Abstract
Background:
Patients with advanced and/or metastatic solid tumors have limited treatment options. Mutations that serve as biomarkers of carcinogenesis can be found in cell-free DNA of patients’ plasma. Analysis of circulating tumor DNA (ctDNA) was developed as a non-invasive, cost-effective alternative to tumor biopsy when such biopsy is not technically feasible or it is associated with high risk for complications. The role of ctDNA in precision oncology is promising but its clinical significance across tumor types remains to be validated. We report a case series of three heavily pretreated patients with advanced solid tumors who received matched targeted therapy based on ctDNA analysis and/or tumor molecular profiling.
Case presentation:
Three patients with advanced, metastatic cancer and the following characteristics are presented: a 71-year-old woman with ovarian cancer and BRCA2 mutation identified in ctDNA and tumor tissue was treated with a PARP inhibitor and achieved partial response by RECIST (Response Evaluation Criteria in Solid Tumors) for 22.6+ months; a 40-year-old woman with adenoid cystic carcinoma of the parotid gland was treated with a MEK/RAF pathway inhibitor on the basis of RAF1 amplification on ctDNA analysis and had stable disease for 20.2 months; and a 56-year-old woman with breast cancer and a BRCA1 mutation identified by ctDNA analysis was treated with a PARP inhibitor and achieved stable disease for 9.1 months. All three patients are alive at the time of this report.
Conclusions:
These results suggest that ctDNA analysis can contribute to selection of targeted therapy in patients with advanced, metastatic cancer. Prospective clinical trials to evaluate and optimize ctDNA biomarkers, as well as the integration of novel and/or alternative targeted therapies, are warranted to fully assess the role of ctDNA analysis in cancer therapy.
Trial registration:
www.clinicaltrials.gov (NCT02152254). Registered May 28, 2014. https://www.clinicaltrials.gov/ct2/show/NCT02152254. MD Anderson protocol # PA12-1161 (approval ID IRB1 FWA00000121) and # PA11-0377 (approval ID IRB4 FWA00005015).
Keywords: cell-free DNA, circulating tumor DNA, genomic profiling, personalized therapy, precision medicine, targeted therapy
Introduction
Genomic alterations play an important role in carcinogenesis, disease progression, resistance and response to targeted therapy. The genetic heterogeneity across tumor types complicates the management of these diseases. Using the genomic profiles of individual patient tumors, precision medicine helps optimize treatment selection to improve clinical outcomes for patients with cancer.1 Mutations that serve as biomarkers of tumor progression and response can be found in cell-free DNA collected from the plasma of individual patients. Analysis of this circulating tumor DNA (ctDNA), therefore, offers a promising, non-invasive, and personalized diagnostic approach to selecting treatment and patients for enrollment in clinical trials.
Tumor biopsy samples are typically analyzed to determine the tumor characteristics, but biopsy is not feasible for some patients because their tumors are not easily accessible or because the biopsy procedure is associated with significant risk. Also, samples obtained from biopsies can be insufficient for tumor analysis. The proportion of patients who are ineligible for molecular screening due to inaccessibility or high risk of tumor biopsy, and therefore are excluded from certain clinical trials offering targeted treatment based on molecular alterations, has not been systematically analyzed. In the BATTLE (Biomarker-integrated Approaches of Targeted Therapy for Lung Cancer Elimination) study, it was estimated that 10% to 15% of patients with lung cancer did not have sufficient material for next-generation molecular testing.2 In an interim analysis of IMPACT2, an ongoing randomized study evaluating genomic profiling and targeted agents in metastatic cancer, of 391 patients who were enrolled in the first part of the study, 19 (4.86%) patients had inadequate tumor cells for analysis and biopsy was not feasible or tumor was not accessible in 15 (3.84%) patients. Overall, 213 (54.48%) of 391 received anticancer therapy (Tsimberidou et al., Npj Precision Oncology, in Press).
CtDNA analysis was developed as a non-invasive, cost-effective alternative to tumor biopsy when such biopsy is associated with significant risk, when tumor tissue is insufficient or inaccessible, and/or when repeated assessment of tumor molecular abnormalities is needed to optimize treatment.3
We previously reviewed the role of ctDNA in guiding targeted therapy in clinical trials that involved ctDNA analysis of specific tumors and across tumor types.3 CtDNA analysis based on epidermal growth factor receptor (EGFR) and v-Ki-ras2 kirsten rat sarcoma viral oncogene homolog (KRAS) has been well established for selecting treatment for patients with advanced non-small cell lung cancer (NSCLC) and colorectal cancer (CRC), respectively.4,5 However, the assessment of ctDNA and clinical trials that involve ctDNA analysis in other tumor types are still needed. Herein, we describe encouraging clinical outcomes of three patients with advanced metastatic cancer who received matched targeted therapy based on the results of ctDNA analysis.
