Abstract
Purpose
Colorectal cancer (CRC) is the third most common cancer in males and the second in females worldwide with very poor prognosis. Extracellular matrix proteins like collagens play important roles in cancer progression. Collagen type V α2 (COL5A2) is increased in several cancers but its role in cancer remains unclear.
Methods
COL5A2 expression was evaluated by interrogation of public Oncomine gene microarray datasets and immunohistochemistry (IHC) analyses of two tissue microarrays containing 180 paired CRC cases. Survival analysis was performed using Kaplan–Meier survival curve and Cox proportional hazards regression methods. COL5A2 was ectopically expressed in CRC cells, and the cell proliferation was measured using the methylthiazolyldiphenyl-tetrazolium bromide (MTT) method.
Results
COL5A2 gene was significantly upregulated in the most types of CRC comparing with the normal counterparts. The mRNA expression of COL5A2 was associated with cancer stages, gender, recurrence, microsatellite instability and KRAS status of CRC. COL5A2 protein increased in the cancer epithelial cells comparing with the normal counterpart and associated with age and T stage of CRC, whereas stromal expression of COL5A2 has no significant change between cancerous and normal tissues. COL5A2 gene and protein (epithelial expression) are independent risk factors and predict poor prognosis of CRC. Ectopic expression of COL5A2 drives colon cancer cell growth and upregulates WNT/β-catenin and PI3K/mTOR signaling via binding DDR1.
Conclusion
COL5A2 is a potential prognostic marker of CRC.
Keywords: colorectal cancer, COL5A2, secreted protein, molecular marker, tissue microarray
Introduction
Colorectal cancer (CRC) is the second most common cancer in men worldwide. According to the latest GLOBOCAN 2018 statistics, there were 1006 and 795 thousand estimated cases in 2018 for men and women, respectively.1 The overall mortality of CRC is 47.2% (475 thousand) in men and 48.7% (387 thousand) in women, indicating a very poor prognosis. Despite the achievements have been made to improve the diagnosis and treatments of CRC patients, the identification of suitable diagnostic or prognostic molecular biomarkers for CRC still remains a challenge.
Previously, we have performed proteomic analyses of the secretome of colon fibroblasts and colon cancer cells and identified several extracellular matrix (ECM) proteins which were associated with colon cancer progression or were prognostic markers of CRC, including follistatin-related protein 1 (FSTL1),2 collagen type VI α3 (COL6A3),3 spondin-2 (SPON2)4 and COL3A1.5 COL5A2 was also identified as one of the secreted proteins which are abundant in CRC cell secretome.3 The human genome encodes 47 collagen genes including 3 pseudogenes. Traditionally, collagens are recognized as secretory ECM proteins which are deposited in cancer stromal region forming a microenvironment scaffold structure for tumor growth and development. Some collagen proteins are upregulated in cancer and associate with cancer progression and prognosis.6 Targeting microenvironmental COL5A3 and COL4A2 have been shown to attenuate mammary tumor cell growth and progression.7,8
Human COL5A2 gene locates at 2q32.2 and spans 55 exons and encodes an alpha chain of fibrillar collagen type V. Heterozygous mutations of COL5A2 and COL5A1 were related to Ehlers-Danlos syndrome.9 COL5A2 mutation was relevant to genetically complex autoimmune disease, like diffuse cutaneous systemic sclerosis.10 COL5A2 gene is upregulated in CRC11 and osteosarcoma,12 and associated with drug resistance of ovarian cancer13 and Muscle-Invasive Bladder Cancer.14 COL5A2 protein was upregulated in CRC patient sera and represented a potential serum marker of CRC.15 These observations suggest that COL5A2 might play an important role in carcinogenesis or progression. However, its clinic and prognosis significance and function in human cancers remain further elucidation.
In the current study, we analyzed COL5A2 mRNA and protein expressions in CRC by interrogation of publicly-available gene expression microarray datasets and immunohistochemistry (IHC) of two tissue microarrays (TMAs). The relationship between COL5A2 expression and the clinical parameters of CRC patients was also analyzed. We also examined the function of COL5A2 in cancer progression.
Materials and Methods
Cell Lines and Cell Culture
CRC cell line SW620 was a gift from Prof. Qiqun Tang’s lab in the Fudan University Shanghai Medical College, Fudan University, China. HEK293T, Hela and CRC cell lines SW480 and HCT116 cells were brought from the Cell Bank of Shanghai Institutes for Biological Sciences, China. SW480 and SW620 cells were cultured in DMEM media. HCT-116 cells were cultivated with McCoy’s 5A medium (Gibco Life Technologies, Shanghai, China). All culture media were supplemented with 10% FBS and 1% Penicillin/streptomycin (Hyclone Laboratories, China) and cells were all cultured in a humidified incubator at 37°C and 5% CO2. The study was approved by the Research Ethics Committee of Fudan University.
Antibodies
COL5A2 antibody (PA5-14245) was purchased from ThermoFisher Scientific, and COL5A2 antibody (TA313657, OriGene) was used for immunohistochemistry (IHC) analysis. Antibodies for human phospho-p70S6K (Thr389) (#9234), phospho-mTOR (Ser2448) (#2971), phospho-GSK-3α/β (Ser21/9) (#8566), GSK-3α/β (#5676), phospho-Akt (Ser473) (#4060), phospho-Akt (Thr308) (#4056), β-catenin (#8480), phospho-PDK1 (Ser241) (#3061), APC (#2504), AXIN1 (#3323), CK1 (#2655), phospho-β-cateninS33/S37/T41 (#9561), phospho-β-cateninS45 (#9564), non-phospho (active) β-cateninS33/37/T41 (#8814), C-MYC (#9402), DDR1 (#3917) and pY792 DDR1 (#11994) were purchased from Cell Signaling Technology. β-actin antibody (60008-1-Ig) and Cyclin D1 antibody (60186-1-Ig) were purchased from Proteintech Group. E-cadherin antibody (no. 610181) was purchased from BD Transduction Laboratories. VEGF antibody (sc-152) was purchased from Santa Cruz Biotechnology. GAPDH antibody was purchased from Kangwei Century Co. LTD, Beijing, China.
Protein Extraction, Western Blot and Densitometry
Cellular protein extraction and Western blot were performed as described previously.4 Western densitometry was performed using Multi Gauge V3.0. The intensities of the paired bands and an adjacent background region were calculated automatically. The band intensities were background-subtracted and a ratio was calculated using the vector transfection as the dividend.
