Abstract
Objectives
In December 2020, Italy began a national immunization campaign using the BNT162b2 coronavirus disease 2019 (COVID-19) mRNA vaccine, prioritizing healthcare workers (HCWs). Immune serum from vaccinated subjects seems (largely) to retain titres of neutralizing antibodies, even against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) VOC 202012/01-lineage B.1.1.7. Here, we describe an outbreak of SARS-CoV-2 lineage B.1.1.7 infection in three HCWs in a hospital setting; two of the HCWs were fully vaccinated (i.e. had received two doses).
Methods
Two physicians and one nurse working on the same shift on 20th February 2021 were involved in the outbreak. Real-time PCR, antigen tests, and serological tests for the IgG anti-spike protein of SARS-CoV-2 were performed, along with whole-genome sequencing (WGS).
Results
SARS-CoV-2 infection was confirmed in all three HCWs; all presented with mild symptoms of COVID-19. The two physicians were fully vaccinated with BNT162b2 vaccine, with the second dose administered 1 month before symptom onset. Both had high titres of IgG anti-spike antibodies at the time of diagnosis. WGS confirmed that all virus strains were VOC 202012/01-lineage B.1.1.7, suggesting a common source of exposure. Epidemiological investigation revealed that the suspected source was a SARS-CoV-2-positive patient who required endotracheal intubation due to severe COVID-19. All procedures were carried out using a full suite of personal protective equipment (PPE).
Conclusions
This mini-outbreak highlights some important issues about the efficacy of vaccines against transmission of SARS-CoV-2 variants, the high risk of exposure among HCWs, and the need for optimized implementation of PPE in hospitals. The wide circulation of VOC 202012/01 in Europe and Italy highlights the need to improve surveillance and genetic sequencing.
Keywords: COVID-19, Healthcare workers, Outbreak, SARS-CoV-2 VOC 202012/01-LINEAGE B.1.1.7, Vaccine, Whole-genome sequencing
Introduction
In December 2020, Italy began a national immunization campaign, prioritizing healthcare workers (HCWs) [1]. Initially, the campaign used the BNT162b2 coronavirus disease 2019 (COVID-19) mRNA vaccine. Clinical trial data demonstrate that two doses of the BNT162b2 vaccine confer 95% protection against COVID-19 caused by wild-type severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [2]. A recent study showed that immune serum from subjects vaccinated with BNT162b2 largely retains titres of neutralizing antibodies against SARS-CoV-2 VOC 202012/01-lineage B.1.1.7 [3], despite a lack of specific effectiveness estimates for the B.1.1.7 variant. SARS-CoV-2 VOC 202012/01 has been circulating in Italy since December 2020 [4]. A flash survey conducted in Italy in February 2021 suggested that the prevalence of this variant was 54.0%, i.e., three-fold higher than that reported 2 weeks previously (17.8%) [5].
Here, we describe an outbreak of SARS-CoV-2 VOC 202012/01-lineage B.1.1.7 infection in three HCWs in a hospital setting in Italy; two of these HCWs were fully vaccinated and working in the intensive care unit of a COVID-19 hospital.
Patients and methods
Two physicians and one nurse working on the same shift on 20th February 2021 were involved in the outbreak. The clinical presentation of the HCWs was classified according to the National Institute of Health (NIH) clinical staging of COVID-19 disease [6]. Nasopharyngeal swabs were collected from the three HCWs after symptom onset and were processed at the Laboratory of Molecular Epidemiology and Public Health of the Hygiene Unit (A.O.U.C. Policlinico Bari), which is the coordinator of the Regional Laboratory Network for SARS-CoV-2 diagnosis in the Apulia region. Samples were subjected to molecular test using a three-target commercial multiplex real-time PCR assay targeting the N, ORF1ab, and S genes (TaqPath RT-PCR COVID-19 Assay; Thermo Fisher Scientific). An antigen test was also performed using the Lumipulse SARS-CoV-2 Ag kit (Fujirebio, Tokyo, Japan). Moreover, to assess responses to vaccination, serological tests for IgG antibodies specific for the spike (S) protein of SARS-CoV-2 were performed (SARS-CoV-2 IgG II Quant assay; Abbott Architect iSystem, Abbott Diagnostics, Chicago, USA). Whole-genome sequencing (WGS) was performed using the Ion Torrent platform (Thermo Fisher Scientific). The aligned reads were used for both reference-guided assembly and variant calling. Assembly was performed using the Iterative Refinement Meta-Assembler (IRMA) v.1.3.0.2.
Ethical approval
Approval by a research ethics committee was not required since the activities described here were conducted as part of the national and regional COVID-19 surveillance effort. All procedures were carried out in accordance with the Declaration of Helsinki, as revised in 2013, for research on human subjects. Informed written consent for publication was obtained from the subjects who provided samples.
