Abstract
Background
Non‐small cell lung carcinoma (NSCLC) is a malignancy with the highest mortality rate. Currently, surgery combined with radiotherapy is the first choice in the clinical treatment of lung carcinoma (LC); however, long‐term radiotherapy leads to radiation resistance in patients, resulting in treatment failure.
Methods
In this study, a new microRNA‐218‐5p (miRNA‐218‐5p) was identified, and its function in LC was investigated.
Results
Reverse transcription quantitative polymerase chain reaction (RT‐qPCR) results revealed that miRNA‐218‐5p was downregulated in LC. Overexpression or inhibition of miRNA‐218‐5p in LC and targeted binding of protein kinase, DNA‐activated, catalytic polypeptide (PRKDC) to miRNA‐218‐5p were confirmed by comprehensive bioinformatic analysis. Exosomes from A549 and H1299 cells were cocultured with miRNA‐218‐5p and then cotransfected into radiation‐resistant A549R and H1299R cells; the proliferation of radiation‐resistant LC cells was found to be effectively inhibited and apoptosis was induced. Overexpression of miRNA‐218‐5p and X‐irradiation could enhance the radiosensitivity of LC cells. Exogenous miRNA‐218‐5p derived from A549 and H1299 cells could be transfected into radiation‐resistant LC cells and could inhibit PRKDC expression, thus accelerating DNA damage, apoptosis, and radiation sensitization of LC cells.
Conclusions
miRNA‐218‐5p could induce apoptosis and enhance the radiosensitivity of LC cells through regulatory activities, thus suggesting its application as a potential target for LC treatment.
Keywords: exosomes, lung carcinoma, miRNA‐218‐5p, PRKDC, radiation resistance
This study explored the role of microRNA‐218‐5p in the radiosensitivity of lung carcinoma cells. microRNA‐218‐5p induced apoptosis and enhanced the radiosensitivity of lung carcinoma cells through regulation, thus suggesting its application as a potential target for the treatment of lung carcinoma.
INTRODUCTION
Lung carcinoma (LC) has become an increasingly serious public health concern worldwide. 1 Statistics indicate that in 2018 there were more than 2 million new LC patients globally 2 and 1.7 million deaths. LC can be grouped into small cell LC (SCLC) and non‐small cell lung carcinoma (NSCLC); NSCLC accounts for over 80% of LCs. Radiotherapy is an important treatment option for malignant thoracic tumors. 3 , 4 , 5 At present, radiotherapy combined with surgery is known to exert potent synergistic effects, 6 , 7 which can induce cell death by breaking the DNA of tumor cells. However, carcinoma treatment is a lengthy process. Patients often show adverse reactions during long‐term radiotherapy, and an increasing number of studies have revealed that NSCLC can acquire resistance to radiotherapy, reducing its efficacy, and tumors can recur and metastasize. 8 In addition, for epidermal growth factor receptor‐mutant lung adenocarcinoma, the radiotherapeutic effects of tyrosine kinase inhibitors are also not ideal. 9 Therefore, exploring radiotherapy tolerance is crucial.
microRNAs (miRNAs), which have emerged as a subject of extensive research, are short‐chain non‐coding RNAs comprising 19–25 nucleotide bases. Recent studies have confirmed that miRNAs can target downstream genes through mRNA 3′‐untranslated regions (UTRs) and participate in tumorigenesis. 10 , 11 , 12 The key role of miRNAs in the pathogenesis and response to treatment has been confirmed in different carcinomas. 13 For example, Meng et al. 14 reported that miR‐183‐5p plays a tumor‐inhibitory role in LC by inhibiting PIK3CA. The levels of miRNA‐218‐5p, an miRNA that was discovered early on, were found to be low in LC 15 ; studies have revealed that exosomes containing miRNA‐218‐5p play a role in osteoblast differentiation as well as oxaliplatin resistance in colon carcinoma. 16 , 17 However, there are few studies on the role of miRNA‐218‐5p in tumor tolerance to radiotherapy in LC.
Cells can communicate directly via cell‐to‐cell contacts and synapses through chemical receptors. 18 Recent studies have shown that exosomes also act in cell‐to‐cell communication. 19 Exosomes can carry lipids, proteins, mRNA, and various miRNAs. 20 It has been reported that exosomes engage recipient cells by fusion with target cell membranes, suggesting that exosomes may act in transporting radiation‐induced serum miRNAs and thus affect the proliferation and radio resistance of cells. 21
In this study, we found that miRNA‐218‐5p reduced the tolerance of LC cells to radiotherapy, thus suggesting a potential therapeutic strategy for tumor tolerance to radiotherapy in LC.
