Abstract
Background
Lung cancer is a malignant tumor with the highest morbidity and mortality rates worldwide, of which lung adenocarcinoma (LUAD) is the most common subtype. Overall, current treatments of LUAD are not satisfactory; therefore, novel targets need to be explored. Let-7b-3p is an important member of the let-7 family of microRNAs (miRNAs), and has not been studied separately in LUAD. This study aimed to investigate the role and molecular mechanism of let-7b-3p in LUAD.
Methods
Herein, let-7b-3p expression was detected by quantitative real-time polymerase chain reaction (qRT-PCR) and fluorescence in situ hybridization (FISH) assays. MTT, colony formation assay, flow cytometry analysis, wound-healing, Transwell and in vivo experiments were conducted to assess let-7b-3p’s function in LUAD. The downstream target TFIIB-related factor 2 (BRF2) was predicted using bioinformatics analyses and confirmed by dual-luciferase reporter assay and rescue experiments. Additionally, BRF2 was found to affect the MAPK/ERK pathway through transcriptome sequencing analysis and western blot (WB) assay.
Results
Let-7b-3p is downregulated in LUAD cells and tissue samples and low let-7b-3p expression is correlated with a poor prognosis in LUAD patients. Let-7b-3p suppresses the proliferation and metastasis of LUAD cells both in vivo and in vitro by directly targeting the BRF2-mediated MAPK/ERK pathway.
Conclusions
Let-7b-3p inhibits the development of LUAD and is an ideal novel therapeutic target for the treatment of LUAD.
Keywords: Lung adenocarcinoma (LUAD), let-7b-3p, TFIIB-related factor 2 (BRF2), MAPK/ERK pathway
Introduction
Lung cancer is the most common and fatal malignancy in humans with lung adenocarcinoma (LUAD) being the predominant form (1,2). Despite significant improvements being made in targeted therapy in recent years, the clinical prognosis of LUAD patients remains dismal (3), and thus further investigations into more effective therapeutic targets for LUAD are required.
MicroRNAs (miRNAs) are about 20-nucleotide-long single-stranded noncoding RNAs (4). The hallmark function of miRNAs is to downregulate the expression of their target genes by binding to their mRNAs’ 3' untranslated region (3'UTR) (5). Studies have suggested that the majority of coding genes are potential targets of miRNAs (6). In addition, miRNAs are aberrantly altered in many diseases including a wide variety of cancers (4,7). Let-7 is among the first miRNAs that were discovered and widely accepted as a tumor suppressor miRNA family (8,9). Several let-7 family members have been progressively used for clinical diagnosis and prognostic evaluation of lung cancer (10-12). Let-7b-3p, previously known as let-7b, was conventionally regarded as the passenger strand was considered to have no biological function. In recent years, however, relevant studies have revealed let-7b-3p to play a significant role in various of diseases (13-15). Despite this, the specific role and mechanism of let-7b-3p in LUAD remains unclear.
RNA polymerase III (RNA Pol III) is an important component of the eukaryotic transcriptional machinery that participates in transcription of small untranslated RNAs (16). Abnormally expressed Pol III products have been found in multiple cancer types (17). TFIIB-related factor 2 (BRF2) is a subunit of the RNA Pol III transcription factor (TFIIIB) complex and critical for RNA Pol III to initiate transcription accurately (18,19). BRF2 acts as an oncogene in numerous malignancies such as lung, gastric, kidney, and esophageal cancers (20-22). Although some studies have indicated that BRF2 can promote the progression of lung cancer cells (23-25), the role of BRF2 in vivo and the mechanisms of action of BRF2 in lung cancer, especially in LUAD remain largely unknown.
In this study, we detected the expression of let-7b-3p in LUAD tissues and cell lines and examined the functions of let-7b-3p in LUAD both in vitro and in vivo. Let-7b-3p expression was found to be downregulated in both LUAD cells and tissues. Moreover, low let-7b-3p expression was associated with a poorer prognosis of LUAD patients. Both in vitro and in vivo experiments showed that let-7b-3p suppressed the development of LUAD cells by directly targeting BRF2. Transcriptome sequencing analysis and Western blotting (WB) assay also confirmed that BRF2 could affect MAPK/ERK pathway. Overall, this study provides insight into a new therapeutic miRNA target for the clinical treatment of LUAD. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-21-299).
Methods
Clinical tissue specimen collection and ethical approvals
Fifty LUAD tissues and corresponding adjacent normal lung tissues were collected from patients undergoing surgery in the Thoracic Surgery Department, Qilu Hospital of Shandong University from January 2018 to July 2019. All tissue samples were immediately placed into liquid nitrogen after surgical resection. Ten pairs of LUAD cancer tissue and their corresponding adjacent tissue blocks, prepared in triplicate separately for hematoxylin and eosin (HE) staining and immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) assays, were obtained from the pathology department of our hospital. This study was reviewed and approved by the Medical Ethics Committee of Qilu Hospital of Shandong University (No. KYLL-2016-097) and conformed to the tenets of the Declaration of Helsinki (as revised in 2013). Each patient or their family signed an informed consent form.
Cell cultures, transfection, and lentivirus transduction
LUAD cell lines (A549, PC9, H1299, H157, H1975), human bronchial epithelial (HBE) cell line, and HEK293 cell line were obtained from Shanghai Academy of Science (Shanghai, China). PPMI-1640 with 10% fetal bovine serum (FBS; Gibco, USA) was used as medium, and the cells were incubated under 5% CO2 at 37 °C. Mimic-let-7b-3p, inhibitor-let-7b-3p, pcDNA3.1-BRF2, si-BRF2 (5'-GCCUACUACCAACAGGCAUTT-3'), and their corresponding negative controls were purchased from GenePharma Co., Ltd. (Shanghai, China) for in vitro assays. They were separately transfected into H1299 and A549 cells using Lipofectamine 3000 (Thermo Fisher Scientific, USA). The anti-let-7b-3p lentivirus (LV-let-7b-3p-inhibitor) and the corresponding negative control (LV-NC-inhibitor) were sourced from Jikai Co. (Shanghai, China) and transduced into A549 cells for in vivo experiments. The stably transduced cells were selected using puromycin (2 µg/mL) for 7 days.
