Abstract
Bacterial DNA methylation occurs at diverse sequence contexts and plays important functional roles in cellular defense and gene regulation. Existing methods for detecting DNA modification from nanopore sequencing data do not effectively support de novo study of unknown bacterial methylomes. In this work, we observed that nanopore sequencing signal displays complex heterogeneity across methylation events of the same type. To enable nanopore sequencing for broadly applicable methylation discovery, we generated a training dataset from an assortment of bacterial species and developed a method, named nanodisco (https://github.com/fanglab/nanodisco), that couples the identification and fine mapping of the three forms of methylation into a multi-label classification framework. We applied it to individual bacteria and mouse gut microbiome for reliable methylation discovery. In addition, we demonstrated the use of DNA methylation for binning metagenomic contigs, associating mobile genetic elements with their host genomes, and identifying misassembled metagenomic contigs.
Editor Summary:
This work describes nanodisco that is a tool for de novo identifying DNA methylations in bacterial species and microbiomes using nanopore sequencing, as well as performing metagenomic binning using microbial DNA methylation pattern.
Introduction
Single Molecule Real-Time (SMRT) and nanopore sequencing provide a great opportunity for the direct detection of DNA modifications1. SMRT sequencing monitors the pulse fluorescence associated with each nucleotide and records the time it takes for the DNA polymerase to translocate from one nucleotide to the next, termed inter-pulse duration (IPD). Deviation of IPD, calculated by comparing native DNA with methylation free DNA (e.g. produced by whole genome amplification, WGA), is correlated with the presence of DNA modifications2. SMRT sequencing has helped to study bacterial methylomes1, 3 which contain three primary forms of DNA methylation: N6-methyladenine (6mA), N4-methylcytosine (4mC), and 5-methylcytosine (5mC). Great progress has also been made in methods development for DNA modification detection using nanopore sequencing. Two early studies showed differences in current at multiple consecutive positions near the modified base when comparing nanopore sequencing signals from the same genomic regions with or without DNA methylation4, 5. Since then, multiple detection methods were published including Nanopolish6, signalAlign7, mCaller8, DeepSignal9, DeepMod10, NanoMod11, and Tombo12. While encouraging, existing methods were either trained for detecting a specific type of DNA methylation from one of few specific sequence contexts (e.g. 5mC at CpG, and 6mA at GATC) or allow more general detection without effectively differentiating between different forms of DNA methylation13. To date, none of these methods have been applied to characterize unknown bacterial methylomes without prior knowledge, which includes de novo identification of methylation type (i.e. assigning methylation type: 4mC, 5mC, or 6mA), and de novo fine mapping of the methylated nucleotide.
In this work, by examining the three types of DNA methylation in diverse sequence contexts, we observed large variation and complex heterogeneity in terms of their impact on ionic current levels captured in nanopore sequencing. This observation suggests that detection methods are best developed using a diverse collection of species. Bacterial epigenomes are highly motif driven, that is, nearly every occurrence ( >95%) of a methylation sequence motif is methylated1, 14, 15. Following this rationale, we built a comprehensive training dataset and developed a multi-label classification framework for de novo methylation typing and fine mapping of the three forms of DNA methylation at constitutively methylated motifs.
Results
Heterogeneous signal variation induced by DNA methylation in nanopore sequencing
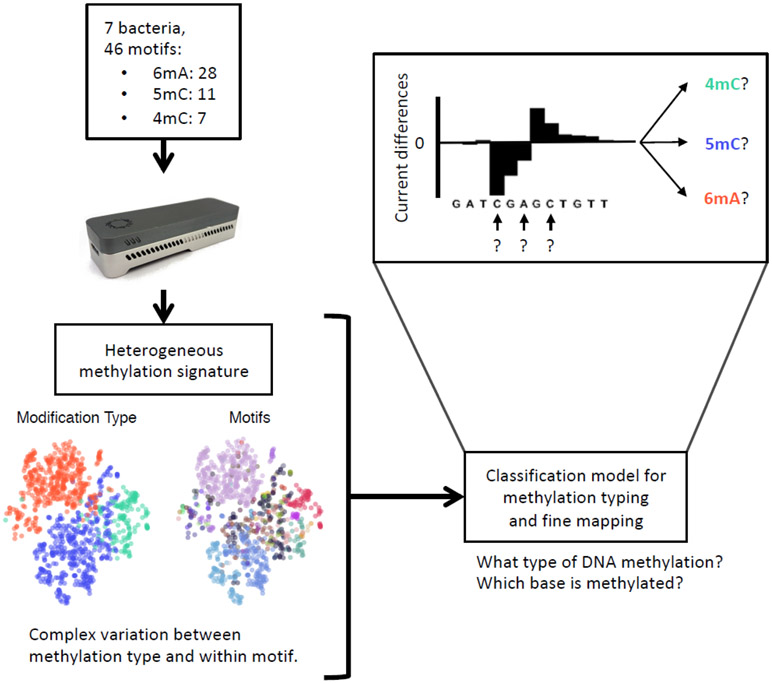
In Bacteria, 6mA, 4mC and 5mC events occur in a highly motif-driven manner. On average, each bacterial genome contains three methylation motifs, and nearly every occurrence of the target motifs is methylated1, 3. To examine the variation of different types of DNA methylation across sequence contexts, we collected seven bacterial species with diverse methylation motifs and genomic GC contents (28.4 to 69.1%; Supplementary Table 1; Methods). According to a previous study3 and REBASE16 (Methods), these strains have a total of 46 unique methylation motifs (6mA: 28; 4mC: 7; 5mC: 11; 308,773 methylation sites in total; Fig. 1; Supplementary Table 2). We conducted nanopore sequencing for both the native and WGA samples on the MinION with R9.4 flow cells achieving 175x coverage on average (Supplementary Table 3).
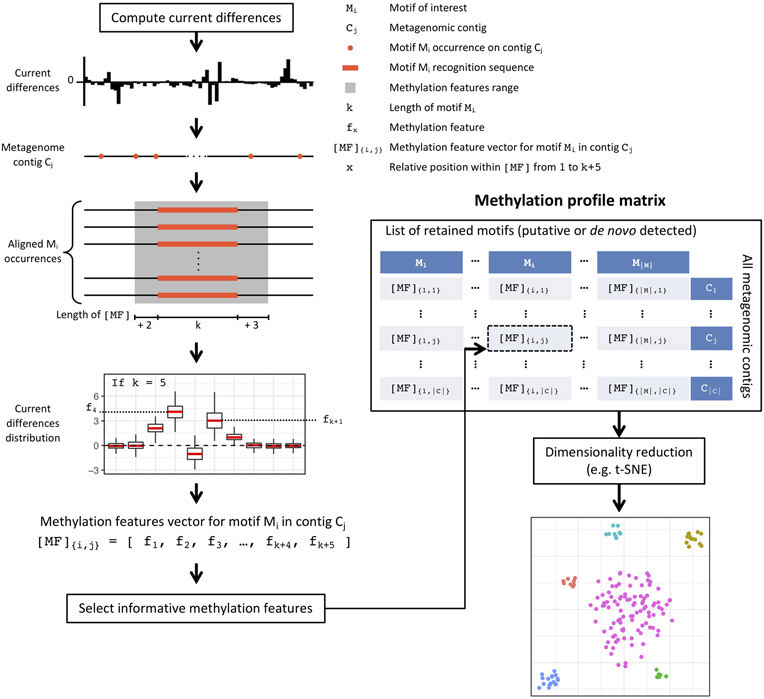
Figure 1:
Schematics for the method design and applications.
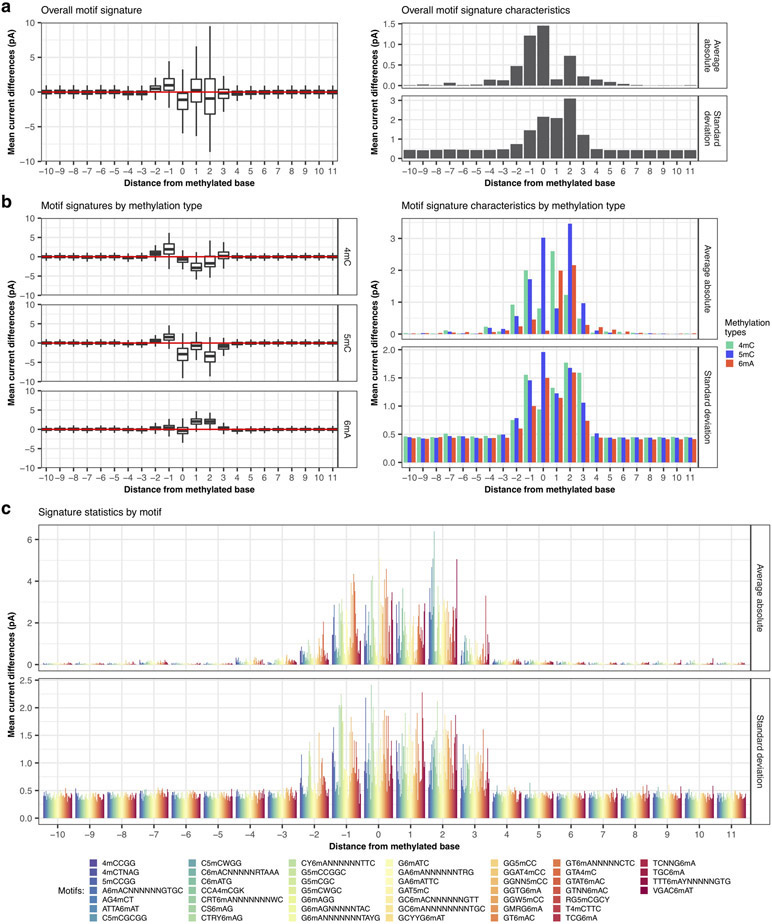
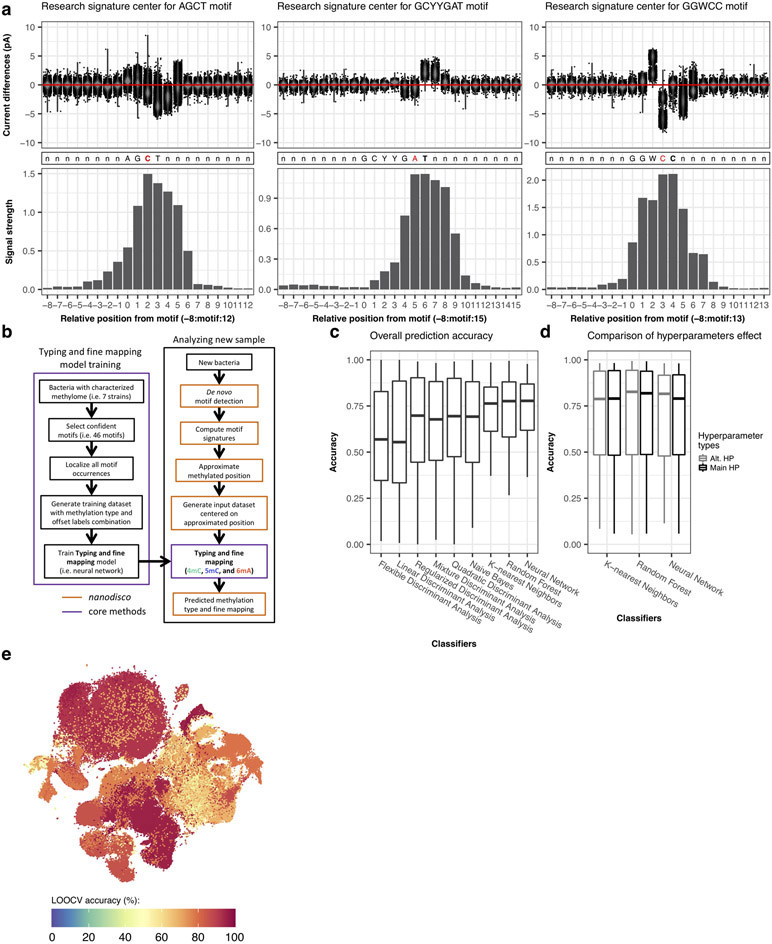
Read events and associated current levels (picoampere, pA) were aligned to reference genomes using Nanopolish6. After normalization and filtering, current differences between native and WGA datasets were computed for each genomic position (Methods). To examine the variation of current differences across different DNA methylation types and motifs, we extracted current differences around each methylated base ([−6 bp, +7 bp]) and computed the methylation motif signatures (i.e. distribution of current differences at relative distance flanking the methylated bases; Fig. 2a). Generally, the widths and amplitudes of perturbation in the methylation motif signatures vary between motifs and methylation types (Extended Data Fig. 1a-c).
