Abstract
With advances in the design and fabrication of nanofluidic devices during the last decade, there have been a few reports on nucleic acid analysis using nanoscale electrophoresis. The attractive nature of nanofluidics is the unique phenomena associated with this length scale that are not observed using microchip electrophoresis. Many of these effects are surface-related and include electrostatics, surface roughness, van de Waals interactions, hydrogen bonding, and the electric double layer. The majority of reports related to nanoscale electrophoresis have utilized glass-based devices, which are not suitable for broad dissemination into the separation community because of the sophisticated, time consuming, and high-cost fabrication methods required to produce the relevant devices. In this study, we report the use of thermoplastic nanochannels (110 nm x 110 nm, depth x width) for the free solution electrokinetic analysis of ribonucleotide monophosphates (rNMPs). Thermoplastic devices with micro- and nanofluidic networks were fabricated using nanoimprint lithography (NIL) with the structures enclosed via thermal fusion bonding of a cover plate to the fluidic substrate. Unique to this report is that we fabricated devices in cyclic olefin copolymer (COC) that was thermally fusion bonded to a COC cover plate. Results using COC/COC devices were compared to poly(methyl methacrylate), PMMA, devices with a COC cover plate. Our results indicated that at pH = 7.9, the electrophoresis in free solution resulted in an average resolution of the rNMPs >4 (COC/COC device range = 1.94 – 8.88; PMMA/COC device range = 1.4 – 7.8) with some of the rNMPs showing field-dependent electrophoretic mobilities. Baseline separation of the rNMPs was not possible using PMMA- or COC-based microchip electrophoresis. We also found that COC/COC devices could be assembled and UV/O3 activated after device assembly with the dose of the UV/O3 affecting the magnitude of the electroosmotic flow, EOF. In addition, the bond strength between the substrate and cover plate of unmodified COC/COC devices was higher compared to PMMA/COC devices. The large differences in the electrophoretic mobilities of the rNMPs afforded by nanoscale electrophoresis will enable a new single-molecule sequencing platform we envision, which uses molecular-dependent electrophoretic mobilities to identify the constituent rNMPs generated from an intact RNA molecule using a processive exonuclease. With optimized nanoscale electrophoresis, the rNMPs could be identified via mobility matching at an accuracy >99% in both COC/COC and PMMA/COC devices.
Keywords: Nanofluidics, electrophoresis, ribonucleotides, thermoplastics
1. Introduction
Analysis of the transcriptome has garnered significant attention recently as it offers information not readily obtainable from the genome. Whole-genome sequencing provides a static view of an organism’s genetic and regulatory information, but transcriptomic analyses allows for assessment of dynamic changes in gene expression in response to various stimuli [1, 2]. In addition, identification of unique transcripts can enhance the understanding of underlying mechanisms governing pathophysiological conditions. Moreover, with the development of precision medicine, RNA analysis will increasingly be relied upon to serve as molecular signatures that define various disease subtypes that allow for predicting pharmacological responses to certain therapies (i.e., precision medicine) [3–5].
The advent of next generation sequencing (NGS) has revolutionized the understanding of the complex and dynamic nature of the transcriptome by allowing RNA analysis through complementary DNA (cDNA) sequencing known as RNA sequencing (RNA-seq) [6]. RNA-seq provides a detailed and quantitative view of gene expression and alternative splicing patterns [7]. With recent advances in the RNA-seq workflow, it has provided deep profiling of the transcriptome [8].
Even though the accomplishments are impressive for RNA-seq, there are still numerous challenges with this platform, including: (i) The propensity of various reverse transcriptases (RT) to generate spurious secondary cDNA strands due to their DNA-dependent polymerase activities; (ii) artifactual cDNA generation due to template switching or contaminating DNA and primer independent cDNA synthesis; (iii) low amounts of cDNA due to the inefficient nature of RT; and (iv) biases introduced during amplification steps [9]. Therefore, it is apparent that new sequencing platforms that can address the aforementioned limitations are necessary, especially those that are capable of sequencing full length transcripts without amplification.
Recently, single-molecule sequencing (SMS), some of which are based on nanopore readout, have become an attractive alternative to ensemble-based sequencing such as NGS as it can eliminate the need for RT and polymerase amplification as well as providing longer reads [10, 11]. Unlike NGS, nanopore sequencing does not require fluorescence labelling [12, 13]. Nanopore sequencing can be accomplished using two different approaches, strand sequencing [14, 15] or exosequencing [11, 12, 16]. Ayub et al. [11] showed that polynucleotide phosphorylase (PNPase), which processively cleaves RNA in the 3′ → 5′ direction releasing inorganic phosphate (Pi) and ribonucleotide diphosphates (rNDPs), sequentially produces rNDPs that can pass through an αHL (HL = hemolysin) pore containing cyclodextrin adapters non-covalently bound to the pore. However, diffusional misordering is a limitation of the exosequencing approach [12, 17]. Moreover, single nucleotide detection approaches are limited by high error rates as nucleotide bases are identified using current blockage event amplitudes only [10].
To overcome these challenges, we are developing an innovative SMS strategy that is based on the exosequencing approach and consists of enzymatically cleaving intact DNAs or RNAs using a processive enzyme to generate individual nucleotide monophosphates [18–20]. For RNA, this can be achieved via a processive enzyme such as exoribonuclease 1 (XRN1). We have recently shown that this enzyme can be tethered to a solid support and processively clip an RNA strand in the 5’ → 3’ direction and generate rNMPs when activated by the cofactor, Mg2+ [21]. The released rNMPs are electrokinetically transported through a nanochannel with the travel time (i.e., Time-of-Flight, TOF) through the nanochannel used to efficiently identify the constituent rNMPs based on their molecular-dependent electrophoretic mobilities. Therefore, a thorough understanding of the electrophoretic behavior of the rNMPs through nanochannels along with the electrophoretic parameters, for example carrier electrolyte pH, field strength, and channel material, that determine their molecular-dependent mobilities will allow for high identification accuracy using our SMS approach.
Separation of rNMPs has been achieved using conventional electrophoresis, most commonly using glass capillaries and alkaline solutions as the carrier electrolyte [22–24]. Sodium borate is the most commonly used alkaline buffer for rNMP electrophoresis [22, 25]. In addition to sodium borate, others have used sodium and ammonium carbonate buffers [26].
Nanofluidics, which use channels with dimensions (width and/or depth) ≤100 nm, have garnered attention recently due to the unique properties compared to microscale electrophoresis arising from scaling effects. For example, electrophoresis in nanochannels can depend on ion valence, ζ (zeta potential), ion mobility, and thickness of the electric double layer (EDL) [27–30]. The increased surface area-to-volume ratio in nanofluidics can also allow for a host of solute/wall interactions, which in turn provide hydrophobic, electrostatic, or van der Waals interactions [31]. However, the majority of the reported nanoscale electrophoresis were performed in glass-type devices, which make these channels not ideal for broad community-based applications as the fabrication of these devices is associated with time-consuming and sophisticated methods, such as direct focused ion beam milling [32, 33].
Thermoplastic nanofluidic devices are considered an attractive alternative to glass/silicon devices due to their diverse physiochemical properties and the many fabrication techniques available to generate the prerequisite structures [34, 35]. Thermoplastic nanofluidic devices can be fabricated using nanoimprint lithography (NIL), which takes advantage of the deformability of the plastic at temperatures above their glass transition temperatures (Tg) to produce multi-scale structures in a relatively high production mode at moderate cost [1, 35]. The diversity in their surface chemistry, which is determined by the identity of the monomer units comprising the polymer chains, is another advantage of thermoplastics for nanoscale electrophoresis. In addition, a diverse range of simple activation techniques, such as O2 plasma or UV/O3 irradiation, can be employed to generate groups that alter the surface chemistry of the plastic nanochannels as well as their wettability [36–38].
Unfortunately, there have been a limited number of studies on nanoscale electrophoresis using thermoplastic nanofluidic devices [20, 39, 40]. Recently, we showed the identification of deoxynucleotide monophosphates (dNMPs) through thermoplastic nanochannels made with PMMA as the substrate and COC as the cover plate [20]. We observed field-dependent mobilities of dNMPs at low electric field strengths due to intermittent motion arising from nanometer surface roughness. Moreover, increasing the pH of the carrier electrolyte increased differences in the electrophoretic mobilities while low ionic strength conditions, where the EDL is thicker, led to poorer differences. However, we observed the delamination of the PMMA-COC nanofluidic device at higher pHs (pH >10.3), which limited the pH that could be used to improve the differences in the electrophoretic mobilities to enhance identification efficiency using mobility matching.
In this work, we report the electrokinetic identification of rNMPs using thermoplastic nanochannels. The major goal of this work was to under the electrophoretic conditions, such as carrier electrolyte composition (i.e., pH), electric field strength, and material effects, on the performance of nanoscale electrophoresis of the rNMPs. While our previous work on nanoscale electrophoresis used exclusively PMMA/COC hybrid devices [40, 41], herein we fabricated COC/COC thermoplastic nanochannel devices with the ability to control the EOF through post-assembly UV/O3 surface activation. Furthermore, these COC/COC devices showed higher bond strengths compared to PMMA/COC devices, which will improve the use of different electrophoresis operating conditions to optimize the identification accuracy of the rNMPs without device failure. We were able to efficiently identify all of the rNMPs using free solution nanoelectrophoresis via their molecular-dependent mobilities with efficiencies >99% in COC/COC and PMMA/COC nanofluidic devices.
2. Materials and Methods
2.1. Reagents and Materials
Silicon (Si) 〈100〉wafers were purchased from University Wafers (Boston, MA). Non-impact modified PMMA was received from ePlastics (San Diego, CA). Cyclic olefin copolymer (COC 8007 and COC 5010) were purchased from TOPAS Advanced Polymers (Florence, KY). COC 6015 was obtained from Knightsbridge Plastics Inc. (Fremont, CA). UV curable polyurethane resin was purchased from Chansang Co. ATTO-532 was secured from Atto-Tec (Siegen, Germany). Uridine 5’-monophophate disodium salt, cytidine 5’-monophosphate disodium salt, adenosine 5’-monophosphate disodium salt, and guanosine 5’-monophosphate disodium salt were all obtained from Sigma-Aldrich (St. Louis, MO). Molecular biology grade water was secured from Thermo Fisher (Waltham, MA).
