva samples are 85.3% and 100% and 100% and 86.5%, respectively.
Keywords: SARS-CoV-2, diagnostics, electrochemistry, COVID-19, point of care, biosensors, angiotensin-converting enzyme 2
Graphical abstract
Progress and potential
To address the need for rapid, inexpensive, and scalable point-of-care diagnostics for COVID-19, we created RAPID, a biosensor that can detect SARS-CoV-2 within 4 min using minimal sample volume through electrochemical impedance spectroscopy. Our test detects SARS-CoV-2 and UK variant B.1.1.7 at extremely low concentrations (1.16 PFU mL−1) with high specificity, sensitivity, and accuracy in both saliva and nasopharyngeal/oropharyngeal samples.
RAPID is a low-cost electrochemical biosensor that diagnoses COVID-19 within 4 min in clinical saliva and nares swab samples.
Introduction
SARS-CoV-2, the virus that causes COVID-19, continues to kill people at a staggering pace,1 threatening the global economy, health care systems, and our lives. Despite the urgency of the pandemic, the high cost of production, lack of scalable technologies, and slow detection time hinder the widespread use of currently available diagnostics. Therefore, low-complexity point-of-care diagnostic tests with a rapid turnaround time are urgently needed.
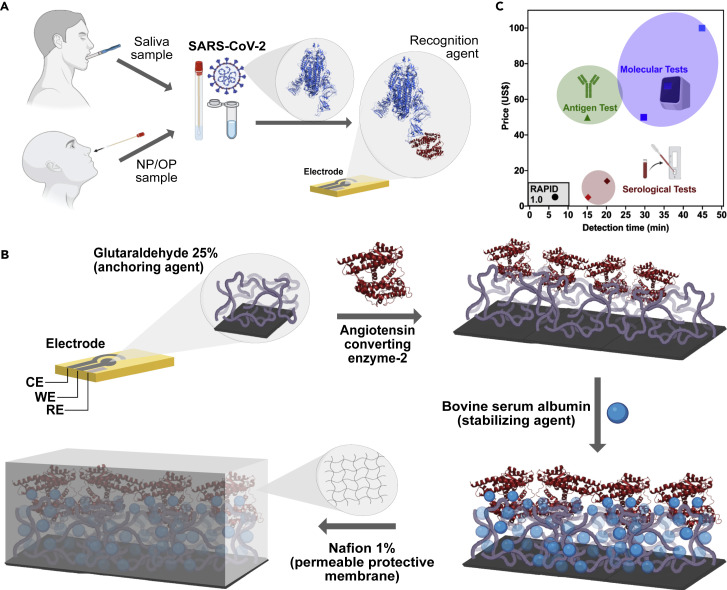
Here, we describe a simple, inexpensive, and rapid test for the detection of SARS-CoV-2 (Figure 1 A). RAPID 1.0 (real-time accurate portable impedimetric detection prototype 1.0) transforms biochemical information from a specific molecular binding event between the SARS-CoV-2 spike protein (SP) and angiotensin-converting enzyme-2 (ACE2) into an electrical signal that can easily be detected.
Figure 1.
Point-of-care detection of SARS-CoV-2 using RAPID 1.0
(A) RAPID 1.0 enables diagnosis using neat saliva and nasopharyngeal/oropharyngeal (NP/OP) swab samples infected with SARS-CoV-2.
(B) Schematic for the preparation of the electrodes. Briefly, screen-printed electrodes in a three-electrode configuration cell (CE, counter electrode; WE, working electrode; and RE, reference electrode) were printed on phenolic paper circuit board or filter paper with conductive carbon and Ag/AgCl inks. The WE was functionalized with glutaraldehyde to enable anchoring of ACE2, which was stabilized by the addition of bovine serum albumin. Detection was improved by adding a Nafion permeable membrane, enabling chemical preconcentration of cation species and protecting the electrode's surface against biofouling with proteins, lipids, and other macromolecules present in the biological sample matrix.
(C) Cost and detection time comparison between RAPID 1.0 and existing FDA-approved antigen, serological, and molecular tests.2, 3, 4 Note that comparisons were made for a single test of each of the different technologies.
RAPID 1.0 (henceforth referred to as RAPID) uses electrochemical impedance spectroscopy (EIS), an electrochemical technique extensively utilized for the characterization of functionalized electrode surfaces and the transduction of biosensors.5 , 6 In our test, the EIS transducer signal reports the selective interaction and binding between the biological receptor immobilized on the electrode surface (i.e., ACE2) and its binding element (i.e., SP).7 The binding between these two molecules causes a change in interfacial electron transfer kinetics between the redox probe, ferricyanide/ferrocyanide in solution, and the conducting electrode sites. This electrochemical change is then detectable by monitoring the charge-transfer resistance (RCT), the diameter of the semi-arc on the Nyquist plot, which correlates with the number of targets bound to the receptive surface.5 The selectivity of an EIS biosensor relies mostly on the specificity between the target and the recognizing bioelement immobilized on the electrode surface and its robustness through the designed architecture surfaces to minimize non-specific binding of the analyte or adsorption of other biomolecules in solution.
