Abstract
YAP1 is a transcriptional co-activator whose activity is controlled by the Hippo signaling pathway. In addition to important functions in normal tissue homeostasis and regeneration, YAP1 has also prominent functions in cancer initiation, aggressiveness, metastasis, and therapy resistance. In this review we are discussing the molecular functions of YAP1 and its roles in cancer, with a focus on the different mechanisms of de-regulation of YAP1 activity in human cancers, including inactivation of upstream Hippo pathway tumor suppressors, regulation by intersecting pathways, miRNAs, and viral oncogenes. We are also discussing new findings on the function and biology of the recently identified family of YAP1 gene fusions, that constitute a new type of activating mutation of YAP1 and that are the likely oncogenic drivers in several subtypes of human cancers. Lastly, we also discuss different strategies of therapeutic inhibition of YAP1 functions.
Keywords: YAP1, Gene fusions, YAP1 fusion, Cancer, Hippo signaling pathway
Introduction
The Hippo Signaling Pathway is a key regulator of cell growth, tissue homeostasis, and organ size. It can be divided into a tumor suppressive upstream core kinase cascade (including the serine/threonine kinases MST1/2 and LATS1/2) that phosphorylates and inhibits the activity of the downstream effector proteins YAP1 (also known as YAP) and TAZ (also known as WWTR1), two transcriptional activators that interact with a series of transcription factors, most notably TEADs. While it is believed that YAP1 and TAZ have similar functions and are both implicated in development and cancer, since the majority of research efforts in this area concentrated on YAP1, this review will predominantly focus on YAP1.
YAP1 is a potent transcriptional co-activator that has essential functions in development, stem cell maintenance, normal tissue homeostasis and regeneration. Deregulation and constitutive activation of YAP1 is implicated in cancer initiation, progression, invasion, and therapy resistance, functions that are mostly attributed to a pro-survival and pro-proliferative transcriptional program elicited via its interaction with TEAD transcription factors. Elevated nuclear YAP1 staining can be detected in many cancers. The causes responsible for YAP1 activation in these tumors are versatile and include loss-of-function mutations in core Hippo pathway upstream regulators, such as LATS1/2, MST1/2, NF2, or mutations in genes encoding proteins that can impact the core Hippo signaling pathway, such as G protein-coupled receptors or viral proteins. By contrast, activating point mutations in the YAP1 coding sequence are rare. Recent whole genome sequencing studies have identified several recurrent YAP1 gene fusion events in various cancer types. These YAP1 gene fusions are oncogenic when expressed in mice and constitute the likely tumor-initiating events and oncogenic drivers in the cancers in which they are found.
In this review we will highlight the oncogenic functions of YAP1 and discuss the different mechanisms responsible for de-regulation of YAP1 activity in human cancers with a specific emphasis on the recently discovered role and mechanisms of YAP1 fusion proteins.
Linking YAP1 to the Hippo signaling pathway
YAP1 was initially identified as an interaction partner of the SRC family member YES, hence its name YES-associated protein 1 (Sudol 1994; Sudol et al. 1995b). Subsequently, YAP1 was identified as a transcriptional co-activator that does not directly bind DNA itself but rather functions via the interaction with several transcription factors (Yagi et al. 1999; Strano et al. 2001; Ferrigno et al. 2002; Komuro et al. 2003; Omerovic et al. 2004). At that time, the role of YAP1 as the principle effector of the Hippo signaling pathway remained unknown. Independently from these findings, the function of the Hippo signaling pathway, as well as its role in regulating organ size during development in Drosophila melanogaster was unraveled by a series of studies that identified the critical members of its core kinase cascade Wts, Hpo, Salvador/Sav, and Mats. These works demonstrated that these genes were working in the same signaling pathway and mutations in these genes resulted in organ overgrowth and a “big-headed” phenotype (resembling Hippos) (Justice et al. 1995; Xu et al. 1995; Tapon et al. 2002; Harvey et al. 2003; Jia et al. 2003; Udan et al. 2003; Wu et al. 2003). The newly identified pathway negatively regulating the growth of Drosophila organs during normal development was named Hippo signaling pathway. Further studies showed that the core Hippo pathway genes are highly conserved between Drosophila and mammals and the expression of LATS1 (mammalian orthologue of Drosophila Wts) was able to rescue the phenotype of Drosophila wts mutants (Tao et al. 1999). However, the actual effector protein of the Hippo pathway remained unknown until Huang and co-authors identified YAP1 and its Drosophila orthologue Yorkie as the principle effectors of the Hippo signaling pathway and demonstrated that YAP1 is inactivated by the Hippo pathway core kinase cascade via direct phosphorylation by LATS/Wts (Huang et al. 2005). Overexpression of Yorkie during fly development lead to tissue overgrowth that resembled the phenotypes observed by inactivation of the Hippo core kinase members Hpo, Sav, or Wts, whereas deletion of Yorkie in eye disc cells lead to smaller eyes. Interestingly, overexpression of human YAP1 was able to rescue the effect of Drosophila Hippo pathway hyperactivation, suggesting a strong evolutionary conservation and importance of the Hippo signaling (Huang et al. 2005).
YAP1 functions in normal tissues and cancer
YAP1 has important functions in both normal tissue development, homeostasis, and regeneration. Drosophila melanogaster Yorkie (yki) null mutants are homozygous lethal and die as late embryos and early first instar larvae; moreover, targeted deletion of Yorkie in eye disc cells results in a significantly reduced organ size (Huang et al. 2005). Similarly, homozygous deletion of Yap1 in mice is also embryonically lethal, as early as E8.5, due to severe developmental defects (Morin-Kensicki et al. 2006). Conversely, deletion of the Hippo pathway tumor suppressors Stk3/4 (also known as Mst1/2) and Sav1 (also known as Ww45) leads to hypoplasia, immature differentiation and embryonic death, whereas targeted deletion of these genes in several organs leads to organ overgrowth due to de-regulated YAP1 activation (reviewed in (Zhao et al. 2010a)). Taken together, these results highlight the important roles of YAP1/Yorkie proteins during normal development. In addition, several studies have highlighted a role for YAP1 and the Hippo Pathway in the regeneration of intestine, skin, liver, heart, and nervous tissues following injury and damage (reviewed in (Wang et al. 2017)). Overall, during normal tissue homeostasis, the Hippo pathway suppresses YAP1 function to maintain stem cell quiescence, but is inhibited upon injury, resulting in YAP1 de-repression and activation to promote stem cell self-renewal and generation of new cells necessary for tissue repair. YAP1 interacts with several signaling pathways, including Wnt signaling, BMP and TGFbeta signaling, as well as EGF and HGF signaling, to promote cell proliferation and in some cases differentiation (Wang et al. 2017).
In addition to its role in normal tissue development and homeostasis, YAP1 has also been shown to play important roles in cancers, where it can act as either a tumor suppressor or oncogene depending on the cellular context (reviewed in (Zhang et al. 2018)). YAP1 is important in regulation of cell apoptosis, but it can have dichotomous roles by promoting either pro- or anti-apoptotic functions. Initial evidence showed that YAP1 acts as a tumor suppressor that binds to P53 family members P63 and P73 via its WW domain and enhances their proapoptotic function (Strano et al. 2001; Strano et al. 2005). In addition, diminished YAP1 expression has been reported in some tumor types and was linked to shorter patient survival (Yuan et al. 2008; Cottini et al. 2014). By contrast, a multitude of studies revealed the pro-oncogenic functions of YAP1, largely attributed to the pro-proliferative and pro-survival transcriptional program elicited by its interaction with the family of TEAD transcription factors (TEAD1–4) (Zhao et al. 2007; Liu-Chittenden et al. 2012). Moreover, elevated and nuclear YAP1 staining and inactivating mutations in upstream Hippo pathway members are frequently found in a variety of cancers (discussed in detail below) (Visser and Yang 2010; Deel et al. 2015; Petrilli and Fernandez-Valle 2016).
YAP1-mediated regulation of proliferation and migration of tumor cells
High YAP activity has been shown to positively affect proliferation, survival, stemness, invasiveness and metastatic behavior, as well as therapy resistance of tumor cells in different cancers (Zanconato et al. 2016; Thompson 2020). The growth and proliferation-promoting functions of YAP1 are exemplified by the organ overgrowth observed in Drosophila upon Yorkie overexpression and/or deletion of Hippo tumor suppressors (Justice et al. 1995; Xu et al. 1995; Tapon et al. 2002; Harvey et al. 2003; Huang et al. 2005). Similarly, expression of hyperactivated S127/397A-YAP1 resulted in increased cell proliferation and growth of cell lines in vitro as well as tumor growth in mice in vivo (Zhao et al. 2010b; Szulzewsky et al. 2020). De-regulation of YAP activity has also been shown to cause excess proliferation in multiple tissues, including liver, gastrointestinal tissues, skin, and heart (Camargo et al. 2007; Dong et al. 2007; Heallen et al. 2011; Schlegelmilch et al. 2011). In addition, YAP1 knockdown reduced the growth and proliferation in vitro and/or the ability to form tumors upon transplantation into mice of several cancer cell lines (Muramatsu et al. 2011; Nallet-Staub et al. 2014; Hiemer et al. 2015; Pei et al. 2015; Zhang et al. 2015; Vigneswaran et al. 2020), whereas overexpression of hyperactivated YAP1 in cancer cells with low endogenous YAP1 expression led to an increase in proliferation (Hiemer et al. 2015; Yu et al. 2018). TEAD-dependent YAP1 functions have also been linked to invasive and metastatic behavior of tumor cells (Lamar et al. 2012) and knockdown of YAP1 expression has also been shown to decrease the migration of several tumor cell lines in vitro and/or the metastatic phenotype in mice in vivo (Chen et al. 2012; Hiemer et al. 2015; Pei et al. 2015; Kim et al. 2017; Choi et al. 2018).
Roles of YAP1 in in cancer therapeutic resistance
YAP activity was linked to therapy resistance in different cancer types. For example, increased YAP activity in esophageal cancer induced EGFR expression and was positively correlated with resistance to 5-fluorouracil and docetaxel, whereas knockdown of YAP1 sensitized cells to these drugs (Song et al. 2015). Furthermore, YAP1 expression correlated with resistance of urothelial cell carcinomas and oral squamous cell carcinomas to cisplatin treatment and knockdown of YAP1 sensitized cells from both cancer types to cisplatin treatment (Yoshikawa et al. 2015; Ciamporcero et al. 2016). YAP1 expression and activity was also linked to resistance to radiation therapy in Glioblastoma and Medulloblastoma, two different types of brain cancer (Fernandez et al. 2012; Alexander et al. 2020). YAP1 expression was shown to confer radio-resistance in Sonic Hedgehog medulloblastoma by promoting ongoing proliferation after radiation (Fernandez et al. 2012). YAP1 enabled the tumor cells to enter mitosis with un-repaired DNA through driving insulin-like growth factor 2 (IGF2) expression and AKT activation. Similarly, YAP activity was also linked to treatment resistance and recurrence in a mouse model of glioblastoma (Alexander et al. 2020). In these tumors YAP1 was expressed in a stem-like and highly radio-resistant cell population that was enriched and proliferating 72 hours post-radiation and contributed to tumor-relapse following radiation therapy. By contrast, another study has shown that inactivation of LATS2 or NF2 or forced expression of hyperactivated YAP1 actually sensitizes several pancreatic cancer cell lines to gemcitabine treatment, indicating that the effect of YAP1 on chemoresistance might be context (in vitro 2D, in vitro 3D, in vivo xenograft or in vivo autochthonous), cell type or cancer type specific (Gujral and Kirschner 2017). This study found that inhibitory Hippo pathway signaling enhanced gemcitabine metabolism and export and Hippo inactivation sensitizes a diverse panel of cell lines and human tumors to gemcitabine in 3D spheroid and mouse xenograft tumors. Taken together, these studies highlight a pro-tumorigenic and also context dependent pro- and anti-therapy resistance functions of YAP1 in a large variety of cancers.
Roles of YAP1 in metabolic reprogramming of cancer cells
Dysregulation of metabolic pathways is observed in a variety of cancers. Cancer cells often display an increased uptake of glucose yet use less glucose for the TCA cycle and oxidative phosphorylation and instead favor the glycolytic pathway even under aerobic conditions (called aerobic glycolysis or Warburg effect). To compensate for these changes and to satisfy the demand for biosynthetic precursors and NADPH while maintaining the functional TCA cycle, cancer cells often rely on elevated rates of glutaminolysis, in which glutamine is metabolized to generate ATP and necessary biosynthetic precursors (Jin et al. 2016). YAP1 has been shown to directly induce the expression of several key enzymes involved in both aerobic glycolysis and glutaminolysis thereby modulating the activity of these pathways (reviewed in (Koo and Guan 2018; Yamaguchi and Taouk 2020)). In addition, the activity of YAP1 can itself be regulated by several of these metabolic cues (discussed further below).
YAP1 directly up-regulates the expression of several genes involved in glycolysis in different cancer types, including GLUT3, HK1, HK2, PFKFB4, PFKP, GAPDH, PGK1, PGAM1, LDHA, PDHA1, and PDHB (Cosset et al. 2017; White et al. 2019). Furthermore, a subset of patient-derived glioma cell lines exhibits an addiction to GLUT3. These cells display high levels of GLUT3 expression that is dependent on YAP1 and are vulnerable to YAP1 inhibition (Cosset et al. 2017).
YAP1 has also been shown to enhance both the uptake and the metabolism of different amino acids, most prominently glutamine. YAP1 up-regulates the expression of several glutamine transporters important for glutamine uptake, such as SLC1A5, SLC7A5, and SLC38A1 (Hansen et al. 2015b; Park et al. 2016; Edwards et al. 2017). YAP1 also promotes glutaminolysis by directly enhancing the expression of GLS1 (responsible for the conversion of glutamine to glutamate), as well as GOT1 and PSAT1, two enzymes that convert glutamate to α-ketoglutarate by transamination (Bertero et al. 2016; Yang et al. 2018). Notably, treatment with a transaminase inhibitor was able to inhibit the growth of YAP1-driven breast cancer cells in vitro (Yang et al. 2018). Other studies have also linked YAP1 to an activation of serine metabolism (Wu et al. 2017) and increased leucin uptake (Hansen et al. 2015b).
Roles of YAP1 in tumor cell immune escape
YAP1 has been shown to contribute to the immune escape of tumor cells in various cancers (reviewed in (Pan et al. 2019)). One mechanism by which tumor cells escape immune surveillance is by inhibiting activation of cytotoxic T cells by expressing the immune checkpoint protein PD-L1. Cytotoxic T cells entering the tumor express PD-1, which upon binding to its ligand PD-L1 attenuates T-cell activation and function. YAP1 contributes to the immune escape of tumor cells by directly binding to the enhancer and activating the expression of the PD-L1 gene (Miao et al. 2017; Hsu et al. 2018; Janse van Rensburg et al. 2018; Kim et al. 2018).
YAP1 is also important for the function of immune-suppressive FOXP3+ T-regulatory cells (Tregs) (Fan et al. 2017; Ni et al. 2018). YAP1 expression in Tregs results in an increase in activin signaling and TGFβ/SMAD-dependent activation of these cells. Pharmacological inhibition of either YAP1 or the activin receptor enhanced anti-tumor immunity and survival in a mouse model of melanoma (Ni et al. 2018). Similarly, another study found that in hepatocellular carcinoma YAP1 expression in peripheral blood mononuclear cells is positively correlated with the abundance of Tregs in the tumor tissue and negatively correlated with patient survival (Fan et al. 2017). This effect was due to the direct transcriptional up-regulation of TGFBR2 by YAP1 in these cells.
Lastly, two recent studies have shown that YAP1 expression is up-regulated in activated CD4+ and CD8+ T cells and that YAP1 functions as an immunosuppressive factor and inhibitor of effector differentiation (Lebid et al. 2020; Stampouloglou et al. 2020). Loss of YAP1 expression in T cells results in an improved ability of T cells to infiltrate and repress tumors; however, the exact mechanisms by which YAP1 attenuates activation of these cells is still unknown.
YAP1 structure and transcriptional control of YAP1 target genes by the YAP1-TEAD complex
YAP1 is a transcriptional co-activator that does not contain a DNA binding domain and its function in regulation of transcription relies on the interaction with several other proteins, such as transcription factors (such as TEADs, RUNX2, SMADs) and chromatin remodeling proteins (Varelas 2014) (Figure 1).
