Abstract
Purpose:
Neurodevelopmental disorders (NDDs) encompass a spectrum of genetically heterogeneous disorders with features that commonly include developmental delay, intellectual disability, and autism spectrum disorders. We sought to delineate the molecular and phenotypic spectrum of a novel neurodevelopmental disorder caused by variants in the GNAI1 gene.
Methods:
Through large cohort trio-based exome sequencing and international data-sharing, we identified 24 unrelated individuals with NDD phenotypes and a variant in GNAI1, which encodes the inhibitory Gαi1 subunit of heterotrimeric G-proteins. We collected detailed genotype and phenotype information for each affected individual.
Results:
We identified 16 unique variants in GNAI1 in 24 affected individuals; 23 occurred de novo and one was inherited from a mosaic parent. Most affected individuals have a severe neurodevelopmental disorder. Core features include global developmental delay, intellectual disability, hypotonia, and epilepsy.
Conclusion:
This collaboration establishes GNAI1 variants as a cause of NDDs. GNAI1-related NDD is most often characterized by severe to profound delays, hypotonia, epilepsy that ranges from self-limiting to intractable, behavior problems, and variable mild dysmorphic features.
Introduction
Neurodevelopmental disorders (NDDs) are heterogeneous disorders, often with a broad and overlapping range of features that commonly include developmental delay, intellectual disability, and autism spectrum disorder. This group of disorders also has an increased incidence of comorbidities such as epilepsy. Advances in genomic technologies have led to an exponential increase in the number of genes associated with NDDs. However, up to half of those affected do not have an identified genetic etiology1, presenting challenges in understanding the long-term prognosis and accessing appropriate support. Phenotype-based genetic investigations of NDDs are hampered by the highly variable clinical manifestations, the phenotypic overlap with other closely related disorders, and the rarity of particular genetic subtypes2. Instead, more recently, large-scale trio-based exome sequencing of clinically heterogeneous populations coupled with international data-sharing has proven a powerful strategy for discovering NDD-associated genes1, 2.
G-protein subunits belong to a family of proteins that have previously been associated with NDDs3, 4. Heterotrimeric G proteins, composed of α, β and γ subunits, transmit the signals of extracellular ligands bound to G-protein-coupled receptors (GPCRs) to intracellular signaling pathways5. G-protein signaling has been implicated in a diverse range of biological functions including neuronal development and synaptic function6. GNAI1 (MIM 139310) encodes the inhibitory Gαi1 subunit of heterotrimeric G-proteins. A recent study found a significant enrichment of de novo variants in GNAI1 in a diverse cohort of individuals with NDDs2, 7. Here, we describe 24 unrelated individuals with GNAI1 variants (23 de novo variants and one inherited from a mosaic parent) and delineate the associated phenotypic features which include global developmental delay with intellectual disability, hypotonia, and seizures.
Methods
Individuals with pathogenic and likely pathogenic variants in GNAI1 (NM_002069.5) were identified via Deciphering Developmental Disorders study (DDD) research study7 and international collaboration facilitated by GeneMatcher8 and MyGene2. Variants were classified using American College of Medical Genetics guidelines9. Individuals were identified via trio exome sequencing by the Deciphering Developmental Disorders study7 (individuals 11, 14, 16, 18-20, and 24), clinical trio exome sequencing at GeneDx as previously described10 (1-3, 5, 7-10, 15, 17, 22-23), trio exome sequencing via clinical practice (4, 6, 12), or by research-based trio exome sequencing (13, 21). The genetic details from thirteen of these individuals was previously reported (2, 3, 7-11, 14, 16, 18-20, 24) with minimal clinical details2, 7 ; we obtained additional, detailed clinical information for these individuals for this study.
Results
We identified 27 unrelated individuals (16 female) with rare variants in GNAI1 (GenBank: NM_002069.6). For three individuals with GNAI1 variants, parental samples were unavailable for testing (Table S1); clinical information for these individuals has not been included in this report. The remaining twenty-four variants would meet the criteria to be classified as likely pathogenic or pathogenic according to ACMG guidelines if GNAI1 were an established disease gene9 (Figure 1, Table 1). Of these, twenty-three variants were de novo in the affected individual (somatic mosaic in Individual 2), while one variant was inherited from a mother with low-level mosaicism (Individual 3, 6.0% alternate allele frequency). Where applicable, de novo status of variants and parental relationships were confirmed. .
