Abstract
Background
The efficacy of immune checkpoint inhibitors (ICIs) remains unexpected and in some patients the resistance to anti-programmed death-1 (anti-PD-1) and anti-programmed death ligand 1 (anti-PD-L1) agents is observed. One of possible explanation may be PD-L2 activity. PD-1 ligands: PD-L1 and PD-L2 are present on cancer cells but also, not without significance, on alveolar macrophages (AMs) contributing to immune-suppression in the tumor microenvironment. The aim of this study was to analyse PD-L2, PD-L1 expression on AMs in bronchoalveolar lavage fluid (BALF) in relation to PD-1 positive T lymphocytes.
Methods
Seventeen patients with lung cancer were investigated. BALF cells from the lung with cancer (clBALF) and from the opposite “healthy” lung (hlBALF) and peripheral blood (PB) lymphocytes were investigated. Flow cytometry method was used.
Results
We found that 100% of CD68+ AMs from the clBALF were PD-L1 and PD-L2-positive. Unexpectedly, fluorescence minus one (FMO) PD-L1 and PD-L2 stained controls and isotype controls also showed strong autofluorescence. The hlBALF AMs exhibited a similar PD-L1 and PD-L2 autofluorescence. The median proportion of PD-1+ T lymphocytes was higher in the clBALF than the hlBALF and PB (28.9 vs. 23.4% vs. 15.6%, P=0.0281).
Conclusions
We discussed the opportunities of exploring the PD-1-PD-L1/PD-L2 pathway in the lung cancer environment, which may help to find new potential biomarkers for immunotherapy. We concluded that precise identification by flow cytometry of macrophages in the BALF is possible, but our study showed that the autofluorescence of macrophages did not allow to assess a real expression of PD-L2 as well as PD-L1 on AMs.
Keywords: Macrophages, bronchoalveolar lavage fluid (BALF), PD-L1, PD-L2, lung cancer microenvironment
Introduction
Lung cancer
Lung cancer is the leading cause of cancer mortality in both women and men worldwide (1,2). Non-small cell lung cancer (NSCLC) accounts for the majority of all lung cancer cases. With the onset of immune checkpoint inhibitors (ICIs) the outlook for patients with advanced NSCLC improved, but the benefit does not exceed 40% of patients (3,4). The results of immunotherapy in lung cancer are not so spectacular as in other tumors, like melanoma (5). It could be explained by very heterogenous character of lung tumors in histology, molecular biology and immunity of respiratory tract. The lack of defined biomarkers leads to no possibility of proper qualification to immunotherapy. The more the group of patients qualified to immunotherapy are patients, which differ in clinical and pathological stage of the disease and performance status. In the ongoing efforts to stimulate non-responders to achieve therapeutic benefits, numerous additional immunomodulatory pathways are being explored. More accurate biomarkers for individual therapy and understanding of the mechanisms of primary and acquired resistance to ICIs are needed.
The programmed death-1/programmed death ligand 1 (PD-1/PD-L1) and PD-L2 checkpoint pathway
In NSCLC, the PD-1/PD-L1 blocking therapy has become a major pillar of immunotherapy. During cancer progression, immune function is impaired, including progressive loss of T-cell effector responses and immunosuppression. The PD-1 pathway has been reported to play a key role in the depletion of T cells in the course of cancer progression, and its blockade may restore many T cell functions (6). PD-1 and its ligands: PD-L1 (B7-H1, CD274) and PD-L2 (B7-H2, B7-DC, CD273), belong to the B7:CD28 family and regulate T cell functions and peripheral immune response (7).
The reaction between PD-1 and its ligands provides an inhibitory signal for T lymphocytes, influencing cytokine production and cell proliferation (8). The two PD-ligands (PD-Ls) display a different expression pattern and are varied in function. PD-L1 is expressed on epithelial and non-epithelial cells, PD-L2 on non-epithelial cells: dendritic cells (DCs) and macrophages, and depending on the local immunity, on other immune cells (9-11). PD-L2 is a cell surface protein that competes with PD-L1 for binding to PD-1 and has a higher affinity for PD-1, but it is expressed at lower levels (12). It is not excluded that PD-L2 expression is more dependent on environmental stimuli [mainly Th2 stimulation and interferon gamma (13)] than PD-L1, which appears to be expressed in a more constitutive way, although this can be further upregulated by pro-inflammatory factors (11). Although the role of PD-1-related pathways has been broadly described (14) and the usefulness of detection of PD-L1 expression on tumor cells was approved as a biomarker for ICIs (15-17) the significance of PD-L1 and especially of PD-L2 detected on non-epithelial cells are not understood completely. Recent data indicate that the role of PD-L2 in malignancy is noteworthy (10,13,18-20). This molecule could be a target for ICIs in patients with low or no PD-L1 expression (15). A high expression of PD-L2 in tumors and relation with molecular alterations (KRAS, EGFR mutations) were observed (13,19). There are some reports on the role of PD-L2 as prognostic factor in lung cancer (20-22). Finally, one can assume that PD-L2 may contribute to lower frequency of immune related adverse events of ICIs (23,24).
This study aimed to evaluate PD-L2 and PD-L1 expression on alveolar macrophages (AMs) in relation to T cell in lung cancer patients. We investigated AMs from bronchoalveolar lavage fluid (BALF) from the lung with cancer (clBALF) and from the opposite “healthy” lung (hlBALF). Our choice of BAL as an investigation method arises from our experience and from the possibility of adopting it in practice (25). We previously showed that the constituents of BALF from the lung with tumor reflect the character of the local immune status (26-28). Macrophages from the main population of BALF cells, up to half a million cells could be available for analysis. Flow cytometry seems to be a method of choice for analyzing BALF cells and their phenotype identification.