Figure 1 –
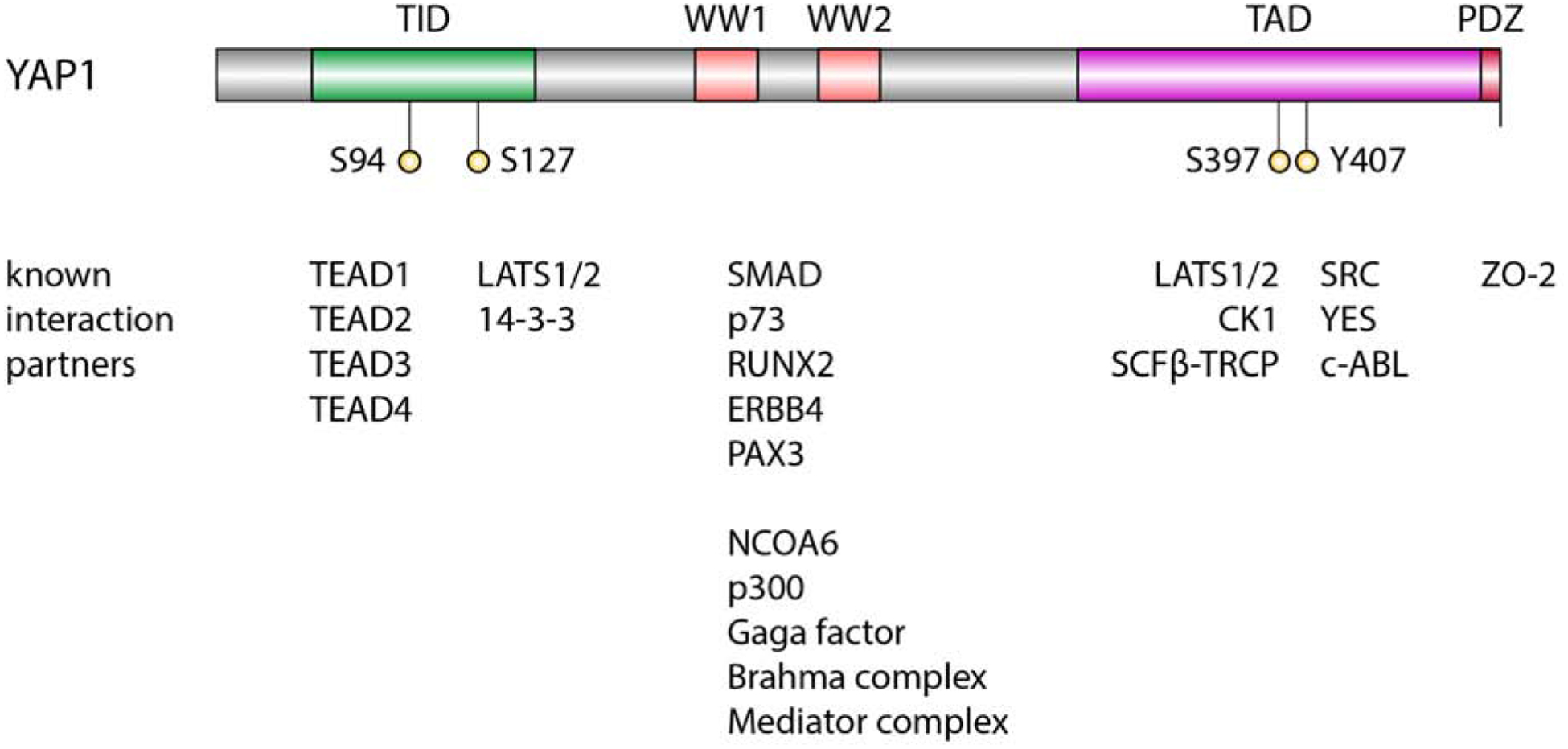
Overview YAP1 protein domains and interaction partners
The transcriptional programs activated by YAP1 and its strong growth promoting functions largely depend on its binding to TEAD transcription factors (Vassilev et al. 2001; Zhao et al. 2008; Zhang et al. 2009). The interaction between YAP1 and TEAD transcription factors is mediated by the TEAD interaction domain (TID), located in the N-terminal region of YAP1 (Vassilev et al. 2001; Li et al. 2010). Serine 94 of TID forms two hydrogen bonds with E240 and Y406 of TEAD1 and mediates a direct interaction between YAP1 and TEAD1 (Li et al. 2010). Furthermore, the TID region contains two short helices with an extended loop containing a PXXΦP motif important for the interaction with TEADs (Chen et al. 2010). TEADs themselves do not contain an activation domain and are thought to function in gene activation mostly through their interaction with YAP1. ChIP-Seq experiments demonstrated a large co-occupancy of YAP1 and TEAD peaks (Zhao et al. 2008; Galli et al. 2015; Stein et al. 2015). Ablation of YAP1-TEAD interaction by S94A mutation of YAP1 resulted in the inability to induce the gene expression changes elicited by wild type YAP1 (Zhao et al. 2008). Furthermore, expression of S94A-YAP1 in NIH-3T3 cells was unable to recapitulate the growth promoting effects of wild type YAP1 (Zhao et al. 2008). Functionally, TEADs direct YAP1 predominantly to distal enhancers and super-enhancers, where YAP1 then recruits the p300 acetyltransferase to induce H3K27 acetylation (Galli et al. 2015; Stein et al. 2015). The YAP1-TEAD complex has also been shown to interact with AP-1 to activate the expression of its target genes (Zanconato et al. 2015; Koo et al. 2020). YAP1 stimulates cell proliferation either directly by controlling the expression of genes involved in cell cycle control, or indirectly by inducing the expression of other transcriptional regulators (Nicolay et al. 2011; Mizuno et al. 2012; Kapoor et al. 2014; Zanconato et al. 2015; Totaro et al. 2018). Prominent YAP1-TEAD target genes include CTGF, CYR61, NPPB, CCND1, AXL, DKK1, ITGB2, WWC1, AMOTL2 and ANKRD1 (Stein et al. 2015; Wang et al. 2018). Importantly, the YAP1-TEAD complex has also been shown to coordinately regulate a network of proproliferative genes together with MYC (Xiao et al. 2013; Croci et al. 2017). YAP1 binds to promoters pre-bound by both TEAD and MYC and MYC-driven cell cycle entry depends on YAP activity.
Located in close proximity to the TID is the YAP1 14-3-3 interaction domain, that is a target of LATS1/2-mediated phosphorylation of YAP1 at S127 and, when phosphorylated, drives interaction with 14-3-3 proteins and cytoplasmic sequestration of YAP1 (Zhao et al. 2007; Zhao et al. 2010b).
Neighboring the 14-3-3-binding domain are WW protein domains (one or two depending on the YAP isoform) that are involved in protein-protein interactions and recognize a PPxY motif in their protein targets. These WW domains are responsible for the interaction of YAP1 with several other proteins, including transcription factors, such as RUNX2 (Yagi et al. 1999), SMADs (Ferrigno et al. 2002), ERBB4 (Komuro et al. 2003; Omerovic et al. 2004; Haskins et al. 2014), p63/p73 (Strano et al. 2001; Strano et al. 2005; Levy et al. 2007; Levy et al. 2008), and PAX3 (Manderfield et al. 2014). The WW domains also mediate the interaction with several chromatin-remodeling proteins. Both YAP1 and Yorkie, the Drosophila orthologue of YAP1, interact with a PPxY motif in Ncoa6, a subunit of the Trithorax-related (Trr) histone H3 lysine 4 (H3K4) methyltransferase complex, which results in increased H3K4 methylation and active transcription (Oh et al. 2014; Qing et al. 2014). In addition, Yorkie was also shown to use WW domains to interact with GAGA factor (GAF), the Brahma complex, and the Mediator complex to induce H3K4 trimethylation and activate transcription of target genes (Oh et al. 2013; Galli et al. 2015). Similarly, human TAZ was shown to interact with BRM, the catalytic subunit of the SWI/SNF chromatin remodeling complex to activate the transcription of TAZ target genes (Skibinski et al. 2014). Again, this interaction was mediated via the WW domains of TAZ and a PPxY motif in BRM. As mentioned earlier, YAP1 has also been shown to recruit p300 to induce H3K27 acetylation (Stein et al. 2015; LeBlanc et al. 2018).
The extended C-terminal region of YAP1 contains an unstructured transactivation domain (TAD) that is rich in serine, threonine and acidic amino acids (Yagi et al. 1999). The exact biochemical mechanisms of how this acidic C-terminal TAD activates gene expression are unknown, however it resembles the acidic activation domain of herpes simplex virus VP16, suggesting that YAP1, like VP16, can directly interact with components of the transcriptional machinery including TFIIB, TBP, TFIIA, and TFIIH (Vassilev et al. 2001). Interestingly, Yorkie, the Drosophila orthologue of YAP1, does not contain the C-terminal TAD, indicating that the exact mechanism of how YAP1 and Yorkie activate transcription differ at least in part (Zhu et al. 2015). In addition, phosphorylation of YAP1 at Y407 located in the TAD by c-ABL, SRC, or YES can influence the function of YAP1 and its interaction with other proteins, such as β-catenin or p73 (Sudol et al. 1995a; Levy et al. 2008; Rosenbluh et al. 2012). Finally, a PDZ binding motif located at the very C-terminus of YAP1 mediates the interaction with ZO-2 and is necessary for the nuclear localization and activity of YAP1 (Oka and Sudol 2009; Oka et al. 2010).
YAP1 and TAZ exert overlapping but not identical functions
The Hippo Pathway is highly conserved across species and YAP1/Yorkie and other key Hippo cascade components were already present in unicellular ancestors (Sebe-Pedros et al. 2012). By contrast, the YAP1 paralogue TAZ is evolutionary much younger and is only found in vertebrates (Pappalardo et al. 2015). YAP1 and TAZ are both required for embryonic development and it is thought that their functions are largely overlapping but not redundant. Homozygous deletion of Yap1 in mice is embryonically lethality as early as E8.5 (Morin-Kensicki et al. 2006), whereas around one-fifth of mice deficient for Taz are viable (Makita et al. 2008), although it is not known if this is due to different tissue expression patterns or differences in their actual transcriptional functions. Recent studies have shown that the transcriptional profiles induced by both YAP1 and TAZ in HEK293 cells largely overlap, however knockout of YAP1 had a bigger influence on cellular physiology (such as proliferation, cell volume, migration) when compared to knockout of TAZ, and YAP1 knockout cells behaved more similar to YAP1/TAZ double knockout cells (Plouffe et al. 2018). Another study performing ChIP-Seq for both YAP1 and TAZ in breast cancer cells also demonstrated a large overlap in the function of both proteins (Zanconato et al. 2015). Around 7,100 peaks (92% of YAP1 peaks and 73% of TAZ peaks) were shared between both proteins, indicating that their functions are similar, however not identical. It is unknown what causes the differences in the functionalities of YAP1 and TAZ. Both proteins share around 60% similarity, however there are also prominent differences. YAP1’s longest isoform contains two WW domains, whereas TAZ only contains one (Hong et al. 2005). In addition, YAP1 contains an SH3-binding motif (necessary for its interaction with YES) and a proline-rich N-terminal region, both of which are absent in TAZ and TAZ was unable to bind the Yes SH3 domain in vitro (Kanai et al. 2000). Furthermore, TAZ contains a second, N-terminal phosphodegron that is not present in YAP1, contributing to the fact that TAZ activity is much more dynamically regulated by protein degradation compared to YAP1 (Huang et al. 2012). Finally, while both YAP1 and TAZ predominantly function through binding to TEADs (Zanconato et al. 2015) and residues necessary for the interaction with TEADs are conserved in TAZ, it does not contain YAP1’s PXXΦP motif and can form both heterodimers and heterotetramers with TEADs (Chen et al. 2010; Li et al. 2010; Kaan et al. 2017). In conclusion, YAP1 and TAZ exert largely overlapping, however not identical functions and further studies are necessary to explain these differences.
Regulation of YAP1 by the Hippo Signaling Pathway
The activity of YAP1 is regulated and inhibited by the upstream core kinase cascade of the Hippo signaling pathway (Figure 2). Serine/threonine kinases MST1/2 (orthologues of Drosophila Hippo) interact with SAV1 (orthologue of Drosophila Salvador) and phosphorylate MOB1 (ortholog of Drosophila Mats) and serine/threonine kinases LATS1/2 (orthologues of Drosophila Warts), a critical event resulting in the activation of LATS1/2 (Chan et al. 2005; Hergovich et al. 2006; Praskova et al. 2008). In turn, LATS1/2-MOB1 complex inactivates YAP1 by direct phosphorylation at a series of serine residues (S61, S109, S127, S164, S397 (S381 depending on the YAP1 isoform)), with serine 127 and 397 seeming to be the most important residues for the Hippo Pathway-mediated regulation of YAP1 (Huang et al. 2005; Zhao et al. 2007). Phosphorylation of YAP1 ultimately leads to its inactivation by both nuclear exclusion via binding of 14-3-3 proteins at phosphorylated S127 and proteasomal degradation initiated by phosphorylation of S397 (Zhao et al. 2007; Liu et al. 2010a; Zhao et al. 2010b).
Figure 2 –
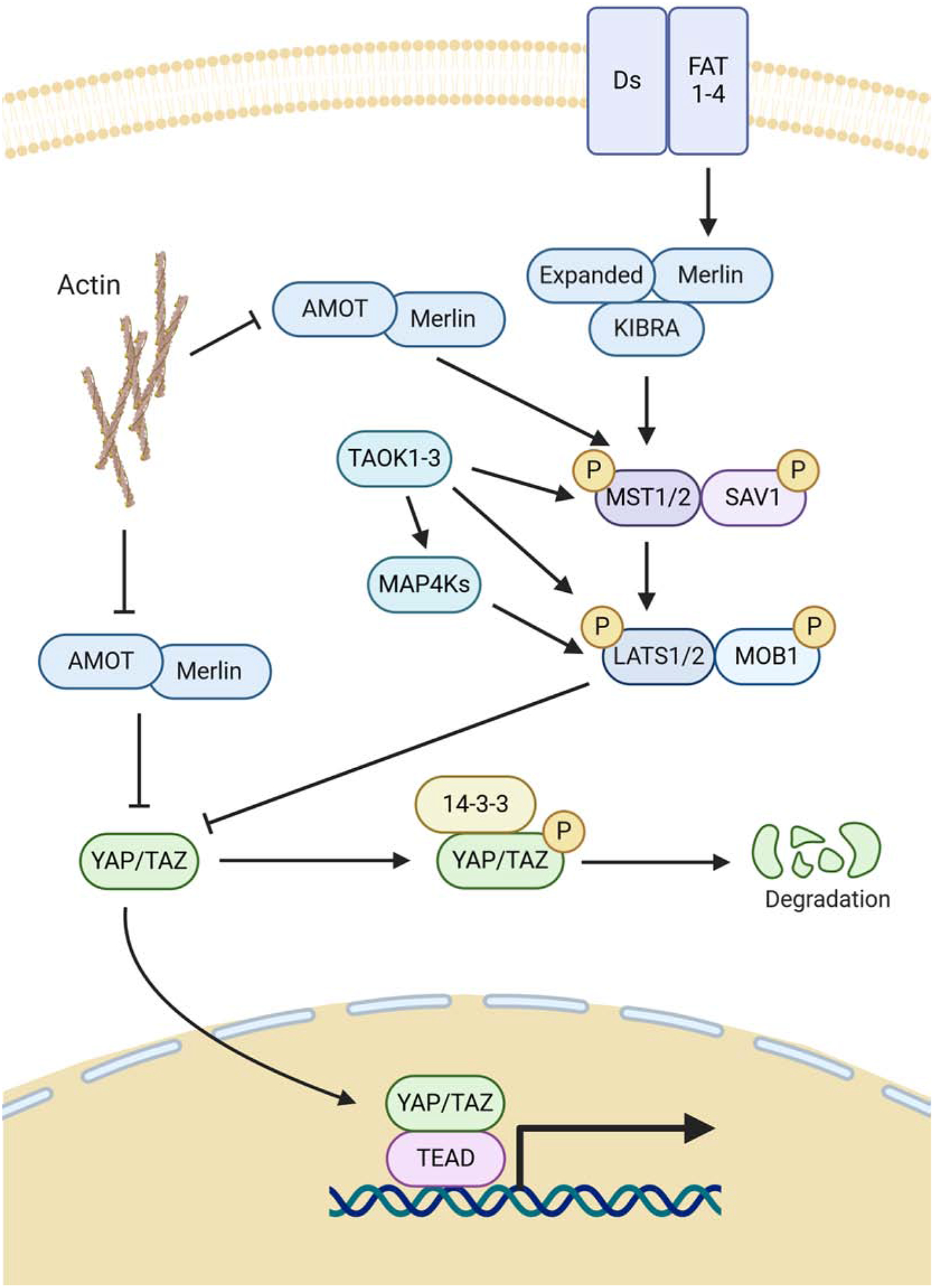
Hippo Core kinases and resulting regulation of YAP1. Figure was created with BioRender.com.
While the above-mentioned core participants of the Hippo pathway play very important roles in regulation of YAP1 and TAZ, mammalian Hippo signaling is very complex and several additional proteins are involved. For example, in addition to MST1/2, LATS1/2 can be also directly phosphorylated and activated by serine-threonine kinases of the MAP4K and TAOK families (Praskova et al. 2008; Meng et al. 2015; Zheng et al. 2015; Meng et al. 2016). In addition to LATS1/2, YAP1 can be also phosphorylated and inhibited by the nuclear kinase PRP4K, cell-cycle kinase CDK1 and AKT (Basu et al. 2003; Zhao et al. 2014; Cho et al. 2018).
Significant information has been accumulated regarding the mechanisms responsible for the upstream regulation of Mst1/2-LATS1/2 cascade (for recent review see (Ma et al. 2019)). One of the most unique aspects of the Hippo signaling pathway is its prominent regulation by extracellular mechanical forces acting on cells exerted by the tissue architecture, extracellular matrix, cellular shapes, cell-cell adhesion and liquid shear stress. In tissue culture, the role of cell contacts and confluency (contact inhibition) in activation of the Hippo cascade and inhibition of YAP1 is especially well documented (Zhao et al. 2007). Inhibition of cell-cell adhesion by loss of adherens junction proteins E-cadherin or alpha-catenin results in prominent YAP1 activation (Kim et al. 2011; Schlegelmilch et al. 2011; Silvis et al. 2011; Karaman and Halder 2018). Gigantic cadherin proteins FAT and Dachsous positively regulate Hippo signaling and inhibit Yorkie activity in Drosophila, and, while the exact mechanism is not conserved in mammals, mouse FAT family proteins have been implicated in the regulation of YAP1 (Silva et al. 2006; Willecke et al. 2006; Willecke et al. 2008; Ragni et al. 2017). The tight junctional and apical-basal cell polarity protein complex CRB3-LIN7cC-PATJ-PALS-MPDZ-AMOT is a prominent negative regulator of YAP1/TAZ (Varelas et al. 2010b). In contrast to cell-cell adhesion structures, cell-substratum adhesion and integrins are strong positive regulators of YAP1/TAZ (Dupont et al. 2011; Elbediwy et al. 2016; Wang et al. 2016b). How exactly the mechanical forces and cell adhesion structures regulate YAP1/TAZ is not completely understood. This regulation may be dependent and independent from the canonical MST1/2-LATS1/2 Hippo signaling pathway (Ma et al. 2019; Zheng and Pan 2019). F-actin cytoskeleton and Rho-family GTPases appear to play a critical role in connecting mechanical forces to YAP1 signaling (Dupont et al. 2011; Aragona et al. 2013). Cell polarity proteins Angiomotins (AMOT, AMOTL1, AMOTL2), Merlin (NF2), Expanded (FRMD6), and the Kibra complex play an important role in the regulation of Hippo signaling and YAP1 (Pan 2010). The Merlin/Kibra/Expanded complex physically associates with both MST1/2 and LATS1/2 and recruits the Hippo kinases to the plasma membrane and the apical membrane domain promoting their activation and phosphorylation of YAP1 (Yin et al. 2013). Interestingly, both Merlin and Angiomotins can directly bind to YAP1 and sequester it from the nucleus (Varelas et al. 2010b; Furukawa et al. 2017). In addition, Angiomotins promote the Hippo signaling pathway by binding to and activation of Merlin (Li et al. 2015). While Angiomotins are not present in the Drosophila genome, in mammals these proteins may play an important role in connecting mechanotransductive F-actin-mediated signaling to YAP1 activity. Angiomotins associate with F-actin through a conserved F-actin-binding domain, which is also used for binding to YAP1. Thus, F-actin sequesters Angiomotins and blocks their interaction and inhibition of YAP1 (Mana-Capelli et al. 2014).