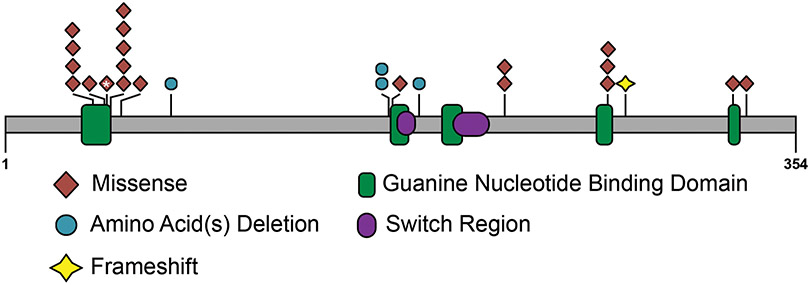
Figure 1: Distribution of disease-causing variants across GNAI1.
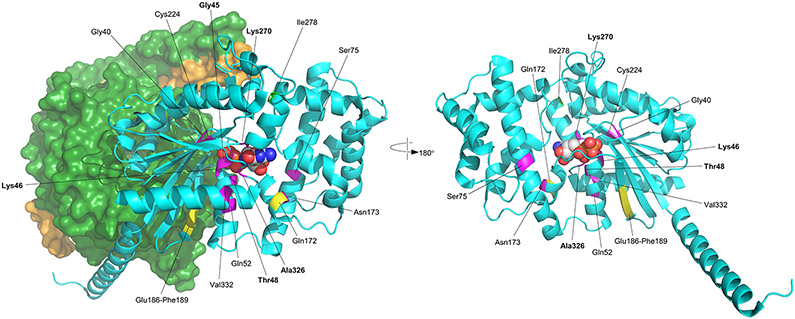
A) Schematic showing the pathogenic and likely pathogenic variants identified in GNAI1, including one previously reported variant (*) (Kaplanis et al. 2020). Variants cluster within the first guanine nucleotide-binding domain (green box). Missense variants are represented as brown diamonds, coding deletion variants as blue circles, and the truncating frameshift variant as a yellow star. Each symbol represents one individual. B) 3D structure of GNAi1. The left figure shows the structure of GNAi1 as part of the trimeric G-protein complex (PDB accession 6crk); GNAi1 is shown as a cyan ribbon, except for positions of novel variants which are coloured as follows: missense, magenta; in-frame deletions, yellow; frameshifting insertion at Ile278, light green; bound GDP is shown as space-filling spheres, coloured by atom type (white, carbon; blue, nitrogen; red, oxygen; orange, phosphorus); the molecular surface is shown for the βγ dimer, with the β1 and γ2 chains coloured dark green and orange respectively. The right figure shows GNAi1 only, rotated around the vertical axis; Gly45 is obscured by the GDP ligand in this view. In both parts, labelling in bold font indicates residues making direct contact with GDP (Gly45, Thr48, Lys270, Ala326).
Table 1:
de novo variants in GNAI1
| Ind. | Variant | Protein Domain | CADD | M-CAP score (prediction) |
phastCons 100way vertebrate |
gnomAD AC |
||
|---|---|---|---|---|---|---|---|---|
| Genomic Coordinates (GRCh37/hg19) |
cDNA Position (NM_002069) |
Protein Position | ||||||
| 1 2 |
chr7:g.79764594G>C | c.118G>C | p.(Gly40Arg) | GDP Binding, GoLoco Binding | 33 | 0.910 (PP) | 9.041 | 0 |
| 3 4 |
chr7:g.79764594G>T | c.118G>T | p.(Gly40Cys) | GDP Binding, GoLoco Binding | 34 | 0.938 (PP) | 9.041 | 0 |
| 5 | chr7:g.79818282G>A | c.134G>A | p.(Gly45Asp) | GDP Binding | 29.2 | 0.549 (PP) | 9.75805 | 0 |
| 6 7 8 |
chr7:g.79818291C>A | c.143C>A | p.(Thr48Lys) | GDP Binding | 32 | 0.432 (PP) | 7.6889 | 0 |
| 9 10 |
chr7:g.79818291C>T | c.143 C>T | p.(Thr48Ile) | GDP Binding | 32 | 0.329 (PP) | 7.6889 | 0 |
| 11 | chr7:g.79818303A>C | c.155A>C | p.(Gln52Pro) | none | 27.4 | 0.534 (PP) | 9.29824 | 0 |
| 12 | chr7:g.79818466_79818468del | c.222_224del | p.(Ser75del) | GoLoco Binding | NA | NA | 9.29824 | 0 |
| 13 14 |
chr7:g.79833072_79833074del | c.514_516del | p.(Gln172del) | none | NA | NA | 9.1759 | 0 |
| 15 | chr7:g. 79833076A>T | c.518A>T | p.(Asp173Val) | GDP Binding | 29.8 | 0.255 (PP) | 9.1759 | 0 |