Total RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA from the cells and tissues was isolated using TRIzol reagent (Takara, Japan) and the complement DNA (cDNA) was subsequently generated by a reverse transcription kit (Takara, Japan). qRT-PCR amplification was then done using the Bio-Rad IQ 5 real-time PCR system with SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). GAPDH and U6 were used as endogenous controls. The primers were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). The oligonucleotides sequences were as follows: BRF2 (F: 5'-GTGAAGCTCCTGGGACTGGAT-3', R: 5'-GTATTTGGCTGGCACAGAAGG-3'); GAPDH (F: 5'-AGGTCGGTGTGAACGGATTTG-3', R: 5'-TGTAGACCATGTAGTTGAGGTCA-3'); Has-let-7b-3p (F: 5'-CCTCCACCCTATACAACCTACTGC-3', R: 5'-CTCGACCCTGGCACCT-3'); U6 (F: 5'-CGCTTCGGCAGCACATATAC-3', R: 5'-TTCACGAATTTGCGTGTCAT-3'). The tests were performed in triplicate, with data analysis being performed using the 2–∆∆CT method.
WB assay
Total protein was isolated using RIPA lysis buffer (Beyotime, Jiangsu, China), and the concentration was measured using a bicinchoninic acid (BCA) kit. Subsequently, the protein was added into 10% SDS-PAGE gels for separation and then transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were then blocked with 5% skim milk for 1 hour at room temperature. After being washed in tris-buffered saline with Tween 20 (TBST) (3 times for 10 minutes), the membranes were incubated with the corresponding primary antibody at 4 °C for 24 hours. They were then washed with TBST (3 times for 10 minutes) and incubated with secondary antibodies for 1 hour at room temperature. All antibodies were purchased from GeneTex (Irvine, CA, USA). Finally, the membranes were detected using the enhanced chemiluminescence (ECL) system, and GAPDH was used as an internal control.
HE staining and IHC
For HE staining, sections were deparaffinized in xylene and then hydrated with decreasing concentrations of alcohol. The sections were then stained with HE and imaged under an optical microscope. For IHC, the sections were first dewaxed and rehydrated. They were then incubated with primary antibodies overnight at 4 °C. Subsequently, they were incubated with secondary antibodies at room temperature for 1 hour after 3 washes with phosphate-buffered saline (PBS). They were finally viewed, and images were captured using an optical microscope.
MTT cell proliferation assay
A549 and H1299 cells were cultured in 96-well microplates (3,000 cells/well). After transfection, the cells were cultured for 24, 48, 72, and 96 hours, and then, 20 µL of MTT solution (5 mg/mL; Solarbio, China) was added into each well. The cells were subsequently treated with 200 µL dimethyl sulfoxide (DMSO; Solarbio, China) after incubation for 4 hours in absolute darkness. Finally, the absorbance of the cells was detected at 490 nm using a microplate reader. These experiments were repeated 3 times.
Colony formation assay
For colony formation assay, 1,000 transfected A549 or H1299 cells were cultured in each well in 6-well plates for 15 days. The cells were stained with 0.1% crystal violet solution for 15 minutes after 30 minutes of fixation in 4% paraformaldehyde. They were then washed twice with PBS, and their images taken using a digital camera. These experiments were repeated 3 times.
Cell cycle and apoptosis analysis
For cell cycle analysis, harvested cells were fixed in 70% ethanol at 4 °C overnight. They were then suspended in 250 µL of RNase A buffer (100 ng/mL) and labeled with propidium iodide (PI, 25 mg/mL) for 30 minutes at 37 °C. Subsequently, a FACSCalibur Flow Cytometer (BD Biosciences, San Jose, CA, USA) was used to count different cell cycle phases. An Annexin-V-FITC apoptosis detection kit (Beyotime, Shanghai, China) was used for cell apoptosis assay according to the instruction manual. Each experiment was conducted in triplicate.
Wound-healing assay
Cells were evenly seeded into 6-well plates and incubated until they were completely fused. After 3 PBS washes, the plates were scratched in the center using sterile 200 µL pipette tips. The cells were then incubated in serum-free PPMI-1640 medium. The wounds were measured and the micrographs captured at 0 and 24 hours under a microscope. The experiment was repeated 3 times.
Transwell assay
For the Transwell assay, 5×104 transfected A549 or H1299 cells in 200 µL of serum-free PPMI-1640 medium were seeded into the upper chambers of Transwell inserts (Corning, Corning, NY, USA) with or without Matrigel. Then, 600 µL of PPMI-1640 medium containing 20% FBS was added into the lower chambers. After 24-hour incubation, cells remaining in the upper chambers were removed with a cotton swab while those that had invaded the bottom surface were fixed with 75% ice-alcohol for 30 minutes and stained with 1% crystal violet solution for 20 minutes. An inverted microscope was then used for imaging. All assays were independently repeated 3 times.
FISH
Ten pairs of tissue slides were used for the FISH assay using a FISH kit sourced from RiboBio (Guangzhou, China). Briefly, the slides were dewaxed, air-dried, and dehydrated. They were then incubated at 37 °C in protease solution for 2 hours. Next, tissue sections were added with let-7b-3p probe RiboBio (Guangzhou, China) and incubated overnight at 37 °C. After hybridization, the slides were washed (0.4× SSC, 0.3% NP-40) and then counterstained with DAPI. Finally, the slides were visualized, and micrographs taken using a confocal fluorescence microscope.
Dual luciferase assay
HEK-293 cells cultured in 24-well plates were transiently co-transfected with the wild-type (WT) or mutated (MUT) 3‘UTR of BRF2 mRNA together with the mimic-let-7b-3p or mimic-NC. Then, 48 hours after transfection, the cells were collected, and the luciferase activity was measured using a dual luciferase assay kit (GeneCopoeia, Guangzhou, China). These experiments were repeated 3 times.
Experiments in vivo
Six-week-old nude mice were purchased from Shanghai Laboratory Animal Center, Laboratory Animal Company (Shanghai, China). For the tumor xenograft experiment, 8 nude mice were randomly divided into two groups. Each nude mouse was subcutaneously injected in the right armpit with 5×106 A549 cells which stably expressed LV-let-7b-3p-inhibitor or LV-NC-inhibitor. Tumor formation was monitored every 4 days up to 24 days post inoculation. The volumes of tumors were estimated using the following equation: V = (length × width2)/2. The mice were euthanized and the tumors were harvested at the end of the study (day 24). Then, 6 other nude mice were randomly divided into two groups to establish the lung metastasis experimental model. Each nude mouse received a tail vein injection with 2×106 A549 cells that expressed LV-let-7b-3p-inhibitor or LV-NC-inhibitor. The lungs of the mice were harvested after 2 months, and their metastatic nodes were counted. All the specimens were further subjected to HE staining and IHC. The animal experiments were performed under a project license (SCXK Lu 20090001) granted by the Shandong University Animal Research Ethics Committee, in compliance with institutional guidelines for the care and use of animals.
