Abstract
Background
Epidemiological studies have found that hyperglycemia, is an independent risk factor for colorectal cancer (CRC), increasing colon cancer incidence and affecting the recurrence, metastasis, and prognosis in colon cancer patients. However, the intercorrelation between hyperglycemia and CRC risk is still unknown, In the present study, we sought to determine whether gene markers, which act in CRC with hyperglycemia, are silenced in CRC without hyperglycemia.
Methods
In order to characterize the mechanism of functional genes associated with CRC with hyperglycemia, A total 24 CRC and matched controls were sequenced. Through bioinformatics analysis includes differential expression analysis, functional enrichment, new isoform prediction and alternative splicing event identification to found biomarker genes related to CRC development.
Results
CRC patients with hyperglycemia were compared with patients without hyperglycemia, and we found that 21 genes were upregulated and 27 were downregulated. Further study showed that these genes are possibly of key genes involved in CRC development with hyperglycemia, such as mannan-binding lectin-associated serine protease 3 (MASP3), which has an immunological role in the activation of the complement system. Based on our comprehensive analysis, a cis-regulatory network for hyperglycemic CRC was reconstructed.
Conclusions
Protein-protein interactions revealed the mechanisms of molecules involved in the interaction of hyperglycemia and cancer development. Our results provide further information on the metabolic pathway interaction with cancer pathways and elucidated the mechanisms of hyperglycemic factors function in cancer development from a transcriptomic perspective.
Keywords: Transcriptome, hyperglycemia, colorectal cancer (CRC)
Introduction
The incidence of colorectal cancer (CRC) is steadily increasing worldwide. Epidemiological data collected over the past decade indicate that the risk of CRC is elevated in those with metabolic syndrome (e.g., obesity, type 2 diabetes, and hypertension) or disorder of plasma or serum components (hypertriglyceridemia, hyperglycemia, and low-density lipoprotein cholesterol), and elevated markers of hyperinsulinemia or insulin resistance [(IR) insulin and C-peptide] (1,2). The mechanisms underlying these associations with CRC are unknown, but may involve the influence of hyperinsulinemia with stimulation of the insulin-like growth factor 1 (IGF-1) axis, which enhances free or bioavailable concentrations of IGF-1, as well as other factors, such as increased cytokine production (3).
As both hyperglycemia and cancer are major diseases that spread on a global scale and develop rapidly, these diseases are influenced by many factors, both genetic and environmental. Therefore, an understanding of the complex interactions between these 2 diseases is of paramount importance for their prevention and treatment. Type 2 diabetes mellitus is commonly characterized by prolonged hyperinsulinemia, IR, and progressive hyperglycemia (4). Hyperinsulinemia and IR are correlated concepts during the pathogenesis of diabetes mellitus. IR is a condition of type 2 diabetes mellitus. Signs and symptoms of IR include high blood sugar. IR is when cells fail to respond to the normal actions of the hormone insulin and can further lead to hyperglycemia. Beta cells in the pancreas subsequently increase their production of insulin, further contributing to hyperinsulinemia. This often remains undetected and can contribute to type 2 diabetes (5).
Hyperglycemia, or high blood sugar, is another progressive abnormal condition, in which an excessive amount of glucose circulates in the blood plasma with concentrations >11.1 mmol/L (200 mg/dL). Patients with a consistent range between ~5.6 and ~7 mmol/L (100–126 mg/dL), according to American Diabetes Association guidelines, are considered hyperglycemic, while patients with >7 mmol/L (126 mg/dL) are generally considered to have diabetes (6). Chronic hyperglycemia that persists even in fasting states is most commonly caused by diabetes mellitus. Certain medications (e.g., corticosteroids, octreotide, and antipsychotic agents) and other conditions, including stroke or myocardial infarction; dysfunction of the thyroid, adrenal, and pituitary glands; pancreatic diseases; sepsis and certain infections; intracranial diseases, encephalitis, and brain tumors, can cause hyperglycemia (7,8).
Many previously published studies have attempted to determine the association between hyperglycemia and CRC risk using epidemiological methods. Blood glucose concentrations have been studied in relation to CRC and adenoma, and it has been indicated that serum glucose levels may be a potential marker of CRC. Early detection and intervention for controlling elevated glucose levels may be indicated as a way to prevent carcinogenesis. In a prospective study in Italy, elevated glucose was associated with a significantly elevated cancer risk for both men and women [combined risk ratio (RR): 1.80] (9). In another large study of Norwegian men and women, women with elevated blood glucose concentrations had a significantly elevated relative RR of 1.98 for CRC, but no significant association was seen in men (10). In the Cardiovascular Health Study, individuals in the top quartile of fasting glucose (RR: 1.8) or 2-h glucose (RR: 2.4) had an elevated risk of CRC relative to those in the bottom quartiles (11).
Glycosylated hemoglobin is a marker of average glycemia, and elevated glycosylated hemoglobin is used as an indicator of chronic hyperglycemia. In the II cohort, higher glycosylated hemoglobin was associated with an elevated risk of CRC (12). In an Italian study, Giovanni et al. assessed the relationship between fructosamine and colorectal adenomas (a precursor to colorectal cancer) in a case-control study. Individuals with higher concentrations of fructosamine had a 2.3-fold elevated risk of adenoma compared with those with lower concentrations (13). In another previously published study, elevated insulin and glucose were found to be associated with increased adenoma risk and decreased apoptosis in normal rectal mucosa. Those in the highest quartile of insulin (odds ratio: 2.2) and glucose (OR: 1.8) were more likely to have an adenoma compared with the lowest quartiles (14). These findings suggest that insulin may act early in the adenoma-carcinoma sequence to promote the development of colorectal adenoma by decreasing apoptosis in normal mucosa. Moreover, based on 4 human CRC cell lines (SW480, SW620, LoVo, and HCT116), high glucose, compared with physiological normal glucose concentration (5 mM), leads to increased cell proliferation. This enhances the hyperglycemic affects, which includes the antiproliferative effect of fluorouracil (5-FU) on CRC cells (15).
For metastatic CRC, an examination of 121 CRC patients showed that diabetes in CRC was associated with higher blood hemoglobin A1C and higher renin–angiotensin–aldosterone concentrations in primary tumors, and a higher incidence of liver metastasis than in non-diabetic cases (16).
Hyperglycemia shows a predominating consistent correlation with high CRC risk. However, epidemiological studies do not indicate how hyperglycemia or diabetes and predisposes to CRC development or whether some common mechanisms exist. To the best of our knowledge, there is a lack of studies elucidating whether cancer itself renders patients more prone to the development of diabetes.
In the present study, we utilized advanced transcriptome technique based on high-throughput next-generation sequencing to elucidate gene regulations in CRC, and investigated the intercorrelation between hyperglycemia and CRC risk. We sought to determine whether gene markers, which act in CRC with hyperglycemia, are silenced in CRC without hyperglycemia. We analyzed the differences between CRC with and without hyperglycemia and elucidated the mechanisms of hyperglycemic factors function in cancer development from a transcriptomic perspective. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-587).
Methods
Sample collection
A total 24 CRC and matched controls were enrolled, including 2 subtypes of CRC, which were allocated to the hyperglycemia and non-hyperglycemia group. Four libraries were created for the 2 CRC groups (6 patients were merged in the same group in each tumor or matched control subgroup). 6 CRC patients with normal glycemia level (T) and 6 matched controls (N), 6 CRC patients (DM-T) with hyperglycemia and 6 matched controls (DM-N).
The study conformed to the provisions of the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee board of The First Affiliated Hospital of Huzhou University (No. 20170118004). Informed consent was taken from all the patients.
