Abstract
Background
Oliver–McFarlane syndrome is characterised by trichomegaly, congenital hypopituitarism and retinal degeneration with choroidal atrophy. Laurence–Moon syndrome presents similarly, though with progressive spinocerebellar ataxia and spastic paraplegia and without trichomegaly. Both recessively inherited disorders have no known genetic cause.
Methods
Whole-exome sequencing was performed to identify the genetic causes of these disorders. Mutations were functionally validated in zebrafish pnpla6 morphants. Embryonic expression was evaluated via in situ hybridisation in human embryonic sections. Human neurohistopathology was performed to characterise cerebellar degeneration. Enzymatic activities were measured in patient-derived fibroblast cell lines.
Results
Eight mutations in six families with Oliver–McFarlane or Laurence–Moon syndrome were identified in the PNPLA6 gene, which encodes neuropathy target esterase (NTE). PNPLA6 expression was found in the developing human eye, pituitary and brain. In zebrafish, the pnpla6 curly-tailed morphant phenotype was fully rescued by wild-type human PNPLA6 mRNA and not by mutation-harbouring mRNAs. NTE enzymatic activity was significantly reduced in fibroblast cells derived from individuals with Oliver–McFarlane syndrome. Intriguingly, adult brain histology from a patient with highly overlapping features of Oliver–McFarlane and Laurence–Moon syndromes revealed extensive cerebellar degeneration and atrophy.
Conclusions
Previously, PNPLA6 mutations have been associated with spastic paraplegia type 39, Gordon–Holmes syndrome and Boucher–Neuhäuser syndromes. Discovery of these additional PNPLA6-opathies further elucidates a spectrum of neurodevelopmental and neurodegenerative disorders associated with NTE impairment and suggests a unifying mechanism with diagnostic and prognostic importance.
INTRODUCTION
Oliver–McFarlane syndrome (MIM 275400) is a rare congenital disorder characterised by trichomegaly, severe chorioretinal atrophy and multiple pituitary hormone deficiencies, including growth hormone (GH), gonadotrophins and thyroid stimulating hormone (TSH).1,2 Thyroid and GH abnormalities may be present at birth and, if untreated, result in intellectual impairment and profound short stature. Congenital hypogonadism occurs in half of patients, and nearly all have documented hypogonadotropic hypogonadism during puberty, with subsequent reproductive dysfunction. Chorioretinal atrophy is typically noted in the first five years of life. Half of reported cases have spinocerebellar involvement, including ataxia, spastic paraplegia and peripheral neuropathy. Fourteen cases have been reported to date, including two sibships, suggesting autosomal-recessive inheritance.1,3–11 However, no genetic cause has been identified.
Laurence–Moon syndrome (MIM 245800) has a clinical presentation similar to Oliver–McFarlane syndrome, including chorioretinopathy and pituitary dysfunction, though with childhood onset of ataxia, peripheral neuropathy and spastic paraplegia.12 Hair findings have not been previously reported. Historically, Laurence–Moon syndrome has been associated with Bardet–Biedl syndrome (BBS).13–19 No link between Oliver–McFarlane and Laurence–Moon syndromes has been reported.
Here, we identify the genetic aetiology for six patients with a diagnosis of Oliver–McFarlane syndrome and four previously reported patients with Laurence–Moon syndrome. Using whole-exome sequencing, we identified eight mutations in the PNPLA6 gene (MIM 603197), encoding neuropathy target esterase (NTE) that plays critical roles in phosphatidylcholine metabolism, membrane phospholipid trafficking and axonal integrity. PNPLA6 mutations alter the activity of the patatin-like domain that hydrolyses phosphatidylcholine into glycerophosphocholine, important for stabilising membrane integrity.20,21
PNPLA6 alleles have been previously implicated in a broad spectrum of neurodegenerative conditions, including spastic paraplegia type 39 (SPG39 (MIM 612020)),22 Gordon–Holmes syndrome (GHS (MIM 212840)) and Boucher–Neuhäuser syndrome (BNHS (MIM 215470)).23,24 Oliver–McFarlane and Laurence–Moon syndromes are distinct from SPG39, GHS and BNHS due to the infantile pituitary dysfunction, typically resulting in small anterior pituitary size, short stature, hypogonadism and intellectual disability if untreated. A remarkable heterogeneity of hair and neurological findings was found between patients and within families with Oliver–McFarlane and Laurence–Moon syndromes. Taken together with previous studies, the mutant alleles of PNPLA6 subserve a myriad of neurodegenerative disorders that include chorioretinopathy, spinocerebellar ataxia, spastic paraplegia, hypopituitarism and trichomegaly.
METHODS
Whole-exome sequencing and Sanger confirmation
Whole genomic DNA was extracted from whole blood by standard methods. For patients A:1 and A:2, library construction was performed on dsDNA, sheared by sonication to an average size of 200 bp in an automated fashion on an IntegenX Apollo324. After nine cycles of PCR amplification using the Clontech Advantage II kit, 1 μg of genomic library was recovered for exome enrichment using the NimbleGen EZ Exome V2 kit. Libraries were sequenced on an Illumina HiSeq2500, generating approximately 30 million paired end reads, each 100 bases long. Data analysis used the Broad Institute’s Genome Analysis Toolkit (GATK).25 Reads were aligned with the Illumina Chastity Filter with the Burrows Wheeler Aligner.26 Variant sites were called using the GATK UnifiedGenotyper module. Single-nucelotide variant calls were filtered using the variant quality score recalibration method.25 Equivalent methods were employed for patients D:1, E:1 and family F. Variant filtering methods for A, D and E are listed in online supplementary table S1.
For patients B:1, C:1 and variant confirmation for other families, Sanger sequencing was performed by standard methods on the 34 exons of the PNPLA6 gene (NM_001166111.1). Primers were designed using Primer3 and listed in online supplementary table S2. PCR amplification and DNA sequencing was performed as described previously.27 Mutation nomenclature was assigned in accordance with GenBank accession number NM_001166111.1 with nucleotide position 1 corresponding to the A of the ATG initiation codon.
Human neurohistopathology
Postmortem brain tissue donation was approved and provided for this project by the Queen Square Brain Bank, University College London. Paraffin blocks from representative areas of the brain were available for histological and immunohistochemical investigation. In brief, 8-mm-thick sections of paraffin embedded tissue blocks were deparaffinised and rehydrated through graded alcohols. Sections were placed in 0.3% H2O2/methanol to block endogenous peroxidase activity. Antigen retrieval was undertaken to unmasked antigen epitopes and was achieved either by pressure cooking in citrate buffer (pH 6.0) for 10 min, incubation either in 99% formic acid for 10 min or in proteinase K solution (Dako) for 10 min. The sections were washed with tris-buffered saline and incubated in 10% non-fat milk solution to prevent non-specific antibody binding. The appropriate primary antibody was applied (see online supplementary table S4), followed by an antimouse or antirabbit secondary antibody (Dako, Ely, UK) at 1/200 dilution, as appropriate. Subsequently, avidinbiotin complex Elite kit (Vector, Peterborough, UK) was applied and colour was developed by diaminobenzidine/H2O2. Sections were finally counterstained with Mayer’s haematoxylin. Routine H&E histological staining was also carried out.
In situ hybridisation
The full-length purified PNPLA6 cDNA was obtained from Source Bioscience (IRATp970G0676D) in a pcmvSPORT6 plasmid. Histological sections of human embryos (Carnegie stages 19 and 23) were obtained from the Human Developmental Biology Resource. In situ hybridisation was performed as described.28
Zebrafish morpholino and rescue
Wild-type (WT) zebrafish (Danio rerio) were maintained under standard laboratory conditions. Embryos were obtained by natural cross. A human PNPLA6 cDNA clone was purchased (Thermo, BC051768) and was subcloned in T7TS vector. Point mutations were introduced with the Quik Change Lightning Site Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, California, USA). Sanger sequencing was completed to verify each construct. RNA was produced using the mMessage mMachine Kit (Life Technologies) and linearised plasmid constructs.
