Abstract
Background
Myocardial infarction (MI), caused by temporary or permanent coronary artery occlusion, poses a serious threat to patients’ lives. Circular RNAs (circRNAs), a new kind of endogenous noncoding RNAs, have been widely studied recently. This study was designed to illustrate and potential molecular mechanisms of circRNA 010567 in hypoxia-induced cardiomyocyte injury in vitro, so as to provide new strategies for the therapy of MI.
Methods
H9c2 cells were cultured in anoxic conditions with 94% N2, 5% CO2, and 1% O2 to establish the in vitro MI model. Cell viability and apoptosis were checked using MTT and flow cytometry assay, respectively, Moreover, the levels of circRNA 010567, miR-141, and DAPK1 was determined using qRT-PCR. The putative targets of circRNA 010567 and miR-141 were confirmed by dual-luciferase reporter system and the RNA immunoprecipitation (RIP) assay. The release of creatine kinase-MB (CK-MB), cardiac troponin I (cTnI), and the viability of mitochondria were detected using assay kits.
Results
The current study revealed that circRNA 010567 and DAPK1 were over-expressed, and miR-141 was low-expressed in hypoxia-induced MI. circRNA 010567 sponges miR-141 and DAPK1 was a direct target of miR-141. Mechanistic investigations revealed that circRNA 010567-siRNA impaired the release of CK-MB and cTnI, and promoted the viability of mitochondria in hypoxia-induced H9c2 cells, while these findings were reversed by the miR-141 inhibitor. In addition, the miR-141 mimic markedly reduced the release of CK-MB and cTnI, and promoted the viability of mitochondria, and these results were reversed by the DAPK1-plasmid. Subsequently, functional experiments revealed that hypoxia-stimulated decreases in H9c2 cell viability, as well as increases in apoptosis and caspase-3 activity, were induced by the miR-141 mimic and circRNA 010567-siRNA. However, these results were reversed by the miR-141 inhibitor and DAPK1-plasmid.
Conclusions
Our results demonstrated that circRNA 010567-siRNA played a protective role in hypoxia-induced cardiomyocyte damage via regulating the miR-141/DAPK1 axis, indicating that circRNA 010567-siRNA may be a promising target for MI therapy.
Keywords: Myocardial infarction (MI), circRNA 010567, miRNA-141/DAPK1
Introduction
Myocardial infarction (MI), caused by temporary or permanent blockage of major coronary arteries, has a high global morbidity and mortality all over the world (1,2). Previous studies have suggested that MI manifests as chest pain, fatigue, sweating, and dyspnea amongst other symptoms, and eventually, the contractility of the myocardium is reduced, leading to heart failure (3,4). Previous studies have shown that thrombolytic drug reperfusion therapy can effectively restore blood supply, but may also result in secondary myocardial injury and limiting the therapeutic effect (5). Therefore, it is urgent to develop more effective therapeutic drugs to decrease the burden of MI.
Various studies have demonstrated that circular RNAs (circRNAs) are associated with the physiological and pathological processes of various diseases, including gastric cancer (6), gallbladder cancer (7), and breast cancer (8). Zhang et al. revealed that the circRNA circNRIP1 promoted gastric cancer development by targeting microRNA-149-5p through the AKT1/mTOR pathway (6). Moreover, Li et al. highlighted the roles of circRNAs in breast cancer (8). Recent studies have also reported that circRNA molecules are rich in microRNA (miRNA) binding sites, which can regulate gene expression by acting as miRNA sponges (9,10). In recent years, the role of circRNA in MI has received more and more attention (11,12). For example, circular RNA CircFndc3b has been reported to modulate cardiac repair after myocardial infarction via FUS/VEGF-A axis (13). Circ-Ttc3 has been confirmed to play the cardioprotective role in MI (14). Besides, circRNA CDYL could induce myocardial regeneration by ceRNA after MI (15). CircRNA 010567, a newly identified circRNA, has been shown to promote myocardial fibrosis through suppression of miR-141 by targeting TGF-beta1 (16). It has also been reported that circRNA 010567 improved MI in rats by inhibiting TGF-beta1 (17). However, the underlying mechanism of circRNA 010567 in the development of MI has not yet been elucidated.
MiRNAs are a kind of endogenous noncoding RNAs found in eukaryotes, with regulatory functions (18). Studies have shown that miRNAs play important roles in cell growth and apoptosis, cell differentiation, insulin secretion, brain morphogenesis, and cardiovascular health and disease, including MI (19). For example, miRNA-145 inhibited MI-treated apoptosis by autophagy via AKT3/mTOR signaling pathway in vitro and in vivo (20). Furthermore, miRNA-141 can regulate BMP-2-mediated calcification in aortic stenosis (21). Nevertheless, whether and how miRNA-141 is involved in the progression of MI has not yet been fully explored.
Death-associated protein kinase 1 (DAPK1), a Ca2+/calmodulin (CaM)-dependent serine/threonine protein kinase, is reported to be involved in different apoptosis pathways, including tumor suppression and neuronal cell death (22-24). Study has reported that disruption of DAPK1 related cell death pathway may be considered as a promising treatment for stroke (25). Another study indicated that DAPK1 is involved in the protective effect of miR-98-5p in cerebral ischemia/reperfusion injury (26). Through the analysis of bioinformatics software, we found that there is a potential binding site between miR-141 and DAPK1. Therefore, we hypothesized that miR-141 might regulate hypoxia-induced cardiomyocyte apoptosis by regulating the expression of DAPK1.
Therefore, tour report was designed to explore the roles and potential molecular mechanisms of circRNA 010567 in hypoxia-induced cardiomyocyte injury in vitro, so as to provide new strategies for the treatment of MI. Our investigations showed that circRNA 010567 was up-regulated in hypoxia-induced cardiomyocytes. CircRNA 010567-siRNA played a protective role in hypoxia-induced cardiomyocyte injury by promoting cell proliferation and suppressing apoptosis via the miRNA-141/DAPK1 axis, providing a novel potential biomarker and therapeutic target for MI.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jtd-21-212).
