Abstract
Aims
Coronavirus disease 2019 (COVID-19), which is a highly contagious disease, is an ongoing outbreak worldwide with high morbidity and mortality. The approaches targeting the autophagy processes might have promising diagnostic and therapeutic values against Coronavirus infection. Here, we aimed to investigate the relationship of Beclin-1 (BECN1), an autophagy-related protein, with blood parameters and the clinical severity in patients with COVID-19.
Materials and methods
We enrolled 108 patients with COVID-19 and 21 healthy controls in this study, from September 2020 to January 2021 and divided all patients into two groups according to the severity of the disease: The non-severe group and the severe group. BECN1 levels and blood parameters were measured with Enzyme-Linked Absorbent Assay and routine techniques, respectively.
Key findings
Serum BECN1 levels were increased in patients with COVID-19 compared to the healthy controls, and its concentrations were significantly higher in the severe group than in the non-severe group (p < 0.001). BECN1 levels showed a significantly positive correlation with coagulation markers such as D-dimer and Fibrinogen (FIB) and inflammation markers such as C-reactive protein (CRP), Procalcitonin (PCT), Ferritin and biochemical markers such as Blood urea nitrogen and Lactate dehydrogenase (p < 0.001). We detected that areas under the ROC curve for BECN1, D-dimer, FIB, PCT, CRP and Ferritin were 0.8662, 0.9110, 0.8278, 0.9996 and 0.9284, respectively (p < 0.0001).
Significance
BECN1 may serve as a predictive biomarker in evaluating the disease severity of COVID-19. Our data suggest that BECN1 mediated-autophagy modulation might have a promising value in improving the clinical outcomes of COVID-19.
Keywords: COVID-19, Beclin 1, Autophagy, Predictive marker, Disease severity
1. Introduction
Coronavirus disease 2019 (COVID-19) is an unprecedented global health problem that mainly affects the respiratory and immune systems, and the ongoing outbreak of this highly contagious disease continues to cause an enormous burden on health care and economic systems worldwide [1]. The clinical spectrum of COVID-19 is broad, and most patients infected with Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have a range of common symptoms, such as fever, cough, shortness of breath, and fatigue, while some of the COVID-19 patients are asymptomatic [2,3]. Severe illness and higher mortality rates are seen among older adults 60 and over age and patients with comorbidities including cardiovascular disease and diabetes [4]. Although recent efforts of scientists around the world increase our knowledge so far, many important questions about early diagnosis and effective treatment of COVID-19 are not responded yet. Therefore, the development of novel therapeutic and diagnostic approaches is urgently required to improve the clinical management of COVID-19.
SARS-CoV-2, a recently identified single-strand RNA virus, is a highly pathogenic agent responsible for a current pandemic of COVID-19 [4]. Previous studies revealed that coronaviruses use various cellular mechanisms, such as autophagy to enhance their replication [5,6]. Autophagy is an essential mechanism in physiological homeostasis that acts as a clearance system by removing and degrading unfolded proteins and damaged organelles. Recent studies emphasize that autophagy-related processes may play a critical role in the action mechanisms of SARS-CoV-2, and therapeutic approaches targeting autophagy modulation would be beneficial in improving the clinical outcomes of COVID-19 [[6], [7], [8]]. In the course of the coronavirus infection, autophagy may act as an anti-viral mechanism to inhibit viral replication [6]. On the other hand, some viruses may use the autophagy mechanisms as a viral replication tool [6]. Unfolded protein response (UPR) is a critical intracellular pathway that can be activated by endoplasmic reticulum (ER) stress arise from the accumulation of unfolded proteins during viral infection [9]. Activated UPR may induce the degradation of unfolded proteins through activation of autophagy in alleviating ER stress and restoring homeostasis [10]. Moreover, UPR and autophagy are closely related mechanisms, and SARS-CoV-2 infection might activate autophagy through UPR induction [8,10]. Beclin-1 (BECN1) is an essential regulator protein that controlling the autophagy process at the nucleation stage, and its deregulation is associated with some diseases, including cancer and diabetic kidney disease [[11], [12], [13]]. However, there is currently no study examining the potential role of serum BECN1 and its clinical value in COVID-19. Given the papers mentioned above on autophagy, we speculate that BECN1, an autophagy-related protein, might be worth investigating in patients with COVID-19. Also, clarifying the relationship of BECN1 as a serum marker with the clinical severity of COVID-19 would be beneficial for physicians to improve the management of the disease. Therefore, we aimed to investigate the relationship of BECN1 with biochemical parameters and the clinical severity in patients with COVID-19.
2. Material and methods
2.1. Study design and participants
We recruited 108 COVID-19 patients who were admitted to Hatay Mustafa Kemal University Hospital and 21 healthy individuals in the present study. Our study was reviewed and approved by the clinical research ethics committee of Hatay Mustafa Kemal University (Approval number: 2020/103), and we performed the study per the Declaration of Helsinki during the period September 2020 and January 2021. All participants, which consisted of COVID-19 patients and healthy individuals, provided an informed consent form. All patients were diagnosed according to recommendations of the World Health Organization, and the diagnosis was confirmed in nasal and pharyngeal swab specimens collected from all patients at admission to the hospital using the real-time quantitative polymerase chain reaction method (RT-qPCR). We detected the presence of SARS-CoV-2 RNA using an RT-qPCR kit (Bioeksen R&D Tech. Ltd., Istanbul, Turkey) and included only RT-qPCR confirmed cases in the current study.