2.2. Conjugation of ATTO-532 to the ribonucleotide monophosphates (rNMPs)
The fluorescence labeling of rNMPs with the ATTO-532 reporter was conducted following a similar procedure reported by our group, but with slight modifications; see Figure S1 [20]. Details can be found in the SI material.
2.3. Fabrication of microchannel thermoplastic devices
Fabrication of microchannel devices were carried out following a method similar to what we previously published [42–47]. Briefly, T-shaped (50 μm depth x 100 μm width and 5 cm long) microfluidic devices were hot embossed into PMMA using a brass master mold, which was fabricated utilizing high precision micromilling. Embossed devices were diced with a bandsaw, reservoirs were drilled using a mechanical drill, and were cleaned with 10% Micro-90, IPA, and nanopure water. The substrate containing the fluidic network and cover plate (150 μm thick PMMA sheet) were UV/O3 modified at 22 mW/cm2 for 16 min prior to thermal fusion bonding. Microchannel dimensions were measured before and after bonding by rapid laser-scanning optical profilometry (VK-X250, Keyence, IL, USA).
2.4. Microscale electrophoresis of the rNMPs
Free solution electrophoresis was carried out following a method reported by our group [40]. Briefly, the T-shaped microfluidic device was primed with 50% methanol/water, filled with 1X NEB buffer at pH 7.9 before carrying out the electrophoresis. A positive voltage was applied to the sample reservoir to initiate injection while grounding the sample waste reservoir until the cross channel was completely filled. The remaining reservoirs were allowed to float during injection. After injection, a positive voltage was applied to the electrophoresis buffer reservoir and the electrophoresis waste reservoir was grounded for the separation. The detector consisted of a laser-induced fluorescence system equipped with a single-photon counting module as we have reported previously [20]. For a schematic of the laser-induced fluorescence detector for microchip electrophoresis and a picture of the microchip see Figure S2 in the SI.
2.5. Fabrication of nanochannel devices
Nanofluidic devices were fabricated in thermoplastics using a method described by our group [20]. Briefly, microchannels were fabricated in a Si wafer (master) via optical lithography followed by wet Si etching. Then, nanochannels were fabricated into the same Si master using focused-ion beam milling. Next, resin stamps were produced from the Si master using a UV curable polyurethane (PUA) resin that covered the Si master and exposing to UV light. COC 6015, which was used as the back plate for the resin stamp, was coated with a NOA72 adhesive. Nanochannels were imprinted into a plastic substrate using a Nanonex 2500 nanoimprint lithography (NIL) machine. The details on the use of the Nanonex 2500 instrument for device fabrication can be found in the SI material (see Figure S3). Preliminary tests were conducted to optimize the imprinting pressure, temperature, and time. The imprinting temperature was kept higher than the Tg of the thermoplastic substrate. For PMMA devices, we used the same imprinting conditions reported previously by our group (140°C, 300 psi, and 5 min) [40]. Imprinting of nanofluidic structures into COC 5010 substrates was done by keeping the imprinting pressure (300 psi) and time (5 min) constant while optimizing the imprinting temperature. The optimized conditions for the imprinting are given in Table S1 for both COC 5010 and PMMA substrates. Imprinted nanofluidic devices were then characterized using SEM and atomic force microscopy, AFM (see Figure 1).
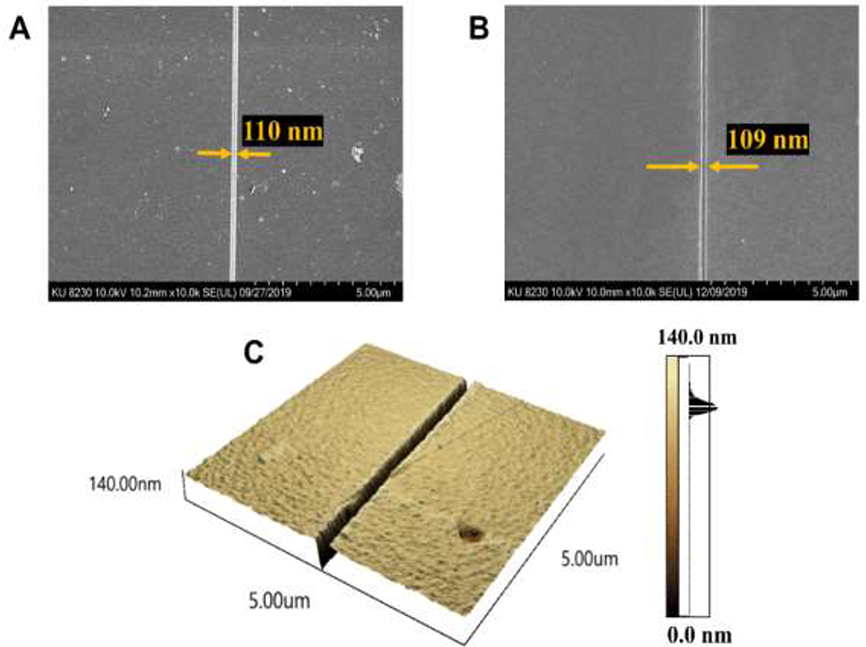
Figure 1.
(A) SEM of a nanochannel replicated via UV-NIL into a polyurethane (PUA) resin to produce the resin stamp. In this cases, the resin stamp has the reverse polarity compared to the Si master from which it was replicated from. (B) SEM of a nanochannel thermally imprinted into a COC 5010 substrate. (C) AFM of a nanochannel thermally imprinted into COC 5010 with a depth determined to be 110 nm.
It is necessary to seal the nanofluidic device after thermal imprinting using a cover plate. Thermal fusion bonding was used for sealing nanochannel devices. Bonding of PMMA/COC devices were done according to a method described by Uba et al. [48] with 1 min O2 plasma treatment for both the substrate and cover plate at 50 W. We attempted a similar protocol to assemble COC/COC devices by modifying both the substrate and cover plate by oxygen plasma, but we were unsuccessful in obtaining a well-sealed device. Therefore, we tried bonding COC 5010 and 8007 without plasma treatment. Successful bonding was achieved at 110 psi bonding pressure, 70°C for 15 min using the NIL machine (see Table S2). The assembled COC/COC devices could then be UV/O3 treated to increase the wettability of the nanochannels, which created surface confined −COOH groups.
2.6. Cover plate/substrate bond strength measurements of nanochannel devices
The crack opening method was used to evaluate the bond strength between the cover plate and substrate [49, 50]. Here, a razor blade of known thickness, tb, is inserted between the bonded substrate and cover plate to generate an interfacial fracture with a length (L) from the edge of the razor blade. The bond strength is represented by equation (1) where ts and tp are the thickness and Ep and Es are the elastic moduli of cover plate and the substrate, respectively.
| (1) |
The elastic moduli of COC 5010 and COC 8007 were 3.0 GPa and 2.6 GPa, respectively, as provided by the manufacturer. In this work, all tests were performed using a stainless-steel single edge razor blade with a thickness of 0.009” and the crack lengths were measured using a calibrated upright microscope with a 10X objective lens. All measurements were replicated five times and the average bond strength was determined along with the standard deviation.
2.7. Evaluation of pH stability and structural integrity of fabricated nanochannel devices
To make sure there was no channel deformation after bonding, COC/COC and PMMA/COC nanochannel devices were filled with a Rhodamine B solution in ultrapure water at pH 7.0 and fluorescence images were captured. Afterwards, to evaluate the pH stability of the nanochannel devices, Rhodamine B solution in ultrapure water was replaced with a Rhodamine B dye solution in pH 10.3 TBE buffer and the fluorescence images were captured at t = 0, 0.5 h, and 3 h to observe any channel deformation or leaking by delamination of the cover plate.
2.8. Electroosmotic flow (EOF) measurements
The EOF in the nanochannel devices was measured using the current monitoring method [51]. A device possessing a single nanochannel (107 μm long, 110 nm deep, and 110 nm wide) connecting two access microchannels was fabricated as described earlier. The entire chip was primed with 50% (v/v water/methanol), drained, and flushed with nuclease free water. After that, the device was filled with 1X NEB (New England Biolabs) buffer 3 (composition of NEB buffer 3 is given in Table S3) and allowed to equilibrate for 4 min under a 500 mV DC bias. After confirming equilibrium by a constant current trace, one reservoir was replaced by 0.94X NEB buffer 3. Pt electrodes were placed into the reservoirs across the channels under a 500 mV DC bias. pClamp10 software and Digidata 1440B low noise digitizer were used for data acquisition.
2.9. Contact Angle measurements
The contact angles of COC surface before and after UV/O3 treatment were measured using VCA optima instrument. Nanopure water was used for all of these measurements.
2.10. Detection system for nanoscale electrokinetics
The translocation of rNMPs through the thermoplastic nanochannels was monitored using a fluorescence imaging system we have reported previously [20, 40].
2.11. Electrokinetic identification of rNMPs in nanochannels
Nanochannel devices were primed with 50% methanol/water mixture for 5 min. Using a vacuum pump, the methanol/water solution was removed from the nanofluidic device. After that, it was filled with 1X NEB buffer 3 (pH 7.9) and allowed to equilibrate for 10 min. ATTO-532 labeled rNMPs (200 nM) were prepared in 1X NEB buffer 3. Next, the rNMP solution was added to one of the reservoirs connecting microchannels by replacing the carrier electrolyte. Afterwards, the microchannel was filled with rNMP solution by applying a vacuum through the opposite side reservoir of the same microchannel. Thereafter, the same volume of carrier electrolyte was added to all other reservoirs. Finally, the dye-labeled rNMPs were injected continuously into the nanochannels by applying a square wave voltage (2 −20 Vpp) for a period of 20 s (50 × 10−3 Hz frequency) using an ATTEN ATF200B function waveform generator allowing multiple injections. Events were recorded for 6,000 frames.
