Abstract
In many areas of oncology, we lack sensitive tools to track low burden disease. While cell-free DNA (cfDNA) shows promise in detecting cancer mutations, we found that the combination of low tumor fraction (TF) and limited number of DNA fragments restricts low disease burden monitoring through the prevailing deep targeted sequencing paradigm. We reasoned that breadth may supplant depth of sequencing to overcome the barrier of cfDNA abundance. Whole genome sequencing (WGS) of cfDNA allowed ultra-sensitive detection, capitalizing on the cumulative signal of thousands of somatic mutations observed in solid malignancies, with TF detection sensitivity as low as 10-5. The WGS approach enabled dynamic tumor burden tracking and post-operative residual disease detection, associated with adverse outcome. Thus, we present an orthogonal framework for cfDNA cancer monitoring via genome wide mutational integration, enabling ultra-sensitive detection, overcoming the limitation of cfDNA abundance, and empowering treatment optimization in low-disease burden oncology care.
Keywords: Circulating tumor DNA, cell free DNA, whole genome sequencing, cancer monitoring
Liquid biopsy technology is a transformative force in cancer care1. While different analytes are being actively explored for cancer surveillance2–7, detection of somatic mutations in circulating tumor DNA (ctDNA) in plasma cell-free DNA (cfDNA) has been shown to enable non-invasive characterization of the somatic malignant genome8,9,10,11,12. Current applications have often been developed in the context of metastatic high tumor-burden disease13,14. In this context, cfDNA tumor fraction (TF) is high, providing for effective mutational characterization with next-generation sequencing (e.g., whole exome sequencing or targeted panels) with minimal modifications10.
One of the major areas of future promise for cfDNA-based cancer studies is the detection of minimal residual disease (MRD) to guide clinical interventions15,16. However, in the MRD context, cfDNA TF is significantly lower as it is correlated with disease burden17. To enable mutation detection of low TF cfDNA, the prevailing paradigm has been to increase the depth of sequencing of a limited high yield target set (e.g., common cancer drivers or patient specific panels sequenced to a depth of ten-to-hundreds of thousands reads per base)18,19,20. While these state-of-the-art methods provide detection with high accuracy, many patients with radiographically evident disease do not show detectable ctDNA by deep targeted sequencing17,18. These data suggest that ultra-deep sequencing currently underperforms imaging, and may be even more challenged in post-operative MRD detection, where significant debulking likely reduces TF further. To overcome this challenge, we present here genome-wide mutational integration for ctDNA detection, whereby breadth supplants depth of sequencing for sensitive detection of low burden cancer.
Results
Small number of DNA fragments in plasma samples poses a fundamental challenge to deep targeted sequencing in low burden disease
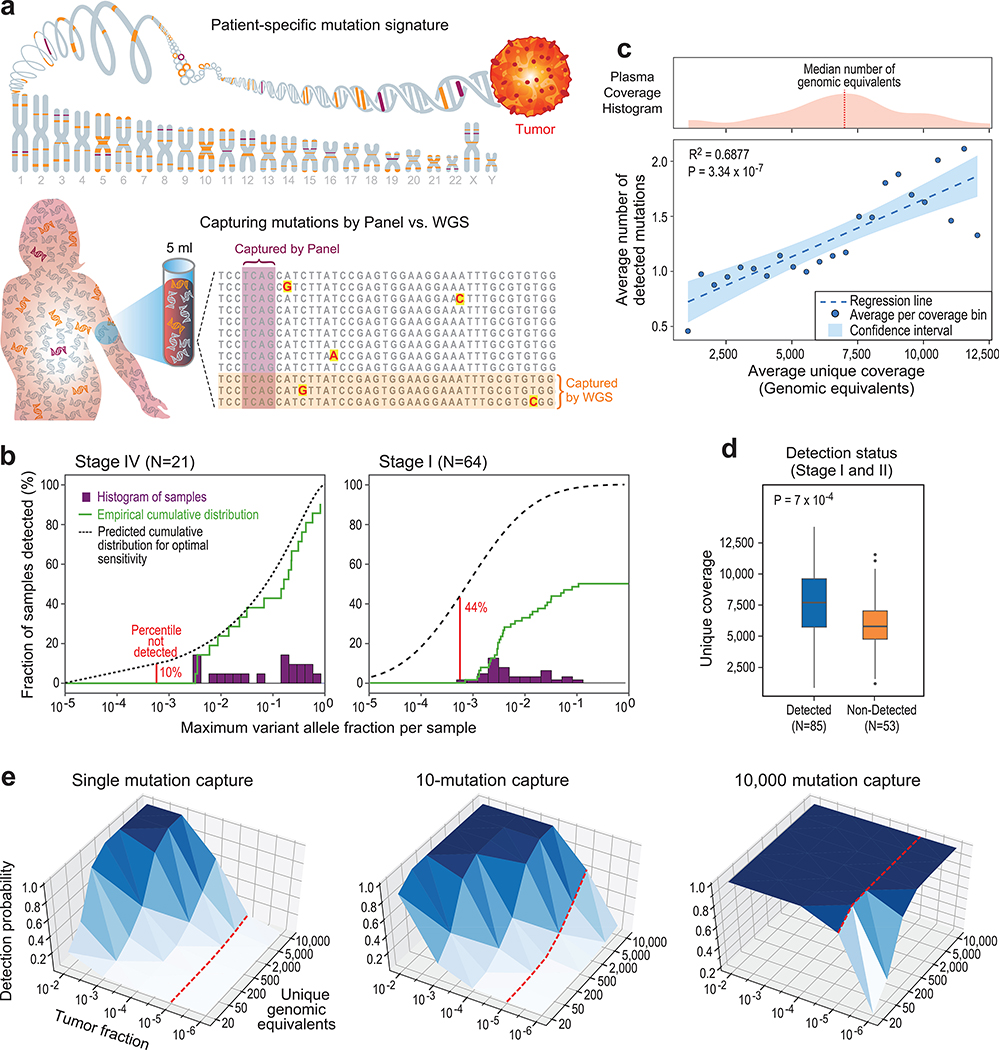
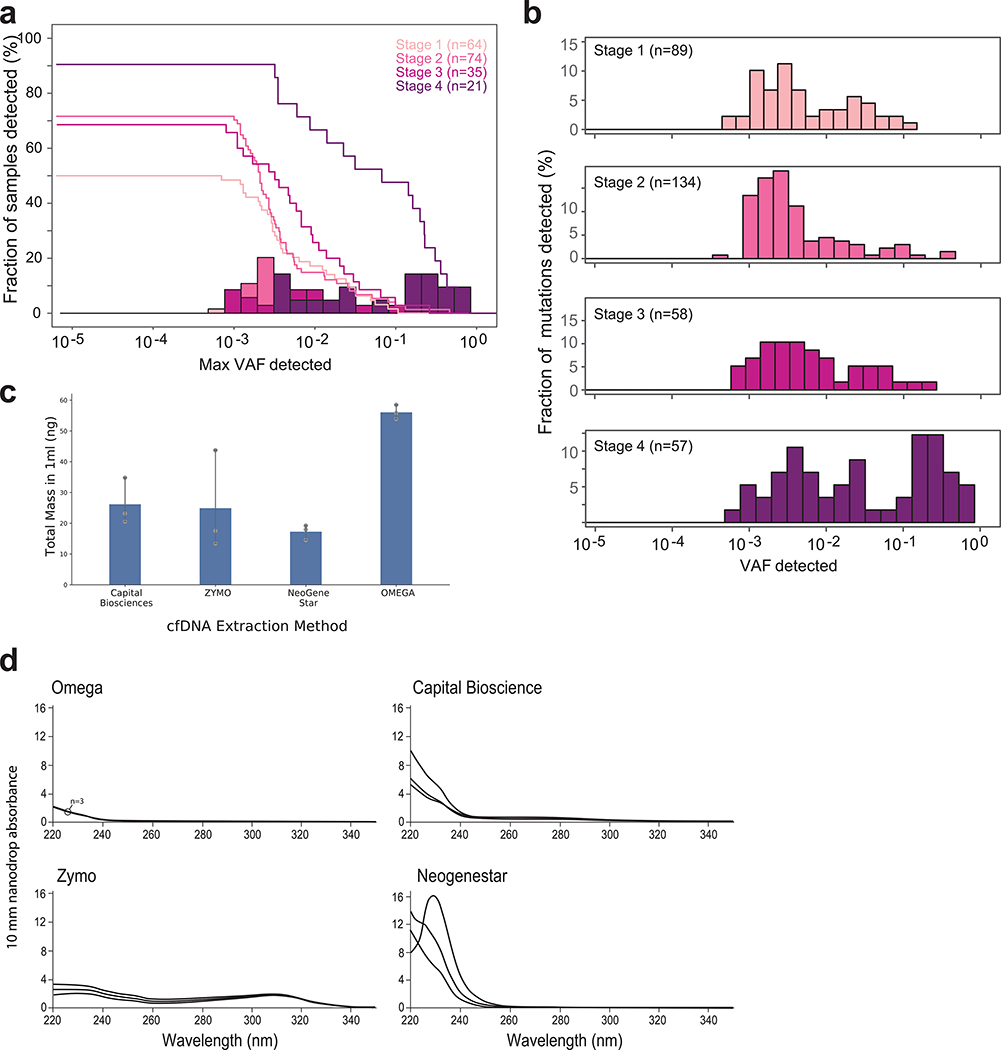
We hypothesized that deep-targeted approaches may be hindered by a fundamental barrier to detection sensitivity – limited input material (the number of cfDNA fragments as measured by genomic equivalents [GEs]). Specifically, in the setting of early cancer or MRD detection, cfDNA abundance may be limiting to deep targeted sequencing, as it imposes a ceiling on effective depth of sequencing (Fig. 1a). To test this hypothesis, we reanalyzed a recent landmark study of early-stage cancer detection using deep targeted sequencing18. Mutation detection was largely limited to 10−3 variant allelic-fraction (VAF) in cfDNA for all cancer stages (Extended Data Fig. 1a,b, consistent with VAF cut-offs implemented in variant detection18), despite ~40,000X coverage depth of sequencing, and the application of an advanced molecular error suppression21. This sensitivity limitation manifested in a stage-dependent decrease in the proportion of patients with detectable ctDNA (Fig. 1b; Extended Data Fig. 1a,b). Consistent with the hypothesis that available input material limits detection, we observed a positive correlation between the number of unique cfDNA reads (representative of the number of genomic equivalents; GEs) in the sample and the number of mutations detected (Fig. 1c), as well as higher unique read coverage in samples with detected ctDNA mutations (Fig. 1d).
Figure 1: Low cfDNA input material limits sensitive ctDNA mutation detection with deep targeted sequencing, but can be overcome by genome-wide mutational integration.
(a) Illustrative schematic showing that mutations captured by targeted panel deep sequencing are only a small fraction of genome-wide somatic tumor mutations (upper panel). The low input and random sampling nature of cfDNA collection by blood samples results in higher probability for mutation capture by WGS than by targeted panels (lower panel). (b-d) constitute a re-analysis of previously published data18. (b) Histogram of samples detected as a function of the maximum variant allele fraction (VAF) in stage IV (left) and stage I (right) cancer. Maximum VAF serves as an estimation of tumor fraction (TF). Patient histogram (solid bars), empirical cumulative distribution (solid line) and predicted “optimal-sensitivity” detection (dashed line, see Methods) are shown. Lower limit of detection (red solid line) shows a residual of 44% of stage I patients that are not detected by the deep targeted sequencing panel (10% for stage IV). Most of these cancers are predicted to have been detected if the VAF lower-limit-of-detection was 10-5. Re-analysis across 64 stage I and 21 stage IV patient samples across tumor types. (c) Plasma input material (depth of coverage) correlates to the number of detected tumor mutations (correlation and statistical significance calculated with a two-sided Pearson correlation.) Upper panel: distribution (histogram) of unique genomic equivalents (GEs, unique coverage, x-axis) across all patient samples (n = 194 patients, across different tumor types and disease stages) sequenced with exhaustive deep sequencing (median 40,729X). A median of 6,000 (range 400–14,000) unique reads (GEs) was found in a median 4.2mL of plasma (range 1.6–9.8). Lower panel: the average number of mutations detected per patient shows a positive correlation with the number of unique GEs in the patient plasma, indicating that input material is a potential limiting factor for mutation detection with deep targeted sequencing (two-sided Pearson correlation). Dashed line shows regression line, shaded area represents 95% confidence interval. (d) Patient samples with higher number of unique GEs (coverage) have higher probability for cancer detection by deep targeted sequencing (P value = 7*10−4, two-sided Wilcoxon rank-sum test). Analysis includes all detected (n = 85) and non-detected (n = 53) stage I/II patients across tumor types. Boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR. (e) Predicted probability (analytic derivation from binomial distribution of Bernoulli trials, see Methods) of detecting at least one mutated (SNV supporting) read as a function of the number of unique GEs (coverage) and the fraction of ctDNA in cfDNA (tumor fraction, TF). Left: targeting only a single SNV (e.g., with digital droplet PCR), the probability for detection decreases rapidly with tumor fraction (e.g., probability to detect at 10−5 with 5000 GEs is <0.05, red line). Middle: targeting 10 patient-specific mutations (e.g., 1Mbp panel for patient with 10/Mbp mutation load or a typical exome-based patient specific panel17) shows low probability of detection in low TF (e.g., probability to detect at 10−5 with 5000 GE is <0.4 , red line). Right: targeting 10,000 patient-specific mutations (e.g., whole genome sequencing for patient with >4/Mbp mutation load) shows significant probability of detection in low tumor fraction even with modest coverage (e.g., probability to detect at 10−5 is > 0.99 with 50X coverage, red line).
To infer a putative underlying TF distribution, we integrated the distribution of the maximal VAF (an approximation of TF) across samples with the fraction of samples that were not detected, with the assumption that detection failures are due to TF below the limit of assay sensitivity (see Online Methods). In advanced disease, higher TF values (median 4% estimated vs. 6% observed in stage IV patients) enabled robust detection in 90% of samples despite the 10−3 sensitivity limitation (Fig. 1b, left panel). However, our analysis suggested that lower TFs in early stage disease (median TF 0.02% estimated vs. 0.4% observed, in stage I patients) result in TF values that are often below the 10−3 sensitivity limitation (Fig. 1b, right panel), and anticipated a substantially increased detection sensitivity through methods capable of detecting TFs as low as 10-5.
These results demonstrated that in the setting of low-burden disease detection – where TF is low – limited input material likely constitutes a major barrier to the effective application of deep targeted sequencing. As the limited cfDNA input cannot be overcome through sample processing improvement (see Online Methods, Extended Data Fig. 1c,d), we require a rethinking of library and analysis design to maximize the utilization of available cfDNA fragments (Fig. 1a).
Breadth of sequencing may supplant depth of sequencing to overcome the limitation of low number of DNA fragments in plasma
Detection of a single somatic single nucleotide variant (SNV) in a plasma sample results from two consecutive sampling processes, each with its own statistical probability. The first process provides the probability that the mutated fragment is sampled in the limited number of GEs present in a typical plasma sample. The second process is the detection of the mutated fragment in the sample given its abundance, sequencing depth and technical variables such as sequencing error rate. While the latter process has been the focus of intensive investigation and technology development (e.g., ultra-deep error free sequencing protocols18,19,20), the former stochastic process is infrequently addressed. However, if no physical fragment that supports a targeted SNV is present in the sample, even ideal ultra-deep targeted sequencing will fail to discover a signal. In practice, this problem is further compounded by the fact that a single observation (mutated read) is rarely sufficient for confident detection.
Mathematical modeling of the probability of sampling mutant fragments in a given cfDNA sample (see Online Methods) predicts that the detection probability in TFs relevant to the low-burden cancer regime (TF < 1%, ~5000 GEs; Fig. 1b,c) will show rapid decline in detection at TFs below 10−3 (detection probability as low as 5% for TF = 10−5, Fig. 1e, left panel). We note that these limitations are observed even under idealized conditions of: (i) exhaustive sequencing; (ii) efficient utilization of all GEs, assuming perfect recovery; and (iii) detection based on a single supporting DNA fragment with no sequencing errors. These results show that the plasma sampling process imposes a formidable barrier to mutation detection at low TF regimes, such as in residual disease.
Conversely, our model also shows that the sampling limitation can be effectively overcome by increasing the number of detected sites (SNVs) through increased breadth. While the detection probability of a large targeted panel (1Mbp total coverage, and patient mutation load of 10/Mbp, corresponding to 10 SNVs), still showed rapid decline in detection at TFs below 10−3 (detection probability lower than 40% for TF = 10−5, Fig. 1e, middle panel), integrating over 10,000 SNVs (mutation load of >4/Mbp found in over a quarter of human cancer22) can provide high detection probability even at TFs as low as 10−5, at a modest sequencing effort readily achieved with standard WGS (detection probability of >85% with 20X depth of coverage, or >99% with 50X; Fig. 1e, right panel).
