Abstract
Flavonoids have been shown to have antioxidant, anti-inflammatory, anti-proliferative, antibacterial and antiviral efficacy. Therefore, in this study, we choose 85 flavonoid compounds and screened them to determine their in-silico interaction with protein targets crucial for SARS-CoV-2 infection. The five important targets chosen were the main protease (Mpro), Spike receptor binding domain (Spike-RBD), RNA - dependent RNA polymerase (RdRp or Nsp12), non-structural protein 15 (Nsp15) of SARS-CoV-2 and the host angiotensin converting enzyme-2 (ACE-2) spike-RBD binding domain. The compounds were initially docked at the selected sites and further evaluated for binding free energy, using the molecular mechanics/generalized Born surface area (MMGBSA) method. The three compounds with the best binding scores were subjected to molecular dynamics (MD) simulations. The compound, tribuloside, had a high average binding free energy of −86.99 and −88.98 kcal/mol for Mpro and Nsp12, respectively. The compound, legalon, had an average binding free energy of −59.02 kcal/mol at the ACE2 spike-RBD binding site. The compound, isosilybin, had an average free binding energy of −63.06 kcal/mol for the Spike-RBD protein. Overall, our results suggest that tribuloside, legalon and isosilybin should be evaluated in future studies to determine their efficacy to inhibit SARS-CoV-2 infectivity.
Keywords: Docking, Molecular dynamics (MD), Binding free energy, MMGBSA, SARS-CoV-2, Flavonoids
List of abbreviations
- Mpro,
Main protease;
- SGp-RBD,
Spike glycoprotein-receptor binding domain;
- ACE2,
Angiotensin-converting enzyme-2;
- RdRp,
RNA dependent RNA polymerase;
- non-structural protein 12,
Nsp12;
- non-structural protein 15 or Endoribonuclease,
Nsp15;
- SARS-CoV-2,
Severe acute respiratory syndrome coronavirus 2;
- COVID-2,
Coronavirus disease;
- Molecular Mechanics/Generalized Born Surface Area,
MM-GBSA;
- root-mean-square deviation,
RMSD
1. Introduction
Currently, numerous research groups are developing compounds and vaccines for the treatment of patients infected with SARS-CoV-2 [1]. Several strategies have been used to develop drugs to treat SARS-CoV-2. Unfortunately, due to restrictions regarding the use of SARS-CoV-2 (i.e. biosafety level III requirements) and an ongoing pandemic, laboratory research has been greatly limited. Computational tools have been widely used for development of variety of drugs including this study by screening libraries to discover compound that could affect the attachment, maturation and replication of SARS-CoV-2 [2,3]. The major focus of these studies was to obtain information about the SARS-CoV-2 proteins and target them with natural phytochemicals. The major targets studied were the SARS-CoV-2 main protease (Mpro) [4], spike glycoprotein-receptor binding domain (Spike-RBD) [5], angiotensin-converting enzyme-2 (ACE2) [6], RNA-dependent RNA polymerase (RdRp; also known as non-structural protein 12(Nsp12)) [7] and non-structural protein 15 (Nsp15) [8]. The protein, Mpro, also known as the 3C-like protease, is required for the proteolytic maturation of the SARS-CoV-2 [9]. It is also essential for the regulation and cleavage of the polyproteins, pp1a and pp1ab, that generates functional proteins, such as Nsp12, Nsp15 and exoribonuclease [10,11]. The inhibition of this enzyme prevents viral maturation and increases the host immune response to SARS-CoV-2 [12,13]. The spike protein, or S protein of SARS-CoV-2, mediates viral infection and is involved in viral pathogenesis [14]. The S1 subunit of the S protein recognizes and binds to the host receptor, angiotensin-converting enzyme 2 (ACE2), which induces the cleavage of the spike protein that produces a conformational change in S2 region to facilitate the fusion of the viral envelope with the host cell membrane [1,15]. The region of the S protein that binds to ACE2 receptor is the receptor-binding domain (RBD) or Spike-RBD [16]. The ACE2 receptor is a transmembrane metalloproteinase that is a functional receptor for SARS-CoV-2 [17,18]. The interaction between the viral S protein and ACE2 trimerizes the S glycoprotein, producing the internalization of the virus by the host cell [19]. Therefore, the ACE2 and S proteins are important drug targets for preventing the entry of SARS-CoV-2 into the host cell [20]. The non-structural protein 12, in combination with the non-structural proteins 7 and 8, catalyzes the synthesis of a positive-stranded RNA molecule that is required for viral translation and replication and thus, infectivity [21,22]. A number of studies have reported that the inhibition of the Nsp12 by compounds such as favipiravir and remdesivir, inhibits the in vitro and in vivo replication of SARS-CoV-2, thereby significantly decreasing infectivity [23,24]. Finally, the coronavirus uridylate-specific endoribonuclease, Nsp15, a highly conserved protein among vertebrate nidoviruses, has been suggested to be a target for the development of drugs for SARS-CoV-2 [25]. Nsp15 mediates replication and the processing of sub-genomic RNAs during the replication cycle and it biodegrades viral polyuridine sequences to decrease the detection of the virus by the immune system of the host cell [26].