Control and PNPLA6 translation blocking (5′-ctgtgtccgatgtgc tctgtcccat-3′)29 morpholinos (MOs) were injected in WT zebrafish embryos at one-to-two-cell stage. Rescue experiments were conducted with 100 pg human PNPLA6 mRNA and 2.5 ng MO. Embryos were examined with a Leica MZ 16 F microscope and images were captured with a Leica DFC 480 camera.
NTE activity assay
Human skin fibroblasts were obtained from patients by 2 mm punch skin biopsies with appropriate topical analgesia. Human skin fibroblasts from a control subject were obtained from Invitrogen (Carlsbad, California, USA) and were maintained minimal essential medium (GIBCO, Invitrogen) supplemented with fetal bovine serum (Hyclone, Thermo Fisher Scientific, Waltham, Massachusetts, USA) and antibiotics at 37°C with 5% CO2.30
NTE enzymatic activity assay was performed in patient fibroblasts as described.31 NTE activity was defined as the difference in phenyl valerate hydrolysis activity inhibited by paraoxon (inhibits background esterase activity) and paraoxon+mipafox (inhibits NTE in addition to background esterase activity). Endpoint absorbance was measured at 486 nm using a SpectraMax 340 microplate reader (Molecular Devices, Sunnydale, California, USA). Protein concentration was determined colorimetrically using the Bio-Rad Protein Assay Dye Reagent (Hercules, California, USA) using standard dilutions of bovine serum albumin (New England BioLabs, Ipswich, Massachusetts, USA). We define one unit of NTE specific activity as 1 nmol phenol produced per minute per milligram protein. Activity levels were normalised to two control fibroblast samples.
Statistical analysis
Statistical analyses performed included Student t test and one-way analysis of variance (ANOVA) with post hoc Tukey’s test (α value 0.05). Calculations were performed using Excel 2010 (Microsoft, Redmond, Washington, USA) and GraphPad Prism 6d (GraphPad Software, La Jolla, California).
RESULTS
PNPLA6 alleles associated with Oliver–McFarlane and Laurence–Moon syndromes
Patient phenotypes (A–F) are summarised in online supplementary results, table 1 and figure 1, and pedigrees are depicted in online supplementary figure S1. Oliver–McFarlane and Laurence–Moon syndromes were distinguished from BNHS and GHS by the presence of congenital or infantile anterior hypopituitarism (hypothyroidism or GH deficiency). Oliver–McFarlane was distinguished from Laurence–Moon syndrome by the presence of trichomegaly or alopecia. Patient E:1 and family F have been described in detail.11,18 Postmortem neuropathology was performed on brain of patient E:1 to characterise the histological features of cerebellar degeneration in Oliver–McFarlane syndrome. The major neuropathological findings included an overall reduction in size of the brain (981 g), frontal white matter and midbrain, and severe cerebellar cortical atrophy involving both cerebellar vermis and hemispheres. Microscopy demonstrated severe cerebellar cortical degeneration with loss of Purkinje and granule cells, numerous empty baskets and prominent Bergmann gliosis (figure 1O–R).
Table 1.
Clinical phenotype and genotype summary
| Phenotype | A:1 | A:2 | B:1 | C:1 | D:1 | E:1 | F:1 | F:2 | F:3 | F:4 |
|---|---|---|---|---|---|---|---|---|---|---|
| Sex | F | M | M | M | M | M | M | F | M | F |
| Age (last Wt, L) | 6.8 | 5.7 | 2.7 | 28.4 | 8.0 | 21.8 | 19 | 42.3 | 41.3 | 15 |
| Birth Wt (SDS) | −0.79 | −0.97 | −1.62 | NR | +0.91 | −3.84 | NR | NR | NR | NR |
| Birth L (SDS) | +1.47 | +2.26 | −3.09 | NR | −1.47 | NR | NR | NR | NR | NR |
| Current Wt | −1.35 | −1.65 | −2.26 | −1.39 | 0.12 | −1.46 | NR | +1.84 | −0.39 | NR |
| Current height | −2.03 | −1.23 | −2.11 | −3.97 | −1.55 | −1.64 | NR | −2.50 | −2.36 | NR |
| Chorioretinopathy | + | + | + | + | + | + | + | + | + | + |
| Trichomegaly | + | +/− | + | +/− | + | + | NR | − | + | NR |
| Alopecia | − | − | − | + | − | + | NR | + | − | NR |
| Facial dysmorphisms | + | + | − | + | − | NR | NR | + | + | NR |
| Intellectual disability | − | − | − | − | − | + | + | + | + | + |
| Short stature | + | + | + | + | + | + | + | + | + | + |
| Delayed bone age | + | + | + | NR | + | + | NR | NR | NR | NR |
| Small A pituitary | + | + | + | + | + | + | NR | + | NR | NR |
| GH deficiency | + | + | + | + | + | + | + | + | + | + |
| Hypothyroidism | + | + | +/− | − | +/− | + | − | − | − | − |
| Micropenis | NA | − | + | + | + | + | + | NA | + | NA |
| Hypogonadism | NA | NA | NA | + | NA | + | + | + | + | + |
| Ataxia | − | − | − | − | − | + | + | + | + | + |
| Spastic paraplegia | − | − | − | − | − | + | + | + | + | + |
| Peripheral neuropathy | − | − | + | − | − | + | + | + | + | + |
| Polydactyly | − | − | − | − | − | − | − | − | − | − |
| Obesity | − | − | − | − | − | − | − | + | − | − |
| PNPLA6 alleles | p.Arg1099Gln p.Gly1176Ser |
p.Arg1031fs*38 p.Gly1129Arg |
c.1973+2T>G p.Val1215Ala |
dup(Ex14–20) p.Val1215Ala |
p.Arg1031fs*38 p.Gly1129Arg |
p.Gly726Arg p.Arg1031fs*38 |
||||
A, anterior; GH, growth hormone; L, length; NA, information not applicable; NR, information not recorded; Wt, weight.
Figure 1.