Results and discussion
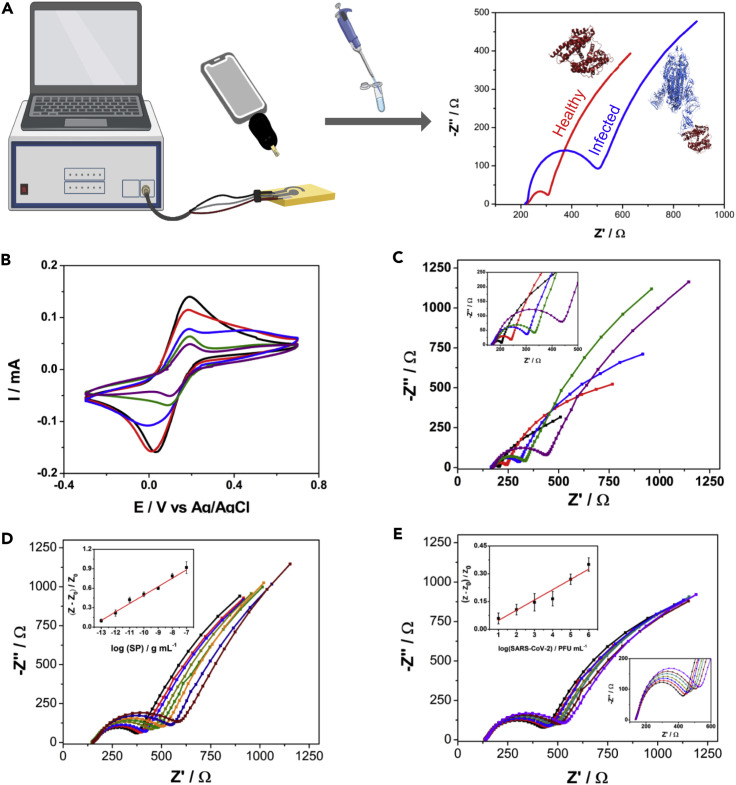
We designed the electrochemical device to explore the remarkable binding affinity of SARS-CoV-2 SP to ACE2, its receptor in the human body7 , 8 (Figure 2 A). The working electrode (WE), where the (electro)chemical reaction/interaction takes place and is converted to a detectable analytical signal, was functionalized by a drop-casting method. Enzyme immobilization was achieved by cross-linking ACE2 using the bifunctional chemical cross-linker glutaraldehyde (GA).9 This dialdehyde reacts mainly with the primary amino groups of proteins, for example, the ε-amino group of lysine residues or the N-terminal group of the protein chain.10 We used bovine serum albumin (BSA) to block the electrode's surface after immobilization of ACE2. BSA is a functionally inert protein with a high density of superficial lysine residues that is commonly used for biosensor development.10
Figure 2.
Characterization and calibration of RAPID 1.0
(A) Schematic representation of the RAPID diagnostic process.
(B and C) (B) Cyclic voltammetry and (C) Nyquist plots (inset shows the zoomed region of the curve with the semi-arc) of all functionalization steps showing progressively increased resistivity between the bare electrode (in black) and the four modification steps: addition of glutaraldehyde (in red), functionalization of ACE2 (in blue), addition of the blocking agent bovine serum albumin (in green), and addition of the Nafion permselective membrane (in purple).
(D) Nyquist plots for different SP concentrations ranging from 100 fg mL−1 to 100 ng mL−1 with 10-fold increments in neat saliva from a healthy donor (negative result by qRT-PCR). The inset shows the linearized correlation between normalized RCT values and the concentration of SP exposed to the electrode.
(E) Nyquist plots for titered inactivated virus solutions at concentrations ranging from 101 to 106 PFU mL−1 with 10-fold increments. The upper left inset shows the linearized correlation between the normalized RCT values and the concentration of inactivated virus in solution. The lower right inset shows a zoomed region of the curve with the Nyquist plots' semi-arc (RCT). The analytical curves presented in (D) and (E) were based on triplicate measurements. All data were recorded using the eChip version of RAPID.
Using these well-established protocols for bioelectrode development, we first added GA for 1 h at 37°C to fully cover the carbon electrode surface, generating a cross-linked polymer that enables the covalent anchoring of ACE2 at 37°C for 1.5 h (Figure 1B). Next, BSA was added to the surface of the electrode for 30 min at 37°C to block possible remaining active sites (i.e., WE's surface areas that were not functionalized with ACE2), thus preventing nonspecific adsorption to the GA layer by other proteins. We also incorporated an additional functionalization step using a 1.0% Nafion solution (Figure S2) to create a protective polymeric membrane, enhancing the robustness of the biosensor.11 Interestingly, Nafion increased the sensitivity of the biosensor up to 2-fold, particularly when used at a concentration ranging between 1.0% and 1.5% (Figure S2). Given these results, we selected 1.0% Nafion (wt %) for subsequent optimization steps because of its optimal analytical response to a low reagent usage ratio.11 This anionic membrane enables small positively charged species to cross and preconcentrate close to the biosensing surface. The Nafion layer also enhanced the robustness of RAPID by protecting against biofouling of the electrodic surface when exposed to the sample's complex matrix (e.g., proteins, lipids, and other macromolecules present in biological samples), which may interfere with detection.11 , 12
To ensure that the biological activity of ACE2 was preserved upon immobilization onto the electrode's surface, we evaluated the response of the sensor when exposed to its natural substrate angiotensin II (Figure S1). A sensitive linear response was observed in the range of 1 pg mL−1 to 10 μg mL−1 of angiotensin II, demonstrating that our anchoring and stabilization strategies maintained the functionality of ACE2's active sites and revealing that the biosensor architecture did not obstruct ACE2.
Next, we characterized the effect of each modifier layer (GA, ACE2, BSA, and Nafion) on the electrochemical response of our modified electrode, recording cyclic voltammetry (CV) and EIS measurements in the presence of 5 mmol L−1 of the redox probe potassium ferricyanide/ferrocyanide (Figures 2B and 2C). These results demonstrated that the peak current signal of the redox probe decreased when using CV and the resistance to charge transfer increased after each functionalization step. The decrease in the peak current signal occurs because of the addition of nonconductive materials (e.g., proteins) that block the active sites of the electrodic surface, hindering the kinetics of charge transfer of the redox probe. Our optimized protocol generated the best analytical signal for the detection of SARS-CoV-2 in human biofluid samples (Figure 1A). It consists of the following four steps: (1) modifying the WE with the immobilizing agent (GA); (2) covalent attachment of the recognition agent ACE2; (3) addition of the stabilization and active-site blocking agent BSA; and (4) incorporating the permselective membrane (Nafion). A detailed protocol describing biosensor preparation, including the production of the screen-printed devices and functionalization, is provided in the supplemental information.
Our test can be performed at room temperature with minimal equipment and reagents, and costs $4.67 to produce ($0.07 to produce the bare electrode, $4.50 to functionalize the electrode with the recognition agent ACE2, and $0.10 to coat the electrode with GA, BSA, and Nafion used; Figure 1C). The overall cost of RAPID may be further reduced through recombinant production of ACE2 and ACE2 variants.13 Our technology is also highly scalable, as the electrodes can be rapidly mass produced by using commercially available screen printers. One laboratory-sized unit is able to produce 35,000 electrodes daily (1.05 M electrodes/month) and this could scaled up to 10.5 billion electrodes monthly with only 10,000 screen printers (Table S1). These estimates take into account both the time needed to print the electrodes and all functionalization steps (i.e., 1 h for GA functionalization, 1.5 h to incorporate ACE2, 0.5 h for BSA, and 1 h for Nafion; total of 4 h). However, it must be noted that these steps can be fully automated into a production line for industrial purposes, drastically reducing time requirements.