Case presentation
Case presentation 1: Patient (ID 001)
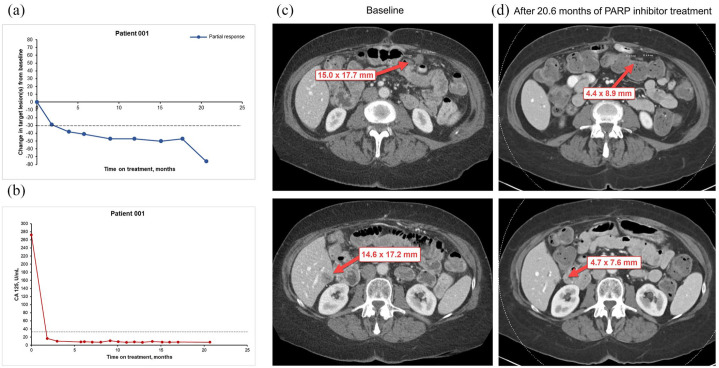
A 71-year-old woman was diagnosed with metastatic ovarian cancer in October 2010. She had been experiencing symptoms of vaginal dryness, dyspareunia, and abdominal and lower back pain. Magnetic resonance imaging (MRI) of the pelvis demonstrated a complex adnexal mass and computed tomography (CT) imaging studies demonstrated a lesion in the upper lobe of the left lung, and an adenoma of the left adrenal gland. Transvaginal ultrasonography confirmed a multilocular mass (6.0 × 5.0 × 5.6 cm) centered in the right adnexal region. The mass had multiple irregular thick septa and a soft tissue component with definite vascular flow in the regions of echogenic soft tissue and was highly suggestive of cystadenocarcinoma. The patient underwent a laparoscopic bilateral salpingo-oophorectomy, and the surgical pathology report demonstrated ovarian high-grade serous carcinoma with extension to the fallopian tube surface and fimbrial end in the right ovary and tube, and metastatic to the omentum. In November 2010, the patient was treated with six cycles of carboplatin and paclitaxel followed by 11 cycles of paclitaxel consolidation therapy. CT imaging studies demonstrated no evidence of disease at 5 months after treatment and annually for 3 years. Three years after treatment, she presented to the clinic with increasing pain and pressure in the lower abdomen, including pain with urination. CT scans demonstrated diffuse peritoneal carcinomatosis, evidenced by multiple nodular masses noted within the peritoneum and the supracolic omentum, and small volume ascites. She was retreated with six cycles of carboplatin and paclitaxel, and restaging scans demonstrated improvement in the peritoneal carcinomatosis and no gross CT evidence of residual disease. She had no evidence of disease for 17 months, and then CT scans showed interval development of a heterogeneous soft tissue lesion in the pelvis compatible with recurrent disease. She underwent tumor reductive surgery followed by six cycles of carboplatin and paclitaxel. Molecular analysis using formalin-fixed paraffin-embedded (FFPE) tissue of the most recently resected pelvic mass demonstrated tumor protein p53 (TP53) mutation and breast cancer type 2 (BRCA2) somatic mutation (Table 1). The disease responded to treatment with carboplatin and paclitaxel but recurred after 20 months and the patient was referred to the Department of Investigational Cancer Therapeutics in July 2017. She was enrolled on a clinical trial that included carboplatin, paclitaxel and immunotherapy. Her best response was immune-related partial response (PR). On cycle 19, day 22, CT images demonstrated increasing size and number of nodules within the peritoneum consistent with increasing metastatic disease; a growing metastasis was also noted in segment five of the liver. ctDNA analysis performed 13 months prior to disease progression demonstrated the following alterations: neurofibromin 1 (NF1), splice site SNV 1.9, TP53 V157F 0.2, guanine nucleotide binding protein, alpha stimulating activity polypeptide (GNAS) R201H 0.2, BRCA2 R3052W 0.1 (Table 1). On the basis of this information and evidence of a BRCA2 somatic mutation in the pelvic mass, the patient was enrolled on a clinical trial of a poly (ADP-ribose) polymerase (PARP) inhibitor administered daily. She underwent genetic counseling, and testing for germline BRCA1 and BRCA2 mutations was negative. Thus far, she has received 21 cycles of the PARP inhibitor, and her best RECIST response was PR (76% decrease from baseline per RECIST 1.1) (Figure 1). Her treatment is ongoing [progression-free survival (PFS) = 22.6+ months] (Table 2). The patient experienced grade 3 anemia that started during cycle 3 and was treated with transfusion of red blood cells (average, 2 units monthly). On cycle 7, day 1, she required a dose adjustment consisting of a 25% reduction in the PARP inhibitor dose and a decrease of days of treatment to 3 weeks on, 1 week off, after which she tolerated the treatment without the need for transfusion.
Table 1.
Genomic analysis using tissue and ctDNA samples.
| Pt. ID | Collection date | Sample source | Genomic alterations | Panel, no. of clinically relevant genes |
|---|---|---|---|---|
| 001 | 16 June 2015 | Pelvic mass | TP53 V157F | Solid tumor genomic assay V2, 146 genes |
| BRCA2 R3052W | ||||
| 20 July 2017 | Blood | BRCA2 R3052W | Guardant360, 73 genes | |
| NF1 splice site | ||||
| TP53 V157F | ||||
| GNAS R201H | ||||
| 002 | 21 July 2017 | Blood | RAF1 amplification | Guardant360, 73 genes |
| 25 September 2017 | Liver | None | Solid tumor genomic assay V1, 134 genes | |
| 22 November 2019 | Liver | BRAF V600dup | Tempus × T assay, 596 genes | |
| MYB-NFIB chromosomal rearrangement | ||||
| KMT2D Q2696 Stop gain – LOF | ||||
| 003 | 29 January 2015 | Lung | ATM loss exons 57–63 | FoundationOne, 315 genes |
| NOTCH2 A3F | ||||
| ESR1 Y537S | ||||
| CCND1 amplification | ||||
| EMSY amplification | ||||
| FGF19 amplification | ||||
| FGF3 amplification | ||||
| FGF4 amplification | ||||
| MCL1 amplification - equivocal | ||||
| 15 October 2018 | Blood | ATM M3011T | Liquid biopsy panel, 70 genes | |
| BRCA1 Q356* | ||||
| ESR1 Y537S | ||||
| TP53 A159V | ||||
| TP53 P152L |
ATM, ATM serine/threonine kinase; BRAF, rapidly accelerated fibrosarcoma homolog B; BRCA1, breast cancer type 1; BRCA2, breast cancer type 2; CCND1, Cyclin D1; EMSY, BRCA2 interacting transcriptional repressor; ESR1, estrogen receptor 1; FGF, fibroblast growth factor; GNAS, guanine nucleotide binding protein, alpha stimulating activity polypeptide; KMT2D, histone-lysine N-methyltransferase 2D; LOF, loss of function; MCL1, myeloid cell leukemia 1; MYB-NFIB, myeloblastosis virus oncogene – nuclear factor 1 B-type; NF1, neurofibromatosis type 1; NOTCH2, notch receptor 2; RAF1, rapidly accelerated fibrosarcoma 1; TP53, tumor protein p53.
Molecular alterations identified by ctDNA analysis and used to select matched therapy are presented in bold text.
Figure 1.
Changes in tumor measurement from baseline in patient 001, who underwent treatment with PARP inhibitor on the basis of ctDNA analysis showing BRCA2 R3052W mutation. (a) Scatter plot illustrates changes in lesion tumor burden. Computed tomography (CT) scan data at the most recent time prior to cycle 1, day 1 is chosen as baseline (time 0). The horizontal (x) axis shows time at restaging CT scans as months from baseline. The vertical (y) axis shows percentage of change in tumor measurement from baseline. Each dot represents a data point collected at each restaging CT scan. The blue line represents PR (⩾30% decrease in tumor measurements from baseline, based on RECIST 1.1). (b) Scatter plot illustrates changes in tumor marker levels over time. (c) Representative CT images from the patient at baseline. (d) Representative CT images from the patient after 20.6 months of targeted therapy with PARP inhibitor.
