Abstract
Given the highly polymorphic nature of Human Leukocyte Antigen (HLA) molecules, it is not surprising that they function as key regulators of the host immune response to almost all invading pathogens, including SARS-CoV-2, the etiological agent responsible for the recent COVID-19 pandemic. Several correlations have already been established between the expression of a specific HLA allele/haplotype and susceptibility/progression of SARS-CoV-2 infection and new ones are continuously emerging. Protective and harmful HLA variants have been described in both mild and severe forms of the disease, but considering the huge amount of existing variants, the data gathered in such a brief span of time are to some extent confusing and contradictory. The aim of this mini-review is to provide a snap-shot of the main findings so far collected on the HLA-SARS-CoV-2 interaction, so as to partially untangle this intricate yarn. As key factors in the generation of antigenic peptides to be presented by HLA molecules, ERAP1 and ERAP2 role in SARS-CoV-2 infection will be revised as well.
Keywords: SARS-CoV-2, HLA, ERAPs, Polymorphisms
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MHC, major histocompatibility complex; HLA, between Human Leukocyte Antigen; ERAP, endoplasmic reticulum aminopeptidase; WHO, World Health Organization; COVID-19, coronavirus disease 2019; MERS-CoV, Middle East respiratory syndrome coronavirus; ACE2, angiotensin-converting enzyme-2 receptor; ARDS, acute respiratory distress syndrome; IRAP, insulin-regulated aminopeptidase; RDB, receptor-binding domain; TMPRSS2, type II transmembrane serine protease; TAP, transporter associated with antigen processing; GWAS, genome-wide association study; SNP, single nucleotide polymorphism, NMD, nonsense-mediated-decay; ERGIC, ER Golgi Intermediate Compartment; ER, endoplasmic reticulum; CTL, Cytotoxic T Lymphocytes
1. Introduction
Infectious diseases are still a significant challenge for public health worldwide, as they are responsible for millions of deaths, mainly, but not only, among older adults and immunosuppressed or chronically ill people. The virulence of the pathogen and the efficacy of the host immune response are the key factors conditioning the onset and progression of infectious diseases. In the context of adaptive immunity, a pivotal role in protection and recovery from infections is played by CD8+ T lymphocytes [1]. These cells detect antigenic peptides bound by major histocompatibility complex (MHC) class I molecules, known as the human leukocyte antigen (HLA) class I in humans, and work through different pathways in order to eradicate the pathogen and the infected cells.
Given the major role played by CD8+ T cells in host immunity, it is conceivable that plenty of studies have tried to decipher the consequences of a jam in CD8+ T cell activation in their response to pathogens. In this setting, since the very first report establishing a correlation between HLA-B27 and Ankylosing Spondylitis [2], [3], the MHC has been recognized as the region of the genome that is associated with the highest number of human diseases [4]. For this same reason, allelic variants and altered MHC expression have been associated with disease severity following infection with several microbes. In parallel, aminopeptidases – in particular endoplasmic reticulum aminopeptidase 1 (ERAP1), ERAP2 and partly insulin-regulated aminopeptidase (IRAP) – have drawn the scientific attention as well. Indeed, they are responsible for antigenic peptide trimming within the ER, thus conditioning the antigen processing pathway [5] in both physiological and pathological contexts including those mediated by infectious agents, among which severe acute respiratory disease coronavirus 2 (SARS-CoV-2) [6].
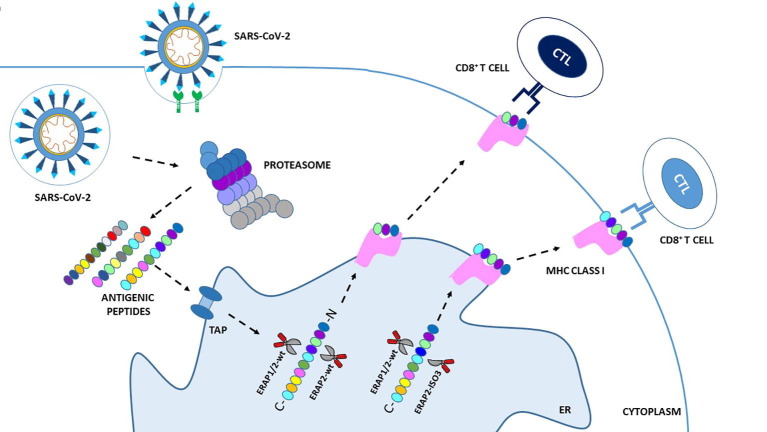
Since SARS-CoV-2 outbreak, the pathogen responsible for COronaVIrus Disease 19 (COVID-19), the scientific community has tried to identify those factors controlling the susceptibility/outcome of the disease and playing a major role in determining the appearance of acute respiratory distress syndrome (ARDS), the life-threatening form of infection [7]. This virus belongs to the family of Coronaviruses, responsible for other two epidemics over the last twenty years: SARS-CoV (Severe Acute Respiratory Syndrome) in Asia in 2003 and MERS-CoV (Middle East Respiratory Syndrome Coronavirus) in Arabian Peninsula in 2012. As an obligate intracellular parasite, SARS-CoV-2 replicates inside the host cells exploiting nucleic acid and protein synthesis mechanisms to facilitate its spreading from one individual to another. The biology of the virus, its infectious and replicative cycle, as well as the human host factors directly or indirectly contributing to its maintenance/annihilation within cells have been exhaustively described elsewhere [8], [9]. Throughout these phases, viral proteins can be unfolded, degraded and further processed by cytosolic and nuclear proteasomes inside host cells; the resulting peptides, 8–16 amino acid long, are then transported into the endoplasmic reticulum (ER) by transporter associated with antigen processing (TAP). Herein, they are further trimmed by ERAP1 and ERAP2 proteins in order to achieve the optimal length to be loaded onto the clefts of MHC class I molecules. In turn, MHC class I molecules, by loading intracellular compartment-derived antigens, provide a recapitulation of the events occurring inside the cell, thus allowing CD8+ T cells to monitor possible ongoing infections (Fig. 1 ).
Fig. 1.
Schematic overview of the MHC-I antigen processing and presentation pathway in SARS-CoV-2 infection. Following SARS-CoV-2 infection rs2248374-A ERAP2 expressing cells produce wild type ERAP2 (ERAP2-wt) which can homodimerize or heterodimerize with ERAP1-wt (ERAP2-wt + ERAP2-wt; ERAP1-wt + ERAP2-wt), in order to process viral antigens to be presented on cell surface for recognition by specific CD8+ cytotoxic T lymphocyte (CTL) clones. Rs2248374-G ERAP2 expressing cells may also produce an alternative spliced isoform: ERAP2-ISO3. This variant, unlike ERAP2-wt, lack the catalytic domain but can still heterodimerize with both ERAP2-wt and ERAP1-wt. As a result, these unconventional heterodimers (ISO3 + ERAP2-wt; ISO3 + ERAP1-wt) may process viral antigens differently from the canonical ones, generating an alternative antigenic repertoire. This in turn may activate other CTL clones possibly triggering a more or less protective immune system response. ER: Endoplasmic reticulum; TAP:
Given the direct participation of ERAP proteins in the antigen presentation pathway of MHC class I, this manuscript primarily aims to review the data correlating antigen presentation by MHC class I molecules and COVID-19 susceptibility/severity so far reported in the scientific literature by in silico and genetic-association studies. Noteworthy, data concerning the role displayed by MHCII and non-classical HLA in SARS-CoV-2 infection will be reported as well. Although this topic has already been reviewed by other authors [10], [11], due to the rising global attention on the current pandemic, researchers are rapidly accumulating an incredible amount of data and an updated revision of the literature is constantly necessary.
2. Antigen presentation by HLA class I in SARS-CoV-2 infection
HLA are highly polymorphic molecules that play a key role in individual genetic susceptibility to human diseases including those provoked by infectious agents [6], [12]. For example, in HIV-1 infection different HLA – mainly HLA-A*29, HLA-B*27, HLA-B*35 and HLA-B*57 – were shown to be correlated with both susceptibility/progression of the disease [13]. Notably, as reviewed in early June 2020 in a work by Ovsyannikova and colleagues, a collection of data provides evaluations of the role of host genetics - including variation in HLA genes - in the immune response to Coronaviruses, among others SARS-CoV-2 [14]. In contrast to what had emerged in studies concerning SARS-CoV [15], [16] and MERS-CoV [17], [18], correlations between SARS-CoV-2 and HLA are still not entirely defined and results so far obtained are sometimes contradictory. Herein we report the so far collected data on this issue (Table 1 ).
Table 1.
HLA variants involved in susceptibility to SARS-CoV-2 infection and/or in disease severity.