We present the study in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1103).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Military Institute of Medicine Ethics Committee (No. 47/WIM/2017) and informed consent was taken from all the patients.
Patients
BALF and PB were obtained from 21 patients during diagnostic bronchoscopy. The exclusion criteria were: anti-cancer therapy, chronic obstructive pulmonary disease (COPD), infection, immunosuppressive therapy, and autoimmune diseases, bloody or purulent BALF. Finally, 17 patients with histologically confirmed primary NSCLC were included. Written informed consent was signed by each patient before each diagnostic procedure (the Military Institute of Medicine Ethics Committee, number 47/WIM/2017). There were 11 women and 6 men, in the mean age 66.5±6.6 years, in I–IIIA stage of the disease in the study group. There were no control group; taking BALF from a healthy person without any lung disease is impossible for ethical reasons.
Materials
BALF was collected from the lung affected by cancer and from the opposite site: “healthy” lung of the same patient during one procedure. One hundred mL of 0.9% NaCl solution was applied to each lung and then removed. BALF processing has been performed as recommended. Cell viability was each time assessed by trypan blue and 7-AAD (Becton Dickinson BD, USA, catalog number: 559925, lot: 7061885) and BALF had a lifetime of over 90% (29). Additionally, 2 mL of PB were obtained.
Flow cytometry analysis
Monoclonal antibodies (Becton Dickinson BD, USA) targeting the cell-surface expression of CD45-V500 (catalog number: 655873, lot: 0164828, clone number: 2D1), HLA-DR-V450 (catalog number: 655874, lot: 0035775, clone number: L243) and intracellular CD68-PE-Cy7 (catalog number: 565595, lot: 7333702, clone number: Y1/82A) were used for AMs identification. AMs were defined as CD45+ bright/CD68+ bright/HLA-DR+ bright and high on SSC/FSC. The subpopulations of lymphocytes T and CD4+ or CD8+ in PB and BALF were analyzed by a panel of multicolor monoclonal antibodies: anti-CD45-V500 (catalog number: 655873, lot: 0164828, clone number: 2D1), anti-CD3-APC-H7(catalog number: 333771, lot: 9324872, clone SK7), anti-CD8-V450 (catalog number: 560347, lot: 9298615, clone RPA-T8) or anti-CD8-APC (catalog number: 345775, lot: 8225759, clone SK1), anti-CD4-PerCP-Cy5.5 (catalog number: 332772, lot: 0064229, clone SK3). The anti-PD-L1-PE (catalog number: 557924, lot: 9273698, clone MIH1) and anti-PD-L2-PerCP-Cy5.5 (catalog number: 564256, lot: 9120940, clone MIH18) antibodies were used to evaluate the expression of these molecules on AMs in BALF and the anti-PD-1-FITC (catalog number: 564494, lot: 9136884, clone EH12.1) was applied to assess the presence of this molecule on the subpopulations of lymphocytes T. Fluorescence minus one (FMO) control for PD-L1-PE and PD-L2-PerCP-Cy5.5 and isotype control (PE Mouse IgG1, κ Isotype Control catalog number: 555749, lot: 5225509, clone: MOPC-21, PerCP-Cy™5.5 Mouse IgG1 κ Isotype Control catalog number: 552834, lot: 7158535 clone MOPC-21) were applied. To extend the use of controls, in addition to the FMO panel for PD-L1 and PD-L2, and isotype control antibodies were placed of the markers of interest. This control was then serve two purposes, the first one to ensure that the isotypes do not cause any background staining in the channels and the second one, to monitor as compensation/autofluorescence overlap.
The FACS Canto II flow cytometer (BD, USA) was used to process the samples. The intensity of geometric mean fluorescence (GMF) and cell proportion (%) were the subject of our analysis. Examinations were performed using BD FACSDiva™ Software 8.0.1 (BD, USA) and Infinicyt 1.8 Flow Cytometry (Cytognos, Salamanca, Spain).
Statistical analysis
Kruskal-Wallis ANOVA, post-hoc analysis test and the Pearson r coefficient were used. The value P<0.05 was considered as statistically significant. All analyses were performed using the Statistica 13.0 software (TIBCO Software, Palo Alto, CA, USA).
Results
The percentage of macrophages and lymphocytes in the investigated group was presented in Table 1. The median percentage of AMs was higher in clBALF than in hlBALF (38.1 q25–q75 range: 28.1–72.3 vs. 33.4 q25–q75 range: 14.4–45.3). Considering the T cell subpopulations, we observed differences of the main T cell subpopulations between both the BALF and PB. The CD4/CD8 ratio was lower in the clBALF and hlBALF than in the PB.
Table 1. Median proportion of the cells from BALF: lymphocytes, lymphocytes T (CD4+, CD8+) and macrophages in the tumor environment (clBALF), hlBALF and PB.
| WBC and cells | clBALF | hlBALF | PB |
|---|---|---|---|
| WBC (k/μL) | 91 [52–114] | 79 [66–187] | 8,920 [8,250–9,420] |
| Lymphocytes | 7.3 (5.0–10.5) | 9.6 (6.6–13.3) | 38.9 (32.2–44.5) |
| Lymphocytes T CD3+ | 4.7 (2.9–6.3) | 7.0 (5.6–11.1) | 19.7 (14.4–27.4) |
| T CD3+ CD4+ | 1.2 (1.0–1.8) | 2.2 (1.4–2.3) | 8.2 (4.2– 11.6) |
| T CD3+ CD8+ | 2.5 (2.1–5.2) | 3.8 (2.7–8.7) | 11.5 (7.6–11.5) |
| CD4/CD8 | 0.4 (0.3–1.0) | 0.4 (0.3–1.0) | 0.9 (0.2–1.5) |
| AMs | 38.1 (28.1–72.3) | 33.4 (14.4–45.3) |
Data expressed as median (q25–q75). BALF, bronchoalveolar lavage fluid; clBALF, BALF from the lung with cancer; hlBALF, BALF from the opposite “healthy” lung; PB, peripheral blood; AMs, alveolar macrophages.