Development and validation of ultra-sensitive ctDNA detection via genome-wide mutational integration (MRDetect)
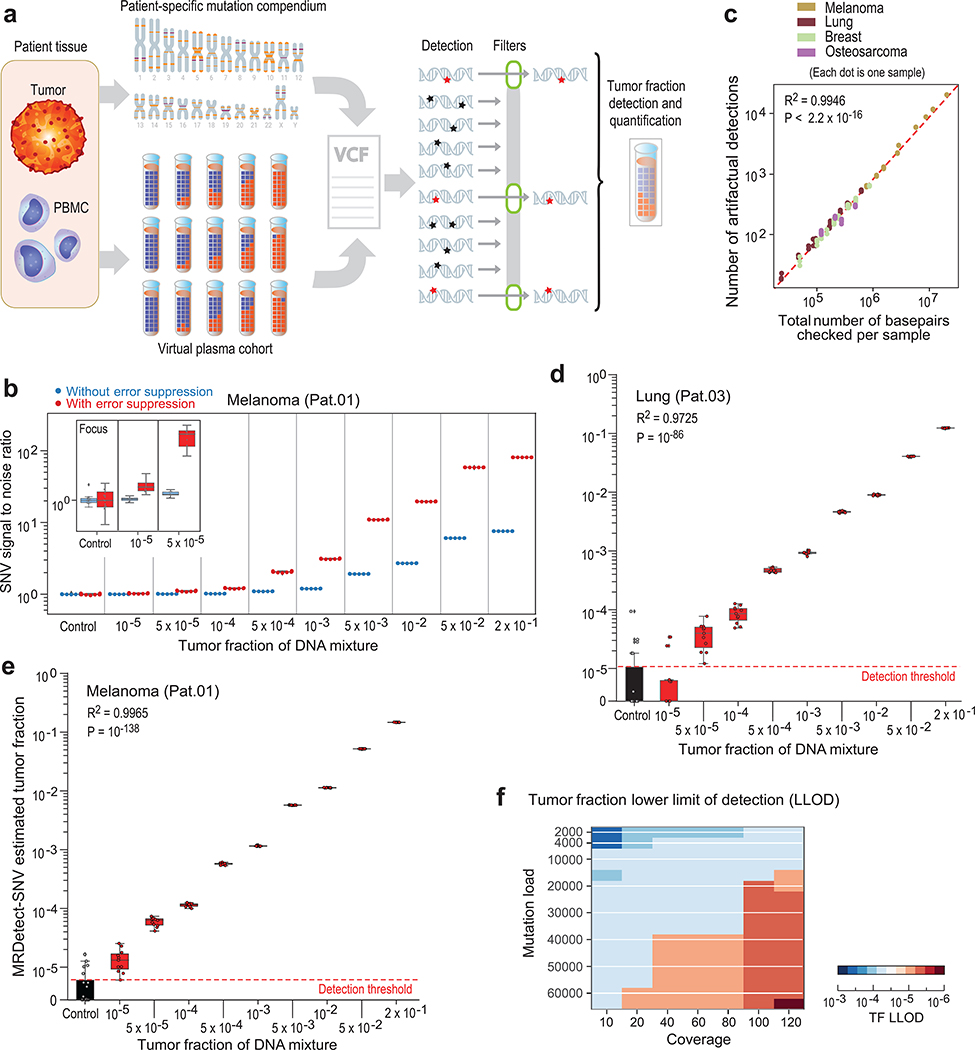
Our model predicts the TF as a function of number of detected sites, mutation load, and sequencing coverage depth. To validate this model, we simulated ctDNA detection using in silico admixtures of tumor and normal WGS data from eight patients with various cancers (Supplementary Table 1), generating over 700 in silico admixtures varying the TF mixtures (range 10−5-0.2) in 35X coverage with replicates (n = 11; see Online Methods; Fig. 2a; Supplementary Table 1).
Figure 2: Patient-specific genome-wide SNV integration provides ultra-sensitive ctDNA detection and precision tumor fraction (TF) estimation.
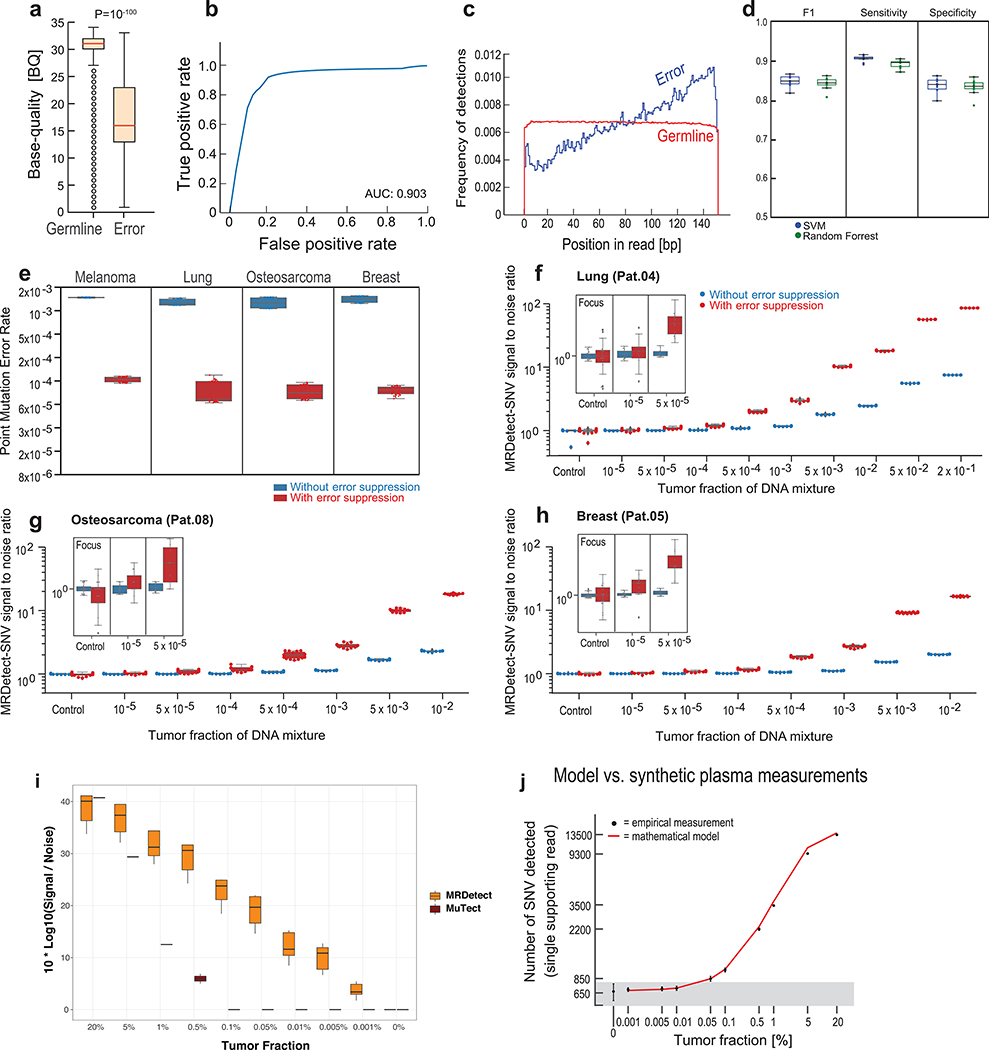
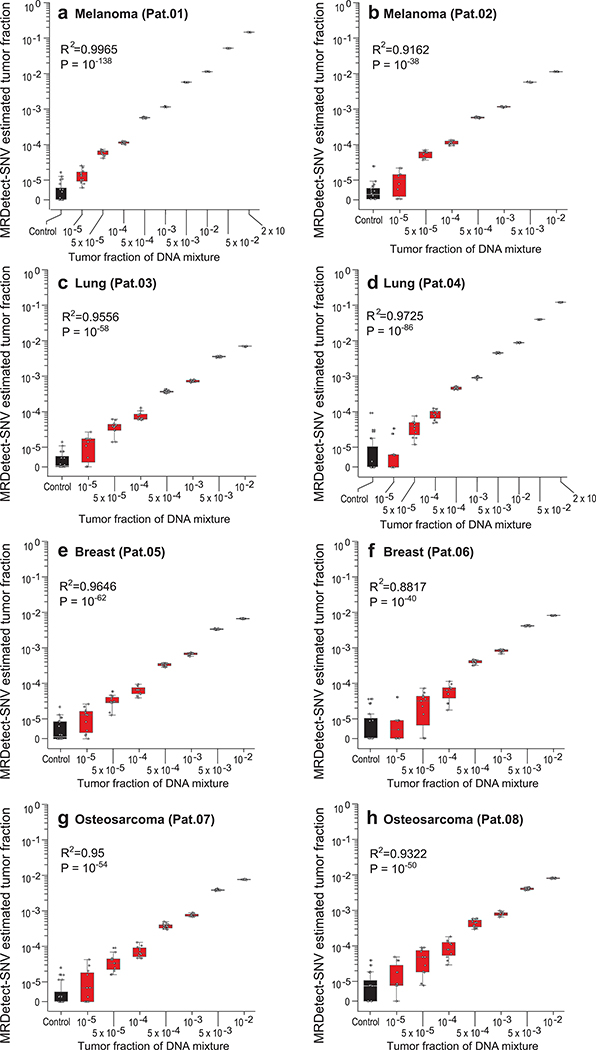
(a) Illustration of genome wide SNV integration for inference of plasma TF, and analytical validation scheme. Patient-specific SNV mutations are detected from matched tumor and peripheral blood mononuclear cells (PBMC) germline whole genome sequencing (WGS). Tumor somatic SNVs are then used to calculate the genome-wide tumor signal in the patient’s plasma sample. To test this approach, for each cancer, tumor and PBMC WGS reads were mixed to generate a range of TFs (10−5-0.2) in 35X coverage with multiple replicates (n = 11, see Methods). Across eight patient samples with four different tumor types (lung, melanoma, breast and osteosarcoma), we generated over 700 in silico admixture samples. Detection and filtering were applied on each patient sample to benchmark the tumor fraction detection sensitivity and accuracy. (b) Patient-specific SNV signal-to-noise quantification over a range of TFs (10−5-0.2) compared to basal noise signal detected in control (TF = 0) samples (left column), estimated using melanoma sample (Pat.01). Signal-to-noise was estimated by calculating the log difference between the number of detections in each plasma sample (TF > 0) and the mean number of detections in the controls (TF = 0). Inset panel shows discrimination of tumor and control samples down to tumor fraction 10−5 after utilizing machine-learning-based sequencing error suppression (red) vs. reduced sensitivity with the raw unfiltered data (blue). For TFs > 0, n = 11 independent admixture samples. For the control (TF = 0), n = 20 independently down-sampled PBMC replicates. (c) Correlation between the number of base-pairs evaluated for SNVs and the number of mismatches detected (artefactual detections), measured over all synthetic control plasma (no tumor DNA, TF = 0, n = 341) from eight patients and mutational compendia from four tumor types (lung, breast, melanoma and osteosarcoma). Results show a constant error rate, independent of tumor type, corresponding to previously published21,40 Illumina sequencing error-rate estimates (~1/1000 bps) (correlation and statistical significance calculated with a two-sided Pearson correlation). (d-e) Tumor fraction (TF) inference using genome-wide SNV integration for lung cancer (Pat.03) (d) and melanoma (Pat.01) (e) samples, shows accurate TF estimation as low as 5*10−5 and 10−5, respectively, discriminated from control (TF = 0) samples (left box-plot). High Pearson correlation (two-sided test) between the input TF mixture (x-axis) and the SNV-based estimated TF prediction, confirms accurate inference based on genome-wide mutational integration. For TFs > 0, n = 11 independent admixture samples. For the control (TF = 0), n = 20 independently down-sampled PBMC replicates. (f) The lower-limit-of-detection (LLOD) was empirically measured as a function of the input mutational load and the WGS coverage showing that for high mutational loads (~60,000 mutations) achieving sensitivity nearing 10−6 is feasible. Analysis was done over 21,420 in silico admixtures, varying TF (10−3-10−6), coverage (10–120X), mutation load (2,000–63,000), and across 20 replicates (random read downsampling) for each admixture. Lower-limit-of-detection (LLOD) was defined by the lowest tumor fraction that show significant Z-score separation from the control (TF = 0) cohort, for the same context (mutation load and coverage depth, see Methods). Throughout the figure, boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR.
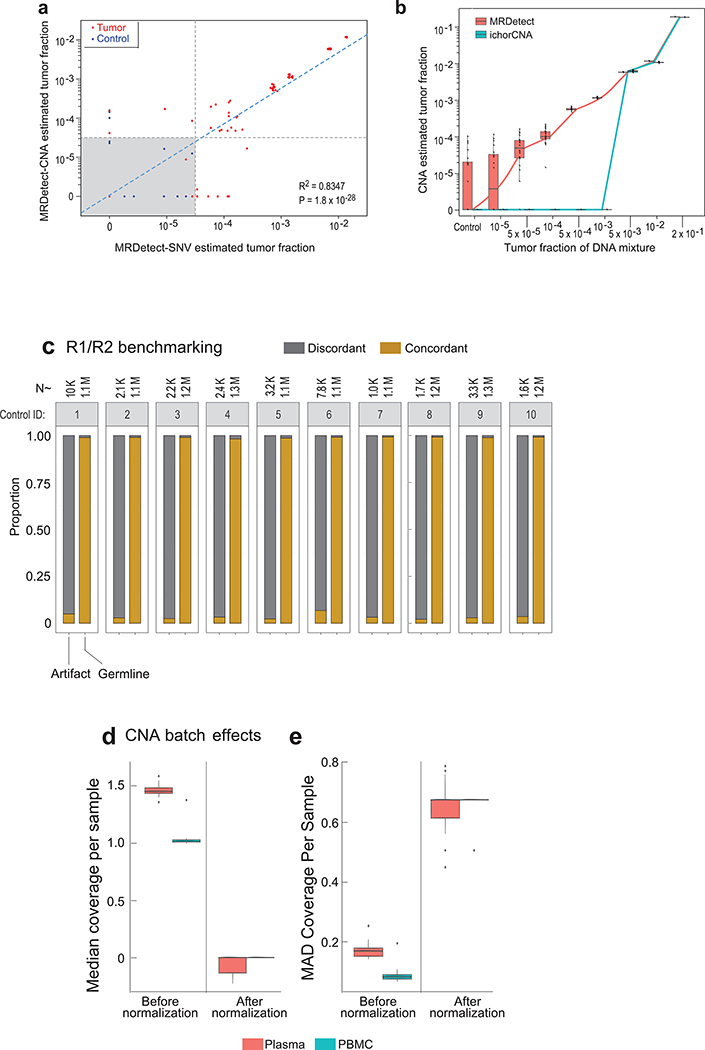
We designed a tumor-informed detection approach for the MRD setting – MRDetect – that utilizes the primary tumor mutational compendium as a prior. In silico plasma mixtures are then evaluated by searching all sites from the matched patient tumor compendium for corresponding mutations. Since integrating many sites also results in accumulation of sequencing errors, we estimated the extent of noise in WGS-based cfDNA detection in a complementary dataset from matched germline WGS ( TF = 0, varying sequencing depth [5–35X], in subsampling replicates [n = 20], n = 341 in total). Signal-to-noise was measured by comparing the rate of detection (sites with a matching variant out of all sites in the patient-specific compendium) in plasma admixtures (TF > 0) compared to controls (TF = 0), with detection of TFs as low as 10−3 even with only 35X depth of coverage WGS (Fig. 2b).
Further increase of the sensitivity of genome-wide SNV detection required reduction of the noise caused by sequencing errors (~1/1000 bases21; Fig. 2c). Lower sequencing quality metrics are associated with sequencing errors, suggesting the possibility to optimize the tradeoff between the true positive and false positive rates. (Extended Data Fig. 2a,b,c). To integrate multiple quality metrics to inform the probability of error in a single read, we implemented a support vector machine (SVM) classification framework (see Online Methods, Extended Data Fig. 2d). We note that this method was specifically designed to address the challenging scenario where the TF is far lower than the inverse of the depth of coverage, such that, at best, one supporting read is expected for each detected somatic mutation (hence read-classification rather than the standard locus-classification frameworks23,24). Applying this filtering strategy further reduced the sequencing noise by a median 14.4-fold (range 11–17) to lower than 1/10,000 bases (Extended Data Fig. 2e).
Importantly, this machine learning approach improved the sensitivity of the MRDetect-SNV down to TF of 10−5 (Fig. 2b; Extended Data Fig. 2f-h), with assay level specificity of 95% as evaluated across 160 control WGS. As anticipated, this read-centric method showed superior performance compared to a leading locus-centric method in detecting mutations in this unique context of ultra-low TF samples (Extended Data Fig. 2i). We further note the high concordance between the known synthetic TF admixtures and our model TF predictions (Fig. 2d,e; Extended Data Fig. 2j, 3a-h). Thus, MRDetect also offers TF inference that is not limited by the number of genomic equivalents and is therefore orthogonal to current prevailing methods that utilize mutational VAF.
As detection sensitivity using genome-wide SNV integration is a function of the mutation load and the sequencing depth, we systematically evaluated the relationship between TF’s lowest limit of detection (LLOD) and these parameters. Using synthetic admixtures of a melanoma tumor/normal pair to generate plasma-like samples with varying depth of coverage (10–120X), mutation load (2,000–63,000 through subsampling of the mutations detected in the tumor) and TFs (10−3-10−6), we demonstrated that at high mutational load, TF detection as low as 10−6 is feasible with 120X sequencing depth (Fig. 2f). Collectively, these results suggest that genome-wide mutational integration offers an orthogonal route to ultra-sensitive and quantitative TF monitoring in the low disease burden setting.