2.12. Data analysis
The nanochannel electrophoresis data were analyzed using a previously reported method by our group [20, 40]. Briefly, the collected videos from the imaging microscope’s EMCCD camera were imported into Image J software and two detection windows of 1 μm2 were placed at the nanochannel entrance and exit. The fluorescence intensity over time recorded from these detection windows were exported into Origin 8.5. Then, the first derivative of each data set was taken to produce two peaks indicating the time the fluorescently labeled rNMPs reached the entrance and exit of the nanochannel. The time difference was taken as the migration time of the rNMPs to travel a fixed distance and the apparent velocity was calculated. The apparent mobility of each rNMP was generated by normalizing the apparent velocity with respect to the electric field strength.
3. Results and Discussion
3.1. Bond strength and pH stability of COC/COC and PMMA/COC devices
In a typical nanofluidic device production pipeline, the final step is enclosure of the nanochannels with a cover plate with the preferred assembly step that does not alter the underlying nanostructures. There are several methods to obtain enclosed fluidic nanochannels such as thermal fusion bonding, ultrasonic bonding, or solvent-assisted bonding [52]. Solvent-assisted bonding can result in bond strengths between the substrate and cover plate that cannot withstand high pressures and voltages for extended periods of time [34]. Also, solvent-assisted bonding can cause dimensional instability of the thermoplastic device due to softening of the plastic substrate leading to changes in channel dimensions and/or collapse of the nanochannel resulting in device failure [52]. Our group previously reported a thermal fusion bonding technique with a high process yield rate (>90%), which consists of bonding a low Tg cover plate to a higher Tg substrate using a bonding temperature close to the Tg of the cover plate [48]. Because the substrate’s Tg is much higher than the bonding temperature used, minimal changes in the dimensions of the nanochannels result. In addition, thermal fusion bonding of the low Tg cover plate to the higher Tg substrate is less time consuming compared to the aforementioned methods, and does not require additional reagents or pre-cleaning in RCA solutions or organic solvents. Thus, we used thermal fusion bonding to assemble all devices herein.
We studied the bond strength of thermally fusion bonded COC/COC devices using the crack opening method. Preliminary tests were conducted to optimize the bonding pressure and time at a bonding temperature of 70°C, which is close to the Tg of the cover plate. The optimized bonding parameters (see Table S2) were utilized to prepare sealed nanofluidic devices. The bond strength of COC/COC devices was 8.5 ±1.1 mJ/cm2. The bond strength of COC/COC devices were significantly higher than that of PMMA/COC devices, which had a bond strength of 0.086 ±0.014 mJ/cm2. This suggests that interfacial adhesion of similar materials is greater than that of dissimilar materials. Therefore, COC devices can be operated at higher pressures and tolerate higher electric field strengths compared to PMMA/COC hybrid devices.
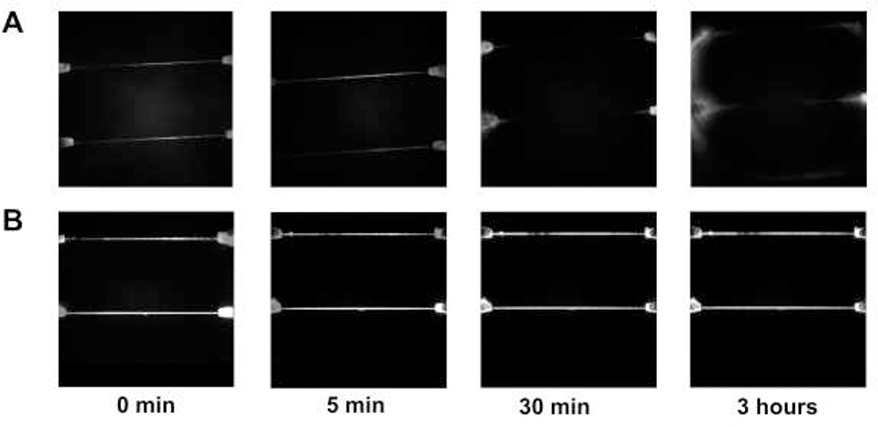
pH stability is another important parameter to evaluate in these devices because high pH may result in improved identification of the rNMPs [22, 24]. However, under high pH conditions, thermoplastics may swell and/or delaminate according to their base resistance. Thus, the base resistance of PMMA/COC nanochannels and COC/COC nanochannels were evaluated prior to nanoscale electrophoresis. The nanofluidic device was flooded with different pH solutions containing Rhodamine B as the reporter and fluorescence imaging was carried out to determine if leaking of the dye solution occurred due to swelling and/or delamination of the cover plate/substrate assembly. A fluorescence image of a dye-filled chip was first taken at pH = 7.0 to make sure that the thermal fusion bonded device had no leakage prior to adding the higher pH (pH = 10.0) buffer. After 30 min, the PMMA/COC devices leaked as fluorescence appeared near the inlets of the nanochannels indicating delamination of the cover plate from the substrate. However, the COC/COC devices even after 3 h did not show signs of leaking or delamination (see Figures 2A and B). Therefore, PMMA/COC devices showed lower base resistance compared to COC/COC devices [53, 54] meaning that COC/COC devices can be used in higher pH carrier electrolytes that may produce better electrophoresis results.
Figure 2.
(A) pH stability of PMMA/COC devices with time at pH 10.3. (B) pH stability of COC/COC devices with time at pH 10.3. All fluorescence images were adjusted to the same intensity scale. In both cases, a Rhodamine B dye solution was infused into the nanofluidic device and the fluorescence monitored using a single-molecule fluorescence microscope equipped with an EMCCD camera.
3.2. Water contact angle and EOF of COC/COC devices
For PMMA/COC devices, O2 plasma treatment was carried out before thermal fusion bonding to increase the bond strength between the PMMA substrate and COC cover plate and increase the wettability of the surfaces to minimize issues with bubble formation in the device. However, for COC/COC devices O2 plasma treatment prior to thermal fusion bonding produced poor adhesion of the cover plate to the substrate. Therefore, we evaluated the ability to assemble COC/COC devices in their native states followed by UV/O3 activation to increase the wettability of the nanochannel by increasing the surface charge density.
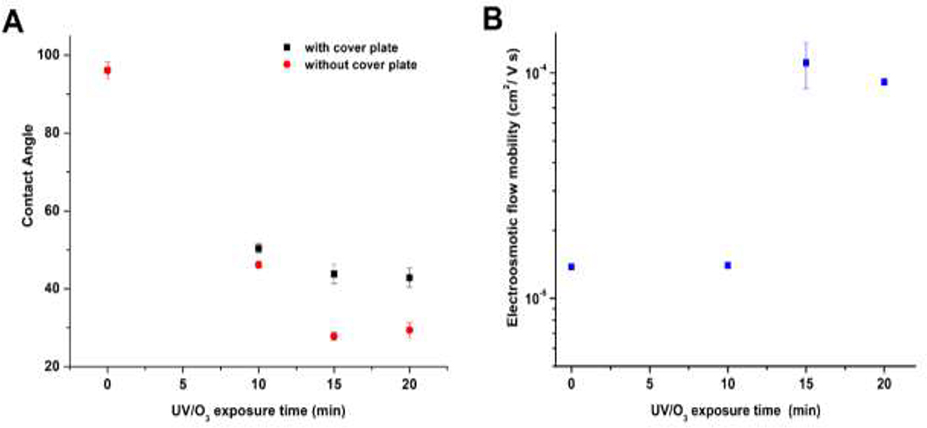
Water contact angle measurements can serve as a measure of the hydrophilicity/hydrophobicity of a surface, although it does not provide insight into the functional groups comprising that surface. Figure 3A shows the water contact angle measured at different post-assembly UV/O3 exposure times for a COC 5010 substrate that was covered with COC 8007. In these experiments, the cover plate was removed following UV/O3 exposure to allow performing the contact angle measurements, which was possible by eliminating the thermal fusion bonding step. Reductions in the water contact angle indicated that the COC surface became more hydrophilic due to polar functional groups generated from UV/O3 activation. The water contact angle did not change for UV/O3 exposure times >15 min as shown in Figure 3A (p = 0.3474, n = 5, contact angle data for 15 min UV vs. 20 min UV exposure).
Figure 3.
(A) Water contact angle of COC 5010 measured at different UV/O3 exposure times using a power density of 22 mW/cm2. The data points named as with cover plate were obtained by keeping a cover plate on top of the substrate and then exposing it with UV/O3 through the cover plate. After exposure, the contact angle of the underlying substrate was measured by removing the cover plate, which was not thermally bonded to the underlying substrate. (B) EOF mobility of COC/COC nanochannel devices as a function of UV/O3 exposure time. The dimensions of the nanochannels were 110 nm x 110 nm (depth x width). The substrate was COC 5010 that was sealed with a COC 8007 cover plate. The UV/O3 activation was done through the cover plate following device assembly. Error bars represent the standard deviations (n = 5).
Another parameter we investigated was the EOF in COC/COC nanochannel devices. We measured the EOF with UV/O3 exposure time in COC/COC nanochannel devices exposed to UV/O3 through the COC 8007 cover plate following thermal fusion bonding. According to the data presented in Figure 3B, the EOF was 1.40 × 10−5 cm2/Vs after 10 min exposure time. However, upon increasing the exposure time to 15 min, the EOF increased by ~10-fold. Further increasing the exposure time to 20 min did not change the EOF significantly (p = 0.87 for 15 min vs 20 min exposure time). We should note that for PMMA/COC devices, post-assembly activation of the device with UV/O3 did not affect the EOF as was seen for COC/COC devices (data not shown). This result is not surprising, because in a previous publication we noted that PMMA is not as UV transparent as COC and the channel side walls for PMMA can reduce penetration of the UV light into the channel [55]. For the PMMA/COC devices, three of the four walls are PMMA while in COC/COC devices all of the surfaces are COC and thus, more transparent to UV light (PMMA has ~1.5% UV transparency and COC is ~53%).