We evaluated the incubation time (i.e., time of exposure of the sample to the biosensor to enable sensitive detection) and whether a centrifugation/dilution step was needed to detect SARS-CoV-2 in complex biological samples such as saliva.14 Next, we built dose-response curves using free SP to assess the effect of incubation time (Figure S4), centrifugation (Figure S3), and sample dilution (Figure S3) on the performance of RAPID. Our optimization steps revealed that an additional centrifugation step was not needed (Figure S3), since the use of neat saliva yielded results similar to those obtained using centrifuged samples. These results demonstrated that our approach is robust and can directly use human samples (nasopharyngeal/oropharyngeal [NP/OP] or saliva) without a prior pretreatment step, thus allowing the application of RAPID for streamlined and rapid point-of-care diagnosis. We selected 2 min as the optimal incubation period of the sample on the WE's surface for sensitive SARS-CoV-2 detection in samples considering the detectability and analytical frequency of the tests (Figure S4). Our very minimal incubation time requirement (2 min) confirms the favorable configuration of the modified electrode that allows rapid interaction kinetics between the SP and immobilized ACE2 (kinetics constant rate of 104 M−1 s−1 in its natural environment7). Overall, RAPID provides a result in 4 min (2 min of sample incubation +2 min to perform the EIS analysis), which is vastly faster than methods currently available for diagnosing COVID-19 (Figure 1C). It is important to note that the total time required to run each blank is an additional 4 min. However, we did not take this into account in our testing time calculations because the blanking step can be done before analyzing clinical samples, and we can use the RCT values obtained for the blanks (PBS or virus transportation medium [VTM]) to compare with the patient sample values.
Taking into account the optimal analytical conditions evaluated (Table 1 ), we built calibration curves for free SP (Figures 2D, S5A, and S5B) and heat-inactivated virus (Figure 2E) using the normalized RCT response, defined by the following equation:
where Z is the RCT of the sample and Z0 is the RCT of the respective blank solution: PBS, VTM, or healthy saliva. The normalization process of RCT aims to correct eventual fluctuations in the sensor operation, such as the temperature at the testing point or variations due to analyst operation. The dose-response curve for the free SP in PBS solution ranged from 1 fg mL−1 to 10 μg mL−1 (Figure S5A). A linear concentration range of 10 fg mL−1 to 100 ng mL−1 was obtained (R2 = 0.993), and limits of detection (LOD) and quantification (LOQ) were calculated as 2.18 and 7.26 fg mL−1 SP based on the signal-to-noise ratio (S/N = 3 and S/N = 10, respectively). We built a dose-response curve for the free SP in VTM at a concentration range of 10 fg mL−1 to 100 pg mL−1 (Figure S5B). A linear concentration range of 10 fg mL−1 to 1 ng mL−1 was obtained (R2 = 0.995) and LOD and LOQ were calculated as 6.29 and 20.96 fg mL−1 SP based on the signal-to-noise ratio (S/N = 3 and S/N = 10, respectively). When performed in neat saliva, the calibration curve was built at a concentration ranging from 100 fg mL−1 to 100 ng mL−1 (Figure 2D). The calculated LOD and LOQ were 1.39 and 4.63 pg mL−1, respectively. The higher LODs obtained in saliva and VTM are consistent with the increased sample complexity compared with PBS solution.
Table 1.
Analytical parameters of RAPID 1.0
| Parameter | Value |
|---|---|
| Linear concentration range (SP in PBS) | 10 fg mL−1 to 100 ng mL−1 |
| Linear concentration range (SP in VTM) | 10 fg mL−1 to 1 ng mL−1 |
| Linear concentration range (SP in saliva) | 100 fg mL−1 to 100 ng mL−1 |
| Limit of detection (SP in PBS) | 2.18 fg mL−1 |
| Limit of detection (SP in VTM) | 6.29 fg mL−1 |
| Limit of detection (SP in saliva) | 1.39 pg mL−1 |
| Limit of quantification (SP in PBS) | 7.26 fg mL−1 |
| Limit of quantification (SP in VTM) | 20.96 fg mL−1 |
| Limit of quantification (SP in saliva) | 4.63 pg mL−1 |
| Working concentration range (IV in VTM) | 101–106 PFU mL−1 |
| Limit of detection (IV in VTM) | 1.16 PFU mL−1 |
| Limit of quantification (IV in VTM) | 3.87 PFU mL−1 |
IV, inactivated virus.
The RCT values of Nyquist plots were extracted by the application of an equivalent circuit (Figure S6). The equivalent circuit comprises two semi-arc regions observed in the Nyquist plots, where the first is a nondefined semi-arc at a high frequency range due to inhomogeneity or defects in the electrode modification step (during drop-casting functionalization) and considerably small (RCT ∼10 Ω).15 , 16 The second parallel component of the equivalent circuit comprises an RCT whose signal intensity was proportional to the logarithm of the SP/virus concentration, and also presented a Warburg element to describe the mass transport (diffusional control).
The concentration range of SP detected by our device was 10–1,000 times lower than that reported in previous studies,17 , 18 thus emphasizing the sensitivity of our approach. To assess the diagnostic capability of RAPID, we calibrated our biosensor using titered solutions of inactivated SARS-CoV-2 ranging from 101 to 106 plaque-forming units (PFU) mL−1 (Figure 2E). RAPID exhibited high sensitivity, presenting an LOD of 1.16 PFU mL−1, which corresponds to the order of 100 RNA copies μL−1,19 , 20 a viral load that correlates with the initial stages of COVID-19 (i.e., 2 to 3 days after onset of symptoms).21 Thus, RAPID's LOD and LOQ values are comparable to those of gold-standard approaches such as RealStar SARS-CoV-2, CDC COVID-19, and ePlex SARS-CoV-2,19 with the advantage of detecting symptomatic and asymptomatic individuals at the earliest stages of the infection allowing for rapid decision-making and the subsequent use of more appropriate and effective countermeasures. To ensure the repeatability, stability, and reproducibility of the results, we carried out three different experiments. First, 21 successive EIS measurements of the medium (PBS) were performed using the same device to verify the drift of the EIS response, yielding a relative standard deviation (RSD) of 5.3% (Figure S7). These results demonstrated that the device exhibits a repeatable and stable response. Next, the reproducibility of RAPID was evaluated by analytical curves in the range of 1 pg mL−1 to 1 ng mL−1 of SP and the analytical sensitivity of 10 electrodes from different batches was assessed (Figure S8). An RSD of 6.8% was obtained, indicating that the electrochemical device fabrication and functionalization protocols display high reproducibility.
