Abstract
Background
In 1957, Francis Crick drew a linear diagram on a blackboard. This diagram is often called the “central dogma.” Subsequently, the relationships between different steps of the “central dogma” have been shown to be considerably complex, mostly because of the emerging world of small molecules. It is noteworthy that metabolites can be generated from the diet through gut microbiome metabolism, serve as substrates for epigenetic modifications, destabilize DNA quadruplexes, and follow Lamarckian inheritance. Small molecules were once considered the missing link in the “central dogma”; however, recently they have acquired a central role, and their general perception as downstream products has become reductionist. Metabolomics is a large-scale analysis of metabolites, and this emerging field has been shown to be the closest omics associated with the phenotype and concomitantly, the basis for all omics.
Aim of review
Herein, we propose a broad updated perspective for the flux of information diagram centered in metabolomics, including the influence of other factors, such as epigenomics, diet, nutrition, and the gut- microbiome.
Key scientific concepts of review
Metabolites are the beginning and the end of the flux of information.
Keywords: Metabolomics, Central dogma, Small molecules, Metabolites
Introduction
Sixty-three years ago, Francis Crick gave a lecture in which he presented the diagram called the “central dogma.” This dogma states that the transfer of information from DNA to DNA/RNA, or from nucleic acid to protein, may be possible, but the transfer from protein to protein or protein to nucleic acid is impossible (Cobb, 2017; CRICK, 1957).
The flux of information attained another dimension when small molecules advanced to the forefront of chemical biology. In 2005, Dr. Schreiber was the first to describe small molecules as the missing link in the central dogma and as the central elements of life (Schreiber, 2005). Consequently, the perception of small molecules as only downstream products became reductionist, as they have been shown to modulate all steps in the flux of information, including the phenotype (Chang et al., 2008; Guijas et al., 2018; Kondratov et al., 2001; Mathew & Padmanaban, 2013; Patti et al., 2012; Rabinowitz & Silhavy, 2013; Yugi & Kuroda, 2018). In 2016, for the first time, a white paper provided recommendations to include metabolomics in precision medicine (Beger et al., 2016). In recent years, several papers have described an integrative data analysis with metabolomics (Ahmed et al., 2021; Damiani et al. 2020; Hou et al., 2020; Long et al., 2020; Mussap et al., 2021; Xie et al., 2021). In the last 20 years, the advance of important analytical tools, such as nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS), has led to the advocacy of small molecules, by the large-scale analysis called metabolomics. These exceptional omics techniques have made it possible to build the Human Metabolome Database (Wishart et al., 2018) and the Biological Magnetic Resonance Data Bank (Ulrich et al., 2007). The assignment of several metabolites has been extremely important for revealing distinct metabolic pathway shifts related to the phenotype, such as the polyol pathway detour under vitamin D intake (Santos et al., 2017), enhanced lipid metabolism in oral cancer (Sant’Anna-Silva, 2018), glycerol metabolism in osteoarthritis (de Sousa et al., 2017a, 2017b, 2019), glutaminolysis in cancer (DeBerardinis et al., 2007; Koczula et al., 2016; Rodrigues et al., 2016; Yang et al., 2016), tumor microenvironments (Costa Dos Santos et al., 2021), and COVID-19 (Costa dos Santos Junior et al., 2020).
Metabolites are the substrates and products of molecular mechanisms linked to the steps in the flux of information. Molecular modulation can start outside the host organism, as metabolites can come from the metabolism of microorganisms, diet, and other exogenous sources (Johnson et al., 2012; Martin et al., 2007; Tang et al., 2015). One good example of a metabolite is trimethylamine N-oxide (TMAO), commonly found in urine; it can be generated directly from the diet by ingestion of fish, or indirectly from gut-microbiome choline metabolism (Thøgersen et al., 2020; Thomas & Fernandez, 2021; Ufnal et al., 2015).
TMAO can affect the proteome, as MAP kinase and nuclear factor-κB (NF-κB) proteins can be activated by TMAO in endothelial cells, and high levels of this metabolite are associated with cardiovascular diseases (Bennett et al., 2013; Gibson et al., 2020; Seldin et al., 2016; Zeisel & Warrier, 2017). The mechanism of interaction of TMAO with proteins is still unknown; however, there is evidence that it can influence enzyme activity and protein stability (Hu et al., 2010; Mashino & Fridovich, 1987). TMAO can affect the genome, epigenome, and transcriptome by stabilizing DNA G-quadruplex structures (Knop et al., 2018; Ueda et al., 2016), altering levels of DNA methylation (Knight et al., 2018), and stabilizing RNA tertiary structures (Denning et al., 2013; Lambert et al., 2010), respectively. In addition, other factors can affect TMAO levels, such as age and sex (Rossner et al., 2017).
Herein, we discuss various pieces of evidence to shed light on the main role of metabolites shaping the other omics levels, such as the epigenome, genome, transcriptome, and proteome (Adamski & Suhre, 2013; Anders et al., 2014; Bhaduri et al., 2018; Choi et al., 2018; Dervan, 2001; Etchegaray & Mostoslavsky, 2016; Francisco & Paulo, 2018; Gao et al., 2016; Kimball & Vondriska, 2020; Neidle, 2001; Padmanabhan et al., 2013; Pogribny et al., 2008; Reid et al., 2017; Rodriguez & Miller, 2014; Tzika et al., 2018; Usanov et al., 2018; Wilson & Li, 2012), as shown in Fig. 1. In addition, this evidence is particularly important in the multiomics era, because it can reveal the modes of action of small molecules, accelerate drug development (Patel-Murray et al., 2020), reveal gut-microbiome alterations (Li et al., 2020a, 2020b), and link with machine learning for diagnoses, such as COVID-19 (Delafiori et al., 2021).
Fig. 1.
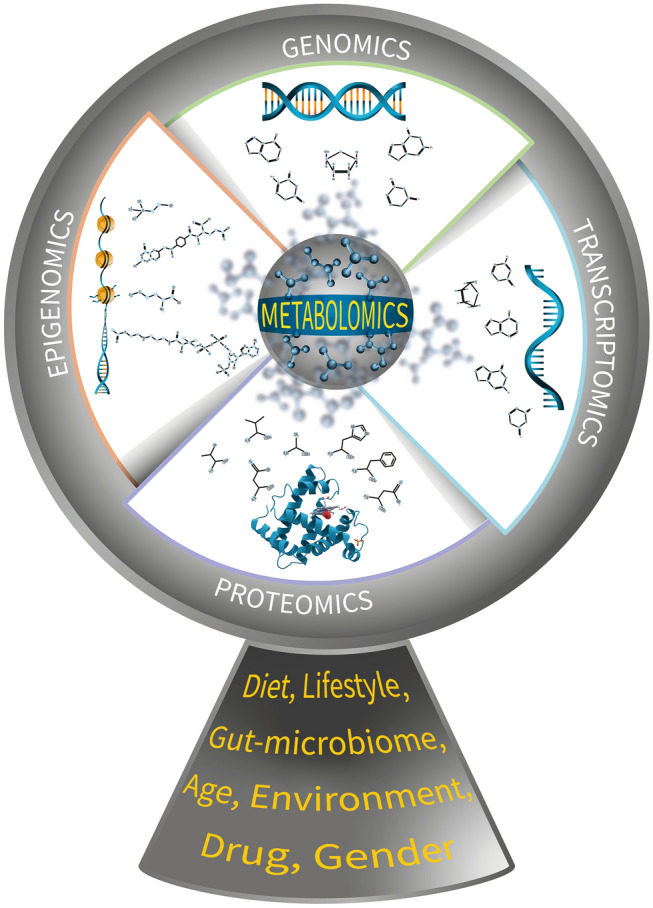
Small molecules are the center of life. A multiinteraction dynamic perspective of the flux of information centered on metabolomics. The small molecules drive all other omics, as a rotor drives the blades in the turbine of a windmill. External factors such as diet, lifestyle, gut-microbiome, age, drug, and sex will affect the metabolomics, e.g. the center of the flux of the information, and consequently affect all other omics
Metabolite–macromolecules interactions and modifications
Metabolites can interact with macromolecules through competitive and allosteric binding to the active site of enzymes, leading to alterations in enzyme activity. This is not exclusive to enzymes; it occurs in other signaling proteins, such as G protein-coupled receptors (GPCRs). The succinate receptor (SUCNR1/GPR91), a cell-surface sensor for extracellular succinate, was shown to be upregulated in mice-activated macrophages, which function as an autocrine and paracrine sensor for extracellular succinate to enhance interleukin 1β (IL-1β) production (Littlewood-Evans et al., 2016). The GPCR (GPR132), which can sense and respond to lactate, was shown to promote an M2-like protumoral phenotype in mice macrophages, stimulating cancer metastasis (Chen et al., 2017). Lactate controls muscle regeneration by inducing M2-like macrophage polarization (Zhang et al., 2020). Some GPCRs (GPR40 and GPR120), which are activated by several fatty acids, including palmitic acid esters of hydroxystearic acids (PAHSAs) in mice and human models, are targets of type II diabetes therapies (Syed et al., 2018; Yore et al., 2014). In neutrophils, short-chain fatty acids engage with GPR43, inducing chemotaxis (Vinolo et al., 2011). Thus, the binding of metabolites to proteins, such as receptors, induces specific cellular responses, leading to the activation of specific signaling pathways.