METHODS
Clinical data
A total of 42 patients with NSCLC who received radiotherapy between January 2013 and January 2015 were selected as participants. The patient received 60 Gy/30 min of radiotherapy. Peripheral venous blood was obtained 24 h before treatment, and peripheral venous blood was collected again after the patient completed the course of treatment. In addition, 30 healthy individuals during the same period were included in a control group. Peripheral blood samples were collected from both groups, held at room temperature for 60 min, and centrifuged at 3000 × g for 10 min. The supernatant was then obtained and stored at −80°C. The patients had not received chemotherapy before this study but were diagnosed with NSCLC. All subjects agreed to participate in the study and signed consent forms voluntarily. This research was approved by the Shanghai Chest Hospital, Shanghai Jiao Tong University and was carried out in compliance with the Declaration of Helsinki. 22
Cell culture and irradiation
HEK‐293 T, A549, H1299, HCC827, and BEAS‐2B cell lines were procured from the American Type Culture Collection and cultured in Dulbecco's modified Eagle's medium. A549 and H1299 cell lines were irradiated for radiation‐resistance analysis. When cell confluence reached 50%, A549 and H1299 cells were subjected to X‐ray irradiation (2 Gy), cultured, digested with trypsin at a 90% convergence rate, and then reirradiated at a 50% convergence rate, with a 2 Gy daily fraction size being administered 30 times for a total dose of 60 Gy. Parental cells were cultured without radiation under the same conditions. In addition, there was an interval of four or five months between the last two partial exposures. Cells were replated for colony formation after recovering for 2–3 weeks after the last irradiation. After 2–3 weeks, the progenies of the clonogenic survivors were obtained.
Cell transfection
miRNA‐218‐5p mimic, miRNA‐218‐5p inhibitor, inhibitor‐NC, and protein kinase, DNA‐activated, catalytic polypeptide (PRKDC) small interfering RNA (si‐PRKDC) were synthesized by GenePharma Co. Ltd. The synthesized RNA was transfected using Lipofect 3000.
Reverse transcription quantitative polymerase chain reaction (RT‐qPCR)
Total RNA was obtained using TRIzol reagent, and reverse transcription was performed to obtain cDNA. TransStart SYBR Green qPCR Supermix (TransGen Biotech) was used for PCR amplification using an ABI 7500 system, and the reaction systems were configured according to the manufacturer's instructions. Three replicate wells were employed, and the test was conducted three times. U6 and GAPDH were used as internal references for miRNAs and mRNAs, respectively, with 2‐△△qct for data analysis. 23 The PCR primer sequences are listed in Table 1.
TABLE 1.
PCR primer sequences
| Gene | Upstream primer | Downstream primer |
|---|---|---|
| miRNA‐218‐5p | 5′‐TTGCGGATGGTTCCGTCA AGCA‐3′ | 5′‐ATCCAGTGCAGGGTCCGAGG‐3′ |
| PRKDC | 5′‐GAAAGGAGGTTCTAAACTAC‐3′ | 5′‐GCTACGTGAATATAGACCATA‐3′ |
| U6 | 5′‐CTCGCTTCGGCAGCACA‐3′ | 5′‐AACGCTTCACGAATTTGCGT‐3′ |
| GAPDH | 5′‐CCATCTTCCAGGAGCGAGAT‐3′ | 5′‐TGCTGATGATCTTGAGGCTG‐3′ |
Colony formation
After 48 h of transfection, the cells were treated with X‐rays at 0, 2, 4, 6, or 8 Gy. Cells were cultured and evaluated for survival using a colony forming assay. Cells were reinoculated into six‐well plates, cultured overnight to allow cell adhesion, treated with 0–8 Gy X‐rays, cultured at 37°C for two weeks, fixed with methanol for 20 min, and stained with 0.1% crystal violet.
3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl‐2H4‐tetrazolium bromide (MTT) assay
MTT was used to evaluate cell proliferation. After 48 h of transfection, the cells were resuspended and the density‐adjusted cells (2 × 103 cells/well) were seeded into 96‐well plates. After 24 h of culture, 20 μl MTT solution (5 mg/ml) was added at 24, 48, and 72 h, followed by incubation at 37°C for another 4 h. Next, the medium was aspirated and 150 μl of dimethyl sulfoxide (DMSO) was added. The optical density (OD) was determined at 492 nm.
Cells in DNA synthesis assay
Cell‐Light EdU DNA cell proliferation kit (Ribo Bio) was used to determine the proliferation rate of A549R and H1299R cells according to the manufacturer's instructions. Briefly, cells were incubated with 50 μM EdU for 2 h, then fixed, permeabilized, and stained with EdU. The nuclei were stained with Hoechst 33342 at a concentration of 5 μg/ml for 30 min, and the cells were then inspected using a fluorescence microscope (Olympus). The viability index was calculated as follows: Viability index = experimental OD value/control OD value. In addition, 5‐ethynyl‐20‐deoxyuridine (EdU) was used to evaluate cell proliferation.
Apoptosis
Cells were obtained after 48 h of transfection, and apoptosis was evaluated using annexin V‐fluorescein isothiocyanate (FITC). Transfected cells were harvested, rinsed, and resuspended. Next, the cells were stained with FITC and propidium iodide (PI) for 15 min. Cytek Aurora and FlowJo were used for the analysis.
Western blotting (WB)
Total protein was extracted, and exosome protein levels were determined using BCA (Bicinchoninic acid, Beyotime), followed by quantification. The proteins were transferred to a polyvinylidene fluoride membrane, which was sealed with phosphate buffer and mixed with diluted anticleaved PARP, anticleaved caspase 3, anti‐CD63, and anti‐β‐actin antibodies. Secondary antibody labeled with horseradish peroxidase was added, followed by incubation for 1 h. Enhanced chemiluminescence was employed to determine protein levels using Bio‐Rad ChemiDoc XRS densitometry and Image Lab version 4.1.