| HLA variants | Associated to | Mechanism of action |
|---|---|---|
| Genetic association on case-control and cohort studies | ||
| HLA‐A*24:02 | SARS-CoV-2 susceptibility | Unknown [19] |
| HLA-A*02:01 | Increased risk for severe COVID-19 outcome | Lower capacity to present SARS-CoV-2 antigens [20] |
| HLA-A*11:01, HLA-A*24:02 | Protection against COVID-19 | More efficient T cell-mediated antiviral responses to SARS-CoV-2 [20] |
| HLA-B*22 | Susceptibility marker for SARS-CoV-2 | Unknown [21] |
| HLA-A*01:01 g-B*08:01 g-C*07:01 g-DRB1*03:01 g | Susceptibility marker of SARS-CoV-2 infection and severe COVID-19 outcome | Unknown [22] |
| HLA-A*02:01 g-B*18:01 g-C*07:01 g-DRB1*11:04 g | Protection against SARS-CoV-2 infection | Unknown [22] |
| HLA-B*27:07, HLA-DRB1*15:01, HLA-DQB1*06:02 | Worst COVID-19 outcome | Unknown [23] |
| HLA-DRB1, HLA-DQB1 | Higher susceptibility to COVID-19 | Unknown [24] |
| HLA-B*15:03 | Severe evolution of COVID-19 | Unknown [25] |
| HLA-A*25:01 | Moderate evolution of COVID-19 | Unknown [25] |
| HLA-A*32 | Higher frequency in healthy controls | Unknown [26] |
| HLA-B*39, HLA-C*16 | More represented in COVID-19 patients | Unknown [26] |
| HLA-C*05 | Higher mortality | Unknown [27] |
| HLA-DRB1*08 | Higher risk of and death | Unknown [36] |
| HLA-E*0101 | High severity of COVID-19 | Lower NKG2C + NK cell response [66] |
| Bioinformatic in silico epitope prediction studies | ||
| HLA‐B*15:03 | Protection against SARS-CoV-2 | High presentation of SARS-CoV-2 immunogenic epitopes [47] |
| HLA‐B*46:01 | Severe symptoms | Reduced presentation of SARS-CoV-2 peptides [47] |
| HLA-A*02:02, HLA-A*11:01, HLA-B*40:01, HLA-B*35:01 | Lower risk for severe COVID-19 | High capacity to present SARS-CoV-2 antigens [49] |
| HLA‐DRB1*01 | Fatality rate in hospitalized patients | Unknown [51] |
| HLA-A*01:01, HLA-A*02:01, HLA-A*03:01, HLA-A*23:01, HLA-A*24:02, HLA-A*26:01, HLA-A*30:02, HLA-A*31:01, HLA-A*68:01, HLA-B*07:02, HLA-B*18:01, HLA-B*35:03, HLA-B*38:01, HLA-B*44:02, HLA-B*44:03, HLA-B*51:01, HLA-C*05:01, HLA-C*07:01, HLA-C*07:02, HLA-C*08:02, HLA-C*15:02, HLA-C*17:01 | Protection against COVID-19 | Strong binding to SARS-CoV-2 peptides [52] |
2.1. Genetic associations on case-control and cohort studies
Because of the relatively recent occurrence of SARS-CoV-2, quite limited published studies have explored the relative frequencies of HLA alleles in case-control, cohort, and observational studies. The results so far obtained are not univocal and limited by relatively small sample size and by heterogeneity in study design. Moreover, in most cases the reported results are merely associative and the mechanism of action possibly displayed by different HLA alleles in SARS-CoV-2 antigen presentation remains elusive. Nonetheless, comparison with previous coronavirus infections and correlation with clinical symptoms may provide essential information prior to a specific mandatory multicenter study to be performed, in the attempt to identify biomarkers of susceptibility/progression of SARS-CoV-2 infection.
One of the first studies in the field documented HLA‐A*24:02 to be correlated with SARS-CoV-2 susceptibility. This allele was found to be expressed in four out of five patients from Wuhan, during the first stages of the pandemic [19]. HLA-A*24:02 frequency in Chinese population is typically 17.2%, a value that is significantly lower than the observed frequency (80%) reported in the infected patients enrolled in the study. As it is conceivable, considering the extremely small sample size, the study lacks statistical power and requires validation in larger sample cohorts. However, soon after the publication of these results, Tomita et al. reported an association between HLA-A*02:01 and an increased risk for COVID-19. HLA-A*02:01, indeed, showed a relatively lower capacity to present SARS-CoV-2 antigens compared with other frequent HLA class I molecules, mainly HLA-A*11:01 or HLA-A*24:02. Therefore, the authors suggest that subjects carrying HLA-A*11:01 or HLA-A*24:02 genotypes may trigger a more efficient T cell-mediated antiviral responses to SARS-CoV-2 compared to HLA-A*02:01 [20]. A subsequent study conducted by Yung et al. determined a positive association between HLA-B*22 serotype with SARS-CoV-2 susceptibility in 190 Hong Kong Chinese patients [21].
In order to define the HLA haplotypes associated with susceptibility to COVID-19 disease, a group of Italian researchers – through a geographical epidemiological analysis – focused on describing the pattern of distribution of the two most common HLA haplotypes (HLA-A*01:01g-B*08:01g-C*07:01g-DRB1*03:01g and HLA-A*02:01g-B*18:01g-C*07:01g-DRB1*11:04g) in the Italian population [22]. They discovered that the huge incidence of infection and mortality rate in the northern regions of Italy correlates with high frequency of HLA-A*01:01g-B*08:01g-C*07:01g-DRB1*03:01g haplotype, suggesting that such haplotype is a potential ‘susceptibility’ marker of the disease. Contrariwise, a lower incidence and mortality for COVID-19 were observed in the central-southern regions of the country, where higher frequency values of the HLA-A*02:01g-B*18:01g-C*07:01g-DRB1*11:04g were reported, allowing to speculate on a defensive mechanism towards SARS-CoV-2 infection elicited in subjects carrying this haplotype. Other possible explanations to such a differing mortality rate from north to south of Italy – e.g. climatic differences, migration and pollution – were considered by the authors. Nevertheless, none of these factors seemed to be that significant. Indeed, no substantial climatic differences distinguish the northern and southern regions, and atmospheric emissions of PM10, PM2.5 and NO2 are actually higher in the southern areas. Finally, the uncontrolled north–south exoduses, which took place just before the beginning of the lockdown, did not result into the spreading of the infection in the southern Italian regions as expected.
Another study conducted by Novelli and colleagues [23], on a small sample of 99 Italian patients affected by a severe or extremely severe form of COVID‐19, investigated the HLA allele frequency distribution, in order to identify variants possibly associated to a worst COVID-19 outcome. Despite the restricted sample size, the researchers found a strong correlation for HLA B*27:07, DRB1*15:01, DQB1*06:02 alleles after comparing the results to a reference group of 1017 Italian individuals. Notably, these data are in line with those published by Kachuri et al. recently identifying DRB1 and DQB1 as key genetic factors controlling host susceptibility to viral infections [24]. Even more recently, HLA class I typing was performed within a pilot study on 45 Spanish patients with different COVID-19 symptoms severity [25]. The results obtained suggest that patients exhibiting a mild form of the disease presented HLA class I molecules characterized by a higher binding affinity to SARS-CoV-2 peptides and showed a higher percentage of heterozygous HLA molecules compared to patients exhibiting moderate and severe symptoms. In addition, the authors stressed the fact that theoretically protective alleles of HLA, such as HLA-B*15:03, were found in patients who died because of a severe evolution of the disease; while, on the other hand, alleles showing low affinity for the viral peptides, such as HLA-A*25:01, were present in patients with a moderate evolution of the disease. Their observation suggests that studying the affinity of the entire HLA genotype for SARS-CoV-2 may be more fruitful than focusing on the specific positive or negative roles of different HLA alleles.
Lorente et al. analysed a total of 3886 healthy controls and 72 COVID-19 patients (10 non-survivor and 62 survivor patients at 30 days) and showed that there was a higher frequency of HLA-A*32 alleles in healthy controls than in COVID-19 patients. conversely, HLA-B*39 and HLA-C*16 were more represented in COVID-19 patients compared to healthy controls. However, the correlation did not reach statistical significance after correction for multiple parameters [26].
Sakuraba et al., by analyzing the frequency of HLA allele in 74 countries from the Allele Frequency Net Database and worldometer.info, in order to investigate the association between class I MHC, HLA-A, -B and -C, and the risk of death due to SARS-CoV-2 infection, found HLA-C*05 allele to be potentially correlated with mortality at a global level [27]. HLA-C molecules work as killer cell immunoglobulin-like receptor (KIR) ligands [28]. The latter are highly polymorphic receptors expressed on natural killer (NK) and T-cells membranes and are distinguished into activating or inhibitory ones, depending on the length of their cytoplasmatic tail [29]. KIRs indeed control the inhibition and activation of cell responses by recognizing polymorphic motifs on HLA I molecules (i.e. HLA-A*03, HLA-A*11, HLA-Bw4, HLA-C1, HLA-C2) expressed on target cells, thus playing a crucial role in regulating the innate immune defense against cancerous cells, adaptive immune responses as well as viral infections [30]. An example is represented by the interaction between KIR3DL1/S1 and HLA-Bw4 and different outcome of HIV-infection as reviewed in [31]. In Sakuraba work, the authors hypothesized that this HLA-KIR combination could lead to immune over-activation, subsequently causing negative selection. Indeed, populations with the highest mortalities (France, Italy and Spain) were demonstrated to present the greatest number of carriers for HLA-C*05 and its receptor KIR2DS4fl. Therefore, patients with a HLA-C*05 and KIR2DS4fl pair may be predisposed to develop an excessive cytokine response and suffer from hyper-cytokinemia, strongly associated to severe forms of the disease and COVID-19 mortality [27]. Incidentally, the central role of HLA-C in virus immune-escape has already been documented, as reported for example by Fredj and colleagues in a study showing an increase in human herpesviruses (HHV) risk of infection in KIR2DL2 and HLA-C1 positive multiple sclerosis (MS) patients [32]. HLA-C1 molecules may induce inhibitory signals in KIR2DL2 positive NK cells from these patients, thus creating an anergic environment with very low levels of IFNγ and consequent lack of NK cell activation and of innate protection to virus infection [33]. Such correlation was more recently associated with an increased susceptibility to HHV-6A infection in patients with a severe Alzheimer’s Disease (AD) status [34].
As was to be expected, a more complete picture of the features of the immune response to SARS-CoV-2 was gained through large-scale genome-wide association studies (GWAS) and biological validation studies based on larger cohorts of patients. Ellinghaus et al. performed a GWAS on 1980 COVID-19 patients enrolled in Spain and Italy. The authors found an association between two regions in the human genome and the virus-induced respiratory failure [35]. One of them is located on chromosome 3 in an area which includes six genes, namely SLC6A20, LZTFL1, FYCO1, CXCR6, XCR1, CCR9. Some of them encode chemokine receptors, while SLC6A20 is translated into a protein with transportation functions, which has been demonstrated to interact with ACE2. The other identified locus is situated within the ABO blood groups locus on chromosome 9, assigning to blood type O a protective role against the disease and, on the other hand, linking group A to a more severe form of the disease. This finding is consistent with other studies reporting similar results [36], [37], [38]. However, any link between HLA alleles and disease susceptibility or severity was found in Ellinghaus’ study. As genetic hyper-polymorphism across MHC locus differentiates thousands of HLA alleles and the clinical manifestations may significantly differ between affected patients, it is possible that the sample size analyzed in this study was not large enough to identify any statistically significant association.