Expanding the genome-wide mutational integration approach to copy number alterations (CNAs)
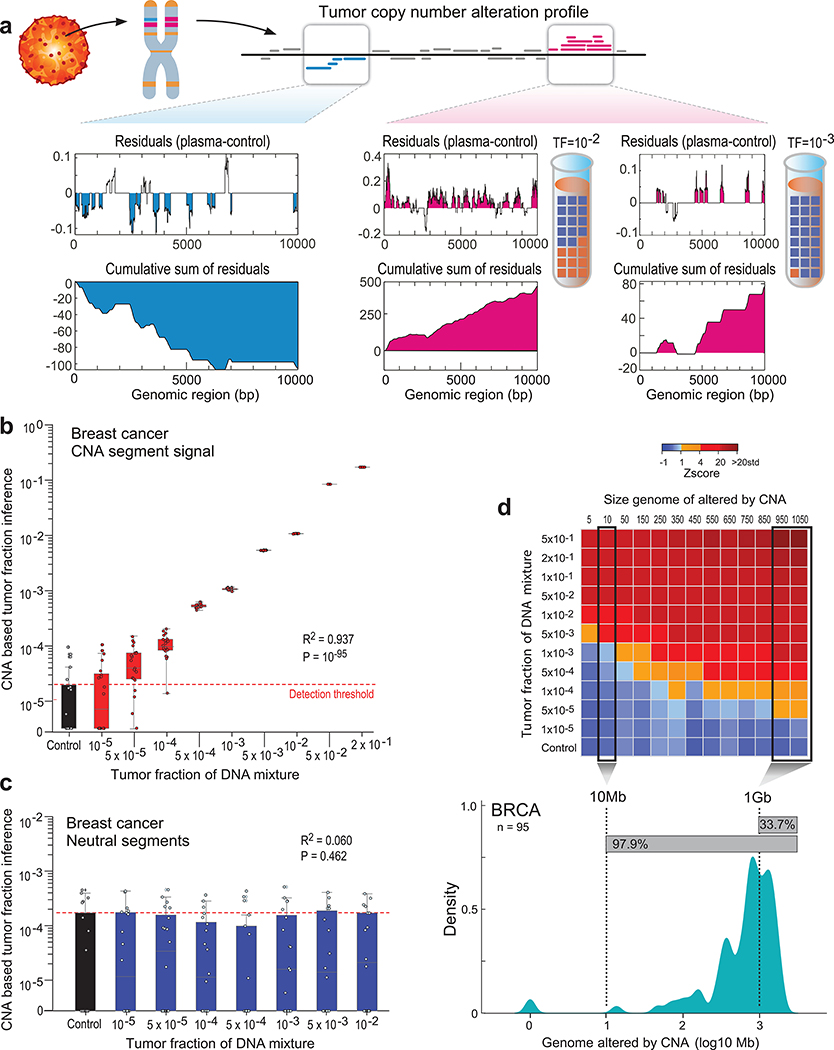
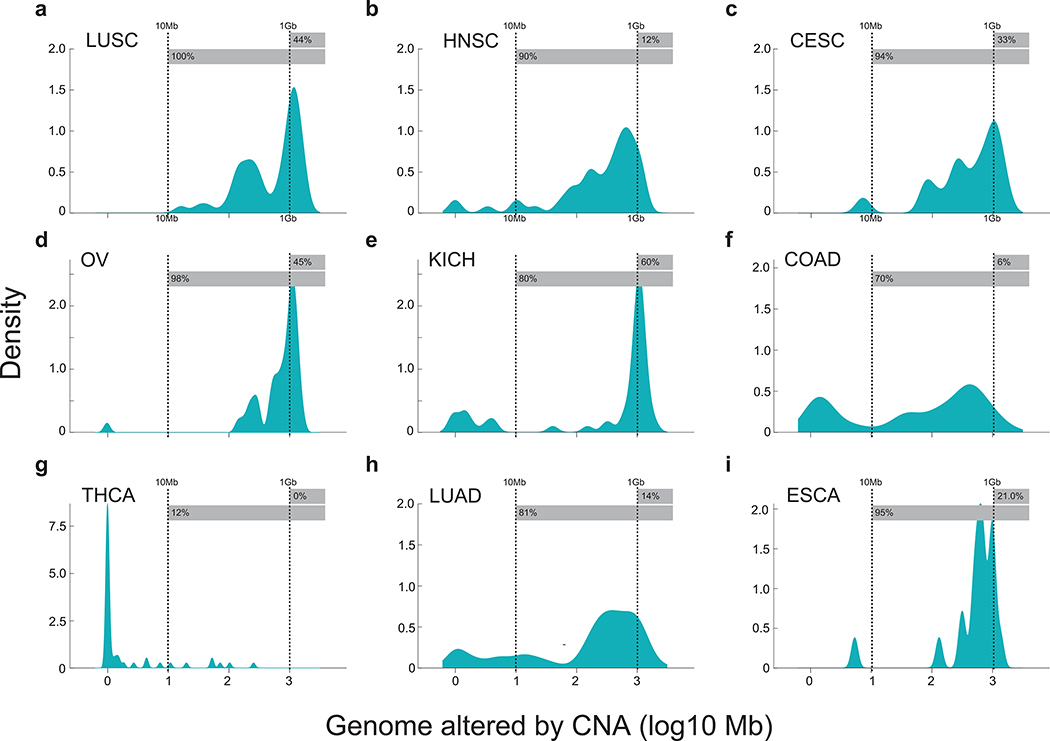
Substantial aneuploidy is observed in 88% of human cancer25 (Extended Data Fig. 4), through which large swaths of the genome undergo amplification and deletion, yielding a potentially robust signal for ctDNA detection. We therefore developed MRDetect-CNA to integrate changes in read depth at patient-specific amplification and deletion segments across the genome. At these CNA segments, the coverage in the plasma represents a mixture of the diploid normal cfDNA, together with ctDNA with directional coverage skews (i.e., greater read depth in amplified regions and lower read depth in deleted regions). Even at low TF admixtures we observed sparse read depth skews, that are biased towards the known patient-specific CNA segment directionality (i.e. amplification or deletion, Fig. 3a). We therefore integrated read depth skews in tumor-specific CNAs across the genome to achieve enhanced ctDNA detection.
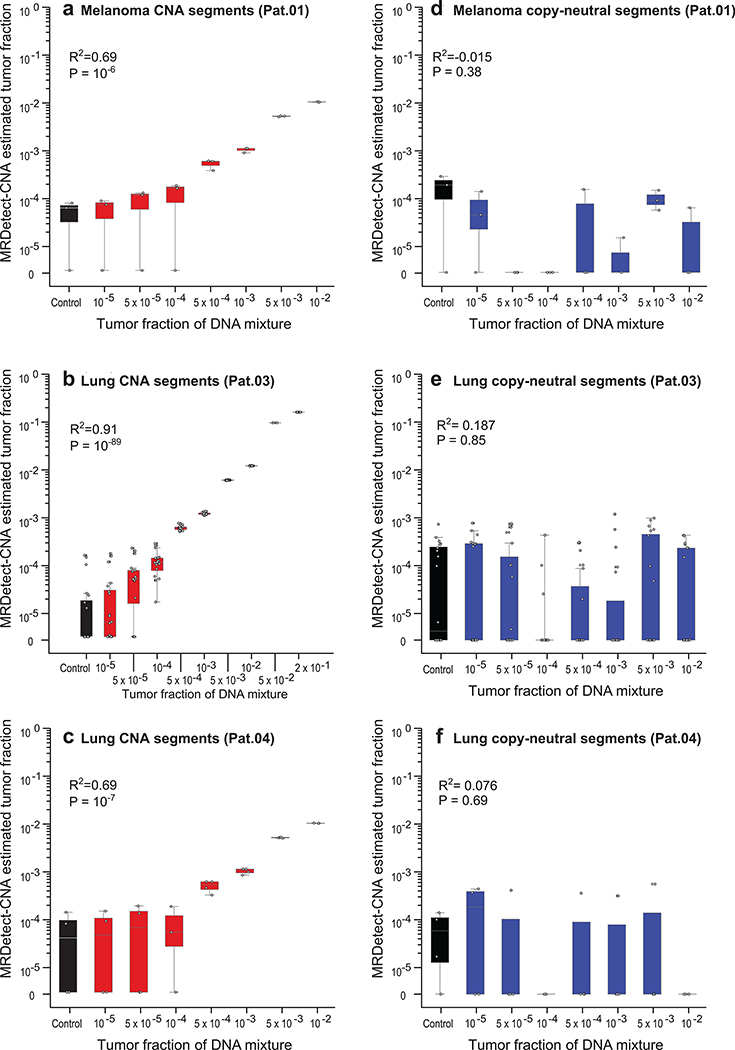
Figure 3: Patient-specific genome-wide CNA integration provides ultra-sensitive ctDNA detection and precision tumor fraction (TF) estimation.
(a) Illustration of the genome-wide copy number alteration (CNA) integration for the inference of plasma tumor fraction (TF). Patient-specific CNA segments are detected from matched tumor and germline WGS. Tumor CNA segments are then used to calculate the genome-wide accumulated signal in the patient’s plasma sample. Amplification segments (right panels, pink) in synthetic plasma with TF of 1% and 0.1% show sparse TF correlated signal that can be integrated due to the positive read coverage bias. Deletion segments (left panel, blue) show similar sparse signal, which is negatively biased. Upper panel: normalized coverage difference between plasma and control (TF = 0) across a 10Kbp region. Lower panel: sum of coverage differences between plasma and control. (b) Utilizing patient-specific CNA mixture model allows for the integrated CNA signal to be directly converted to TF estimates in a breast cancer (Pat.05) in silico synthetic plasma sample, with accurate TF estimation as low as 5*10−5, discriminated from the control samples (TF = 0, left box-plot). High Pearson correlation (two-sided test) between the input TF mixture (x-axis) and the CNA-based estimated TF (y-axis) shows accurate inference based on CNA patterns. N = 20 for each TF > 0 sample, as well as the control (TF = 0) samples. (c) Utilizing the CNA mixture model on copy-neutral regions of the same tumor genome (i.e., regions that do not show differential read-coverage between tumor and germline WGS), do not show correlation with TF (two-sided Pearson correlation). N = 20 for each TF > 0 sample, as well as the control (TF = 0) samples. (d) CNA method lower-limit-of-detection (LLOD) was empirically measured as a function of the CNA load showing high sensitivity (5X10−5) for tumors with copy number variation footprint of 1Gb or more.Upper panel: ctDNA detection at various TF (10−5 - 5*10−1 and control (TF=0)) and CNA load (5Mb - 1050Mb) estimated in a breast cancer (Pat.05) in silico synthetic plasma samples. Detection is observed at TF=5*10−3 at 5Mb and as low as TF=5*10−5 at 1Gb (Z score >= 1). Lower panel: Smoothed histogram (kernel density estimate) of a representative cohort of WGS breast cancer (BRCA) samples from TCGA (n = 95). Majority (97.9%) of the samples have CNA load above 10Mb and 33.7% have CNA load above 1Gb, corresponding with ultra-high ctDNA sensitivity. CNA method lower-limit-of-detection (LLOD) was empirically measured as a function of the CNA load showing high sensitivity (5X10−5) for tumors with copy number variation footprint of 1Gb or more. Upper panel: ctDNA detection at various TF (10−5 - 5*10−1 and control (TF=0); 20 random admixtures each) and CNA load (5Mb - 1050Mb; 3 random subsampling each) estimated in a breast cancer (Pat.05) in silico synthetic plasma samples. Throughout the figure, boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR.
We observed high concordance between input synthetic admixture TF and TF inferred by MRDetect-CNA (Fig. 3b; Extended Data Fig. 5a-c), while tumor copy neutral regions, where no directional coverage skews are expected, showed no change in detection signal in relation to TF (Fig. 3c; Extended Data Fig. 5d-f). A high concordance was also noted between the orthogonal SNV- and CNA-based estimates (Extended Data Fig. 6a). CNA-based detection thus enabled sensitive detection at TF as low as 5*10−5 (range 10−4-10−5, Fig. 3b; Extended Data Fig. 5a-c), extending the detection range by two orders of magnitude compared to a leading CNA ctDNA algorithm10 (Extended Data Fig. 6b). To systematically evaluate the relationship between lowest limit of detection (LLOD) and the footprint of the aggregated CNAs, we down-sampled the cumulative size of segments affected by CNA used for detection. This analysis shows the expected relationship whereby a larger cumulative size of genome affected by CNAs translates into greater sensitivity, with highest sensitivity (TF < 5*10−5) with 1Gbp CNA footprint (Fig. 3d), applicable to a substantial proportion of patients across tumor types (Extended Data Fig. 4).
Application of the genome-wide mutational integration approach to clinical plasma samples
To evaluate MRDetect in the clinical setting, we tested the method on clinical cohorts comprising lung adenocarcinoma patients (LUAD; n = 39), colorectal cancer (CRC; n = 19) and melanoma (n = 2) undergoing surgery (LUAD and CRC patients) or immunotherapy (melanoma patients). We also collected plasma from controls with no known malignancy (n = 38), with similar age and tobacco exposure, to characterize the noise background and estimate clinical specificity (Supplementary Table 2 and 3).
Sequencing noise in cfDNA WGS was comparable to noise rates obtained from the in silico DNA mixtures (median error rate = 1.03*10−3 [1/bp]; Fig. 4a, unfiltered), allowing for facile adaptation of our SVM de-noising strategy using a training set of control plasma samples (n = 8), with comparable suppression performance across test control samples (n = 30, Fig. 4a,). We further leveraged the short size of cfDNA fragments (~165bp), to detect discordance between read pairs (150bp paired end protocol) indicative of potential sequencing errors (Extended Data Fig. 6c). This combined filtering scheme showed reduction of sequencing error rate to median 4.96*10−5 (median 21-fold reduction, Fig. 4a, filtered), with analytic specificity (probability of a true negative detection per locus sequenced at 30X) of 99.85%. The application of MRDetect-CNA to this cohort required additional adaptations (see Online Methods), to account for differences in read depth profiles in WGS cfDNA compared with WGS from genomic DNA (Extended Data Fig. 6d,e), possibly due to differences in library preparation (PCR vs. PCR-free protocols) and non-uniformity of cfDNA coverage driven by DNA degradation and epigenetic features (e.g., chromatin accessibility)6.
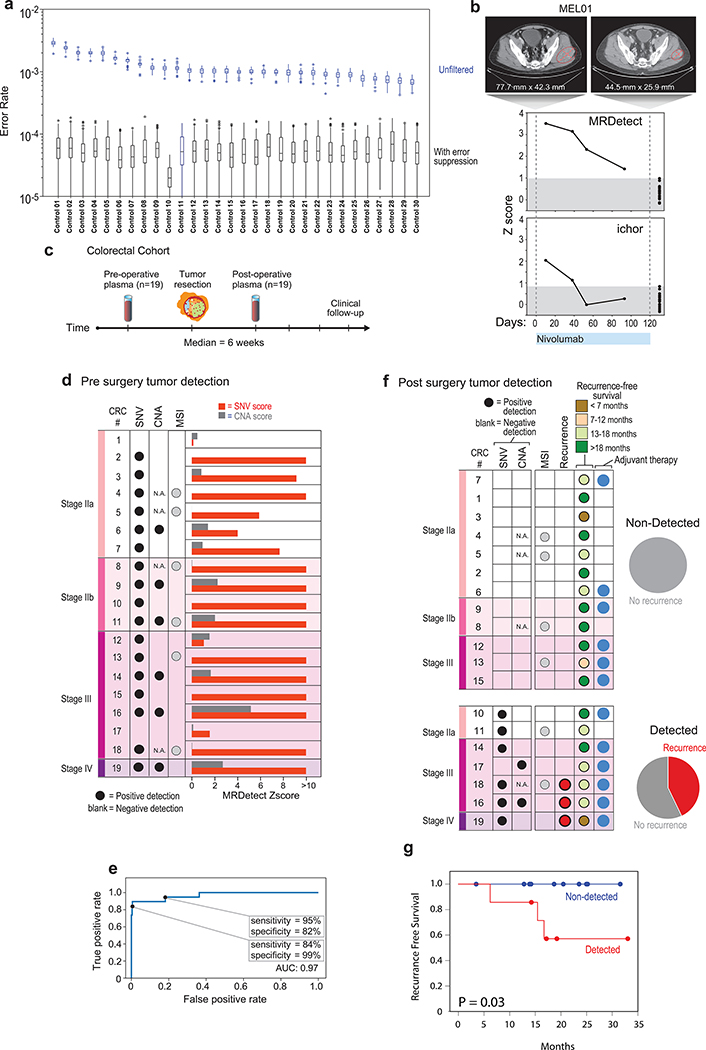
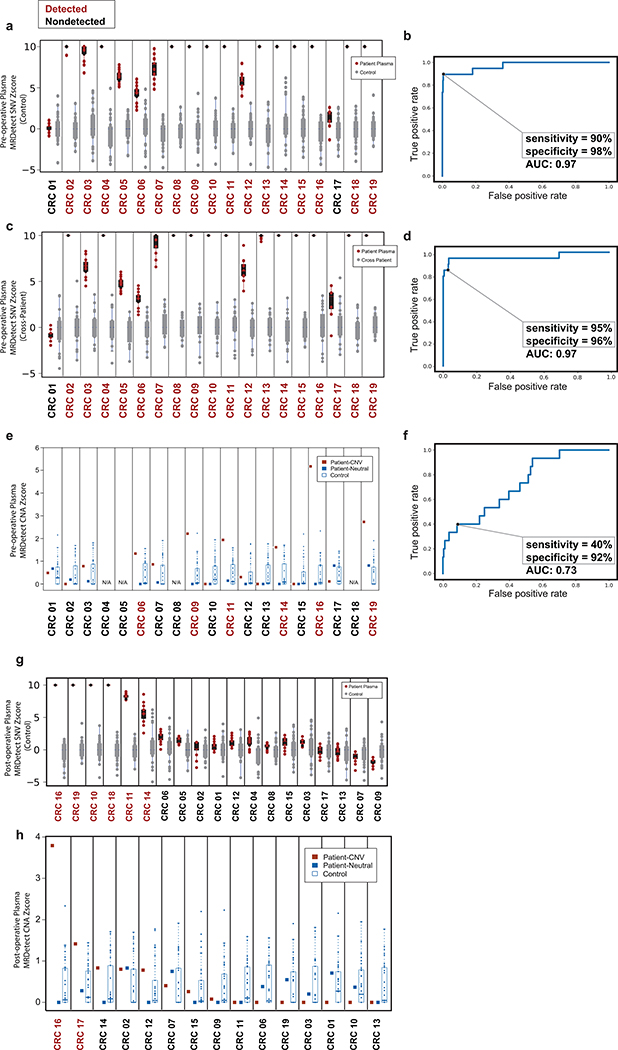
Figure 4: Detection of ctDNA using MRDetect in Melanoma during immunotherapy and colon cancer post-operatively.