RNA sample preparation
Experimental protocols for RNA sequence were performed according to the manufacturer’s instructions. Briefly, RNA was isolated from about tumor tissue with TRIzol reagent (Invitrogen, Grand Island, NY, USA), and total RNA was purified using the Qiagen RNeasy mini kit (Qiagen, Valencia, CA, USA). Purified RNA was analyzed on a ND-8000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA), and agarose electrophoresis using a 2100-Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), to determine the quantity. RNA samples were used if there was no smear on the agarose. RNA samples were also used if the 260/280 ratio was >2.0 and the RNA integrity number was >7. Single- and double-stranded cDNA were synthesized from mRNA samples using SuperScript II (Invitrogen, Carlsbad, CA, USA). High-quality total RNA (1 µg) was used as the starting material. The Truseq RNA sample preparation kit (Baoli Medical Technology Co. LTD., Beijing, China) was used for the mRNA purification and fragmentation, the first-strand cDNA synthesis, and the second-strand cDNA synthesis. The double-stranded cDNA was then purified for end repair, dA tailing, adaptor ligation, and DNA fragment enrichment. The size of DNA was checked using a DNA specific chip, such as the Agilent DNA-1000 on the Agilent Technologies 2100 Bioanalyzer. The libraries were quantified using Qubit (Invitrogen, Carlsbad, CA, USA) according to the Qubit user guide.
Read mapping, differential expression analysis, and functional enrichment
The raw paired end reads were trimmed and quality controlled by SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) with default parameters. Clean reads were then separately aligned to then reference genome with orientation mode using TopHat software (http://tophat.cbcb.umd.edu/). To identify differentially expressed genes (DEGs) between the 2 samples, the expression level for each transcript was calculated using the fragments per kilobase of exon per million mapped reads method. Cufflinks (http://cufflinks.cbcb.umd.edu/) was used for the differential expression analysis (17). DEGs between the 2 samples were selected using the following criteria: (I) the logarithmic of fold change was >1; and (II) the false discovery rate (FDR) was <0.05. To understand the functions of the DEGs, gene oncology (GO) functional enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were done using Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do) (18). DEGs were significantly enriched in GO terms and metabolic pathways when their Bonferroni-corrected P value was <0.05.
New isoform prediction and alternative splicing event identification
The TopHat-Cufflinks pipeline was used to predict gene isoforms from our RNA-seq data. In TopHat (version 2.0.0, http://tophat.cbcb.umd.edu/), the option “min-isoform-fraction” was disabled (19); instead, “coverage-search”, “butterfly-search”, and “microexon-search” were used. The expected fragment length was set to 200 bp, and “small-anchor-fraction” was set to 0.08, which requires at least 8 bp on each side of an exon junction for our 100-bp RNA-seq data. Cuffcompare was used to compare and merge the reference annotation and the isoform predictions. All the alternative splicing events that occurred in our sample were identified using the in-house perl scripts, which were obtained from Keren et al. (20). Only isoforms that were similar to the reference or comprised novel splice junctions were considered, and we detected 7 types of alternative splicing events as follows: (I) exon skipping (ES); (II) intron retention; (III) alternative 5’ splice site (5S); (IV) alternative 3’ splice site (3S); (V) alternative first exon [3 prime untranslated region (3’UTR)]; (VI) alternative last exon (5’UTR); and (VII) others.
Gene cis-regulatory network analysis
DEGs and isoform transcripts were used to extract their promoter sequences. HOMER was used to discover the enriched motifs in the promoter sequences. Significant de novo motifs and known motifs were detected and used to reconstruct the gene cis-regulatory network.
Protein-protein interaction (PPI) analysis
Human PPI was downloaded from the Human Protein Reference Database (HPRD). For each comparison of significantly expressed genes, we mapped these genes on the PPI network and obtained the subnetwork for each CRC subtype. Visualization of the network was performed using Cytoscape software.
Statistical analysis
The differences between groups were tested by Tukey’s Multiple Comparison Test. P values less than 0.05 and 0.01 were considered significant. Statistical test was performed using GraphPad PRISM version 5.0 (La Jolla, CA, USA). P<0.05 was designated as the cut-off for statistical significance. DEGs were significantly enriched in GO terms and metabolic pathways when their Bonferroni-corrected P value was <0.05.
Results
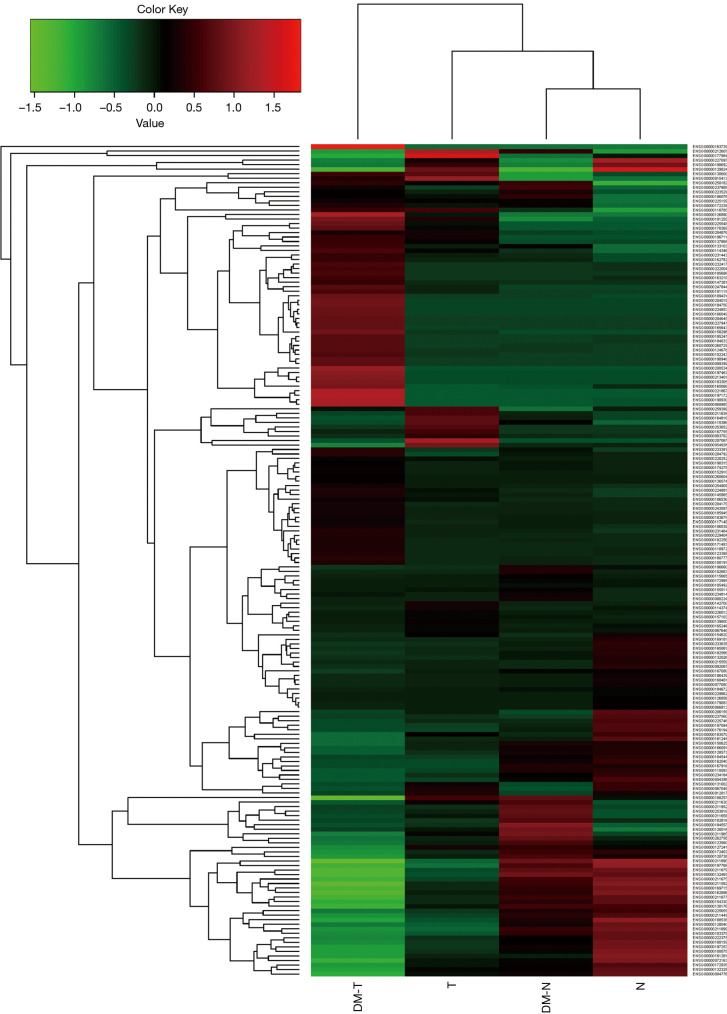
To study hyperglycemic function in CRC risk, 24 cancer patients were enrolled in the present study; 6 CRC patients with normal glycemia level (T) and 6 matched controls (N), 6 CRC patients (DM-T) with hyperglycemia and 6 matched controls (DM-N). Before performing high-throughput sequencing, we pooled 6 samples in the same group together, and created 4 libraries representing the T and N and T-DM and N-DM groups. We then conducted transcriptome library and performed sequencing using a Hiseq2500 sequencer for each group library. We used human genome in the Ensembl database as the reference (21), and mapped each filtered read on it using TopHat (22). We obtained mapped transcriptome bam files for gene and isoform expression downstream analyses via SAM tools (23). In general, 90.99%, 92.55%, 92.39%, and 85.81% mapped rates were obtained for DM-N, DM-T, N, and T, respectively. Therefore, most of the high-quality reads can be mapped to the human genome to ensure downstream analysis of the transcriptome. Aligned reads were further processed to compute gene and isoform transcripts expression as FPKM via Cufflinks software (24). The clustered gene profiling is shown in Figure 1.
Figure 1.
Clustering of gene expression profile pattern for colorectal cancer with matched controls.
CRC-related expression genes in patients with normal blood sugar level
After comparing the N and T groups, we obtained 15 upregulated genes and 7 downregulated genes (Table 1), as well as 37 upregulated isoform transcripts and 10 downregulated isoforms transcripts in tumors with normal blood sugar level (without hyperglycemia, FDR <0.05 and |logfold change| >1) (Table S1). The Pearson correlation between N and T among the global gene expression profile was 0.9184 for isoforms transcripts and 0.9183 for genes, respectively. We introduced the biologic functions for these DEGs and found that enriched functional GO terms related to metallodipeptidase activity, γ-aminobutyric acid:sodium symporter activity, short-chain fatty acid import, short-chain fatty acid import, cellular amide catabolic process, monocarboxylic acid transport, response to calcium ion, and response to purine-containing compound. Similarly, we performed a pathway enrichment analysis using multiple pathway databases to categorize the significant DEGs in tumors without hyperglycemia. We found that leukotriene biosynthesis (P=0.004), alternative complement pathway activation (P=0.007), Wnt lrp6 pathway signaling, integrin cell surface interactions, and extracellular matrix-receptor interactions are functionally active during tumor development without hyperglycemia.
