Abstract
Purpose
CircRNA circ_POLA2 has been reported as an oncogene in lung cancer, while its role in other malignancies is unknown. This study aimed to explore the role of circ_POLA2 in acute myeloid leukemia (AML).
Methods
The expression levels of circ_POLA2, mature miR-34a and miR-34a precursor in bone marrow mononuclear cells (BMMNCs) from AML patients (n = 50) and healthy controls (n = 50) were determined by RT-qPCR. Correlations among them were analyzed by Pearson’s correlation coefficient. Overexpression of circ_POLA2 was achieved in AML cell lines, followed by the measurement of the expression levels of mature miR-34a and miR-34a precursor. The role of circ_POLA2 and miR-34a in regulating AML cell proliferation was assessed by CCK-8 assay.
Results
Circ_POLA2 was upregulated in AML and inversely correlated with mature miR-34a, but not miR-34a precursor. In AML cells, overexpression of circ_POLA2 decreased the expression levels of mature miR-34a, but not miR-34a precursor. Cell proliferation analysis showed that the overexpression of miR-34a attenuated the effects of overexpression of circ_POLA2 on cell proliferation.
Conclusion
Circ_POLA2 is upregulated in AML and promotes cell proliferation by suppressing the production of mature miR-34a.
Keywords: circ_POLA2, acute myeloid leukemia, miR-34a, maturation
Introduction
Acute myeloid leukemia (AML), also known as acute non-lymphocytic leukemia, is a type of malignancy that originates from white blood cells in the bone marrow.1 AML is rare and only accounts for less than 1% of all cancer cases.2 However, due to its highly aggressive and rapidly growing nature, AML is a common cause of cancer deaths in clinical practice.3,4 It is estimated that only less than 24% of AML patients can survive more than 5 years.3,4 AML mainly affects adults, especially people older than 60 years old. Although there are standard treatments for elderly AML patients, such as chemotherapy and stem cell transplant, the treatment outcomes are generally poor and side effects are common.5,6
Novel anti-AML therapeutic approaches, such as molecular targeted therapies, are still under research.7–9 Specifically, safe and effective targets for targeted AML therapy remain lacking, mainly due to the unclear molecular mechanism of the initiation, development and progression of AML.10 Circular RNAs (circRNAs, covalently closed) have identified as emerging novel critical players in cancer biology.11,12 CircRNAs participate in cancer biology mainly by regulating the expression of related genes.11,12 Therefore, circRNAs may serve as potential targets for molecular targeted therapy.13 Unfortunately, the functions of most circRNAs in cancer biology are unknown. CircRNA circ_POLA2 has been identified as an oncogene in lung cancer,14 while its role in other malignancies is unknown. Our preliminary microarray analysis observed an altered expression of circ_POLA2 and its close correlation with miR-34a, which plays a tumor suppressive role in AML.15,16 Therefore, this study was carried out to investigate the interaction between circ_POLA2 and miR-34a in AML.
Materials and Methods
AML Patients and Healthy Controls
This study was carried out in accordance with the Declaration of Helsinki. This study was approved by the Ethics Committee of Shandong Qianfoshan Hospital. Both AML patients (n = 50) and healthy controls (n = 50) who were admitted to the aforementioned hospital from May 2018 to May 2020 were enrolled in this study. The AML group included 30 males and 20 females, with a mean age of 61.2 ± 5.3 years old. The control group also included 30 males and 20 females, with a mean age of 61.3 ± 5.1 years old. These two groups had similar distributions of gender and age. Patients with other severe diseases or initiated therapies within 3 months prior to admission were excluded. The healthy controls showed normal physiological functions in systemic physiological exams carried out at the aforementioned hospital. Informed consent was provided by all participants. Bone marrow (BM) biopsy was performed on both AML patients and healthy controls to collect BM samples. BM mononuclear cells (BMMNCs) were isolated from all BM samples using lymphocyte separation medium (TBD Sciences). Samples of BMMNCs were kept in liquid nitrogen prior to the subsequent experiments.