Dual‐luciferase reporter gene constructs
Potential targets of miRNA‐216‐5p were identified, and PRKDC was the target of miRNA‐216‐5p. To verify that PRKDC was a target of miRNA‐216‐5p, pCMV‐REPORT‐PRKDC‐3′UTR‐WT was established by inserting the PCMV‐reporter vector into the DNA sequence of PRKDC 3′UTR. The binding site of miRNA‐216‐5p was confirmed. A pCMV‐REPORT‐PRKDC‐3′UTR‐mut with a mutant miRNA‐216‐5p binding site was also constructed. miRNA‐216‐5p mimic and control were transfected into HEK‐293 T cells, and the luciferase activity was tested.
Tumor formation in nude mice
A total of 12 male BALB/c nude mice (5‐weeks‐old) were procured from Charles River Laboratories. A normal diet was fed to the mice. This study conformed to the Animal Ethics Committee and the Laboratory Animal Guidelines. Overexpressed miRNA‐216‐5p (A549R cells) were injected into the mice, and X‐ray irradiation (12 Gy) was conducted when tumor diameters reached 5 mm. The mice were grouped into an miRNA‐NC group, miRNA‐216‐5p‐mimics group, miRNA‐NC + 12 Gy group, and miRNA‐216‐5p‐mimics+2 Gy group, with three mice in each group. Two days after X‐ray irradiation, the tumor volume (V) was measured using a caliper. The spherical volume was calculated as follows: spherical volume = 4/3 π r̂3 or (4/3) π (length/2) × (width/2) × (depth/2).
Statistical analysis
SPSS v.24.0 was applied for analysis, and GraphPad 8 was used for visualization. Counting data were represented in (%) and compared using the Chi‐squared test. Measurement data were represented as means ± SEM and compared using Student's t‐test. Single‐factor analysis of variance (ANOVA) and least significant difference (LSD) post‐hoc test were applied for multiple groups. Repeated measures ANOVA and Bonferroni post‐hoc test were applied for multiple time points. The diagnostic predictive value was tested using the receiver operating characteristic curve. At p < 0.05, the difference was considered to be statistically significant.
RESULTS
Role of radiotherapy in improving miRNA‐218‐5p levels in LC
First, miRNA‐218‐5p levels in LC from The Cancer Genome Atlas (TCGA) data were determined, and RT‐qPCR results revealed that the miRNA‐218‐5p level in the serum of LC patients was evidently lower than that in the normal control group (Figure 1a,b ). Further comparison showed that the miRNA‐218‐5p level in the serum of LC patients after radiotherapy was evidently higher than that before intervention (Figure 1c ). In addition, the miRNA‐218‐5p level in LC cell lines was evaluated using RT‐qPCR, and the results revealed that the miRNA‐218‐5p levels in the LC cell lines after X irradiation declined (Figure 1d ). This suggests that miRNA‐218‐5p may be a latent observation index for radiotherapy in LC.
FIGURE 1.
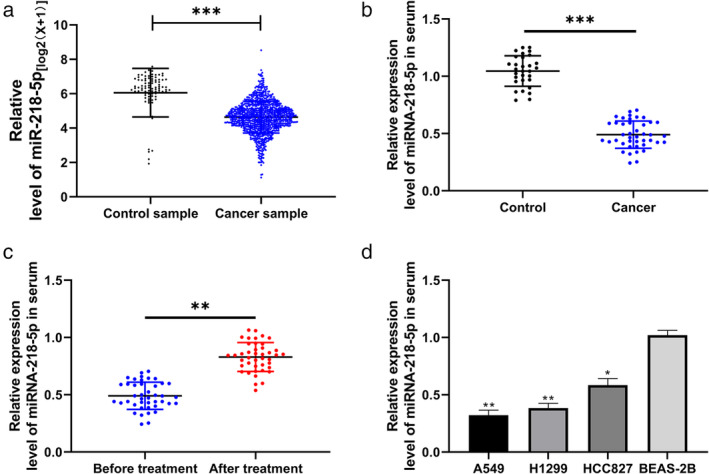
miRNA‐218‐5p expression in lung carcinoma patients after radiotherapy. (a) Relative expression of miRNA‐218‐5p in lung carcinoma in TCGA. (b) Reverse transcription quantitative polymerase chain reaction (RT‐qPCR) was used to determine miRNA‐218‐5p expression in the serum of control and patient groups. ***p < 0.001, lung carcinoma groups compared with the control. (c) RT‐qPCR was used to determine miRNA‐218‐5p expression in serum before and after radiotherapy. ***p < 0.001 after radiotherapy compared with before radiotherapy. (d) RT‐qPCR was used to determine miRNA‐218‐5p expression in lung carcinoma cell lines. **p < 0.01 and *p < 0.05 carcinoma cell lines compared with the BEAS‐2B cell line
Elevation of miRNA‐218‐5p levels on proliferation and apoptosis of radiation‐resistant LC cells
A549 and H1299 cells were exposed to X‐rays to establish radiation‐resistant LC cells. Cloning experiments showed that the radiation‐resistant cell lines (A549R and H1299R) had enhanced resistance to X‐rays compared with the parental cells (Figure 2a ). RT‐qPCR revealed that the miRNA‐218‐5p abundance in A549R and H1299R cells was evidently lower than that in parental cells, suggesting that miRNA‐218‐5p may play a role in the radiation resistance of LC cells (Figure 2b,c ). Therefore, we established an miRNA‐218‐5p‐mimic plasmid and transferred it into A549R and H1299R cells to further observe any changes in cellular biological functions (Figure 2d ). MTT (Figure 2e) assays, cloning assays (Figure 3a), and EdU experiments (Figure 3b) revealed that the metabolic survival (Figure 2e), clonogenic survival (Figure 3a), and DNA synthesis activity (Figure 3b), respectively, of A549R and H1299R cells declined after transfection with the miRNA‐218‐5p‐mimic, whereas apoptosis of A549R and H1299R cells was elevated after transfection of the miRNA‐218‐5p‐mimic, as revealed by flow cytometry (Figure 3c ).
