Abstract
In this communication, we report the presence of RNA of bovine viral diarrhea virus (BVDV) as a contaminant of different biological products used in Mexico for veterinary vaccine production. For this purpose, six batches of monovalent vaccines, eight cell line batches used for vaccine production, and 10 fetal bovine serum lots (FBS) commercially available in Mexico from different suppliers were tested by reverse transcription polymerase chain reaction (RT-PCR). Viral RNA was detected in 62.5% of the samples analyzed. Phylogenetic analysis revealed the presence of the subgenotypes BVDV-1a, 1b, and BVDV-2a in the tested samples. Collectively, these findings indicate that contamination by BVDV RNA occurs in commercial vaccines and reagents used in research and production of biological products. The ramifications of this contamination are discussed.
The viruses associated with the clinical presentation referred to as bovine viral diarrhea (BVD) belong to three species in the genus Pestivirus: Pestivirus A (bovine viral diarrhea virus 1, BVDV1), Pestivirus B (bovine viral diarrhea virus 2, BVDV2), and Pestivirus H (HoBi-like pestivirus, an atypical ruminant pestivirus) [1]. Within these three species, subgenotypes have been suggested based on phylogenetic analysis. At least 21 subgenotypes have been reported for BVDV-1 (1a-1u), four for BVDV-2 (2a-2d), and four for HoBi-like viruses (a-d) [2]. Regional differences have been observed in the prevalence of these species and subgenotypes. In Mexico, BVDV subgenotypes 1a, 1b, 1c, and 2a have been found, while HoBi-like viruses have not been detected [3].
Clinical manifestations of BVD in cattle include respiratory disease, hemorrhagic syndrome, gastrointestinal disorders, and reproductive problems (abortions, infertility, and congenital malformations). In addition, transplacental infection of the fetus can result in the birth of an immunotolerant, persistently infected (PI) animal [4].
Due to fetal infections, fetal bovine serum (FBS) is frequently contaminated with BVD-associated viruses [5–10]. Contamination of FBS leads to contamination of biological products that use FBS in production, including cell cultures [11–14] and vaccines for animal [15, 16] and human use [17, 18].
Vaccine contamination may not only influence the results of vaccination but may also lead to new infections, causing serious BVD outbreaks [16, 19]. BVDV contamination can affect the outcome of cell-culture-based research and diagnostic procedures, resulting in misinterpretation of research data or an incorrect diagnosis [20]. Reliable FBS screening is crucial for the safety of biological products used in cattle populations. Therefore, BVDV contamination remains a major concern and a continuous challenge for biological product safety [16, 17, 21, 22].
In this study, we investigated BVDV contamination in FBS and cell lines used in vaccine production and in live viral vaccines for veterinary use that are commercially available in Mexico. Furthermore, BVDV-positive samples were genotyped.
A total of six batches of two modified live vaccines, one against parainfluenza virus 3 (PI3) and infectious rhinotracheitis virus 1 (IBR) and the other against rabies virus, produced by different Mexican manufacturers, eight batches of MDCK, MDBK, and BHK-21 cells, kindly provided by several national vaccine manufacturers, and 10 gamma-irradiated lots of FBS used in animal vaccine production in Mexico were tested for BVDV RNA contamination by RT-PCR assay (Table 1).
Table 1.
Vaccines and cell lines used in the current study
| Type of sample | Sample identifier |
|---|---|
| Vaccines | |
| Modified live PI3 and IBR vaccine | Vaccine D |
| Modified live PI3 and IBR vaccine | Vaccine E |
| Modified live rabies virus vaccine for cattle | Vaccine F |
| Modified live rabies virus vaccine for cattle | Vaccine G |
| Modified live PI3 and IBR vaccine | Vaccine H |
| Modified live rabies virus vaccine for cattle | Vaccine I |
| Cell lines | |
| BHK-1 | Cell line 1 |
| MDBK | Cell line 2 |
| MDBK | Cell line 3 |
| MDCK | Cell line 4 |
| MDCK | Cell line 5 |
| MDBK | Cell line 6 |
| MDBK | Cell line 7 |
| MDBK | Cell line 8 |
Positive samples are shown in boldface
BHK-1 bovine hamster kidney, MDBK Madin-Darby bovine kidney, MDCK Madin-Darby canine kidney
Ten different lots of commercially available FBS were used in this study. The BVDV NADL reference strain was used as a positive control and a supernatant of mock-infected BVDV-free MDBK cells was used as a negative control.