AML Cell Lines and Transient Transfections
Human bone marrow stromal HS-5 cells and two human AML cell lines Kasumi-6 and Kasumi-3 purchased from ATCC (USA) were used as the cell model of AML. Cells were cultivated in an incubator at 37°C with 95% humidity and 5% CO2. Cell culture medium was composed of 10% FBS and 90% RPMI-1640 medium. Cells were cultivated up to about 80% confluence prior to the subsequent transient transfections.
To overexpress miR-34a and circ_POLA2, mimic of miR-34a and miRNA negative control (NC) were purchased from Sigma-Aldrich, and circ_POLA2 expression vector was established using pcDNA3.1(+) CircRNA Mini Vector (Addgene) as backbone. Lipofectamine 2000 (Invitrogen) was used to transfect cells with either 1 μg circ_POLA2 expression vector or 40 nM miR-34a mimic. NC experiments (empty vector- or miRNA NC-transfected cells) and C (untransfected cells) were included in each experiment. Prior to the subsequent experiments, fresh medium was used to cultivate cells for another 48 h.
RNA Isolation and Genomic DNA Removal
Ribozol reagent (VWR) was used to isolate total RNAs from both BMMNCs and two AML cell lines. RNA samples were incubated with DNase I (Invitrogen) at 37°C for 75 min to achieve removal of genomic DNAs. RNA integrity was checked with electrophoresis using 5% urea-PAGE gel. RNA purity was determined by checking the OD 260/280 ratio.
RT-qPCR Assay
The synthesis of cDNA samples was performed using RNA samples with satisfactory quality (good integrity and OD260/280 ratios close to 2.0) as template through reverse transcriptions (RTs). RTs were completed using SS-IV-RT system (Invitrogen). Primers were from the kit. To determine the expression of circ_POLA2 and PD-L1, SYBR Green Master Mix (Bio-Rad) was used to perform qPCRs with 18S rRNA as the internal control.
The expression of mature miR-34a and miR-34a precursor were determined using the All-in-One™ miRNA qRT-PCR Detection Kit (Genecopoeia). To analyze the expression of mature miR-34a, mature miR-34a was added with poly (A), followed by using poly (T) as reverse primer to perform both RTs and qPCRs. To analyze the expression of miR-34a precursor, sequence-specific primers were used to perform RTs and qPCRs. U6 was used as the internal control of miR-34a.
Primer sequences were as follows: 5ʹ-TGAGCTTGTGAGTGAGTGGT-3ʹ (forward) and 5ʹ-GCAAGGAGAATGGCGAGATG-3ʹ (reverse) for circ_POLA2; 5ʹ-CTACCACATCCAAGGAAGCA-3ʹ (forward) and 5ʹ-TTTTTCGTCACTACCTCCCCG-3ʹ (reverse) for human 18S rRNA; 5ʹ-CACGGTTCCCAAGGACCTAT-3ʹ (forward) and 5ʹ-TGGAGGATGTGCCAGAGGTA-3ʹ (reverse) for PD-L1; 5ʹ-GGCCAGCTGTGAGTGTTTC-3ʹ (forward) and 5ʹ-CCACAACGTCAGCACTTCT-3ʹ (reverse) for miR-34a precursor. 5ʹ-CTCGCTTCGGCAGCACA-3ʹ (forward) and 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ (reverse) for U6; Forward primer for mature miR-34a was 5ʹ-TGGCAGTGTCTTAGCTGG-3ʹ. Reverse primer of miR-34a and U6 primers were from the kit. It is worth noting that multiple primer pairs were tried and only the ones that reached about 100% amplification efficiency were used. All qPCRs were performed on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). The 2−ΔΔCT method was used to normalize Ct values of target genes to internal controls.
Cell Counting Kit-8 (CCK-8) Kit
Both cell lines were collected at 48 h post-transfection and the proliferation of both cell lines was analyzed by cell proliferation assay using a CCK-8 kit (Dojindo). Briefly, 3000 cells in 0.1 mL medium were added into each well of a 96-well plate, and cells were cultivated at 37°C. To monitor the cell proliferation, OD values at 450 nm were measured every 24 h until 96 h. Prior to the measurement of the OD values, 10 μL CCK-8 solution was added into each well.
