Abstract
There is growing evidence that measurement of SARS-CoV-2 viral copy number can inform clinical and public health management of SARS-CoV-2 carriers and COVID-19 patients. Here we show that quantification of SARS-CoV-2 is feasible in a clinical setting, using a duplex RT-qPCR assay which targets both the E gene (Charité assay) and a human RNA transcript, RNase P (CDC assay) as an internal sample sufficiency control.
Samples in which RNase P is not amplified indicate that sample degradation has occurred, PCR inhibitors are present, RNA extraction has failed or swabbing technique was insufficient. This important internal control reveals that 2.4 % of nasopharyngeal swabs (15/618 samples) are inadequate for SARS-CoV-2 testing which, if not identified, could result in false negative results.
We show that our assay is linear across at least 7 logs and is highly reproducible, enabling the conversion of Cq values to viral copy numbers using a standard curve. Furthermore, the SARS-CoV-2 copy number was independent of the RNase P copy number indicating that the per-swab viral copy number is not dependent on sampling- further allowing comparisons between samples.
The ability to quantify SARS-CoV-2 viral copy number will provide an important opportunity for viral burden-guided public health and clinical decision making.
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; VTM, virus transport medium; Cq, quantification cycle
Keywords: SARS-CoV-2, COVID-19, RT-qPCR, Viral burden, RNase P
1. Introduction
Viral burden correlates with morbidity and mortality in many viral diseases. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is no exception: per-swab SARS-CoV-2 copy number may predict severity of symptoms and mortality in symptomatic hospitalized patients (Pujadas et al., 2020), and varies over >7 logs between individuals, with infectious virus reported to be recoverable only from samples with >1,000,000 copies/mL virus transport medium (VTM) (Wölfel et al., 2020).
Although the factors which determine transmission of SARS-CoV-2 are at present poorly understood, viral burden in the upper respiratory tract likely contributes to transmission potential. Within most individuals SARS-CoV-2 viral copy number in nasopharyngeal swabs appears to peak at the onset of symptoms and decline to undetectable levels one to two weeks later, coinciding with convalescence (Wölfel et al., 2020; Walsh et al., 2020). A considerable fraction of individuals remain asymptomatic on infection, with younger people more likely to remain asymptomatic and less likely to experience severe disease than older people (Verity et al., 2020; Docherty et al., 2020). Critically, viral burden does not correlate with age (Jones et al., 2020), and the viral burden of asymptomatic carriers is comparable to those with symptoms (Ra et al., 2020). Thus, within an individual or group of individuals (e.g. schoolchildren) mild or absent symptoms does not infer a low viral burden, or indeed lower transmission potential. Currently, SARS-CoV-2 laboratory results are reported qualitatively and the number of copies of the virus per swab is not considered in the U.K. national test and trace program. There is a clear clinical and public health need to move to standardized, quantitative tests.
This report describes an in-house method which uses primers and probes from the Charité and CDC protocols to detect sarbecovirus viral RNA in nasopharyngeal samples using real-time quantitative PCR (Corman et al., 2020). The Charité E gene primers and probe are duplexed with primers and a probe which amplify and detect a human RNA target (RNase P) (Centre for Disease Control and Prevention, 2020). This serves as an internal positive control which provides evidence of sample sufficiency, successful RNA extraction, successful reverse transcription and lack of PCR inhibition within a reaction. Here, we report the sensitivity and reproducibility of this assay and compare performance of the assay with testing for the E gene alone or the N gene in nasopharyngeal swabs, which will allow others to implement this assay in their lab with a substantial reduction in the amount of work required to validate the assay locally. We report the range of viral burden observed, the proportion of samples which were inadequate for SARS-CoV-2 testing, and the efficiency of sampling in samples collected by healthcare professionals.
2. Materials and methods
2.1. Specimen collection
Data in this manuscript was generated using nose or throat swab samples in virus transport media donated, following written informed consent, to the Communicable Diseases Research Tissue Bank (NRES reference 20/SC/0226) and using the anonymized excess virus transport media from diagnostic samples. Upper respiratory tract specimens (nasal swabs, throat swabs and nasopharyngeal swabs) were collected from staff and hospitalized patients with suspected COVID-19 disease or a history of exposure to SARS-CoV-2, for routine clinical investigation at Imperial College Healthcare NHS Trust. Unless otherwise stated, swabs were stored for less than 24 h ‘wet’, immersed in virus transport medium (VTM, MWE Σ-Virocult) at 2−4 °C.