The role of MHC class II molecules, which intervene in antigen presentation to helper CD4+ T cells to facilitate the humoral immune response, has been investigated as well, in relation to SARS-CoV-2 infection. Some of the results obtained have already been reported in the above paragraphs [22], [23], however the most encouraging findings seem to be related to HLA-DR expression levels, as its dowregulation – mainly in monocytes – is often associated with a dysregulated immune response, [39], [40] even in SARS-CoV-2 infection as recently reviewed in [41].
In this perspective, Giamarellos-Bourboulis et al. [42] performed a well-designed study on 54 COVID-19 patients, who showed hyper-inflammatory reactions in the form of either macrophage activation syndrome (MAS) or immune dysregulation. The latter was characterized by lower expression of HLA-DR on CD14+ monocytes, secondary to monocyte hyperactivation, excessive release of interleukin-6 (IL-6), and severe lymphopenia. The leading hypothesis was that IL-6 was responsible for the reduced level of HLA-DR on CD14+ monocytes. The increase of circulating HLA-DR+ cells during the healing period of one patient with a moderately severe infection from SARS-CoV-2 further endorsed this hypothesis [43]. In conclusion, they identified a new feature of immune dysregulation in SARS-CoV-2 patients, which supports the rationale of clinical trials – which were ongoing at the time this work was published – based on the use of Anakinra, Sarilumab, Siltuximab, and Tocilizumab to hamper the production of inflammatory cytokines in these patients. In another study conducted by Amoroso and colleagues [36], they analyzed HLA-A, B, and DRB1 frequencies on a sample of 40 904 individuals (32294 transplant recipients and 8610 waitlisted patients) and found a higher frequency of HLA-DRB1*08 in COVID-19 patients and a significant correlation with an increased risk of death. Consistently, the peptide binding prediction analyses demonstrated that the DRB1*08 allele is unable to bind any viral peptide with high affinity. Although the obtained data needs further confirmation, the present study provides promising results, especially if the consistency with previous works is taken into account.
2.2. Bioinformatic in-silico epitope prediction studies
Because of the highly polymorphic nature of HLA molecules and the limited quantity of biological data gathered in roughly a year of pandemics, some authors focused on the use of predictive algorithms to find which HLA alleles are associated with viral peptide epitope recognition [44], [45], [46]. In particular, Nguyen et al. analyzed peptides from SARS‐CoV‐2 proteome across more than a hundred HLA I alleles with the aim of mapping susceptibility loci for COVID‐19 [47]. Results of these analyses showed that HLA‐B*15:03 is highly capable of presenting peptides from SARS‐CoV‐2, suggesting a possible protective role for this allele against SARS-CoV-2 infection. On the other hand, HLA‐B*46:01 was predicted to bind to the fewest number of peptides from the virus, suggesting that immune response in carriers of this allele may be weaker, resulting in more severe symptoms. Consistently with that, HLA‐B*46:01 has already been reported to be significantly associated with the severity of SARS-CoV infection in Asian populations [48].
A recent study by La Porta and colleagues compared the different binding affinities between coronavirus-derived peptides and a series of HLA class I molecules for SARS-CoV-2, SARS-CoV, and HCoV-OC43. The first two share 80% of the genome and are both responsible of the potential onset of severe symptoms following infection, while HCoV-OC43 is a coronavirus associated with mild respiratory symptoms [49]. Specifically, using two epitope prediction algorithms – both based on artificial neural networks – the authors evaluated binding affinities between SARS-CoV-2 peptides and 79 HLA class I molecules; results were compared with the ones from analogous predictions made for SARS-CoV and HCoV-OC43. What emerged was a strong similarity in the binding patterns for SARS-CoV-2 and SARS-CoV. Importantly, HCoV-OC43 was characterized by peptides with a stronger HLA binding ability compared to peptides belonging to the other two viruses. The authors individuated two sets of haplotypes respectively correlated to weak and strong binding capacity towards SARS-CoV-2 peptides, the latter including HLA-A*02:02, HLA-A*11:01, HLA-B*40:01 and HLA-B*35:01. They then investigated the heterogeneous responses to SARS-CoV-2 infection in human populations by measuring the prevalence of the haplotypes in different human populations, finding that strongly binding haplotypes are more represented in Asian populations. Despite no clinical or immunological consequences related to this haplotypes were investigated in this work, these data could be relevant to study the diffusion of the disease across the world and could represent the basis to develop individualized tests in order to identify the immune susceptibility to COVID-19 among different populations. This study may be considered as another significant step towards the possibility to perform population screening and to predict individual severity scores of infection, in order to develop personalized therapeutic strategies against COVID-19.
A recent report described peptide-binding affinities between 438 HLA class I and class II proteins and the proteomes of seven pandemic viruses, including coronaviruses, influenza viruses and the immunodeficiency virus [50]. In this work HLA alleles were examined in relation to peptide-binding affinities and then grouped into four categories, namely strong, regular, weak or nonbinding, based on the different kind of affinity they show towards various peptides. Notably the authors observed that the frequencies of the strongest and weakest HLA molecules are influenced by geographical location. Indeed, among native Americans the frequencies are higher for the strongest and lower for the weakest HLA binders, possibly as a consequence of previous selective pressure applied by historical infectious agents. However, results demonstrated that the majority of HLA proteins are not specific binders of SARS-CoV-2 peptides, as they bind viral peptides in an aspecific way.
Romero-Lòpez et al. performed a bioinformatic prediction of which epitopes of the SARS‐CoV‐2 spike protein are significantly immunogenic and could be presented by HLA Class I and II in different populations [51]. They also established an ecological correlation of HLA allele frequency with the predicted fatality rate in hospitalized patients of 28 states in Mexico. The only negative significant correlation observed was between the frequency of HLA‐DRB1*01 and the fatality rate in hospitalized patients in Mexico. Remarkably, this correlation was weak, suggesting that other key factors, apart from HLA, could be involved in COVID-19 outcome and further experimental studies are needed to reinforce these results.
Using a different approach based on panHLA analysis, Campbell et al. were able to recognize 368,145 unique combinations of peptide-HLA complexes (pMHCs) with a strong binding affinity and an overlap between class I and II predicted pMHCs [44]. Though highly informative this kind of approach surely presents some limitations: 1) the analysis focused on pMHC complexes wizth predicted binding affinities of less than 500 nM, which could lead to underestimate the number of alleles correlated to the predicted antigenic peptides; 2) the research was restricted to 9-mers and 15-mers, which represent the length of most but not all reported HLA class I and class II binding peptides; 3) the data does not provide any measure of the quantity and timing of viral protein expression in host cells; and 4) the research of global population frequencies was conducted only on a restricted number of HLA alleles and countries. Overall, however, this valuable pan-HLA approach allowed identification of new possible interactions between HLA molecules and peptides, while leading to the recognition of other peptides from less prevalent HLA types. Besides, the overlap between class I and II predicted pMHCs makes conceivable that some epitopes may be presented to both CD4+ and CD8+ T lymphocytes.
De Moura et al. described 24 epitopes derived from the SARS-CoV-2 S protein that could interact with 17 different MHC-I alleles in the Brazilian population [52]. These epitopes can elicit an effective CD8+ T cells immune response and may be useful to develop strategic methods for vaccines against COVID-19. The immunoinformatic approach reveals that the protective MHC class I alleles include HLA-A*01:01, HLA-A*02:01, HLA-A*03:01, HLA-A*11:01, HLA-A*23:01, HLA-A*24:02, HLAA*26:01, HLA-A*30:02, HLA-A*31:01, HLA-A*68:01, HLA-B*07:02, HLA-B*18:01, HLA-B*35:01, HLA-B*35:03, HLA-B*38:01, HLA-B*44:02, HLAB*44:03, HLA-B*51:01, HLA-C*05:01, HLA-C*07:01, HLA-C*07:02, HLA-C*08:02, HLA-C*15:02 and HLA-C*17:01.
Since the very first identification of allele specific motifs for T-cell antigens by HLA class I molecules, in 1991, epitope identification and characterization has been significantly simplified by in silico prediction strategies [53]. Indeed, many ongoing vaccines designs, targeting infectious pathogens, have been exploiting prediction algorithms, whose accuracy is progressively improving. Nevertheless, these tools present some limitations, mainly consisting in: differences between the real and the Protein Data Bank structure, accuracy of the methods for simulate proteasome cleavage, imperfections in the reproduction of molecular modelling, docking and dynamics and others, which have been recently revised in [54]. Such considerations should be taken into account to properly use the innovative programs for T-cell immunogen design for epitope-based therapies and future epidemiological investigations.
3. Antigen presentation by non-classical HLA in SARS-CoV-2 infection
HLA-G and HLA-E are both non classical molecules with tolerogenic and immunosuppressive properties which influence the onset of autoimmune and infectious diseases [55], [56], [57]. In particular, HLA-G was shown to be upregulated following HIV, HCMV and HCV infections leading to the suggestion that this could represent an immune evasion strategy ([58], [59]. Actually, HLA-G can bind immune inhibitory receptors such as ILT2 and ILT4, thus preventing the generation of optimal immune responses and facilitating virus immune escape [60]. Based on these premises, HLA-G and HLA-E were considered by different authors as possible biomarkers to monitor SARS-CoV-2 infection, as exhaustively revised by Zidi [61].
In particular, Zhang and colleagues published a case report aimed to analyze the expressions of HLA-G and its receptors (ILT2, ILT4 and KIR2DL4) in peripheral immune cells of a patient critically infected with SARS-CoV-2, during the 23-day hospitalization [62]. In this study, HLA-G expression in peripheral immune cells was shown to follow a high–low–high pattern, which may reflect the three stages of infection, indicating that the status of SARS-CoV-2 infection may influence the regulation of HLA-G expression. However, questioning the real relevance of this observation, results showing that the expression of the HLA-G receptors, ILT4 and KIR2DL4, remained relatively stable during the disease were reported. As being a single case-report, the relevance of this findings should be confirmed by further independent studies.