Data acquisition and transcriptome sequencing
Gene expression data and clinical information of LUAD patients in The Cancer Genome Atlas (TCGA) database were downloaded from UCSC Xena Browser (https://xenabrowser.net/). The downstream targets of let-7b-3p were predicted by MiRDB (www.mirdb.org), MiRbase (www.mirbase.org), and TargetScan (www.targetscan.org) bioinformatics software. Library preparation and transcriptome sequencing was performed by the HuaDa Corporation (Beijing, China).
Statistical analysis
All experiments were independently repeated at 3 times. Data analyses were conducted using the GraphPad Prism (GraphPad, San Diego, CA, USA) and R (version 3.6.1) software (R Foundation for Statistical Computing, Vienna, Austria). Experimental results are expressed as mean ± SD. Student’s t-test (two-sided), Kaplan-Meier survival analysis, and Pearson’s correlation analysis were performed as indicated. One-way ANOVA was used to compare the differences among more than two groups. A P value less than 0.05 (P<0.05) indicated a significant difference between groups.
Results
Let-7b-3p expression was downregulated in LUAD
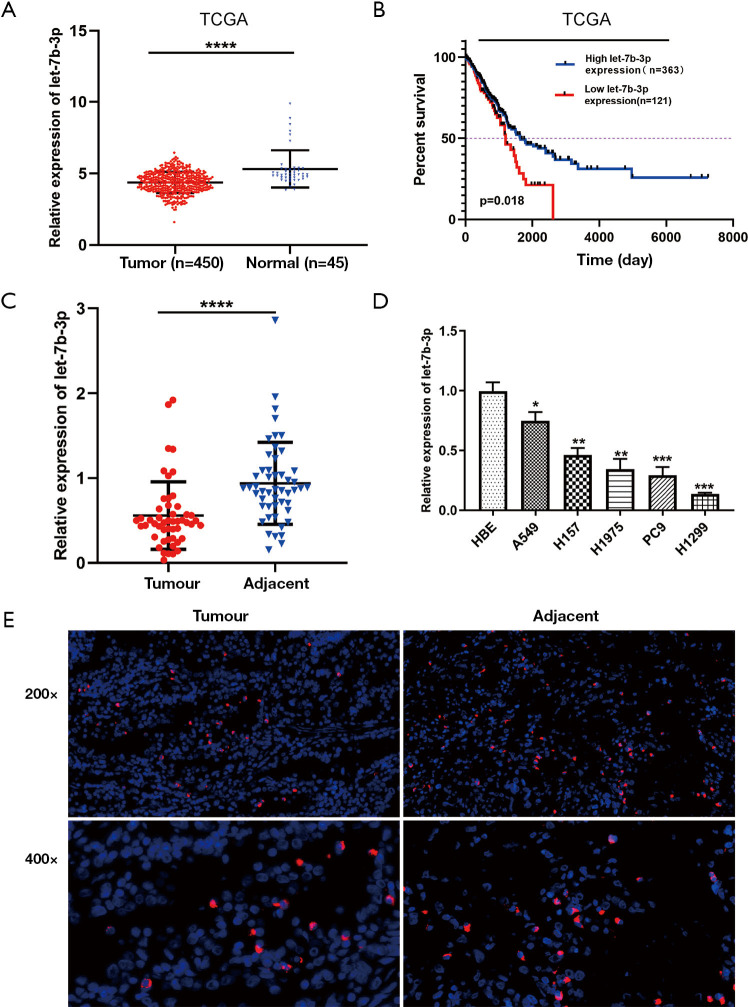
Let-7b-3p expression was first analyzed in TCGA database. Compared with normal lung tissues, decreased let-7b-3p expression was observed in LUAD tissues (P<0.0001; Figure 1A). The Kaplan-Meier survival analysis showed that patients with low let-7b-3p expression had a lower overall survival percentage (P=0.018; Figure 1B). Then, qRT-PCR assay was used to detect let-7b-3p expression in 50 pairs of LUAD tissues and matched paracancerous tissues. Significantly low expression levels of let-7b-3p were observed in LUAD tissues (P<0.0001; Figure 1C). At the cellular level, let-7b-3p expression was significantly decreased in the 5 LUAD cell lines compared to that in the HBE cell line according to qRT-PCR assay (Figure 1D). FISH assay further revealed that adjacent normal lung tissues had a higher proportion of let-7b-3p expressed cells than LUAD tissues (Figure 1E). In conclusion, let-7b-3p expression was decreased, and low let-7b-3p expression was correlated with a poor prognosis in LUAD patients.
Figure 1.
Let-7b-3p expression was decreased in LUAD tissues and cell lines. (A) Let-7b-3p expression in TCGA database. (B) Kaplan-Meier analysis of overall survival according to TCGA database. (C) Let-7b-3p expression in clinical specimens was detected by qRT-PCR (n=50). (D) Let-7b-3p expression in cell lines was assessed by qRT-PCR. (E) FISH of LUAD tumor tissues and corresponding adjacent nontumorous tissues. The probes were labeled by Cy3 (red) and the nuclei are counter-stained with DAPI (blue) (*, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001). LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas; qRT-PCR, quantitative real-time polymerase chain reaction; FISH, fluorescence in situ hybridization.