Next, we evaluated the stability of RAPID at different temperature storage conditions (25°C, 8°C, and −20°C) over 10 days (Figure S9). Analytical curves were generated with SP at a concentration ranging from 1 pg mL−1 to 1 ng mL−1 and the sensitivity was normalized by the mean value of the three different biosensors used immediately after the functionalization steps. The biosensors stored at room temperature did not detect the SP after 24 h due to loss of enzymatic activity (Figure S9). The sensors stored at 8°C were stable after 24 h, but after 48 h presented decreased sensitivity (around 50% of the initial response), keeping this low sensitivity for 7 days (Figure S9). Biosensors stored at −20°C exhibited the most promising results, since they were as sensitive as those used right after functionalization even after 96 h and retained 50% of their sensitivity after 10 days of storage (Figure S9).
Next, the performance of RAPID was assessed using both SARS-CoV-2-positive and negative clinical samples from the Hospital of the University of Pennsylvania (Tables S2 and S3), including the highly contagious SARS-CoV-2 UK B.1.1.7 variant (Tables 2 and S2). All samples were heat inactivated at 56°C for 1 h. The effect of heat inactivation of SARS-CoV-2 samples on the analytical response of our biosensor was evaluated through measurements taken before and after sample inactivation at 56°C for 1 h (Figure S10. The results indicated that thermal inactivation affected the ability of SP to bind to ACE2, since a decrease of up to 60% was detected in the analytical response for sample 2 after heat inactivation (Figure S10). These results indicate that heat-inactivated clinical samples with very low viral titers may fall below our current LOD. Rath and Kumar22 demonstrated using molecular dynamics simulations that temperatures >50°C trigger the closing of the spike receptor binding motif, which buries the receptor-binding residues, preventing contacts between the SP and the ACE2 receptor. These insights may help explain the results obtained upon thermal inactivation of our biosensor (Figure S10). However, despite this decrease in SP binding to ACE2 upon heat inactivation, the sensitivity of our method still enabled accurate viral detection in clinical samples containing a range of viral titers (Figure 2E).
Table 2.
Positive and negative values obtained by qRT-PCR and sensitivity, specificity, and accuracy of RAPID 1.0 using NP/OP and saliva samples
| RAPID (NP/OP) | qRT-PCR |
Sensitivity | Specificity | Accuracy | ||
|---|---|---|---|---|---|---|
| Positive (n = 109a) | Negative (n = 30) | Total (n = 139) | ||||
| Positive | 91 | 0 | 91 | 91/109 (83.5%) | 121/139 (87.1%) | |
| Negative | 18 | 30 | 48 | 30/30 (100%) | ||
| RAPID (saliva) | qRT-PCR |
Sensitivity | Specificity | Accuracy | ||
|---|---|---|---|---|---|---|
| Positive (n = 13) | Negative (n = 37) | Total (n = 50) | ||||
| Positive | 13 | 5 | 18 | 13/13 (100.0%) | 45/50 (90.0%) | |
| Negative | 0 | 32 | 32 | 32/37 (86.5%) | ||
Clinical sample set includes a highly contagious SARS-CoV-2 UK variant B.1.1.7 from a patient.
We also observed that centrifuging the samples did not lead to increased impedimetric detection of the SP (Figure S3). Therefore, the NP/OP and saliva samples were used in VTM and PBS, respectively, following the US FDA recommendation for regulatory applications. Of note, the detectability of impedimetric measurements after 2 min of incubation of the sample on the WE's surface was as high as that for longer incubation times of 5 and 10 min (Figure S4), thus demonstrating RAPID's fast interaction kinetics between the SP and the functionalized WE, as discussed above. Thus, we selected 2 min of incubation and set as a cut-off value a 10% change in the RCT compared with the blank solution. Such a cut-off threshold takes into account the LOQ obtained for inactivated virus analysis (Figure 2E), thus allowing discrimination between SARS-CoV-2-negative and SARS-CoV-2-positive samples (Tables S2 and S3).
In blinded tests, we analyzed 139 NP/OP swab samples (in VTM) obtained from patients after heat inactivation, 109 of which were COVID-19 positive and 30 COVID-19 negative as determined by qRT-PCR and clinical assessment (Table S2). RAPID demonstrated high sensitivity, specificity, and accuracy for NP/OP (83.5%, 100%, and 87.1%, respectively; Table 2) and saliva (100%, 86.5%, and 90.0%, respectively; Table 2) samples. RAPID missed a single sample, which presented a viral count lower than its LOD (10−1 RNA copies μL−1). It is worth noting that although the heat-inactivation protocol decreased the response of our biosensor due to the inactivation of SP (Figure S10), the outstanding sensitivity of RAPID (Table 1) enabled high detectability (Table 1) and hit rate (Table 2). Of the 12 negative NP/OP swab samples present in our sample set, 100% were confirmed as SARS-CoV-2 negative by RAPID (Table 2). In addition, the highly contagious SARS-CoV-2 UK variant B.1.1.7 was obtained from a government testing site in Philadelphia (Tables 2 and S3). RAPID successfully identified this sample as positive with a normalized RCT value of 1.10 (Table S2), thus highlighting its ability to detect emerging mutant variants of SARS-CoV-2.