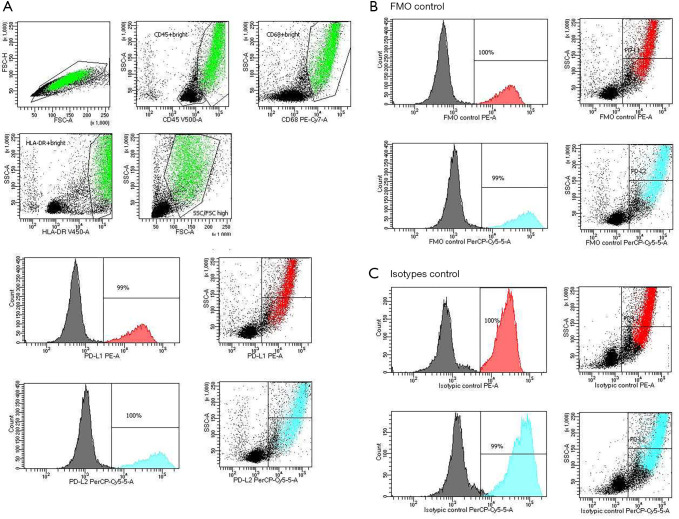
Prior to beginning the experiment, it was important to set up and evaluate FMO and isotype controls. Figure 1 shows a representative BALF macrophages gating strategy. First to removed clumps, we gated the cells that have height and an equal area [greater FSC-A compared to FSC-H and debris (very low FSC) (FSC-A vs. FSC-H plot)]. Next, we selected AMs based on their SSC/CD45, SSC/CD68, SSC/HLA-DR and SSC/FSC properties: high expression of CD45, CD68, HLA-DR and FSC. Finally, the expression of PD-L1, PD-L2 on AMs in the clBALF and hlBALF were examined. HLA-DR expression was high in both clBALF and hlBALF on macrophages. The HLA-DR expression was used mainly in the gating strategy of macrophages in clBALF and hlBALF.
Figure 1.
FACS analysis of BALF cells with specific antibodies for AMs and PD-L1 or PD-L2 expression: (A) CD45+ bright/CD68+ bright/HLA-DR+ bright on SSC/FSC macrophage population was gated (green population) and “PD-L1 positive population AMs” (red population: approximately 100%) and “PD-L2 positive population AMs” (blue population: approximately 100%) were determined. (B) FMO controls for PD-L1-PE minus and PD-L2-PerCp-Cy5.5 minus were analyzed. The percentages of autofluorescent population were shown in the SSC/FL1 (PE) (red population 100%) and in the SSC/FL1 (PerCp-Cy5.5) (green population 100%) plots and histograms. (C) Isotypes controls for PD-L1-PE and PD-L2-PerCp-Cy5.5 were evaluated. The percentage of autofluorescent population were shown in the SSC/FL1 (PE isotypes controls) (red population 100%) and in the SSC/FL1 (PerCp-Cy5.5 isotypes control) (green population 100%) plots and histograms. BALF, bronchoalveolar lavage fluid; AM, alveolar macrophage; FMO, fluorescence minus one.
AMs from the clBALF as well as hlBALF were PD-L1, PD-L2-positive approximately in 100%. (Figure 1A with representative macrophages gating strategy and PD-L1, PD-L2 expressions). Unexpectedly, FMO stained control for PD-L1 and PD-L2 also showed strong autofluorescence and similar proportions (approximately 100%) of the PD-L1 and PD-L2 as stained population (Figure 1B). Isotype controls for PD-L1 and PD-L2 also showed strong autofluorescence (Figure 1C). GMF intensity of PD-L1 and PD-L2 on AMs was also difficult to assess being covered by autofluorescence, however, it was slightly higher for PD-L2 (Table 2). T lymphocytes did not express PD-L1 and PD-L2 and did not show autofluorescence.
Table 2. Proportion of PD-L1 and PD-L2 on macrophages in clBALF and hlBALF.
| PD-L1/PD-L2 macrophages | clBALF | hlBALF |
|---|---|---|
| Macrophages PD-L1 (%) | 98.10 (97.10–99.60) | 98.3 (97.6–99.6) |
| Macrophages PD-L2 (%) | 99.20 (99.10–99.80) | 99.6 (9.1–100.0) |
| Macrophages PD-L1 (GMF) | 28,215 (12,283–36,457) | 36,414 (7,392–41,022) |
| Macrophages PD-L2 (GMF) | 60,359 (36,917–87,419) | 83,224 (25,479–95,810) |
Data expressed as median (q25–q75) of % and GMF intensity. BALF, bronchoalveolar lavage fluid; clBALF, BALF from the lung with cancer; hlBALF, BALF from the opposite “healthy” lung; GMF, geometric mean fluorescence.