Figure 2:
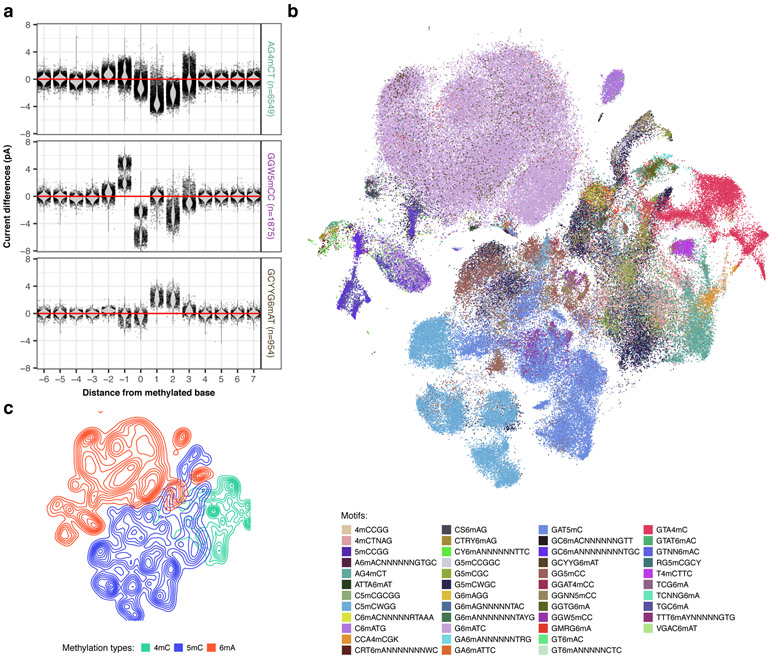
Systematic examination of three main types of DNA methylation with nanopore sequencing. (a) Variation of current differences across methylation occurrences as illustrated by motif signatures from three motifs, AG4mCT (n=6549 occurrences), GGW5mCC (n=1875 occurrences), and GCYYG6mAT (n=954 occurrences). For each motif, current differences near methylated bases ([− 6 bp, + 7 bp]) from all isolated occurrences are plotted with conservation of relative distances to methylated bases. Distributions of current differences for each relative distance are displayed as a violin plot. (b) Variation of current differences across methylation occurrences as illustrated by projection with t-SNE for 46 well-characterized motifs (Supplementary Table 2). Each dot represents one isolated motif occurrence colored by methylation motif. For each motif occurrence, current differences from 22 positions near methylated bases ([− 10 bp, + 11 bp]) were used. (c) Similar to b but colored by DNA methylation type with cluster density indicated by relief.
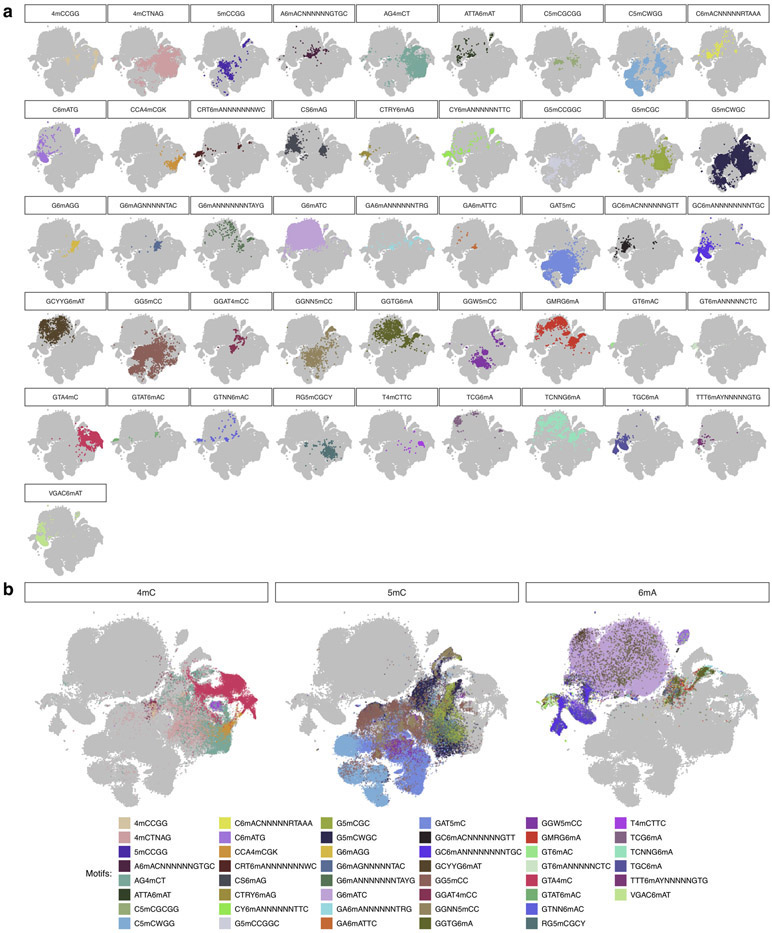
To obtain an overall view of the current differences across all the methylation types and methylation motifs, we subjected the 14 bp vectors ([−6 bp, +7 bp]) capturing current differences across 183,818 non-overlapping methylation motif occurrences to t-distributed stochastic neighbor embedding (t-SNE)17 (Fig. 2b,c, Extended Data Fig. 2). While methylation motif occurrences from the same methylation type tend to cluster together (Fig. 2c and Extended Data Fig. 2b), some individual motifs form distinct sub-clusters (e.g. T4mCTTC and GTA4mC; Fig. 2c and Extended Data Fig. 2b), likewise between methylation sites within the same methylation motif (e.g. GGW5mCC, Fig. 2a,b and Extended Data Fig. 2a). Further analysis of the motif signatures suggests that the across-motif and within-motif variations can be largely explained by sequence variation from degenerated position in the motifs (e.g. GGW5mCC, Fig. 3a,b) or by their flanking sequences (e.g. GAT5mC, Fig. 3c). We found these observations are robust after examining potential sources of variations such as base callers, signal processing workflows, and genome assembly (Supplementary Text, Extended Data Fig. 3, and Supplementary Fig. 1).
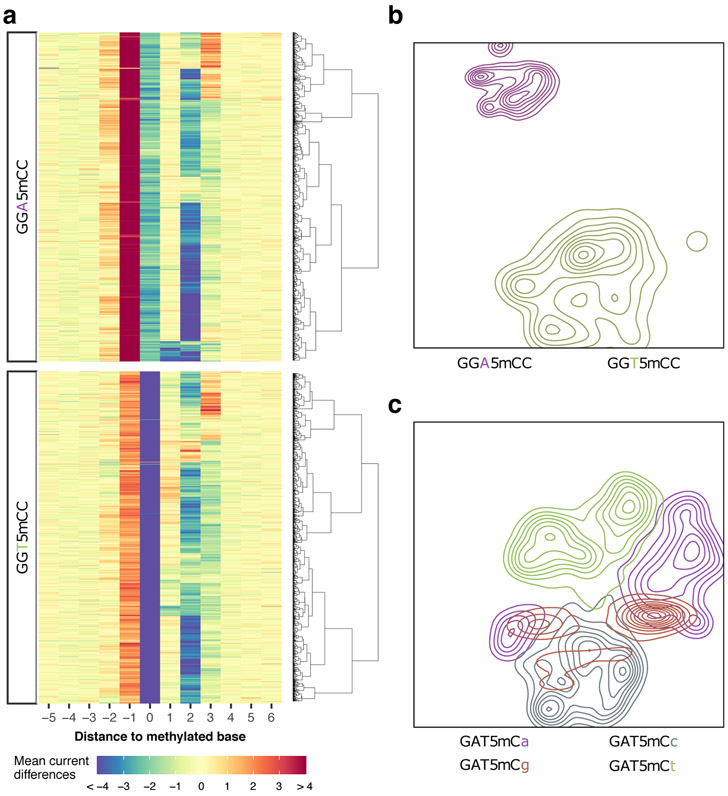
Figure 3:
Local sequence context effect on motif signatures. (a) Current differences from violin plots of GGW5mCC in Fig. 2a were plotted as a heatmap with each row representing current differences flanking a methylation occurrence ([−5, +6] relative to methylation). GGW5mCC motif occurrences were split into two groups according to degenerated base (W=[A∣T]; n=933 for GGACC and n=942 for GGTCC) and ordered, within groups, using hierarchical clustering to highlight current difference patterns. (b) Independent t-SNE projection of GGW5mCC motif occurrences from a with cluster density displayed as relief. Clusters are colored according to degenerated base within the methylation motif. (c) Another example of sequence-dependent variation for GAT5mC motif occurrences displayed after independent t-SNE projection with cluster density displayed as relief. Clusters are colored according to the first base following GAT5mC motif.
de novo methylation typing and fine mapping
The above analyses suggested great signature diversity exists in methylation induced current differences across sequence contexts. To account for this diversity, we developed a method to identify the type of DNA methylation (i.e., methylation typing) and to identify the position of the methylated base (i.e., fine mapping).
Methylation motif enrichment.
The methylation detection and motif enrichment procedure are built on existing methods6, 12, 18. In brief, 1) current levels are compared between native and WGA datasets for each genomic position; 2) p-values are combined locally with a sliding window-based approach followed by peak detection; 3) flanking sequences around the center of peaks are used as input for MEME motif discovery analysis (Methods). Overall, 45 of the total 46 well-characterized methylation motifs from seven bacteria were successfully re-discovered (Supplementary Table 2). The only undetected motif, GT6mAC from H. pylori, has much fewer occurrences (i.e. 198) than other 4-mer motifs (7169 occurrences on average). The motif discovery analysis also found six additional motifs not among the 46 well-characterized motifs (Methods, Supplementary Text), which were not included in the training dataset described below.
de novo methylation typing and fine mapping.
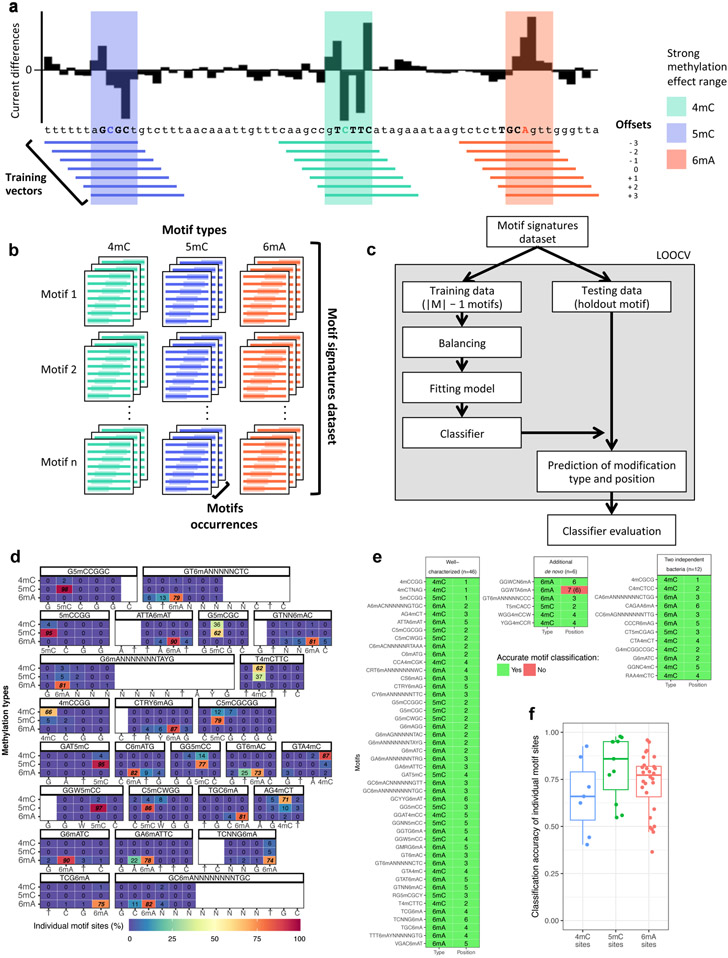
The t-SNE analysis shows that DNA methylation events of the same type generally cluster well (Fig. 2c). We hypothesized that a classification model trained using diverse methylation types and motifs may serve as a reliable approach for categorizing de novo detected methylation. While both methylation type and methylation position are known for the well-characterized training samples (i.e. feature vectors can be consistently defined relatively to the methylated base for classifier training), features vector for the test samples cannot be aligned consistently because the methylated position is yet to be predicted in a de novo discovery setting. Essentially, methylation type classification and methylation fine mapping are coupled problems that need to be approached simultaneously. Building on the observation that no more than +/− 3 bp offsets from peak centers across the 46 well-characterized motifs (Supplementary Table 2; Extended Data Fig. 4a), we designed a multi-label classifier training strategy. For training, each methylation occurrence from a wide range of sequence context is learned 7 times by the classifier, each time using current differences at a specific offset from the methylated base; for a given test sample with unknown methylation type and unknown methylated position, the classifier will first use the center of current differences as an approximation of the methylated position and then predict the methylation type and the exact methylated position (Methods; Fig. 4a-c).
