Summary
The capacity of antibodies to engage with immune cells via the Fc region is important in preventing and controlling many infectious diseases. The evolution of such antibodies during convalescence from coronavirus disease 2019 (COVID-19) is largely unknown. We develop assays to measure Fc-dependent antibody functions against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S)-expressing cells in serial samples from subjects primarily with mild-moderate COVID-19 up to 149 days post-infection. We find that S-specific antibodies capable of engaging Fcγ receptors decay over time, with S-specific antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent phagocytosis (ADP) activity within plasma declining accordingly. Although there is significant decay in ADCC and ADP activity, they remain readily detectable in almost all subjects at the last time point studied (94%) in contrast with neutralization activity (70%). Although it remains unclear the degree to which Fc effector functions contribute to protection against SARS-CoV-2 re-infection, our results indicate that antibodies with Fc effector functions persist longer than neutralizing antibodies.
Keywords: SARS-CoV-2, COVID-19, antibody, decay, Fc effector functions, ADCC, ADP, phagocytosis, trogocytosis, convalescence
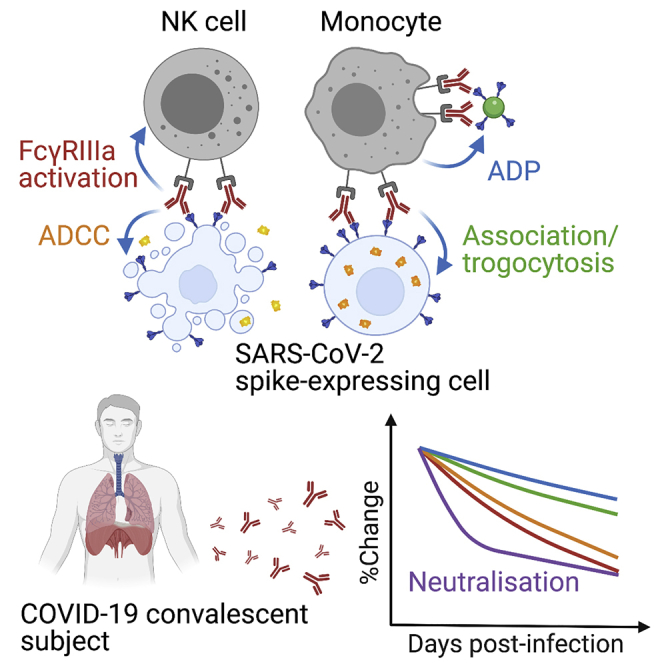
Graphical abstract
Highlights
SARS-CoV-2 spike-specific Fc effector functions may contribute to COVID-19 control
Spike-specific ADCC and ADP responses decline in the 4 months post-infection
Compared to neutralization, ADCC and ADP are detectable longer post-infection
Cross-reactive antibodies against human coronavirus spike increase post-infection
Lee et al. report the decline of Fc-dependent antibody functions against SARS-CoV-2 spike in COVID-19 convalescent subjects up to 149 days post-infection. Unlike neutralization activity, plasma ADCC and ADP responses are sustained in the majority of subjects at the last time point measured.
Introduction
Most individuals who recover from coronavirus disease 2019 (COVID-19) develop binding and neutralizing antibody responses against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein,1,2 with neutralizing antibody responses generally targeted to the receptor-binding domain (RBD) of S.3 Passive transfer of neutralizing monoclonal antibodies (mAbs) can protect animal models from subsequent SARS-CoV-2 challenge,4, 5, 6 suggesting neutralization is likely to be a correlate of protection in humans.7 However, the duration of protection from re-infection in humans conferred by neutralizing antibodies is not known. Several studies now show neutralizing antibodies decline rapidly during early convalescence,2,8,9 with the magnitude of the antibody response positively correlating with disease severity.10,11 Following mild COVID-19, many subjects mount modest neutralizing antibody responses that decline to undetectable levels within 60 days, despite the maintenance of S- and RBD-specific immunoglobulin G (IgG) binding antibodies.10 Given that reported cases of SARS-CoV-2 re-infection have been rare to date, it is likely that immune responses beyond neutralization, including T cell responses,12 contribute to SARS-CoV-2 protective immunity. Apart from direct virus neutralization, antibodies can also mediate antiviral activity, such as antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent phagocytosis (ADP), by engaging Fc gamma receptors (FcγR) on NK cells or phagocytes. Fc effector functions contribute to the prevention and control of other viral infections, including HIV-1, influenza, and Ebola.13, 14, 15 Butler et al.16 recently showed that SARS-CoV-2 RBD-specific antibodies within plasma could crosslink Fcγ receptors and mediate ADP and antibody-dependent complement deposition. Importantly, two recent challenge studies demonstrated that certain RBD-specific mAbs rely on Fc effector functions to mediate protection against SARS-CoV-2 in mice.17,18
We previously reported that binding antibodies to SARS-CoV-2 S exhibit substantially longer half-lives than the neutralizing antibody response,8 suggesting that Fc-mediated antibody function may extend the protective window beyond that inferred from neutralizing activity alone. At present, analyses of Fc-mediated functions of SARS-CoV-2 antibodies within COVID-19 convalescent subjects have focused upon cross-sectional analyses or short-term longitudinal studies up to 1 to 2 months post-symptom onset.16,19,20 We extend these findings and analyze Fc effector functions mediated by S-specific antibodies in a cohort of 53 convalescent individuals up to 149 days post-symptom onset. We developed functional assays using SARS-CoV-2 S-expressing cells to comprehensively analyze plasma ADCC and ADP activity against SARS-CoV-2 S. Our results show that plasma ADCC and ADP activity decays over the first 4 months post-infection, mirroring the decline in S-specific IgG titers. Importantly, however, S-specific antibodies capable of Fc-mediated antiviral activity remain readily detectable in almost all donors up to 4 months post-infection, even in donors whose neutralizing antibody responses have waned to undetectable levels. Although the protective potential of antibody Fc effector functions against SARS-CoV-2 re-infection remains to be determined, our results suggest that ADCC and ADP activity outlasts neutralizing activity within plasma following convalescence from COVID-19.
Results
Decay of dimeric FcγR-binding S and RBD-specific antibodies
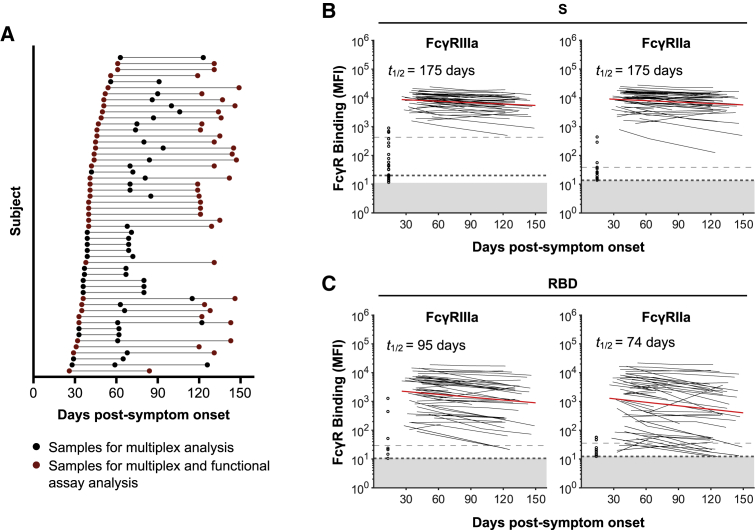
We collected repeated (2–4) longitudinal samples from a cohort of 53 subjects after recovery from COVID-19 (Figure 1A; Table S1). The first sample was collected at a median of 41 days post-symptom onset (interquartile range [IQR] 36–48), and the last sample was collected at a median of 123 days post-symptom onset (IQR 86–135). The engagement of dimeric recombinant soluble FcγRIIIa and FcγRIIa proteins by antibodies mimics the immunological synapse required for FcγR activation of innate immune cells and is a surrogate measure of ADCC and ADP, respectively.21,22 To determine the dynamics of Fc-mediated function in plasma samples over time, we measured the capacity of dimeric FcγRIIIa and FcγRIIa receptors to engage antibodies specific for SARS-CoV-2 S antigens (trimeric S, S1, or S2 subunits or the RBD; Table S2) with a multiplex bead array. Using mixed-effects modeling, we assessed the fit of single-phase or two-phase decay in FcγR binding between the time points analyzed. We found that dimeric FcγRIIIa (V158)-binding antibodies against SARS-CoV-2 trimeric S and RBD both had single-phase decay kinetics with half-lives (t1/2) of 175 and 95 days, respectively (Figures 1B and 1C). Dimeric FcγRIIa (H131) binding antibodies against SARS-CoV-2 trimeric S and RBD also decayed constantly with t1/2 of 175 and 74 days, respectively. Kinetics of decay for dimeric FcγR-binding antibodies against S and RBD for the lower affinity polymorphisms of FcγRIIIa (F158) and FcγRIIa (R131) were broadly similar to their higher affinity counterparts (Figure S1A), with dimeric FcγR-binding antibodies against RBD decaying faster than for S. Consistent with our previous report that S1-specific IgG decays faster than S2-specific IgG,8 FcγR binding activity with antibodies against the S1 subunit decayed faster than that of S2 (FcγRIIIa, t1/2 of 84 versus 227 days; FcγRIIa, t1/2 of 65 versus 317 days; Figure S1B).