2.2. Inactivation
Infectious material was inactivated in a containment level 3 laboratory by incubation in lysis buffer containing guanidinium thiocyanate, guanidinium chloride and/or sodium dodecyl sulfate (SDS) from the relevant RNA extraction kit. Effective concentrations for inactivation are described in Pastorino et al. (2020). For automated extraction this was achieved by adding 200 μl of VTM to 300 μl (1–10 % SDS, 38 % guanidinium chloride, Maxwell HT Viral TNA Kit, Promega) or 600 μl (2.5–10 % SDS, innuPREP Virus DNA/RNA Kit Analytik Jena) lysis buffer and incubating for 10 min at room temperature. Manual RNA extractions required the addition of 200 μl of VTM to 800 μl (50–70 % guanidinium thiocyanate, QIAamp Viral RNA mini kit, Qiagen) AVL buffer. Buffer AVL significantly reduces the titre of infectious SARS-CoV-2, however does not completely inactivate the virus, thus samples prepared using this method should be handled in appropriate containment conditions (Pastorino et al., 2020).
2.3. RNA extraction
The RNA extraction of SARS-CoV-2 was performed on the CyBio FeliX liquid handling robot (Analytik Jena), as previously described (Crone et al., 2020), using the Maxwell HT Viral TNA Kit (Promega), or InnuPREP Virus DNA/RNA Kit (Analytik Jena). Manual RNA extractions were carried out using QIAamp Viral RNA mini kit (Qiagen) as per manufacturer’s protocol. All samples were eluted into 50 μl of kit buffer.
2.4. In house RT-qPCR
RNA was reverse transcribed and subjected to quantitative PCR in a 20 μL reaction containing 10 μL of RNA, 5 μL of 4× TaqMan Fast virus one step Master Mix (ThermoFisher). Primer and probe sequences, as well as optimized concentrations are shown in Table 1 . Thermal cycling was performed at 55 °C for 10 min for reverse transcription, followed by 94 °C for 3 min and then 40 cycles of 94 °C for 15 s, 58 °C for 30 s using a Bio-Rad CFX real time PCR system. Where indicated, primers and a FAM-labelled hydrolysis probe for the E gene (Corman et al., 2020) (supplied by TIB-Molbiol, Germany) were duplexed with primers and a HEX-labelled hydrolysis probe for RNase P (Centre for Disease Control and Prevention, 2020). The working concentration of the RNase P primers was deliberately restricted to avoid exhaustion of reagents preventing amplification of the E gene. A SARS-CoV-2 PCR positive control was included in each run to monitor assay performance. This consisted of purified viral RNA obtained from culture supernatant (Public Health England) or a stabilized synthetic RNA construct containing the E gene sequence (SARSCOV2, Exact Diagnostics, Bio-Rad). An ‘extraction’ positive control which consisted of a frozen (−80 °C) aliquot of a high copy number patient sample diluted in VTM, was inactivated, extracted and amplified in every batch of samples processed. A PCR negative control (nuclease free water with or without 1 μg/μl Poly A carrier RNA (1017647, Qiagen)) was also included in all runs.
Table 1.
Primers and probes used in this study.
| Sequence (5′ to 3′) | Working concentration | |
|---|---|---|
| E_Sarbeco_F (Corman et al., 2020) | 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′ | 400 nM |
| E_Sarbeco_R (Corman et al., 2020) | 5′-ATATTGCAGCAGTACGCACACA-3′ | 400 nM |
| E_Sarbeco_P1 (Corman et al., 2020) | 5′-FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ-3′ | 200 nM |
| RNaseP_F (Centre for Disease Control and Prevention, 2020) | 5′-AGATTTGGACCTGCGAGCG-3′ | 100 nM |
| RNaseP_R (Centre for Disease Control and Prevention, 2020) | 5′-GAGCGGCTGTCTCCACAAGT-3′ | 100 nM |
| RNaseP_Pr (Centre for Disease Control and Prevention, 2020) | 5′-HEX-TTCTGACCTGAAGGCTCTGCGCG-BBQ-3′ | 100 nM |
| 2019-nCoV_N1-F (Centre for Disease Control and Prevention, 2020) | 5′-GACCCCAAAATCAGCGAAAT-3′ | 400 nM |
| 2019-nCoV_N1-R (Centre for Disease Control and Prevention, 2020) | 5′-TCTGGTTACTGCCAGTTGAATCTG-3′ | 400 nM |
| 2019-nCoV_N1-P (Centre for Disease Control and Prevention, 2020) | 5′-FAM-ACCCCGCATTACGTTTGGTGGACC-BBQ-3′ | 100 nM |
2.5. Data processing
On completion of thermal cycling, qPCR data was processed using Bio-Rad CFX maestro software (version 1.1). A fluorescence drift correction was applied, and the background was subtracted using the ‘curve fit’ option. The baseline was set manually to 800 for the required detectors (FAM alone or in combination with HEX).
