Abstract
Tight junctions (TJs) are composed of a claudin-based anastomosing network of TJ strands at which plasma membranes of adjacent epithelial cells are closely attached to regulate the paracellular permeability. Although the TJ proteins occludin and tricellulin have been known to be incorporated in the TJ strand network, their molecular functions remain unknown. Here, we established tricellulin/occludin-double knockout (dKO) MDCK II cells using a genome editing technique and evaluated the structure and barrier function of these cells. In freeze-fracture replica electron microscopy, the TJ strands of tricellulin/occludin-dKO cells had fewer branches and were less anastomosed compared with the controls. The paracellular permeability of ions and small tracers was increased in the dKO cells. A single KO of tricellulin or occludin had limited effects on the morphology and permeability of TJs. Mathematical simulation using a simplified TJ strand network model predicted that reduced cross-links in TJ strands lead to increased permeability of ions and small macromolecules. Furthermore, overexpression of occludin increased the complexity of TJ strand network and strengthened barrier function. Taken together, our data suggest that tricellulin and occludin mediate the formation and/or stabilization of TJ-strand branching points and contribute to the maintenance of epithelial barrier integrity.
INTRODUCTION
Tight junctions (TJs) control the paracellular movement of ions and molecules in the epithelial and endothelial tissues to maintain barrier functions (Farquhar and Palade, 1963; Turner et al., 2014; Van Itallie and Anderson, 2014; Zihni et al., 2016; Piontek et al., 2020). TJs are cell–cell junction structures at the apical part of the cell, where the plasma membranes of the adjacent cells are closely apposed to each other with multiple tight attachments (so-called kissing points) in the cross-sections and provide a physical barrier to prevent free diffusion of molecules (Farquhar and Palade, 1963). This structure appears as a continuous anastomosing network of fibrils (TJ strands) on freeze-fracture replica electron microscopy (FFEM) (Chalcroft and Bullivant, 1970; Wade and Karnovsky, 1974).
TJs consist of various transmembrane proteins, such as claudins, TJ-Associated MARVEL (MAL and related proteins for vesicle trafficking and membrane link) Proteins (TAMPs), angulins, and Junctional Adhesion Molecules (JAMs) (Chiba et al., 2008; Higashi and Chiba, 2020). Claudins are considered major integral membrane proteins of TJ strands (Furuse et al., 1998a; Piontek et al., 2020) and consist of a protein family of more than 20 members, which are differentially expressed in most epithelial tissues in different combinations. Some claudins can form charge-selective paracellular pores and determine the physiological property of permeability (Furuse et al., 2001; Yu et al., 2009; Krug et al., 2012). When a claudin gene product is exogenously expressed in mouse L-fibroblasts, which lack claudin expression and TJ structure, well-developed TJ strand–like structures were reconstituted on the cell–cell contact sites (Furuse et al., 1998b), suggesting that claudins constitute the TJ strands. Recently, it was shown that claudin family-knockout MDCK (Madin-Darby canine kidney) II cells lack TJ strands (Otani et al., 2019), which demonstrated that claudins are essential for TJ-strand formation and further supported the view that claudins are the backbone of TJ strands.
Although the roles of claudins in the formation of TJ strands have been well understood, the molecular function of TAMPs remains relatively less characterized. The TAMP family is composed of three members with four transmembrane domains, occludin (encoded by Ocln), tricellulin (encoded by Tric/Marveld2), and MarvelD3 (encoded by Marveld3) (Steed et al., 2009; Raleigh, Marchiando, Zhang, et al., 2010). Occludin was discovered as the first integral membrane TJ protein (Furuse et al., 1993). Occludin appears to be copolymerized with claudins in TJ strands when expressed in fibroblasts (Furuse et al., 1998b; Cording et al., 2013; Van Itallie et al., 2017); however, it is dispensable for the formation of TJ strands, because the epithelial cells in the Ocln-knockout (KO) embryoid body and Ocln-KO mice have mature TJ strands (Saitou et al., 1998, 2000). Ocln-knockdown did not alter the steady-state barrier function for ions and uncharged solutes in cultured epithelial cells (Yu et al., 2005; Raleigh, Marchiando, Zhang, et al., 2010; Van Itallie et al., 2010), suggesting that occludin alone does not play a crucial role in the steady-state barrier function of epithelial cells. The occludin-related protein tricellulin was identified as being uniquely localized at tricellular tight junctions (tTJs) (Ikenouchi et al., 2005). However, the contribution of tricellulin to barrier function is still under debate. Tric knockdown significantly reduced the barrier for ions and low molecular weight macromolecules in EpH4 cells (Ikenouchi et al., 2005), and Tric-overexpression improved the barrier for macromolecules in MDCK II cells (Krug et al., 2009), supporting the view that tricellulin contributes to the establishment of a strong epithelial barrier especially for macromolecules. However, Tric knockdown in MDCK II cells and Caco-2 cells did not affect the barrier for ions and macromolecules (Raleigh, Marchiando, Zhang, et al., 2010; Van Itallie et al., 2010), implying that tricellulin does not have essential roles in epithelial barrier establishment. The discrepancy might arise from the varied contribution of tTJs to the overall epithelial barrier among different solutes and different cell types (Krug, 2017).
The paracellular permeability has size-and charge-selectivity. It has been proposed that TJs allow passage of solutes through two distinct routes with different selectivities (Watson et al., 2001; Anderson and Van Itallie, 2009): pore and leak pathways (Shen, Weber, et al., 2011). The pore pathway is a charge-selective and high-capacity route for ions and small molecules (∼4 Å or less), which is presumed to be based on the “pore-forming claudins” (Furuse et al., 2001; Yu et al., 2009; Krug et al., 2012). The leak pathway has less size- and charge-selectivity with relatively lower capacity compared with the pore pathway. The well-accepted model for the leak pathway is based on the dynamic TJ-strand remodeling (Claude, 1978; Sasaki, Matsui, et al., 2003). Because the TJ strand–like structure observed in the fluorescent protein–labeled, claudin-expressing fibroblasts is highly dynamic and exhibits frequent break and annealing (Sasaki, Matsui, et al., 2003; Van Itallie et al., 2017) and the TJ-forming claudins (and occludin) in epithelial cells also undergo rapid and continuous turnover (Shen et al., 2008; Van Itallie et al., 2019), it would be plausible to consider that the TJ-strand network in epithelial cells also dynamically remodels its structure and allows the permeation of solutes during the process of break and repair of the anastomosing network structure.
It has been assumed that the number and pattern of the TJ-strand network, as well as its components, determine the tightness of TJs and are dynamically regulated in response to physiological and pathological stimuli (Claude and Goodenough, 1973; Claude, 1978; Madara and Dharmsathaphorn, 1985; Tsukita et al., 2001; Schneeberger and Lynch, 2004; Zihni et al., 2016; Varadarajan, Stephenson, and Miller, 2019; Lynn, Peterson, and Koval, 2020). However, the molecular mechanisms regulating the TJ-strand network structure have yet to be uncovered. It was reported that occludin is preferentially concentrated at the strand-to-strand junction points (branching points) of the TJ strands when exogenously expressed with claudin-2 in Rat-1 fibroblasts (Van Itallie et al., 2017) and that tricellulin affects the morphology of claudin-1–based TJ strands in L-fibroblasts (Ikenouchi et al., 2008), suggesting that occludin and tricellulin could be key molecules for the regulation of TJ-strand network architecture. However, it remains unclear whether occludin and tricellulin modulate the complexity of the TJ-strand network in epithelial cells because of a lack of loss-of-function experiments.
Here, we established Tric/Ocln-double knockout (dKO) MDCK II cells and explored the roles of tricellulin and occludin in the structure and function of TJs. Our results showed that tricellulin and occludin are required for the formation of anastomosing meshwork of TJ strands and contribute to the maintenance of epithelial barrier function. Furthermore, we simulated the effect of reduced cross-links in the TJ-strand meshwork on the permeability using a mathematical model of the simplified TJ-strand network. Our results and simulation suggest that TAMP family proteins affect the molecular architecture of TJs and regulate epithelial permeability.