Data Mining of Oncomine Gene Expression Microarray Datasets
The expression values of COL5A2 gene across different microarray datasets of CRC, expressed as Log2 median-centered intensity or Log2 median-centered ratio, were retrieved directly from the graphical user interface of the Oncomine microarray database (https://www.oncomine.org/resource/login.html). COL5A2 expression of each dataset were dichotomized into higher-than-median and lower-than-median groups, and clinicopathological parameter and prognosis analyses were performed as previously described.4,5
IHC and Quantitative Staining Analysis
IHC was performed with the COL5A2 antibody using two commercial TMA (catalog no. HCol-Ade180Sur-04 (Sur-04) and HCol-Ade180Sur-08 (Sur-08), Shanghai Outdo Biotech, China) as described previously.4,5 For both TMAs, each case has one core with a diameter of 1.5 mm. The TMA HCol-Ade180Sur-04 contains 90 pairs of CRC and the matched normal mucosa counterparts. The surgical time was from July 2006 to May. 2007 and the follow-up information was available from November 2006 to Aug. 2013. The survival time was 3~ 85 months with a median survival time of 56 months. Follow-up records were unavailable for 7 cases. The TMA HCol-Ade180Sur-08 contains 90 pairs of CRC and the matched normal mucosa counterparts. The surgical time was from June 2007 to April. 2008 and the follow-up information was available from 2007 to July. 2015. The survival time was 2~ 97 months with a median survival time of 88 months. Follow-up records were unavailable for one case. The IHC of the TMAs was performed using Dako REALTM EnVisionTM Detection System, Peroxidase/DAB, Rabbit/Mouse kit (Dako Denmark A/S) according to the manufactory’s instructions. First, the paraffin-sealed TMAs were baked in a 63°C oven for one hour. The TMAs were de-vaxed automatically in the Dako Autostainer (Dako Denmark A/S) with a sequential procedure: dimethylbenzene 15 min × 2 times; water-free alcohol, 7 min × 2 times; 90% alcohol, 7 min; 80% alcohol, 7 min; 70% alcohol, 7 min. The chips were triple-rinsed with water for three minutes each time. Antigen retrieval was performed for 5 min in boiled citric acid solution. Next, the chips were incubated in blocking solution (76.8% methanol and 7.2% H2O2) for 10 min, followed by triple-rinsing with 1 × PBS for 5 min each time. Then the TMAs were stained with the rabbit polyclonal COL5A2 antibody (OriGene) diluted with Dako REAL™ Antibody Diluent (Dako S3022) at a ratio of 1:1000. This dilution was optimized and determined by preliminary experiments. The incubation was performed overnight in a humid container in a 4°C refrigerator. After the first antibody incubation, the chips were allowed to recovered to room temperature for one hour. The chips were triple-rinsed with 1 × PBS for 5 min each time before a further incubation with Dako EnVision™+/HRP second antibody reagent (Dako SM802) for 30 min. This is dextran coupled with peroxidase molecules and goat secondary antibody molecules against rabbit immunoglobulins. The chips were washed with PBS and stained with substrate 3,3ʹ-diaminobenzidine (DAB) (Dako EnVision™) DAB+ Chromogen (Dako DM827) in EnVision™ Substrate Buffer (Dako SM803) for 5 min for color developing, followed by washing with water for 15 min. The nuclei were counterstained with Dako Hematoxylin for the Dako Autostainer for 2 min before being treated with 0.25% HCl/alcohol for 2 seconds and rinsed with water for 2 min. The chips were dehydrated automatically in the immunostaining instrument with the sequential steps: 75% alcohol, 3 min; 85% alcohol, 3 min; 95% alcohol, 3 min; water-free alcohol, 5 min × 2 times; dimethylbenzene 15 min × 2 times. The chips were sealed with dimethylbenzene-diluted resin and scanned using Scanscope XT (Aperio Technologies).
To get a precise measure of COL5A2 protein expression, the staining images were subjected to a quantitative work flow as described previously.5 Briefly, for “whole tissue spot” analysis, the mucosa area of each tissue spot was selected with the Pen Tool using Aperio ImageScope v12.1 and the muscularis mucosae and submucosa area were excluded for analysis. To selectively quantify the expression level of the epithelial cells of the cancerous or normal tissue, these areas were firstly selected using the Pen Tool and then the unwanted stromal areas were excluded using the Negative Pen Tool. Similarly, to specifically analyze the stromal region, the epithelial area was excluded. The intensities of the selected regions were measured in an automated fashion using an integrated algorithm of Aperio ImageScope, Positive Pixel Count v9.1. The measure results were exported and parsed into Microsoft Excel for further analysis. The Positivity value (Number of Positive/Total Number (Positive + Negative)) was used as an expression measurement.16 To merge the results from the two TMA staining, the Positivity values from TMA Sur-08 were normalized using the formula: Positivity(Sur-08)normalized = Positivity(Sur-08)/[Median(Sur-08)/Median(Sur-04)]. After normalization, the median values of TMA Sur-08 and Sur-04 were equal. For survival curve and COX regression analysis, the most optimal cutpoint to distinguish the high and low expression of COL5A2 was determined using X-Tile v 3.6.1.17 For clinicopathological parameter analysis, the median value was used to dichotomized the cases into low and high groups. All of the clinical information and the Positivity values were deposited in Table S1.