Additionally, metabolites are related to prions. The cellular prion protein, PrPc is a cell-surface glycoprotein anchored to the plasma membrane via glycosylphosphatidylinositol (GPI). PrPc is found in mammalian tissues, mostly in neuronal cells, despite some evidence of its involvement in signal transduction (Bate et al., 2016; Mouillet-Richard et al., 2000), memory formation, peripheral myelin maintenance, circadian rhythm neuroprotection, and immune system activation (Dearmond et al., 1999; Ermonval, Mouillet-Richard, Codogno, Kellermann, & Botti, 2003). Although the exact role of PrPc is not well defined, it is well known that PrPc is the pathology of its misfolded isoform related to disease, PrPsc. This isoform can aggregate with other PrP molecules, causing a fatal neurodegenerative process (Ermonval et al., 2003). Glycosylation is strongly related to glucose and glutamine metabolism (Carvalho-cruz et al., 2018): it is the most important posttranslational modification of PrP. Unglycosylated PrP has been reported to induce apoptosis in oral squamous cell carcinoma and colon adenocarcinoma (Yap & Say, 2011). Another study reported that glycosylation is important for determining the PrP location on the membrane, to inhibit its aggregation and diminish its cytotoxicity by lowering the intracellular levels of reactive oxygen species (ROS) (Yi et al., 2018).
In 2012, Koonin refuted the “central dogma” idea that information cannot be transferred from proteins to the genome. This refutation is based on the genetic assimilation of prion-dependent phenotypic heredity, which is mediated by epigenetic mechanisms. He named it the “general look-ahead effect” (Koonin, 2012). Conventionally, epigenetic modifications comprise all aspects of the chromatin structure, histone posttranslational modifications, RNA, DNA methylation, or hydroxymethylation, and metabolites act as the substrates for these modifications (Carrer & Wellen, 2014; Gerhäuser, 2012; Janke et al., 2015; Johnson et al., 2015; Petersen et al., 2014; Roberti et al., 2021). In 1942, Waddington introduced the concept of epigenetics. He defined internal and external interactions between genes and their products, leading to the development of phenotypes (Waddington, 1942). In recent years, this concept has been slightly updated to the most accepted definition: “the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence” (Robertson, 2005).
Posttranslational modifications of histones are involved in the repression or activation of gene transcriptions and can be linked to DNA methylation. One good example is histone acetylation, through an acetyl donor, the metabolite acetyl-CoA, which modifies the chromatin structure and creates an accessible structure that is beneficial for transcription. The availability of the metabolite acetate and its pathway is important for maintaining acetylation levels (Di Cerbo & Schneider, 2013; Pogo et al., 1966; Turner & O’Neill, 1995; Wolfe, 2005). In addition, histone acetylation is responsible for generating binding sites that are recognized by bromodomain-containing regulators, promoting gene activation (Filippakopoulos & Knapp, 2014; Shogren-Knaak et al., 2006). Acetate supplementation restored chromatin accessibility in a neuroblastoma differentiation model (Li et al., 2020a, 2020b). Lauterbach et al. observed that in macrophages, lipopolysaccharide (LPS) signaling promotes the incorporation of acetyl-CoA into histones through an increased glycolytic flux and ATP-citrate lyase activity. This histone acetylation induces specific gene sets associated with proinflammatory responses (Lauterbach et al., 2019).
In addition to acetyl-CoA, there are other metabolites responsible for posttranslational modifications, such as succinyl-CoA (succinylation) and sugar molecules, such as uridine 5′-diphosphoglucose (UDP-glucose) (glycosylation and GlcNAcylation). Succinylation was observed to mediate the mitochondrial translocation of pyruvate kinase M2 (PKM2), thereby playing a role in switching the cellular machinery from proliferation to cell survival mode and vice versa in cancer cells (Qi et al., 2019). In macrophages, Lys311 is a key succinylated site in the regulation of PKM2 activity, promoting inflammation (Wang et al., 2017). O-GlcNAcylation plays an important role in the regulation of cellular signaling, translation, and transcription in response to nutrients and stresses (Li et al., 2019; Ong et al., 2018); glycosylation is important in the correct folding and trafficking of proteins, such as Relmα, CD206, and CD301, which are destined for secretion or exportation to the cell surface in anti-inflammatory macrophages (Jha et al., 2015).
In addition, α-ketoglutarate and flavin adenine dinucleotide (FAD) from the tricarboxylic acid cycle (TCA) increase DNA demethylase, while succinate and fumarate function as antagonists to inhibit such demethylases, thus demonstrating the pivotal role of TCA in epigenetic reprogramming (Xiao et al., 2012) and tumorigenesis (Kaelin & McKnight, 2013; Martínez-Reyes & Chandel, 2020; Soga, 2013). Previous studies have demonstrated that mutations in metabolic enzymes, such as succinate dehydrogenase, fumarate hydratase, and NADP+-dependent isocitrate dehydrogenase (both cytosolic and mitochondrial) favor the generation of oncometabolites through the accumulation of succinate and fumarate, triggering hypermethylation of DNA (Eijkelenkamp et al., 2020; Mohammad et al., 2020; Parsons et al., 2008; Ward et al., 2010; Zhu et al., 2020).
Methylation and demethylation are key modifications of histones, DNA, and RNA; they play an important role in the regulation of gene expression and are essential for cell differentiation and embryonic development (Bachman et al., 2014; Petkovich et al., 2017). In this process, the methyl group of the metabolite, S-adenosylmethionine (SAM), is donated to the methyl transferase enzymes (Warth et al., 2017). DNA methylation is strongly associated with histone deacetylases (HDACs), leading to chromatin inactivation (Tiwari et al., 2008). In addition, methylation turnover can differ within the genome (Ginno et al., 2020).
Epitranscriptomics has been shown to be extremely important. Among 170 epigenetic modifications in coding and noncoding RNAs (Boccaletto et al., 2018), N6-methyladenosine methylation (m6A) is the most studied (Zaccara et al., 2019b). Some proteins recognize this RNA modification and mediate several functions, such as mRNA splicing, stability, microRNA processing, and translation (Meyer & Jaffrey, 2017; Shi et al., 2019; Zaccara et al., 2019a). In addition, m6A is linked to diverse biological functions, such as cancer (Sun et al., 2019), T cell differentiation (Furlan et al., 2019), and skin morphogenesis (Xi et al., 2020).
The association of metabolites with metabolic states can enable the formation of metabolic signatures. Furthermore, the identification of the relationships between metabolites and enzyme activity, nutrient import, and posttranslational modifications (Table 1) reinforces their role in phenotype acquisition.
Table 1.
Metabolites as modulators of enzymes, posttranslational modifications, and nutrient transport expressions
| Metabolites as modulators of enzymes | |||||
|---|---|---|---|---|---|
| Metabolite | Effect/modulated enzyme | Product | References | ||
| Accumulation of 2-oxoglutarate (2OG) with a concomitant decrease in citrate |
Activation of isocitrate dehydrogenase (IDH) toward the reductive carboxylation of 2OG HBO |
Generation of citrate | Mullen et al. (2014) | ||
| Fructose-1,6- bisphosphate (Fru-1,6-BP) and high levels of serine | Activation of the M2 isoform of pyruvate kinase (PKM2); increase in the activity of PKM2 for phosphoenolpyruvate (PEP) | Conversion of phosphoenolpyruvate to pyruvate with the generation of ATP | Chaneton et al. (2012), Israelsen and Vander Heiden (2015) | ||
| PEP | Major allosteric regulation of PKM | Regulation of PKM activity promoting tetramerization and stabilization in the active state | Chaneton et al. (2012), Israelsen and Vander Heiden (2015) | ||
| Low levels of serine | Allosteric activation of PKM2 generating reduced activity | Shuttle of glucose-derived carbons to serine biosynthesis | Chaneton et al. (2012) | ||
| AMP | Allosteric activation of AMP-activated protein kinase (PKM2) favoring its phosphorylation | AMPK phosphorylates its targets modulating metabolic fluxes: induction of energy-producing processes and repression of energy-consuming processes. Exphosphorylation of Acetyl-CoA-carboxykinase (ACC) generating malonyl-CoA, which in turn inhibits carnitine palmitoyltransferase (CPT) | Gowans et al. (2013), Wegner et al. (2015) | ||
| High concentrations of succinate, fumarate, 2OG, and 2-hydroxyglutarate (2HG) | Prolyl hydroxylase 2 (PHD2) | Hydroxylation of hypoxia-inducible factor-1α (HIF-1α) inhibiting its proteasomal degradation and stabilization, independent of the oxygen supply | Chaneton et al. (2012), Wegner et al. (2015), Xu et al. (2011) | ||
| Ceramides | Allosteric activation of protein phosphatase 2A (PP2A) | Dephosphorylation of AKT at Thr308 and Ser473 triggering insulin resistance | Blouin et al. (2010) | ||
| Resveratrol metabolites: resveratrol-3′-O-sulfate (R3S) and resveratrol-4′-O-sulfate (R4S) | Inhibition of COX-1 | Anti-inflammatory effects | Luca et al. (2020) | ||
| R4S | Activation of SIRT1 | Deacetylation of histones | Luca et al. (2020) | ||
| Tetrahydrocurcumin | Activation of glutathione peroxidase (GPX), glutathione S-transferase (GST), and NADPH: quinone reductase | Antioxidant effects | Luca et al. (2020) | ||