Methods
Cell culture and MI model establishment
The rat cardiomyocyte cell line H9c2 was obtained from ATCC(Manassas, VA, USA) and seeded in Dulbecco’s modified Eagle medium (DMEM, Gibco, MD, USA) containing 10% fetal calf serum (FBS, Gibco, MD, USA) and 1% penicillin-streptomycin, and cultured in an incubator at 37 °C with 5% CO2. In order to establish an in vitro model of MI, H9c2 cells were exposed to a hypoxic incubator (94% N2, 5% CO2, and 1% O2) for 48 h (27).
qRT-PCR analysis
CircRNA 010567, miR-141, and DAPK1 mRNA expression were measured by qRT-PCR. The isolation of RNA from H9c2 cells was carried out using the miRNeasy Mini Kit (Life Technologies, Carlsbad, California, USA) referring to the manufacturer’s manual, and cDNA was synthesized by PrimeScript RT kit (Takara, Shiga, Japan). PCR amplification was then performed on an ABI PRISM 7900 sequence detection system (Applied Biosystems, Carlsbad, California, USA) with SYBR Premix Ex-Taq (TaKaRa). Primers were obtained from Sangon Biotech (Shanghai, China) and presented as following: circRNA 010567 forward, 5'-CAGCGTCCTTTCTCAAGGGA-3'; Reverse 5'-GACCTGATTGGCCACTCAGTA-3'; miR-141 forward 5'-GTCCATCTCCATCAGTACAGGTTG-3'; Reverse 5'-AGCCATCTTACTCACAGAGTGTG-3'; DAPK1 forward 5'-AATCCTAGACGTGGTCCGGTAT-3'; Reverse 5'-GTCCTCGGTGCGTATCCTTTCG3'; U6 forward 5'-GCTTGCTTCACACACACATA-3'; Reverse 5'-AAAAAACATCGACTCADG-3'; GAPDH forward 5'-CGGAGTCAACGGATTTGGTCGTAT-3'; Reverse 5'-AGCCTTCTCCATGGTGGTGAAGAC-3'. Target gene expression was calculated using the 2−ΔΔCt method.
Dual-luciferase reporter assay
Bioinformatics software (StarBase: http://starbase.sysu.edu.cn/, TargetScan: http://www.targetscan.org/vert_72/) was applied to identify the relationship between circRNA 010567 and miR-141, or miR-141 and DAPK1. Taking circRNA 010567 and miR-141 as an example, The 3'UTR of miR-141, which contains the circRNA 010567 binding site or mutated target site were amplified by RT-PCR and cloned into pMIR vectors (Ambion, USA) to construct the reporter vector circRNA 010567 wild-type (circRNA 010567-WT) or circRNA 010567 mutated-type (circRNA 010567-MUT) Our data revealed that DAPK1 was the potential target of miR-141. MiR-141-WT or miR-141-MUT and DAPK1 mimics or negative control (NC) were co-transfected into HEK293 cells by Lipofectamine 2000 (Life Technologies Corporation, Carlsbad, California, USA) and incubated for 48 h. Then, the relative luciferase activity was detected using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) referring to the manufacturer’s instructions.
RNA immunoprecipitation (RIP) assay
The RIP assay is a powerful tool to study the binding of RNA to proteins in cells, and to understand the dynamic processes of post-transcriptional regulatory networks. In brief, H9c2 cells transfected with the circRNA 010567 mimics and circRNA 010567-NC were lysed using RIP buffer. Magnetic beads were incubated with anti-ago2 or IgG antibody, and then co-incubated with cell lysate referring to the manufacturer’s protocol. Antibodies for RIP assays against anti-Ago2 and IgG were obtained from Abcam (Cambridge, MA, USA).
Cell transfection
CircRNA 010567-siRNA (circ-siRNA), control-siRNA, miR-141 inhibitor, inhibitor control, miR-141 mimic, mimic control, control-plasmid, or DAPK1-plasmid were transfected into hypoxia-induced H9c2 cells using Lipofectamine 2000 (Life Technologies Corporation, Carlsbad, California, USA) for 48 h, following the manufacturer’s manual. Subsequently, qRT-PCR was applied to evaluate the efficiency of cell transfection.
Detection of biochemical cardiac markers
The concentrations of cTnI and CK-MB, and mitochondrial activity in H9c2 cells were determined using corpuscle chemiluminescence (Beckman Coulter, Shanghai, China) following the manufacturer’s protocol.
MTT assay
After treatment, H9c2 cells were seeded into 96-well plates and incubated for 24 h at 37 °C. After that, H9c2 cells were induced by 20 µL MTT (5 mg/mL) solution and incubated for a further 4 h. After incubation, we removed the solution, added 100 µL DMSO to each well and kept the plate in the dark for 15 min. Finally, the optical density (OD) at the wavelength of 570 nm was determined by a multifunctional plate reader (BioTek, Richmond, Florida, USA) in accordance with the manufacturer’s protocol.
Flow cytometry analysis
After transfection for 48 h, H9c2 cells were trypsinized and stained using Annexin V-FITC/PI Apoptosis Detection Kit for 15 min in the dark following the manufacturer’s direction. The apoptotic cells were calculated by flow cytometry (BD, Biosciences, Franklin Lakes, NJ, USA), and the results were analyzed using CellQuest software.
Detection of caspase-3 activity
Caspase-3 activity in H9c2 cells was assessed using the Caspase-3 Assay Kit (Abcam, China) referring to the manufacturer’s direction. Briefly, H9c2 cells were lysed with buffer solution. Cell supernatants were obtained and centrifuged for 10 min at 1,500 ×g. The supernatants were then treated with the caspase-3 reagent and incubated at 37 °C for 2 h. After treatment, a microplate reader was used to measure the OD of caspase-3 activity at 405 nm.
Western blot assay
Total proteins from H9c2 cells were quantified using the BCA Protein Assay Kit (Invitrogen, CA, USA), separated by 10% SDS-PAGE and transferred to PVDF membranes (BioRad, USA). The membranes were blocked, then treated with primary antibodies against GAPDH (Cat no. G5262-1VL; Sigma) and DAPK1 (Cat no. SAB2109107; Sigma) overnight at 4 °C. After that, the membranes were incubated in secondary antibody (Cat no. 7074; Cell signaling technology), and bands were analyzed using the ECL detection system (Millipore, MA, USA) referring to the manufacturer’s direction.