Western Blot
RIPA solution was used to isolate total proteins. BCA assay (Sigma-Aldrich) was used to quantify protein samples. After denaturation, proteins were separated using SDS-PAGE gels (8%). Gel transfer to PVDF membranes was performed and blocking was carried out in 5% non-fat milk. The primary antibodies were GAPDH (1:1000, Abcam) and PD-L1 (1:1000, Abcam), CDK4 (1:1000; Cell Signaling Technology), CDK6 (1:1000; Cell Signaling Technology), and β-tubulin (1:2000; Wanleibio, Beijing, China). The secondary antibody was HRP (IgG) goat antibody (1:1000; ab6721; Abcam). ECL (Sigma-Aldrich) was used to develop signals. Quantity One software was used to quantify data.
5-Bromo-2-deoxyUridine (BrdU) Assay
The BrdU assay was used to investigate cell proliferation as described in previous studies.17 Each group of AML cells was seeded into 24-well plates at a density of 2×104 cells per well, followed by incubation with 10 mM BrdU (BD Pharmingen) for 2 h. After that, cells were fixed in 70% ethanol for 20 min, followed by incubation with a peroxidase-coupled anti-BrdU-antibody (Sigma-Aldrich) for 1 h. Peroxidase substrate (tetramethylbenzidine) was then used to incubate with the cells for 30 min. Finally, OD values were measured at 450 nm to reflect cell proliferation.
Cell Cycle Analysis
For cell cycle analysis, cells were taken from the test tube and washed twice with 4 mL PBS. Pre-cooled 75% ethanol was added and incubate overnight at 4°C. The cells were washed twice to remove all the ethanol. Each test tube sample was stained with 1×106 cells. For PI/RNase staining, the cells were resuspended in 0.5 mL PI/RNase staining solution (BD Biosciences) and incubated for 15 min at 25°C in the dark. Analysis of flow data was performed using FlowJo version X.10.0.7–1 (FlowJo LLC).18,19
Statistical Analyses
ANOVA Tukey’s test was used to compare multiple independent cell transfection groups. Unpaired t test was used to compare independent control and AML groups. The overall survival rate estimates over time were calculated using the Kaplan–Meier method with Log rank test. Correlations were analyzed by Pearson’s correlation coefficient. P < 0.05 was statistically significant.
Results
AML Group Exhibited Altered Expression of circ_POLA2, Mature miR-34a and miR-34a Precursor
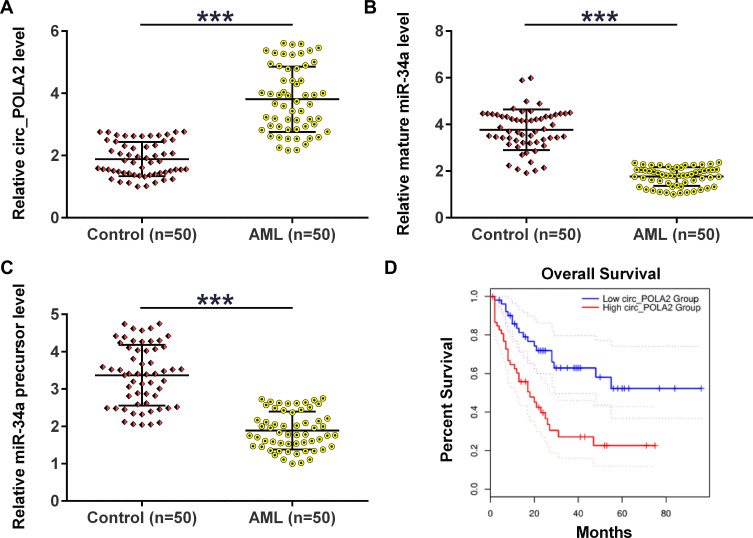
The expression levels of circ_POLA2, mature miR-34a and miR-34a precursor in BM samples collected from AML patients (n = 50) and healthy controls (n = 50) were analyzed using RT-qPCRs. The results showed that circ_POLA2 was significantly upregulated (Figure 1A, p < 0.01) in AML group, while mature miR-34a (Figure 1B, p < 0.001) and miR-34a precursor (Figure 1C, p < 0.001) were significantly downregulated in AML group compared to that in the control group. The relationship between circ_POLA2 levels and survival time in AML patients was then evaluated. The Kaplan–Meier survival curve showed that the patients with higher expression levels of circ_POLA2 had lower overall survival rate (Figure 1D).