Red arrows indicate target lesions.
Table 2.
Clinical outcomes of three patients with advanced solid tumors who received matched therapy based on ctDNA analysis.
| Pt. ID | ctDNA biomarker | Matched therapy | Best RECIST response | PFS*, months | Progr.status | Subsequent Rx | Survival status | OS†, months |
|---|---|---|---|---|---|---|---|---|
| 001 | BRCA2 R3052W | PARP inhibitor | PR | 22.6+ | N/A | N/A | Alive | 22.6+ |
| 002 | RAF1 amplification | MEK/RAF pathway inhibitor | SD | 20.2 | PD | Investigational | Alive | 28.4+ |
| 003 | BRCA1 Q356* | PARP inhibitor | SD | 9.1 | PD | Doxorubicin; radiotherapy | Alive | 14.8+ |
N/A, non-applicable; OS, overall survival; PD, progressive disease; PFS, progression-free survival; PR, partial response; Progr., progression; Rx, therapy; SD, stable disease.
BRAC1, breast cancer type 1; BRAC2, breast cancer type 2; MEK, mitogen-activated protein kinase kinase; PARP, poly (ADP-ribose) polymerase; RAF1, rapidly accelerated fibrosarcoma 1.
Progression-free survival is measured in months from cycle 1, day 1 to time of radiologic scan showing progressive disease.
Overall survival is measured in months from cycle 1, day 1 to the most recent time of this report.
Case presentation 2: Patient (ID 002)
A 40-year-old woman was first diagnosed in 2007 with adenoid cystic carcinoma of the left parotid gland and bone metastases. She was a non-smoker and had a pertinent family history of multiple cancers, notably of the brain, stomach, and skin. The patient underwent superficial parotidectomy with apparently negative margins followed by post-operative adjuvant radiotherapy. Subsequent imaging revealed a lytic lesion at the L3 vertebral body; biopsy did not demonstrate metastatic tumor. She had no evidence of disease until 2013, when a positron emission tomography (PET)/CT revealed increased activity in the L3 vertebra. The patient experienced back pain while lifting heavy objects or when the lesion was percussed, post-operative paresthesia in the lobule and tragus of the left ear and stiffening of the left neck. In October 2013, she received stereotactic radiation therapy to the L3 vertebra (24 Gy to the gross tumor volume and 16 Gy to the clinical target volume). Three months later, PET/CT imaging studies demonstrated no evidence of FDG-avid metastasis.
She had no evidence of active disease for 33 months, until October 2016, when MRI and PET/CT imaging studies demonstrated left pulmonary metastasis, an enlarging lesion at L3–L4 consistent with progressive disease. She was referred to the Department of Investigational Cancer Therapeutics in February 2017. She was treated with an investigational combination therapy comprising a STAT3 inhibitor and nivolumab for eight cycles. Her best RECIST response was stable disease (SD), with a 13% decrease in tumor measurements from baseline to cycle 3, day 26 (RECIST 1.1). She was taken off protocol in July 2017 owing to disease progression that included liver metastases. Possibly related adverse events included grade 1 diarrhea and abdominal cramps.
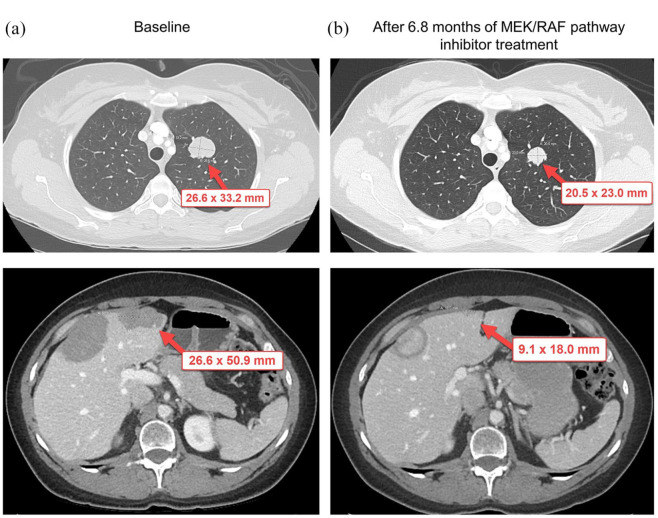
Molecular diagnostic solid tumor genomic assay using FFPE slides from a liver biopsy performed in September 2017 did not identify any somatic mutations (Table 1). However, ctDNA analysis using the patient’s blood sample demonstrated a strongly positive rapidly accelerated fibrosarcoma 1 (RAF1) amplification, with a magnitude in the 50th to 90th percentile (Table 1). On the basis of this finding, the patient was started on a clinical trial with a mitogen-activated protein kinase kinase/ rapidly accelerated fibrosarcoma (MEK/RAF) pathway inhibitor in January 2018. Her best RECIST response was SD with an 18% decrease in tumor measurements (RECIST 1.1) after 10 cycles (Figure 2). She received 29 cycles of the study drug and was taken off study owing to progressive disease confirmed by CT imaging studies. PFS duration was 20.2 months and overall survival duration was 28.4+ months. Adverse events possibly associated with MEK/RAF pathway inhibitor were all grade 1 and included intermittent diarrhea and vomiting; maculopapular and acneiform rash; neutropenia; blurry vision; and hair thinning.
Figure 2.
Changes in tumor measurement from baseline in patient 002, who underwent treatment with MEK/RAF pathway inhibitor on the basis of ctDNA analysis showing RAF1 amplification. (a) Representative computed tomography (CT) images from the patient at baseline. (b) Representative CT images from the patient after 6.8 months of treatment with MEK/RAF pathway inhibitor.
Red arrows indicate target lesions.