Table 1. Statistics of transcriptome library and RNA-seq data production.
| Library | Raw reads | Raw bases (bp) | ≥ Q20 (%) | Filtered reads | Filtered bases (bp) | ≥ Q20 (%) | Mapping rate (%) |
|---|---|---|---|---|---|---|---|
| DM-N | 143,488,864 | 14,492,375,264 | 92.27 | 137,834,871 | 13,191,087,921 | 96.075 | 90.99 |
| DM-T | 121,649,882 | 12,286,638,082 | 92.69 | 117,055,765 | 11,240,687,930 | 96.17 | 92.55 |
| N | 113,419,514 | 11,455,370,914 | 91.74 | 108,992,176 | 10,361,165,082 | 96.155 | 92.39 |
| T | 123,049,788 | 12,428,028,588 | 92.83 | 118,605,931 | 11,399,230,908 | 96.225 | 85.81 |
| Mean | 125,402,012 | 12,665,603,212 | 92.38 | 120,622,186 | 11,548,042,960 | 96.156 | 90.40 |
| Median | 122,349,835 | 12,357,333,335 | 92.48 | 117,830,848 | 11,319,959,419 | 96.163 | 91.69 |
CRC-related expression genes in patients with hyperglycemia
As with the case-control comparison method, we directly conducted a differential gene expression analysis between CRC with matched controls (all with hyperglycemia) (DM_N5 vs. DM_T5). Twenty-six genes were found to be upregulated and 29 were found to be downregulated (Table 2); isoform transcripts were also found (Table S2). Using enrichment analysis, most of these significantly expressed genes were found to participate in the regulation of gene expression, macromolecule metabolic process, metabolic process, RNA biosynthetic process, and RNA metabolic process, complement activation, the cellular macromolecule biosynthetic process, antigen binding, and high-affinity sodium:dicarboxylate symporter activity. In significantly enriched pathways, we found that genes had an active function in the sodium-coupled sulfate, di- and tri-carboxylate transporters (P=0.004), acetylcholine neurotransmitter release cycle (P=0.008), interleukin (IL)-4-mediated signaling event (P=0.014), γ-interferon signaling, lectin-induced complement pathway, α-linolenic acid metabolism, linoleic acid metabolism, mitogen-activated protein kinase (MAPK) signaling, type 2 diabetes mellitus (ENSG00000184557), fat digestion and absorption (ENSG00000188257), inflammation mediated by chemokine and cytokine signaling, notch-mediated HES/HEY network, vascular endothelial growth factor signaling, adipocytokine signaling pathway [suppressor of cytokine signaling 3 (SOCS3), ENSG00000184557], and insulin signaling pathways (SOCS3, ENSG00000184557). From the phenotypes of CRC and hyperglycemia, the differential genes are related to pathways related to the progression of type 2 diabetes and IR. IR is the main step toward the progression of type 2 diabetes, and has been linked to increased circulating levels of cytokines, leading to chronic low-grade inflammation (25). The increased expression of SOCS3 mediates the inhibitory effects of IL-6 on insulin signaling and glucose metabolism (26,27). Chronically elevated IL-6 levels lead to the increased expression of SOCS3 proteins in skeletal muscle, liver, and adipose tissue (28). The role of elevated IL-6 levels on insulin signaling include the activation of Adenosine activates protein kinases (AMPK) signaling pathway and the involvement of leptin and SOCS3. Specifically, in chronic disease states, increased IL-6 levels are thought to play a critical role in the regulation of IR in peripheral tissue, and has been used as a marker of IR.
Table 2. Differentially expressed genes in the N vs. T comparison group.
| Gene name | log2FC (T/N) | log2FC (N/T) | P value | FDR | Upregulation/downregulation (T4/N2) |
|---|---|---|---|---|---|
| DPEP1 | 6.426717452 | –6.426717452 | 5.00E−05 | 0.0265652 | Upregulation |
| EPYC | 1.546131404 | –1.546131404 | 5.00E−05 | 0.0265652 | Upregulation |
| AQP8 | –4.307850642 | 4.307850642 | 5.00E−05 | 0.0265652 | Downregulation |
| REG1A | 4.366525052 | –4.366525052 | 5.00E−05 | 0.0265652 | Upregulation |
| SPP1 | 5.553699867 | –5.553699867 | 5.00E−05 | 0.0265652 | Upregulation |
| DKK2 | 2.108899091 | –2.108899091 | 5.00E−05 | 0.0265652 | Upregulation |
| SLC6A1 | 2.47446979 | –2.47446979 | 5.00E−05 | 0.0265652 | Upregulation |
| HS3ST6 | –2.618363183 | 2.618363183 | 5.00E−05 | 0.0265652 | Downregulation |
| KLK6 | 2.447624156 | –2.447624156 | 5.00E−05 | 0.0265652 | Upregulation |
| GSG1L | –1.101038179 | 1.101038179 | 5.00E−05 | 0.0265652 | Downregulation |
| ATOH1 | 1.404973948 | –1.404973948 | 5.00E−05 | 0.0265652 | Upregulation |
| FAM150A | 1.922802968 | –1.922802968 | 5.00E−05 | 0.0265652 | Upregulation |
| CFD | –5.887352053 | 5.887352053 | 5.00E−05 | 0.0265652 | Downregulation |
| AC021218.2 | 1.654944152 | –1.654944152 | 5.00E−05 | 0.0265652 | Upregulation |
| SNORA72 | 5.507864395 | –5.507864395 | 5.00E−05 | 0.0265652 | Upregulation |
| IGLV4-60 | 2.991968927 | –2.991968927 | 5.00E−05 | 0.0265652 | Upregulation |
| IGHA2 | –5.693133995 | 5.693133995 | 5.00E−05 | 0.0265652 | Downregulation |
| RP11-40H20.2 | 2.329947348 | –2.329947348 | 5.00E−05 | 0.0265652 | Upregulation |
| AP001434.2 | 1.910467888 | –1.910467888 | 5.00E−05 | 0.0265652 | Upregulation |
| RP11-505P4.7 | –1.88179381 | 1.88179381 | 5.00E−05 | 0.0265652 | Downregulation |
| NKX6-3 | –3.152243462 | 3.152243462 | 1.00E−04 | 0.0465128 | Downregulation |
| RP11-887P2.3 | 1.839536389 | –1.839536389 | 1.00E−04 | 0.0465128 | Upregulation |
FDR, false discovery rate; N, normal group; T, Tumour group.
Hyperglycemic-specific genes and function in CRC development
To analysis how hyperglycemia specifically functions in CRC development, we compared the active genes in tumor patients with hyperglycemia with those active only in tumor patients without hyperglycemia from transcriptome. We found that there were no overlapping gene and isoform transcript functions in tumor patients with hyperglycemia versus tumor patients without. Therefore, total DM-T upregulated or downregulated genes and isoform transcripts are only significantly expressed in tumors with hyperglycemia, but not in tumors with normal blood sugar level. In contrast, total T upregulated or downregulated genes and isoform transcripts are only differentially expressed in tumors with normal sugar level, but not in tumors with hyperglycemia. This suggests that DEGs in tumors with hyperglycemia mostly function in relation to hyperglycemic-specific regulatory modules/pathways and are associated with CRC risk. Different molecular gene profiles can be identified as classifiers for the 2 subtypes: CRC with and CRC without hyperglycemia.