Another correlation between SARS-CoV-2 and a non-classical HLA was investigated in a study by Bortolotti and colleagues with the aim of evaluating the effect of SARS-CoV-2 spike (SP1) protein expression in the control of NK cell activation [63]. Results showed that SP1 expression provoked: 1) HLA-E induction, whose expression was stabilized by the interaction with a SP1-derived HLA-E-binding peptide; 2) increased levels of the inhibitory receptor NKG2A/CD94); and 3) NK cell-reduced degranulation. In view of deepen new aspects and therapeutic strategies against COVID-19, this study highlights the potential of targeting the S1 protein or using the anti-NKG2A monoclonal antibody – already in use against rheumatoid arthritis and some neoplastic diseases [64] – in an attempt to enhance the innate immune response at the early stage of SARS-CoV-2 infection [65].
Vietzen et al. performed an analysis on a total of 361 Austrian COVID-19 patients to assess the influence of host genetic variants on the severity of COVID-19. Considering the significant reduction of NK cells observed in patients with a severe clinical outcome [66], they focused on NKG2C – an activating NK cell receptor encoded by the KLRC2 gene – which binds to HLA-E on infected cells allowing NK cell activation. The study confirmed that NKG2C+ NK are potent antiviral effector cells, even in SARS-CoV-2 infection. Consistently, the deletion of KLRC2 which naturally occurs, together with a higher degree of the heterozygous HLA-E*0101/0103 variant and the HLA-E*0101 allele were registered in hospitalized patients, especially those who required intensive care in comparison to patients with mild symptoms. Other in vitro studies had already highlighted a lower cell surface expression levels for HLA-E*0101/0101 than for HLA-E*0103/0103 [66], which likely results in decreased NKG2C+ NK cell response, thus influencing the severity of COVID-19. These antiviral mechanisms driven by NK cell could play a critical role in gaining new insight about the protective immune responses against SARS-CoV-2 and further studies are needed to support and clarify these findings.
4. ERAP1 and ERAP2 role in SARS-CoV-2 infection
ERAP1 and ERAP2 are two aminopeptidases which share 50% homology. They belong to the M1 family of zinc-dependent aminopeptidases and play key roles in the activation of the human adaptive immune response [67]. All of them are characterized by the presence of four functional domains: domain I comprises 3 beta sheets and contains the antigenic peptide binding site; domain II constitutes the catalytic site; domain III acts as a hinge between domain II and IV, thus allowing conformational changes of the protein; domain IV forms an arc with domain II and determines the closed state of the protein [47], [68].
Within the Endoplasmic Reticulum (ER), ERAP1 and ERAP2 cleave the N-terminus of precursor peptides, previously processed in the cytoplasm by the proteasome, so as to generate antigenic peptides of 8–9 amino acid residues, which perfectly accommodate within the binding groove of MHC I molecules.
These two proteins act in concert but their activity is not redundant as they maintain marked differences in their enzymatic specificity. ERAP1 preferentially cleaves peptides with lysine, leucine, asparagine and tyrosine residues [69] and shows a strong tendency to cut 9–16 amino acids peptides into pieces of 8–9 amino acids, the optimal length for loading onto MHC I molecules [70], [71], [72], [73]. Contrariwise, ERAP2 presents a striking predilection for the positively charged arginine and lysine residues situated at the N-terminal and shows a greater efficiency toward shorter peptides, that ERAP1 processes poorly [74].
These aminopeptidases are, therefore, essential for creating the appropriate immunopeptidome, capable of activating a suitable immune response by CD8+ T cells. For this same reason, ERAPs polymorphisms altering their functionality and/or expression level have been demonstrated to influence the onset and progression of several diseases. Indeed, ERAP1 has been associated with Ankylosing spondylitis (AS), psoriasis and Behçet's disease (BD) in epistasis with the risk HLA-B*27[75] and -B*40:01 [76], -C*06:02, [77] and HLA-B*51 alleles [78], respectively. ERAP2 has been associated with AS, psoriasis and natural resistance to HIV-1 infection [79], [80] as well, but only in this latter it seems to be in epistasis with HLA-B*57.
As expected, an altered functioning of these aminopeptidases, due to specific mutations, has evident consequences even on susceptibility/progression of infectious diseases included COVID-19 [6].
A work by Stamatakis et al. was pivotal in understanding how these enzymes within the ER process SARS-CoV-2 antigens and suggested the possibility of modulating ERAPs efficacy to improve and boost the effectiveness of antiviral responses [81]. In particular, they utilized a novel approach to investigate the trimming activity of ERAP1, ERAP2 and IRAP, focusing on S1 spike glycoprotein, known to be the largest antigen of the virus. The authors incubated a mixture of synthetic peptides derived from the sequence of the SARS-CoV-2 S1 spike glycoprotein with either ERAP1, ERAP2, or IRAP alone, or with a mixture of ERAP1 and ERAP2. All three aminopeptidases generated shorter peptides with sequences appropriate for binding onto HLA alleles, even if with different trimming specificities. ERAP1 was the most efficient in generating peptides 8-11aa long – the correct length for HLA binding – ERAP2 and the ERAP1/ERAP2 mixture followed, while IRAP was the less efficient. The conclusions of these elegant analyses confirmed the hypothesis that, as long as peptide trimming is mediated by ERAP1, HLA-B*15:03 is likely to present more SARS-CoV-2 epitopes than HLA-B*46:01; notably, ERAP1 is considered to have the leading trimming activity in the ER. The authors also discovered that only 7% of the SARS-CoV-2 antigenic peptides potentially presented by the above-mentioned HLA alleles were produced by either ERAP1, ERAP2 or IRAP. This finding reveals that, through their trimming activity, aminopeptidases can markedly filter and determine which antigens can be presented by MHC I molecules. Thus, ERAPs genes and their allelic variants, which encode for proteins that establish a bottleneck for the fitting of processed antigens into the binding pocket of MHC I molecules, represent a further variable which should be taken into account in the context of peptide prediction algorithms. In line with this, one of the main functions recently ascribed to ERAP1 is to limit the peptides available for MHC I [82]. Therefore, this innovative approach could be useful in optimizing bioinformatic predictions of potential MHC I epitopes.
In a subsequent study, Lu et al. correlated ERAP2 genotype to COVID-19 severity. The authors examined 193 deaths from 1412 infections in a group of 5871 UK Biobank SARS-CoV-2 positive patients and found rs150892504 variant in ERAP2 gene to be one – out of the 5 novel risk variants in 4 genes discovered – of the genetic risk factors associated with survival from infection in SARS-CoV-2-infected individuals [83].
Even more recently, our group conducted a study based on the consistent amount of data giving ERAP2 a pivotal role in viral infections [6]. The work was built on the 2018 results of Ye and colleagues showing the presence of two novel genetic variants – ERAP2/Iso3 and ERAP2/Iso4 – transcribed following influenza viral stimuli by monocyte-derived dendritic cells isolated from homozygous HapB-carrying individuals [84]. These latter carry the G allele for rs2248374, a single nucleotide polymorphism (SNP) previously demonstrated to prime only the transcription of a spliced ERAP2 variant (ERAP2/Iso2), further degraded by nonsense-mediated-decay (NMD) [85]. Conversely, the A allele for this SNP leads to the transcription of ERAP2/Iso1, the wild type (WT) form of the enzyme, which is thoroughly functioning. The two short isoforms diverge from each other by alternative splicing at a secondary splice site at exon 15, but while ERAP2/Iso4 harbors a premature termination codon leading to its NMD, ERAP2/Iso3 may contribute to shape the antigen repertoire. Indeed, although it loses the catalytic domain, its capacity to dimerize is preserved. The interaction of ERAP2/Iso3 with the WT-forms of ERAPs could result into an alternative trimming efficacy of viral peptides, thus suggesting its indirect, yet crucial involvement in the anti-microbial response (Fig. 1). The results reported in the paper answered three main issues: first, they demonstrated that the newly characterized ERAP2/Iso3 mRNA expression is not flu-specific, as being also prompted by other pathogens including HIV, SARS-CoV-2, CMV and Bacteria (LPS); second, they proved that ERAP2/Iso3mRNA can be translated into a protein in response to microbial infections; third, they showed that ERAP2/Iso3 mRNA is expressed in a dose-dependent manner following viral infections [86]. Indeed, ERAP2/Iso3 expression was found to be directly proportional to the multiplicity of infection (MOI) of SARS-CoV-2 or HIV-1 used to in vitro infect cells, leading to the conclusion that ERAP2/Iso3 expression is strictly dependent on the viral dose of exposure. Finally, these results also showed that SARS-CoV-2 exposure provokes the expression of ERAP1 and ERAP2/Iso1 in a dose-dependent way, once again suggesting ERAPs participation in the handling of the anti-viral response during SARS-CoV-2 infection.
Besides their direct participation in the antigen presentation machinery ERAPs may be released in the extracellular milieu by immunocompetent cells triggered by inflammatory stimuli [87], [88], possibly conditioning SARS-CoV-2 infection through at least two alternative mechanisms. Firstly, once released ERAPs can modulate innate immunity besides acquired one, by promoting inflammasome activation, monocyte differentiation and phagocytic activity of THP-1–derived macrophages [89], [90], thus altering the immunological microenvironment in which the virus is growing. Secondly, ERAPs are two renin-angiotensin system (RAS) regulators, a pathway which is often altered in patients showing a severe form of COVID-19 [91]. In particular, ERAP1 cleaves angiotensin II into angiotensin (Ang) III and IV while ERAP2 cuts Ang III into Ang IV [92], [93]. Thus, loss-of-function variants of these aminopeptidases impair Ang III and Ang IV production, contributing to the increase of circulating Ang II levels resulting in hypertension [94]. As following SARS-CoV-2 infection the ACE2 receptor is depleted from cellular surface causing an accumulation of Ang II [95], ERAP1 and ERAP2 dysfunctional status may accentuate the clinical manifestations of the disease, thus further worsening the effects of SARS-CoV-2 infection [96].