The total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Briefly, a 400-µl aliquot from each sample was mixed with 300 µl of TRIzol and incubated at room temperature for 5 min. Then, 150 µl of chloroform was added, and the tubes were vigorously shaken and then chilled on ice for 7 min, followed by centrifugation at 13,800 g for 20 min. The aqueous phase was collected, and 500 µl of isopropanol was added. Tubes were incubated at room temperature for 10 min and centrifuged at 18,800 g at 4°C for 20 min. The resulting pellet was washed twice with 1 ml of 75% ethanol in RNase-free water. The pellet was air-dried at room temperature, dissolved in 20 µl of RNase-free water, and stored at − 70 °C.
The RNA obtained from the samples and the positive and negative controls were subjected to reverse transcription polymerase chain reaction (RT-PCR). RT-PCR was performed using the primers 5UTR and START as described previously [23] in order to amplify a fragment of the 5’UTR region. The PCR products were detected by electrophoresis in a 1% agarose gel stained with GelRed Nucleic Acid® and then purified according to the manufacturer’s recommendations using a QIAquick Gel Extraction Kit (QIAGEN GmbH).
We determined the detection limit of the PCR by performing tenfold dilutions of a titrated sample of the BVDV1a-NADL reference strain using minimal essential medium (MEM, Gibco, Grand Island, NY) as the diluent. RNA extraction and RT-PCR of each dilution were carried out as described above. The detection limit of the PCR was defined as the highest dilution at which a positive amplification signal was obtained.
Sequencing of 5’UTR amplicons was performed at the Molecular Biology Unit of the Cell Physiology Institute of the Universidad Nacional Autonoma de México (IFC-UNAM) using the Sanger dideoxy method on a ABI Prism 3500xl Genetic Analyzer (Applied Biosystems/Hitachi, Forest City, CA). Nucleotide sequences were aligned using the Clustal W [24] program with BioEdit software [25]. Phylogenetic analysis was performed in MEGA 7 [26] by the maximum-likelihood method with the Kimura 2-parameter model. Bootstrap analysis was carried out on 1000 replicates, and phylogenetic trees were drawn in MEGA 7. Nucleotide sequences obtained from this study were submitted to the GenBank database, and the corresponding accession numbers of the 5´UTR sequences are KC252579-KC252582 for vaccine samples, KC252583-KC252590 for FBS samples, and KC252591-KC252597 for cell line samples.
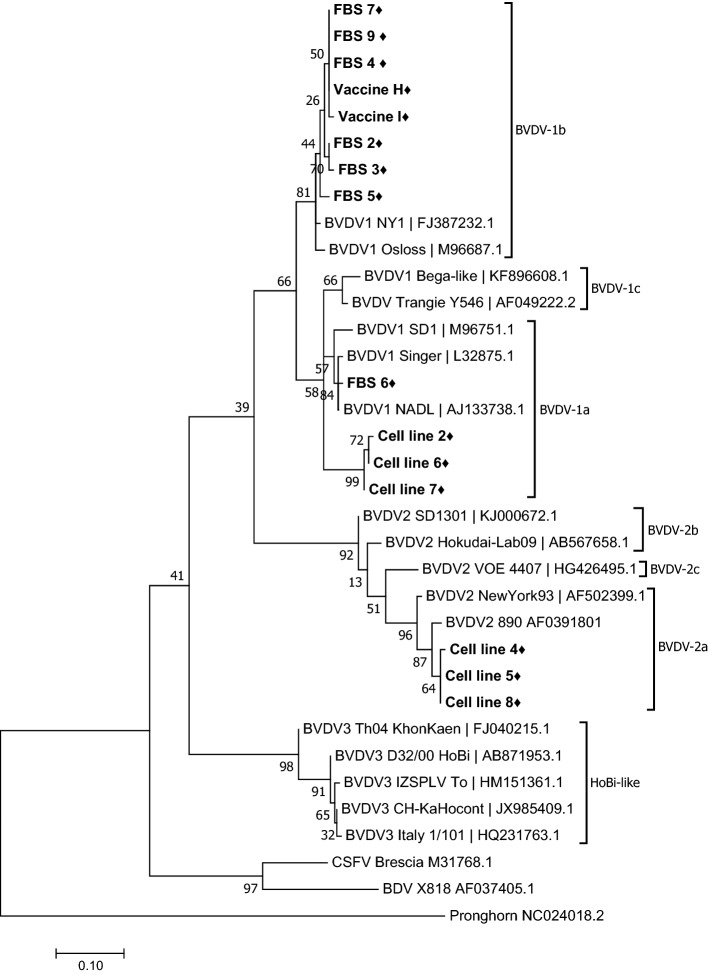
Six commercial veterinary vaccines, eight cell lines, and 10 FBS lots from different manufacturers were tested for the presence of BVDV contamination. Fifteen samples out of 24 were positive by RT-PCR using the primers 5UTR and START, which have a detection limit of 2.4 × 103 TCID50/mL for the BVDV1a-NADL reference strain. In order to determine the genotypes of the viruses that were detected, phylogenetic analysis was performed based on comparisons of a 280-nucleotide region of the 5’UTR (Fig. 1). This analysis segregated the amplified sequences into three BVDV subgenotypes. Sequences obtained from samples of FBS lots 2, 3, 4, 5, 7, and 9 and vaccines H and I belonged to BVDV genotype 1b. Sequences from FBS lot 6 and cell lines 2, 6, and 7 were grouped within clade BVDV 1a, and the sequences in cell line samples 4, 5, and 8 were grouped within BVDV genotype 2a.
Fig. 1.