Figure 1.
AML group exhibited altered expression of circ_POLA2, mature miR-34a and miR-34a precursor. BM samples were collected from AML patients (n = 50) and healthy controls (n = 50), followed by RNA preparations and RT-qPCRs to analyze the expression of circ_POLA2 (A), mature miR-34a (B) and miR-34a precursor (C). Unpaired t test was used to compare two groups. The Kaplan-Meier survival curve showed the survival rate in patients with high expression levels of circ_POLA2 and those with low expression levels of circ_POLA2 (D). ***p < 0.001.
Circ_POLA2 Was Only Inversely Correlated with Mature miR-34a
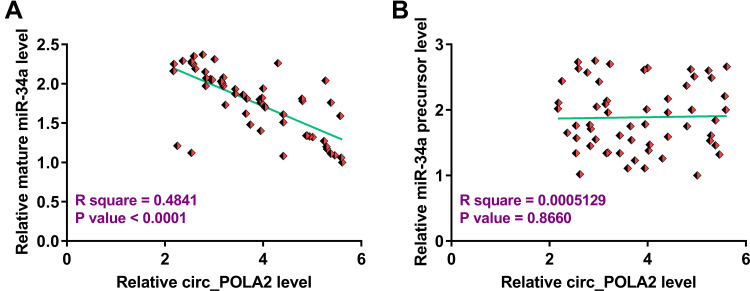
To explore the relationship between circ_POLA2 and mature miR-34a or miR-34a precursor, Pearson’s correlation coefficient was used and it was observed that circ_POLA2 was significantly and inversely correlated with mature miR-34a (Figure 2A), but not miR-34a precursor (Figure 2B). Therefore, circ_POLA2 may be involved in the maturation of miR-34a from precursor to mature miRNA.
Figure 2.
Circ_POLA2 was only inversely correlated with mature miR-34a. To analyze the relationship between circ_POLA2 and mature miR-34a (A) or miR-34a precursor (B), Pearson’s correlation coefficient was used to analyze the correlations among them.
Overexpression of circ_POLA2 Suppressed the Maturation of miR-34a in AML Cells
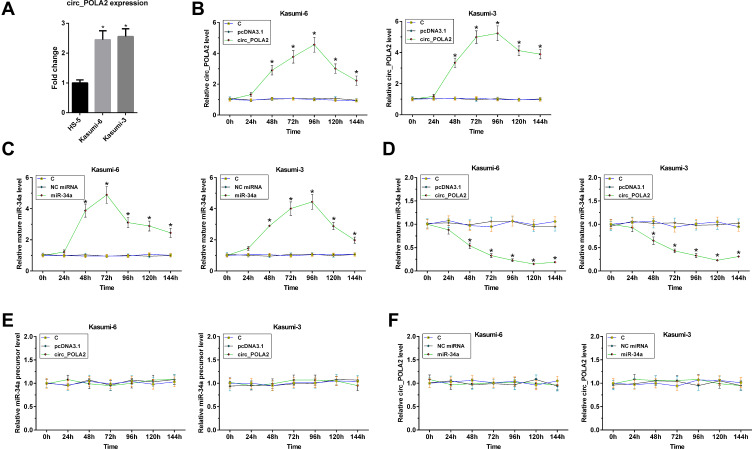
To investigate the biological relevance of circ_POLA2 in AML, we analyzed baseline expression of circ_POLA2 in two human AML cell lines (Kasumi-6 and Kasumi-3) and one human bone marrow stromal HS-5 cell line. The results showed that the expression levels of circ_POLA2 were higher in Kasumi-6 and Kasumi-3 cells than that in HS-5 cells (Figure 3A, p < 0.05). To explore the role of circ_POLA2 in the maturation of miR-34a, Kasumi-6 and Kasumi-3 cells were transfected with either circ_POLA2 expression vector or miR-34a mimic. The expression of circ_POLA2 and mature miR-34a were determined every 24 h until 144 h. It was observed that circ_POLA2 and miR-34a were significantly upregulated from 48 to 144 h (Figure 3B and C, p < 0.05). In addition, the overexpression of circ_POLA2 significantly decreased the expression levels of mature miR-34a between 48 h and 144 h (Figure 3D, p < 0.05), but did not affect the expression of miR-34a precursor at each time point (Figure 3E). Moreover, the transfection of miR-34a mimic did not affect the expression of circ_POLA2 at each time point (Figure 3F). Therefore, circ_POLA2 may suppress the maturation of miR-34a in AML cells.
