Abstract
The pandemic of coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) is continuously worsening globally, herein we have proposed an electrochemical biosensor for the sensitive monitoring of SARS-CoV-2 RNA. The presence of target RNA firstly triggers the catalytic hairpin assembly circuit and then initiates terminal deoxynucleotidyl transferase-mediated DNA polymerization. Consequently, a large number of long single-stranded DNA products can be produced, and these negatively charged DNA products will bind a massive of positively charged electroactive molecular of Ru(NH3)63+ due to the electrostatic adsorption. Therefore, significantly amplified electrochemical signals can be generated for sensitive analysis of SARS-CoV-2 RNA in the range of 0.1–1000 pM with the detection limit as low as 26 fM. Besides the excellent distinguishing ability for SARS-CoV-2 RNA against single-base mismatched RNA, the proposed biosensor can also be successfully applied to complex matrices, as well as clinical patient samples with high stability, which shows great prospects of clinical application.
Keywords: SARS-CoV-2, COVID-19, Catalytic hairpin assembly, TdT-mediated DNA polymerization
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a newly discovered human virus, has caused the global pandemic of coronavirus disease 2019 (COVID-19), leading to extremely high morbidity and severe mortality worldwide (Huang et al., 2020; Lee et al., 2021; Li et al., 2020; Wang et al., 2021). In addition to the incubation period up to 24 days, a fact that cannot be ignored is that 40%–45% of infected people have no symptoms (Oran et al., 2020). The unknown transmission routes and transmission capabilities of these asymptomatic infectors pose a huge risk for virus prevention and control (Lai et al., 2020; Zou et al., 2020). Therefore, thoroughly screening of suspicious people through accessible, fast and accurate molecular techniques is one of the most effective means to contain virus spreads and threats.
Real-time reverse transcription polymerase chain reaction (RT-PCR) as the most wide detection method has made tremendous contributions to the epidemic prevention and control of COVID-19 since the SARS-CoV-2 genome sequence was released (Corman et al., 2020). However, some technical shortcomings such as long processing time, laborious, involving specialized instruments, skilled personnel and insurmountable false negatives encourage the further efforts to develop more reliable diagnosis systems for thoroughly screening of suspicious patients (Ali et al., 2021; Baek et al., 2020; Pang et al., 2020; Zhu et al., 2020). Electrochemical biosensors are considered to be one of the most powerful alternative tools for the real-time monitoring of COVID-19 in the clinical diagnosis (Ji et al., 2020; Mahshid et al., 2021), for instance, Chaibun et al. have reported a rapid electrochemical biosensor for detection of SARS-CoV-2 RNA as low as 1 copy/μL within 2 h, showing its high sensitivity, rapidity and robustness (Chaibun et al., 2021). Moreover, electrochemical biosensors possess the advantages of portability, miniaturization, and low-cost, which are beneficial to their widespread and rapid deployment in remote regions or outbreak areas where professional testing laboratories are difficult to access (Chen et al., 2019; Masson et al., 2017; Soler et al., 2019; Zheng et al., 2020). For example, Zhao et al. have proposed a portable electrochemical smartphone based on alixarene functionalized graphene oxide to detect SARS-CoV-2 RNA (Zhao et al., 2021), which shows great application potentials in point-of-care testing (POTC). However, most of these methods rely on nanomaterials or cumbersome modifications of probes to improve detection performance. Further endeavors are still demanded to achieve more convenient and effective analysis of COVID-19.