It has been reported that natural phytochemicals, such as certain bioflavonoids, have efficacy against viruses such as the dengue virus, herpes simplex virus (HSV), Japanese encephalitis virus and Ebola [[27], [28], [29], [30], [31], [32]]. In addition, certain bioflavonoid compounds have in vitro and in vivo anti-inflammatory and anti-oxidant efficacy [33]. Thus, it is possible that bioflavonoids could have efficacy in the treatment of SARS-CoV-2 by decreasing viral infectivity and replication and/or decreasing the inflammatory response. The identification of naturally-derived anti-SARS-CoV-2 compounds can be done by screening various databases/libraries using computational approaches, such as molecular docking, pharmacophore mapping, 3D-QSAR and molecular dynamics (MD) simulations [[34], [35], [36], [37], [38], [39], [40], [41]]. The computational screening approach is advantageous at this time due to the limited access to laboratories and researchers can focus on the outcome of these studies, which could represent a platform for the design and development of novel compounds within a relatively short period of time [42].
In this study, we screened 85 flavonoids that were previously reported to have antimicrobial and antioxidant efficacy based on docking scores and molecular mechanics/generalized Born surface area (MMGBSA) [43]. The purpose of the high-throughput screening was to identify compounds that have high docking scores for the active site of the SARS-CoV-2 proteins, Mpro, SpikeRBD, Nsp12 and NSP15 in SARS-CoV-2 and the host ACE-2 receptor.
2. Material and methods
2.1. Platform for molecular modeling
All computational studies and visualization of the protein-ligand complexes were performed using the Schrodinger drug discovery suite (Release 2020-4: Schrödinger, LLC, New York, NY, 2020) and Pymol version 2.4.1 [44].
2.2. Flavonoid compounds library preparation
A dataset of 85 flavonoid molecules, with known antiviral and antimicrobial efficacy, were selected from the PubChem database (www.pubchem.ncbi.nlm.nih.gov) and their in silico interaction with the SARS-CoV-2 target proteins was determined for binding energy [Suppl file Table 1 ]. The SDF files of the compounds were imported to the Maestro molecular modeling suite. The LigPrep tool of Maestro (Schrödinger Release 2020-4) was used to generate optimized, low-energy conformers of the ligands using the OPLS3e force field [45]. Possible tautomeric states were generated for each ligand at a pH of 7.0 ± 2.0 using the Epik [46,47]. The specified chirality was retained and a maximum of 32 stereoisomers were generated per ligand. This approach yielded 843 structures generated from the 85 selected compounds and were used for docking and to estimate binding free energy.
Table 1.
Docking score and binding free energy values for the SARS-CoV-2 Mpro active site.
2.3. Protein preparation
Based on the available 3D structures of the target proteins, high resolution structures were retrieved from the protein data bank (PDB) [48]. The structures of the SARS-CoV-2 main protease (Mpro) (PDB ID: 6LU7) [49], the non-structural protein 12 (nsp12) (PDB ID: 7BV2) [42] and non-structural protein 15 (nsp15) (PDB ID: 6WXC) [43], were complexed with a co-crystallized inhibitor. The structure of the spike receptor binding domain was complexed with the angiotensin-converting enzyme-2 (ACE2) without a co-crystallized inhibitor (PDB ID: 6M0J) [44]. Using the Protein Preparation Wizard of Maestro, the selected structures were processed as as per the general protocol. Briefly, the missing hydrogen atoms were added to the structure and the hydrogen bond network was optimized using PROPKA at pH 7.0. All water molecules beyond 3 Å were removed and a restrained minimization was conducted using the OPLS3e force field with a convergence criterion of 0.3 Å root-mean-square deviation (RMSD) for all the heavy atoms. The histidine protonation state for the structures of Mpro and Nsp15 were given specific attention. The histidine protonation network for SARS-CoV-2 Mpro was optimized as per our previous protocol [50]. While processing the structure of Nsp15, the protonation state was optimized using an interactive H-bond optimiser panel usinf PROPKA at pH 7.0. The His-250 residue was kept delta protonated (HID-250), whereas His-235 was eta protonated (HIE-235) as previously reported [11].
2.4. Docking protocol
For docking with the SARS-CoV-2 main protease, Nsp12 and Nsp15, the coordinates of the co-crystallized ligands were used to prepare the receptor grid. For the spike RBD and ACE2, the binding site was defined based on the interactions between the interfaces of spike and ACE2 protein. The receptor grid generation tool of the docking program Glide in Maestro was used to define the docking site for the target proteins [[51], [52], [53]]. The partial charge limit was 0.25 and the van der Waals scaling factor was 0.80. While preparing the grid for the target proteins, the size of the outer box of the docking grid was fixed at 30 Å × 30 Å × 30 Å and the inner box dimensions were 10 Å × 10 Å × 10 Å. The size of the ligands to be docked was ≤20 Å. The docking was conducted using the Glide extra precision (XP) modes with flexible ligand sampling. The Epik state penalties were added to the final docking scores. For each ligand, ten poses were used for post-docking minimization. A maximum one pose per ligand was generated. The docking protocol for Mpro, Nsp12 and Nsp15 was validated by redocking the co-crystallized ligand with their respective crystal positions.
2.5. Binding free energy estimation
The output pose viewer file from the Glide XP docking was used as an input to re-rank the compounds using the Prime MM-GBSA binding energy [54,55] values calculated with implicit solvation model (VSGB2.1) in the OPLS3e force field. The binding free energy was calculated using the following equation:
where is the difference in energy between the complex structure and the sum of the energies of the ligand and unliganded receptor. The is the difference in the GBSA solvation energy of the complex and the sum of the solvation energies for the ligand and unliganded receptor. The is the difference in the surface area energy for the complex and the sum of the surface area energies for the ligand and uncomplexed receptor. The Prime/MM-GBSA energies after MD simulation were calculated from the last 50 ns of the simulation trajectories.