2.2. The Classifications of COVID-19 severity
Clinical classification of all participants was performed according to the World Health Organization (WHO) classification criteria for COVID-19 [14] and divided all patients into four groups: 1) mild group, patients who present with symptoms of COVID-19 disease without signs of viral pneumonia or hypoxia; 2) moderate group, patients who have signs of non-severe pneumonia without signs of severe pneumonia on clinical examination, a saturation of oxygen (SpO2) higher than 90% on room air; 3) severe group, patients who have clinical signs of severe pneumonia (fever, cough, dyspnea, fast breathing) plus a respiratory frequency higher than 30 breaths/min and/or have SpO2 lower than 90% on room air; and 4) critically group, patients who have acute respiratory distress syndrome (ARDS) and/or septic shock and/or multiple organ dysfunction, with requiring mechanical ventilation and intensive care unit admission.
2.3. Data and sample collection
We collected and analyzed the demographic and clinical data of all patients, including age, sex, comorbidities, clinical symptoms, medications and hospitalization days. We also evaluated the results of laboratory tests performed according to the clinical needs of patients, including hematological parameters (white blood cells [WBC, 103/μL], lymphocyte [LYM, 103/μL], monocytes [MON, 103/μL], eosinophil [EOS, 103/μL], red blood cells [RBC, 106/μL], hemoglobin [HMG, g/dL], hematocrit [HCT, %], mean corpuscular volume [MCV, fL], platelet [PLT, 103/μL], mean platelet volume [MPV, fL], platelet distribution width [PWD, ratio]), coagulation parameters (D-dimer [ng/L], fibrinogen [FIB, mg/L]), inflammatory markers (Erythrocyte sedimentation rate [ESR, mm/h] and C-reactive protein [CRP, mg/L]) and biochemical markers (alanine aminotransferase [ALT, U/L], aspartate aminotransferase [AST, U/L], creatinine [CREA, mg-dL]), Blood Urea Nitrogen [BUN, mg/dL], Creatine kinase [CK, U/L], lactate dehydrogenase [LDH, U/L], Pro-calcitonin [PCT, ng/mL] and Ferritin [ng/mL]. All laboratory tests were conducted in the central laboratory of Hatay Mustafa Kemal University Hospital using routine standard methods. Venous blood samples were collected via venipuncture from healthy individuals and COVID-19 patients, and blood samples were transferred into regular test tubes containing gel for serum separation and tubes containing Ethylenediaminetetraacetic acid and sodium citrate for other analysis. The blood samples in biochemistry tubes containing gel were allowed to clot at room temperature for 30–50 min, and then, all samples were centrifuged at 1500 ×g for 10 min in a pre-cooled (4 °C) centrifuge, and serum samples were aliquoted into tubes and store at −80 °C until BECN1 concentrations were measured.
2.4. BECN1 concentrations
BECN1 concentrations were analyzed from the serum of COVID-19 patients and healthy individuals using commercial Enzyme-linked Immunosorbent Assay (ELISA) kits. The detection range for BECN1 was 0.16–10 ng/mL, and the intra-assay and inter-assay coefficients of variation for BECN1 were 5% and 5.14%, respectively.
2.5. Statistical analysis
We first performed the Kolmogorov-Smirnov test to detect the normal distribution of the data and presented categorical variables as percentages and number and continuous variables as medians (interquartile range). The Kruskal Wallis H test was used for multiple comparisons of continuous data, and Mann Whitney U test was utilized to compare two groups. Analysis of categorical data was conducted using the chi-square test. Spearman's rho correlation analysis was conducted to evaluate the association between BECN1 and biochemical parameters. p values <0.05 was considered as statistical significant. We performed receiver operating characteristic (ROC) analysis to evaluate the potential diagnostic value of BECN1 and other biochemical parameters in COVID-19 patients. All data were analyzed using SPSS version 23.0 (IBM Corporation, Armonk, NY, USA), and graphs were drawn using GraphPad Prism 8.0.
3. Results
3.1. Demographic and clinical characteristics of patients with COVID-19
A total of 108 patients with COVID-19 whose SARS-CoV-2 RNA was confirmed by qRT-PCR and 21 healthy individuals were enrolled in the present study. There were 70 COVID-19 positive cases in the non-severe group (16 mild and 54 moderate), while there were 38 COVID-19 positive cases in the severe group (24 severe and 14 critically ill). No significant difference in age and sex were determined between healthy subjects and non-severe and severe groups. The most common clinical symptoms of all patients with COVID-19 were fever, cough, fatigue and dyspnea. The most common clinical symptoms of patients with COVID-19 in the non-severe group were cough, fever and fatigue, while common clinical symptoms of patients with COVID-19 in the severe group were dyspnea, cough, fatigue, and chest tightness. The majority (79.6%) of COVID-19 patients had one or more complications, and hypertension (22.2%) and diabetes (19.44%) in the patient groups were more common comorbidities (p < 0.05). Hospitalization times of patients in the severe group were significantly higher than the patients in the non-severe groups (p < 0.001). A total of 7 patients from the severe group died (18.4%) during hospitalization, and as expected, the severe group had a higher mortality rate than the non-severe group (p < 0.001). Demographic and clinical data of patients with COVID-19 and healthy subjects were presented in Table 1 .
Table 1.