RESULTS
Establishment of Tric-KO, Ocln-KO, and Tric/ Ocln-dKO epithelial cell lines
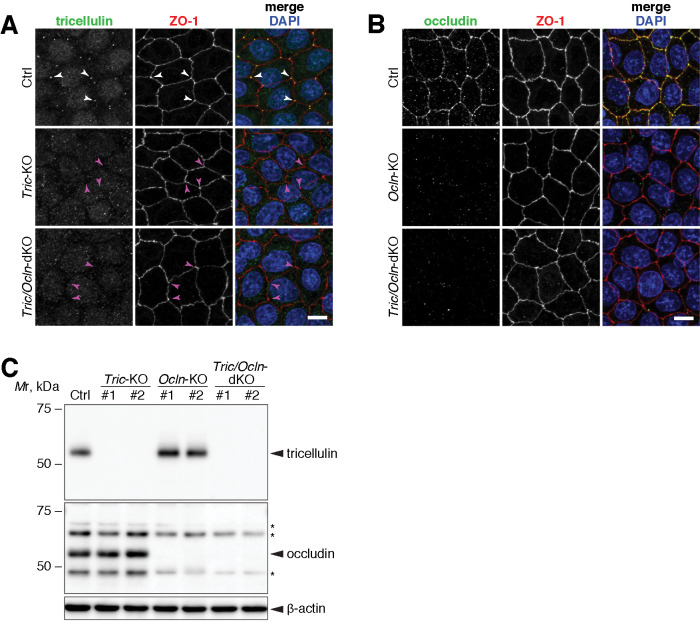
Although MDCK II cells have been used for the studies of TJs for many years, it has been difficult to precisely evaluate the changes in barrier function, especially for ions using MDCK II cells because they express a highly permeable claudin, claudin-2 (Furuse et al., 2001; Tokuda and Furuse, 2015). To overcome this problem, first, we generated a Cldn2-KO MDCK II cell line using CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9-based genome editing (Supplemental Figure S1). There was no significant change in the expression of TJ and AJ proteins in Cldn2-KO MDCK II cells (Supplemental Figure S1E). FFEM analysis revealed that TJ strands of Cldn2-KO cells were associated with the protoplasmic (P)-face, and were mostly continuous, which is in good contrast to the discontinuous appearance of TJ strands in the parental MDCK II cells (Supplemental Figure S2A), as reported previously (Furuse et al., 2001). The complexity of the TJ-strand network pattern in Cldn2-KO cells was significantly decreased compared with that in parental MDCK II cells (Supplemental Figure S2, A–H). Transepithelial electric resistance (TER) measurement showed that Cldn2-KO MDCK II cells exhibited increased resistance by 17.6-fold on day 6 after seeding (Supplemental Figure S2I), which is similar to the results of a previous study using TALEN-mediated Cldn2-KO cells (Tokuda and Furuse, 2015). Throughout this article, we use these Cldn2-KO MDCK II cells as parental cells, and we call this line “Control (Ctrl)” cells. From these Ctrl cells, we established two independent clones of Tric-KO cells (Tric-KO #1 and Tric-KO #2) (Supplemental Figure S3). Likewise, two clones of Ocln-KO cells were established (Ocln-KO #1 and Ocln-KO #2) (Supplemental Figure S3). Next, we generated Tric/Ocln-double knockout cell clones (Tric/Ocln-dKO #1 and Tric/Ocln-dKO #2). Because the gene loci of Tric and Ocln are next to each other, we introduced the guide RNAs (gRNAs) targeting Tric and Ocln together to induce a large deletion (Supplemental Figure S3). Genome sequencing validated a large deletion and frame shifts at both loci (Supplemental Figure S3). The knockout of the genes was further confirmed by immunofluorescence staining (Figure 1, A and B) and immunoblotting (Figure 1C) analyses. Because two clones of each KO cell line exhibited essentially the same phenotype, we present data for #1 cells as a representative of each knockout phenotype unless otherwise specified.
FIGURE 1:
Establishment of Tric-KO, Ocln-KO, and Tric/Ocln-dKO cells. (A, B) Ctrl, Tric-KO #1, and Tric/Ocln-dKO #1 cells (A) and Ctrl, Ocln-KO #1, and Tric/Ocln-dKO #1 cells (B) were immunostained with rat anti-tricellulin mAb (A) or rat anti-occludin mAb (B) (green) and rabbit anti-ZO-1 pAb (red). All cells were stained with DAPI (blue). Note that tricellulin signal (white arrowheads) is absent in the Tric-KO and Tric/Ocln-dKO cells (magenta arrowheads) in A. Scale bars, 20 µm. (C) Immunoblotting of the total cell lysates of Ctrl, Tric-KO, Ocln-KO, and Tric/Ocln-dKO cells using rabbit anti-tricellulin mAb and rabbit anti-occludin pAb. Asterisks on the occludin panel indicate nonspecific bands. β-Actin served as a loading control.
Loss of tricellulin and occludin does not change the expression and localization of other TJ proteins
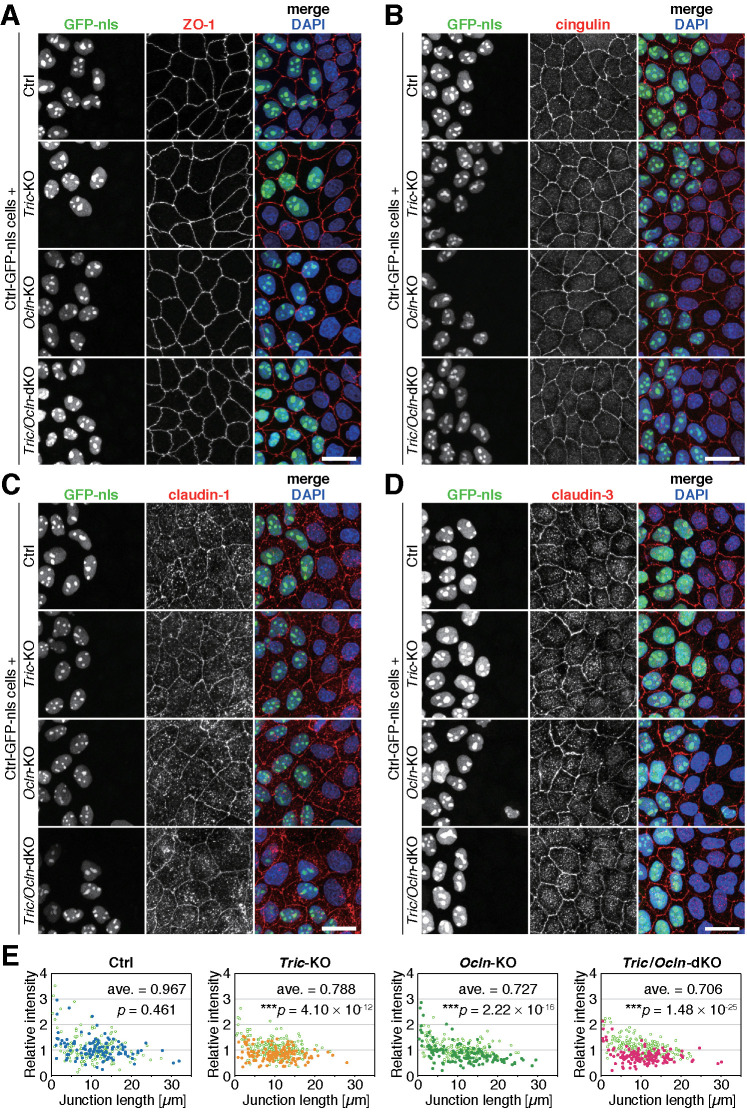
We examined the localization of TJ components, including ZO-1, cingulin, claudin-1, claudin-3, and claudin-4, by immunofluorescence staining (Figure 2, A–D, and Supplemental Figure S4A). To compare the signal intensity of TJ proteins with that of the internal control, we established a Ctrl cell clone expressing 3× nuclear localization signal–conjugated GFP (Ctrl-GFP-nls) and mixed it with Ctrl, Tric-KO, Ocln-KO, or Tric/Ocln-dKO cells. The localization and intensity of ZO-1, cingulin, claudin-1, and claudin-4 in Tric-KO, Ocln-KO, and Tric/Ocln-dKO cells, as well as Ctrl cells, appeared indistinguishable from those in the Ctrl-GFP-nls cells (Figure 2, A–C, and Supplemental Figure S4A). Claudin-3 is localized at TJs and cytoplasmic puncta in all cell clones, although the fluorescent signal at TJs was significantly weaker in the Tric-KO, Ocln-KO, and Tric/Ocln-dKO cells compared with the Ctrl-GFP-nls cells (Figure 2, D and E). We also examined the protein expression levels of TJ and adherens junction (AJ) proteins by immunoblotting and found that there were no significant changes in the amount of junctional proteins, including claudin-3 (Supplemental Figure S4B). These data suggest that tricellulin and occludin do not have essential roles in the expression or localization of TJ proteins except for claudin-3.