In this regard, we have proposed an electrochemical biosensor for the monitoring of SARS-CoV-2 RNA by combining the signal amplification capabilities of catalytic hairpin assembly (CHA) (Dai et al., 2018; Li et al., 2011, 2018; Liu et al. 2017a, 2019; Xu et al., 2021) and terminal deoxynucleotidyl transferase (TdT) (Liu et., 2017b; Yuan et al., 2017). In a typical CHA reaction, two complementary DNA strands are pre-designed to be locked into two metastable hairpin structures that cannot interact with each other. Nevertheless, the addition of the catalytic single strand DNA or RNA can lead the two hairpins to be opened one after another, generating remarkably enhanced signal for target detection (Peng et al., 2018). On the other hand, as a template-free polymerase, TdT can catalyze the addition of multiple deoxyribonucleotides to the 3′-end of a DNA strand, thereby polymerizing into a long single-stranded structure, which may be used as a powerful tool for biological analysis. In our design, the presence of target sequence can trigger CHA to generate a double-stranded product accompanying with the recycle of target. The double-stranded product will then hybridize with the DNA hairpin structure modified on the electrode surface and form a Y-shaped structure DNA with three protruding 3′-end, which can be extended by the TdT enzyme to form long single-stranded DNA products. Thus, after adding Ru(NH3)6 3+, a remarkably enhanced signal for sensitive quantification of SARS-CoV-2 RNA is generated. In addition to the advantages of high signal and excellent anti-interference ability, the proposed electrochemical biosensor can be successfully performed in clinical samples, demonstrating its great clinical application potential for efficient and thorough diagnosis of COVID-19.
2. Experimental
2.1. Chemicals and materials
The purified DNA sequences and RNA sequences involved in the experiment (Table S1) were synthesized by Sangon Biotech. Co. Ltd. (Shanghai, China) and Invitrogen Biotechnology Co., Ltd. (Shanghai, China), respectively. The sequence of target gene is according to GenBank, SARS-CoV-2 NC_045512. The terminal deoxynucleotidyl transferase (TdT) enzyme and deoxyribonucleotides mixture (dNTPs) were ordered from New England Biolabs Co.,Ltd (Beijing, China). Hexaammineruthenium (III) chloride (Ru(NH3)6Cl3) was purchased from Aladdin (Shanghai, China). Human serum samples and the total RNA clinical specimens were provided by the Second Affiliated Hospital of Southeast University (Nanjing, China). Other reagents required in the experiment were provided by Sigma-Aldrich (Shanghai, China). All reagents were of analytical grades. The research was approved by the ethical committee of the Second Hospital of Nanjing and Nanjing University, and informed consent was obtained in all cases.
2.2. Electrode treatment
The bare gold electrode (AuE) was treated according to the previous reported protocol to obtain a mirror-like surface (Li et al., 2016). Before the experiment, the HP, HP1 as well as HP2 were annealed (maintained at 95 °C for 5 min, and then gradually cooled to room temperature) to ensure respective hairpin structures. The 10 μL HP (0.5 μM) treated with TCEP for 30 min for reducing the S–S bonds was subsequently dropped on the surface of AuE for 2 h. After that, a drop of 10 μL MCH (1 mM) was added onto the resulting AuE for another 2 h to block non-specific sites.
2.3. Detection protocol for SARS-COV-2 RNA
According to the “WHO interim guidance for laboratory testing for COVID-19 in humans”, a 26-nt-long ORF1ab fragment of SARS-CoV-2 RNA is selected as the target sequence for the following tests, which has been demonstrated to be a highly conserved region and has been widely used in recent studies [Qiu et al., 2020]. First, the mixture of HP1 (1 μM), HP2 (1 μM) and different concentrations of the target RNA was dropped on the sensing electrode (HP/MCH/AuE) and incubated together for 2 h at room temperature. Then, the mixture of dNTP (1 mM) and TdT (10 U) was dropped onto the sensing electrode and incubated for another 1 h. Finally, the electrode was immersed into Tris-HCl buffer containing Ru(NH3)6 3+ (5 μM) for 5 min to measure the electrochemical signal. In order to protect RNA from RNase degradation, all solutions involved in the experiments were prepared with DEPC-treated Milli-Q water and autoclaved in an RNase-free environment. Moreover, all experiments were performed at a clean bench.
2.4. Electrochemical measurements
All electrochemical measurements were performed on a CHI660D Potentiostat (CH Instruments) containing a three-electrode system (working electrode: the modified sensing electrode; reference electrode: a saturated calomel electrode; counter electrode: a platinum wire). The electrochemical impedance spectra (EIS) were scanned in the mixture solution of KCl (1 M) and [Fe(CN)6]3-/4- (5 mM) with parameter settings as follows: amplitude, 5 mV; potential, 0.21 V; frequency range, 0.1 Hz to 10 kHz. The differential pulse voltammetry (DPV) was scanned between the potential from-0.15 V to −0.4 V in Tris-HCl buffer containing Ru(NH3)6 3+ (5 μM) with parameter settings as follows: sample width, 0.0167 s; pulse width, 0.05 s; pulse amplitude, 0.05 V.