Figure 4:
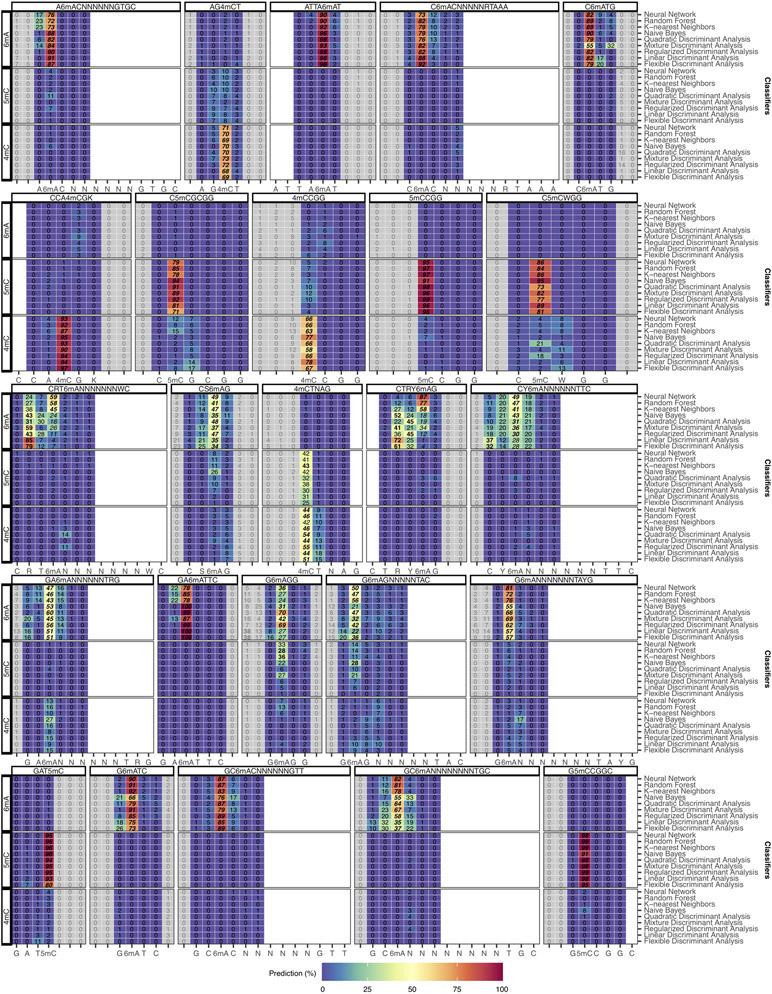
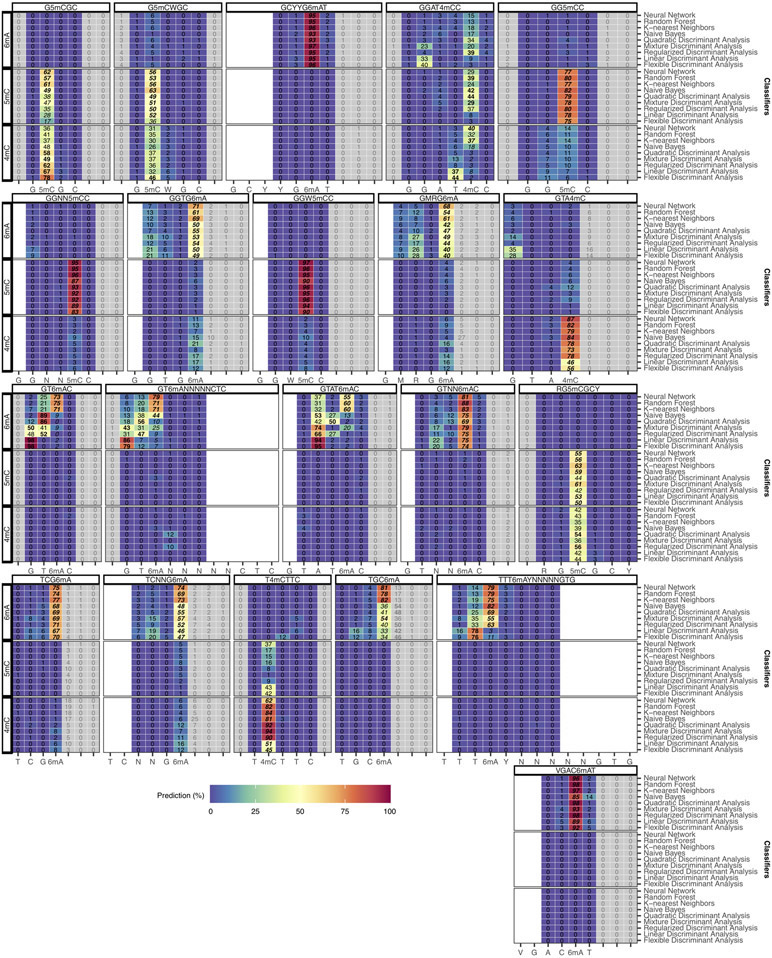
Classification and fine mapping of three types of DNA methylation. (a) For each motif occurrence, we produced 7 training vectors of length 12 with +/− offsets from 0 to 3 position(s) relative to current differences core defined as [−2, +3] (Extended Data Fig. 1a-c). (b) Each training vector is labeled with methylation type and offset used. They are gathered into a training dataset of current differences flanking 183,818 methylated bases from 46 distinct motifs (Methods). (c) Description of the classifier performance evaluation using leave-one-out cross-validation (LOOCV). (d) Detailed classifier evaluation results for neural network model from the LOOCV evaluation for a subset of the 46 well-characterized methylation motifs are displayed for illustration. Filling colors correspond to percentage of occurrences classified to a specific class: blue (0%) to red (100%). Prediction percentages of expected classes are displayed in italics and fine mapped methylated positions in each motif are displayed in bold. (e) Summary of methylation motifs typing and fine mapping results from the neural network model. Green shows accurately typed and/or fine mapped methylation in motif, while red shows inaccurate prediction with the expected result in parentheses. LOOCV results are used for the “Well-characterized” motifs (n=46), while classification results from the final neural network model trained on the 46 well-characterized motifs are used for both “Additional de novo (n=6)” and “Two independent bacteria (n=12)” motifs. (f) Classification accuracy for individual motifs sites (n=46 motifs including 6mA: 28, 4mC: 7, 5mC: 11) from the neural network model. The lower and upper hinges correspond to the 25th and 75th percentiles while the lower and upper whisker extends to the minima and maxima respectively (capped at 1.5 time the inter-quartile range).
A set of nine different classifiers was separately trained using current differences flanking known methylated bases following the offset strategy described above (Methods; Fig. 4a-c; Supplementary Table 4; Extended Data Fig. 4b,d). For evaluation, we used leave-one-out cross-validation (LOOCV) strategy where one motif is held out for testing while all the other 45 motifs are used for training. With all held out individual methylation sites belonging to a single methylation motif classified, predicted methylated type and position within motif was determined from the consensus across tested occurrences (Methods). Overall results for k-nearest neighbors, random forest, and neural network are largely consistent in terms of accuracy for classifying individual methylation sites (Extended Data Fig. 4c) and methylation motifs with at least 95.7% of motifs correctly typed and fine mapped (Extended Data Fig. 5 and 6). For simplicity, only results from the neural network model are used for the remainder of the study (Fig. 4d,e). Methylation site classification accuracy varies widely ranging from 36% for G6mAGG to 98% for G5mCCGGC (median accuracy of 78%; Extended Data Fig. 4c, 4e, 5, and 6), which is consistent with the observation that motifs of the same methylation type can have different signatures (Fig. 2c and Extended Data Fig. 2,3,7d-e). Furthermore, breaking down individual methylation sites per methylation type shows balanced accuracy results for 4mC, 6mA, and 5mC motifs, while all the 46 well-characterized motifs are correctly typed and fine mapped (Fig. 4f, Extended Data Fig. 5 and 6).
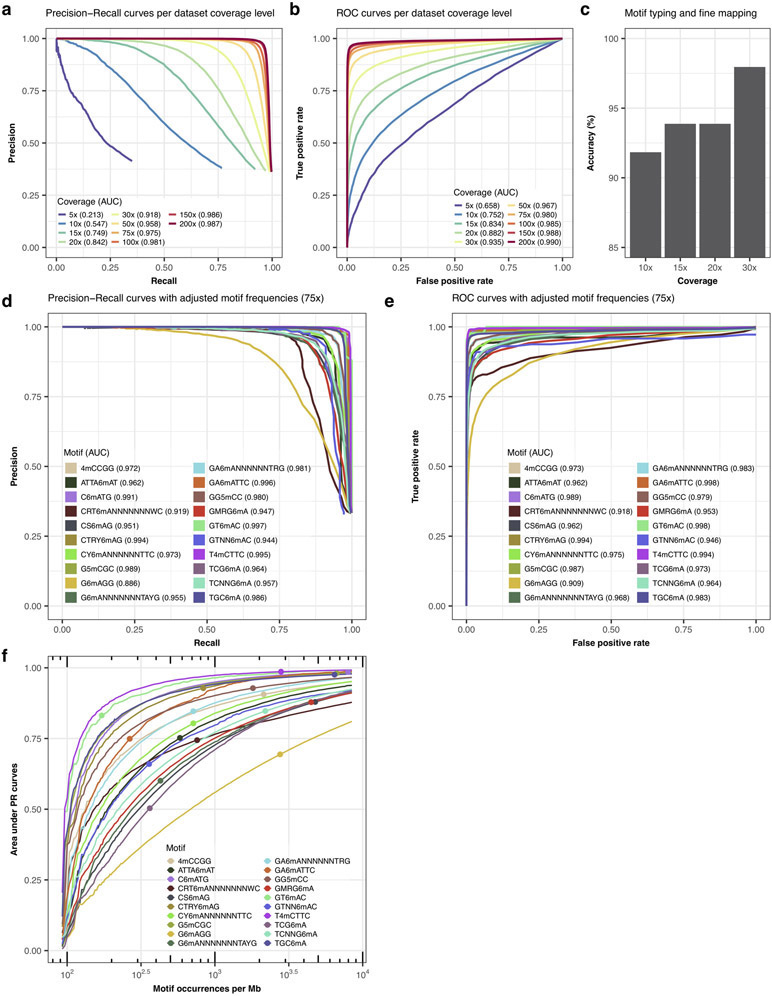
After training of a final model from the 46 well-characterized motifs, motif typing and fine mapping performances were further assessed on two independent bacterial samples: N. otitidiscaviarum and T. phaeum. All the 12 known methylation motifs were de novo re-discovered as well as accurately typed and fine mapped (11 were not among the 46 motifs in the final training; Supplementary Table 5). We also applied the classification method on all de novo discovered motifs including the six additional motifs, which resulted in an overall accuracy of 98.1% (51/52 motifs well classified combining LOOCV results for the 46 well-characterized motifs and using the final model on the 6 additional motifs). We further evaluate the impacts of de novo genome assembly (Extended Data Fig. 3m,n), genome coverage (Extended Data Fig. 7a-c), and motif frequency (Extended Data Fig. 7f).
