Abstract
Background
Discriminating the subtypes of non-small cell lung cancer (NSCLC) based on computed tomography (CT) images is a challenging task for radiologists. Although several machine learning methods such as radiomics, and deep learning methods such as convolutional neural networks (CNNs) have been proposed to explore the problem, large sample sizes are required for effective training, and this may not be easily achieved in single-center datasets.
Methods
In this study, an automated subtype recognition model with capsule net (CapsNet) was developed for the subtype discrimination of NSCLC. CapsNet utilizes an activity vector to record the relative spatial relationship of image elements that can subsequently better delineate the global image characteristics. CT images of 72 adenocarcinoma (AC) and 54 squamous cell carcinoma (SCC) patients were retrospectively collected from a single clinical center. The cancer region on the CT image was manually segmented for every subject by an experienced radiologist, and CapsNet, CNN, and four radiomics models were then used to construct the recognition model.
Results
The study demonstrated that CapsNet achieved the best discriminative performance (accuracy 81.3%, specificity 80.7%, sensitivity 82.2%) although its area under the curve was just marginally better than that of the optimal random forest (RF) based radiomics model. Not surprisingly, the performance of the CNN was only comparable to the other radiomics models.
Conclusions
This study demonstrated that CapsNet is a viable potential framework for discriminating the subtypes of NSCLC, and its use could be extended to the recognition of other diseases especially in limited single-center datasets.
Keywords: Capsule net (CapsNet), convolutional neural network (CNN), radiomics, non-small cell lung cancer (NSCLC), computed tomography imaging (CT imaging)
Introduction
Lung cancer is the leading cause of cancer-related death in the world (1). Non-small cell lung cancer (NSCLC) accounts for about 89% of all types of lung cancers, and adenocarcinoma (AC) and squamous cell carcinoma (SCC) are the two major histological subtypes of NSCLC (2). Current advancements in the precision therapy for NSCLC are dependent on the accurate differential diagnosis of the histopathological subtypes of NSCLC (3,4). Histopathological examination of tumor tissue via biopsy or surgery is the golden criterion for diagnosing AC and SCC in routine clinical practice. However, both procedures are invasive and costly. Specifically, biopsies and cytological examinations of small pieces of tumor may be not representative of the entire tumor (5,6). In addition, to evaluate the efficacy of treatments through repeated biopsies are inconvenient and not feasible. Therefore, a non-invasive, low-cost, and convenient method for the accurate differential diagnosis of NSCLC subtypes is highly desirable for clinical application.
Computed tomography (CT) imaging serves as the initial screening tool for lung cancer. Although AC and SCC lesions are obviously different under microscopic examination of pathology sections, these lesions display similar visual characteristics on CT images, which makes the discrimination between AC and SCC a highly challenging task for radiologists. Radiomics is an alternative technique that employs high-throughput quantitative imaging features for clinical diagnosis and prognosis (7-10). Previous studies have explored the potential of radiomics and machine learning methods in the histological subtype classification of NSCLC using CT images (11-16). However, these prediction models rely upon predefined radiomics feature sets for training the model. As different studies have adopted different radiomics feature sets (17-19), it is difficult for radiologists to choose the appropriate feature sets, making the application of these prediction models in clinical practice not straightforward (20). Thus, a more intelligent and expert-like automatic feature detection method is essential for improving these types of diagnostic tasks.
Convolutional neural networks (CNNs) are popular deep learning methods that have been widely used in medical images processing. They are capable of discovering the informative feature representations in a self-taught manner and thus can function independently from prior knowledge and human input (21,22). CNNs have been used to discriminate the subtypes of NSCLC (23). However, large amounts of data are usually needed for effective training, and this requirement is not easily satisfied for most medical imaging datasets. Moreover, CNNs are sensitive to any changes in the viewpoint of the image element, and cannot interpret images in terms of objects and their parts. Therefore, the performance of CNNs may be weakened in situations where the sample size is limited.