Similarly, we also compared CRC with hyperglycemia to CRC without hyperglycemia and obtained total 48 significantly expressed genes, including 21 upregulated genes and 27 downregulated genes (Table S3). A total of 56 isoforms were detected as significantly expressed transcripts; 20 were upregulated and 36 were downregulated (Table S4). Most of these genes were involved in the functions of ribonucleoprotein granule and the regulation of the nuclear-transcribed mRNA catabolic process, positive regulation of mRNA 3’-end processing, and deadenylation-dependent decay; as well as in the Wnt lrp6 signaling (P=0.01), γ-interferon signaling (P=0.02), type 2 diabetes mellitus (P=0.03), and Erythropoietin (EPO) signaling pathways by utilizing GO and pathway analysis. Some DEGs are related to the pathogenesis of diabetes. As reported in previously published studies, SOCS3 involvement in the type 2 diabetes mellitus pathway is related to the pathogenesis of obesity and is associated with metabolic abnormalities (28). It is a major negative regulator of both leptin and insulin signaling. Muscle SOCS3 overexpression can suppresses leptin-regulated genes involved in fatty acid oxidation and mitochondrial function, and mediate insulin and leptin resistance in obesity. Another previously published study also revealed a functional and mechanistic link between miR-185 and SOCS3 in the pathogenesis of diabetes; miR-185 level was found to be inversely correlated with SOCS3 expression in diabetic patients (29).
To understand the background transcriptome of normal tissues in the hyperglycemic or non-hyperglycemic group, we compared the DM-N with N. Ten upregulated and 17 downregulated genes were detected (Figure 1). We obtained a total of 168 significantly expressed transcripts; 31 were upregulated and 137 were downregulated.
We created a Venn plot to represent the relationship between each group of comparisons, including DM-T versus T, DM-N versus N, DM-T versus DM-N, and T versus N (Figure 2); tumor subgroup gene comparisons and gene expression trends are shown. The clustering of overlapping genes and hyperglycemic-specific genes are shown in Figure 3A. The DM-T versus DM-N group had the most significantly expressed gene set, which was hyperglycemic-specific gene function modules associated with CRC development Figure 3B. After comparison with the DM-T versus T group, we found that there was a total of 13 genes in both the DM-T versus T group and the DM-T versus DM-N group, including Melanoma Antigen Genes B2 (MAGEB2), homeobox-C 11 (HOXC11), Human t-Complex Protein 11 (TCP11), Manna-binding lectin-associated serine protease 1 (MASP1), GATA-binding protein 4 (GATA4), Melanoma Antigen-A2B (MAGEA2B), Melanoma Antigen-A11 (MAGEA11), Phospholipase A2-IIA (PLA2G2A), CSAG3, CSAG1, CXorf61, RP11-543D5.1, and AC009133.21. These genes are possibly of key genes involved in CRC development with hyperglycemia. For example, little is known about the role of MASP3 in tumor development. MASP3 has an immunological role through the activation of the complement system (30). Two different human MASP proteins have been described. MASP1 and MASP3 are encoded by the same gene, generated by alternative splicing (31). Functional analyses have confirmed that MASP3 is a bona fide candidate antitumor protease, as it is either mutated or downregulated in tumor samples, and it plays a role in the regulation of cell proliferation and subcutaneous tumor formation (32). MASP1 is active in immune response, and was also identified as a novel candidate high-risk gene for CRC in an analysis of whole-exome sequencing data in 16 high-risk CRC families (33 3’UTR) (33).
Figure 2.
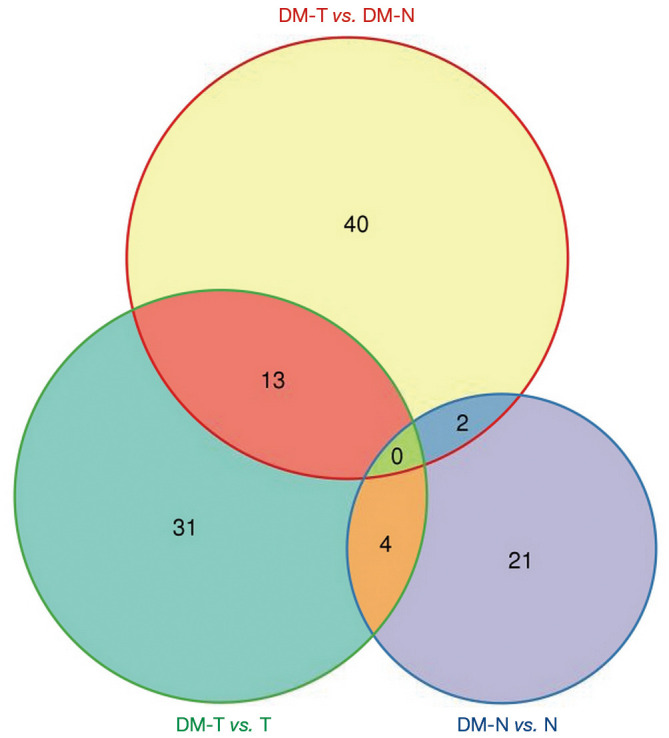
Venn plot for multiple comparisons. DM-N, diabetes mellitus-normal; DM-T, diabetes mellitus-tumour; N, normal group; T, tumour group.
Figure 3.
Scatter plot and volcano plot for genes and isoforms in the comparison of the N and T group. (A) Gens scatter plot of (N vs. T), (B) isoforms scatter plot of (N vs. T). N, normal group; T, tumour group.
We also found that the DM-N versus the N group had DEGs (e.g., SOCS3, ENSG00000184557) that participate in type 2 diabetes mellitus pathways; those DEGs are related to the metabolic regulation of blood sugar. This suggests that, even in normal tissue with a transcriptome background, hyperglycemia must affect basic transcriptome of the human body, and then influence CRC tissue via dysfunctional mechanisms.
Alternative splicing in CRC with normal sugar level
For each gene, we tested 6 subtypes of alternative splicing (AS) events, including 3S, 3’UTR, 5S, 5’UTR, ES, and IR. Intergenic and non-intergenic AS events were all considered.
For CRC without hyperglycemia, we found that 4,333 AS events in 2,475 gene transcripts had at least 1 subtype of AS events in CRC without hyperglycemia in non-intergenic regions, as well as 1,867 intergenic AS events. We found that 3’UTR (18.28%), 3S (17.76%), and 5S (14.89%) were the 3 most frequent AS events occurring in CRC tissues without hyperglycemia.
A total of 2,855 AS events in 1,742 gene transcripts were found to have at least 1 subtype of AS events in CRC-matched controls without hyperglycemia in non-intergenic regions, as well as 1,224 intergenic AS events alternatively. The combined AS event (which may contain multiple AS events in the same gene) types and corresponding 1,560 genes are shown in Table 3. 3’UTR (22.69%), 3S (21.09%), and 5S (12.56%) were also the 3 most frequent AS events occurring in CRC-matched control tissues without hyperglycemia. We noticed that all 3 subtypes’ fractions were larger than those of tumor tissues.
Table 3. Differentially expressed genes in the DM-N vs. DM-T comparison group.