At low EDL thicknesses (~0.8 nm for buffer used), μeof can be represented in terms of the bulk solvent viscosity, ηo, and ζ by the Helmholtz-Smoluchowski equation where ϵ0 ϵr are the permittivity of air and the relative permittivity of the buffer, respectively, through the use of equation (2).
| (2) |
Furthermore, ζ can be represented in terms of σs (surface charge) and the Debye length (λD) for different electrolyte solutions using equation (3) [38].
| (3) |
In equation (3), ϵ0 and ϵr are constants for a given carrier electrolyte and if the ionic strength remains constant, λD will not change. In our experiments, only σs is changing as a function of UV/O3 dose and therefore, EOF changes arise from changes in the number of −COOH groups on the plastic surface. Similar to the contact angle data, the EOF remained constant for exposure times longer than 15 min. O’Neil et al., whom mapped the distribution and number density of −COOH groups in COC by super-resolution microscopy, observed a reduction of −COOH groups at 20 min UV/O3 exposure times compared to 15 min exposure [37]. This would indicate that the EOF may be reduced with UV/O3 doses >15 min (see equation (3)). While we noticed a slight reduction in the EOF at 20 min exposure compared to 15 min, it was not significantly different.
3.3. Microchip electrophoresis of the rNMPs
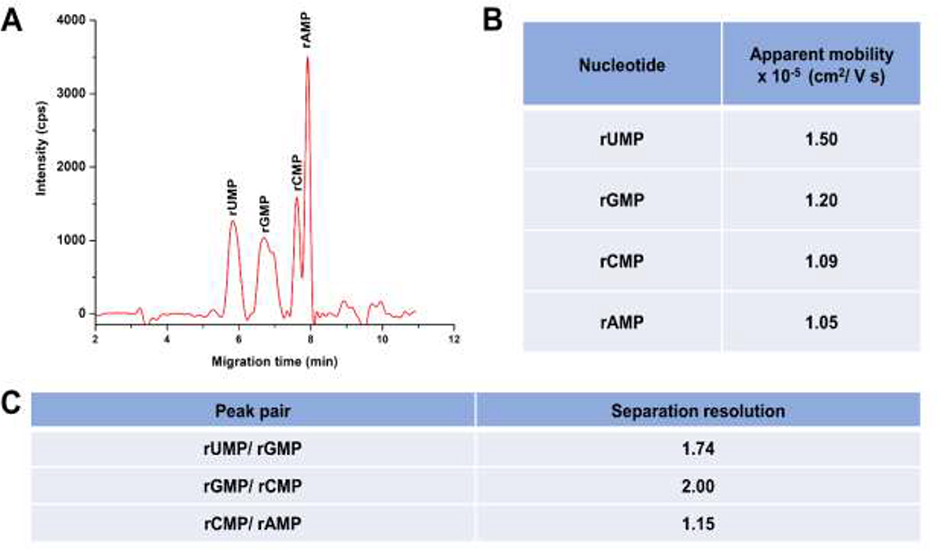
We carried out microchip electrophoretic separations of the rNMPs using a T-shaped microchip fabricated in PMMA (depth = 50 μm; width = 100 μm; total column length = 5 cm) to understand the effects of scaling on the rNMP identification. The free solution electrophoretic separation of the rNMPs was performed in 1X NEB buffer 3 at pH 7.9 (see Table S3 for composition of this buffer). During the microchip electrophoresis, rNMPs migrated from anode to cathode in spite of their anionic nature. As shown in Figure 4A, at pH 7.9 we could separate all four rNMPs. The apparent mobilities of the rNMPs were calculated using equation (4) and are presented in Figure 4B. In this case, l is the distance from the injection point to the detector (4.0 cm), tm is the migration time of each rNMP, and E is the electric field strength (1,000 V/ cm). The apparent mobility order of the rNMPs was rAMP < rCMP < rGMP < rUMP.
Figure 4.
(A) Microchip electropherogram of the rNMPs in PMMA microchannels having dimensions of 50 μm x 100 μm (depth and width, respectively) with a 5 cm total length channel (effective length = 4 cm). (B) Calculated apparent mobilities of the rNMPs using equation (4). (C) Resolutions (R) calculated for adjacent peak pairs using the electropherogram shown in (A). R = 1.18(tm2-tm1)/ (w1+w2), where tm1 and tm2 are migration times and w1 and w2 correspond to the peak widths at the base of the peaks.
| (4) |
Uhrova et al. [22] has shown the separation of rNMPs in a fused silica capillary without suppressing the EOF by free solution electrophoresis using a borate buffer at pH 8 and observed a similar migration order except for rUMP, which showed a higher migration time in their data. Baseline separation (resolution >1.5) was obtained for the rGMP/rUMP pair, but for the rAMP/rCMP pair, no baseline separation was observed (see Figure 4C). We should note that the shoulder for the rGMP peak could be due to 8-oxo-G, because guanine is highly susceptible to oxidation [56]. Moreover, rNMPs were separated in a COC/COC microfluidic chip and in this device, we obtained a similar migration order, but lower separation resolution (see Figure S4).
3.4. Effect of material type on the nanoscale electrophoresis of ATTO-532 labeled rNMPs
In Sections 3.1 and 3.2, we compared the mechanical and chemical properties of nanochannel devices made from COC/COC and PMMA/COC, where the first material listed is the substrate containing the nanochannel and the second material is the cover plate. In this section we will present results for the identification of rNMPs in PMMA/COC and COC/COC nanochannel devices to understand material effects on nanoscale electrophoresis. The EOF for PMMA/COC devices was reported in [40], which was found to be 4.2 × 10−4 cm2/Vs, approximately 4 times higher than that of COC/COC devices.
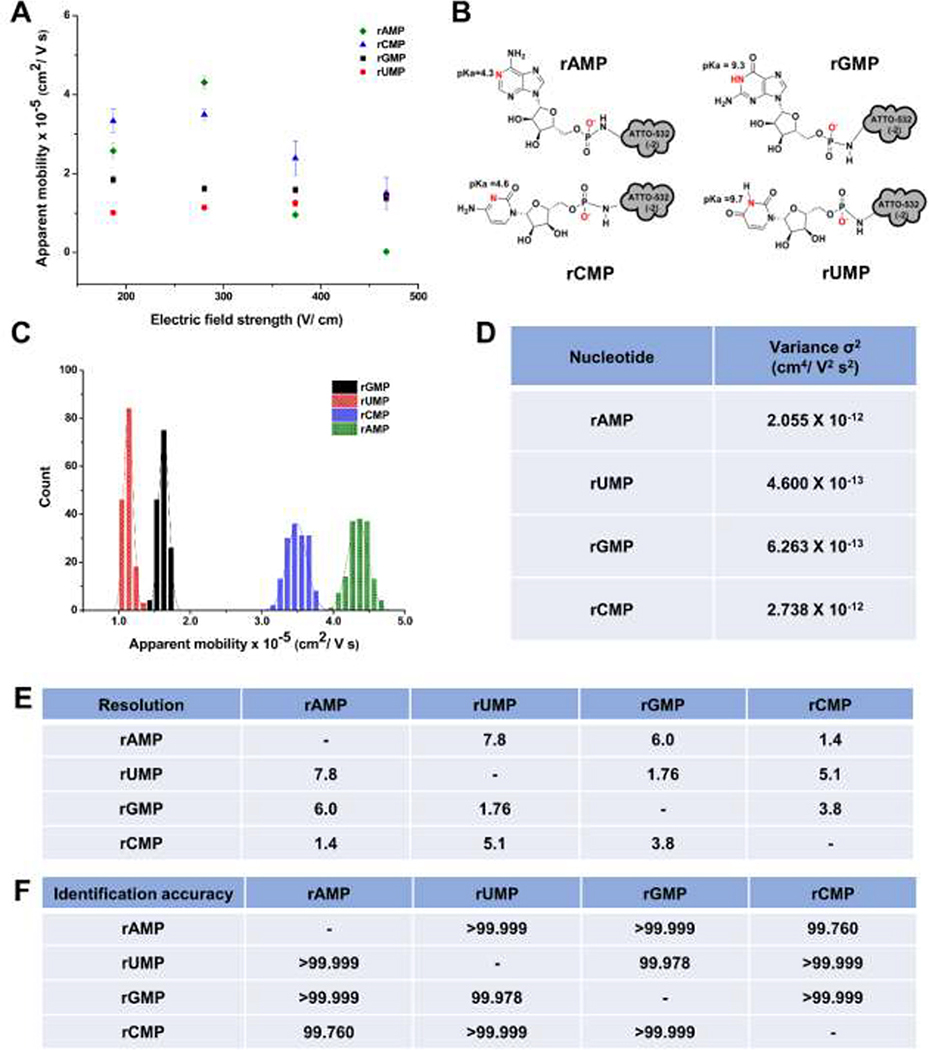
Figure 5A shows the apparent mobility versus electric field strength for the four rNMPs using a 110 nm x 110 nm channel fabricated in PMMA/COC with a carrier electrolyte consisting of 1x NEB buffer 3 (ionic strength of 145 mM) at pH 7.9. At pH 7.9, all ATTO-532 labeled rNMPs have a net charge of −3; the ATTO-532 dye contributes a −2 charge and the phosphate group contributes −1 (see Figure 5B) with all nucleobases not carrying a charge at this pH. Because the same dye reporter was covalently attached to each rNMP using the same linker, differences in the apparent mobilities of the rNMPs were assumed to arise from the nucleobase.
Figure 5.