FIGURE 2.
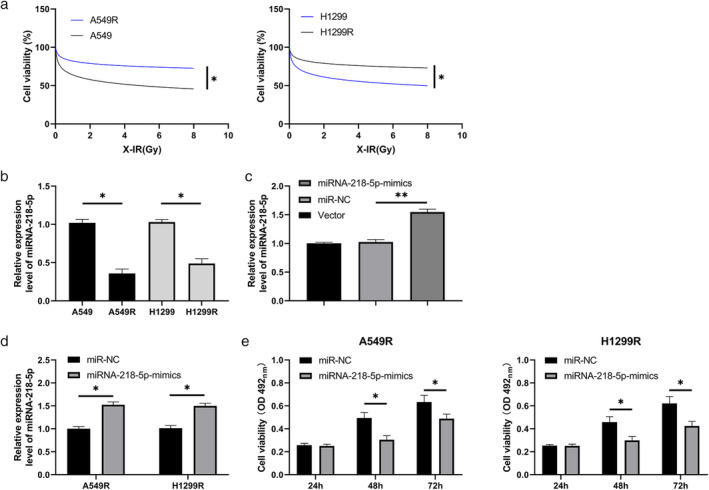
Role of miRNA‐218‐5p in the growth of radiation‐induced lung carcinoma cells. (a) MTT assay was used to evaluate metabolic survival of parental cells and radiation‐resistant cell lines under different doses of X‐rays. (b) RT‐qPCR was used to determine miRNA‐218‐5p expression in radiation‐resistant cell lines. (c) RT‐qPCR was used to determine miRNA‐218‐5p expression in miRNA‐218‐5p‐mimic plasmid. (d) RT‐qPCR was used to determine miRNA‐218‐5p expression after transfection of miRNA‐218‐5p‐mimic into radiation‐resistant cell lines. (e) MTT assay was used to evaluate the proliferation of irradiated cell lines after transfection of the miRNA‐218‐5p‐mimic. (b), p < 0.05, radiation‐resistant cell lines compared with the control cell lines. (c–e), *p < 0.05, miRNA‐218‐5p‐mimic compared with the NC
FIGURE 3.
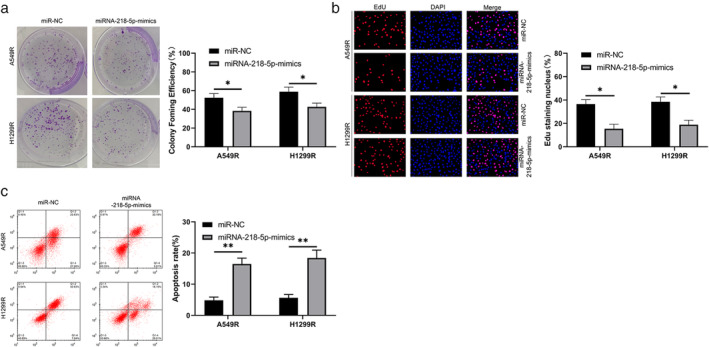
Role of miRNA‐218‐5p in the apoptosis of radiation‐induced lung carcinoma cells. (a) Colony formation was used to evaluate changes in the cloning ability of irradiated cell lines after transfection of the miRNA‐218‐5p‐mimic. (b) EdU was used to evaluate the proliferation of irradiated cell lines after the transfection of the miRNA‐218‐5p‐mimic. (c) Flow cytometry was used to evaluate changes in the apoptosis rate of irradiated cell lines after transfection of the miRNA‐218‐5p‐mimic. *p < 0.05; **p < 0.01
miRNA‐218‐5p exhibits targeted binding to PRKDC
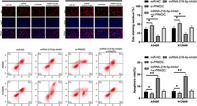
It has been proven that miRNAs regulate tumor growth by combining with downstream target genes. PRKDC and miRNA‐218‐5p have latent binding targets, identified through online website prediction analysis (Figure 4a ). To verify the correlations among them, we first tested PRKDC in LC in TCGA database and found that PRKDC expression in LC is elevated (Figure 4b ). We then tested PRKDC expression in LC patients using RT‐qPCR, and the results were consistent with those obtained from the database (Figure 4c ). Through correlation analysis, PRKDC expression was negatively correlated with miRNA‐218‐5p levels in the serum of LC patients (Figure 4d ), suggesting that miRNA‐218‐5p may exhibit targeted binding to PRKDC. To verify this speculation, we conducted dual‐luciferase reporter gene construct experiments, and the results revealed that the miRNA‐218‐5p‐mimic could hinder the fluorescence activity of PRKDC‐WT (Figure 4e ), whereas the miRNA‐218‐5p‐inhibitor enhanced the fluorescence activity of PRKDC‐WT. In addition, RT‐qPCR and WB revealed that PRKDC mRNA and protein levels in cells transfected with the miRNA‐218‐5p‐mimic or inhibitor were obviously hindered and accelerated, respectively (Figure 4f,g ). This suggests that miRNA‐218‐5p exhibits targeted binding to PRKDC.