Figure 1.
Dynamics of SARS-CoV-2 S and RBD-specific dimeric FcγR-binding antibodies in COVID-19 convalescent individuals
(A) Timeline of sample collection for each COVID-19 convalescent subject (n = 53). Subjects with 2 samples at least 60 days apart were chosen for functional assay analysis (n = 36).
(B and C) Kinetics of SARS-CoV-2 S and RBD-specific dimeric FcγRIIIa (V158) and dimeric FcγRIIa (H131) binding antibodies over time measured using the bead-based multiplex assay. The best-fit decay slopes (red lines) and estimated half-lives (t1/2) are indicated for COVID-19 convalescent individuals. Uninfected controls (n = 33) are shown in open circles, with the median and 90% percentile responses presented as thick and thin dashed lines, respectively. The limit of detection is shown as the shaded area.
See also Figure S1.
Decay of S-specific ADCC
ADCC could play a role in eliminating cells infected with SARS-CoV-2. We generated Ramos- and A549-derived cell lines as model target cells that stably express membrane-localized S with either mOrange2 or luciferase reporters (Figures S2A and S2B). The capacity of plasma IgG to recognize S was measured in 36 subjects in our cohort who had at least 60 days between the first and last visits (median of 89 days between first and last visits; Table S1) and 8 seronegative controls. Using a Ramos cell line expressing high levels of S (Ramos S-Orange), we find IgG binding to cell-surface-displayed S proteins decayed significantly between the first and last visits (p < 0.0001; Figure S2D; gating in Figure S2C) with a half-life of 97 days (Figure S2E). These results are consistent with the decay of S-specific IgG titers we observed previously8 and the decay of dimeric FcγR-binding antibodies against S in Figure 1B.
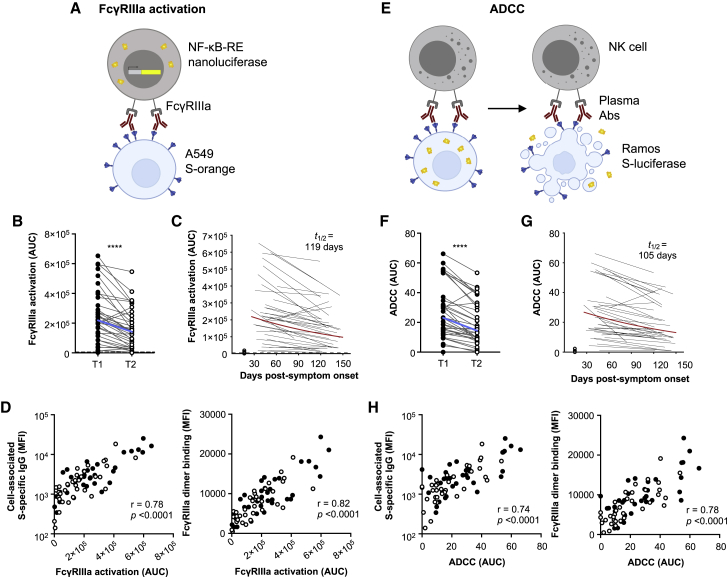
As a surrogate measure of ADCC, we next used FcγRIIIa reporter cells to quantify the capacity of S-specific antibodies in plasma to engage cell-surface FcγRIIIa and activate downstream nuclear factor κB (NF-κB) signaling (measured by induced nanoluciferase expression in the FcγRIIIa reporter cells co-cultured with S-expressing A549 cells; Figures 2A and S3A). FcγRIIIa activity decayed significantly over time (p < 0.0001; Figure 2B), with a half-life of 119 days (Figure 2C), and was correlated with S-specific IgG titers measured using stably transduced cells or by binding to dimeric FcγRIIIa (Figure 2D). Because FcγRIIIa crosslinking and activation may not necessarily reflect downstream target lysis, we next performed an ADCC assay to confirm antibody recognition could mediate killing of S-expressing cells. We quantified the loss of cellular luciferase signal in Ramos S-luciferase target cells in the presence of convalescent plasma and primary human NK cells (Figures 2E and S3B). S-specific ADCC decayed significantly over time (p < 0.0001; Figure 2F), with a half-life of 105 days (Figure 2G), and correlated with both cell-associated S-specific IgG and dimeric FcγRIIIa-binding antibodies against S (Figure 2H). These positive correlations demonstrate that S-specific IgG and FcγRIIIa-binding antibodies are important factors for S-specific ADCC. S-specific FcγRIIIa-activating antibodies also correlated strongly with S-specific ADCC (Figure S3C), showing that FcγRIIIa-mediated activation is a surrogate for NK-cell-mediated ADCC.
Figure 2.
ADCC responses in COVID-19 convalescent individuals over time
(A) Schematic of the FcγRIIIa NF-κB activation assay. IIA1.6 cells expressing FcγRIIIa V158 and a NF-κB response-element-driven nanoluciferase reporter were co-incubated with A549 S-orange target cells and plasma from COVID-19 convalescent individuals or uninfected controls. The engagement of FcγRIIIa by S-specific antibodies activates downstream NF-κB signaling and nanoluciferase expression.
(B) S-specific FcγRIIIa-activating plasma antibodies in COVID-19 convalescent individuals in the first (T1; filled) and last (T2; open) time points available. Blue lines indicate the median responses of COVID-19 convalescent individuals (N = 36), and dashed lines indicate median responses of uninfected controls (N = 8).
(C) The best-fit decay slopes (red lines) and estimated half-life (t1/2) for FcγRIIIa-activating plasma antibodies in COVID-19 convalescent individuals. Uninfected controls are shown in open circles, with the median response presented as a dashed line.
(D) Correlation of S-specific FcγRIIIa-activating antibodies to cell-associated S-specific IgG and S-specific dimeric FcγRIIIa-binding antibodies.
(E) Schematic of the luciferase-based ADCC assay. Purified NK cells from healthy donors were co-incubated with Ramos S-luciferase target cells and plasma. ADCC is measured as the loss of cellular luciferase.
(F) S-specific ADCC mediated by plasma antibodies from COVID-19 convalescent individuals in the first (T1; filled) and last (T2; open) time points available. Blue lines indicate the median responses of COVID-19 convalescent individuals (N = 36), and dashed lines indicate median responses of uninfected controls (N = 8).
(G) The best-fit decay slopes (red lines) and estimated half-life (t1/2) for FcγRIIIa-activating plasma antibodies in COVID-19 convalescent individuals. Uninfected controls are shown in open circles, with the median response presented as a dashed line.
(H) Correlation of S-specific ADCC to cell-associated S-specific IgG and S-specific dimeric FcγRIIIa-binding antibodies. Statistical analyses were performed with the Wilcoxon signed-rank test (∗∗∗∗p < 0.0001). Correlations were performed with the non-parametric Spearman test.
See also Figures S2 and S3.
Decay of S-specific ADP
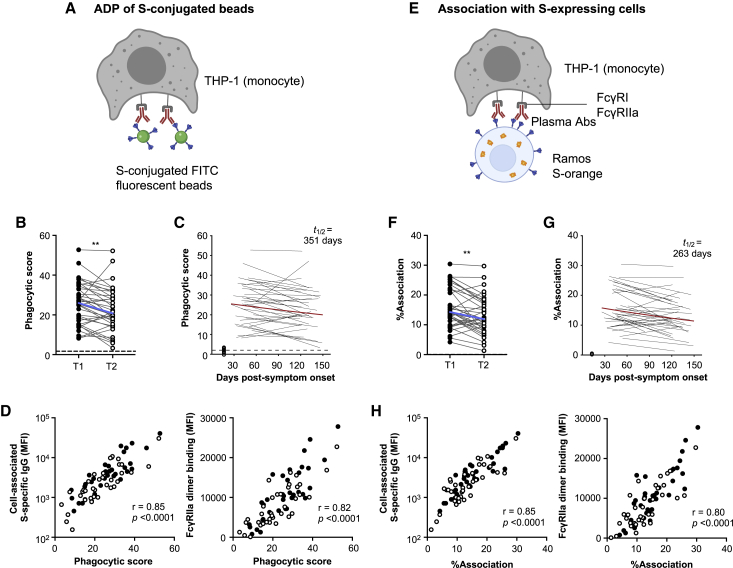
As has been suggested for SARS-CoV, ADP could play a role in eliminating antibody-opsonized virions.23 We first used a well-established ADP assay24 to measure antibody-mediated uptake of S-conjugated fluorescent beads into THP-1 monocytes (Figure 3A; gating in Figures S4A and S4B and optimization in Figures S5A–S5C). ADP of S-conjugated beads was detected in all 36 subjects at the first time point studied but decayed significantly over time (p < 0.01; Figure 3B), with a half-life of 351 days (Figure 3C). ADP of S-conjugated beads correlated with cell-associated S-specific IgG and S-specific dimeric FcγRIIa-binding antibodies (Figure 3D).