In November 2019, the patient underwent a core needle biopsy of the liver for completion of molecular profiling, which demonstrated rapidly accelerated fibrosarcoma homolog B (BRAF) V600dup and myeloblastosis virus oncogene – nuclear factor 1 B-type (MYB-NFIB) chromosomal rearrangement, biologically relevant histone-lysine n-methyltransferase 2D (KMT2D) genomic variant, negative programmed death-ligand 1 (PDL1) expression, normal DNA mismatch repair protein expression, and RNA sequencing demonstrating fibroblast growth factor receptor 2 (FGFR2) and fibroblast growth factor receptor 1 (FGFR1) overexpression (Table 1). Two months later, she underwent transcatheter arterial hepatic embolization of the larger liver metastases. The patient was alive at the time of this report (Table 2). She did not experience any serious adverse events and did not require any blood transfusions.
Case presentation 3: Patient (ID 003)
A 56-year-old woman with a history of breast cancer was referred to the Department of Investigational Cancer Therapeutics in January 2015. She had no pertinent medical history and was in good health until early 2005, when she felt a mass in her left breast. She was a non-smoker and had a family history of breast cancer and prostate cancer. Physical examination demonstrated a 3 × 3 cm mass in the 10 to 11-o’clock position in the superior aspect of the left breast and a 2 cm lymph node in the left axilla. Ultrasound-guided core needle biopsy performed in February 2005 demonstrated an estrogen receptor- and progesterone receptor-positive, human epidermal growth factor receptor 2 (HER-2/neu)-negative invasive mammary carcinoma of the left breast [modified Black’s nuclear grade 1 (well differentiated)], carcinoma in situ, low grade, solid type, without necrosis). CT scans indicated no evidence of metastasis; however, fine-needle aspiration of the left axilla lymph nodes demonstrated metastatic carcinoma consistent with the primary breast tumor.
In March 2005, the patient was enrolled on a clinical trial that included weekly paclitaxel chemotherapy for 12 cycles followed by six cycles of 5-fluorouracil, epirubicin, and cyclophosphamide. Four months later, imaging studies indicated a reduction in tumor size. In September 2005, she underwent a left skin-sparing total mastectomy with axillary lymph node dissection followed by delayed reconstruction using a submuscular tissue expander. Then, from October to December 2005, she underwent post-operative adjuvant radiation therapy (50 Gy in 25 fractions to the left central chest and left lateral chest wall with an additional 10 Gy boost to the tumor bed). She also intermittently received Tamoxifen between 2005 and 2008. In December 2006, imagining studies demonstrated bone metastasis and the patient was treated with radiation therapy and anastrozole plus fulvestrant from December 2006 to March 2014. The bone disease stabilized, but she had disease progression in her lungs and lymph nodes. Although her tumor did not harbor an alteration in the PI3K/Akt/mTOR pathway, she was treated with the standard-of-care exemestane and everolimus from March 2014 to January 2015.
In January 2015, CT scans demonstrated increasing thoracic lymphadenopathy and pulmonary metastatic disease, and a nuclear medicine bone scan demonstrated bone metastasis involving the left ilium, acetabulum, and ischium. The patient was referred to the Department of Investigational Cancer Therapeutics in January 2015 and underwent biopsy of a lung tumor. Molecular profiling of the tumor demonstrated the following genomic alterations: ATM serine/threonine kinase (ATM) loss in exons 57 to 63, notch receptor 2 (NOTCH2) A3F, estrogen receptor 1 (ESR1) Y537S, and amplification of the following genes: cyclin D1 (CCND1), myeloid cell leukemia 1 (MCL1), BRCA2 interacting transcriptional repressor (EMSY), fibroblast growth factor 19 (FGF19), fibroblast growth factor 3 (FGF3), and fibroblast growth factor 4 (FGF4) (Table 1). Early assessments suggested that preclinical models with increased cyclin D1 or cyclin-CDK-Rb pathway activation may have increased sensitivity to CDK4/6 inhibitors.6,7 However, the patient did not receive a CDK4/6 inhibitor because clinical trials were not available at that time. She was presented at the multidisciplinary conference for optimization of treatment selection. On the basis of tumor molecular profiling demonstrating multiple mutations in FGF (FGF19, FGF3, and FGF4), the patient was offered an investigational therapy comprising FGFR inhibitor, but she pursued treatment with eribulin; after four cycles she developed disease progression. Subsequently, she was treated with 11 cycles of fulvestrant and palbociclib and was taken off study owing to progressive disease in August 2017.
On the basis of her tumor’s NOTCH2 A3F alteration, in October 2017, the patient was enrolled on a clinical trial consisting of a NOTCH inhibitor, cisplatin, and gemcitabine. Her best RECIST response was PR per RECIST 1.1 criteria on cycle 12, day 19. She received 22 cycles of the investigational drug combination for 19 months and was taken off study owing to disease progression. Adverse events included impaired hearing and thrombocytopenia.
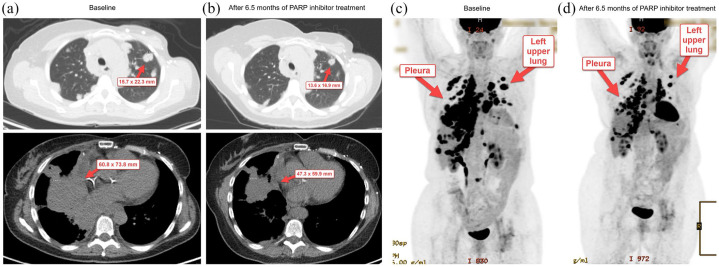
The patient underwent genetic counseling, and testing for germline BRCA1 and BRCA2 mutations was negative. CtDNA analysis in October 2018 demonstrated multiple somatic mutations which included a breast cancer type 1 (BRCA1) nonsense mutation on exon 10 (Table 1). On the basis of these findings, the patient was enrolled on a clinical trial with a PARP inhibitor. Her best RECIST response was SD with a 14% decrease in tumor measurement from baseline per RECIST 1.1, and the PFS duration was 9.1 months (Figure 3 and Table 2). Imaging studies in January 2020 demonstrated evidence of progressive disease and she was taken off study. Subsequently, she received radiotherapy and doxorubicin. At the time of this report, the patient was still alive, and the overall survival duration was 14.8+ months (Table 2).
Figure 3.
Changes in tumor measurement from baseline in patient 003, who underwent treatment with PARP inhibitor on the basis of ctDNA analysis showing BRCA1 Q356* mutation. (a) Representative computed tomography (CT) images from the patient at baseline. (b) Representative CT images from the patient after 6.5 months of treatment with PARP inhibitor. (c) Representative positron emission tomography (PET) overview image from the patient at baseline. (d) Representative PET overview image from the patient after 6.5 months of treatment with PARP inhibitor.