Demographic and clinic characteristics of COVID-19 patients and healthy controls.
| Variables | Healthy controls (n = 21) | COVID-19 patients |
p value | ||
|---|---|---|---|---|---|
| Non-severe group (n = 70) | Severe group (n = 38) | All patients (n = 108) | |||
| Age | 45.52 (41.5–50) | 47.98 (36–59) | 50.82 (47.5–56) | 48.94 (40–58) | 0.053 |
| Sex (n, %) | |||||
| Male | 12 (57.1) | 38 (54.3) | 27 (71.1) | 65 (60.2) | 0.229 |
| Female | 9 (42.9) | 32 (45.7) | 11 (28.9) | 43 (39.8) | |
| Hospitalization (day) | 8.64 ± 3.18 | 15.71 ± 9.82 | 11.15 ± 7.19 | <0.001 | |
| Comorbidities (n, %) | |||||
| Hypertension | 14 (20) | 10 (26.3) | 24 (22.2) | 0.041 | |
| Diabetes mellitus | 7 (10) | 14 (36.8) | 21 (19.44) | <0.001 | |
| Chronic kidney disease | 1 (1.4) | 5 (13.2) | 6 (5.5) | 0.012 | |
| Coronary heart disease | 2 (2.9) | 4 (10.5) | 6 (5.5) | 0.106 | |
| Cerebrovascular disease | 0 (0) | 4 (10.5) | 4 (3.7) | 0.007 | |
| Chronic liver disease | 2 (2.9) | 2 (5.3) | 4 (3.7) | 0.528 | |
| Pulmonary disease | 4 (5.7) | 4 (10.5) | 8 (7.4) | 0.267 | |
| Thyroid disease | 3 (4.3) | 3 (7.9) | 6 (5.5) | 0.378 | |
| Malignancy | 3 (4.3) | 4 (10.5) | 7 (6.48) | 0.191 | |
| Clinical symptoms (n, %) | |||||
| Fever | 26 (37.1) | 5 (13.2) | 31 (28.7) | <0.001 | |
| Cough | 33 (47.1) | 14 (36.8) | 47 (43.5) | <0.001 | |
| Loss of appetite | 1 (1.4) | 3 (7.9) | 4 (3.7) | 0.121 | |
| Expectoration | 6 (8.6) | 0 (0) | 6 (5.5) | 0.071 | |
| Chest tightness | 5 (7.1) | 7 (18.4) | 12 (11.1) | 0.043 | |
| Fatigue | 25 (35.7) | 8 (21.1) | 33 (30.5) | 0.003 | |
| Dyspnea | 6 (8.6) | 24 (63.2) | 30 (27.7) | <0.001 | |
| Diarrhea | 2 (2.9) | 1 (2.6) | 3 (2.7) | 0.740 | |
| Sore throat | 7 (10) | 0 (0) | 7 (6.48) | 0.044 | |
| Myalgia | 3 (4.3) | 0 (0) | 3 (2.7) | 0.274 | |
| Nausea or vomiting | 2 (2.9) | 3 (7.9) | 5 (4.62) | 0.261 | |
| Abdominal pain | 3 (4.3) | 1 (2.6) | 4 (3.7) | 0.598 | |
| Loss of taste or smell | 1 (1.4) | 0 (0) | 1 (0.9) | 0.654 | |
| Deaths (n, %) | 0 (0) | 7 (18.4) | 7 (6.48) | <0.001 | |
The comparisons of continuous data were performed using the Kruskal Wallis H test followed by the Mann-Whitney U test, and analysis of categorical data was conducted using the chi-square test. Continuous values were expressed as medians (interquartile range [IQR]), while categorical variables were presented as numbers (percentages). Statistically significant p values less than 0.05 were indicated as bold.
3.2. The laboratory findings of patients with COVID-19 and healthy controls
We presented the laboratory findings of COVID-19 patients and healthy controls in Table 2 . Analysis of hematological parameters demonstrated that WBC count was significantly higher in the severe group than in the non-severe group and healthy controls (p < 0.001), and no significant difference was found in WBC between healthy controls and the non-severe group (p = 0.091). LYM, EOS and HGB levels were significantly reduced in the non-severe and the severe group compared to the healthy controls (p < 0.05). LYM counts were significantly decreased among patients who had died from SARS-CoV-2 infections than survival patients (p < 0.001). HCT levels were significantly lower in the severe group than in the non-severe group and healthy controls (p < 0.001), while no significant difference was observed in HCT levels between healthy controls and the non-severe group (p = 0.107). PLT levels were significantly higher in the severe group than in the non-severe group and healthy controls. No significant difference in MON and MCV was found between healthy controls and the non-severe and the severe groups (p > 0.05). Serum D-dimer and FIB levels were significantly elevated in the non-severe and the severe groups compared to the healthy controls (p < 0.001). Also, serum D-dimer and FIB levels were significantly higher in the severe group than in the non-severe group (p < 0.001). ESR, CRP, PCT and Ferritin levels were significantly increased in the non-severe and severe group compared to the healthy controls (p < 0.001). These parameters were significantly higher in the severe group than in the non-severe group (p < 0.001). Biochemical analyses showed a significant increase in serum ALT, AST, BUN, CK and LDH levels in the non-severe and the severe group compared to the healthy controls. No significant differences were found in CREA among all groups (p > 0.05).
Table 2.