| 16 | chr7:g.79833114_79833125del | c.556_567del | p.(Glu186_Phe189del) | none | NA | NA | 9.1759 | 0 |
| 17 18 |
chr7:g.79840365G>A | c.671G>A | p.(Cys224Tyr) | none | 29.2 | 0.309 (PP) | 9.818 | 0 |
| 19 20 |
chr7:g.79842120A>G | c.809A>G | p.(Lys270Arg) | GDP Binding | 32 | 0.611 (PP) | 9.24465 | 0 |
| 21 | chr7:79842121G>C | c.810G>C | p.(Lys270Asn) | GDP Binding | 27.5 | 0.596 (PP) | 6.72193 | 0 |
| 22 | chr7:g.79842143dup | c.832dupA | p.(Ile278Asnfs*20) | NA | NA | NA | 6.26325 | 0 |
| 23 | chr7:g.79846720G>C | c.976G>C | p.(Ala326Pro) | GDP Binding | 28.6 | 0.606 (PP) | 9.818 | 0 |
| 24 | chr7:g.79846739T>A | c.995T>A | p.(Val332Glu) | none | 27.6 | 0.540 (PP) | 7.98329 | 0 |
Abbreviations: AC, allele count; GDP, Guanosine-diphosphate; NA, not applicable; PP, possibly pathogenic
Among the 24 individuals with variants in GNAI1, there were 16 unique variants, with seven recurrent variants identified in two or three individuals each. At three residues (Gly40, Thr48, Lys270), there were two different pathogenic variants resulting in different amino acid changes. Of the 16 variants, 12 were missense variants, three were small in-frame deletions, and one was a protein-truncating variant. The majority of missense and coding deletion variants (9/15; 60%) affect amino acids within the guanine nucleotide binding motifs of GNAI1 (Figure 1). Variants predominantly cluster in the first GDP binding motif (also known as the Walker A motif or P-loop), where five variants at three sites (Gly40, Gly45, and Thr48) accounted for 10/24 individuals (42%), while variants at Arg270 and Ala326 affect residues in the fourth and fifth guanine nucleotide binding motifs, respectively.
We performed detailed phenotyping of the 24 affected individuals with de novo GNAI1 variants (Table 2, Table S1). Age at last medical review ranged from 3 years 10 months to 18 years (median age 11 years). All participants have global developmental delays ranging in severity from mild to profound. Speech is significantly affected with language delays reported in 21/23 (91%) individuals; 16 individuals are nonverbal (at ages 3–18 years), and only one individual has achieved fluent speech. Gross motor delays are also common. Delayed sitting was reported in 18/21 (86%) individuals, five of whom cannot yet sit independently (at ages 18 months –18 years). Delayed walking was reported in 19/23 (83%) individuals; nine individuals remain nonambulatory (ages 18 months – 18 years). Intellectual disability was reported in all individuals for whom data was available (20/20) and ranged from mild (3/20; 15%) to severe/profound (11/20; 55%).