(A) Apparent mobility vs electric field strength of rNMPs in 110 nm x 110 nm nanochannels fabricated in PMMA/COC nanofluidic devices using 1X NEB buffer 3 at pH 7.9 as the carrier electrolyte. (B) Structures of the rNMPs with ATTO-532 label and the pKa of the nucleobases. (C) Histogram of apparent mobilities of rNMPs at 280 V/cm in 110 × 110 nm PMMA/COC nanochannel devices using 1X NEB buffer 3 at pH 7.9 as the carrier electrolyte. The histograms were fit to a Gaussian function and each bin represented 2 × 10−6 cm2/Vs. (D) The variance (σ2) of peaks estimated from the Gaussian fits to the histograms. (E) The resolution of the Gaussian fits was calculated using R= 1.18 (Δμ/w0.5 + w0.5), where w0.5 corresponds to the full width at half maximum of the Gaussians. F) Identification accuracies of rNMPs calculated from Gaussian fit overlap. Identification accuracy = area of non-overlapped/total peak area.
The apparent mobility is a sum of the EOF (μeof) and the electrophoretic mobility of the rNMP (μep). In this case, the electrophoretic mobility of the dye/rNMP conjugate is opposite in direction to the EOF and thus, a larger μep value results in a lower apparent mobility. Because all rNMPs have the same charge at this pH, differences in the apparent mobility are due to differences in the size of the nucleobase if the identification mechanism is solely electrophoretic. We could only monitor the mobilities of the rNMPs at electric field strengths <460 V/cm in the PMMA/COC devices due to the high mobilities of the rNMPs at electric field strengths >460 V/cm that generated significant slurring of the images that resulted from the limited framerate we could operate the EMCCD camera to provide sufficient signal-to-noise ratio to track the rNMP motion. As seen in Figure 5A, the apparent mobility for rCMP and rAMP showed electric field dependency, which was not observed in the case of rUMP and rGMP. In our previous report for the nanoscale electrophoresis of the dNMPs in PMMA/COC devices, all of the nucleotides showed a field dependent apparent mobility [20]. For the dNMPs, we surmised that wall interactions occurred at low electric field strengths but were absent at higher electric field strengths. Here, we surmised potential wall interactions for rCMP and rAMP due to their field-dependent mobilities. Inspection of Figure 5A indicated that for these two nucleotides, the apparent mobility decreased at higher electric field strengths indicating an increase in the electrophoretic mobility of rCMP and rAMP, which would indicate less wall interactions at higher electric field strengths.
Figure 5C shows histograms of the apparent mobility for the ATTO-532 labeled rNMPs at 280 V/cm in PMMA/COC devices. This electric field was selected as it provided the optimal resolution between the rNMPs with minimum standard deviations in the histograms (i.e., the standard deviations in these histograms is directly related to peak dispersion). These histograms were fit to a Gaussian function and the variance (σ2), resolution, and identification accuracies were calculated (Figure 5D and E). The apparent mobility order at this electric field strength was rUMP < rGMP < rCMP < rAMP. The observed apparent mobility order here was different than the apparent mobility order we observed for microchip electrophoresis of the rNMPs (see Figure 4A).
Differences in the apparent mobility order between microchip electrophoresis and nano-electrophoresis could be due to scaling effects. When the channel dimensions are significantly reduced, the surface area-to-volume ratio increases and thus, surface interactions of solutes with channel walls can become more prominent. These wall interactions can arise from surface roughness that is comparable to the channel dimensions (width and depth) giving rise to intermittent motion and/or hydrophobic/hydrophilic, electrostatic, or van der Waals interactions [20]. In addition, because of the amorphous nature of thermoplastics resulting in a heterogeneous distribution of surface charges, recirculation can occur at low electric fields giving rise to intermittent motion [37]. For rAMP and rCMP, which showed electric-field dependent mobilities at lower electric fields indicative of wall effects on the mobility, their peak variances were larger than those for rUMP and rGMP, which did not show electric field dependent mobilities. In spite of these considerations, the average resolution between Gaussian fits to the apparent mobility histograms was 4.3 (ranged from 1.4–7.8). Another important metric for the potential utilization of mobility matching for the identification of ribonucleotides is the identification accuracy. We defined the identification accuracy as the amount of overlap between two adjacent Gaussian fits to the histograms of the ATTO-532 rNMPs’ apparent mobilities. The percent overlap of the Gaussian peaks was calculated using a previously described method [57]. The average identification accuracy for the rNMPs in PMMA/COC devices was >99.955% (see Figure 5F).
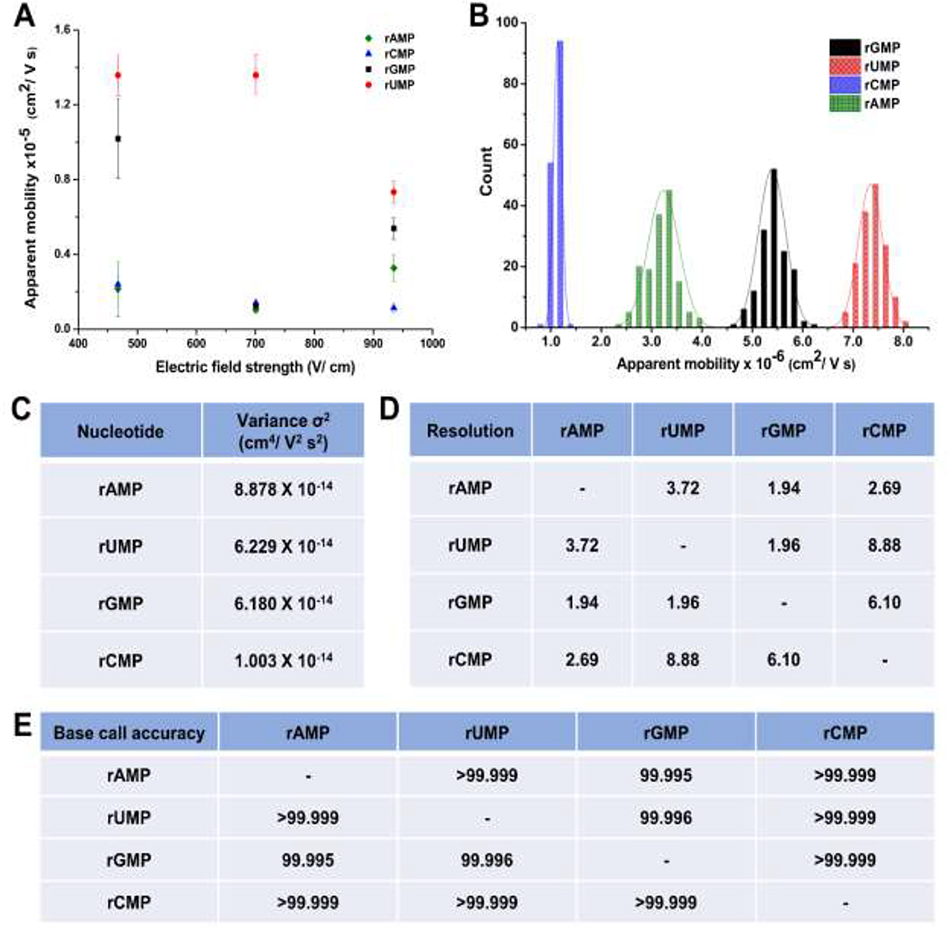
The EOF for COC/COC devices was 1.1 × 10−4 cm2/Vs, which was ~4 times smaller than that of PMMA/COC devices as noted above [40]. Therefore, the overall magnitude of μapp should be smaller in COC/COC devices compared to PMMA/COC devices (see Figure 6A), which allowed us to investigate the nanoscale electrophoresis at higher electric field strengths in the absence of image slurring compared to the PMMA/COC devices. Figure 6A shows the apparent mobility vs. electric field strength of the rNMPs in COC/COC nanochannel devices. In contrast to the data shown in Figure 5A, we could only perform the identification at high electric field strengths because at field strengths <450 V/cm, we observed trapping at the surface resulting from dielectrophroesis generated from surface roughness [38]; COC channels did show higher surface roughness compared to PMMA channels (data not shown). In the COC/COC devices, we observed reductions in the apparent mobility for rUMP above electric field strengths of 700 V/cm while for rGMP, the apparent mobility decreased going from 470 to 700 V/cm and then showed an increase at 930 V/cm. In addition, the other two rNMPs only showed a slight electric field dependent mobility. These trends for the most part are different than those observed in the PMMA/COC nanochannels most likely due to differences in surface chemistry between these two devices as well as surface roughness, which generates intermittent motion and recirculation that are significantly reduced at high electric field strengths [37].
Figure 6.
(A) Apparent mobility vs electric field strength of rNMPs in 110 nm x 110 nm nanochannels fabricated in COC/COC nanochannels using 1X NEB buffer 3 at pH 7.9 as the carrier electrolyte. (B) Histogram of apparent mobilities of rNMPs at 930 V/cm in 110 × 110 nm COC/COC nanochannel devices using 1X NEB buffer 3 at pH 7.9 as the carrier electrolyte. The histograms were fit to Gaussian functions and each bin width represented 2 × 10−6 cm2/Vs. (C) The variance (σ2) of peaks estimated from the Gaussian fits to the histogram data shown in (B). (D) The resolution of Gaussian fits calculated using R = 1.18 (Δμ/w0.5 + w0.5), where w0.5 correspond to the full width at half maximum of the Gaussian fits. (E) Identification accuracies of rNMPs calculated from Gaussian fit overlap.
Figure 6B shows histograms of the apparent mobilities for the ATTO-532 labeled rNMPs at 930 V/cm, which was selected because it provided optimal resolution between the rNMPs. The resolution was determined from Gaussian fits to the histograms. The variance, σ2, was obtained from standard deviations associated with the Gaussian fits to the histogram data (see Figure 6C). Figure 6D shows the resolution values between rNMP Gaussian fit pairs using 1X NEB buffer 3 at pH 7.9. The apparent mobility order here was rCMP < rAMP < rGMP < rUMP and is different than the apparent mobility order observed in PMMA/COC nanochannel devices. At a carrier electrolyte pH of 7.9 and a field strength of 930 V/cm (λd = 0.3 nm), the average identification resolution of the rNMPs was 4.2 (range = 1.94 – 8.88, see Figure 6D). Figure 6E shows the calculated identification accuracies for the rNMPs. As can be seen, the average identification accuracy for the rNMPs in COC/COC devices was >99.997%, slightly higher than that seen for PMMA/COC devices. But, the average identification accuracy was higher for the rNMPs in both materials compared to our previous nanoscale electrophoresis analysis of the dNMPs (deoxynucleotide monophosphates), where the average identification accuracy was >95% [58].