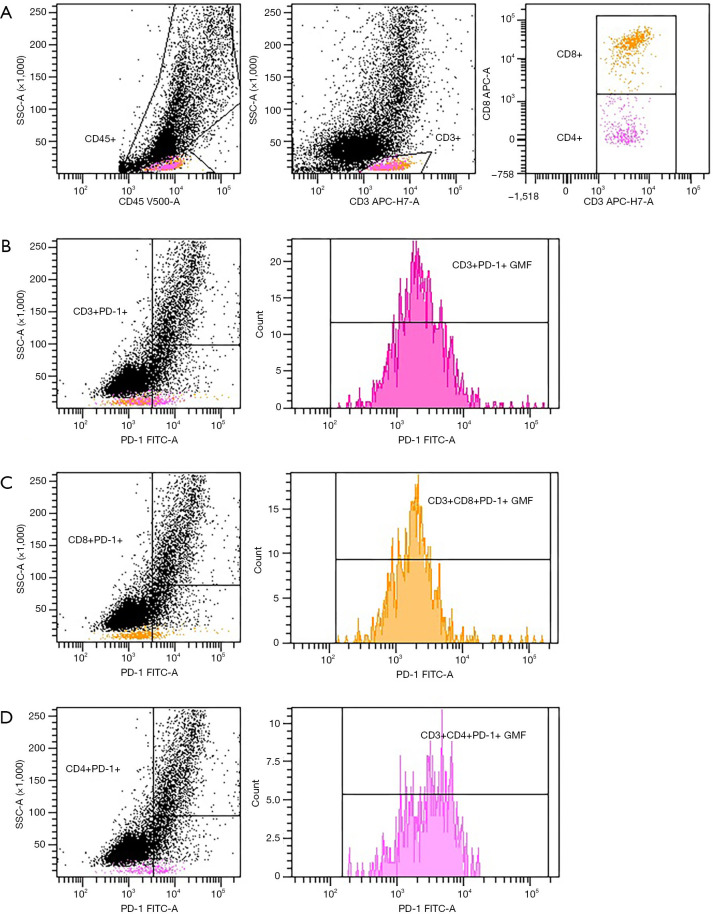
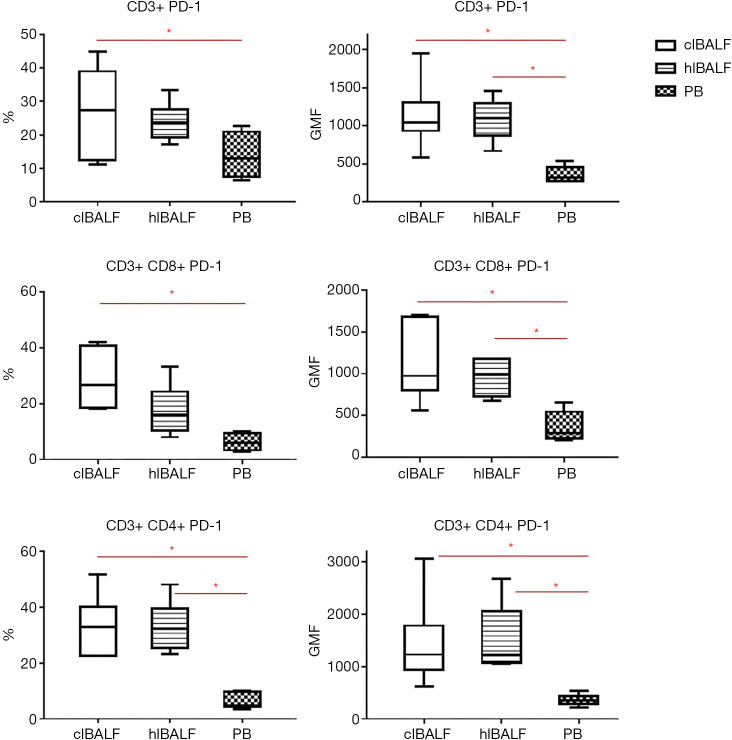
The evaluation of expression of PD-1 on lymphocytes gave much clearer results than for AMs (Figure 2). In general, the median proportion of T cells and T cell subpopulations with the expression of PD-1 was higher in the clBALF than hlBALF and PB (Table 3). The GMF intensity of PD-1 on the BALF lymphocytes was higher than on the PB cells, but did not differ between the clBALF and hlBALF (Figure 3).
Figure 2.
FACS analysis of PD-1 expression on T lymphocytes in clBALF from example patient with lung cancer. (A) Gating strategy of T cells. (B) PD-1 expression on CD3+ T cells read as % and GMF intensity. (C) PD-1 expression on CD8+ T cells read as % and GMF intensity. (D) PD-1 expression on CD4+ T cells read as % and GMF intensity. BALF, bronchoalveolar lavage fluid; clBALF, BALF from the lung with cancer; hlBALF, BALF from the opposite “healthy” lung; GMF, geometric mean fluorescence.