YAP1 activity can be regulated by several different mechanisms
The first mechanism of YAP1 inactivation is mediated by the exclusion of the YAP1 protein from the nucleus. Recent studies have suggested that even at steady-state conditions YAP1 is continuously shuttled between the nucleus and cytoplasm and the switch between a more nuclear or cytoplasmic localization is a dynamic process that is regulated by several factors and processes (reviewed in (Manning et al. 2020)). Phosphorylation of YAP1 by LATS1/2 results in its binding to 14-3-3 proteins, followed by nuclear exclusion and cytoplasmic sequestration (Oh and Irvine 2008; Ren et al. 2010). In addition, a nuclear exclusion sequence (NES) was identified within the sequence encompassed by leucine 308 to leucine 320 of YAP1 that can be bound by exportin-1 (XPO1) (Wang et al. 2016a). Treatment with the XPO1 inhibitor Leptomycin B resulted in a decrease in XPO1-mediated YAP1 nuclear exclusion and a more prominent nuclear localization of YAP1 (Wang et al. 2016a). The upstream Hippo pathway member NF2/Merlin has also been shown to directly interact with YAP1 and facilitate its nuclear exclusion (Furukawa et al. 2017). Furthermore, Kofler and colleagues identified a nuclear localization sequence (NLS) within amino acids 413 and 427 of human YAP1, as well as an NES in its TEAD binding domain (Kofler et al. 2018). Upon binding to TEAD transcription factors the NES is masked, resulting in nuclear retention of YAP1.
The second mechanism leading to YAP1 inactivation is mediated by proteasomal degradation. Phosphorylation of YAP1 at S397 by LATS1/2 primes YAP1 for additional phosphorylation by CK1δ/ɛ, resulting in the formation of a phosphodegron that primes YAP1 for ubiquitination by the SCFβ-TRCP E3 ubiquitin ligase and subsequent degradation (Zhao et al. 2007; Zhao et al. 2010b; Azzolin et al. 2014). Interestingly, both mechanisms, nuclear exclusion and proteasomal degradation, seem to independently regulate YAP1 activity. Even though S127A mutation of YAP1 results in a more prominent nuclear localization of YAP1, this does not result in the complete de-regulation of YAP activity and expression of S127A-YAP1 is not sufficient to cause tumor formation in many organs (Zhao et al. 2010b; Chen et al. 2015; Szulzewsky et al. 2020). This is likely due to a compensatory regulation by proteasomal degradation, mediated via phosphorylation at S397 (Zhao et al. 2010b; Chen et al. 2015). In turn, inhibition of both nuclear exclusion and proteasomal degradation by combined S127/397A mutation (2SA-YAP1) or mutation of all five phosphorylated by LATS1/2 serine residues (5SA-YAP1) renders YAP1 fully oncogenic (Zhao et al. 2010b; Zhang et al. 2019; Eder et al. 2020; Szulzewsky et al. 2020). These results indicate that the activity of YAP1 is regulated by several compensatory mechanisms and a single point mutation - such as a K-Ras G12V mutation that results in oncogenic Ras - is insufficient to de-regulate YAP1 and render it oncogenic.
De-regulation of YAP1 activity in cancer
Elevated YAP1 activity and nuclear staining are observed in several human cancer types and this is often correlated with worse prognostic outcome (Zanconato et al. 2016; Dey et al. 2020). Amplifications of genomic regions harboring YAP1 are frequent events in certain cancers, such as head and neck squamous cell carcinomas (~14% of tumors), esophageal squamous cell carcinomas (~15% of tumors), lung squamous carcinomas (~16% of tumors), and cervical squamous cell carcinomas (~17% of tumors) (Lorenzetto et al. 2014; Sanchez-Vega et al. 2018; Dey et al. 2020). However, it is unclear if YAP1 gene locus amplifications are the tumor initiating events and oncogenic drivers in these tumors or if they are mostly passengers that can potentially contribute to tumor aggressiveness but are not the key genetic changes responsible for malignant transformation. Indeed, simple overexpression of wild type YAP1 in mice is insufficient to cause tumor formation (Szulzewsky et al. 2020). Additional inactivation of upstream Hippo tumor suppressors is potentially necessary to initiate transformation. Similarly, activating point mutations in YAP1 are generally rare in human cancer (Wang et al. 2018). This may be potentially attributed to the fact that LATS1/2 phosphorylate YAP1 at several serine residues and a minimum of two co-occurring point mutations (S127A and S397A) are necessary to fully de-regulate YAP1 and render it insensitive to both Hippo-mediated nuclear exclusion and degradation (Zhao et al. 2007; Zhao et al. 2010b; Szulzewsky et al. 2020). Indeed, while clearly an extremely rare event, a hyperactivated YAP mutant containing several co-occurring serine-to-alanine miss-sense mutations - including S127A and S397A - has been observed in cutaneous melanoma (Zhang et al. 2019).
Recent pan-cancer studies by The Cancer Genome Atlas Research (TCGA) Network identified several recurrent alterations in Hippo Pathway members in human cancers, such as amplifications of YAP1 and/or deletions, loss-of-function mutations, or epigenetic silencing of Hippo Pathway tumor suppressor genes NF2/Merlin, FAT1–4, TAOK1–3, WW45, and LATS1/2. (Sanchez-Vega et al. 2018; Wang et al. 2018). In addition to loss-of-function events directly involving Hippo Pathway tumor suppressors, several studies have shown that other factors can contribute to de-regulation of YAP1 activity in cancers. These involve miRNAs, mutations in non-Hippo up-stream regulators (e.g. G protein-coupled receptors), viral oncoproteins, mechanical stimuli, and gene fusion events involving YAP1 and TAZ (Figure 3). Below, we will briefly discuss and summarize these findings.
Figure 3 –
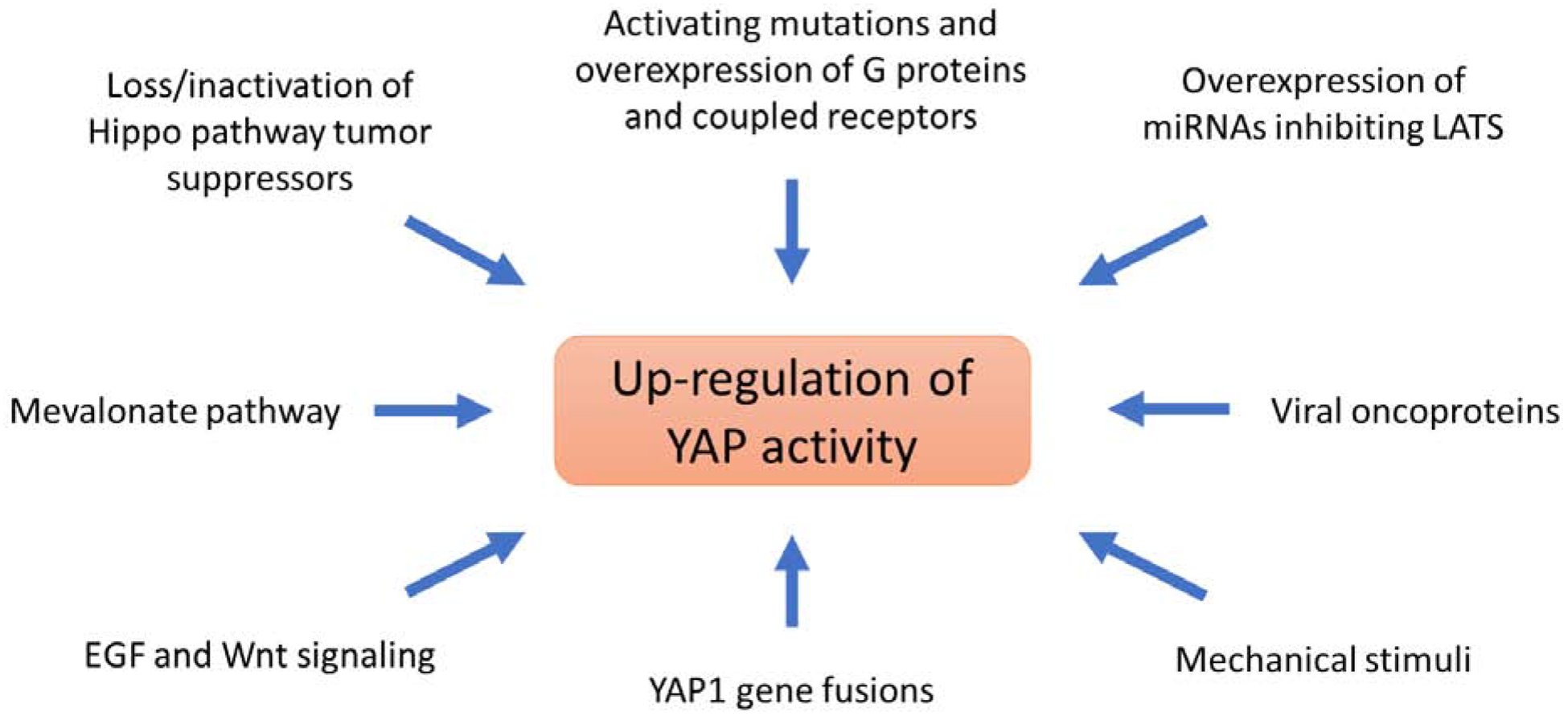
Mechanisms of YAP1 de-regulation in cancer
Mutations in Hippo pathway tumor suppressor genes in cancer
FAT proteins
FATs are giant Cadherin proteins and upstream activators of NF2/Merlin and the Hippo pathway. FAT1–4 loss-of-function events are frequently observed in several human cancer types. The TCGA pan-cancer study reported inactivating FAT1 mutations or gene deletions in head and neck squamous cell carcinoma, lung squamous cell carcinoma, stomach and esophageal cancer, colorectal cancer, uterine corpus endometrial carcinoma, cervical cancer, and sarcoma tumors (Sanchez-Vega et al. 2018). Interestingly, FAT1 inactivation in head and neck squamous cell carcinoma was predominantly observed in HPV-negative cases, indicating that HPV-positive tumors might rely on alternative ways of YAP1 activation (discussed in more detail below). In addition, FAT2–4 genes were also deleted in several colorectal cancers and uterine corpus endometrial carcinoma tumors (Sanchez-Vega et al. 2018). Furthermore, FAT1 was homozygously deleted in ~80% of primary oral cancer cases and FAT1 gene expression was down-regulated or lost in oral cancer cell lines due to either homozygous deletion or promoter methylation (Nakaya et al. 2007; Katoh 2012). Loss of heterozygosity of FAT1 occurred in ~42% of low grade diffuse astrocytoma and ~63% of glioblastoma samples (Chosdol et al. 2009). Lastly, FAT1 expression was significantly decreased in invasive breast cancer samples compared to ductal carcinoma in situ and FAT1 knockdown promoted the progression from ductal carcinoma in situ to invasive breast cancer (Katoh 2012; Lee et al. 2012).
NF2
NF2/Merlin is a potent positive regulator of the Hippo pathway, which can also directly bind to YAP1 and sequester it from the nucleus. Deletion or loss-of-function mutation of the NF2 gene (most commonly frameshift or nonsense mutation) causes Neurofibromatosis Type 2 (NF2), a rare genetic disorder that results in the frequent formation of several central nervous system tumors, such as schwannomas, meningiomas, and spinal ependymomas (Rubio et al. 1994; Ebert et al. 1999; Ahronowitz et al. 2007; Petrilli and Fernandez-Valle 2016; Lee et al. 2019). YAP1 function is necessary for the proliferation, survival, and in vivo tumor growth of NF2 null Schwann cells (Guerrant et al. 2016). In addition, somatic NF2 mutations have also been found in a large percentage of malignant mesothelioma patients (Sekido et al. 1995; Cheng et al. 1999; Baser et al. 2002; Sanchez-Vega et al. 2018). The prevalence of functional NF2/Merlin inactivation in other cancers is still not fully understood. Somatic NF2 mutations have been found in approximately 4.5% of breast and colorectal cancer cases (Sjoblom et al. 2006). Two studies found only a low prevalence of somatic NF2 mutations in human colorectal carcinoma (2 out of 24 samples and 2 out of 44 samples, respectively), whereas another study found NF2 loss-of-heterozygosity in 20% of sporadic colorectal cancers and it was found to be more frequent in larger and less differentiated tumors (Arakawa et al. 1994; Rustgi et al. 1995; Cacev et al. 2014). In addition to somatic mutations resulting in the expression of a truncated or inactive NF2/Merlin protein, elevated levels of Merlin phosphorylation at serine 518, which causes inactivation of its tumor suppressive function, may present an alternative route of NF2/Merlin loss-of-function, and this was observed in several cancer cell lines, including melanoma and prostate cancer (Horiguchi et al. 2008; Murray et al. 2012).
TAOK proteins
TAOK1–3 are serine/threonine kinases that can directly phosphorylate and activate LATS1/2 independently of MST1/2. The pan-cancer TCGA studies observed loss-of-function events involving TAOK genes predominantly in stomach and esophageal cancer, colorectal cancer, uterine corpus endometrial carcinoma, sarcomas, and breast invasive carcinomas (Sanchez-Vega et al. 2018; Wang et al. 2018).
MST1/2
MST1/2 are serine/threonine kinases that directly phosphorylate and activate LATS1/2. The pan-cancer TCGA studies identified recurrent loss-of-function events of MST1 (STK4) and MST2 (STK3) genes mostly in colorectal cancer and uterine corpus endometrial carcinoma, whereas they were relatively uncommon in other cancers (Sanchez-Vega et al. 2018; Wang et al. 2018). These loss-of-function events were predominantly caused by point-mutations and gene fusions. Decreased expression of MST1/STK4 is furthermore observed in several hematopoietic malignancies, including myelodysplastic syndrome, classic myeloproliferative neoplasm, and acute leukemias (Stoner et al. 2019). These cancers often exhibit heterozygous deletion of chromosome 20 (harboring the MST1/STK4 gene) and MST1/STK4 was one of nine genes that were down-regulated to subhaploinsufficient levels, and this contributed to malignancy via chronic innate immune activation (Stoner et al. 2019).
LATS1/2
LATS1/2 are serine/threonine kinases that directly phosphorylate YAP1 and inhibit its function. The TCGA pan-cancer study reported LATS1 and/or LATS2 loss-of-function events in several cancers. Inactivating mutations, copy number loss, or epigenetic silencing of LATS2 was observed in low-grade gliomas, EBV-positive esophagogastric cancer, and diffuse large B cell lymphoma (Sanchez-Vega et al. 2018). By contrast, loss of LATS1 function was observed predominantly in uveal melanoma, colorectal cancer, and uterine corpus endometrial carcinoma. Another study found that LATS1 was mutated in stomach adenocarcinoma (~6% of cases), uterine corpus endometrial carcinoma (~4%), and bladder urothelial carcinoma (~3%), whereas LATS2 mutations occurred in uterine corpus endometrial carcinoma (~5% of cases), stomach adenocarcinoma (~4%), and lung adenocarcinoma (~4%) (Yu et al. 2015b).
In summary, pan-cancer TCGA studies indicate that inactivating mutations in Hippo pathway members predominantly occur in LATS (especially LATS2) and FAT (especially FAT1) genes, whereas mutations in other Hippo pathway core cascade members were more rare. In addition, very little is known about inactivating events in MAP4K genes. The relative rarity of inactivating events in MST1/2 (STK4/3) and TAOK1–3 genes might be explained by a potential functional redundancy between these two families of proteins, that independently regulate LATS1/2 activity. Therefore, loss-of-function of MST proteins might be compensated by MAP4K and TAOK proteins and vice versa, and therefore not sufficient to de-regulate YAP1 activity. Recurrent NF2/Merlin loss-of-function events are predominantly found in cancers associated with hereditary NF2 mutations, such as schwannomas, meningiomas, and spinal ependymomas. In addition, somatic NF2 loss-of-function mutations are present in a large percentage of malignant mesothelioma patients but are rare in other cancers.
Additional regulators of YAP1 activity in cancer
G protein-coupled receptors
The Hippo Pathway intersects with several other signaling pathways and the activity of different Hippo Pathway proteins is directly regulated by several non-Hippo Pathway proteins. Genetic or epigenetic changes affecting these important regulators are frequently observed in several cancer types.
G protein-coupled receptors (GPCRs) constitute a large family of cell surface receptors that transmit diverse extracellular signals. Several studies demonstrated that activation of GPCRs coupled with G proteins Gα12/13, Gαq/11, or Gαi/o upregulates YAP1 signaling (reviewed in (Luo and Yu 2019)). These GPCRs include protease-activated receptors (PARs, thrombin signaling) (Mo et al. 2012), LPA receptors (Yu et al. 2012; Cai and Xu 2013), S1P receptors (Yu et al. 2012), and GPER estrogen receptors (Zhou et al. 2015). They were linked to increased YAP1 activation in such cancer types as ovarian, prostate, breast, lung, pancreatic, colon cancer, and melanoma (Luo and Yu 2019). The direct mechanism how GPCRs modulate the Hippo Pathway and YAP1 activation remains unknown, however, it has been demonstrated that GPCRs activate Rho GTPases which in turn modulate the F-actin cytoskeleton, which can then be sensed by the Hippo Pathway and leads to activation of YAP1 (Yu et al. 2015a; Luo and Yu 2019).