Phylogenetic analysis based on the 5’UTR region. Phylogenetic inference was conducted in MEGA 7 using the maximum-likelihood method. Distances were computed using the Kimura 2-parameter model. Reference sequences are identified by GenBank accession number. The nucleotide sequences from this work are shown in bold font with the symbol “filled diamond”.
FBS is the most commonly used growth-supporting factor in cell culture and production of biological products such as vaccines. Even with irradiation, it is difficult to absolutely guarantee elimination of all viral contaminants, particularly if those contaminants are present in large numbers [27]. Thus, there is an inherent risk of the introduction of bovine viruses via the use of FBS. Because of the high rate of BVDV contamination of FBS, these viruses have been detected in research reagents such as cell lines and vaccines [13–18]. Contamination of vaccines with live BVDV can result in immunosuppression, which can reduce the effectiveness of the vaccines and may make animals more vulnerable to opportunistic infections [9, 15]. In laboratory diagnosis and research assays, BVDV contamination can potentially affect the results of assays performed using cell culture and infectious agents [13, 28].
While the incidence of BVDV infection and the genotypes circulating in Mexican cattle populations have begun to be analyzed [3], little is known about the prevalence of BVDV contamination of biological products. There are no surveys for the presence of BVDV as an adventitious virus in vaccines and cell lines used in Mexico, despite the report of HoBi virus RNA in an FBS lot obtained from Mexico [8]. The results obtained in this study provide evidence for the presence of BVDV in biological products commercially available in Mexico, including live veterinary vaccines, cell lines, and FBS used nationally in veterinary vaccine production. In addition, genotyping of these contaminants showed that they belong to BVDV subgenotypes found previously in the USA and Canada [29, 30]. Furthermore, no evidence was found for the presence of HoBi-like viruses, which have not been found in the USA or Canada.
In our study, around 62.5% of the samples tested were contaminated with BVDV RNA, suggesting the potential contamination with BVDV of some of the vaccines used in the cattle industry in Mexico to prevent PI3, IBR, and rabies virus infections. In this context, one of the main limitations of this survey was the lack of a certified BVDV-free cell line at the time of our study, which prevented us from assessing the presence of live BVDV and thus confirming the relevance of our findings.
Analysis based on the nucleotide sequence of the 5’UTR indicated that the sequences obtained belonged to BVDV subgenotypes 1a, 1b, and 2a. These subgenotypes have been detected in cattle in the USA, Canada [29, 30], and Mexico. Thus, the BVDV subgenotypes detected in these biological samples are similar, and this correlates with the diversity detected in the animal population of Mexico [3]. These results were consistent with a study conducted in Argentina, where an analysis of FBS samples resulted in the identification of BVDV-1a, 1b, 2a, and HoBi-like viruses in that country [31]. This suggests that analysis of lots of FBS produced in Mexico on a regular basis may be a valuable tool for detecting the circulation of new pestiviruses in this country.