5. Conclusions
The aim of this brief review is to collect and present the results actually available on the role of antigen presentation in SARS-CoV-2 infection. At the time of writing this paper, the COVID-19 pandemic is continuing its course, with a total of 155 million cases and more than 3 million deaths to date registered [97]. The scientific community is relentlessly focusing on identifying the features of the immune response against the virus and the role of genetics in influencing susceptibility and degree of severity of the disease. More and more studies are deepening the mechanisms of T cell response to SARS-CoV-2, considering that – unlike vaccines based on the formation of antibodies against the surface spike glycoprotein – T cell vaccines have the capacity to generate immune responses against viral proteome in its entirety [98]. In addition, a recent study states that robust cellular immunity against SARS-CoV-2 is likely to be present within the great majority of adults at six months following asymptomatic and mild to moderate infection. This also lowers the level of concern that immune responses following natural infection may not be lasting, thus predisposing to recurrent infection [99]. Even if different vaccines are currently available and distributed worldwide, it is important to highlight how the clarification of the complex interactions between this devastating coronavirus and our immune system is a matter of strong need, to learn how to control the ill-fated progression of COVID-19.
Author contributions
I.S. and C.V are responsible for investigation, and writing the original draft. M.C. and M.B conceived the project supervised the work and reviewed the manuscript.
Funding
This work was supported by own funding.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Woon A.P., Purcell A.W. The use of proteomics to understand antiviral immunity. Semin. Cell Dev. Biol. 2018;84:22–29. doi: 10.1016/j.semcdb.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Brewerton D.A., Hart F.D., Nicholls A., Caffrey M., James D.C.O., Sturrock R.D. Ankylosing spondylitis and HL-A 27. Lancet. 1973;301(7809):904–907. doi: 10.1016/S0140-6736(73)91360-3. [DOI] [PubMed] [Google Scholar]
- 3.Schlosstein L., Terasaki P.I., Bluestone R., Pearson C.M. High association of an HL-A antigen, W27, with ankylosing spondylitis. N Engl J Med. 1973;288(14):704–706. doi: 10.1056/NEJM197304052881403. [DOI] [PubMed] [Google Scholar]
- 4.Trowsdale J., Knight J.C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genom. Hum. Genet. 2013;14(1):301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saveanu L., Carroll O., Hassainya Y., van Endert P. Complexity, contradictions, and conundrums: studying post-proteasomal proteolysis in HLA class I antigen presentation. Immunol Rev. 2005;207(1):42–59. doi: 10.1111/j.0105-2896.2005.00313.x. [DOI] [PubMed] [Google Scholar]
- 6.I. Saulle, C. Vicentini, M. Clerici, M. Biasin, An overview on ERAP roles in infectious diseases, Cells. 9 (2020) 720. https://doi.org/10.3390/cells9030720. [DOI] [PMC free article] [PubMed]
- 7.F.A. Rabi, M.S. Al Zoubi, G.A. Kasasbeh, D.M. Salameh, A.D. Al-Nasser, SARS-CoV-2 and Coronavirus Disease 2019: What We Know So Far, Pathog. Basel Switz. 9 (2020). https://doi.org/10.3390/pathogens9030231. [DOI] [PMC free article] [PubMed]
- 8.Schurr T.G. Host genetic factors and susceptibility to SARS-CoV-2 infection. Am. J. Hum. Biol. 2020;32 doi: 10.1002/ajhb.23497. [DOI] [PubMed] [Google Scholar]
- 9.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J.Y. Paik, R. Rakosi-Schmidt, M.D. Jun Liu, The Role of MHC System in COVID-19 Susceptibility: A Qualitative Review of Current Literature, North Am. J. Med. Sci. 13 (2020). https://najms.com/index.php/najms/article/view/541 (accessed April 12, 2021).
- 11.Tavasolian F., Rashidi M., Hatam G.R., Jeddi M., Hosseini A.Z., Mosawi S.H., Abdollahi E., Inman R.D. HLA, immune response, and susceptibility to COVID-19. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.601886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dendrou C.A., Petersen J., Rossjohn J., Fugger L. HLA variation and disease. Nat. Rev. Immunol. 2018;18(5):325–339. doi: 10.1038/nri.2017.143. [DOI] [PubMed] [Google Scholar]
- 13.L. Piacentini, M. Biasin, C. Fenizia, M. Clerici, Genetic correlates of protection against HIV infection: the ally within, J. Intern. Med. 265 (2009) 110–124. https://doi.org/10.1111/j.1365-2796.2008.02041.x. [DOI] [PubMed]
- 14.Ovsyannikova I.G., Haralambieva I.H., Crooke S.N., Poland G.A., Kennedy R.B. The role of host genetics in the immune response to SARS‐CoV‐2 and COVID‐19 susceptibility and severity. Immunol. Rev. 2020;296(1):205–219. doi: 10.1111/imr.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y.-M.- A., Liang S.-Y., Shih Y.-P., Chen C.-Y., Lee Y.-M., Chang L., Jung S.-Y., Ho M.-S., Liang K.-Y., Chen H.-Y., Chan Y.-J., Chu D.-C. Epidemiological and genetic correlates of severe acute respiratory syndrome coronavirus infection in the hospital with the highest nosocomial infection rate in Taiwan in 2003. J. Clin. Microbiol. 2006;44(2):359–365. doi: 10.1128/JCM.44.2.359-365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hajeer A.H., Balkhy H., Johani S., Yousef M.Z., Arabi Y. Association of human leukocyte antigen class II alleles with severe Middle East respiratory syndrome-coronavirus infection. Ann. Thorac. Med. 2016;11(3):211. doi: 10.4103/1817-1737.185756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Josset L., Menachery V.D., Gralinski L.E., Agnihothram S., Sova P., Carter V.S., Yount B.L., Graham R.L., Baric R.S., Katze M.G., Buchmeier M.J., Buchmeier M.J. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio. 2013;4(3) doi: 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menachery V.D., Schäfer A., Burnum-Johnson K.E., Mitchell H.D., Eisfeld A.J., Walters K.B., Nicora C.D., Purvine S.O., Casey C.P., Monroe M.E., Weitz K.K., Stratton K.G., Webb-Robertson B.-J., Gralinski L.E., Metz T.O., Smith R.D., Waters K.M., Sims A.C., Kawaoka Y., Baric R.S. MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc. Natl. Acad. Sci. USA. 2018;115(5):E1012–E1021. doi: 10.1073/pnas.1706928115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren R.L., Birol I. HLA predictions from the bronchoalveolar lavage fluid samples of five patients at the early stage of the wuhan seafood market COVID-19 outbreak. ArXiv. 2020 [Google Scholar]
- 20.Tomita Y., Ikeda T., Sato R., Sakagami T. Association between HLA gene polymorphisms and mortality of COVID‐19: An in silico analysis. Immun. Inflamm. Dis. 2020;8(4):684–694. doi: 10.1002/iid3.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yung Y.-L., Cheng C.-K., Chan H.-Y., Xia J.T., Lau K.-M., Wong R.S.M., Wu A.K.L., Chu R.W., Wong A.C.C., Chow E.Y.D., Yip S.-F., Leung J.N.S., Lee C.-K., Ng M.H.L. Association of HLA-B22 serotype with SARS-CoV-2 susceptibility in Hong Kong Chinese patients. HLA. 2020 doi: 10.1111/tan.14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pisanti S., Deelen J., Gallina A.M., Caputo M., Citro M., Abate M., Sacchi N., Vecchione C., Martinelli R. Correlation of the two most frequent HLA haplotypes in the Italian population to the differential regional incidence of Covid-19. J. Transl. Med. 2020;18(1) doi: 10.1186/s12967-020-02515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novelli A., Andreani M., Biancolella M., Liberatoscioli L., Passarelli C., Colona V.L., Rogliani P., Leonardis F., Campana A., Carsetti R., Andreoni M., Bernardini S., Novelli G., Locatelli F. HLA allele frequencies and susceptibility to COVID-19 in a group of 99 Italian patients. HLA. 2020;96:610–614. doi: 10.1111/tan.14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kachuri L., Francis S.S., Morrison M.L., Wendt G.A., Bossé Y., Cavazos T.B., Rashkin S.R., Ziv E., Witte J.S. The landscape of host genetic factors involved in immune response to common viral infections. Genome Med. 2020;12(1) doi: 10.1186/s13073-020-00790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iturrieta-Zuazo I., Rita C.G., García-Soidán A., de Malet Pintos-Fonseca A., Alonso-Alarcón N., Pariente-Rodríguez R., Tejeda-Velarde A., Serrano-Villar S., Castañer-Alabau J.L., Nieto-Gañán I. Possible role of HLA class-I genotype in SARS-CoV-2 infection and progression: a pilot study in a cohort of Covid-19 Spanish patients. Clin. Immunol. 2020;219:108572. doi: 10.1016/j.clim.2020.108572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorente L., Martín M.M., Franco A., Barrios Y., Cáceres J.J., Solé-Violán J., Perez A., Marcos y Ramos J.A., Ramos-Gómez L., Ojeda N., Jiménez A., Lorente L., Franco A., Barrios Y., Perez A., Jiménez A., Pérez-Cejas A., Pérez-Llombet A., Uribe L., González L., Alvarez R., Martín M.M., Alcoba-Flórez J., Estupiñan A., Cáceres J.J., Vega P., Gonzalez L., Solé-Violán J., Ojeda N., López S., Rodríguez-Pérez A., Domínguez C., Marcos y Ramos J.A., Zapata M.F., Ramos-Gómez L., Ortiz-López R. HLA genetic polymorphisms and prognosis of patients with COVID-19. Med. Intensiva. 2021;45(2):96–103. doi: 10.1016/j.medin.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.A. Sakuraba, H. Haider, T. Sato, Population Difference in Allele Frequency of HLA-C*05 and Its Correlation with COVID-19 Mortality, Viruses. 12 (2020). https://doi.org/10.3390/v12111333. [DOI] [PMC free article] [PubMed]