Red arrows indicate target lesions.
Discussion and conclusions
The clinical significance of ctDNA analysis is evolving. The FDA has approved the use of next-generation sequencing (NGS) analysis as a companion diagnostic device for multiple biomarkers detected by ctDNA analysis using plasma specimens of patients with NSCLC (EGFR mutations for the use of gefitinib, osimertinib and erlotinib; and ALK rearrangements for treatment with alectinib), prostate (BRCA1 and BRCA2 alterations for the use of rucaparib and olaparib; and ATM alterations for the use of olaparib), ovarian (BRCA1 and BRCA2 alterations for the use of rucaparib), and breast cancer (PI3K mutations for the use of alpelisib). CtDNA analysis is also used for KRAS in CRC5 and in the management of resistance to ALK inhibitors in lung cancer.8 However, ctDNA analysis has not been systematically validated for patients with other tumor types. If the specific genomic alterations associated with these FDA-approved drugs are not detected in the blood, then tumor biopsies should be performed to identify these alterations.
In this case series, the use of ctDNA analysis alone or in combination with tumor molecular profiling was associated with encouraging clinical outcomes in three patients with advanced, metastatic cancer treated with targeted therapy. One patient had a durable PR of 22.6+ months as evidenced by CT imaging and tumor marker monitoring (Figure 1); the other two patients had SD lasting for 20.2 months and 9.1 months, respectively (Table 2). Our data demonstrate that in heavily pretreated patients with advanced, metastatic cancer, targeted therapy selected on the basis of ctDNA and/or tumor genomic analysis was associated with encouraging results. Our observation emphasizes the need to systematically analyze ctDNA in correlation with tumor genomic analysis. Further, the role of ctDNA analysis should be assessed in prospective clinical trials for selection of personalized therapy in individual patients.
In our study, patient ID 002, who received MEK/RAF pathway inhibitor on the basis of ctDNA analysis demonstrating a strongly positive RAF1 amplification, had disease stabilization for 20.2 months and overall survival (OS) >28.4+ months. Genomic analysis using tumor biopsy also demonstrated BRAF_V600dup alteration. Tumor harboring both RAF amplification and BRAF V600dup may have sensitivity to treatment with MEK/RAF inhibitors, and our results warrant further investigation. There is minimal evidence on RAF1 amplification and enhanced tumor response to targeted therapies. Overexpression of RAF1 promotes c-Raf signaling in vitro, and preclinical studies have suggested that tumors with activating c-Raf signaling may be more sensitive to MEK inhibition.9–11 Tumors with RAF1 amplification therefore might benefit from MEK inhibitor therapy. Studies conducted by other investigators in a phase III, randomized clinical trial demonstrated that patients with melanoma harboring either RAF1, KRAS or CCND1 amplification who received carboplatin, paclitaxel, and sorafenib, had a longer OS and PFS compared with only carboplatin and paclitaxel.12 The outcomes were attributed to targeting of RAF1 by sorafenib. Hence, it is possible that RAF1 amplification in this tumor type might sensitize cells to sorafenib and other multi-tyrosine kinase inhibitors.
BRAF_V600dup has been reported to be an activating alteration. This alteration is a duplication of codon V600, a “hotspot” codon located within the BRAF kinase domain (amino acids 457–717), which is commonly mutated in cancer. Codon V600 is important for maintaining the Asp-Phe-Gly (DFG) motif of BRAF in an inactive state.13 Alterations at codon V600 increase BRAF activity by stabilizing the active conformation of BRAF. Several studies conducted by other investigators demonstrated that multiple missense mutations at V600 promote constitutive activation of BRAF and alterations at K601 and T599I promote BRAF kinase activity.13–17 These alterations and other overlapping deletion and/or insertion events have been reported to be activating in vitro as evidenced by the activation of BRAF kinase activity, increase in MEK and ERK signaling, and transformation in NIH-3T3 cells.18
The effect of BRAF inhibitors on tumors harboring activating non-BRAF V600 mutations is unclear. Limited preclinical data suggest that the majority of non-BRAF V600 mutations are associated with decreased response or resistance to BRAF inhibitors, vemurafenib and dabrafenib.19,20 In contrast, other investigators reported that activating non-V600 variants may retain some sensitivity to dabrafenib.21 Some preclinical data indicate that MEK and ERK inhibitors may be effective for tumors harboring non-BRAF V600 mutations.21–23
Plasma ctDNA analysis may address tissue heterogeneity by capturing somatic alterations that represent multiple metastatic sites; therefore, it may be more informative than tissue molecular profiling. These alterations may indicate tumor sensitivity to targeted therapies or resistance to treatment. ctDNA analysis is associated with a shorter turnaround time compared with tumor tissue molecular analysis and eliminates the risks associated with invasive biopsies. Identification of tumor genomic alterations using ctDNA analysis also may help select patients for enrollment in clinical trials.
The use of different panels and genomic assays in our study may contribute to the differences in molecular alterations found in tumor biopsy versus ctDNA. Other factors may include the methodology used for genomic assays, tumor heterogeneity, and tumor burden at the time of diagnosis and at the time of disease progression. Notably, in patient ID 002, RAF1 amplification detected using ctDNA analysis was not found in tumor analysis (Table 1). The results are consistent with the concept that ctDNA analysis may be more representative of the overall tumor molecular profiling, regardless of the location of the tumor.