Table 3. Proportion of PD-1 lymphocytes in clBALF, hlBALF and PB.
| PD-1 positive lymphocytes | clBALF (A) | hlBALF (B) | PB (C) | P (group A-B-C) | P (groups A-C, B-C) |
|---|---|---|---|---|---|
| T CD3+ PD-1 (%) | 28.9 (21.7–37.0) | 23.4 (17.3–25.6) | 15.6 (7.9–20.4) | P=0.0281* | A-C: 0.0244*; B-C: – |
| T CD3+ PD-1 (GMF) | 1,039.0 (945–1,481) | 1,074.0 (924–1,291) | 317 (277–453) | P=0.0033* | A-C: 0.0074*; B-C: 0.0148* |
| T CD3+ CD8+ PD-1 (%) | 26.7 (18.5–40.2) | 16.1. (11.0–21.4) | 6.0 (3.5–9.2) | P=0.0023* | A-C: 0.0016*; B-C: – |
| T CD3+ CD8+ PD-1 (GMF) | 974.0 (886–1,672) | 985.5 (744–1,179) | 292.5 (237–513) | P=0.0046* | A-C: 0.0105*; B-C: 0.0175* |
| T CD3+ CD4+ PD-1 (%) | 32.9 (22.7–36.5) | 32.3 (26.2–36.7) | 4.7 (4.6–9.5) | P=0.0034* | A-C: 0.0125*; B-C: 0.0089* |
| T CD3+ CD4+ PD-1 (GMF) | 1,234.0 (1,049–1,358) | 1,227.5 (1,072–1,854) | 354.5 (310–397) | P =0.0033* | A-C: 0.0148*; B-C: 0.0074* |
Data expressed as median (q25–q75) of percentage (%) and GMF intensity. Group A-B-C: ANOVA, Kruskal-Wallis; groups A-C, B-C: post-hoc analysis. *, P<0.05. BALF, bronchoalveolar lavage fluid; clBALF, BALF from the lung with cancer; hlBALF, BALF from the opposite “healthy” lung; PB, peripheral blood; GMF, geometric mean fluorescence.
Figure 3.
The differences between PD-1 expression on T lymphocytes (CD4+ and CD8+) in clBALF, hlBALF and PB patients with lung cancer. (A) PD-1 expression on T cells read as % and GMF intensity. (B) PD-1 expression on CD8+ T cells read as % and GMF. (C) PD-1 expression on CD4+ T cells read as % and GMF. The median values and quartile q25–q75 were shown on graphs (*, P<0.05). BALF, bronchoalveolar lavage fluid; clBALF, BALF from the lung with cancer; hlBALF, BALF from the opposite “healthy” lung; PB, peripheral blood; GMF, geometric mean fluorescence.
Discussion
There are many studies assessing the PD-1/PD-L1 pathway based mainly on the analysis of PD-1 on lymphocytes and PD-L1 on tumor cells (16,30). Researches focus on the detection of PD-1 ligands on cancer cells not on the non-epithelial ones. Whereas it can be assumed that lymphocytes have more contact with macrophages, DCs and other stromal cells in the tumor milieu than with tumor cells. Furthermore, the latter are very different in their vitality and the expression of surface markers importantly differs between fragments of cancer tissue (17). The reported range of proportion of PD-L1 positive cells is wide (31). These reasons prompted us to focus on a rich population of BALF AMs, which is relatively uniform. We could expect to find measurable expression of PD-1 ligands on AMs modulated by the local immunity. Unfortunately, our study shows that the analysis of PD-Ls on BALF AMs by flow cytometry is problematic.
PD-L1/PD-L2 expression on AMs by flow cytometry
Using a combination of the panel of surface and intracellular markers (CD45+ bright/CD68+ bright/HLA-DR+ bright), granularity parameter (high on SSC/FSC) and gating strategy allowed us to identify AMs in BALF by flow cytometry. Other researchers have also confirmed that macrophages can be assessed by flow cytometry using an appropriate analysis strategy (32-35). We found that AMs from the clBALF and from the hlBALF were PD-L1, PD-L2-positive approximately in 100%. FMO stained control for PD-L1 and PD-L2 and isotypes control also showed strong autofluorescence and similar percentages of PD-L1 and PD-L2 as stained population. Thus, the false-positive results for PD-Ls on AMs may be associated with the occurrence of autofluorescence, which may depend on the microenvironment, the type of molecule being tested, depending on the biological and physiological condition of cells. Autofluorescence is a very common phenomenon and can interfere studies using fluorescence labeling. In some samples with low expression of tested antigens and dim dyes, there may be a problem with masking the tested targets in flow cytometry (36). Duan et al. (37) have shown that AMs exhibit high autofluorescence, which often decreases the resolution between “positive” and “negative” populations, leading to false positivity for a given antigen or fluorochrome and that the use of different techniques is helpful (spectral scanning). Garn et al. (38) and Daigneault et al. (39) suggest that a high content of the granularity is associated with a significant number of cell organelles in the macrophage cytoplasm, which is likely to cause autofluorescence. Moreover, in the conditions when some antigens with low baseline expression, which are not bright enough, autofluorescence may completely hinder their detection. After activation in a specific environment their detection could be improved. In addition, for low-expressed antigens, researchers suggest that cytometry may be insufficient (40). There are some ways for improving their detection by flow cytometry. Since autofluorescence is lower at longer light wavelengths, fluorophores emitting above 600 nm will have less autofluorescence interference. The use of a very bright fluorophore will lower the effects of autofluorescence. There are some potential approaches to analyze cells that are naturally autofluorescent: using fluorophores that emit in channels that are not known for autofluorescence or using very bright fluorophores in the channels that exhibit autofluorescence. The use of Alexa488 instead of FITC, PerCP-Cy5.5 instead of PerCP could be helpful (41). Li et al. analysed expression of Foxp3 in macrophages and concluded that the expression is likely an artefact and more specific methods need to be implemented to identify the antigen or a combination of several methods, for example immunofluorescence, immunohistochemistry (IHC) and molecular methods (40,42). However, there are some reports on the possibility of assessing PD-1 ligands by flow cytometry. These studies are mostly carried out on mouse material after in vitro stimulation, which significantly changes this expression (43,44) or on human macrophages stimulated in culture, not in material directly from the patient (45). The other method of identification of checkpoint molecules is IHC, which is commonly used in PD-L1 expression tests in practice. However, there are some discrepancies in the reports of IHC quantitative methods which, in addition, are time consuming and subjective. The differentiation between cancer cells and immune cells in simple IHC staining could be burdened with an error (20). Unlike IHC and histological techniques, flow cytometry enables rapid sampling of the cell population and detailed analysis. Furthermore, the fact that most standard flow cytometry analyzers are capable of measuring 8 or more fluorescent signals together in a single sample means that cells and their subpopulations can be defined by multiple surface and/or intracellular specific cell markers (46).