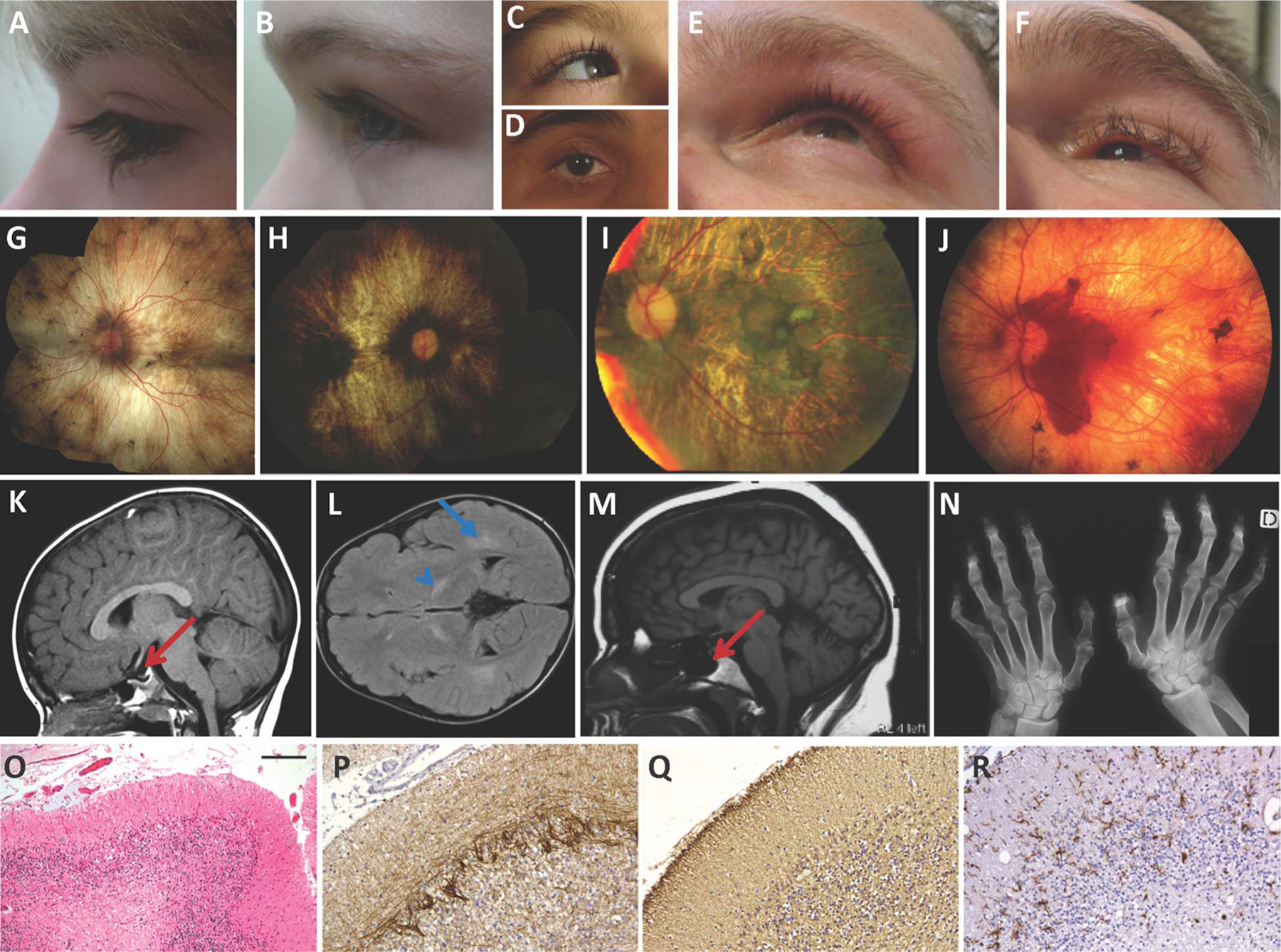
Clinical phenotypes of Oliver–McFarlane and Laurence–Moon syndromes. (A–F) Variability in eyelash length and eyebrow and scalp hair quality for patients A:1 (A), A:2 (B), B:1 (C), C:1 (D), F:1 (E) and F:2 (F). (G–J) Chorioretinal atrophy with variable pigment findings including macular and peripapillary sparing and peripheral pigment clumping in patients A:1 (G), C:1 (H), D:1 (I) and F:1 (J). (K–M) Brain MRI findings including small anterior pituitary noted in Oliver–McFarlane syndrome patient A:1 (K) and Laurence–Moon syndrome patient F:1 (M), along with signal abnormalities of the posterior internal capsule and deep white matter bilaterally in patient B:1 (L). (N) Spastic paraplegia findings on hand X-ray of patient F:1. (O–R) Neuropathology findings for patient E:1, including severe depletion of Purkinje and granule cells in cerebellar cortex (O), numerous empty baskets (P), severe astrogliosis (Q) and an increased activated microglial cells (R). (O) H&E stain, (P) α-internexin immunohistochemistry, (Q) glial fibrillary acidic protein immunohistochemistry and (R) IBa1 immunohistochemistry. Scale bar represents 90 μm on (A) and 45 μm on (B–D).
To determine the genetic cause for Oliver–McFarlane and Laurence–Moon syndromes, whole-exome sequencing was performed independently for patients A:1–2, D:1, E:1 and family F. Autosomal-recessive inheritance, both homozygous and compound heterozygous, was assumed during the filtering stages (see online supplementary table S1). Sanger sequencing of PNPLA6 was performed for patients B:1 and C:1 and for segregation analysis (see online supplementary figure S1). Familial carrier testing to demonstrate disease allele segregation was possible for families A, D, E and F. Carriers were asymptomatic for all phenotypes.
Compound heterozygous mutations were found in the PNPLA6 gene for each patient (table 1). PNPLA6 is composed of 34 exons, encoding a 1375 amino acid polypeptide with three cyclic nucleotide monophosphate (cNMP) binding domains and one patatin-like domain (figure 2A, B). In total, five missense (p.[Gly726Arg], p.[Arg1099Gln], p.[Gly1176Ser], p.[Gly1129Arg], p.[Val1215Ala]), one splice site mutation (c.[1973+2T>G]), one insertion mutation (c.[3091–3092_insAGCC]) and an inline duplication of exons 14–20 of PNPLA6 (see online supplementary figure S2A) were observed in our Oliver–McFarlane and Laurence–Moon affected individuals (figure 2A, B and table 1). Duplication of exons 14–20 would result in a frameshift at residue 832 and early truncation of the protein (see online supplementary figure S2B).
Figure 2.
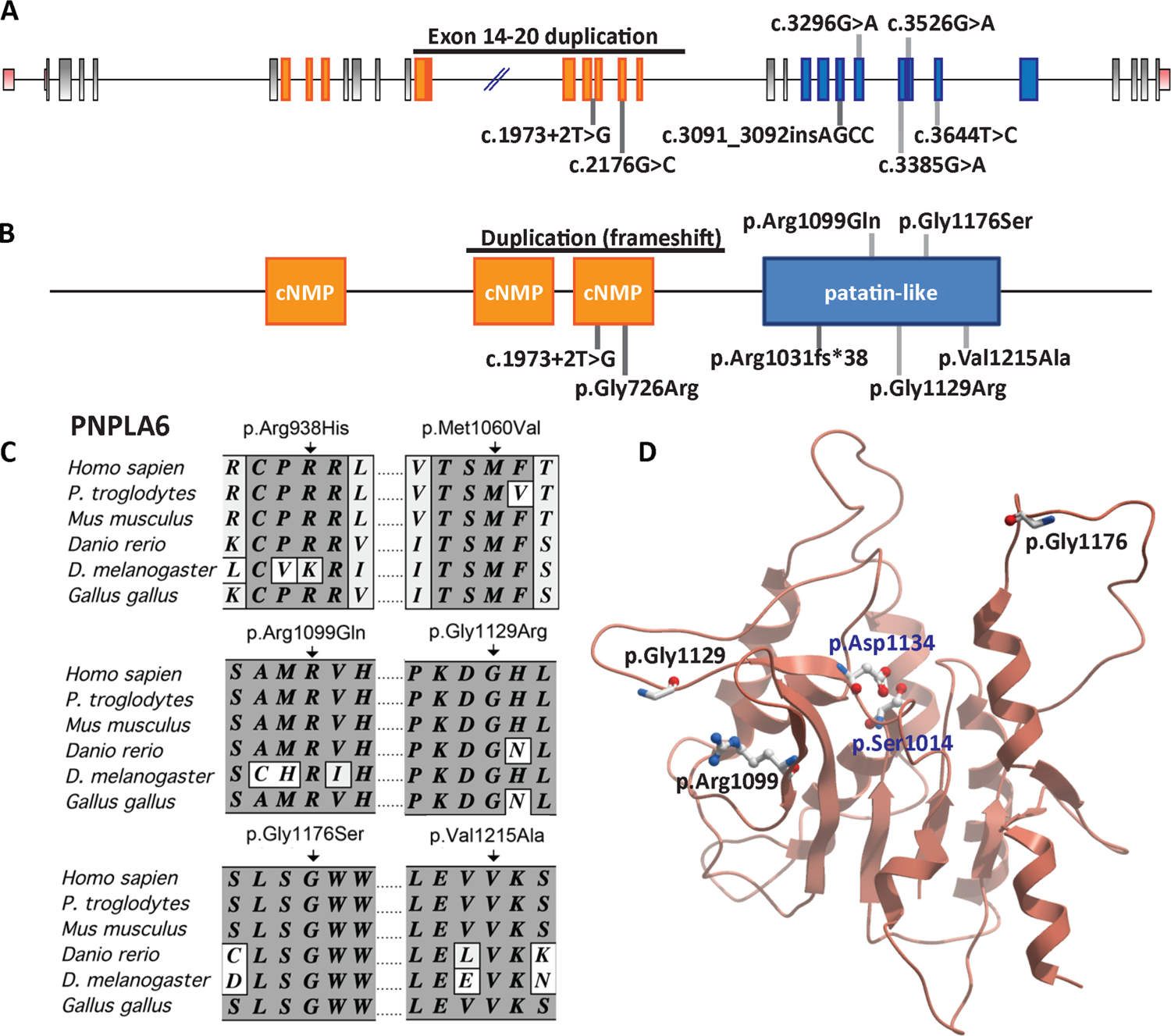
PNPLA6 mutations are associated with Oliver–McFarlane and Laurence–Moon syndromes. (A) An illustration of PNPLA6 gene structure along with the mutations. Exons highlighted corresponding to protein domains. (B) Predicted PNPLA6 protein structure includes three cNMP domains and one patatin-like domain. (C) Alignment of portions of PNPLA6 proteins from various species, showing conservation of the neuropathy target esterase domain residues mutated in patients with Oliver–McFarlane and SPG39 syndromes. (D) A homology model of the catalytic domain of PNPLA6 constructed using SWISS-MODEL,52 and 10XW.PDB as a template. The location of mutations (red) and the enzymatically active residues (blue) are shown. cNMP, cyclic-nucleotide monophosphate.