2.5. Detection of SARS-COV-2 RNA in serum and saliva
Prior to experiments, the collected saliva and serum samples were treated with RNase inhibitor under a clean condition to prevent RNA from degrading by RNase. Afterward, different concentrations of target RNA were spiked into human serum and saliva to be detected according to our detection protocol described above.
2.6. Extraction and detection of SARS-COV-2 RNA from clinical samples
The total RNA was extracted from 200 μL of each clinical sample using a Nucleic Acid Isolation Kit (Magnetic Beads) from Bioperfectus Technologies (Taizhou, China), according to the manufacturer. The RNA sample was stored in nuclease-free water at −80 °C for use. 10 μL of the extracted RNA was further used as the sample input for the detection of SARS-CoV-2 RNA in accordance with our detection protocol described above.
3. Results and discussions
3.1. Principle of the designed biosensor
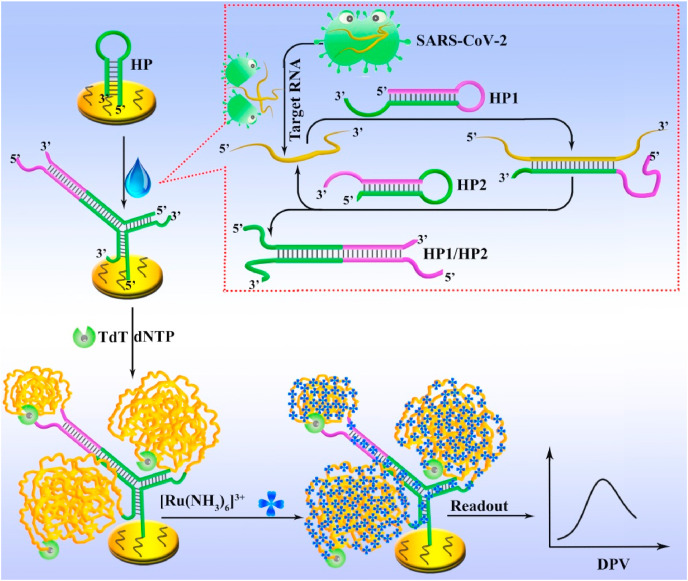
The principle of the proposed biosensor is depicted in Scheme 1 . The complementary HP1 and HP2 responsible for the CHA reaction are designed into metastable hairpin structures. The HP modified with thiol group at the 5′-end is fixed on the electrode through thiol-gold interaction. In the absence of the target RNA, all probes can coexist without significant cross-hybridization. In such circumstances, the TdT-mediated DNA polymerization on the electrode is inhibited due to the steric hindrance effect of the hairpin structure of HP. In addition, the blunt 3′-end of HP will further weaken the function of TdT because the catalytic activity of TdT enzyme requires a protruding 3′-end of more than 3 bases as an initiator (Michelson et al., 1982). On the contrary, in the presence of target RNA, the hairpin structure of HP1 will be unfolded to expose the stem region due to the hybridization process between the target and HP1. The newly exposed single-stranded stem region can further hybridize with HP2 to form a duplex complex (HP1/HP2) along with the release of the target RNA to bind with another HP1, thereby achieving the recycle of the target. Meanwhile, the two functional tails (green region) of HP1/HP2 duplex can simultaneously bind to the loop region of HP to form a Y-shaped DNA structure with three free 3′-end, which can further activate TdT-induced polymerization reaction in a dNTP pool for generation of numerous long single-stranded DNA products. As a result, a mass amount of Ru(NH3)6 3+ can be adsorbed to the negatively charged DNA phosphate backbone through the strong electrostatic interaction. Thus, significantly amplified electrochemical signals can be obtained for sensitive monitoring of target RNA.
Scheme 1.
Principle of the proposed electrochemical biosensor for sensitive analysis of SARS-CoV-2 RNA.