| Metabolites and posttranslational modifications | |||||
|---|---|---|---|---|---|
| Metabolite | Effect | Product | References | ||
| Saturated fatty acids (SFA) | Provide substrates for the synthesis of lipids, such as diacylglycerol (DAG) and ceramides; TLR4 mediated activation of inhibitors of nuclear factor-kβ (NF- kβ) kinase subunit-β (IKKβ) and Jun N-terminal kinase (JNK) | Impairment of insulin sensitivity through IKKβ and JNK-mediated phosphorylation of IRS1 on Ser307; increased production of proinflammatory cytokines | Yang et al. (2018) | ||
| Polyunsaturated fatty acids (PUFAs) | Activation of GPCR120 | Recruitment of β-arrestin 2, which sequesters TAK1-binding protein (TAB1), triggering the inhibition of MAP3K7 to prevent JNK and IKKβ activity and resulting in anti-inflammatory effects | Yang et al. (2018) | ||
| Palmitic acid esters of hydroxystearic acid (PAHSAs) | Activation of the long form of GPCR120 | Enhanced insulin-stimulated GLUT4 translocation to membrane | Yore et al. (2014) | ||
| Acetyl-CoA | Acetylation of IRS proteins in lysine residues by the histone acetyltransferase, p300 | Impaired insulin signaling | Cao et al. (2017) | ||
| Acetylation of AKT at lys14 and lys20 by p300 and the p300/CBP-associated factor (P/CAF) | Blockage of AKT binding to phosphatidylinositol (3,4,5)-triphosphate (PIP3) at the plasma membrane | Sundaresan et al. (2011) | |||
| Deacetylation of AKT at lys14 and lys20 by SIRT1 | AKT activation through AKT-PIP3 binding | Sundaresan et al. (2011) | |||
| Acetylation of glucosamine-6-phosphate | Synthesis of uridine-diphosphate-GlcNac | Yang and Qian (2017) | |||
| Palmitate | Palmitoylation of GLUT4 at Cy223 | Insulin-stimulated GLUT4 translocation to membrane | Du et al. (2017) | ||
| β-OHB | Inhibition of HDAC1 and HDAC2 activity | Increase in histone acetylation at promoters of FOXO3 and metallothionein 2 resulting in protection against oxidative stress | Shimazu et al. (2013) | ||
| Metabolites and solute carrier transporters | |||||
|---|---|---|---|---|---|
| Metabolite condition | Transporter modulation | Cellular effect | References | ||
| Glucose starvation | Induction of SLC7A11 expression | Uptake of cystine by SLC7A11 transporters depleting intracellular NADPH and inducing ROS, triggering cancer cell death under glucose starvation | Koppula et al. (2017), Shin et al. (2017) | ||
| Cystine deprivation, oxidative stress | Induction of SLC7A11 expression via nuclear factor erythroid 2-related factor 2 (NRF2) and activating transcription factor 4 (ATF4) | Import of cystine and reestablishment of redox balance | Koppula et al. (2018) | ||
| Loss of lactate transport | Decreased expression of SLC5A8 | Decreased transport of butyrate | Zhang et al. (2019) | ||
| Low levels of butyric acid in the gut | Downregulation of SLC16A1 | Decreased transport of butyric acid | Thibault et al. (2007) | ||
| mTORC complex | Increased GLUT1 expression | Downstream aerobic glycolysis in T cells to support their proliferation and effector function | Song et al. (2020) | ||
Diet microbiome, metabolites, and epigenetic changes
Changes in the extracellular environment, such as food deprivation, caloric restriction, and intense exercise, modify intracellular nutrient-responsive pathways that promote adaptation and epigenetic changes (Dai et al., 2020; Gut & Verdin, 2013; Pizzorusso & Tognini, 2020).
Diet and lifestyle are important external factors that can affect metabolomics and, consequently, the phenotype. Vitamin D is mainly produced through endogenous production; however, through the lifestyle factor, solar ultraviolet-B radiation (UV-B) irradiates 7-dehydrocholesterol present in the skin to generate cholecalciferol (Pilz et al., 2013), which is subsequently activated in the liver and kidney. The other source of vitamin D is dietary intake, which includes supplementation with ergocalciferol or cholecalciferol. Most of the biological functions of the active metabolite of vitamin D are mediated through the regulation of gene expression. The 1,25 dihydroxyvitamin D (1,25(OH)2D3) binds to its nuclear receptor (nVDR) with high affinity and specificity, forming a heterodimer with the retinoid X receptor. Thus, this complex can repress or amplify the transcription of target genes. This occurs through its binding to vitamin D-responsive elements in DNA (Pilz et al., 2013). nVDR is found in some immune cells, such as monocytes and macrophages (Neve et al., 2014). Thus, the active metabolite of vitamin D has an anti-inflammatory effect on macrophages, modifying the phenotype, and downregulating the expression and production of several proinflammatory cytokines, including TNF-α, IL-1β, IL-6, and IL-8 (Giulietti et al., 2007), in different diseases, such as COVID-19 (Jain et al., 2020), hepatic inflammation and steatosis (Dong et al., 2020).
Folate, choline, and methionine are essential metabolites related to methylation through the conversion of homocysteine to SAM. These metabolites are dietary requirements for maintaining methylation levels (Elango, 2020; Niculescu & Zeisel, 2002; Pizzorusso & Tognini, 2020). A methyl-deficient diet reduces one-third of the remethylation of one-carbon cycle metabolites (Farias et al., 2015; Ferrari & Pasini, 2013; Gerhäuser, 2012; Townsend, Davis, Mackey, & Gregory, 2004) and leads to aberrant DNA-methyltransferase activity, abnormal DNA methylation in liver tumors (Lopatina, 1998), and RNA methylation (Mosca et al., 2019). In addition to dietary sources, folate can be synthesized by small intestine flora, and most of it is absorbed by the host (Camilo et al., 1996). Betaine supplementation can enhance the levels of mRNA methylation and decrease fat mass and obesity-associated (FTO) expression in mice on a high-fat diet (Zhou et al., 2015). The presence of selenium in the culture media affects the methylation levels of selenocysteine tRNAs (Diamond et al., 1993).
Among the metabolites, β-hydroxybutyrate (β-OHB), a ketone body delivered by the liver during fasting or prolonged exercise, reshapes DNA by inhibiting HDACs (Mikami et al., 2020; Shimazu et al., 2013). After 12 h of fasting, the intake of ketogenic diets, or prolonged intense exercises, high circulation levels of β-OHB reach 1–2 mM (Koeslag, Noakes, & Sloan, 1980). β-OHB acts through GPR109, a GPCR that binds short-chain fatty acids. Such actions inhibit class I HDACs, which regulate gene expression through the deacetylation of lysine residues on histone and nonhistone proteins (Newman & Verdin, 2014).
β-OHB increases the intracellular content of acetyl-CoA, indirectly altering histone acetylation by promoting acetyltransferase activity via an acetyl-CoA flux (Ku et al., 2020; Xie et al., 2012). Furthermore, histone hyperacetylation triggered by β-OHB results in the increased expression of forkhead box O3 (FOXO3) (Kenyon, 2010). The increase in FOXO3 and metallothionein promoted by the hyperacetylation of histone H3 on lysine 9 (H3K9) offers resistance to oxidative stress (Shimazu et al., 2013; Xie et al., 2016).
Tanegashima et al. demonstrated that fasting induces H3K9 acetylation in the enhancer region of the Slc2a1 gene in neuronal and endothelial cells, due to increased levels of β-OHB. This further maintains a constant glucose concentration in the brain during fasting (Tanegashima et al., 2017). In contrast, supplementation with ketone esters has recently been shown to inhibit glycolysis in the brains of nonfasted mice (Suissa et al., 2021).
In addition to β-OHB, other metabolites are required for enzymes capable of modifying DNA or histones, and during fasting or energy scarcity, the intracellular levels of nicotinamide adenine nucleotide (NAD+), an energy carrier, increase. This metabolite is a cofactor for sirtuin and poly (ADP-ribose) polymerases (PARP), which regulate cellular functions ranging from gene expression to fatty acid metabolism. Once NAD+ levels increase during fasting or energy scarcity, this reflects the potential of such metabolites to regulate intracellular processes in response to environmental changes (Katsyuba et al., 2020; Verdin, 2015).
It was previously demonstrated that food bioactive compounds, such as sulforaphane, a thiocyanate particularly found in broccoli, increase VDR expression (Apprato et al., 2020), inhibit HDAC activity (Tortorella et al., 2015), and increase histone H3 and H4 acetylation (Juge et al., 2007). Furthermore, sulforane cysteine, sulforane N-acetyl-cysteine, allyl mercaptan, and diallyl disulfide, the metabolites produced by microbial metabolism of cruciferous vegetables and garlic induce epigenetic changes by inhibiting histone deacetyl transferase enzymes (Kim et al., 2010).
It is well known that the intestinal microbiota is sensitive to environmental changes, such as high-fat diets (David et al., 2014; Peng et al., 2021), micronutrient deficiency (Hibberd et al., 2017), obesity (Aron-Wisnewsky et al., 2021; Turnbaugh et al., 2009), and chronic inflammation (Couto et al., 2020; Round & Mazmanian, 2009). Therefore, modulating the microbiome maintains and/or improves health through the metabolites produced by intestinal microorganisms in response to the components of the diet, thus resulting in the production of short-chain fatty acids (SCFA), such as butyrate, acetate, and propionate (Guilloteau et al., 2010). In addition, SCFAs are produced through the fermentation of nondigestible carbohydrates, such as dietary fibers, which activate GPCRs, such as GPR41 and GPR43, leading to the inhibition of HDACs (Bhat & Kapila, 2017).