The practice of ordering ctDNA analysis when response to ongoing therapy starts to wane may allow for optimized treatment selection without delay at the time of disease progression. Patients with advanced, metastatic cancer typically have a higher total systemic tumor burden compared with that of patients with early stage disease and, therefore, higher ctDNA levels.24 For instance, in CRC, the ctDNA levels decreased after tumor resection and generally increased with the presentation of new lesions evidenced by imaging scans.24
CtDNA analysis and selected targeted treatments have been well established4,5,25,26 in CRC and NSCLC, and the concordant rates between ctDNA and matched tissue biopsies across other advanced/metastatic solid tumors range from 48% to 100%. The respective published data include patients with breast cancer,27 castration-resistant prostate cancer,28 pancreatic cancer,29 neuroblastoma,30 and melanoma.31
Analysis of ctDNA and tumor tissue samples in patients with metastatic breast cancer using droplet digital polymerase chain reaction analysis reported an overall concordance rate of 74.3% in ESR1 mutation status,27 indicating acquired resistance to endocrine therapy.27 In patients with metastatic, castration-resistant prostate cancer, the concordance between ctDNA and matched metastatic tissue biopsies was 100% in somatic mutations and 88.9% for individual copy number calls of the driver genes androgen receptor (AR), BRCA2, ATM, phosphatase and tensin homolog (PTEN), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta (PIK3CB), phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1), TP53, and RB transcriptional corepressor 1 (RB1).28 In patients with advanced pancreatic ductal adenocarcinoma, the concordance rates between ctDNA and tumor tissue DNA alterations were 61% for TP53 and 52% for KRAS mutations.29 For KRAS alterations, concordance between ctDNA and tumor tissue DNA was significantly higher for metastatic than for primary tumors.29
One 83-year-old male patient with advanced pancreatic ductal adenocarcinoma (PDAC) treated with trametinib based on evidence of MEK pathway (GNAS, KRAS, and NF1) mutations, remained on treatment for 6+ months; although he was unable to undergo CT due to renal insufficiency, the levels of total ctDNA and CA19-9 significantly decreased.29
In patients with neuroblastoma, the concordance rate between ctDNA and tumor tissue for one of three anaplastic lymphoma kinase (ALK) mutational hotspots, the ALK F1174L mutation, was 100%.30 In patients with metastatic melanoma, the concordance rate between ctDNA from plasma compared with tumor tissue ranged from 75% to 100% (average, 89%).31
Serial monitoring of ctDNA can be used to assess clonal evolution, predict progressive disease, and identify resistant clones. Other investigators have reported on ctDNA analysis in diverse solid tumors including advanced breast, ovarian, lung, and colorectal cancers.32–35 CtDNA clonal mutations were assessed to monitor tumor burden and disease progression in patients with advanced gastric cancer.36 For instance, serial monitoring of ctDNA samples from patients with gastric cancer and evaluation of the molecular tumor burden index (mTBI) identified progressive disease a mean of 18 weeks before it was seen on radiographic results. In patients who received anti-HER2 therapy, ctDNA analysis identified 32 expanding mutations potentially related to trastuzumab resistance. In patients who received chemotherapy, prediction of progressive disease using mTBI was validated with 94% sensitivity. Higher mTBI (⩾1%) in pre-treatment ctDNA was associated with shorter PFS.36
Microsatellite instability (MSI) and high tumor mutation burden (TMB-High) are promising pan-tumor biomarkers for the selection of patients to receive checkpoint blockade immunotherapy. Studies conducted by other investigators demonstrated the feasibility of using ctDNA analysis to assess MSI and TMB-high status.37,38 However, sensitive detection of MSI using ctDNA is still in early phases of development owing to technical and bioinformatics challenges including efficient molecular capture, sequencing, mapping, variant calling, error correction at microsatellite loci, the highly repetitive genomic context and low tumor fraction in circulation, high level of normal cell contamination, and technical noise due to polymerase slippage. Other factors affecting the accuracy of MSI detection using ctDNA and associated with discrepancies between studies include the number and type of microsatellite markers used, tumor cell fraction, and intratumor and intertumor heterogeneity.39 In patients with gastric cancer, high MSI status, as assessed by ctDNA analysis, was associated with favorable clinical response to immunotherapy.38 MSI detection using ctDNA sequencing demonstrated high specificity, precision, and sensitivity up to a limit of 0.1% tumor content. CtDNA analysis detected 87% of tissue with high MSI and 99.5% of microsatellite stable tissue, with an overall accuracy of 98.4% and a 95% positive predictive value. Objective responses were noted in 63% of these patients.38 Ongoing studies are investigating the role of ctDNA in assessment of MSI and TMB-high status as predictive biomarkers for the selection of checkpoint inhibitors.
Encouraging results have been reported using ctDNA analysis to select targeted therapy across tumor types. The TARGET (Tumour chARacterisation to Guide Experimental Targeted therapy) study, which was initiated in 2015, matches patients with diverse advanced tumors to early-phase clinical trials based on the tumor molecular abnormalities identified by ctDNA analysis.40 In 100 patients (Part A) with advanced colorectal cancer, breast cancer, NSCLC, small-cell lung carcinoma, sarcoma, prostate cancer, cholangiocarcinoma, small bowel cancer, melanoma, adrenal cancer, solitary fibrous tumor, cancer of unknown primary, or other cancers, the concordance rate between mutations identified in ctDNA and in the tumor tissue was 70%. Genomic profiling of ctDNA using NGS identified actionable mutations in 41 patients, of whom 11 patients received a matched therapy and 17 patients received a non-matched therapy.40 Four of the 11 patients treated with targeted therapy had a PR lasting for 8, 12, 18, and 20 months, respectively; and four of the 17 patients treated with non-matched therapy had SD (no objective response was noted). The study is accruing patients in Part B with a planned enrollment of 450 patients over 3 years to compare clinical outcomes between matched and non-matched therapies.40 The TARGET study indicates the feasibility of using ctDNA analysis to guide targeted therapy in early-phase clinical trials. Notably, in August 2020, the FDA approved Guardant360 CDx, the first liquid biopsy companion diagnostic approved to provide information on multiple solid tumor biomarkers.41 In October and November 2020, the FDA also approved FoundationOne Liquid CDx test (Foundation Medicine, Inc.) as a companion diagnostic device for multiple additional biomarkers detected in cell-free DNA isolated from plasma specimens.42
Clinical trials that allow ctDNA analysis for selection of targeted therapy are in development, including the TAPUR (Targeted Agent and Profiling Utilization Registry) study, a non-randomized, open-label, multi-basket, pragmatic phase II precision oncology trial that integrates tumor genomic profiling using tumor biopsy and/or liquid biopsy to match patients with diverse advanced cancers to targeted anticancer agents outside of their FDA-approved indications (NCT02693535).43 The NEXT-2 [Next Generation pErsonalized tX (Therapy) with plasma DNA] trial is another ongoing prospective study being conducted in Korea that evaluates the role of ctDNA in refractory solid tumors (NCT02140463).44 The investigators reported that 30 (15.5%) of 195 patients who underwent comprehensive ctDNA genomic profiling were enrolled on clinical trials with matched therapy and noted objective responses in six of nine patients with gastric cancer, 13 of 15 patients with NSCLC, one of two patients with melanoma, and zero of one patient in the other tumor types category.44
Limitations to ctDNA analysis are associated with discordance between ctDNA and tumor tissue genomic analysis due to biologic and technical differences (sensitivity and specificity), metastatic sites, histology, time of treatment administration prior to blood/plasma collection, low tumor burden, and absence of a primary tumor.25,45–47 CtDNA is secreted by tumor cells, phagocyte-engulfed tumor cells, and necrotic or apoptotic tumor cells, and therefore reflects tumor from all sites of disease.3 In addition, genomic analysis using only ctDNA may exclude other biomarkers. Analysis of circulating RNAs, circulating tumor cells, and exosomes may overcome this challenge.45
In conclusion, ctDNA and tumor profiling for treatment optimization for patients with advanced solid tumors requires greater precision. Although the role of ctDNA in guiding targeted treatments is well established in NSCLC and in CRC,4,5 the assessment of ctDNA and its clinical significance in other tumor types remains to be validated, despite some encouraging results. Prospective clinical trials involving ctDNA analysis and targeted agents are ongoing and/or awaiting results. ctDNA analysis at the time of diagnosis, or early evidence of progressive disease during ongoing treatment, will yield significant insights into the evolution of the patient’s tumor biology, accelerate drug development, and contribute to the implementation of precision medicine to improve clinical outcomes.