FIGURE 2:
TJs of Tric-KO, Ocln-KO, and Tric/Ocln-dKO cells appear normal in fluorescence microscopy. (A–D) Ctrl, Tric-KO #1, Ocln-KO #1, or Tric/Ocln-dKO #1 cells were mixed and cocultured with Ctrl-GFP-nls cells (green). Cells were immunostained with rat anti-ZO-1 mAb (A), mouse anti-cingulin mAb (B), mouse anti–claudin-1 mAb (C) or rabbit anti–claudin-3 pAb (D) (red) and stained with DAPI (blue). Scale bars, 20 µm. (E) Quantification of fluorescence intensity of claudin-3 at cell–cell junctions. The intensity at each cell–cell junction in the Ctrl (dark blue closed circles), Tric-KO #1 (orange closed circles), Ocln-KO #1 (green closed circles), and Tric/Ocln-dKO #1 cells (magenta closed circles) were normalized to the averaged intensity in the Ctrl-GFP-nls cells (lime-green open circles) and plotted against the length of the cell–cell junctions. n = 158 and 120 (Ctrl), 202 and 134 (Tric-KO), 160 and 139 (Ocln-KO), and 209 and 136 (Tric/Ocln-dKO). ***p < 0.001 in Welch’s t test of junction-length-weighted average of the fluorescence intensity.
Tricellulin is not exclusively concentrated at tTJs in Ocln-KO cells
Because it has been reported that tricellulin is mislocalized to bicellular TJs in Ocln-knockdown MDCK II cells (Ikenouchi et al., 2008) and Ocln-KO mice (Kitajiri et al., 2014), we examined the localization of tricellulin in Ocln-KO cells. Although the majority of tricellulin is still found at tTJs in Olcn-KO cells, the intensity of tricellulin at bicellular TJs is increased in Ocln-KO cells compared with the internal control, Ctrl-GFP-nls cells (Supplemental Figure S5A). Quantification analysis revealed that the intensity of tricellulin at bicellular junctions is higher in Ocln-KO cells compared with that in the Ctrl cells (Supplemental Figure S5, C and D), suggesting that occludin may regulate the exclusive localization of tricellulin at tTJs. Because the tricellular localization of tricellulin is mediated by angulins (Masuda et al., 2011; Higashi et al., 2013), we examined the localization of angulin-1 in each cell line. The relative confinement of angulin-1 was not significantly changed in either Ocln-KO, Tric-KO, or Tric/Ocln-dKO cells compared with the Ctrl-GFP-nls cells (Supplemental Figure S5, B and E), suggesting that occludin removes tricellulin from the bicellular TJs in an angulin-1–independent manner.
Tric-KO, Ocln-KO, and Tric/ Ocln dKO cells appear to have TJs in the ultrathin sections
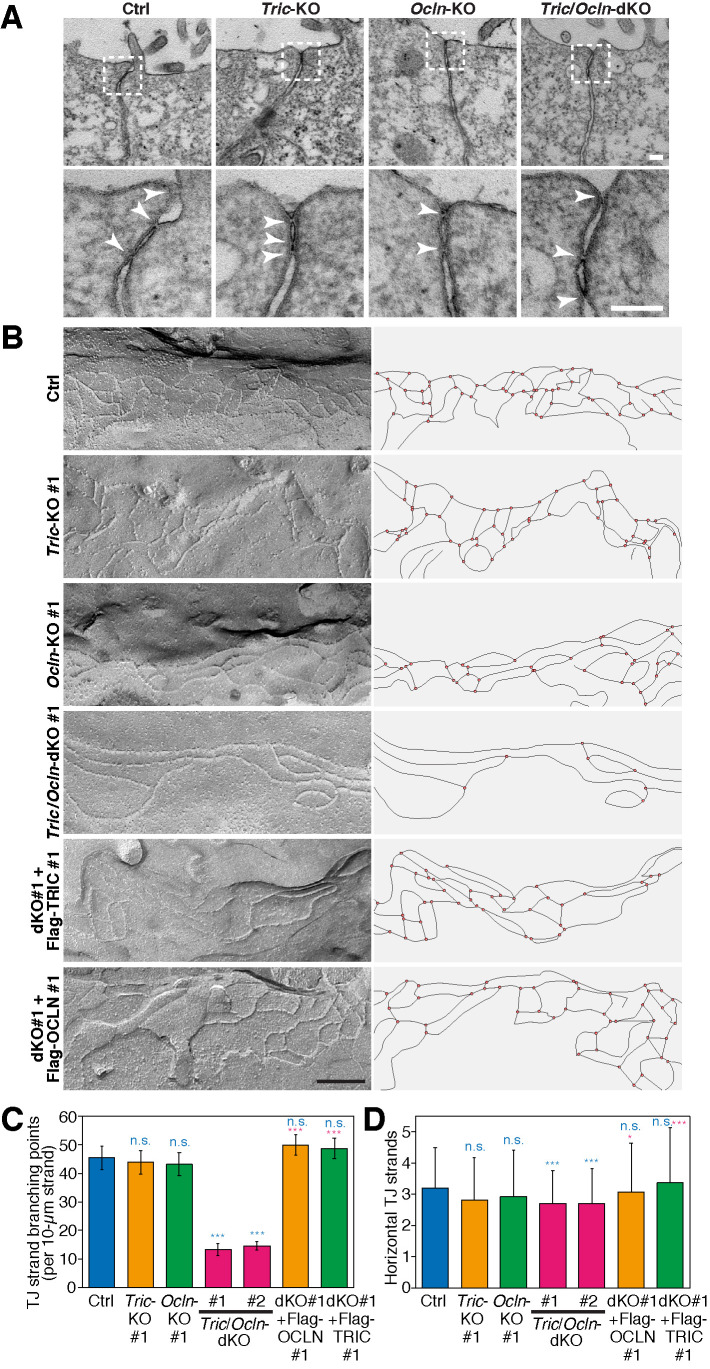
We examined the morphology of TJs in the KO cell lines using transmission electron microscopy (TEM). At the TJs of Ctrl cells, the plasma membranes of adjacent cells were apposed to each other and eliminated the paracellular space to restrict the passage of the fluid and solutes (Figure 3A). The TJs of Tric/Ocln-dKO cells, as well as those of Tric-KO and Ocln-KO cells, also appeared to have membrane apposition similar to that of the Ctrl cells (Figure 3A) and exhibited no differences in the gross morphology of TJs, suggesting that tricellulin and occludin are not required for TJ formation.