All mutations identified in this cohort are novel, except the frameshift mutation p.Arg1031fs*38 previously associated with GHS.23 The c.3526G>A (rs142422525) variant was observed once in over 13 000 individual alleles listed in the National Heart, Lung, and Blood Institute Exome Sequencing Project variant (EVS) database. However, it was not found in over 300 ethnically matched controls32 and was predicted pathogenic by MutationTaster,33 Polyphen-2,34 SIFT35 and PROVEAN.36 None of the other variants were found in the control samples, in the 1000 Genome Project, or in the EVS databases, and all were predicted to be deleterious.
PNPLA6 expression during human central nervous system and retina development
In order to further understand the effect of PNPLA6 mutations leading to congenital and childhood onset of pituitary and retinal degeneration, expression analyses were conducted in these tissues at embryonic and adult stages. First, PNPLA6 expression was evaluated in human embryonic eye, pituitary and brain. As organogenesis occurs during the first trimester, human embryonic tissue sections were evaluated for PNPLA6 expression at Carnegie stages 19 and 23 by in situ hybridisation (figure 3). As anticipated, PNPLA6 expression was noted throughout the neural retina, retinal pigment epithelium (RPE), choroid, anterior and posterior pituitary, cerebellum and ventricular zones of the developing brain (figure 3A–F). Expression was also detected in the epidermis, lens, extraocular muscles, nasal epithelium, trigeminal ganglion and diencephalon (figure 3A, D, F). We also observed consistent expression of PNPLA6 expression in all adult human and mouse tissues examined (see online supplementary figure S3A,B), in the developing and adult mouse brain and eye (see online supplementary figure S3B,C), and in dissected segments of the postnatal mouse eye and brain (see online supplementary figure S3D,E).
Figure 3.

Human embryonic expression of PNPLA6. PNPLA6 in situ hybridisation of human embryos as Carnegie stage 19 (E) and stage 23 (A–D and F). (A and D) Horizontal sections with expression in the retina and periventricular zones of the lateral and third ventricles. (B) Eye expression observed in the retina, fovea (arrow), retinal pigment epithelium (arrowhead), choroid, external epithelium and lens (L). (C) Anterior and posterior pituitary expression in the epithelium and parenchyma. (E) PNPLA6 expression in the developing cerebellum (arrow) and hindbrain (arrowhead). (F) PNPLA6 is expressed in the nasal epithelium (arrow), eyelid epithelium (arrowhead), extraocular muscles (EOM) and lens (L). Scale bar represents 400 μm in (A), (D) and (E) 100 μm in (B) and (C), and 200 μm in (F).
Mutant alleles are clustered in the Patatin-like domain of NTE
We found one missense mutation in the cNMP domain. Mutations in cNMP domain have been reported in human patients and in Drosophila swiss cheese mutants.23,37 Furthermore, the inline duplication and splice site mutation were predicted to alter transcription of the terminal cNMP domain. However, most of our disease-associated mutations were located in or predicted to disrupt transcription of the patatin-like domain, critical for the esterase activity of NTE. Notably, the two previously reported SPG39 mutations (p.-Arg938His and p.Met1060Val) are also located in the patatin-like domain, as are the majority of mutations reported for GHS and BNHS.22–24 These results suggest that esterase activity of NTE is essential for physiological function in retina and pituitary.
Therefore, we analysed structural effects of different mutations in the patatin-like domain (figure 2C, D). ClustalW analysis of the two SPG39 and five Oliver–McFarlane and Laurence–Moon alleles indicated a high level of evolutionary conservation for all mutated residues (figure 2C and data not shown). We also visualised the location of the amino acid substitutions in relation to the two enzymatically active residues for phosphatidylcholine hydrolysis, p.Asp1134 and p.Ser1014. Residues p.Gly1176, p.Gly1129 and p.Arg1099 are not in close proximity to the catalytic residues or within the funnel proposed to be the enzymatic site (figure 2D), similar to reported SPG39, BNHS and GHS alleles.22,24,38 Therefore, given the early and severe onset of Oliver–McFarlane and Laurence–Moon syndromes compared with other PNPLA6-associated diseases, we reasoned that differential impairment of NTE activity might underlie the mechanism for this spectrum of disorders. Because molecular modelling demonstrated that these mutations lie outside the esterase active site and thus limit our ability to predict the effect on enzymatic function of the encoded protein, we investigated disease allele pathogenicity in zebrafish and patient cells.
Impaired activity of human PNPLA6 alleles during development
To determine the potential deleterious effects of PNPLA6 mutations associated with Oliver–McFarlane or Laurence–Moon syndromes during normal development, MO knockdown of pnpla6 was performed in the zebrafish model system. Injection of pnpla6 MO in single cells postfertilisation resulted in larvae with curved tails as well as small heads and eyes as previously described.29 The phenotypes were grossly scored in 48 h posthatching zebrafish embryos into four categories: (i) normal, (ii) mild with small head and eyes, (iii) intermediate additionally had distal tail curvature and (iv) severe had full tail and body curvature in addition to small head and eyes (figure 4A–D). Injection of a control scrambled MO or WT human PNPLA6 mRNA alone showed no difference from WT embyros (figure 4E and not shown). Coinjection of MO and WT PNPLA6 mRNA significantly rescued the morphant phenotype and were indistinguishable from WT mRNA alone (figure 4E).
Figure 4.