| Gene name | log2FC (DM-T/DM-N) | log2FC (DM-T/DM-N) | P value | FDR | Upregulation/downregulation (DM-T/DM-N) |
|---|---|---|---|---|---|
| MAGEB2 | 3.345509924 | –3.345509924 | 5.00E−05 | 0.0265652 | Upregulation |
| VGLL1 | 2.962611949 | –2.962611949 | 5.00E−05 | 0.0265652 | Upregulation |
| SGCG | –1.109193378 | 1.109193378 | 5.00E−05 | 0.0265652 | Downregulation |
| POU6F2 | 0.451340372 | –0.451340372 | 5.00E−05 | 0.0265652 | Upregulation |
| SMYD1 | –2.232835711 | 2.232835711 | 5.00E−05 | 0.0265652 | Downregulation |
| SLC5A7 | –2.799472802 | 2.799472802 | 5.00E−05 | 0.0265652 | Downregulation |
| FGF23 | 0.81775422 | –0.81775422 | 5.00E−05 | 0.0265652 | Upregulation |
| EGR1 | –5.237626829 | 5.237626829 | 5.00E−05 | 0.0265652 | Downregulation |
| HOXC11 | 1.232838781 | –1.232838781 | 5.00E−05 | 0.0265652 | Upregulation |
| PLP1 | –6.259429956 | 6.259429956 | 5.00E−05 | 0.0265652 | Downregulation |
| TCP11 | 7.039365022 | –7.039365022 | 5.00E−05 | 0.0265652 | Upregulation |
| CTAG2 | 6.5014694 | –6.5014694 | 5.00E−05 | 0.0265652 | Upregulation |
| MASP1 | –5.957919737 | 5.957919737 | 5.00E−05 | 0.0265652 | Downregulation |
| SPINK2 | –2.933219475 | 2.933219475 | 5.00E−05 | 0.0265652 | Downregulation |
| FOXP2 | –4.750570574 | 4.750570574 | 5.00E−05 | 0.0265652 | Downregulation |
| H19 | 5.003969292 | –5.003969292 | 5.00E−05 | 0.0265652 | Upregulation |
| IGJ | –7.55777445 | 7.55777445 | 5.00E−05 | 0.0265652 | Downregulation |
| GATA4 | 2.194796245 | –2.194796245 | 1.00E−04 | 0.0465128 | Upregulation |
| STRA6 | 6.84810149 | –6.84810149 | 5.00E−05 | 0.0265652 | Upregulation |
| MAGEA4 | 1.92730864 | –1.92730864 | 5.00E−05 | 0.0265652 | Upregulation |
| GPM6A | –4.878738746 | 4.878738746 | 5.00E−05 | 0.0265652 | Downregulation |
| SLC13A3 | 6.48502215 | –6.48502215 | 1.00E−04 | 0.0465128 | Upregulation |
| PIGR | –5.97822265 | 5.97822265 | 5.00E−05 | 0.0265652 | Downregulation |
| CMTM5 | –2.054181662 | 2.054181662 | 5.00E−05 | 0.0265652 | Downregulation |
| KRT24 | –2.145879581 | 2.145879581 | 5.00E−05 | 0.0265652 | Downregulation |
| LGI3 | –1.428533704 | 1.428533704 | 5.00E−05 | 0.0265652 | Downregulation |
| SSX6 | 0.861915675 | –0.861915675 | 5.00E−05 | 0.0265652 | Upregulation |
| SYNPO2 | –4.973711063 | 4.973711063 | 5.00E−05 | 0.0265652 | Downregulation |
| ARPP21 | –1.661585441 | 1.661585441 | 5.00E−05 | 0.0265652 | Downregulation |
| KRTAP13-2 | –3.152855114 | 3.152855114 | 5.00E−05 | 0.0265652 | Downregulation |
| MAGEA2B | 4.451765 | –4.451765 | 5.00E−05 | 0.0265652 | Upregulation |
| CTAG1B | 3.170383385 | –3.170383385 | 5.00E−05 | 0.0265652 | Upregulation |
| DHRS7C | –1.120212575 | 1.120212575 | 5.00E−05 | 0.0265652 | Downregulation |
| SOCS3 | –5.154826771 | 5.154826771 | 1.00E−04 | 0.0465128 | Downregulation |
| RALYL | –1.371770721 | 1.371770721 | 5.00E−05 | 0.0265652 | Downregulation |
| MAGEA11 | 3.075833803 | –3.075833803 | 5.00E−05 | 0.0265652 | Upregulation |
| PRAME | 6.712358666 | –6.712358666 | 5.00E−05 | 0.0265652 | Upregulation |
| PLA2G2A | –8.002392563 | 8.002392563 | 5.00E−05 | 0.0265652 | Downregulation |
| SLC30A10 | –1.376796016 | 1.376796016 | 5.00E−05 | 0.0265652 | Downregulation |
| CSAG3 | 4.810566494 | –4.810566494 | 5.00E−05 | 0.0265652 | Upregulation |
| CSAG1 | 5.811948767 | –5.811948767 | 5.00E−05 | 0.0265652 | Upregulation |
| SSX4B | 3.260329825 | –3.260329825 | 5.00E−05 | 0.0265652 | Upregulation |
| CXorf61 | 3.878558603 | –3.878558603 | 5.00E−05 | 0.0265652 | Upregulation |
| IGKV1D-13 | –3.569381719 | 3.569381719 | 5.00E−05 | 0.0265652 | Downregulation |
| IGLC2 | –5.965995712 | 5.965995712 | 5.00E−05 | 0.0265652 | Downregulation |
| IGLC3 | –6.956741988 | 6.956741988 | 5.00E−05 | 0.0265652 | Downregulation |
| IGHV3-49 | –5.603130774 | 5.603130774 | 5.00E−05 | 0.0265652 | Downregulation |
| SNORA45 | –4.402360639 | 4.402360639 | 1.00E−04 | 0.0465128 | Downregulation |
| MAGEA12 | 4.532958984 | –4.532958984 | 5.00E−05 | 0.0265652 | Upregulation |
| LINC00393 | 3.694022119 | –3.694022119 | 5.00E−05 | 0.0265652 | Upregulation |
| RP11-143M1.2 | –3.013890985 | 3.013890985 | 5.00E−05 | 0.0265652 | Downregulation |
| RP11-543D5.1 | 3.593712308 | –3.593712308 | 5.00E−05 | 0.0265652 | Upregulation |
| RP11-124L5.7 | 0.980610305 | –0.980610305 | 5.00E−05 | 0.0265652 | Upregulation |
| CT5A3 | 2.576638226 | –2.576638226 | 5.00E−05 | 0.0265652 | Upregulation |
| AC009133.21 | –4.738573303 | 4.738573303 | 5.00E−05 | 0.0265652 | Downregulation |
Alternative splicing in CRC with hyperglycemia
AS event counts and type distribution for CRC with hyperglycemia was also determined. In the DM-T group (CRC tissues with hyperglycemia), there were 7,944 AS events in 4,147 gene transcripts with at least 1 subtype of AS in non-intergenic regions, as well as 3,465 Intergenic AS events alternatively. We found that 3’UTR (16.83%), 5S (16.27%), and 3S (14.83%) were the 3 most frequent AS events occurring in CRC with hyperglycemia (DM-T), which were all lower than the T group (CRC without hyperglycemia).
A total of 7,943 AS events in 4,190 gene transcripts with at least 1 subtype of AS events in CRC controls (DM-N) with hyperglycemia in non-intergenic regions were detected, as well as 3,218 intergenic AS events alternatively. We found that 5S (16.4%), 3’UTR (16.28%), and 3S (15.77%) were the 3 most frequent AS events occurring in the DM-T group, which was similar to those of the DM-N group (CRC with hyperglycemia). Based on our observations of AS events, we found differences in AS usage frequency between various CRC subtypes.
Hyperglycemic-specific gene cis-regulatory network
To investigate the role of cancer biomarker genes in cancer development, we performed cis-regulation network analysis for the 2 CRC subtypes. We used a novel motif discovery algorithm that was designed for regulatory element analysis in genomics applications, such as gene promoters. We first obtained 2-kb promoter sequences near the TSS for significantly expressed genes and isoforms in the T and N comparison groups, respectively. We then used de novo motif discovery method, in which 2 sets of sequences are used to identify the regulatory elements that are specifically enriched in a set relative to the other using ZOOPS scoring (zero-or-one occurrence per sequence), together with hypergeometric enrichment calculations, to determine motif enrichment in significantly expressed genes. We obtained 2 known motifs and 16 de novo motifs. Using the JASPAR database, the known motifs on each gene promoter were found. Ten known transcription factor (TF)-gene target associations and 17 de novo motif-gene target associations were reconstructed as the T group’s gene cis-regulatory network (Tables 4 and 5). In the network, TATA-Box can regulate multiple genes, such as glyoxalase 1 (GLO1), SMARCB1, Dickkopf2 (DKK2), ATOH1, high-density lipoprotein binding protein (HDLBP), CFD, EPYC, zinc finger protein 259 (ZNF259), and nuclear autoantigenic sperm protein (NASP).