- 28.P. Parham, Immunogenetics of killer-cell immunoglobulin-like receptors, Tissue Antigens. 62 (2003) 194–200. https://doi.org/10.1034/j.1399-0039.2003.00126.x. [DOI] [PubMed]
- 29.M. Yawata, N. Yawata, M. Draghi, A.-M. Little, F. Partheniou, P. Parham, Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function, J. Exp. Med. 203 (2006) 633–645. https://doi.org/10.1084/jem.20051884. [DOI] [PMC free article] [PubMed]
- 30.Caroll M.C., Prodeus A.P. Linkages of innate and adaptive immunity. Curr. Opin. Immunol. 1998;10(1):36–40. doi: 10.1016/S0952-7915(98)80028-9. [DOI] [PubMed] [Google Scholar]
- 31.Kӧrner C., Altfeld M. Role of KIR3DS1 in human diseases. Front. Immunol. 2012;3 doi: 10.3389/fimmu.2012.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ben Fredj N., Rizzo R., Bortolotti D., Nefzi F., Chebel S., Rotola A., Frih-Ayed M., Di Luca D., Aouni M. Evaluation of the implication of KIR2DL2 receptor in multiple sclerosis and herpesvirus susceptibility. J. Neuroimmunol. 2014;271(1-2):30–35. doi: 10.1016/j.jneuroim.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 33.Rizzo R., Gentili V., Casetta I., Caselli E., De Gennaro R., Granieri E., Cassai E., Di Luca D., Rotola A. Altered natural killer cells' response to herpes virus infection in multiple sclerosis involves KIR2DL2 expression. J. Neuroimmunol. 2012;251(1-2):55–64. doi: 10.1016/j.jneuroim.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Rizzo R., Bortolotti D., Gentili V., Rotola A., Bolzani S., Caselli E., Tola M.R., Di Luca D., Fülöp T. KIR2DS2/KIR2DL2/HLA-C1 haplotype is associated with Alzheimer’s disease: implication for the role of herpesvirus infections. JAD. 2019;67(4):1379–1389. doi: 10.3233/JAD-180777. [DOI] [PubMed] [Google Scholar]
- 35.Severe Covid-19 GWAS Group, D. Ellinghaus, F. Degenhardt, L. Bujanda, M. Buti, A. Albillos, P. Invernizzi, J. Fernández, D. Prati, G. Baselli, R. Asselta, M.M. Grimsrud, C. Milani, F. Aziz, J. Kässens, S. May, M. Wendorff, L. Wienbrandt, F. Uellendahl-Werth, T. Zheng, X. Yi, R. de Pablo, A.G. Chercoles, A. Palom, A.-E. Garcia-Fernandez, F. Rodriguez-Frias, A. Zanella, A. Bandera, A. Protti, A. Aghemo, A. Lleo, A. Biondi, A. Caballero-Garralda, A. Gori, A. Tanck, A. Carreras Nolla, A. Latiano, A.L. Fracanzani, A. Peschuck, A. Julià, A. Pesenti, A. Voza, D. Jiménez, B. Mateos, B. Nafria Jimenez, C. Quereda, C. Paccapelo, C. Gassner, C. Angelini, C. Cea, A. Solier, D. Pestaña, E. Muñiz-Diaz, E. Sandoval, E.M. Paraboschi, E. Navas, F. García Sánchez, F. Ceriotti, F. Martinelli-Boneschi, F. Peyvandi, F. Blasi, L. Téllez, A. Blanco-Grau, G. Hemmrich-Stanisak, G. Grasselli, G. Costantino, G. Cardamone, G. Foti, S. Aneli, H. Kurihara, H. ElAbd, I. My, I. Galván-Femenia, J. Martín, J. Erdmann, J. Ferrusquía-Acosta, K. Garcia-Etxebarria, L. Izquierdo-Sanchez, L.R. Bettini, L. Sumoy, L. Terranova, L. Moreira, L. Santoro, L. Scudeller, F. Mesonero, L. Roade, M.C. Rühlemann, M. Schaefer, M. Carrabba, M. Riveiro-Barciela, M.E. Figuera Basso, M.G. Valsecchi, M. Hernandez-Tejero, M. Acosta-Herrera, M. D’Angiò, M. Baldini, M. Cazzaniga, M. Schulzky, M. Cecconi, M. Wittig, M. Ciccarelli, M. Rodríguez-Gandía, M. Bocciolone, M. Miozzo, N. Montano, N. Braun, N. Sacchi, N. Martínez, O. Özer, O. Palmieri, P. Faverio, P. Preatoni, P. Bonfanti, P. Omodei, P. Tentorio, P. Castro, P.M. Rodrigues, A. Blandino Ortiz, R. de Cid, R. Ferrer, R. Gualtierotti, R. Nieto, S. Goerg, S. Badalamenti, S. Marsal, G. Matullo, S. Pelusi, S. Juzenas, S. Aliberti, V. Monzani, V. Moreno, T. Wesse, T.L. Lenz, T. Pumarola, V. Rimoldi, S. Bosari, W. Albrecht, W. Peter, M. Romero-Gómez, M. D’Amato, S. Duga, J.M. Banales, J.R. Hov, T. Folseraas, L. Valenti, A. Franke, T.H. Karlsen, Genomewide Association Study of Severe Covid-19 with Respiratory Failure, N. Engl. J. Med. 383 (2020) 1522–1534. https://doi.org/10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed]
- 36.A. Amoroso, P. Magistroni, F. Vespasiano, A. Bella, S. Bellino, F. Puoti, S. Alizzi, T. Vaisitti, S. Boros, P.A. Grossi, S. Trapani, L. Lombardini, P. Pezzotti, S. Deaglio, S. Brusaferro, M. Cardillo, Italian Network of Regional Transplant Coordinating Centers, HLA and AB0 Polymorphisms May Influence SARS-CoV-2 Infection and COVID-19 Severity, Transplantation. 105 (2021) 193–200. https://doi.org/10.1097/TP.0000000000003507. [DOI] [PubMed]
- 37.Zhao J., Yang Y., Huang H., Li D., Gu D., Lu X., Zhang Z., Liu L., Liu T., Liu Y., He Y., Sun B., Wei M., Yang G., Wang X., Zhang L., Zhou X., Xing M., Wang P.G. Relationship between the ABO Blood Group and the COVID-19 Susceptibility. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020 doi: 10.1093/cid/ciaa1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M. Zietz, J. Zucker, N.P. Tatonetti, Associations between blood type and COVID-19 infection, intubation, and death, Nat. Commun. 11 (2020) 5761. https://doi.org/10.1038/s41467-020-19623-x. [DOI] [PMC free article] [PubMed]
- 39.F. Venet, J. Demaret, M. Gossez, G. Monneret, Myeloid cells in sepsis-acquired immunodeficiency, Ann. N. Y. Acad. Sci. n/a (n.d.). https://doi.org/10.1111/nyas.14333. [DOI] [PubMed]
- 40.M.S. Winkler, A. Rissiek, M. Priefler, E. Schwedhelm, L. Robbe, A. Bauer, C. Zahrte, C. Zoellner, S. Kluge, A. Nierhaus, Human leucocyte antigen (HLA-DR) gene expression is reduced in sepsis and correlates with impaired TNFα response: A diagnostic tool for immunosuppression?, PloS One. 12 (2017) e0182427. https://doi.org/10.1371/journal.pone.0182427. [DOI] [PMC free article] [PubMed]
- 41.Benlyamani I., Venet F., Coudereau R., Gossez M., Monneret G. Monocyte HLA-DR measurement by flow cytometry in COVID-19 patients: an interim review. Cytometry A. 2020;97:1217–1221. doi: 10.1002/cyto.a.24249. [DOI] [PubMed] [Google Scholar]
- 42.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.-E., Katsaounou P., Ntaganou M., Kyriakopoulou M., Dimopoulos G., Koutsodimitropoulos I., Velissaris D., Koufargyris P., Karageorgos A., Katrini K., Lekakis V., Lupse M., Kotsaki A., Renieris G., Theodoulou D., Panou V., Koukaki E., Koulouris N., Gogos C., Koutsoukou A. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michot J.-M., Albiges L., Chaput N., Saada V., Pommeret F., Griscelli F., Balleyguier C., Besse B., Marabelle A., Netzer F., Merad M., Robert C., Barlesi F., Gachot B., Stoclin A. Tocilizumab, an anti-IL-6 receptor antibody, to treat COVID-19-related respiratory failure: a case report. Ann. Oncol. 2020;31(7):961–964. doi: 10.1016/j.annonc.2020.03.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell K.M., Steiner G., Wells D.K., Ribas A., Kalbasi A. Prediction of SARS-CoV-2 epitopes across 9360 HLA class I alleles. BioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.03.30.016931. [DOI] [Google Scholar]
- 45.Lee C.H., Koohy H. In silico identification of vaccine targets for 2019-nCoV. F1000Res. 2020;9:145. doi: 10.12688/f1000research.22507.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.A. Nguyen, J.K. David, S.K. Maden, M.A. Wood, B.R. Weeder, A. Nellore, R.F. Thompson, Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2, J. Virol. 94 (2020). https://doi.org/10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed]