2.6. Molecular dynamics (MD) simulation
The highest scoring ligand-protein complexes, with high binding free energies (ΔGBind) and/or Glide docking scores, were submitted to a 100ns molecular dynamics (MD) simulation using Desmond (Schrödinger Release 2020-4: Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, USA, 2020) [56]. The simulation was used to refine the docked poses and evaluate the stability of binding at the active sites on the target proteins. In order to compare the efficiency of these screened compounds, the native crystal complexes of Mpro, Nsp12 and Nsp15 were simulated using the same protocol. The simulation systems were prepared using the System Builder tool of the Desmond module. The single point charge (SPC) water [57] was chosen as the explicit solvation model. Each system was neutralized using an appropriate number of Na+ or Cl-counter ions. An orthorhombic simulation box, with Periodic Boundary Conditions (PBC) and a 10 Å buffer space between the solute and the box edge, was used for each system. The simulation systems were minimized and equilibrated before the actual simulations and before production simulation in a stepwise manner. After system relaxation, the production simulation was performed using the NPT ensemble for 100 ns, using a reversible reference system propagator algorithms (RESPA) integrator [56]. The temperature (300 K) was set using the Nosé–Hoover chain thermostat [[58], [59], [60]], with a relaxation time of 1.0 ps. The pressure was set at 1.01325 bar with the Martyna–Tobias–Klein barostat [61], using isotropic coupling and a relaxation time of 2.0 ps. Long-range interactions were handled using the U-series method [62] and for short range interactions, a cut-off radius of 9.0 Å was used. The MD trajectories were analyzed using the Maestro built-in Simulation Interactions Diagram tool and Microsoft Excel360.
2.7. Docking validation
The docked poses of co-crystallized complexes were redocked at their respective binding sites to determine the precision of the docking protocol we used for Mpro, Nsp12 and Nsp15.
3. Results
3.1. Computational screening of the therapeutic targets of SARS-CoV-2
The ligprep generated 843 structures of the flavonoid's compounds from the original 85 entries that were subjected to brief molecular docking and a binding free energy estimation protocol. The compounds were then selected based on binding free energy, docking scores and individual observation of each pose at the binding sites. For each target, the 10 compounds with best docking pose were identified. The molecular dynamics (MD) simulation of the best 3 compounds at each of the targets were selected and subjected to 100 ns MD simulation. The RMSD and RMSF graphs for Cα carbon atoms were plotted. The interaction of the ligand with each protein was performed. The average binding free energy calculated by Prime-MM/GBSA method was based on the last 50 ns MD simulation.
3.1.1. SARS-CoV-1 main protease (Mpro) site
The docking and subsequent binding free energy estimation yielded the top 10 flavonoids (Table 1). The monoxerutin shows the highest docking score among all the compounds at the Mpro site. The MMGBSA-based binding free energy for monoxerutin was −78.88 kcal/mol, which is comparable to that of peptide inhibitor, N3. Furthermore, the compounds, tribuloside and panasenoside, had a greater interaction with mPro compared to co-crystallized N3. All of the top 10 compounds had a binding free energy > −70 kcal/mol (Table 1).
3.1.2. The spike -receptor binding domain (Spike-RBD) site
The glycosylated spike protein (S-protein) directly interacts with the human angiotensin-converting enzyme 2 (ACE2) receptor via the receptor-binding domain (RBD) of the S-protein [63]. As the S-protein is exposed to the surface and is essential for viral entry into the host cells, the S-protein is considered to be a first-line therapeutic target for the treatment of SARS-CoV-2 infection [64]. The MMGBSA - based binding free energy for isosilybin was −68.39 kcal/mol and this value was the highest compared to the other flavonoids for the Spike RBD. Table 2 shows the 10 compounds with the highest scores for the RBD of spike, with binding free energy > −50 kcal/mol before MD simulation.
Table 2.
Docking score and binding free energy values for the SARS-CoV-2 Spike RBD active site.
3.1.3. Angiotensin converting enyme-2(ACE-2) spike protein binding site
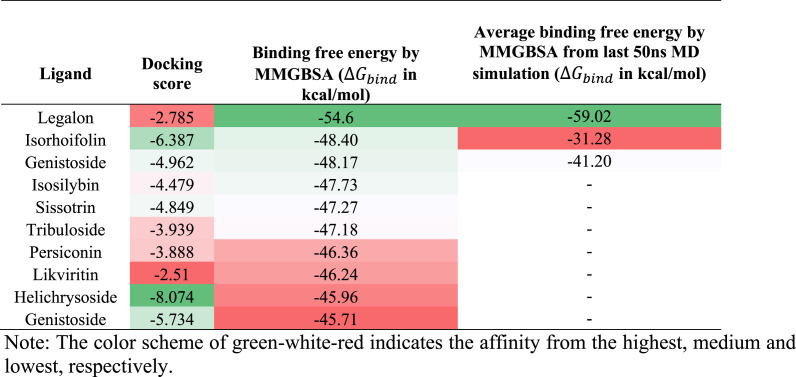
The angiotensin-converting enzyme-2 (ACE-2) is a receptor that binds the spike protein of SARS-CoV-2, which activates the host cell protease and produces the fusion of the spike protein with host cell membrane [1]. Consequently, the inhibition of the binding of the spike protein with the ACE-2 protein could be an effective strategy to prevent SARS-CoV-2 infeciton [1]. The flavonoids with the highest docking scores are shown in Table 3 . For the ACE-2 spike protein binding site, isorhoifolin had the highest docking score, whereas kikviritin had the lowest docking score. The MMGBSA based binding free energy for legalon was −54.6 kcal/mol.