Activating mutations and/or overexpression of GPCRs and G-coupled proteins are frequent events in cancer and there is evidence that these mutations contribute to YAP1 activation in the affected tumors (reviewed in (Nieto Gutierrez and McDonald 2018)). This includes overexpression of PAR1 and GPER in breast cancer (Hernandez et al. 2009; Zhou et al. 2015), activating point mutations in GNAQ and GNA11 in uveal melanoma and blue naevi (Van Raamsdonk et al. 2009; Van Raamsdonk et al. 2010; Feng et al. 2014; Yu et al. 2014; Lyubasyuk et al. 2015), and inactivating mutations in GNAS in medulloblastoma and basal-cell carcinoma (He et al. 2014; Kool et al. 2014; Iglesias-Bartolome et al. 2015).
Rho GTPases and the mevalonate pathway
The mevalonate signaling pathway is an essential metabolic pathway that utilizes acetyl-CoA to generate sterol and non-sterol isoprenoids necessary for production of thousands of important biomolecules, such as cholesterol, all steroid hormones, dolichol, heme-A, isopentenyl tRNA, and ubiquinone. The mevalonate pathway has been shown to positively affect the activity of Rho GTPases, which in turn promote the nuclear localization and activity of YAP1 (Sorrentino et al. 2014). In turn, inhibition of HMG-CoA reductase, the rate-limiting enzyme of the mevalonate pathway, resulted in the inhibition of YAP1. Importantly, Sorrentino et al. demonstrated that mutant p53 activates the expression of sterol regulatory element-binding proteins SREBP1 and SREBP2, which in turn positively regulate the expression of the enzymes of the mevalonate pathway (Sorrentino et al. 2014). Breast cancer cells expressing mutant p53 exhibited activation of both mevalonate signaling and YAP1, whereas knockdown of mutant p53 in MDA-MB-231 cells inhibited YAP1 activity.
O-GlcNAcylation
High glucose uptake and aerobic glycolysis are hallmarks of cancer (discussed in detail above). High glucose levels positively correlate with increased global protein O-GlcNAcylation, a post-translational modification consisting of the addition of an N-acetyl-glucosamine residue (GlcNAc) to serine or threonine amino acids. Increased O-GlcNAcylation is observed in many cancer cells and this can activate YAP1 signaling. O-GlcNAcylation of YAP1 at several residues inhibits its interaction with LATS1 as well as with the SCFβ-TRCP E3 ubiquitin ligase, thereby increasing its stability and activity (Peng et al. 2017; Zhang et al. 2017).
Wnt pathway
Wnt signaling is a signal transduction pathway playing a pivotal role in regulation of development, normal tissue homeostasis and cancer. Beta-catenin, an essential effector of the canonical Wnt signaling can directly interact with the YAP1 protein (Imajo et al. 2012). The exact mechanisms and the significance of the interaction between YAP1/Hippo and Wnt signaling pathways are very complex and even the functional outcome of this interaction (cooperation versus competition) is sometimes opposite and context dependent (Jiang et al. 2020). YAP1 forms a complex with β-catenin and the transcription factor TBX5 and subsequent phosphorylation of YAP1 by YES1 leads to localization of this complex to the promoters of antiapoptotic genes (Rosenbluh et al. 2012). In turn, cytoplasmic YAP1 inhibits Wnt signaling by facilitating both the degradation and sequestration of β-catenin (Varelas et al. 2010a; Imajo et al. 2012; Azzolin et al. 2014) (reviewed in (Hansen et al. 2015a). YAP1 expression is also positively regulated by Wnt signaling and is a direct target gene of β-catenin (Konsavage et al. 2012). In addition, Protein kinase C zeta (PKCζ) has been shown to inhibit the function of both YAP1 and β-catenin through direct interaction (Llado et al. 2015). PKCζ phosphorylates YAP1 at several residues, including S109 and T110, leading to increased degradation of the YAP1 protein. Importantly, functional inactivation of PKCζ has been reported in colorectal cancer via down-regulation and inactivating mutations, clinically implicating PKCζ as a tumor suppressor (Wood et al. 2007; Ma et al. 2013).
Receptor and non-receptor type tyrosine kinases-PI3K-AKT signaling
Constitutive activation of Tyrosine kinase-PI3K-AKT signaling pathway is one of the hallmarks and a major tumor initiating and promoting factor in human cancer (Biscardi et al. 1999; Fruman et al. 2017). Significant literature implicates Hippo and YAP1/TAZ as important downstream targets of this oncogenic pathway. Epidermal growth factor (EGF) signaling activates YAP1 function by inhibiting LATS (reviewed in (Hansen et al. 2015a)). EGF treatment of immortalized mammary cells inhibits LATS function via a PI3K-PDK1 pathway. PDK1 associates with the Hippo Pathway scaffolding protein SAV1, which in turn leads to the dissociation of the Hippo Core kinase complex, the inhibition of LATS and to the dephosphorylation and nuclear translocation of YAP1 (Fan et al. 2013). Similarly, EGF signaling also activates a EGFR-RASMAPK-AJUBA pathway that likewise activates YAP1 through inhibiting LATS (Reddy and Irvine 2013). The non-receptor type tyrosine kinases of the SRC and FAK families are heavily implicated in human cancer and also activate YAP1 via both Hippo dependent and Hippo independent pathways (Rosenbluh et al. 2012; Kim and Gumbiner 2015; Taniguchi et al. 2015; Elbediwy et al. 2016; Li et al. 2016; Si et al. 2017; Lamar et al. 2019). YAP1 is a functionally essential downstream target of PI3K-AKT signaling pathway activated by either expression of oncogenic PIK3CA or deletion of PTEN (Chen et al. 2019; Roy et al. 2019).
ARRDC3
The ARRDC3 tumor suppressor protein is involved in controlling signaling and trafficking of PAR1. It is down-regulated in several cancer types, including invasive breast carcinomas and renal cell carcinoma (Arakaki et al. 2018; Xiao et al. 2018). ARRDC3 inhibits YAP1 and TAZ activity by directly facilitating their degradation (Xiao et al. 2018; Arakaki et al. 2020). Interestingly, Arakaki et al demonstrated that in invasive breast cancer the tumor suppressive functions of ARRDC3 were mostly related to the inhibition of TAZ, whereas YAP1 function was dispensable for the invasive behavior of these tumor cells (Arakaki et al. 2020).
Leukemia inhibitory factor receptor
Leukemia inhibitory factor receptor (LIFR) has been identified as a breast cancer metastasis suppressor and low LIFR expression correlates with poor breast cancer prognosis (Chen et al. 2012). Loss of LIFR expression leads to YAP1 activation and a YAP1-dependent increase of invasive behavior of breast cancer cells. Re-expression of LIFR resulted in an increase of p-S127 (inactivated) YAP1 levels, accompanied by an increase in the levels of phosphorylated (activated) MST1/2 and LATS1; however, the direct mechanism of LIFR-mediated activation of Hippo Pathway signaling is unknown.
MicroRNAs
Several miRNAs have been shown to either positively or negatively influence the activity of YAP1 and/or tumor suppressive Hippo Pathway proteins (reviewed in (Li et al. 2017; Han 2019). Several miRNAs such as miR-550a-3–5p, miR-195, miR-874–3p negatively regulate YAP1 (Li et al. 2017; Han 2019). The tumor-suppressive miR-375 directly represses YAP1, TEAD4, and CTGF expression and was down-regulated in several gastric cancer samples (Kang et al. 2018). By contrast, miR-31 and miR-135b are overexpressed in several cancers and have been shown to inhibit the translation of LATS2 (Liu et al. 2010b; Lin et al. 2013; Mitamura et al. 2014). Similarly, miR-624–5p and miR-665 are overexpressed in osteosarcoma and hepatocellular carcinoma, respectively, and enhance YAP1/TAZ activity by inhibiting LATS function via down-regulating PTPRB (Hu et al. 2018b; Luo et al. 2019).
The Hippo signaling pathway and its effector YAP1 have a profound impact on biogenesis on many miRNAs, including important regulators of MYC (Mori et al. 2014). Activated YAP1 binds p72 (DDX17), a regulatory component of the miRNA-processing machinery, and prevents its interaction with the microprocessor resulting in widespread inhibition of miRNA production (Mori et al. 2014). In addition, YAP1 can also upregulate expression of miR-29 and miR-130 that inhibit the biosynthesis of the tumor suppressor PTEN (Tumaneng et al. 2012; Shen et al. 2015). High expression of miR-130 family members has been observed in several cancer types, including bladder cancer and gastric cancer (Duan et al. 2016; Egawa et al. 2016).
Viral oncogenes
Historically, research on the biology of oncogenic viruses (or Oncoviruses) has led to important findings on the biology of cancer and oncogenes. The transforming activity of most of these oncogenic viruses arises from incorporated activated cellular oncogenes, such as the Rous sarcoma virus that harbors a truncated and constitutively active form of Src. By contrast, some viruses are oncogenic because they developed their own viral proteins that are able to disrupt cellular signaling pathways. This includes the Merkel cell polyomavirus (MCV or MCPyV) in Merkel cell carcinoma (Feng et al. 2008) and the human papillomavirus in cervical cancer (Bosch et al. 1995). Viral proteins from both of these viruses can positively affect YAP1 function, either indirectly by inhibiting the function of Hippo tumor suppressor proteins or by directly interacting with YAP1 itself. Abundant YAP1 and TAZ positivity has been observed in cervical carcinoma samples (Buglioni et al. 2016) and, in addition, low expression of MST1 was observed in cervical carcinoma cell lines (Morgan et al. 2020). The HPV E6 and E7 proteins have been shown to up-regulate the expression of miR-18a, which in turn down-regulates the expression of the Hippo tumor suppressor MST1, ultimately leading to increased YAP1 activity (Morgan et al. 2020). Concurrently, miR-18a knockdown increased the expression of MST1 and significantly reducing cervical cancer cell proliferation Similarly, both MCV and SV40 small T (ST) antigens have been shown to enhance YAP1 activity via PAK1-dependent inhibition of NF2 (Nguyen et al. 2014). Expression of ST lead to both an increase of PAK1 protein levels as well as active phospho-T423 PAK1, which in turn lead to phosphorylation and inhibition of NF2. Furthermore, knockdown of YAP1 was sufficient to inhibit the transforming properties of ST (Nguyen et al. 2014). In addition, the murine polyomavirus small t antigen (PyST) has been shown to directly bind and stabilize the YAP1 protein (Hwang et al. 2014). Binding to PyST enhanced YAP1 association with the protein phosphatase PP2A, leading to a decrease of phospho-S397 YAP1 and a subsequent reduction of YAP1 protein degradation. A subsequent study from the same lab later showed that also the murine polyomavirus middle T (MT) antigen binds to the WW domains of both YAP1 and TAZ (Rouleau et al. 2016). YAP1 was necessary for MT-mediated transformation of NIH3T3 cells and binding to MT resulted in a dephosphorylation of S397 and stabilization of the YAP1 protein, as well as a localization of YAP1 at the cell membrane. Taken together, these findings demonstrate that activation of YAP1 is an important factor for the transformation function of various viral oncoproteins.
Mechanotransduction
Contact inhibition is a major factor in regulating tissue architecture and cell growth in vitro. Cells can sense external forces and changes in the surrounding tissues and react to these stimuli by changing the tension and organization of their F-actin cytoskeleton (Halder et al. 2012; Zanconato et al. 2016). The architecture of normal healthy tissues has tumor suppressive functions and altered tissue composition, stiffness, and extracellular matrix (ECM) might contribute to oncogenic signaling in tumors (Lee and Vasioukhin 2008). The activity of YAP1 is subject to regulation by these mechanical stimuli, as exemplified by the nuclear exclusion of the YAP1 protein at high cell confluency conditions or when cells are grown on a soft ECM in vitro (Dupont et al. 2011; Aragona et al. 2013). Importantly, while LATS1/2 function was dispensible for the mechanotransduction-mediated controls YAP1 activity and localization, it primarily relied on F-actin architecture and Rho activity (Dupont et al. 2011; Aragona et al. 2013). In line with these findings, it has been shown that increased actin polymerization and extra F-actin, caused by the overexpression of the actin nucleation factor Diaphanous (DiaCA), lead to YAP1/Yorkie-dependent tissue overgrowth in vivo in Drosophila (Sansores-Garcia et al. 2011). In turn, overexpression of actin capping proteins, that inhibit the accumulation of F-actin, lead to an inhibition of YAP1/Yorkie activity in Drosophila (Fernandez et al. 2011). A recent study by Panciera et al. has shown that the transformation of primary cells by activated RTK-Ras signaling requires additional mechanical stimuli that resulted in activated YAP1 signaling (Panciera et al. 2020).
YAP1 gene fusions
Recent cancer genome sequencing studies have identified a series of gene fusions involving the N-terminal regions of YAP1 in different tumor types, including Supratentorial (ST) Ependymoma (YAP1-MAMLD1, YAP1-FAM118B) (Pajtler et al. 2015; Pajtler et al. 2019), Epithelioid Hemangioendothelioma (YAP1-TFE3) (Antonescu et al. 2013), Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma (YAP1-SS18) (Hu et al. 2018a), Poroma/Porocarcinoma (YAP1-MAML2, YAP1-NUTM1) (Sekine et al. 2019), NF2-wild type Meningioma (YAP1-MAML2, YAP1-FAM118B, YAP1-PYGO1, YAP1-LMO1) (Schieffer et al. 2020; Sievers et al. 2020), and Sclerosing Epithelioid Fibrosarcoma (YAP1-KMT2A) (Kao et al. 2020; Puls et al. 2020) (Table 1, Figure 4). Several of these YAP1 gene fusions are recurrent and their presence defines a separate tumor subtype, such as ST-EPN-YAP1 ependymoma (YAP1-MAMLD1) (Pajtler et al. 2015) or Epithelioid Hemangioendothelioma (YAP1-TFE3) (Antonescu et al. 2013). In addition, gene fusions involving TAZ have also been recurrently identified in different cancer subtypes, such as WWTR1(TAZ)-CAMTA1 in a subtype of Epithelioid Hemangioendothelioma (Errani et al. 2011; Tanas et al. 2011; Tanas et al. 2016).
Table 1:
Summary of recurrent YAP1 and TAZ fusions identified in human cancers.