Methylation discovery from microbiome and methylation-enhanced metagenomic analyses
A number of methods have been developed to group metagenomic contigs (i.e. binning) using composition features19-22, contig coverages23-26, and chromosome interaction maps27-29. Recent work by Beaulaurier et al. demonstrates that microbial DNA methylation can be exploited as complementary features to enhance metagenomic binning (i.e. methylation binning) using SMRT sequencing30. We hypothesized that methylation binning of metagenomic contigs with nanopore sequencing holds great promise by utilizing all three DNA methylations types (6mA, 4mC, and 5mC) beyond the scope of Beaulaurier et al. that focused on 6mA (SMRT sequencing does not effectively detect 5mC at diverse sequence contexts)30.
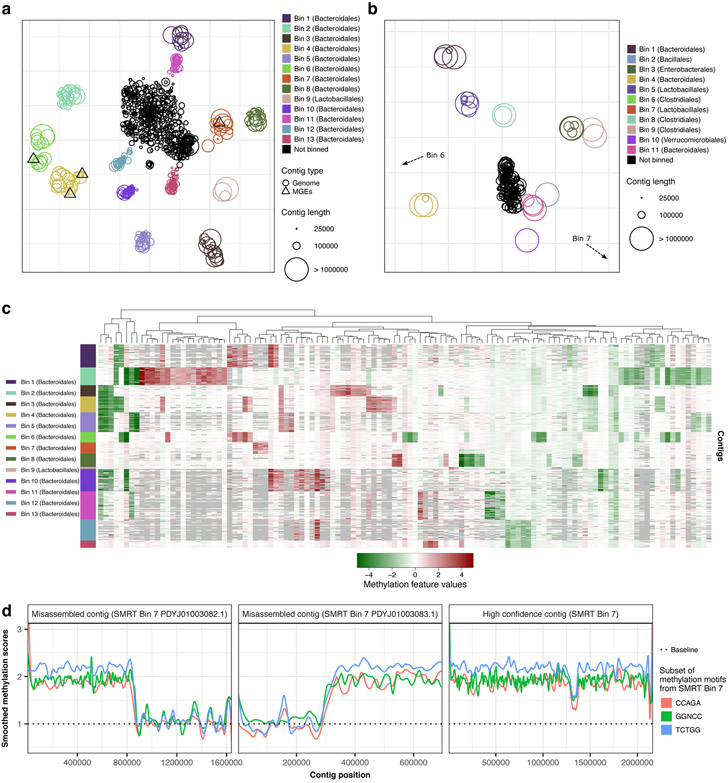
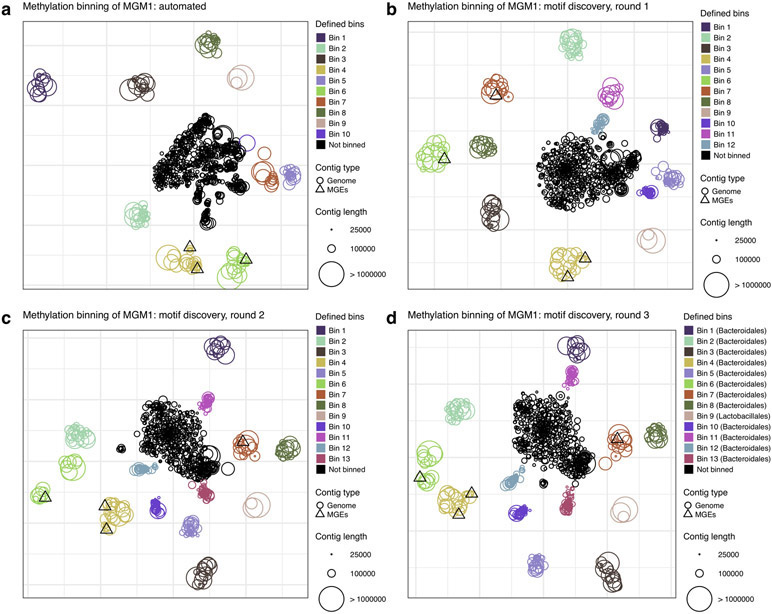
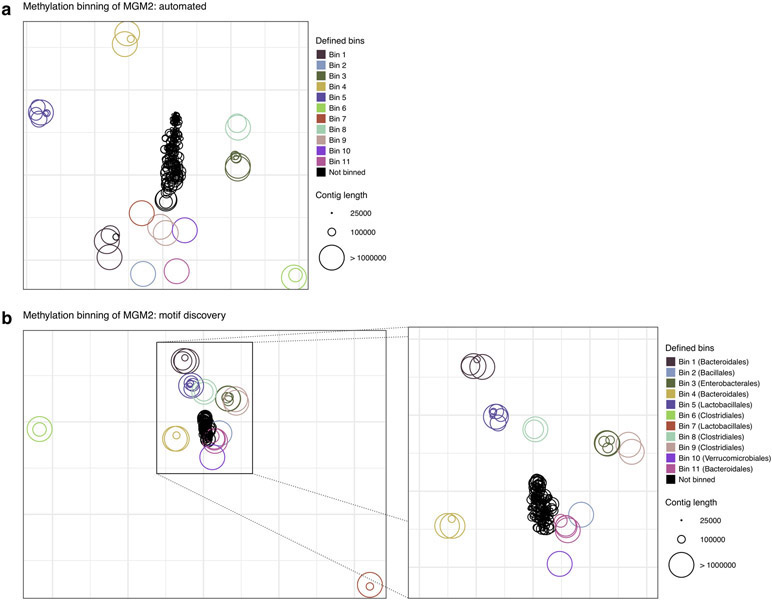
We developed a methylation binning approach with nanopore sequencing data considering the fundamental differences from SMRT sequencing (Methods; Extended Data Fig. 8). In a nutshell, we had to address the observation that current differences associated with methylation are spanning multiple events near methylated bases (Fig. 2a, Fig. 3a, and Extended Data Fig. 1) rather than confined to a single base for 6mA or 4mC as in SMRT sequencing. After prototyping and evaluation on a mock community (Supplementary Text; Supplementary Fig. 2), we applied the method on nanopore sequencing data of the same mouse fecal sample used in the SMRT sequencing study (MGM1; Supplementary Table 6). To summarize, after the de novo metagenome assembly, we computed methylation feature vectors for a large set of candidate methylation motifs (n=210,176; Methods). Motifs with informational feature (i.e. significant current differences) were first selected based on large contigs, and methylation feature vectors were then computed in remaining contigs. Methylation feature vectors are then arranged in a methylation profile matrix, which is subjected to clustering analysis based on similarity among contigs (Methods). This initial automated binning resulted in ten bins (Extended Data Fig. 9a), which were further refined by per-bin motif detection and binning guided by discovered motifs (Methods; Extended Data Fig. 9b-d). The final methylation binning of MGM1 contigs was performed using the 80 de novo detected methylation motifs (Supplementary Table 7),which revealed thirteen bins containing from 3 to 43 contigs in each (Fig. 5a; Extended Data Fig. 9d; Supplementary Table 8). The unique methylation profiles for each bin are displayed in Figure 5c. Among contigs with length >50kb and with average coverage >5x, 85% can be binned based on methylation information, which correspond to 91% of the contig cumulative lengths. The method was further tested with a second microbiome sample, MGM2 (Supplementary Table 6), in which eleven bins with unique methylation profiles were identified (Fig. 5b; Extended Data Fig. 10; Supplementary Table 9). We observe that bins from nanopore sequencing data closely matched those from SMRT sequencing data30, and none of the nanopore sequencing bins contained misclassified contigs (Methods; Supplementary Fig. 3a and Supplementary Table 10). Consistent between the two technologies, methylation binning effectively separated the multiple Bacteroidales species that are usually hard to distinguish from each other due to their highly similar genome sequence composition and abundance30.
Figure 5:
Methylation analysis of mouse gut microbiome samples. (a) Methylation binning of MGM1 contigs using de novo discovered motifs (after three rounds of binning followed by motif discovery; Methods, Extended Data Figs. 8, 9). Methylation features computed from de novo discovered motifs with t-SNE analysis. Contigs are colored based on bin identities with point sizes matching contig length. (b) Same as a but for MGM2 contigs (one round of binning followed by motif discovery; Methods, Extended Data Figs. 8, 10). Non-zoomed plot (with visible Bins 6, 7) in Extended Data Fig. 10b. (c) Heatmap of methylation feature values (all de novo discovered motifs) across binned contig from MGM1 sample (n=309 contigs). Only the significant features with absolute values above 1.5 pA in the bin of origin (where the motif was discovered) were selected (n=119 methylation features). Missing methylation features from contigs (less than 5 motif occurrences) are colored in grey. (d) Detection of misassemblies using methylation signal along contigs. Left and middle panels: misassembled contigs mislabeled as Bin 7 in SMRT analysis (PDYJ01003082.1 and PDYJ01003083.1, contigs marked with an asterisk in Supplementary Fig. 3a. Right panel: an example of a properly assembled contig from SMRT Bin 7 (PDYJ01000763.1). We selected three de novo detected motifs from SMRT Bin 7 and scored their methylation sites along the three contigs. Methylation scores were smoothed and displayed with one color per motif. Methylation scores are consistent in the contig in the right panel, but not in the misassembled contigs. A switch of methylome occurs near 800 kbp and 300 kbp in the left two panels respectively, supporting misassemblies (detailed in Supplementary Fig. 4a,b).
Through the methylation binning analysis, 80 methylation motifs were detected using MEME from the thirteen bins from MGM1 sample (Supplementary Table 7). We applied the methylation typing and fine mapping method and made confident prediction of methylation types and modified positions for 64 motifs (6mA: 18; 5mC: 46; Supplementary Table 11). The de novo detection of a large number of 5mC motifs is encouraging because previous large-scale bacterial methylome studies were almost exclusively based on SMRT sequencing, which is known to be ineffective for detecting 5mC methylation across diverse motifs. However, not every 6mA motif found with SMRT sequencing was detected in the analysis of nanopore sequencing data. The missing ones are mostly bipartite 6mA motifs, which are usually not frequent and thus more challenging to detect using nanopore sequencing. This is probably due to the diffuse nature of current differences around 6mA (Fig. 2a and Extended Data Fig. 1) in contrast to the highly specific signal right on top of 6mA in SMRT sequencing.
We further attempted to link mobile genetic elements (MGEs) to their host genome based on their methylation profiles. Using the SMRT metagenomic assembly with de novo discovered methylation motifs, we were able to bin 11 of the 19 annotated MGEs from this microbiome sample according to their methylation profiles (five plasmids and six conjugative transposons; Supplementary Fig. 3b; Supplementary Table 12), while nine were binned with the SMRT analysis30. With eight MGEs binned as with SMRT analysis and three newly binned MGEs, nanopore sequencing increased MGEs linking potential compared to SMRT methylation binning likely owing to its better sensitivity to 5mC motifs. From our nanopore-only de novo metagenome assembly, fewer MGEs were identified (eight), although similar results were obtained in terms of linking MGEs to their host genomes, i.e. four out of the eight MGEs identified were binned correctly (Fig. 5a).