Table 3.
Docking score and binding free energy values for the SARS-CoV-2 SpikeRBD active site of the human ACE2 receptor.
3.1.4. The non-structural protein-12 (Nsp12)
The RNA-dependent RNA polymerase (RdRp), also known as Nsp12, catalyzes the synthesis of viral RNA, in combination with cofactors Nsp-7 and Nsp-8, within the host cell and plays a major role in the replication and transcription of SARS-CoV-2 [65]. In this study, we docked the compounds at Nsp12 using co-ordinates for co-crystallized remdesivir (Table 4 ). The flavonoids, isoorientin and tribuloside, had significantly greater docking scores of −10.96 and −11.571 kcal/mol, respectively, compared to −6.244 kcal/mol for remdesivir (it is important to note that it can form a covalent bond at this site). Similarly, the binding free energy of isoorientin and tribuloside for Nsp12 were appreciable compared to remdesivir.
Table 4.
Docking score and binding free energy values for t remdesivir at the SARS-CoV-2 protein, Nsp12.
3.1.5. Molecular docking with endoribonuclease (NSP15)
The non-structure protein, Nsp15, is essential in the life cycle and virulence of SARS-CoV-2 [66]. We performed docking and MMGBSA analysis of our compounds at the tipiracil binding site and the results are presented in Table 5 . The 10 compounds with the highest docking scores compounds at the tipiracil binding site were > - 4 kcal/mol, except for tribuloside. The docking scores and binding free energy value of Persiconin and Neosakuranin were comparable to that of Tipiracil co-crystallized at the tipiracil binding site on Nsp15.
Table 5.
Docking scorse and binding free energy values at the tipiracil binding site on SARS-CoV-2 Nsp15.
3.1.6. Validation of docking
Before performing the docking analysis at these selected sites, the redocking of co-crystallized inhibitors was performed where the structure of the co-crystallized inhibitor was available. Unfortunately, we could not validate the docking performed at the ACE2 and Spike RBD site due to unavailability of the co-crystalized inhibitor complex at the interface between ACE2 and the Spike RBD. At the N3 and remdesivir binding sites, redocking was performed by cleaving the covalent bond using the 3D builder tool of maestro, which does not change the actual orientation of the groups after regenerating the original structure of ligand and amino acid side chain. Our results indicated a slight deviation in the orientation of N3 during redocking that was due to the larger size of this peptide ligand as we were able to generate very accurate redocked poses at the other two sites (Fig. 1 ).
Fig. 1.
a The redocked N3 peptide (cyancolour ball and stick representation), in grey, at the active site of SARS-CoV-2 Mpro (PDB ID: 6LU7) with co-crystallized. covalently bonded N3 peptide (represented by and orange ball and stick). b. Co-crystallized tipiracil (represented by a cyan colour ball and stick) and redocked tipiracil (represented by a pink ball and stick) showing key interactions (green sticks representing amino acids, phosphate groups represented as red sticks and interactions represented by cyan dash lines) with Nsp15 (grey cartoon representation). c. Co-crystallized and redocked remdesivir (represented by white and green balls and sticks, respectively) with Nsp12 protein (the interacting amino acids and RNA chains are represented by thin grey sticks). Nonpolar hydrogens are hidden for clarity.
4. Discussion
Numerous studies have reported that flavonoids and their derivatives are efficacious in vitro in several viral strains, including avian influenza strain H5N1, HIV, HSV and Ebola [67]. In this study, we used an in silico approach to screen a library of 85 flavonoids to identify compounds that have a high binding affinity for the critical SARS-CoV-2 proteins, main protease (Mpro), receptor binding domain of spike protein- (Spike-RBD), angiotensin-converting enzyme-2 (ACE-2), non-structural protein 12 and non-structural protein 15.
The active site of SARS-CoV-2 Mpro has been reported to have 4 sub-sites: S1′, S1, S2 and S4 [49]. The catalytic Cys-145 and His-163 are located in the S1’ site. The Glu-166 residue is in the S1 pocket, whereas Met-165 and Val-186 are in the S2 pocket and Pro-168 and Gln-189 are located in the S4 site [49]. As compared to other receptor sites in the SARS-Cov-2 proteins, the Mpro site is comparatively flexible and can accommodate a wide variety of ligands, including peptides and small molecules [68]. This characteristic is important as it makes the docking critical because many compounds could have appreciable docking scores at this site. Consequently, the use of MD simulation is critical for determining stable interactions between the screened compounds and the docking site. As shown in Fig. 2 , certain flavonoid compounds formed an efficient hydrogen bond network with key amino acids and the details of these interactions are shown in the supplementary results [see suppl file- Figure S1, S6, S11, S16].
Fig. 2.
The 3D structure of SARS-CoV-2 Mpro (PDB ID:6LU7) and its interaction with Tribuloside (left) and the ligand interaction in 2D (right side). The protein is represented in grey, tribuloside is represented by a cyan ball and sticks, amino acids are represented by green sticks (right) and donor and acceptor hydrogen bonds interactions are represented by pink arrows (left).