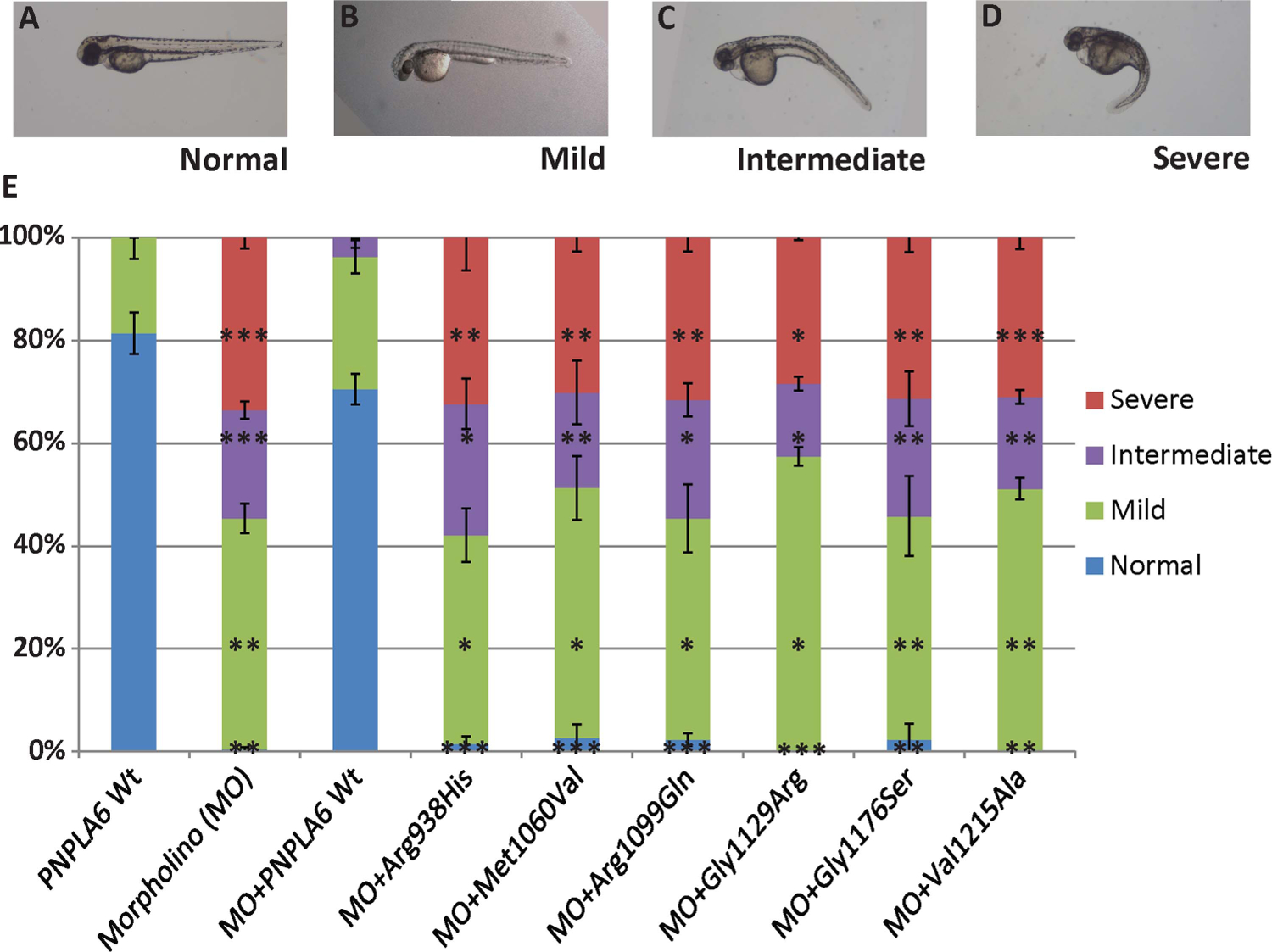
Suppression of pnpla6 expression produces developmental defects in zebrafish embryos. (A) Normal embryos class. (B–D) Embryos injected with morpholino (MO) against pnpla6 have developmental defects, curved head (mild), curved head and distal tail (intermediate), and curved, small head with full tail curvature (severe). (E) Percentage of embryos categorised in various phenotypic classes after injection with pnpla6 MO, wild-type mRNA or coinjection of PNPLA6 harbouring mutant alleles (N>100 embyros per condition; error bars=SEM; *p<0.05, **p<0.01, ***p<0.001).
Next, coinjections were performed with MO and PNPLA6 mRNA encoding one of the missense alleles associated with SPG39 (p.Arg938His, p.Met1012Val) or the novel patatin-like domain mutations (p.Arg1099Gln, p.Gly1129Arg, p.Gly1176Ser, p.Val1215Ala). Importantly, none of the six human mutant PNPLA6 mRNAs were sufficient to restore normal embryogenesis (figure 4E and see online supplementary figure S4). For each of the six mutations, the distribution of hatching zebrafish among the four phenotypes was indistinguishable from MO alone and significantly different from controls and WT rescue in all categories (figure 4E). Thus, the four novel patatin-like domain missense alleles appear to affect protein function.
The pnpla6 morphant gross phenotypes resulting from coinjection of SPG39 mutations or Oliver–McFarlane/Laurence–Moon mutations were not significantly different between alleles (figure 4E and see online supplementary figure S4), suggesting either that there are subtle differences that would require more detailed histology or that the effect of these mutations on NTE protein was similar, specifically reduced enzymatic function as previously described for SPG39 alleles.31,39
To ensure that Oliver–McFarlane/Laurence–Moon mutations did not cause mistargeting of NTE protein from the proper location in the endoplasmic reticulum,40 we transfected HEK293T cells with N-terminus or C-terminus GFP-tagged PNPLA6 cDNAs constructs (see online supplementary figure S5). We observed similar potentially artifactual perinuclear GFP aggregates as reported previously with PNPLA6 transfection in COS cells.40 Confocal imaging of transfected cells did not reveal any obvious changes in the expression or targeting of mutant NTE (see online supplementary figure S5). Therefore, we predicted that the inability of the novel mutations to rescue the zebrafish morphant phenotype is due to NTE loss of function rather than protein degradation or misrouting.
Severe loss of NTE activity in congenital PNPLA6 disease
In order to elucidate whether subtle differences exist between loss of NTE function in Oliver–McFarlane/Laurence–Moon syndromes and SPG39, we compared enzymatic activity between disease states. Given that Oliver–McFarlane and Laurence–Moon syndromes have earlier onset compared with SPG39, we predicted the hydrolase activity would be significantly reduced in these childhood-onset diseases compared with adult-onset SPG39. We compared the esterase activity among fibroblast cells from WT controls, a homozygous affected patient with SPG39 (p.Met1012Val/p.Met1012Val) and carrier, and patients A:1 and A:2 (p.Arg1099Gln/p.Gly1176Ser) and their carrier parents (figure 5). ANOVA was significantly different between groups (p value=1.15E–15; figure 5). Cells from heterozygous carrier parents and compound heterozygous affected patients from family A had reduced hydrolase activity compared with controls (figure 5A, B). Phenol production was significantly less in affected patient cells than cells with heterozygous mutations for p.Arg1099Gln and p.Gly1176Ser, and total reductions appeared to be allele dose dependent.
Figure 5.
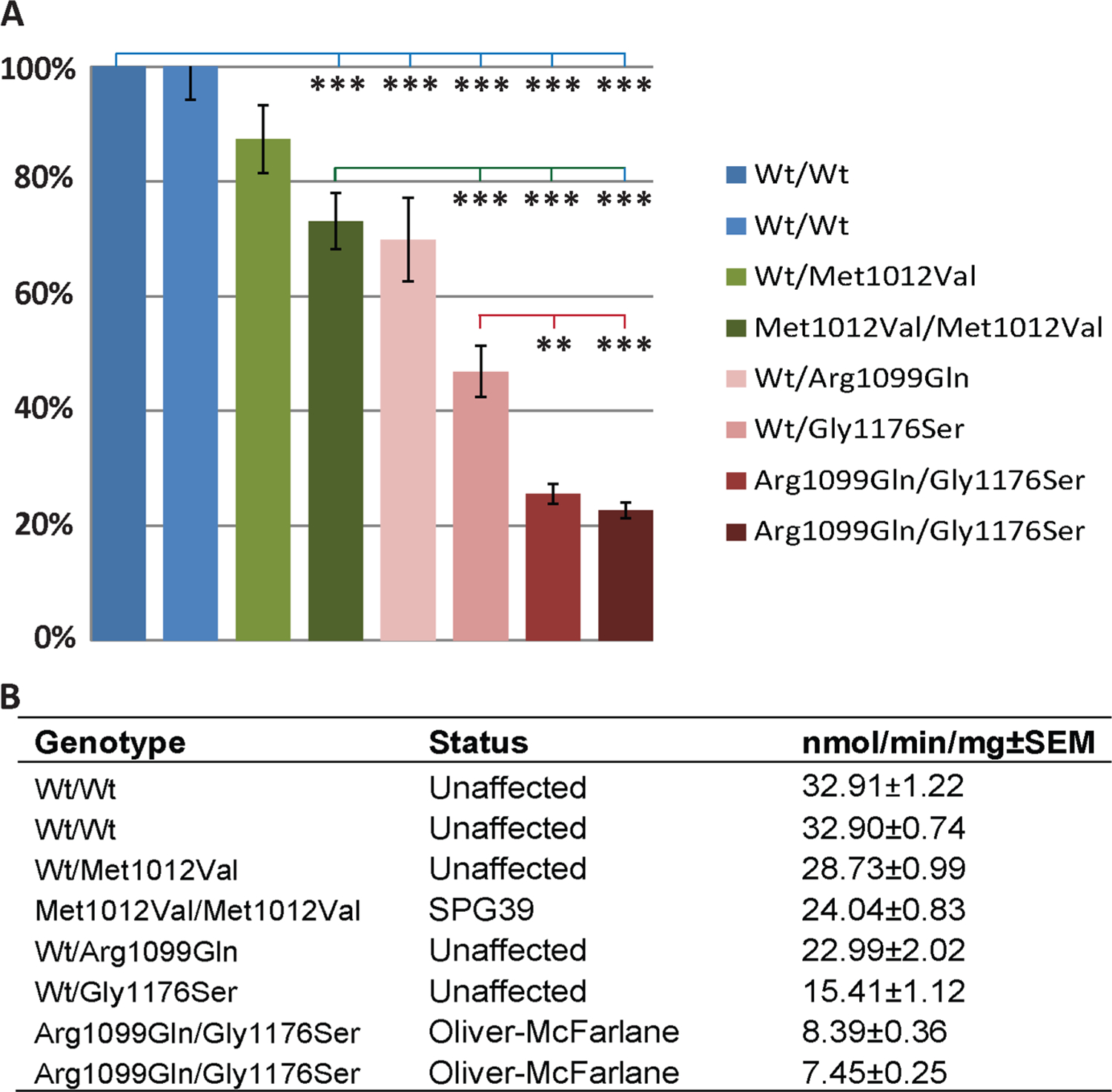
Differential neuropathy target esterase (NTE) hydrolase activity among PNPLA6-associated diseases. (A) Phenol production normalised to wild-type control cells. Statistical significance of analysis of variance post hoc Tukey’s test comparing wild type (blue line), SPG39 patient cells (green line) or family A carrier cells (red line). (B) Values for phenol production (±SEM) for each cell line along with genotype and affected status. Error bars=SEM; *p<0.05, **p<0.01, ***p<0.001.