Judicious choice of nanochannel material is critical for optimizing identification accuracy. For example, COC/COC have similar chemistries except for slight differences in the norbornene content of the copolymer [59]. But hybrid devices, such as PMMA/COC, can demonstrate distortions in the plug flow due to large differences in the EOF that can introduce dispersion [60].
3.5. Effect of pH on the nanoscale electrophoresis of the ATTO-532 labeled rNMPs in COC/COC devices
We investigated the nanoscale electrophoresis of the rNMPs at pH 10.3, because a majority of microscale electrophoresis of rNMPs showed better resolution at higher pH (pH >9) [22, 24]. In addition, we noted better identification accuracies using nanoscale electrophoresis for the dNMPs at high pH. For the high pH experiments, the same carrier electrolyte as that used for pH = 7.9 was used, but with slight modifications. Tris-HCl in 1X NEB buffer 3 was replaced with glycine, however, the ionic strength at both pH values was the same. Glycine has a high buffer capacity at pH 10.3 (pKa2 9.6) and therefore, the pH of the carrier electrolyte is maintained properly during the course of the experiment. In these experiments the electrophoresis was carried out at 930 V/cm.
The apparent mobility order at this pH was rAMP < rCMP < rGMP < rUMP with the apparent mobilities about one-order of magnitude higher compared to the electrophoresis performed at pH 7.9. In addition, at pH 10.3 the identification accuracy was reduced compared to pH 7.9 partly due to the increased peak dispersion that was observed. For example, the peak variance ranged from 1.3 × 10−11 cm4/V2s2 to 4.1 × 10−13 cm4/V2s2 for rAMP; these values were about 2-orders of magnitude larger than observed at pH 7.9. The identification accuracy for the rNMPs performed at this pH ranged from 0.78 to 2.55.
Increasing the carrier electrolyte pH had a two-fold effect: (1) Increasing the EOF due to increased ionization of the surface functional groups (pKa of the surface −COOH groups are ~ 6.9 [38]). The extent of ionization of the surface −COOH groups can be calculated using the Henderson-Hasselbalch equation and the pH value for the carrier electrolyte [61]. At pH 7.9, 90% ionization of the surface −COOH groups would be expected while at pH 10.3, 99.99% of these groups are deprotonated. The EOF mobility measured at pH 10.3 was 7.5 ×10−4 cm2/Vs, which was approximately 7-times higher compared to the EOF observed at pH 7.9 (1.1 ×10−4 cm2/Vs). (2) Changing the charge state on some rNMPs. In this case, we would expect rGMP and rUMP to have a lower μapp compared to the other two rNMPs due to increases in their negative charge at pH = 10.3 yielding a larger μep (see Figure 5B).
While one would surmise increased dispersion arising from increased wall interactions, electrostatically we would not expect this because of the larger negative charge on the wall of the nanochannel as well as two rNMPs at this pH. Because these identifications were done at 930 V/cm and the larger EOF at this high pH, we suspect that the increased dispersion was due to image slurring resulting from the limited framerate that we could operate the EMCCD to secure sufficient signal-to-noise ratios to track the motion of the ATTO-532 labeled rNMPs. Irrespective of these observations, the mobility order was different at pH 10.3 compared to the mobility order at 7.9.
3.6. Nanoscale electrophoresis of ATTO-532 labeled methylated rNMPs in COC/COC devices
The “epitranscriptome” is a term that refers to biochemical modifications of the transcriptome, which can play a major role in disease progression. However, studying the epitranscriptome is challenged by a lack of tools to interpret the entire portfolio of RNA modifications (>170). Most of these epitranscriptome modifications occur in ribosomal (rRNA) and transfer RNA (tRNA), which influence tRNA stability and cellular stress response (5-mC) and microRNA stability (2’-O-methlyation). Moreover, RNA base modifications of messenger RNAs (mRNA), such as N6-methyladenosine (m6A), 5-methylcytidine (5-mC), inosine (I), pseudouridine (ψ), N1-methyladenosine (m1A), and 5-hydroxylmethylcytidine (5-hmC), can alter its function and metabolism [62]. To provide additional tools to investigate the epitranscriptome, the nanoscale electrophoresis of methylated rNMPs and their corresponding rNMPs was undertaken.
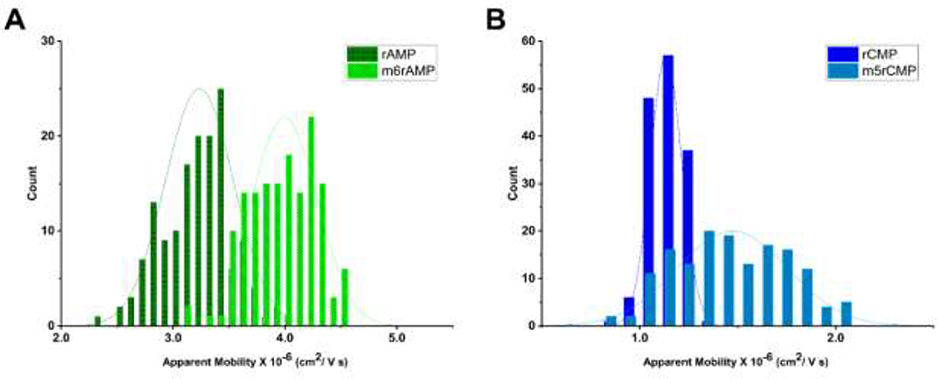
Figure 7A shows histograms of the apparent mobilities of rAMP and m6rAMP and Figure 7B displays the histograms of apparent mobilities for rCMP and m5rCMP at a field strength of 930 V/cm in pH 7.9 NEB buffer using COC/COC nanochannel devices. A higher apparent mobility was observed for both methylated rNMPs (m6rAMP and m5rCMP) compared to their non-methylated counterparts. We calculated the resolution for the methylated from non-methylated forms and found that it was 0.59 for rAMP/m6rAMP and 0.73 for rCMP/m5rCMP. While baseline resolution was not achieved, further investigation into various electrophoresis conditions can improve these results. For example, one can optimize the electric field strength, carrier electrolyte conditions (pH, ionic strength), and/or wall chemistry (increasing/decreasing the dose of UV/O3 irradiation, switching the choice of polymer substrate). In addition, because more than 170 post-transcriptional RNA modifications have been identified to date [63], we will also be investigating multi-dimensional nanoscale electrophoresis to efficiently identify the entire complement of modifications.
Figure 7.
(A) Histograms of apparent mobilities for rAMP and m6rA MP. (B) Histograms of apparent mobilities of rCMP and m5rCMP. Electrokinetic separation was carried out at 930 V/cm in 110 × 110 nm COC/COC nanochannel devices using 1X NEB buffer 3 at pH 7.9 as the carrier electrolyte.
4. Conclusions
Our proposed SMS approach addresses the limitations of moderate base call accuracy associated with current nanopore sequencing by generating mononucleotides using a processive enzyme from an intact RNA molecule with the individual rNMPs identified using their molecular-dependent electrophoretic mobility; concept termed Time-of-Flight (ToF) identification. What makes ToF identification attractive is that a variety of experimental conditions can be altered to optimize the identification, such as channel material type, electric field strength, and pH to name a few. In this work, PMMA/COC and COC/COC nanofluidic devices were investigated. The advantage of COC/COC devices was the ability to perform post-assembly surface activation using UV/O3 light to control the EOF depending on the dose, which was not possible in PMMA/COC devices. In addition, the bond strength between the cover plate and the substrate for COC/COC devices was much higher when using post-assembly UV/O3 activation to allow a diverse range of electrophoresis conditions to be employed to optimize the identification accuracies. However, both materials resulted in high ToF identification accuracy of the rNMPs.
There are, however, scale-dependent processes that can affect the performance of nanoscale electrophoresis, such as dielectrophoretic trapping generated by surface roughness. This surface roughness can create inhomogeneous electric fields within the channel when its dimensions are on the same order as the surface roughness and can result in intermittent motion of molecules that can increase peak dispersion [34, 38]. We found that high electric fields can largely mitigate this issue. Others scale-dependent effects include EDL overlap, concentration polarization, and increased surface interactions due to the high surface area-to-volume ratio associated with nanoscale electrophoresis. In our previous report on the identification of dNMPs using nanoscale electrophoresis using PMMA/COC devices [20], we found that partial EDL overlap can degrade identification accuracy. This artifact was overcome by using high carrier electrolyte concentrations to compress the EDL and the use of channels that are relatively large in cross section (110 × 110 nm, width and depth). Minimal EDL overlap also reduces issues associated with concentration polarization.
While organic modifiers can be used to improve electrophoretic performance if wall interactions are present (i.e., capillary electrochromatography, CEC), they are not practical for nanoscale electrophoresis using thermoplastic channels due to polymer swelling when placed in organic solvents that may block the nanochannel making the device fail. In addition, addition of organic modifiers would not be practical for our SMS approach as the presence of organic solvents may denature the surface-immobilized ribo-exonuclease.
In both COC/COC and PMMA/COC nanochannels, the average resolution was similar when the pH was 7.9 generating ToF identification accuracies >99%. The successful identification of rNMPs in free solution using thermoplastic nanochannels will enable development of our SMS approach for RNA. Our recent report on using surface immobilized XRN1 to plastic pillars serves as another foundational piece to deliver our SMS approach [21]. Our SMS approach will provide high read lengths (determined by the processivity of the exonuclease) and high call accuracies (determined by the apparent mobility differences in the rNMPs). The utility of RNA sequencing is becoming even more paramount in light of the COVID-19 pandemic because understanding the structure of the SARS-CoV-2 RNA genome will provide helpful guidance on generating and assigning proper vaccines to the population based on sequence variations the virus may/may not possess.