Tribuloside had the highest magnitude of interaction among all of the compounds for Mpro, based on the binding free energy before and after MD simulation. There was a slight decrease in initial interaction energy from −93.55 to −86.99 kcal/mol during the simulation. However, it is important to note that the latter value is the average binding score, which suggests that it could inhibit Mpro if tested in vitro, although this remains to be determined.
As with the ACE2 site, the binding pocket of RBD is also relatively shallow and the MD simulation for the ligand is important to determine the stability of the interactions of the compounds at this site. Furthermore, this will allow for the detection of false positive ligand poses during docking. Isosilybin had hydrogen bond interactions with Gly-202 and Gly-493, in addition to water mediated interactions with Thr-505, Arg-403 and Gly-498 (Fig. 3 ). Isosilybin also forms pi-pi interactions with Tyr-505, which increases the stability of its interaction with the binding pocket of RBD compared to panasenoside. Compared to panasenoside and helichrysoside, whose interaction decreased after MD simulation, isosilybin had a better docking pose. [see suppl file- Figure S2, S7, S12, S17].
Fig. 3.
The 3D structure of the interaction of SARS-CoV-2 Spike RBD protein (PDB ID:6MOJ) with isosilybin (left) and the ligand interaction in 2D(right side). The proteinis represented in grey, isosilybin represented by cyan balls and stick and the amino acids are represented by green sticks (right). The hydrogen bond interactions are represented in pink and the pi-pi interactions are represented by green arrows (left).
In this study, we selected a novel binding site when we conducted the docking poses at ACE2. The rationale behind the selection of this site was based on the fact that this site is in close proximity to the N-glycosylation site of ACE2, which occurs at the conserved residue, Asn-90 [[69], [70], [71], [72]]. It has been reported that N-glycosylation of ACE2 and its inhibition by chloroquine and its derivatives could affect the binding of the Spike-RBD to the ACE2 receptor [[51], [52], [53], [54]].
Fig. 4 shows the binding interactions of legalon at the ACE2 site. The major interactions between legalon and the ACE2 site involved Asn-90, Lys-26, Asp-30, His-34, Gly-37, Gly-96, Ala-387 and Arg-393. Additional details of the interactions are presented in the supplemental results (Figure S3, S8, S13, S18 in Suppl file). The interactions of legalon could affect SpikeRBD binding at this site and future in vitro studies should be done to verify this possibility.
Fig. 4.
The 3D structure of SpikeRBD binding domain of human ACE2 receptor (PDB ID:6MOJ) with Legalon (left) and ligand interaction in 2D view (right side). The protein is in grey cartoon representation, Legalon in cyan ball and sticks, amino acids in green sticks (right). The hydrogen bond interactions in pink and pi-pi interactions in green arrows (left).
The protein, Nsp12, in combination with the proteins, Nsp7 and Nsp8, is involved in the replication of the SARS-CoV-2 RNA genome [73]. Previous studies have reported that various flavonoids, including the compounds, C4, C8, C12, C16, C17, C20, C22, C23, calophyllolide, genistein, quercetin, tangeritin, arbutin and liquiritin, interact with the β-hairpin residues of Nsp12 [74]. The compounds we chose for MD simulation, tribuloside, isoorientin and silybin, had stable interactions at the remdesivir binding site is shown in Fig. 5 . The rank order of the docking scores was tribuloside > isoorientin [see Suppl file-Figure S4, S9, S14, S19]. The root mean square fluctuation (RMSF) for the three compounds was convergent and was stable thought the MD simulation. The per residue interaction calculated using RMSF remained stable and was similar to the RMSF for the covalently bound, co-crystallized ligand, remdesivir.
Fig. 5.
The 3D structure of SARS-CoV-2 Nsp12 (PDB ID:7BV2), with Isoorientin (left) and the ligand interaction in 2D (right side). The protein is represented in grey, representation, isoorientin is represented by cyan balla and sticks, amino acids are represented by green sticks (right and the donor and acceptor hydrogen bond interactions are represented as pink arrows (left).
Our results indicated that the active binding site of Nsp15 is highly specific for the uridine moiety of tipiracil and consequently, tipiracil inhibits Nsp15 and thus, it catalytic activity [57]. The uracil ring of tipiracil can stack against Tyr-341 and forms hydrogen bond interactions with Ser-294 and His-250. Our docking and binding free energy analysis indicated that certain flavonoid had high docking and binding free energy values at the active site of Nsp12. In addition, the compounds with the highest values were subjected to MD simulation but none of them maintained the initial interactions before simulation. The compounds, persiconin and isorhoifolin, had moderate interactions, whereas as biorobin lost significant interactions for Nsp12 after MD simulation. Furthermore, the RMSF did not converge well for biorobin. The detail binding interactions are shown in Fig. 6 and in the supplemental results [Suppl file-Figure S5, S10, S15, S20].
Fig. 6.
The 3D structure of SARS-CoV-2 Nsp15 (PDB ID:6WXC) with Persiconin and (left) Isorhoifolin (right) and ligand interaction in 2D view (right side). The protein is in grey cartoon representation, Persiconin and Isorhoifolin in cyan ball and sticks, amino acids in green sticks (right), donor and acceptor hydrogen bond interactions in pink arrows (left), Non-polar hydrogens are hidden for clarity.