Capsule net (CapsNet) is an improved type of neural network with a structural basis called the “capsule”. The capsule is a group of neurons whose activity vector represents the instantiation parameters of an object in the image (24). The two key features of CapsNet are layer-based squashing and dynamic routing. Unlike CNNs with individual neurons squashed through nonlinearities, the CapsNet output is squashed as an entire vector. Dynamic routing iteratively tunes the weight coefficients and routes the outputs of low-level capsules to the appropriate high-level capsule, thus determining the pose of the objects more accurately. Taken together, CapsNet is both rotation invariant and spatially aware, and can encode the relative relationships (e.g., locations, scales, orientations) between local parts and the whole object, resulting in a satisfactory performance within small datasets.
Thus far, few studies have compared the three above-mentioned models in discriminating the subtypes of NSCLC within the same dataset. It is unclear whether CapsNet can achieve better performances relative to the CNN and radiomics models in a limited dataset. Therefore, in this study, we retrospectively collected NSCLC CT imaging data from a single clinical center, and proposed and evaluated three classification models: CapsNet, CNN, and radiomics (with four different machine learning methods). We hypothesized that CapsNet would achieve the best performance among the three models.
Methods
Subjects
We retrospectively collected the CT data of suspected lung cancer patients from the picture archiving and communication system (PACS) of the Third Medical Center of PLA General Hospital between January 2014 and June 2018. The study was approved by the institutional ethics board of the Third Medical Center of PLA General Hospital. Individual patient consent for this retrospective analysis was waived. The inclusion criteria were as follows: (I) the non-enhanced CT images were obtained by the same scanner with identical scanning parameters; (II) all lesions showed a solid-appearing mass; (III) the tumor short-axis diameters were at least 10 mm to guarantee sufficient volume of interests (VOIs); (IV) histopathological evidence of AC or SCC was obtained by tumor resection or biopsy after the CT scans; and (V) there were no treatments prior to the CT scan. Patients who had accepted any treatments, or patients with a diameter of lesion less than 10 mm were not enrolled in the study. Patients showing ground-glass opacities or part-solid nodules were excluded. Patients presenting with other types of lung cancer identified by histopathology including small cell lung cancer and adeno-squamous carcinoma were also excluded. Finally, 126 patients were enrolled in this study, including 72 patients with AC and 54 patients with SCC.
The demographic data and semi-quantitative clinical features of all patients in the AC and SCC cohorts were summarized and listed in Table 1. Tumors detected in the segmental or more proximal bronchi were classified as centrally located, while tumors found in the subsegmental bronchi or more distal airway were classified as peripherally located. The grade of tissue adhesion was based on the surface area of the tumor adhering to neighboring tissues. The differences in the patient’s age, tumor density, and tissue adhesion grade between the AC and SCC cohorts were assessed using an independent two sample t-test, and the difference in gender and tumor location were assessed by the χ2 test. The reported statistical significance levels were all two-tailed with P<0.05.
Table 1. The demographic and clinical characteristics of all the patients.
| Category | Details | AC (n=72) | SCC (n=54) | P value |
|---|---|---|---|---|
| Demographic information | Gender (M/F) | 44/28 | 48/6 | 0.0005 |
| Age | 61.5 [35–85] | 62 [45–76] | 0.4240 | |
| Radiographic features | Location (central/peripheral) | 15/57 | 24/30 | 0.0045 |
| Volume (cm3) | 44.6 [0.4–404.2] | 74.6 [0.8–325.5] | 0.0153 | |
| Tissue adhesion (no/yes) | 16/56 | 11/43 | 0.8025 | |
| Density (Hu) | 33 [12–57] | 33.5 [16–62] | 0.3478 |
Age, volume, and density are displayed as mean [range]. AC, adenocarcinoma; SCC, squamous cell carcinoma; M/F, male/female; Hu, Hounsfield unit.
Imaging protocol and data acquisition
All patients were scanned with a 64-slice CT system (Discovery CT750 HD, GE Healthcare, USA). The non-enhanced CT images were acquired encompassing the entire thorax with the identical scanning protocol (120 kV; 40–200 mA; scan type: helical; rotation time: 0.6 s; detector coverage: 40 mm; interval: 5 mm; reconstruction width: 1.25 mm; matrix: 512×512).