- 47.Nguyen T.T., Chang S.-C., Evnouchidou I., York I.A., Zikos C., Rock K.L., Goldberg A.L., Stratikos E., Stern L.J. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat. Struct. Mol. Biol. 2011;18(5):604–613. doi: 10.1038/nsmb.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin M., Tseng H.-K., Trejaut J.A., Lee H.-L., Loo J.-H., Chu C.-C., Chen P.-J., Su Y.-W., Lim K.H., Tsai Z.-U., Lin R.-Y., Lin R.-S., Huang C.-H. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med. Genet. 2003;4(1) doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.La Porta C.A.M., Zapperi S. Estimating the binding of Sars-CoV-2 Peptides to HLA Class I in human subpopulations using artificial neural networks. Cell Systems. 2020;11(4):412–417.e2. doi: 10.1016/j.cels.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barquera R., Collen E., Di D., Buhler S., Teixeira J., Llamas B., Nunes J.M., Sanchez-Mazas A. Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA. 2020;96:277–298. doi: 10.1111/tan.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romero‐López J.P., Carnalla‐Cortés M., Pacheco‐Olvera D.L., Ocampo‐Godínez J.M., Oliva‐Ramírez J., Moreno‐Manjón J., Bernal‐Alferes B., López‐Olmedo N., García‐Latorre E., Domínguez‐López M.L., Reyes‐Sandoval A., Jiménez‐Zamudio L. A bioinformatic prediction of antigen presentation from SARS‐CoV‐2 spike protein revealed a theoretical correlation of HLA‐DRB1*01 with COVID‐19 fatality in Mexican population: An ecological approach. J. Med. Virol. 2021;93(4):2029–2038. doi: 10.1002/jmv.26561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Moura R.R., Agrelli A., Santos-Silva C.A., Silva N., Assunção B.R., Brandão L., Benko-Iseppon A.M., Crovella S. Immunoinformatic approach to assess SARS-CoV-2 protein S epitopes recognised by the most frequent MHC-I alleles in the Brazilian population. J. Clin. Pathol. 2020 doi: 10.1136/jclinpath-2020-206946. [DOI] [PubMed] [Google Scholar]
- 53.Falk K., Rötzschke O., Stevanovié S., Jung G., Rammensee H.-G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351(6324):290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 54.S. Silva-Arrieta, P.J.R. Goulder, C. Brander, In silico veritas? Potential limitations for SARS-CoV-2 vaccine development based on T-cell epitope prediction, PLoS Pathog. 16 (2020) e1008607. https://doi.org/10.1371/journal.ppat.1008607. [DOI] [PMC free article] [PubMed]
- 55.Morandi F., Rizzo R., Fainardi E., Rouas-Freiss N., Pistoia V. Recent advances in our understanding of HLA-G biology: lessons from a wide spectrum of human diseases. J. Immunol. Res. 2016;2016:1–14. doi: 10.1155/2016/4326495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morandi F., Pistoia V. Interactions between HLA-G and HLA-E in physiological and pathological conditions. Front. Immunol. 2014;5:394. doi: 10.3389/fimmu.2014.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rizzo R., Bortolotti D., Bolzani S., Fainardi E. HLA-G molecules in autoimmune diseases and infections. Front. Immunol. 2014;5:592. doi: 10.3389/fimmu.2014.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amiot L., Vu N., Rauch M., L’Helgoualc’h A., Chalmel F., Gascan H., Turlin B., Guyader D., Samson M. Expression of HLA-G by mast cells is associated with hepatitis C virus-induced liver fibrosis. J. Hepatol. 2014;60(2):245–252. doi: 10.1016/j.jhep.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 59.C. Li, I. Toth, J. Schulze Zur Wiesch, F. Pereyra, J. Rychert, E.S. Rosenberg, J. van Lunzen, M. Lichterfeld, X.G. Yu, Functional characterization of HLA-G+ regulatory T cells in HIV-1 infection, PLoS Pathog. 9 (2013) e1003140. https://doi.org/10.1371/journal.ppat.1003140. [DOI] [PMC free article] [PubMed]
- 60.A. Lin, W.-H. Yan, Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges, Front. Immunol. 9 (2018) 2164. https://doi.org/10.3389/fimmu.2018.02164. [DOI] [PMC free article] [PubMed]
- 61.Zidi I. Puzzling out the COVID-19: therapy targeting HLA-G and HLA-E. Hum. Immunol. 2020;81(12):697–701. doi: 10.1016/j.humimm.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang S., Gan J., Chen B.-G., Zheng D., Zhang J.-G., Lin R.-H., Zhou Y.-P., Yang W.-Y., Lin A., Yan W.-H. Dynamics of peripheral immune cells and their HLA‐G and receptor expressions in a patient suffering from critical COVID‐19 pneumonia to convalescence. Clin. Transl. Immunol. 2020;9(5) doi: 10.1002/cti2.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D. Bortolotti, V. Gentili, S. Rizzo, A. Rotola, R. Rizzo, SARS-CoV-2 Spike 1 Protein Controls Natural Killer Cell Activation via the HLA-E/NKG2A Pathway, Cells. 9 (2020) 1975. https://doi.org/10.3390/cells9091975. [DOI] [PMC free article] [PubMed]
- 64.van Hall T., André P., Horowitz A., Ruan D.F., Borst L., Zerbib R., Narni-Mancinelli E., van der Burg S.H., Vivier E. Monalizumab: inhibiting the novel immune checkpoint NKG2A. J. Immunotherapy Cancer. 2019;7(1) doi: 10.1186/s40425-019-0761-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Masselli E., Vaccarezza M., Carubbi C., Pozzi G., Presta V., Mirandola P., Vitale M. NK cells: A double edge sword against SARS-CoV-2. Adv. Biol. Regul. 2020;77:100737. doi: 10.1016/j.jbior.2020.100737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vietzen H., Zoufaly A., Traugott M., Aberle J., Aberle S.W., Puchhammer-Stöckl E. Deletion of the NKG2C receptor encoding KLRC2 gene and HLA-E variants are risk factors for severe COVID-19. Genet. Med. 2021;23(5):963–967. doi: 10.1038/s41436-020-01077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neefjes J., Jongsma M.L.M., Paul P., Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011;11(12):823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 68.Kochan G., Krojer T., Harvey D., Fischer R., Chen L., Vollmar M., von Delft F., Kavanagh K.L., Brown M.A., Bowness P., Wordsworth P., Kessler B.M., Oppermann U. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc. Natl. Acad. Sci. 2011;108(19):7745–7750. doi: 10.1073/pnas.1101262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Serwold T., Gaw S., Shastri N. ER aminopeptidases generate a unique pool of peptides for MHC class I molecules. Nat. Immunol. 2001;2(7):644–651. doi: 10.1038/89800. [DOI] [PubMed] [Google Scholar]
- 70.Chen H., Li L., Weimershaus M., Evnouchidou I., van Endert P., Bouvier M. ERAP1-ERAP2 dimers trim MHC I-bound precursor peptides; implications for understanding peptide editing. Sci. Rep. 2016;6(1) doi: 10.1038/srep28902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Evnouchidou I., Weimershaus M., Saveanu L., van Endert P. ERAP1–ERAP2 Dimerization Increases Peptide-Trimming Efficiency. J. Immunol. 2014 doi: 10.4049/jimmunol.1302855. [DOI] [PubMed] [Google Scholar]
- 72.Hattori A., Tsujimoto M. Endoplasmic reticulum aminopeptidases: biochemistry, physiology and pathology. J. Biochem. (Tokyo) 2013;154:219–228. doi: 10.1093/jb/mvt066. [DOI] [PubMed] [Google Scholar]
- 73.Saveanu L., Carroll O., Lindo V., Del Val M., Lopez D., Lepelletier Y., Greer F., Schomburg L., Fruci D., Niedermann G., van Endert P.M. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat. Immunol. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 74.Stratikos E., Stern L.J. Antigenic peptide trimming by ER aminopeptidases–insights from structural studies. Mol. Immunol. 2013;55:212–219. doi: 10.1016/j.molimm.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Evans D.M., Spencer C.C.A., Pointon J.J., Su Z., Harvey D., Kochan G., Oppermann U., Dilthey A., Pirinen M., Stone M.A., Appleton L., Moutsianas L., Leslie S., Wordsworth T., Kenna T.J., Karaderi T., Thomas G.P., Ward M.M., Weisman M.H., Farrar C., Bradbury L.A., Danoy P., Inman R.D., Maksymowych W., Gladman D., Rahman P., Morgan A., Marzo-Ortega H., Bowness P., Gaffney K., Gaston J.S.H., Smith M., Bruges-Armas J., Couto A.-R., Sorrentino R., Paladini F., Ferreira M.A., Xu H., Liu Y., Jiang L., Lopez-Larrea C., Díaz-Peña R., López-Vázquez A., Zayats T., Band G., Bellenguez C., Blackburn H., Blackwell J.M., Bramon E., Bumpstead S.J., Casas J.P., Corvin A., Craddock N., Deloukas P., Dronov S., Duncanson A., Edkins S., Freeman C., Gillman M., Gray E., Gwilliam R., Hammond N., Hunt S.E., Jankowski J., Jayakumar A., Langford C., Liddle J., Markus H.S., Mathew C.G., McCann O.T., McCarthy M.I., Palmer C.N.A., Peltonen L., Plomin R., Potter S.C., Rautanen A., Ravindrarajah R., Ricketts M., Samani N., Sawcer S.J., Strange A., Trembath R.C., Viswanathan A.C., Waller M., Weston P., Whittaker P., Widaa S., Wood N.W., McVean G., Reveille J.D., Wordsworth B.P., Brown M.A., Donnelly P. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cortes A., Pulit S.L., Leo P.J., Pointon J.J., Robinson P.C., Weisman M.H., Ward M., Gensler L.S., Zhou X., Garchon H.-J., Chiocchia G., Nossent J., Lie B.A., Førre Ø., Tuomilehto J., Laiho K., Bradbury L.A., Elewaut D., Burgos-Vargas R., Stebbings S., Appleton L., Farrah C., Lau J., Haroon N., Mulero J., Blanco F.J., Gonzalez-Gay M.A., Lopez-Larrea C., Bowness P., Gaffney K., Gaston H., Gladman D.D., Rahman P., Maksymowych W.P., Crusius J.B.A., van der Horst-Bruinsma I.E., Valle-Oñate R., Romero-Sánchez C., Hansen I.M., Pimentel-Santos F.M., Inman R.D., Martin J., Breban M., Wordsworth B.P., Reveille J.D., Evans D.M., de Bakker P.I.W., Brown M.A. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat. Commun. 2015;6:7146. doi: 10.1038/ncomms8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2, A. Strange, F. Capon, C.C.A. Spencer, J. Knight, M.E. Weale, M.H. Allen, A. Barton, G. Band, C. Bellenguez, J.G.M. Bergboer, J.M. Blackwell, E. Bramon, S.J. Bumpstead, J.P. Casas, M.J. Cork, A. Corvin, P. Deloukas, A. Dilthey, A. Duncanson, S. Edkins, X. Estivill, O. Fitzgerald, C. Freeman, E. Giardina, E. Gray, A. Hofer, U. Hüffmeier, S.E. Hunt, A.D. Irvine, J. Jankowski, B. Kirby, C. Langford, J. Lascorz, J. Leman, S. Leslie, L. Mallbris, H.S. Markus, C.G. Mathew, W.H.I. McLean, R. McManus, R. Mössner, L. Moutsianas, A.T. Naluai, F.O. Nestle, G. Novelli, A. Onoufriadis, C.N.A. Palmer, C. Perricone, M. Pirinen, R. Plomin, S.C. Potter, R.M. Pujol, A. Rautanen, E. Riveira-Munoz, A.W. Ryan, W. Salmhofer, L. Samuelsson, S.J. Sawcer, J. Schalkwijk, C.H. Smith, M. Ståhle, Z. Su, R. Tazi-Ahnini, H. Traupe, A.C. Viswanathan, R.B. Warren, W. Weger, K. Wolk, N. Wood, J. Worthington, H.S. Young, P.L.J.M. Zeeuwen, A. Hayday, A.D. Burden, C.E.M. Griffiths, J. Kere, A. Reis, G. McVean, D.M. Evans, M.A. Brown, J.N. Barker, L. Peltonen, P. Donnelly, R.C. Trembath, A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1, Nat. Genet. 42 (2010) 985–990. https://doi.org/10.1038/ng.694. [DOI] [PMC free article] [PubMed]