Food bioactive compounds can interact directly with DNA, and dietary behavior can affect DNA structure, leading to a different phenotype. For example, three flavonoids, quercetin, kaempferol, and delphinidin can bind to adenine and guanine (major groove), and thymine (minor groove) (Kanakis et al., 2005), and saffron derivate metabolites can interact with DNA guanine-quadruplexes (G4) (Hoshyar et al., 2012). These G4 motifs are stable structures related to gene promoter regulation and DNA methylation (Hardin et al., 1993; Mao et al., 2018); they can be dysregulated by aberrant DNA methylation due to folate deprivation (Tavakoli Shirazi et al., 2018). Upon interfering with the genome, all other steps of the flux of information will be affected, including the phenotype.
Conclusion
Metabolites can induce macromolecule activity and control phenotypes. It has been shown that omics-scale techniques provide better correlation than single approaches, which indicates that the “central dogma” is a wide integration of information (Piras et al., 2012). Recently, Bar et al. analyzed 1251 metabolites from the serum of 491 individuals and by machine learning deduced that the diet and microbiome both represent 50% of an individual’s metabolic profile (Bar et al., 2020). This concept is a paradigm shift in that it reshapes the conventional thinking about the molecular linear “central dogma,” placing metabolomics at the center, not only providing a simple readout to other omics, but also acting as a master regulator of the whole system (Guijas et al., 2018).
Acknowledgements
The authors thank Professor Tatiana El-Bacha for the rich discussion in this article. We would like to thank Editage (www.editage.com) for English language editing.
Author contributions
GCS conceived and designed the ideas of this study. MRM and NMB developed some parts of the text and reviewed the paper.
Funding
Not applicable.
Data availability
Not applicable.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Research involving human participants and/or animals
GCS, MRM and NMB complied with Springer’s ethical policies. This article does not contain any studies with human or animal subjects performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gilson Costa dos Santos, Jr., Email: gilson.junior@uerj.br
Natália Mesquita de Brito, Email: nataliamesquitadebrito@gmail.com.
References
- Adamski J, Suhre K. Metabolomics platforms for genome wide association studies—Linking the genome to the metabolome. Current Opinion in Biotechnology. 2013 doi: 10.1016/j.copbio.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Ahmed Z, Zeeshan S, Foran DJ, Kleinman LC, Wondisford FE, Dong XQ. Integrative clinical, genomics and metabolomics data analysis for mainstream precision medicine to investigate COVID-19. BMJ Innovations. 2021 doi: 10.1136/bmjinnov-2020-000444. [DOI] [Google Scholar]
- Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, Loven J, Sigova AA, Smith WB, Lee TI, Bradner JE. Genome-wide localization of small molecules. Nature Biotechnology. 2014;32(1):92–96. doi: 10.1038/nbt.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apprato G, Fiz C, Fusano I, Bergandi L, Silvagno F. Natural epigenetic modulators of vitamin D receptor. Applied Sciences (Switzerland) 2020 doi: 10.3390/APP10124096. [DOI] [Google Scholar]
- Aron-Wisnewsky J, Warmbrunn MV, Nieuwdorp M, Clément K. Metabolism and metabolic disorders and the microbiome: The intestinal microbiota associated with obesity, lipid metabolism, and metabolic health—Pathophysiology and therapeutic strategies. Gastroenterology. 2021;160(2):573–599. doi: 10.1053/j.gastro.2020.10.057. [DOI] [PubMed] [Google Scholar]
- Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrell A, Balasubramanian S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nature Chemistry. 2014;6(12):1049–1055. doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar N, Korem T, Weissbrod O, Zeevi D, Rothschild D, Leviatan S, Kosower N, Lotan-Pompan M, Weinberger A, Le Roy CI, Menni C. A reference map of potential determinants for the human serum metabolome. Nature. 2020 doi: 10.1038/s41586-020-2896-2. [DOI] [PubMed] [Google Scholar]
- Bate C, Nolan W, Williams A. Sialic acid on the glycosylphosphatidylinositol anchor regulates PrP-mediated cell signaling and prion formation. Journal of Biological Chemistry. 2016;291(1):160–170. doi: 10.1074/jbc.M115.672394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beger RD, Dunn W, Schmidt MA, Gross SS, Kirwan JA, Cascante M, Brennan L, Wishart DS, Oresic M, Hankemeier T, Broadhurst DI. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics. 2016;12(10):149. doi: 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BJ, Vallim TQDA, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA. Trimethylamine-N-Oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metabolism. 2013;17(1):49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri S, Ranjan N, Arya DP. An overview of recent advances in duplex DNA recognition by small molecules. Beilstein Journal of Organic Chemistry. 2018;14:1051–1086. doi: 10.3762/bjoc.14.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat MI, Kapila R. Dietary metabolites derived from gut microbiota: Critical modulators of epigenetic changes in mammals. Nutrition Reviews. 2017;75(5):374–389. doi: 10.1093/nutrit/nux001. [DOI] [PubMed] [Google Scholar]
- Blouin CM, Prado C, Takane KK, Lasnier F, Garcia-Ocana A, Ferré P, Dugail I, Hajduch E. Plasma membrane subdomain compartmentalization contributes to distinct mechanisms of ceramide action on insulin signaling. Diabetes. 2010;59(3):600–610. doi: 10.2337/db09-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, MacHnicka MA, Purta E, Pitkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Research. 2018;46(D1):D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilo E, Zimmerman J, Mason JB, Golner B, Russell R, Selhub J, Rosenberg IH. Folate synthesized by bacteria in the human upper small intestine is assimilated by the host. Gastroenterology. 1996;110(4):991–998. doi: 10.1053/gast.1996.v110.pm8613033. [DOI] [PubMed] [Google Scholar]
- Cao J, Peng J, An H, He Q, Boronina T, Guo S, White MF, Cole PA, He L. Endotoxemia-mediated activation of acetyltransferase P300 impairs insulin signaling in obesity. Nature Communications. 2017;8(1):131. doi: 10.1038/s41467-017-00163-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer A, Wellen KE. Metabolism and epigenetics: A link cancer cells exploit. Current Opinion in Biotechnology. 2014;34:23–29. doi: 10.1016/j.copbio.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-cruz P, Alisson-Silva F, Todeschini AR, Dias WB. Cellular glycosylation senses metabolic changes and modulates cell plasticity during epithelial to mesenchymal transition. Developmental Dynamics. 2018;247(3):481–491. doi: 10.1002/dvdy.24553. [DOI] [PubMed] [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, Martin ACL, Maddocks ODK, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, Frezza C. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491(7424):458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Bray SM, Li Z, Zarnescu DC, He C, Jin P, Warren ST. Identification of small molecules rescuing fragile X syndrome phenotypes in Drosophila. Nature Chemical Biology. 2008;4(4):256–263. doi: 10.1038/nchembio.78. [DOI] [PubMed] [Google Scholar]
- Chen P, Zuo H, Xiong H, Kolar MJ, Chu Q, Saghatelian A, Siegwart DJ, Wan Y. Gpr132 sensing of lactate mediates tumor-macrophage interplay to promote breast cancer metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(3):580–585. doi: 10.1073/pnas.1614035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YK, Park JH, Yun JA, Cha JH, Kim Y, Won MH, Kim KW, Ha KS, Kwon YG, Kim YM. Heme oxygenase metabolites improve astrocytic mitochondrial function via a Ca2+-dependent HIF-1α/ERRα circuit. PLoS One. 2018;13(8):e0202039. doi: 10.1371/journal.pone.0202039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb M. 60 years ago, Francis Crick changed the logic of biology. PLOS Biology. 2017;15(9):e2003243. doi: 10.1371/journal.pbio.2003243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa dos Santos Junior G, Pereira CM, da Silva K, Fidalgo T, Valente AP. Saliva NMR-based metabolomics in the war against COVID-19. Analytical Chemistry. 2020;92(24):15688–15692. doi: 10.1021/acs.analchem.0c04679. [DOI] [PubMed] [Google Scholar]
- Couto MR, Gonçalves P, Magro F, Martel F. Microbiota-derived butyrate regulates intestinal inflammation: Focus on inflammatory bowel disease. Pharmacological Research. 2020 doi: 10.1016/j.phrs.2020.104947. [DOI] [PubMed] [Google Scholar]
- CRICK, F. H. (1957). On protein synthesis. Symposia of the Society for Experimental Biology, 11, 138–163. http://libgallery.cshl.edu/items/show/52220. Accessed 21 August 2020. [PubMed]
- Dai Z, Ramesh V, Locasale JW. The evolving metabolic landscape of chromatin biology and epigenetics. Nature Reviews Genetics. 2020 doi: 10.1038/s41576-020-0270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani C, Gaglio D, Sacco E, Alberghina L, Vanoni M. Systems metabolomics: From metabolomic snapshots to design principles. Current Opinion in Biotechnology. 2020;63:190–199. doi: 10.1016/j.copbio.2020.02.013. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa EB, de Farias GC, dos Santos Junior GC, Almeida FC, Duarte ML, Neto VM, Aguiar DP. Normal and osteoarthritic synovial fluid present different metabolomic profile. Osteoarthritis and Cartilage. 2017;25(2017):S384. doi: 10.1016/j.joca.2017.02.657. [DOI] [Google Scholar]