Laboratory findings of 108 patients with COVID-19 and 21 healthy controls.
| Parameters | Normal range | Healthy controls (n = 21) | COVID-19 patients |
p value | ||
|---|---|---|---|---|---|---|
| Non-severe group (n = 70) | Severe group (n = 38) | All patients (n = 108) | ||||
| WBC, 103/μL | 4–10 | 6.80 (6.19–7.40) | 5.70 (4.43–7.45) | 10.47 (6.51–12.06) | 6.57 (4.66–10.32) | <0.001a, c, d,=0.091b |
| LYM, 103/μL | 0.8–4 | 2.22 (1.65–2.51) | 1.47 (0.99–1.85) | 0.82 (0.64–1.34) | 1.23 (0.85–1.81) | <0.001a, b, c, d |
| MON, 103/μL | 0.12–1.2 | 0.45 (0.40–0.50) | 0.42 (0.31–0.55) | 0.47 (0.32–0.72) | 0.45 (0.32–0.61) | =0.103a,=0.278b, =0.311c,=0.047d |
| EOS, 103/μL | 0.02–0.5 | 0.16 (0.13–0.27) | 0.02 (0.005–0.07) | 0.01 (0.00–0.052) | 0.02 (0.00–0.06) | <0.001a, b, c,=0.266d |
| RBC, 106/μL | 4–5.5 | 4.49 (4.29–4.88) | 4.8 (4.44–5.18) | 4.38 (3.78–4.77) | 4.60 (4.27–5.06) | =0.001a,=0.078b, =0.095c,<0.001d |
| HGB, g/dL | 12–16 | 14.4 (13–15.3) | 13.2 (12.4–14.3) | 11.85 (9.4–13.3) | 12.85 (11.82–14.1) | =0.051a,=0.029b,<0.001c, d |
| HCT, % | 40–54 | 41.9 (38.4–44.25) | 39.1 (37.5–43.35) | 35.8 (29.37–39.35) | 38.65 (35.92–41.9) | <0.001a, c, d,=0.107b |
| MCV, fL | 80–100 | 87.5 (82.05–90.75) | 84.8 (80.90–88.25) | 85.35 (80.6–89.3) | 84.85 (80.92–88.3) | =0.217a,=0.091b, =0.137c,=0.908d |
| PLT, 103/μL | 150–450 | 247 (208–288) | 207 (177–273) | 322.5 (250–369) | 249 (186–306.75) |
<0.001a, d,=0.082b =0.009c |
| MPV, fL | 6.5–12 | 9.8 (8.8–11.4) | 9.6 (9.3–10.5) | 10.7 (10.25–11.1) | 10.1 (9.4–11) | =0.001a,=0.565b, =0.05c,<0.001d |
| PDW, ratio | 15–17 | 16.1 (15.85–16.4) | 16 (15.7–16.4) | 16.3 (16.1–16.42) | 16.1 (15.82–16.4) | =0.023a,=0.461b, =0.168c,=0.006d |
| D-dimer, ng/mL | 0–500 | 207 (170.5–254) | 377 (259,5-650,68) | 1040 (590–2640) | 560 (300–1050) | <0.001a, b, c, d |
| FIB, mg/dL | 200–400 | 213 (202–239.5) | 365 (302–474) | 519 (417.75–567.5) | 421.5 (314.25–533) | <0.001a, b, c, d |
| ESR, mm/h | 0–20 | 8 (6.5–11) | 16 (10. 50–35.5) | 38 (19.5–51.25) | 22.5 (12.25–42) | <0.001a, b, c, d |
| CRP, mg/L | 0–5 | 1.05 (0.94–1.88) | 17.4 (5.99–52.35) | 52.52 (20.17–78.8) | 29.15 (9.25–61.45) | <0.001a, b, c,=0.001d |
| PCT, ng/mL | 0.05–0.09 | 0.012 (0.01–0.018) | 0.03 (0.01–0.05) | 0.135 (0.04–0.33) | 0.04 (0.02–0.12) | <0.001a, b, c, d |
| Ferritin, ng/mL | 10–291 | 23 (10.5–40) | 122.4 (47,5–309) | 664.7 (246–1189.92) | 263.75 (70.3–647.65) | <0.001a, b, c, d |
| LDH, U/L | 120–246 | 132 (121.5–145) | 241.5 (200.75–311) | 312 (203.5–437.75) | 256 (201–356.75) | <0.001a,b,c,<0.049d |
| ALT, U/L | 0–40 | 19 (15–24) | 29 (21–43.25) | 38.5 (18.75–78.25) | 30.5 (20.25–52) | =0.001a,<0.01b, c,=0.239d |
| AST, U/L | 0–49 | 19 (16–23.5) | 31 (25–43.25) | 29.5 (21.75–52.25) | 30.5 (23–44.75) | <0.001a, b, c,=0.662d |
| CREA, mg/dL | 0.5–1.1 | 0.76 (0.62–0.81) | 0.77 (0.6–0.96) | 0.87 (0.64–1.18) | 0.80 (0.60–0.99) | =0.135a,=0.291b, =0.062c,=0.166d |
| BUN, mg/dL | 8–23 | 9 (7.5–9.5) | 13 (10–16.25) | 22 (14–31) | 15 (11−22) | <0.001a, b, c, d |
| CK, U/L | 30–200 | 63 (49.5–88) | 92 (56.75–157.25) | 208 (127.5–350.25) | 112 (78–229.75) | <0.001a, c, d,=0.014b |
The comparisons of data were performed using the Kruskal Wallis H test followed by the Mann-Whitney U test, and the values were expressed as medians (interquartile range [IQR]). Statistically significant p values less than 0.05 were indicated as bold. Abbreviations: WBC: White blood cells, LYM: Lymphocyte, MON: Monocytes, EOS: Eosinophil, RBC: Red blood cells, HGB: Hemoglobin, HCT: Hematocrit, MCV: Mean corpuscular volume, MPV: Mean platelet volume, PDW: Platelet distribution width, FIB: Fibrinogen, ESR: Erythrocyte sedimentation rate, CRP: C-reactive protein, PCT: Procalcitonin, LDH: Lactate dehydrogenase, ALT: Alanine aminotransferase, AST: Aspartate aminotransferase, CREA: Creatinine, BUN: Blood urea nitrogen, CK: Creatine kinase.