Table 4. Known motifs discovered in isoform transcripts in colorectal cancer without hyperglycemia.
| Motif | Promoter ID | Name | Chr | Description |
|---|---|---|---|---|
| FOXP1 (Forkhead) | NM_001195193 | NASP | 1p34.1 | Nuclear autoantigenic sperm protein (histone-binding) |
| TBP | NM_006708 | GLO1 | 6p21.3-p21.1 | Glyoxalase I |
| TATA-Box (TBP) | NM_001007468 | SMARCB1 | 22q11.23|22q11 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily b, member 1 |
| TATA-Box (TBP) | NM_001007468 | SMARCB1 | 22q11.23|22q11 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 |
| TATA-Box (TBP) | NM_001007468 | SMARCB1 | 22q11.23|22q11 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily b, member 1 |
| TATA-Box (TBP) | NM_014421 | DKK2 | 4q25 | Dickkopf 2 homolog (Xenopus laevis) |
| TATA-Box (TBP) | NM_005172 | ATOH1 | 4q22 | Atonal homolog 1 (Drosophila) |
| TATA-Box (TBP) | NM_005172 | ATOH1 | 4q22 | Atonal homolog 1 (Drosophila) |
| TATA-Box (TBP) | NM_005172 | ATOH1 | 4q22 | Atonal homolog 1 (Drosophila) |
| TATA-Box (TBP) | NM_005172 | ATOH1 | 4q22 | Atonal homolog 1 (Drosophila) |
| TATA-Box (TBP) | NM_001243900 | HDLBP | 2q37.3 | High-density lipoprotein binding protein |
| TATA-Box (TBP) | NM_001928 | CFD | 19p13.3 | Complement factor D (adipsin) |
| TATA-Box (TBP) | NM_004950 | EPYC | 12q21 | Epiphycan |
| TATA-Box (TBP) | NM_003904 | ZNF259 | 11q23.3 | Zinc finger protein 259 |
| TATA-Box (TBP) | NM_001195193 | NASP | 1p34.1 | Nuclear autoantigenic sperm protein (histone-binding) |
| TATA-Box (TBP) | NM_001195193 | NASP | 1p34.1 | Nuclear autoantigenic sperm protein (histone-binding) |
NASP, nuclear autoantigenic sperm protein; SNF, sucrose non fermentable; SWI, SWItch.
Table 5. De novo motifs discovered in isoform transcripts in colorectal cancer without hyperglycemia.
| Motif name | Name | Promoter ID | Chr | Description |
|---|---|---|---|---|
| 1-TYAGGCTTCA | MRGPRF | NM_001098515 | 11q13.3 | MAS-related GPR, member F |
| 2-ABCCCACACT | HNRNPK | NM_002140 | 9q21.32-q21.33 | Heterogeneous nuclear ribonucleoprotein K |
| 3-GGGACCTCGT | MRGPRF | NM_001098515 | 11q13.3 | MAS-related GPR, member F |
| 3-GGGACCTCGT | ATP1A1 | NM_000701 | 1p21 | ATPase, Na+/K+ transporting, α1 polypeptide |
| 4-CCATGCAGCA | ATOH1 | NM_005172 | 4q22 | Atonal homolog 1 (Drosophila) |
| 1-ATCTGCAAGCCT | SMARCB1 | NM_001007468 | 22q11.23|22q11 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily b, member 1 |
| 2-TCCTTCTCCTTC | OTUD5 | NM_001136157 | Xp11.23 | OTU domain containing 5 |
| 7-CGCCTTTGGTCT | MRGPRF | NM_001098515 | 11q13.3 | MAS-related GPR, member F |
| 14-GGGTTGTAGGGC | STMN1 | NM_001145454 | 1p36.11 | Stathmin 1 |
| 15-GGGGCTGGAAAA | HNRNPK | NM_002140 | 9q21.32-q21.33 | Heterogeneous nuclear ribonucleoprotein K |
| 15-GGGGCTGGAAAA | MMP11 | NM_005940 | 22q11.23 | Matrix metallopeptidase 11 (stromelysin 3) |
| 16-AAAAACCTCAAA | EPYC | NM_004950 | 12q21 | Epiphycan |
| 22-GCCATGGCAGCC | ZNF259 | NM_003904 | 11q23.3 | Zinc finger protein 259 |
| 1-AGCACGCA | DKK2 | NM_014421 | 4q25 | Dickkopf 2 homolog (Xenopus laevis) |
| 1-AGCACGCA | ATP1A1 | NM_000701 | 1p21 | ATPase, Na+/K+ transporting, α1 polypeptide |
| 1-AGCACGCA | NASP | NM_001195193 | 1p34.1 | Nuclear autoantigenic sperm protein (histone binding) |
| 4-GATCTCCG | ATOH1 | NM_005172 | 4q22 | Atonal homolog 1 (Drosophila) |
GPR, G-protein coupled receptor; OTU, ovarian tumor; SNF, Sucrose non fermentable; SWI, SWItch.
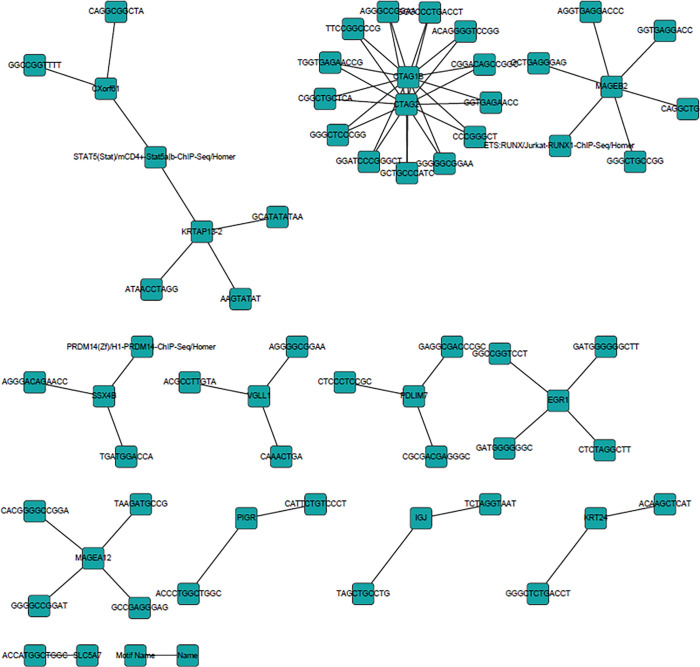
Similarly, for CRC with hyperglycemia, we also reconstructed the gene cis-regulatory network in DM-T group (Figure 4). Further differences in the network structure were found, including the discovery of less-known TF-target gene associations (4 edges) and de novo motif-target gene associations (94 edges) (Table 6). In total, 98 TF motif-target gene associations were included in the gene cis-regulatory network (Figure 4). We found that multiple de novo motifs may co-regulate CTAG1B and CTAG2, while 6 motifs co-regulate MAGEB2; STAT5 regulates CXorf61 and KRTAP13-2. Other singular motif-gene association precisions require further validation. These motifs and their regulations to their target genes may constitute CRC development with hyperglycemia, which is different to the CRC without hyperglycemia.
Figure 4.
Cis-regulatory network of colorectal cancer with hyperglycemia.