Statistical analysis
Statistical analysis was conducted using SPSS 20.0. All measurement data were presented as mean ± standard deviation (SD) from three independent experiments. Mean differences among groups were assessed by the unpaired Student’s t-test or one-way ANOVA. P<0.05 and P<0.01 indicated a significant difference.
Results
CircRNA 010567 was up-regulated and miR-141 was down-regulated in hypoxia-induced H9c2 cells
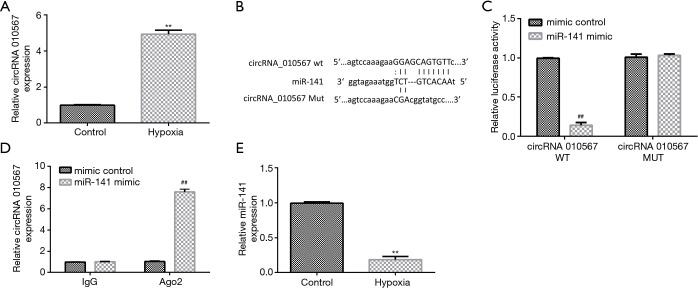
H9c2 cells were cultured under hypoxic conditions to establish the in vitro MI model. Firstly, the level of circRNA 010567 in hypoxia-induced H9c2 cells was determined using qRT-PCR. We found that circRNA 010567 was remarkably elevated in hypoxia-stimulated H9c2 cells, as compared to control (Figure 1A). Moreover, putative circRNA 010567 targets were predicted using bioinformatics tools. Our data indicated that miR-141 was a latent target of circRNA 010567 (Figure 1B). The dual-luciferase reporter gene system (Figure 1C) and the RIP assay (Figure 1D) confirmed that miR-141 was a direct target of circRNA 010567. Furthermore, we detected the expression of miR-141 in hypoxia-induced H9c2 cells using qRT-PCR. Down-regulation of miR-141 was found in hypoxia-induced H9c2 cells (Figure 1E). These results demonstrated that circRNA 010567 was up-regulated and miR-141 was down-regulated in hypoxia-induced H9c2 cells.
Figure 1.
Expression of circRNA 010567 and miR-141 in hypoxia-induced H9c2 cells. (A) The expression of circRNA 010567 was evaluated using qRT-PCR. (B) The binding site of miR-141 and circRNA 010567. The dual-luciferase reporter gene system (C) and the RIP assay (D) were conducted to verify the relationship between circRNA 010567 and miR-141. (E) qRT-PCR analysis of miR-141 levels in hypoxia-induced H9c2 cells. Results were presented as mean ± SD. **, P<0.01 vs. control; ##, P<0.01 vs. mimic control.
MiR-141 inhibitor reversed the effects of circRNA 010567-siRNA on hypoxia-induced cardiomyocyte injury
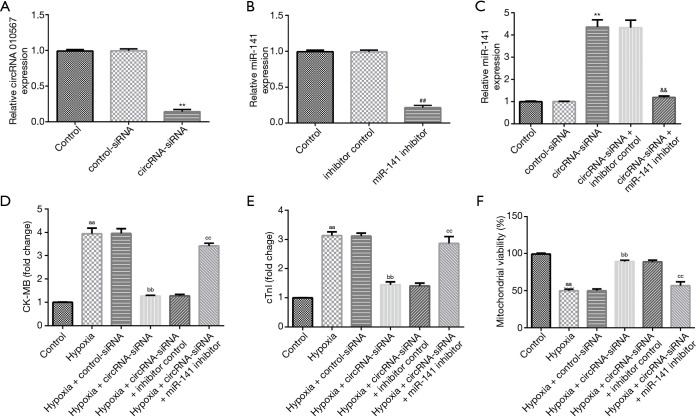
To verify the roles of circRNA 010567 on hypoxia-induced cardiomyocyte damage, circRNA 010567-siRNA, control-siRNA, miR-141 inhibitor, or inhibitor control were transfected in H9c2 cells for 48 h under hypoxic conditions. qRT-PCR analysis revealed that the circRNA 010567 level was significantly decreased in H9c2 cells after circRNA 010567-siRNA transfection compared to the control-siRNA group (Figure 2A). Furthermore, the miR-141 level was significantly reduced in miR-141 inhibitor-transfected H9c2 cells compared with the inhibitor control group (Figure 2B). In addition, the level of miR-141 was significantly enhanced in circRNA 010567-siRNA-transfected H9c2 cells, while this increase was reversed by the miR-141 inhibitor (Figure 2C). Our data revealed that circRNA 010567 could interfere with miR-141 expression.
Figure 2.
CircRNA 010567-siRNA protected H9c2 cells against hypoxia-induced cell damage. CircRNA 010567-siRNA, control-siRNA, miR-141 inhibitor, or inhibitor control were transfected into H9c2 cells under hypoxic conditions. (A) qRT-PCR analysis of circRNA 010567 levels in H9c2 cells after circRNA 010567-siRNA treatment. (B) The expression of miR-141 in miR-141 inhibitor-transfected cells. (C) MiR-141 expression in H9c2 cells after circ-siRNA and miR-141 inhibitor treatment. The release of CK-MB (D), cTnI (E), and mitochondrial viability (F) in different groups. Results were shown as mean ± SD. **, P<0.01 vs. control-siRNA; ##, P<0.01 vs. inhibitor control; &&, P<0.01 vs. circRNA-siRNA + inhibitor control; aa, P<0.01 vs. control; bb, P<0.01 vs. hypoxia + control-siRNA; cc, P<0.01 vs. hypoxia + circ-siRNA + inhibitor control.
Moreover, we detected the release of CK-MB and cTnI, and evaluated mitochondrial viability. Our data revealed that hypoxia obviously promoted the release of CK-MB (Figure 2D) and cTnI (Figure 2E), and inhibited mitochondrial viability (Figure 2F). However, we found that the release of CK-MB and cTnI were reduced, and that mitochondrial viability was promoted in the hypoxia + circRNA 010567-siRNA group, and these results were reversed by the miR-141 inhibitor.