Acknowledgments
We thank the patients, their families, and caregivers for participating in the study.
Footnotes
Authors’ contributions: MFN and HHC contributed to literature search, data acquisition, data analysis, data interpretation and writing the manuscript. DV contributed to data acquisition, data analysis, data interpretation, and review of the manuscript. AMT (corresponding author) conceptualized the manuscript, contributed to patient enrollment, patient treatment and assessment, data analysis, data interpretation and writing the manuscript. All authors read and approved the final manuscript.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Supported by donor funds from Jamie’s Hope, Mr. and Mrs. Zane W. Arrott, and Mr. and Mrs. Steven McKenzie for Dr. Tsimberidou’s Personalized Medicine Program. This work was also supported in part by the National Institutes of Health/National Cancer Institute award number P30 CA016672 (University of Texas MD Anderson Cancer Center).
Conflict of interest statement: Apostolia M. Tsimberidou: Clinical Trial Research Funding/Grant Support: IMMATICS, Parker Institute for Cancer Immunotherapy, Tempus, OBI Pharma, EMD Serono, Baxalta, ONYX, Bayer, Boston Biomedical, Placon Therapeutics, Karus Therapeutics, Tvardi, CPRIT; Travel: ASCO, Japanese Society of Medical Oncology.
Consulting or Advisory Role: Covance, Genentech, Tempus
David J. Vining: Other ownership interests: VisionSR, Majority owner and CEO, multimedia structured reporting for use in advancing medical research; Bracco Diagnostics, Inventor, virtual colonoscopy-related products for colorectal cancer imaging.
The remaining authors have no competing interests.
Ethics approval and consent to participate: The protocol was approved by the FDA and the Institutional Review Board at The University of Texas MD Anderson Cancer Center. The study was conducted in accordance with the Declaration of Helsinki and the International Conference on Harmonization Good Clinical Practice guidelines. All the study participants provided written informed consent before enrollment on the individual clinical trials in which they participated.
Consent for publication: Patient consent for publication is not required. Patients consented to participate in the study.
ORCID iD: Apostolia-Maria Tsimberidou  https://orcid.org/0000-0003-2713-233X
https://orcid.org/0000-0003-2713-233X
Availability of data and material: Data are available upon reasonable request. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request and approval from study sponsor and institution according to available guidelines at time of request.
Contributor Information
Mohammad Faraz Naqvi, Department of Investigational Cancer Therapeutics, Phase I Clinical Trials Program, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Henry Hiep Vo, Department of Investigational Cancer Therapeutics, Phase I Clinical Trials Program, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
David Vining, Department of Abdominal Imaging, Division of Diagnostic Imaging, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Apostolia-Maria Tsimberidou, Department of Investigational Cancer Therapeutics, Phase I Clinical Trials Program, The University of Texas MD Anderson Cancer Center, Unit 455, 1515 Holcombe Boulevard, Houston, TX 77030, USA.
References
- 1. Said R, Tsimberidou AM. Basket trials and the MD Anderson precision medicine clinical trials platform. Cancer J 2019; 25: 282–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tam AL, Papadimitrakopoulou V, Wistuba II, et al. The value of interventional radiology in clinical trial teams: experience from the BATTLE lung cancer trials. Clin Radiol 2021; 76: 155.e25-155.e34. [DOI] [PubMed] [Google Scholar]
- 3. Said R, Guibert N, Oxnard GR, et al. Circulating tumor DNA analysis in the era of precision oncology. Oncotarget 2020; 11: 188–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol 2016; 34: 3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014; 20: 430–435. [DOI] [PubMed] [Google Scholar]
- 6. Finn RS, Dering J, Conklin D, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res 2009; 11: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cabrera MC, Diaz-Cruz ES, Kallakury BV, et al. The CDK4/6 inhibitor PD0332991 reverses epithelial dysplasia associated with abnormal activation of the cyclin-CDK-Rb pathway. Cancer Prev Res (Phila) 2012; 5: 810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dagogo-Jack I, Rooney M, Lin JJ, et al. Treatment with next-generation ALK inhibitors fuels plasma ALK mutation diversity. Clin Cancer Res 2019; 25: 6662–6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Imielinski M, Greulich H, Kaplan B, et al. Oncogenic and sorafenib-sensitive ARAF mutations in lung adenocarcinoma. J Clin Invest 2014; 124: 1582–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li D, Mrsny RJ. Oncogenic Raf-1 disrupts epithelial tight junctions via downregulation of occludin. J Cell Biol 2000; 148: 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoyle PE, Moye PW, Steelman LS, et al. Differential abilities of the Raf family of protein kinases to abrogate cytokine dependency and prevent apoptosis in murine hematopoietic cells by a MEK1-dependent mechanism. Leukemia 2000; 14: 642–656. [DOI] [PubMed] [Google Scholar]
- 12. Wilson MA, Zhao F, Khare S, et al. Copy number changes are associated with response to treatment with carboplatin, paclitaxel, and sorafenib in melanoma. Clin Cancer Res 2016; 22: 374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004; 116: 855–867. [DOI] [PubMed] [Google Scholar]
- 14. Houben R, Becker JC, Kappel A, et al. Constitutive activation of the Ras-Raf signaling pathway in metastatic melanoma is associated with poor prognosis. J Carcinog 2004; 3: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feng YZ, Shiozawa T, Miyamoto T, et al. BRAF mutation in endometrial carcinoma and hyperplasia: correlation with KRAS and p53 mutations and mismatch repair protein expression. Clin Cancer Res 2005; 11: 6133–6138. [DOI] [PubMed] [Google Scholar]