Table 6. Known motifs discovered in isoform transcripts in colorectal cancer with hyperglycemia.
| Motif name | Name | Ref-seq | Name | Chr | Description |
|---|---|---|---|---|---|
| PRDM14 (Zf)/H1-PRDM14-ChIP-Seq | SSX4B | NM_001034832 | SSX4B | Xp11.23 | Synovial sarcoma, X breakpoint 4B |
| STAT5 (Stat)/mCD4+-Stat5a|b-ChIP-Seq | CXorf61 | NM_001017978 | CXorf61 | Xq23 | Chromosome X open reading frame 61 |
| STAT5 (Stat)/mCD4+-Stat5a|b-ChIP-Seq | KRTAP13-2 | NM_181621 | KRTAP13-2 | 21q22.1 | Keratin-associated protein 13-2 |
| ETS:RUNX/Jurkat-RUNX1-ChIP-Seq | MAGEB2 | NM_002364 | MAGEB2 | Xp21.3 | Melanoma antigen family B, 2 |
Protein-protein interaction network in CRC with hyperglycemia
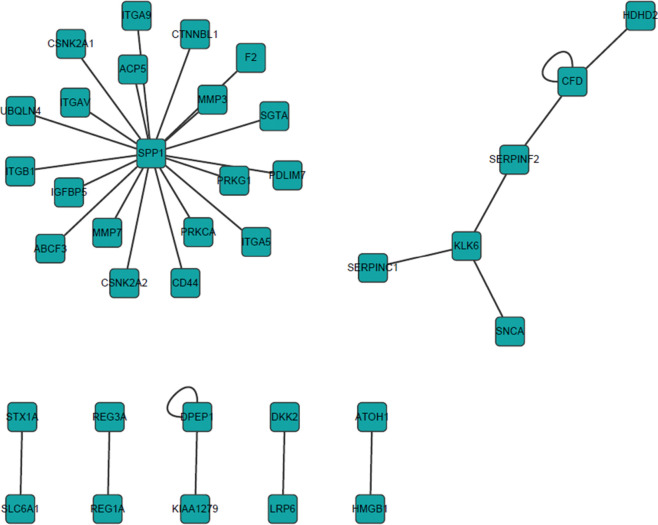
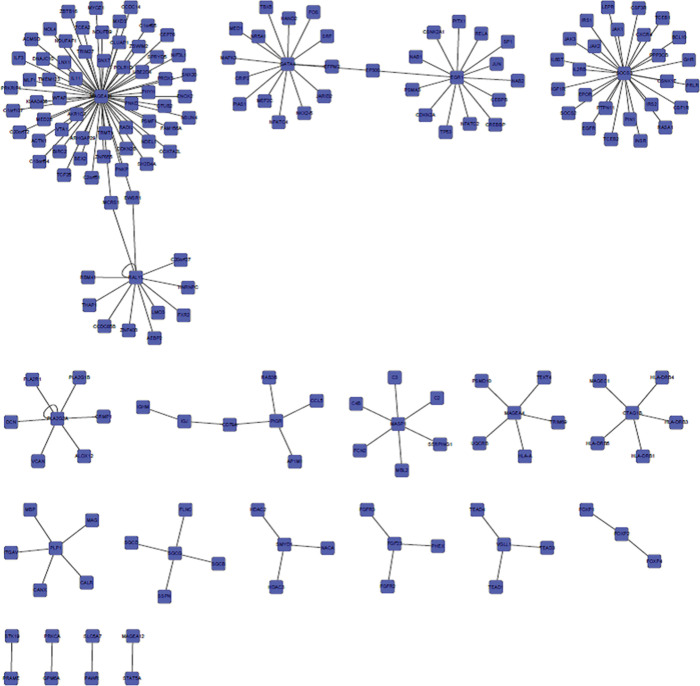
Human PPI was analyzed in gene functions. Therefore, the Protein InteraCtion KnowLedge BasE (PICKLE) global human PPI network generated from the HPRD was mapped using above identified CRC associated genes. For CRC without hyperglycemia (T group), we found that 36 proteins were involved in 31 PPIs in the reconstructed PPI network (Figure 5). Similarly, we identified 194 genes in 179 PPIs in CRC with hyperglycemia (DM-T group) (Figure 6). We found that there were no overlapping interactions between the 2 CRC subtypes. MAGEA11, GATA4, EGR1, SOCS3 and RALYL were closely related to DM-T CRC. Those pathways are substantially involved in sugar metabolism regulations. As well as metabolic-related pathways, immune-related and signaling pathways associated with cancer development were also identified, such as the T-cell receptor signaling pathway, IL-3/IL-4/IL-6/IL-7/IL-22 signaling pathway, cytokine-cytokine receptor interaction, complement and coagulation cascades, Jak-STAT signaling pathway, Epidermal growth factor (EGF) signaling pathway, MAPK signaling pathway, adherens junction, Wnt signaling pathway, and transforming growth factor-β signaling pathway, ErbB signaling pathway. Certain genes (e.g., PRKCA, EGFR, CDKN2A, MAPK3, TP53) are also reported in other cancer pathways, such as non-small cell lung cancer, and EGFR, CDKN2A, RELA, MAPK3, TP53, JAK1 are reported in CRC, pancreatic cancer, prostate cancer, glioma, and melanoma. Cell cycle associated with cancer development are expected in results, too.
Figure 5.
Protein-protein interaction network associated with colorectal cancer without hyperglycemia.
Figure 6.
Protein-protein interaction network associated with colorectal cancer with hyperglycemia.
For CRC without hyperglycemia, ECM-receptor interaction, Wnt signaling pathway, focal adhesion, complement and coagulation cascades, and the regulation of actin cytoskeleton are significantly enriched. Those pathways and functional modules mostly occur in cancer development, similar to CRC with hyperglycemia. As expected, blood glucose metabolic-associated pathways have been found to be enriched in CRC with hyperglycemia.
Discussion
CRC is one of the most preventable cancers, yet it is still the 3rd most common cancer among both men and women in the world. Epidemiological studies have revealed that hyperglycemia or diabetes is associated with CRC etiology. Diabetes has been known to increase the risk of cancer, and hyperglycemia (high blood sugar) has been reported in people with diabetes, type 2 diabetes, high blood pressure, depression, and bipolar disorder. Cancer and diabetes frequently co-exist, even after adjusting for other factors, such as disease stage, body weight, and smoking habits. It has been reported that the risk for CRC can be lowered in the same ways as for type 2 diabetes. Both diseases are complex and have multiple subtypes.
Diabetes is typically divided into 2 major subtypes: types 1 and 2 diabetes, while cancer is typically classified by its anatomic origin in which there may be multiple subtypes. Further, the pathophysiology underlying both cancer and diabetes is incompletely understood. Mechanisms postulated for increased cancer risk in diabetes include hyperglycemia, hyperinsulinemia, and obesity. However, the molecular mechanisms underlying the association between hyperglycemia and CRC have not been well studied. Therefore, in the present study, we discuss the possible biologic links between hyperglycemia and cancer risk using transcriptome sequencing technique. We compared CRC with hyperglycemia with CRC without hyperglycemia.
Carcinogenesis is a complex process in which normal cells must undergo multiple genetic mutations before the full neoplastic phenotype of growth, invasion, and metastasis occurs. From a transcriptome level, gene function effects that are caused by genetic mutation changes and various random factors, including known environmental and unknown factors that affect 1 or more steps of this pathway, are associated with cancer incidence or mortality. Diabetes may influence the neoplastic process by several mechanisms, including hyperinsulinemia (either endogenous due to IR or exogenous due to administered insulin or insulin secretagogues), hyperglycemia, or chronic inflammation.
Epidemiological studies indicate that elevated circulating IGF-1 and IR, and associated complications, such as elevated fasting plasma insulin, glucose and free fatty acids, glucose intolerance, increased body mass index, and visceral adiposity, are linked with an increased risk of CRC. However, the role of insulin and markers of glucose control in the development of adenomas, which are precursors to CRC, has not been fully explored. Based on the findings from the transcriptome analysis, we found that DEGs in CRC with hyperglycemic function in the IGF-I pathway, including IGF1R, FOS, CSNK2A1, JUN, MAPK3, SRF, IRS1, RASA1, and PTPN11. This pathway is not significantly enriched in CRC without hyperglycemia. With regard to the insulin signaling pathway, we also found that several genes, such as FOS, CSNK2A1, JUN, MAPK3, SRF, INSR, IRS1, RASA1, and PTPN11, are related to cancer development. Type 2 diabetes mellitus (IRS2, SOCS2, SOCS3, MAPK3, INSR, IRS1) and type I diabetes mellitus (HLA-DRB1, HLA-DRB3, HLA-A, HLA-DRB4, HLA-DRB5) are both enriched only in CRC with hyperglycemia. This suggests that the functions of these genes are related to the association of CRC with hyperglycemia.