VOI segmentation
The segmentation of lung tumor is essential for subsequent image analysis. In this study, a semi-automatic active contour segmentation method was used to segment the VOIs with ITK-SNAP software (http://www.itksnap.org) by a radiologist with more than 10 years of experience in thoracic diagnosis. To confirm the accuracy of the VOI segmentation, all segmentation results were reviewed by another radiologist with more than 15 years of experience in thoracic diagnosis. In the case of disagreements in segmentation, a consensus would be reached by discussions and modifications between two experts. All Images were resampled into isotropic voxels of unit dimension to ensure comparability, with one voxel corresponding to 1 mm3. This was achieved using linear and nearest neighbor interpolations for the image and annotations, respectively.
The radiomics framework
The radiomics features were extracted from the VOIs of the CT images by using Pyradiomics 1.2.0, and included 14 shape-based features, 18 first-order statistic features, 24 gray level co-occurrence matrix features, 14 gray level dependence matrix features, 16 gray level run-length matrix features, 16 gray level size zone matrix features, and 5 neighboring gray tone difference matrix features. The details of these radiomics features are described in http://www.radiomics.io/pyradiomics.html. Finally, a total of 107 radiomics features were obtained for every subject. In our study, the demographic information and semiquantitative features (six features in total, see Table 1) obtained from the CT reports were incorporated with the radiomics features.
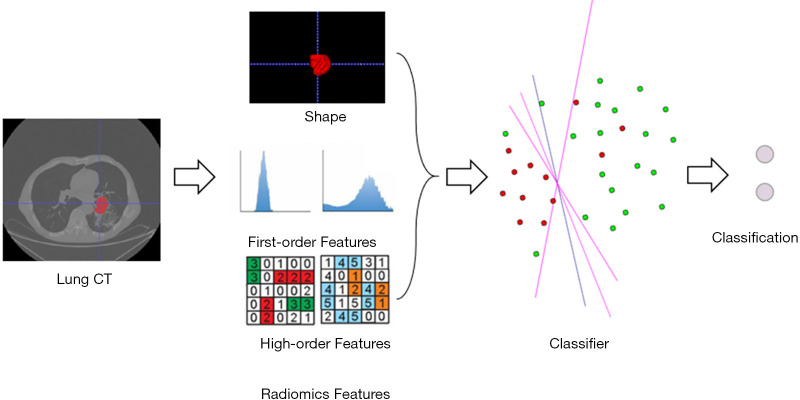
Fisher score (14,25) was then used to select the effective feature sets; because there was no optimal threshold for the Fisher score, the numbers of feature sets were incrementally increased by 10% of the total features. The best performing feature sets were used in the radiomics model. Four machine learning methods were then used on the extracted features (after normalization) to discriminate the subtypes of NSCLC, including random forest (RF), logistic regression (LR), logistic regression with L1 regularization (LR-L1), and logistic regression with principal component analysis (LR-PCA). All of these methods were conducted using the widely used machine learning tool, Scikit-Learn (www.scikit-learn.org). For RF and LR, the default parameter settings were used in respective functions within Scikit-Learn. For LR-PCA and LR-L1, an inner grid search cross-validation was conducted to determine the optimal dimensionality of PCA and the sparse coefficient C. The whole flowchart of the radiomics framework is shown in Figure 1.
Figure 1.
The flowchart of the radiomics framework for the subtype discrimination of NSCLC. NSCLC, non-small cell lung cancer; CT, computed tomography.
The CapsNet framework
In contrast to alternative deep neural networks, each node of the capsule layers in the CapsNet is a capsule containing a series of neurons. The activity of each capsule is represented by an activation vector (activation values of a certain number of neurons inside). The norm of this vector represents the probability that an object of interest possesses a certain property. The classification layer consists of classification capsules, and the classification output norm represents the probability that an instance belongs to a certain class.