3.2. Feasibility investigation
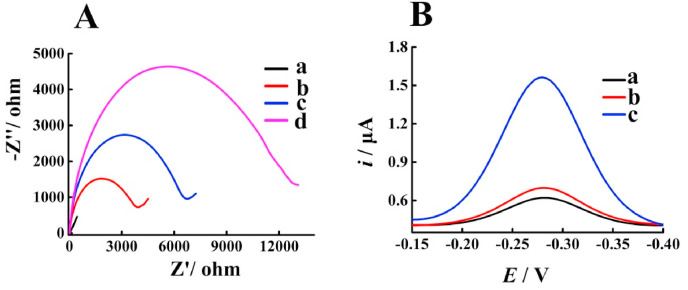
The performance of the sensor has been first verified by the electrochemical impedance spectroscopy (EIS) in Fe(CN)6 3−/4- solution containing KCl. As shown in Fig. 1 A, bare electrode shows extremely weak resistance (curve a), as the semicircular diameter shows a positive correlation with the resistance of charge transfer. However, the modification of HP and MCH on the electrode can significantly increase the semicircle area (curve b), which is probably due to the shielding ability of the negative charge of DNA on the electron transfer. The addition of the mixture of HP1, HP2 and target RNA to the sensing electrode leads to an increase in the semicircular area (curve c) since the Y-shaped DNA structure induced by the double-stranded product of CHA increases the electronegativity and steric hindrance. Furthermore, subsequent incubation with TdT causes a further enlarged semicircular area (curve d), because the generation of long single-stranded DNA triggered by TdT can further enhance the electronegativity and steric hindrance to hinder the charge transfer. After that, the signal responses under different conditions have been recorded to further evaluate the feasibility of the proposed electrochemical biosensor. As exhibited in Fig. 1B, the sensing electrode shows the weakest current peak (curve a), which is because the HP on the electrode can adsorb a small amount of Ru(NH3)6 3+. Moreover, the incubation with HP1 and HP2 results in a slightly increased current intensity (curve b), presumably due to the weak non-specific cross-hybridization between the probes. However, after the introduction of the target RNA, an obvious enhancement in signal intensity can be observed (curve c), owing to the generation of the long single-stranded DNA polymers catalyzed by TdT. These above results have demonstrated the excellent feasibility of our biosensor for the analysis of SARS-CoV-2 RNA.
Fig. 1.
(A) EIS characterizations corresponding to different treatments of the electrode: (a) AuE, (b) MCH/HP/AuE, (c) (HP1 + HP2 + target RNA)/MCH/HP/AuE, and (d) (HP1 + HP2 + target RNA+TdT)/MCH/HP/AuE. (B) DPV responses under different conditions: (a) (TdT + Ru(NH3)63+) MCH/HP/AuE, (b) (HP1 + HP2 + TdT + Ru(NH3)63+)/MCH/HP/AuE, (c) (HP1 + HP2 + target RNA + TdT + Ru(NH3)63+)/MCH/HP/AuE.
3.3. Sensitivity investigation
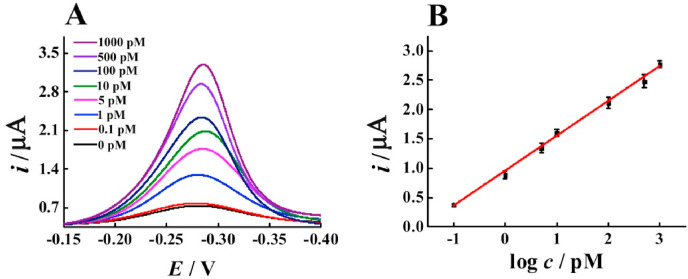
Under the optimal experimental conditions (Fig. S1), the sensitivity of the proposed electrochemical biosensor has been tested by using various abundances of the target RNA. It can be noticed from Fig. 2 A that the signal response is highly dependent on the target RNA concentration. The increased concentration of the target RNA ranging from 0.1 pM to 1000 pM leads to a corresponding enhancement in current intensity. Moreover, as shown in Fig. 2B, the peak current exhibits a good linear relationship to the logarithm of concentration of the target RNA. The obtained regression equation is I (μA) = 0.5933 lgc (pM) + 0.9422 (R2 = 0.9963), and the detection limit estimated by the 3σ rule is 26 fM, which is better than several methods currently reported (Jiao et al., 2020; Qiu et al., 2020).
Fig. 2.
(A) DPV currents of the biosensor with various concentrations of the target RNA. (B) The linear relationship between the value of peak current and the logarithm of the target RNA concentrations. Error bars: SD, n = 3.