- de Sousa, E. B., dos Santos Junior, G. C., Duarte, M. E. L., Moura Neto, V., & Aguiar, D. P. (2017b). Metabolomics as a promising tool for early osteoarthritis diagnosis. Brazilian Journal of Medical and Biological Research, 50(11). 10.1590/1414-431x20176485 [DOI] [PMC free article] [PubMed]
- de Sousa EB, dos Santos Junior GC, Aguiar RP, da Costa Sartore R, de Oliveira ACL, Almeida FCL, Neto VM, Aguiar DP. Osteoarthritic synovial fluid modulates cell phenotype and metabolic behavior in vitro. Stem Cells International. 2019;2019:1–14. doi: 10.1155/2019/8169172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearmond SJ, Qiu Y, Sànchez H, Spilman PR, Ninchak-Casey A, Alonso D, Daggett V. PrP(c) glycoform heterogeneity as a function of brain region: Implications for selective targeting of neurons by priori strains. Journal of Neuropathology and Experimental Neurology. 1999;58(9):1000–1009. doi: 10.1097/00005072-199909000-00010. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delafiori J, Navarro LC, Siciliano RF, De Melo GC, Busanello ENB, Nicolau JC, Sales GM, De Oliveira AN, Val FF, De Oliveira DN, Eguti A. Covid-19 automated diagnosis and risk assessment through metabolomics and machine learning. Analytical Chemistry. 2021;93(4):2471–2479. doi: 10.1021/acs.analchem.0c04497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning EJ, Thirumalai D, Mackerell AD. Protonation of trimethylamine N-oxide (TMAO) is required for stabilization of RNA tertiary structure. Biophysical Chemistry. 2013;184:8–16. doi: 10.1016/j.bpc.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervan PB. Molecular recognition of DNA by small molecules. Bioorganic & Medicinal Chemistry. 2001;9(July):2215–2235. doi: 10.1016/S0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- Di Cerbo V, Schneider R. Cancers with wrong HATs: The impact of acetylation. Briefings in Functional Genomics. 2013;12(3):231–243. doi: 10.1093/bfgp/els065. [DOI] [PubMed] [Google Scholar]
- Diamond AM, Choi IS, Crain PF, Hashizume T, Pomerantz SC, Cruz R, Steer CJ, Hill KE, Burk RF, McCloskey JA. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec) Journal of Biological Chemistry. 1993;268(19):14215–14223. doi: 10.1016/S0021-9258(19)85229-8. [DOI] [PubMed] [Google Scholar]
- Dong B, Zhou Y, Wang W, Scott J, Kim KH, Sun Z, Guo Q, Lu Y, Gonzales NM, Wu H, Hartig SM. Vitamin D receptor activation in liver macrophages ameliorates hepatic inflammation, steatosis, and insulin resistance in mice. Hepatology. 2020;71(5):1559–1574. doi: 10.1002/hep.30937. [DOI] [PubMed] [Google Scholar]
- Dos Santos GC, Saldanha-Gama R, De Brito NM, Renovato-Martins M, Barja-Fidalgo C. Metabolomics in cancer and cancer-associated inflammatory cells. Journal of Cancer Metastasis and Treatment. 2021;2021:1–19. doi: 10.20517/2394-4722.2020.86. [DOI] [Google Scholar]
- Du K, Murakami S, Sun Y, Kilpatrick CL, Luscher B. DHHC7 palmitoylates glucose transporter 4 (Glut4) and regulates Glut4 membrane translocation. Journal of Biological Chemistry. 2017;292(7):2979–2991. doi: 10.1074/jbc.M116.747139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelenkamp K, Osinga TE, Links TP, van der Horst-Schrivers ANA. Clinical implications of the oncometabolite succinate in SDHx-mutation carriers. Clinical Genetics. 2020 doi: 10.1111/cge.13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango R. Methionine nutrition and metabolism: Insights from animal studies to inform human nutrition. Journal of Nutrition. 2020;150(Supplement_1):2518S–2523S. doi: 10.1093/jn/nxaa155. [DOI] [PubMed] [Google Scholar]
- Ermonval M, Mouillet-Richard S, Codogno P, Kellermann O, Botti J. Evolving views in prion glycosylation: Functional and pathological implications. Biochimie. 2003 doi: 10.1016/S0300-9084(03)00040-3. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Mostoslavsky R. Interplay between metabolism and epigenetics: A nuclear adaptation to environmental changes. Molecular Cell. 2016 doi: 10.1016/j.molcel.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias N, Ho N, Butler S, Delaney L, Morrison J, Shahrzad S, Coomber BL. The effects of folic acid on global DNA methylation and colonosphere formation in colon cancer cell lines. The Journal of Nutritional Biochemistry. 2015;26(8):818–826. doi: 10.1016/j.jnutbio.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Ferrari, K. J., & Pasini, D. (2013). Regulation and function of DNA and histone methylations. Current Pharmaceutical Design, 19(4), 719–733. http://www.ncbi.nlm.nih.gov/pubmed/23016854 [PubMed]
- Filippakopoulos P, Knapp S. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nature Reviews Drug Discovery. 2014 doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- Francisco AP, Paulo A. Oncogene expression modulation in cancer cell lines by DNA G-quadruplex-interactive small molecules. Current Medicinal Chemistry. 2018;24(42):4873–4904. doi: 10.2174/0929867323666160829145055. [DOI] [PubMed] [Google Scholar]
- Furlan M, Galeota E, de Pretis S, Caselle M, Pelizzola M. M6A-dependent RNA dynamics in T cell differentiation. Genes. 2019;10(1):28. doi: 10.3390/genes10010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Lin SH, Ren F, Li JT, Chen JJ, Yao CB, Yang HB, Jiang SX, Yan GQ, Wang D, Wang Y. Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nature Communications. 2016;7(1):1–14. doi: 10.1038/ncomms11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhäuser C. Cancer cell metabolism, epigenetics and the potential influence of dietary components—A perspective. Biomedical Research. 2012;23(SPEC. ISSUE):69–89. [Google Scholar]
- Gibson R, Lau CHE, Loo RL, Ebbels TMD, Chekmeneva E, Dyer AR, Miura K, Ueshima H, Zhao L, Daviglus ML, Stamler J. The association of fish consumption and its urinary metabolites with cardiovascular risk factors: The International Study of Macro-/Micronutrients and Blood Pressure (INTERMAP) American Journal of Clinical Nutrition. 2020;111(2):280–290. doi: 10.1093/ajcn/nqz293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Gaidatzis D, Feldmann A, Hoerner L, Imanci D, Burger L, Zilbermann F, Peters AH, Edenhofer F, Smallwood SA, Krebs AR. A genome-scale map of DNA methylation turnover identifies site-specific dependencies of DNMT and TET activity. Nature Communications. 2020;11(1):1–16. doi: 10.1038/s41467-020-16354-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulietti A, van Etten E, Overbergh L, Stoffels K, Bouillon R, Mathieu C. Monocytes from type 2 diabetic patients have a pro-inflammatory profile. 1,25-Dihydroxyvitamin D3 works as anti-inflammatory. Diabetes Research and Clinical Practice. 2007;77(1):47–57. doi: 10.1016/j.diabres.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Gowans GJ, Hawley SA, Ross FA, Hardie DG. AMP is a true physiological regulator of amp-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metabolism. 2013;18(4):556–566. doi: 10.1016/j.cmet.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijas C, Montenegro-Burke JR, Warth B, Spilker ME, Siuzdak G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nature Biotechnology. 2018;36(4):316–320. doi: 10.1038/nbt.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilloteau P, Martin L, Eeckhaut V, Ducatelle R, Zabielski R, Van Immerseel F. From the gut to the peripheral tissues: The multiple effects of butyrate. Nutrition Research Reviews. 2010;23(2):366–384. doi: 10.1017/S0954422410000247. [DOI] [PubMed] [Google Scholar]
- Gut P, Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature. 2013 doi: 10.1038/nature12752. [DOI] [PubMed] [Google Scholar]
- Hardin CC, Corregan M, Brown BA, Frederick LN. Cytosine—Cytosine+ base pairing stabilizes DNA quadruplexes and cytosine methylation greatly enhances the effect. Biochemistry. 1993;32(22):5870–5880. doi: 10.1021/bi00073a021. [DOI] [PubMed] [Google Scholar]
- Hibberd MC, Wu M, Rodionov DA, Li X, Cheng J, Griffin NW, Barratt MJ, Giannone RJ, Hettich RL, Osterman AL, Gordon JI. The effects of micronutrient deficiencies on bacterial species from the human gut microbiota. Science Translational Medicine. 2017;9(390):eaal4069. doi: 10.1126/scitranslmed.aal4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshyar R, Bathaie SZ, Kyani A, Mousavi MF. Is there any interaction between telomeric DNA structures, G-quadruplex and I-motif, with Saffron active metabolites? Nucleosides, Nucleotides and Nucleic Acids. 2012;31(11):801–812. doi: 10.1080/15257770.2012.730164. [DOI] [PubMed] [Google Scholar]