2.6. Data interpretation
Samples in which amplified product was detected in the FAM channel (amplification curve crossed the threshold) before 36.5 cycles were called as ‘SARS-CoV-2 detected’. All other samples were called as ‘SARS-CoV-2 not detected’ provided RNase P was detected in the HEX channel before 37 cycles. If RNase P was not detected before 37 cycles, the sample was called as ‘inadequate’, and a repeat sample was requested. Before releasing results, each run was checked for any evidence of product in the PCR negative control (before 36.5 cycles) and for any evidence of deviation from the acceptable range for the SARS-CoV-2 PCR positive control. To determine the acceptable quantification cycle (Cq) range for each batch of SARS-CoV-2 PCR positive control material, the material was assayed in 4 independent runs, and the average Cq was calculated. The acceptable range was set as the mean plus or minus 1 Cq of the mean. Results were released if the PCR negative control was classed as ‘not detected’, and the PCR positive control was in the acceptable range. When required (Results section 3.9), the 2^(-ΔΔCt) method was used to normalize E and RNase P Cq values in test samples to the Cq values of E and RNase P in the Exact positive control. As the ratio of both E to RNase P in the Exact positive control is known (200 copies E:75 copies RNase P), the resulting value was converted into copies of E per copy RNase P by multiplying by 200/75. Further details of the assay are included in Table 1- Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) (Bustin et al., 2009). Probit analysis was performed by calculating detection probability units for each replicate sample set tested using the Excel function 5+NORMSINV(P), following which a linear regression line was plotted and used to calculate C95, the 95 % detection rate, corresponding to a probit value of 6.64.
2.7. Assay accreditation
The assay was accredited by UKAS on the basis of validation data which included analysis of 251 clinical specimens which had previously been characterized using the Coronavirus typing (8-well assay, ref 20619 V1, AusDiagnostics which detects ORF1ab) provided by Paul Randell and Pinglawathee Madona, Virology, North West London Pathology, Charing Cross Hospital.
3. Results
3.1. Efficiency and linear range of the duplex E/RNase P RT-qPCR assay
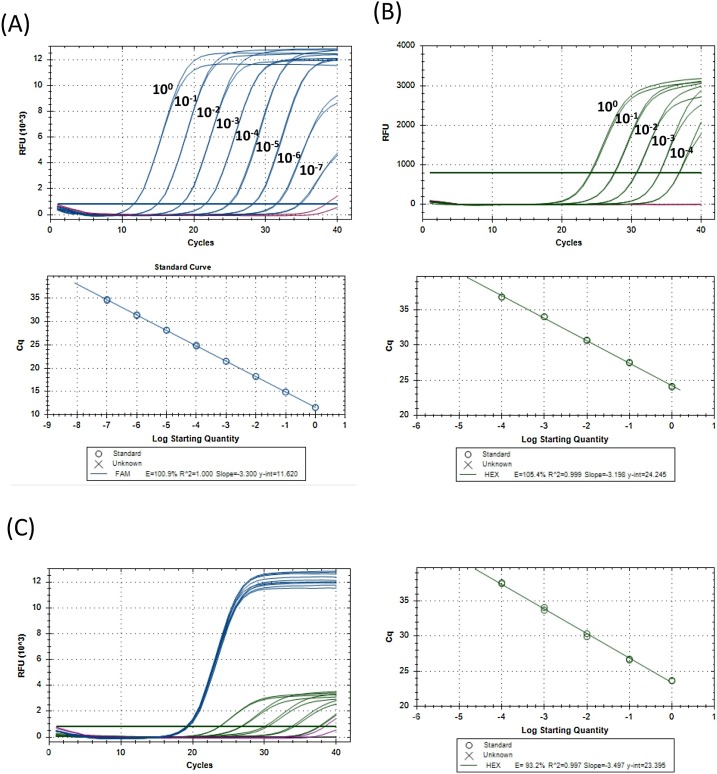
As SARS-CoV-2 is an emerging pathogen, at the time this assay was established, there was no independently verified copy number controlled material available commercially. Thus, we were compelled to conduct the initial assay validation work using cellular RNA extracted from Vero cells which had been infected with SARS-CoV-2 in vitro, kindly provided by Prof. Wendy Barclay. Serial dilutions of this material were assayed with the E/RNase P duplex assay to evaluate PCR efficiency. E gene Cq values were linear over 7 log dilutions, with a nominal PCR efficiency of 100.9 % (Fig. 1 A). RNase P expression was not detected in RNA from infected Vero cells, thus, RNA extracted from human peripheral blood mononuclear cells (PBMCs) was also analyzed to evaluate the efficiency of RNAse P amplification. This revealed a nominal efficiency of 105 % for RNase P (Fig. 1B). To determine whether RNase P amplification was inhibited in the presence of SARS-CoV-2 RNA, PBMC RNA was mixed with RNA from infected Vero cells, the same dilution series of PBMC RNA shown in Fig. 1B was prepared, and spiked with a uniform amount of RNA from SARS-Cov-2-infected Vero cells (Fig. 1C). As expected, the E gene Cq was consistent for all dilutions tested (average Cq 19.19, range 19.07–19.39), RNase P Cq values were comparable to those observed in Fig. 1B, and were linear across the 4 log dilutions tested, with a nominal efficiency of 93 % (96.4 % if the lowest concentration sample which had a Cq of 38 is omitted).