Let-7b-3p inhibited LUAD cell proliferation
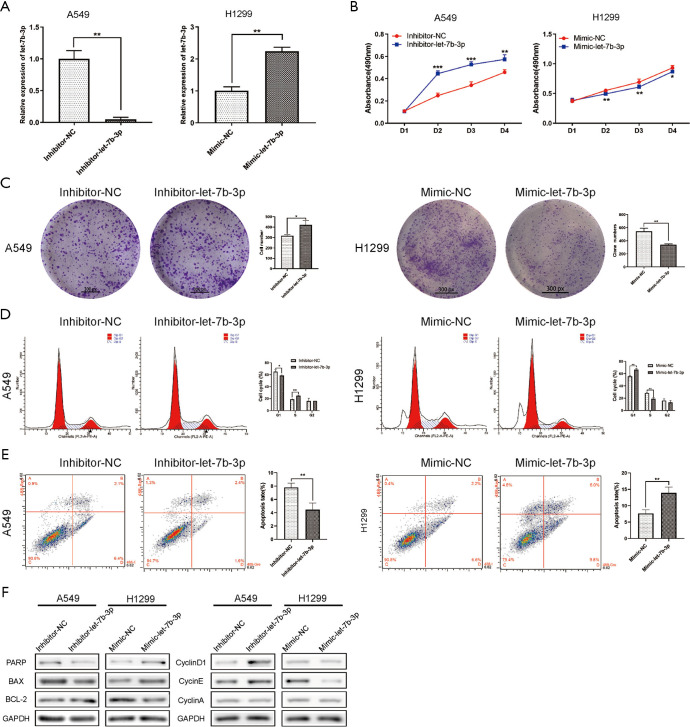
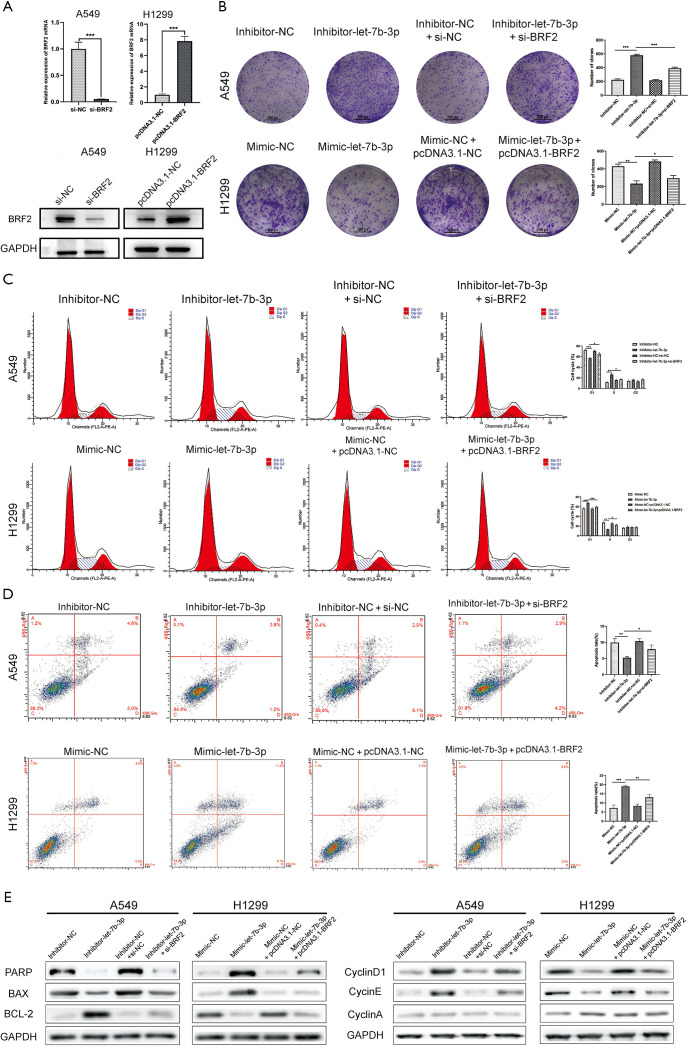
Among the LUAD cell lines, A549 cells had the highest let-7b-3p expression level while H1299 cells had the lowest expression level (Figure 1D). These two cell lines were therefore selected for the follow-up experiments. A549 and H1299 cells were transfected with inhibitor-let-7b-3p, mimic-let-7b-3p, and their corresponding negative controls. qRT-PCR assay was then used for detecting the transfection efficiency. Let-7b-3p expression was significantly decreased in A549 cells (P<0.01) while H1299 cells had a markedly upregulated let-7b-3p expression (P<0.01) compared to the negative controls (Figure 2A). When MTT and colony formation assays were performed, A549 cells in the inhibitor-let-7b-3p group were found to have a higher cell growth activity than the cells in the negative control group. In the H1299 cell line, the growth activity was significantly decreased in mimic-let-7b-3p H1299 cells compared to mimic-NC H1299 cells (Figure 2B,C). Flow cytometry results revealed that relatively high let-7b-3p expression significantly increased the G1-phase cell population but in turn reduced the cell population in the S phase (Figure 2D). This indicated that let-7b-3p induced G1/S cell cycle arrest. Besides this, relatively high let-7b-3p expression groups had more apoptotic A549 and H1299 cells (Figure 2E). We then explored the expression of proteins associated with cell cycle and cell apoptosis. When let-7b-3p expression was downregulated in A549 cells, the expression of ribose polymerase (PARP) and BAX was decreased while that of BCL-2, cyclin D1, and cyclin E was increased. The opposite trend was observed when let-7b-3p expression was upregulated in H1299 cells. Cyclin A showed no change in both A549 and H1299 cells (Figure 2F). Overall, let-7b-3p was found to inhibit LUAD cell proliferation.
Figure 2.
Let-7b-3p inhibited LUAD cell proliferation. (A) The transfection efficiency in A549 and H1299 cells was verified by qRT-PCR. (B) Cell proliferation activity was assessed by MTT assay. (C) The clonogenic capacity of transfected cells was detected by clone formation assay. The cells were stained with crystal violet. (D,E) The effect of let-7b-3p on cell cycle and cell apoptosis was detected by flow cytometry. (F) The expression of cell cycle and cell apoptosis-related proteins were detected by WB. Data are presented as mean ± SD according to Student’s t-test. (*, P<0.05; **, P<0.01; ***, P<0.001). LUAD, lung adenocarcinoma; qRT-PCR, quantitative real-time polymerase chain reaction; WB, Western blot.