FIGURE 4.
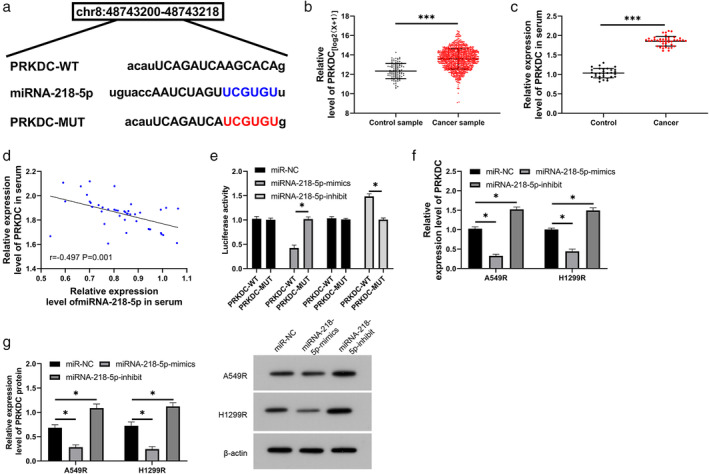
Targeted binding of miRNA‐218‐5p and protein kinase, DNA‐activated, catalytic polypeptide (PRKDC). (a) StarBase predictions were used to identify the binding and mutated sites of miRNA‐218‐5p and PRKDC. (b) Expression of PRKDC in lung carcinoma in TCGA database. (c) Reverse transcription quantitative polymerase chain reaction (RT‐qPCR) was used to evaluate the relative expression of PRKDC in the serum of patients with lung carcinoma. (d) Pearson's test was applied to identify any correlation involving miRNA‐218‐5p and PRKDC in patients with lung carcinoma. (e) Dual‐luciferase reporter gene analysis of miRNA‐218‐5p and PRKDC target binding. (f) and (g) RT‐qPCR and WB were used to evaluate PRKDC mRNA and protein levels, respectively, in radiation‐resistant lung carcinoma cells transfected with miRNA‐218‐5p‐mimic/inhibitor. *p < 0.05; ***p < 0.001
Decreased PRKDC expression can inhibit the growth promotion of miRNA‐218‐5p‐inhibitor on radiation‐resistant LC cells
A cotransfection experiment was performed to test the role of miRNA‐218‐5p and PRKDC in radiation‐resistant LC. The metabolic survival of radiation‐resistant cells was increased (Figure 5a), the clone survival rate (Figure 5b) was enhanced, and apoptosis was hindered (Figure 6a,b). However, the metabolic and clonal survival of radiation‐resistant cells declined after transfection of si‐PRKDC. Compared with miRNA‐NC + si‐NC cells, miRNA‐218‐5p‐inhibitor and si‐PRKDC cotransfected cells showed no evident difference in metabolic survival, clonal survival, or apoptosis.
FIGURE 5.
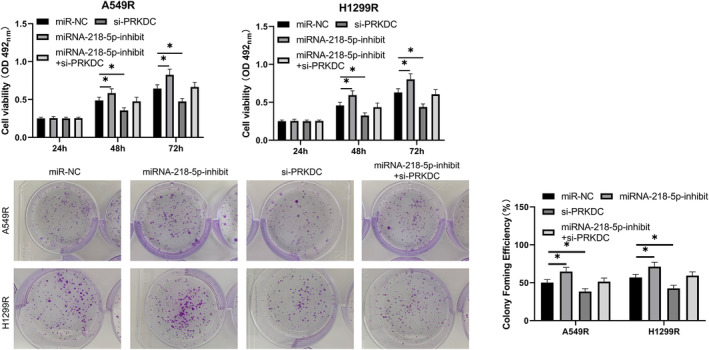
Decrease in protein kinase, DNA‐activated, catalytic polypeptide (PRKDC) levels can inhibit promotion of miRNA‐218‐5p‐inhibitor on growth of radiation‐resistant lung carcinoma cells. (a) MTT assay was used to evaluate changes in the number of cells surviving metabolically after cotransfection of the miRNA‐218‐5p‐inhibitor and si‐PRKDC. (b) Colony‐forming experiments were performed to evaluate changes in cell survival of reproductive integrity after cotransfection of the miRNA‐218‐5p‐inhibitor and si‐PRKDC. *p < 0.05; **p < 0.01
FIGURE 6.