In addition to contig binning, we hypothesized that the microbial DNA methylation pattern can also be used to discover misassembled contigs. The methylation pattern is expected to be largely consistent across different regions of an authentic metagenomic contig. Following this rationale, we discovered two contigs from SMRT sequencing based metagenomic assembly of the MGM1 sample (marked by an asterisk in Supplementary Fig. 3a) showing inconsistent intra-contig methylation status (Fig. 5d). By comparing methylation patterns from methylation motif sets from the other bins, we found that the contigs in question are chimeric contigs representing two Bacteroidales species (Supplementary Fig. 4, SMRT Bins 2 and 7). This is consistent with the previous examination of coverage uniformity and contamination through single-copy gene count30, confirming that those contigs annotated as Bin 7 were misassembled by HGAP2 combining parts of Bin 2 and Bin 7 genomes. Generally, this analysis highlights the benefit of incorporating DNA methylation status (ideally all three types: 6mA, 4mC, and 5mC), which not only help better distinguishing microbial species but also help assess contig homogeneity revealing eventual misassemblies.
Discussion
In this work, we developed a method for de novo discovery (methylation typing and fine mapping) of three forms of bacterial DNA methylation (4mC, 5mC, and 6mA). We also developed a method for nanopore sequencing-based methylation binning of metagenomic contigs and MGEs-to-host mapping, building on the method reported for SMRT sequencing data30. In addition, we demonstrated that examining the methylation pattern along assembled metagenomic contigs could help identify chimeric contigs. While methylation binning provides additional discriminative power for species with highly similar genome sequences, other binning features (e.g. sequence composition and coverage binning) have advantages when contigs are short or when two organisms have the same methylome, although this latter case is not common based on the diversity and variation of bacterial methylomes30.
Our comparative methylation binning analysis between SMRT and Nanopore sequencing from the same microbiome sample provided important insights. First, nanopore sequencing provides reliable 5mC detection across diverse sequence contexts, addressing a challenge faced by SMRT sequencing. The large number of 5mC motifs discovered from the mouse gut microbiome sample using nanopore sequencing suggests the prevalence and diversity of 5mC motifs could have been largely underestimated in the >2,700 bacterial methylome analyses that were almost exclusively based on SMRT sequencing16, 30. Second, we found that multiple long and rare methylation motifs well detected by SMRT sequencing in the metagenome analysis were missed by nanopore sequencing, which can be explained by the diffuse current differences associated with methylation in contrast to the high IPD ratios confined to a single methylation site (4mC or 6mA) for SMRT sequencing2, 31-34. Collectively, it shows that SMRT sequencing and nanopore sequencing have their own strengths and limitations, hence the two technologies are expected to complement each other in various applications.
For individual methylation sites, we would like to highlight that the accuracy of the current method for methylation typing and fine mapping varies across different motifs, which calls for development of more accurate methods in future work. In practice, existing tools such as Tombo allow the estimation of partial methylation at individual methylation sites once motifs are de novo discovered and characterized, thus are complementary to our method.
Lastly, while this study focused on three types of DNA methylation, similar design could be extended for the detection for other forms of DNA modification35, 36, as well as RNA modifications37, 38 owing to nanopore technology direct RNA sequencing39, 40.
Online Methods
Samples collection and DNA extraction
A set of nine bacteria was selected using a previous study3 and REBASE16 to provide a large diversity of methylation motifs: Bacillus amyloliquefaciens H, Bacillus fusiformis 1226, Clostridium perfringens ATCC 13124, Escherichia coli K-12 substr. MG1655 ATCC 47076, Methanospirillum hungatei JF-1, Helicobacter pylori JP26, Neisseria gonorrhoeae FA 1090, Nocardia otitidiscaviarum NEB252, and Thermacetogenium phaeum DSM 12270.
B. amyloliquefaciens H, B. fusiformis 1226, and N. otitidiscaviarum NEB252 DNA samples were obtained from New England Biolabs (NEB, Ipswich, MA). Those for C. perfringens ATCC 13124, M. hungatei JF-1, H. pylori JP26, N. gonorrhoeae FA 1090 and T. phaeum DSM 12270 were obtained from the Human Health Therapeutics Research Area at National Research Council Canada, the Department of Microbiology, Immunology, and Molecular Genetics at University of California Los Angeles, the Department of Medicine at New York University Langone Medical Center (NYUMC), the University of Oklahoma Health Sciences Center, and the Department of Biology at the University of Konstanz (Germany), respectively. Finally, we obtained E. coli K-12 substr. MG1655 ATCC 47076 from the American Type Culture Collection (ATCC, Manassas, VA).
The adult mouse gut microbiome DNA samples (MGM1 and MGM2) were obtained from the Department of Medicine at NYUMC. MGM1 DNA sample was extracted from the fecal pellets used in the SMRT sequencing study30 while MGM2 DNA sample comes from fecal pellets of the same mouse after antibiotic treatment with tylosin. Fecal DNA extraction was performed using QIAamp DNA Microbiome Kit (QIAGEN, Hilden, Germany) followed by cleanup with DNA Clean & Concentrator – 5 elution buffer (ZYMO Research, Irvine, CA) and final elution in 10 mM Tris-HCl, pH 8.5, 0.1 mM EDTA.
Library preparation and sequencing
The quality of input DNA was controlled with Nanodrop 2000 and concentration measured using Qubit 3.0 (Thermo Fisher Scientific, Waltham, MA). Native libraries were prepared following 1D Genomic DNA by ligation protocol (SQK-LSK108; version GDE_9002_v108_revT_18Oct2016) with minor modifications described below. Whole genome amplification samples were prepared using REPLI-g Mini Kits (QIAGEN, Hilden, Germany) according to the protocol with 12.5 ng of input DNA and 16 h incubation. Next, WGA samples were treated with T7 endonuclease I (NEB) to maximize nanopore sequencing yield according to ONT documentation. WGA libraries were prepared following Premium whole genome amplification protocol from T7 step (version WAL_9030_v108_revJ_26Jan2017) with minor modifications described below. Bacteria (other than E. coli and H. pylori) and mouse gut microbiome DNA samples, native and WGA, were RNase A treated (FEREN0531, Thermo Fisher Scientific) then fragmented at 8 kbp with g-TUBEs (Covaris, Woburn, MA) to homogenized DNA fragments lengths increasing accuracy of input DNA molarity calculation to maximize yields. Final fragment length distributions were determined using Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Samples were sequenced on R9.4 and R9.4.1 MinION flow cells using MinKNOW (versions 1.5.12 and 1.10.11; Supplementary Table 3 and 6).
E. coli and H. pylori libraries (native and WGA) were prepared without fragmentation or Formalin-Fixed, Paraffin-Embedded (FFPE) DNA repair. E. coli and H. pylori WGA input DNA was increased to 3 μg in T7 step with 20 min incubation. Remaining steps were performed according to corresponding ONT protocol and final libraries sequenced on 3 flow cells with a maximum of two consecutive runs per flow cell. Flow cells were washed between runs using the Flow Cell Wash Kit (EXP-WSH002) from ONT. An additional WGA was produced for H. pylori and referred to as independent WGA. Sequencing of native and WGA libraries for E. coli and H. pylori generated from 289 to 2630x genomic coverage but were down sampled at 200x to more accurately represent common yield targets.
DNA samples for the additional bacteria (B. amyloliquefacien, B. fusiformis, C. perfringens, M. hungatei, N. gonorrhoeae, N. otitidiscaviarum, and T. phaeum) were pooled in equimolar quantity for library preparation. Pooling possibility was confirmed by mapping mock nanopore reads datasets generated using Nanosim41 (version 1.0.0; simulator.py linear -r <path_to_fasta> -c <error_model> -o <path_output> -n 50000 --min_len 200 --max_len 50000 using the E. coli error model provided by the authors on 03/23/17) on the combined references and verifying accurate separation of reads into genome of origin. Any reads mapping on more than one genome were discarded from all the analysis presented in our study, independently of the mapping type. Native and WGA library preparations were performed using aforementioned ONT protocol and sequenced on separate flow cells (Supplementary Table 3). Sequencing of native and WGA generated datasets with coverage ranging from 65 to 297x.
Finally, mouse gut microbiome libraries (MGM1 and MGM2) were generated according to the One-pot ligation protocol for Oxford Nanopore libraries (dx.doi.org/10.17504/protocols.io.k9acz2e) including the FFPE DNA repair step with exception for the room temperature incubation times that were increased from 10 to 20 minutes. 300 fmol of input DNA were used in FFPE DNA repair steps. Native and WGA libraries were sequenced on separate flow cells for 48 h (Supplementary Table 6).
Nanopore sequencing signal processing
Nanopore sequencing reads are base called using ONT Albacore Sequencing Pipeline Software (version 2.3.4). Reads are mapped to corresponding references using BWA-MEM (version 0.7.15 with –x ont2d option)42. The following steps are performed using R (version 3.5.3). Reads are separated by strand according to the initial alignment (package Rsamtools; version 1.34.1)43, and both groups are processed as forward strand reads by mapping reverse strand reads on the reverse complement of the reference genome using BWA-MEM. Supplementary and reverse strand alignments are then filtered out with samtools (version 1.3; flags 2048 and 16)44. Next, events are associated to genomic positions according to alignment coordinates from reads and expected current levels with Nanopolish eventalign (version 0.11.0)6. Event levels are normalized across reads by correcting signal scaling and shifting. Both normalization factors are computed for each read by fitting events level to ONT 6-mer model (nanopolish configuration file r9.4_450bps.nucleotide.6mer.template.model) using robust regression (rlm function). Event level outliers are removed using Tukey’s fences methods based on interquartile range (IQR=1.5) for each genomic position. Finally, mean event current differences (pA) were computed by comparing event levels between native sample (maintained methylation state) and WGA sample (essentially methylation free) at each genomic position for both strands separately. This metric is referred to as current differences in our manuscript. Associated p-values from two-sided Mann-Whitney U test are also computed (wilcox.test function) which was proposed in Stoiber et al.12. Only genomic positions with sufficient coverage are considered in later analysis (min_cov=5).
Motif enrichment analysis
DNA methylation affects nanopore sequencing signal at multiple positions around the methylated base (Fig. 2a and Extended Data Fig. 1a-c)4 meaning detection of methylated sites can be reinforced by combining information from consecutive genomic positions. As in Stoiber et al., consecutive p-values are combined with Fisher’s method (sumlog function) in sliding windows (5 bp) smoothing statistical signal along the genome12. It combines the methylation related signal near methylated bases and reduces signal noises from spurious genomic positions. Resulting smoothed statistical signals form peaks near methylated positions. Detected peaks are ranked according to their smoothed p-value and the top 2000 peaks are then selected for motif discovery. An alternative strategy is to randomly sample peaks from more than the top-2000 positions as described below. Corresponding genomic sequences are then extracted (22 bp, at the peak position, which was defined to encompass the complete motif recognition sequence for the subsequent motif discovery according to the H. pylori dataset) and used as input for de novo motifs discovery with MEME software (version 4.11.4; parameters: -dna -mod zoops -nmotifs 5 -minw 4 -maxw 14 -maxsize 1000000)18. The selection of region of interest based on combined p-values followed by motif detection using MEME was initially proposed in a preprint by Stoiber et al.12. However, we enhanced the motif discovery potential by closely integrating MEME in our pipeline as described in next paragraphs.
Running time for motif discovery with MEME rapidly increases with size of the sequence dataset to such extent that we had to limit the number of input sequences used. To address this constraint, we adopt a repeated procedure of back and forth between peak detection and motif discovery steps3, 31. For each pass, a limited number of input sequences are analyzed with MEME and motifs achieving a sufficient confidence (E-value <= 10−30) are reported. After each motif discovery step, peaks explained by discovered motifs, whose corresponding genomic sequence contains at least one of the de novo detected motifs, are removed making it possible to discover less frequent motifs and ones with weaker signals. This motif discovery procedure is automatically stopped when no additional motif can be found enriched in the input sequences (i.e. no motifs are significantly more frequent in the input sequence than in the background). This repeated procedure is adapted for detecting any number of methylated motifs while decreasing processing time.