PD-1 expression on lymphocytes
To have an insight into the local immune status and perform a comparative analysis we evaluated lymphocyte T subtyping and PD-1 expression on lymphocytes. We observed significant differences between the cell proportion in the BALF and PB. These results are consistent with our previous findings and with these presented by others (27,47,48). The immune system plays a protective role against cancer, mainly through the ability of cytotoxic CD8+ T cells to recognize and destroy cancer cells. Cancer cells often develop escape mechanisms from the immune system by activating the PD-1 receptor, which is an inhibitory checkpoint, mainly expressed on T cells, and activation of the PD-1/PD-L1 pathway (27,49,50). Therefore, in our study we decided to confirm the PD-1 expression on lymphocytes T subpopulation (CD4+ and CD8+) in three difference compartments in patients with NSCLC: clBALF, hlBALF and PB and assess their relationship with PD-L1 and PD-L2 expression on AMs. The proportion of CD8+ and CD4+ cells bearing PD-1 were the highest in clBALF. CD8+ cells are involved in anticancer response in a full extend. It was confirmed mainly on the basis of tumor infiltrating lymphocytes (TILs) evaluation but also in the BALF analyses (23). PD-1 may also be a cell activation marker, but there is no conclusive data on this (51). Duraiswamy et al. suggested that PD-1 expression may be involved in regulating the anti-tumor response in both activated and depleted cells (52). Thus PD-1 expression can also be considered as a marker of activated tumor-reactive T cells. The uncountable proportion of PD-L1 and PD-L2 positive AMs did not allow us to perform a correlation test with PD-1 T cells. There are reports on relation of cells expressing PD-L2 detected by IHC with TIL CD8+ cells in oesophageal cancer tissue (53) and hepatocellular carcinoma (18).
In our study we analysed also the GMF of PD-1 T cells. The GMF was used to describe and define the expression level and mean intensity of PD-1 on T cells. We found higher GMF of PD-1 on T cells (both CD4 and CD8) in clBALF and hlBALF than in PB. In our opinion the use of the GMF parameter is better to show the difference between BALF and PB. However, in our study GMF of PD-1 does not show differences between the lungs (clBALF and hlBALF).
Conclusions
Conclude the counts of macrophages in the lung environment suggests a significant contribution these cells to the development of lung cancer. However, plasticity for theirs functions: with on the one hand tumor suppression on the other hand tumor growth and demonstration of high autofluorescence, underline the challenges of attacking these cells in anti-cancer therapies. The above-presented data of another studies and our results confirm that the pathway associated with PD-1 on lymphocytes may plays important role in lung cancer patients and it can be examined by flow cytometry. We focused on the need of examination the PD-1/PD-1 ligands pathway not only on lymphocytes but also with participation of macrophages, including analysing also the PD-L2 ligand. We conclude that precise macrophages identification in BALF using flow cytometry is possible. In contrary to expression of PD-1 on lymphocytes, the quantitative analysis of the expression of PD-1 ligands (PD-L1, PD-L2) on AMs by flow cytometry is disturbed by autofluorescence and needs further elaboration.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by grants from Military Institute of Medicine number: 488.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Military Institute of Medicine Ethics Committee (No. 47/WIM/2017) and informed consent was taken from all the patients.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1103
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1103
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1103). The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 2019;144:1941-53. 10.1002/ijc.31937 [DOI] [PubMed] [Google Scholar]
- 3.Costantini A, Grynovska M, Lucibello F, et al. Immunotherapy: a new standard of care in thoracic malignancies? A summary of the European Respiratory Society research seminar of the Thoracic Oncology Assembly. Eur Respir J 2018;51:1702072. 10.1183/13993003.02072-2017 [DOI] [PubMed] [Google Scholar]
- 4.Dudnik E, Moskovitz M, Daher S, et al. Effectiveness and safety of nivolumab in advanced non-small cell lung cancer: the real-life data. Lung Cancer 2018;126:217-23. 10.1016/j.lungcan.2017.11.015 [DOI] [PubMed] [Google Scholar]
- 5.Daud AI, Loo K, Pauli ML, et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest 2016;126:3447-52. 10.1172/JCI87324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong H, Strome SE, Salomao DR, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 2002;8:793-800. 10.1038/nm730 [DOI] [PubMed] [Google Scholar]
- 7.Keir ME, Butte MJ, Freeman GJ, et al. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol 2008;26:677-704. 10.1146/annurev.immunol.26.021607.090331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharpe AH, Wherry EJ, Ahmed R, et al. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol 2007;8:239-45. 10.1038/ni1443 [DOI] [PubMed] [Google Scholar]