| Fusion | Tumor type | Retained YAP1/TAZ amino acids | Retained C-terminal fusion partner amino acids | References |
|---|---|---|---|---|
| YAP1-FAM118B | ST-Ependymoma; NF2-wild type Meningioma | aa1-aa388 (exons 1–7) | aa1-aa351 (entire sequence) | (Pajtler et al. 2015; Schieffer et al. 2020) |
| YAP1-KMT2A | Sclerosing Epithelioid Fibrosarcoma; Fibromyxoid Sarcoma | 1) aa1-aa267 (exons 1–4) 2) aa1-aa328 (exons 1–5) |
1) aa1112-aa3969 (exons 5–36) 2) aa1053-aa3969 (exons 4–36) |
(Kao et al. 2020; Puls et al. 2020) |
| YAP1-LMO1 | NF2-wild type Meningioma | aa1-aa267(exons 1–4) | aa9-aa156 (exons 2–4) | (Sievers et al. 2020) |
| YAP1-MAMLD1 | ST-Ependymoma | 1) aa1-aa328 (exons 1–5) 2) aa1-aa344 (exons 1–6) |
1) aa58-aa774 (exons 3–7) 2) aa33-aa774 (exons 2–7) |
(Pajtler et al. 2015) |
| YAP1-MAML2 | Head and Neck Carcinoma; Nasopharyngeal Carcinomas; NF2-wild type Meningioma; Ovarian Carcinoma; Poroma/Porocarcinoma; Retiform and Composite Hemangioendothelioma | 1) aa1-aa107 (exon 1) 2) aa1-aa328 (exons 1–5) |
1) aa172-aa1152 (exons 2–5) 2) aa172-aa1152 (exons 2–5) |
(Picco et al. 2019; Sekine et al. 2019; Antonescu et al. 2020; Sievers et al. 2020) |
| YAP1-NUTM1 | Poroma/Porocarcinoma | 1) aa1-aa229 (exons 1–3) 2) aa1-aa267 (exons 1–4) |
1) aa6-aa1132 (exons 2–7) 2) aa6-aa1132 (exons 2–7) |
(Sekine et al. 2019) |
| YAP1-PYGO1 | NF2-wild type Meningioma | aa1-aa267(exons 1–4) | aa17-aa419(exons 2–3) | (Sievers et al. 2020) |
| YAP1-SS18 | Cervical Squamous Cell Carcinoma; Endocervical Adenocarcinoma | aa1-aa107 (exon 1) | aa1-aa418 (entire sequence) | (Szulzewsky et al. 2020) |
| YAP1-TFE3 | Epithelioid Hemangioendothelioma | aa1-aa107 (exon 1) | aa179-aa575 (exons 4–10) | (Antonescu et al. 2013) |
| TAZ-CAMTA1 | Epithelioid Hemangioendothelioma | 1) exons 1–2 2) exons 1–4 |
1) exons 9–23 2) exons 8/9–23 |
(Errani et al. 2011; Tanas et al. 2011) |
Figure 4 –
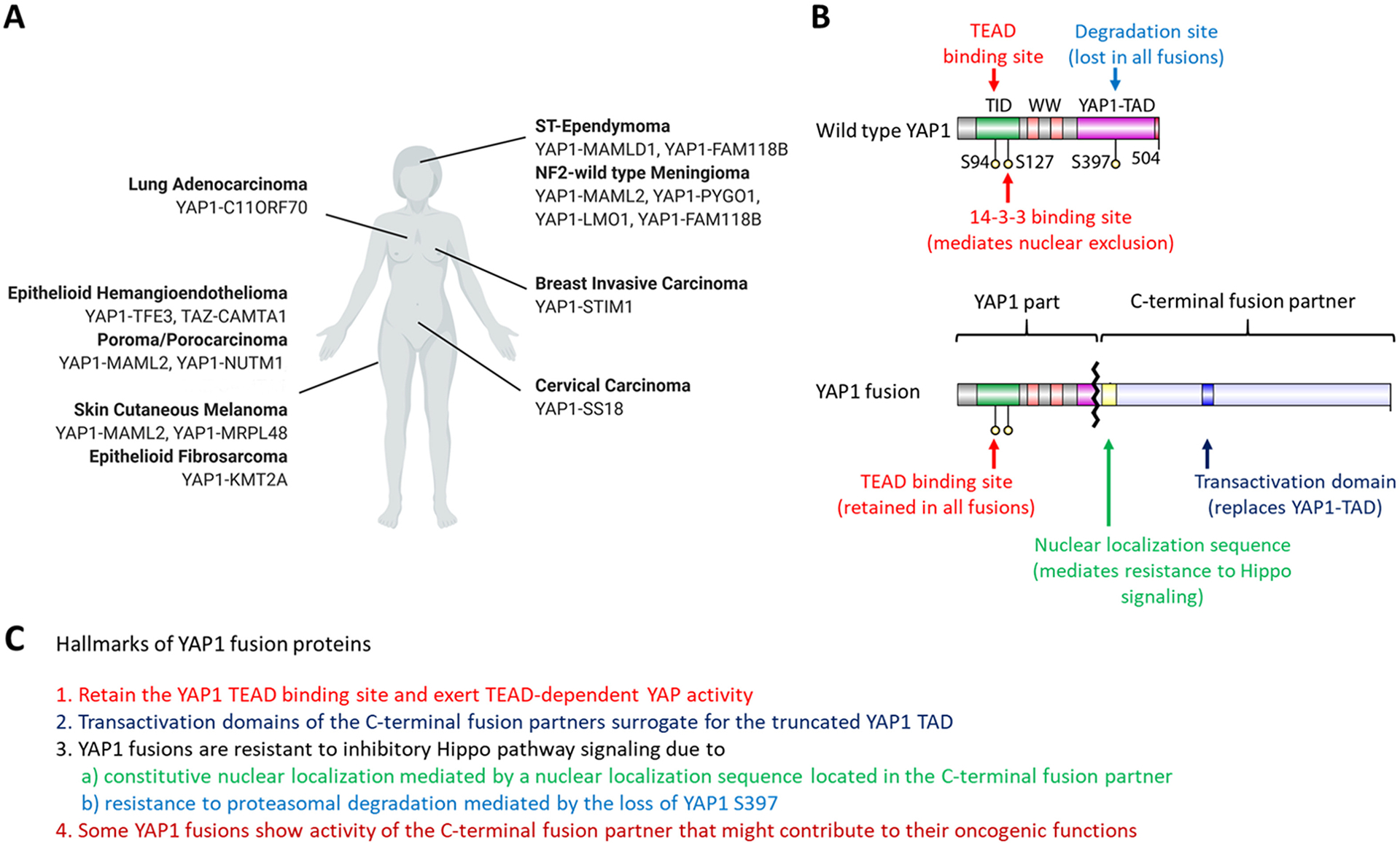
YAP1 and TAZ fusions in different cancer types. A: Overview of known YAP1 and TAZ gene fusions and the cancer types they are found in. B: Structural features that are shared between known YAP1 fusion proteins. All known YAP1 fusions retain the YAP1 TID domain necessary for binding to TEADs and lose the S397 residue of wild type YAP1 important for Hippo-mediated proteasomal degradation. In addition, the C-terminal fusion partner contains a nuclear localization sequence that mediates the nuclear localization of the YAP1 fusion proteins. The C-terminal fusion partner also contains a transactivation domain (TAD) that surrogates for the YAP1 TAD that is lost in the YAP1 fusions. C) Hallmarks of known YAP1 fusions. Figure was created with BioRender.com
These YAP1 gene fusions occur in tumors that generally show a low overall mutational burden (Pajtler et al. 2015; Rosenbaum et al. 2020), suggesting that these fusion transcripts might be the tumor initiating events and oncogenic drivers in these tumors, as well as potential therapeutic targets. We and others (Pajtler et al. 2019; Szulzewsky et al. 2020; Takadera et al. 2020) have recently shown that several of these YAP1 fusion transcripts (YAP1-MAMLD1, YAP1-FAM118B, YAP1-TFE3, and YAP1-SS18) are sufficient to induce tumor formation when expressed either in the brain or in the muscles of prenatal or neonatal mice using somatic cell gene transfer mouse models, strongly supporting the notion that these fusions are the likely oncogenic drivers in their respective tumors.
All examined YAP1 fusion proteins exert TEAD-dependent YAP activity and were able to both activate YAP1-responsive reporter systems and induce the expression of known YAP1 target genes (Pajtler et al. 2019; Sekine et al. 2019; Szulzewsky et al. 2020). The sequence of these YAP1 fusions consists of an N-terminal part of the YAP1 protein fused to the C-terminal part of another protein. Depending on the particular YAP1 gene fusion, different lengths of the N-terminal YAP1 sequence are retained. YAP1-TFE3, YAP1-SS18, and some variants of YAP1-MAML2 only retain the first 107 amino acids of YAP1, that are missing the 14-3-3 binding domain, the WW and the YAP1 transactivation domains (TAD). By contrast, YAP1-MAMLD1 and YAP1-FAM118B retain large parts of YAP1 sequence (up to amino acid 344 and 388, respectively), YAP1-FAM118B retains a part of the YAP1 TAD, although it is unclear if it is still functional. What all N-terminal YAP1 fusions have in common is that they retain the TEAD binding domain (including the S94 residue) but are truncated upstream of the S397 residue, which is necessary for the Hippo Pathway-mediated proteasomal degradation of YAP1. Most of the known C-terminal fusion partners possess putative transactivation or p300 interaction domains that might surrogate for the YAP1 TAD that is missing in fusions. In fact, truncation experiments have shown that the MAMLD1 TAD is necessary and sufficient for the YAP1-like transcriptional activity of YAP1-MAMLD1 (Szulzewsky et al. 2020).
As stated earlier, the activity of wild type YAP1 is regulated through phosphorylation by LATS1/2, which in turn leads to both nuclear exclusion and proteasomal degradation (Zhao et al. 2007; Zhao et al. 2010b). Phosphorylation of a series of five serine residues is important for this regulation (S61, S109, S127, S164, S397), with S127 (nuclear exclusion) and S397 (proteasomal degradation) being the functionally most important residues (Zhao et al. 2007; Zhao et al. 2010b).
By contrast, the YAP activity of YAP1 fusion proteins is resistant to inhibitory Hippo Pathway signaling (Figure 5). While the YAP activity of wild type YAP1 was significantly reduced upon overexpression of the Hippo proteins LATS1, MOB1, and MST1, the YAP activity of YAP1 fusions was either unaffected (YAP1-MAMLD1, YAP1-FAM118B, YAP1-TFE3) or significantly less affected compared to wild type YAP1 (YAP1-SS18) (Szulzewsky et al. 2020). This is likely mediated by two factors. On the one hand, the YAP1 sequence of all known YAP1 fusion transcripts is truncated upstream of S397 (important for wild-type YAP1 protein degradation) and in vitro experiments at different cell densities indicate that, in contrast to wild type YAP1, YAP1 fusion proteins are not degraded at high cell densities (Szulzewsky et al. 2020). On the other hand, YAP1 fusion proteins displayed a constitutively nuclear localization even at high cell densities and computational mapping and truncation experiments revealed NLS in the different C-terminal fusion partners (such as MAMLD1, FAM118B, TFE3, and SS18) that were responsible for the constitutive nuclear localization of the corresponding YAP1 fusion proteins (Sekine et al. 2019; Szulzewsky et al. 2020). Deletion of this NLS from the YAP1-MAMLD1 sequence led to a purely cytoplasmic localization of the fusion protein and a loss of its ability to induce tumor formation (Pajtler et al. 2019; Szulzewsky et al. 2020). Similarly, WWTR1(TAZ)-CAMTA1 has also been shown to be resistant to inhibitory Hippo Pathway signaling, attributed at least in part to an NLS near the C-terminus of CAMTA1, deletion of which resulted in cytoplasmic localization and a reduced ability to transform NIH3T3 cells in vitro (Tanas et al. 2016).
Figure 5 -.
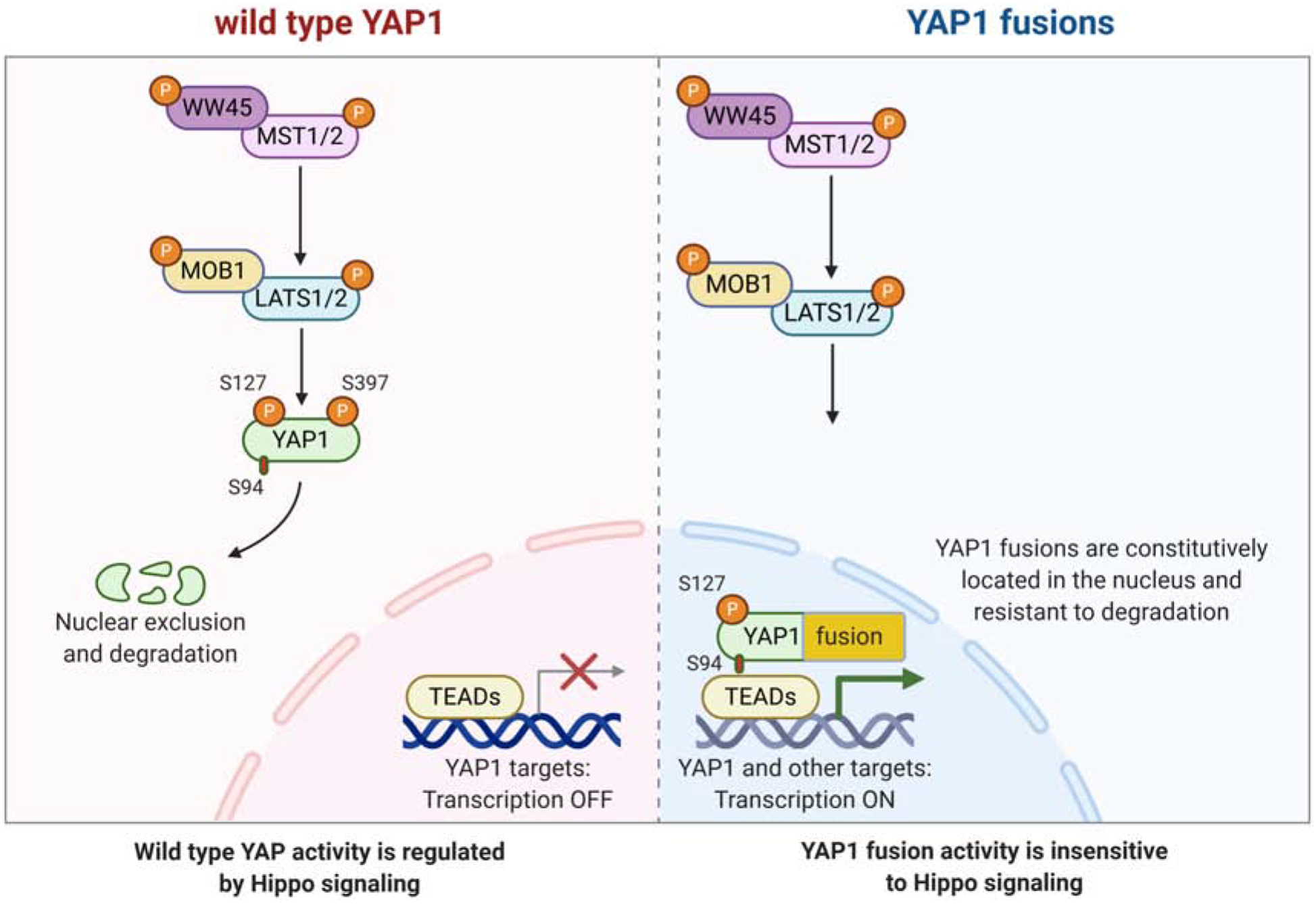
Oncogenic mechanisms of YAP1 fusions. Left: Regulation of wild type YAP1 activity by the Hippo pathway. Right: YAP1 fusion proteins are resistant to Hippo pathway-mediated regulation, due to both constitutive nuclear localization (mediated by the nuclear localization sequence present in the C-terminal fusion partner sequence) and resistance to proteasomal degradation (mediated by the loss of YAP1 S397 in the fusion protein). Figure was created with BioRender.com
Moreover, constitutive activation of YAP1 signaling by expression of a two point-mutant (S127/397A-YAP1), that was resistant to both nuclear exclusion and proteasomal degradation, was sufficient to induce tumor formation upon intracranial expression in new born pups, whereas single point-mutants (S127A-YAP1 or S397A-YAP1) were unable to cause cancer (Szulzewsky et al. 2020). In addition, targeted combined knockout of Lats1 and Lats2 in Neurod6-positive brain cells resulted in the formation of brain tumors shortly after birth (Eder et al. 2020). These results suggest that YAP1 fusion events represent activating mutations that generated a deregulated YAP1, insensitive to Hippo Pathway-mediated functional inhibition.
RNA-Seq experiments indicated that YAP1-MAMLD1, YAP1-FAM118B, and YAP1-TFE3 shared a core transcriptional signature that overlapped with wild type YAP1. Moreover, Cut&Run experiments demonstrated that all three YAP1 fusions occupied YAP1 target regions. In addition, YAP1-TFE3 also occupied TFE3 target regions, indicating that at least some YAP1 fusion proteins exert activity of both fusion partners. This was further supported by the fact that intracranial tumors caused by expression of YAP1-TFE3 showed a drastically different histomorphology compared to tumors generated by other YAP1 fusions, suggesting that the TFE3 activity contributed to the biology of the tumor cells (Szulzewsky et al. 2020).
Several studies have shown that the YAP activity of several YAP1 fusion proteins (YAP1-MAMLD1, YAP1-FAM118B, YAP1-TFE3, YAP1-SS18), as well as WWTR1(TAZ)-CAMTA1 largely relies on the interaction with TEAD transcription factors (Tanas et al. 2016; Pajtler et al. 2019; Szulzewsky et al. 2020). TEAD binding motifs were enriched in Cut&Run and ChIP-Seq peaks of both human ST-EPN-YAP1 tumors and in vitro-transduced cells expressing YAP1-MAMLD1, YAP1-FAM118B, or YAP1-TFE3 (Pajtler et al. 2019; Szulzewsky et al. 2020) and TEAD1–4 knockdown and/or S94A mutation of the YAP1 fusion sequences both resulted in reduced YAP1 activity (Pajtler et al. 2019; Szulzewsky et al. 2020). Similar results have been observed for WWTR1(TAZ)-CAMTA1 (Tanas et al. 2016).
The ability of YAP1 fusions to form tumors was either completely abolished (YAP1-FAM118B and YAP1-SS18) or significantly reduced (YAP1-MAMLD1 and YAP1-TFE3) by S94A point mutation, which ablates YAP1-TEAD interaction (Szulzewsky et al. 2020). Moreover, treatment with Verteporfin decreased the YAP transcriptional activity of YAP1 fusion proteins as well as inhibited the growth of YAP1-FAM118B-driven mouse tumor cells in vitro (Szulzewsky et al. 2020). These results suggest that inhibition of YAP activity via pharmacological YAP1-TEAD disruption might be a feasible approach for the treatment of tumors displaying activated YAP1 (or TAZ) as well as YAP1 fusion-driven cancers, however further studies will be necessary to address this point.
In summary, all YAP1 gene fusions analyzed to date were oncogenic and functioned by exerting TEAD-dependent YAP activity that was resistant to inhibitory Hippo Pathway signaling, while the activity of the C-terminal fusion partner was able to contribute to the functions of some YAP1 fusions.
Anti-YAP1 therapy
Several small molecule inhibitors against YAP1 function have been reported (reviewed in (Deel et al. 2015)). Some of these inhibitors have been designed to activate the tumor suppressive Hippo Core kinase pathway, e.g. by inhibiting PP2A (Swingle et al. 2009) or by directly activating MST/LATS kinases (Basu et al. 2014). However, these drugs would likely be ineffective in tumors that harbor inactivating mutations in Hippo pathway members, tumors that achieve YAP1 overactivation through Hippo-independent pathways, or YAP1 fusion positive tumors, since YAP1 fusion proteins are resistant to inhibitory Hippo signaling (Szulzewsky et al. 2020). A second class of molecules could attenuate YAP1 function independently of Hippo signaling, by inhibiting the transduction of mechanical stimuli, e.g. by inhibiting the function of RHO/ROCK or destabilizing F-actin (Dupont et al. 2011).
Lastly, the dependence of the YAP1 transforming functions on the interaction with TEAD transcription factors might be an Achilles’ heel of this potent oncogene that can be therapeutically exploited. Verteporfin is a small molecule inhibitor that is presently used as a photosensitizer for photodynamic therapy to treat macular degeneration (Verteporfin In Photodynamic Therapy Study 2001), but it has also been shown to inhibit the function of YAP1 by disrupting the interaction between YAP1 and TEADs in a photosensitizer-independent manner (Liu-Chittenden et al. 2012). Treatment with Verteporfin reduced the growth of YAP1-driven tumors in vivo (Brodowska et al. 2014) and recently several other small molecule inhibitors targeting YAP1-TEAD interactions have been developed (Pobbati et al. 2015; Song et al. 2018; Bum-Erdene et al. 2019). The importance of the interaction with TEAD transcription factors is also shared by all analyzed YAP1 fusion proteins and this could make it possible to treat different YAP1 fusion-positive tumors in a similar manner, however, further studies will be necessary to analyze if this could result in regression of established tumors in vivo.
It remains to be seen if anti-YAP1 therapy in general will be sufficient to inhibit the growth of human tumors that display activated YAP signaling. Tumors in which aberrant YAP activity is the underlying tumor-initiating event and the main oncogenic driver, such as YAP1 fusion-driven tumors or potentially also tumors caused by inactivation of Merlin/NF2, are likely to be sensitive to anti-YAP1 therapy. In turn, targeting YAP1 function may or may not be an effective treatment for tumors in which aberrant YAP activity is not a causal event, but rather the result of oncogenic signaling mediated by other oncogenic drivers. These tumors may not exclusively depend on aberrant YAP activity and might use alternative pathways to support their growth. However, recent studies demonstrated that YAP1 contributes to the resistance of melanoma and breast cancer cells to BRAF and HER2 inhibitors, respectively, indicating that anti-YAP1 therapies might be a feasible concomitant treatment to overcome this resistance (Lin et al. 2015; Kim et al. 2016).