To evaluate RAPID's diagnostic efficacy in a more complex biological environment, we tested saliva samples from 50 patients (Table S3) under the same conditions used for the NP/OP swab samples. The greater complexity of saliva, compared with swab samples, is known to hinder the accurate detection of infectious agents.14 , 21 Saliva is a biofluid that is susceptible to large variations in composition depending on different factors such as the ingestion of food and drinks prior (30–60 min) to sample collection, which can lead to the dilution of the saliva matrix, and the insertion of exogenous molecular species that may interfere with accurate detection. Even using highly heterogeneous saliva samples, the sensitivity of RAPID remained high (100%); however, false positives led to decreased specificity (86.5%) and an accuracy of 90.0% (Table 2). These results may be explained by potential interactions between ACE2, which is a carboxypeptidase and amino acid transporter, and other biomolecules that can be found in neat biofluids, such as regulatory peptides and peptide hormones (e.g., angiotensin, bradykinin, ghrelin, apelin, neurotensin, and dynorphin).23 Thus, we believe the performance of RAPID will improve when using fresh saliva samples at the point-of-care. It is worth noting that among the SARS-CoV-2-positive saliva samples, our test identified as positive two samples that had been previously erroneously detected as negative by qRT-PCR, therefore indicating that RAPID may help correctly diagnose COVID-19 in samples previously misdiagnosed by other methods.
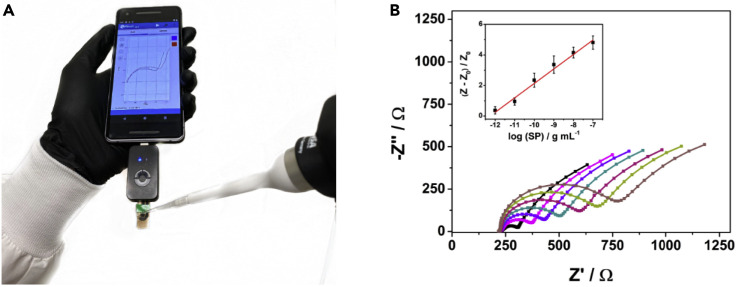
Several key analytical features were used to compare the performance of RAPID with respect to other electrochemical methods reported in the literature (Table 3 ). Our method provides the highest sensitivity (LOD of 2.8 fg mL−1) for the detection of SARS-CoV-2 SP with excellent time of detection and overall cost (Table 3). In addition, the robustness of RAPID was evaluated in a large clinical sample set (Tables S2 and S3), and all results were compared with those obtained by qRT-PCR (Table 2), thus highlighting the reliability of our method. All experiments described thus far (e.g., detection of SARS-CoV-2 SP and clinical samples) were performed using the eChip version of the electrode (e.g., Figure 2, Table 2, Figures S2 and S3). After successfully applying the eChip (composed of printed circuit board) to clinical samples (Tables S2 and S3) and obtaining robust and sensitive results (Table 1), we sought to construct an optimized electrode composed of a material that was more accessible and less expensive to enable scale-up production of RAPID. We selected filter paper as the main component of the electrochemical paper-based analytical device (ePAD) as it is easy to handle (malleable), accessible, and inexpensive (paper filter costs $0.50/m2, whereas printed circuit board costs $40.00/m2).34 , 35 We adapted and demonstrated the applicability of ePAD in a portable potentiostat connected to a smart device (Figure 3 A). We used the screen-printing method to fabricate the electrodes and combined wax-printing technology to pattern the electrochemical cell onto the paper filter. Thus, the ePAD is composed of more accessible and low-cost material, enabling scalable manufacturing and on-demand testing at the point-of-care.34 , 35
Table 3.
Comparison of methods reported for COVID-19 diagnosis
| Sensor | Technique | Biological target | Lowest concentration detected | Number of clinical samples | Pricea (US$) | Time (min) | Reference |
|---|---|---|---|---|---|---|---|
| RAPID 1.0 | EIS | SARS-CoV-2 spike protein | 2.8 fg mL−1 | 151 | 4.67 | 4 | this work |
| SARS-CoV-2 RapidFlex | DPV and OCP-EIS | Viral antigen nucleocapsid protein | 500 pg mL−1 | 16 | – | 10 | 24 |
| SARS-CoV-2 RapidFlex | DPV and OCP-EIS | IgM and IgG antibodies | 250 ng mL−1 | 16 | – | 10 | 24 |
| SARS-CoV-2 RapidFlex | DPV and OCP-EIS | C-reactive protein | 50 ng mL−1 | 16 | – | 10 | 24 |
| SCC | SARS-CoV-2 RNA | 231 RNA copies μL−1 | 48 | 10 | 5 | 25 | |
| DPV | SARS-CoV-2 RNA | 200 RNA copies μL−1 | 33 | – | <5 | 26 | |
| EIS | SARS-CoV-2 spike protein | 0.1 mg mL−1 | 4 | – | 3 | 18 | |
| SWV | IgM and IgG antibodies | 1 μg mL−1 | 17 | – | 45 | 27 | |
| DETECTR | CRISPR technology | E gene and N gene | 10 RNA copies μL−1 | 11 | – | 40 | 28 |
| colorimetric assay | N gene | 0.18 ng μL−1 | 1 | – | 30 | 29 | |
| localized surface plasmon resonance | RdRp | 2.26 × 104 RNA copies μL−1 | 5 | – | 2 | 30 | |
| DNA nanoscaffold-based hybrid chain reaction | synthetic RNA conserved region | 0.96 pmol L−1 | 0 | – | 10 | 31 | |
| RT-LAMP | orf1ab | 20–200 RNA copies μL−1 | 130 | – | 60 | 32 | |
| RT-LAMP | N gene | 100 RNA copies μL−1 | 27 | – | 30 | 33 |
DPV, differential pulse voltammetry; EIS, electrochemical impedance spectroscopy; OCP-EIS, open-circuit potential-electrochemical impedance spectroscopy; RT-LAMP, reverse transcription loop-mediated isothermal amplification; SCC, signal conditioning circuit; SWV, square-wave voltammetry.
Prices reported correspond to a single test for each of the technologies and do not account for the equipment required in each case.
Figure 3.
Use of miniaturized and portable RAPID 1.0 for rapid point-of-care diagnosis of COVID-19
(A) Photo of mobile device-compatible handheld RAPID 1.0 during real-time sample analysis.