Let-7b-3p inhibited LUAD cell invasion and migration
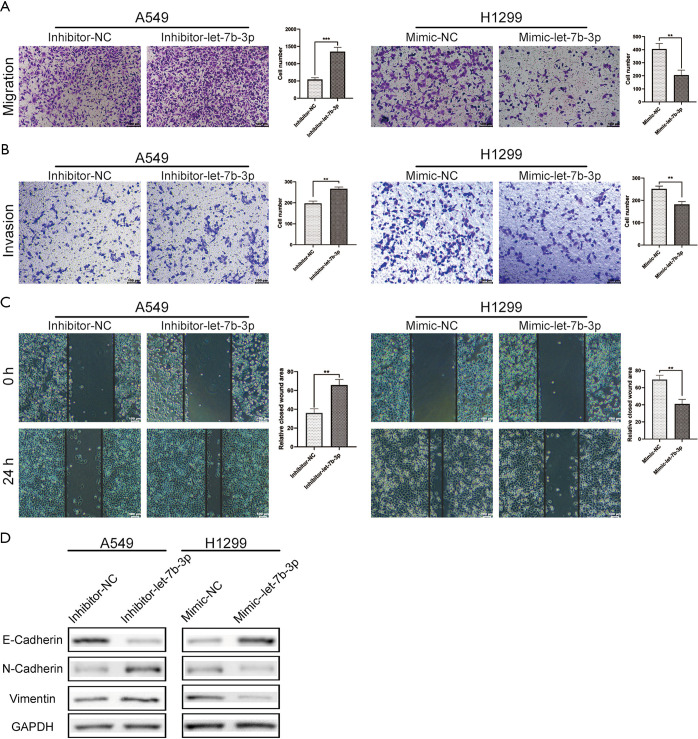
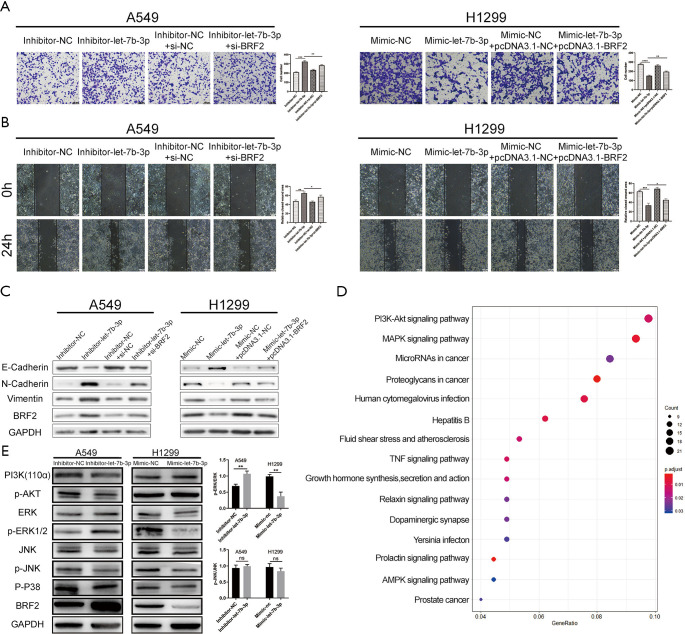
Cell migration and invasion abilities were assessed using Transwell and scratch wound-healing assays. As shown in Figure 3A,B,C, let-7b-3p knockdown significantly strengthened cell migration and invasion abilities of A549 cells, while let-7b-3p overexpression dramatically inhibited these abilities in H1299 cells. We further examined epithelial-mesenchymal transition (EMT)-associated marker proteins in transfected A549 and H1299 cells. Knockdown of let-7b-3p expression in A549 cells suppressed the expression of E-cadherin and promoted the expression of N-cadherin and Vimentin. Conversely, the expression of E-cadherin was upregulated while the expression of N-cadherin and Vimentin was downregulated in mimic-let-7b-3p H1299 cells (Figure 3D). Thus, let-7b-3p was found to inhibit LUAD cell invasion and migration.
Figure 3.
Let-7b-3p inhibited LUAD cell invasion and migration. (A,B) Migration and invasion ability of transfected cells was evaluated by Transwell assay (without or with Matrigel). Scale bars in (A) and (B) represent 100 µm. (C) Migration ability of transfected cells was detected by wound-healing experiment. Scale bars represent 100 µm. (D) The expression of EMT-related proteins was detected by WB. Data are presented as mean ± SD according to Student’s t-test (**, P<0.01; ***, P<0.001). LUAD, lung adenocarcinoma; EMT, epithelial-mesenchymal transition; WB, Western blot.
BRF2 was a direct target of let-7b-3p
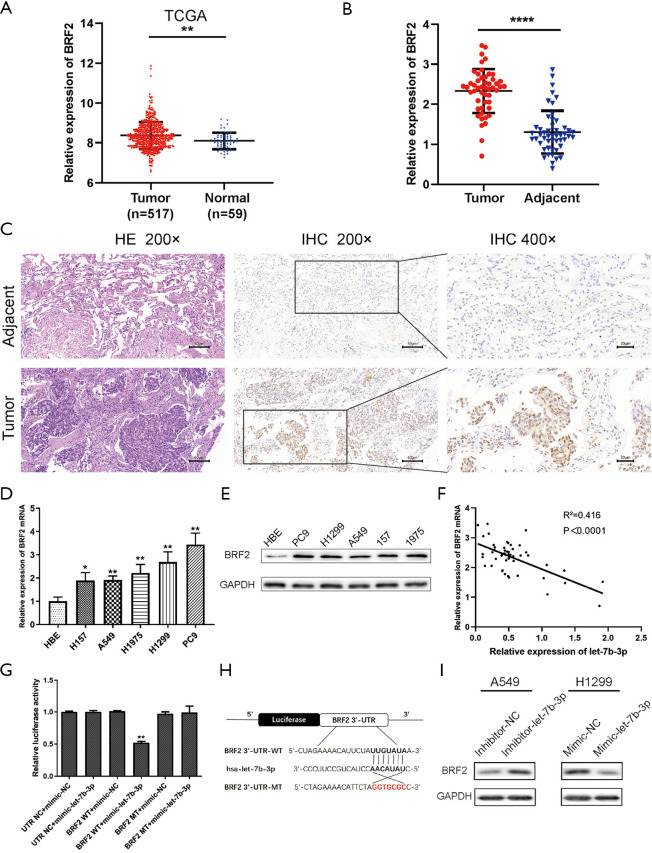
A search was conducted in MiRDB, MiRbase, and TargetScan prediction programs to investigate the downstream targets of let-7b-3p. BRF2 was finally confirmed to be a potential target of let-7b-3p. Compared with normal lung tissues, BRF2 expression level was much higher in LUAD tissues in TCGA database (P<0.01; Figure 4A). The expression of BRF2 was also determined at both the cell and tissue level by qRT-PCR, IHC, and WB tests, and it was found to be increased in both LUAD tissues and cells (Figure 4B,C,D,E). In addition, the expression of BRF2 mRNA and let-7b-3p in LUAD tissues presented a negative correlation (Figure 4F; r2=0.4164; P<0.0001). Dual-luciferase reporter assay was then conducted to verify whether there is a direct relationship between let-7b-3p and BRF2 mRNA. Let-7b-3p decreased luciferase activity in the WT 3'UTR group whereas it had no effect on the MUT 3'UTR group (P<0.01, Figure 4G). The possible binding site of let-7b-3p and BRF2 3'UTR is presented in Figure 4H. WB results showed BRF2 expression was dramatically upregulated when let-7b-3p expression was inhibited in A549 cells, while it was significantly reduced when the expression of let-7b-3p was upregulated in H1299 cells (Figure 4I). All the above results confirmed that BRF2 is a direct downstream target of let-7b-3p.