Conclusions
In addition to its pivotal roles in normal tissue development, homeostasis, and regeneration, YAP1 also exerts potent pro-oncogenic functions and high nuclear YAP1 staining or elevated expression of YAP1 has been detected in several human cancer types. The mechanisms by which YAP1 activity is de-regulated in human cancers are versatile and differ between cancer (sub)-types, which may have important implications for the efficacy of anti-YAP therapy. In some cancer types de-regulated YAP activity is the actual tumor-initiating event and sustained YAP activity is required for tumor maintenance, such as in tumors harboring YAP1 gene fusions, especially since these tumors generally harbor very few additional mutations; and it is highly plausible that in these tumors anti-YAP therapy might be a feasible strategy. By contrast, in other cancers, such as tumors harboring several co-occurring mutations and/or gains and losses of whole chromosomes, de-regulation of YAP activity may not be a bona fide oncogenic driver but rather a consequence of aberrant oncogenic signaling. In these tumors YAP activity might rather contribute to aggressiveness and/or resistance to therapy, but inhibition of YAP activity itself might not be sufficient to inhibit the growth of these tumors. Further studies will be necessary to determine if targeting YAP1 in these cancers could be a possible approach in combination with other drugs to overcome treatment resistance.
Highlights:
De-regulation of YAP1 activity plays important roles in the progression, aggressiveness, and therapy resistance of several human cancers
The mechanisms that facilitate the de-regulation of YAP1 activity in human cancers are versatile and include inactivation of tumor suppressive upstream Hippo Pathway regulators (such as LATS, NF2, or FAT), activation by intersecting pathways (such as GPCRs, the Wnt pathway, or the mevalonate pathway), miRNAs, or viral oncogenes
YAP1 gene fusions are the likely oncogenic drivers in several subtypes of human cancers and constitute a novel type of activating YAP1 mutation, that renders YAP activity resistant to inhibitory signaling from the Hippo pathway
Therapeutic targeting of YAP activity by inhibiting the YAP1-TEAD interaction might be a feasible approach, but efficacy might depend on whether YAP activity is an actual oncogenic driver and required for the maintenance of a particular tumor
Acknowledgements
Research in the V.V. laboratory is supported by the U.S. National Cancer Institute grants R01CA188452 and R01CA234050. Research in the E.C.H. laboratory is supported by the U.S. National Cancer Institute grant U54CA243125-01 and the Ivy Foundation Translational Adult Glioma Grant Award from The Ben and Catherine Ivy Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Ahronowitz I, Xin W, Kiely R, Sims K, MacCollin M, Nunes FP. 2007. Mutational spectrum of the NF2 gene: a meta-analysis of 12 years of research and diagnostic laboratory findings. Hum Mutat 28: 1–12. [DOI] [PubMed] [Google Scholar]
- Alexander J, LaPlant QC, Pattwell SS, Szulzewsky F, Cimino PJ, Caruso FP, Pugliese P, Chen Z, Chardon F, Hill AJ et al. 2020. Multimodal single-cell analysis reveals distinct radioresistant stem-like and progenitor cell populations in murine glioma. Glia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR, Dickson BC, Sung YS, Zhang L, Suurmeijer AJH, Stenzinger A, Mechtersheimer G, Fletcher CDM. 2020. Recurrent YAP1 and MAML2 Gene Rearrangements in Retiform and Composite Hemangioendothelioma. Am J Surg Pathol 44: 1677–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR, Le Loarer F, Mosquera JM, Sboner A, Zhang L, Chen CL, Chen HW, Pathan N, Krausz T, Dickson BC et al. 2013. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer 52: 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. 2013. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154: 1047–1059. [DOI] [PubMed] [Google Scholar]
- Arakaki AKS, Pan W-A, Trejo J. 2020. Regulation of GPCR activation of the Hippo pathway in metastatic breast cancer. The FASEB Journal 34: 1–1. [Google Scholar]
- Arakaki AKS, Pan WA, Lin H, Trejo J. 2018. The alpha-arrestin ARRDC3 suppresses breast carcinoma invasion by regulating G protein-coupled receptor lysosomal sorting and signaling. J Biol Chem 293: 3350–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H, Hayashi N, Nagase H, Ogawa M, Nakamura Y. 1994. Alternative splicing of the NF2 gene and its mutation analysis of breast and colorectal cancers. Hum Mol Genet 3: 565–568. [DOI] [PubMed] [Google Scholar]
- Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V et al. 2014. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell 158: 157–170. [DOI] [PubMed] [Google Scholar]
- Baser ME, De Rienzo A, Altomare D, Balsara BR, Hedrick NM, Gutmann DH, Pitts LH, Jackler RK, Testa JR. 2002. Neurofibromatosis 2 and malignant mesothelioma. Neurology 59: 290–291. [DOI] [PubMed] [Google Scholar]
- Basu D, Lettan R, Damodaran K, Strellec S, Reyes-Mugica M, Rebbaa A. 2014. Identification, mechanism of action, and antitumor activity of a small molecule inhibitor of hippo, TGF-beta, and Wnt signaling pathways. Mol Cancer Ther 13: 1457–1467. [DOI] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. 2003. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell 11: 11–23. [DOI] [PubMed] [Google Scholar]
- Bertero T, Oldham WM, Cottrill KA, Pisano S, Vanderpool RR, Yu Q, Zhao J, Tai Y, Tang Y, Zhang YY et al. 2016. Vascular stiffness mechanoactivates YAP/TAZ-dependent glutaminolysis to drive pulmonary hypertension. J Clin Invest 126: 3313–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscardi JS, Tice DA, Parsons SJ. 1999. c-Src, receptor tyrosine kinases, and human cancer. Adv Cancer Res 76: 61–119. [DOI] [PubMed] [Google Scholar]
- Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst 87: 796–802. [DOI] [PubMed] [Google Scholar]
- Brodowska K, Al-Moujahed A, Marmalidou A, Meyer Zu Horste M, Cichy J, Miller JW, Gragoudas E, Vavvas DG. 2014. The clinically used photosensitizer Verteporfin (VP) inhibits YAP-TEAD and human retinoblastoma cell growth in vitro without light activation. Exp Eye Res 124: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buglioni S, Vici P, Sergi D, Pizzuti L, Di Lauro L, Antoniani B, Sperati F, Terrenato I, Carosi M, Gamucci T et al. 2016. Analysis of the hippo transducers TAZ and YAP in cervical cancer and its microenvironment. Oncoimmunology 5: e1160187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bum-Erdene K, Zhou D, Gonzalez-Gutierrez G, Ghozayel MK, Si Y, Xu D, Shannon HE, Bailey BJ, Corson TW, Pollok KE et al. 2019. Small-Molecule Covalent Modification of Conserved Cysteine Leads to Allosteric Inhibition of the TEADYap Protein-Protein Interaction. Cell Chem Biol 26: 378–389 e313. [DOI] [PubMed] [Google Scholar]
- Cacev T, Aralica G, Loncar B, Kapitanovic S. 2014. Loss of NF2/Merlin expression in advanced sporadic colorectal cancer. Cell Oncol (Dordr) 37: 69–77. [DOI] [PubMed] [Google Scholar]
- Cai H, Xu Y. 2013. The role of LPA and YAP signaling in long-term migration of human ovarian cancer cells. Cell Commun Signal 11: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. 2007. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol 17: 2054–2060. [DOI] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH. 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24: 2076–2086. [DOI] [PubMed] [Google Scholar]
- Chen D, Sun Y, Wei Y, Zhang P, Rezaeian AH, Teruya-Feldstein J, Gupta S, Liang H, Lin HK, Hung MC et al. 2012. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med 18: 1511–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chan SW, Zhang X, Walsh M, Lim CJ, Hong W, Song H. 2010. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev 24: 290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Zhao D, Li J, Liang X, Li J, Chang A, Henry VK, Lan Z, Spring DJ, Rao G et al. 2019. Symbiotic Macrophage-Glioma Cell Interactions Reveal Synthetic Lethality in PTEN-Null Glioma. Cancer Cell 35: 868–884 e866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang N, Xie R, Wang W, Cai J, Choi KS, David KK, Huang B, Yabuta N, Nojima H et al. 2015. Homeostatic control of Hippo signaling activity revealed by an endogenous activating mutation in YAP. Genes Dev 29: 1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JQ, Lee WC, Klein MA, Cheng GZ, Jhanwar SC, Testa JR. 1999. Frequent mutations of NF2 and allelic loss from chromosome band 22q12 in malignant mesothelioma: evidence for a two-hit mechanism of NF2 inactivation. Genes Chromosomes Cancer 24: 238–242. [PubMed] [Google Scholar]
- Cho YS, Zhu J, Li S, Wang B, Han Y, Jiang J. 2018. Regulation of Yki/Yap subcellular localization and Hpo signaling by a nuclear kinase PRP4K. Nat Commun 9: 1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JS, Kim CS, Berdis A. 2018. Inhibition of Translesion DNA Synthesis as a Novel Therapeutic Strategy to Treat Brain Cancer. Cancer Res 78: 1083–1096. [DOI] [PubMed] [Google Scholar]
- Chosdol K, Misra A, Puri S, Srivastava T, Chattopadhyay P, Sarkar C, Mahapatra AK, Sinha S. 2009. Frequent loss of heterozygosity and altered expression of the candidate tumor suppressor gene ‘FAT’ in human astrocytic tumors. BMC Cancer 9: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciamporcero E, Shen H, Ramakrishnan S, Yu Ku S, Chintala S, Shen L, Adelaiye R, Miles KM, Ullio C, Pizzimenti S et al. 2016. YAP activation protects urothelial cell carcinoma from treatment-induced DNA damage. Oncogene 35: 1541–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosset E, Ilmjarv S, Dutoit V, Elliott K, von Schalscha T, Camargo MF, Reiss A, Moroishi T, Seguin L, Gomez G et al. 2017. Glut3 Addiction Is a Druggable Vulnerability for a Molecularly Defined Subpopulation of Glioblastoma. Cancer Cell 32: 856–868 e855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottini F, Hideshima T, Xu C, Sattler M, Dori M, Agnelli L, ten Hacken E, Bertilaccio MT, Antonini E, Neri A et al. 2014. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat Med 20: 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croci O, De Fazio S, Biagioni F, Donato E, Caganova M, Curti L, Doni M, Sberna S, Aldeghi D, Biancotto C et al. 2017. Transcriptional integration of mitogenic and mechanical signals by Myc and YAP. Genes Dev 31: 2017–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deel MD, Li JJ, Crose LE, Linardic CM. 2015. A Review: Molecular Aberrations within Hippo Signaling in Bone and Soft-Tissue Sarcomas. Front Oncol 5: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Varelas X, Guan KL. 2020. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat Rev Drug Discov 19: 480–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Zhang H, Qu Y, Deng T, Huang D, Liu R, Zhang L, Bai M, Zhou L, Ying G et al. 2016. Onco-miR-130 promotes cell proliferation and migration by targeting TGFbetaR2 in gastric cancer. Oncotarget 7: 44522–44533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183. [DOI] [PubMed] [Google Scholar]
- Ebert C, von Haken M, Meyer-Puttlitz B, Wiestler OD, Reifenberger G, Pietsch T, von Deimling A. 1999. Molecular genetic analysis of ependymal tumors. NF2 mutations and chromosome 22q loss occur preferentially in intramedullary spinal ependymomas. Am J Pathol 155: 627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder N, Roncaroli F, Dolmart MC, Horswell S, Andreiuolo F, Flynn HR, Lopes AT, Claxton S, Kilday JP, Collinson L et al. 2020. YAP1/TAZ drives ependymoma-like tumour formation in mice. Nat Commun 11: 2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DN, Ngwa VM, Wang S, Shiuan E, Brantley-Sieders DM, Kim LC, Reynolds AB, Chen J. 2017. The receptor tyrosine kinase EphA2 promotes glutamine metabolism in tumors by activating the transcriptional coactivators YAP and TAZ. Sci Signal 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa H, Jingushi K, Hirono T, Ueda Y, Kitae K, Nakata W, Fujita K, Uemura M, Nonomura N, Tsujikawa K. 2016. The miR-130 family promotes cell migration and invasion in bladder cancer through FAK and Akt phosphorylation by regulating PTEN. Sci Rep 6: 20574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbediwy A, Vincent-Mistiaen ZI, Spencer-Dene B, Stone RK, Boeing S, Wculek SK, Cordero J, Tan EH, Ridgway R, Brunton VG et al. 2016. Integrin signalling regulates YAP and TAZ to control skin homeostasis. Development 143: 1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errani C, Zhang L, Sung YS, Hajdu M, Singer S, Maki RG, Healey JH, Antonescu CR. 2011. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer 50: 644–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R, Kim NG, Gumbiner BM. 2013. Regulation of Hippo pathway by mitogenic growth factors via phosphoinositide 3-kinase and phosphoinositide-dependent kinase-1. Proc Natl Acad Sci U S A 110: 2569–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Gao Y, Rao J, Wang K, Zhang F, Zhang C. 2017. YAP-1 Promotes Tregs Differentiation in Hepatocellular Carcinoma by Enhancing TGFBR2 Transcription. Cell Physiol Biochem 41: 1189–1198. [DOI] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319: 1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, Zaidi MR, Ksander BR, Merlino G, Sodhi A et al. 2014. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell 25: 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez BG, Gaspar P, Bras-Pereira C, Jezowska B, Rebelo SR, Janody F. 2011. Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development 138: 2337–2346. [DOI] [PubMed] [Google Scholar]
- Fernandez LA, Squatrito M, Northcott P, Awan A, Holland EC, Taylor MD, Nahle Z, Kenney AM. 2012. Oncogenic YAP promotes radioresistance and genomic instability in medulloblastoma through IGF2-mediated Akt activation. Oncogene 31: 1923–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrigno O, Lallemand F, Verrecchia F, L’Hoste S, Camonis J, Atfi A, Mauviel A. 2002. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene 21: 4879–4884. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. 2017. The PI3K Pathway in Human Disease. Cell 170: 605–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa KT, Yamashita K, Sakurai N, Ohno S. 2017. The Epithelial Circumferential Actin Belt Regulates YAP/TAZ through Nucleocytoplasmic Shuttling of Merlin. Cell Rep 20: 1435–1447. [DOI] [PubMed] [Google Scholar]
- Galli GG, Carrara M, Yuan WC, Valdes-Quezada C, Gurung B, Pepe-Mooney B, Zhang T, Geeven G, Gray NS, de Laat W et al. 2015. YAP Drives Growth by Controlling Transcriptional Pause Release from Dynamic Enhancers. Mol Cell 60: 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrant W, Kota S, Troutman S, Mandati V, Fallahi M, Stemmer-Rachamimov A, Kissil JL. 2016. YAP Mediates Tumorigenesis in Neurofibromatosis Type 2 by Promoting Cell Survival and Proliferation through a COX-2-EGFR Signaling Axis. Cancer Res 76: 3507–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujral TS, Kirschner MW. 2017. Hippo pathway mediates resistance to cytotoxic drugs. Proc Natl Acad Sci U S A 114: E3729–E3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Dupont S, Piccolo S. 2012. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol 13: 591–600. [DOI] [PubMed] [Google Scholar]
- Han Y 2019. Analysis of the role of the Hippo pathway in cancer. J Transl Med 17: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CG, Moroishi T, Guan KL. 2015a. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol 25: 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CG, Ng YL, Lam WL, Plouffe SW, Guan KL. 2015b. The Hippo pathway effectors YAP and TAZ promote cell growth by modulating amino acid signaling to mTORC1. Cell Res 25: 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. 2003. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114: 457–467. [DOI] [PubMed] [Google Scholar]
- Haskins JW, Nguyen DX, Stern DF. 2014. Neuregulin 1-activated ERBB4 interacts with YAP to induce Hippo pathway target genes and promote cell migration. Sci Signal 7: ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang L, Chen Y, Remke M, Shih D, Lu F, Wang H, Deng Y, Yu Y, Xia Y et al. 2014. The G protein alpha subunit Galphas is a tumor suppressor in Sonic hedgehog-driven medulloblastoma. Nat Med 20: 1035–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. 2011. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332: 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Schmitz D, Hemmings BA. 2006. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem Biophys Res Commun 345: 50–58. [DOI] [PubMed] [Google Scholar]
- Hernandez NA, Correa E, Avila EP, Vela TA, Perez VM. 2009. PAR1 is selectively over expressed in high grade breast cancer patients: a cohort study. J Transl Med 7: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiemer SE, Zhang L, Kartha VK, Packer TS, Almershed M, Noonan V, Kukuruzinska M, Bais MV, Monti S, Varelas X. 2015. A YAP/TAZ-Regulated Molecular Signature Is Associated with Oral Squamous Cell Carcinoma. Mol Cancer Res 13: 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA et al. 2005. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309: 1074–1078. [DOI] [PubMed] [Google Scholar]
- Horiguchi A, Zheng R, Shen R, Nanus DM. 2008. Inactivation of the NF2 tumor suppressor protein merlin in DU145 prostate cancer cells. Prostate 68: 975–984. [DOI] [PubMed] [Google Scholar]
- Hsu PC, Miao J, Wang YC, Zhang WQ, Yang YL, Wang CW, Yang CT, Huang Z, You J, Xu Z et al. 2018. Inhibition of yes-associated protein down-regulates PD-L1 (CD274) expression in human malignant pleural mesothelioma. J Cell Mol Med 22: 3139–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Wang Q, Tang M, Barthel F, Amin S, Yoshihara K, Lang FM, Martinez-Ledesma E, Lee SH, Zheng S et al. 2018a. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res 46: D1144–D1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yang C, Yang S, Cheng F, Rao J, Wang X. 2018b. miR-665 promotes hepatocellular carcinoma cell migration, invasion, and proliferation by decreasing Hippo signaling through targeting PTPRB. Cell Death Dis 9: 954. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. 2005. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122: 421–434. [DOI] [PubMed] [Google Scholar]