Results
The first case was a healthy 41-year-old male physician. The date of symptom onset (conjunctivitis) was 23rd February 2021. On 25th February he presented with strong asthenia, low-grade fever (Tmax 37.2°C), and coryza. On the following day, he developed anosmia and ageusia. A molecular test of a nasopharyngeal swab collected on 27th February was positive for SARS-CoV-2. The second case was a healthy 34-year-old female physician who presented with a cough on 28th February. The diagnosis was confirmed by a molecular test on a nasopharyngeal swab collected on 2nd March. Serological tests were performed on both patients on the day of diagnosis. Antibody titres in the male and female patients were 1774 AU/mL and 2500 AU/mL, respectively. The third case was a 51-year-old male nurse who developed a fever (Tmax 38°C) on 25th February; there were no respiratory symptoms. The diagnosis was confirmed on 27th February. This case showed high-grade fever for 8 days from symptom onset. The clinical presentation of the three cases was classified as mild (Table 1 ).
Table 1.
Laboratory and vaccination data of the healthcare workers involved in the outbreak
| Case | Date of sample collection | Clinical presentation | Antigenic test (pg/mL) | Real-time PCR (Ct) |
Vaccine (BNT162b2 mRNA COVID-19 vaccine) |
|||
|---|---|---|---|---|---|---|---|---|
| N gene | ORF1ab gene | S gene | Dose I | Dose II | ||||
| 1 | 27th Feb 2021 | Mild | >5000 | +(17) | +(17) | — | 12/31/2020 | 01/24/2021 |
| 2 | 2nd Mar 2021 | Mild | >5000 | +(21) | +(22) | — | 01/01/2021 | 01/24/2021 |
| 3 | 27th Feb 2021 | Mild | >5000 | +(15) | +(16) | — | — | — |
Ct, cycle threshold.
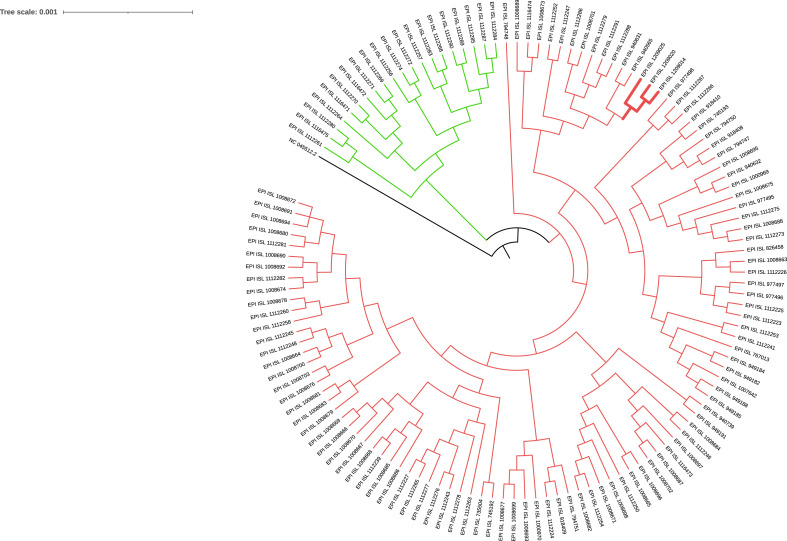
The two physicians had been fully vaccinated with the BNT162b2 COVID-19 mRNA vaccine, having received two doses; the second dose was administered 21 days after the first, in accordance with the recommended schedule. The second dose was administered 1 month before symptom onset (Table 1). The nurse refused the vaccination. The multiplex real-time PCR assay performed on nasopharyngeal swabs from all three HCWs identified an S gene dropout, a good proxy for VOC 202012/01 (Table 1). All three samples tested positive in the antigen test. WGS confirmed that all strains were VOC 202012/01-lineage B.1.1.7, suggesting a common source of exposure (Fig. 1 ). Epidemiological investigation revealed that it is likely that the three HCWs were infected by exposure to aerosols while conducting an invasive procedure on a patient with COVID-19. No other risk factors or exposure to SARS-CoV-2 were reported. The suspected source was a 50-year-old male patient with a confirmed diagnosis of SARS-CoV-2 infection who presented at the emergency department with respiratory failure and pulmonary oedema; the severity of his condition required endotracheal intubation. The patient died 2 h later. No post-mortem samples were available for molecular characterization of SARS-CoV-2. All procedures were carried out using a full set of personal protective equipment (PPE): particulate filter respirators (P3), two pairs of gloves, face shields, and a single-use coverall. No eye glasses were used under the face shields.
Fig. 1.
Phylogenetic tree of 127 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) full genome sequences from Apulia, including the three genomes identified in this study. The green clade includes genomes assigned to the B.1.177 lineage (dubbed ‘the Spanish lineage’), while the red clade includes all Apulian genomes assigned to the B.1.1.7 lineage (VOC 202012/01). The red subclade in bold includes the three genomes examined in this study. The reference SARS-CoV-2 genome from Wuhan (AC:NC_045512.2) was included to root the tree. Multiple alignment of the sequences was performed using Multiple Alignment using Fast Fourier Transform (MAFFT, v7.475), with the following options: --6merpair, --keeplength, --addfragments. The resulting multiple alignment was used to build the phylogenetic tree in FastTreeMP v2.1.10, with the -gamma option.