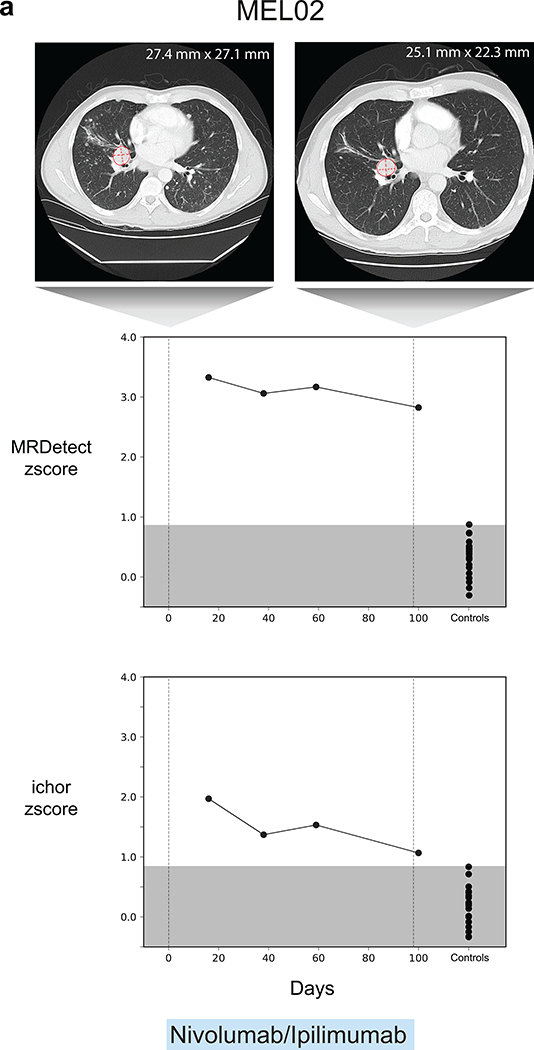
(a) Error rate estimation in a cohort the test control plasma samples (n = 30) with and without error suppression. Applying support vector machine (SVM) error suppression and paired-end read concordance allow sequencing error reduction by a median 21 fold and increase the uniformity between samples (2-fold decrease in coefficient of variation). Boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR. (b) Melanoma treatment response during immunotherapy (Nivolumab) is monitored by blood samples. Treatment monitoring by computed tomography (CT) shows response to therapy but residual disease after 3 months of therapy (upper panel). MRDetect Z-scores effectively track tumor responses, matching radiographic changes, in higher temporal resolution than that feasible with imaging (middle panel). Ichor-CNA sensitivity captures initial treatment response dynamics but does not detect residual disease after 3 months of treatment. Z-score is calculated from a single plasma sample for each timepoint compared to a panel of control samples (n = 30). (c) Illustration of the colon cancer clinical cohort and potential clinical utility for liquid biopsy residual disease detection post-operatively. Upon surgical resection of tumor, the primary tumor is sequenced to define the tumor mutational compendium. Pre-operative plasma is used to verify tumor mutational integration detection in cfDNA and establish detection performance metrics. Post-operative plasma (median 6 weeks) is used to establish the presence of residual disease. (d) Colorectal cancer (CRC) detection on pre-operative plasma samples (n = 19 patients), using SNV (red) and CNA (gray) MRDetect methods. Z-score estimation (bar plots) was used to calculate mutation signature detection in the patient plasma in comparison to detection in the control plasma test cohort (n = 30). Z-score (> 4 and >1.3) was used for the SNV or CNA methods, respectively, to define a positive tumor detection based on ROC optimization on each method (e) Receiver operating curve (ROC) analysis on a combined detection model of SNV and CNA mutational compendia. Pre-operative plasma samples (n = 19) were used as the positive label, and the panel of control plasma samples against all patient mutational compendia (n = 570, nineteen mutational compendia assessed across thirty control samples) was used as the negative label. Two operational points (Z-score thresholds) are shown that correspond with two different specificity regimes. (f) Detection of ctDNA post-operative (6 weeks after surgery) in plasma samples (n = 19) was performed using Z-score estimation in comparison to the cohort of control plasma samples (n = 30). Patients with clinical recurrence, are indicated by red dots. Post-operative ctDNA detection (lower panel) is associated with early disease recurrence as shown in the pie chart. None of patients with the non-detected post-operative ctDNA (upper panel) showed clinical recurrence after median clinical follow-up of 15 (range 4–32) months. (g) Kaplan Meier disease free survival analysis was done over all patients with detected (n = 7) and non-detected (n = 12) post-operative ctDNA. Post-operative ctDNA detection shows association with shorter recurrence-free survival (RFS) (two-sided log-rank test).
We first applied MRDetect to the challenge of monitoring immunotherapy response in high disease burden metastatic melanoma. Patient-specific mutational compendia were obtained from baseline (pre-treatment) plasma WGS, and applied to plasma samples during therapy. The Z-score between the patient plasma signal and the test control plasma noise distribution (n = 30) was used to measure the significance of discrimination between ctDNA detection in patient vs. control samples (Fig. 4b and Extended Data Fig. 7). These data demonstrated that MRDetect effectively track tumor responses, matching radiographic changes, but with higher temporal resolution. Notably, while disease burden estimated by MRDetect decreased with therapy, our data supported residual disease detection, consistent with imaging results and in contrast to benchmarked methods where ctDNA burden decreased below the limits of detection (Fig. 4b, lower panel).
Genome-wide mutational integration enables detection of post-operative residual disease
One of the foremost challenges for residual disease monitoring is detection shortly after surgery to aid with the application of adjuvant therapy in patients with high risk of relapse (Fig. 4c). In this clinical scenario, resected tumors are sequenced to define the patient tumor-specific mutational profiles, which can then be used to detect minute amounts of ctDNA in the patient plasma post-operatively.
To evaluate MRDetect in the post-operative MRD setting, we tested CRC cfDNA, where post-operative ctDNA detection was shown to predict adverse outcome26. We collected plasma from a cohort of 19 operable CRC patients prior to surgery and at a median of 43 days after surgery (Supplementary Table 2 and 3), including 6 CRCs with micro-satellite instability (MSI) and 13 micro-satellite stable (MSS) tumors (Fig. 4d, Supplementary Table 4). To further ensure the robustness of the ctDNA detection, we bootstrapped the patient plasma sample by down-sampling the WGS data (down to 80% coverage, n = 20 random down-sampling replicates), confirming that detection is not driven by isolated sites (Extended Data Fig. 8a). MRDetect-SNV detection performance was evaluated using a ROC analysis showing AUC±SE = 0.97 ± 0.025 (sensitivity±SE = 90% ± 0.069%, specificity±SE = 98% ± 0.006%, Extended Data Fig. 8b). An orthogonal noise model can be generated by applying the patient-specific mutational compendium to other CRC patient plasma samples, which may reveal artifactual non patient-specific detection due to matching mutational sequence context preference (e.g., MSI signature). This cross-patient analysis showed similar detection profiles and performance (Extended Data Fig. 8c, d), supporting detection that is highly specific to an individual malignancy.
MRDetect-CNA showed lower performance (AUC = 0.73 ± 0.064, sensitivity = 40% ± 0.126%, specificity = 92% ± 0.013%; Extended Data Fig. 8e, f), consistent with the lower CNA load in CRC (Extended Data Fig. 4f). However, as the MRDetect-SNV and -CNA signals are independent, they can be combined to a single statistical detection score (see Online Methods), showing robust performance in operable CRC detection (Fig. 4e).
Following the validation of sensitive and specific pre-operative plasma detection, we tested the more challenging task of detecting ctDNA post-operatively. Post-operative detection was evaluated in the same CRC cohort (n = 19, Fig. 4f; Extended Data Fig. 8g,h), and was found to be associated with shorter disease-free survival (Fig. 4g, see Supplementary Table 5 for analysis accounting for additional clinical features) over a median post-operative follow-up of 15 months (range 4–32; Fig. 4g). Four patients with post-operative detection did not show evidence of recurrence, of which three have received adjuvant therapy which may have eliminated residual disease, consistent with prior reports showing that adjuvant therapy may reduce recurrence even in patients with detectable post-operative ctDNA26. The remaining patient had MSI tumor which has been shown to be associated with extended course to relapse27 and even rarely with spontaneous regression28,29. Alternatively, this individual may reflect a false positive classification.
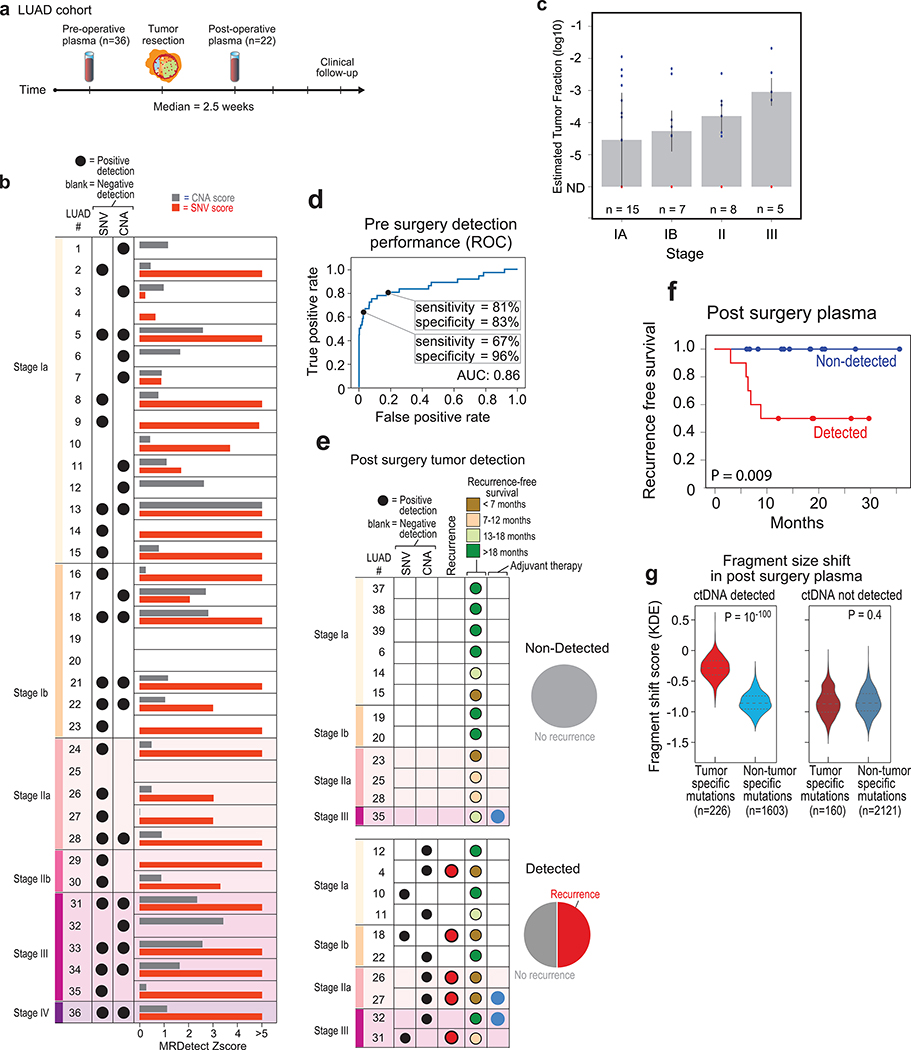
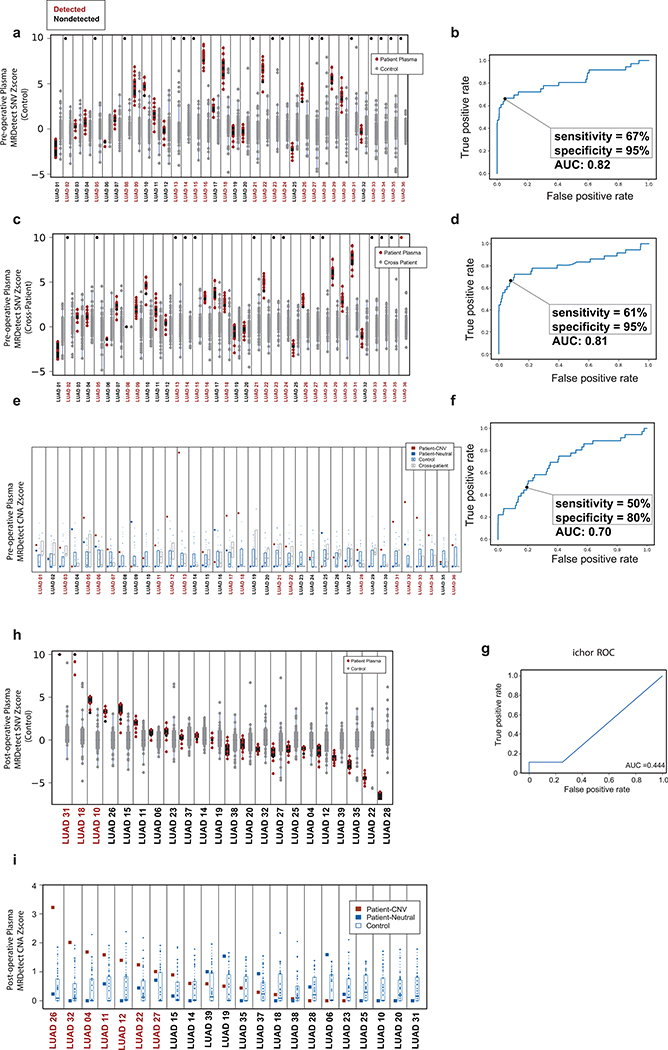
In addition, we tested ctDNA detection in the challenging case of low-burden disease LUAD, where even tumor-informed targeted panel detection showed limited pre- and post-operative sensitivity17. We collected plasma from a cohort of early-stage LUAD patients (78% stage I-IIa LUAD) prior to surgery and at a median of 2.5 weeks after surgery (Fig. 5a, Supplementary Table 2 and 3). MRDetect-SNV applied to pre-operative plasma (Fig. 5b, Extended Data Fig. 9a, Supplementary Table 4) showed AUC = 0.82 ± 0.049 (Extended Data Fig. 9b). Tumor fraction estimates using our mathematical model showed the expected correlation between ctDNA TF and disease stage (Fig. 5c), as well as confirmed that ctDNA TF in early stage LUAD can be often < 10−3, in agreement with the above-described modeling result (Fig. 1b,c). Performance was maintained in the cross-patient analysis (Extended Data Fig. 9c,d), validating MRDetect high specificity even in a more challenging setting. MRDetect-CNA showed comparable performance to CRC (Fig. 5b and Extended Data Fig. 9e, f), with integration of SNV and CAN detection leading to improved performance in early stage LUAD detection (Fig. 5d). Increasing the detection threshold to achieve higher clinical specificity of 96%± 0.006% still maintained high LUAD detection sensitivity of 67% ± 0.079% (Fig. 5d). These data show that genome-wide integration for disease monitoring can perform as well, or better, than deep targeted approaches18,17 or other CNA-based methods10 (Extended Data Fig. 9g), while overcoming the limitation of input genomic equivalents abundance.
Figure 5: Detection of ctDNA using MRDetect in lung adenocarcinoma (LUAD) pre- and post-operative.