MiR-141 inhibitor abolished the effects of circRNA 010567-siRNA on cell growth and apoptosis in hypoxia-induced cardiomyocytes
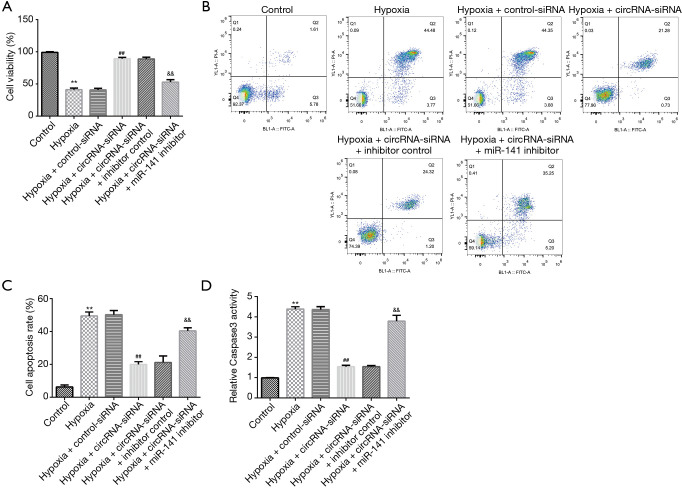
To further explore the potential underlying regulatory mechanism between miR-141 and circRNA 010567, an miR-141 inhibitor was used to evaluate the effects of circRNA 010567-siRNA on cell proliferation and apoptosis. The MTT assay and flow cytometry analysis demonstrated that hypoxia dramatically suppressed the viability of H9c2 cells, induced apoptosis, and enhanced the activity of caspase-3. In addition, we observed increased cardiomyocyte viability (Figure 3A), reduced apoptotic cells (Figure 3B,C), and inhibited caspase-3 activity (Figure 3D) in circRNA 010567-siRNA transfected cells. Moreover, these effects were abolished when the miR-141 inhibitor was co-transfected with circRNA 010567-siRNA in hypoxia-induced cardiomyocytes. These findings indicated that circRNA 010567-siRNA protected against hypoxia-induced cell damage by up-regulating miR-141 expression.
Figure 3.
CircRNA 010567-siRNA protected H9c2 cells against hypoxia-induced apoptosis. CircRNA 010567-siRNA, control-siRNA, miR-141 inhibitor, or inhibitor control were transfected into hypoxia-induced H9c2 cells. (A) The viability of cardiomyocytes was assessed using the MTT assay; (B) Fow cytometry analysis of apoptotic cardiomyocytes; (C) statistical analysis of apoptotic cells; (D) detection of caspase-3 activity. Results were presented as mean ± SD. **, P<0.01 vs. control; ##, P<0.01 vs. hypoxia + control-siRNA; &&, P<0.01 vs. hypoxia + circ-siRNA + inhibitor control.
DAPK1 directly interacted with miR-141
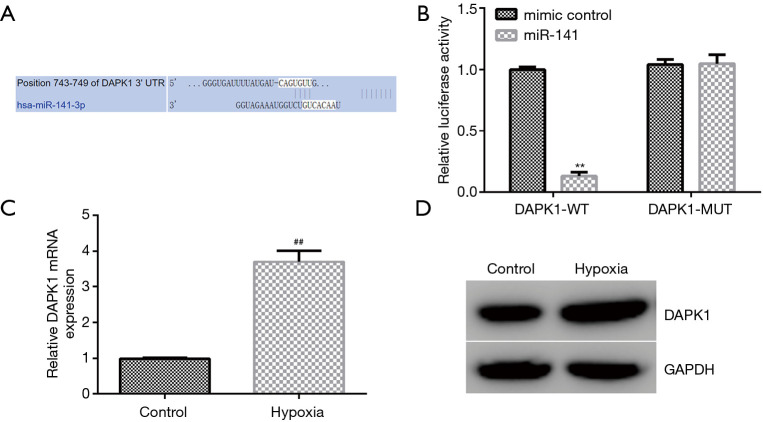
Furthermore, we demonstrated the molecular mechanism behind the involvement of miR-141 in hypoxia-induced cardiomyocyte injury. TargetScan was applied to illustrate the correlation between DAPK1 and miR-141. Our observations revealed that DAPK1 was a latent target of miR-141 (Figure 4A). Moreover, the dual-luciferase reporter system verified the binding site of miR-141 on DAPK1. We observed that miR-141 mimics significantly suppressed DAPK1-WT reporter luciferase activity, while there were no changes in DAPK1-MUT luciferase activity (Figure 4B). We also evaluated the level of DAPK1 in hypoxia-induced cardiomyocytes using qRT-PCR and western blot. As presented in Figure 4C,D, DAPK1 was significantly enhanced in hypoxia-induced cardiomyocytes. These observations demonstrated that miR-141 might be involved in hypoxia-induced cardiomyocyte injury through regulating DAPK1.
Figure 4.
MiR-141 binds to the 3-prime untranslated region (3’-UTR) of DAPK1. (A) TargetScan was applied to predict the binding of miR-141 to the 3’-UTR of DAPK1. (B) Dual-luciferase reporter gene system was applied to verify the relationship between miR-141 and DAPK1. (C) qRT-PCR and (D) western blot analysis of DAPK1 levels Results were presented as mean ± SD. **, P<0.01 vs. mimic control; ##, P<0.01 vs. control.