Figure 3.
ADP responses in COVID-19 convalescent individuals over time
(A) Schematic of the bead-based ADP assay. THP-1 cells were incubated with S-conjugated fluorescent beads and plasma from COVID-19 convalescent individuals or uninfected controls. The uptake of fluorescent beads was measured by flow cytometry.
(B) ADP of S-conjugated beads mediated by plasma antibodies from COVID-19 convalescent individuals in the first (T1) and last (T2) time points available. Blue lines indicate the median responses of COVID-19 convalescent individuals (N = 36), and dashed lines indicate median responses of uninfected controls (N = 8).
(C) The best-fit decay slopes (red lines) and estimated half-life (t1/2) for plasma ADP activity in COVID-19 convalescent individuals. Uninfected controls are shown in open circles, with the median response presented as a dashed line.
(D) Correlation of ADP to cell-associated S-specific IgG and S-specific dimeric FcγRIIa-binding antibodies.
(E) Schematic of the THP-1 FcγR-dependent cell association assay. Ramos S-orange cells were pre-incubated with plasma prior to co-incubation with THP-1 cells. The association of THP-1 cells with Ramos S-orange cells was measured by flow cytometry.
(F) FcγR-dependent association of THP-1 cells with Ramos S-orange cells mediated by plasma antibodies from COVID-19 convalescent individuals in the first (T1) and last (T2) time points available. Blue lines indicate the median responses of COVID-19 convalescent individuals (N = 36), and dashed lines indicate median responses of uninfected controls (N = 8).
(G) The best-fit decay slopes (red lines) and estimated half-life (t1/2) for THP-1 association in COVID-19 convalescent individuals. Uninfected controls are shown in open circles, with the median response presented as a dashed line.
(H) Correlation of association of THP-1 cells with Ramos S-orange cells to cell-associated S-specific IgG and S-specific dimeric FcγRIIIa-binding antibodies. Red lines indicate the median responses of COVID-19 convalescent individuals (N = 36), and dashed lines indicate median responses of uninfected controls (N = 8). Statistical analyses were performed with the Wilcoxon signed-rank test (∗∗p < 0.01). Correlations were performed with the non-parametric Spearman test.
See Figure S4 for gating strategies and Figure S5 for assay optimization.
In addition to uptake of antibody-opsonized virions, phagocytes could also potentially mediate clearance of infected cells expressing SARS-CoV-2 S on the cell surface. THP-1 cells have been shown to mediate both trogocytosis (sampling of plasma membrane fragments from target cells that can lead to cell death) and phagocytosis via antibody Fc-FcγR interactions with target cells.25, 26, 27 As such, we measured the FcγR-dependent association of THP-1 cells with Ramos S-orange cells following incubation with plasma from convalescent individuals or uninfected controls (Figure 3E; gating in Figure S4C and optimization in Figures S5D–S5F). Association of THP-1 cells with Ramos S-orange cells was detected in all subjects at the first time point but decayed significantly over time (p < 0.01; Figure 3F), with a half-life of 263 days (Figure 3G), correlating with IgG binding to cell-associated S and S-specific dimeric FcγRIIa-binding antibodies (Figure 3H). These strong positive correlations indicate S-specific IgG and FcγRIIa-binding antibodies are important for THP-1-mediated ADP and cell association with S-expressing cells.
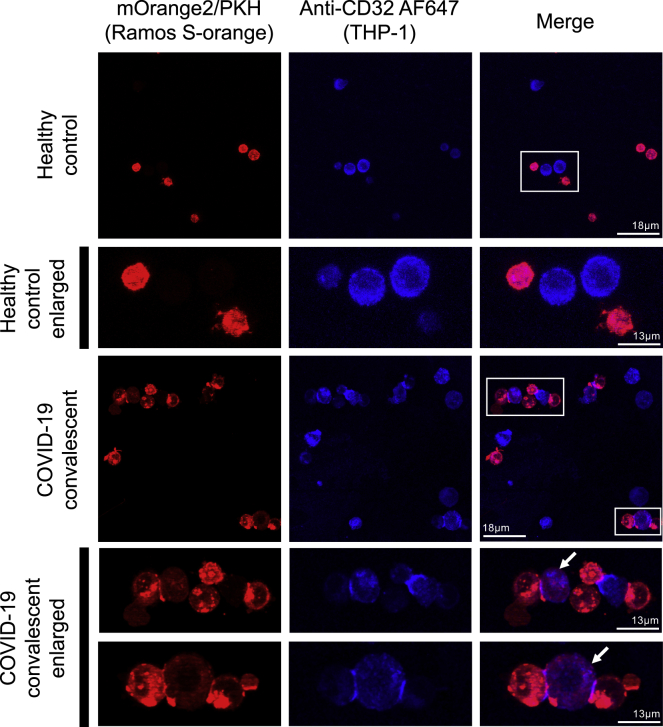
To confirm that Fc-dependent cell association can lead to trogocytosis, we performed confocal microscopy to visualize the interaction of THP-1 cells and target Ramos S-orange cells (Figure 4). We confirmed that close association of THP-1 cells (blue) and Ramos S-orange cells (red) only occurred in the presence of COVID-19 convalescent plasma. Further, we observed that THP-1 cells acquired PKH-26 dye from Ramos S-orange cells, indicative of trogocytosis of the S-expressing Ramos cell membrane by the THP-1 cells.
Figure 4.
Confocal microscopy visualization of Fc-mediated association and trogocytosis
THP-1 monocytes were incubated with Ramos S-orange target cells in the presence of healthy control plasma or COVID-19 convalescent plasma. Cells were fixed on slides and visualized using a confocal microscope (60× objective). THP-1 cells were stained with CD32-AF647 (blue), and Ramos S-orange cells (expressing SARS-CoV-2 spike and mOrange2) were labeled with the membrane dye PKH-26 (red). Examples of trogocytosis in the COVID-19 convalescent plasma sample (white boxes) are enlarged and shown in the fourth and fifth rows. The white arrows indicate THP-1 cells (blue) that have received plasma membrane proteins from Ramos S-orange cells (red). No trogocytosis occurred in the healthy control sample (white box, enlarged in second row). Scale bars are 13 μM for the first and third rows and 18 μM for the enlarged images in the second, fourth, and fifth rows.