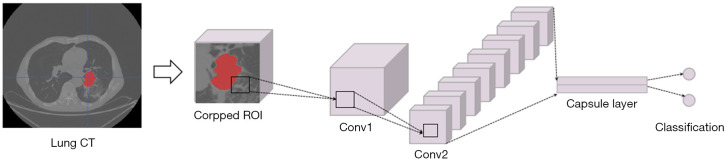
In our proposed CapsNet for histological subtype identification, the cropped image patches were firstly filtered by two convolution layers. Then, a capsule layer which contains two identification capsules (representing AC and SCC) was introduced to obtain identification results.
As illustrated in Figure 2, the CapsNet used in this study contains two convolution layers which acted to obtain feature representation F, and a capsule layer for identifying histologic subtypes. For convolution layer 1, the number of input channels is 16, and the number of output channels is 256. For convolution layer 2, the number of input channels is 256, and the number of output channels is 64. The kernel size of these two convolution layers is set as 9 and the stride is set as 2.
Figure 2.
The flowchart of the CapsNet framework for the subtype discrimination of NSCLC. CapsNet, capsule net; NSCLC, non-small cell lung cancer; CT, computed tomography; ROI, region of interest; Conv, convolutional layer.
For the capsule layer, F is first reshaped to form a series of 8-dimension capsules [f1,…,fi,…,fM]. Then, [f1,…,fi,…,fM] are connected to identification capsules that model the probability that an instance is AC or SCC. The length of each identification capsule is 8.
The connections between [f1,…,fi,…,fM] and the identification capsules are optimized via the dynamic routing by agreement algorithm. Denoting the output of capsule i in representation capsules with fi, its parent capsule j among the identification capsules is computed by:
| [1] |
where Wij is a trainable weight matrix between fi and its parent capsule j (AC or SCC). A coupling cij between them is defined as follows:
| [2] |
Where bij represents the probability that fi is coupled with capsule j. The probability is initialized with a value of 0, and k =2 is the number of capsules in the identification capsules. The parent capsule j has an input, sj, as computed by the following:
| [3] |
Then, a squashing function, as formulated by Eq. [4], is applied to restrict the norm of output vector vj which is obtained from the capsule j to the range of [0, 1]. Therefore, the norm of this vector can act as a probability for classification.
| [4] |
For the identification capsules, the norm of vj represents the probability that a subject belongs to AC or SCC.
An agreement factor aij between fi and its parent capsule j is defined as follows:
| [5] |
The agreement factor aij is then added to bij in the next iteration step of the dynamic routing to enhance or weaken the coupling between these two capsules. The loss function LD of CapsNet is defined as follows:
| [6] |
where Tc=1 iff an instance belongs to class c (AC or SCC), vc is the output of the capsule, representing subtype c, λ is a weight set to 0.5, m+=0.9, and m−=0.1.
The CNN framework
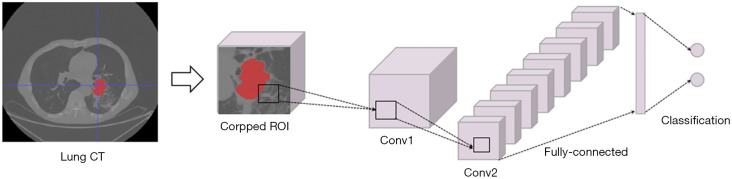
As shown in Figure 3, the comparative CNN model shared the same convolutional layers as the CapsNet described above, which contained a 16-to-256 convolutional layer and a 256-to-64 convolutional layer to obtain feature representation from the cropped region of interest (ROI) input. Then, a fully connected network layer was placed to achieve the final classification results instead of the identification capsule layer (Figure 2). The number of neurons in this fully connected layer shared the same value as that in the identification capsule layer for fair comparison.
Figure 3.
The flowchart of the CNN framework for the subtype discrimination of NSCLC. CNN, convolutional neural network; NSCLC, non-small cell lung cancer; CT, computed tomography; ROI, region of interest; Conv, convolutional layer.