DAPK1-plasmid reversed the effects of miR-141 on hypoxia-induced cardiomyocyte injury
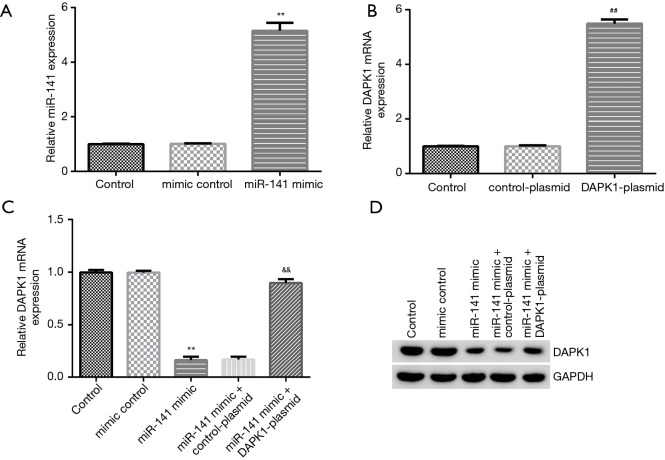
To demonstrate the relative roles of DAPK1 and miR-141 in hypoxia-induced cardiomyocyte injury, DAPK1-plasmid, control-plasmid, miR-141 mimic, or mimic control were transfected into cardiomyocytes for 48 h under hypoxic conditions. qRT-RCR and western blot were performed to evaluate cell transfection efficiency. Our data indicated that the miR-141 mimic markedly enhanced miR-141 expression in hypoxia-induced cardiomyocytes compared to the mimic control group (Figure 5A). Additionally, DAPK1-plasmid significantly promoted DAPK1 mRNA expression in cardiomyocytes compared to the control-plasmid group (Figure 5B). As displayed in Figure 5C and 5D, the miR-141 mimic significantly decreased DAPK1 mRNA and protein expression, while these results were abolished by DAPK1-plasmid.
Figure 5.
MiR-141 negatively regulated the expression of DAPK1 in hypoxia-induced cardiomyocytes. MiR-141 mimic, mimic control, control-plasmid, or DAPK1-plasmid were transfected into hypoxia-induced H9c2 cells for 48 h. (A) MiR-141 levels in the mimic control and miR-141 mimic groups. (B) Detection of DAPK1 mRNA in DAPK1-plasmid and control-plasmid groups. DAPK1 mRNA (C) and protein expression (D) in different groups were evaluated using qRT-PCR and western blot, respectively. Results were presented as mean ± SD. **, P<0.01 vs. mimic control; ##, P<0.01 vs. control-plasmid; &&, P<0.01 vs. miR-141 mimic + control-plasmid.
DAPK1-plasmid reversed the effects of the miR-141 mimic on hypoxia-induced cardiomyocyte damage
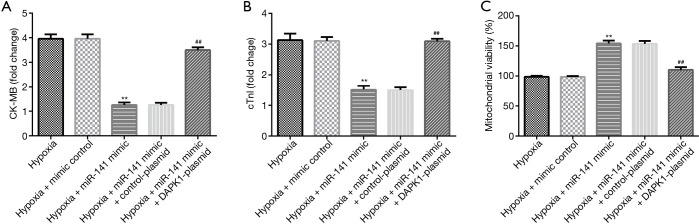
To further probe the roles of miR-141 and DAPK1 in hypoxia-induced cardiomyocytes, mimic control, miR-141 mimic, control-plasmid, or DAPK1-plasmid were transfected into H9c2 cells under hypoxic conditions. We then detected CK-MB and cTnI release, and mitochondrial viability in hypoxia-induced H9c2 cells. The release of CK-MB (Figure 6A) and cTnI (Figure 6B) were decreased, and mitochondrial viability (Figure 6C) was enhanced in the hypoxia + miR-141 mimic group compared to the hypoxia + mimic control group. Nevertheless, these findings were reversed in the hypoxia + miR-141 mimic + DAPK1-plasmid group. Based on these observations, we verified that the DAPK1-plasmid abolished the protective effects of the miR-141 mimic on H9c2 cell injury.
Figure 6.
DAPK1-plasmid reversed the roles of miR-141 in hypoxia-induced cardiomyocyte injury. MiR-141 mimic, mimic control, control-plasmid, or DAPK1-plasmid were transfected into hypoxia-induced H9c2 cells for 48 h. The secretion of (A) CK-MB and (B) cTnI in hypoxia-induced cardiomyocytes was determined. (C) Mitochondrial activity was evaluated in different groups. Results were presented as mean ± SD. **, P<0.01 vs. hypoxia + mimic control; ##, P<0.01 vs. hypoxia + miR-141 mimic + control-plasmid.
DAPK1-plasmid reversed the effects of the miR-141 mimic on cell growth and apoptosis in hypoxia-induced cardiomyocytes
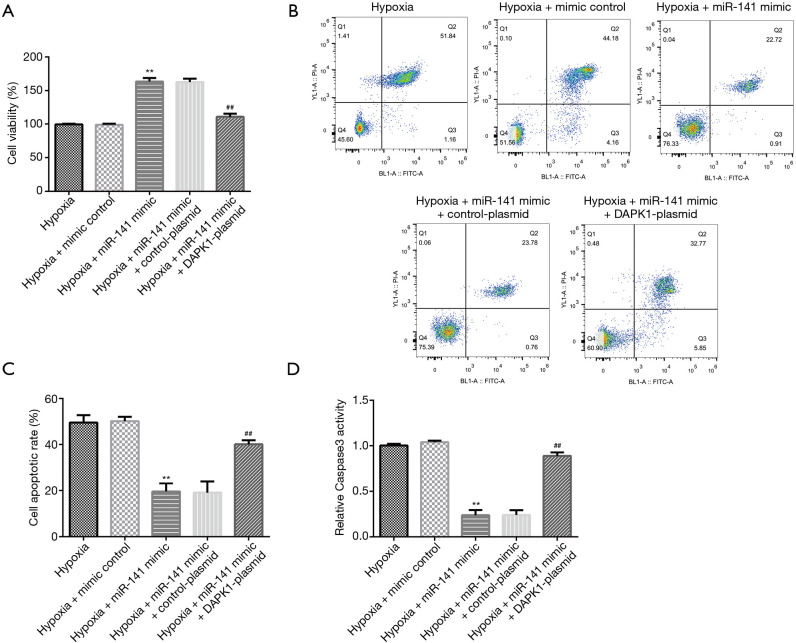
To lucubrate the responses of H9c2 cells co-regulated by miR-141 and DAPK1, we checked the effects of the miR-141 mimic and DAPK1-plasmid on cell proliferation and apoptosis using MTT and flow cytometry. As displayed in Figure 7A, compared to control group, the miR-141 mimic signally enhanced H9c2 cell viability (Figure 7A), reduced apoptosis (Figure 7B,C), and decreased caspase-3 activity (Figure 7D). However, these effects were reversed in the hypoxia + miR-141 mimic + DAPK1-plasmid group. In summary, our study revealed that circRNA 010567-siRNA played a protective role in hypoxia-treated myocardial cell damage through regulating the miR-141/DAPK1 axis.