Figure 3.
Overexpression of circ_POLA2 suppress the maturation of miR-34a in AML cells. The expression of circ_POLA2 was detected in human bone marrow stromal HS-5 cells and two human AML cell lines (A). To explore the role of circ_POLA2 in the maturation of miR-34a, Kasumi-6 and Kasumi-3 cells were transfected with either circ_POLA2 expression vector or miR-34a mimic. Expression of circ_POLA2 and mature miR-34a expression was determined every 24 h until 144 h (B and C). The effects of circ_POLA2 expression vector transfection on the expression of mature miR-34a (D) and the expression of miR-34a precursor at each time point (E), as well as the effects of miR-34a mimic transfection on the expression of circ_POLA2 (F) were analyzed by RT-qPCR. *p < 0.05.
Overexpression of circ_POLA2 Increased the Proliferation of AML Cells Through miR-34a
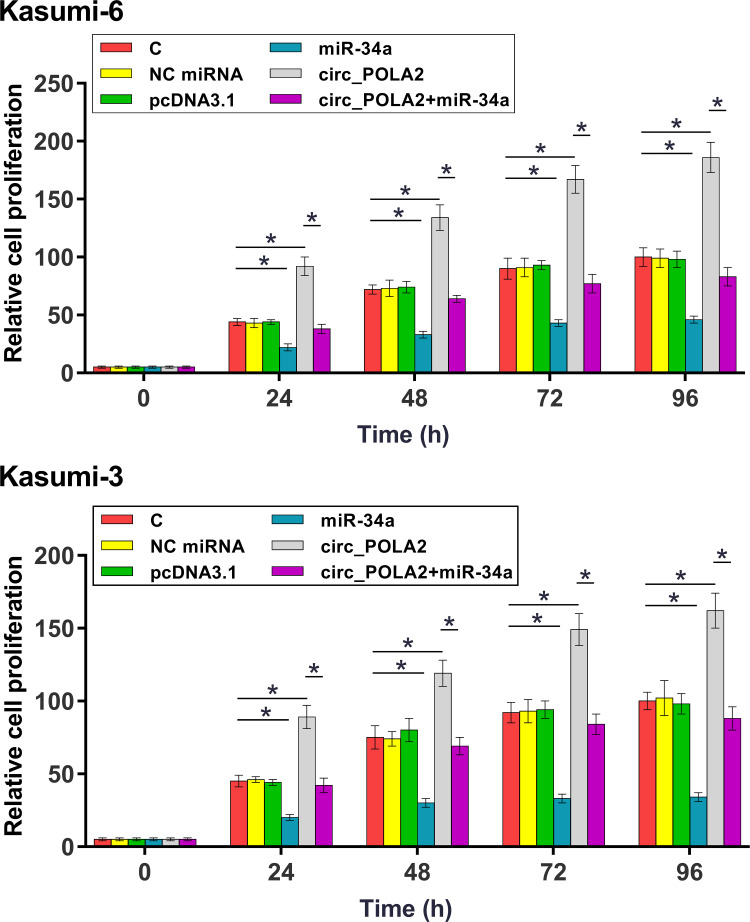