- 16. Matsuse M, Mitsutake N, Tanimura S, et al. Functional characterization of the novel BRAF complex mutation, BRAF(V600delinsYM), identified in papillary thyroid carcinoma. Int J Cancer 2013; 132: 738–743. [DOI] [PubMed] [Google Scholar]
- 17. Cantwell-Dorris ER, O’Leary JJ, Sheils OM. BRAFV600E: implications for carcinogenesis and molecular therapy. Mol Cancer Ther 2011; 10: 385–394. [DOI] [PubMed] [Google Scholar]
- 18. Eisenhardt AE, Olbrich H, Roring M, et al. Functional characterization of a BRAF insertion mutant associated with pilocytic astrocytoma. Int J Cancer 2011; 129: 2297–2303. [DOI] [PubMed] [Google Scholar]
- 19. Yang H, Higgins B, Kolinsky K, et al. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res 2010; 70: 5518–5527. [DOI] [PubMed] [Google Scholar]
- 20. Joshi M, Rice SJ, Liu X, et al. Trametinib with or without vemurafenib in BRAF mutated non-small cell lung cancer. PLoS One 2015; 10: e0118210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Noeparast A, Teugels E, Giron P, et al. Non-V600 BRAF mutations recurrently found in lung cancer predict sensitivity to the combination of Trametinib and Dabrafenib. Oncotarget 2017; 8: 60094–60108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anastasaki C, Estep AL, Marais R, et al. Kinase-activating and kinase-impaired cardio-facio-cutaneous syndrome alleles have activity during zebrafish development and are sensitive to small molecule inhibitors. Hum Mol Genet 2009; 18: 2543–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pratilas CA, Hanrahan AJ, Halilovic E, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res 2008; 68: 9375–9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008; 14: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grasselli J, Elez E, Caratu G, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol 2017; 28: 1294–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bando H, Kagawa Y, Kato T, et al. A multicentre, prospective study of plasma circulating tumour DNA test for detecting RAS mutation in patients with metastatic colorectal cancer. Br J Cancer 2019; 120: 982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, et al. Comparison of ESR1 mutations in tumor tissue and matched plasma samples from metastatic breast cancer patients. Transl Oncol 2017; 10: 766–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wyatt AW, Annala M, Aggarwal R, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst 2017; 109: djx118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patel H, Okamura R, Fanta P, et al. Clinical correlates of blood-derived circulating tumor DNA in pancreatic cancer. J Hematol Oncol 2019; 12: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Combaret V, Iacono I, Bellini A, et al. Detection of tumor ALK status in neuroblastoma patients using peripheral blood. Cancer Med 2015; 4: 540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Calapre L, Giardina T, Robinson C, et al. Locus-specific concordance of genomic alterations between tissue and plasma circulating tumor DNA in metastatic melanoma. Mol Oncol 2019; 13: 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov 2015; 5: 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schiavon G, Hrebien S, Garcia-Murillas I, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med 2015; 7: 313ra182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Russo M, Siravegna G, Blaszkowsky LS, et al. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer Discov 2016; 6: 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013; 497: 108–112. [DOI] [PubMed] [Google Scholar]
- 36. Wang Y, Zhao C, Chang L, et al. Circulating tumor DNA analyses predict progressive disease and indicate trastuzumab-resistant mechanism in advanced gastric cancer. EBioMedicine 2019; 43: 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Georgiadis A, Durham JN, Keefer LA, et al. Noninvasive detection of microsatellite instability and high tumor mutation burden in cancer patients treated with PD-1 blockade. Clin Cancer Res 2019; 25: 7024–7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Willis J, Lefterova MI, Artyomenko A, et al. Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin Cancer Res 2019; 25: 7035–7045. [DOI] [PubMed] [Google Scholar]
- 39. Wang L, Ajani JA. Ushering in liquid biopsy for the microsatellite status: advantages and caveats. Clin Cancer Res 2019; 25: 6887–6889. [DOI] [PubMed] [Google Scholar]
- 40. Rothwell DG, Ayub M, Cook N, et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med 2019; 25: 738–743. [DOI] [PubMed] [Google Scholar]
- 41. U. S. Food and Drug Administration. FDA approves first liquid biopsy next-generation sequencing companion diagnostic test, https://www.fda.gov/news-events/press-announcements/fda-approves-first-liquid-biopsy-next-generation-sequencing-companion-diagnostic-test (2020, accessed 11 January 2021).
- 42. U. S. Food and Drug Administration. FDA approves liquid biopsy NGS companion diagnostic test for multiple cancers and biomarkers, https://www.fda.gov/drugs/fda-approves-liquid-biopsy-ngs-companion-diagnostic-test-multiple-cancers-and-biomarkers (2020, accessed 11 January 2021).
- 43. Targeted Agent and Profiling Utilization Registry (TAPURTM) study, https://www.tapur.org/ (2020, accessed 30 July 2020).
- 44. Kim ST, Banks KC, Lee S-H, et al. Prospective feasibility study for using cell-free circulating tumor DNA–guided therapy in refractory metastatic solid cancers: an interim analysis. JCO Precis Oncol 2017; 1: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakamura Y, Yoshino T. Clinical utility of analyzing circulating tumor DNA in patients with metastatic colorectal cancer. Oncologist 2018; 23: 1310–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vidal J, Muinelo L, Dalmases A, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol 2017; 28: 1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bachet JB, Bouche O, Taieb J, et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: the AGEO RASANC prospective multicenter study. Ann Oncol 2018; 29: 1211–1219. [DOI] [PubMed] [Google Scholar]