Fig. 1.
Linear range and PCR efficiency for E and RNase P.
RNA was extracted from SARS-CoV-2-infected Vero cells (A), serially diluted (10 fold) in nuclease-free water with carrier RNA and assayed in duplicate using the E/RNase P RT-qPCR assay. Relative fluorescence units (RFU) detected in the FAM channel for the E gene for samples containing viral RNA (blue) or carrier RNA alone (purple) are shown. (B) RNA was extracted from human peripheral blood mononuclear cells (PBMC), serially diluted (10 fold) in nuclease-free water with carrier RNA and assayed in duplicate using the E/RNase P RT-qPCR assay. RFU detected in the HEX channel for the RNase P for samples containing PBMC RNA (blue) or carrier RNA alone (purple) are shown. (C) RNA from SARS-CoV-2-infected Vero cells was spiked into serial dilutions of PBMC RNA. RFU from samples with PBMC and infected Vero cells RNA is shown in blue (FAM) and green (HEX), and carrier RNA alone is shown in purple (FAM) and black (HEX). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.2. Estimation of viral copy number
In April 2020, a known copy number product became available (EXACT standard, Bio-Rad, product code SARSCOV2), which consisted of RNA which had been tested by digital droplet PCR (SARS-CoV-2 RNA Transcripts: Envelope (E) Gene, Nucleocapsid (N) Gene, ORF1ab Gene, RdRP Gene, and the Spike Protein (S) Gene each at 200,000 copies/mL). A standard curve which could be used to estimate the SARS-CoV-2 RNA copy number was established by plotting the E gene Cq values of serial dilutions of synthetic RNA on a log/linear scale, followed by linear regression analysis. In runs in which the positive control was within the acceptable range for the assay, the following calculation was performed to estimate the copy number:
| E gene copies/10 μl RNA = 10^((Cq(E gene)-38.332)/(-3.114)) |
To estimate the copy number per ml of VTM, this value was multiplied by 25 to correct for the volume of VTM analyzed. This equation assumes a PCR efficiency of 109 %, observed empirically in serial dilutions of synthetic RNA. As the highest concentration of standardized material tested was 2000 copies/10 μL RNA, higher copy numbers are extrapolated.
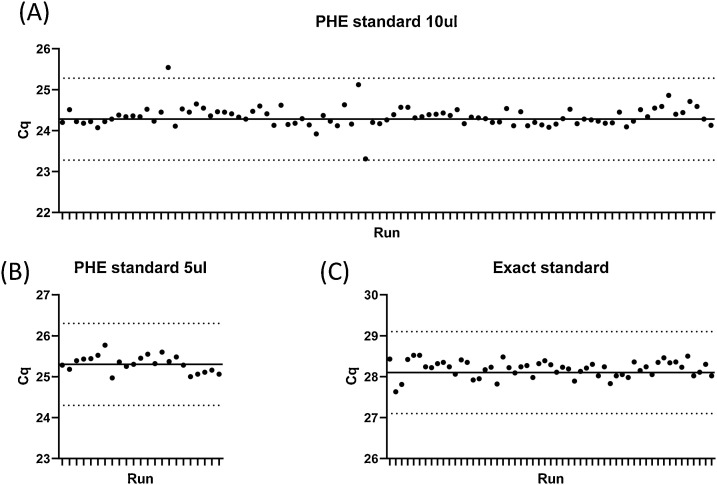
3.3. Assay reproducibility
Data on assay reproducibility were collected from 171 runs (Fig. 2 ). For three separate batches of control material, the standard deviation was between 0.20-0.25.
Fig. 2.
Assay reproducibility.
Aliquots of viral RNA (A, B) extracted from culture supernatant or a commercial synthetic RNA construct (C) were assayed using the E/RNase P duplex PCR. Each dot represents the E Cq of an independent run. The solid line represents the mean Cq of four runs (not included in the graph), and the dotted lines indicate the mean plus or minus 1 cycle.