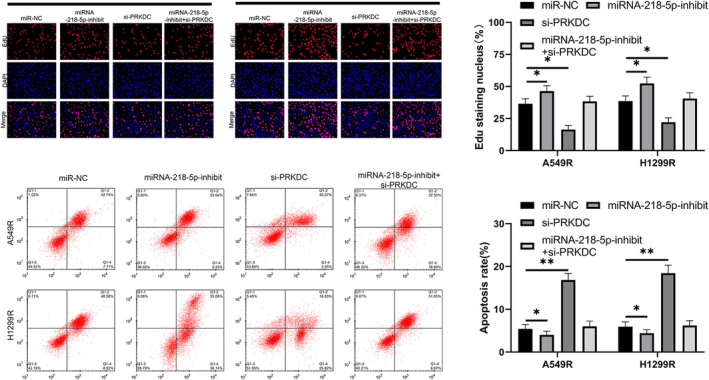
Decreasing the level of protein kinase, DNA‐activated, catalytic polypeptide (PRKDC) can reverse miRNA‐218‐5p‐inhibitor radiation‐resistant lung cancer cell apoptosis. (a) An EdU DNA cell proliferation test was used to evaluate changes in the number of cells undergoing DNA synthesis after cotransfection of the miRNA‐218‐5p‐inhibitor and si‐PRKDC. (b) Flow cytometry was used to evaluate changes in the apoptosis rates after cotransfection of the miRNA‐218‐5p‐inhibitor and si‐PRKDC. *p < 0.05; **p < 0.01
miRNA‐218‐5p‐mimics increases the radiosensitivity of lung cancer nude mice
In the above experiment, we determined the function of miRNA‐218‐5p in radiation‐resistant lung cancer cells. In order to further determine whether miRNA‐218‐5p can be a potential target in clinical radiotherapy, we established a xenograft in a nude mouse model. Through measurement, we found that the tumor volume of nude mice after 12 Gy X‐ray irradiation was significantly lower than that of miRNA‐NC, while the tumor volume in nude mice injected with miRNA‐218‐5p‐mimics combined with 12 Gy X‐ray irradiation was significantly lower than that of miRNA‐NC (Figure 7a,b). In addition, our research observed that the miRNA‐218‐5p in the tumor tissue of nude mice after 12 Gy X‐ray irradiation was significantly higher than that of miRNA‐NC, while miRNA‐218‐5p in tumors of nude mice after injection of miRNA‐218‐5p‐mimics combined with 12 Gy X‐ray irradiation was significantly higher than that of nude mice after miRNA‐NC and 12 Gy X‐ray irradiation. This suggests that miRNA‐218‐5p can increase the radio sensitivity in a lung cancer nude mouse model.
FIGURE 7.
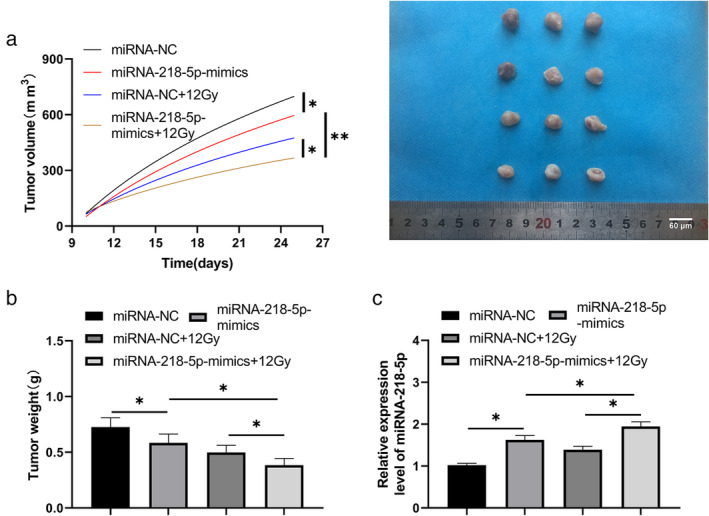
miRNA‐218‐5p‐mimics increases the radiosensitivity of lung cancer cells in a nude mice model. (a) The tumor volume changes in the nude mice over 25 days. (b) After 25 days, the nude mice were sacrificed and the tumor tissue mass was compared. *p < 0.05, **p < 0.01
DISCUSSION
Radiation exposure increases the levels of free radicals in cells, accompanied by DNA strand breakage and dysfunction of the mitochondria, endoplasmic reticulum, and other organelles, eventually leading to apoptosis. 24 , 25 At present, research concerning the mechanism involved in the radiation resistance in LC shows that P53 mutations, an X‐linked inhibitor of apoptosis protein, and survivin have positive effects on radiotherapy. 26 However, the mechanisms and latent targets of radiotherapy remain unclear.