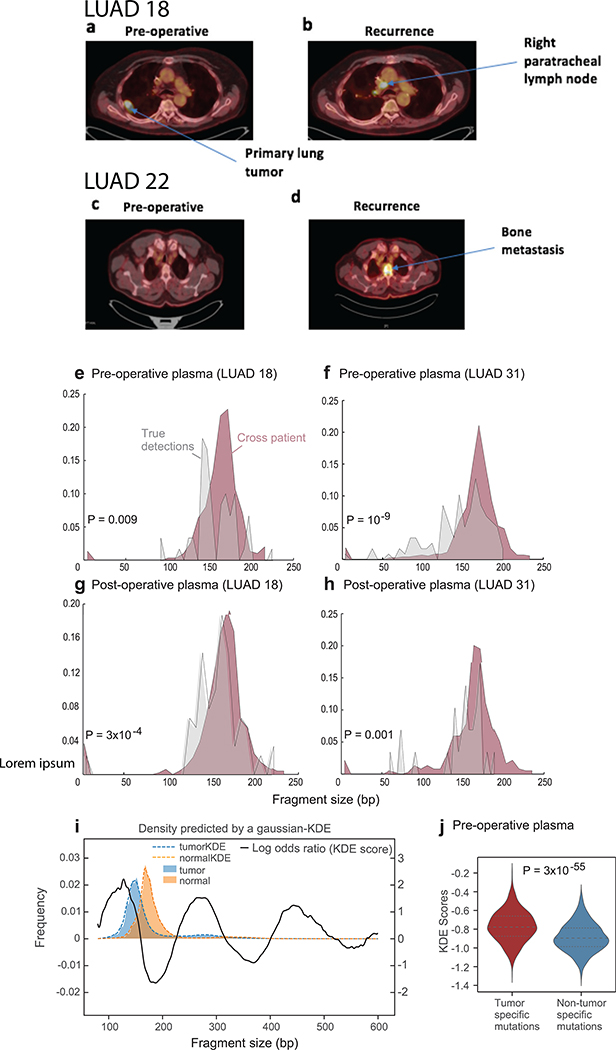
(a) Illustration of the LUAD clinical cohort and potential clinical utility for liquid biopsy residual disease detection post-operatively. Upon surgical resection of tumor, the primary tumor is sequenced to define the tumor mutational compendium. Pre-operative plasma is used to verify tumor mutational integration detection in cfDNA and establish detection performance metrics. Post-operative plasma (median 2.5 weeks) is used to establish the presence of residual disease. Patients with detectable ctDNA in their post-operative plasma samples show higher probability for disease recurrence even when there is no other clinical or imaging confirmation at that time. (b) Early stage lung adenocarcinoma (LUAD) detection on pre-operative plasma samples (n = 36 patients), using SNV (red) and CNA (gray) MRDetect methods. Z-score estimation (bar plots) was used to calculate the mutation signature detection in the patient plasma in comparison to detection in the control plasma test cohort (n = 30). Z-score was used for the SNV or CNA methods to define a positive tumor detection, indicated by black dots. (c) Early stage lung adenocarcinoma (LUAD) tumor fraction estimation per stage. All pre-operative plasma samples were considered for tumor fraction estimation by MRDetect, non-detected samples were assigned a 0 value. Bar plot represent the median TF value per stage, black line represents the confidence intervals (iqr) and the specific patient TF values are overlaid for the detected (blue dots) and non-detected (red dots) patients. Estimated tumor fraction shows association with cancer stage, with stage III samples showing the highest TF values. (d) Receiver operating curve (ROC) analysis on a combined detection model of SNV and CNA mutational compendia. Pre-operative plasma samples (n = 36) were used as the positive label, and the panel of control plasma samples against all patient mutational compendia (n = 1,080, thirty-six mutational compendia assessed across thirty control samples) was used as the negative label. Two operational points (Z-score thresholds) are shown that correspond with two different specificity regimes. (e) Detection of ctDNA post-operative (median 2.5 weeks after surgery) in plasma samples (n = 22) was performed using Z-score estimation in comparison to the cohort of control plasma test cohort (n = 30). Z-score was used for the SNV or CNA methods to define a positive tumor detection, indicated by black dots. Patients with clinical recurrence are indicated by red dots. Post-operative ctDNA detection (lower panel) is associated with early disease recurrence, as shown in the pie chart. None of patients with the non-detected ctDNA (upper panel) showed clinical recurrence after median clinical follow-up of 18 (range 6–36) months. (f) Kaplan Meier disease free survival analysis was done over all patients with detected (n = 10) and non-detected (n = 12) post-operative ctDNA. Post-operative ctDNA detection shows association with shorter recurrence-free survival (RFS) (two-sided log-rank test). (g) Fragments containing tumor mutations show fragment size shift in comparison to non-tumor (cross-patient) mutations in the same patient plasma. The fragment size shift was estimated on detected post-operative patients and non-detected post-operative patients using a kernel-density-estimation method (see Methods). Patients that showed ctDNA detection post-operative also exhibit significant shift (two-sample t-test) in their tumor-specific mutation detections in the patient plasma (light red, n = 226), compared to non-tumor (cross-patient) detected mutations on the same samples (light blue, n = 1603). Patients that do not show ctDNA detection post-operative also do not exhibit significant fragment size shift in tumor-specific mutation detections (dark red, n = 160) compared to non-tumor (cross-patient) detected mutations on the same samples (dark blue, n = 2121). Violin plots depict kernel density estimates of the density distribution. Center dashed lines represent the median and dashed lines represent the interquartile range.
Post-operative detection was evaluated for a cohort of early stage post-operative samples (n = 22, note that these patients were from the same cohort evaluated in the pre-operative detection, except 3 patients [LUAD37–39] where a pre-operative sample was not available). ctDNA was detected in 10 post-operative plasma samples by MRDetect (SNV or CNA method; Fig. 5e; Extended Data Fig. 9h, i), associated with shorter disease-free survival (Fig. 5e, f, see Supplementary Table 5 for analysis accounting for additional clinical features). Thus, MRD ctDNA detection anticipated recurrence shortly post-operatively, compared with delayed identification with imaging (Extended Data Fig. 10a-d). The cohort of patients with ctDNA detected post-operatively also included five patients with no evidence of recurrence, including a patient that was treated with adjuvant chemoradiation after plasma collection. The additional four patients with no evidence of recurrence may reflect shorter follow up time than needed to demonstrate recurrence in these early stage malignancies (e.g., LUAD11 with only 12 months follow up, below median time for recurrence30), with longer follow-up potentially reveal that residual disease detected anticipates additional recurrences, or alternatively may reflect false positive testing. In contrast, the 12 patients where ctDNA was not detectable in post-operative plasma showed no recurrence over a median follow-up of 18 months post-operatively (range 6–36; Fig. 5e).
Notably, it has been shown that ctDNA fragments are shorter than other cfDNA fragments6 and thus this feature may even be used for detection of high burden ctDNA30. Consistent with this notion, tumor-specific mutations detected in patient plasma showed a shorter fragment size compared to non-patient-specific (cross-patient) mutations that are detected in the same samples (Extended Data Fig. 10e-h). We further developed a kernel-density-estimator (KDE) trained on patient derived xenograft (PDX) samples to discriminate between tumor-derived and normal-derived cfDNA based on the fragment size signature (see Online Methods, Extended Data Fig. 10i). Applying this KDE method to the detected pre and post-operative samples showed significant shift towards tumor fragment size signature in the tumor-specific mutations vs. non patient-specific (cross-patient) mutations (Fig. 5g and Extended Data Fig. 10j), providing an orthogonal confirmation that the detected mutations are indeed ctDNA derived, and further suggesting the potential of future integration of fragment length into machine learning de-noising approaches.
Discussion
Deep targeted sequencing approaches are limited in the context of low disease burden due to (i) low fraction of ctDNA, often below 0.1%, and (ii) limited numbers of available DNA fragments in a typical plasma sample, often in the range of only hundreds to several thousand genomic equivalents. The limiting number of genomic equivalents effectively places a ceiling on the depth of sequencing beyond which available distinct fragments are exhausted. Thus, in low disease-burden cancer, deep targeted sequencing is limited to ctDNA detection in frequency above 10−3, resulting in detection in only 20–70% of cases with radiographically demonstrated early stage cancer18,19,17. Residual disease (e.g., post-operative) detection is anticipated to pose an even greater challenge to current technologies26. To overcome the fundamental limitation of low input of cfDNA, we integrated genome-wide mutational signal to allow ultra-sensitive detection, as well as quantitative dynamic monitoring of disease burden. Our results show that through genome-wide mutational integration, we enable accurate and sensitive ctDNA detection in fractions as low 10-5. As this was achieved with a modest sequencing depth (35X), the genome-wide approach severs the limiting tie between available genomic equivalents and detection sensitivity.
WGS also enables effective integration across orthogonal data dimensions such as SNV and CNA, allowing clinical application to a wide range of tumor types that have either high mutation load22 or aneuploidy25. WGS provides an abundance of additional information sources to further increase sensitivity, including shorter fragment length of ctDNA compared with normal cfDNA and nucleosome position information6,27,29,31,32. All these data dimensions could be potentially extracted from the same WGS data, allowing for versatile and cost-effective use of a simple experimental workflow.
We note that while our approach provides sensitive detection of ctDNA it provides limited confidence in the sensitivity to detect any individual site (e.g., a driver mutational event). This suggests that in scenarios where therapeutic targeting requires such information, deep targeted approaches are more appropriate. However, in the context of the broad cancer detection challenge, this may serve as a potential advantage. Genome-wide detection is often based on the majority of events that are ancestral (i.e., preceding transformation30,32), and thus less affected by clonal diversification which may hinder detection via personalized targeting panels due to spatial subsampling or clonal tides. Moreover, targeted cancer gene panel mutation detection may be confounded by clonal growths of hematopoietic origin18,33,34. This finding may present a larger limitation to panel strategies, given the emerging challenge to distinguish pervasive clonal outgrowths across tissues in physiological aging35–37, from true malignancies. In contrast, MRDetect leads to exquisite specificity driven by the patient-specific mutational compendia, such that even when applied cross-patient to samples from individuals with a known malignancy, specificity is maintained.
MRDetect further obviates the need for optimizing the design of capture panels and molecular de-noising, enabling detection with standard WGS. The simplified workflow enabled by WGS based ctDNA detection may thus pave the path to wide-scale clinical application, given the anticipated decline in raw sequencing costs. In addition, the approach presented here requires lower amount of input material (1ml of plasma), in contrast to deep sequencing approaches, where sensitivity hinges on completely saturating the available genomic equivalents. This aspect is an important consideration in clinical applicability as sampling related variability of the amount of available plasma (and genomic equivalents) may hinder real-world application of deep targeted approaches, leading to fluctuating sensitivity.
Notably, the application to clinical care will also be closely informed by the negative and positive predictive values. For example, for stage Ib LUAD the estimated recurrence rate may be as high as 40–50% 38, 39. In this scenario the positive predictive value (PPV) and negative predictive value (NPV) of MRDetect (with sensitivity 81% and specificity 83%) are 0.8 and 0.84, respectively. With the higher thresholds (sensitivity 67% and specificity 96%) PPV and NPV are 0.93 and 0.78, respectively. These performance metrics are in line with current risk-benefit assessment which translates to practice guidelines offering adjuvant therapy for stage IIa NSCLC with recurrence rate of 0.55. While further validation in large cohort studies is required, these proof-of-principle data highlight the potential of MRDetect to deliver precision stratification of patients for adjuvant therapy, and more broadly to address the urgent unmet need of informing the complex residual disease therapy decision-making.
Methods
Human subjects, sample collection and processing.
The study was approved by the local ethics committee and by the Institutional Review Board (IRB) and conducted in accordance to the Declaration of Helsinki protocol. Blood samples were collected in Blood Collection Tubes (BCT) (Streck, La Vista, Nebraska) from patient and healthy adult volunteers enrolled on clinical research protocols at the NewYork-Presbyterian/Weill Cornell Medical Center (NYP/WCMC), Memorial Sloane Kettering Cancer Center (MSKCC) or the Massachusetts General Hospital (MGH) approved by the respective Institutional Review Boards. Tumor tissue were collected from resected lung and colorectal cancer biopsies. The diagnosis of lung adenocarcinoma and colorectal cancer was established according to the World Health Organization (WHO) criteria and confirmed in all cases by an independent pathology review. Informed consent on IRB-approved protocols for genomic sequencing of patients’ samples was obtained prior to the initiation of sequencing studies.
Germline and tumor DNA processing.
Tumor tissue and matched germline DNA from peripheral blood mononuclear cells (PBMC) or adjacent normal tissue were collected and stored in −80oC until they were processed for extraction (see Supplementary Table 3). Genomic DNA was extracted from tumor tissue using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Genomic DNA was extracted from PBMC using QIAamp DNA Blood kit (Qiagen, Hilden, Germany). Libraries were prepared using the TruSeq DNA PCR-free Library Preparation Kit (Illumina, San Diego, CA) with 1μg DNA input following the recommended protocol41 with minor modifications as described below. Intact genomic DNA was concentration normalized and sheared using the Covaris LE220 sonicator (Woburn, MA) to a target size of 450bp. After cleanup and end-repair, an additional double-sided bead-based size selection was added to produce sequencing libraries with highly consistent insert sizes. This was followed by A-tailing, ligation of Illumina DNA Adapter Plate (DAP) adapters and two post-ligation bead-based library cleanups. These stringent cleanups resulted in a narrow library size distribution and the removal of remaining unligated adapters. Final libraries were run on a Fragment Analyzer (Agilent, Santa Clara, CA) to assess their size distribution and quantified by qPCR with adapter specific primers (Kapa Biosystems, Wilmington, MA). Libraries were pooled together based on expected final coverage and sequenced across multiple flow cell lanes to reduce impact of lane-to-lane variations in yield. Whole genome sequencing was performed on the HiSeq X (HCS HD 3.5.0.7; RTA v2.7.7) at 2×150 bp read length, using SBS v3 reagents.
Patient Derived Xenograft (PDX) plasma collection.
Mouse PDX studies were reviewed and approved by Institutional Animal Care and Use Committee (IACUC) at Weill Cornell Medicine (protocol number 2014–0024). PDX were established using fresh pathological tissue fragments from a patient with triple negative breast cancer, implanted subcutaneously into eight-week-old anesthetized NSG mouse. Implant growth was assessed by palpation and when required tumor masses were harvested (< 1.5 cm3). Recipient animals were checked regularly and sacrificed at early sign of distress. Serial propagation were performed implanting tissue seed, as described before42. Blood collection were obtained via tail vein. Blood was collected into Eppendorf tubes contained 5μl of Streck solution. Samples were span for 15 minutes at 14,000rpm. Plasma was collected and stored at −80oC. cfDNA were extracted and sequenced to shallow WGS (1X) to obtain human (Tumor) circulating DNA and mouse (Normal) circulating DNA.
Plasma DNA processing.
At the same day of blood collection, BCT tubes were centrifuged at 2000rpm for 10 minutes to separate plasma. Cell free (cfDNA) was then extracted from human blood plasma by using Mag-Bind® cfDNA Kit (Omega Bio-tek, Norcross, GA). Protocol was optimized and modified in order to optimize yield. Elution time was increased to 20 minutes on thermomixer at 1600rpm at room temperature and eluted in 35ul elution buffer. The concentration of the samples was quantified by Qubit Fluorometer (ThermoFisher, Waltham, MA) and they were run on fragment analyzer by using High Sensitivity Genomic DNA Analysis Kit (Agilent, Santa Clara, CA), to define the size of cfDNA extracted and genomic DNA contamination.
Extraction protocol optimization.
Given the limited amount of cfDNA in the setting of low tumor burden, we examined the potential of cfDNA extraction optimization. To decrease variation derived from sample acquisition and inter-individual variation, we compared available extraction methods on uniform cfDNA material generated through large-volume plasma collections (~300cc) through plasmapheresis of healthy subjects and patients with blood cancer in remission undergoing hematopoietic stem cell collection. The large volume of plasma allows the testing of multiple methods and protocol parameters on the same cfDNA input, enabling accurate measurement of subtle differences in yield and quality. We compared cfDNA extraction methods from Capital Biosciences (Gaithersburg, MD), Zymo Research (Irvine, CA), Omega Bio-tek (Norcross, GA), and NeoGeneStar (Somerset, NJ). Extractions were performed on 1ml of the large-volume plasma sample according to the manufacturer’s instructions and multiple plasma aliquots were processed in parallel to assess both inter- and intra-method variability. The yield and purity of each recovered cfDNA sample was determined using Qubit Fluorometer (total mass), Nanodrop (ThermoFisher, Waltham, MA) via UV absorbance (detection of salt and protein contaminants), and on-chip electrophoresis (size distribution and gDNA contamination). We found that the Mag-Bind cfDNA Extraction Kit from Omega Bio-tek displayed overall somewhat better performance (Extended Data Fig. 1c,d) and ease of use compared to other tested methods. We therefore elected to perform Omega Bio-tek extraction for patient and control plasma samples.
Plasma cfDNA library preparation and sequencing.
Two library preparation methods were used to sequence cfDNA: (i) KAPA Hyper Library Preparation and (ii) NEXTflex Cell Free DNA-Seq Library Preparation. We have transitioned to the KAPA method due to ability to generate library on variable input material and somewhat lower duplication rate. We note no significant differences between the methods in terms of MRDetect performance and coverage variation across the genome. For KAPA Hyper Library Preparation, samples having a mass above 1ng were prepared for next generation sequencing on Illumina’s HiSeq X’s by using modified manufacturer’s protocol. The protocol was scaled down to half reaction by using 25ul of extracted cfDNA. IDT for Illumina TruSeq Unique Dual Indexes were used by diluting 1:15 with EB and ligation reaction was adjusted to 30 minutes. Additional 0.8x SPRISelect Magnetic beads (Beckman Coulter, Pasadena, CA) clean-up was included to the protocol after post-ligation clean-up in order to remove excess adapters and adapter dimers. cfDNA from 1mL of plasma was used for all of the plasma samples in this study. For samples with low concentration (5 samples in the entire study), an additional 1mL of plasma was extracted and the DNA aliquot with the highest mass was used for library preparation (see Supplementary Table 3, for the total plasma extracted for each sample). The number of PCR cycles was dependent on initial cfDNA total mass. For samples with >5ng of total cfDNA, 5 PCR cycles were performed. For samples with <5ng of total cfDNA, 7–10 PCR cycles were performed. Quality metrics were performed on the libraries by Qubit Fluorometer, High Sensitivity NGS DNA Analysis Kit, and KAPA SYBR Fast qPCR Kit (Kapa Biosystems, Wilmington, MA).
For NEXTflex Cell Free DNA-Seq Library Preparation, 1–5ng of the extracted cfDNA were prepared to make libraries for next generation sequencing on Illumina’s HiSeq X’s by using manufacturer’s protocol for Illumina Sequencing. NEXTflex™ DNA Barcode was diluted 1:8 and ten PCR cycles were performed on the libraries. Additional 0.8x SPRISelect Magnetic beads clean-up was included to the protocol after post PCR clean-up in order to remove excess adapter dimers. Quality check was performed by Qubit Fluorometer, High Sensitivity NGS DNA Analysis Kit by AATI, and KAPA SYBR Fast qPCR Kit. Whole genome sequencing was performed on the HiSeq X (HCS HD 3.5.0.7; RTA v2.7.7) at 2×150 bp read length, using SBS v3 reagents.