FIGURE 3:
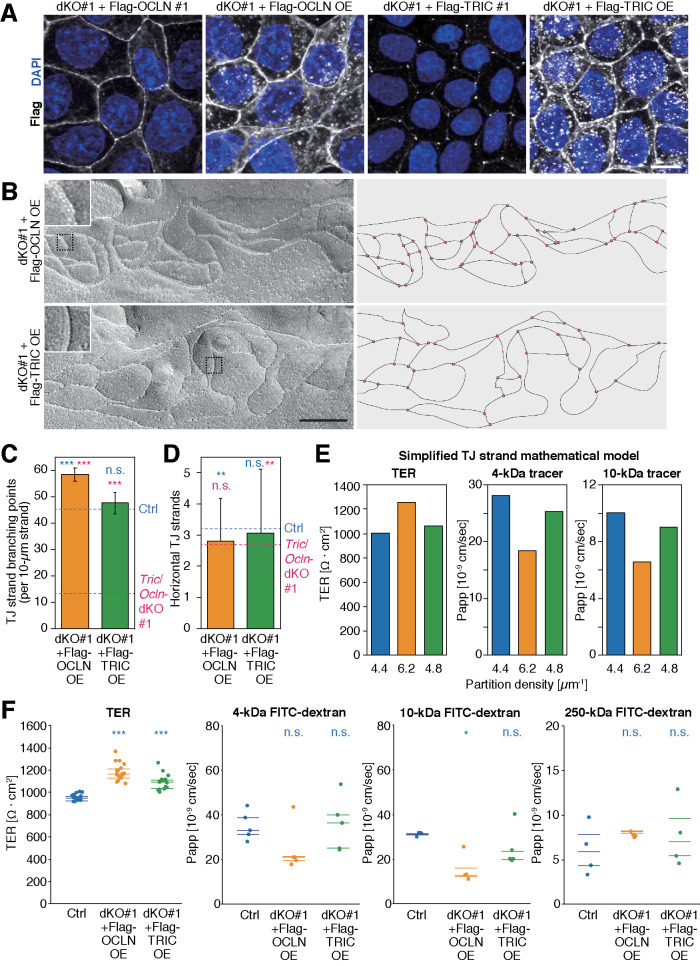
The TJ-strand network of Tric/Ocln-dKO cells is less branched. (A) TEM images of Ctrl, Tric-KO #1, Ocln-KO #1, and Tric/Ocln-dKO #1 cells. The magnified images of the TJ regions in the top panels (dashed rectangles) are shown in the bottom panels. Kissing points between the adjacent cells are shown with white arrowheads in the bottom panels. Scale bars, 100 nm. (B) Freeze-fracture replica electron microscopy of Ctrl, Tric-KO #1, Ocln-KO #1, Tric/Ocln-dKO #1, dKO #1+Flag-OCLN #1, and dKO #1+Flag-TRIC #1 cells are shown (left panels). Additional images are shown in Supplemental Figure S6. Scale bars, 200 nm. TJ strands on the replica images were traced, and the network organization is shown (right panels). (C) Numbers of TJ-strand junction points (branching points). Number of three-way junction points were divided by total length of TJ strands (>100 µm from more than 20 images for each clone). Four-way junctions were regarded as two three-way junctions. Error bars indicate 95% confidence intervals. Total lengths of the measured TJ strands were 101 µm (Ctrl), 103 µm (Tric-KO #1), 103 µm (Ocln-KO #1), 127 µm (Tric/Ocln-dKO #1), 248 µm (Tric/Ocln-dKO #2), 155 µm (dKO #1+Flag-OCLN #1), 146 (dKO #1+Flag-TRIC #1). ***p < 0.001; n.s., p ≥ 0.05 in Poisson’s exact rate test. (D) Number of horizontal TJ strands. n = 188 (Ctrl), 184 (Tric-KO #1), 208 (Ocln-KO #1), 198 (Tric/Ocln-dKO #1), 244 (Tric/Ocln-dKO #2), 356 (dKO #1-Flag-OCLN #1), 310 (dKO #1-Flag-TRIC #1). ***p < 0.001; *p < 0.05; n.s., p ≥ 0.05 in Welch’s t test with Bonferroni’s correction. See Supplemental Figure S2, B and C, for detailed quantification methods for C and D.
The TJ-strand junction points are reduced in the Tric/ Ocln-dKO cells
Because the TAMP family proteins, including tricellulin and occludin, have been implicated in the regulation of TJ-strand network morphology (Ikenouchi et al., 2008; Cording et al., 2013), we conducted FFEM to visualize the TJ strands of Ctrl, Tric-KO, Ocln-KO, and Tric/Ocln-dKO cells (Figure 3B). As expected, Tric-KO, Ocln-KO, and Tric/Ocln-dKO cells still retained the TJ strands at the most apical part of the cell–cell interfaces (Figure 3B). This result is in clear contrast to ZO-1/2 dKO MDCK II cells and claudin-null MDCK II cells, both of which are devoid of TJ strands (Otani et al., 2019). The number and pattern of TJ strands in Tric-KO and Ocln-KO cells were indistinguishable from those in the Ctrl cells (Figure 3B and Supplemental Figure S6). Interestingly, the TJ strands of Tric/Ocln-dKO #1 and #2 cells were less branched compared with the Ctrl, Tric-KO, and Ocln-KO cells, and the complexity of the TJ strand network was drastically reduced (Figure 3B and Supplemental Figure S6). Quantification analysis revealed that the number of TJ-strand junction points (branching points) in the Tric/Ocln-dKO cells was less than one-third compared with that of the Ctrl cells (Ctrl, 45.4 branching points per 10-µm TJ-strand length; Tric/Ocln-dKO #1, 13.3; Tric/Ocln-dKO #2, 14.7) (Figure 3C and Supplemental Figure S7A). The branch angles were similar among the Ctrl, Tric-KO, Ocln-KO, and Tric/Ocln-dKO cells (Supplemental Figures S2B and S7C). The average number of horizontal TJ strands was significantly reduced by 15% in the Tric/Ocln-dKO cells compared with Ctrl cells (Figure 3D and Supplemental Figures S2C and S7B). To confirm that the changes in the TJ-strand network pattern were caused by gene knockout, we established cell lines expressing Flag-tricellulin or Flag-occludin exogenously using Tric/Ocln-dKO #1 cells as parental cells and designated them dKO #1+Flag-TRIC cells and dKO #1+Flag-OCLN cells, respectively (Supplemental Figure S8). In the dKO #1+Flag-TRIC cells and dKO #1+Flag-OCLN cells, the number of TJ-strand junction points and average number of horizontal TJ strands were restored (Figure 3, C and D, and Supplemental Figure S7, A and B), indicating that the loss of tricellulin and occludin resulted in an altered TJ-strand network pattern in the Tric/Ocln-dKO cells. These results indicate that tricellulin and occludin are required for the formation or maintenance of the TJ-strand junction points and regulate the complexity of the TJ-strand network.
Tric/ Ocln-dKO cells exhibit increased permeability for ions and macromolecules
To examine whether the loss of tricellulin and occludin affects the epithelial barrier development, we measured TER of Ctrl and KO cell lines (Figure 4A and Supplemental Table S1). The TER value of Tric-KO #1 cells was comparable to that of the Ctrl cells, whereas the Tric-KO #2 cells showed slightly decreased TER. Both of the Ocln-KO cell clones exhibited a slight, but significant, reduction in TER by 15–18% compared with the Ctrl cells, suggesting that occludin may contribute to the tightness of the barrier. In contrast, the TER values of both of the Tric/Ocln-dKO cell lines decreased by ∼50% compared with that of the Ctrl cells, indicating that tricellulin and occludin are required for the establishment of permeability barrier for ions. The TER values of the dKO #1+Flag-TRIC cells and dKO #1+Flag-OCLN cells were restored to values comparable to those of the Ctrl cells (Figure 4A), confirming that the deletion of tricellulin and occludin caused the decreased TER values in the Tric/Ocln-dKO cells.