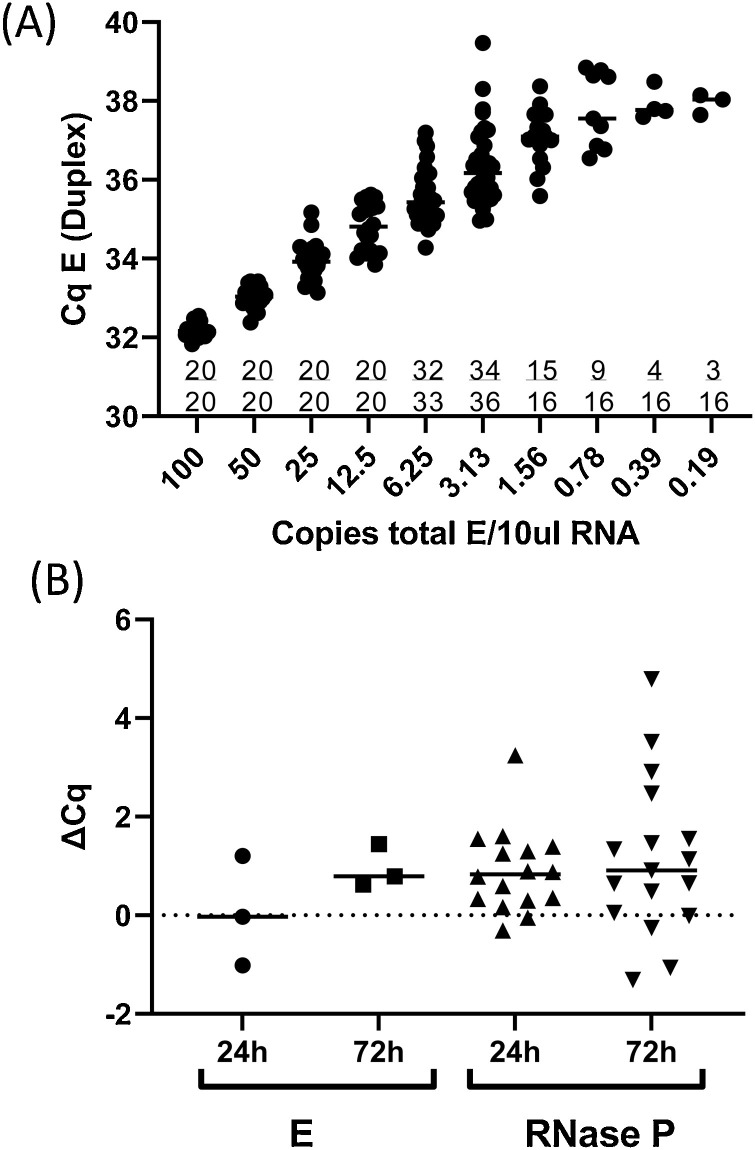
3.4. Sensitivity and limit of detection
The synthetic RNA standard was diluted in nuclease free water with 1 μg/μl PolyA carrier RNA (Qiagen) to a range of concentrations from 100 copies/10 μl- 0.19 copies/10 μl and assayed using the E/RNase P duplex assay (Fig. 3 a). To determine the limit of detection samples were analyzed in replicate sets, to identify the copy number at which 95 % of replicates were detected. At 12.25 copies/10 μl and above, products of which the E gene probe/primer set were detectable in 20/20 wells. At a concentration of 6.25 copies/10 μl, 32/33 wells had detectable E gene signal (97 % detection rate), with a median Cq of 35.4 (range, 34.28–37.20; standard deviation, 0.67). At 3.125 copies/10 μL RNA, 34/36 wells had detectable E gene signal (94 % detection rate). Probit analysis gave a similar result of 3.3 copies/10 μL RNA. To maximize assay specificity, 6 copies was taken to be the limit of detection of the assay, which equates to 150 copies/mL VTM. A cut off for calling a sample as “detected” was set a Cq = 36.5, and Cq values greater than 36.5 are called as “not detected”.
Fig. 3.
Limit of detection and effect of sample storage.
(A) The EXACT synthetic RNA standard was diluted in nuclease free water with 1 μg/μl PolyA carrier RNA (Qiagen) to a range of concentrations from 100 copies/10 μl- 0.19 copies/10 μl. Replicates of each concentration were assayed using the E/RNase P duplex assay. Cqs for E are shown for each replicate within each concentration, and the number of replicates in which a signal was detected is indicated on the graph as a fraction. (B) RNA was extracted from clinical samples immediately on receipt or after 24 h (n = 16) or 72 h (n = 17) storage at room temperature. The difference in Cq values (ΔCq) was calculated by subtracting the E or RNase P Cq observed in the sample processed and assayed on receipt from the E/ RNase P Cq observed in the sample processed after storage.