Furthermore, we observed that with some genomes, top peaks (based on smoothed p-value) could be enriched in specific motif combinations (i.e. motifs in close proximity) preventing MEME from discovering individual motifs in favor of the specific motif combination. This is due to larger than average smoothed p-value happening when two motif occurrences are near each other, which affects current in a broader genomic region. This phenomenon was observed for genomes with multiple frequent motifs such as H. pylori. To limit this bias when observed, we provide an option to randomly select sequences among top peaks (i.e. smoothed p-values above a threshold resulting in more than 2000 peaks), effectively avoiding the enrichment of specific motif combinations.
Raw motifs called by MEME were further refined by leveraging current difference information. The rationale is that if the initial motif found with MEME is not precise (e.g. GATCH instead of GATC or CCAGG instead of CCWGG) then we can refine it by looking at the motif signature of related motifs, which is expected to stay flat when the motif is not methylated (i.e. current differences distributed around 0 for all positions). For each motif reported by MEME, we generated a set of related motifs by introducing substitutions, one substitution at a time. For example, the refinement of GATC will give 12 related motifs with substituted nucleotide in bold: AATC, CATC, TATC, GCTC, GGTC, GTTC, GAAC, GACC, GAGC, GATA, GATG, GATT. We then compute each related motif signature (see Motif typing and fine mapping) with associated scores representing total divergence from non-methylated signature (i.e. sum of absolute average current differences).
Parameter tuning for signal processing and motif detection
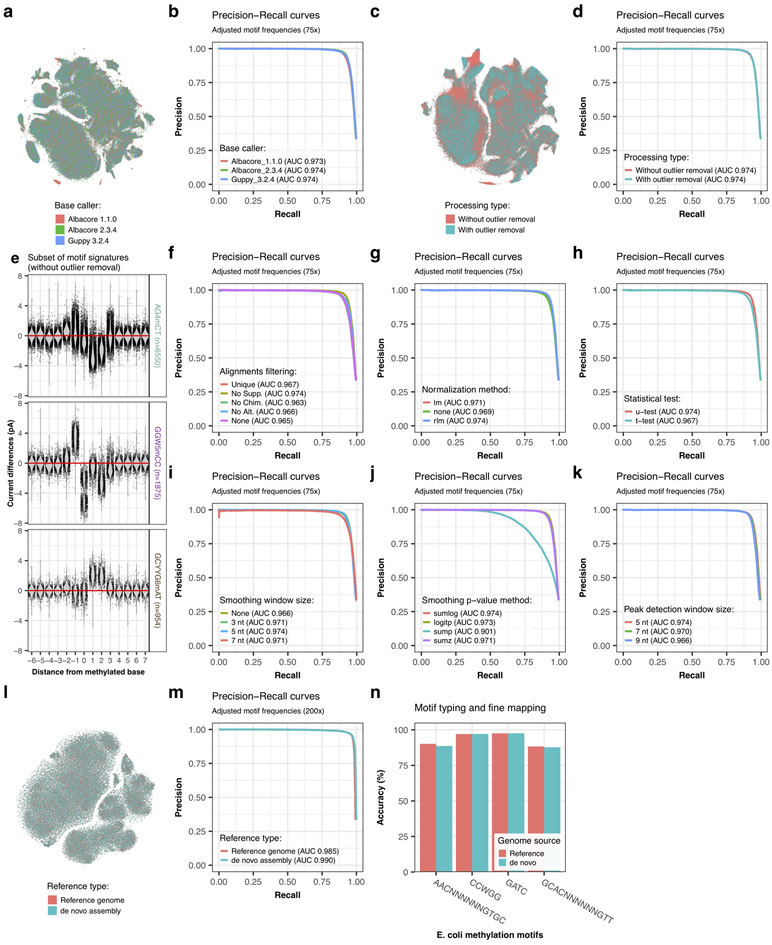
To assess our methods performance for de novo motif discovery and tune parameters, we evaluated the enrichment of MEME input sequences for expected motifs as the chosen smoothed p-value threshold varies. Method development and choice of default parameters was guided by evaluating various metrics including Precision-Recall (PR) curves, Receiver Operating Characteristic (ROC) curves and area under curves (AUC). We used the following two comparisons to define contingency table classes (i.e. two current differences datasets): native versus WGA, and independent WGA versus WGA. The independent WGA versus WGA comparison is used to improve the true negative (TN) and the false positive (FP) estimation by including information at unmethylated motif occurrences, which is absent from the native versus WGA comparison. True positives (TP) and false negatives (FN) are respectively defined as motif occurrences with or without signal peaks above a threshold in native versus WGA. False positives (FP) are genomic regions without motifs and with signal peaks above a threshold in native versus WGA as well as motif occurrences with signal peaks above a threshold in independent WGA versus WGA. Finally, true negatives (TN) are defined as genomic regions without motifs and without signal peaks above a threshold in native versus WGA as well as motif occurrences without signal peaks above a threshold in independent WGA versus WGA. State of motif occurrences were defined whether a peak was detected above the chosen threshold in a 22 bp window encompassing expected methylated base of motif occurrences. For genomic regions devoid of motif, those were split in 22 bp consecutive units, and used in the computation of FP and TN with similar status definition. Performances were computed on the first 500 kbp of the reference genome only. When comparing performances for de novo detection between individual motifs, we took into consideration variation in frequencies (i.e. a rare motif will be more difficult to detect). Therefore, in order to make the evaluation more generally applicable, we fixed the ratio of positive regions (22 bp windows from motif occurrences in native versus WGA) over all queried regions to one third by random subsampling either the motif occurrences or the genomic regions without motifs depending on the natural motif frequency (i.e. the original ratio of motif occurrences over all queried regions), effectively avoiding variation in frequencies across the set of H. pylori motifs. In the opposite, we also evaluate the impact of motif frequency on de novo detection by creating in silico datasets with a wide range of motif frequencies using a similar random subsampling strategy. Note that this method evaluation design, which assumes that all motif occurrences are methylated, could be slightly underestimated FP and TP, while FN could be slightly overestimated.
Using the aforementioned method, we evaluated parameter performances for de novo methylation detection for the following steps or parameters: read mapping (Extended Data Fig. 3f), event current normalization (Extended Data Fig. 3g), outlier removal (Extended Data Fig. 3c,d), statistical test (Extended Data Fig. 3h), smoothing window size (Extended Data Fig. 3i), p-value combining function (Extended Data Fig. 3j), and peaks window size (Extended Data Fig. 3k). We also evaluated the impact of coverage by subsampling at 10 depths ranging from 5x to 200x as well as the impact of motif frequency and the motif specific context (i.e. how methylation type and sequence context affect detection potential; Extended Data Fig. 7).
Validation of methylation motifs used for classification
E. coli and H. pylori were sequenced with SMRT sequencing in order to confirm 4mC and 6mA methylation motifs using the RS_Modification_and_Motif_Analysis protocol from SMRT Analysis Server (v2.3.0). Methylation status summaries for the remaining bacterial species (modifications.csv and motif_summary.csv files) were obtained from the U.S. Department of Energy Joint Genome Institute and NEB. We confirmed effective methylation of 4mC and 6mA motifs individually by checking if IPD ratio (IPDnative over IPDcontrol, which is either obtained from a WGA sample or an in silico model) consistently peaked on expected methylated bases. Finally, REBASE annotation was used as a gold standard for 5mC motifs. Methylation motifs with an ambiguous status (e.g. weak or partial IPD ratio peaks) or not reported in REBASE annotation were not used for the classifier training and the performance evaluation.
Motif typing and fine mapping
For each bacterial genome, we list methylated genomic positions from each strand based on motif recognition sequences. Methylated positions in close proximity are discarded to avoid introducing unwanted complexity (at least 22 bp apart, each strand considered independently as current signal is strand specific). Ambiguous motifs are removed from downstream analysis (see Validation of methylation motifs used for classification in Methods). We extract current differences in [− 10 bp, + 11 bp] range relative to methylated base positions allowing for the subsequent creation of the offsetted dataset used for the classifier training. Each occurrence is labeled with genome of origin, recognition sequence, methylation type, methylation position within motif, and genomic coordinates. This dataset constitutes our methylation motif signatures for motif typing and fine mapping, while we use a subset of it, [− 6 bp, + 7 bp], to examine the variation of current differences across different DNA methylation types and motifs. Note that for de novo detected methylation motif and refinement function, signatures are generated considering every position in the motif as potentially methylated, which produced a longer signature not necessarily centered on the methylated base.
The training dataset for classification is generated from methylation motif signatures to permit labeling of methylation type and position within motifs simultaneously (Fig. 4a). For each vector of current differences from a methylated site, we generate 7 smaller vectors, lengths 12, offsetted by one position so that each of them still contains the [− 2 bp, + 3 bp] range relative to the methylated base (range with the most current differences, Extended Data Fig. 1). In other words, those 7 vectors contain current differences from the [− 2 bp, + 3 bp] range with up to 3 additional position(s) before or after it (i.e. [− 5 bp, + 6 bp] +/− 0 to 3 bp). Each of those vectors is labeled with the type of DNA methylation from corresponding motifs as well as corresponding offset used (from − 3 to + 3) resulting in 21 different labels (7 offsets x 3 DNA methylation types).
For the testing datasets, methylated base position is unknown and current difference vectors cannot be defined in the same way. However, methylated base position can be approximate by computing the center of current differences from a motif signature. For that, we average absolute current differences from a motif signature using a sliding window of length 5 and the position with the largest variation is used as an approximation of methylation position within the motif (Extended Data Fig. 4a). In practice, approximations are not further than 3 bp from the methylated position meaning that the vectors of current differences centered on those approximations will match one type of vector offset used for training because they are generated with − 3 to + 3 bp offsets.
Prior to any model fitting, the training dataset is balanced by random sampling to contain a similar number of vectors for each label in order to avoid bias toward the more common methylation type. In addition, we also attempted to balance the training dataset according to the local sequence context near the methylation (i.e. [− 1, + 1] range relative to the methylated base) by downsampling common context in priority instead of random sampling. However, while we observe an overall improvement of motif occurrences classification compared to the default balancing (average LOOCV accuracy +3.8%), not all motifs benefit from it. While the context balancing method does not currently improve the motif typing and fine mapping, we note that it could be helpful when the methylation motif signature database becomes larger. Classifier hyperparameters (Supplementary Table 4) were tuned on the balanced training dataset containing all motifs using repeated 10-fold cross-validation (n=3) with balanced accuracy (mean and standard deviation) as the main metric. Robustness of chosen hyperparameters was confirmed by comparing performances from three classifiers (k-nearest neighbors, random forest, and neural network) when using parameters either tuned on a dataset containing all motifs (as described above) or a dataset only containing H. pylori motifs only. Both sets of hyperparameters gave similar results when tested on a dataset without H. pylori motifs (Extended Data Fig. 4d).
Classifier performance evaluation was performed using leave-one-out cross-validation strategy (LOOCV) by holding out current difference vectors from one motif and training on remaining vectors (from all motifs except one). The resulting model is then used to predict the label of held out vectors from the tested motif. The LOOCV strategy simulates models’ behavior when faced with an unseen motif signature. For testing, we only used the set of vectors corresponding to the approximated methylation position found as described previously. Predicted methylated base type and position for a motif are defined using consensus across all tested motif occurrences. Note that the classifier prognosticates the offset between the approximated methylation position chosen as input and the predicted methylation position, which is then converted into a position within tested motifs. The confidence of the final prediction is defined as percentage of motif occurrences assigned to the type-position combination with the highest number of assignment. Alternatively, the p-value associated with the classifier prediction can be incorporated into the calculation of prediction confidence. One can leverage the p-value distribution generated from all occurrences of the same motifs to estimate the confidence of the prediction or define a confidence threshold.
Nanopore sequencing based de novo assembly
Genome assembly for E. coli was performed using Canu45 (version 1.8; -nanopore-raw genomeSize=4.7m overlapper=mhap utgReAlign=true) with the native nanopore reads (200x dataset). Next, we generated the genomic consensus with Racon46 (version 1.3.3; default parameters) to correct raw contigs, and correct contig ends using nucmer47 (version 4.0.0beta; --maxmatch –nosimplify and show-coords -lrcTH) to identify and trim remaining overlaps. Then, we polished the assembly consensus using Nanopolish6 (version 0.11.0; variants --min-candidate-frequency 0.1 for five consecutive times) with the native nanopore reads. Finally, we performed another polishing step with Nanopolish using nanopore WGA reads (methylation free) to correct remaining assembly error caused by DNA methylation signal in the native reads (same parameters for five consecutive times).