- Hou YCC, Yu HC, Martin R, Cirulli ET, Schenker-Ahmed NM, Hicks M, Cohen IV, Jönsson TJ, Heister R, Napier L, Swisher CL. Precision medicine integrating whole-genome sequencing, comprehensive metabolomics, and advanced imaging. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(6):3053–3062. doi: 10.1073/pnas.1909378117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CY, Lynch GC, Kokubo H, Montgomery Pettitt B. Trimethylamine N-oxide influence on the backbone of proteins: An oligoglycine model. Proteins: Structure, Function and Bioinformatics. 2010;78(3):695–704. doi: 10.1002/prot.22598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen WJ, Vander Heiden MG. Pyruvate kinase: Function, regulation and role in cancer. Seminars in Cell and Developmental Biology. 2015 doi: 10.1016/j.semcdb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Chaurasia R, Sengar NS, Singh M, Mahor S, Narain S. Analysis of vitamin D level among asymptomatic and critically ill COVID-19 patients and its correlation with inflammatory markers. Scientific Reports. 2020;10(1):1–8. doi: 10.1038/s41598-020-77093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke R, Dodson AE, Rine J. Metabolism and epigenetics. Annual Review of Cell and Developmental Biology. 2015;31:473–496. doi: 10.1146/annurev-cellbio-100814-125544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha AK, Huang SCC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, Pearce EJ. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42(3):419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic metabolomics: Major impact on the metabolome. Annual Review of Pharmacology and Toxicology. 2012;52:37–56. doi: 10.1146/annurev-pharmtox-010611-134748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Warmoes MO, Shen X, Locasale JW. Epigenetics and cancer metabolism. Cancer Letters. 2015;356(2 Pt A):309–314. doi: 10.1016/j.canlet.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juge N, Mithen RF, Traka M. Molecular basis for chemoprevention by sulforaphane: A comprehensive review. Cellular and Molecular Life Sciences. 2007 doi: 10.1007/s00018-007-6484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013 doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanakis CD, Tarantilis PA, Polissiou MG, Diamantoglou S, Tajmir-Riahi HA. Dna interaction with naturally occurring antioxidant flavonoids quercetin, kaempferol, and delphinidin. Journal of Biomolecular Structure and Dynamics. 2005;22(6):719–724. doi: 10.1080/07391102.2005.10507038. [DOI] [PubMed] [Google Scholar]
- Katsyuba E, Romani M, Hofer D, Auwerx J. NAD+ homeostasis in health and disease. Nature Metabolism. 2020 doi: 10.1038/s42255-019-0161-5. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010 doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kim G-W, Gocevski G, Wu C-J, Yang X-J. Dietary, metabolic, and potentially environmental modulation of the lysine acetylation machinery. International Journal of Cell Biology. 2010;2010:1–14. doi: 10.1155/2010/632739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball TH, Vondriska TM. Metabolism, epigenetics, and causal inference in heart failure. Trends in Endocrinology and Metabolism. 2020 doi: 10.1016/j.tem.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight AK, Park HJ, Hausman DB, Fleming JM, Bland VL, Rosa G, Kennedy EM, Caudill MA, Malysheva O, Kauwell GP, Sokolow A. Association between one-carbon metabolism indices and DNA methylation status in maternal and cord blood. Scientific Reports. 2018;8(1):1–9. doi: 10.1038/s41598-018-35111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop JM, Patra S, Harish B, Royer CA, Winter R. The deep sea osmolyte trimethylamine N-oxide and macromolecular crowders rescue the antiparallel conformation of the human telomeric G-quadruplex from urea and pressure stress. Chemistry-A European Journal. 2018;24(54):14346–14351. doi: 10.1002/chem.201802444. [DOI] [PubMed] [Google Scholar]
- Koczula KM, Ludwig C, Hayden R, Cronin L, Pratt G, Parry H, Tennant D, Drayson M, Bunce CM, Khanim FL, Günther UL. Metabolic plasticity in CLL: Adaptation to the hypoxic niche. Leukemia. 2016;30(1):65–73. doi: 10.1038/leu.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeslag JH, Noakes TD, Sloan AW. Post-exercise ketosis. The Journal of Physiology. 1980;301(1):79–90. doi: 10.1113/jphysiol.1980.sp013190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Komarov PG, Becker Y, Ewenson A, Gudkov AV. Small molecules that dramatically alter multidrug resistance phenotype by modulating the substrate specificity of P-glycoprotein. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):14078–14083. doi: 10.1073/pnas.241314798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Does the central dogma still stand? Biology Direct. 2012;7:1–7. doi: 10.1186/1745-6150-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppula P, Zhang Y, Shi J, Li W, Gan B. The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate. Journal of Biological Chemistry. 2017;292(34):14240–14249. doi: 10.1074/jbc.M117.798405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppula P, Zhang Y, Zhuang L, Gan B. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Communications (London, England) 2018 doi: 10.1186/s40880-018-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku JT, Chen AY, Lan EI. Metabolic engineering design strategies for increasing acetyl-coa flux. Metabolites. 2020 doi: 10.3390/metabo10040166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert D, Leipply D, Draper DE. The osmolyte TMAO stabilizes native RNA tertiary structures in the absence of Mg2+: Evidence for a large barrier to folding from phosphate dehydration. Journal of Molecular Biology. 2010;404(1):138–157. doi: 10.1016/j.jmb.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbach MA, Hanke JE, Serefidou M, Mangan MSJ, Kolbe CC, Hess T, Rothe M, Kaiser R, Hoss F, Gehlen J, Engels G. Toll-like receptor signaling rewires macrophage metabolism and promotes histone acetylation via ATP-citrate lyase. Immunity. 2019;51(6):997–1011.e7. doi: 10.1016/j.immuni.2019.11.009. [DOI] [PubMed] [Google Scholar]
- Li Y, Xie M, Men L, Du J. O-GlcNAcylation in immunity and inflammation: An intricate system (Review) International Journal of Molecular Medicine. 2019 doi: 10.3892/ijmm.2019.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Huang X, Liang X, Su M, Lai KP, Chen J. Integrated omics analysis reveals the alteration of gut microbe–metabolites in obese adults. Briefings in Bioinformatics. 2020 doi: 10.1093/bib/bbaa165. [DOI] [PubMed] [Google Scholar]
- Li Y, Gruber JJ, Litzenburger UM, Zhou Y, Miao YR, LaGory EL, Li AM, Hu Z, Yip M, Hart LS, Maris JM. Acetate supplementation restores chromatin accessibility and promotes tumor cell differentiation under hypoxia. Cell Death and Disease. 2020;11(2):1–17. doi: 10.1038/s41419-020-2303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood-Evans A, Sarret S, Apfel V, Loesle P, Dawson J, Zhang J, Muller A, Tigani B, Kneuer R, Patel S, Valeaux S. GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. Journal of Experimental Medicine. 2016;213(9):1655–1662. doi: 10.1084/jem.20160061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long NP, Nghi TD, Kang YP, Anh NH, Kim HM, Park SK, Kwon SW. Toward a standardized strategy of clinical metabolomics for the advancement of precision medicine. Metabolites. 2020 doi: 10.3390/metabo10020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopatina N. Elevated expression and altered pattern of activity of DNA methyltransferase in liver tumors of rats fed methyl-deficient diets. Carcinogenesis. 1998;19(10):1777–1781. doi: 10.1093/carcin/19.10.1777. [DOI] [PubMed] [Google Scholar]
- Luca SV, Macovei I, Bujor A, Miron A, Skalicka-Woźniak K, Aprotosoaie AC, Trifan A. Bioactivity of dietary polyphenols: The role of metabolites. Critical Reviews in Food Science and Nutrition. 2020 doi: 10.1080/10408398.2018.1546669. [DOI] [PubMed] [Google Scholar]
- Mao SQ, Ghanbarian AT, Spiegel J, Martínez Cuesta S, Beraldi D, Di Antonio M, Marsico G, Hänsel-Hertsch R, Tannahill D, Balasubramanian S. DNA G-quadruplex structures mold the DNA methylome. Nature Structural and Molecular Biology. 2018;25(10):951–957. doi: 10.1038/s41594-018-0131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F-PJ, Dumas M-E, Wang Y, Legido-Quigley C, Yap IKS, Tang H, Zirah S, Murphy GM, Cloarec O, Lindon JC, Sprenger N. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Molecular Systems Biology. 2007;3(1):112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nature Communications. 2020 doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashino T, Fridovich I. Effects of urea and trimethylamine-N-oxide on enzyme activity and stability. Archives of Biochemistry and Biophysics. 1987;258(2):356–360. doi: 10.1016/0003-9861(87)90355-9. [DOI] [PubMed] [Google Scholar]
- Mathew AK, Padmanaban VC. Metabolomics: The apogee of the omics trilogy. International Journal of Pharmacy and Pharmaceutical Sciences. 2013 doi: 10.1038/nrm3314. [DOI] [Google Scholar]
- Meyer KD, Jaffrey SR. Rethinking m6A readers, writers, and erasers. Annual Review of Cell and Developmental Biology. 2017 doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami D, Kobayashi M, Uwada J, Yazawa T, Kamiyama K, Nishimori K, Nishikawa Y, Nishikawa S, Yokoi S, Taniguchi T, Iwano M. β-Hydroxybutyrate enhances the cytotoxic effect of cisplatin via the inhibition of HDAC/survivin axis in human hepatocellular carcinoma cells. Journal of Pharmacological Sciences. 2020;142(1):1–8. doi: 10.1016/j.jphs.2019.10.007. [DOI] [PubMed] [Google Scholar]
- Mohammad N, Wong D, Lum A, Lin J, Ho J, Lee CH, Yip S. Characterisation of isocitrate dehydrogenase 1/isocitrate dehydrogenase 2 gene mutation and the d-2-hydroxyglutarate oncometabolite level in dedifferentiated chondrosarcoma. Histopathology. 2020;76(5):722–730. doi: 10.1111/his.14018. [DOI] [PubMed] [Google Scholar]
- Mosca P, Leheup B, Dreumont N. Nutrigenomics and RNA methylation: Role of micronutrients. Biochimie. 2019 doi: 10.1016/j.biochi.2019.07.008. [DOI] [PubMed] [Google Scholar]
- Mouillet-Richard S, Ermonval M, Chebassier C, Laplanche JL, Lehmann S, Launay JM, Kellermann O. Signal transduction through prion protein. Science. 2000;289(5486):1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, Boriack R, Rakheja D, Sullivan LB, Linehan WM, Chandel NS. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Reports. 2014;7(5):1679–1690. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussap M, Noto A, Piras C, Atzori L, Fanos V. Slotting metabolomics into routine precision medicine. Expert Review of Precision Medicine and Drug Development. 2021 doi: 10.1080/23808993.2021.1911639. [DOI] [Google Scholar]