miRNAs play a role in the radiation resistance and radiosensitivity of various tumor cells. For example, Zhu et al. 27 reported that miRNA‐145 antagonizes the dryness and radiation resistance of colorectal carcinoma mediated by SNAI1; another study showed that miR‐99a elevates the radiosensitivity of NSCLC via mTOR. 28 miRNA‐218‐5p, an miRNA that was discovered early on, is located on human chromosome 4p15.31. A previous study revealed that miRNA‐218‐5p levels are low in LC cells, 29 as confirmed by our results. In addition, a study has found that miRNA‐218‐5p plays a role in the sensitivity to ionizing radiation in breast carcinoma; however, there are relatively few studies on the role on miRNA‐218‐5p in radiation resistance of LC. 30 miRNA‐218‐5p levels in radiation‐resistant LC cells were found to be decreased; the metabolic and clonogenic survival of radiation‐resistant LC cells were obviously reduced, and apoptosis was induced by elevating miRNA‐218‐5p levels. In addition, the transfection of the miRNA‐218‐5p‐mimic increased the sensitivity of radiation‐resistant cells to X‐irradiation, suggesting that miRNA‐218‐5p is a latent therapeutic target for radiation‐resistant LC cells.
It has been reported that miRNAs influence downstream target genes to participate in tumor radiation resistance mechanisms. 31 , 32 To further investigate the mechanisms underlying the effect of miRNA‐218‐5p on the radiation resistance of LC cells, we predicted the downstream target genes of miRNA‐218‐5p and found that there were targeted binding sites for PRKDC with miRNA‐218‐5p. This suggests that miRNA‐218‐5p may be a latent target for radiation resistance. PRKDC belongs to the non‐homologous end joining pathway of the DNA double‐strand break (DSB) repair response and can be recruited by DNA double‐strand breaks to form DNAPK complexes with DNA‐bound Ku70/80 heterodimers to promote the repair of DNA DSBs. 33 , 34 PRKDC levels in patients with LC are evidently elevated. Furthermore, miRNA‐218‐5p levels in the serum of LC patients showed a negative correlation with PRKDC abundance. In addition, miRNA‐218‐5p exhibited targeted binding to PRKDC. Knockdown of PRKDC could reverse the promotion of the miRNA‐218‐5p‐inhibitor on the proliferation of radiation‐resistant LC cells and inhibit apoptotic activity. It is suggested that miRNA‐218‐5p regulates the growth and apoptosis of radiation‐resistant LC cells through PRKDC.
However, this study had certain limitations. First, we did not detect miRNA‐218‐5p in the serum exosomes of patients with LC. Previous studies have shown that miRNAs in serum exosomes can also be applied as a latent diagnostic marker for tumors. Second, owing to the short collection time of the samples, we did not conduct patient follow‐up for a long period. In the future, we aim to perform additional clinical experiments to further verify the significance of miRNA‐218‐5p in serum exosomes in LC, in order to supplement our experimental results.
This study explored the role of miRNA‐218‐5p in the radiosensitivity of LC cells. miRNA‐218‐5p could induce apoptosis and enhance the radiosensitivity of LC cells through regulation, thus suggesting its application as a potential target for LC treatment.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
ACKNOWLEDGMENT
This research was funded by National Natural Science Fundation of China Project No. 8187070347.
Chen X, Xu Y, Jiang L, Tan Q. miRNA‐218‐5p increases cell sensitivity by inhibiting PRKDC activity in radiation‐resistant lung carcinoma cells. Thorac Cancer. 2021;12:1549–1557. 10.1111/1759-7714.13939
Funding information National Natural Science Fundation of China, Grant/Award Number: 8187070347
REFERENCES
- 1. Watanabe SI, Nakagawa K, Suzuki K, Takamochi K, Ito H, Okami J, et al. Neoadjuvant and adjuvant therapy for stage III non‐small cell lung cancer. Jpn J Clin Oncol. 2017;47:1112–8. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 3. Romaszko AM, Doboszyńska A. Multiple primary lung cancer: a literature review. Adv Clin Exp Med. 2018;27:725–30. [DOI] [PubMed] [Google Scholar]
- 4. Villalobos P, Wistuba II. Lung cancer biomarkers. Hematol Oncol Clin North Am. 2017;31:13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Steven A, Fisher SA, Robinson BW. Immunotherapy for lung cancer. Respirology. 2016;21:821–33. [DOI] [PubMed] [Google Scholar]
- 6. Chua GWY, Chua KLM. Which patients benefit most from stereotactic body radiotherapy or surgery in medically operable non‐small cell lung cancer? An in‐depth look at patient characteristics on both sides of the debate. Thorac Cancer. 2019;10:1857–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lemjabbar‐Alaoui H, Hassan OU, Yang YW, Buchanan P. Lung cancer: biology and treatment options. Biochim Biophys Acta. 1856;2015:189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD‐1 blockade in non‐small cell lung cancer. Science. 2015;348:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peng S, Wang R, Zhang X, Ma Y, Zhong L, Li K, et al. EGFR‐TKI resistance promotes immune escape in lung cancer via increased PD‐L1 expression. Mol Cancer. 2019;18:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu Y, Wang Y, Xiao X, Cheng W, Hu L, Yao W, et al. MiR‐204 inhibits hepatocellular cancer drug resistance and metastasis through targeting NUAK1. Biochem Cell Biol. 2019;97:563–70. [DOI] [PubMed] [Google Scholar]