Comparison among all groups.
Comparison between Healthly controls and Non-severe group.
Comparison between Healthly controls and Severe group.
Comparison between Non-severe group and Severe group.
3.3. Serum BECN1 levels in patients with COVID-19 and healthy controls
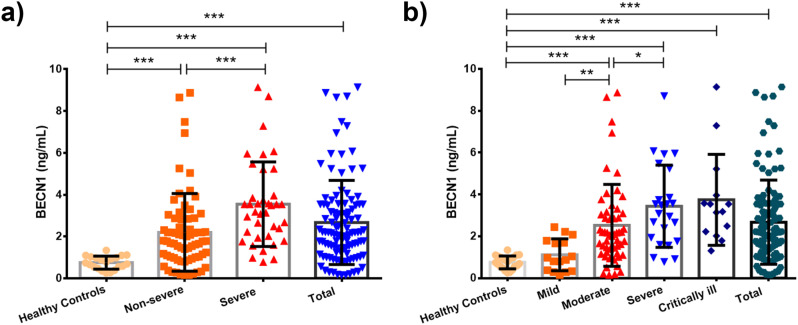
We measured the serum BECN1 concentrations in COVID-19 patients and healthy controls to detect the possible effect of COVID-19 on autophagy-related processes at the systemic level. Our results showed that serum BECN1 levels were significantly increased in all COVID-19 patients when compared to the healthy controls (p < 0.001, Fig. 1a). Also, BECN1 levels were significantly higher in the severe group than in the non-severe group (p < 0.001, Fig. 1a). We also analyzed the levels of BECN1 in clinical subgroups of COVID-19 to compare whether the effect of clinical severity on this protein level (Fig. 1b). Our findings indicated that BECN1 levels were increased with concomitantly clinical severity of the COVID-19 disease. Furthermore, no significant differences were observed in BECN1 levels between patients with and without comorbidities.
Fig. 1.
The BECN1 protein levels in patients with COVID-19 and healthy controls (a). Comparison of BECN1 protein levels in COVID-19 patients according to the classification of mild, moderate, severe and critically ill disease (b). BECN1: Beclin-1. Data were expressed as mean ± SD. p < 0.05 values were considered as significant. ***p < 0.001, **p < 0.01, *p < 0.05.
3.4. Correlations between BECN1 and biochemical parameters
We analyzed the association of serum BECN1 levels with hematological, inflammatory and biochemical parameters in COVID-19 patients and healthy controls. As presented in Table 3 , our results revealed that there is a significant negative correlation between BECN1 and LYM, EOS, HGB, HCT and MCV, while there is a significant positive correlation between BECN1 and WBC and PLT. Furthermore, our findings unveiled that BECN1 has a significant positive correlation with coagulation markers, such as D-dimer and FIB and inflammatory parameters, such as ESR, CRP, Ferritin and PCT, and other biochemical markers, such as LDH, ALT, AST, CREA, BUN and CK (Table 3).
Table 3.
Correlation of serum BECN1 and biochemical parameters in patients with COVID-19 and healthy controls.
| Variables | BECN1 |
|
|---|---|---|
| r | p value | |
| WBC, 103/μL | 0.183 | 0.038 |
| LYM, 103/μL | −0.322 | 0.001 |
| MON, 103/μL | 0.098 | 0.272 |
| EOS, 103/μL | −0.424 | <0.001 |
| RBC, 106/μL | −0.012 | 0.895 |
| HGB, g/dL | −0.234 | 0.008 |
| HCT, % | −0.224 | 0.011 |
| MCV, fL | −0.206 | 0.019 |
| PLT, 103/μL | 0.217 | 0.013 |
| MPV, fL | 0.058 | 0.515 |
| PDW, ratio | 0.012 | 0.896 |
| D-dimer, ng/mL | 0.483 | <0.001 |
| FIB, mg/dL | 0.473 | <0.001 |
| ESR, mm/h | 0.245 | 0.005 |
| CRP, mg/L | 0.464 | <0.001 |
| PCT, ng/mL | 0.452 | <0.001 |
| Ferritin, ng/mL | 0.516 | <0.001 |
| ALT, U/L | 0.214 | 0.015 |
| AST, U/L | 0.246 | 0.005 |
| CREA, mg/dL | 0.273 | 0.002 |
| BUN, mg/dL | 0.487 | <0.001 |
| CK, U/L | 0.394 | <0.001 |
| LDH, U/L | 0.463 | <0.001 |
The correlation of BECN1 with biochemical parameters was analyzed using Spearman's rho test, and statistically significant p values less than 0.05 were indicated as bold. Abbreviations: BECN1:Beclin-1WBC: White blood cells, LYM: Lymphocyte, MON: Monocytes, EOS: Eosinophil, RBC: Red blood cells, HGB: Hemoglobin, HCT: Hematocrit, MCV: Mean corpuscular volume, MPV: Mean platelet volume, PDW: Platelet distribution width, FIB: Fibrinogen, ESR: Erythrocyte sedimentation rate, CRP: C-reactive protein, PCT: Procalcitonin, LDH: Lactate dehydrogenase, ALT: Alanine aminotransferase, AST: Aspartate aminotransferase, CREA: Creatinine, BUN: Blood urea nitrogen, CK: Creatine kinase.