Gene Cloning, Ectopic Expression and Methylthiazolyldiphenyl-Tetrazolium Bromide (MTT) Assay
Human full length COL5A2 cDNA clone plasmid was purchased from Sino Biological Inc., China. The coding sequence (CDS) of COL5A2 was amplified from the plasmid and sub-cloned into pcDNA3.1/myc-His(-)A vector for overexpression assay using primers COL5A2-F-NheI: CTAGCTAGCGTATTTAGACATGATGGCAAACTGGG and COL5A2-R-NotI: ATAAGAATGCGGCCGCCACAAAACAAACTGGCCCA. The transfection using Lipofectamine 2000 (Life Technologies - Invitrogen) and MTT proliferation assay were performed as described previously.4
RNA Extraction, Reverse Transcription and Quantitative PCR (qPCR)
Cellular total RNA was extracted using TRIzol Reagent (Invitrogen) according to the manufactory’s instructions. Reverse transcription was performed using the PrimeScript™ RT Master Mix (Takara). Real-time PCR was performed using LightCycler®480 SYBR Green I Master (Roche) on LightCycler® 480 System (Roche). The cycling parameters were as follows: 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s for 40 cycles. All reactions were performed in duplicate. The relative expression levels of the target genes were determined using the comparative Ct method equation: 2-∆∆Ct. ACTB was used as the endogenous control. The value was normalized to the average fold change in the negative control cell lines, which was defined as 1.0. The primers used were: COL5A2-F, CAGGCTCCATAGGAATCAGAGG, COL5A2-R, CCAGCATTTCCTGCTTCTCCAG; β-catenin-F, CACAAGCAGAGTGCTGAAGGTG, β-catenin-R, GATTCCTGAGAGTCCAAAGACAG; c-MYC-F, CCTGGTGCTCCATGAGGAGAC, c-MYC-R, CAGACTCTGACCTTTTGCCAGG; CyclinD1-F, TCTACACCGACAACTCCATCCG, CyclinD1-R, TCTGGCATTTTGGAGAGGAAGTG; VEGF-F, TTGCCTTGCTGCTCTACCTCCA, VEGF-R, GATGGCAGTAGCTGCGCTGATA; APC-F, AGGCTGCATGAGAGCACTTGTG, APC-R, CACACTTCCAACTTCTCGCAACG; AXIN1-F, GTATGTGCAGGAGGTTATGCGG, AXIN1-R, CACCTTCCTCTGCGATCTTGTC; CK1-F, GAAGATGTCCACGCCTGTTGAAG, CK1-R, GCGGAATAGCTGCCTCAGATAC; GSK3β-F, CCGACTAACACCACTGGAAGCT, GSK3β-R, AGGATGGTAGCCAGAGGTGGAT; E-cadherin-F, GCCTCCTGAAAAGAGAGTGGAAG, E-cadherin-R, TGGCAGTGTCTCTCCAAATCCG; PDK1-F, CATGTCACGCTGGGTAATGAGG, PDK1-R, CTCAACACGAGGTCTTGGTGCA; AKT1-F, TGGACTACCTGCACTCGGAGAA, AKT1-R, GTGCCGCAAAAGGTCTTCATGG; AKT2-F, CATCCTCATGGAAGAGATCCGC, AKT2-R, GAGGAAGAACCTGTGCTCCATG, mTOR-F, AGCATCGGATGCTTAGGAGTGG, mTOR-R, CAGCCAGTCATCTTTGGAGACC; GAPDH-F, GTCTCCTCTGACTTCAACAGCG, GAPDH-R, ACCACCCTGTTGCTGTAGCCAA; β-Actin-F, CACCATTGGCAATGAGCGGTTC, β-Actin-R, AGGTCTTTGCGGATGTCCACGT.
Statistical Analysis
The significance was calculated using two-tailed Student’s t test. The correlation between COL5A2 expression and the clinicopathological parameters was evaluated by the Chisq test using PASW Statistics 18. The Kaplan-Meier survival curves were calculated using the Log rank test with GraphPad Prism 6.01. Univariate and multivariate Cox proportional hazard regression analyses using enter and forward stepwise (Likelihood ratio) method, respectively, were carried out with PASW Statistics 18. A p value < 0.05 was considered as statistically significant.
Results
COL5A2 Gene and Epithelial-Expressed COL5A2 Protein Was Significantly Upregulated in CRC
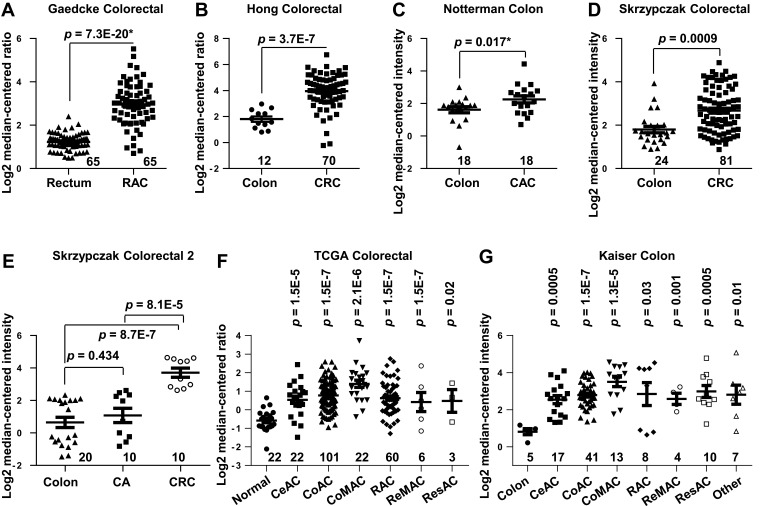
We analyzed COL5A2 mRNA expression in cancer tissues and normal counterparts by interrogation of the Oncomine microarray datasets. COL5A2 was significantly upregulated in CRC. This is supported by 7 out of all 8 independent datasets containing normal control data, which were generated by Gaedcke et al,18 Hong et al,19 Notterman et al,20 Skrzypczak et al,21 The Cancer Genome Atlas (TCGA) (http://tcga-data.nci.nih.gov/tcga/) and Kaiser et al22 (Figure 1). Furthermore, the upregulation of COL5A2 is evident in almost all subtypes of CRCs, as revealed using TCGA and Kaiser Colon datasets22 (Figure 1F and G). These datasets contain 558 cancerous and 166 normal cases.
Figure 1.
Upregulation of COL5A2 gene and protein expression in colorectal cancer (CRC). Gene expression of COL5A2 was upregulated in CRC tissues comparing with the normal tissues revealed using (A) the Gaedcke Colorectal dataset (GSE20842) (Oncomine database, https://www.oncomine.org/resource/login.html)18, (B) Hong Colorectal dataset (GSE9348),19 (C) Notteman colon dataset,20 (D) Skrzypczak Colorectal dataset (GSE20916),21 (E) Skrzypczak Colorectal 2 dataset (GSE20916),21 (F) TCGA colorectal dataset (http://tcga-data.nci.nih.gov/tcga/) and (G) Kaiser Colon dataset (GSE5206).22 The total number of cases were shown under each category. Other type of colon cancers includes colon signet ring cell adenocarcinoma, colon small cell carcinoma, rectal signet ring cell adenocarcinoma, rectosigmoid mucinous adenocarcinoma. The p values were calculated using two-tailed and unpaired Student’s t test. The p value indicated with an asterisk was calculated using paired Student’s t test.
Abbreviations: CA, colon adenoma; CeAC, cecum adenocarcinoma; RAC, rectal adenocarcinoma; CoAC, colon adenocarcinoma; CoMAC, colon mucinous adenocarcinoma; ReMAC, rectal mucinous adenocarcinoma; ResAC, rectosigmoid adenocarcinoma.