As anticipated, cells from patients A:1 and A:2 had significantly reduced NTE specific activity compared with SPG39 cells (p<0.001). Surprisingly, significant reductions in hydrolysis activity were measured in cells from unaffected carrier parents in family A. Compared with the SPG39 patient cells (p.Met1012Val/p.Met1012Val), cells with heterozygous p.Arg1099Gln mutation had similar activity (p=0.65) and cells with heterozygous p. Gly1176Ser mutation had significantly reduced activity (p=0.001). The carrier parents of patients A:1 and A:2 have no neurological symptoms, gait abnormalities or peripheral neuropathy, suggesting biallelic mutations may be required for phenotypic expression. In summary, biallelic mutations confer dose-dependent reductions in hydrolysis activity, and disease severity and congenital onset of Oliver–McFarlane and Laurence–Moon syndromes correlate with reduction of NTE activity compared with SPG39.
DISCUSSION
We describe the clinical phenotype of 10 patients with Oliver–McFarlane and Laurence–Moon syndromes and identify 8 NTE loss-of-function alleles of PNPLA6 as the underlying genetic aetiology. Further, we add congenital disease and developmental phenotypes including anterior hypopituitarism, trichomegaly, alopecia, and facial dysmorphisms to the spectrum caused by NTE dysfunction due to PNPLA6 mutations. This study also provides the first insights into the relationship between Oliver–McFarlane and Laurence–Moon syndromes. The combination of growth retardation, pigmentary degeneration of the retina and trichomegaly was first described by Oliver and McFarlane in 1965 and subsequently 13 other cases including two sibling pairs.1,3–11 Laurence–Moon syndrome, described a century earlier, has nearly complete overlap with Oliver–McFarlane syndrome with the exceptions of absent hair findings and earlier-onset neurological signs (see online supplementary table S5).1
Laurence–Moon syndrome has historically been included in the Bardet–Biedl spectrum, though distinct given the marked choroidal atrophy, hypopituitarism with short stature, early neurological involvement and the absence of polydactyly and renal disease.13–19 The retinal degeneration is quite distinct, with Laurence–Moon and Oliver–McFarlane syndromes resembling choroideremia, and BBS resembling cone-rod dystrophy or retinitis pigmentosa with choroidal sparing. The overlap between Laurence–Moon, Oliver–McFarlane and BBS is not surprising as cilia require membrane specialisations that rely on proper trafficking of membrane phospholipids. This may represent an important interface between the larger classes of ciliopathies and phospholipase disorders.
Marked variability of hair and neurological features was noted within families A and F (figure 1). This raises the possibility that, in specific cases, Laurence–Moon and Oliver–McFarlane syndromes are part of a heterogenic spectrum, and clinical diagnosis is based on predominant hair or neurological signs. Laurence–Moon and Oliver–McFarlane syndromes may also be allelic. With the exception of the frameshift insertion, the mutations found in our patients have not been reported among the PNPLA6 disease-associated alleles.22–24,41 Patients with early neurological involvement (patients B:1, E:1 and family F), characteristic of Laurence–Moon syndrome and infrequently reported in Oliver–McFarlane syndrome, have the same frameshift allele, p.Arg1031fs*38, observed in patients with recessive spastic ataxia and GHS.23 Notably, like E:1, both previously reported patients had cerebellar atrophy, suggesting that important prognostic information may lie in genotype–phenotype correlations. Therefore, combinations of disease-causing alleles may also underlie differences among PNPLA6-opathies.
Our data further enhance the spectrum of congenital and childhood phenotypes across tissues involved in PNPLA6-associated diseases. The extent of chorioretinal atrophy and retinal pigment migration was variable (figure 1), though peripapillary sparing of disease was typically present. Pituitary hormone deficiencies were also heterogeneous. Patients A:1 and A:2 had marked hypothyroidism and GH deficiency, while patients C:1 and D:1 had GH deficiency and hypogonadotropic hypogonadism with thyroid sparing. Small anterior pituitary size is typically noted after hormonal deficiencies are detected, and while it remains unclear whether this represents hypoplasia or atrophy, patients A:1 and E:1 had interval decreases in anterior pituitary size, suggesting a degenerative process. We add eyelash and eyebrow hyperpigmentation as well as mild facial dysmorphisms to the clinical descriptions for certain patients, including prominent chin and frontal bone. The distinct difference in ocular, trichologic and pituitary involvement in Oliver–McFarlane and Laurence–Moon syndromes compared with SPG39, BNHS and GHS further implies that individual tissues require differential levels of NTE activity.
We found significant reduction in NTE-mediated hydrolysis in cells from patients with Oliver–McFarlane syndrome compared with those with SPG39. This implies similar pathophysiology, namely a requirement for hydrolysis of phospholipids at the endoplasmic reticulum to maintain the cell membrane lipid bilayer and the choline-recycling pathway.20,21,42 Spinocerebellar features in humans and NTE-deficient mice appear to result from degeneration of long axons, correlating with increased phosphatidylcholine levels in the mouse brain.20,42–45 In the pituitary, reduced vesicle release from gonadotropes may underlie gonadotropin deficiency and pubertal failure, as shown in vitro.24 This mechanism may apply to the release of other anterior pituitary hormones as well, for example, GH release from somatotropes and TSH release from thyrotropes. Trichomegaly results from increased prostaglandin production due to increased phosphatidylethanolamine in follicular cells.46 Retinal degeneration may relate to abnormalities of multiple membrane phospholipid functions, including outer segment disc membrane formation in photoreceptor cells, lipid second messengers in visual transduction, degeneration of optic nerve axons and vitamin A analogue processing or melanosome trafficking in RPE.47,48 Notably, carriers of alleles associated with Oliver–McFarlane/Laurence–Moon appear completely unaffected, though their cells exhibit similar or reduced function compared with SPG39 patients, suggesting that biallelic mutations are required to confer disease status. Similarly, heterozygous Pnpla6 knockout mice have 50% NTE activity and are phenotypically normal, while homozygous mutations are embryonic lethal due to failed placenta formation.49 Future studies will explore the relative NTE activity and mechanistic differences among tissue-specific phenotypic expression of PNPLA6 mutations causing this spectrum of neurodegenerative disorders.