3.5. Predictive value of BECN1 levels for evaluating disease severity
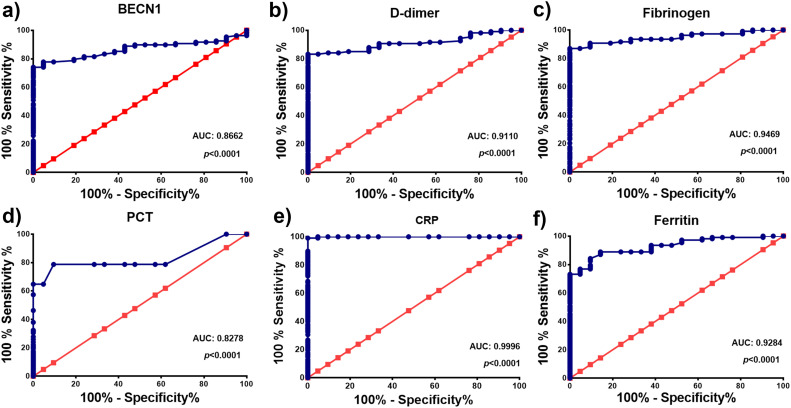
We utilized ROC analyses to evaluate the diagnostic potential and detect the optimal cut-off values of BECN1 and other biochemical parameters in COVID-19 patients. As presented in Table 4 and Fig. 2 , we detected that areas under the ROC curve for BECN1, D-dimer, Fibrinogen, PCT, CRP and Ferritin were 0.8662, 0.9110, 0.8278, 0.9996 and 0.9284, respectively (p < 0.0001). 95% confidence intervals for BECN1, D-dimer, Fibrinogen, PCT, CRP and Ferritin were 0.8058–0.9266, 0.8620–0.9600, 0.9105–0.9833, 0.7572–0.8984, 0.9981–1.001 and 0.8831–0.9736, respectively. Our data suggest that BECN1, CRP, Ferritin, D-dimer, Fibrinogen and PCT have clinical values for the management of COVID-19.
Table 4.
Area under the receiver operating characteristic curve and optimal cut-off values of BECN1, D-dimer, Fibrinogen, PCT, CRP and Ferritin in COVID-19 patients.
| Variables | AUC (95%CI) | Sensitivity % | Specificity % | Cut-off | p value |
|---|---|---|---|---|---|
| BECN1 | 0.8662 (0.8058–0.9266) | 79.6 | 80.9 | 1.03 ng/mL | <0.0001 |
| D-dimer | 0.9110 (0.8620–0.9600) | 84.1 | 85.7 | 258 ng/mL | <0.0001 |
| Fibrinogen | 0.9469 (0.9105–0.9833) | 90.7 | 90.5 | 252 mg/dL | <0.0001 |
| PCT | 0.8278 (0.7572–0.8984) | 78.7 | 61.9 | 0.01 ng/mL | <0.0001 |
| CRP | 0.9996 (0.9981–1.001) | 99.1 | 95.2 | 3.04 mg/mL | <0.0001 |
| Ferritin | 0.9284 (0.8831–0.9736) | 87.0 | 85.7 | 44.8 ng/mL | <0.0001 |
Abbreviations: AUC: Area under the receiver operating characteristic curve, BECN1: Beclin-1, PCT: Procalcitonin, CRP: C-reactive Protein.
Fig. 2.
ROC curve analyses representing the potential diagnostic values of BECN1 (a), D-dimer (b), Fibrinogen (c), PCT (d), CRP (e) and Ferritin (f) in COVID-19 patients. Abbreviations: BECN1: Beclin-1, PCT: Procalcitonin, CRP: C-reactive Protein.
4. Discussion
COVID-19, which is a highly contagious disease, is an ongoing outbreak worldwide with high morbidity and mortality, and there is no available effective therapy for this deadly disease. Therefore, the understanding of the molecular pathogenesis of COVID-19 and identification of diagnostic and therapeutic targets are crucial in improving the treatment outcomes of SARS-CoV-2 infected patients and timely control of disease progression. The existing evidence suggests that autophagy mechanisms might be part of combat strategies against SARS-CoV-2 infections [[5], [6], [7], [8]]. However, the precise role of autophagy in COVID-19 remains unclear.
To the best of our knowledge, we are the first to examine the relationship of BECN1 with biochemical parameters and the clinical severity in patients with COVID-19. Here, we report that serum BECN1 levels were significantly increased in the severe and non-severe COVID-19 patients. Furthermore, our ROC curve analyses reveal that BECN1 might have a potential value in improving the clinical outcomes of COVID-19. Previous studies revealed that viral infections may induce autophagy through several mechanisms, and autophagy could counteract viral replication by regulating certain anti-viral mechanisms and survival functions in infected and immune cells [6]. Furthermore, viral infection and replication stimulate UPR, which is a crucial intracellular mechanism by leading to the accumulation of unfolded proteins in the ER [15,16]. The UPR pathway is associated with autophagy, and induced UPR could promote autophagy processes, suggesting that SARS-CoV-2 infection might possibly trigger autophagy mechanisms in patients with COVID-19 [8,15,17]. Our data suggest that increased BECN1 levels in COVID-19 patients may result from UPR mediated-ER stress induced by viral replication. Considering existing evidence, we may speculate that BECN1 mediated-autophagy stimulation might have anti-viral effects against Coronaviruses infection, notwithstanding further detailed studies are undoubtedly needed.