The deep learning models were implemented based on the widely used deep learning framework PyTorch (https://pytorch.org/). 3D isotropic patches of 64×64×64 in size were extracted around manually labeled lesions and normalized to act as input to our deep learning models. The CNN and CapsNet shared the same training strategy, the applied optimizer was Adam, and the parameter setting of the optimizer was set to learning rate =0.001, betas = (0.9, 0.999), and eps =1e–08. An early stopping strategy was not used during the model training. Several data augmentation strategies (such as translation and rotation) were also tested for the CNN, but without any improvement in performance, so only results without data augmentation are reported in the following sections. Moreover, the demographic and semiquantitative features (six features in total, see Table 1) were not adopted in either CapsNet or the CNN.
The patients were proportionally appointed to the training data group (75% of all patients, n=94) and the testing data group (the remaining 25% of patients, n=32). The appointing process was repeated four times. All three classification models were evaluated using the mean value obtained from the four repetitions. Sensitivity, specificity, accuracy, area under the curve (AUC), F1_score, Matthews correlation coefficient (MCC), and log_loss were calculated.
Results
For the demographic and semi-quantitative clinical features, there were significant statistical differences in the patient’s gender (P<0.01) and tumor location (P<0.05), with SCC more prone to occur in males and AC more likely to occur in the peripheral lung field (Table 1). No significant statistical differences were found in age, tissue adhesion, or tumor density (all P>0.05; Table 1).
Figure 4 illustrates the classification performances with different radiomics features (increment: 10% of total features), and it was clear that model performances fluctuated with the number of input features. A comparison of the accuracies of the different methods is shown in Table 2. Based on the results, CapsNet outperformed CNN and the four radiomics methods, and obtained an 81.3% accuracy, 82.2% sensitivity, and 80.7% specificity. Notably, the AUC of CapsNet was only marginally better than the AUC of the optimal radiomics model with RF. The CNN, the radiomics with RF, and the radiomics with LR-PCA models achieved suboptimal performances with accuracy of 75% (sensitivity: 68.3%, specificity: 80.7%), 75.9% (sensitivity: 72.6%, specificity: 79.1%) and 74.1% (sensitivity: 64.4%, specificity: 82.4%), respectively. Among the four radiomics models, the LR and LR-L1 penalty performed the worst (both with accuracy <0.7). Figure 5 displays the receiver operating characteristic (ROC) curves for the three models.
Figure 4.
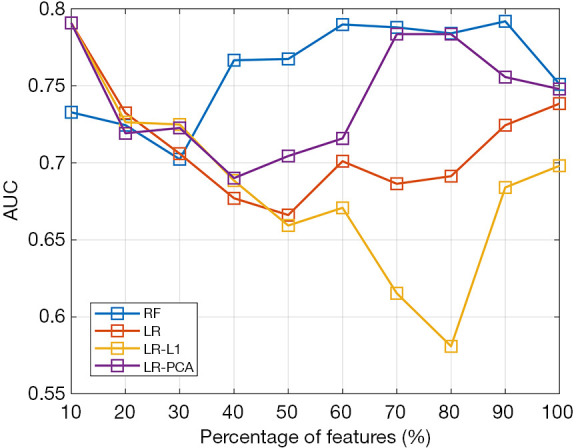
The classification of performances with different numbers of radiomics features. RF, random forest; LR, logistic regression; PCA, principal component analysis.
Table 2. The classification results of all the models.
| Classifiers | ACC | Sen | Spe | AUC | F1_score | MCC | Log_loss |
|---|---|---|---|---|---|---|---|
| RF | 0.759 (0.074) | 0.726 (0.018) | 0.791 (0.061) | 0.84 | 0.727 | 0.511 | 8.325 |
| LR | 0.643 (0.016) | 0.619 (0.029) | 0.657 (0.045) | 0.669 | 0.53 | 0.272 | 12.335 |
| LR-L1 | 0.670 (0.014) | 0.667 (0.020) | 0.675 (0.014) | 0.656 | 0.641 | 0.336 | 11.41 |
| LR-PCA | 0.741 (0.034) | 0.644 (0.014) | 0.824 (0.078) | 0.777 | 0.688 | 0.473 | 8.943 |
| CNN | 0.750 (0.029) | 0.683 (0.103) | 0.807 (0.048) | 0.725 | 0.708 | 0.492 | 8.635 |
| CapsNet | 0.813 (0.018) | 0.822 (0.070) | 0.807 (0.048) | 0.852 | 0.796 | 0.624 | 6.476 |
ACC, Sen, and Spe are displayed as mean (standard deviation). ACC, accuracy; Sen, sensitivity; Spe, specificity; AUC, area under the curve; MCC, Matthews correlation coefficient; RF, random forest; LR, logistic regression; PCA, principal component analysis; AUC, area under curve; CNN, convolutional neural network; CapsNet, capsule network.