Several events of vaccine contamination have been documented in the past. In studies in Switzerland [18], Japan, and Italy [17], contamination with subgenotypes 1a, 1b, 1c, and 1d were reported in human vaccines against mumps, measles, rubella, and poliomyelitis. In animals, contamination of vaccines against classical swine fever virus, bovine syncytial respiratory virus, and infectious bovine rhinotracheitis virus was responsible for clinical signs of BVD in vaccinated animals [15, 16].
In previous surveys of BVDV contamination in animal vaccines, the genotypes of the contaminating viruses were not determined [19, 32–34]. In this study, contaminants found in vaccines against parainfluenza virus 3 (PI3) and infectious rhinotracheitis virus 1 (IBR) and against rabies virus, identified as vaccines H and I, respectively, were found to belong to the BVDV 1b group (Fig. 1), which is one of the subgenotypes that has been reported in vaccines for human use. So far, the impact of BVDV contamination of human vaccines is unknown, but possible detrimental effects on human health should not be dismissed. In both animals and humans, BVDV contamination represents a biohazard that has to be taken into account by vaccine manufacturers.
Moreover, in this study, eight batches of FBS were found to be positive for BVDV RNA. The sequences of these contaminants were very similar to those of BVDV 1a and 1b. These results reveal the presence of at least two subgenotypes in commercially available FBS in Mexico and are similar to those obtained in studies by Miroslaw et al., in which BVDV 1a, 1b, and 1c were identified but not isolated [9]. Similarly, several studies have reported contamination of FBS with different BVDV subgenotypes with a prevalence up to 100% in the samples tested [27, 35, 36].
The presence of BVDV in FBS is known to be the main cause of contamination in cell culture, because the use of contaminated FBS in growing cultured cells promotes dissemination of the virus through serial passages over time. Previous studies have shown that around 24% of bovine, swine, mouse, monkey, rabbit, and horse cell cultures tested were contaminated with BVDV RNA [36, 37]. In our survey, six out of eight cell lines used in animal vaccine production were found to be positive for BVDV 1a and 2a. Furthermore, the BVDV RNA contamination was present at relatively high levels in the samples. While detection of BVDV RNA in vaccines does not prove that live BVDV is present, the calculated amount of BVDV RNA detected in these products is close to that found in BVDV vaccines.
The results obtained in this work highlight the need for the implementation of continuous screening processes, not only for the vaccine production sector but also for research laboratories and regulatory authorities overseeing livestock production, in order to assure the safety of products used for vaccine development and the reliability of reagents used in diagnosis and research.
Funding
Funding for this project was provided by Programa de Apoyo a Proyecto de Investigación e Innovación Tecnológica (PAPIIT Project no. IN217919).
Declarations
Conflict of interest
The authors declare no conflict of interest regarding the authorship and/or publication of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Smith DB, Meyers G, Bukh J, Gould EA, Monath T, Muerhoff AS. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J Gen Virol. 2017;98:2106–2112. doi: 10.1099/jgv.0.000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeşilbağ K, Alpay G, Becher P. Variability and global distribution of subgenotypes of bovine viral diarrhea virus. Viruses. 2017;9:128. doi: 10.3390/v9060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gómez-Romero N, Basurto-Alcántara FJ, Verdugo-Rodríguez A, Bauermann FV, Ridpath JF. Genetic diversity of bovine viral diarrhea virus in cattle from Mexico. J Vet Diagn Invest. 2017;29(3):362–365. doi: 10.1177/1040638717690187. [DOI] [PubMed] [Google Scholar]