Cross-reactivity with HCoV S-specific antibodies
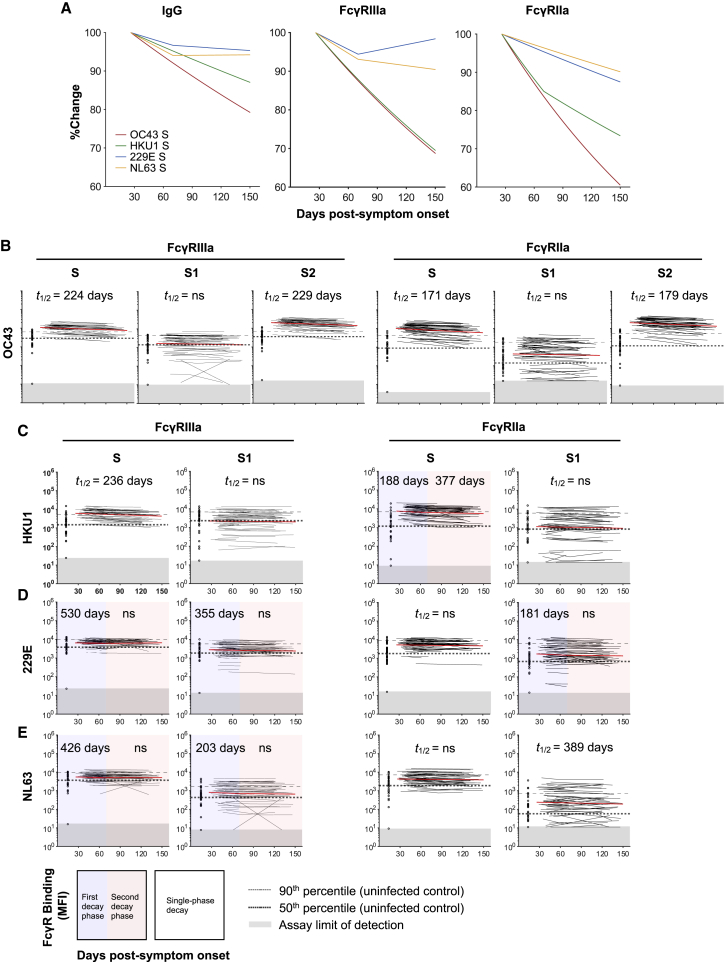
Cross-reactive antibodies between endemic human coronaviruses (HCoVs) and SARS-CoV-2 have been widely reported,28,29 suggesting past exposure to HCoVs may prime ADCC and ADP immunity against SARS-CoV-2. In addition, several studies have shown back boosting of antibodies against endemic HCoVs following infection with SARS-CoV-2,30,31 likely due to the recall of pre-existing B cell responses against conserved regions of S. We thus determined whether IgG antibody levels against S from four HCoV strains (OC43, HKU1, 229E, and NL63; Table S2) were higher in COVID-19 convalescent subjects compared to uninfected healthy controls. Using a multiplex bead array, we found that COVID-19 convalescent subjects had increased IgG antibodies against S from the betacoronaviruses OC43 and HKU1 (which are more closely related to SARS-CoV-2) at the first time point sampled compared to uninfected controls (Figure S6), while there was no difference in IgG levels against S from the alphacoronaviruses 229E and NL63. Correspondingly, the elevated IgG against OC43 and HKU1 S decayed over time while IgG against 229E and NL63 S remained stable (Figure 5A). We then measured whether dimeric FcγR-binding antibodies against HCoV S antigens in COVID-19 convalescent individuals declined over time. Dimeric FcγR-binding antibodies against OC43 and HKU1 S were much higher in COVID-19 convalescent individuals than in healthy controls (Figures 5B and 5C) and decayed more rapidly over time compared to that against 229E and NL63 (Figures 5D and 5E). Although there was an overall decay of dimeric FcγR-binding antibodies against OC43 S (Figure 5B; FcγRIIIa t1/2 = 224; FcγRIIa t1/2 = 171 days), this was largely due to a decay in antibodies against the more conserved S2 subunit (FcγRIIIa t1/2 = 229; FcγRIIa t1/2 = 179 days). FcγR-binding antibodies against the S1 subunit were not increased compared to healthy controls and did not change over time (Figure 5B). This was also the case for HKU1, where dimeric FcγR-binding antibodies against S decayed over time, but antibodies against the S1 subunit did not change (Figure 5C).
Figure 5.
Dynamics of dimeric FcγR-binding antibodies against HCoV S antigens in COVID-19 convalescent individuals
(A) Best-fit decay slopes of IgG and dimeric FcγR-binding antibodies against S from HCoV strains OC43, HKU1, 229E, and NL63. The responses at time point 1 for each parameter are set to 100%, and the %change over time is shown.
(B–E) Kinetics of dimeric FcγRIIIa (V158) and FcγRIIa (H131) binding antibodies against S, S1, or S2 from HCoV strains (B) OC43, (C) HKU1, (D) 229E, and (E) NL63 over time in COVID-19 convalescent individuals (N = 53) measured using the bead-based multiplex assay. The best-fit decay slopes (red lines) and estimated half-lives (t1/2) are indicated for COVID-19 convalescent individuals. Uninfected controls (N = 33) are shown in open circles, with the median and 90% percentile responses presented as thick and thin dashed lines, respectively. The limit of detection is shown as the shaded area.
See also Figure S6.
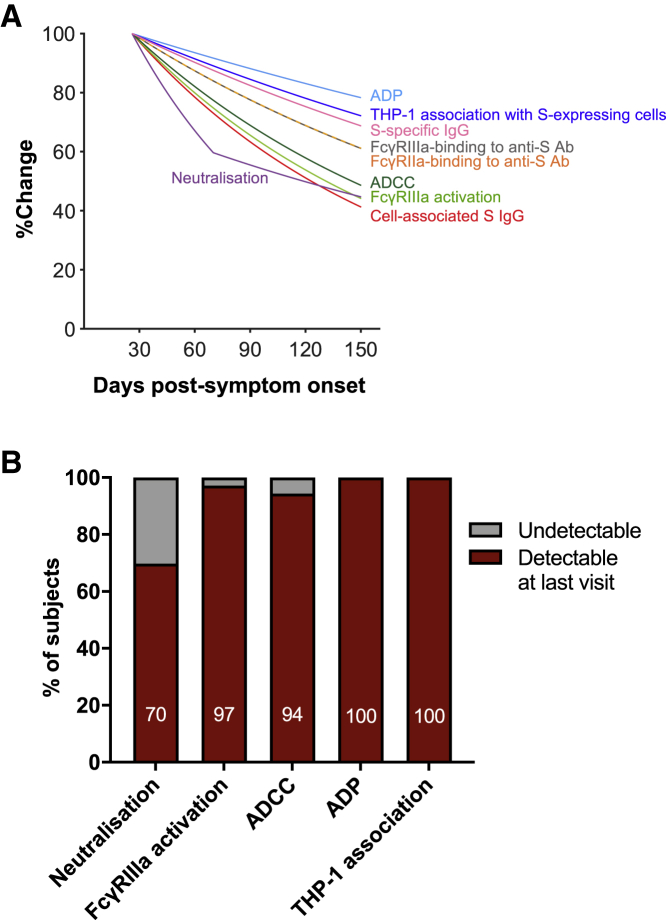
Decay kinetics of S-specific antibodies, neutralization, and Fc effector functions
To compare the decay kinetics of S-specific antibodies, neutralization, and Fc effector functions, we plotted the best-fit decay slopes over time as a percentage of the response measured at time point 1 (Figure 6A). The best-fit decay slopes of S-specific IgG and plasma neutralization titers (Figure S7) were obtained from a previous dataset that encompasses the same subjects analyzed for dimeric FcγR-binding antibodies and Fc effector functions.8 The general decline in plasma S-specific IgG titers and dimeric FcγR-binding activity was similarly reflected in reductions in Fc effector functions during convalescence from COVID-19. Importantly, Fc effector functions at the last time point sampled were still readily detectable above baseline activity observed in uninfected controls (97% for FcγRIIIa activation, 94% for ADCC, 100% for ADP, and 100% for THP-1 association). This contrasted with plasma neutralization activity, which was detectable above background for only 70% of subjects (Figure 6B). The longer persistence of S-specific IgG and dimeric FcγR-binding antibodies against S has important implications for the durability of SARS-CoV-2 immunity following the decline of neutralizing antibodies.
Figure 6.
Decay kinetics of binding antibodies, neutralization, and Fc effector functions following SARS-CoV-2 infection
(A) Best-fit decay slopes of various antibody parameters against SARS-CoV-2 S over time. The responses at time point 1 for each parameter are set to 100%, and the %change over time is shown.
(B) The percentage of subjects having detectable responses above (red) and below (gray) background levels at the last visit are shown. Background levels for each assay were the median responses of uninfected controls.
Discussion
Using a multiplex bead array and assays measuring Fc effector functions against SARS-CoV-2 S, we find that FcγR-binding, ADCC, and ADP activities of S-specific antibodies decay during convalescence from COVID-19. The decline of plasma ADCC and ADP activity correlated with the decay of S-specific IgG and FcγR-binding antibodies. Importantly, Fc effector functions were readily detectable above uninfected controls in 94% of subjects for all assays at the last time point sampled, in contrast with neutralization activity, which remained detectable above background for only 70% of subjects. Although neutralizing antibodies are likely to form a correlate of protection for SARS-CoV-2,7,32 several studies find that neutralizing antibodies in convalescent donors with mild COVID-19 wane rapidly.2,8,9 The rapid decline of plasma neutralization activity in the early weeks following infection is likely in part explained by the rapid decline of plasma IgM and IgA titers against S and RBD,20,33 which substantially contribute to neutralization of SARS-CoV-2.34, 35, 36 Given the relative scarcity of re-infection cases reported to date, it is likely that immune responses beyond neutralization, including antibody Fc effector functions and T cell responses, contribute to long-term protection from SARS-CoV-2. Indeed, a recent study demonstrated that cellular immunity in convalescent macaques, mainly CD8+ T cells, contribute to protection against re-challenge after neutralizing antibodies have waned.37
Our results demonstrate that FcγR-binding antibodies against betacoronaviruses OC43 and HKU1 are much higher in COVID-19 convalescent individuals compared to uninfected controls. This could either be due to the back boosting of pre-existing HCoV antibodies that are cross-reactive with SARS-CoV-228,29 or the de novo generation of SARS-CoV-2 antibodies that are cross-reactive with conserved HCoV epitopes. Cross-reactive S antibodies were largely directed against the more conserved S2 subunit, in line with other reports,28,29 which also likely explains the longer half-life of S2 antibodies that we observed relative to S1 antibodies. A recent study found cross-reactive binding and neutralizing antibodies against SARS-CoV-2 S2 in uninfected children and adolescents,28 suggesting prior infections with OC43 or HKU1 can elicit cross-reactive antibodies against the S2 subunit of SARS-CoV-2 S. These findings raise the interesting question of whether cross-reactive antibodies are recalled rapidly during early SARS-CoV-2 infection and can contribute to Fc effector functions against conserved epitopes within the S2 subunit. The presence of cross-reactive S2-specific antibodies capable of mediating Fc effector functions in early infection could potentially ameliorate disease symptoms and severity. Follow-up studies to dissect the influence of S1 or S2 antibody epitope localization on FcγR engagement and the impact on Fc effector functions are also warranted.