- 78.Kirino Y., Bertsias G., Ishigatsubo Y., Mizuki N., Tugal-Tutkun I., Seyahi E., Ozyazgan Y., Sacli F.S., Erer B., Inoko H., Emrence Z., Cakar A., Abaci N., Ustek D., Satorius C., Ueda A., Takeno M., Kim Y., Wood G.M., Ombrello M.J., Meguro A., Gül A., Remmers E.F., Kastner D.L. Genome-wide association analysis identifies new susceptibility loci for Behçet’s disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet. 2013;45:202–207. doi: 10.1038/ng.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.M. Biasin, M. Sironi, I. Saulle, M. de Luca, F. la Rosa, R. Cagliani, D. Forni, C. Agliardi, S. lo Caputo, F. Mazzotta, D. Trabattoni, J. Macias, J.A. Pineda, A. Caruz, M. Clerici, Endoplasmic reticulum aminopeptidase 2 haplotypes play a role in modulating susceptibility to HIV infection, AIDS Lond. Engl. 27 (2013) 1697–1706. https://doi.org/10.1097/QAD.0b013e3283601cee. [DOI] [PubMed]
- 80.International Genetics of Ankylosing Spondylitis Consortium (IGAS), A. Cortes, J. Hadler, J.P. Pointon, P.C. Robinson, T. Karaderi, P. Leo, K. Cremin, K. Pryce, J. Harris, S. Lee, K.B. Joo, S.-C. Shim, M. Weisman, M. Ward, X. Zhou, H.-J. Garchon, G. Chiocchia, J. Nossent, B.A. Lie, Ø. Førre, J. Tuomilehto, K. Laiho, L. Jiang, Y. Liu, X. Wu, L.A. Bradbury, D. Elewaut, R. Burgos-Vargas, S. Stebbings, L. Appleton, C. Farrah, J. Lau, T.J. Kenna, N. Haroon, M.A. Ferreira, J. Yang, J. Mulero, J.L. Fernandez-Sueiro, M.A. Gonzalez-Gay, C. Lopez-Larrea, P. Deloukas, P. Donnelly, Australo-Anglo-American Spondyloarthritis Consortium (TASC), Groupe Française d’Etude Génétique des Spondylarthrites (GFEGS), Nord-Trøndelag Health Study (HUNT), Spondyloarthritis Research Consortium of Canada (SPARCC), Wellcome Trust Case Control Consortium 2 (WTCCC2), P. Bowness, K. Gafney, H. Gaston, D.D. Gladman, P. Rahman, W.P. Maksymowych, H. Xu, J.B.A. Crusius, I.E. van der Horst-Bruinsma, C.-T. Chou, R. Valle-Oñate, C. Romero-Sánchez, I.M. Hansen, F.M. Pimentel-Santos, R.D. Inman, V. Videm, J. Martin, M. Breban, J.D. Reveille, D.M. Evans, T.-H. Kim, B.P. Wordsworth, M.A. Brown, Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci, Nat. Genet. 45 (2013) 730–738. https://doi.org/10.1038/ng.2667. [DOI] [PMC free article] [PubMed]
- 81.Stamatakis G., Samiotaki M., Mpakali A., Panayotou G., Stratikos E. Generation of SARS-CoV-2 S1 spike glycoprotein putative antigenic epitopes in vitro by intracellular aminopeptidases. J. Proteome Res. 2020;19:4398–4406. doi: 10.1021/acs.jproteome.0c00457. [DOI] [PubMed] [Google Scholar]
- 82.Komov L., Kadosh D.M., Barnea E., Milner E., Hendler A., Admon A. Cell Surface MHC class I expression is limited by the availability of peptide-receptive “empty” molecules rather than by the supply of peptide ligands. Proteomics. 2018;18 doi: 10.1002/pmic.201700248. [DOI] [PubMed] [Google Scholar]
- 83.C. Lu, R. Gam, A.P. Pandurangan, J. Gough, Genetic risk factors for death with SARS-CoV-2 from the UK Biobank, MedRxiv. (2020) 2020.07.01.20144592. https://doi.org/10.1101/2020.07.01.20144592.
- 84.Ye C.J., Chen J., Villani A.-C., Gate R.E., Subramaniam M., Bhangale T., Lee M.N., Raj T., Raychowdhury R., Li W., Rogel N., Simmons S., Imboywa S.H., Chipendo P.I., McCabe C., Lee M.H., Frohlich I.Y., Stranger B.E., De Jager P.L., Regev A., Behrens T., Hacohen N. Genetic analysis of isoform usage in the human anti-viral response reveals influenza-specific regulation of ERAP2 transcripts under balancing selection. Genome Res. 2018;28:1812–1825. doi: 10.1101/gr.240390.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Andrés A.M., Dennis M.Y., Kretzschmar W.W., Cannons J.L., Lee-Lin S.-Q., Hurle B., Schwartzberg P.L., Williamson S.H., Bustamante C.D., Nielsen R., Clark A.G., Green E.D. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saulle I., Vanetti C., Goglia S., Vicentini C., Tombetti E., Garziano M., Clerici M., Biasin M. A new ERAP2/Iso3 isoform expression is triggered by different microbial stimuli in human cells. Could it play a role in the modulation of SARS-CoV-2 infection? Cells. 2020;9 doi: 10.3390/cells9091951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goto Y., Ogawa Y., Tsumoto H., Miura Y., Nakamura T.J., Ogawa K., Akimoto Y., Kawakami H., Endo T., Yanoshita R., Tsujimoto M. Contribution of the exosome-associated form of secreted endoplasmic reticulum aminopeptidase 1 to exosome-mediated macrophage activation. Biochim. Biophys. Acta BBA - Mol. Cell Res. 2018;1865:874–888. doi: 10.1016/j.bbamcr.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 88.Saulle I., Ibba S.V., Torretta E., Vittori C., Fenizia C., Piancone F., Minisci D., Lori E.M., Trabattoni D., Gelfi C., Clerici M., Biasin M. Endoplasmic reticulum associated aminopeptidase 2 (ERAP2) is released in the secretome of activated MDMs and reduces in vitro HIV-1 infection. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.01648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goto Y., Ogawa K., Nakamura T.J., Hattori A., Tsujimoto M. TLR-Mediated secretion of endoplasmic reticulum aminopeptidase 1 from macrophages. J. Immunol. 2014;192:4443–4452. doi: 10.4049/jimmunol.1300935. [DOI] [PubMed] [Google Scholar]
- 90.Saulle I., Marventano I., Saresella M., Vanetti C., Garziano M., Fenizia C., Trabattoni D., Clerici M., Biasin M. ERAPs reduce in vitro HIV infection by activating innate immune response. J. Immunol. Baltim. Md. 2021;1950(206):1609–1617. doi: 10.4049/jimmunol.2000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gathiram P., Mackraj I., Moodley J. The renin-angiotensin system, hypertension, and SARS-CoV-2 infection: a review. Curr. Hypertens. Rep. 2021;23:17. doi: 10.1007/s11906-021-01134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sato Y. Role of aminopeptidase in angiogenesis. Biol. Pharm. Bull. 2004;27:772–776. doi: 10.1248/bpb.27.772. [DOI] [PubMed] [Google Scholar]
- 93.Tanioka T., Hattori A., Masuda S., Nomura Y., Nakayama H., Mizutani S., Tsujimoto M. Human Leukocyte-derived Arginine Aminopeptidase the third member of the oxytocinase subfamily of aminopeptidases. J. Biol. Chem. 2003;278:32275–32283. doi: 10.1074/jbc.M305076200. [DOI] [PubMed] [Google Scholar]
- 94.S. Ranjit, J.Y. Wong, J.W. Tan, C. Sin Tay, J.M. Lee, K. Yin Han Wong, L.H. Pojoga, D.L. Brooks, A.E. Garza, S.A. Maris, I.A. Katayama, J.S. Williams, A. Rivera, G.K. Adler, G.H. Williams, J.R. Romero, Sex-specific differences in endoplasmic reticulum aminopeptidase 1 modulation influence blood pressure and renin-angiotensin system responses, JCI Insight. 4 (2020). https://doi.org/10.1172/jci.insight.129615. [DOI] [PMC free article] [PubMed]
- 95.Wang K., Gheblawi M., Oudit G.Y. Angiotensin converting enzyme 2: a double-edged sword. Circulation. 2020;142:426–428. doi: 10.1161/CIRCULATIONAHA.120.047049. [DOI] [PubMed] [Google Scholar]
- 96.D’Amico S., Tempora P., Lucarini V., Melaiu O., Gaspari S., Algeri M., Fruci D. ERAP1 and ERAP2 enzymes: a protective shield for RAS against COVID-19? Int. J. Mol. Sci. 2021;22:1705. doi: 10.3390/ijms22041705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.WHO Coronavirus Disease (COVID-19) Dashboard | WHO Coronavirus Disease (COVID-19) Dashboard, (n.d.). https://covid19.who.int/ (accessed December 7, 2020).
- 98.Gilbert S.C. T-cell-inducing vaccines - what’s the future. Immunology. 2012;135:19–26. doi: 10.1111/j.1365-2567.2011.03517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zuo J., Dowell A.C., Pearce H., Verma K., Long H.M., Begum J., Aiano F., Amin-Chowdhury Z., Hallis B., Stapley L., Borrow R., Linley E., Ahmad S., Parker B., Horsley A., Amirthalingam G., Brown K., Ramsay M.E., Ladhani S., Moss P. Robust SARS-CoV-2-specific T cell immunity is maintained at 6 months following primary infection. Nat. Immunol. 2021:1–7. doi: 10.1038/s41590-021-00902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]