- 9.Latchman Y, Wood CR, Chernova T, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol 2001;2:261-8. 10.1038/85330 [DOI] [PubMed] [Google Scholar]
- 10.Arrieta O, Montes-Servin E, Hernandez-Martinez JM, et al. Expression of PD-1/PD-L1 and PD-L2 in peripheral T-cells from non-small cell lung cancer patients. Oncotarget 2017;8:101994-2005. 10.18632/oncotarget.22025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rozali EN, Hato SV, Robinson BW, et al. Programmed death ligand 2 in cancer-induced immune suppression. Clin Dev Immunol 2012;2012:656340. 10.1155/2012/656340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Youngnak P, Kozono Y, Kozono H, et al. Differential binding properties of B7-H1 and B7-DC to programmed death-1. Biochem Biophys Res Commun 2003;307:672-7. 10.1016/S0006-291X(03)01257-9 [DOI] [PubMed] [Google Scholar]
- 13.Shibahara D, Tanaka K, Iwama E, et al. Intrinsic and extrinsic regulation of PD-L2 expression in oncogene-driven non-small cell lung cancer. J Thorac Oncol 2018;13:926-37. 10.1016/j.jtho.2018.03.012 [DOI] [PubMed] [Google Scholar]
- 14.Salmaninejad A, Valilou SF, Shabgah AG, et al. PD-1/PD-L1 pathway: basic biology and role in cancer immunotherapy. J Cell Physiol 2019;234:16824-37. 10.1002/jcp.28358 [DOI] [PubMed] [Google Scholar]
- 15.Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol 2019;16:341-55. 10.1038/s41571-019-0173-9 [DOI] [PubMed] [Google Scholar]
- 16.Kerr KM, Tsao MS, Nicholson AG, et al. Programmed death-ligand 1 immunohistochemistry in lung cancer: in what state is this art? J Thorac Oncol 2015;10:985-9. 10.1097/JTO.0000000000000526 [DOI] [PubMed] [Google Scholar]
- 17.Hirsch FR, McElhinny A, Stanforth D, et al. PD-L1 Immunohistochemistry assays for lung cancer: results from phase 1 of the blueprint PD-L1 IHC assay comparison project. J Thorac Oncol 2017;12:208-22. 10.1016/j.jtho.2016.11.2228 [DOI] [PubMed] [Google Scholar]
- 18.Liao H, Chen W, Dai Y, et al. Expression of programmed cell death-ligands in hepatocellular carcinoma: correlation with immune microenvironment and survival outcomes. Front Oncol 2019;9:883. 10.3389/fonc.2019.00883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calles A, Liao X, Sholl LM, et al. Expression of PD-1 and its ligands, PD-L1 and PD-L2, in smokers and never smokers with KRAS-mutant lung cancer. J Thorac Oncol 2015;10:1726-35. 10.1097/JTO.0000000000000687 [DOI] [PubMed] [Google Scholar]
- 20.Shinchi Y, Komohara Y, Yonemitsu K, et al. Accurate expression of PD-L1/L2 in lung adenocarcinoma cells: a retrospective study by double immunohistochemistry. Cancer Sci 2019;110:2711-21. 10.1111/cas.14128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsubara T, Takada K, Azuma K, et al. A clinicopathological and prognostic analysis of PD-L2 expression in surgically resected primary lung squamous cell carcinoma. Ann Surg Oncol 2019;26:1925-33. 10.1245/s10434-019-07257-3 [DOI] [PubMed] [Google Scholar]
- 22.Takamori S, Takada K, Azuma K, et al. Prognostic impact of programmed death-ligand 2 expression in primary lung adenocarcinoma patients. Ann Surg Oncol 2019;26:1916-24. 10.1245/s10434-019-07231-z [DOI] [PubMed] [Google Scholar]
- 23.Khunger M, Rakshit S, Pasupuleti V, et al. Incidence of pneumonitis with use of programmed death 1 and programmed death-ligand 1 inhibitors in non-small cell lung cancer: a systematic review and meta-analysis of trials. Chest 2017;152:271-81. 10.1016/j.chest.2017.04.177 [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Zhou S, Yang F, et al. Treatment-related adverse events of PD-1 and PD-L1 inhibitors in clinical trials: a systematic review and meta-analysis. JAMA Oncol 2019;5:1008-19. 10.1001/jamaoncol.2019.0393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domagala-Kulawik J. The relevance of bronchoalveolar lavage fluid analysis for lung cancer patients. Expert Rev Respir Med 2020;14:329-37. 10.1080/17476348.2020.1708720 [DOI] [PubMed] [Google Scholar]
- 26.Kwiecien I, Stelmaszczyk-Emmel A, Polubiec-Kownacka M, et al. Elevated regulatory T cells, surface and intracellular CTLA-4 expression and interleukin-17 in the lung cancer microenvironment in humans. Cancer Immunol Immunother 2017;66:161-70. 10.1007/s00262-016-1930-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwiecien I, Skirecki T, Polubiec-Kownacka M, et al. Immunophenotype of T cells expressing programmed death-1 and cytotoxic T cell antigen-4 in early lung cancer: local vs. systemic immune response. Cancers (Basel) 2019;11:567. 10.3390/cancers11040567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osińska I, Stelmaszczyk-Emmel A, Polubiec-Kownacka M, et al. CD4+/CD25(high)/FoxP3+/CD127- regulatory T cells in bronchoalveolar lavage fluid of lung cancer patients. Hum Immunol 2016;77:912-15. 10.1016/j.humimm.2016.07.235 [DOI] [PubMed] [Google Scholar]
- 29.Chciałowski A, Chorostowska-Wynimko J, Fal A, et al. Recommendation of the Polish Respiratory Society for bronchoalveolar lavage (BAL) sampling, processing and analysis methods. Pneumonol Alergol Pol 2011;79:75-89. [PubMed] [Google Scholar]
- 30.Ahmadzadeh M, Johnson LA, Heemskerk B, et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009;114:1537-44. 10.1182/blood-2008-12-195792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dietel M, Savelov N, Salanova R, et al. Real-world prevalence of programmed death ligand 1 expression in locally advanced or metastatic non-small-cell lung cancer: the global, multicenter EXPRESS study. Lung Cancer 2019;134:174-9. 10.1016/j.lungcan.2019.06.012 [DOI] [PubMed] [Google Scholar]