The coumarin ring of isorhoifolin and phenyl rings formed pi-pi interactions with Trp-333 and His-235 respectively. Isorhoifolin formed hydrogen bonds with Gly-285, Gly-151, Gly-189 and Thr-286. Similarly, periconin had significant interactions with Ser-316, Val-318, Trp-333, Leu-346 and Gly-347. However, none of the compounds formed the key interactions as observed for co-crystalized tipiracil. Also, none of the flavonoids interacted with key residues, such as Ser-294 and His-250.
5. Conclusion
In the present in silico study, 85 flavonoids, with known antiviral efficacy, were selected to determine their interactions at five proteins that are known to play a role in the infectivity of SARS-CoV-2. Our results indicated that tribuloside formed significant interactions with the SARS-CoV-2 proteins, Mpro and Nsp12. In contrast, our results suggested that legalon could be a potential inhibitor of ACE2 spike RBD binding site. Compared to all of the compounds, isosilybin had the highest interaction with the Spike-RBD protein. The interaction of all of the compounds with the active site of Nsp15 was significantly weaker than that of tipiracil, which has been reported to have a low to moderate inhibitory efficacy for Nsp15. It is possible that further investigation of the effects of these compounds for the protein targets could help elucidate their exact mechanism of binding. Finally, our results could prove useful for exploring and developing novel natural compounds that have anti-SARS-CoV-2 efficacy.
Availability of data and material
During this project, all data is stored on the local backed-up storage servers utilizing the Biocentre Finland storage facility (RB's laboratory is the provider of that service). In addition, after the project was completed, all data were archived and made available through the IDA research data storage service (produced by CSC, the IT Center for Science Ltd, Finland, http://www.csc.fi), so that when needed these data can be retrieved for usage and sharing.
Author's contributions
All authors contributed equally to writing the paper and approved the final manuscript.
Funding
R.B. gratefully acknowledges the financial support of the Academy of Finland (mobility grant) and the Joe, Pentti and Tor Borg Memorial Fund.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The Sigrid Jusélius Foundation, Biocenter Finland Bioinformatics and Drug Discovery and Chemical Biology networks, CSC IT Center for Science. Joe, Pentti and Tor Borg Memorial Fund and Prof. Mark Johnson and Dr. Jukka Lehtonen are gratefully acknowledged for the excellent computational infrastructure at the Åbo Akademi University. We would also like to thank Dr. Outi M. H. Salo-Ahen for constructive feedback on the computational part.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104464.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Amawi H., Abu Deiab G.a.I., A Aljabali A.A., Dua K., Tambuwala M.M. COVID-19 pandemic: an overview of epidemiology, pathogenesis, diagnostics and potential vaccines and therapeutics. Ther. Deliv. 2020;11:245–268. doi: 10.4155/tde-2020-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pandey P., Rane J.S., Chatterjee A., Kumar A., Khan R., Prakash A., Ray S. Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: an in silico study for drug development. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1796811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhardwaj V.K., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput. Biol. Med. 2021;128 doi: 10.1016/j.compbiomed.2020.104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tariq A., Mateen R., Sohail Afzal S., Saleem M. 2020. Paromomycin: A Potential Dual Targeted Drug Effectively Inhibits Both Spike (S1) and Main Protease of COVID-19. Available at SSRN 3575839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng X., Hackbart M., Mettelman R.C., O'Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. Unit. States Am. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohammad T., Shamsi A., Anwar S., Umair M., Hussain A., Rehman M.T., AlAjmi M.F., Islam A., Hassan M.I. Identification of high-affinity inhibitors of SARS-CoV-2 main protease: towards the development of effective COVID-19 therapy. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198102. 198102-198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.-K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science (New York, N.Y.) 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubey K., Dubey R. Computation screening of narcissoside a glycosyloxyflavone for potential novel coronavirus 2019 (COVID-19) inhibitor. Biomed. J. 2020;43(4):363–367. doi: 10.1016/j.bj.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das P., Majumder R., Mandal M., Basak P. In-Silico approach for identification of effective and stable inhibitors for COVID-19 main protease (Mpro) from flavonoid based phytochemical constituents of Calendula officinalis. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1796799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall D.C., Jr., Ji H.-F. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Trav. Med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vankadari N. Arbidol: a potential antiviral drug for the treatment of SARS-CoV-2 by blocking the trimerization of viral spike glycoprotein? Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang J., Wan Y., Liu C., Yount B., Gully K., Yang Y., Auerbach A., Peng G., Baric R., Li F. Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., Aziz B. Structural variations in human ACE2 may influence its binding with SARS‐CoV‐2 spike protein. J. Med. Virol. 