Figure 5.
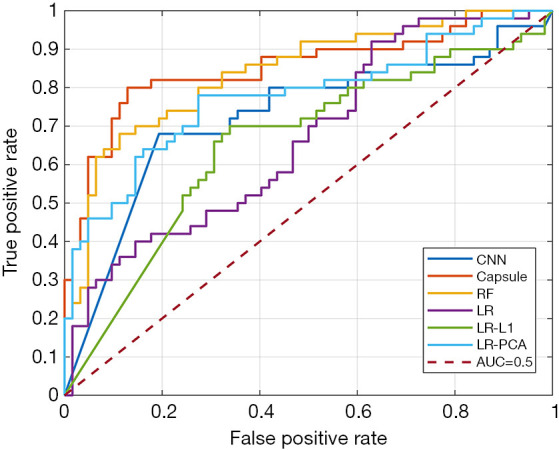
The ROC curve of the three classification models. ROC, receiver operating characteristic; CNN, convolutional neural network; RF, random forest; LR, logistic regression; PCA, principal component analysis; AUC, area under curve.
Discussion
We developed and compared three frameworks for the classification of AC and SCC using CT images. The investigations revealed that CapsNet could better classify AC from SCC compared with the CNN and radiomics models using our dataset. Although the highest accuracy obtained by CapsNet was just 81.3%, it displayed great potential in future studies especially for limited single-center datasets.
Although radiomics has been widely used in the diagnosis of various cancers (26), it only had an adequate performance among the three models and obtained a maximum 75.9% accuracy. In addition, the radiomics performance was also dependent on the classifier type, and RF and LR-PCA performed better than the other two models. Several factors may limit the performance of the radiomics models. First, the feature sets of radiomics are predefined before the model construction. However, if there exist certain features that do not belong to the radiomics feature sets but in fact contribute to discrimination, then the radiomics model will miss these features. Second, the principal source of radiomics features is texture feature, which is predominantly the first-order or second-order statistical description for voxel-level image characteristics. In comparison, deep learning models may dig further into the hierarchical features through the multi-layer network structure. The four machine learning methods including RF, LR, LR-L1, and LR-PCA performed differently from each other but with accuracy all lower than 80%. One possible explanation for this is that the radiomics feature sets might not have fully delineated the differences between SCC and AC, thereby limiting the performances of these machine learning methods. Although there are other kinds of radiomics feature sets with more features, high correlation has been shown to exist in radiomics feature sets (11). Lastly, it is important to note that feature selection and the selection of classifier types are both important to the performances of the radiomics models.
CNNs have been developed and successfully used in various medical imaging tasks, such as tumor segmentation, classification, and evaluation of prognosis (22). When applied in this study, CNN did not achieve sufficiently satisfactory results compared with CapsNet. One possible reason may be the relatively small training dataset, which is a common condition for most medical imaging studies. The current solution for this problem is to collect large datasets through multicenter accumulation. However, multicenter datasets may have larger heterogeneity than single datasets, and few studies have developed the specific algorithm to eliminate these multicenter differences. It is still not clear exactly which factors contribute to multicenter differences, and hence, even if CNNs could achieve good results in large samples, it would be difficult to interpret the biological significance of the features detected by the CNNs. Future studies are needed to enhance the interpretability of CNNs and to incorporate brain-inspired structures into CNNs.