Preprocessing, quality control analysis and sample identification/concordance.
Whole genome sequencing (WGS) reads for primary tumor, matched germline and plasma samples were demultiplexed using Illumina’s bcl2fastq (v2.17.1.14) to generate FASTQ files. The primary tumor and matched germline WGS was submitted to the NYGC somatic preprocessing pipeline which includes alignment to the GRCh37 reference (1000 Genomes version) with BWA-ALN (v0.6.2)43. We used a modified alignment pipeline for plasma cfDNA to accommodate adapter trimming. We did this after observing increased adapter contaminated reads in cfDNA samples as compared to tumor samples, presumably due to the fact that cfDNA has shorter fragment size which can lead to R1/R2 overhang (data not shown). For cfDNA samples, we used Skewer44 for adapter trimming (default settings), and subsequently aligned samples using BWA-MEM (default settings) to the GRCh37 reference (1000 Genomes version). For all samples, duplicate marking and sorting was done using Novosort MarkDuplicates (v1.03.01, a multi-threaded bam sort/merge tool by Novocraft technologies45) followed by indel realignment (done jointly for the tumor and matched germline) and base quality score recalibration (BQSR) using GATK (v3.4.0)46 resulting in a final coordinate sorted bam file per sample. Alignment quality metrics were computed using Picard (v1.83; QualityScoreDistribution, MeanQualityByCycle, CollectBaseDistributionByCycle, CollectAlignmentSummaryMetrics, CollectInsertSizeMetrics, CollectGcBiasMetrics, CollectOxoGMetrics) and GATK (average coverage, percentage of mapped and duplicate reads). To specifically assess for potential sample contamination we applied Conpair47, which validated genetic concordance between the matched germline, tumor and plasma samples, as well as evaluated any inter-individual contamination in the samples. Samples that showed low concordance (< 0.99) were excluded from further analysis. Specifically, three pre-operative plasma samples from LUAD patients #37, #38 and #39 were rejected from analysis due to low concordance score.
Generation of synthetic-plasma DNA admixtures.
We generated in silico admixtures of tumor and matched germline DNA from eight patients with various cancer types, including LUAD, breast cancer, melanoma and osteosarcoma (Supplementary Table 1). Admixtures were generated by downsampling and mixing, using SAMtools (v1.1, view -s and merge commands), of tumor and germline reads sequenced with the same protocol and sequencing device (see tumor DNA processing above) used for the lung adenocarcinoma cohort reported in our study. The ratio of tumor vs. germline reads was defined by the tumor fraction (TF) taking into account the tumor-specific purity and ploidy using the following model. In the tumor sample, the fraction of DNA that is actually coming from the tumor (TTF) can be characterized by the following equation:
| Eq. 1 |
Where PU denotes purity, PL denotes ploidy in the tumor sample, Treads denote the number of tumor-tissue reads sequenced from the tumor sample, Nreads denote the number of healthy-tissue reads (normal tissue in the tumor sample or contamination) sequenced from the tumor sample. Down-sampling the original tumor to a lower TF (so as to represent highly tumor diluted plasma samples) will require the downsampling ratio S:
| Eq. 2 |
Lastly scaling down the coverage of each sample, tumor and germline, and mixing will take the following form
| Eq. 3 |
Where covreq is the required read depth coverage for the admixture sample and covT, covN are the read depth coverage of the tumor and germline samples, respectively.
Tumor / Normal somatic mutation calling.
The primary tumor and matched germline bam files were processed through somatic variant calling pipeline which consists of MuTect (v1.1.7)23, Strelka (v1.0.14)48 and LoFreq (2.1.3a)49 for single nucleotide variants (SNVs). We note that for the two patients with metastatic melanoma, no primary tumor was available for sequencing. We therefore performed mutation calling comparing the pre-treatment plasma WGS to PBMC WGS. To achieve stringent somatic variant calling we enforced intersection between callers. We further removed triallelic sites and common germline variants (minor allele fraction [MAF] > 5% in cancer genes DNMT3A, TET2, JAK2, ASXL1, TP53, GNAS, PPM1D, BCORL1 and SF3B1, and with MAF ≥ 1% elsewhere in the genome, as reported in the 1000 Genomes Project release 3 and the Exome Aggregation Consortium [ExAC] server). Finally, we removed a subset of artifactual calls by the use of a “blacklist” generated by calling somatic variants on 16 random pairings of 80x/40x in-house sequenced HapMap WGS data. Small deletions and insertions (indels) were not used in our methodology, due to smaller frequency and somewhat greater challenges in calling accuracy. Copy number alterations (CNA), including deletions, amplifications and copy-neutral loss of heterozygosity, were called using NBIC-seq (v0.7)50. Segments with log2 > 0.2 were categorized as amplifications, and segments with log2 < −0.235 were categorized as deletions (corresponding to a single copy gain or loss, respectively, at 30% purity genome).
Plasma cfDNA single nucleotide variant (SNV) identification.
Sensitive detection of patient-specific compendia of SNVs is performed by searching the plasma WGS for all sites from the matched patient-tumor compendium with corresponding mutations in the same genomic site and the same exact substitution. To efficiently identify variants present in the sequencing data, we used a custom python script (python version 3.6.2), which utilizes the pysam module to efficiently extracts alignments harboring variants, and extracted any read that both uniquely maps to a variant of interest and was in an aligned portion of the read (no clipping or soft-masking at the position of the variant). All extracted reads were then subjected to the support vector machine (SVM, see below) model for further classification.
Sequencing error suppression.
The detection of mutations in cfDNA WGS required a novel conceptualization of the mutation calling process. Current mutation calling is typically “locus-centric”23,48,49 whereby the presence of multiple supporting reads for the same mutation is incorporated into statistical models. However, in highly underrepresented ctDNA detection in low burden disease, the tumor fraction is orders of magnitude lower than the inverse of depth of sequencing, such that, at best, only a single supporting read is available. We therefore generated a novel analytic framework that is “read-centric”51 using a machine learning models to scan through individual reads and provide a measure of confidence to classify them as likely true somatically mutated ctDNA vs. sequencing artifacts. These data are then integrated across the genome using analytic models (see below) to measure the tumor fraction representation in cfDNA.
Our read-centric denoising method applies a support vector machine (SVM) framework. Five features were included in the model training, which are known to represent sequencing error patterns, and which showed association to artifactual detections in our training control plasma cohort (n = 8). Variant base quality (VBQ) indicates our trust in the particular mismatch, which showed significant enrichment with sequencing error (P value < 10−100 , two sample t-test, Extended Data Fig. 2b,c). Mean read base quality (MRBQ) represents the overall quality of sequencing within a particular read pair. Position in read (PIR) captures cycle specific errors, as 3’ showed higher association with sequencing error (Extended Data Fig. 2d). R1/R2 allow us to test concordance between overlapping read-pair sequences, where discordance is associated with sequencing error (Extended Data Fig. 5a). Mapping quality (MQ) is an alignment-provided metric corresponding to the trust the aligner has in the particular alignment.
In order to train the read-centric SVM model, we first focused on building a high-quality truth set of confident alterations and likely errors. For confident alterations we aimed for variants with a high degree of support. We used GATK (v3.4.0) to call variants on each of the training control plasma sample, using the -L flag to specify only returning sites in dbSNP (build 151) variants. For error mutations, we searched for mutations with a low degree of support by performing pileup using SAMtools mpileup to identify mismatches genome wide. These were then filtered for coverage (coverage > 10X) while enforcing that the variant has low support of <= 0.1 VAF. We sampled 10,000 reads randomly from the training set of 8 control patients’ cfDNA for each class for training.
For model training we utilized sklearn’s SVM package, using linear SVM with a C: 1.0, tol 1e-7, hinge loss, and l2 regularization. Using 0 as a cutoff we evaluated model performance by F1, sensitivity, and specificity using 10-fold cross validation (Extended Data Fig. 2e). Throughout development we considered the decision boundaries when various features were fixed. Visualizations were interpreted for expected relationships between features: VBQ correlates with PIR, R2 has a more stringent bias, MRBQ correlates with VBQ and when both VBQ and MRBQ are high we make a confident decision. As a comparison to our SVM method, we evaluated the performance of a random forest fit using the same dataset and features. The number of trees was set to 50. The SVM model outperformed the random forest model across several metrics (Extended Data Fig. 2e).
Additional de-noising methods.
Paired-end sequencing of the short cfDNA fragments (read length 150bp) result in overlapping R1 and R2 reads which provide an additional means of error suppression due to overlapping information from the same DNA fragment. Given the low probability of a sequencing error occurring in both of the overlapping reads, pairs that contained the same alternative base in their overlaps (concordant mutation) were considered as more likely to represent true mutations as opposed to pairs in which only one read differed (discordant), which are more likely to reflect sequencing errors. To test this notion, we evaluated the proportion of concordant read pairs at dbSNP sites and at likely artifactual sites, which were defined by read pairs with variants overlapping the union of all patient somatic SNVs compendia across all LUAD patients, that were observed with the same variant in the control plasma samples (Extended Data Fig. 5a). This analysis revealed that pairs are far more likely to be discordant at artifactual sites, and mostly concordant at sites containing true germline variant. This analysis was conducted using a custom python script (python version 3.6.2) which utilizes the pysam module. Briefly, for each variant, reads containing the variant of interest and their read mates were compared to evaluate whether both (concordant) or only one (discordant) contained the variant of interest, and discordant read pairs were discarded. Additionally, we generated a “blacklist” of regions in the genome that are more likely to contain artifactual variants. To do this, we performed artifact detection mode on the training control plasma samples (n = 8), any variant detected in at least two controls was included in the blacklist and removed from subsequent patient plasma analysis.
Plasma SNV-based ctDNA detection and quantification.
We reasoned that the fraction of the patient-specific SNVs that are observed in the plasma cfDNA WGS follows a binomial distribution over N independent Bernoulli trials, where N is the number of SNVs in the patient-specific mutational compendium (identified through standard SNV detection on matched tumor and germline DNA WGS data). Each such trial includes multiple rounds of random sampling that depends on the local coverage, where the probability of sampling a DNA fragment containing a given variant in each round is defined as the tumor fraction (the fraction of circulating tumor DNA in the cfDNA pool). We note that we have not explicitly modeled heterozygosity or lower VAF due to subclonal events, and thus our Bernoulli model likely somewhat underestimates the true TF. However, these aspects will likely lead to a 2–3 fold underestimation at most (given depth of sequencing, purity for subclonal SNVs), which is anticipated to have limited impact considering the heavy dilution of ctDNA in low burden disease (< 1%).
Therefore, the relationship between coverage, mutation load (SNV/tumor), number of detected variants in cfDNA WGS, and the tumor fraction corresponds to the following equation:
| Eq. 4 |
Where M denotes the number of SNVs detected in the plasma sample, N denotes the number of SNVs (mutation load) in the patient-specific mutational compendium, TF denotes the tumor fraction, cov denote the local coverage in sites with a tumor-specific SNV, μ denoted the mean noise rate (number of_errors/number of reads evaluated) that corresponds to the patient-specific SNV compendium evaluated in control plasma WGS data (see below), and R denote the total number of reads covering the patient-specific mutational compendium. This relationship allows the calculation of the plasma TF from the mutation detection rate, even in extremely low allele fraction where the mutation allele fraction itself is not informative (random sampling between 0 and 1 supporting read at best).
To address variation in sequencing artifact noise (μ) across patients with different mutational compendia, we apply the patient-specific mutational compendium to calculate the expected noise distribution across the cohort of control plasma samples. The process described above is performed to detect the patient-specific SNVs in control plasma samples or other patients (cross-patient analysis). These detections represent the background noise model for which we calculate the mean and standard-deviation (μ,σ) of artifactual mutation detection rate. Confident cfDNA tumor detection can then be defined by converting the patient-specific detection rate (det_rate = number of SNVs detected in cfDNA/number of reads checked = M/R) to a Z-score = , and define a threshold that will keep the specificity above 80% (Z-score > 1.2). Specificity and sensitivity performance values were further validated using receiver operating characteristic (ROC) curve using the Python (version 2.7.10) module numpy.metrics.roc_curve and numpy.metrics.roc_auc_score.
Calculating the patient tumor fraction from point mutation detection was then carried out by the following equation (which is an inversion of Eq.4):
| Eq. 5 |
Where M denotes the number of SNVs detected in the plasma sample, N denotes the number of SNVs (mutation load) in the patient-specific mutational compendium, TF denotes the tumor fraction, cov denote the local coverage in sites with a tumor-specific SNV, μ denoted the noise rate (number of errors/number of reads evaluated) that corresponds to the patient-specific SNV compendium, and R denote the total number of reads covering the patient-specific mutational compendium.
Plasma copy number alteration (CNA)-based quantification.
To generate a patient-specific CNA compendium (amplifications and deletions), we applied NBIC-seq50 to the primary tumor and generated an interval list of amplifications, deletions and copy-neutral loss of heterozygosity. This interval list was then interrogated in cfDNA WGS to calculate an integrated signal (score) for representation of ctDNA in the cfDNA. Average coverage per base-pair was calculated in plasma samples using GATK (v3.4.0) DepthOfCoverage. The genomic region of interest (CNA or Neutral segments) was divided into non-overlapping 500bp bins and median depth of coverage per-bin was calculated. We note general stability of performance for bin size between 200bp and 1Kbp with lower performance with increasing bin size (not shown). To compare across samples with varying depths of coverages, we normalized the depth of coverage in each bin by dividing the coverage by the average coverage of the sample. To further correct for sample-specific variation in depth of coverage, a robust Z-score normalization was applied to each plasma sample separately. Median and median absolute deviation (MAD) were calculated per sample over the entire genomic region of interest (over the aggregate of all the patient’s CNA segments), subsequently all genomic bins were normalized by subtracting the median and dividing the result by the MAD. Specifically, every 500bp bin is converted to a normalized_coverage = .
Generating plasma reference sample.
Read depth profiles from cfDNA and from gDNA (tumor or PBMC) showed significant differences (Extended Data Fig. 5b,c), possibly due to differences in library preparation (PCR vs. PCR-free protocols) and non-uniformity of cfDNA coverage driven by DNA degradation and epigenetic features (e.g., chromatin accessibility). To ensure that the signal calculated is not driven by these effects, we generated a control plasma reference for plasma differential coverage measurement. Control plasma training set samples (n = 8) were randomly down-sampled and admixed, using SAMtools (v1.1, view -s, merge), to generate a reference cfDNA plasma sample with 25X depth of coverage. The robust Z-score normalized coverage of this reference sample was used as the reference plasma for all subsequent analysis, in place of patient-specific PBMC reference. We further note that detection results with the plasma reference and with patient-specific PBMC WGS showed high concordance confirming the robustness of these results (Supplementary Table 4).
Removing artifactual CNA events.
We observed that some genomic regions show recurrent amplification and deletion patterns in control samples; these include centromeres and other regions of poor coverage known as “gap regions”. To remove genomic bins (500bp) with recurrent and significant high or low coverage compared to the rest of the genome, we removed bins with extreme coverage values (), on the reference sample, after robust Z-score normalization. Given that each sample is diluted in the plasma reference (composed of downsampled 8 samples) this likely results in removal of only recurrent artifactual amplification or deletion. Somatic CNA events originating from possible clonal hematopoiesis can also create biases in plasma cfDNA CNA analysis, as most cfDNA is derived from blood cells. To assess for this potential artifactual, CNA matched PBMC WGS were assessed for CNAs using NBIC-seq (v0.7) against a contemporary normal sample, NA1287852, sequenced using the same protocol as the PBMC. Copy-neutral LOH events were also examined using B-allele frequency (BAF) analysis. Copy number alterations (CNA) were called using NBIC-seq (v0.7)11 on PBMC vs contemporary normal. Segments (>1Mb in length) with log2 > 0.2 were categorized as amplifications, and segments with log2 < −0.235 were categorized as deletions (corresponding to a single copy gain or loss, respectively, at 30% purity genome) in the PBMC. These regions were then excluded from the patient-specific CNA interval list. Detected events were removed from the CNA and neutral interval lists to prevent biases in MRDetect CNA signal. Three patients had a detectable somatic-PBMC events, LUAD10 (amp Chr12:60138–133841502), LUAD26 (CN-LOH Chr4:50400000–191044164), and CRC03 (del Chr3:234305–80851349; del Chr5:75605307–180877637; del Chr7:95649215–159128563; del Chr10:50003039–108417985; del Chr15:36365636–63901029; del Chr15:36365636–63901029; del Chr17:7602691–20374289; del Chr18:24227106–78017148).
Plasma cfDNA CNA signal measurement.
Differential coverage between plasma sample (pre-surgery or post-surgery) and the reference showed a tendency for a positive skew in bins overlapping patient-specific amplifications and a negative skew in bins overlapping patient-specific deletions. Therefore, multiplying the differential (Plasma_sample – PON_reference) depth of coverage by the directionality of the CNA segment (amplification multiplied by +1, deletion multiplied by −1) and summing these values across the CNA regions provides the CNA signal, while neutral regions (diploid regions in the matched tumor) provides the background noise, in which random directionality in depth of coverage is expected to result in low integrated signal. This is represented by the following equation:
| Eq. 6 |
Where M is the number of non-overlapping 500bp bins covering the region of interest (CNA segment). P(i) and N(i) are robust Z-score normalized depth of coverage values in the window i for the plasma sample and reference, respectively. Sign(T(i)-N(i)) represents the direction of the tumor CNA segment (amplification multiplied by +1, deletion multiplied by −1) that overlap with window i.