Metagenome methylation binning
While methylation motif detection could be performed as for individual bacteria, metagenome assemblies often result in many contigs from multiple organisms with various lengths making individual contig analysis lacking power. Instead, we propose to first bin contigs with similar methylation profiles then perform the motif detection. Nanopore sequencing native and WGA datasets are processed in the same way as for individual bacteria (except that supplementary alignment were conserved) generating current differences alongside metagenome contigs using the nanopore sequencing-only de novo metagenome assembly.
De novo metagenome assemblies for MGM1 and MGM2 were performed using Flye48 (version 2.4.2; --meta –nano-raw –genome-size 100M) with the native nanopore reads. Next, the metagenome consensus was computed using Racon46 for four consecutive rounds (default parameters). Then, the resulting metagenome assemblies were polished using Nanopolish6 with first the native, then with the WGA nanopore reads (variants --min-candidate-frequency 0.1 for five rounds with each set of reads).
For a candidate motif, an associated methylation feature vector is computed by averaging current differences from aggregated occurrences on a metagenomic contig (Extended Data Fig. 8). Unlike well-characterized methylation motifs, the methylated position in a candidate motif is unknown. Therefore, we consider every position in motifs as potentially methylated by including all potentially affected current differences in the methylation feature vector calculation. For a motif of length k, we compute a methylation feature vector of length k + (2 + 3), which corresponds to the length of current differences that are possibly affected by a methylated base in a k-mer motif (the core current differences is defined as [− 2 bp, + 3 bp] range flanking a methylated base, Extended Data Fig. 1). This procedure results in a methylation feature vector of average current differences of length k + 5, which effectively capture methylation signal flanking a motif of interest for a contig, and discriminate between different modification types of the same motif. This step represents a major difference from SMRT sequencing based methylation binning method where a single methylation score is generated for a motif on a contig30.
The next step is to create a methylation profile matrix comprising methylation feature vectors for each motif of interest in each metagenomic contig, which will be used for methylation binning (Extended Data Fig. 8). A set of 210,176 candidate motifs is generated according to common structures (4-, 5-, and 6-mers, as well as bipartite motifs with 3 to 4 bp specificity part separated by 5 to 6 bp gaps). In order to select motifs of interest, an initial round of motif evaluation is performed on a subset of longer contigs (100 kbp using nanopore sequencing de novo assembly) with sufficient coverage (10x; Supplementary Fig. 2) with the rationale that results will have a higher statistical power. Uninformative methylation features are filtered out by discarding the ones with small absolute current difference values across the initial contig set (< 1.5 pA; chosen based on our mock metagenome analysis) as well as the ones computed from fewer than 20 motif occurrences. Next, we additionally filtered out uninformative methylation features from bipartite motifs by removing methylation feature vectors with fewer than two significant features across the initial contig set (significant features if absolute value >= 1.5 pA) to account for the longer vector and generally lower motif frequency. Finally, methylation features from bipartite motifs that overlap with any remaining 4 to 6-mer motifs are also discarded. The resulting list of informative methylation features is then evaluated in each contig of the metagenome assembly to construct a methylation profile matrix (Extended Data Fig. 8). This two-step approach effectively reduces the initial research space on the set of large contigs speeding up the analysis, and reduces noise by only considering methylation features selected from contigs with higher statistical power. The resulting methylation profile matrix (significant methylation features computed across all contigs) is then processed using t-SNE dimensionality reduction method to visualize contig clusters (Extended Data Fig. 8). Missing methylation features and ones computed from fewer than 5 motifs occurrences are set to small random pseudovalues in the [− 0.2, + 0.2] range (reducing correlation from missing methylation features; random number generation seeds are set at 2, 3, and 4 for MGM1, MGM2, and the SMRT assemblies respectively). Small contigs are not considered for methylation binning (<25 kbp for the nanopore sequencing de novo assembly analysis), and remaining ones are weighted according to their length. Weighting factors are defined as quotient of contig length divided by 50,000 and capped at a percentage of the number of remaining contigs to avoid extreme imbalance (only contigs with coverage >= 10x for both native and WGA are weighted). We set the capping value at 5% for metagenome with high diversity (large number of metagenome contigs, MGM1) and 10% for simpler metagenome (<500 contigs, MGM2). Finally, bins are defined after t-SNE dimension reduction using DBSCAN (package dbscan version 1.1-4; size of the epsilon neighborhood, eps, set to 5 and number of minimum points in the eps region, minPts, set to 3), an automated clustering method, with additional manual annotation of visible bins that can be missed by DBSCAN.
The analysis using the SMRT metagenome assembly (GCA_002754755.1) is performed as described previously using thresholds of 500 kbp and 10x of coverage for initial methylation feature selection (contigs from Bin 3, Bin 4, and Bin 9 are not covered sufficiently due to the use of a different DNA extraction kit than the SMRT study). Contigs smaller than 10 kbp are not considered.
Motif detection from bins is performed the same way as for individual bacteria. With de novo detected motifs, methylation feature vectors used for binning are not filtered, keeping the full-length methylation feature vectors. Missing methylation features from individual contigs are handled as described previously and contigs are also weighted. We performed three consecutive rounds of binning and motif detection for MGM1 as new bins and therefore new methylation motifs are identified in the first two rounds (Extended Data Fig. 9), while one round was sufficient for MGM2 (Extended Data Fig. 10). Confirmation of de novo discovered motifs in MGM1 sample (potential 6mA and 4mC motifs) from nanopore sequencing analysis were realized with per bin motif detection from SMRT sequencing data using the SMRT portal pipeline (RS_Modification_and_Motif_Analysis.1).
Binning focused on associating mobile genetic elements (MGEs) to host genome (Supplementary Fig. 3b) was performed using metagenome reference from the SMRT study where binned contigs were replaced by per-bin reassemblies6. MGEs contigs from the nanopore-only de novo metagenome assemblies were identified according to the alignment of MGEs sequences from the SMRT study using minimap2 (version 2.15; -ax asm20)49.
Detection of metagenome contigs misassemblies
The rationale is to examine the consistency of methylation signal for a motif across different occurrence of the motif along a metagenomic contig. For every single motif occurrence, we calculate a score by taking the average of absolute current differences from six consecutives positions with the most perturbation. Then, these individual scores are averaged using a sliding window across the contig to examine the continuity. Motif occurrences from both strands are used in this analysis. However, if a motif occurrence overlaps with another motif site being examined (<15 bp) then both are discarded.
Data availability
All sequencing data generated for this study are available at Sequence Read Archive (SRA) under the BioProjects PRJNA559199 for individual bacteria and PRJNA559386 for the mouse gut microbiomes samples. NCBI reference sequences used for the individual bacteria analysis are available under the accession codes: CP041693, CP041696, NC_008261.1, CP014225.1, CP023448.1, NC_007796.1, NC_002946.2, CP041695, and CP003732.1 (Supplementary Table 1). Information related to methylation motifs are available from REBASE database (http://rebase.neb.com)16. Data from the SMRT sequencing metagenomic study can be found under the BioProject PRJNA404082.
Code availability
Software nanodisco and a detailed tutorial with supporting data are available at http://github.com/fanglab/nanodisco.
Extended Data
Extended Data Fig. 1. General statistics of motif signatures.
(a) Distribution of current differences are shown for all confident motifs altogether (n=46 motifs) as well as average absolute differences and associated standard deviations near methylated bases ([− 10 bp, + 11 bp]). The lower and upper hinges correspond to the 25th and 75th percentiles while the lower and upper whisker extends to the minima and maxima respectively (capped at 1.5 time the inter-quartile range). (b) Same as a with distinction between DNA methylation types (n=28 6mA motifs, n=7 4mC motifs, n=11 5mC motifs). (c) Same as a but for individual methylation motifs.
Extended Data Fig. 2. Systematic examination of three main DNA methylation types with nanopore sequencing.
(a) t-SNE projection of isolated methylation motif occurrences separated per motif. The same dataset as Fig. 2b was used with occurrences colored per motif. Other motifs are colored in grey. (b) Same as a but grouped by methylation type.
Extended Data Fig. 3. Nanopore sequencing signal processing variable.
(a) Comparison of current differences across methylation occurrences between datasets base called with Albacore 1.1.0, Albacore 2.3.4, and Guppy 3.2.4 illustrated by projection with t-SNE from for 46 well-characterized motifs (Supplementary Table 2). Each dot represents one isolated motif occurrence colored by base caller versions. 100, 000 motif occurrences were randomly selected from each dataset to reduce the scatter plot density and ease the visualization. For each motif occurrence, current differences from 22 positions near methylated bases ([− 10 bp, + 11 bp]) were used. (b) Performance for de novo methylated site detection between datasets base called with Albacore 1.1.0, Albacore 2.3.4, and Guppy 3.2.4. We evaluated individual motif occurrences detection using Precision-Recall curves for H. pylori at 75x coverage. Precision-Recall curves and area under the curves (AUC) were computed as described in the Method section. Only confident H. pylori motifs were considered for the evaluation. (c) Comparison of current differences across methylation occurrences (same as a) between datasets produced with or without outlier removal step (Methods). (d) Performance for de novo methylated site detection (similar than b) with datasets produced with or without outlier removal step. (e) Variation of current differences across methylation occurrences without outlier removal step as illustrated by motif signatures from three motifs, AG4mCT (n=6550 occurrences), GGW5mCC (n=1875 occurrences), and GCYYG6mAT (n=954 occurrences). For each motif, current differences near methylated bases ([− 6 bp, + 7 bp]) from all isolated occurrences are plotted with conservation of relative distances to methylated bases. Distributions of current differences for each relative distance are displayed as a violin plot. Current differences axis is limited to −8 to 8 pA range. (f) Performance for de novo methylated site detection across current difference datasets generated with different read alignment type filtering: remove alternative alignments (filtered out XA bam flags; named No Alt.), remove supplementary alignments (filtered out 2048 bam flags; named No Supp.), remove chimeric alignments (filtered out SA bam flags; named No Chim.), only conserve unique mapping (filtered out XA and SA bam flags; named Unique), and keep all alignments (named None). (g) Performance for de novo methylated site detection across datasets normalized with linear regression (lm function), robust regression (rlm function) or no additional normalization (annotated as none). (h) Performance for de novo methylated site detection across datasets generated using two-sided Mann-Whitney U test or Student’s t-test. (i) Performance for de novo methylated site detection across datasets generated using different p-value smoothing window size: no smoothing (named None), 3 nt, 5 nt, and 7 nt. (j) Performance for de novo methylated site detection across datasets generated using different function for combining consecutives p-values: Fisher’s method (named sumlog), logit method (named logitp), sum p method (named sump), and sum z method (named sumz). (k) Performance for de novo methylated site detection across peaks datasets generated using different peak detection window size: 5 nt, 7 nt, and 9 nt. Plots f, g, h, i, j, and k show Precision-Recall curves and area under the curves (AUC) for various signal processing steps and were computed as described in the Method section. (l) Comparison of current differences across methylation occurrences (same as a) with E. coli datasets (200x) produced using either the reference genome or the de novo assembly (Methods). (m) Performance for de novo methylated site detection in E. coli datasets (200x) using either the reference genome or the de novo assembly. (n) Performance of methylation motif typing and fine mapping on E. coli datasets (200x) produced using either the reference genome or the de novo assembly (motif occurrences: n=458 for AACNNNNNNGTGC, n=18451 for CCWGG, n=28110 for GATC, n=463 for GCACNNNNNNGTT). Only results for k-nearest neighbors, neural network, and random forest are displayed.
Extended Data Fig. 4. Additional information for classification of methylation motif occurrences.