- Huang W, Lv X, Liu C, Zha Z, Zhang H, Jiang Y, Xiong Y, Lei QY, Guan KL. 2012. The N-terminal phosphodegron targets TAZ/WWTR1 protein for SCFbeta-TrCP-dependent degradation in response to phosphatidylinositol 3-kinase inhibition. J Biol Chem 287: 26245–26253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JH, Pores Fernando AT, Faure N, Andrabi S, Adelmant G, Hahn WC, Marto JA, Schaffhausen BS, Roberts TM. 2014. Polyomavirus small T antigen interacts with yes-associated protein to regulate cell survival and differentiation. J Virol 88: 12055–12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Bartolome R, Torres D, Marone R, Feng X, Martin D, Simaan M, Chen M, Weinstein LS, Taylor SS, Molinolo AA et al. 2015. Inactivation of a Galpha(s)-PKA tumour suppressor pathway in skin stem cells initiates basal-cell carcinogenesis. Nat Cell Biol 17: 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imajo M, Miyatake K, Iimura A, Miyamoto A, Nishida E. 2012. A molecular mechanism that links Hippo signalling to the inhibition of Wnt/beta-catenin signalling. EMBO J 31: 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse van Rensburg HJ, Azad T, Ling M, Hao Y, Snetsinger B, Khanal P, Minassian LM, Graham CH, Rauh MJ, Yang X. 2018. The Hippo Pathway Component TAZ Promotes Immune Evasion in Human Cancer through PD-L1. Cancer Res 78: 1457–1470. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. 2003. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev 17: 2514–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Li J, Zhang C, Shang Y, Lin J. 2020. YAPmediated crosstalk between the Wnt and Hippo signaling pathways (Review). Mol Med Rep 22: 4101–4106. [DOI] [PubMed] [Google Scholar]
- Jin L, Alesi GN, Kang S. 2016. Glutaminolysis as a target for cancer therapy. Oncogene 35: 3619–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. 1995. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 9: 534–546. [DOI] [PubMed] [Google Scholar]
- Kaan HYK, Chan SW, Tan SKJ, Guo F, Lim CJ, Hong W, Song H. 2017. Crystal structure of TAZ-TEAD complex reveals a distinct interaction mode from that of YAP-TEAD complex. Sci Rep 7: 2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC et al. 2000. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J 19: 6778–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Huang T, Zhou Y, Zhang J, Lung RWM, Tong JHM, Chan AWH, Zhang B, Wong CC, Wu F et al. 2018. miR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesis. Cell Death Dis 9: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao YC, Lee JC, Zhang L, Sung YS, Swanson D, Hsieh TH, Liu YR, Agaram NP, Huang HY, Dickson BC et al. 2020. Recurrent YAP1 and KMT2A Gene Rearrangements in a Subset of MUC4-negative Sclerosing Epithelioid Fibrosarcoma. Am J Surg Pathol 44: 368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, Zhong Y, Wu CJ, Sadanandam A, Hu B et al. 2014. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman R, Halder G. 2018. Cell Junctions in Hippo Signaling. Cold Spring Harb Perspect Biol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M 2012. Function and cancer genomics of FAT family genes (review). Int J Oncol 41: 1913–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Kim CG, Kim SK, Shin SJ, Choe EA, Park SH, Shin EC, Kim J. 2018. YAP-Induced PD-L1 Expression Drives Immune Evasion in BRAFi-Resistant Melanoma. Cancer Immunol Res 6: 255–266. [DOI] [PubMed] [Google Scholar]
- Kim MH, Kim J, Hong H, Lee SH, Lee JK, Jung E, Kim J. 2016. Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. EMBO J 35: 462–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NG, Gumbiner BM. 2015. Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J Cell Biol 210: 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NG, Koh E, Chen X, Gumbiner BM. 2011. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci U S A 108: 11930–11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Hwang D, Lee D, Kim JH, Kim SY, Lim DS. 2017. MRTF potentiates TEAD-YAP transcriptional activity causing metastasis. EMBO J 36: 520–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler M, Speight P, Little D, Di Ciano-Oliveira C, Szaszi K, Kapus A. 2018. Mediated nuclear import and export of TAZ and the underlying molecular requirements. Nat Commun 9: 4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Nagai M, Navin NE, Sudol M. 2003. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem 278: 33334–33341. [DOI] [PubMed] [Google Scholar]
- Konsavage WM, Jr., Kyler SL, Rennoll SA, Jin G, Yochum GS. 2012. Wnt/beta-catenin signaling regulates Yes-associated protein (YAP) gene expression in colorectal carcinoma cells. J Biol Chem 287: 11730–11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo JH, Guan KL. 2018. Interplay between YAP/TAZ and Metabolism. Cell Metab 28: 196–206. [DOI] [PubMed] [Google Scholar]
- Koo JH, Plouffe SW, Meng Z, Lee DH, Yang D, Lim DS, Wang CY, Guan KL. 2020. Induction of AP-1 by YAP/TAZ contributes to cell proliferation and organ growth. Genes Dev 34: 72–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M et al. 2014. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25: 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO. 2012. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci U S A 109: E2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar JM, Xiao Y, Norton E, Jiang ZG, Gerhard GM, Kooner S, Warren JSA, Hynes RO. 2019. SRC tyrosine kinase activates the YAP/TAZ axis and thereby drives tumor growth and metastasis. J Biol Chem 294: 2302–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebid A, Chung L, Pardoll DM, Pan F. 2020. YAP Attenuates CD8 T Cell-Mediated Anti-tumor Response. Front Immunol 11: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc L, Lee BK, Yu AC, Kim M, Kambhampati AV, Dupont SM, Seruggia D, Ryu BU, Orkin SH, Kim J. 2018. Yap1 safeguards mouse embryonic stem cells from excessive apoptosis during differentiation. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Vasioukhin V. 2008. Cell polarity and cancer--cell and tissue polarity as a non-canonical tumor suppressor. J Cell Sci 121: 1141–1150. [DOI] [PubMed] [Google Scholar]
- Lee S, Karas PJ, Hadley CC, Bayley VJ, Khan AB, Jalali A, Sweeney AD, Klisch TJ, Patel AJ. 2019. The Role of Merlin/NF2 Loss in Meningioma Biology. Cancers (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Stewart S, Nagtegaal I, Luo J, Wu Y, Colditz G, Medina D, Allred DC. 2012. Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Res 72: 4574–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Adamovich Y, Reuven N, Shaul Y. 2007. The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ 14: 743–751. [DOI] [PubMed] [Google Scholar]
- Levy D, Adamovich Y, Reuven N, Shaul Y. 2008. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell 29: 350–361. [DOI] [PubMed] [Google Scholar]
- Li N, Xie C, Lu N. 2017. Crosstalk between Hippo signalling and miRNAs in tumour progression. FEBS J 284: 1045–1055. [DOI] [PubMed] [Google Scholar]
- Li P, Silvis MR, Honaker Y, Lien WH, Arron ST, Vasioukhin V. 2016. alphaE-catenin inhibits a Src-YAP1 oncogenic module that couples tyrosine kinases and the effector of Hippo signaling pathway. Genes Dev 30: 798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou H, Li F, Chan SW, Lin Z, Wei Z, Yang Z, Guo F, Lim CJ, Xing W et al. 2015. Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway. Cell Res 25: 801–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, Guan KL, Xu Y. 2010. Structural insights into the YAP and TEAD complex. Genes Dev 24: 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Pelissier FA, Zhang H, Lakins J, Weaver VM, Park C, LaBarge MA. 2015. Microenvironment rigidity modulates responses to the HER2 receptor tyrosine kinase inhibitor lapatinib via YAP and TAZ transcription factors. Mol Biol Cell 26: 3946–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM et al. 2013. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat Commun 4: 1877. [DOI] [PubMed] [Google Scholar]
- Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. 2012. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev 26: 1300–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, Li T, Chan SW, Lim CJ, Hong W et al. 2010a. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem 285: 37159–37169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, Demidenko E, Korc M, Shi W, Preis M et al. 2010b. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest 120: 1298–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llado V, Nakanishi Y, Duran A, Reina-Campos M, Shelton PM, Linares JF, Yajima T, Campos A, Aza-Blanc P, Leitges M et al. 2015. Repression of Intestinal Stem Cell Function and Tumorigenesis through Direct Phosphorylation of beta-Catenin and Yap by PKCzeta. Cell Rep 10: 740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzetto E, Brenca M, Boeri M, Verri C, Piccinin E, Gasparini P, Facchinetti F, Rossi S, Salvatore G, Massimino M et al. 2014. YAP1 acts as oncogenic target of 11q22 amplification in multiple cancer subtypes. Oncotarget 5: 2608–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Yu FX. 2019. GPCR-Hippo Signaling in Cancer. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Liu W, Tang P, Jiang D, Gu C, Huang Y, Gong F, Rong Y, Qian D, Chen J et al. 2019. miR-624–5p promoted tumorigenesis and metastasis by suppressing hippo signaling through targeting PTPRB in osteosarcoma cells. J Exp Clin Cancer Res 38: 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubasyuk V, Ouyang H, Yu FX, Guan KL, Zhang K. 2015. YAP inhibition blocks uveal melanogenesis driven by GNAQ or GNA11 mutations. Mol Cell Oncol 2: e970957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Tao Y, Duran A, Llado V, Galvez A, Barger JF, Castilla EA, Chen J, Yajima T, Porollo A et al. 2013. Control of nutrient stress-induced metabolic reprogramming by PKCzeta in tumorigenesis. Cell 152: 599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Meng Z, Chen R, Guan KL. 2019. The Hippo Pathway: Biology and Pathophysiology. Annu Rev Biochem 88: 577–604. [DOI] [PubMed] [Google Scholar]
- Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, Mitani A, Nagase T, Yatomi Y, Aburatani H et al. 2008. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am J Physiol Renal Physiol 294: F542–553. [DOI] [PubMed] [Google Scholar]
- Mana-Capelli S, Paramasivam M, Dutta S, McCollum D. 2014. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol Biol Cell 25: 1676–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manderfield LJ, Engleka KA, Aghajanian H, Gupta M, Yang S, Li L, Baggs JE, Hogenesch JB, Olson EN, Epstein JA. 2014. Pax3 and hippo signaling coordinate melanocyte gene expression in neural crest. Cell Rep 9: 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning SA, Kroeger B, Harvey KF. 2020. The regulation of Yorkie, YAP and TAZ: new insights into the Hippo pathway. Development 147. [DOI] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Guan KL. 2016. Mechanisms of Hippo pathway regulation. Genes Dev 30: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Mottier-Pavie V, Plouffe SW, Hansen CG, Hong AW, Park HW, Mo JS, Lu W, Lu S et al. 2015. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun 6: 8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Hsu PC, Yang YL, Xu Z, Dai Y, Wang Y, Chan G, Huang Z, Hu B, Li H et al. 2017. YAP regulates PD-L1 expression in human NSCLC cells. Oncotarget 8: 114576–114587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitamura T, Watari H, Wang L, Kanno H, Kitagawa M, Hassan MK, Kimura T, Tanino M, Nishihara H, Tanaka S et al. 2014. microRNA 31 functions as an endometrial cancer oncogene by suppressing Hippo tumor suppressor pathway. Mol Cancer 13: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Murakami H, Fujii M, Ishiguro F, Tanaka I, Kondo Y, Akatsuka S, Toyokuni S, Yokoi K, Osada H et al. 2012. YAP induces malignant mesothelioma cell proliferation by upregulating transcription of cell cycle-promoting genes. Oncogene 31: 5117–5122. [DOI] [PubMed] [Google Scholar]
- Mo JS, Yu FX, Gong R, Brown JH, Guan KL. 2012. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs). Genes Dev 26: 2138–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan EL, Patterson MR, Ryder EL, Lee SY, Wasson CW, Harper KL, Li Y, Griffin S, Blair GE, Whitehouse A et al. 2020. MicroRNA-18a targeting of the STK4/MST1 tumour suppressor is necessary for transformation in HPV positive cervical cancer. PLoS Pathog 16: e1008624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, Gregory RI. 2014. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 156: 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O’Neal W, Milgram SL. 2006. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol 26: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T, Imoto I, Matsui T, Kozaki K, Haruki S, Sudol M, Shimada Y, Tsuda H, Kawano T, Inazawa J. 2011. YAP is a candidate oncogene for esophageal squamous cell carcinoma. Carcinogenesis 32: 389–398. [DOI] [PubMed] [Google Scholar]
- Murray LB, Lau YK, Yu Q. 2012. Merlin is a negative regulator of human melanoma growth. PLoS One 7: e43295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya K, Yamagata HD, Arita N, Nakashiro KI, Nose M, Miki T, Hamakawa H. 2007. Identification of homozygous deletions of tumor suppressor gene FAT in oral cancer using CGH-array. Oncogene 26: 5300–5308. [DOI] [PubMed] [Google Scholar]
- Nallet-Staub F, Marsaud V, Li L, Gilbert C, Dodier S, Bataille V, Sudol M, Herlyn M, Mauviel A. 2014. Proinvasive activity of the Hippo pathway effectors YAP and TAZ in cutaneous melanoma. J Invest Dermatol 134: 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Hong X, Tan S, Chen Q, Chan L, Fivaz M, Cohen SM, Voorhoeve PM. 2014. Viral small T oncoproteins transform cells by alleviating hippo-pathway-mediated inhibition of the YAP protooncogene. Cell Rep 8: 707–713. [DOI] [PubMed] [Google Scholar]
- Ni X, Tao J, Barbi J, Chen Q, Park BV, Li Z, Zhang N, Lebid A, Ramaswamy A, Wei P et al. 2018. YAP Is Essential for Treg-Mediated Suppression of Antitumor Immunity. Cancer Discov 8: 1026–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Islam AB, Lopez-Bigas N, Frolov MV. 2011. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev 25: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto Gutierrez A, McDonald PH. 2018. GPCRs: Emerging anti-cancer drug targets. Cell Signal 41: 65–74. [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD. 2008. In vivo regulation of Yorkie phosphorylation and localization. Development 135: 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Slattery M, Ma L, Crofts A, White KP, Mann RS, Irvine KD. 2013. Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep 3: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Slattery M, Ma L, White KP, Mann RS, Irvine KD. 2014. Yorkie promotes transcription by recruiting a histone methyltransferase complex. Cell Rep 8: 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Remue E, Meerschaert K, Vanloo B, Boucherie C, Gfeller D, Bader GD, Sidhu SS, Vandekerckhove J, Gettemans J et al. 2010. Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem J 432: 461–472. [DOI] [PubMed] [Google Scholar]
- Oka T, Sudol M. 2009. Nuclear localization and pro-apoptotic signaling of YAP2 require intact PDZ-binding motif. Genes Cells 14: 607–615. [DOI] [PubMed] [Google Scholar]
- Omerovic J, Puggioni EM, Napoletano S, Visco V, Fraioli R, Frati L, Gulino A, Alimandi M. 2004. Ligand-regulated association of ErbB-4 to the transcriptional co-activator YAP65 controls transcription at the nuclear level. Exp Cell Res 294: 469–479. [DOI] [PubMed] [Google Scholar]
- Pajtler KW, Wei Y, Okonechnikov K, Silva PBG, Vouri M, Zhang L, Brabetz S, Sieber L, Gulley M, Mauermann M et al. 2019. YAP1 subgroup supratentorial ependymoma requires TEAD and nuclear factor I-mediated transcriptional programmes for tumorigenesis. Nat Commun 10: 3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C, Johann P et al. 2015. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 27: 728–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D 2010. The hippo signaling pathway in development and cancer. Dev Cell 19: 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Tian Y, Cao C, Niu G. 2019. The Emerging Role of YAP/TAZ in Tumor Immunity. Mol Cancer Res 17: 1777–1786. [DOI] [PubMed] [Google Scholar]
- Panciera T, Citron A, Di Biagio D, Battilana G, Gandin A, Giulitti S, Forcato M, Bicciato S, Panzetta V, Fusco S et al. 2020. Reprogramming normal cells into tumour precursors requires ECM stiffness and oncogene-mediated changes of cell mechanical properties. Nat Mater 19: 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappalardo A, Porreca I, Caputi L, De Felice E, Schulte-Merker S, Zannini M, Sordino P. 2015. Thyroid development in zebrafish lacking Taz. Mech Dev 138 Pt 3: 268–278. [DOI] [PubMed] [Google Scholar]
- Park YY, Sohn BH, Johnson RL, Kang MH, Kim SB, Shim JJ, Mangala LS, Kim JH, Yoo JE, Rodriguez-Aguayo C et al. 2016. Yes-associated protein 1 and transcriptional coactivator with PDZ-binding motif activate the mammalian target of rapamycin complex 1 pathway by regulating amino acid transporters in hepatocellular carcinoma. Hepatology 63: 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei T, Li Y, Wang J, Wang H, Liang Y, Shi H, Sun B, Yin D, Sun J, Song R et al. 2015. YAP is a critical oncogene in human cholangiocarcinoma. Oncotarget 6: 17206–17220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Zhu Y, Zhang W, Liao Q, Chen Y, Zhao X, Guo Q, Shen P, Zhen B, Qian X et al. 2017. Regulation of the Hippo-YAP Pathway by Glucose Sensor O-GlcNAcylation. Mol Cell 68: 591–604 e595. [DOI] [PubMed] [Google Scholar]
- Petrilli AM, Fernandez-Valle C. 2016. Role of Merlin/NF2 inactivation in tumor biology. Oncogene 35: 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picco G, Chen ED, Alonso LG, Behan FM, Goncalves E, Bignell G, Matchan A, Fu B, Banerjee R, Anderson E et al. 2019. Functional linkage of gene fusions to cancer cell fitness assessed by pharmacological and CRISPR-Cas9 screening. Nat Commun 10: 2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouffe SW, Lin KC, Moore JL 3rd, Tan FE, Ma S, Ye Z, Qiu Y, Ren B, Guan KL. 2018. The Hippo pathway effector proteins YAP and TAZ have both distinct and overlapping functions in the cell. J Biol Chem 293: 11230–11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobbati AV, Han X, Hung AW, Weiguang S, Huda N, Chen GY, Kang C, Chia CS, Luo X, Hong W et al. 2015. Targeting the Central Pocket in Human Transcription Factor TEAD as a Potential Cancer Therapeutic Strategy. Structure 23: 2076–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova M, Xia F, Avruch J. 2008. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol 18: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls F, Agaimy A, Flucke U, Mentzel T, Sumathi VP, Ploegmakers M, Stoehr R, Kindblom LG, Hansson M, Sydow S et al. 2020. Recurrent Fusions Between YAP1 and KMT2A in Morphologically Distinct Neoplasms Within the Spectrum of Low-grade Fibromyxoid Sarcoma and Sclerosing Epithelioid Fibrosarcoma. Am J Surg Pathol 44: 594–606. [DOI] [PubMed] [Google Scholar]