Table 2:
Summary of clinical features of individuals with de novo variants in GNAI1
| Individual | Sex | Variant | DD | Delayed Sitting |
Delayed Walking |
Language Delays |
Intellectual Disabilities |
Autism | Seizures | Tone | MRI anomalies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | p.(Gly40Arg) | + | nr | nr | nr | nr | + | + | Hypotonia | + |
| 2 | F | p.(Gly40Arg)^ | + | nr | − | − | − | + | − | Hypotonia | nr |
| 3 | M | p.(Gly40Cys) | + | nr | + | Nonverbal | S | + | + | Normal | + |
| 4 | F | p.(Gly40Cys) | P | Not achieved | Not achieved | Nonverbal | S | nr | + | Hypertonia | + |
| 5 | M | p.(Gly45Asp) | + | + | + | Nonverbal | nr | + | + | Hypotonia | − |
| 6 | F | p.(Thr48Lys) | + | + | + | Nonverbal | P | − | + | Hypotonia | + |
| 7 | M | p.(Thr48Lys) | + | + | Not achieved | Nonverbal | nr | − | + | Hypotonia | nr |
| 8 | M | p.(Thr48Lys) | P | Not achieved | Not achieved | Nonverbal | P | nr | + | Hypotonia | + |
| 9 | M | p.(Thr48Ile) | + | + | − | Nonverbal | S | + | − | Hypotonia | − |
| 10 | F | p.(Thr48Ile) | + | − | + | + | nr | − | − | Hypotonia | − |
| 11 | M | p.(Gln52Pro) | + | + | Not achieved | Nonverbal | S | + | + | Hypotonia | + |
| 12 | F | p.(Ser75del) | S | + | Not achieved | Nonverbal | S-P | − | + | Hypotonia | − |
| 13 | M | p.(Gln172del) | P | Not achieved | Not achieved | Nonverbal | S-P | − | + | Hypotonia | + |
| 14 | F | p.(Gln172del) | + | + | − | Nonverbal | S | − | − | nr | nr |
| 15 | F | p.(Asp173Val) | + | − | + | + | Mi | − | + | Hypotonia | − |
| 16 | F | p.(Glu186_Phe189del) | + | − | + | Nonverbal | + | − | nr | Hypotonia | nr |
| 17 | F | p.(Cys224Tyr) | Mo-S | + | − | + | Mo-S | − | + | Hypotonia | − |
| 18 | F | p.(Cys224Tyr) | + | + | + | − | Mo | + | − | Hypotonia | − |
| 19 | F | p.(Lys270Arg) | + | + | + | + | Mo | − | + | Hypotonia | − |
| 20 | M | p.(Lys270Arg) | + | + | + | Nonverbal | S | − | + | Hypotonia | − |
| 21 | M | p.(Lys270Asn) | + | Not achieved | Not achieved | Nonverbal | Mo | − | + | Hypotonia | + |
| 22 | F | p.(Ile278AsnfsX20) | P | Not achieved | Not achieved | Nonverbal | S-P | − | + | Hypotonia | + |
| 23 | F | p.(Ala326Pro) | + | + | + | + | Mi | − | − | Hypotonia | − |
| 24 | F | p.(Val332Glu) | + | + | Not achieved | Nonverbal | + | nr | + | Hypertonia | + |
| Totals | 24/24 | 18/21 | 19/23 | 21/23 | 20/20 | 7/21 | 17/23 | 22/23 | 10/20 | ||
Abbreviations: DD, Developmental Delay; F, female; M, male; Mi, mild; Mo, moderate; nr, not reported; S, severe; P, profound; +, present; −, absent
somatic mosaic variant.
Other prominent phenotypic features include hypotonia (20/23 individuals, 87%), and epilepsy (17/23 individuals, 74%) individuals. For many patients, hypotonia was severe and had a significant effect on daily functioning. Median age of seizure onset was 5 months (range 36 hours to 7 years), with seizures beginning in the first six months of life in 10/15 (67%) individuals for whom data were available. Seizure types were variable with the most common being absence seizures (n=5), generalized tonic-clonic seizures (n=5), and focal onset impaired awareness seizures (n=4).
While behavioral anomalies were present in 17/19 individuals (89%), they were variable across the cohort. The most common behavioral features included aggression and temper tantrums (n=7), autism (n=7), hypersensitivity (n=5), and hand stereotypies (n=6). Other variable features included feeding difficulties (n=9) and obesity (n=7). MRI abnormalities were reported for 10/20 (50%) individuals, with the most common finding being brain atrophy seen in four individuals.