Figure 7.
MiR-141 protected H9c2 cells against hypoxia-induced cell apoptosis by regulating DAPK1. MiR-141 mimic, mimic control, control-plasmid, or DAPK1-plasmid were transfected into hypoxia-induced H9c2 cells for 48 h. (A) MTT assay was conducted to assess the cardiomyocyte viability. (B,C) Detection of apoptotic cells using flow cytometry analysis. (D) Measurement of caspase-3 activity. Results were presented as mean ± SD. **, P<0.01 vs. hypoxia + mimic control; ##, P<0.01 vs. hypoxia + miR-141 mimic + control-plasmid.
Discussion
MI, defined as myocardial cell death caused by coronary artery occlusion, is the major cause of death and disability all over the world (28). Previous studies have revealed that MI decreases blood supply to the left ventricular myocardium, leading to the formation of fibrous scar tissues and remodeling of ventricular tissues (29,30). Hypoxia, a state of oxygen deficiency, is one of the main risks of MI (31). Under hypoxia, energy metabolism changes from mitochondrial respiration to anaerobic glycolysis, which causes a large number of physiological and pathological responses, leading to acidosis and cell necrosis. Therefore, it is considered that protecting myocardial cells from hypoxia is a promising strategy for MI treatment (32). H9c2 is a commercial myogenic cell line derived from embryonic rat ventricles. H9c2 cells displayed almost the same hypertrophic responses as primary cardiomyocytes and showed various characteristics of skeletal muscles (33). Therefore, H9c2 cells are commonly used for in vitro models. In this report, H9c2 cells were induced by hypoxia for 48 h.
CircRNAs, characterized by covalent 3’ and 5’ termini, are stable transcripts from different genomic locations (34). CircRNAs are abundant in the transcriptome of eukaryotes, and diverse genomic regions can produce different types of RNA loops, suggesting that circRNAs may play a crucial role in cellular processes (35). Recently, several reports have suggested that circRNAs are involved in many diseases, especially cardiovascular diseases. For example, Zhou et al. confirmed that circRNAs may be novel biomarkers for cardiovascular diseases (36). Previous reports have also confirmed that inhibition of circRNA 010567 inhibits myocardial fibrosis and improves MI infarction in rats (17). Unfortunately, up to now, the specific roles and mechanisms of circRNA 010567 in MI have not been fully elucidated.
In this study, we detected circRNA 010567 expression in hypoxia-induced H9c2 cardiomyocytes. We found that circRNA 010567 was memorably increased in hypoxia-treated H9c2 cells, compared to the control group. Increasing evidence has revealed the relationship between circRNAs and miRNAs, and their key roles in tumorigenesis (37,38). Our study demonstrated the circRNA-miRNA relationship using the dual-luciferase reporter gene system and the RIP assay. Our data revealed that there is a binding site between miR-141 and circRNA 010567. MiR-141, belongs to miR-200 family, is implicated in many cancers, while its function in MI has not yet been clarified (39). Up-regulation of miR-141 has been evidenced to suppress the invasion and migration of hepatocellular carcinoma and pancreatic cancer cells (40,41). In this report, we observed that the level of miR-141 was lower in hypoxia-induced H9c2 cardiomyocytes than that in control group. Our data indicated that circRNA 010567 was involved in MI by regulating miR-141.
Previous studies have revealed that 24 h of hypoxic treatment memorably promoted apoptosis and impaired H9c2 cell growth, suggesting that cell injury occurred in MI via hypoxia in vitro (42). We speculated that altered circRNA 010567 or miR-141 levels might influence the functions of H9c2 cells. We then evaluated the effects of circRNA 010567 on cell injury in hypoxia-induced H9c2 cells. Inhibitor control, circRNA 010567-siRNA, control-siRNA, or miR-141 inhibitor were transfected into H9c2 cells for 48 h, and qRT-PCR was used to measure transfection efficiency. We found that circRNA 010567-siRNA inhibited the level of circRNA 010567 in H9c2 cells. In addition, the miR-141 inhibitor dramatically reduced the miR-141 level, and circRNA 010567-siRNA significantly enhanced the miR-141 level in H9c2 cells, compared to the control-siRNA group. However, this effect was abolished in the circ-siRNA + miR-141 inhibitor group. We then examined the release of CK-MB and cTnI, which are important markers of MI (43), along with mitochondrial viability. We observed a obvious reduction in the release of CK-MB and cTnI, and increased mitochondrial viability in the hypoxia + circRNA 010567-siRNA group, and these results were reversed by the miR-141 inhibitor. It has also been confirmed that circRNAs often interact with miRNAs to mediate their biological processes, including cell viability, differentiation, and apoptosis (16). Our results were consistent with previous literature which revealed that circRNA 010567-siRNA increased cell viability and reduced cell apoptosis, and inhibited the activity of caspase-3, suggesting that inhibition of circRNA 010567 may alleviate hypoxia-induced H9c2 cell damage. Moreover, these results were reversed by the miR-141 inhibitor. Thus, circRNA 010567-siRNA can mitigate hypoxia-induced cardiomyocyte injury by up-regulating the expression of miR-141.