Discussion
This mini outbreak highlights some important issues about the efficacy of vaccines against transmission of SARS-CoV-2 variants, the higher risk of exposure among HCWs, and the need for optimized implementation of PPE in hospitals. Data on the efficacy of BNT162b2 were confirmed after a mass vaccination campaign in Israel, in which the estimated efficacy at ≥7 days after the second dose was 92% for preventing documented infection, and 94% for preventing symptomatic COVID-19 infection; these data were robust, despite wide circulation of VOC 202012/01 [7]. However, our findings suggest that the increased transmissibility of this variant raises concerns about such strains causing possible symptomatic post-vaccination infections. It is hypothesized that SARS-CoV-2 variants are capable of at least partially evading vaccine-induced immunity, and that they replicate more efficiently [8]; in fact, all vaccinated cases described herein had a high viral load (Ct < 21). This is worrisome because they may also be a possible source of infection. The high antibody titres found in the two HCWs on the day of diagnosis suggests a vaccine-induced immune response rather than a natural response to SARS-CoV-2. Despite the presence of anti-spike IgG antibodies, the two individuals developed a symptomatic infection; this may not be as rare as once thought [7]. In vaccinated patients, a breakthrough infection must be differentiated from vaccine-associated enhancement disease (VAED) [9]. VAED is defined as a modified clinical presentation in individuals exposed to a pathogen after having received a prior vaccination against the same pathogen [10]. Assessment of this adverse event following immunization is of particular interest, especially in the context of SARS-CoV-2 vaccine development, given the urgent global need for safe and effective vaccines. In the two cases here described, a breakthrough infection could be hypothesized since the clinical presentation was mild, and no biomarkers suggestive of VAED were identified.
Vaccines are a crucial long-term strategy for controlling SARS-CoV-2; as such, vaccination of HCWs should be mandatory because this population is at high risk of infection. Frequent and comprehensive routine screening of HCWs, even if vaccinated, will help to identify asymptomatic carriage and protect patients and hospital staff. The wide circulation of VOC 202012/01 in Europe, coupled with its high transmissibility, highlight the need for preventive measures in hospital settings. Finally, improved surveillance and a more coordinated global effort regarding genetic sequencing are mandatory to investigate outbreaks and to monitor the spread and circulation of all possible VOCs.
Author contributions
MC designed the study. DL drafted the manuscript. AL and ASang collected data. ASal and MA performed experiments and data curation. MC and AP supervised the study and revised the manuscript. All authors read and approved the final version of the manuscript.
Transparency declaration
The authors declare that they have no conflicts of interest. No external funding was received for this study.
Editor: L. Leibovici
References
- 1.European Centre for Disease Prevention and Control (ECDC) 2020. COVID-19 vaccination and prioritisation strategies in the EU/EEA.https://www.ecdc.europa.eu/sites/default/files/documents/COVID-19-vaccination-and-prioritisation-strategies.pdf Available from: [Google Scholar]
- 2.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muik A., Wallisch A.K., Sänger B., Swanson K.A., Mühl J., Chen W. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loconsole D., Sallustio A., Accogli M., Centrone F., Capozzi L., Del Sambro L. Genome sequence of a SARS-CoV-2 VUI 202012/01 strain identified from a patient returning from London, England, to the Apulia Region of Italy. Microbiol Resour Announc. 2021;10 doi: 10.1128/MRA.01487-20. e01487-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Istituto Superiore di Sanita' (ISS) 2021. Prevalenza delle varianti VOC 202012/01 (lineage B.1.1.7), P.1, e 501.V2 (lineage B.1.351) in Italia Indagine del.https://www.iss.it/documents/20126/0/Relazione+tecnica+terza+indagine+flash+per+le+varianti+del+virus+SARS-CoV-2+%282%29.pdf?fbclid=IwAR2yoTbzwM-fU8K2KIwpM4Gnk76qTHD4zihFwEf8zYF1nwsG8T0tNpRlVIo Available from: [Google Scholar]
- 6.National Institute of Health (NIH) Coronavirus disease 2019 (COVID-19) treatment guidelines. https://files.covid19treatmentguidelines.nih.gov/guidelines/covid19treatmentguidelines.pdf Available from: [PubMed]
- 7.Dagan N., Barda N., Kepten E., Miron O., Perchik S., Katz M.A. BNT162b2 mRNA Covid-19 vaccine in a nationwide mass vaccination setting. N Engl J Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kidd M., Richter A., Best A., Cumley N., Mirza J., Percival B. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-qPCR. J Infect Dis. 2021:jiab082. doi: 10.1093/infdis/jiab082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munoz F.M., Cramer J.P., Dekker C.L., Dudley M.Z., Graham B.S., Gurwith M. Vaccine-associated enhanced disease: case definition and guidelines for data collection, analysis, and presentation of immunization safety data. Vaccine. 2021;S0264–410X doi: 10.1016/j.vaccine.2021.01.055. 00094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham B.S. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]