Even though transcriptome data provide further information on gene regulation and the pathways involved in CRC with hyperglycemia, there is less information on CRC without hyperglycemia and cancer prognosis or cancer-specific mortality. It is unclear whether the association between hyperglycemia and cancer is direct (e.g., due to hyperglycemia), whether hyperglycemia is a marker of underlying biologic factors that alter cancer risk (e.g., IR and hyperinsulinemia), or whether the cancer–hyperglycemia association is indirect and due to common risk factors, such as obesity. Whether cancer risk is influenced by the duration of diabetes is a critical and complex issue that be further complicated by the multidrug therapy that is often necessary for diabetes treatment. A better understanding of whether hyperglycemia influences cancer prognosis is needed.
Acknowledgments
Funding: The Research Project of Science and Technology of Zhejiang Province (2017C37177); The Medical Scientific Research Project of Health commission of Zhejiang Province (2018KY775); The Research Project of Science and Technology of Huzhou City (2020GYB13); The Key Research Project of Science and Technology of Huzhou City (2018GZ34); The Development and Application of South Taihu Science and Technology Innovation Leading Talents Project.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study conformed to the provisions of the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee board of The First Affiliated Hospital of Huzhou University (No. 20170118004). Informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-587
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-587
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-587). The authors have no conflicts of interest to declare.
(English Language Editor: R. Scott)
References
- 1.Hsing AW, Sakoda LC, Chua S, Jr. Obesity, metabolic syndrome, and prostate cancer. Am J Clin Nutr 2007;86:s843-57. 10.1093/ajcn/86.3.843S [DOI] [PubMed] [Google Scholar]
- 2.Ishino K, Mutoh M, Totsuka Y, et al. Metabolic syndrome: a novel high-risk state for colorectal cancer. Cancer Lett 2013;334:56-61. 10.1016/j.canlet.2012.10.012 [DOI] [PubMed] [Google Scholar]
- 3.Pandey A, Forte V, Abdallah M, et al. Diabetes mellitus and the risk of cancer. Minerva Endocrinol 2011;36:187-209. [PubMed] [Google Scholar]
- 4.Onitilo AA, Stankowski RV, Berg RL, et al. Type 2 diabetes mellitus, glycemic control, and cancer risk. Eur J Cancer Prev 2014;23:134-40. 10.1097/CEJ.0b013e3283656394 [DOI] [PubMed] [Google Scholar]
- 5.Gawlinska K, Gawlinski D, Przegalinski E, et al. Maternal high-fat diet during pregnancy and lactation provokes de pressive-like behavior and influences the irisin/brain-derived neurotrophic factor axis and inflammatory factors in male and female offspring in rats. Journal of Physiology and Pharmacology 2019;70:407-17. [DOI] [PubMed] [Google Scholar]
- 6.Valpione S, Gremel G, Mundra P, et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur J Cancer 2018;88:1-9. 10.1016/j.ejca.2017.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cetin M, Yetgin S, Kara A, et al. Hyperglycemia, ketoacidosis and other complications of L-asparaginase in children with acute lymphoblastic leukemia. J Med 1994;25:219-29. [PubMed] [Google Scholar]
- 8.Luna B, Feinglos M. Drug-induced hyperglycemia. JAMA 2001;286:1945-8. 10.1001/jama.286.16.1945 [DOI] [PubMed] [Google Scholar]
- 9.Trevisan M, Liu J, Muti P, et al. Markers of insulin resistance and colorectal cancer mortality. Risk Factors and Life Expectancy Research Group. Cancer Epidemiol Biomarkers Prev 2001;10:937-41. [PubMed] [Google Scholar]
- 10.Nilsen TI, Vatten LJ. Prospective study of colorectal cancer risk and physical activity, diabetes, blood glucose and BMI: exploring the hyperinsulinaemia hypothesis. Br J Cancer 2001;84:417-22. 10.1054/bjoc.2000.1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoen RE, Tangen CM, Kuller LH, et al. Increased blood glucose and insulin, body size, and incident colorectal cancer. J Natl Cancer Inst 1999;91:1147-54. 10.1093/jnci/91.13.1147 [DOI] [PubMed] [Google Scholar]
- 12.Kono S, Honjo S, Todoroki I, et al. Glucose intolerance and adenomas of the sigmoid colon in Japanese men (Japan). Cancer Causes Control 1998;9:441-6. 10.1023/A:1008879920140 [DOI] [PubMed] [Google Scholar]
- 13.Misciagna G, Michele GD, Guerra V, et al. Serum fructosamine and colorectal adenomas. Eur J Epidemiol 2004;19:425-32. 10.1023/B:EJEP.0000027359.95727.24 [DOI] [PubMed] [Google Scholar]
- 14.Keku TO, Lund PKL, Galanko J, et al. Insulin resistance, apoptosis, and colorectal adenoma risk. Cancer Epidemiol Biomarkers Prev 2005;14:2076-81. 10.1158/1055-9965.EPI-05-0239 [DOI] [PubMed] [Google Scholar]
- 15.Ma YS, Yang IPY, Tsai HL, et al. High glucose modulates antiproliferative effect and cytotoxicity of 5-fluorouracil in human colon cancer cells. DNA Cell Biol 2014;33:64-72. 10.1089/dna.2013.2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimomoto T, Ohmori H, Luo Y, et al. Diabetes-associated angiotensin activation enhances liver metastasis of colon cancer. Clin Exp Metastasis 2012;29:915-25. 10.1007/s10585-012-9480-6 [DOI] [PubMed] [Google Scholar]
- 17.Trapnell C, Hendrickson DG, Sauvageau M, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013;31:46-53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie C, Mao X, Huang JJ, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011;39:W316-22. 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105-11. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 2010;11:345-55. 10.1038/nrg2776 [DOI] [PubMed] [Google Scholar]
- 21.Hubbard T, Barker D, Birney E, et al. The Ensembl genome database project. Nucleic Acids Res 2002;30:38-41. 10.1093/nar/30.1.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105-11. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Handsaker B, Wysoker A, et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009;25:2078-9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562-78. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mauer J, Bhagirath C, Goldau J, et al. Signaling by IL-6 promotes alternative activation of macrophages to limit endotoxemia and obesity-associated resistance to insulin. Nat Immunol 2014;15:423-30. 10.1038/ni.2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ueki K, Kondo T, Kahn CR. Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol Cell Biol 2004;24:5434-46. 10.1128/MCB.24.12.5434-5446.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi H, Cave B, Inouye K, et al. Overexpression of suppressor of cytokine signaling 3 in adipose tissue causes local but not systemic insulin resistance. Diabetes 2006;55:699-707. 10.2337/diabetes.55.03.06.db05-0841 [DOI] [PubMed] [Google Scholar]
- 28.Ueki K, Kadowaki T, Kahn CR. Role of suppressors of cytokine signaling SOCS-1 and SOCS-3 in hepatic steatosis and the metabolic syndrome. Hepatol Res 2005;33:185-92. 10.1016/j.hepres.2005.09.032 [DOI] [PubMed] [Google Scholar]
- 29.Yang Z, Hulver M, McMillan RP, et al. Regulation of Insulin and Leptin Signaling by Muscle Suppressor of Cytokine Signaling 3 (SOCS3). PLoS One 2012;7:e47493. 10.1371/journal.pone.0047493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao L, Fu XD, Si MW, et al. MicroRNA-185 Targets SOCS3 to Inhibit Beta-Cell Dysfunction in Diabetes. PLoS One 2015;10:e0116067. 10.1371/journal.pone.0116067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwaeble W, Dahl MRD, Thiel S, et al. The mannan-binding lectin-associated serine proteases (MASPs) and MAp19: four components of the lectin pathway activation complex encoded by two genes. Immunobiology 2002;205:455-66. 10.1078/0171-2985-00146 [DOI] [PubMed] [Google Scholar]
- 32.Fraile JM, Ordóñez GR, Quirós PM, et al. Identification of novel tumor suppressor proteases by degradome profiling of colorectal carcinomas. Oncotarget 2013;4:1919-32. 10.18632/oncotarget.1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeRycke MS, Gunawardena SR, Middha S, et al. Identification of Novel Variants in Colorectal Cancer Families by High-Throughput Exome Sequencing. Cancer Epidemiol Biomarkers Prev 2013;22:1239-51. 10.1158/1055-9965.EPI-12-1226 [DOI] [PMC free article] [PubMed] [Google Scholar]