3.4. Selectivity investigation
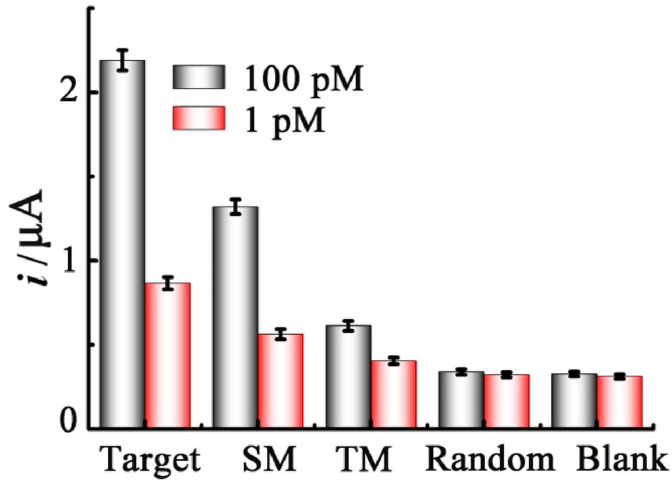
The selectivity of our electrochemical sensor has also been investigated using challenging several control sequences, including single-base mismatched and two-base mismatched RNA, as well as the random sequence at the concentrations of 100 pM and 1 pM. As illustrated in Fig. 3 , it is evident that the current response induced by the SM RNA decreases significantly as compared to that of the target RNA, and the TM RNA leads to a further weaker signal. Moreover, the presence of the random sequence produces a signal similar to the blank, indicating the excellent selectivity of the biosensor to the target RNA.
Fig. 3.
Selective study of the proposed biosensor for the target RNA, single-base mismatched (SM) RNA, two-base mismatched (TM) RNA and random sequence at the concentrations of 100 pM and 1 pM, respectively. Error bars: SD, n = 3.
3.5. Detection of the target RNA in serum and saliva
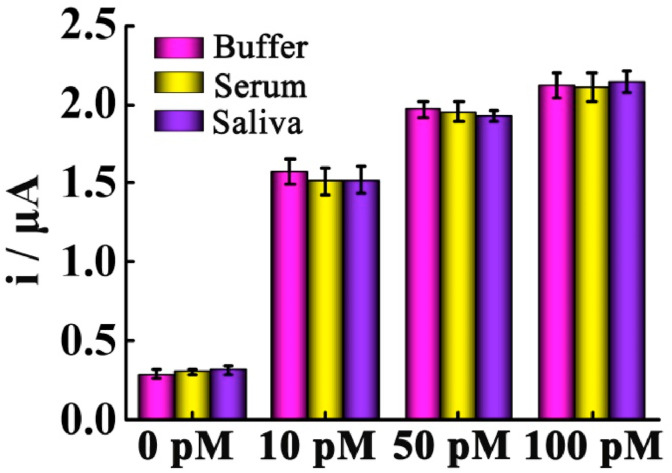
To further assess the analytical performance of the as-proposed electrochemical biosensor in complex biological samples, we have dispersed different concentrations of the target RNA in buffer, 10% serum and saliva for recovery tests, respectively. As shown in Fig. 4 , the variations of the signal responses between the buffer, 10% serum and saliva are negligible, suggesting the satisfactory stability of the biosensor. In addition, the recovery of the spiked RNA in the serum is between 93.0% and 101.2% with an appropriate RSD ranging from 3.2% to 5.5%, while the recovery of the spiked RNA in the saliva is between 95.0% and 105.8% with an RSD of 1.9%–5.3% (Table S2). Thus, these results have further demonstrated that the proposed biosensor can be applied to complicated biological matrix with excellent practicality potential.
Fig. 4.
Current responses for different concentrations of the target RNA in buffer, serum and saliva. Error bars: SD, n = 3.