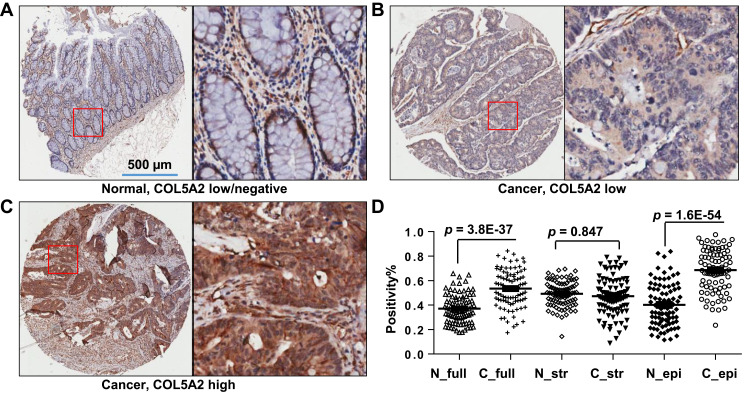
COL5A2 protein expression was analyzed by IHC using two commercial TMAs (Figure 2, Table S1). Low or high expression of COL5A2 staining in the normal or cancerous tissues was shown (Figure 2A–C). We found that COL5A2 was significantly increased in CRC tissues comparing with the normal mucosa tissues (full tissue spot, p = 3.8E-37) (Figure 2D). COL5A2 was mainly increased in cancer epithelial cells comparing with the normal tissue (p = 1.6E-54). No significant change was noted in the stromal region (p = 0.847) (Figure 2D).
Figure 2.
Expression of COL5A2 protein in colon cancer tissues. Immunohistochemistry (IHC) analysis was performed using two commercial tissue microarrays (TMAs) containing a total of 180 cases. (A) The image shows the negative or low expression of COL5A2 in normal colonic mucosa tissue. The bar represents 500 µm. The boxed area is enlarged to the right side. (B) Low and (C) high expression of COL5A2 in CRC tissue. (D) A statistic of COL5A2 expression between the normal and cancer tissues based on the scanned staining intensity values.
Abbreviations: N_full, normal tissue (full tissue spot on the TMA); C_full, cancer tissue (full tissue spot on the TMA); N_str, normal tissue, stromal part; C_str, cancer tissue, stromal part; N_epi, normal tissue, epithelial part; C_epi, cancer tissue, epithelial part.
Correlation of COL5A2 mRNA and Protein Expression with CRC Clinicopathological Characteristics
By data mining of Oncomine datasets, we found that expression of COL5A2 mRNA expression significantly (p < 0.05) associates with higher Dukes stage (Jorissen Colorectal23 and Bittner Colon (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2109)), overall stage (Tsukamoto Colorectal24) and T stage (Bittner Colon and TCGC Colorectal) (Table S2). In addition, COL5A2 expression significantly associates with sex (Gaedcke Colorectal18 and Tsuji Colorectal25), recurrence (Jorissen Colorectal23), microsatellite instability (Watanabe Colon26) and KRAS status (Gaedcke Colorectal18) (Table S2).
The IHC analysis of CRC TMAs revealed that the expression of COL5A2 protein was significantly related to age (p = 0.042) and T stage (p = 0.014 for full tissue spot and p = 0.0373 for epithelial expression) (Table 1). The stromal expression of COL5A2 does not correlate to any clinicopathological parameters.
Table 1.
Correlation Between the Epithelial Expression of COL5A2 and Clinicopathological Variables in Patients with Colorectal Cancer Revealed by Immunohistochemistry Analysis of a Tissue Microarray
| Clinicopathological Parameters | Full Tissue Spot | χ2 | p | Epithelia | χ2 | p | Stroma | χ2 | p | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| < M | > M | < M | > M | < M | > M | |||||||
| Gender | ||||||||||||
| Male | 47 | 44 | 0.049 | 0.826 | 47 | 44 | 0.611 | 0.4344 | 48 | 44 | 0.3644 | 0.5461 |
| Female | 44 | 44 | 37 | 44 | 40 | 44 | ||||||
| Age | ||||||||||||
| <70 | 47 | 41 | 0.458 | 0.499 | 48 | 38 | 4.137 | 0.042 | 44 | 44 | 0.0486 | 0.8255 |
| ≥70 | 44 | 47 | 36 | 53 | 44 | 47 | ||||||
| Grade | ||||||||||||
| I | 4 | 4 | 1.437 | 0.488 | 5 | 3 | 1.748 | 0.4173 | 3 | 5 | 0.5961 | 0.7423 |
| II, I–II | 58 | 63 | 51 | 50 | 50 | 53 | ||||||
| III, I–III, II–III | 29 | 21 | 28 | 38 | 35 | 33 | ||||||
| T | ||||||||||||
| T1-T2 | 11 | 4 | 8.491 | 0.014 | 11 | 4 | 4.337 | 0.0373 | 7 | 8 | 1.331 | 0.5139 |
| T3 | 72 | 64 | 71 (T3+T4) | 86 (T3+T4) | 71 | 65 | ||||||
| T4a T4b | 7 | 18 | 10 | 15 | ||||||||
| N | ||||||||||||
| N0 | 55 | 55 | 0.192 | 0.909 | 52 | 56 | 0.052 | 0.9744 | 50 | 60 | 1.82 | 0.4025 |
| N1a N1b N1c | 26 | 25 | 23 | 26 | 29 | 22 | ||||||
| N2a N2b | 10 | 8 | 9 | 9 | 9 | 9 | ||||||
| AJCC 7th stage | ||||||||||||
| 1 | 10 | 3 | 4.432 | 0.109 | 9 | 4 | 2.894 | 0.2352 | 7 | 6 | 1.871 | 0.3924 |
| 2A 2B 2C | 44 | 51 | 41 | 52 | 43 | 52 | ||||||
| 3A 3B 3C 4A 4B | 36 | 32 | 32 | 36 | 38 | 30 | ||||||
| Tumor size | ||||||||||||
| <50 cm2 | 64 | 61 | 0.34 | 0.844 | 60 | 61 | 0.12 | 0.9417 | 62 | 63 | 0.1201 | 0.9417 |
| 50–150 cm2 | 11 | 9 | 10 | 10 | 10 | 10 | ||||||
| ≥150 cm2 | 16 | 18 | 14 | 20 | 16 | 18 | ||||||
| Positive lymph node | ||||||||||||
| 0 | 53 | 48 | 9E-04 | 0.976 | 52 | 48 | 0.292 | 0.5887 | 48 | 53 | 0.6207 | 0.4308 |
| 1~12 | 35 | 32 | 31 | 34 | 36 | 31 | ||||||
Note: The protein expression of COL5A2 was dichotomized into low and high groups using the median (M) value as the cutoff.