FIGURE 4:
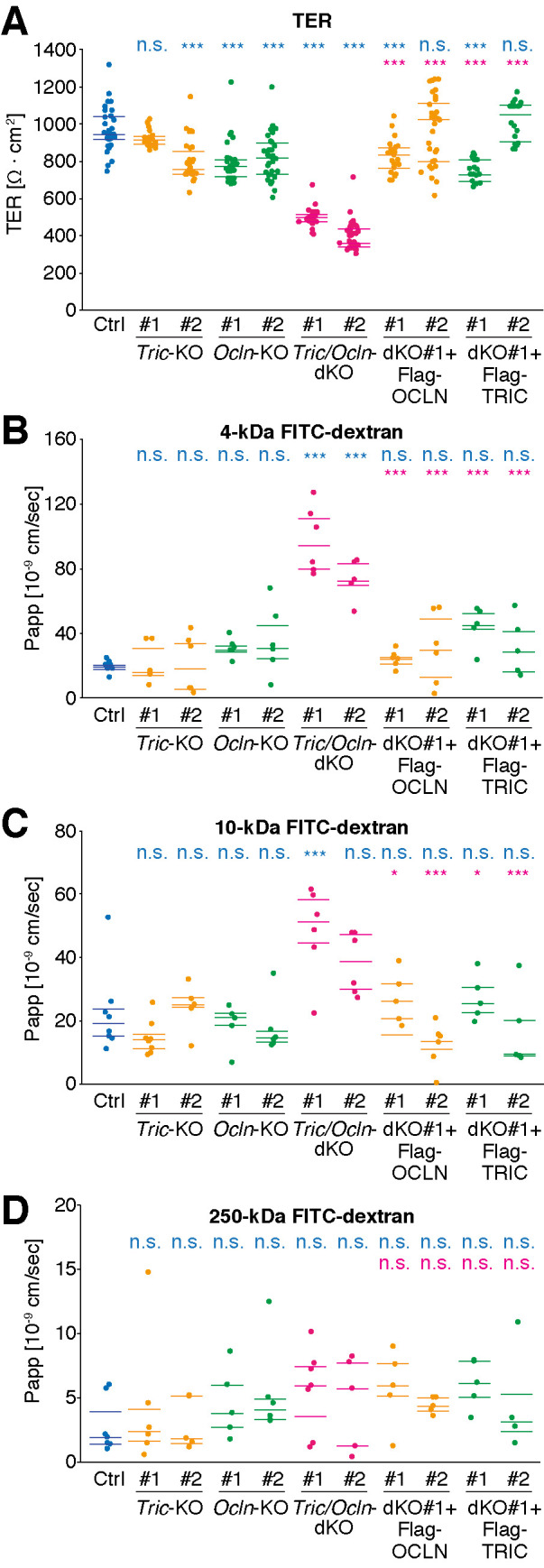
Loss of tricellulin and occludin leads to increased permeability of ions and macromolecules. (A) TER of Ctrl, Tric-KO, Ocln-KO, Tric/Ocln-dKO, dKO #1+Flag-OCLN, and dKO #1+Flag-TRIC cell sheets was measured on day 5 after seeding onto the filter cups. Two independent clones for each genotype were analyzed (#1 and #2). n = 17–35. (B–D) Apparent permeability (Papp) of 4-kDa (B), 10-kDa (C), or 250-kDa (D) FITC-dextran through Ctrl, Tric-KO, Ocln-KO, Tric/Ocln-dKO, dKO #1+Flag-OCLN, and dKO #1+Flag-TRIC cell sheets was measured on day 8 after seeding onto the filter cups. n = 4–8. 25; 50, and 75 percentiles are shown (lines). *p < 0.05; ***p < 0.001; n.s., p ≥ 0.05 in the Tukey–Kramer test. Top and bottom rows indicate the comparison with Ctrl cells (cyan) and Tric/Ocln-dKO #1 cells (magenta), respectively. The p values in Tukey–Kramer test are shown in Supplemental Table S1.
Next, we measured the macromolecular permeability by the tracer-flux method using fluorescein isothiocyanate (FITC)-dextran with 4, 10, and 250 kDa. For the 4-kDa tracer, the apparent permeability coefficient (Papp) significantly increased in the Tric/Ocln-dKO cells compared with the Ctrl cells (Figure 4B and Supplemental Table S1). Similarly, the Papp for the 10-kDa tracer was greater in the Tric/Ocln-dKO cells (Figure 4C), although the difference was statistically significant only for the Tric/Ocln-dKO #1 cells in comparison with the Ctrl cells (Supplemental Table S1). The Papp for the 250-kDa tracer was unaltered (Figure 4D and Supplemental Table S1). The Tric-KO cells and Ocln-KO cells showed no significant change in Papp of any size. The permeability barrier for macromolecules was restored in the dKO #1+Flag-TRIC cells and dKO #1+Flag-OCLN cells (Figure 4, B–D).
Taken together, these results indicate that at least either of tricellulin or occludin is required for the establishment of a tight permeability barrier against ions and macromolecules.
Complexity of the TJ-strand network affects the permeability of ions and macromolecules in a mathematical model
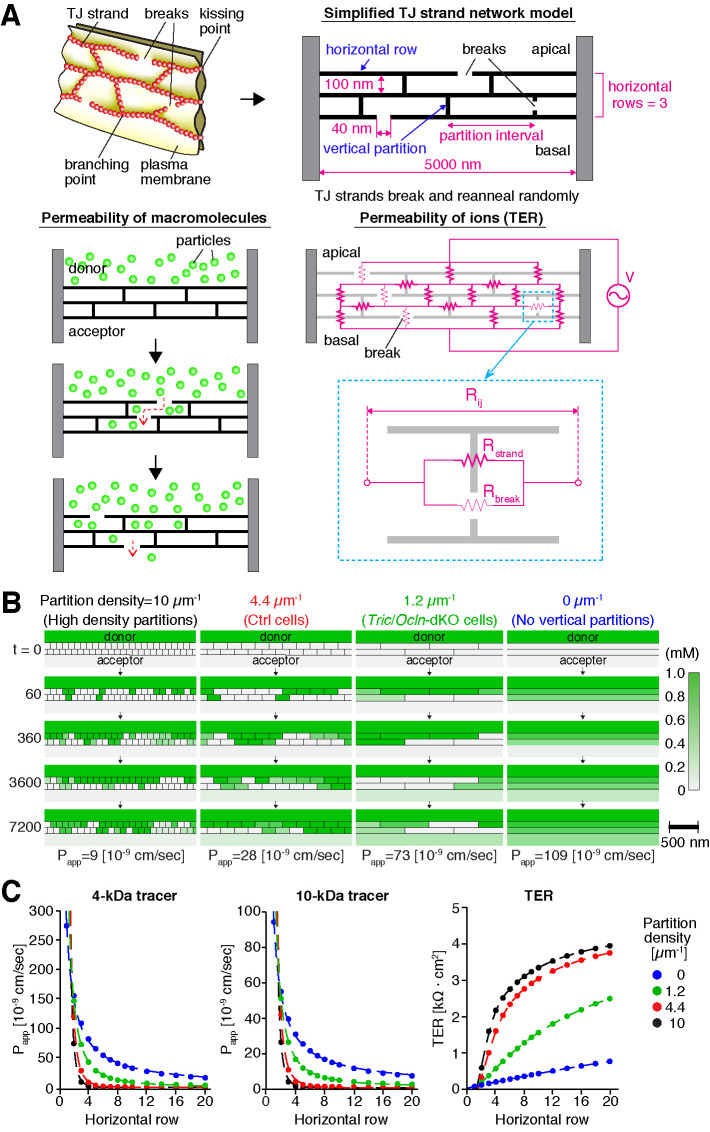
We have shown that loss of both tricellulin and occludin reduces TJ-strand branching and increases the permeability of epithelial sheets. To integrate these findings, we next asked whether the complexity of the meshwork affects the permeability of ions and macromolecules using a mathematical model of the TJ-strand network. To assess the contribution of TJ-strand cross-links, we used a simplified TJ-strand model (Tervonen et al., 2019) in which straight TJ strands run horizontally (horizontal rows) and vertical TJ strands connect the horizontal rows to divide the spaces between the rows into smaller compartments (vertical partitions) (Figure 5A). These TJ strands of horizontal rows and vertical partitions stochastically undergo breaks and reannealing over time to allow passage of ions and macromolecules, mimicking the leak pathway. The branching points of the TJ strand are fixed and unchanged during simulation.
FIGURE 5:
Computational simulation of paracellular permeability using a simplified TJ network model. (A) Schematic diagram of the simulation. The complex network of TJ strands was simplified to two-dimensional grids with multiple compartments. The TJ strands of each compartment stochastically undergo a break and reanneal. In the macromolecular permeability model (left), macromolecules can pass only when the TJ strand has a break, and the diffusion of macromolecules among the grids was simulated over time. In the TER model (right), overall the resistance between the apical and basal sides was calculated. The break in the TJ strand drastically reduced the electric resistance between the compartments. (B) Typical heat maps of macromolecular concentrations in each compartment at 0, 60, 360, 3600, and 7200 [s] in the three-horizontal-row model of 4-kDa tracer permeability. Examples of models with partition density of 10, 4.4, 1.2, and 0 µm–1 are shown. The donor and acceptor compartments are depicted in a smaller size than actual because of space limitations. The Papp values are an average of 512 simulations. See Supplemental Video 1 for the entire heat maps of macromolecular concentrations throughout the simulation (0–7200 [s]). (C) Averaged Papp of 4- and 10-kDa macromolecule tracers and TER values in the models with different partition densities (10, 4.4, 1.2, and 0 µm–1) and horizontal row numbers (1–20 rows). The Papp and TER values are an average of 512 and 106 simulations, respectively.