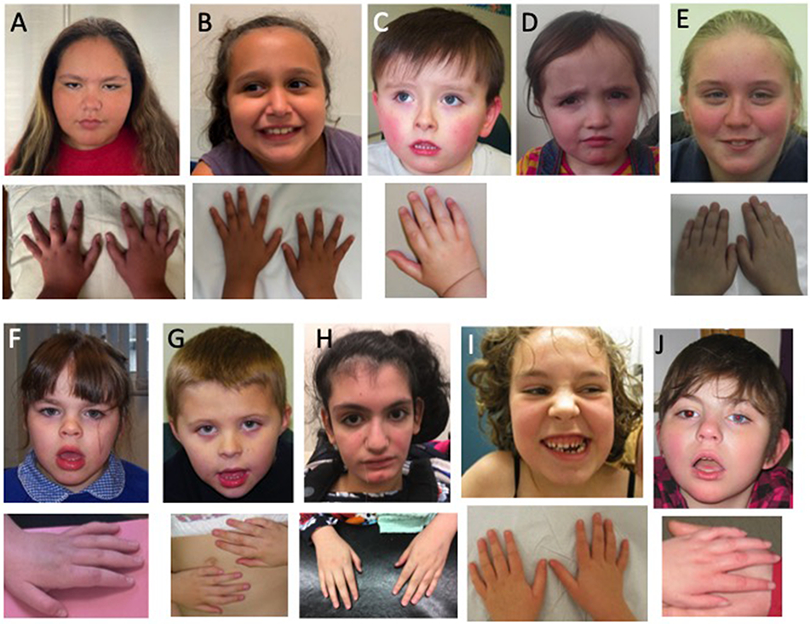
Dysmorphic features were reported in 16/21 (76%) individuals (Figure 2); however, the described physical features were variable. The most commonly reported features included tapered fingers (n=9), a markedly long hallux (n=5), and a short, upturned nose (n=8). Other common facial features included a tented upper lip or open mouth appearance (n=5), and a thin upper lip or prominent lower lip (n=4).
Figure 2: Photographs of affected individuals.
A) individual 2; B) individual 10; C) individuals 11; D) individual 16; E) individual 18; F, individual 19; G) individual 20; H) individual 22; I) individual 23; J) individual 24. Affected individuals have variable minor dysmorphic features and tend to have tapering fingers.
Discussion
We describe a novel, severe neurodevelopmental disorder due to de novo variants in the GNAI1 gene, which encodes Gαi1, a member of the Gi/o inhibitory family of G-protein α-subunits. Heterotrimeric G-proteins act as a molecular switch. The GDP-bound Gα subunit binds the Gβγ dimer, maintaining the heterotrimeric protein in an inactive state. In response to an extracellular stimulant, bound GDP is replaced by GTP, resulting in a conformational change leading to the disassociation of the Gα subunit from the Gβγ dimer. Once separated, the Gα subunit and the Gβγ dimer are able to activate (or inhibit) downstream signaling pathways via modulation of cAMP levels. The intrinsic GTPase activity of the Gα subunit will eventually result in GTP hydrolysis, returning the protein to its GDP-bound inactive heterotrimeric state5.
Gαi1 is part of the Gi/o inhibitory family of α-subunits named for their ability to inhibit adenylyl cyclase activity. In the central nervous system, Gαi1 has been shown to mediate major signaling pathways Akt-mTORC1 and Erk–MAPK11, 12 and control the gating of G protein-activated potassium channels13. Other G-protein subunits have also been implicated in neurological disease including GNAQ (MIM: 600998) associated with Sturge-Weber syndrome, GNAL (MIM: 139312) associated with dystonia, GNAO1 (MIM: 139311) in which loss of function variants are associated with developmental and epileptic encephalopathy while gain of function variants are associated with movement disorders14, and GNB1 (MIM: 139380) associated with developmental delay. Individuals with loss of function variants in GNB1, a β-subunit of heterotrimeric G-proteins, have a strikingly similar phenotype to individuals with de novo GNAI1 variants, including profound developmental delay commonly accompanied by hypotonia and seizures4, suggesting there may be a common pathogenetic disease mechanism between GNB1 and GNAI1.
Gα proteins contain five highly conserved guanine nucleotide binding motifs that fold to form a single deep pocket for binding guanine nucleotides10. Of the 16 pathogenic or likely pathogenic variants in GNAI1 reported in this study and one previously reported variant (p.Lys46Glu)2, nine variants at six sites (Gly40, Gly45, Lys46, Thr48, Lys270, and Ala326) are located in the guanine nucleotide binding pocket of GNAI1 (Figure 1).Gly40, Gly45, Lys46, and Thr48 reside in the highly conserved first GDP binding motif. Gly40 lies at the mouth of the nucleotide binding pocket immediately N-terminal to a series of GDP-interacting residues, including Gly45 and Thr48. Notably, although Arg270 and Ala326 are distant in the linear sequence to Gly45 and Thr48, they lie in close spatial proximity on the opposite face of the GDP binding pocket 11. Gly45, Thr48, and Lys270 make direct contacts with the GDP ligand and therefore the Gly45Asp, Thr48Lys, Thr48Ile, Lys270Asn, and Lys270Arg substitutions are likely to disrupt these interactions. While Gly40 does not directly interact with the GDP ligand, structural modeling predicts that the Gly40Arg and Gly40Cys substitutions will have a significant destabilizing effect on the GDP binding pocket (increases in free energy compared to the native structure of ~9.4 kcal/mol and ~27.6 kcal/mol respectively; values >3 kcal/mol are generally regarded as strongly destabilizing15). As such, the pathogenic variants in guanine nucleotide binding motifs of Gαi1 are all predicted to have adverse effects on Gαi1 function through the disruption of Gαi1 ability to bind GDP and GTP and/or hydrolyze GTP.