Alternate mechanisms may also underlie differences among the PNPLA6-opathies. First, different enzymatic activities may exist within the patatin-like domain that may be selectively affected by disease-specific mutations. Relevant to this possibility is our observation that the novel mutations do not localise to the putative active site, yet affect NTE activity. It is possible that they differentially affect inter-domain interactions or other properties such as substrate recognition or binding affinity.20 Second, there may be tissue-specific phospholipid substrates for NTE that modulate these phenotypes. Other phospholipases are involved in ocular, pituitary and spinocerebellar disease. For example, mutations in PLA2G5 and PLA2G6, both of which are calcium-independent phospholipases, cause two very distinct conditions, benign fleck retinopathy ((MIM 228980)) and neurodegeneration with brain iron accumulation ((MIM603604)), respectively.50,51 Further investigations into common mechanisms of phospholipase disorders will improve our understanding of this disease spectrum.
In conclusion, PNPLA6 mutations cause a broad spectrum of ocular, trichologic, pituitary and cerebellar phenotypes. Our findings support that the timing and severity of disease is related to the absolute hydrolase activity of the patatin-like domain, though unknown modifying factors within tissues or alternate enzymatic activities of NTE likely contribute to the phenotypic heterogeneity. We propose that disease phenotypes might correlate with NTE activity in a tissue-specific manner. These disorders overlap clinically with more common diseases such as BBS and other ciliopathies, though the prognostic, mechanistic and future treatment implications vary drastically. Given the congenital onset of many features of Oliver–McFarlane syndrome and PNPLA6-associated Laurence–Moon syndrome, early intervention with hormone replacement and, in the future, gene or enzyme replacement therapy may improve visual, developmental and neurological outcomes.
Supplementary Material
Acknowledgements
We would like to thank the participating patients and their families, and the healthcare professionals involved in their care. We are grateful to E Richard and A Giese for technical assistance, M Keddache for exome sequencing, K Kaufman for variant frequency data in local controls, S Dai for clinical information and S Hufnagel for critical reading of this manuscript. We would like to thank the PHRC programme for the French Health ministry, API programme of the Hôpitaux Universitaires de Strasbourg, RETINA France, FORMICOEUR and UNADEV.
Funding
This study was sponsored by the National Institute on Deafness and Other Communication Disorders (NIDCD/NIH) research grants R01 DC012564 to ZMA and R01 DC011803 to SR. HDBR is supported by the Medical Research Council Grant G0700089 and the Wellcome Trust Grant 082557. MTD is funded by Great Ormond Street Hospital Children’s Charity. GA, SH and ATM are supported by the National Institute for Health Research UK (Moorfields BRC), Fight for Sight and The Rosetrees trust.
Footnotes
Competing interests None.
Patient consent This study was conducted under institutional review board-approved protocols in accordance with the Declaration of Helsinki for the release of clinical information, family history, blood draw and skin biopsies. Informed consent was obtained after the explanation of the study risks and benefits.
Ethics approval This study followed the tenets of the Declaration of Helsinki and was approved by all local ethics committees.
Provenance and peer review Not commissioned; externally peer reviewed.
Additional material is published online only. To view please visit the journal online (http://dx.doi.org/10.1136/jmedgenet-2014-102856)
REFERENCES
- 1.Oliver GL, McFarlane DC. Congenital trichomegaly: with associated pigmentary degeneration of the retina, dwarfism, and mental retardation. Arch Ophthalmol 1965;74:169–71. [DOI] [PubMed] [Google Scholar]
- 2.Chang TS, McFarlane DC, Oliver G, Willis NR. Congenital trichomegaly, pigmentary degeneration of the retina and growth retardation (Oliver-McFarlane syndrome): 28-year follow-up of the first reported case. Can J Ophthalmol 1993;28:191–3. [PubMed] [Google Scholar]
- 3.Corby DG, Lowe RS Jr, Haskins RC, Hebertson LM. Trichomegaly, pigmentary degeneration of the retina, and growth retardation. A new syndrome originating in utero. Am J Dis Child 1971;121:344–5. [DOI] [PubMed] [Google Scholar]
- 4.Haritoglou C, Rudolph G, Kalpadakis P, Boergen KP. Congenital trichomegaly (Oliver-McFarlane syndrome): a case report with 9 years’ follow up. Br J Ophthalmol 2003;87:119–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathieu M, Goldfarb A, Berquin P, Boudailliez B, Labeille B, Piussan C. Trichomegaly, pigmentary degeneration of the retina and growth disturbances. a probable autosomal recessive disorder. Genet Couns 1991;2:115–8. [PubMed] [Google Scholar]
- 6.Sheng X, Zhang S, Peter Boergen K, Li H, Liu Y. Oliver-McFarlane Syndrome in a Chinese Boy: Retinitis Pigmentosa, Trichomegaly, Hair Anomalies and Mental Retardation. Ophthalmic Genet 2013. [DOI] [PubMed]
- 7.Sonmez S, Forsyth RJ, Matthews DS, Clarke M, Splitt M. Oliver-McFarlane syndrome (chorioretinopathy-pituitary dysfunction) with prominent early pituitary dysfunction: differentiation from choroideremia-hypopituitarism. Clin Dysmorphol 2008;17:265–7. [DOI] [PubMed] [Google Scholar]
- 8.Zaun H, Stenger D, Zabransky S, Zankl M. [The long-eyelash syndrome (trichomegaly syndrome, Oliver-McFarlane)]. Hautarzt 1984;35:162–5. [PubMed] [Google Scholar]
- 9.Haimi M, Gershoni-Baruch R. Autosomal recessive Oliver-McFarlane syndrome: retinitis pigmentosa, short stature (GH deficiency), trichomegaly, and hair anomalies or CPD syndrome (chorioretinopathy-pituitary dysfunction). Am J Med Genet A 2005;138A:268–71. [DOI] [PubMed] [Google Scholar]
- 10.Cant JS. Ectodermal dysplasia. J Pediatr Ophthalmol 1967;4:13–7. [Google Scholar]
- 11.Patton MA, Harding AE, Baraitser M. Congenital trichomegaly, pigmentary retinal degeneration, and short stature. Am J Ophthalmol 1986;101:490–1. [DOI] [PubMed] [Google Scholar]
- 12.Laurence JZ, Moon RC. Four cases of “retinitis pigmentosa” occurring in the same family, and accompanied by general imperfections of development. 1866. Obes Res 1866;3:400–3. [DOI] [PubMed] [Google Scholar]
- 13.Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA. New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet 1999;36:437–46. [PMC free article] [PubMed] [Google Scholar]
- 14.Farag TI, Teebi AS. Bardet-Biedl and Laurence-Moon syndromes in a mixed Arab population. Clin Genet 1988;33:78–82. [DOI] [PubMed] [Google Scholar]
- 15.Green JS, Parfrey PS, Harnett JD, Farid NR, Cramer BC, Johnson G, Heath O, McManamon PJ, O’Leary E, Pryse-Phillips W. The cardinal manifestations of Bardet-Biedl syndrome, a form of Laurence-Moon-Biedl syndrome. N Engl J Med 1989;321:1002–9. [DOI] [PubMed] [Google Scholar]
- 16.Hutchinson J Slowly progressive paraplegia and diseases of the choroids. Arch Surg 1900;11:118–22. [Google Scholar]
- 17.Bowen P, Ferguson-Smith MA, Mosier D, Lee CS, Butler HG. The Laurence-Moon syndrome. Association with hypogonadotrophic hypogonadism and sex-chromosome aneuploidy. Arch Intern Med 1965;116:598–604. [DOI] [PubMed] [Google Scholar]
- 18.Chalvon-Demersay A, Tardieu M, Crosnier H, Benichou JJ, Pienkowski C, Rochiccioli P, Labrune B. [Laurence-Moon (Bardet-Biedl) syndrome with growth hormone deficiency]. Arch Fr Pediatr 1993;50:859–62. [PubMed] [Google Scholar]