(B) Nyquist plots were obtained using ePAD coupled to a smart device for different concentrations of SP ranging from 1 pg mL−1 to 100 ng mL−1. The inset shows the calibration curve for the normalized RCT values of the different concentrations of SP recorded in triplicate.
To enhance the detectability (i.e., the LOD) of RAPID, we added 2.5-fold increased volumes of the modifiers (GA, ACE2, BSA, and Nafion) on the surface of the WE during the fabrication process. This approach allowed higher sensitivity toward the detection of SP, which was used to generate a calibration curve (Figure 3B). We attribute the enhanced detection (7-fold increase) of the paper-based version of RAPID compared with the phenolic-based electrode (eChip) to the higher amount of recognition element (ACE2) used on the WE's surface. However, it is worth noting that the eChip version already demonstrated excellent performance at detecting SARS-CoV-2 (Tables 1 and 2) and, although its sensitivity can be further increased by using a higher concentration of ACE2 (Figure 3B), this would increase the cost of the test since recombinant ACE2 accounts for 95% of the final cost of RAPID 1.0 (Figure 1C).
RAPID presented higher accuracy, specificity, and selectivity than most existing electrochemical methods available for SARS-CoV-2 detection (Table 3).19 We also assessed RAPID's specificity in cross-reactivity assays by exposing our sensor to the following seven viruses: three coronaviruses (MHV, murine hepatitis virus; HCoV-OC43, human coronavirus OC43; and human coronavirus 229E; Table S4) and four non-coronavirus viral strains (H1N1, A/California/2009; H3N2, A/Nicaragua; influenza B, B/Colorado; HSV2, herpes simplex virus 2; Table S4). We did not detect cross-reactivity events against any of the viruses tested (RCT < 10%) (Table S4), thus further highlighting the translatability of our diagnostic test. Our biosensor is inexpensive and portable, enabling decentralized diagnosis at the point of care. The time of detection of our approach (4 min) is significantly lower than existing diagnostic tests18 , 19 , 36 and could potentially be lowered even more by using engineered versions of human ACE2 with enhanced selective binding toward SARS-CoV-2 SP.13 The use of such ACE2 variants would also help reduce the rate of false positives in complex biofluids such as saliva.13 , 37 , 38
RAPID can also be multiplexed to allow detection of other emerging biological threats such as bacteria, fungi, and other viruses. Thus, our technology serves as a platform for the rapid diagnosis of COVID-19 and future endemic/pandemic outbreaks at the point-of-care. Its low cost, speed of detection, scalability, and implementation using smart devices and telemedicine platforms may facilitate much needed population-wide deployment and express use in hospitals, private companies, and public events.
Experimental procedures
Key resources table
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Chemicals | ||
| Human angiotensin converting enzyme 2 | GenScript | Z03484-1 |
| Sulfuric acid (H2SO4) | Sigma | 258105-1L-PC |
| Potassium chloride (KCl) | Sigma | P3911-1KG |
| Potassium ferricyanide K3 (Fe(CN)6) | Sigma | 244023-5G |
| Potassium ferrocyanide K4 (Fe(CN)6) | Sigma | P3289-5G |
| Bovine serum albumin | Sigma | A2153-10G |
| Glutaraldehyde | Fisher | S25341 |
| Nafion | Sigma | 527084-25ML |
| Phosphate-buffered saline | VWR | P32200 |
| Software and algorithms | ||
| Squidstat | Admiral Instruments | |
| PSTrace | PalmSens | |
| PSTouch | PalmSens | |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Cesar de la Fuente-Nunez (cfuente@upenn.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Test results used for analysis will be provided upon reasonable request.
Experimental model and subject details
Clinical sample preparation
Patient saliva and NP/OP swab samples were collected and processed according to the appropriate biosafety procedure (2020 CDC COVID-19 test protocol for details on specimen collection) at the Hospital of the University of Pennsylvania. Clinical matrices were heated at 56°C for 1 h for viral inactivation. Saliva used for optimization steps was collected from healthy donors (attested by negative result by qRT-PCR) and used promptly for experiments. The samples obtained are derived from patients with a wide range of symptoms (i.e., from negative cases to patients with very low viral loads to severe COVID-19 cases), thus providing a highly representative COVID-19 population sample set. All the steps were approved and performed under conditions detailed in University of Pennsylvania IRB 844145.
Method details
Fabrication of electrochemical devices
The electrochemical sensors were screen-printed in a three-electrode configuration cell (dimensions: 1.8 × 1.2 cm) on two accessible substrates: (1) a qualitative filter paper and (2) phenolic paper circuit board material. First, specific patterns were wax printed on A4 size filter paper using a commercial Xerox ColorQube 8570 printer (Xerox, Brazil). The patterns consisted of small white rectangles (1.1 × 1.7 cm) to delimit the electrochemical cell on paper substrates. In a single A4 size paper, 80 patterns were printed, thus affording 80 disposable ePADs. Following this, the screen-printing process was performed in the previously patterned paper using electrically conductive carbon and Ag/AgCl inks (Creative Materials, USA) to fabricate the working/auxiliary electrodes and reference electrodes, respectively. The printed filter paper sheets were then placed in a thermal oven for 30 min at 100°C. The heating process induces the curing step of the conductive tracks and melts the deposited wax layer that then penetrates the cellulosic structure, forming a 3D hydrophobic barrier around the hydrophilic patterns (electrochemical cell). Finally, the electrochemical paper-based analytical devices (ePADs) were cut with scissors, and the backside of the devices was covered with a transparent tape to prevent solution leakage through the device and to add structural integrity. The phenolic paper is a material largely used as a printed circuit board substrate. The boards were washed thoroughly with deionized water and isopropyl alcohol. The screen-printing process on the paper phenolic resin was performed using the same design and dimensions reported for the filter paper platform. The electrochemical circuit board-based devices (eChip) present a rigid substrate and low wettability that dispenses with the use of a hydrophobic barrier. After the curing step on the printed electrodes, they were cut into small pieces (2 × 2 cm) and a nonconductive layer was applied to delimit the electrode area.