Our laboratory results showed the significant abnormalities of hematological parameters in COVID-19 patients, consistent with previous studies. We found that the severe COVID-19 patients had higher WBC counts than in the non-severe patients. Our findings revealed that LYM counts were significantly decreased in the severe group compared with the non-severe group, and lymphopenia is highly correlated with the clinical severity of SARS-CoV-2 infections, consistent with previous studies [18]. Moreover, BECN1 levels were highly correlated with LYM, EOS and HGB. We also evaluated the coagulation-related factors, including D-dimer and Fibrinogen, in the current study. D-dimer, which serves as a serum biomarker of coagulation, is a fibrinolysis degradation product and increased D-dimer concentrations are related to the clinical severity and poor prognosis of COVID-19 [19,20]. Fibrinogen is a crucial coagulation factor that plays a role in maintaining homeostasis, and its levels were increased in COVID-19 patients [20,21]. Consistent with the studies mentioned above [20,21], our analyses reveal that D-dimer and FIB levels were increased in the non-severe and severe COVID-19 patients. Our findings unveiled that there are abnormalities of coagulation profiles in COVID-19 patients, and BECN1 levels were highly correlated with D-dimer and Fibrinogen concentrations. We also investigated the inflammation-related markers [22,23], including CRP, ESR, PCT and Ferritin in COVID-19 patients and the relationship of these parameters with BECN1. C-reactive protein (CRP), which is synthesized by the liver, is an inflammatory biomarker, and its expressions were increased in SARS-CoV-2 infected cases [24,25]. Our results revealed that CRP is significantly elevated in COVID-19 patients, and CRP is related to the clinical severity of COVID-19. These findings are consistent with previous studies [24,25]. Like CRP, ESR, which is known as an acute-phase reactant, is an inflammation marker, and we here show that ESR levels were significantly elevated in COVID-19 patients, consistent with previous studies [20]. Accumulating evidence demonstrates that Ferritin and PCT are inflammatory markers, and their levels are increased due to cytokine storms in COVID-19 [20]. Similarly, our results revealed that Ferritin and PCT levels are increased among COVID-19 patients. Our correlation analyses unveiled that BECN1 is highly correlated with inflammatory markers such as CRP, ESR, Ferritin and PCT. ROC curve analyses revealed that CRP, Ferritin and PCT might have potential clinical values in improving treatment outcomes of COVID-19 patients.
Considering the potentially detrimental effects of SARS-CoV-2 infection on many organs [22,23], we also investigated the biochemical parameters such as LDH, ALT, AST, CK, CREA and BUN and the association of these parameters with BECN1. Lactate dehydrogenase (LDH), which serves as an indicator of organ damage, is a cytoplasmic enzyme, and its levels were increased in COVID-19 patients [20]. As expected, our findings showed that LDH levels were higher in the severe cases than in the non-severe patients. We also analyzed the ALT and AST levels which are liver function indicators in COVID-19 patients, and our results revealed a significant increase in these parameters, consistent with previous studies [20,21]. We analyzed the CREA and BUN levels as renal function indicators in Covid-19 patients, and our results showed that the BUN levels were significantly higher in severe infection patients than in the non-severe patients, consistent with previous studies [22,26].
The current study is not without any limitations, and our findings might not represent all COVID-19 patients since it is a single-center cross-sectional study. The sample size, which consists of patients who were admitted to Hatay Mustafa Kemal University at a certain time, was small in our study. Also, the patients enrolled in our study were receiving treatment and some of them had one or more comorbid conditions. The confounder effect of these treatments and comorbidities on serum BECN1 concentrations is unknown.
5. Conclusion
In conclusion, we report for the first time that serum BECN1 levels were elevated in patients with COVID-19 and increased BECN1 concentrations were associated with hematological, coagulation, inflammatory and biochemical parameters and clinical severity in COVID-19. Furthermore, our results suggest that BECN1 may serve as a biomarker for the prediction of disease severity of COVID-19, and BECN1 mediated-autophagy modulation might have a promising value in improving the clinical outcomes of COVID-19. More importantly, our results highlight the potential importance of autophagy mechanisms in the pathogenesis of COVID-19 at the systemic level. However, further mechanistic studies are required to clarify the precise role of BECN1 in the molecular pathogenesis of COVID-19.
Funding
This study no received specific grant from any funding organization.
Author contributions
HMO and SD designed the study. SD, TB, and MC collected the samples and clinical data. HMO and SD performed the laboratory and statistical analysis. HMO, SD, TB and MC wrote, edited and revised the manuscript and interpreted the results. HMO and SD created tables, graphs and figures.
All authors listed in the present paper have made significant contributions to the study and all authors read and approved the final version of the paper.
Declaration of competing interest
We all declare that we do not have any conflicts of interest.