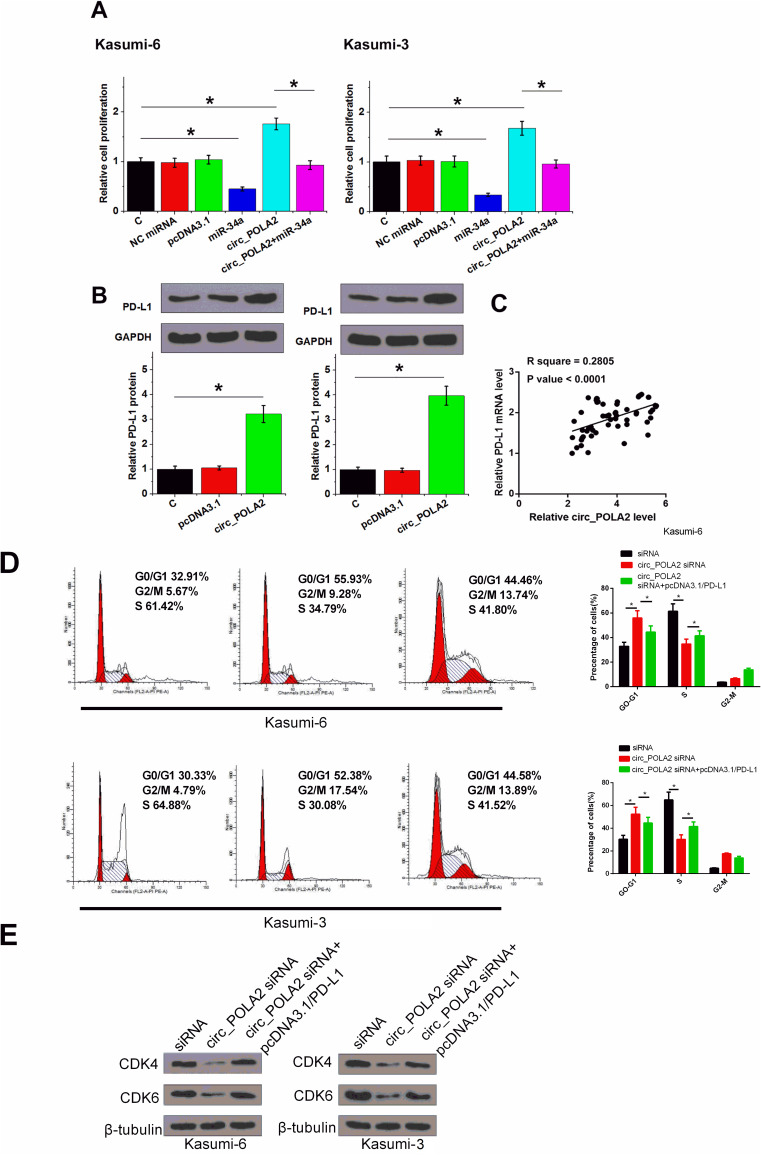
The role of circ_POLA2 and miR-34a in the proliferation of Kasumi-6 and Kasumi-3 cells were analyzed by CCK-8 assay. It was observed that overexpression of circ_POLA2 increased cell proliferation, and the overexpression of miR-34a reduced cell proliferation. In addition, the overexpression of miR-34a attenuated the effects of the overexpression of circ_POLA2 on cell proliferation (Figure 4, p < 0.05). BrdU assay was performed to further confirm the role of circ_POLA2 and miR-34a. As shown in Figure 5A, consistent results were obtained (p < 0.05).
Figure 4.
Overexpression of circ_POLA2 increased the proliferation of AML cells through miR-34a. The role of circ_POLA2 and miR-34a in the proliferation of Kasumi-6 and Kasumi-3 cells were analyzed by CCK-8 assay. OD values at 450 nm were measured every 24 h until 96 h to monitor cell proliferation. *p < 0.05.