2020;92(9):1580–1586. doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 20.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 21.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhury S., Moulick D., Saikia P., Mazumder M.K. Evaluating the potential of different inhibitors on RNA-dependent RNA polymerase of severe acute respiratory syndrome coronavirus 2: a molecular modeling approach. Med. J. Armed Forces India. 2020 doi: 10.1016/j.mjafi.2020.05.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017;93:449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konkolova E., Dejmek M., Hřebabecký H., Šála M., Böserle J., Nencka R., Boura E. Remdesivir triphosphate can efficiently inhibit the RNA-dependent RNA polymerase from various flaviviruses. Antivir. Res. 2020;182 doi: 10.1016/j.antiviral.2020.104899. 104899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hackbart M., Deng X., Baker S.C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johari J., Kianmehr A., Mustafa M.R., Abubakar S., Zandi K. Antiviral activity of baicalein and quercetin against the Japanese encephalitis virus. Int. J. Mol. Sci. 2012;13:16785–16795. doi: 10.3390/ijms131216785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zandi K., Teoh B.-T., Sam S.-S., Wong P.-F., Mustafa M.R., AbuBakar S. Antiviral activity of four types of bioflavonoid against dengue virus type-2. Virol. J. 2011;8:560. doi: 10.1186/1743-422X-8-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amoros M., Simõs C., Girre L., Sauvager F., Cormier M. Synergistic effect of flavones and flavonols against herpes simplex virus type 1 in cell culture. Comparison with the antiviral activity of propolis. J. Nat. Prod. 1992;55:1732–1740. doi: 10.1021/np50090a003. [DOI] [PubMed] [Google Scholar]
- 30.Qiu X., Kroeker A., He S., Kozak R., Audet J., Mbikay M., Chrétien M. Prophylactic efficacy of quercetin 3-β-Od-glucoside against Ebola virus infection. Antimicrob. Agents Chemother. 2016;60:5182–5188. doi: 10.1128/AAC.00307-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma J., Kumar Bhardwaj V., Singh R., Rajendran V., Purohit R., Kumar S. An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against non structural protein-15 of SARS-CoV-2. Food Chem. 2021;346 doi: 10.1016/j.foodchem.2020.128933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amawi H., Ashby C.R., Tiwari A.K. Cancer chemoprevention through dietary flavonoids: what's limiting? Chin. J. Canc. 2017;36:1–13. doi: 10.1186/s40880-017-0217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.da Silva Hage-Melim L.I., Federico L.B., de Oliveira N.K.S., Francisco V.C.C., Correa L.C., de Lima H.B., Gomes S.Q., Barcelos M.P., Francischini I.A.G. Virtual screening, ADME/Tox predictions and the drug repurposing concept for future use of old drugs against the COVID-19. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Shan Y., Pan X., Wang C., Xu W., He L. Molecular docking, 3D‐QSAR Studies, and in silico ADME prediction of p‐aminosalicylic acid derivatives as neuraminidase inhibitors. Chem. Biol. Drug Des. 2011;78:709–717. doi: 10.1111/j.1747-0285.2011.01179.x. [DOI] [PubMed] [Google Scholar]
- 36.Kumar Bhardwaj V., Purohit R., Kumar S. Himalayan bioactive molecules as potential entry inhibitors for the human immunodeficiency virus. Food Chem. 2021;347 doi: 10.1016/j.foodchem.2020.128932. [DOI] [PubMed] [Google Scholar]
- 37.Aier I., Varadwaj P.K., Raj U. Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci. Rep. 2016;6 doi: 10.1038/srep34984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yadav G., Rao R., Raj U., Varadwaj P.K. Computational modeling and analysis of prominent T-cell epitopes for assisting in designing vaccine of ZIKA virus. J. Appl. Pharmaceut. Sci. 2017;7:116–122. [Google Scholar]
- 39.Kumar H., Raj U., Srivastava S., Gupta S., Varadwaj P.K. Identification of dual natural inhibitors for chronic myeloid leukemia by virtual screening, molecular dynamics simulation and ADMET analysis. Interdiscipl. Sci. Comput. Life Sci. 2016;8:241–252. doi: 10.1007/s12539-015-0118-7. [DOI] [PubMed] [Google Scholar]
- 40.Kumar A., Mehta V., Raj U., Varadwaj P.K., Udayabanu M., Yennamalli R.M., Singh T.R. Computational and in-vitro validation of natural molecules as potential acetylcholinesterase inhibitors and neuroprotective agents. Curr. Alzheimer Res. 2019;16:116–127. doi: 10.2174/1567205016666181212155147. [DOI] [PubMed] [Google Scholar]
- 41.Mahanta S., Chowdhury P., Gogoi N., Goswami N., Borah D., Kumar R., Chetia D., Borah P., Buragohain A.K., Gogoi B. Potential anti-viral activity of approved repurposed drug against main protease of SARS-CoV-2: an in silico based approach. J. Biomol. Struct. Dynam. 2020:1–10. doi: 10.1080/07391102.2020.1768902. [DOI] [PubMed] [Google Scholar]
- 42.Kant K., Lal U.R., Kumar A., Ghosh M. A merged molecular docking, ADME-T and dynamics approaches towards the genus of Arisaema as herpes simplex virus type 1 and type 2 inhibitors. Comput. Biol. Chem. 2019;78:217–226. doi: 10.1016/j.compbiolchem.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X., Wong S.E., Lightstone F.C. 2014. Toward Fully Automated High Performance Computing Drug Discovery: a Massively Parallel Virtual Screening Pipeline for Docking and Molecular Mechanics/generalized Born Surface Area Rescoring to Improve Enrichment, ACS Publications. [DOI] [PubMed] [Google Scholar]
- 44.Giardina S.F., Werner D.S., Pingle M., Feinberg P.B., Foreman K.W., Bergstrom D.E., Arnold L.D., Barany F. Novel, self-assembling dimeric inhibitors of human β tryptase. J. Med. Chem. 2020;63:3004–3027. doi: 10.1021/acs.jmedchem.9b01689. [DOI] [PubMed] [Google Scholar]