References
- 1.Pollard C.A., Morran M.P., Nestor-Kalinoski A.L. The COVID-19 pandemic: a global health crisis. Physiol. Genomics. 2020;52:549–557. doi: 10.1152/physiolgenomics.00089.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cevik M., Bamford C.G.G., Ho A. COVID-19 pandemic-a focused review for clinicians. Clin. Microbiol. Infect. 2020;26:842–847. doi: 10.1016/j.cmi.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim G.U., Kim M.J., Ra S.H., Lee J., Bae S., Jung J., Kim S.H. Clinical characteristics of asymptomatic and symptomatic patients with mild COVID-19. Clin. Microbiol. Infect. 2020;26:948.e1–948.e3. doi: 10.1016/j.cmi.2020.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J., Qi T., Liu L., Ling Y., Qian Z., Li T., Li F., Xu Q., Zhang Y., Xu S., Song Z., Zeng Y., Shen Y., Shi Y., Zhu T., Lu H. Clinical progression of patients with COVID-19 in Shanghai, China. J. Inf. Secur. 2020;80:e1–e6. doi: 10.1016/j.jinf.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi Y., Bowman J.W., Jung J.U. Autophagy during viral infection - a double-edged sword. Nat. Rev. Microbiol. 2018;16:341–354. doi: 10.1038/s41579-018-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmona-Gutierrez D., Bauer M.A., Zimmermann A., Kainz K., Hofer S.J., Kroemer G., Madeo F. Digesting the crisis: autophagy and coronaviruses. Microb. Cell. 2020;7:119–128. doi: 10.15698/mic2020.05.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Z., Lu K., Mao B., Liu S., Trilling M., Huang A., Lu M., Lin Y. The interplay between emerging human coronavirus infections and autophagy. Emerg. Microbes Infect. 2021;10:196–205. doi: 10.1080/22221751.2021.1872353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shojaei S., Suresh M., Klionsky D.J., Labouta H.I., Ghavami S. Autophagy and SARS-CoV-2 infection: a possible smart targeting of the autophagy pathway. Virulence. 2020;11:805–810. doi: 10.1080/21505594.2020.1780088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung T.S., Huang M., Liu D.X. Coronavirus-induced ER stress response and its involvement in regulation of coronavirus-host interactions. Virus Res. 2014;194:110–123. doi: 10.1016/j.virusres.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooper K.M., Barlow P.G., Henderson P., Stevens C. Interactions between autophagy and the unfolded protein response: implications for inflammatory bowel disease. Inflamm. Bowel Dis. 2019;25:661–671. doi: 10.1093/ibd/izy380. [DOI] [PubMed] [Google Scholar]
- 11.Salminen A., Kaarniranta K., Kauppinen A., Ojala J., Haapasalo A., Soininen H., Hiltunen M. Impaired autophagy and APP processing in Alzheimer’s disease: the potential role of Beclin 1 interactome. Prog. Neurobiol. 2013;106-107:33–54. doi: 10.1016/j.pneurobio.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Naguib M., Rashed L.A. Serum level of the autophagy biomarker Beclin-1 in patients with diabetic kidney disease. Diabetes Res. Clin. Pract. 2018;143:56–61. doi: 10.1016/j.diabres.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 13.Fu L.L., Cheng Y., Liu B. Beclin-1: autophagic regulator and therapeutic target in cancer. Int. J. Biochem. Cell Biol. 2013;45:921–924. doi: 10.1016/j.biocel.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization. Clinical Management of COVID-19: Interim Guidance, (WHO/2019-nCoV/clinical/2020.5).
- 15.Banerjee A., Czinn S.J., Reiter R.J., Blanchard T.G. Crosstalk between endoplasmic reticulum stress and anti-viral activities: a novel therapeutic target for COVID-19. Life Sci. 2020;255 doi: 10.1016/j.lfs.2020.117842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oakes S.A., Papa F.R. The role of endoplasmic reticulum stress in human pathology. Annu. Rev. Pathol. 2015;10:173–194. doi: 10.1146/annurev-pathol-012513-104649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi Z., Chen L. Endoplasmic reticulum stress and autophagy. Adv. Exp. Med. Biol. 2019;1206:167–177. doi: 10.1007/978-981-15-0602-4_8. [DOI] [PubMed] [Google Scholar]
- 18.Henry B.M., de Oliveira M.H.S., Benoit S., Plebani M., Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin. Chem. Lab. Med. 2020;58:1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 19.Halaby R., Popma C.J., Cohen A., Chi G., Zacarkim M.R., Romero G., Goldhaber S.Z., Hull R., Hernandez A., Mentz R., Harrington R., Lip G., Peacock F., Welker J., Martin-Loeches I., Daaboul Y., Korjian S., Gibson C.M. D-dimer elevation and adverse outcomes. J. Thromb. Thrombolysis. 2015;39:55–59. doi: 10.1007/s11239-014-1101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skevaki C., Fragkou P.C., Cheng C., Xie M., Renz H. Laboratory characteristics of patients infected with the novel SARS-CoV-2 virus. J. Inf. Secur. 2020;81:205–212. doi: 10.1016/j.jinf.2020.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Zheng L., Liu L., Zhao M., Xiao J., Zhao Q. Liver impairment in COVID-19 patients: a retrospective analysis of 115 cases from a single centre in Wuhan city, China. Liver Int. 2020;40:2095–2103. doi: 10.1111/liv.14455. [DOI] [PubMed] [Google Scholar]
- 22.Cheng Y., Luo R., Wang K., Zhang M., Wang Z., Dong L., Li J., Yao Y., Ge S., Xu G. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020;97:829–838. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pourbagheri-Sigaroodi A., Bashash D., Fateh F., Abolghasemi H. Laboratory findings in COVID-19 diagnosis and prognosis. Clin. Chim. Acta. 2020;510:475–482. doi: 10.1016/j.cca.2020.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G., Wu C., Zhang Q., Wu F., Yu B., Lv J., Li Y., Li T., Zhang S., Wu C., Wu G., Zhong Y. C-reactive protein level may predict the risk of COVID-19 aggravation. Open Forum Infect. Dis. 2020;7:ofaa153. doi: 10.1093/ofid/ofaa153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W., Zheng K.I., Liu S., Yan Z., Xu C., Qiao Z. Plasma CRP level is positively associated with the severity of COVID-19. Ann. Clin. Microbiol. Antimicrob. 2020;19 doi: 10.1186/s12941-020-00362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao M., Li X., Liu F., Tian T., Luo J., Yang Y. Acute kidney injury is associated with severe infection and fatality in patients with COVID-19: a systematic review and meta-analysis of 40 studies and 24,527 patients. Pharmacol. Res. 2020;161 doi: 10.1016/j.phrs.2020.105107. [DOI] [PMC free article] [PubMed] [Google Scholar]