To simulate the movement of macromolecules, we calculated the apparent permeability (Papp) of particles over time in the simplified TJ-strand network model (Figure 5A, bottom left). Because the tracer of 250 kDa (∼206 Å diameter) is too large to pass the intercellular space between the apposed plasma membranes at TJs (∼60–70 Å) (Otani et al., 2019), we decided to test only 4-kDa (∼28 Å diameter) and 10-kDa (∼46 Å diameter) particles in the simulation. We simulated the movement of the tracer particles among the compartments over time and recorded the concentration of tracers in each grid (Figure 5A). Because Ctrl and Tric/Ocln-dKO cells possessed three horizontal rows of TJ strands on average in our observation using FFEM samples (Figure 3D), we used a three-horizontal-row model and tested the permeability of the 4-kDa particles first (Figure 5B and Supplemental Video 1). On the basis of the quantification of TJ-strand junction point frequency (Figure 3C), the densities of the vertical partitions of the Ctrl cell model and Tric/Ocln-dKO cell model were determined to be 4.4 and 1.2 µm–1, respectively (see Materials and Methods for details). The Papp of the Ctrl cell model (2.81 × 10–8 cm/s) was comparable with the experimental data (1.94 × 10–8 cm/s) (Figure 4B). Interestingly, the Papp value of the Tric/Ocln-dKO cell model was increased ∼2.6 times (7.29 × 10–8 cm/s) compared with that of the Ctrl cell model (Figure 5B and Supplemental Video 1), which is in good agreement with the increased permeability observed in the Tric/Ocln-dKO cells (7.31 × 10–8 cm/s). We also examined the permeability of the models with different densities of the vertical partitions. In the models with higher-density partitions (10 µm–1) and without vertical partitions (0 µm–1), the Papp values were 32% and 387%, respectively, compared with the Ctrl cell model. Next, we tested the models of different numbers of horizontal rows and vertical partitions and calculated the Papp by averaging the results from 512 simulations for each parameter setting (Figure 5C). In the models of any horizontal row numbers, higher density of the vertical partitions resulted in decreased permeability of the particles (Figure 5C). Using the same models, we also tested the permeability of 10-kDa particles (Figure 5C). The Papp values were ∼1/3 compared with the 4-kDa simulation in each model, showing size selectivity. The permeability of the 10-kDa particles was dependent on the density of partitions, which was similar to the 4-kDa results. As expected, an increased number of horizontal rows of the TJ strand resulted in the decreased Papp in both sizes of particles with 4 and 10 kDa, and the single horizontal strand model without cross-links exhibited the highest Papp value among all other conditions (Figure 5C).
Video S1.
Computational simulation of continuous flow of macromolecules through broken TJ strands in simplified TJ network model Image sequences of typical heat map of macromolecular concentrations (4 kDa) in each compartment from 0-7200 [sec] in the three‐horizontal‐row model. The movie in the order from top to bottom indicates partition density = 0, 1.2, 4.4 and 10 μm‐1, respectively. The display sizes of donor and acceptor were reduced. The images were constructed using Matlab's (command “heat map”), were animated using ImageJ software (“Import”‐“Image Sequence”), and were converted to mp4 filename extension using FFmpeg.
Next, we simulated the permeability of ions (transepithelial current). Similar to the simulation for macromolecule permeability, we used the break-repair model of the TJ-strand network (Figure 5A, bottom right). We hypothesized that the intact TJ strands have a high electric resistance and the breaks in TJ strands drastically reduce the resistance. We calculated the overall electric resistance between apical and basal sides as TER (Figure 5A). Similar to the macromolecule simulation, decreased density of the vertical of partitions (fewer cross-links) caused decreased TER values (Figure 5C). The TER value of the Ctrl cell model (1002 Ω·cm2) was comparable with the experimental data (970 Ω·cm2) (Figure 4A), and the TER of the Tric/Ocln-dKO cell model was decreased to 35% (354 Ω·cm2) of the Ctrl cell model (Figure 5C), which reasonably corresponds to the experimental TER (395 Ω·cm2) (Figure 4A).
This simplified TJ-strand mathematical model predicted that the reduced complexity of the TJ-strand meshwork results in an increased permeability of ions and macromolecules, which fits well with the obtained experimental data.
Overexpression of occludin in the Tric/ Ocln-dKO cells increases the network complexity of TJ strands and improves barrier function
To further examine the relationship between the complexity of the TJ-strand network and barrier function, we established cell clones overexpressing OCLN and TRIC using the Tric/Ocln-dKO #1 cells as a parental cell line and designated them dKO #1+Flag-OCLN OE and dKO #1+Flag-TRIC OE, respectively (Figure 6A and Supplemental Figure S8). Although Flag-OCLN was localized at the bicellular TJs in the dKO #1+Flag-OCLN #1, dKO #1+Flag-OCLN OE cells exhibited a broader distribution of Flag-OCLN at the entire lateral membrane and cytoplasmic puncta in addition to the TJs (Figure 6A). Flag-TRIC was also localized at the lateral membrane and cytoplasmic puncta in the dKO #1+Flag-TRIC OE cells, which is in good contrast to the dKO #1+Flag-TRIC #1 cells where Flag-TRIC localization is confined to the tTJs (Figure 6A). FFEM analysis revealed that the TJ-strand network of the dKO #1+Flag-OCLN OE cells was more complex (Figure 6B) and that the TJ-strand branching points were significantly increased by ∼30% compared with the Ctrl cells (Figure 6C and Supplemental Figure S7A). The horizontal TJ-strand number of the dKO #1+Flag-OCLN OE cells was indistinguishable from that of the Tric/Ocln-dKO cells and was significantly smaller than that of the Ctrl cells (Figure 6D and Supplemental Figure S7B). The TJ strands in these cells appeared to be thicker than in the Ctrl and any other MDCK II cells (Figure 6B, inset). On the other hand, the TJ-strand morphology of the dKO #1+Flag-TRIC OE cells was similar to that of the Ctrl cells (Figure 6B), although thick TJ strands were occasionally observed (Figure 6B, inset). The complexity of the TJ-strand network was slightly, but not significantly, higher (Figure 6C and Supplemental Figure S7A), and horizontal strand number was similar, compared with the Ctrl cells (Figure 6D and Supplemental Figure S7B). We simulated the permeability of ions and macromolecules in these cells using the simplified TJ-strand mathematical model with corresponding TJ network complexity. The partition densities in the models of the dKO #1+Flag-OCLN OE cells (OCLN-OE model) and dKO #1-Flag-TRIC OE cells (TRIC-OE model) were determined to be 6.2 and 4.8 µm–1, respectively, based on the branching frequency shown in Figure 6C. The TER value was predicted to be higher in the OCLN-OE and TRIC-OE models by 26% and 6%, respectively (Figure 6E). The predicted Papp values were lower than that in the Ctrl model (OCLN-OE model, 34%; TRIC-OE model, 10% in both 4- and 10-kDa simulations) (Figure 6E). Next, we examined actual barrier function of these cells by TER measurement and tracer-flux assay. The TER values of the dKO #1+Flag-OCLN OE cells and dKO #1+Flag-TRIC OE cells were significantly higher than that of the Ctrl cells (OCLN, 24%; TRIC, 14%) (Figure 6F). In the dKO #1+Flag-OCLN OE cells, the Papp value of 10-kDa dextran was significantly decreased by 50% compared with the Ctrl cells and there was no change in the flux of 4- and 250-kDa tracers. No significant differences were detected in the Papp values of any tracer size between the Ctrl cells and the dKO #1+Flag-TRIC OE cells (Figure 6F).