- 45.Roos K., Wu C., Damm W., Reboul M., Stevenson J.M., Lu C., Dahlgren M.K., Mondal S., Chen W., Wang L., Abel R., Friesner R.A., Harder E.D. OPLS3e: extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theor. Comput. 2019;15:1863–1874. doi: 10.1021/acs.jctc.8b01026. [DOI] [PubMed] [Google Scholar]
- 46.Greenwood J.R., Calkins D., Sullivan A.P., Shelley J.C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput. Aided Mol. Des. 2010;24:591–604. doi: 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- 47.Shelley J.C., Cholleti A., Frye L.L., Greenwood J.R., Timlin M.R., Uchimaya M. Epik: a software program for pK( a ) prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 48.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 50.Alamri M.A., Tahir Ul Qamar M., Mirza M.U., Bhadane R., Alqahtani S.M., Muneer I., Froeyen M., Salo-Ahen O.M.H. Pharmacoinformatics and molecular dynamics simulation studies reveal potential covalent and FDA-approved inhibitors of SARS-CoV-2 main protease 3CL(pro) J. Biomol. Struct. Dynam. 2020:1–13. doi: 10.1080/07391102.2020.1782768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision Glide: docking and scoring incorporating a model of hydrophobic enclosure for Protein−Ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 52.Halgren T.A., Murphy R.B., Friesner R.A., Beard H.S., Frye L.L., Pollard W.T., Banks J.L. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 53.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 54.Jacobson M.P., Pincus D.L., Rapp C.S., Day T.J., Honig B., Shaw D.E., Friesner R.A. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 55.Jacobson M.P., Friesner R.A., Xiang Z., Honig B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 56.Bowers K.J., Chow E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., Klepeis J.L., Kolossvary I., Moraes M.A., Sacerdoti F.D., Salmon J.K., Shan Y., Shaw D.E. Association for Computing Machinery; Tampa, Florida: 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters; p. 84. (Proceedings of the 2006 ACM/IEEE Conference on Supercomputing). es. [Google Scholar]
- 57.H.J. Berendsen, J.P. Postma, W.F. van Gunsteren, J. Hermans, INteraction Models for Water in Relation to Protein Hydration, Intermolecular Forces, Springer1981, pp. 331-342.
- 58.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 59.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 60.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 61.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 62.Predescu C., Lerer A.K., Lippert R.A., Towles B., Grossman J.P., Dirks R.M., Shaw D.E. The u-series: a separable decomposition for electrostatics computation with improved accuracy. J. Chem. Phys. 2020;152 doi: 10.1063/1.5129393. [DOI] [PubMed] [Google Scholar]
- 63.Hasan A., Paray B.A., Hussain A., Qadir F.A., Attar F., Aziz F.M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1754293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bourgonje A.R., Abdulle A.E., Timens W., Hillebrands J.-L., Navis G.J., Gordijn S.J., Bolling M.C., Dijkstra G., Voors A.A., Osterhaus A.D., van der Voort P.H., Mulder D.J., van Goor H. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19) J. Pathol. 2020;251:228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahmad J., Ikram S., Ahmad F., Rehman I.U., Mushtaq M. SARS-CoV-2 RNA Dependent RNA polymerase (RdRp) – a drug repurposing study. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vardhan S., Sahoo S.K. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput. Biol. Med. 2020;124 doi: 10.1016/j.compbiomed.2020.103936. 103936-103936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.da Silva F.M.A., da Silva K.P.A., de Oliveira L.P.M., Costa E.V., Koolen H.H., Pinheiro M.L.B., de Souza A.Q.L., de Souza A.D.L. Flavonoid glycosides and their putative human metabolites as potential inhibitors of the SARS-CoV-2 main protease (Mpro) and RNA-dependent RNA polymerase (RdRp) Mem. Inst. Oswaldo Cruz. 2020:115. doi: 10.1590/0074-02760200207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ngo S.T., Nguyen H.M., Thuy Huong L.T., Quan P.M., Truong V.K., Tung N.T., Vu V.V. Assessing potential inhibitors of SARS-CoV-2 main protease from available drugs using free energy perturbation simulations. RSC Adv. 2020;10:40284–40290. doi: 10.1039/d0ra07352k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kristiansen T.Z., Bunkenborg J., Gronborg M., Molina H., Thuluvath P.J., Argani P., Goggins M.G., Maitra A., Pandey A. A proteomic analysis of human bile. Mol. Cell. Proteomics : MCP. 2004;3:715–728. doi: 10.1074/mcp.M400015-MCP200. [DOI] [PubMed] [Google Scholar]
- 71.Chen R., Jiang X., Sun D., Han G., Wang F., Ye M., Wang L., Zou H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J. Proteome Res. 2009;8:651–661. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- 72.Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., Patane M.A., Pantoliano M.W. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vijayakumar B.G., Ramesh D., Joji A., Kannan T. In silico pharmacokinetic and molecular docking studies of natural flavonoids and synthetic indole chalcones against essential proteins of SARS-CoV-2. Eur. J. Pharmacol. 2020;886 doi: 10.1016/j.ejphar.2020.173448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
During this project, all data is stored on the local backed-up storage servers utilizing the Biocentre Finland storage facility (RB's laboratory is the provider of that service). In addition, after the project was completed, all data were archived and made available through the IDA research data storage service (produced by CSC, the IT Center for Science Ltd, Finland, http://www.csc.fi), so that when needed these data can be retrieved for usage and sharing.