- Neidle S. DNA minor-groove recognition by small molecules. Natural Product Reports. 2001 doi: 10.1039/a705982e. [DOI] [PubMed] [Google Scholar]
- Neve A, Corrado A, Cantatore FP. Immunomodulatory effects of vitamin D in peripheral blood monocyte-derived macrophages from patients with rheumatoid arthritis. Clinical and Experimental Medicine. 2014;14(3):275–283. doi: 10.1007/s10238-013-0249-2. [DOI] [PubMed] [Google Scholar]
- Newman JC, Verdin E. Ketone bodies as signaling metabolites. Trends in Endocrinology and Metabolism. 2014 doi: 10.1016/j.tem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu MD, Zeisel SH. Diet, methyl donors and DNA methylation: Interactions between dietary folate, methionine and choline. The Journal of Nutrition. 2002;132(8):2333S–2335S. doi: 10.1093/jn/132.8.2333S. [DOI] [PubMed] [Google Scholar]
- Ong Q, Han W, Yang X. O-GlcNAc as an integrator of signaling pathways. Frontiers in Endocrinology. 2018 doi: 10.3389/fendo.2018.00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan N, Jia D, Geary-Joo C, Wu X, Ferguson-Smith AC, Fung E, Bieda MC, Snyder FF, Gravel RA, Cross JC, Watson ED. Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell. 2013;155(1):81–93. doi: 10.1016/j.cell.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JCH, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-Murray NL, Adam M, Huynh N, Wassie BT, Milani P, Fraenkel E. A multi-omics interpretable machine learning model reveals modes of action of small molecules. Scientific Reports. 2020;10(1):954. doi: 10.1038/s41598-020-57691-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: The apogee of the omics trilogy. Nature Reviews Molecular Cell Biology. 2012 doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Xu X, He Z, Li N, Ouyang Y, Zhu Y, Lu N, He C. Helicobacter pylori infection worsens impaired glucose regulation in high-fat diet mice in association with an altered gut microbiome and metabolome. Applied Microbiology and Biotechnology. 2021;105(5):2081–2095. doi: 10.1007/s00253-021-11165-6. [DOI] [PubMed] [Google Scholar]
- Petersen A-K, Zeilinger S, Kastenmüller G, Römisch-Margl W, Brugger M, Peters A, Meisinger C, Strauch K, Hengstenberg C, Pagel P, Huber F. Epigenetics meets metabolomics: An epigenome-wide association study with blood serum metabolic traits. Human Molecular Genetics. 2014;23(2):534–45. doi: 10.1093/hmg/ddt430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovich DA, Podolskiy DI, Lobanov AV, Lee SG, Miller RA, Gladyshev VN. Using DNA methylation profiling to evaluate biological age and longevity interventions. Cell Metabolism. 2017;25(4):954–960.e6. doi: 10.1016/j.cmet.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz S, Kienreich K, Rutters F, De Jongh R, Van Ballegooijen AJ, Grübler M, Tomaschitz A, Dekker JM. Role of vitamin D in the development of insulin resistance and type 2 diabetes. Current Diabetes Reports. 2013;13(2):261–270. doi: 10.1007/s11892-012-0358-4. [DOI] [PubMed] [Google Scholar]
- Piras V, Tomita M, Selvarajoo K. Is central dogma a global property of cellular information flow? Frontiers in Physiology. 2012;3(November):1–8. doi: 10.3389/fphys.2012.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorusso T, Tognini P. Interplay between metabolism, nutrition and epigenetics in shaping brain dna methylation, neural function and behavior. Genes. 2020 doi: 10.3390/genes11070742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogo BGT, Allfrey VG, Mirsky AE. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 1966;55:805–812. doi: 10.1073/pnas.55.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP, Karpf AR, James SR, Melnyk S, Han T, Tryndyak VP. Epigenetic alterations in the brains of Fisher 344 rats induced by long-term administration of folate/methyl-deficient diet. Brain Research. 2008;1237:25–34. doi: 10.1016/j.brainres.2008.07.077. [DOI] [PubMed] [Google Scholar]
- Qi H, Ning X, Yu C, Ji X, Jin Y, McNutt MA, Yin Y. Succinylation-dependent mitochondrial translocation of PKM2 promotes cell survival in response to nutritional stress. Cell Death & Disease. 2019;10(3):170. doi: 10.1038/s41419-018-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, Silhavy TJ. Metabolite turns master regulator. Nature. 2013;500(7462):283–284. doi: 10.1038/nature12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid MA, Dai Z, Locasale JW. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nature Cell Biology. 2017 doi: 10.1038/ncb3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti A, Fernández AF, Fraga MF. Nicotinamide N-methyltransferase: At the crossroads between cellular metabolism and epigenetic regulation. Molecular Metabolism. 2021 doi: 10.1016/j.molmet.2021.101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K.D. (2005) Epigenetic Mechanisms of Gene Regulation. In: DNA Methylation and Cancer Therapy. Medical Intelligence Unit. Springer, Boston, MA. 10.1007/0-387-27443-X_2
- Rodrigues MF, Obre E, de Melo FHM, Santos GC, Galina A, Jasiulionis MG, Rossignol R, Rumjanek FD, Amoêdo ND. Enhanced OXPHOS, glutaminolysis and β-oxidation constitute the metastatic phenotype of melanoma cells. The Biochemical Journal. 2016;473(6):703–15. doi: 10.1042/BJ20150645. [DOI] [PubMed] [Google Scholar]
- Rodriguez R, Miller KM. Unravelling the genomic targets of small molecules using high-throughput sequencing. Nature Reviews Genetics. 2014 doi: 10.1038/nrg3796. [DOI] [PubMed] [Google Scholar]
- Rossner R, Kaeberlein M, Leiser SF. Flavin-containing monooxygenases in aging and disease: Emerging roles for ancient enzymes. Journal of Biological Chemistry. 2017 doi: 10.1074/jbc.R117.779678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature Reviews Immunology. 2009 doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant’Anna-Silva ACB, Santos GC, Campos SPC, Oliveira Gomes AM, Pérez-Valencia JA, Rumjanek FD. Metabolic profile of oral squamous carcinoma cell lines relies on a higher demand of lipid metabolism in metastatic cells. Frontiers in Oncology. 2018;8:13. doi: 10.3389/fonc.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos GC, Zeidler JD, Pérez-Valencia JA, Sant’Anna-Silva ACB, Da Poian AT, El-Bacha T, Almeida FCL. Metabolomic analysis reveals vitamin D-induced decrease in polyol pathway and subtle modulation of glycolysis in HEK293T cells. Scientific Reports. 2017;7(1):9510. doi: 10.1038/s41598-017-10006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SL. Small molecules: The missing link in the central dogma. Nature Chemical Biology. 2005;1(2):64–66. doi: 10.1038/nchembio0705-64. [DOI] [PubMed] [Google Scholar]
- Seldin MM, Meng Y, Qi H, Zhu WF, Wang Z, Hazen SL, Lusis AJ, Shih DM. Trimethylamine N-oxide promotes vascular inflammation through signaling of mitogen-activated protein kinase and nuclear factor-κb. Journal of the American Heart Association. 2016;5(2):e002767. doi: 10.1161/JAHA.115.002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wei J, He C. Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Molecular Cell. 2019 doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD, Newgard CB. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339(6116):211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin CS, Mishra P, Watrous JD, Carelli V, D’Aurelio M, Jain M, Chan DC. The glutamate/cystine xCT antiporter antagonizes glutamine metabolism and reduces nutrient flexibility. Nature Communications. 2017;8(1):1–11. doi: 10.1038/ncomms15074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311(5762):844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Soga T. Cancer metabolism: Key players in metabolic reprogramming. Cancer Science. 2013 doi: 10.1111/cas.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Li D, Tao L, Luo Q, Chen L. Solute carrier transporters: The metabolic gatekeepers of immune cells. Acta Pharmaceutica Sinica B. 2020;10(1):61–78. doi: 10.1016/j.apsb.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suissa L, Kotchetkov P, Guigonis JM, Doche E, Osman O, Pourcher T, Lindenthal S. Ingested ketone ester leads to a rapid rise of acetyl-coa and competes with glucose metabolism in the brain of non-fasted mice. International Journal of Molecular Sciences. 2021;22(2):1–17. doi: 10.3390/ijms22020524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomedicine and Pharmacotherapy. 2019 doi: 10.1016/j.biopha.2019.108613. [DOI] [PubMed] [Google Scholar]
- Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V, Raghuraman H, Cunningham JM, Gupta M, Gupta MP. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Science Signaling. 2011;4(182):ra46–ra46. doi: 10.1126/scisignal.2001465. [DOI] [PubMed] [Google Scholar]
- Syed I, Lee J, Moraes-Vieira PM, Donaldson CJ, Sontheimer A, Aryal P, Wellenstein K, Kolar MJ, Nelson AT, Siegel D, Mokrosinski J. Palmitic acid hydroxystearic acids activate GPR40, which is involved in their beneficial effects on glucose homeostasis. Cell Metabolism. 2018;27(2):419–427.e4. doi: 10.1016/j.cmet.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanegashima K, Sato-Miyata Y, Funakoshi M, Nishito Y, Aigaki T, Hara T. Epigenetic regulation of the glucose transporter gene Slc2a1 by β-hydroxybutyrate underlies preferential glucose supply to the brain of fasted mice. Genes to Cells. 2017;22(1):71–83. doi: 10.1111/gtc.12456. [DOI] [PubMed] [Google Scholar]