Both results suggest that COL5A2 expression might correlate with the carcinogenesis and development process of CRC.
Upregulation of COL5A2 mRNA and Protein Predicts Poor Prognosis of CRC
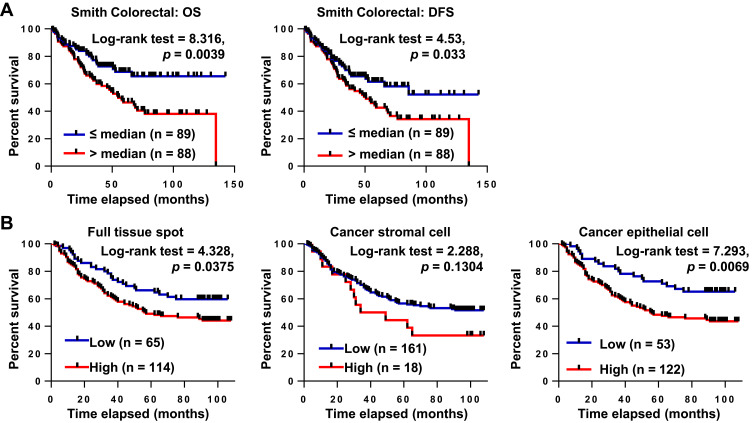
We analyzed the relationship of COL5A2 mRNA expression with CRC prognosis by reanalyzing the Oncomine Colorectal dataset GSE17536 by Smith et al.27 We concluded that high expression of COL5A2 mRNA predicted poor overall survival (OS) and disease free survival (DFS) of CRC patients (Figure 3A). In univariate Cox regression analysis, COL5A2 expression, grade, recurrence and stage are significant hazard variables for CRC DFS and OS (Table 2). By adjusting the impact for the co-variants in the multivariate analysis, COL5A2 expression (p = 0.049), relapse (p = 0.000) and stage (p = 0.041) are independent worse predictors for both DFS and OS (Table 2).
Figure 3.
COL5A2 gene and protein expressions are prognostic markers of CRC. (A) Kaplan-Meier analysis of the overall survival (OS) and disease free survival (DFS) of CRC patients based on COL5A2 gene expression using Smith Colorectal dataset (GSE17536) from Oncomine database.27 The low or high expression of COL5A2 was defined as lower-than the median (≤ median) or higher-than the median (> median). The case numbers of each group are indicated in the blankets. The p value was calculated using the Log-rank (Mantel-Cox) test. (B) COL5A2 protein expression was analysed in CRC using two commercial TMAs containing 180 CRC and paired normal tissues. The “full tissue spot”, the “stromal” region and the “epithelial” region were defined manually and the staining intensities were calculated automatically using the Positive Pixel Count algorithm v9.1 with Aperio ImageScope. The Positivity value was used to represent the expression level of COL5A2 protein. The optimal cutoff to dichotomize the cases was determined dynamically using X-title v3.6.1.17
Table 2.
Univariate and Multivariate Analyses of Clinicopathological Variables for the Disease-Free Survival and Overall Survival of CRC Patients
| Univariate | Multivariate | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | SE | Wald | Sig. | Exp (β) | 95.0% CI for Exp (β) | β | SE | Wald | Sig. | Exp (β) | 95.0% CI for Exp (β) | |||
| Lower | Upper | Lower | Upper | |||||||||||
| 1. Oncomine gene expression dataset analysisa | ||||||||||||||
| DFS | ||||||||||||||
| Age | 0.003 | 0.008 | 0.116 | 0.733 | 1.003 | 0.986 | 1.020 | |||||||
| COL5A2 mRNA | 0.475 | 0.226 | 4.432 | 0.035 | 1.608 | 1.033 | 2.503 | |||||||
| Grade | 0.692 | 0.230 | 9.062 | 0.003 | 1.997 | 1.273 | 3.133 | |||||||
| Recurrence | 2.272 | 0.293 | 60.110 | 0.000 | 9.703 | 5.463 | 17.235 | 2.113 | 0.299 | 49.839 | 0.000 | 8.276 | 4.603 | 14.882 |
| Sex | 0.051 | 0.222 | 0.052 | 0.819 | 1.052 | 0.681 | 1.626 | |||||||
| Stage | 0.985 | 0.141 | 48.820 | 0.000 | 2.677 | 2.031 | 3.528 | 0.429 | 0.197 | 4.742 | 0.029 | 1.536 | 1.044 | 2.259 |
| OS | ||||||||||||||
| Age | 0.006 | 0.009 | 0.472 | 0.492 | 1.006 | 0.988 | 1.025 | |||||||
| COL5A2 mRNA | 0.697 | 0.247 | 7.984 | 0.005 | 2.008 | 1.238 | 3.257 | 0.677 | 0.344 | 3.876 | 0.049 | 1.969 | 1.003 | 3.863 |
| Grade | 0.695 | 0.241 | 8.311 | 0.004 | 2.004 | 1.249 | 3.215 | |||||||
| Recurrence | 1.569 | 0.308 | 25.958 | 0.000 | 4.801 | 2.625 | 8.778 | 1.293 | 0.317 | 16.592 | 0.000 | 3.644 | 1.956 | 6.788 |
| Sex | 0.100 | 0.237 | 0.176 | 0.675 | 1.105 | 0.694 | 1.759 | |||||||
| Stage | 1.049 | 0.154 | 46.482 | 0.000 | 2.854 | 2.111 | 3.859 | 0.448 | 0.219 | 4.180 | 0.041 | 1.565 | 1.019 | 2.404 |
| 2. Immunohistochemistry of a TMAb | ||||||||||||||
| OS | ||||||||||||||
| Age | 0.520 | 0.240 | 4.706 | 0.030 | 1.682 | 1.051 | 2.689 | 0.663 | 0.252 | 6.912 | 0.009 | 1.94 | 1.184 | 3.181 |
| Sex | −0.068 | 0.212 | 0.102 | 0.750 | 0.935 | 0.617 | 1.416 | |||||||
| Positive lymph node | 0.940 | 0.213 | 19.406 | 0.000 | 2.560 | 1.685 | 3.888 | |||||||
| T | 0.429 | 0.189 | 5.160 | 0.023 | 1.535 | 1.061 | 2.222 | |||||||
| N | 0.833 | 0.143 | 33.770 | 0.000 | 2.301 | 1.737 | 3.048 | 0.964 | 0.157 | 37.850 | 0.000 | 2.623 | 1.929 | 3.566 |
| M | 1.415 | 0.590 | 5.749 | 0.016 | 4.115 | 1.295 | 13.077 | |||||||
| AJCC 7th staging | 0.901 | 0.172 | 27.368 | 0.000 | 2.462 | 1.756 | 3.450 | |||||||
| Grade | 0.864 | 0.274 | 9.940 | 0.002 | 2.373 | 1.387 | 4.062 | 0.583 | 0.247 | 5.565 | 0.018 | 1.791 | 1.104 | 2.905 |
| Tumor size | 0.555 | 0.234 | 5.643 | 0.018 | 1.742 | 1.102 | 2.755 | 0.578 | 0.245 | 5.577 | 0.018 | 1.782 | 1.103 | 2.879 |
| Epithelial COL5A2 | 0.687 | 0.260 | 6.962 | 0.008 | 1.987 | 1.193 | 3.309 | 0.574 | 0.262 | 4.792 | 0.029 | 1.776 | 1.062 | 2.969 |
Notes: aThe analysis was performed using the Smith Colorectal dataset from the Oncomine cancer gene expression microarray DB (https://www.oncomine.org/resource/login.html). bThe analysis was based on the immunohistochemistry assay of a tissue microarray (HCol-Ade180Sur-04) containing 90 pairs of CRC and matched normal tissues.