Modification of the eChips and ePADs
The electrodes were washed with deionized water and cleaned and activated electrochemically by CV recorded in sulfuric acid solution (0.1 mol L−1) in the potential range −1.3 to 1.5 V at the scan rate of 100 mV s−1 for 5 cycles. The eChips were dried at room temperature and 4 μL of GA solution (25% in water) was added on the surface of the WE using the drop-casting method. After 1 h, 4 μL of ACE2 solution (0.32 μg mL−1) prepared in PBS medium was added on top of the WE and left to dry at room temperature for 1.5 h. Subsequently, 4 μL of BSA solution (1 mg mL−1) was added on the surface of the WE to stabilize the protein and block nonspecific sites of the electrode. After 30 min, 4 μL of Nafion solution (1.0% in PBS) was added to the WE's surface and left for 1 h before the final washing with deionized water. The ePADs were modified using the same protocol but applying 2.5-fold higher volume of the modifying agent solutions.
Electrochemical measurements
Squidstat Plus (Admiral Instruments) and Sensit Smart (PalmSens) potentiostats controlled by a laptop running the software Squidstat and a smartphone running the software PSTouch, respectively, were used to record all electrochemical data. The electrodes were characterized by a CV technique using a mixture of 5 mmol L−1 potassium ferricyanide/ferrocyanide in the medium of 0.1 mol L−1 KCl solution before and after electrode modification using a potential range of 0.7 to −0.3 V at the scan rate of 50 mV s−1. EIS was used to characterize the biosensor and for SARS-CoV-2 detection. The EIS measurements were performed using 200 μL of a mixture of 5 mmol L−1 ferricyanide/ferrocyanide prepared in 0.1 mol L−1 KCl solution added after the sample incubation on the electrode (10 μL of OP/NP or saliva samples) and the gentle washing process in PBS solution to remove the unbound SP/SARS-CoV-2. A sinusoidal signal was applied in the frequency range between 105 and 10−1 s−1 using a typical open-circuit potential of 0.15 V and an amplitude of 10 mV at room temperature.
Optimization tests
We evaluated the main experimental parameters and processes that affect the efficiency of the developed biosensor. For modification steps, both GA and BSA were used at high concentration levels to ensure the complete recovery of the electrode surface, providing the best conditions for the covalent attachment of ACE2 and its stabilization. The formation of the permselective membrane was evaluated by using different Nafion concentrations in the range of 0.5–3.0 wt %. After the biosensor preparation, we evaluated its response to different concentrations (1 pg mL−1 to 10 μg mL−1) of angiotensin II, the natural substrate of ACE2, to verify if the anchoring and stabilization strategies maintain the biological activity of ACE2. To assess the kinetics of interaction between SP and the architecture of the modified electrode, we carried out calibration curves ranging from 1 pg mL−1 to 1 ng mL−1 SP using different times of incubation (from 1 to 10 min) to obtain the best analytical response to RAPID1.0. Finally, the need for pretreatment of saliva samples was evaluated using three different approaches: (1) direct use of raw saliva, (2) 2 min of centrifugation at 10,000 rpm, and (3) simple dilution of sample 1:1 (v/v) with PBS. We performed this study with saliva samples because they present a greater matrix complexity (high viscosity and content of proteins, lipids, and other biomolecules that can cause biofouling of the electrodic surface) compared with NP/OP swab samples.
Cross-reactivity experiments
Cross-reactivity assays were carried out by exposing the sensor to three coronaviruses (MHV at 108 PFU mL−1, HCoV-OC43 at 104 PFU mL−1, and human coronavirus 229E at 107 PFU mL−1) and four non-coronavirus viral strains (H1N1, H3N2, influenza B, and HSV2, all at 105 PFU mL−1) to assess the specificity of our biosensor. The conditions used were the same as those used for all SARS-CoV-2 samples: incubation time of 5 min, 10 μL of virus sample, and EIS measurements as specified above (“electrochemical measurements”).
Quantification and statistical analysis
CV and electrochemical impedimetric spectroscopy measurements are presented as an average of three or seven different replicates for each condition and described in each figure caption. Graphs were created and statistical tests conducted in GraphPad Prism 9.
Acknowledgments
C.d.l.F.-N. holds a Presidential Professorship at the University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation, and acknowledges funding from the Institute for Diabetes, Obesity, and Metabolism, the Penn Mental Health AIDS Research Center of the University of Pennsylvania, the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM138201, and the Defense Threat Reduction Agency (DTRA; HDTRA11810041 and HDTRA1-21-1-0014). Research reported in this publication was supported by the Nemirovsky Prize and by funds provided by the Dean's Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania. W.R.A. acknowledges funding from the Brazilian funding agencies CAPES (grant 88887.479793/2020-00), FAPESP (grant 2018/08782-1), FAEPEX/PRP/UNICAMP (grant 3374/19), and CNPq (grants 438828/2018-6, 401256/2020-0) for supporting the research. All figures were prepared using the Biorender drawing toolkit. We thank Jonathan A. Epstein for his support and for revising the manuscript. We also thank Scott Hensley, Sara Cherry, Susan Weiss, Ronald Collman, Michael Feldman, Benjamin Abella, and Paul Calahan for donating samples. A provisional patent application has been filed on the technology described in this article (Invention Disclosure 21-9515). This study was performed under the University of Pennsylvania IRB protocol 844145.