Although we utilized dye labeled rNMPs in this study to allow tracking the molecules during their transport through nanochannels, our envisioned SMS platform will not require labeling of the rNMPs [64]. We will employ a label free approach for determining the ToF of the rNMPs. This is accomplished by fabricating a thermoplastic nanochannel device with two in-plane nanopores poised at each end of the nanochannel to measure the ToF [65]. The detection therefore uses resistive pulse sensing of single molecules.
Finally, we are employing NIL to make the nanofluidic chips, which is convenient due to the lack of need of specialized equipment to make each device, such as FIB or EBL. Currently, >10 imprinting runs can be carried out with a single resin stamp. Because the Si master can produce >100 resin stamps from a single Si mold master, we can make >1,000 chips from a single Si master. Currently we are working on producing the nanofluidic devices using nano-injection molding [66, 67], which uses a mold insert made from Ni via electroplating. The Ni mold insert can produce many more parts compared to resin stamps used in NIL and injection molding can produce parts at higher rate (~1000 chips per day per injection molding machine).
Supplementary Material
HIGHLIGHTS.
Greater than 99% identification efficiency of the ribonucleotide monophosphates (rNMPs) through mobility matching using a 110 × 110 nm (width x depth) plastic channels that are 100 μm in length using free solution nanoscale electrophoresis.
The nanochannels can be made in a thermoplastic using Nanoimprint Lithography (NIL) to produce nanofluidic devices in plastics at a much higher rate and lower cost compared to glass-based nanofluidic devices. We are currently working on fabricating the nanofluidic devices using nano-injection molding to further reduce cost and increase production rate.
The high identification efficiency afforded by nanoscale electrophoresis will support a new single-molecule RNA sequencing strategy being developed by our group.
5. Acknowledgements
The authors would like to thank the NIH for funding of this work (NIBIB: P41 EB020594; NCI: P30 CA168524; P20 GM130423, P20 GM103638). The authors would also like to thank the National Science Foundation for partial support of this work as well (1507577). Finally, the authors would like to thank the University of Kansas Nanofabrication Facility for assistance in making the nanofluidic devices used in this work. We would like to thank Dr. Prem S.Thapa from Microscopy and Imaging Laboratory for his support on SEM imaging. The authors would like to thank Chemical Biology of Infectious Disease CoBRE (CBID), National Institute of General Medical Sciences of the National Institutes of Health Award Number P20GM113117 and Center for Molecular Analysis of Disease Pathways (CMADP) National Institute of General Medical Sciences of the National Institutes of Health Award Number P20GM103638 for labelling rNMPs with ATTO-532 dye reporter. We would also like to acknowledge Dr. Mateusz Hupert for assistance in collecting the microchip electrophoresis data and Dr. Malgorzata Witek for helpful discussions on the data interpretation.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- [1].Abgrall P, Low L-N, Nguyen N-T, Fabrication of planar nanofluidic channels in a thermoplastic by hot-embossing and thermal bonding, Lab on a Chip 7(4) (2007) 520–522. [DOI] [PubMed] [Google Scholar]
- [2].Hoeijmakers WA, Bártfai R, Stunnenberg HG, Transcriptome analysis using RNA-Seq, Malaria, Springer; 2012, pp. 221–239. [DOI] [PubMed] [Google Scholar]
- [3].Whitley SK, Horne WT, Kolls JK, Research techniques made simple: methodology and clinical applications of RNA sequencing, Journal of Investigative Dermatology 136(8) (2016) e77–e82. [DOI] [PubMed] [Google Scholar]
- [4].Grada A, Weinbrecht K, Next-generation sequencing: methodology and application, The Journal of investigative dermatology 133(8) (2013) e11. [DOI] [PubMed] [Google Scholar]
- [5].Ray M, Horne W, McAleer JP, Ricks DM, Kreindler JL, Fitzsimons MS, Chan PP, Trevejo-Nunez G, Chen K, Fajt M, RNA-seq in pulmonary medicine: how much is enough?, American journal of respiratory and critical care medicine 192(3) (2015) 389–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B, Mapping and quantifying mammalian transcriptomes by RNA-Seq, Nature methods 5(7) (2008) 621. [DOI] [PubMed] [Google Scholar]
- [7].Wang Z, Gerstein M, Snyder M, RNA-Seq: a revolutionary tool for transcriptomics, Nat Rev Genet 10(1) (2009) 57–63. 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sudhagar A, Kumar G, El-Matbouli M, Transcriptome analysis based on RNA-seq in understanding pathogenic mechanisms of diseases and the immune system of fish: a comprehensive review, International journal of molecular sciences 19(1) (2018) 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ari Ş, Ari an M, Next-generation sequencing: advantages, disadvantages, and future, Plant omics: Trends and applications, Springer; 2016, pp. 109–135. [Google Scholar]
- [10].Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, The potential and challenges of nanopore sequencing, Nanoscience and technology: A collection of reviews from Nature Journals, World Scientific 2010, pp. 261–268. [Google Scholar]
- [11].Ayub M, Hardwick SW, Luisi BF, Bayley H, Nanopore-based identification of individual nucleotides for direct RNA sequencing, Nano letters 13(12) (2013) 6144–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Astier Y, Braha O, Bayley H, Toward Single Molecule DNA Sequencing: Direct Identification of Ribonucleoside and Deoxyribonucleoside 5’-Monophosphates by Using an Engineered Protein Nanopore Equipped with a Molecular Adapter, Journal of the American Chemical Society 128(5) (2006) 1705–1710. 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- [13].Clarke J, Wu H-C, Jayasinghe L, Patel A, Reid S, Bayley H, Continuous base identification for single-molecule nanopore DNA sequencing, Nature nanotechnology 4(4) (2009) 265. [DOI] [PubMed] [Google Scholar]
- [14].Cockroft SL, Chu J, Amorin M, Ghadiri MR, A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution, Journal of the American Chemical Society 130(3) (2008) 818–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lieberman KR, Cherf GM, Doody MJ, Olasagasti F, Kolodji Y, Akeson M, Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase, Journal of the American Chemical Society 132(50) (2010) 17961–17972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bayley H, Sequencing single molecules of DNA, Current opinion in chemical biology 10(6) (2006) 628–637. [DOI] [PubMed] [Google Scholar]
- [17].Reiner JE, Balijepalli A, Robertson JW, Drown BS, Burden DL, Kasianowicz JJ, The effects of diffusion on an exonuclease/nanopore-based DNA sequencing engine, The Journal of chemical physics 137(21) (2012) 214903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Novak BR, Moldovan D, Nikitopoulos DE, Soper SA, Distinguishing Single DNA Nucleotides Based on Their Times of Flight Through Nanoslits: A Molecular Dynamics Simulation Study, Journal of Physical Chemistry B 117(12) (2013) 3271–3279. 10.1021/jp309486c. [DOI] [PubMed] [Google Scholar]
- [19].Xia K, Novak BR, Weerakoon-Ratnayake KM, Soper SA, Nikitopoulos DE, Moldovan D, Electrophoretic Transport of Single DNA Nucleotides through Nanoslits: A Molecular Dynamics Simulation Study, Journal of Physical Chemistry B 119 (2015) 11443–11448. [DOI] [PubMed] [Google Scholar]
- [20].O’Neil C, Amarasekara CA, Weerakoon-Ratnayake KM, Gross B, Jia Z, Singh V, Park S, Soper SA, Electrokinetic transport properties of deoxynucleotide monophosphates (dNMPs) through thermoplastic nanochannels, Anal Chim Acta 1027 (2018) 67–75. 10.1016/j.aca.2018.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Athapattu US, Amarasekara CA, Nagel AC, Immel JR, Bloom S, Barany F, Soper SA, Solid-Phase XRN1 Reactions for RNA Cleavage: Application in Single-Molecule Sequencing, Nucleic Acids Research (under review) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Uhrová M, Deyl Z, Suchánek M, Separation of common nucleotides, mono-, di- and triphosphates, by capillary electrophoresis, Journal of Chromatography B: Biomedical Sciences and Applications 681(1) (1996) 99–105. [DOI] [PubMed] [Google Scholar]
- [23].Mateos-Vivas M, Rodríguez-Gonzalo E, Domínguez-Álvarez J, García-Gómez D, Ramírez-Bernabé R, Carabias-Martínez R, Analysis of free nucleotide monophosphates in human milk and effect of pasteurisation or high-pressure processing on their contents by capillary electrophoresis coupled to mass spectrometry, Food chemistry 174 (2015) 348–355. [DOI] [PubMed] [Google Scholar]
- [24].Geldart SE, Brown PR, Analysis of nucleotides by capillary electrophoresis, Journal of Chromatography A 828(1–2) (1998) 317–336. [DOI] [PubMed] [Google Scholar]
- [25].Liu H, Qi S, Zhang Y, Huang A, Sun Y, Determination of inosine 5′-monophosphate and guanosine 5′-monophosphate in pig feed by capillary zone electrophoresis, Journal of High Resolution Chromatography 20(4) (1997) 242–244. [Google Scholar]
- [26].Geldart SE, Brown PR, Separation of purine and pyrimidine bases by capillary zone electrophoresis with carbonate buffers, Journal of Chromatography A 831(1) (1999) 123–129. [Google Scholar]
- [27].Pennathur S, Santiago JG, Electrokinetic Transport in Nanochannels. 1. Theory, Analytical Chemistry 77(21) (2005) 6772–6781. 10.1021/ac050835y. [DOI] [PubMed] [Google Scholar]