Initial concerns for antibody-dependent enhancement (ADE) of COVID-19 were driven by the reported association of higher SARS-CoV-2 antibody titers with severe disease.38 However, this could simply be the result of prolonged antigen exposure due to higher viral loads. Importantly, Zohar et al.33 showed that, in subjects with severe COVID-19, those who survived had higher levels of S-specific antibodies and Fc-mediated effector functions compared to those who died. Notably, numerous trials of convalescent plasma (CP) therapy for COVID-19 have been safely conducted,39, 40, 41 with no enhancement of disease reported to date.42, 43, 44 Because RBD-specific IgG1 antibodies in severe COVID-19 are more likely to have afucosylated Fc regions and trigger hyper-inflammatory responses from monocytes and macrophages,45,46 there could be implications for ADE in people who are re-infected with SARS-CoV-2 after initial neutralizing antibodies have waned but non-neutralizing antibodies remain. Excessive Fc-mediated effector functions and immune complex formation in the absence of neutralization could potentially trigger a hyper-inflammatory response and lead to ADE of disease, as observed for respiratory syncytial virus and measles infections.47,48 Although ADE during re-infection remains only a theoretical risk, there have been two reported cases of re-infection where the second infection resulted in worse disease.49,50 However, antibody levels after the first infection were not measured for one case49 and only IgM was detectable after the first infection for the second case,50 arguing against Fc-mediated effector functions as the cause of increased pathogenicity.
Overall, we find that mild to moderate COVID-19 generates robust FcγR-binding, ADCC, and ADP antibody functions that decay at a slower rate than plasma neutralization activity. Further dissecting the protective potential of antibody Fc effector functions will be critical for defining the durability of immunity generated by infection or vaccination.
Limitations of study
We acknowledge several limitations in this study. (1) Our cohort of COVID-19 convalescent subjects is composed primarily of individuals who exhibited mild disease symptoms. Although this is representative of the typical spectrum of COVID-19 across this age group, the low number of subjects with moderate and severe disease limited our ability to make any meaningful comparisons between disease-severity groups. (2) Due to the more laborious nature of the functional ADCC and ADP assays, they were only performed with a subset of 36 donors at the first and last time points. Nevertheless, this is still a significant number of donors and spanned a median of 89 days between time points. (3) The target cells we used for the FcγRIIIa activation, ADCC, and THP-1 cell association and trogocytosis assays express high levels of S, which may differ from the physiological levels of S expressed on infected cells. However, these cells provide a potentially more reproducible target than infected cells. (4) The bead-based ADP assay used fluorescent beads conjugated with S trimer as a surrogate and as such may also differ physiologically from actual SARS-CoV-2 virions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human IgG Fc PE | SouthernBiotech | 9040-09; RRID:AB_2796601 |
| Mouse anti-human IgG1 (hinge) PE | SouthernBiotech | 9052-09; RRID:AB_2796621 |
| Mouse anti-human IgG2 Fc PE | SouthernBiotech | 9070-09; RRID:AB_2796639 |
| Mouse anti-human IgG3 (hinge) PE | SouthernBiotech | 9210-09; RRID:AB_2796701 |
| Mouse anti-human IgG4 Fc PE | SouthernBiotech | 9200-09; RRID:AB_2796693 |
| Mouse anti-human IgA1 PE | SouthernBiotech | 9130-09; RRID:AB_2796656 |
| Mouse anti-human IgA2 PE | SouthernBiotech | 9140-09; RRID:AB_2796664 |
| Mouse anti-human IgM PE | Mabtech | 3880-6-250 |
| Streptavidin PE | ThermoFisher Scientific | S866 |
| Mouse anti-human IgG APC (HP6017) | BioLegend | 409306; RRID:AB_11149491 |
| Mouse anti-human CD32 AF647 (FUN-2) | BioLegend | 303212; RRID:AB_2262705 |
| Mouse anti-human CD64 BV510 (10.1) | BioLegend | 305027; RRID:AB_2562512 |
| Mouse anti-human CD89 APC (A59) | BioLegend | 354106; RRID:AB_2565257 |
| Mouse anti-human CD32 FITC (FUN-2) | BioLegend | 303204; RRID:AB_314336 |
| Biological samples | ||
| Whole blood samples and derivatives (peripheral blood mononuclear cells (PBMCs), plasma and serum) from COVID-19 convalescent donors and uninfected controls | The Peter Doherty Institute for Infection and Immunity, The University of Melbourne | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 3,3′,5,5′-Tetramethylbenzidine (TMB) Liquid Substrate System for ELISA | Sigma | T0440-1L |
| SARS-CoV-2 RBD | BEI | NR-52366 |
| SARS-CoV-2 Spike | Juno et al.1 | N/A |
| SARS-CoV-2 S1 subunit | Sino Biological | 40591-V08H |
| SARS-CoV-2 S2 subunit | Sino Biological | S2N-C52H5 |
| HCoV-229E Spike | Sino Biological | 40605-V08B |
| HCoV-229E S1 subunit | Sino Biological | 40601-V08H |
| HCoV-HKU1 Spike | Sino Biological | 40606-V08B |
| HCoV-HKU1 S1 subunit | Sino Biological | 40021-V08H |
| HCoV-NL63 Spike | Sino Biological | 40604-V08B |
| HCoV-NL63 S1 subunit | Sino Biological | 40600-V08H |
| HCoV-OC43 Spike | Sino Biological | 40607-V08B |
| HCoV-OC43 S1 subunit | Sino Biological | 40607-V08H1 |
| HCoV-OC43 S2 subunit | Sino Biological | 40607-V08B1 |
| Clostridium Tetani Tetanus Toxin | Sigma-Aldrich | T3194 |
| SIV gp120 | Sino Biological | 40415-V08H |
| Influenza A H1N1 (A/Cali/07/2009) Hemagglutinin | Sino Biological | 11085-V08H |
| G418 Geneticin (neomycin analog) | ThermoFisher/GIBCO | Cat# 10131-027 |
| Hygromycin B | ThermoFisher/Invitrogen | Cat# 10687010 |
| Critical commercial assays | ||
| EasySep Human NK Cell Enrichment Kit | StemCell Technologies, Inc. | 19055 |
| Nano-Glo® Luciferase Assay System | Promega | N1120 |
| britelite plus Reporter Gene Assay System, 100 mL | Perkin Elmer | 6066761 |
| Amaxa cell line nucleofector kit T | Lonza Bioscience | Cat# VCA-1002 |
| Deposited data | ||
| SARS-CoV-2 S-specific IgG and microneutralisation data | Wheatley et al.8 | N/A |
| Experimental models: cell lines | ||
| THP-1 | ATCC | TIB-202 |
| IIA1.6 | Jan G van de Winkel | N/A |
| Phoenix Packaging line | Garry Nolan Lab | N/A |
| Ramos S-orange | This paper | N/A |
| Ramos S-luciferase | This paper | N/A |
| A549 S-orange | This paper | N/A |
| Recombinant DNA | ||
| FcγRIIIa V158 cDNA/pMXneo | Mark Hogarth Lab. | N/A |
| NanoLuc® Reporter Vector with NF-κB Response Element/ pNL3.2.NF-κB-RE[NlucP/NF-κB-RE/Hygro] | Promega | Cat# N1111 |
| Software and algorithms | ||
| FlowJo v10 | Tree Star | https://www.flowjo.com/ |
| GraphPad Prism v8 | GraphPad | https://www.graphpad.com/ |
| R: A language and environment for statistical computing v4.0.2 | The Comprehensive R Archive Network | https://cran.r-project.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Stephen Kent (skent@unimelb.edu.au).
Materials availability
All unique reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
This study did not generate any unique datasets or code.