Abbreviations: OS, overall survival; DFS, disease-free survival; b, beta coefficients; SE, standard error for each parameter estimate; Wald, Wald statistic; Sig., significance; Exp, the exponentiated parameter estimates; CI, confidence interval.
By IHC analysis, we found that the high expression of COL5A2 significantly correlated with survival of CRC patients (Log rank test, p = 0.0375 for full tissue spot analysis and p = 0.0069 for epithelial region analysis) (Figure 3B). However, stromal expression of COL5A2 has no significant relationship with prognosis (p = 0.1304). Univariate analysis reveals that the epithelial expression of COL5A2, age, positive lymph node, T/N/M stage, grade, tumor size and American Joint Committee on Cancer (AJCC) staging are the significant risk factors of CRC prognosis (Table 2). The multivariate analysis demonstrates that the epithelial expression of COL5A2 (p = 0.029), age, N stage, grade and tumor size are the independent danger factors of CRC outcome (Table 2).
The above analyses suggest that both COL5A2 mRNA and protein expression are independent risk factors of CRC survival after surgery.
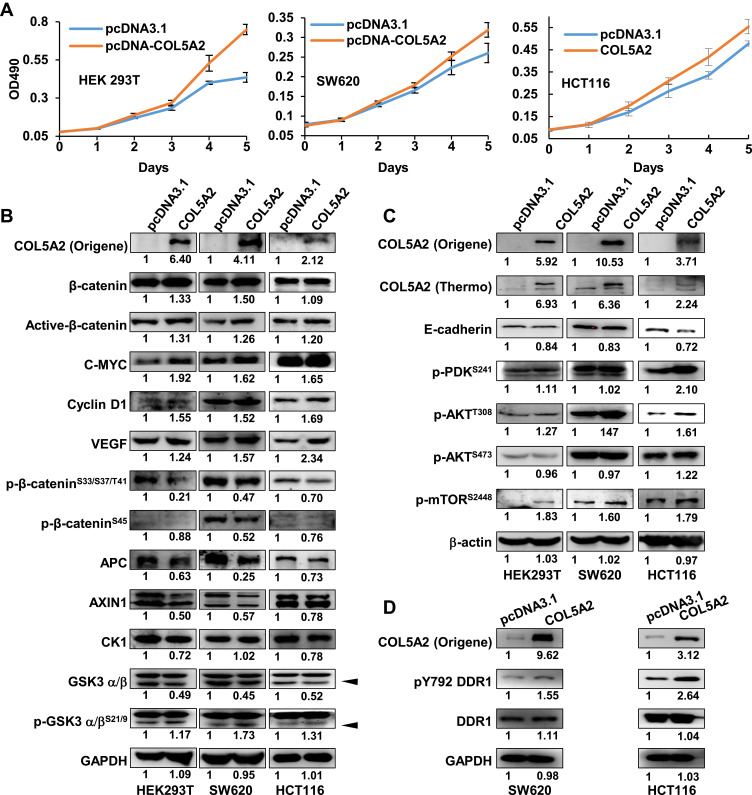
COL5A2 Promotes Cancer Cell Proliferation and Activates PI3K/mTOR Signaling
To address the function of COL5A2 gene, we cloned the full-length gene of COL5A2 and performed overexpression analysis. COL5A2 expression vector and blank vector were transfected into HEK293T, colon cancer SW620 and HCT116 cells, and cell growth was measured using the MTT method. Interestingly, ectopic expression of COL5A2 promoted the proliferation of HEK293T, SW620 and HCT116 cells (Figure 4A). Then we measured the WNT/β-catenin and PI3K/AKT/mTOR phosphorylation signaling pathway that is commonly activated and involved in colon cancer cell carcinogenesis and progression. We observed that the active form of β-catenin, as well as its downstream targets C-MYC, Cyclin D1 and VEGF, were slightly upregulated by COL5A2 overexpression (Figure 4B). Statistics of the changes across the three biological replicates were shown in Figure S1A. The stability of β-catenin was known to be controlled by its N-terminal phosphorylation and subsequent ubiquitination and proteasome-dependent degradation.28 We found that the phosphorylation of the four N-terminal residues of β-catenin was slightly decreased in both cell lines after COL5A2 transfection, which was accompanied with a reduction of APC and AXIN1, two components of its destruction complex. Whereas the increase of phosphorylation of GSK3β might attenuate its kinase activity for β-catenin phosphorylation. Besides the protein and phosphorylation changes, we also analyzed the mRNA expression of these pathway-related molecules. The downstream molecules of WNT/β-catenin signaling, C-MYC and VEGF, were consistently upregulated across the three cell lines after COL5A2 overexpression (Figure S2). CTNNB1 (β-catenin) was increased, suggesting it was regulated at both post-translational and transcriptional level by COL5A2 expression. The gene expression of AXIN1 was downregulated by COL5A2 expression in all three cell lines, while APC was downregulated in 293T and SW620 cell lines (Figure S2).
Figure 4.