Figure 4.
BRF2 was a direct target of let-7b-3p. (A) The expression of BRF2 in TCGA database. (B) BRF2 expression in clinical specimens was detected by qRT-PCR (n=50). (C) HE staining of LUAD tumor tissues and adjacent normal tissues and the expression of BRF2 was detected by IHC. (D,E) BRF2 expression in cell lines was assessed by qRT-PCR and WB. (F) Pearson’s correlation analysis between let-7b-3p and BRF2 mRNA expression in LUAD tumor tissues. (G) BRF2 3'UTR luciferase reporter assays in HEK293 cells. (H) The predicted binding site between let-7b-3p and BRF2 3'UTR. (I) BRF2 protein expression in transfected A549 and H1299 cells was assessed by WB (*, P<0.05; **, P<0.01; ****, P<0.0001). BRF2, TFIIB-related factor 2; TCGA, The Cancer Genome Atlas; qRT-PCR, quantitative real-time polymerase chain reaction; HE, hematoxylin and eosin; LUAD, lung adenocarcinoma; IHC, immunohistochemistry; 3'UTR, 3' untranslated region.
Let-7b-3p exerted its tumor-suppressive functions via targeting the BRF2-mediated MAPK/ERK pathway
Rescue experiments were conducted to further confirm if BRF2 was responsible for the function of let-7b-3p on LUAD cells. pcDNA3.1-BRF2 was used to upregulate BRF2 expression in mimic-let-7b-3p H1299 cells while the inhibitor-let-7b-3p A549 cells were transfected with si-BRF2 to downregulate its expression. The knockdown efficiency of si-BRF2 and overexpression efficiency of pcDNA3.1-BRF2 at both protein and mRNA levels was showed in Figure 5A. Colony formation and flow cytometry assays revealed that BRF2 reversed let-7b-3p-induced inhibition of LUAD cell proliferation (Figure 5B,C,D). At the same time, the expression of the proteins related to cell cycle and cell apoptosis changed correspondingly (Figure 5E) Further to this, Transwell and wound-healing assays revealed that BRF2 reversed the migratory and invasive features induced by let-7b-3p (Figure 6A,B). Meanwhile, the expression of the proteins related to EMT changed correspondingly (Figure 6C). We further performed RNA-sequencing (RNA-seq) transcriptome analyses after knocking down the expression of BRF2 by si-RNA in H1299 cells to investigate the signaling pathways that were affected by BRF2. R software was then used to conduct Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Figure 6D). It was found that, when BRF2 was knocked down, PI3K/AKT and MAPK/ERK pathways were significantly changed. As such, PI3K/AKT and MAPK/ERK signaling pathway-related proteins were detected in both transfected A549 and H1299 cells lines using WB. Among those proteins, the expression of PI3K and p-AKT changed slightly, while p-ERK1/2 was remarkedly changed accordingly when BRF2 expression was altered by the up-or downregulation of let-7b-3p. Beyond this, the bands of p-ERK, total ERK, p-JNK and total JNK were quantified and the ratios of p-ERK to total ERK and p-JNK to total JNK were shown. In A549 cells, the ratio of p-ERK to total ERK in the Inhibitor-let-7b-3p group was notably higher than that in the Inhibitor-NC group (P<0.01). In H1299 cells, this ratio was significantly lower in the Mimic-let-7b-3p group than that in the Mimic-NC group (P<0.01). However, the ratio of p-JNK to total JNK showed no appreciable difference among the groups in both A549 cells and H1299 cells (Figure 6E). The above results strongly indicated that let-7b-3p exerts its tumor-suppressive functions mainly by regulating the BRF2-mediated MAPK/ERK pathway.
Figure 5.
BRF2 reversed let-7b-3p-induced inhibition of LUAD cell proliferation. (A) The knockdown efficiency of si-BRF2 in A549 cells and the overexpression efficiency of pcDNA3.1-BRF2 in H1299 cells were measured by q-PCR and WB assays. (B) Upregulation of BRF2 reversed the inhibitory effect of let-7b-3p on cell proliferation, while downregulation of BRF2 enhanced this effect. The cells were stained with crystal violet. (C,D) The effects of inhibiting cell cycle progression and accelerating apoptosis of let-7b-3p was reversed by high expression of BRF2, while knockdown of BRF2 enhanced these effects. (E) BRF2 reversed the expression changes of cell cycle and cell apoptosis-related proteins induced by let-7b-3p. Data are presented as mean ± SD according to Student’s t-test (*, P<0.05; **, P<0.01; ***, P<0.001). BRF2, TFIIB-related factor 2; LUAD, lung adenocarcinoma; q-PCR, quantitative polymerase chain reaction; WB, Western blot.
Figure 6.