3.6. Clinical sample analysis
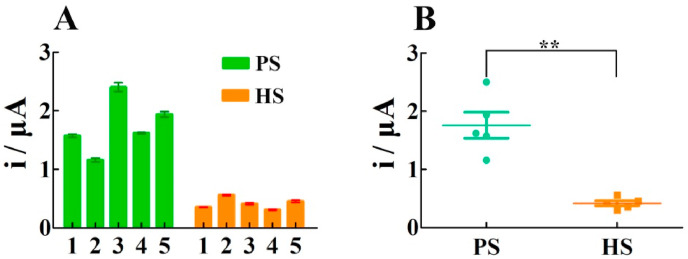
To further demonstrate the potential of our electrochemical biosensor in clinical diagnosis, RNA isolated from the oropharyngeal swabs of 5 healthy samples and 5 patient samples with COVID-19 have been collected for tests. As exhibited in Fig. 5 A, the current signals triggered by the RNA from the patients are much stronger than that from the healthy persons. In addition, the scattered dot plots in the statistical analysis shown in Fig. 5B can further reflect the significant difference between the healthy population and the patient population, which is consistent with the results of clinical standard PCR test (Table S3), demonstrating the applicability of our method in clinical diagnosis.
Fig. 5.
Clinical sample analysis of the SARS-COV-2 RNA. (A) DPV responses and (B) scattered dot plots of SARS-COV-2 RNA from healthy samples (HS) and patient samples (PS). The statistical significances are calculated by t-test (**, p < 0.01). Error bars: SD, n = 3.
4. Conclusion
In conclusion, we have proposed an electrochemical biosensor for sensitive monitoring of the SARS-CoV-2 RNA. By coupling the signal amplification capabilities of CHA and TdT-induced polymerization, a significantly amplified current signal can be obtained to achieve a detection limit as low as 26 fM. Meanwhile, the successful discrimination of target RNA from single-base mismatched RNA, two-base mismatched RNA, as well as random sequence has verified the strong anti-interference ability and accuracy of this biosensor. Moreover, the proposed approach can be applied to detect the SARS-CoV-2 RNA in serum, saliva, and throat test paper of infected patients with excellent stability, showing great clinical application potential. Compared with the previously reported methods, the proposed electrochemical approach avoids sophisticated instrumental techniques, tedious experimental steps, and complicated probe modification, greatly simplifying the detection procedure and saving experimental costs (Table S4). Therefore, this proposed electrochemical biosensor may provide a simple, low-cost and easy-to-operate analysis platform to facilitate more efficient and thorough diagnoses of COVID-19 in the near future.
CRediT authorship contribution statement
Ying Peng: Conceptualization, Methodology, Software, Writing – review & editing. Yanhong Pan: Investigation, Data Collection and Curation. Zhaowei Sun: Investigation. Jinlong Li: Data Collection and Curation. Yongxiang Yi: Data curation, Validation. Jie Yang: Data curation. Genxi Li: Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by the National Natural Science Foundation of China (Grant No. 81772593), and the Fundamental Research Funds for the Central Universities (Grant No. 14380131).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113309.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ali M.A., Hu C., Jahan S., Yuan B., Saleh M.S., Ju E., Gao S.J., Panat R. Adv. Mater. 2021;33 doi: 10.1002/adma.202006647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek Y.H., Um J., Antigua K.J.C., Park J.H., Kim Y., Oh S., Kim Y.I., Choi W.S., Kim S.G., Jeong J.H., Chin B.S., Nicolas H.D.G., Ahn J.Y., Shin K.S., Choi Y.K., Park J.S., Song M.S. Emerg. Microb. Infect. 2020;9:998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A.P., Vongpunsawad S., Poovorawan Y., Lee S.Y., Lertanantawong B. Nat. Commun. 2021;12:802. doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.Y., Liu Z.J., Wang X.W., Ye C.L., Zheng Y.J., Peng H.P., Zhong G.X., Liu A.L., Chen W., Lin X.H. Anal. Chem. 2019;91:4552–4558. doi: 10.1021/acs.analchem.8b05692. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Euro Surveill. 2020;25:2000045. [Google Scholar]