Experimental model and subject details
Human subjects
People who recovered from COVID-19 and healthy controls were recruited to provide serial whole blood samples. Convalescent donors either had a PCR+ test during early infection or clear exposure to SARS-CoV-2, and were confirmed to have SARS-CoV-2 S- and RBD-specific antibodies via ELISA as previously reported1. Contemporaneous uninfected controls who did not experience any COVID-19 symptoms were also recruited and confirmed to be seronegative via ELISA. For all subjects, whole blood was collected with sodium heparin or lithium heparin anticoagulant. The plasma fraction was then collected and stored at −80°C. A subset of 36 donors with at least 60 days between the first and last visits were chosen to proceed with the more labor-intensive functional ADCC and ADP assays. Plasma was heat-inactivated at 56°C for 30 minutes prior to use in functional assays. Characteristics of the COVID-19 convalescent and uninfected donors are described in Table S1. The study protocols were approved by the University of Melbourne Human Research Ethics Committee (#2056689). All subjects provided written informed consent in accordance with the Declaration of Helsinki.
Cell lines
As target cells for the functional antibody assays, Ramos and A549 cells stably expressing full-length SARS-CoV-2 S and the reporter proteins mOrange2 or luciferase were generated by lentiviral transduction (Figure S2A). To stain for S-expression, transduced cells were incubated with convalescent plasma (1:100 dilution) prior to staining with a secondary mouse anti-human IgG-APC antibody (1:200 dilution; clone HP6017, BioLegend). S-luciferase cells were bulk sorted on high S expression while S-orange cells were bulk sorted on high S- and mOrange2-expression. Following a week of outgrowth, the bulk sorted cells were single-cell sorted to obtain clonal populations of S-orange and S-luciferase cells (Figure S2B). The Ramos cell lines were grown in complete RPMI medium (10% fetal calf serum (FCS) with 1% penicillin strepytomycin glutamine (PSG)) while the A549 cell lines were grown in complete DMEM medium (10% FCS with 1% PSG).
FcγRIIIa-NF-κB-RE nanoluciferase reporter cells were used as effector cells for the FcγRIIIa activation assay. IIA1.6 cells expressing the Fc receptor gamma subunit (FcR-γ) were maintained in RPMI containing 10% FCS, 2.5 mM L-glutamine, 55 μM 2-mercaptoethanol, 100 units penicillin and 100 units streptomycin (Sigma Aldrich). These were further transduced as described previously51 using a FcγRIIIa V158 cDNA in pMX-neo and the packaging line Phoenix. IIA1.6/FcR-γ/FcγRIIIa V158 cells were transfected with a NF-κB response element driven nanoluciferase (NanoLuc) reporter construct (pNL3.2.NF-κB-RE[NlucP/NF-κB-RE/Hygro] (Promega) by nucleofection (Amaxa Kit T, Lonza) and selected in the presence of 200 μg/ml hygromycin. Reporter cells were maintained in media containing 400 μg/ml neomycin and 50 μg/ml hygromycin (ThermoFisher).
THP-1 monocytes (ATCC) were cultured in complete RPMI medium and maintained below a cell density of 0.3 × 106/ml. Flow cytometry was used to confirm stable expression of FcγRIIa (CD32), FcγRI (CD64) and FcαR (CD89) on THP-1 monocytes prior to use in assays.
Method details
Luminex bead-based multiplex assay
A custom bead array was designed using SARS-CoV-2 S trimer, S1 subunit (Sino Biological), S2 subunit (ACRO Biosystems) and RBD (BEI Resources), as well as HCoV (OC43, HKU1, 229E, NL63) S and S1 subunit (Sino Biological) (as described in Table S2)52. Tetanus toxoid (Sigma-Aldrich), influenza hemagglutinin (H1Cal2009; Sino Biological) and SIV gp120 (Sino Biological) were also included in the array as positive and negative controls respectively. These antigens were covalently coupled to magnetic carboxylated beads (Bio Rad) using a two-step carbodiimide reaction and blocked with 0.1% BSA, before being resuspended and stored in PBS 0.05% sodium azide till use.
Using the respective antigen-coupled beads, a custom CoV multiplex assay was formed to investigate the dimeric recombinant soluble FcγR-binding capacity of pathogen-specific antibodies present in COVID-19 convalescent plasma samples and uninfected controls52. Briefly, 20μl of working bead mixture (1000 beads per antigen-coupled bead region) and 20μl of diluted plasma (final dilution 1:200) were added per well and incubated overnight at 4°C on a shaker. Different detectors were used to assess pathogen-specific antibodies. Single-step detection was done using phycoerythrin (PE)-conjugated mouse anti-human pan-IgG (Southern Biotech; 1.3μg/ml, 25μl/well). For the detection of FcγR-binding, recombinant soluble FcγR dimers (higher affinity polymorphisms FcγRIIIa-V158 and FcγRIIa-H131, lower affinity polymorphisms FcγRIIIa-F158 and FcγRIIa-R131; 1.3μg/ml, 25μl/well) were first added to the beads, washed, and followed by the addition of streptavidin R-PE (Thermo Fisher Scientific). Assays were read on the Flexmap 3D and performed in duplicates.
FcγRIIIa activation assay
A549 S-orange cells were plated (2 × 105/ml, 100 μl/well) in 96-well white flat-bottom plates (Corning). The next day, COVID-19 convalescent and uninfected plasma were serially diluted and 50 μl aliquots transferred to the aspirated A549 S-orange cells and incubated at 37°C, 60 min, 5% CO2. Unbound antibody was removed by aspirating the wells and refilling with RPMI (200 μl) four times. FcγRIIIa-NF-κB-RE nanoluciferase reporter cells (4 × 105/ml, 50 μl/well) were added to the aspirated wells containing the opsonised A549 S-orange cells. After incubation (37°C, 4h, 5%CO2) cells were lysed by adding 50 μl/well of 10 mM Tris-pH 7.4, containing 5 mM EDTA, 0.5 mM DTT, 0.2% Igepal CA-630 (Sigma Aldrich), and Nano-Glo luciferase assay substrate (1:1000). Induction of nanoluciferase was measured using a 1 s read on a Clariostar Optima plate reader (BMG Labtech) with background luminescence from control wells without agonist subtracted from test values.
Luciferase-based ADCC assay
A luciferase-based ADCC assay was performed to examine ADCC against S-expressing cells. NK cells from healthy donors were first enriched from freshly isolated PBMCs using the EasySep Human NK Cell Enrichment Kit (StemCell Technologies, Inc.). In a 96-well V-bottom cell culture plate, purified NK cells (20,000/well) were mixed with Ramos S-luciferase cells (5,000/well) in the presence or absence of plasma from convalescent or uninfected donors at 1:100, 1:400 and 1:1600 dilutions. Each condition was tested in duplicate and “no plasma” and “target cell only” controls were included. Cells were centrifuged at 250 g for 4 min prior to a 4-hour incubation at 37°C with 5% CO2. Cells were then washed with PBS and lysed with 25μl of passive lysis buffer (Promega). Cell lysates (20μl) were transferred to a white flat-bottom plate and developed with 30μl of britelite plus luciferase reagent (Perkin Elmer). Luminescence was read using a FLUOstar Omega microplate reader (BMG Labtech). The relative light units (RLU) measured were used to calculate %ADCC with the following formula: (“no plasma control” – “plasma sample”) ÷ “target cell only control” × 100. For each plasma sample, %ADCC was plotted against log10(plasma dilution-1) and the area under curve (AUC) was calculated using Graphpad Prism.
Bead-based THP-1 ADP assay
To examine ADP mediated by COVID-19 convalescent plasma, a previously published bead-based ADP assay was adapted for use in the context of SARS-CoV-224. SARS-CoV-2 S trimer was biotinylated using EZ-Link Sulfo-NHS-LC biotinylation kit (Thermo Scientific) with 20mmol excess according to manufacturer’s instructions and buffer exchanged using 30kDa Amicon centrifugal filters (EMD millipore) to remove free biotin. The binding sites of 1 μm fluorescent NeutrAvidin Fluospheres beads (Invitrogen) were coated with biotinylated S at a 1:3 ratio overnight at 4°C. S-conjugated beads were washed four times with 2% BSA/PBS to remove excess antigen and incubated with plasma (1:100 dilution) for 2 hours at 37°C in a 96-well U-bottom plate (see Figure S5 for optimization). THP-1 monocytes (10,000/well) were then added to opsonized beads and incubated for 16 hours under cell culture conditions. Cells were fixed with 2% formaldehyde and acquired on a BD LSR Fortessa with a HTS. The data was analyzed using FlowJo 10.7.1 (see Figure S4 for gating strategy) and a phagocytosis score was calculated as previously described53 using the formula: (%bead-positive cells × mean fluorescent intensity)/103. To account for non-specific uptake of S-conjugated beads, the phagocytosis scores for each plasma sample were subtracted with that of the “no plasma” control.