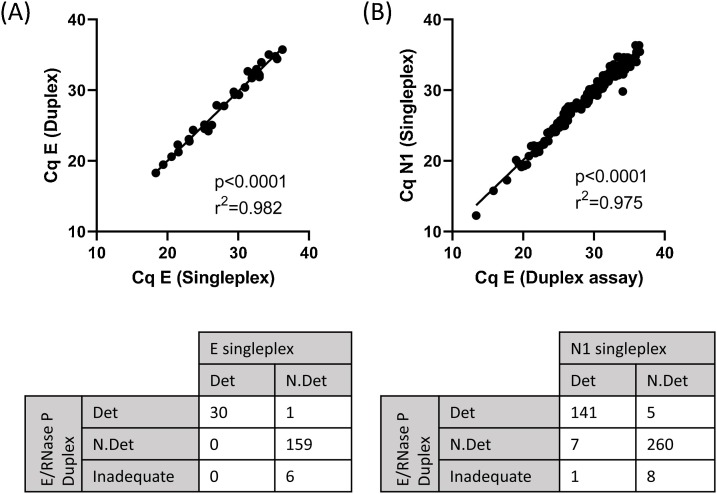
3.5. Concordance of duplex E and RNase P RT-qPCR with E singleplex RT-qPCR using clinical specimens
To compare the sensitivity of the E/RNase P duplex assay with the E singleplex assay, duplicate assays were run, testing 196 clinical samples in total (Fig. 4 A). The duplex assay was highly concordant with the singleplex assay. Thirty samples had detectable SARS-CoV2 sequences (Cq <36.5) in the E singleplex assay and the E/RNase P duplex assay. One additional sample was classed as ‘SARS-CoV-2 detected’ by the E/RNase P duplex assay, with Cq of 34.7, close to the limit of detection for the E gene. Cq values recorded for E gene in the duplex assay were not significantly different from the singleplex assay (p = 0.23, Wilcoxon matched pairs signed rank test) and displayed a strong linear correlation across Cq = 18 to Cq = 36, approximately 6 logs. Only samples which were close to the limit of detection were variably detected between the two assays. One hundred and sixty six samples were classed as “not detected” in the singleplex assay. Of these, 159 were also classed as “not detected”, in the duplex assay with six classed as “inadequate”. Thus, the duplex assay adds the additional benefit of classifying 3% of samples as inadequate, ensuring that these are reported correctly and are flagged for re-testing.
Fig. 4.
Comparison of E and N1 Singleplex and Duplex E/RNase P assays.
(A) Clinical samples (n = 196) were assayed using the E primers and FAM-labelled probe alone (E singleplex) or in combination with RNase P primers and HEX-labelled probe (E/RNase P Duplex) (B) Clinical samples (n = 422) were assayed using the CDC N1 primers and FAM-labelled probe alone (N1 singleplex) or with a combination with E and RNase P primers and probes (E/RNase P Duplex).
3.6. Comparison of duplex E assay and CDC singleplex N1 assay using clinical specimens
We also compared the sensitivity of the E/RNase P duplex assay with the widely used CDC N1 assay (Centre for Disease Control and Prevention, 2020), which is run in singleplex with a separate control well in which RNase P is run in singleplex as a sample sufficiency control. We tested 422 clinical samples with the E/RNase P duplex assay or the N1 singleplex assay (Fig. 4B), and observed that the E/RNase P duplex assay was highly concordant with the N1 singleplex assay. Using the same limit of detection cut-off for N1 (Cq<36.5), one hundred and forty one samples had detectable SARS-CoV2 sequences in both assays. Eight samples had detectable SARS-CoV-2 sequences in the N1 assay but not the E/RNase P duplex assay (one of which was classed as inadequate), and five samples had detectable SARS-CoV-2 sequences in the E/RNase P duplex assay but were not detected by the N1 assay. Samples which were variably detected between the two assays had Cq values which were at the limit of detection: range 34.0–36.4 for N1, and 34.7–36.1 for E in the duplex assay. Although the Cq values recorded for N1 were significantly lower than those recorded for the E gene in the duplex assay (p = 0.0015, Wilcoxon matched pairs signed rank test), the difference was small (0.2 cycles). There was also a strong linear correlation across Cq = 13 to Cq = 36, approximately 7 logs. In this cohort 2.1 % of samples (9/422) were classified as inadequate by RNase P Cq, approximately reproducing the rate observed in the cohort above.
3.7. Effect of sample storage on sensitivity of the duplex E/RNase P RT-qPCR
RNA was extracted from three aliquots of clinical samples collected in VTM immediately on receipt or after 24 h (n = 16) or 72 h (n = 17) incubation at room temperature (Fig. 3b). Three of 16 samples extracted at 0 and 24 h had detectable SARS-CoV-2 at both timepoints. One additional sample was classed as “not detected” when processed immediately, but had a Cq of 36.18 when extracted after 24 h incubation, consistent with stochastic detection of low copy number samples. A 24 h delay in RNA extraction was associated with an average increase of 0.05 in Cq values for the E gene in positive samples and an increase of 0.89 the Cq values for RNase P. When processed after 72 h incubation at room temperature, the same three samples were also classified as “detected”, and the mean Cq increased by 0.85 for the E gene in positive samples and by 1.13 for RNase P.