- [28].Pennathur S, Santiago JG, Electrokinetic Transport in Nanochannels. 2. Experiments, Analytical Chemistry 77(21) (2005) 6782–6789. 10.1021/ac0508346. [DOI] [PubMed] [Google Scholar]
- [29].Xuan X, Li D, Electrokinetic transport of charged solutes in micro-and nanochannels: The influence of transverse electromigration, Electrophoresis 27(24) (2006) 5020–5031. [DOI] [PubMed] [Google Scholar]
- [30].Garcia AL, Ista LK, Petsev DN, O’Brien MJ, Bisong P, Mammoli AA, Brueck SR, López GP, Electrokinetic molecular separation in nanoscale fluidic channels, Lab on a Chip 5(11) (2005) 1271–1276. [DOI] [PubMed] [Google Scholar]
- [31].Baldessari F, Santiago JG, Electrophoresis in nanochannels: brief review and speculation, Journal of Nanobiotechnology 4(1) (2006) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang L, Shao P, van Kan J, Ansari K, Bettiol A, Pan X, Wohland T, Watt F, Fabrication of nanofluidic devices utilizing proton beam writing and thermal bonding techniques, Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms 260(1) (2007) 450–454. [Google Scholar]
- [33].Duan C, Wang W, Xie Q, Fabrication of nanofluidic devices, Biomicrofluidics 7(2) (2013) 026501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chantiwas R, Hupert ML, Pullagurla SR, Balamurugan S, Tamarit-López J, Park S, Datta P, Goettert J, Cho Y-K, Soper SA, Simple replication methods for producing nanoslits in thermoplastics and the transport dynamics of double-stranded DNA through these slits, Lab on a Chip 10(23) (2010) 3255–3264. [DOI] [PubMed] [Google Scholar]
- [35].Chantiwas R, Park S, Soper SA, Kim BC, Takayama S, Sunkara V, Hwang H, Cho Y-K, Flexible fabrication and applications of polymer nanochannels and nanoslits, Chemical Society Reviews 40(7) (2011) 3677–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chai J, Lu F, Li B, Kwok DY, Wettability interpretation of oxygen plasma modified poly (methyl methacrylate), Langmuir 20(25) (2004) 10919–10927. [DOI] [PubMed] [Google Scholar]
- [37].Oneil CE, Jackson JM, Shim S-H, Soper SA, Interrogating Surface Functional Group Heterogeneity of Activated Thermoplastics Using Super-Resolution Fluorescence Microscopy, Analytical Chemistry 88(7) (2016) 3686–3696. 10.1021/acs.analchem.5b04472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Uba FI, Pullagurla SR, Sirasunthorn N, Wu J, Park S, Chantiwas R, Cho Y-K, Shin H, Soper SA, Surface charge, electroosmotic flow and DNA extension in chemically modified thermoplastic nanoslits and nanochannels, Analyst 140(1) (2015) 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Weerakoon-Ratnayake KM, Uba FI, Oliver-Calixte NJ, Soper SA, Electrophoretic separation of single particles using nanoscale thermoplastic columns, Analytical chemistry 88(7) (2016) 3569–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Amarasekara CA, Athapattu US, Rathnayaka C, Choi J, Park S, Soper SA, Open-tubular Nanoelectrochromatography (OT-NEC): Gel-free Separation of Single Stranded DNAs (ssDNAs) in Thermoplastic Nanochannels, Electrophoresis 41(18–19) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].O’Neil Colleen, Amarasekara CA, Weerakoon-Ratnayake KM, Gross Bethany, Jia Z, Singh V, Park Sunggook, Soper SA, Electrokinetic transport properties of deoxynucleotide monophosphates (dNMPs) through thermoplastic nanochannels, Anal Chim Acta 1027 (2018) 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hu W, Soper SA, Jackson JM, Time-Delayed Integration–Spectral Flow Cytometer (TDI-SFC) for Low-Abundance-Cell Immunophenotyping, Analytical chemistry 91(7) (2019) 4656–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ford SM, Kar B, McWhorter S, Davies J, Soper SA, Klopf M, Calderon G, Saile V, Microcapillary electrophoresis devices fabricated using polymeric substrates and X-ray lithography, J. Microcolumn Sep. 10(5) (1998) 413–422. [Google Scholar]
- [44].Sinville R, Soper SA, High resolution DNA separations using microchip electrophoresis, Journal of Separation Science 30(11) (2007) 1714–1728. 10.1002/jssc.200700150. [DOI] [PubMed] [Google Scholar]
- [45].Obubuafo A, Balamurugan S, Shadpour H, Spivak D, McCarley RL, Soper SA, Poly(methyl methacrylate) microchip affinity capillary gel electrophoresis of aptamer-protein complexes for the analysis of thrombin in plasma, Electrophoresis 29(16) (2008) 3436–3445. 10.1002/elps.200700854. [DOI] [PubMed] [Google Scholar]
- [46].Njoroge SK, Witek MA, Hupert ML, Soper SA, Microchip electrophoresis of Alu elements for gender determination and inference of human ethnic origin, Electrophoresis 31(6) (2010) 981–990. 10.1002/elps.200900641. [DOI] [PubMed] [Google Scholar]
- [47].Albrecht JC, Kotani A, Lin JS, Soper SA, Barron AE, Simultaneous detection of 19 K-ras mutations by free-solution conjugate electrophoresis of ligase detection reaction products on glass microchips, Electrophoresis 34(4) (2013) 590–597. 10.1002/elps.201200462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Uba FI, Hu B, Weerakoon-Ratnayake K, Oliver-Calixte N, Soper SA, High process yield rates of thermoplastic nanofluidic devices using a hybrid thermal assembly technique, Lab on a Chip 15(4) (2015) 1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ramm P, Lu JJ-Q, Taklo MM, Handbook of wafer bonding, John Wiley & Sons; 2011. [Google Scholar]
- [50].Tsao C, Hromada L, Liu J, Kumar P, DeVoe D, Low temperature bonding of PMMA and COC microfluidic substrates using UV/ozone surface treatment, Lab on a Chip 7(4) (2007) 499–505. [DOI] [PubMed] [Google Scholar]
- [51].Huang X, Gordon MJ, Zare RN, Current-monitoring method for measuring the electroosmotic flow rate in capillary zone electrophoresis, Analytical Chemistry 60(17) (1988) 1837–1838. 10.1021/ac00168a040. [DOI] [Google Scholar]
- [52].Cho Y, Park J, Park H, Cheng X, Kim B, Han A, Fabrication of high-aspect-ratio polymer nanochannels using a novel Si nanoimprint mold and solvent-assisted sealing, Microfluidics and Nanofluidics 9(2–3) (2010) 163–170. [Google Scholar]
- [53].Ali U, Karim KJBA, Buang NA, A review of the properties and applications of poly (methyl methacrylate)(PMMA), Polymer Reviews 55(4) (2015) 678–705. [Google Scholar]
- [54].Nunes PS, Ohlsson PD, Ordeig O, Kutter JP, Cyclic olefin polymers: emerging materials for lab-on-a-chip applications, Microfluidics and nanofluidics 9(2–3) (2010) 145–161. [Google Scholar]
- [55].Jackson JM, WItek MA, Hupert ML, Brady C, Pullagurla SR, Kamande JW, Aufforth RD, Tignanelli CJ, Torphy RJ, Yeh JJ, Soper SA, UV Activation of Polymeric High Aspect Ratio Microstructures: Ramifications in Antibody Surface Loading for Circulating Tumor Cell Selection, Lab on a Chip 14 (2014) 106–117. 10.1039/C3LC50618E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kino K, Hirao-Suzuki M, Morikawa M, Sakaga A, Miyazawa H, Generation, repair and replication of guanine oxidation products, Genes and Environment 39(1) (2017) 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bidleman TF, The relationship between resolution and percent band overlap, Journal of Chemical Education 56(5) (1979) 293. [Google Scholar]
- [58].O’Neil C, Amarasekara CA, Weerakoon-Ratnayake KM, Gross B, Jia Z, Singh V, Park S, Soper SA, Electrokinetic transport properties of deoxynucleotide monophosphates (dNMPs) through thermoplastic nanochannels, Anal. Chim. Acta 1027 (2018) 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].O’Neil CE, Taylor S, Ratnayake K, Pullagurla S, Singh V, Soper SA, Characterization of activated cyclic olefin copolymer: effects of ethylene/norbornene content on the physiochemical properties, Analyst 141 (2016) 6521–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rani SD, You BH, Soper SA, Murphy MC, Nikitopoulos DE, Influence of material transition and interfacial area changes on flow and concentration in electro-osmotic flows, Anal Chim Acta 770 (2013) 103–110. 10.1016/j.aca.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Po HN, Senozan N, The Henderson-Hasselbalch equation: its history and limitations, Journal of Chemical Education 78(11) (2001) 1499. [Google Scholar]
- [62].Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S, Mason CE, The birth of the Epitranscriptome: deciphering the function of RNA modifications, Genome biology 13(10) (2012) 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Frye M, Harada BT, Behm M, He C, RNA modifications modulate gene expression during development, Science 361(6409) (2018) 1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Davis LM, Fairfield FR, Harger CA, Jett JH, Keller RA, Hahn JH, Krakowski LA, Marrone BL, Martin JC, Nutter HL, Ratliff RL, Shera EB, Simpson DJ, Soper SA, Rapid DNA sequencing based upon single molecule detection, Genetic Analysis: Biomolecular Engineering 8(1) (1991) 1–7. [DOI] [PubMed] [Google Scholar]
- [65].Jia Z, Choi J, Park S, Surface Charge Density-Dependent DNA Capture through Polymer Planar Nanopores, ACS applied materials & interfaces 10(47) (2018) 40927–40937. [DOI] [PubMed] [Google Scholar]
- [66].Teillet J, Martinez Q, Tijunelyte I, Chami B, Bancaud A, Characterization and minimization of band broadening in DNA electrohydrodynamic migration for enhanced size separation, Soft Matter (2020). [DOI] [PubMed] [Google Scholar]
- [67].Zhang N, Byrne CJ, Browne DJ, Gilchrist MD, Towards nano-injection molding, Materials today 15(5) (2012) 216–221. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.