Author contributions
M.D.T.T., W.R.A., and C.d.l.F.-N. designed the experiments. W.R.A. manufactured the electrochemical devices. M.D.T.T., L.F.L., and A.L.F. performed experiments. M.D.T.T., W.R.A., and C.d.l.F.-N. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: May 9, 2021
Supplemental information
References
- 1.The New York Times Coronavirus map: tracking the global outbreak. https://www.nytimes.com/interactive/2020/world/coronavirus-maps.html
- 2.Government A.C. Information of coronavirus (COVID-19) testing. https://health.arlingtonva.us/covid-19-coronavirus-updates/covid-19-testing/
- 3.Service R. Coronavirus antigen tests: quick and cheap, but too often wrong? Science. 2020 doi: 10.1126/science.abc9586. [DOI] [Google Scholar]
- 4.Administration U.S.F.&D. In vitro diagnostics EUAs. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas
- 5.Muñoz J., Montes R., Baeza M. Trends in electrochemical impedance spectroscopy involving nanocomposite transducers: characterization, architecture surface and Bio-Sensing. TrAC Trends Anal. Chem. 2017;97:201–215. doi: 10.1016/j.trac.2017.08.012. [DOI] [Google Scholar]
- 6.Bahadır E.B., Sezgintürk M.K. A Review on impedimetric biosensors. Artif. Cells Nanomedicine, Biotechnol. 2016;44:248–262. doi: 10.3109/21691401.2014.942456. [DOI] [PubMed] [Google Scholar]
- 7.Yang J., Petitjean S.J.L., Koehler M., Zhang Q., Dumitru A.C., Chen W., Derclaye S., Vincent S.P., Soumillion P., Alsteens D. Molecular interaction and Inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbosa O., Ortiz C., Berenguer-Murcia Á., Torres R., Rodrigues R.C., Fernandez-Lafuente R. Glutaraldehyde in bio-catalysts design: a useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014;4:1583–1600. doi: 10.1039/C3RA45991H. [DOI] [Google Scholar]
- 10.Pereira A.R., Sedenho G.C., Souza J.C.P.de, Crespilho F.N. Advances in enzyme bioelectrochemistry. An. Acad. Bras. Cienc. 2018;90(1 suppl 1):825–857. doi: 10.1590/0001-3765201820170514. [DOI] [PubMed] [Google Scholar]
- 11.Mauritz K.A., Moore R.B. State of understanding of Nafion. Chem. Rev. 2004;104:4535–4586. doi: 10.1021/cr0207123. [DOI] [PubMed] [Google Scholar]
- 12.e Silva R.F., Longo Cesar Paixão T.R., Der Torossian Torres M., de Araujo W.R. Simple and inexpensive electrochemical paper-based analytical device for sensitive detection of Pseudomonas Aeruginosa. Sensors Actuators B Chem. 2020;308:127669. doi: 10.1016/j.snb.2020.127669. [DOI] [Google Scholar]
- 13.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamal A.J., Mozafarihashjin M., Coomes E., Powis J., Li A.X., Paterson A., Anceva-Sami S., Barati S., Crowl G., Faheem A., et al. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2. Clin. Infect. Dis. 2020:ciaa848. doi: 10.1093/cid/ciaa848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uygun Z.O., Ertuğrul Uygun H.D. A short footnote: circuit design for Faradaic impedimetric sensors and biosensors. Sensors Actuators B Chem. 2014;202:448–453. doi: 10.1016/j.snb.2014.05.029. [DOI] [Google Scholar]
- 16.Bertok T., Lorencova L., Chocholova E., Jane E., Vikartovska A., Kasak P., Tkac J. Electrochemical impedance spectroscopy based biosensors: mechanistic principles, analytical examples and challenges towards commercialization for assays of protein cancer biomarkers. ChemElectroChem. 2019;6:989–1003. doi: 10.1002/celc.201800848. [DOI] [Google Scholar]
- 17.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 18.Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., Valdez J., Flynn J., Williams S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021;171:112709. doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhteg K., Jarrett J., Richards M., Howard C., Morehead E., Geahr M., Gluck L., Hanlon A., Ellis B., Kaur H., et al. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J. Clin. Virol. 2020;127:104384. doi: 10.1016/j.jcv.2020.104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao S.N., Manissero D., Steele V.R., Pareja J. A narrative systematic review of the clinical utility of cycle threshold values in the context of COVID-19. Infect. Dis. Ther. 2020;9:573–586. doi: 10.1007/s40121-020-00324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rath S.L., Kumar K. Investigation of the effect of temperature on the structure of SARS-CoV-2 spike protein by molecular dynamics simulations. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.583523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner A.J. The Protective Arm of the Renin Angiotensin System (RAS) Elsevier; 2015. ACE2 cell biology, regulation, and physiological functions; pp. 185–189. [DOI] [Google Scholar]
- 24.Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H., Liu F., Xie W., Zhou T.-C., OuYang J., Jin L., Li H., Zhao C.-Y., Zhang L., Wei J., et al. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensors Actuators B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021;176:112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., et al. CRISPR–Cas12-Based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 31.Jiao J., Duan C., Xue L., Liu Y., Sun W., Xiang Y. DNA nanoscaffold-based SARS-CoV-2 detection for COVID-19 diagnosis. Biosens. Bioelectron. 2020;167:112479. doi: 10.1016/j.bios.2020.112479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baek Y.H., Um J., Antigua K.J.C., Park J.-H., Kim Y., Oh S., Kim Y., Choi W.-S., Kim S.G., Jeong J.H., et al. Development of a Reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. Microbes Infect. 2020;9:998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozer T., McMahon C., Henry C.S. Advances in paper-based analytical devices. Annu. Rev. Anal. Chem. 2020;13:85–109. doi: 10.1146/annurev-anchem-061318-114845. [DOI] [PubMed] [Google Scholar]
- 35.Ataide V.N., Mendes L.F., Gama L.I.L.M., de Araujo W.R., Paixão T.R.L.C. Electrochemical paper-based analytical devices: ten years of development. Anal. Methods. 2020;12:1030–1054. doi: 10.1039/C9AY02350J. [DOI] [Google Scholar]
- 36.Kaushik A.K., Dhau J.S., Gohel H., Mishra Y.K., Kateb B., Kim N.-Y., Goswami D.Y. Electrochemical SARS-CoV-2 sensing at point-of-care and artificial intelligence for intelligent COVID-19 management. ACS Appl. Bio Mater. 2020 doi: 10.1021/acsabm.0c01004. acsabm.0c01004. [DOI] [PubMed] [Google Scholar]
- 37.Glasgow A., Glasgow J., Limonta D., Solomon P., Lui I., Zhang Y., Nix M.A., Rettko N.J., Zha S., Yamin R., et al. Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc. Natl. Acad. Sci. 2020;117:28046–28055. doi: 10.1073/pnas.2016093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorokina M., Teixeira M.C., Barrera-Vilarmau J.S., Paschke R., Papasotiriou I., Rodrigues J.P.G.L.M., Kastritis P.L. Structural models of human ACE2 variants with SARS-CoV-2 spike protein for structure-based Drug design. Sci. Data. 2020;7:309. doi: 10.1038/s41597-020-00652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Test results used for analysis will be provided upon reasonable request.