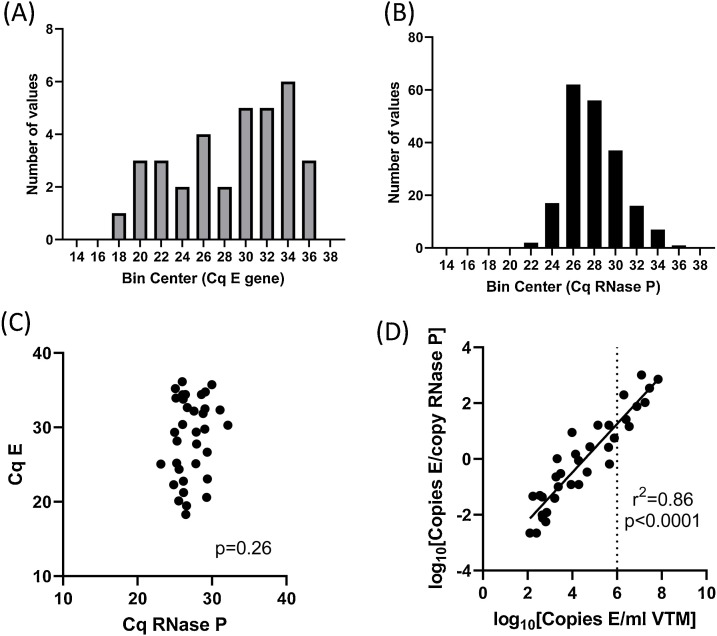
3.8. Relationship between swabbing efficiency and viral copy number
In order to determine how swabbing efficiency relates to the viral copy number detected, we analyzed 204 clinical specimens collected between April 16th to 29th 2020. The viral copy numbers in these samples ranged from 1 × 102 copies/swab to 7 × 107 copies/swab. Histograms of the Cq values for E gene and RNase P are plotted in Fig. 5 . No correlation was observed between E and RNase P Cqs, indicating that, when detected, the copy number of E is likely to be representative of the viral burden in the tissue sampled, rather than a function of how much material is sampled by the swab (Fig. 5C).
Fig. 5.
Distribution of E and RNase P Cq in clinical samples.
Clinical samples (n = 204) were assayed using the E primers and FAM-labelled probe in combination with RNase P primers and HEX-labelled probe (Duplex). N = 34 samples had detectable SARS-CoV-2, 6 were classed as inadequate due to lack of amplification of RNAse P, and the remainder were classified as ‘SARS-CoV-2 not detected’. Histograms of Cq values for E gene in samples with detectable SARS-CoV-2 (A) and Cq values for RNase P in all adequate samples (B). (C) E versus RNAse P Cq values in all samples with detectable SARS-CoV-2. (D) Normalised copy number of E versus per-swab copy number. Dotted line indicates 1,000,000 copies/mL (Cq E gene = 24 in the duplex assay).
3.9. Normalization of E to RNase P
To investigate the feasibility of normalizing viral copy number between swabs (i.e. correcting for sampling due to differences in the amount of material collected per swab), we normalized the number of copies of E to the number of copies of RNase P using the 2^(-ΔΔCt) method. We observed a range of 0.002–1027 copies of E per copy of RNase P in samples with detectable SARS-CoV-2 in this cohort, and the normalized E copy number was significantly correlated with the per-swab copy number (Fig. 5D).
4. Discussion
In this paper we show that, with the appropriate controls, accurate assessment of viral burden of SARS-CoV-2 is achievable in a clinical setting using RT-qPCR. To our knowledge, there are no other reports in the literature which take the same approach we describe here, which builds on the strengths of the Corman protocol (the Charité E primers and probe are extremely sensitive and can detect >99.9 % of variants deposited in GISAID at the time of writing) and the CDC protocol (an internal control to confirm swab efficiency). This will allow clinicians and public health officials to incorporate viral burden into their decision making process. In fact, although the results of most RT-qPCR assays of SARS-CoV-2 are currently reported qualitatively (as “detected”/”not detected”), Cq/Ct values generated could be converted to viral copy numbers without needing to change the assay substantially.
Our data demonstrates that a duplex assay which detects viral and human RNA targets has an efficiency approaching 100 % over 7 logs, is highly reproducible over a broad dynamic range, and has a limit of detection comparable to other available assays (Pujadas et al., 2020; Corman et al., 2020; Centre for Disease Control and Prevention, 2020; Arnaout et al., 2021). We also show how storage affects quantification of viral copy number: as expected, sensitivity of detection in samples with low viral copy numbers is disproportionately negatively affected by storage. Although the clinical significance of these low viral burden carriers remains to be elucidated, assessment of low copy number samples must bear in mind the effect of storage on this subset of samples.
In our assay the RNase P Cq values generated from nasopharyngeal swabs were normally distributed, and did not correlate with the viral copy number detected. We conclude from these data that swabbing efficiency is robust enough to discriminate individuals with high viral burden from those with low viral burden with high confidence. It has been reported in the literature that infectious virus can only be recovered from swabs in which there are >1,000,000 viral copies per swab. In our assays this equates to a Cq of 24 for the E gene, when 10 μL RNA is analyzed (40 μL VTM). In practice, a cut off of RNase P Cq >37 is sufficient to identify all individuals with a viral burden consistent with recovery of infectious virus. Further work would also be required to investigate whether the total viral copy number per swab or the normalized E copy number is a better predictor of the probability of recovering infectious virus, and an individual’s potential to transmit the virus.