- Qing Y, Yin F, Wang W, Zheng Y, Guo P, Schozer F, Deng H, Pan D. 2014. The Hippo effector Yorkie activates transcription by interacting with a histone methyltransferase complex through Ncoa6. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni CV, Diguet N, Le Garrec JF, Novotova M, Resende TP, Pop S, Charon N, Guillemot L, Kitasato L, Badouel C et al. 2017. Amotl1 mediates sequestration of the Hippo effector Yap1 downstream of Fat4 to restrict heart growth. Nat Commun 8: 14582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BV, Irvine KD. 2013. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev Cell 24: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Zhang L, Jiang J. 2010. Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev Biol 337: 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum E, Jadeja B, Xu B, Zhang L, Agaram NP, Travis W, Singer S, Tap WD, Antonescu CR. 2020. Prognostic stratification of clinical and molecular epithelioid hemangioendothelioma subsets. Mod Pathol 33: 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC et al. 2012. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 151: 1457–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau C, Pores Fernando AT, Hwang JH, Faure N, Jiang T, White EA, Roberts TM, Schaffhausen BS. 2016. Transformation by Polyomavirus Middle T Antigen Involves a Unique Bimodal Interaction with the Hippo Effector YAP. J Virol 90: 7032–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Murphy RM, Deng M, MacDonald JW, Bammler TK, Aldinger KA, Glass IA, Millen KJ. 2019. PI3KYap activity drives cortical gyrification and hydrocephalus in mice. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MP, Correa KM, Ramesh V, MacCollin MM, Jacoby LB, von Deimling A, Gusella JF, Louis DN. 1994. Analysis of the neurofibromatosis 2 gene in human ependymomas and astrocytomas. Cancer Res 54: 45–47. [PubMed] [Google Scholar]
- Rustgi AK, Xu L, Pinney D, Sterner C, Beauchamp R, Schmidt S, Gusella JF, Ramesh V. 1995. Neurofibromatosis 2 gene in human colorectal cancer. Cancer Genet Cytogenet 84: 24–26. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S et al. 2018. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 173: 321–337 e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansores-Garcia L, Bossuyt W, Wada K, Yonemura S, Tao C, Sasaki H, Halder G. 2011. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J 30: 2325–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieffer KM, Agarwal V, LaHaye S, Miller KE, Koboldt DC, Lichtenberg T, Leraas K, Brennan P, Kelly BJ, Crist E et al. 2020. YAP1-FAM118B Fusion Defines a Rare Subset of Childhood and Young Adulthood Meningiomas. Am J Surg Pathol. [DOI] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR et al. 2011. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell 144: 782–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe-Pedros A, Zheng Y, Ruiz-Trillo I, Pan D. 2012. Premetazoan origin of the hippo signaling pathway. Cell Rep 1: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido Y, Pass HI, Bader S, Mew DJ, Christman MF, Gazdar AF, Minna JD. 1995. Neurofibromatosis type 2 (NF2) gene is somatically mutated in mesothelioma but not in lung cancer. Cancer Res 55: 1227–1231. [PubMed] [Google Scholar]
- Sekine S, Kiyono T, Ryo E, Ogawa R, Wakai S, Ichikawa H, Suzuki K, Arai S, Tsuta K, Ishida M et al. 2019. Recurrent YAP1-MAML2 and YAP1-NUTM1 fusions in poroma and porocarcinoma. J Clin Invest 129: 3827–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Guo X, Yan H, Lu Y, Ji X, Li L, Liang T, Zhou D, Feng XH, Zhao JC et al. 2015. A miR-130a-YAP positive feedback loop promotes organ size and tumorigenesis. Cell Res 25: 997–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y, Ji X, Cao X, Dai X, Xu L, Zhao H, Guo X, Yan H, Zhang H, Zhu C et al. 2017. Src Inhibits the Hippo Tumor Suppressor Pathway through Tyrosine Phosphorylation of Lats1. Cancer Res 77: 4868–4880. [DOI] [PubMed] [Google Scholar]
- Sievers P, Chiang J, Schrimpf D, Stichel D, Paramasivam N, Sill M, Gayden T, Casalini B, Reuss DE, Dalton J et al. 2020. YAP1-fusions in pediatric NF2-wildtype meningioma. Acta Neuropathol 139: 215–218. [DOI] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. 2006. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol 16: 2081–2089. [DOI] [PubMed] [Google Scholar]
- Silvis MR, Kreger BT, Lien WH, Klezovitch O, Rudakova GM, Camargo FD, Lantz DM, Seykora JT, Vasioukhin V. 2011. alpha-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci Signal 4: ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274. [DOI] [PubMed] [Google Scholar]
- Skibinski A, Breindel JL, Prat A, Galvan P, Smith E, Rolfs A, Gupta PB, LaBaer J, Kuperwasser C. 2014. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Rep 6: 1059–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Honjo S, Jin J, Chang SS, Scott AW, Chen Q, Kalhor N, Correa AM, Hofstetter WL, Albarracin CT et al. 2015. The Hippo Coactivator YAP1 Mediates EGFR Overexpression and Confers Chemoresistance in Esophageal Cancer. Clin Cancer Res 21: 2580–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Xie M, Scott AW, Jin J, Ma L, Dong X, Skinner HD, Johnson RL, Ding S, Ajani JA. 2018. A Novel YAP1 Inhibitor Targets CSC-Enriched Radiation-Resistant Cells and Exerts Strong Antitumor Activity in Esophageal Adenocarcinoma. Mol Cancer Ther 17: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino G, Ruggeri N, Specchia V, Cordenonsi M, Mano M, Dupont S, Manfrin A, Ingallina E, Sommaggio R, Piazza S et al. 2014. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol 16: 357–366. [DOI] [PubMed] [Google Scholar]
- Stampouloglou E, Cheng N, Federico A, Slaby E, Monti S, Szeto GL, Varelas X. 2020. Yap suppresses T-cell function and infiltration in the tumor microenvironment. PLoS Biol 18: e3000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein C, Bardet AF, Roma G, Bergling S, Clay I, Ruchti A, Agarinis C, Schmelzle T, Bouwmeester T, Schubeler D et al. 2015. YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers. PLoS Genet 11: e1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner SA, Yan M, Liu KTH, Arimoto KI, Shima T, Wang HY, Johnson DT, Bejar R, Jamieson C, Guan KL et al. 2019. Hippo kinase loss contributes to del(20q) hematologic malignancies through chronic innate immune activation. Blood 134: 1730–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S, Monti O, Pediconi N, Baccarini A, Fontemaggi G, Lapi E, Mantovani F, Damalas A, Citro G, Sacchi A et al. 2005. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol Cell 18: 447–459. [DOI] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G. 2001. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem 276: 15164–15173. [DOI] [PubMed] [Google Scholar]
- Sudol M 1994. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene 9: 2145–2152. [PubMed] [Google Scholar]
- Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D. 1995a. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem 270: 14733–14741. [DOI] [PubMed] [Google Scholar]
- Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. 1995b. Characterization of a novel protein-binding module--the WW domain. FEBS Lett 369: 67–71. [DOI] [PubMed] [Google Scholar]
- Swingle MR, Amable L, Lawhorn BG, Buck SB, Burke CP, Ratti P, Fischer KL, Boger DL, Honkanen RE. 2009. Structure-activity relationship studies of fostriecin, cytostatin, and key analogs, with PP1, PP2A, PP5, and(beta12-beta13)-chimeras (PP1/PP2A and PP5/PP2A), provide further insight into the inhibitory actions of fostriecin family inhibitors. J Pharmacol Exp Ther 331: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulzewsky F, Arora S, Hoellerbauer P, King C, Nathan E, Chan M, Cimino PJ, Ozawa T, Kawauchi D, Pajtler KW et al. 2020. Comparison of tumor-associated YAP1 fusions identifies a recurrent set of functions critical for oncogenesis. Genes Dev 34: 1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takadera M, Satomi K, Szulzewsky F, Cimino PJ, Holland EC, Yamamoto T, Ichimura K, Ozawa T. 2020. Phenotypic characterization with somatic genome editing and gene transfer reveals the diverse oncogenicity of ependymoma fusion genes. Acta Neuropathol Commun 8: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanas MR, Ma S, Jadaan FO, Ng CK, Weigelt B, Reis-Filho JS, Rubin BP. 2016. Mechanism of action of a WWTR1(TAZ)-CAMTA1 fusion oncoprotein. Oncogene 35: 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, Flanagan J, Luo Y, Fenwick K, Natrajan R et al. 2011. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med 3: 98ra82. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Wu LW, Grivennikov SI, de Jong PR, Lian I, Yu FX, Wang K, Ho SB, Boland BS, Chang JT et al. 2015. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature 519: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Zhang S, Turenchalk GS, Stewart RA, St John MA, Chen W, Xu T. 1999. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat Genet 21: 177–181. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber D, Hariharan IK. 2002. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110: 467–478. [DOI] [PubMed] [Google Scholar]
- Thompson BJ. 2020. YAP/TAZ: Drivers of Tumor Growth, Metastasis, and Resistance to Therapy. Bioessays 42: e1900162. [DOI] [PubMed] [Google Scholar]
- Totaro A, Panciera T, Piccolo S. 2018. YAP/TAZ upstream signals and downstream responses. Nat Cell Biol 20: 888–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan N, Fitamant J, Bardeesy N, Camargo FD, Guan KL. 2012. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol 14: 1322–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. 2003. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol 5: 914–920. [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, Simpson EM, Barsh GS, Bastian BC. 2009. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457: 599–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N et al. 2010. Mutations in GNA11 in uveal melanoma. N Engl J Med 363: 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X 2014. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 141: 1614–1626. [DOI] [PubMed] [Google Scholar]
- Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H et al. 2010a. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell 18: 579–591. [DOI] [PubMed] [Google Scholar]
- Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. 2010b. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev Cell 19: 831–844. [DOI] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. 2001. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev 15: 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verteporfin In Photodynamic Therapy Study G. 2001. Verteporfin therapy of subfoveal choroidal neovascularization in age-related macular degeneration: two-year results of a randomized clinical trial including lesions with occult with no classic choroidal neovascularization--verteporfin in photodynamic therapy report 2. Am J Ophthalmol 131: 541–560. [DOI] [PubMed] [Google Scholar]
- Vigneswaran K, Boyd NH, Oh SY, Lallani S, Boucher A, Neill SG, Olson JJ, Read RD. 2020. YAP/TAZ transcriptional co-activators create therapeutic vulnerability to verteporfin in EGFR mutant glioblastoma. Clin Cancer Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser S, Yang X. 2010. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle 9: 3892–3903. [DOI] [PubMed] [Google Scholar]
- Wang J, Sinnett-Smith J, Stevens JV, Young SH, Rozengurt E. 2016a. Biphasic Regulation of Yes-associated Protein (YAP) Cellular Localization, Phosphorylation, and Activity by G Protein-coupled Receptor Agonists in Intestinal Epithelial Cells: A NOVEL ROLE FOR PROTEIN KINASE D (PKD). J Biol Chem 291: 17988–18005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Luo JY, Li B, Tian XY, Chen LJ, Huang Y, Liu J, Deng D, Lau CW, Wan S et al. 2016b. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature 540: 579–582. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu X, Maglic D, Dill MT, Mojumdar K, Ng PK, Jeong KJ, Tsang YH, Moreno D, Bhavana VH et al. 2018. Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer. Cell Rep 25: 1304–1317 e1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu A, Yu FX. 2017. The Hippo pathway in tissue homeostasis and regeneration. Protein Cell 8: 349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SM, Avantaggiati ML, Nemazanyy I, Di Poto C, Yang Y, Pende M, Gibney GT, Ressom HW, Field J, Atkins MB et al. 2019. YAP/TAZ Inhibition Induces Metabolic and Signaling Rewiring Resulting in Targetable Vulnerabilities in NF2-Deficient Tumor Cells. Dev Cell 49: 425–443 e429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. 2006. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol 16: 2090–2100. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Sansores-Garcia L, Tao C, Halder G. 2008. Boundaries of Dachsous Cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc Natl Acad Sci U S A 105: 14897–14902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J et al. 2007. The genomic landscapes of human breast and colorectal cancers. Science 318: 1108–1113. [DOI] [PubMed] [Google Scholar]
- Wu Q, Li J, Sun S, Chen X, Zhang H, Li B, Sun S. 2017. YAP/TAZ-mediated activation of serine metabolism and methylation regulation is critical for LKB1-deficient breast cancer progression. Biosci Rep 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. 2003. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114: 445–456. [DOI] [PubMed] [Google Scholar]
- Xiao J, Shi Q, Li W, Mu X, Peng J, Li M, Chen M, Huang H, Wang C, Gao K et al. 2018. ARRDC1 and ARRDC3 act as tumor suppressors in renal cell carcinoma by facilitating YAP1 degradation. Am J Cancer Res 8: 132–143. [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Wang J, Ou C, Zhang Y, Ma L, Weng W, Pan Q, Sun F. 2013. Mutual interaction between YAP and c-Myc is critical for carcinogenesis in liver cancer. Biochem Biophys Res Commun 439: 167–172. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. 1995. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121: 1053–1063. [DOI] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. 1999. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J 18: 2551–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Taouk GM. 2020. A Potential Role of YAP/TAZ in the Interplay Between Metastasis and Metabolic Alterations. Front Oncol 10: 928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CS, Stampouloglou E, Kingston NM, Zhang L, Monti S, Varelas X. 2018. Glutamine-utilizing transaminases are a metabolic vulnerability of TAZ/YAP-activated cancer cells. EMBO Rep 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. 2013. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154: 1342–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K, Noguchi K, Nakano Y, Yamamura M, Takaoka K, Hashimoto-Tamaoki T, Kishimoto H. 2015. The Hippo pathway transcriptional co-activator, YAP, confers resistance to cisplatin in human oral squamous cell carcinoma. Int J Oncol 46: 2364–2370. [DOI] [PubMed] [Google Scholar]
- Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, Zhao L, Peyman G, Ouyang H, Jiang W et al. 2014. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell 25: 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Guan KL. 2015a. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 163: 811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H et al. 2012. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Chen Y, Li X, Yang R, Zhang L, Huangfu L, Zheng N, Zhao X, Lv L, Hong Y et al. 2018. YAP1 contributes to NSCLC invasion and migration by promoting Slug transcription via the transcription co-factor TEAD. Cell Death Dis 9: 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Bachman J, Lai ZC. 2015b. Mutation analysis of large tumor suppressor genes LATS1 and LATS2 supports a tumor suppressor role in human cancer. Protein Cell 6: 6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Tomlinson V, Lara R, Holliday D, Chelala C, Harada T, Gangeswaran R, Manson-Bishop C, Smith P, Danovi SA et al. 2008. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell Death Differ 15: 1752–1759. [DOI] [PubMed] [Google Scholar]
- Zanconato F, Cordenonsi M, Piccolo S. 2016. YAP/TAZ at the Roots of Cancer. Cancer Cell 29: 783–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. 2015. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol 17: 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. 2009. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem 284: 13355–13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Gao Y, Li F, Tong X, Ren Y, Han X, Yao S, Long F, Yang Z, Fan H et al. 2015. YAP promotes malignant progression of Lkb1-deficient lung adenocarcinoma through downstream regulation of survivin. Cancer Res 75: 4450–4457. [DOI] [PubMed] [Google Scholar]
- Zhang X, Abdelrahman A, Vollmar B, Zechner D. 2018. The Ambivalent Function of YAP in Apoptosis and Cancer. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Qiao Y, Wu Q, Chen Y, Zou S, Liu X, Zhu G, Zhao Y, Chen Y, Yu Y et al. 2017. The essential role of YAP O-GlcNAcylation in high-glucose-stimulated liver tumorigenesis. Nat Commun 8: 15280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tang JZ, Vergara IA, Zhang Y, Szeto P, Yang L, Mintoff C, Colebatch A, McIntosh L, Mitchell KA et al. 2019. Somatic Hypermutation of the YAP Oncogene in a Human Cutaneous Melanoma. Mol Cancer Res 17: 1435–1449. [DOI] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. 2010a. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev 24: 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. 2010b. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev 24: 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L et al. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM et al. 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Khanal P, Savage P, She YM, Cyr TD, Yang X. 2014. YAP-induced resistance of cancer cells to antitubulin drugs is modulated by a Hippo-independent pathway. Cancer Res 74: 4493–4503. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Pan D. 2019. The Hippo Signaling Pathway in Development and Disease. Dev Cell 50: 264–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wang W, Liu B, Deng H, Uster E, Pan D. 2015. Identification of Happyhour/MAP4K as Alternative Hpo/Mst-like Kinases in the Hippo Kinase Cascade. Dev Cell 34: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang S, Wang Z, Feng X, Liu P, Lv XB, Li F, Yu FX, Sun Y, Yuan H et al. 2015. Estrogen regulates Hippo signaling via GPER in breast cancer. J Clin Invest 125: 2123–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Li L, Zhao B. 2015. The regulation and function of YAP transcription co-activator. Acta Biochim Biophys Sin (Shanghai) 47: 16–28. [DOI] [PubMed] [Google Scholar]