To address variation in noise across patients with different CNA compendia we used the patient specific CNA interval list to calculate patient-specific noise distribution over a cohort of control plasma samples (n = 30). The process described above was done to detect the patient-specific CNA compendia in control plasma samples or in other patients’ plasma (cross-patient). These detections represent the background noise model for which we calculate the mean and standard-deviation (μ,σ) of artifactual background noise. Subsequently, the patient integrated CNA signal and control noise values were converted to Z-scores to define the significance of CNA detection in patient samples compared to background noise.
These detections represent the background noise model for which we calculate the mean and standard-deviation (μ,σ) of artifactual CNA detection signal. Confident cfDNA tumor detection can then be defined by converting the patient-specific CNA signal to a Z-score = , and define a threshold that will keep the specificity above 80% (Z-score > 1.2). Specificity and sensitivity performance values were further validated using receiver operating characteristic (ROC) curve using the Python (version 2.7.10) module numpy.metrics.roc_curve and numpy.metrics.roc_auc_score.
Utilizing the CNA mixture model on copy-neutral regions of the same tumor genome was done as an additional negative control with the same model defined above (Eq.6). In this case, for each 500bp bin, the directionality was defined as the local sign(T-N), meaning the local sign of the difference between the tumor normalized coverage and the germ-line normalized coverage in this specific 500bp bin (in copy-neutral regions this sign takes random +1/−1 values without a defined trajectory).
CNA downsampling.
To demonstrate sensitivity at various TFs and size of genome altered we used the in silico admixtures of tumor and matched germline DNA from breast cancer (described in Methods above) and randomly downsampled the CNAs footprint. We generated 8 replicates across TFs (1×10−5 to 5×10−1 and control) and 3 replicates across CNA footprint sizes (5Mb to 1050Mb). We applied MRDetect CNA (described in methods above) comparing the replicates to control plasma samples to generate CNA Z-scores and finally compute median Z-score across all replicates.
CNA load across tumor types analysis.
We applied a junction balance analysis algorithm (JaBbA)53 on 588 TCGA WGS samples across 10 tumor types to obtain purity and ploidy corrected CNA segments. Briefly, JaBbA53 infers somatic segmental CN simultaneously with rearrangement junction copy numbers from paired tumor-normal WGS. High-density (200 bp bins) coverage data is collected from tumor and normal WGS aligned reads, corrected for GC-content and mappability, and rearrangement junctions are inferred by SvABA (doi:10.1101/gr.221028.117). JaBbA53 reconstructs a junction-balanced genome graph by optimizing a complexity-regularized likelihood function with mixed-integer programming.
Integrating SNV and CNA scores.
For definition of tumor detection (pre and post operatively), we applied thresholds designed based on the ROC analysis. The threshold for SNV detection was defined as the first value to provide > 95% specificity, Z score > 3 for lung (Fig. 5b,e and Extended Data Fig. 9) and Z score > 4 for CRC (Fig. 4d,f and Extended Data Fig. 8). The threshold for CNA detection was defined as the first value to provide > 80% specificity, Z score > 0.9 for lung (Fig. 5b,e and Extended Data Fig. 9) and Z score > 1.3 for CRC (Fig. 4d,f and Extended Data Fig. 8). To further evaluate performance, we have also applied identical Z score thresholds across the cohorts. For example, in SNV analysis, a Z-score of > 3, selected based on specificity>90% in CRC (specificity −98%, sensitivity −90%), yields similar results in LUAD (specificity −95%, sensitivity – 67%). For the CNA analysis, a Z-score of > 1.3, selected based on specificity>90% in CRC (specificity −92%, sensitivity −40%), yields similar results in LUAD (specificity −89%, sensitivity – 27%). These thresholds did not affect the association between post-operative detection and adverse outcome (P value of 0.03 for CRC cohort, P value of 0.024 for LUAD cohort).
Additionally, as the SNV noise model and CNA noise model are largely independent, generated by different noise mechanisms, the SNV and CNA Z-score values of control plasma samples can be combined together and still generate a normal distribution around zero mean. Measuring this combined noise Z-score across all control samples by applying all the different patient mutation compendia (total n = 132 combinations) yielded a mean Z-score of 0.07. In contrast, patient cfDNA CNA and SNV Z-scores are correlated due to the underlying circulating tumor DNA detection signal, which will allow an increased signal-to-noise for combined CNA and SNV detection signal. SNV Z-score and CNA Z-score are combined to get the integrated score (integrated_score = SNV_zscore + CNA_zscore) for all patient plasma and control plasma , the combined metrics performance is shown for lung and CRC (Fig. 4e and 5d).
ichorCNA.
Ichor-CNA10 (version 2.0) was used as an orthogonal CNA-based method for the estimation of plasma tumor fraction (TF) in the in silico admixtures and in patient samples. We optimized input setting for more sensitive detection in low tumor burden disease using the modified flags: --altFracThreshold 0.001, --normal .99, --NORMWIG for using matched normal instead of the ichor panel of normal; all other settings were set to default values. As a matched normal for the in silico admixtures, we used an independent matched germ-line sample. For the patient samples, we used our control plasma PON (see above in the CNA methods) as the matched normal.
Fragment size kernel density estimator (KDE).
Circulating DNA fragments that originate from the tumor show shorter fragment size in comparison to “normal” DNA fragments that originate mainly from apoptosis of hematopoietic cells (immune cells)54,32. We therefore reasoned that shorter fragment length of DNA fragments of somatic variants detected in cfDNA could provide orthogonal support to the fact that these indeed represent ctDNA. To generate a robust model that can quantify the probability of a single DNA fragment to be from tumor or normal origin, we used a joint kernel density estimator (KDE) to characterize the fragment size distribution of circulating DNA. This method allows to rigorously quantitate the fragment size shift across the entire fragment sizes distribution (80–600bp), instead of focusing only on < 150bp fragments30. To train the KDE estimator we used a triple negative breast cancer (TNBC) patient derived xenograft (PDX) to create a purified set of human circulating tumor DNA (human-ctDNA). We collected blood from the mouse and sequenced the cfDNA (see “Patient Derived Xenograft (PDX) plasma collection” section above). Circulating tumor DNA fragments were then isolated by retaining only human-aligned DNA reads, which were then used as a tumor-labeled training set (n = ~3M fragments). Normal cfDNA training set was defined by a random sample of reads from our human control plasma cohort (n = ~6M fragments, from 8 control plasma samples). A KDE model was trained over the tumor and normal cfDNA sets using the Python (version 2.7.10) module scipy.stats.gaussian_kde, using scott-rule to choose optimal bandwidth (bw_method=‘scott’). We then scored collections of cfDNA fragments (e.g., tumor specific detections or cross-patient detection) based on their fragment sizes using the following equation:
| Eq.7 |
where for each specific DNA fragment the logpdf value was calculated using scipy.stats.gaussian_kde().logpdf() for both the tumor KDE and the normal KDE separately.
The KDE score was tested on the aggregated tumor-specific mutation detections and aggregated cross-patient mutation detection in different patient sets (pre-surgery plasma, ctDNA-positive post-surgery plasma and ctDNA-negative post-surgery plasma). We used a permutation test to check the significance of the difference in KDE-score between tumor-specific mutation detections and cross-patient mutation detection. For each mutation set, we randomly sub-sampled 150 DNA fragments 1,000 times with replacement and calculated the KDE-score for each permutation sub-sample. Violin-plot statistics of these permuted KDE-score cohorts is shown (Fig. 5g; Extended Data Fig. 10j), and statistical difference was calculated using two-sample t-test.
Reanalysis of targeted panel cfDNA sequencing data18.
Supplementary tables S3 and S7 from Phallen et al18 were downloaded and, for each patient in table S3, the variant allele frequency (VAF) information was retrieved from table S7. Distributions were fit to the patients’ maximum VAF values using the R package “fitdistrplus” setting the ‘distr’ parameter to ‘beta’ or ‘lnorm’ as appropriate (Fig. 1b). Goodness-of-fit was determined by computing the Mean Squared Error (MSE) between the predicted distributions and the empirical cumulative distributions. The number of genomic equivalents (unique coverage) for each patient was taken from table S4. Linear regression was performed using the ‘lm’ function in R (version 3.4.2) and R2 and P value statistics were calculated (Fig. 1c).
Statistical analysis.
Statistical analysis was performed with Python 2.7.13 and R version 3.4.2. Continuous variables were compared using the Student’s t-test, Wilcoxon rank-sum test or non-parametric permutation test, as appropriate. All P values are two-sided and considered significant at the 0.05 level unless otherwise noted.
Reporting Summary.
Further information regarding research protocol and study design is available in the Life Sciences Reporting Summary associated with this article.
Extended Data
Extended Data Fig. 1. Deep targeted sequencing analysis and DNA extraction optimization.
(a-b) constitute a re-analysis of previously published data18. (a) Histogram of samples detected as a function of the maximum variant allele fraction (VAF) in stage I-IV cancer. Maximum VAF serves as an estimation of tumor fraction (TF). Patient histogram (solid bars) and empirical cumulative distribution (solid line) are shown, displaying stage dependent detection probability from 50% sensitivity in stage I cancer to >90% sensitivity in stage IV. Color coding represents different cancer stages. Number of patients considered per cancer stage is indicated in the figure. (b) Histogram of variants (mutations) detected as a function of the variant allele fraction (VAF) for different cancer stages. Higher cancer stage exhibits an increase in the mutation VAF, associated with higher probability of detection. Number of mutations included in the analysis per cancer stage is indicated in the figure. (c) Comparison of cfDNA yields across various leading commercial extraction kits. Multiple extractions (n = 3) for each kit were performed over 1mL aliquots of the same input plasma sample (from plasmapheresis of a normal donor), \and total mass (per 1mL) was determined with Qubit (ThermoFisher, Waltham, MA). Mean value and confidence interval (standard deviation) is shown. (d) Extracted cfDNA was evaluated with Nanodrop (ThermoFisher, Waltham, MA) to detect impurities such as salt and genomic DNA. Omega Bio-Tek (Norcross, GA) had the lowest amount of contaminant carry over compared to other kits. Multiple tests (n = 3, solid lines) for each kit were performed over 1mL aliquots of the same input plasma sample
Extended Data Fig. 2. Patient-specific genome-wide SNV integration and error suppression.
a) Base quality (BQ) signal for high quality germline single nucleotide polymorphisms (SNPs; n = 9,142,326) vs. low VAF (single supporting read) artifactual variants (n = 407,061) from representative PBMC sample (Pat.01), showing distinct separation between SNP and the low VAF artifacts BQ distributions (P value < 10−100, two-sample t-test), and supporting effective filtration of sequencing artifacts by BQ filtration. (b) Receiver-operating-curve (ROC) analysis for BQ filtering including germline SNPs (true labels) and low VAF artifactual variants (false labels), using the same data set as in (a), and showing high filter performance for this simple quality metric (AUC = 0.9, n = 9,142,326). (c) Variant position-in-read (PIR) shows association between low VAF artifactual variants (n = 407,061) and position at the 3’ of the sequencing read, while germline SNPs (n = 9,142,326) show uniform spread across the sequencing read length. (d) Support vector machine (SVM) classification performance between germline SNPs (random subsampling, n = 100,000) vs. low VAF artifactual variants (random subsampling, n = 100,000) from all PBMC WGS data (n = 8). Performance of SVM and random forest classification was compared over the same sample set with 10-fold cross validation. (e) Box plot of error rate estimations before error suppression (blue) and after SVM-based error suppression (red) over four cancer types and 40 PBMC-derived replicates (2 patients per cancer type, 20 replicates per patient’s PBMC sample). Error rate was calculated as the number of mismatches detected over the number of bp checked. Showing a uniform error reduction (median 14 fold-change reduction, range 11–17). (f-h) Patient-specific SNV signal-to-noise quantification over a range of TFs (10−5-10−2) compared to basal noise signal detected in control (TF = 0, subsampled PBMC DNA fragments) samples (left column). Signal-to-noise was estimated by calculating the log difference between the number of detections in each plasma-like admixture (TF > 0) and the mean number of detections in the controls (TF = 0). Analysis was done separately using tumor and matched germline (PBMC) WGS from lung (f, Pat.04), breast (g, Pat.05) and osteosarcoma (h, Pat.08) patients. Inset panel shows discrimination of tumor and control samples down to tumor fraction 10−5 after utilizing machine-learning-based sequencing error suppression (red) vs. reduced sensitivity with the raw unfiltered data (blue). (i) Benchmarking of mutation detection performance for mutation centric method23 vs. read-centric method (MRDetect). Patient-specific SNV signal-to-noise quantification over a range of TFs (10−5-2X10−1) compared to basal noise signal detected in control (TF = 0, subsampled PBMC DNA fragments) samples (right column). Signal-to-noise was estimated by calculating the log difference between the number of detections in each plasma-like admixture (TF > 0) and the mean number of detections in the controls (TF = 0). (j) Single nucleotide variant (SNV) point mutation detection in plasma mixtures with different tumor fractions (TF > 0) and controls (TF = 0) is shown. Y-axis shows the number of detections (variants observed in tumor WGS and also detected in plasma synthetic admixture) as a function of TF (x-axis). Red line constitutes the number of detections predicted for each TF based on the mutation load, coverage, noise model. Gray area represents the area under the background noise model threshold (1.5std), showing robust discrimination from noise for TF > 10-3. Analysis was done on 35X coverage lung cancer (Pat.04) admixture cohort. Centre values represent mean and error bars represent standard deviation. In (f-j), n = 11 independent admixture samples for TFs > 0 and n = 20 independently down-sampled PBMC replicates for the control (TF=0) of each patient. Throughout the figure, boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR.
Extended Data Fig. 3. Patient-specific genome-wide SNV integration provides accurate read out of tumor fraction.
(a-h) Tumor fraction (TF) inference plots using MRDetect genome-wide SNV integration for two melanomas (a: Pat.01, b: Pat.02), two lung cancers (c: Pat.03, d: Pat.04), two breast cancers (e: Pat.05, f: Pat.06), and two osteosarcomas (g: Pat.07, h: Pat.08). Each plot was generated using in silico admixtures of varying TF (range 10−5-0.2) with 35X depth of coverage by randomly downsampling and mixing tumor reads (mean coverage 97X, range 85X-110X) and germline reads (mean coverage 49X, range 43X-56X) from WGS data (see Methods; Supplementary Table 1). For TFs > 0, n = 11 independent admixture samples. For the control (TF = 0), n = 20 independently down-sampled PBMC replicates. We observed accurate TF estimation as low as 10−5, discriminated from control (TF = 0) samples (left box-plot), with high Pearson correlation (two-sided test) between the input TF mixture (x-axis) and the SNV-based estimated TF prediction, confirming accurate inference based on genome-wide mutational integration. Throughout the figure, boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR.
Extended Data Fig. 4. CNA load across tumor types.
Histogram of CNA load in WGS samples across cancer types from the TCGA cohort. Measured as a function of the size of genome altered by CNA (in log10Mb). Dashed lines represent the percentage of samples that have CNA load of over 10 Mb and 1 Gb respectively, for each cancer type. Cancer types include LUSC: Lung squamous cell carcinoma (n=50), HNSC: Head and Neck squamous cell carcinoma (n=50), CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma (n=18), OV: Ovarian serous cystadenocarcinoma (n=50), KICH: Kidney Chromophobe (n=50), COAD: Colon adenocarcinoma (n=53), THCA: Thyroid carcinoma (n=50), LUAD: Lung adenocarcinoma (N=152), ESCA: Esophageal carcinoma (n=19).
Extended Data Fig. 5. CNA based detection of ctDNA tumor fraction.
(a-c) Tumor fraction (TF) inference using MRDetect genome-wide CNA integration for representative patients, including melanoma (a: Pat.01) and lung cancer (b: Pat.03, c: Pat.04). Each plot was generated using in silico admixtures of varying TF (range 10−5 – 0.2) in 18X coverage, by randomly downsampling and mixing tumor reads (mean coverage 97X, range 85X-110X) and germline reads (mean coverage 49X, range 43X-56X) from WGS data (see Methods; Supplementary Table 1). Twenty replicates used for each TF > 0 sample, and for the control (TF = 0) samples, showing accurate TF estimation as low as 5*10−5, discriminated from control (TF = 0) samples (left box-plot), with high Pearson correlation between the input TF mixture (x-axis) and the CNA-based estimated TF prediction (two-sided test). (d-f) Tumor fraction (TF) inference in neutral regions (no copy number gain or loss in the tumor WGS data) for the same in silico admixtures (d: melanoma, Pat.01; e: lung, Pat.03; f: lung, Pat.04), shows the expected low Pearson correlation between input admixture TF and the signal (two-sided test), consistent with no expected coverage changes in the plasma admixtures in these regions. Throughout the figure, boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR.
Extended Data Fig. 6. CNA and SNV MRDetect correlation and further error suppression.