- 19.Rizzo JF III, Berson EL, Lessell S. Retinal and neurologic findings in the Laurence-Moon-Bardet-Biedl phenotype. Ophthalmology 1986;93:1452–6. [DOI] [PubMed] [Google Scholar]
- 20.Richardson RJ, Hein ND, Wijeyesakere SJ, Fink JK, Makhaeva GF. Neuropathy target esterase (NTE): overview and future. Chem Biol Interact 2013; 203:238–44. [DOI] [PubMed] [Google Scholar]
- 21.Zaccheo O, Dinsdale D, Meacock PA, Glynn P. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J Biol Chem 2004;279:24024–33. [DOI] [PubMed] [Google Scholar]
- 22.Rainier S, Bui M, Mark E, Thomas D, Tokarz D, Ming L, Delaney C, Richardson RJ, Albers JW, Matsunami N, Stevens J, Coon H, Leppert M, Fink JK. Neuropathy target esterase gene mutations cause motor neuron disease. Am J Hum Genet 2008;82:780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Synofzik M, Gonzalez MA, Lourenco CM, Coutelier M, Haack TB, Rebelo A, Hannequin D, Strom TM, Prokisch H, Kernstock C, Durr A, Schols L, Lima-Martinez MM, Farooq A, Schule R, Stevanin G, Marques W Jr, Zuchner S. PNPLA6 mutations cause Boucher-Neuhauser and Gordon Holmes syndromes as part of a broad neurodegenerative spectrum. Brain 2014;137(Pt 1):69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Topaloglu AK, Lomniczi A, Kretzschmar D, Dissen GA, Kotan LD, McArdle CA, Koc AF, Hamel BC, Guclu M, Papatya ED, Eren E, Mengen E, Gurbuz F, Cook M, Castellano JM, Kekil MB, Mungan NO, Yuksel B, Ojeda SR. Loss of function mutations in pnpla6 encoding neuropathy target esterase underlie pubertal failure and neurological deficits in Gordon Holmes syndrome. J Clin Endocrinol Metab 2014:99:E2067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaworek TJ, Kausar T, Bell SM, Tariq N, Maqsood MI, Sohail A, Ali M, Iqbal F, Rasool S, Riazuddin S, Shaikh RS, Ahmed ZM. Molecular genetic studies and delineation of the oculocutaneous albinism phenotype in the Pakistani population. Orphanet J Rare Dis 2012;7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaston-Massuet C, Andoniadou CL, Signore M, Jayakody SA, Charolidi N, Kyeyune R, Vernay B, Jacques TS, Taketo MM, Le Tissier P, Dattani MT, Martinez-Barbera JP. Increased Wingless (Wnt) signaling in pituitary progenitor/stem cells gives rise to pituitary tumors in mice and humans. Proc Natl Acad Sci USA 2011;108:11482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song Y, Wang M, Mao F, Shao M, Zhao B, Song Z, Shao C, Gong Y. Knockdown of Pnpla6 protein results in motor neuron defects in zebrafish. Dis Model Mech 2013;6:404–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vose SC, Fujioka K, Gulevich AG, Lin AY, Holland NT, Casida JE. Cellular function of neuropathy target esterase in lysophosphatidylcholine action. Toxicol Appl Pharmacol 2008;232:376–83. [DOI] [PubMed] [Google Scholar]
- 31.Hein ND, Rainier SR, Richardson RJ, Fink JK. Motor neuron disease due to neuropathy target esterase mutation: enzyme analysis of fibroblasts from human subjects yields insights into pathogenesis. Toxicol Lett 2010;199:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel ZH, Kottyan LC, Lazaro S, Williams MS, Ledbetter DH, Tromp H, Rupert A, Kohram M, Wagner M, Husami A, Qian Y, Valencia CA, Zhang K, Hostetter MK, Harley JB, Kaufman KM. The struggle to find reliable results in exome sequencing data: filtering out Mendelian errors. Front Genet 2014;5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014;11:361–2. [DOI] [PubMed] [Google Scholar]
- 34.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res 2001;11:863–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 2012;7:e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kretzschmar D, Hasan G, Sharma S, Heisenberg M, Benzer S. The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J Neurosci 1997;17:7425–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Synofzik M, Kernstock C, Haack TB, Schols L. Ataxia meets chorioretinal dystrophy and hypogonadism: Boucher-Neuhauser syndrome due to PNPLA6 mutations. J Neurol Neurosurg Psychiatry 2014. [DOI] [PubMed]
- 39.Hein ND, Stuckey JA, Rainier SR, Fink JK, Richardson RJ. Constructs of human neuropathy target esterase catalytic domain containing mutations related to motor neuron disease have altered enzymatic properties. Toxicol Lett 2010;196:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Dinsdale D, Glynn P. Protein domains, catalytic activity, and subcellular distribution of neuropathy target esterase in Mammalian cells. J Biol Chem 2003;278:8820–5. [DOI] [PubMed] [Google Scholar]
- 41.Yoon G, Baskin B, Tarnopolsky M, Boycott KM, Geraghty MT, Sell E, Goobie S, Meschino W, Banwell B, Ray PN. Autosomal recessive hereditary spastic paraplegia-clinical and genetic characteristics of a well-defined cohort. Neurogenetics 2013;14:181–8. [DOI] [PubMed] [Google Scholar]
- 42.Hou WY, Long DX, Wu YJ. Effect of inhibition of neuropathy target esterase in mouse nervous tissues in vitro on phosphatidylcholine and lysophosphatidylcholine homeostasis. Int J Toxicol 2009;28:417–24. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka D Jr, Bursian SJ, Lehning E. Selective axonal and terminal degeneration in the chicken brainstem and cerebellum following exposure to bis(1-methylethyl) phosphorofluoridate (DFP). Brain Res 1990;519:200–8. [DOI] [PubMed] [Google Scholar]
- 44.Classen W, Gretener P, Rauch M, Weber E, Krinke GJ. Susceptibility of various areas of the nervous system of hens to TOCP-induced delayed neuropathy. Neurotoxicology 1996;17:597–604. [PubMed] [Google Scholar]
- 45.Read DJ, Li Y, Chao MV, Cavanagh JB, Glynn P. Neuropathy target esterase is required for adult vertebrate axon maintenance. J Neurosci 2009;29:11594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto K, Taketomi Y, Isogai Y, Miki Y, Sato H, Masuda S, Nishito Y, Morioka K, Ishimoto Y, Suzuki N, Yokota Y, Hanasaki K, Ishikawa Y, Ishii T, Kobayashi T, Fukami K, Ikeda K, Nakanishi H, Taguchi R, Murakami M. Hair follicular expression and function of group X secreted phospholipase A2 in mouse skin. J Biol Chem 2011;286:11616–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhan C, Wang J, Kolko M. Diverse regulation of retinal pigment epithelium phagocytosis of photoreceptor outer segments by calcium-independent phospholipase A(2), group VIA and secretory phospholipase A(2), group IB. Curr Eye Res 2012;37:930–40. [DOI] [PubMed] [Google Scholar]
- 48.Giusto NM, Pasquare SJ, Salvador GA, Ilincheta de Boschero MG. Lipid second messengers and related enzymes in vertebrate rod outer segments. J Lipid Res 2010;51:685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moser M, Li Y, Vaupel K, Kretzschmar D, Kluge R, Glynn P, Buettner R. Placental failure and impaired vasculogenesis result in embryonic lethality for neuropathy target esterase-deficient mice. Mol Cell Biol 2004;24:1667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgan NV, Westaway SK, Morton JE, Gregory A, Gissen P, Sonek S, Cangul H, Coryell J, Canham N, Nardocci N, Zorzi G, Pasha S, Rodriguez D, Desguerre I, Mubaidin A, Bertini E, Trembath RC, Simonati A, Schanen C, Johnson CA, Levinson B, Woods CG, Wilmot B, Kramer P, Gitschier J, Maher ER, Hayflick SJ. PLA2G6, encoding a phospholipase A2, is mutated in neurodegenerative disorders with high brain iron. Nat Genet 2006;38:752–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sergouniotis PI, Davidson AE, Mackay DS, Lenassi E, Li Z, Robson AG, Yang X, Kam JH, Isaacs TW, Holder GE, Jeffery G, Beck JA, Moore AT, Plagnol V, Webster AR. Biallelic mutations in PLA2G5, encoding group V phospholipase A2, cause benign fleck retina. Am J Hum Genet 2011;89:782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 2006;22:195–201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