CapsNet achieved the best performance in this study, and demonstrated its validity in a limited sample set. Most single-center datasets are not large enough, but the clinical criteria for the collective data are easy to control. Therefore, training a medical image prediction model with single-center dataset has distinct clinical values since intercenter and intersubject differences have yet to be clearly explained. There may be multiple reasons why CapsNet displayed the best performance for the discriminative analysis of NSCLC subtypes. First, AC and SCC patients may show different global feature patterns. CNNs and radiomics models generally focus on the local feature patterns, while CapsNet can record and recognize global patterns (that is, the relative spatial relationship of image elements). Second, CapsNet can record the object orientation information for local feature patterns, while both CNN and radiomics lack this function. The orientation information is important for the differential diagnosis of histological images between AC and SCC, and therefore it may also be helpful for the discriminative analysis of CT imaging features. Thus, CapsNet could obtain better classification performance than CNN with the same network structure within limited sample sets, making it popular in disease classification (27,28). Meanwhile, the theory of CapsNet is continuously improving (29), and it can be expected that CapsNet will contribute significantly to the medical imaging domain, especially for datasets with limited samples.
There were several limitations in this study. First, the features used in the three models have not been compared because it is difficult to display the extracted features for the deep learning methods. Moreover, the radiomics models combined the demographic and semiquantitative features, while the CNN and CapsNet models did not include these features. Indeed, incorporating the demographics and semiquantitative features may further improve the performance of the deep learning models. Second, this study only compared a commonly used network design for CapsNet and CNN, and did not identify the optimal design for Capsnet or CNN. Therefore, future studies could be conducted using more complex network architectures so as to achieve the most effective design for the CNN and CapsNet.
Conclusions
In this study, three discrimination models for subtypes of NSCLC were compared, and CapsNet demonstrated the best performance in classification with an accuracy of 81.3%. This may be due to its capacity to discover global feature patterns and orientational local feature patterns. The current study demonstrated the potential of CapsNet in identifying subtypes of NSCLC in a limited single-center dataset, and its use could be extended to the diagnosis of other diseases.
Acknowledgments
Funding: This study was supported by the Beijing Municipal Commission of Education (No. KM202010025025).
Ethical Statement: This study was approved by the institutional ethics board of the Third Medical Center of PLA General Hospital, and individual patient consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-734). The authors have no conflicts of interest to declare.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Socinski MA, Obasaju C, Gandara D, Hirsch FR, Bonomi P, Bunn P, Kim ES, Langer CJ, Natale RB, Novello S, Paz-Ares L, Perol M, Reck M, Ramalingam SS, Reynolds CH, Spigel DR, Stinchcombe TE, Wakelee H, Mayo C, Thatcher N. Clinicopathologic features of advanced squamous NSCLC. J Thorac Oncol 2016;11:1411-22. 10.1016/j.jtho.2016.05.024 [DOI] [PubMed] [Google Scholar]
- 3.Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res 2015;4:36-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shroff GS, Benveniste MF, de Groot PM, Wu CC, Viswanathan C, Papadimitrakopoulou VA, Truong MT. Targeted therapy and imaging findings. J Thorac Imaging 2017;32:313-22. 10.1097/RTI.0000000000000294 [DOI] [PubMed] [Google Scholar]
- 5.Galvin JR, Franks TJ. Lung cancer diagnosis: radiologic imaging, histology, and genetics. Radiology 2013;268:9-11. 10.1148/radiol.13130558 [DOI] [PubMed] [Google Scholar]
- 6.Sabour S. Reproducibility of histopathological diagnosis in poorly differentiated NSCLC: statistical issue. J Thorac Oncol 2015;10:e3-4. 10.1097/JTO.0000000000000354 [DOI] [PubMed] [Google Scholar]