Cell-based THP-1 association assay
To assess the capacity of THP-1 monocytes to associate with S-expressing target cells via Ab-FcγR interactions, an assay using THP-1 cells as effectors and Ramos S-orange cells as targets was performed. THP-1 monocytes were first stained with CellTrace™ Violet (CTV) (Life Technologies) as per manufacturer’s instructions. In a 96-well V-bottom cell culture plate, Ramos S-orange cells (10,000/well) were incubated with plasma from convalescent or uninfected donors (1:2700 dilution) for 30 minutes (see Figure S5 for optimization). Opsonised Ramos S-orange cells were then washed prior to co-culture with CTV-stained THP-1 monocytes (10,000/well) for 1 hour at 37°C with 5% CO2. After the incubation, cells were washed with PBS, fixed with 2% formaldehyde and acquired using the BD LSR Fortessa with a high-throughput sampler attachment (HTS). The data was analyzed using FlowJo 10.7.1 (see Figure S4 for gating strategy). The percentage of Ramos S-orange cells associated with THP-1 monocytes (% association) was measured for each plasma sample and background-subtracted with the “no plasma” control.
Confocal microscopy
Prior to performing the cell-based THP-1 association assay as described above, Ramos S-orange cells were stained with the membrane dye PKH-26 (Sigma-Aldrich; MINI26-1KT). Stained Ramos S-orange cells were incubated with uninfected healthy control plasma or COVID-19 convalescent plasma before THP-1 monocytes were added at a 1:1 ratio for 1 hour. To label THP-1 monocytes, samples were then stained with anti-CD32 AF647 (clone FUN-2, BioLegend) on ice for 60 minutes. Samples were washed and loaded onto poly-L-lysine (Sigma; P4707) coated coverslips. Cells on coverslips were fixed with 2% formaldehyde and rinsed with PBS. Coverslips were mounted onto slides with ProLong Diamond Antifade Mountant (Life Technologies; P36961) and were left in the dark at room temperature for 24 hours to set. The cells were visualized on Zeiss LSM710 laser scanning confocal microscope (Z stack 12 slices, 60 × magnification) and analyzed using ImageJ.
Decay rate estimation
The decay rate was estimated by fitting a linear mixed effect model for each response variable (yij for subject i at time point j) as a function of days post-symptom onset and assay replicate (as a binary categorical variable). The model can be written as below:
The parameter is a constant (intercept), and is a subject-specific adjustment to the overall intercept. The slope parameter is a fixed effect to capture the decay slope before (as a fixed parameter, 70 days); which also has a subject-specific random effect . To fit a model with two different decay rates, an extra parameter (with a subject-specific random effect ) was added to represent the difference between the two slopes. Assay variability between replicates (only for HCoV response variables) was modeled as a single fixed effect , in which we coded the replicate as a binary categorical variable . The random effect was assumed to be normally distributed with zero mean and variance .
We fitted the model to log-transformed data of various response variables (assuming exponential decay), and we censored the data from below if it was less than the threshold for detection. The data for each subject included either two or three time points. The response variables had background levels subtracted by taking the mean of all the background values, and the threshold for detection was set at two standard deviations of the background responses. The model was fitted by using lmec library in R, using the ML algorithm to fit for the fixed effects. We also tested if the response variables can be fitted better by using a single or two different decay slopes (likelihood ratio test – based on the likelihood value and the difference in the number of parameters). These analyses were carried out in R: A language and environment for statistical computing version 4.0.2.
Quantification and statistical analysis
Statistics
Statistical analyses were performed with Graphpad Prism 8. Correlations between functional ADCC and ADP responses with cell-associated S-specific IgG and FcγR-binding S-specific antibodies were assessed using the non-parametric Spearman test. Comparisons of functional ADCC and ADP responses between first and last visits were performed using the Wilcoxon signed-rank test. Comparisons between uninfected individuals and COVID-19 convalescent individuals were performed using the Mann-Whitney test. Statistical details of experiments can be found in the figure legends.
Acknowledgments
We thank the cohort participants for generously providing samples. We thank Francesca Mordant and Kanta Subbarao (University of Melbourne) for performing the SARS-CoV-2 neutralization assays. We acknowledge the Melbourne Cytometry Platform for provision of flow cytometry services. The following reagent was produced under HHSN272201400008C and obtained through BEI Resources, NIAID, NIH: Spike glycoprotein receptor binding domain (RBD) from SARS-related coronavirus 2, Wuhan-Hu-1 with C-terminal histidine tag, recombinant from HEK293F cells, NR-52366. This study was supported by the Victorian Government, an Australian Government Medical Research Future Fund award GNT2002073 (S.J.K., M.P.D., and A.K.W.), the ARC Centre of Excellence in Convergent Bio-Nano Science and Technology (S.J.K.), an NHMRC program grant APP1149990 (S.J.K. and M.P.D.), NHMRC project grants GNT1162760 (A.K.W.) and GNT1145303 (P.M.H. and B.D.W.), an NHMRC-EU collaborative award APP1115828 (S.J.K. and M.P.D.), the European Union Horizon 2020 Research and Innovation Programme under grant agreement 681137 (S.J.K.), and Emergent Ventures Fast Grants (A.W.C.). J.A.J., A.W.C., and S.J.K. are supported by NHMRC fellowships. A.K.W., D.C., and M.P.D. are supported by NHMRC Investigator grants. Figures were created using BioRender.
Author contributions
Conceptualization, W.S.L., A.K.W., M.P.D., A.W.C., and S.J.K.; methodology, W.S.L., K.J.S., S.K.D., and B.D.W.; formal analysis, W.S.L., K.J.S., S.K.D., B.D.W., A.R., D.C., M.P.D., and A.W.C.; investigation, W.S.L., K.J.S., S.K.D., B.D.W., E.R.H., and H.-X.T.; resources, R.E., H.G.K., J.A.J., and A.K.W.; writing – original draft, W.S.L. and S.J.K.; writing – review & editing, W.S.L., K.J.S., S.K.D., B.D.W., A.R., H.-X.T., J.A.J., A.K.W., P.M.H., D.C., M.P.D., A.W.C., and S.J.K.; supervision, A.K.W., A.W.C., and S.J.K.; funding acquisition, A.K.W., P.M.H., M.P.D., A.W.C., and S.J.K.
Declaration of interests
The authors declare no competing interests.
Published: May 9, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100296.
Contributor Information
Amy W. Chung, Email: awchung@unimelb.edu.au.
Stephen J. Kent, Email: skent@unimelb.edu.au.