- Dai W., Zhang J., Meng X., He J., Zhang K., Cao Y., Wang D., Dong H., Zhang X. Theranostics. 2018;8:2646–2656. doi: 10.7150/thno.24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji T., Liu Z., Wang G., Guo X., Akbar Khan S., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q. Biosens. Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J., Duan C., Xue L., Liu Y., Sun W., Xiang Y. Biosens. Bioelectron. 2020;167:112479. doi: 10.1016/j.bios.2020.112479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Liu Y.H., Wang C.Y., Wang Y.H., Hsueh S.C., Yen M.Y., Ko W.C., Hsueh P.R. J. Microbiol. Immunol. Infect. 2020;53:404–412. doi: 10.1016/j.jmii.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Degani I., Cheong J., Lee J., Choi J., Cheon J., Lee H. Biosens. Bioelectron. 2021;178:113049. doi: 10.1016/j.bios.2021.113049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Ellington A.D., Chen X. Nucleic Acids Res. 2011;39:e110. doi: 10.1093/nar/gkr504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Huang Y., Yu Y., Wang Y., Li G. Biosens. Bioelectron. 2016;80:560–565. doi: 10.1016/j.bios.2016.02.012. [DOI] [PubMed] [Google Scholar]
- Li P., Wei M., Zhang F., Su J., Wei W., Zhang Y., Liu S. ACS Appl. Mater. Interfaces. 2018;10:43405–43410. doi: 10.1021/acsami.8b15330. [DOI] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhang Y., Xie H., Zhao L., Zheng L., Ye H. Small. 2019;15 doi: 10.1002/smll.201902989. [DOI] [PubMed] [Google Scholar]
- Liu Y., Wei M., Li Y., Liu A., Wei W., Zhang Y., Liu S. Anal. Chem. 2017;89:3430–3436. doi: 10.1021/acs.analchem.6b04176. [DOI] [PubMed] [Google Scholar]
- Liu Y., Xiong E., Li X., Li J., Zhang X., Chen J. Biosens. Bioelectron. 2017;87:970–975. doi: 10.1016/j.bios.2016.09.069. [DOI] [PubMed] [Google Scholar]
- Mahshid S.S., Flynn S.E., Mahshid S. Biosens. Bioelectron. 2021;176:112905. doi: 10.1016/j.bios.2020.112905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson J.F. ACS Sens. 2017;2:16–30. doi: 10.1021/acssensors.6b00763. [DOI] [PubMed] [Google Scholar]
- Michelson A.M., Orkin S.H.J. Biol. Chem. 1982;257:14773–14782. [PubMed] [Google Scholar]
- Oran D.P., Topol E.J. Ann. Intern. Med. 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B., Xu J., Liu Y., Peng H., Feng W., Cao Y., Wu J., Xiao H., Pabbaraju K., Tipples G., Joyce M.A., Saffran H.A., Tyrrell D.L., Zhang H., Le X.C. Anal. Chem. 2020;92:16204–16212. doi: 10.1021/acs.analchem.0c04047. [DOI] [PubMed] [Google Scholar]
- Peng Y., Li D., Yuan R., Xiang Y. Biosens. Bioelectron. 2018;105:1–5. doi: 10.1016/j.bios.2018.01.017. [DOI] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Soler M., Huertas C.S., Lechuga L.M. Expert Rev. Mol. Diagn. 2019;19:71–81. doi: 10.1080/14737159.2019.1554435. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang Y., Chen J., Wang M., Zhang T., Luo W., Li Y., Wu Y., Zeng B., Zhang K., Deng R., Li W. Anal. Chem. 2021;93:3393–3402. doi: 10.1021/acs.analchem.0c04303. [DOI] [PubMed] [Google Scholar]
- Xu E., Feng Y., Yang H., Li P., Kong L., Wei W., Liu S. Sens. Actuators, B. 2021;337:129745–129751. [Google Scholar]
- Yuan P.X., Deng S.Y., Zheng C.Y., Cosnier S., Shan D. Biosens. Bioelectron. 2017;97:1–7. doi: 10.1016/j.bios.2017.05.033. [DOI] [PubMed] [Google Scholar]
- Zhao H., Liu F., Xie W., Zhou T.C., OuYang J., Jin L., Li H., Zhao C.Y., Zhang L., Wei J., Zhang Y.P., Li C.P. Sens. Actuators, B. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Shi H., Wang M., Duan C., Huang Y., Li C., Xiang Y., Li G. Anal. Chem. 2020;92:2194–2200. doi: 10.1021/acs.analchem.9b04877. [DOI] [PubMed] [Google Scholar]
- Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S., Xing M., Chen H., Wang Y. Biosens. Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.L., Peiris M., Wu J. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.