Our approach of measuring both E and RNase P revealed a sample inadequacy of 2–2.5 % across our clinical samples from hospitalized patients swabbed by healthcare professionals. This information is critical, as otherwise these samples would be reported as ‘SARS-CoV-2 not detected’, and could be false negatives. Measurement of RNase P in a replicate well is standard in the CDC protocols, however there are significant advantages to measuring both E and RNase P in the same PCR reaction: it provides an internal control for sample sufficiency (particularly important in the context of self-swabbing), and will reveal the presence of PCR inhibitors. In addition, setting up a single PCR reaction rather than two reduces the cost of enzymes and the workload, and perhaps most importantly in the current pandemic, allows greater capacity for testing without compromising on test quality.
Funding
This work was supported by the NIHR Imperial Biomedical Research Centre (BRC).
CRediT authorship contribution statement
Aileen G. Rowan: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing - original draft, Writing - review & editing. Philippa May: Conceptualization, Formal analysis, Investigation, Methodology, Writing - review & editing. Anjna Badhan: Conceptualization, Formal analysis, Investigation, Methodology, Writing - review & editing. Carolina Herrera: Conceptualization, Formal analysis, Investigation, Methodology, Writing - review & editing. Patricia Watber: Formal analysis, Investigation, Writing - review & editing. Rebecca Penn: Resources, Writing - review & editing. Michael A. Crone: Conceptualization, Methodology, Software, Writing - review & editing. Marko Storch: Methodology, Software, Writing - review & editing. Jeremy A. Garson: Conceptualization, Methodology, Writing - review & editing. Myra McClure: Conceptualization, Resources, Writing - review & editing. Paul S. Freemont: Conceptualization, Resources, Writing - review & editing. Pinglawathee Madona: Resources, Writing - review & editing. Paul Randell: Conceptualization, Resources, Writing - review & editing. Graham P. Taylor: Conceptualization, Resources, Investigation, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
We are extremely grateful for the generous assistance of volunteers from Imperial College St Mary’s Campus who contributed to sample processing during the early response to the SARS-CoV-2 pandemic, without whom none of this work would have been possible: Marco Biones, Simon Dustan, Silva Hilburn, Maryam Khan, Akif Khawaja, Andrew Lovell, Katie O’Fee, Ana Pedrero-Llamas, Rachael Quinlan, Zainab Saeed, Sophie Sagawe, Bethany Schneiderman, Charlotte-Eve Short, Thilipan Thaventhiran, Frederic Toulza, Jocelyn Turpin and Sonia Wolf. Many thanks also to Dr. Jie Zhou and Professor Wendy Barclay for helpful discussions regarding PCR, and to Leanne Hughes and Tony MacDonald for logistics and IT support.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2021.114174.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Arnaout R., Lee R.A., Lee G.R., Callahan C., Cheng A., Yen C.F., et al. The limit of detection matters: the case for benchmarking severe acute respiratory syndrome coronavirus 2 testing. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciaa1382. 2020.06.02.131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Centre for Disease Control and Prevention . 2020. Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes.https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crone M.A., Priestman M., Ciechonska M., Jensen K., Sharp D.J., Anand A., et al. A role for Biofoundries in rapid development and validation of automated SARS-CoV-2 clinical diagnostics. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-18130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty A.B., Harrison E.M., Green C.A., Hardwick H.E., Pius R., Norman L., et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ. 2020;369:1–12. doi: 10.1136/bmj.m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones Tc, Mühlemann B., Veith T., Biele G., Zuchowski M., Hoffmann J., et al. An analysis of SARS-CoV-2 viral load by patient age. medRxiv. 2020 2020.06.08.20125484. [Google Scholar]
- Pastorino B., Touret F., Gilles M., Luciani L., de Lamballerie X., Charrel R.N. Evaluation of chemical protocols for inactivating SARS-CoV-2 infectious samples. Viruses. 2020;12 doi: 10.3390/v12060624. 131–2, 135–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujadas E., Chaudhry F., McBride R., Richter F., Zhao S., Wajnberg A., et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir. Med. 2020;2:2. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ra S.H., Lim J.S., Kim G., Kim M.J., Jung J., Kim S.-H. Upper respiratory viral load in asymptomatic individuals and mildly symptomatic patients with SARS-CoV-2 infection. Thorax. 2020 doi: 10.1136/thoraxjnl-2020-215042. thoraxjnl-2020-215042. [DOI] [PubMed] [Google Scholar]
- Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P., et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Infect. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.