Supplemental information
References
- 1.Juno J.A., Tan H.X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 2.Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., McMahon M., Meade P., Mendu D.R., Muellers K. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 Spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 6.Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Addetia A., Crawford K.H.D., Dingens A., Zhu H., Roychoudhury P., Huang M.L., Jerome K.R., Bloom J.D., Greninger A.L. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J. Clin. Microbiol. 2020;58:e02107-20. doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheatley A.K., Juno J.A., Wang J.J., Selva K.J., Reynaldi A., Tan H.X., Lee W.S., Wragg K.M., Kelly H.G., Esterbauer R. Evolution of immune responses to SARS-CoV-2 in mild-moderate COVID-19. Nat. Commun. 2021;12:1162. doi: 10.1038/s41467-021-21444-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford K.H.D., Dingens A.S., Eguia R., Wolf C.R., Wilcox N., Logue J.K., Shuey K., Casto A.M., Fiala B., Wrenn S. Dynamics of neutralizing antibody titers in the months after SARS-CoV-2 infection. J. Infect. Dis. 2020 doi: 10.1093/infdis/jiaa618. Published online September 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., Hemmings O., O’Byrne A., Kouphou N., Galao R.P. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L.C., Rattigan S.M., Borgert B.A., Moreno C.A., Solomon B.D., Trimmer-Smith L. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyllie D., Mulchandani R., Jones H.E., Taylor-Phillips S., Brooks T., Charlett A., Ades A.E., Makin A., Oliver I., Moore P., EDSAB-HOME investigators SARS-CoV-2 responsive T cell numbers are associated with protection from COVID-19: a prospective cohort study in keyworkers. medRxiv. 2020 doi: 10.1101/2020.11.02.20222778. [DOI] [Google Scholar]
- 13.Bournazos S., Klein F., Pietzsch J., Seaman M.S., Nussenzweig M.C., Ravetch J.V. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014;158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiLillo D.J., Palese P., Wilson P.C., Ravetch J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Invest. 2016;126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bournazos S., DiLillo D.J., Goff A.J., Glass P.J., Ravetch J.V. Differential requirements for FcγR engagement by protective antibodies against Ebola virus. Proc. Natl. Acad. Sci. USA. 2019;116:20054–20062. doi: 10.1073/pnas.1911842116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler S.E., Crowley A.R., Natarajan H., Xu S., Weiner J.A., Bobak C.A., Mattox D.E., Lee J., Wieland-Alter W., Connor R.I. Distinct features and functions of systemic and mucosal humoral immunity among SARS-CoV-2 convalescent individuals. Front. Immunol. 2021;11:618685. doi: 10.3389/fimmu.2020.618685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:e20201993. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan C.E.Z., Seah S.G.K., Chye D.H., Massey S., Torres M., Lim A.P.C., Wong S.K.K., Neo J.J.Y., Wong P.S., Lim J.H. The Fc-mediated effector functions of a potent SARS-CoV-2 neutralizing antibody, SC31, isolated from an early convalescent COVID-19 patient, are essential for the optimal therapeutic efficacy of the antibody. bioRxiv. 2020 doi: 10.1101/2020.10.26.355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natarajan H., Crowley A.R., Butler S.E., Xu S., Weiner J.A., Bloch E.M. Markers of Polyfunctional SARS-CoV-2 Antibodies in Convalescent Plasma. mBio. 2021;12 doi: 10.1128/mBio.00765-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dufloo J., Grzelak L., Staropoli I., Madec Y., Tondeur L., Anna F., Pelleau S., Wiedemann A., Planchais C., Buchrieser J. Asymptomatic and symptomatic SARS-CoV-2 infections elicit polyfunctional antibodies. Cell Rep. Med. 2021 doi: 10.1016/j.xcrm.2021.100275. Published online April 20, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wines B.D., Vanderven H.A., Esparon S.E., Kristensen A.B., Kent S.J., Hogarth P.M. Dimeric FcγR ectodomains as probes of the Fc receptor function of anti-influenza virus IgG. J. Immunol. 2016;197:1507–1516. doi: 10.4049/jimmunol.1502551. [DOI] [PubMed] [Google Scholar]
- 22.Ana-Sosa-Batiz F., Johnston A.P.R., Hogarth P.M., Wines B.D., Barr I., Wheatley A.K., Kent S.J. Antibody-dependent phagocytosis (ADP) responses following trivalent inactivated influenza vaccination of younger and older adults. Vaccine. 2017;35:6451–6458. doi: 10.1016/j.vaccine.2017.09.062. [DOI] [PubMed] [Google Scholar]
- 23.Yasui F., Kohara M., Kitabatake M., Nishiwaki T., Fujii H., Tateno C., Yoneda M., Morita K., Matsushima K., Koyasu S., Kai C. Phagocytic cells contribute to the antibody-mediated elimination of pulmonary-infected SARS coronavirus. Virology. 2014;454-455:157–168. doi: 10.1016/j.virol.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ackerman M.E., Moldt B., Wyatt R.T., Dugast A.S., McAndrew E., Tsoukas S., Jost S., Berger C.T., Sciaranghella G., Liu Q. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods. 2011;366:8–19. doi: 10.1016/j.jim.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beum P.V., Mack D.A., Pawluczkowycz A.W., Lindorfer M.A., Taylor R.P. Binding of rituximab, trastuzumab, cetuximab, or mAb T101 to cancer cells promotes trogocytosis mediated by THP-1 cells and monocytes. J. Immunol. 2008;181:8120–8132. doi: 10.4049/jimmunol.181.11.8120. [DOI] [PubMed] [Google Scholar]
- 26.Daubeuf S., Lindorfer M.A., Taylor R.P., Joly E., Hudrisier D. The direction of plasma membrane exchange between lymphocytes and accessory cells by trogocytosis is influenced by the nature of the accessory cell. J. Immunol. 2010;184:1897–1908. doi: 10.4049/jimmunol.0901570. [DOI] [PubMed] [Google Scholar]
- 27.Richardson S.I., Crowther C., Mkhize N.N., Morris L. Measuring the ability of HIV-specific antibodies to mediate trogocytosis. J. Immunol. Methods. 2018;463:71–83. doi: 10.1016/j.jim.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 28.Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song G., He W., Callaghan S., Anzanello F., Huang D., Ricketts J. Cross-reactive serum and memory B cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat. Commun. 2021;12:2938. doi: 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aydillo T., Rombauts A., Stadlbauer D., Aslam S., Abelenda-Alonso G., Escalera A., Amanat F., Jiang K., Krammer F., Carratala J. Antibody immunological imprinting on COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.10.14.20212662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westerhuis B.M., Aguilar-Bretones M., Raadsen M.P., de Bruin E., Okba N.M.A., Haagmans B.L., Langerak T., Endeman H., van den Akker J.P.C., Gommers D.A.M.P.J. Severe COVID-19 patients display a back boost of seasonal coronavirus-specific antibodies. medRxiv. 2020 doi: 10.1101/2020.10.10.20210070. [DOI] [Google Scholar]
- 32.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021 doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 33.Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020;183:1508–1519.e12. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gasser R., Cloutier M., Prévost J., Fink C., Ducas É., Ding S., Dussault N., Landry P., Tremblay T., Laforce-Lavoie A. Major role of IgM in the neutralizing activity of convalescent plasma against SARS-CoV-2. Cell Rep. 2021;34:108790. doi: 10.1016/j.celrep.2021.108790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffman H.-H., Oliveira T.Y., Oren D.A. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021;13:eabf1555. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claër L., Quentric P., Fadlallah J., Devilliers H., Ghillani P. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13:eabd2223. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin. Infect. Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. USA. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joyner M., Wright R.S., Fairweather D., Senefeld J., Bruno K., Klassen S. Early safety indicators of COVID-19 convalescent plasma in 5,000 patients. J. Clin. Invest. 2020;130:4791–4797. doi: 10.1172/JCI140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwal A., Mukherjee A., Kumar G., Chatterjee P., Bhatnagar T., Malhotra P., PLACID Trial Collaborators Convalescent plasma in the management of moderate covid-19 in adults in India: open label phase II multicentre randomised controlled trial (PLACID Trial) BMJ. 2020;371:m3939. doi: 10.1136/bmj.m3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L., Zhang W., Hu Y., Tong X., Zheng S., Yang J., Kong Y., Ren L., Wei Q., Mei H. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening COVID-19: a randomized clinical trial. JAMA. 2020;324:460–470. doi: 10.1001/jama.2020.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simonovich V.A., Burgos Pratx L.D., Scibona P., Beruto M.V., Vallone M.G., Vázquez C., Savoy N., Giunta D.H., Pérez L.G., Sánchez M.D.L., PlasmAr Study Group A randomized trial of convalescent plasma in Covid-19 severe pneumonia. N. Engl. J. Med. 2021;384:619–629. doi: 10.1056/NEJMoa2031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chakraborty S., Gonzalez J., Edwards K., Mallajosyula V., Buzzanco A.S., Sherwood R., Buffone C., Kathale N., Providenza S., Xie M.M. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 2021;22:67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoepel W., Chen H.-J., Allahverdiyeva S., Manz X., Aman J., Bonta P., Amsterdam UMC COVID-19 Biobank Anti-SARS-CoV-2 IgG from severely ill COVID-19 patients promotes macrophage hyper-inflammatory responses. Sci. Transl. Med. 2021:eabf8654. doi: 10.1126/scitranslmed.abf8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polack F.P., Teng M.N., Collins P.L., Prince G.A., Exner M., Regele H., Lirman D.D., Rabold R., Hoffman S.J., Karp C.L. A role for immune complexes in enhanced respiratory syncytial virus disease. J. Exp. Med. 2002;196:859–865. doi: 10.1084/jem.20020781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polack F.P. Atypical measles and enhanced respiratory syncytial virus disease (ERD) made simple. Pediatr. Res. 2007;62:111–115. doi: 10.1203/PDR.0b013e3180686ce0. [DOI] [PubMed] [Google Scholar]
- 49.Tillett R.L., Sevinsky J.R., Hartley P.D., Kerwin H., Crawford N., Gorzalski A., Laverdure C., Verma S.C., Rossetto C.C., Jackson D. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 2021;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prado-Vivar B., Becerra-Wong M., Guadalupe J.J., Márquez S., Gutierrez B., Rojas-Silva P., Grunauer M., Trueba G., Barragán V., Cárdenas P. A case of SARS-CoV-2 reinfection in Ecuador. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30910-5. Published online November 23, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wines B.D., Trist H.M., Monteiro R.C., Van Kooten C., Hogarth P.M. Fc receptor gamma chain residues at the interface of the cytoplasmic and transmembrane domains affect association with FcalphaRI, surface expression, and function. J. Biol. Chem. 2004;279:26339–26345. doi: 10.1074/jbc.M403684200. [DOI] [PubMed] [Google Scholar]
- 52.Selva K.J., van de Sandt C.E., Lemke M.M., Lee C.Y., Shoffner S.K., Chua B.Y. Systems serology detects functionally distinct coronavirus antibody features in children and elderly. Nat. Commun. 2021;12:2037. doi: 10.1038/s41467-021-22236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darrah P.A., Patel D.T., De Luca P.M., Lindsay R.W., Davey D.F., Flynn B.J., Hoff S.T., Andersen P., Reed S.G., Morris S.L. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.