- 7.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen B, Zhang R, Gan Y, Yang L, Li W. Development and clinical application of radiomics in lung cancer. Radiat Oncol 2017;12:154. 10.1186/s13014-017-0885-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Chen T, Zhao W, Wei C, Li X, Duan S, Ji L, Lu Z, Shen J. Radiomics prediction model for the improved diagnosis of clinically significant prostate cancer on biparametric MRI. Quant Imaging Med Surg 2020;10:368-79. 10.21037/qims.2019.12.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang N, Cui L, Luo Y, Chang Z, Yu B, Liu Z. Development and multicenter validation of a CT-based radiomics signature for discriminating histological grades of pancreatic ductal adenocarcinoma. Quant Imaging Med Surg 2020;10:692-702. 10.21037/qims.2020.02.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu W, Parmar C, Grossmann P, Quackenbush J, Lambin P, Bussink J, Mak R, Aerts HJ. Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol 2016;6:71. 10.3389/fonc.2016.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen C, Liu Z, Guan M, Song J, Lian Y, Wang S, Tang Z, Dong D, Kong L, Wang M, Shi D, Tian J. 2D and 3D CT radiomics features prognostic performance comparison in non-small cell lung cancer. Transl Oncol 2017;10:886-94. 10.1016/j.tranon.2017.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X, Dong D, Chen Z, Fang M, Zhang L, Song J, Yu D, Zang Y, Liu Z, Shi J, Tian J. Radiomic signature as a diagnostic factor for histologic subtype classification of non-small cell lung cancer. Eur Radiol 2018;28:2772-8. 10.1007/s00330-017-5221-1 [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Jing B, Han W, Long Z, Mo X, Li H. A comparative texture analysis based on NECT and CECT images to differentiate lung adenocarcinoma from squamous cell carcinoma. J Med Syst 2019;43:59. 10.1007/s10916-019-1175-y [DOI] [PubMed] [Google Scholar]
- 15.Digumarthy SR, Padole AM, Gullo RL, Sequist LV, Kalra MK. Can CT radiomic analysis in NSCLC predict histology and EGFR mutation status? Medicine (Baltimore) 2019;98:e13963. 10.1097/MD.0000000000013963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 2015;60:5471-96. 10.1088/0031-9155/60/14/5471 [DOI] [PubMed] [Google Scholar]
- 18.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts H. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017;77:e104-7. 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao B, Tan Y, Tsai WY, Qi J, Xie C, Lu L, Schwartz LH. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 2016;6:23428. 10.1038/srep23428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aerts HJ. The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol 2016;2:1636-42. 10.1001/jamaoncol.2016.2631 [DOI] [PubMed] [Google Scholar]
- 21.Mazurowski MA, Buda M, Saha A, Bashir MR. Deep learning in radiology: An overview of the concepts and a survey of the state of the art with focus on MRI. J Magn Reson Imaging 2019;49:939-54. 10.1002/jmri.26534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng 2017;19:221-48. 10.1146/annurev-bioeng-071516-044442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, Bussink J, Gillies RJ, Mak RH, Aerts H. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med 2018;15:e1002711. 10.1371/journal.pmed.1002711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabour S, Frosst N, Hinton GE. Dynamic routing between capsules. In: Advances in Neural Information Processing Systems 30 (NIPS 2017). 2017:3856-66. [Google Scholar]
- 25.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts H. Machine learning methods for quantitative radiomic biomarkers. Sci Rep 2015;5:13087. 10.1038/srep13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, Wang S, Dong D, Wei J, Fang C, Zhou X, Sun K, Li L, Li B, Wang M, Tian J. The applications of radiomics in precision diagnosis and treatment of oncology: opportunities and challenges. Theranostics 2019;9:1303-22. 10.7150/thno.30309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao Z, Huang P, Kam TE, Hsu LM, Wu Y, Zhang H, Shen D. Dynamic routing capsule networks for mild cognitive impairment diagnosis. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer, 2019:620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afshar P, Mohammadi A, Plataniotis KN. Brain tumor type classification via capsule networks. In: 2018 25th IEEE International Conference on Image Processing (ICIP). IEEE, 2018:3129-33. [Google Scholar]
- 29.Kosiorek A, Sabour S, Teh YW, Hinton GE. Stacked capsule autoencoders. In: Advances in Neural Information Processing Systems 32 (NeurIPS 2019). 2019:15512-22. [Google Scholar]