- 4.Simmonds P, Becher B, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff S, Pletnev A, Rico-Hesse R, Smith D.B, Stapleton, JT; ICTV Report Consortium (2017) ICTV virus taxonomy profile: Flaviviridae. J Gen Virol 98:2–3 [DOI] [PMC free article] [PubMed]
- 5.Bolin SR, Matthews PJ, Ridpath JF. Methods for detection and frequency of contamination of foetal calf serum with bovine viral diarrhoea virus and antibodies against bovine viral diarrhoea virus. J Vet Diagn. Invest. 1991;3:199–203. doi: 10.1177/104063879100300302. [DOI] [PubMed] [Google Scholar]
- 6.Bolin SR, Ridpath JF. Prevalence of bovine viral diarrhoea virus genotypes and antibody against those viral genotypes in foetal bovine serum. J Vet Diagn Invest. 1998;10:135–139. doi: 10.1177/104063879801000203. [DOI] [PubMed] [Google Scholar]
- 7.Nuttal PA, Luther PD, Stott EJ. Viral contamination of bovine foetal serum and cell cultures. Nature. 1977;266:835–837. doi: 10.1038/266835a0. [DOI] [PubMed] [Google Scholar]
- 8.Xia H, Vijayaraghavan B, Belák S, Liu L. Detection and identification of the atypical bovine pestiviruses in comercial foetal serum batches. PLoS ONE. 2011;6(12):1–3. doi: 10.1371/journal.pone.0028553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miroslaw PP, Jerzy R, Zmündzinski J. Contamination of foetal bovine serum with bovine viral diarrhoea virus (BVDV) Bull Vet Inst Pulawy. 2008;52:501–505. [Google Scholar]
- 10.Zabal O, Kobrak AL, Lager IA, Schudel AA, Weber EL. Contamination of bovine fetal serum with bovine viral diarrhea virus. Rev Argent Microbiol. 2000;32(1):27–32. [PubMed] [Google Scholar]
- 11.Bolin SR, Ridpath JF, Black J, Macy M, Roblin R. Survey of cell lines in the American type culture collection for bovine viral diarrhoea virus. J Virol Methods. 1994;48:211. doi: 10.1016/0166-0934(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 12.Büttner M, Oghmig A, Weiland F, Rziha HJ, Pfaff E. Detection of virus specific nucleic acid in food-stuff or bioproducts-hazards and risk assessment. Arch Virol. 1997;13:57–66. doi: 10.1007/978-3-7091-6534-8_6. [DOI] [PubMed] [Google Scholar]
- 13.Woods RD, Kunkle RA, Ridpath JE, Bolin SR. Bovine viral diarrhea virus isolated from fetal calf serum enhances pathogenicity of attenuated transmissible gastroenteritis virus in neonatal pigs. J Vet Diagn Invest. 1999;11(5):400–407. doi: 10.1177/104063879901100503. [DOI] [PubMed] [Google Scholar]
- 14.Schirrmeier H, Strebelow G, Depner K, Hoffmann B, Beer M. Genetic and antigenetic characterization of an atypical pestivirus isolate, a putative member of a novel pestivirus species. J Gen Virol. 2004;85:3647–3652. doi: 10.1099/vir.0.80238-0. [DOI] [PubMed] [Google Scholar]
- 15.Pastoret PP. Human and animal vaccine contaminations. Biologicals. 2010;38:332–334. doi: 10.1016/j.biologicals.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 16.Barkema HW, Bartels CJ, van Wuijckhuise L, Hesselink JW, Holzhauer M, Weber MF, et al. Outbreak of bovine virus diarrhea on Dutch dairy farms induced by a bovine herpesvirus 1 marker vaccine contaminated with bovine virus diarrhea virus type 2. Tijdschr Diergeneeskd. 2001;126:158–165. [PubMed] [Google Scholar]
- 17.Giangaspero M, Vacirca G, Harasawa R, Büttner M, Panuccio A, De Giuli MC, Zanetti A, Belloli A, Verhulst A. Genotypes of pestivirus RNA detected in live virus vaccines for human use. J Vet Med Sci. 2001;63(7):723–733. doi: 10.1292/jvms.63.723. [DOI] [PubMed] [Google Scholar]
- 18.Studer E, Bertoni CU. Detection and characterization of pestivirus contaminations in human live viral vaccines. Biologicals. 2002;30(4):289–296. doi: 10.1006/biol.2002.0343. [DOI] [PubMed] [Google Scholar]
- 19.Falcone E, Tollis M, Conti G (1999) Bovine viral diarrhea disease associated with a contaminated vaccine. Vaccine 14:18 (5–6):387–388 [DOI] [PubMed]
- 20.Pastoret P-P, Blancou J, Vannier P, Verschueren C (eds) (1997) Veterinary vaccinology, 1st edn. Amsterdam