Furthermore, we illustrated the molecular mechanisms of miR-141 in hypoxia-induced cell damage in cardiomyocytes. TargetScan and the dual-luciferase reporter gene system suggested that DAPK1 directly interacted with miR-141. DAPK1, a positive mediator of apoptosis, was used as a tumor suppressor because of its ability to sensitize cells, including death receptors, cytokines, and oncogene-induced proliferation involved in tumorigenesis (44,45). To further explore the functional mechanisms of miR-141 in H9c2 cells, we explored the roles of miR-141 on hypoxia-induced cardiomyocyte damage. MiR-141 mimic, mimic control, control-plasmid, or DAPK1-plasmid were transfected into H9c2 cells for 48 h, and qRT-PCR analysis revealed that the miR-141 mimic markedly enhanced miR-141 expression and suppressed DAPK1 levels. Furthermore, this effect was abolished by the DAPK1-plasmid. Functional analysis demonstrated that the miR-141 mimic significantly inhibited the release of CK-MB and cTnI, and enhanced mitochondrial activity in hypoxia-induced H9c2 cells. Additionally, the miR-141 mimic significantly improved H9c2 cell viability, reduced cell apoptosis, and decreased caspase-3 activity in hypoxia-induced H9c2 cells. However, all these effects were reversed by the DAPK1-plasmid.
Taken together, our data revealed that circRNA 010567-siRNA may play a protective role in hypoxia-induced cardiomyocyte injury by regulating the miR-141/DAPK1 axis, indicating that circRNA 010567 may be a promising therapeutic target for MI treatment. However, this study did not investigate the effect of circRNA 010567 in MI in vivo, and this was a limitation of this study. We will further study the role of circRNA 010567 in MI in vivo in the future.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jtd-21-212
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-21-212
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-21-212). The authors have no conflicts of interest to declare.
(English Language Editor: C. Betlazar-Maseh)
References
- 1.Anand A, Shah A, Beshiri A, et al. Global Adoption of High-Sensitivity Cardiac Troponins and the Universal Definition of Myocardial Infarction. Clin Chem 2019;65:484-9. 10.1373/clinchem.2018.298059 [DOI] [PubMed] [Google Scholar]
- 2.Reindl M, Tiller C, Holzknecht M, et al. Prognostic implications of global longitudinal strain by feature-tracking cardiac magnetic resonance in ST-elevation myocardial infarction. Circ Cardiovasc Imaging 2019;12:e009404. 10.1161/CIRCIMAGING.119.009404 [DOI] [PubMed] [Google Scholar]
- 3.Lataro RM, Imori P, Santos ES, et al. Heart failure developed after myocardial infarction does not affect gut microbiota composition in the rat. Am J Physiol Gastrointest Liver Physiol 2019;317:G342-G348. 10.1152/ajpgi.00018.2019 [DOI] [PubMed] [Google Scholar]
- 4.Mouton AJ, Rivera OJ, Lindsey ML. Myocardial infarction remodeling that progresses to heart failure: a signaling misunderstanding. Am J Physiol Heart Circ Physiol 2018;315:H71-H79. 10.1152/ajpheart.00131.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filippone SM, Samidurai A, Roh SK, et al. Reperfusion therapy with rapamycin attenuates myocardial infarction through activation of AKT and ERK. Oxid Med Cell Longev 2017;2017:4619720. 10.1155/2017/4619720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer 2019;18:20. 10.1186/s12943-018-0935-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Zhang Y, Cai Q, et al. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol Cancer 2019;18:145. 10.1186/s12943-019-1078-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Chen Z, Hu G, et al. Roles of circular RNA in breast cancer: present and future. Am J Transl Res 2019;11:3945-54. [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Q, Zhang W, Wu Z, et al. Construction of a circular RNA-microRNA messengerRNA regulatory network in stomach adenocarcinoma. J Cell Biochem 2020;121:1317-31. 10.1002/jcb.29368 [DOI] [PubMed] [Google Scholar]
- 10.Cao S, Zhang D, Yuan J, et al. MicroRNA and circular RNA expression in affected skin of patients with postherpetic neuralgia. J Pain Res 2019;12:2905-13. 10.2147/JPR.S221615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S, Li X, Zheng H, et al. Loss of Super-Enhancer-Regulated circRNA Nfix Induces Cardiac Regeneration After Myocardial Infarction in Adult Mice. Circulation 2019;139:2857-76. 10.1161/CIRCULATIONAHA.118.038361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altesha MA, Ni T, Khan A, et al. Circular RNA in cardiovascular disease. J Cell Physiol 2019;234:5588-600. 10.1002/jcp.27384 [DOI] [PubMed] [Google Scholar]
- 13.Garikipati VNS, Verma SK, Cheng Z, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun 2019;10:4317. 10.1038/s41467-019-11777-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai L, Qi B, Wu X, et al. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J Mol Cell Cardiol 2019;130:10-22. 10.1016/j.yjmcc.2019.03.007 [DOI] [PubMed] [Google Scholar]
- 15.Zhang M, Wang Z, Cheng Q, et al. Circular RNA (circRNA) CDYL Induces Myocardial Regeneration by ceRNA After Myocardial Infarction. Med Sci Monit 2020;26:e923188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun 2017;487:769-75. 10.1016/j.bbrc.2017.04.044 [DOI] [PubMed] [Google Scholar]
- 17.Bai M, Pan CL, Jiang GX, et al. CircRNA 010567 improves myocardial infarction rats through inhibiting TGF-beta1. Eur Rev Med Pharmacol Sci 2020;24:369-75. [DOI] [PubMed] [Google Scholar]
- 18.Gjorgjieva M, Sobolewski C, Dolicka D, Correia DSM, Foti M. MiRNAs and NAFLD: from pathophysiology to therapy. Gut 2019;68:2065-79. 10.1136/gutjnl-2018-318146 [DOI] [PubMed] [Google Scholar]