FIGURE 6:
Overexpression of occludin results in increased TJ network complexity and reduced permeability. (A) dKO #1+Flag-OCLN #1, dKO #1+Flag-OCLN OE, dKO #1+Flag-TRIC #1, and dKO #1+Flag-TRIC OE cells were stained with mouse anti-Flag mAb (white) and DAPI (blue). Scale bar, 20 µm. (B) Freeze-fracture replicas of dKO #1+Flag-OCLN OE and dKO #1+Flag-TRIC OE cells. Insets are magnified images of the region indicated with black dotted squares. Scale bar, 200 nm. (C) Numbers of TJ-strand junction points (branching points). Branching point numbers of Ctrl (cyan dashed line) and Tric/Ocln-dKO #1 (magenta dashed line) cells shown in Figure 3C are indicated for comparison. Error bars indicate 95% confidence intervals. Total lengths of the measured TJ strands were 100 µm (dKO #1+Flag-OCLN OE) and 284 µm (dKO #1+Flag-TRIC OE). ***p < 0.001; n.s., p ≥ 0.05 in Poisson’s exact rate test (vs. Ctrl cells [cyan] and vs. Tric/Ocln-dKO #1 cells [magenta]). (D) Number of horizontal TJ strands. Horizontal strand numbers of Ctrl (cyan dashed line) and Tric/Ocln-dKO #1 (magenta dashed line) cells shown in Figure 3D are indicated for comparison. n = 286 (dKO #1+Flag-OCLN OE) and 667 (dKO #1+Flag-TRIC OE). **p < 0.01; n.s., p ≥ 0.05 in Welch’s t test with Bonferroni’s correction (vs. Ctrl cells [cyan] and vs. Tric/Ocln-dKO #1 cells [magenta]). (E) Averaged TER values and Papp of 4- and 10-kDa macromolecule tracers in the simplified TJ-strand models with partition densities of 4.4 (Ctrl cell model), 6.2 (OCLN-OE cell model), and 4.8 µm–1(TRIC-OE cell model) and three horizontal rows. The Papp and TER values are an average of 512 and 106 simulations, respectively. (F) TER values and Papp of 4-, 10-, and 250-kDa FITC-dextran in the Ctrl, dKO #1+Flag-OCLN OE, and dKO #1+Flag-TRIC OE cells. n = 16–17 (TER), n = 4–5 (tracer flux); 25, 50, and 75 percentiles are shown (lines). ***p <0 .001; **p < 0.01; *p < 0.05; n.s., p ≥ 0.05 in Tukey–Kramer test.
These data indicate that overexpression of occludin and tricellulin leads to increased complexity and support the idea that occludin and tricellulin strengthen the epithelial barrier by regulating the complexity of the TJ-strand network.
DISCUSSION
In the present study, we showed that double KO of the Tric and Ocln genes reduces the strand-to-strand junction points (branching points, cross-points, T-junction, or bifurcation) of the TJ strands, whereas a single KO of either Tric or Ocln has no drastic effects. TER measurement and tracer-flux assay revealed that the Tric/Ocln-dKO cells exhibited increased permeability for ions and macromolecules of 4–10 kDa compared with the Ctrl cells, Tric-KO cells and Ocln-KO cells. The increased permeability in the Tric/Ocln-dKO cells cannot be explained by reduced TJ localization of claudin-3 (Figure 2, D and E) alone because the localization of claudin-3 was also altered in the Tric-KO and Ocln-KO cells, in which the barrier function was only slightly impaired. Hence, we hypothesized that the reduced complexity of the TJ-strand network is the direct cause of the reduced barrier function. In concordance with the experimental data, our computational simulation using a simplified TJ-strand network model predicted that reduced cross-links in the TJ-strand network resulted in the increased permeability of both ions and small-sized macromolecules. These data collectively demonstrate that tricellulin and occludin coordinately regulate the formation and/or stabilization of TJ-strand branching points, which contributes to the establishment and maintenance of the tight barrier of epithelial cells.
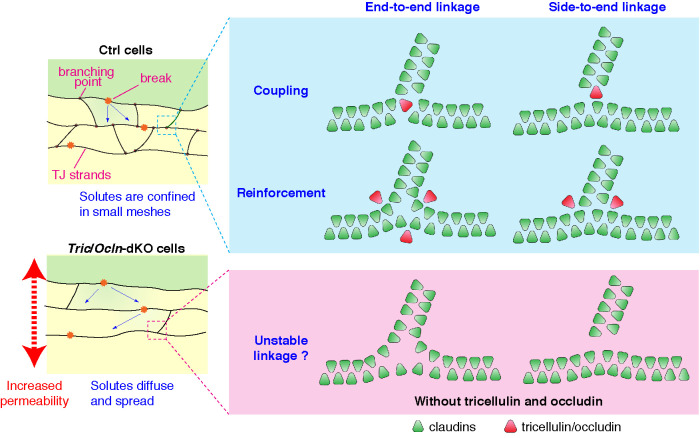
Because occludin is preferentially localized at the free ends and strand-to-strand junction points of the TJ strands (Van Itallie et al., 2017) and the branching points of the TJ strands were reduced by the loss of tricellulin and occludin (Figure 3, B and C, and Supplemental Figures S6 and S7A), one would speculate that tricellulin and occludin serve as connectors (Figure 7, coupling model). We would argue that this would not be likely for the following reasons: 1) TJ strand–like structures in claudin-expressing L-fibroblasts or Rat-1 fibroblasts, in which no expression of TAMP family proteins have been reported, are branched (Sasaki, Matsui, et al., 2003; Van Itallie et al., 2017), suggesting that claudins have an intrinsic ability to form bifurcations; 2) these branching points of TJ strands can break and anneal at all three join points, suggesting that there is no end-to-side attachment mechanism (Van Itallie et al., 2017); and 3) the coupling model demands that TJ strands preferentially make branching points with specific angles; however, there is no change in the distribution of angles between strands at the branching points in the Tric-KO, Ocln-KO, or Tric/Ocln-dKO cells compared with the Ctrl cells (Supplemental Figure S7C). Based on the above, it is most likely that tricellulin and occludin may stabilize the branching points and reduce the propensity for detachment (Figure 7, reinforcement model). This model can explain how loss of tricellulin/occludin leads to fragile connection of branching points (Figure 7, bottom). Although the reduction in the branching points can account for the impaired barrier function observed in the Tric/Ocln-dKO cells, we cannot exclude the possibility that tricellulin and occludin also regulate the stability and/or continuity of TJ strands themselves. Future structural studies of the intramembranous interaction between claudins and tricellulin/occludin would help us understand the exact action of tricellulin/occludin on the TJ strands.
FIGURE 7:
Hypothetical working models for how tricellulin and occludin contribute to the TJ assembly and barrier function. Schematic diagram of branching points of claudin-based TJ strands in the presence or absence of tricellulin/occludin expression. TJ strands are connected to each other in either end-to-end (left) or end-to-side (right) manner at the branching points. Tricellulin and occludin may facilitate the formation of branching points by directly connecting strands (coupling model) or stabilizing the linkage of junctions (reinforcement model). In the Ctrl cells, the solutes entering the strand network through breaks are confined in the compartment. On the other hand, in the absence of tricellulin/occludin, the solutes easily diffuse and spread to interstrand spaces, which results in an increased permeability.
It has been presumed that the number and complexity of TJ strands are important determinants of the TJ barrier properties (Claude and Goodenough, 1973; Claude, 1978; Madara and Dharmsathaphorn, 1985; Tsukita et al., 2001; Schneeberger and Lynch, 2004; Varadarajan, Stephenson, and Miller, 2019). It would be easy to understand that the number of horizontal rows of TJ strands in the network is corelated with the barrier property (Claude and Goodenough, 1973; Claude, 1978; Madara and Dharmsathaphorn, 1985) because the horizontal strands directly prevent the passage of substances on the paracellular pathway. However, the importance of strand complexity has not been demonstrated before because it has been unclear how the complexity of the TJ-strand network is regulated. In the present study, by establishing Tric/Ocln-dKO cells, we discovered one of the mechanisms defining the complexity of the TJ-strand network. Furthermore, using a mathematical model, we clearly demonstrated that the complexity of the TJ-strand network regulates barrier function. A small meshwork structure of the TJ strand may prevent the horizontal diffusion of solutes and reduce the permeability of the paracellular pathway (Figure 7, left). Although our model predicts that the TJ strand complexity regulates the permeability of ions and macromolecules through large breaks in the TJ strands (leak pathway), the strand complexity may also regulate the “pore” pathway, because pore-forming claudin channels are gated and shuttle between “open” and “closed” states (Weber et al., 2015), similar to the stochastic “breaks” on TJ strands in the leak pathway.