Let-7b-3p exerted tumor-suppressor effects via the BRF2-mediated MAPK/ERK pathway in LUAD. (A,B) Restoration of BRF2 rescued the invasive and migratory phenotypes induced by let-7b-3p. The cells were stained with crystal violet. Scale bars in (A) and (B) represent 100 µm. (C) The variance of protein expression induced by let-7b-3p was partly restored by BRF2. (D) KEGG pathway enrichment analysis. (E) The expression of PI3K/AKT and MAPK pathway-related proteins were determined by WB. Data are presented as mean ± SD according to Student’s t-test (*, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001; ns, no significance). BRF2, TFIIB-related factor 2; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Let-7b-3p inhibited tumor growth and metastasis of LUAD in vivo
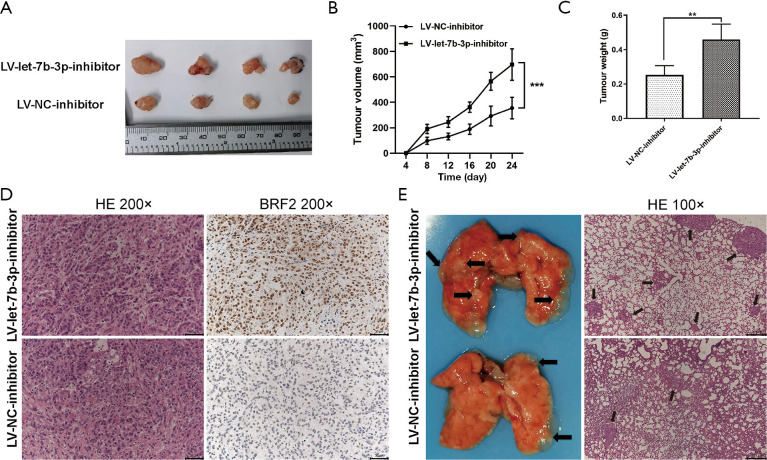
A nude mouse tumorigenesis test was used to evaluate the inhibitory effects of let-7b-3p on tumor growth. Mice in the LV-let-7b-3p-inhibitor group exhibited a significantly faster tumor growth rate compared to the mice in the LV-NC-inhibitor group (Figure 7A,B; P<0.001). Besides this, the weight of the tumor tissues in the LV-let-7b-3p-inhibitor group was much higher (Figure 7C; P<0.01). IHC of the tumor specimens showed that BRF2 expression was significantly increased in the LV-let-7b-3p-inhibitor compared to that in the control group (Figure 7D). We also performed pulmonary metastasis assay to determine the inhibitory effects of let-7b-3p on tumor metastasis. More lung metastatic nodules were observed in the LV-let-7b-3p-inhibitor group than in the LV-NC-inhibitor group (Figure 7E). These results proved that let-7b-3p inhibits LUAD development via targeting BRF2 in vivo.
Figure 7.
Let-7b-3p inhibited tumor growth and metastasis in vivo. (A) An image of subcutaneous tumors. (B) The growth curve of subcutaneous tumors of different groups. (C) The weight of tumors in different groups. (D) HE staining of the subcutaneous tumors and the expression of BRF2 was detected by IHC. (E) Images (left) and HE staining (right) of the lungs obtained from different groups; black arrowheads indicate lung metastasis nodules. (**, P<0.01; ***, P<0.001). HE, hematoxylin and eosin; BRF2, TFIIB-related factor 2; IHC, immunohistochemistry.
Discussion
Let-7b-3p is an indispensable member of the let-7 family. Over the past years, accumulated studies have revealed that let-7b-3p is closely related to malignant diseases. In 2018, it was reported that let-7b-3p inhibits the progression of hepatocellular carcinoma by targeting PLK1 (26). Recently, Min et al. reported that let-7b-3p may be a biomarker of early colon cancer (27). However, thus far, no study has focused on the role of let-7b-3p in LUAD. To our knowledge, this study is the first to report the role and mechanism of let-7b-3p in LUAD both in vitro and in vivo. We found in this study that let-7b-3p expression was obviously downregulated in both LUAD cell lines and tissues compared with normal ones. Moreover, LUAD patients with low let-7b-3p expression had a poorer prognosis. This suggests that let-7b-3p may be a tumor suppressor in LUAD. Results of in vitro and in vivo experiments further revealed that let-7b-3p suppressed the development of LUAD via modulating multiple links, such as cell proliferation, cell cycle, cell apoptosis, and EMT.
Subsequent dual-luciferase and rescue experiments confirmed that BRF2 is directly targeted by let-7b-3p and responsible for the biological function of let-7b-3p. BRF2 is indicated to be a subunit of the RNA Pol III transcription initiation complex and essential for recruiting Pol III to promote transcription initiation (28). BRF2 has been found to be overexpressed in lung cancer and has been associated with a poor prognosis of patients with lung cancer (20). The results of our study also support this view. Therefore, BRF2 inhibition may be a promising new strategy for LUAD therapy. Previous studies have reported that knocking down the expression of BRF2 could suppress the growth and metastasis of lung cancer cells (24,29). However, the above studies lack comprehensive in vivo experiments. In this study, in addition to conducting a series of in vitro experiments, we also carried out tumorigenesis and pulmonary metastasis assays. It was observed that knocking down BRF2 expression by upregulating the expression of let-7b-3p could effectively inhibit the proliferation and metastatic abilities of LUAD cells both in vitro and in vivo. In contrast, exogenous knockdown the expression of let-7b-3p caused the upregulation of BRF2 and promoted the progression of LUAD. These findings further clarify the BRF2-related gene regulatory networks in LUAD.
Tumor development and progression is often accompanied by the excessive activation of many signaling pathways, including the PI3K/AKT and MAPK signaling pathways (30-32). In order to investigate the pathways that are affected by the let-7b-3p/BRF2 axis, we conducted transcriptome sequencing after knocking down the expression of BRF2. KEGG pathway enrichment analysis suggested that silencing of BRF2 affected a series of pathways including PI3K/AKT and MAPK pathways. The ensuing WB experiment indicated that, BRF2 silencing could not inhibit the activation of PI3K/AKT pathway, it obviously suppressed the MAPK/ERK pathway activation. Thus, we concluded that let-7b-3p exerts its tumor suppressor function mainly via a BRF2-mediated MAPK/ERK pathway.
Notably, miRNAs are known to be double edged sword, and depending on the stimulation and context can play different roles. Apart from that, let-7b-3p may exert its function by simultaneously targeting other genes in LUAD. Consequently, more studies about let-7b-3p, especially in the context of different genetic alterations are required.
Conclusions
This is the first study to comprehensively examine let-7b-3p miRNA in LUAD. The expression of let-7b-3p is significantly decreased in both LUAD cell lines and tissues. Functionally, let-7b-3p inhibits the proliferation and metastasis of LUAD cells both in vitro and in vivo. Mechanistically, let-7b-3p suppresses the development of LUAD mainly by directly targeting the BRF2-meditated MAPK/ERK pathway, and is thus a potential therapeutic target of LUAD.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors appreciate the academic support from AME Lung Cancer Collaborative Group.