GNAI1 is predicted to be intolerant to loss of function variants (pLI=0.91; e/o= 0.12 [0.05 - 0.38])16. Of the 16 variants in GNAI1 identified in 24 individuals, only one was a truncating variant: p.(Ile278Asnfs*20) frameshift identified in a single individual with profound developmental delay, axial hypotonia, and seizures; this variant is located within the last 50bp of the penultimate exon and is predicted to escape nonsense-mediated decay. We identified one additional individual with an early truncating variant, but inheritance information was not available (Supplementary Table 1). We also previously identified large, heterozygous deletions encompassing GNAI1 (and additional genes) in two unrelated individuals with epilepsy17. In one case, the deletion segregated in a large family, with at least 5 affected individuals in 3 generations; all had variable types of generalized epilepsy (ranging from mild to severe) and learning difficulties. Additional individuals with truncating variants need to be identified in order to determine if there are differences in phenotypes between individuals with missense and truncating variants in GNAI1.
GNAI1-related NDD is most often characterized by severe to profound delays, hypotonia, epilepsy that ranges from self-limiting to intractable, behavior problems, and variable mild dysmorphic features, though there is range of severity for all phenotypic features associated with pathogenic variants in GNAI1. Like many NDDs that have been recently described, GNAI1-related disorder may not be distinctly recognizable; many features overlap with other single-gene disorders including GNB14, PPP3CA18, TANC219 and others. While many individuals have severe developmental delay accompanied by several additional comorbidities, there are also some more mildly affected individuals. In some cases, additional genetic variants may contribute to the phenotype; for example, individual 9 has a de novo, likely pathogenic variant in ACTA1, which can cause myopathy and may contribute to hypotonia or club feet in this case. Although we identified seven recurrent variants, our cohort is too small to determine whether there are clear genotype-phenotype correlations. The two individuals with p.(Lys270Arg) variants (individuals 19 and 20) have similar presentations, with similar ages at sitting and walking, development of some speech, and seizures. In contrast, for the two individuals with p.(Gln172del) variants, one is nonverbal, nonambulatory and has intractable seizures (individual 13), while the other (individual 14) is ambulatory, has aggressive behaviors and has not had seizures. This lack of clear genotype-phenotype correlation will make it difficult to predict the severity of the disease progression in newly diagnosed individuals.
In summary, we report 24 individuals with de novo variants in GNAI1 and a neurodevelopmental disorder characterized by global developmental delay, intellectual disability, hypotonia, and seizures. While there is a spectrum of severity associated with pathogenic variants in GNAI1, most individuals are profoundly affected. Identification of additional cases as well systematic studies that implement uniform tools to evaluate phenotypes such as behavior and cognitive functioning will provide further insight into the full spectrum of neurological features associated with GNAI1 and potentially elucidate subtle genotype-phenotype correlations not apparent in this cohort.
Supplementary Material
Acknowledgements
We thank all affected individuals and their families for participating in this study. AMM received support from the American Epilepsy Society. HCM received support from the National Institute of Neurological Disorders and Stroke (R01 NS069605). FL and AG received funding from European Union and Région Normandie in the context of Recherche Innovation Normandie (RIN 2018). Europe gets involved in Normandie with the European Regional Development Fund (ERDF). Sequencing and analysis were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung and Blood Institute grants UM1 HG008900, R01 HG009141 and by the Chan Zuckerberg Initiative to the Rare Genomes Project. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003). This study makes use of DECIPHER (http://decipher.sanger.ac.uk), which is funded by Wellcome. See Nature PMID: 25533962 or www.ddduk.org/access.html for full acknowledgement.