- 11. Lin YH. MicroRNA networks modulate oxidative stress in cancer. Int J Mol Sci. 2019;20:4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tuo L, Chu X, Sha S, Zhang X. MicroRNA and lung cancer: a mini review. Zhongguo Fei Ai Za Zhi. 2018;21:727–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu L, Hu B, Zhao B, Liu Y, Yang Y, Zhang L, et al. Circulating microRNA‐422a is associated with lymphatic metastasis in lung cancer. Oncotarget. 2017;8:42173–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meng F, Zhang L. miR‐183‐5p functions as a tumor suppressor in lung cancer through PIK3CA inhibition. Exp Cell Res. 2019;374:315–22. [DOI] [PubMed] [Google Scholar]
- 15. Yang Y, Ding L, Hu Q, Xia J, Sun J, Wang X, et al. MicroRNA‐218 functions as a tumor suppressor in lung cancer by targeting IL‐6/STAT3 and negatively correlates with poor prognosis. Mol Cancer. 2017;16:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iwamoto N, Fukui S, Takatani A, Shimizu T, Umeda M, Nishino A, et al. Osteogenic differentiation of fibroblast‐like synovial cells in rheumatoid arthritis is induced by microRNA‐218 through a ROBO/slit pathway. Arthritis Res Ther. 2018;20:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jin X, Liu X, Zhang Z, Guan Y. lncRNA CCAT1 acts as a microRNA‐218 sponge to increase Gefitinib resistance in NSCLC by targeting HOXA1. Mol Ther Nucleic Acids. 2020;19:1266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koritzinsky EH, Street JM, Star RA, Yuen PS. Quantification of Exosomes. J Cell Physiol. 2017;232:1587–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilligan KE, Dwyer RM. Engineering Exosomes for cancer therapy. Int J Mol Sci. 2017;18:1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qu Y, Zhang Q, Cai X, Li F, Ma Z, Xu M, et al. Exosomes derived from miR‐181‐5p‐modified adipose‐derived mesenchymal stem cells prevent liver fibrosis via autophagy activation. J Cell Mol Med. 2017;21:2491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang Y, Cui Y, Li Z, Jiao Z, Zhang Y, He Y, et al. Radiation‐induced miR‐208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J Exp Clin Cancer Res. 2016;35:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Issue information‐declaration of Helsinki. J Bone Miner Res. 2018;33:BM i–BM ii. [DOI] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C[T]) method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 24. Storch K, Dickreuter E, Artati A, Adamski J, Cordes N. BEMER electromagnetic field therapy reduces cancer cell radioresistance by enhanced ROS formation and induced DNA damage. PLoS One. 2016;11:e0167931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baskar R, Lee KA, Yeo R, Yeoh KW. Cancer and radiation therapy: current advances and future directions. Int J Med Sci. 2012;9:193–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tommelein J, De Vlieghere E, Verset L, Melsens E, Leenders J, Descamps B, et al. Radiotherapy‐activated cancer‐associated fibroblasts promote tumor progression through paracrine IGF1R activation. Cancer Res. 2018;78:659–70. [DOI] [PubMed] [Google Scholar]
- 27. Zhu Y, Wang C, Becker SA, Hurst K, Nogueira LM, Findlay VJ, et al. miR‐145 antagonizes SNAI1‐mediated stemness and radiation resistance in colorectal cancer. Mol Ther. 2018;26:744–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yin H, Ma J, Chen L, Piao S, Zhang Y, Zhang S, et al. MiR‐99a enhances the radiation sensitivity of non‐small cell lung cancer by targeting mTOR. Cell Physiol Biochem. 2018;46:471–81. [DOI] [PubMed] [Google Scholar]
- 29. Yang Q, Li J, Hu Y, Tang X, Yu L, Dong L, et al. MiR‐218‐5p suppresses the killing effect of natural killer cell to lung adenocarcinoma by targeting SHMT1. Yonsei Med J. 2019;60:500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Labbé M, Hoey C, Ray J, Potiron V, Supiot S, Liu SK, et al. microRNAs identified in prostate cancer: correlative studies on response to ionizing radiation. Mol Cancer. 2020;19:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Y, Wang P, Wu LL, Yan J, Pang XY, Liu SJ. miR‐26a‐5p inhibit gastric cancer cell proliferation and invasion through mediated Wnt5a. Onco Targets Ther. 2020;13:2537–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu B, Wang C, Wang YL, Chen SQ, Wu JP, Zhu WD, et al. miR‐143 inhibits renal cell carcinoma cells metastatic potential by suppressing ABL2. Kaohsiung J Med Sci. 2020;36:592–8. [DOI] [PubMed] [Google Scholar]
- 33. Pannunzio NR, Watanabe G, Lieber MR. Nonhomologous DNA end‐joining for repair of DNA double‐strand breaks. J Biol Chem. 2018;293:10512–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brand TM, Iida M, Luthar N, Starr MM, Huppert EJ, Wheeler DL. Nuclear EGFR as a molecular target in cancer. Radiother Oncol. 2013;108:370–7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]