- 32.Misharin AV, Morales-Nebreda L, Mutlu GM, et al. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol 2013;49:503-10. 10.1165/rcmb.2013-0086MA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu YR, Hotten DF, Malakhau Y, et al. Flow cytometric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. Am J Respir Cell Mol Biol 2016;54:13-24. 10.1165/rcmb.2015-0146OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bharat A, Bhorade SM, Morales-Nebreda L, et al. Flow cytometry reveals similarities between lung macrophages in humans and mice. Am J Respir Cell Mol Biol 2016;54:147-9. 10.1165/rcmb.2015-0147LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnston LK, Rims CR, Gill SE, et al. Pulmonary macrophage subpopulations in the induction and resolution of acute lung injury. Am J Respir Cell Mol Biol 2012;47:417-26. 10.1165/rcmb.2012-0090OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hulspas R, O'Gorman MR, Wood BL, et al. Considerations for the control of background fluorescence in clinical flow cytometry. Cytometry B Clin Cytom 2009;76:355-64. 10.1002/cyto.b.20485 [DOI] [PubMed] [Google Scholar]
- 37.Duan M, Li WC, Vlahos R, et al. Distinct macrophage subpopulations characterize acute infection and chronic inflammatory lung disease. J Immunol 2012;189:946-55. 10.4049/jimmunol.1200660 [DOI] [PubMed] [Google Scholar]
- 38.Garn H. Specific aspects of flow cytometric analysis of cells from the lung. Exp Toxicol Pathol 2006;57 Suppl 2:21-4. 10.1016/j.etp.2006.02.009 [DOI] [PubMed] [Google Scholar]
- 39.Daigneault M, Preston JA, Marriott HM, et al. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS One 2010;5:e8668. 10.1371/journal.pone.0008668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li F, Yang M, Wang L, et al. Autofluorescence contributes to false-positive intracellular Foxp3 staining in macrophages: a lesson learned from flow cytometry. J Immunol Methods 2012;386:101-7. 10.1016/j.jim.2012.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flores-Montero J, Kalina T, Corral-Mateos A, et al. Fluorochrome choices for multi-color flow cytometry. J Immunol Methods 2019;475:112618. 10.1016/j.jim.2019.06.009 [DOI] [PubMed] [Google Scholar]
- 42.Menguy S, Prochazkova-Carlotti M, Beylot-Barry M, et al. PD-L1 and PD-L2 are differentially expressed by macrophages or tumor cells in primary cutaneous diffuse large B-cell lymphoma, leg type. Am J Surg Pathol 2018;42:326-34. 10.1097/PAS.0000000000000983 [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez-García M, Porichis F, de Jong OG, et al. Expression of PD-L1 and PD-L2 on human macrophages is up-regulated by HIV-1 and differentially modulated by IL-10. J Leukoc Biol 2011;89:507-15. 10.1189/jlb.0610327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terrazas C, de Dios Ruiz-Rosado J, Amici SA, et al. Helminth-induced Ly6C(hi) monocyte-derived alternatively activated macrophages suppress experimental autoimmune encephalomyelitis. Sci Rep 2017;7:40814. 10.1038/srep40814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wen ZF, Liu H, Gao R, et al. Tumor cell-released autophagosomes (TRAPs) promote immunosuppression through induction of M2-like macrophages with increased expression of PD-L1. J Immunother Cancer 2018;6:151. 10.1186/s40425-018-0452-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore JP, Sakkal S, Bullen ML, et al. A flow cytometric method for the analysis of macrophages in the vascular wall. J Immunol Methods 2013;396:33-43. 10.1016/j.jim.2013.07.009 [DOI] [PubMed] [Google Scholar]
- 47.Domagała-Kulawik J, Hoser G, Droszcz P, et al. T-cell subtypes in bronchoalveolar lavage fluid and in peripheral blood from patients with primary lung cancer. Diagn Cytopathol 2001;25:208-13. 10.1002/dc.2040 [DOI] [PubMed] [Google Scholar]
- 48.Domagała-Kulawik J, Guzman J, Costabel U. Immune cells in bronchoalveolar lavage in peripheral lung cancer--analysis of 140 cases. Respiration 2003;70:43-8. 10.1159/000068414 [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Cao X. Immunosuppressive cells in tumor immune escape and metastasis. J Mol Med (Berl) 2016;94:509-22. 10.1007/s00109-015-1376-x [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Teng F, Kong L, et al. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther 2016;9:5023-39. 10.2147/OTT.S105862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon S, Labarriere N. PD-1 expression on tumor-specific T cells: Friend or foe for immunotherapy? Oncoimmunology 2017;7:e1364828. 10.1080/2162402X.2017.1364828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duraiswamy J, Ibegbu CC, Masopust D, et al. Phenotype, function, and gene expression profiles of programmed death-1(hi) CD8 T cells in healthy human adults. J Immunol 2011;186:4200-12. 10.4049/jimmunol.1001783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohigashi Y, Sho M, Yamada Y, et al. Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin Cancer Res 2005;11:2947-53. 10.1158/1078-0432.CCR-04-1469 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as