(a) Spearman correlation between SNV and CNA TF estimation across TF admixtures for a lung tumor (Pat.03) shows high correlation (two-sided test) between the two orthogonal inference methods. Red dots correspond to cancer plasma (TF > 0) samples and blue dots correspond to control plasma (TF = 0) samples. Detection threshold (dashed lines) were set on TF < 5*10−5 for both methods. Eleven replicates used for each TF > 0 sample, and 20 replicates were used for the control (TF = 0) samples. (b) Comparison to an orthogonal CNA-based TF method- ichor-CNA15. Analyzing the same cohort of breast cancer (Pat.05) in silico synthetic admixture samples shows concordance in TF estimation for high TF (TF > 5*10−3), with extension of detection for MRDetect to lower TFs. The same 20 replicates were used for both MRDetect and ichor-CNA for each of the TF > 0 and control (TF = 0) samples. (c) Proportion of variant concordant (brown) vs. discordant (gray) read pairs (R1 and R2) detected in germline SNPs and artifactual variants. Analysis was done across 10 control (benign lung lesions) plasma samples, comparing read-pairs associated with germline SNPs (right bar) vs. read-pairs associated with artifactual variants (left bar) per plasma sample. The artifactual variants were defined by read pairs with variants overlapping the union of all patient somatic SNVs compendia across all LUAD patients, that were observed with the same variant in the control plasma sample. The number of read-pairs used in the analysis is indicated above each bar. (d) Median genome-wide normalize (divided by mean coverage) coverage from matched germline PBMC WGS samples from patients with LUAD (cyan, n = 15) and control plasma WGS samples from patients with benign lung lesions (red, n = 11) before and after robust Z-score normalization. (e) Median absolute deviation (MAD) calculated over normalized-coverage (i.e., divided by mean coverage) from matched germline PBMC and control plasma WGS samples as in (d) before and after robust Z-score normalization. Throughout the figure, boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR.
Extended Data Fig. 7. Serial application of MRDetect to monitor melanoma response to immunotherapy.
Melanoma treatment response (Patient MEL02) during immunotherapy (Nivolumab) is monitored by blood samples. Upper panels- Treatment monitoring by computed tomography (CT) shows response to therapy but residual disease after 3 months of therapy. Middle panel- MRDetect Z-scores effectively track tumor responses, matching radiographic changes, in higher temporal resolution than that feasible with imaging. Lower panel- ichor-CNA captures treatment response dynamics but showing lower signal to noise ratio compared to the MRDetect method. For both MRDetect and ichor-CNA methods, Z-score is calculated from a single plasma sample for each timepoint compared to a panel of control samples (n = 30). Throughout the figure, boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR.
Extended Data Fig. 8. MRDetect performance in colorectal monitoring.
(a) Robust Z-score discrimination between signal detected across 20 random subsamplings (80% of reads per subsampling iteration) of LUAD patient pre-operative plasma (black, n = 36 patients) and the cohort of control plasma test set (gray, n = 30). The signal was measured on the subsampling set and control test set using the same patient-specific point mutation (SNV) compendium. Z-score was calculated using the noise parameters estimated in the control test cohort (see Methods). (b) Receiver-operating-curve (ROC) analysis was performed over all SNV-based Z-score values calculated on the patients’ pre-operative plasma and control plasma as in (a). (c) Cross patient noise evaluation. Robust Z-score discrimination between signal detected at 20 random subsamplings (80% of reads per subsampling iteration) of LUAD patient pre-operative plasma WGS (black, n = 36 patients), cross-patient noise estimation via application of the patient-specific compendium to all other patient pre-operative plasma (n = 35, gray). Z-score was calculated using the noise parameters estimated in the cross-patient cohort (see Methods). (d) Receiver-operating-curve (ROC) analysis was performed over all SNV-based Z-score values calculated on the matched patients and cross-patient plasma. (e) Z-score discrimination between MRDetect-CNA on LUAD patient pre-operative plasma (red, n = 36 patients) compared to signal detected in neutral regions (as a negative control, blue), control plasma test cohort (n = 30) and cross-patient cohort (n = 35). Cross-patient noise was estimated by applying the patient-specific CNA compendium to other patient plasma samples (n = 35, all other patients). Z-score was calculated using the noise parameters estimated by the control plasma cohort. (f) Receiver-operating-curve (ROC) analysis was performed over all CNA-based Z-score values calculated on the patients’ pre-operative plasma and control patients. (g) ROC analysis was performed over all ichor-CNA4 TF values calculated on the LUAD patients’ pre-operative plasma and control patients (n = 66). Interestingly the two patient plasma samples detected by ichor-CNA included events that do not appear in the tumor, one of them was found to be a PBMC specific somatic event (potentially from clonal hematopoiesis). Throughout the figure, boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR. (h) Z-score discrimination between signal detected in 20 random subsampling (80% of reads per subsampling iteration) of LUAD patient plasma WGS (n = 22 patients) collected at a median of 17 days after surgery and a cohort of control plasma test samples (gray, n = 30). The signal was measured on the matched plasma and control set using the same patient-specific point mutation (SNV) compendium. Z-score was calculated using the noise parameters estimated in the control cohort (see Methods). (i) Z-score discrimination between MRDetect-CNA on LUAD patient plasma (red, n = 22 patients) collected at a median of 17 days after surgery, compared to signal detected in neutral regions (as a negative control, blue) and control plasma cohort (n = 30). Z-score was calculated using the noise parameters estimated by the control plasma cohort (see Methods). Throughout the figure, boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR.
Extended Data Fig. 9. MRDetect performance in LUAD monitoring.
(a) Robust Z-score discrimination between signal detected across 20 random subsamplings (80% of reads per subsampling iteration) of LUAD patient pre-operative plasma (black, n = 36 patients) and the cohort of control plasma test set (gray, n = 30). The signal was measured on the subsampling set and control test set using the same patient-specific point mutation (SNV) compendium. Z-score was calculated using the noise parameters estimated in the control test cohort (see Methods). (b) Receiver-operating-curve (ROC) analysis was performed over all SNV-based Z-score values calculated on the patients’ pre-operative plasma and control plasma as in (a). (c) Cross patient noise evaluation. Robust Z-score discrimination between signal detected at 20 random subsamplings (80% of reads per subsampling iteration) of LUAD patient pre-operative plasma WGS (black, n = 36 patients), cross-patient noise estimation via application of the patient-specific compendium to all other patient pre-operative plasma (n = 35, gray). Z-score was calculated using the noise parameters estimated in the cross-patient cohort (see Methods). (d) Receiver-operating-curve (ROC) analysis was performed over all SNV-based Z-score values calculated on the matched patients and cross-patient plasma. (e) Z-score discrimination between MRDetect-CNA on LUAD patient pre-operative plasma (red, n = 36 patients) compared to signal detected in neutral regions (as a negative control, blue), control plasma test cohort (n = 30) and cross-patient cohort (n = 35). Cross-patient noise was estimated by applying the patient-specific CNA compendium to other patient plasma samples (n = 35, all other patients). Z-score was calculated using the noise parameters estimated by the control plasma cohort. (f) Receiver-operating-curve (ROC) analysis was performed over all CNA-based Z-score values calculated on the patients’ pre-operative plasma and control patients. (g) ROC analysis was performed over all ichor-CNA4 TF values calculated on the LUAD patients’ pre-operative plasma and control patients (n = 66). Interestingly the two patient plasma samples detected by ichor-CNA included events that do not appear in the tumor, one of them was found to be a PBMC specific somatic event (potentially from clonal hematopoiesis). Throughout the figure, boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR. (h) Z-score discrimination between signal detected in 20 random subsampling (80% of reads per subsampling iteration) of LUAD patient plasma WGS (n = 22 patients) collected at a median of 17 days after surgery and a cohort of control plasma test samples (gray, n = 30). The signal was measured on the matched plasma and control set using the same patient-specific point mutation (SNV) compendium. Z-score was calculated using the noise parameters estimated in the control cohort (see Methods). (i) Z-score discrimination between MRDetect-CNA on LUAD patient plasma (red, n = 22 patients) collected at a median of 17 days after surgery, compared to signal detected in neutral regions (as a negative control, blue) and control plasma cohort (n = 30). Z-score was calculated using the noise parameters estimated by the control plasma cohort (see Methods). Throughout the figure, boxplots represent median, bottom and upper quartile; whiskers correspond to 1.5 x IQR.
Extended Data Fig. 10. Imaging of sample monitored LUAD cases and fragment length analysis.
Positron emission tomography–computed tomography (PET-CT) of two patients (LUAD#3 and LUAD#6) confirms no radiographically observable metastatic spread at the time of surgery (a and c, respectively), while metastatic recurrence has been identified approximately six months post-operative by PET-CT (b and d, respectively). (e-h) Representative fragment size histograms showing the distribution of DNA fragments as a function of the fragment length. DNA fragments that are associated with tumor mutations (gray) are showing significantly shorter size compared to DNA fragments that are associated with artifactual non patient-specific detections (red, derived from applying cross-patient mutational compendia; median P value < 10−3, two-sample t-test). Plots include two pre-operative plasma samples (e: LUAD#18, tumor mutation detection n = 35, artefactual detection n = 18802 ; f: LUAD#31, tumor mutation detection n=82, artefactual detection n = 32530) and matching plasma samples after surgery (g: LUAD#18, tumor mutation detection n = 55, artefactual detection n = 22737; h: LUAD#31, tumor mutation detection n = 27, artefactual detection n = 14610). (i) Kernel-density-estimator (KDE) trained to discriminate between tumor-derived (human aligned reads from a patient derived xenograft model, see Methods, blue) and normal-derived (from control plasma samples, orange) cfDNA based on the fragment size signature. The log difference between the tumor and normal density functions (black solid line) was used as a score function that integrates the fragment size shift signal across the entire fragment sizes distribution (80bp-600bp). (j) Pre-operative cfDNA showed significant shift (two-sample t-test) in their tumor-specific mutation detections in the patient plasma (red, n=563), compared to non-tumor (cross-patient) detected mutations in the same samples (blue, n=4184). Violin plots depict kernel density estimates of the density distribution. Center dashed lines represent the median and dashed lines represent the interquartile range.
Supplementary Material
Supplementary Table 1. Patient clinical information and sample QC for the in silico admixture cohort.
Supplementary Table 2. Clinical information of samples and patients for control, melanoma, CRC, and LUAD cohorts.
Supplementary Table 3. Sample sequencing details, metrics, and QC for control, melanoma, CRC, and LUAD cohorts.
Supplementary Table 4. MRDetect SNV and CNA detection metrics for CRC and LUAD cohorts.
Supplementary Table 5. Survival Analysis for CRC and LUAD cohorts.
Acknowledgments
We thank the Landau lab and the NYGC computational biology and sequencing teams for help and feedback throughout this work. A.Z. is supported by an EMBO long-term fellowship (ALTF 140-2016). D.A.L. is supported by the Burroughs Wellcome Fund Career Award for Medical Scientists, Pershing Square Sohn Prize for Young Investigators in Cancer Research, and the National Institutes of Health (NIH) Director’s New Innovator Award (DP2-CA239065). This work was also supported by the Mark Foundation ASPIRE Award, the American Lung Association Cancer Discovery Award, the Daedalus Fund for Innovation and the Meyer Cancer Center.
Competing interests
D.A.L., A.Z., V.A. and S.T.K.H. submitted two patent applications. D.A.L. and A.Z. are co-founders of C2i Genomics. D.A.L. participated in an advisory board for Illumina, Inc and has received research support. J.D.W. consulted for Adaptive Biotech; Advaxis; Amgen; Apricity; Array BioPharma; Ascentage Pharma;Astellas; Bayer; Beigene; Bristol Myers Squibb; Celgene; Chugai; Elucida; Eli Lilly; F Star; Genentech; Imvaq; Janssen; Kleo Pharma; Kyowa Hakko Kirin; Linneaus; MedImmune; Merck; Neon Therapuetics; Northern Biologics; Ono; Polaris Pharma; Polynoma; Psioxus; Puretech; Recepta; Takara Bio; Trieza; Turvax; Sellas Life Sciences; Serametrix; Surface Oncology; Syndax; Syntalogic. JDW also received research support from Bristol Myers Squibb; Medimmune; Merck Pharmaceuticals; Genentech. Equity in: Potenza Therapeutics; Tizona Pharmaceuticals; Adaptive Biotechnologies; Elucida; Imvaq; Beigene; Trieza and Linneaus.
Footnotes
Code availability:
The analytic code used for this work is provided for non-commercial use at: https://ctl.cornell.edu/technology/mrdetect-licence-request
Data availability
Submission of all data described in this work is pending at the European Genome-Phenome Archive.
References
- 1.Wan JCM et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Cohen JD et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadeh R et al. ChIP-seq of plasma cell-free nucleosomes identifies cell-of-origin gene expression programs. doi: 10.1101/638643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moss J et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun 9, 5068 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen SY et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Snyder MW, Kircher M, Hill AJ, Daza RM & Shendure J Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell vol. 164 57–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cristiano S et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S et al. Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin. Cancer Res 16, 1324–1330 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi S et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med 352, 786–792 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Adalsteinsson VA et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun 8, 1324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murtaza M et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Diehl F et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med 14, 985–990 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sozzi G et al. O-297 Quantification of free circulating DNA as a diagnostic marker in lung cancer. Lung Cancer vol. 41 S86–S87 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Bettegowda C et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med 6, 224ra24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y et al. Prognostic Potential of Circulating Tumor DNA Measurement in Postoperative Surveillance of Nonmetastatic Colorectal Cancer. JAMA Oncol (2019) doi: 10.1001/jamaoncol.2019.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Wezel EM et al. Whole-genome sequencing identifies patient-specific DNA minimal residual disease markers in neuroblastoma. J. Mol. Diagn 17, 43–52 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Abbosh C et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phallen J et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman AM et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nature Medicine vol. 20 548–554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman AM et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol 34, 547–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennedy SR et al. Detecting ultralow-frequency mutations by Duplex Sequencing. Nature Protocols vol. 9 2586–2606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell BB et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 171, 1042–1056.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cibulskis K et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spinella J-F et al. SNooPer: a machine learning-based method for somatic variant identification from low-pass next-generation sequencing. BMC Genomics 17, 912 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor AM et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 33, 676–689.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinert T et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol (2019) doi: 10.1001/jamaoncol.2019.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim CG et al. Effects of microsatellite instability on recurrence patterns and outcomes in colorectal cancers. Br. J. Cancer 115, 25–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chida K et al. Spontaneous regression of transverse colon cancer: a case report. Surgical Case Reports vol. 3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karakuchi N et al. Spontaneous regression of transverse colon cancer with high-frequency microsatellite instability: a case report and literature review. World J. Surg. Oncol 17, 19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mouliere F et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang P et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc. Natl. Acad. Sci. U. S. A 115, E10925–E10933 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang P et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. U. S. A 112, E1317–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bauml J & Levy B Clonal Hematopoiesis: A New Layer in the Liquid Biopsy Story in Lung Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research vol. 24 4352–4354 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Razavi P et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med 25, 1928–1937 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martincorena I et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yizhak K et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salk JJ et al. Ultra-Sensitive TP53 Sequencing for Cancer Detection Reveals Progressive Clonal Selection in Normal Tissue over a Century of Human Lifespan. Cell Reports vol. 28 132–144.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstraw P et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J. Thorac. Oncol 11, 39–51 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Systemic therapy in resectable non-small cell lung cancer. in UpToDate (ed. Post TW) (UpToDate, 2019). [Google Scholar]
References (Methods)
- 40.Fox EJ & Reid-Bayliss KS Accuracy of Next Generation Sequencing Platforms. Journal of Next Generation Sequencing & Applications vol. 01 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.TruSeq DNA PCR-free Library Preparation Protocol. https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_truseq/truseq-dna-pcr-free-workflow/truseq-dna-pcr-free-workflow-reference-1000000039279-00.pdf.
- 42.Guerrera F et al. The Influence of Tissue Ischemia Time on RNA Integrity and Patient-Derived Xenografts (PDX) Engraftment Rate in a Non-Small Cell Lung Cancer (NSCLC) Biobank. PLoS One 11, e0145100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang H, Lei R, Ding S-W & Zhu S Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novocraft. http://www.novocraft.com.
- 46.Data Sciences Platform @ Broad Institute. GATK; | Home. https://software.broadinstitute.org/gatk/. [Google Scholar]
- 47.Bergmann EA, Chen B-J, Arora K, Vacic V & Zody MC Conpair: concordance and contamination estimator for matched tumor–normal pairs. Bioinformatics vol. 32 3196–3198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saunders CT et al. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics vol. 28 1811–1817 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Wilm A et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Research vol. 40 11189–11201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xi R, Luquette J, Hadjipanayis A, Kim T-M & Park PJ BIC-seq: a fast algorithm for detection of copy number alterations based on high-throughput sequencing data. Genome Biol. 11, O10 (2010). [Google Scholar]
- 51.Kothen-Hill Steven T., Zviran Asaf, Schulman Rafael C., Deochand Sunil, Gaiti Federico, Maloney Dillon, Huang Kevin Y., Liao Will, Robine Nicolas, Omans Nathaniel D., Landau Dan A.. Deep learning mutation prediction enables early stage lung cancer detection in liquid biopsy. ICLR 2018 (2018). [Google Scholar]
- 52.Data portal | 1000 Genomes. http://www.internationalgenome.org/data-portal/sample/NA12878. [Google Scholar]
- 53.Hadi K et al. Novel patterns of complex structural variation revealed across thousands of cancer genome graphs. doi: 10.1101/836296. [DOI] [PMC free article] [PubMed]
- 54.Underhill HR et al. Fragment Length of Circulating Tumor DNA. PLoS Genet. 12, e1006162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Patient clinical information and sample QC for the in silico admixture cohort.
Supplementary Table 2. Clinical information of samples and patients for control, melanoma, CRC, and LUAD cohorts.
Supplementary Table 3. Sample sequencing details, metrics, and QC for control, melanoma, CRC, and LUAD cohorts.
Supplementary Table 4. MRDetect SNV and CNA detection metrics for CRC and LUAD cohorts.
Supplementary Table 5. Survival Analysis for CRC and LUAD cohorts.
Data Availability Statement
Submission of all data described in this work is pending at the European Genome-Phenome Archive.