Ectopic expression of COL5A2 promotes cancer cell proliferation and upregulates WNT/β-catenin and PI3K/mTOR pathway. (A) COL5A2 was ectopically expressed in HEK293T and colon cancer cell lines SW620 and HCT116 and the cell variability was measured using MTT method in different days. (B and C) Ectopic expression of COL5A2 and selected signaling molecules were measured by Western blot. (D) Western blot analyses of DDR1 and pY792 DDR1 in COL5A2 overexpressing CRC cells. The Western blot densitometry was calculated using Multi Gauge V3.0 and the fold change was normalized using the blank vector transfection. COL5A2 protein was probed using two antibodies. Active β-catenin was the endogenous β-catenin protein when residues Ser33, Ser37, and Thr41 are not phosphorylated. The arrow heads indicate the positions of GSK3β.
We also found that p-AKTT308 and p-mTORS2448 were increased by the ectopic expression of COL5A2, while p-AKTS473 and p-PDK1S241 remained unchanged (Figures 4C and S1B). Interestingly, mTOR mRNA expression was significantly increased by COL5A2 expression across all three cell lines (Figure S2). E-cadherin was consistently upregulated at both protein and mRNA levels by COL5A2 expression (Figures 4C and S2).
In the current study, DDR1 has been identified as a cancer-associated receptor tyrosine kinase that is highly expressed in several malignancies relative to normal tissues.29 As an important receptor, DDR1 can bind all types of collagens and could influence their signalling upon collagen stimulation.30 It has been reported that the phosphorylation of DDR1 could activate the WNT/β-catenin and AKT/mTOR signaling to drive cancer cell metastasis and progression.31,32 We observed that pY792 DDR1, the active form of DDR1, was upregulated by COL5A2 overexpression in HCT116 and SW620 cells (Figure 4D).
These observations indicate that COL5A2 expression upregulates WNT/β-catenin and PI3K/mTOR signaling via binding DDR1 with phosphorylation and activation of this receptor, which might contribute to CRC tumorigenesis or progression.
Discussion
Collagens are commonly recognized as extracellular secreted proteins and implicated in cancerous transformation and progression. COL5A2 is previously identified to associate with Ehlers-Danlos syndrome and autoimmune diseases10,33 and it also upregulated in human cancers.11,12,34 Upregulation of COL5A2 mRNA is associated with increased multiple drug resistance in ovarian cancer cell lines.13 These observations suggest it might involve in neoplastic transformation. However, no study has been performed to address its potential function in cancer.
In current study, COL5A2 mRNA was found to be upregulated in cancer comparing with normal tissues in 87.5% CRC and 100% GC datasets in Oncomine database with available normal control data. COL5A2 mRNA upregulation was observed in almost all subtypes of both cancers. COL5A2 protein was also significantly increased in CRC tissues by IHC analysis, especially in the cancer epithelial cells. These observations, as well as previous reports,11,15,34 suggest that COL5A2 upregulation is common in cancer and it might associate with carcinogenesis and cancer progressing. By meta-analysis, we found that COL5A2 mRNA expression was associated with CRC clinicopathological variables like stages, gender, recurrence, microsatellite instability and KRAS status, while COL5A2 protein expression in cancer epithelial cell was related to age and T stage. The discrepancy between the mRNA and protein analysis may be explained by the difference of the clinicopathological parameters recorded in different cohort and the difference between the genetic features of gene and protein expression.35 Nevertheless, these results indicate that COL5A2 expression is correlated with the carcinogenesis and development process of CRC.
Further analyses reveal that high mRNA expression and the epithelial expression of COL5A2 are associated with poor prognosis of CRC, and both are independent risk factors of CRC outcome. Therefore, high mRNA or epithelial protein expression of COL5A2 might serve as a worse prognostic marker of CRC. Whereas another collagen molecule, COL6A3, is a typical stromal marker for worse prognosis of CRC, as we have addressed previously.3
Interestingly, COL5A2 protein was found to be expressed in the cytosol and nucleus of partial cancer samples by IHC, and immunofluorescence analysis indicated that COL5A2 could be detected in the nucleus of CRC cell lines SW620, HCT116 and SW480 (Figure S3). The nuclear localization of COL5A2 in CRC cells was strengthened by a proteomics report by Albrethsen J.36 These observations suggest that COL5A2 might involve in intracellular or nuclear processes. As a verification, we found that the ectopic expression of COL5A2 gene accelerated colon cancer cell proliferation. These findings indicate that some collagens might function as intracellular cancer drivers, instead of as the extracellular matrix proteins, which have been traditionally recognized.
Conclusion
Our study demonstrates that COL5A2 mRNA and protein are significantly upregulated in CRC. COL5A2 promotes cancer cell proliferation via binding DDR1 and the activation of WNT/β-catenin and AKT/mTOR signaling pathway. COL5A2 is the independent prognostic marker and a potential drug target of colon cancer.
Acknowledgments
This work was funded by the Chinese National Key Program on Basic Research (973) Grant (2011CB910702, 2013CB911202); National Natural Science Foundation of China (81572833); Natural Science Foundation of Shanghai (14ZR1402100); Science and Technology Development Foundation of Pudong New District, Shanghai, China (PKJ2015-S29).