- Tang WH, Wang Z, Kennedy DJ, Wu Y, Buffa JA, Agatisa-Boyle B, Li XS, Levison BS, Hazen SL. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circulation Research. 2015;116(3):448–455. doi: 10.1161/CIRCRESAHA.116.305360.Gut. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakoli Shirazi P, Leifert WR, Fenech MF, François M. Folate modulates guanine-quadruplex frequency and DNA damage in Werner syndrome. Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 2018;826:47–52. doi: 10.1016/j.mrgentox.2017.12.002. [DOI] [PubMed] [Google Scholar]
- Thibault R, De Coppet P, Daly K, Bourreille A, Cuff M, Bonnet C, Mosnier JF, Galmiche JP, Shirazi-Beechey S, Segain JP. Down-regulation of the monocarboxylate transporter 1 is involved in butyrate deficiency during intestinal inflammation. Gastroenterology. 2007;133(6):1916–1927. doi: 10.1053/j.gastro.2007.08.041. [DOI] [PubMed] [Google Scholar]
- Thøgersen R, Rasmussen MK, Sundekilde UK, Goethals SA, Van Hecke T, Vossen E, De Smet S, Bertram HC. Background diet influences TMAO concentrations associated with red meat intake without influencing apparent hepatic TMAO-related activity in a porcine model. Metabolites. 2020;10(2):57. doi: 10.3390/metabo10020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MS, Fernandez ML. Trimethylamine N-oxide (TMAO), diet and cardiovascular disease. Current Atherosclerosis Reports. 2021 doi: 10.1007/s11883-021-00910-x. [DOI] [PubMed] [Google Scholar]
- Tiwari VK, McGarvey KM, Licchesi JDF, Ohm JE, Herman JG, Schübeler D, Baylin SB. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biology. 2008;6(12):2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorella SM, Royce SG, Licciardi PV, Karagiannis TC. Dietary sulforaphane in cancer chemoprevention: The role of epigenetic regulation and HDAC inhibition. Antioxidants and Redox Signaling. 2015 doi: 10.1089/ars.2014.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JH, Davis SR, Mackey AD, Gregory JF. Folate deprivation reduces homocysteine remethylation in a human intestinal epithelial cell culture model: Role of serine in one-carbon donation. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2004;286(4):G588–G595. doi: 10.1152/ajpgi.00454.2003. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, B. M., & O’Neill, L. P. (1995). Histone acetylation in chromatin and chromosomes. Seminars in Cell Biology, 6(4), 229–36. http://www.ncbi.nlm.nih.gov/pubmed/8562915. Accessed 10 April 2014. [DOI] [PubMed]
- Tzika E, Dreker T, Imhof A. Epigenitics and metabolism in health and disease. Frontiers in Genetics. 2018 doi: 10.3389/fgene.2018.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda YM, Zouzumi YK, Maruyama A, Nakano SI, Sugimoto N, Miyoshi D. Effects of trimethylamine N-oxide and urea on DNA duplex and G-quadruplex. Science and Technology of Advanced Materials. 2016;17(1):753–759. doi: 10.1080/14686996.2016.1243000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufnal M, Zadlo A, Ostaszewski R. TMAO: A small molecule of great expectations. Nutrition. 2015;31(11–12):1317–1323. doi: 10.1016/j.nut.2015.05.006. [DOI] [PubMed] [Google Scholar]
- Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E. BioMagResBank. Nucleic Acids Research. 2007;36(Database):D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usanov DL, Chan AI, Maianti JP, Liu DR. Second-generation DNA-templated macrocycle libraries for the discovery of bioactive small molecules. Nature Chemistry. 2018;10(7):704–714. doi: 10.1038/s41557-018-0033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E. NAD+ in aging, metabolism, and neurodegeneration. Science. 2015 doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- Vinolo MAR, Ferguson GJ, Kulkarni S, Damoulakis G, Anderson K, Bohlooly-Y M, Stephens L, Hawkins PT, Curi R. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS One. 2011;6(6):e21205. doi: 10.1371/journal.pone.0021205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- Wang F, Wang K, Xu W, Qin X, Yang P, Correspondence HY. SIRT5 desuccinylates and activates pyruvate kinase M2 to block macrophage IL-1β production and to prevent DSS-induced colitis in mice. Cell Reports. 2017 doi: 10.1016/j.celrep.2017.05.065. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth B, Raffeiner P, Granados A, Huan T, Fang M, Forsberg E, Benton HP, Goetz L, Johnson CH, Siuzdak G. Metabolomics reveals that dietary xenoestrogens alter cellular metabolism induced by palbociclib/letrozole combination cancer therapy. bioRxiv. 2017 doi: 10.1101/188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner A, Meiser J, Weindl D, Hiller K. How metabolites modulate metabolic flux. Current Opinion in Biotechnology. 2015 doi: 10.1016/j.copbio.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Wilson W, Li K. Targeting RNA with small molecules. Current Medicinal Chemistry. 2012;7(1):73–98. doi: 10.2174/0929867003375434. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Research. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. The acetate switch. Microbiology and Molecular Biology Reviews. 2005;69(1):12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi L, Carroll T, Matos I, Luo JD, Polak L, Pasolli HA, Jaffrey SR, Fuchs E. M6a rna methylation impacts fate choices during skin morphogenesis. eLife. 2020;9:1–63. doi: 10.7554/ELIFE.56980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, Zhao S. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes and Development. 2012;26(12):1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y. Lysine succinylation and lysine malonylation in histones. Molecular and Cellular Proteomics. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zhang D, Chung D, Tang Z, Huang H, Dai L, Qi S, Li J, Colak G, Chen Y, Xia C. Metabolic regulation of gene expression by histone lysine β-hydroxybutyrylation. Molecular Cell. 2016;62(2):194–206. doi: 10.1016/j.molcel.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G, Wang L, Chen T, Zhou K, Zhang Z, Li J, Sun B, Guo Y, Wang X, Wang Y, Zhang H. A metabolite array technology for precision medicine. Analytical Chemistry. 2021;93(14):5709–5717. doi: 10.1021/acs.analchem.0c04686. [DOI] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H, Ito S, Yang C, Wang P, Xiao MT, Liu LX. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Qian K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nature Reviews Molecular Cell Biology. 2017;18(7):452–465. doi: 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Achreja A, Yeung TL, Mangala LS, Jiang D, Han C, Baddour J, Marini JC, Ni J, Nakahara R, Wahlig S. Targeting stromal glutamine synthetase in tumors disrupts tumor microenvironment-regulated cancer cell growth. Cell Metabolism. 2016;24(5):685–700. doi: 10.1016/j.cmet.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Vijayakumar A, Kahn BB. Metabolites as regulators of insulin sensitivity and metabolism. Nature Reviews Molecular Cell Biology. 2018 doi: 10.1038/s41580-018-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap YHY, Say YH. Resistance against apoptosis by the cellular prion protein is dependent on its glycosylation status in oral HSC-2 and colon LS 174T cancer cells. Cancer Letters. 2011;306(1):111–119. doi: 10.1016/j.canlet.2011.02.040. [DOI] [PubMed] [Google Scholar]
- Yi CW, Wang LQ, Huang JJ, Pan K, Chen J, Liang Y. Glycosylation significantly inhibits the aggregation of human prion protein and decreases its cytotoxicity. Scientific Reports. 2018;8(1):12603. doi: 10.1038/s41598-018-30770-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, Patel RT, Lee J, Chen S, Peroni OD, Dhaneshwar AS. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159(2):318–332. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yugi K, Kuroda S. Metabolism as a signal generator across trans-omic networks at distinct time scales. Current Opinion in Systems Biology. 2018 doi: 10.1016/j.coisb.2017.12.002. [DOI] [Google Scholar]
- Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nature Reviews Molecular Cell Biology. 2019 doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nature Reviews Molecular Cell Biology. 2019 doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- Zeisel SH, Warrier M. Trimethylamine N-oxide, the microbiome, and heart and kidney disease. Annual Review of Nutrition. 2017;37(1):157–181. doi: 10.1146/annurev-nutr-071816-064732. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang Y, Sun K, Meng Z, Chen L. The SLC transporter in nutrient and metabolic sensing, regulation, and drug development. Journal of Molecular Cell Biology. 2019;11(1):1–13. doi: 10.1093/jmcb/mjy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Muri J, Fitzgerald G, Gorski T, Gianni-Barrera R, Masschelein E, D’Hulst G, Gilardoni P, Turiel G, Fan Z, Wang T. Endothelial lactate controls muscle regeneration from ischemia by inducing M2-like macrophage polarization. Cell Metabolism. 2020;31(6):1136–1153.e7. doi: 10.1016/j.cmet.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Chen J, Chen J, Wu W, Wang X, Wang Y. The beneficial effects of betaine on dysfunctional adipose tissue and N6-methyladenosine mRNA methylation requires the AMP-activated protein kinase α1 subunit. The Journal of Nutritional Biochemistry. 2015;26(12):1678–1684. doi: 10.1016/j.jnutbio.2015.08.014. [DOI] [PubMed] [Google Scholar]
- Zhu H, Lee OW, Shah P, Jadhav A, Xu X, Patnaik S, Shen M, Hall MD. Identification of activators of human fumarate hydratase by quantitative high-throughput screening. SLAS Discovery. 2020;25(1):43–56. doi: 10.1177/2472555219873559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.