Figure 5.
Overexpression of PD-L1 rescued the effect of knockdown of circ_POLA2. BrdU assay was performed to further confirm the role of circ_POLA2 and miR-34a (A). The role of circ_POLA2 in regulating the expression of PD-L1 was analyzed by Western-blot (B). The expression of PD-L1 mRNA in BM samples were also analyzed RT-qPCR. Pearson’s correlation coefficient was performed to analyze the correlation between circ_POLA2 and PD-L1 mRNA across BM tissues from AML patients (C). Cell cycle was measured by flow cytometric analysis (D). CDK4, CDK6 protein levels were measured by Western blot in Kasumi-6 and Kasumi-3 cells (E). *p < 0.05.
Overexpression of PD-L1 Rescued the Effect of Knockdown of circ_POLA2
PD-L1 is a well-established target of miR-34a.16 It was observed that the overexpression of circ_POLA2 significantly increased the expression levels of PD-L1 in both cell lines (Figure 5B, p < 0.05). Pearson’s correlation coefficient analysis showed that circ_POLA2 and PD-L1 mRNA were closely and significantly correlated across BM tissues from AML patients (Figure 5C, p < 0.05). The knockdown of circ_POLA2 caused G1/G0 cell cycle arrest, while the overexpression of PD-L1 significantly reduced the number of cells in G1/G0 (Figure 5D, p < 0.05). Furthermore, the knockdown of circ_POLA2 caused the downregulation of CDK4, CDK6, which were counteracted following the overexpression of PD-L1 (Figure 5E, p < 0.05).
Discussion
The involvement of circ_POLA2 in AML was explored in this study. We found that circ_POLA2 was significantly upregulated in AML. In addition, circ_POLA2 may suppress the maturation of miR-34a to promote AML cell proliferation.
In a recent study, circ_POLA2 was reported to be significantly upregulated in lung cancer and positively regulated the stemness of lung cancer cells.14 Moreover, mechanism studies showed that circ_POLA2 may upregulate G protein subunit beta 1 by serving as an endogenous competing RNA for miR-326.14 This study is the first to explore the involvement of circ_POLA2 in AML. We found that circ_POLA2 was upregulated in AML. Moreover, significantly increased AML cell proliferation was observed after the overexpression of circ_POLA2. Therefore, circ_POLA2 might play an oncogenic role in AML by promoting cell proliferation. However, the role of circ_POLA2 in tumor growth remains to be further explored using animal model experiments.
MiR-34a plays tumor suppressive roles in many types of cancer, including AML.15,16 It is reported that miR-34a is downregulated in AML and it can target HMGB1 to promote cancer cell apoptosis and inhibit autophagy in AML.15 In another study miR-34a was reported to target PD-L1, thereby increasing the sensitivity of AML cells to chemotherapy.16 Consistently, our study confirmed the downregulation of miR-34a in AML. In addition, besides cell apoptosis, our study showed that miR-34a may also suppress the proliferation of AML cells.
Although it is known that miR-34a suppresses multiple types of cancer by targeting different oncogenes, the upstream regulator of miR-34a in cancer remains unclear. In this study, we showed that circ_POLA2 could suppress the formation of mature miR-34a, but not miR-34a precursor in AML cells. It is known that circRNAs can interact with miRNAs by serving as miRNA endogenous competing RNAs.20 Therefore, our study enriched our knowledge of the interaction between circRNAs and miRNAs. We speculated that circ_POLA2 may affect the transportation of miR-34a precursor from the nucleus to the cytoplasm, which is required for the maturation of miR-34a.
In conclusion, circ_POLA2 is upregulated in AML. In addition, circ_POLA2 may suppress the maturation of miR-34a to increase cell proliferation.