Disclosure
The authors reported no conflicts of interest for this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Chen SX, Xu XE, Wang XQ, et al. Identification of colonic fibroblast secretomes reveals secretory factors regulating colon cancer cell proliferation. J Proteomics. 2014;110:155–171. doi: 10.1016/j.jprot.2014.07.031 [DOI] [PubMed] [Google Scholar]
- 3.Qiao J, Fang CY, Chen SX, et al. Stroma derived COL6A3 is a potential prognosis marker of colorectal carcinoma revealed by quantitative proteomics. Oncotarget. 2015;6(30):29929–29946. doi: 10.18632/oncotarget.4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Q, Wang XQ, Wang J, et al. Upregulation of spondin-2 predicts poor survival of colorectal carcinoma patients. Oncotarget. 2015;6(17):15095–15110. doi: 10.18632/oncotarget.3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang XQ, Tang ZX, Yu D, et al. Epithelial but not stromal expression of collagen alpha-1(III) is a diagnostic and prognostic indicator of colorectal carcinoma. Oncotarget. 2016;7(8):8823–8838. doi: 10.18632/oncotarget.6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang M, Yuan J, Peng C, Li Y. Collagen as a double-edged sword in tumor progression. Tumour Biol. 2014;35(4):2871–2882. doi: 10.1007/s13277-013-1511-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.JingSong H, Hong G, Yang J, et al. siRNA-mediated suppression of collagen type iv alpha 2 (COL4A2) mRNA inhibits triple-negative breast cancer cell proliferation and migration. Oncotarget. 2017;8(2):2585–2593. doi: 10.18632/oncotarget.13716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang G, Ge G, Izzi V, Greenspan DS. alpha3 Chains of type V collagen regulate breast tumour growth via glypican-1. Nat Commun. 2017;8(1):14351. doi: 10.1038/ncomms14351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortini F, Villa C, Marinelli B, et al. Ehlers-Danlos Syndrome classical type: a novel COL5A2 missense mutation with possible additive effect of a COL5A1 stop-gain mutation in a strongly correlated phenotype. Meta Gene. 2018;18:132–136. doi: 10.1016/j.mgene.2018.08.012 [DOI] [Google Scholar]
- 10.Mak AC, Tang PL, Cleveland C, et al. Brief report: whole-exome sequencing for identification of potential causal variants for diffuse cutaneous systemic sclerosis. Arthritis Rheumatol. 2016;68(9):2257–2262. doi: 10.1002/art.39721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwong KY, Bloom GC, Yang I, et al. Synchronous global assessment of gene and protein expression in colorectal cancer progression. Genomics. 2005;86(2):142–158. doi: 10.1016/j.ygeno.2005.03.012 [DOI] [PubMed] [Google Scholar]
- 12.Wu D, Chen K, Bai Y, et al. Screening of diagnostic markers for osteosarcoma. Mol Med Rep. 2014;10(5):2415–2420. doi: 10.3892/mmr.2014.2546 [DOI] [PubMed] [Google Scholar]
- 13.Januchowski R, Swierczewska M, Sterzynska K, Wojtowicz K, Nowicki M, Zabel M. Increased expression of several collagen genes is associated with drug resistance in ovarian cancer cell lines. J Cancer. 2016;7(10):1295–1310. doi: 10.7150/jca.15371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng XY, Shi MJ, Zeng ZH, et al. The role of COL5A2 in patients with muscle-invasive bladder cancer: a bioinformatics analysis of public datasets involving 787 subjects and 29 cell lines. Front Oncol. 2019;8. doi: 10.3389/fonc.2018.00659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou X, Feng B, Dong T, et al. Up-regulation of type I collagen during tumorigenesis of colorectal cancer revealed by quantitative proteomic analysis. J Proteomics. 2013;94:473–485. doi: 10.1016/j.jprot.2013.10.020 [DOI] [PubMed] [Google Scholar]
- 16.Safronetz D, Prescott J, Feldmann F, et al. Pathophysiology of hantavirus pulmonary syndrome in rhesus macaques. Proc Natl Acad Sci U S A. 2014;111(19):7114–7119. doi: 10.1073/pnas.1401998111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713 [DOI] [PubMed] [Google Scholar]
- 18.Gaedcke J, Grade M, Jung K, et al. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosomes Cancer. 2010;49(11):1024–1034. doi: 10.1002/gcc.20811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis. 2010;27(2):83–90. doi: 10.1007/s10585-010-9305-4 [DOI] [PubMed] [Google Scholar]
- 20.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61(7):3124–3130. [PubMed] [Google Scholar]
- 21.Skrzypczak M, Goryca K, Rubel T, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5(10). doi: 10.1371/annotation/8c585739-a354-4fc9-a7d0-d5ae26fa06ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser S, Park YK, Franklin JL, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8(7):R131. doi: 10.1186/gb-2007-8-7-r131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan L, Zhan C, Wang S, Wang S, Guo L. Genetic analysis of radiation-specific biomarkers in sinonasal squamous cell carcinomas. Tumour Biol. 2016;37(9):12001–12009. doi: 10.1007/s13277-016-5057-3 [DOI] [PubMed] [Google Scholar]
- 24.Tsukamoto S, Ishikawa T, Iida S, et al. Clinical significance of osteoprotegerin expression in human colorectal cancer. Clin Cancer Res. 2011;17(8):2444–2450. doi: 10.1158/1078-0432.CCR-10-2884 [DOI] [PubMed] [Google Scholar]
- 25.Tsuji S, Midorikawa Y, Takahashi T, et al. Potential responders to FOLFOX therapy for colorectal cancer by Random Forests analysis. Br J Cancer. 2012;106(1):126–132. doi: 10.1038/bjc.2011.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe T, Kobunai T, Toda E, et al. Distal colorectal cancers with microsatellite instability (MSI) display distinct gene expression profiles that are different from proximal MSI cancers. Cancer Res. 2006;66(20):9804–9808. doi: 10.1158/0008-5472.CAN-06-1163 [DOI] [PubMed] [Google Scholar]
- 27.Smith JJ, Deane NG, Wu F, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138(3):958–968. doi: 10.1053/j.gastro.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rammal H, Saby C, Magnien K, et al. Discoidin domain receptors: potential actors and targets in cancer. Front Pharmacol. 2016;7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao Y, Wang R, Lai Q, et al. Targeting of DDR1 with antibody-drug conjugates has antitumor effects in a mouse model of colon carcinoma. Mol Oncol. 2019;13(9):1855–1873. doi: 10.1002/1878-0261.12520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeitany M, Leroy C, Tosti P, et al. Inhibition of DDR1-BCR signalling by nilotinib as a new therapeutic strategy for metastatic colorectal cancer. EMBO Mol Med. 2018;10(4). doi: 10.15252/emmm.201707918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chappell WH, Candido S, Abrams SL, et al. Influences of TP53 and the anti-aging DDR1 receptor in controlling Raf/MEK/ERK and PI3K/Akt expression and chemotherapeutic drug sensitivity in prostate cancer cell lines. Aging (Albany NY). 2020;12(11):10194–10210. doi: 10.18632/aging.103377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park AC, Phillips CL, Pfeiffer FM, et al. Homozygosity and heterozygosity for null Col5a2 alleles produce embryonic lethality and a novel classic Ehlers-Danlos syndrome-related phenotype. Am J Pathol. 2015;185(7):2000–2011. doi: 10.1016/j.ajpath.2015.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer H, Stenling R, Rubio C, Lindblom A. Colorectal carcinogenesis is associated with stromal expression of COL11A1 and COL5A2. Carcinogenesis. 2001;22(6):875–878. doi: 10.1093/carcin/22.6.875 [DOI] [PubMed] [Google Scholar]
- 35.Wilhelm M, Schlegl J, Hahne H, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509(7502):582–587. doi: 10.1038/nature13319 [DOI] [PubMed] [Google Scholar]
- 36.Albrethsen J, Knol JC, Piersma SR, et al. Subnuclear proteomics in colorectal cancer: identification of proteins enriched in the nuclear matrix fraction and regulation in adenoma to carcinoma progression. Mol Cell Proteomics. 2010;9(5):988. doi: 10.1074/mcp.M900546-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]