Funding: This research was funded by the National Natural Science Foundation of China (No. 81672292) and the Taishan Scholar Program of Shandong Province (No. ts201712087).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments related to patients were reviewed and approved by the Medical Ethics Committee of Qilu Hospital of Shandong University (No. KYLL-2016-097) and conformed to the tenets of the Declaration of Helsinki (as revised in 2013). Each patient or their family signed an informed consent form. The animal experiments were performed under a project license (SCXK Lu 20090001) granted by the Shandong University Animal Research Ethics Committee, in compliance with institutional guidelines for the care and use of animals.
Footnotes
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-21-299
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-21-299
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-21-299). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Cheng TY, Cramb SM, Baade PD, et al. The international epidemiology of lung cancer: latest trends, disparities, and tumor characteristics. J Thorac Oncol 2016;11:1653-71. 10.1016/j.jtho.2016.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: a review. JAMA 2019;322:764-74. 10.1001/jama.2019.11058 [DOI] [PubMed] [Google Scholar]
- 4.Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 2016;96:1297-325. 10.1152/physrev.00041.2015 [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105. 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. 10.1073/pnas.0510565103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000;403:901-6. 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- 9.Büssing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med 2008;14:400-9. 10.1016/j.molmed.2008.07.001 [DOI] [PubMed] [Google Scholar]
- 10.Dai X, Fan W, Wang Y, et al. Combined delivery of let-7b microRNA and paclitaxel via biodegradable nanoassemblies for the treatment of KRAS mutant cancer. Mol Pharm 2016;13:520-33. 10.1021/acs.molpharmaceut.5b00756 [DOI] [PubMed] [Google Scholar]
- 11.Leidinger P, Keller A, Meese E. MicroRNAs - important molecules in lung cancer research. Front Genet 2012;2:104. 10.3389/fgene.2011.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamichhane SR, Thachil T, De Ieso P, et al. Prognostic role of microRNAs in human non-small-cell lung cancer: a systematic review and meta-analysis. Dis Markers 2018;2018:8309015. 10.1155/2018/8309015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Z, Bing Z, Shaohuan Q, et al. Expression of miRNAs in plasma exosomes derived from patients with atrial fibrillation. Clin Cardiol 2020;43:1450-9. 10.1002/clc.23461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao R, Dong X, Gong J, et al. hsa-miR-106b-5p participates in the development of chronic thromboembolic pulmonary hypertension via targeting matrix metalloproteinase 2. Pulm Circ 2020;10:2045894020928300. 10.1177/2045894020928300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butler AE, Ramachandran V, Cunningham TK, et al. Increased microRNA levels in women with polycystic ovarian syndrome but without insulin resistance: a pilot prospective study. Front Endocrinol (Lausanne) 2020;11:571357. 10.3389/fendo.2020.571357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White RJ. Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genet 2011;12:459-63. 10.1038/nrg3001 [DOI] [PubMed] [Google Scholar]
- 17.White RJ. RNA polymerase III transcription and cancer. Oncogene 2004;23:3208-16. 10.1038/sj.onc.1207547 [DOI] [PubMed] [Google Scholar]
- 18.Gouge J, Guthertz N, Kramm K, et al. Molecular mechanisms of Bdp1 in TFIIIB assembly and RNA polymerase III transcription initiation. Nat Commun 2017;8:130. 10.1038/s41467-017-00126-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dergai O, Cousin P, Gouge J, et al. Mechanism of selective recruitment of RNA polymerases II and III to snRNA gene promoters. Genes Dev 2018;32:711-22. 10.1101/gad.314245.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabarcas S, Schramm L. RNA polymerase III transcription in cancer: the BRF2 connection. Mol Cancer 2011;10:47. 10.1186/1476-4598-10-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu M, Tian H, Yue W, et al. Overexpression of TFIIB-related factor 2 is significantly correlated with tumor angiogenesis and poor survival in patients with esophageal squamous cell cancer. Med Oncol 2013;30:553. 10.1007/s12032-013-0553-4 [DOI] [PubMed] [Google Scholar]
- 22.Lockwood WW, Chari R, Coe BP, et al. Integrative genomic analyses identify BRF2 as a novel lineage-specific oncogene in lung squamous cell carcinoma. PLoS Med 2010;7:e1000315. 10.1371/journal.pmed.1000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu M, Tian H, Yue W, et al. TFIIB-related factor 2 over expression is a prognosis marker for early-stage non-small cell lung cancer correlated with tumor angiogenesis. PLoS One 2014;9:e88032. 10.1371/journal.pone.0088032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bian Y, Li Q, Li Q, et al. Silencing of BRF2 inhibits the growth and metastasis of lung cancer cells. Mol Med Rep 2020;22:1767-74. 10.3892/mmr.2020.11285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Qu J, Zhou L, et al. MicroRNA-373 inhibits cell proliferation and invasion via targeting BRF2 in human non-small cell lung cancer A549 cell line. Cancer Res Treat 2018;50:936-49. 10.4143/crt.2017.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Z, Deng W, Jiang B, et al. Hsa-let-7b inhibits cell proliferation by targeting PLK1 in HCC. Gene 2018;673:46-55. 10.1016/j.gene.2018.06.047 [DOI] [PubMed] [Google Scholar]
- 27.Min L, Zhu S, Chen L, et al. Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: a comparison with plasma total miRNAs. J Extracell Vesicles 2019;8:1643670. 10.1080/20013078.2019.1643670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larminie CG, Cairns CA, Mital R, et al. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J 1997;16:2061-71. 10.1093/emboj/16.8.2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Han L, Liu Z, et al. Long noncoding RNA MNX1-AS1 contributes to lung cancer progression through the miR-527/BRF2 pathway. J Cell Physiol 2019;234:13843-50. 10.1002/jcp.28064 [DOI] [PubMed] [Google Scholar]
- 30.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 2009;9:537-49. 10.1038/nrc2694 [DOI] [PubMed] [Google Scholar]
- 31.Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer 2020;20:74-88. 10.1038/s41568-019-0216-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bugaj LJ, Sabnis AJ, Mitchell A, et al. Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science 2018;361:eaao3048. 10.1126/science.aao3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as