- 21.Yanagi M, Bukh J, Emerson SU, Purcell RH. Contamination of commercially available fetal bovine sera with bovine viral diarrhea virus genomes: implications for the study of hepatitis C virus in cell cultures. J Infect Dis. 1996;174(6):1324–1327. doi: 10.1093/infdis/174.6.1324. [DOI] [PubMed] [Google Scholar]
- 22.Makoschey B, van gelder PT, Keijsera V, Goovaerts D, Bovine viral diarrhea virus antigen in foetal calf serum batches and consequences of such contamination for vaccine production. Biologicals. 2003;30:203–208. doi: 10.1016/S1045-1056(03)00058-7. [DOI] [PubMed] [Google Scholar]
- 23.Mahony TJ, McCarthy FM, Gravel JL, Corney B, Young PL, Vilcek S. Genetic analysis of bovine viral diarrhea viruses from Australia. Vet Microbiol. 2005;106:1–6. doi: 10.1016/j.vetmic.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 24.Thompson JD, Higgins DG, Gibson TJ. Clustal W; improving the sensitivity of progressive multiple sequence alignment trough sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall TA. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 26.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gauvin G, Nims R. Gamma-irradiation of serum for the inactivation of adventitious contaminants. PDA J Pharm Sci Technol. 2010;64:432–435. [PubMed] [Google Scholar]
- 28.Yanagi M, Bukh J, Emerson SU, Purcell RH (1996) Contamination of commercially available fetal bovine sera with bovine viral diarrhea virus genomes: implications for the study of hepatitis C virus in cell cultures. J Infect Dis 174:1324e7 [DOI] [PubMed]
- 29.Gilbert SA, Burton KM, Prins SE, Deregt D. Typing of bovine viral diarrhea viruses directly from blood of persistently infected cattle by multiplex PCR. J Clin Microbiol. 1999;37(6):2020–2023. doi: 10.1128/JCM.37.6.2020-2023.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridpath JF, Fulton RW, Kirkland PD, Neill JD. Prevalence and antigenic differences observed between Bovine viral diarrhea virus subgenotypes isolated from cattle in Australia and feedlots in the southwestern United States. J Vet Diagn Invest. 2010;22(2):184–191. doi: 10.1177/104063871002200203. [DOI] [PubMed] [Google Scholar]
- 31.Pecora A, Perez Aguirreburualde MS, Ridpath JF, Dus Santos MJ. Molecular characterization of pestiviruses in fetal Bovine sera originating from Argentina: evidence of circulation of HoBi-like viruses. Front Vet Sci. 2019;16(6):359. doi: 10.3389/fvets.2019.00359.PMID:31681812;PMCID:PMC6805694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falcone E, Cordioli P, Tarantino M, Muscillo M, Sala G, La Rosa G, Archetti IL, Marianelli C, Lombardi G, Tollis M. Experimental infection of calves with Bovine viral diarrhoea virus type-2 (BVDV2) isolated from a contaminated vaccine. Vet Res Commun. 2003;27:577–589. doi: 10.1023/A:1026064603630. [DOI] [PubMed] [Google Scholar]
- 33.Wensvoort G, Terpstra C. Bovine viral diarrhea virus infections in piglets born to sows vaccinated against swine fever with contaminated vaccine. Res Vet Sci. 1988;45:143–148. doi: 10.1016/S0034-5288(18)30920-2. [DOI] [PubMed] [Google Scholar]
- 34.Lohr C, Evermann J, Ward A. Investigation of dams and their offspring inoculated with vaccine contaminated by bovine viral diarrhea virus. Vet Med/Small Anim Clin. 1983;78:8. [Google Scholar]
- 35.Giammarioli M, Ridpath JF, Rossi E, Bazzucchi M, Casciari C, De Mia GM. Genetic detection and characterization of emerging HoBi-like viruses in archival foetal bovine serum batches. Biologicals. 2015;43:220–224. doi: 10.1016/j.biologicals.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Pinheiro de Oliveira TF, Fonseca AA, Camargos MF, de Oliveira AM, Pinto Cottorello AC, Souza Ados R, de Almeida IG, Heinemann MB. Detection of contaminants in cell cultures, sera and trypsin. Biologicals. 2013;41(6):407–414. doi: 10.1016/j.biologicals.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Audet SA, Crim RL, Beeler J (2000) Evaluation of vaccines, interferons and cell substrates for pestivirus contamination. Biologicals 28:41e6. 10.1006/biol.1999.0240 [DOI] [PubMed]