Acknowledgments
The authors would like to express our gratitude for those who have critically reviewed this manuscript and those who gave us help during these experiments. This research was supported by no funders.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–1152. doi: 10.1056/NEJMra1406184 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 3.Juliusson G, Antunovic P, Derolf Å, et al. Age and acute myeloid leukemia: real world data on decision to treat and outcomes from the Swedish Acute Leukemia Registry. Blood. 2009;113(18):4179–4187. doi: 10.1182/blood-2008-07-172007 [DOI] [PubMed] [Google Scholar]
- 4.Li G, Zhou Z, Yang W, et al. Long-term cardiac-specific mortality among 44,292 acute myeloid leukemia patients treated with chemotherapy: a population-based analysis. J Cancer. 2019;10(24):6161–6169. doi: 10.7150/jca.36948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crossnohere NL, Richardson DR, Reinhart C, et al. Side effects from acute myeloid leukemia treatment: results from a national survey. Curr Med Res Opin. 2019;35(11):1965–1970. doi: 10.1080/03007995.2019.1631149 [DOI] [PubMed] [Google Scholar]
- 6.Short NJ, Konopleva M, Kadia TM, et al. Advances in the treatment of acute myeloid leukemia: new drugs and new challenges. Cancer Discov. 2020;10(4):506–525. doi: 10.1158/2159-8290.CD-19-1011 [DOI] [PubMed] [Google Scholar]
- 7.Walasek A. The new perspectives of targeted therapy in acute myeloid leukemia. Adv Clin Exp Med. 2019;28(2):271–276. doi: 10.17219/acem/81610 [DOI] [PubMed] [Google Scholar]
- 8.Yu J, Jiang PYZ, Sun H, et al. Advances in targeted therapy for acute myeloid leukemia. Biomark Res. 2020;8:17. doi: 10.1186/s40364-020-00196-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Dijk AD, de Bont ESJM, Kornblau SM. Targeted therapy in acute myeloid leukemia: current status and new insights from a proteomic perspective. Expert Rev Proteomics. 2020;17(1):1–10. doi: 10.1080/14789450.2020.1717951 [DOI] [PubMed] [Google Scholar]
- 10.Martens JHA, Stunnenberg HG. The molecular signature of oncofusion proteins in acute myeloid leukemia. FEBS Lett. 2010;584(12):2662–2669. doi: 10.1016/j.febslet.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 11.Shang Q, Yang Z, Jia R, et al. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. doi: 10.1186/s12943-018-0934-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. doi: 10.1016/j.canlet.2018.03.035 [DOI] [PubMed] [Google Scholar]
- 13.Lei B, Tian Z, Fan W, et al. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301. doi: 10.7150/ijms.28047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan Z, Bai Y, Zhang Q, et al. CircRNA circ_POLA2 promotes lung cancer cell stemness via regulating the miR-326/GNB1 axis. Environ Toxicol. 2020;35(10):16. doi: 10.1002/tox.22980 [DOI] [PubMed] [Google Scholar]
- 15.Liu L, Ren W, Chen K. MiR-34a promotes apoptosis and inhibits autophagy by targeting HMGB1 in acute myeloid leukemia cells. Cell Physiol Biochem. 2017;41(5):1981–1992. doi: 10.1159/000475277 [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Li J, Dong K, et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015;27(3):443–452. doi: 10.1016/j.cellsig.2014.12.003 [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Zhang C, Zhang X. Downregulation of long noncoding RNA ANRIL promotes proliferation and migration in hypoxic human pulmonary artery smooth muscle cells. Mol Med Rep. 2020;21(2):589–596. doi: 10.3892/mmr.2019.10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, Yuan Y, Ma R, Xu B, Zhang R. lncRNA SNHG7 affects malignant tumor behaviors through downregulation of EZH2 in uveal melanoma cell lines. Oncol Lett. 2020;19(2):1505–1515. doi: 10.3892/ol.2019.11240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao H, Hou G, Wang Q, Xu W, Zhao H, Xu Y. LncRNA SPRY4IT1 promotes progression of osteosarcoma by regulating ZEB1 and ZEB2 expression through sponging of miR101 activity. Int J Oncol. 2020;56(1):85–100. doi: 10.3892/ijo.2019.4910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271–73281. doi: 10.18632/oncotarget.19154 [DOI] [PMC free article] [PubMed] [Google Scholar]