- 19.Xu P, Wu Q, Lu D, et al. A systematic study of critical miRNAs on cells proliferation and apoptosis by the shortest path. BMC Bioinformatics 2020;21:396. 10.1186/s12859-020-03732-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan L, Guo N, Cao Y, et al. miRNA145 inhibits myocardial infarctioninduced apoptosis through autophagy via Akt3/mTOR signaling pathway in vitro and in vivo. Int J Mol Med 2018;42:1537-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yanagawa B, Lovren F, Pan Y, at al. miRNA-141 is a novel regulator of BMP-2-mediated calcification in aortic stenosis. J Thorac Cardiovasc Surg 2012;144:256-62. 10.1016/j.jtcvs.2011.10.097 [DOI] [PubMed] [Google Scholar]
- 22.Bialik S, Kimchi A. The death-associated protein kinases: structure, function, and beyond. Annu Rev Biochem 2006;75:189-210. 10.1146/annurev.biochem.75.103004.142615 [DOI] [PubMed] [Google Scholar]
- 23.Cohen O, Inbal B, Kissil JL. DAP-kinase participates in TNF-alpha- and Fas-induced apoptosis and its function requires the death domain. J Cell Biol 1999;146:141-8. 10.1083/jcb.146.999.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelled D, Raveh T, Riebeling C. Death-associated protein (DAP) kinase plays a central role in ceramide-induced apoptosis in cultured hippocampal neurons. J Biol Chem 2002;277:1957-61. 10.1074/jbc.M104677200 [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Shi X, Li H, et al. DAPK1 Signaling Pathways in Stroke: from Mechanisms to Therapies. Mol Neurobiol 2017;54:4716-22. 10.1007/s12035-016-0008-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu S, Zhai J, Yu J, et al. miR-98-5p Protects Against Cerebral ischemia/reperfusion Injury Through Anti-apoptosis and Anti-oxidative stress in Mice. J Biochem 2021;169:195-206. 10.1093/jb/mvaa099 [DOI] [PubMed] [Google Scholar]
- 27.Gong L, Chang H, Zhang J, et al. Astragaloside IV Protects Rat Cardiomyocytes from Hypoxia-Induced Injury by Down-Regulation of miR-23a and miR-92a. Cell Physiol Biochem 2018;49:2240-53. 10.1159/000493827 [DOI] [PubMed] [Google Scholar]
- 28.Reed GW, Rossi JE, Cannon CP. Acute myocardial infarction. Lancet 2017;389:197-210. 10.1016/S0140-6736(16)30677-8 [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Chen Y, Zhao Z, et al. Engineered exosomes with ischemic myocardium-targeting peptide for targeted therapy in myocardial infarction. J Am Heart Assoc 2018;7:e008737. 10.1161/JAHA.118.008737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruparelia N, Digby JE, Jefferson A, et al. Myocardial infarction causes inflammation and leukocyte recruitment at remote sites in the myocardium and in the renal glomerulus. Inflamm Res 2013;62:515-25. 10.1007/s00011-013-0605-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo Y, Tan J, Miao Y, et al. Effects of Microvesicles on Cell Apoptosis under Hypoxia. Oxid Med Cell Longev 2019;2019:5972152. 10.1155/2019/5972152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arriagada C, Silva P, Torres VA. Role of glycosylation in hypoxia-driven cell migration and invasion. Cell Adh Migr 2019;13:13-22. 10.1080/19336918.2018.1491234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Wang H, Zhang Y, et al. Pretreatment of ghrelin protects H9c2 cells against hypoxia/reoxygenation-induced cell death via PI3K/AKT and AMPK pathways. Artif Cells Nanomed Biotechnol 2019;47:2179-87. 10.1080/21691401.2019.1620253 [DOI] [PubMed] [Google Scholar]
- 34.Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell 2018;71:428-42. 10.1016/j.molcel.2018.06.034 [DOI] [PubMed] [Google Scholar]
- 35.Stoll L, Sobel J, Rodriguez-Trejo A, et al. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol Metab 2018;9:69-83. 10.1016/j.molmet.2018.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Q, Zhang Z, Bei Y, et al. Circular RNAs as novel biomarkers for cardiovascular diseases. Adv Exp Med Biol 2018;1087:159-70. 10.1007/978-981-13-1426-1_13 [DOI] [PubMed] [Google Scholar]
- 37.Tu C, Du T, Ye X, et al. Using miRNAs and circRNAs to estimate PMI in advanced stage. Leg Med (Tokyo) 2019;38:51-7. 10.1016/j.legalmed.2019.04.002 [DOI] [PubMed] [Google Scholar]
- 38.Liu S, Xie X, Lei H, et al. Identification of key circRNAs/lncRNAs/miRNAs/mRNAs and pathways in preeclampsia using bioinformatics analysis. Med Sci Monit 2019;25:1679-93. 10.12659/MSM.912801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong H, Weng C, Bai R, et al. The regulatory network of miR-141 in the inhibition of angiogenesis. Angiogenesis 2019;22:251-62. 10.1007/s10456-018-9654-1 [DOI] [PubMed] [Google Scholar]
- 40.Yao B, Xue Y, Liu Z, et al. Over-expression of miR-141 inhibits the proliferation, invasion and migration of hepatocellular carcinoma MHCC-97H cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2016;32:1083-7. [PubMed] [Google Scholar]
- 41.Xu L, Li Q, Xu D, et al. hsa-miR-141 down-regulates TM4SF1 to inhibit pancreatic cancer cell invasion and migration. Int J Oncol 2014;44:459-66. 10.3892/ijo.2013.2189 [DOI] [PubMed] [Google Scholar]
- 42.Sundaram R L, Sali VK, Vasanthi HR. Protective effect of rutin isolated from Spermococe hispida against cobalt chloride-induced hypoxic injury in H9c2 cells by inhibiting oxidative stress and inducing apoptosis. Phytomedicine 2018;51:196-204. 10.1016/j.phymed.2018.09.229 [DOI] [PubMed] [Google Scholar]
- 43.Fan J, Ma J, Xia N, et al. Clinical value of combined detection of CK-MB, MYO, cTnI and Plasma NT-proBNP in diagnosis of acute myocardial infarction. Clin Lab 2017;63:427-33. 10.7754/Clin.Lab.2016.160533 [DOI] [PubMed] [Google Scholar]
- 44.Wu B, Yao H, Wang S, et al. DAPK1 modulates a curcumin-induced G2/M arrest and apoptosis by regulating STAT3, NF-kappaB, and caspase-3 activation. Biochem Biophys Res Commun 2013;434:75-80. 10.1016/j.bbrc.2013.03.063 [DOI] [PubMed] [Google Scholar]
- 45.Li D, Xu D, Xu Y, et al. MicroRNA-141-3p targets DAPK1 and inhibits apoptosis in rat ovarian granulosa cells. Cell Biochem Funct 2017;35:197-201. 10.1002/cbf.3248 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as