Footnotes
Web Resources
gnomAD v2.1.1: https://gnomad.broadinstitute.org/
CADD: http://cadd.gs.washington.edu/
FoldX suite: http://foldxsuite.crg.eu/
GeneMatcher: https://www.genematcher.org/
MyGene2: https://mygene2.org/MyGene2/
Matchmaker Exchange: https://www.matchmakerexchange.org/
M-CAP Score: http://bejerano.stanford.edu/mcap/
OMIM: https://www.omim.org/
Ethics Declaration
This study was approved by local institutional review boards of the participating centers (University of Washington and UK Ethics Research Committee). Informed consent was obtained from all individuals or was provided by a parent or legal guardian in the case of minors or individuals with intellectual disability. Their permission for inclusion in this case series, including photographs, was obtained locally.
Conflict of Interest
IMW, RES, KGM, JJ, and LR are employees of GeneDx, Inc. The other authors declare no conflicts of interest.
Data Availability
All methods and data are available on request.
References
- 1.Deciphering Developmental Disorders S. Large-scale discovery of novel genetic causes of developmental disorders Nature. 2015. March 12;519:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplanis J, Samocha KE, Wiel L, Zhang Z, Arvai KJ, Eberhardt RY, et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data Nature. 2020. October;586:757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura K, Kodera H, Akita T, Shiina M, Kato M, Hoshino H, et al. De Novo mutations in GNAO1, encoding a Galphao subunit of heterotrimeric G proteins, cause epileptic encephalopathy American journal of human genetics. 2013. September 5;93:496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrovski S, Kury S, Myers CT, Anyane-Yeboa K, Cogne B, Bialer M, et al. Germline De Novo Mutations in GNB1 Cause Severe Neurodevelopmental Disability, Hypotonia, and Seizures American journal of human genetics. 2016. May 5;98:1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS. G-protein signaling: back to the future Cellular and molecular life sciences : CMLS. 2005. March;62:551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, et al. The G protein-coupled receptor repertoires of human and mouse Proceedings of the National Academy of Sciences of the United States of America. 2003. April 15;100:4903–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deciphering Developmental Disorders S. Prevalence and architecture of de novo mutations in developmental disorders Nature. 2017. February 23;542:433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene Human mutation. 2015. October;36:928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology Genet Med. 2015. May;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, et al. Clinical application of whole-exome sequencing across clinical indications Genetics in medicine : official journal of the American College of Medical Genetics. 2016. July;18:696–704. [DOI] [PubMed] [Google Scholar]
- 11.Cao C, Huang X, Han Y, Wan Y, Birnbaumer L, Feng GS, et al. Galpha(i1) and Galpha(i3) are required for epidermal growth factor-mediated activation of the Akt-mTORC1 pathway Science signaling. 2009. April 28;2:ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall J, Zhou XZ, Chen G, Yang SQ, Li Y, Wang Y, et al. Antidepression action of BDNF requires and is mimicked by Galphai1/3 expression in the hippocampus Proceedings of the National Academy of Sciences of the United States of America. 2018. April 10;115:E3549–E3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peleg S, Varon D, Ivanina T, Dessauer CW, Dascal N. G(alpha)(i) controls the gating of the G protein-activated K(+) channel, GIRK Neuron. 2002. January 3;33:87–99. [DOI] [PubMed] [Google Scholar]
- 14.Feng H, Sjogren B, Karaj B, Shaw V, Gezer A, Neubig RR. Movement disorder in GNAO1 encephalopathy associated with gain-of-function mutations Neurology. 2017. August 22;89:762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The stability effects of protein mutations appear to be universally distributed J Mol Biol. 2007. June;369:1318–1332. [DOI] [PubMed] [Google Scholar]
- 16.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans Nature. 2016. August 18;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mefford HC, Yendle SC, Hsu C, Cook J, Geraghty E, McMahon JM, et al. Rare copy number variants are an important cause of epileptic encephalopathies Annals of neurology. 2011. December;70:974–985. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers CT, Stong N, Mountier EI, Helbig KL, Freytag S, Sullivan JE, et al. De Novo Mutations in PPP3CA Cause Severe Neurodevelopmental Disease with Seizures Am J Hum Genet. 2017. October;101:516–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo H, Bettella E, Marcogliese PC, Zhao R, Andrews JC, Nowakowski TJ, et al. Disruptive mutations in TANC2 define a neurodevelopmental syndrome associated with psychiatric disorders Nat Commun. 2019. 10;10:4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All methods and data are available on request.