(a) Approximation of DNA methylation position in three motifs, AG4mCT (n=6549 occurrences), GGW5mCC (n=1875 occurrences), and GCYYG6mAT (n=954 occurrences). Signal strength is computed using a sliding window alongside motif signature to choose the best vector positioning to use for classification. (b) Flowchart description of procedure for classifier training and novel motifs dataset annotation. Training the classifier consists of gathering a set of bacteria with characterized methylomes. Confident motifs are selected to assure the robustness of the final classifier, then all motif occurrences are localized in the genome (from corresponding reference genome or de novo assembled and polished genome). Current differences are then computed along the genome. Next, the training dataset is built from the offsetted vector of current differences labelled with the known methylation type and the offset combination. Finally, the classifier is trained using the chosen model(s). Analyzing a new bacterial sample consists of de novo detecting the methylated motif from processed current differences (see Methods). Then methylated motif occurrences are localized and the motif signatures are computed (i.e. distribution of current differences at relative distance from the methylated bases). Next, those signatures are leveraged to approximate the methylated position for each de novo detected motif (see Methods), which is used to define the classifier inputs (i.e. vector of current differences centered on the approximate methylated position). Finally, the trained classifier is used to predict the methylation type and fine map the DNA methylation for each motif. (c) Boxplot of overall prediction accuracy in LOOCV evaluation (n=46 motifs) for each classifier. Classifiers are ordered by average accuracy. The lower and upper hinges correspond to the 25th and 75th percentiles while the lower and upper whisker extends to the minima and maxima respectively (capped at 1.5 time the inter-quartile range). (d) Effect of hyperparameters on classification accuracy. Boxplot of overall prediction accuracy in LOOCV evaluation with classifiers trained on all motifs except the ones from H. pylori (n=27 motifs). Hyperparameters were either tuned on H. pylori motifs only (“Alt. HP”) or on all motifs (“Main HP”). The lower and upper hinges correspond to the 25th and 75th percentiles while the lower and upper whisker extends to the minima and maxima respectively (capped at 1.5 time the inter-quartile range). (e) Relationship between LOOCV accuracy and current difference signal similarities. Current difference signal near methylated bases is visualized by projection with t-SNE for the 46 well-characterized motifs similar to Fig. 2b. Each dot represents one isolated motif occurrence colored by accuracy from LOOCV analysis.
Extended Data Fig. 5. Classification and fine mapping of three types of DNA methylation (part 1).
Similar to Fig. 4d with full set of prediction results for a subset of methylation motifs for k-nearest neighbors, random forest, and neural network. Filling colors correspond to percentage of occurrences classified to a specific class ranging from blue (0%) to red (100%). Greyed out prediction correspond to out of motif position. Blank columns correspond to within-motif positions without prediction. Prediction percentages of expected classes are displayed in italic and selected predictions based on consensus are displayed in bold.
Extended Data Fig. 6. Classification and fine mapping of three types of DNA methylation (part 2).
See Extended Data Fig. 5.
Extended Data Fig. 7. Evaluation of motif enrichment with Precision-Recall curves.
(a) Effect of coverage on de novo methylated site detection. We evaluated individual motif occurrences detection using Precision-Recall curves (PR curves) for H. pylori. Studied datasets with coverage ranging from 5x to 200x were generated by random subsampling of native and WGA datasets. Precision-Recall curves were generated as described in the Method section. We considered only confident H. pylori motifs for evaluation. (b) Same as a but using ROC curves for representation. Motif occurrences without data due to low coverage (<5x) were not considered. (c) Performance of methylation motif typing and fine mapping (n=46 motifs) on datasets with genomic coverage subsampled at 10x, 15x, 20x, and 30x. Only results for k-nearest neighbors, neural network, and random forest are displayed. (d) Precision-Recall curves summarizing the detection performance at 75x coverage of individual methylation sites for each motif in H. pylori with adjusted frequency (Methods). (e) Same as d but using ROC curves for representation. (f) Effect of motif frequency on de novo methylated site detection. For each methylation motif, in silico datasets with a wide range of motif frequencies were created using a random subsampling strategy (either the motif occurrences or the genomic regions without motifs, see Methods). The natural motif frequencies (i.e. the original ratio of motif occurrences over all queried regions) are annotated by a point on each motif curve.
Extended Data Fig. 8. Schematic representation of methylation feature vectors computation and methylation binning of contigs.
The computation of methylation features and the building of the methylation profile matrix is described in the method.
Extended Data Fig. 9. Detailed methylation analysis of MGM1 sample.
(a) Methylation binning using automated methylation features selection (without precise methylation motif discovery; Methods). Methylation features are projected on two dimensions using t-SNE. Contigs are colored per bin defined using DBSCAN, with point sizes matching contig length according to the legend. Two bins with the same methylation motifs were manually merged into Bin 4. (b) Methylation binning using de novo discovered motifs on each bin found in a (Methods). Methylation features computed from de novo discovered motifs are projected on two dimensions using t-SNE. Contigs are colored per bin defined using DBSCAN except Bin 11, which was manually defined. (c) Methylation binning using de novo discovered motifs on each bin found in b. Contigs are colored per bin defined using DBSCAN except for Bin 13, which was manually defined. (d) Methylation binning of MGM1 metagenome contigs using de novo discovered motifs (after three rounds of motif discovery (same as Fig. 5a).
Extended Data Fig. 10. Detailed methylation analysis of MGM2 sample.
(a) Methylation binning using automated methylation features selection (without precise methylation motif discovery; Methods). Methylation features are projected on two dimensions using t-SNE. Contigs are colored per defined bin with point sizes matching contig length according to the legend. Bin 1, 3, 4, and 5 were defined using DBSCAN. The other bins are composed of one or two contigs and were manually defined after de novo methylation motif discovery. (b) Methylation binning using de novo discovered motifs on each bin found in a (Methods). Methylation features computed from de novo discovered motifs are projected on two dimensions using t-SNE. Contigs are colored per bin as described in a.
Supplementary Material
Acknowledgements
We thank Alexey Fomenkov and Sir Richard J. Roberts from New England Biolabs for their help with the bacterial strain selection and for providing us with DNA samples (B. amyloliquefaciens, B. fusiformis, and N. otitidiscaviarum). We also thank Robert Gunsalus from the University of California, Los Angeles (M. hungatei), Susan Logan from the National Research Council Canada (C. perfringens), Lydgia Jackson from the University of Oklahoma Health Sciences Center (N. gonorrhoeae), Bernhard Schink, Nicolai Müller, and Anja Keller from the University of Konstanz, Germany (T. phaeum) for providing us with DNA samples. We thank Yimeng Kong and Mi Ni for providing helpful feedback for early versions of this manuscript. This work was supported by a seed fund from Icahn Institute for Genomics and Multiscale Biology (G.F.), and by R01 GM128955 (G.F.), R35 GM139655 (G.F.) and R56 HG011095 (G.F.) from the National Institutes of Health. G.F. is a Hirschl Research Scholar by Irma T. Hirschl/Monique Weill-Caulier Trust, and a Nash Family Research Scholar. This work was also supported in part through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Footnotes
Ethics declaration
A.T. and G.F. are inventors of two US Provisional patent applications (62/860,952 and 62/851,205) that describe the methods in this manuscript.
Reference
- 1.Beaulaurier J, Schadt EE & Fang G Deciphering bacterial epigenomes using modern sequencing technologies. Nat Rev Genet 20, 157–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flusberg BA et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7, 461–465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blow MJ et al. The Epigenomic Landscape of Prokaryotes. PLoS Genet 12, e1005854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laszlo AH et al. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc Natl Acad Sci U S A 110, 18904–18909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schreiber J et al. Error rates for nanopore discrimination among cytosine, methylcytosine, and hydroxymethylcytosine along individual DNA strands. Proc Natl Acad Sci U S A 110, 18910–18915 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simpson JT et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods 14, 407–410 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Rand AC et al. Mapping DNA methylation with high-throughput nanopore sequencing. Nat Methods 14, 411–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McIntyre ABR et al. Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat Commun 10, 579 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni P et al. DeepSignal: detecting DNA methylation state from Nanopore sequencing reads using deep-learning. Bioinformatics 35, 4586–4595 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Liu Q et al. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat Commun 10, 2449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Q, Georgieva DC, Egli D & Wang K NanoMod: a computational tool to detect DNA modifications using Nanopore long-read sequencing data. BMC Genomics 20, 78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoiber M et al. De novo Identification of DNA Modifications Enabled by Genome-Guided Nanopore Signal Processing. bioRxiv (2017). [Google Scholar]
- 13.Amarasinghe SL et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol 21, 30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wion D & Casadesus J N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol 4, 183–192 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casadesus J & Low D Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev 70, 830–856 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts RJ, Vincze T, Posfai J & Macelis D REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 43, D298–299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Der Maaten L Accelerating t-SNE using tree-based algorithms. J. Mach. Learn. Res. 15, 3221–3245 (2014). [Google Scholar]
- 18.Bailey TL, Johnson J, Grant CE & Noble WS The MEME Suite. Nucleic Acids Res 43, W39–49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saeed I, Tang SL & Halgamuge SK Unsupervised discovery of microbial population structure within metagenomes using nucleotide base composition. Nucleic Acids Res 40, e34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iverson V et al. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science 335, 587–590 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Laczny CC, Pinel N, Vlassis N & Wilmes P Alignment-free visualization of metagenomic data by nonlinear dimension reduction. Sci Rep 4, 4516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laczny CC et al. VizBin - an application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome 3, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albertsen M et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31, 533–538 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Sharon I et al. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res 23, 111–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alneberg J et al. Binning metagenomic contigs by coverage and composition. Nat Methods 11, 1144–1146 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Nielsen HB et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 32, 822–828 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Marbouty M et al. Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms. Elife 3, e03318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burton JN, Liachko I, Dunham MJ & Shendure J Species-level deconvolution of metagenome assemblies with Hi-C-based contact probability maps. G3 (Bethesda) 4, 1339–1346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marbouty M, Baudry L, Cournac A & Koszul R Scaffolding bacterial genomes and probing host-virus interactions in gut microbiome by proximity ligation (chromosome capture) assay. Sci Adv 3, e1602105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beaulaurier J et al. Metagenomic binning and association of plasmids with bacterial host genomes using DNA methylation. Nat Biotechnol 36, 61–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang G et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol 30, 1232–1239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray IA et al. The methylomes of six bacteria. Nucleic Acids Res 40, 11450–11462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schadt EE et al. Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res 23, 129–141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaulaurier J et al. Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes. Nat Commun 6, 7438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song CX, Yi C & He C Mapping recently identified nucleotide variants in the genome and transcriptome. Nat Biotechnol 30, 1107–1116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshihara M, Jiang L, Akatsuka S, Suyama M & Toyokuni S Genome-wide profiling of 8-oxoguanine reveals its association with spatial positioning in nucleus. DNA Res 21, 603–612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S & Mason CE The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet 15, 127–150 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Roundtree IA, Evans ME, Pan T & He C Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garalde DR et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods 15, 201–206 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Workman RE et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat Methods 16, 1297–1305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang C, Chu J, Warren RL & Birol I NanoSim: nanopore sequence read simulator based on statistical characterization. Gigascience 6, 1–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-bio.GN] (2013). [Google Scholar]
- 43.Morgan M, Pagès H, Obenchain V & Hayden N Rsamtools: Binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. (2016). [Google Scholar]
- 44.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koren S et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaser R, Sovic I, Nagarajan N & Sikic M Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27, 737–746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurtz S et al. Versatile and open software for comparing large genomes. Genome Biol 5, R12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolmogorov M, Yuan J, Lin Y & Pevzner PA Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Li H Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data generated for this study are available at Sequence Read Archive (SRA) under the BioProjects PRJNA559199 for individual bacteria and PRJNA559386 for the mouse gut microbiomes samples. NCBI reference sequences used for the individual bacteria analysis are available under the accession codes: CP041693, CP041696, NC_008261.1, CP014225.1, CP023448.1, NC_007796.1, NC_002946.2, CP041695, and CP003732.1 (Supplementary Table 1). Information related to methylation motifs are available from REBASE database (http://rebase.neb.com)16. Data from the SMRT sequencing metagenomic study can be found under the BioProject PRJNA404082.