Although claudins are conserved in both vertebrates and invertebrates (Furuse and Tsukita, 2006), the TAMP family proteins (tricellulin, occludin, and MarvelD3) are conserved only in vertebrates (Steed et al., 2009; Raleigh, Marchiando, Zhang, et al., 2010)., Invertebrates have occluding junctions called septate junctions (SJs) as a functional counterpart of tight junctions (Furuse and Tsukita, 2006). Interestingly, SJs are composed of 5–30 strands with no cross-links (Noirot-Timothee and Noirot, 1980). In our mathematical modeling, a TJ-strand network without cross-links exhibited an increased permeability. Surprisingly, if the network is exclusively composed of parallel strands without cross-links, 12–200 horizontal rows of strands are required to exert the barrier function similar to the cross-linked network with 3–7 rows of horizontal strands. Invertebrates and vertebrates may have adopted distinct strategies to improve the tightness of the paracellular barrier. Invertebrates increased the number of unbranched SJ strands to achieve a tight barrier, while the ancestral vertebrates may have acquired the ability to cross-link the TJ strands to utilize network complexity in improving the tightness of the barrier.
Tricellulin and occludin may not be the only determinants of branching formation in the TJ-strand network. In vertebrates, TJ strands found in the interlamellar TJs of the oligodendrocytes in the brain, “Sertoli junctions” formed between Sertori cells in the testis, and TJs in the basal cells of stria vascularis in the inner ear are scarcely branched and tend to run parallel (Gow et al., 1999, 2004; Morita et al., 1999; Kitajiri, Miyamoto, et al., 2004). There is no detectable expression of occludin and tricellulin at the interlamellar junctions (Morita et al., 1999; Iwamoto et al., 2014). However, in the Sertoli cells of mice and rats (Moroi et al., 1998) and basal cells in stria vascularis (Gow et al., 2004; Kitajiri, Miyamoto, et al., 2004), occludin is expressed and localized at cell–cell junctions. Claudin-11/OSP (oligodendrocyte-specific protein) is the major component of the TJs in these cells (Gow et al., 1999, 2004; Morita et al., 1999; Kitajiri, Miyamoto, et al., 2004), and claudin-11–based TJ strand–like structures in the L-fibroblasts are scarcely branched and tend to run parallel to each other unlike in other classic claudins (Morita et al., 1999). Thus, it is likely that claudin-11 has an intrinsic property to form unbranched TJ strands irrespective of the presence of occludin and tricellulin. Fluorescence resonance energy transfer (FRET) analysis using CFP-claudin-11– and YFP-TAMPs–expressing HEK293 cells revealed that there was no cis-interaction between claudin-11 and TAMPs compared with classic claudins-1–5 (Cording et al., 2013), further supporting the view that occludin does not induce bifurcations of claudin-11–based TJ strands. However, Ocln-KO mice exhibited male sterility (Saitou et al., 2000), suggesting that occludin may have some function other than making cross-links of TJ strands at the Sertoli junctions. Claudin-19 is another example of a claudin that is also reported to induce the formation of scarcely branched parallel TJ strands (Hou et al., 2008), even in the presence of occludin/tricellulin (Gong, Renigunta, et al., 2015). In light of the above, the intrinsic property of claudins, as well as the regulation by tricellulin/occludin, may determine the complexity of the TJ-strand network in the tissues in vivo.
It was reported that loss of occludin expression did not change the steady-state TER values in Ocln-knockdown MDCK II cells (Yu et al., 2005; Van Itallie et al., 2010) or Caco-2 cells (Raleigh, Marchiando, Zhang, et al., 2010) and in the small and large intestines of Ocln-KO mice (Saitou et al., 2000; Schulzke et al., 2005). Our result that Ocln-KO cells exhibit slightly decreased TER is in line with these previous reports. As for tricellulin, it was reported that tricellulin-knockdown in EpH4 cells (Ikenouchi et al., 2005) or HT-29/B6 cells (Krug, 2017) decreased the TER and increased the macromolecular permeability. In contrast, our results and other previous reports using MDCK II cells (Van Itallie et al., 2010) and Caco-2 cells (Raleigh, Marchiando, Zhang, et al., 2010) showed that tricellulin is dispensable for the establishment of a tight barrier. The divergence of results might arise from the difference in cell types or experimental setup. Double-knockdown of tricellulin and occludin was reported to delay the maturation of the TJ barrier but did not alter the steady-state TER (Raleigh, Marchiando, Zhang et al., 2010), which is also in clear contrast to our results. This difference could be due to the cell type. Alternatively, the residual proteins might still function in knockdown cells, which was the case for ZO proteins (Umeda, Ikenouchi, et al., 2006; Otani et al., 2019). The other TAMP family protein, MarvelD3, was also suggested to have redundant roles with tricellulin and occludin in barrier establishment (Raleigh, Marchiando, Zhang, et al., 2010). A triple knockout study in the future would reveal the common function of the TAMP family.
Although tricellulin and occludin have similar domain organizations, these proteins are differentially localized. Tricellulin is exclusively localized at tricellular junctions, where three epithelial cells meet (Higashi and Miller, 2017; Higashi and Chiba, 2020), whereas occludin is distributed along bicellular TJ, suggesting that tricellulin and occludin function at tTJs and bicellular TJs, respectively. Previous reports (Ikenouchi et al., 2008; Kitajiri et al., 2014) and our data (Supplemental Figure S5) showed that occludin is responsible for the confinement of tricellulin at tricellular contacts. In the absence of occludin, tricellulin may ectopically localize at bicellular TJs through an angulin-1–independent mechanism (Supplemental Figure S5) and act as a substitute for occludin in Ocln-KO cells. Although it would be interesting to investigate the tTJ structure in the Tric-KO, Ocln-KO, and Tric/Ocln-dKO cells, we could not find a sufficient number of tTJs in order to discuss the changes in structure, because tTJs are quite rare and difficult to find in freeze-fracture replica samples. How tricellulin and occludin contribute to the formation of tTJs is an interesting future question to be addressed.
In summary, we found that tricellulin and occludin play an important role in forming TJ-strand branching points and regulate the complexity of the anastomosing TJ-strand network. We believe that our mathematical modeling will help clarify how the complexity of the TJ-strand network affects the paracellular permeability. It would be of great interest for future studies to investigate whether the tricellulin/occludin-mediated regulatory mechanism functions in physiological and pathological situations.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture
MDCK II, HEK293T, and SP2 cells were cultured in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich) and were maintained in a 5% CO2 incubator (Panasonic Healthcare, Tokyo, Japan) at 37°C. All KO cell clones of the MDCK II cells were established from a parental Cldn2-KO MDCK II cell line. Thus, the Cldn2-KO cell line is written as “Ctrl” cell in this article.
Antibodies
Rat anti-occludin monoclonal antibody (mAb) (clone MOC37) (Saitou et al., 1997) and rabbit anti–angulin-1 polyclonal antibody (pAb) (Oda, Sugawara, et al., 2020) were previously described. Rabbit anti–occludin pAb (#LS-B2187) was purchased from Lifespan (USA). Rat anti–ZO-1 (alpha+) mAb (clone R40.76; sc-33725), mouse anti–claudin-1 mAb (clone XX7; sc-81796), and mouse anti-cingulin mAb (clone G-6; sc-365264) were obtained from Santa Cruz Biotechnology (USA). Rabbit anti–ZO-1 pAb (#61-7300), rabbit anti-tricellulin mAb (clone 54H19L38; #700191), rabbit anti–claudin-2 pAb (#51-6100), rabbit anti–claudin-3 pAb (#34-1700), and mouse anti–claudin-4 mAb (#32-9400) were from Thermo Fisher Scientific (USA). Rabbit anti–claudin-1 pAb (#18815) and rabbit anti–claudin-7 pAb (#18875) were from Immuno-Biological Laboratories (Gunma, Japan). Mouse anti–E-cadherin mAb (clone 36; #610182) was from BD Biosciences (USA). Rabbit anti–α-catenin mAb (clone 23B2; #3240) and rabbit anti–E-cadherin mAb (clone 24E10; #3195T) were from Cell Signaling Technology (USA). Mouse anti–β-actin mAb (clone AC-15; #A1978) and mouse anti-Flag M2 horseradish peroxidase (HRP)-linked mAb (#A8592) were obtained from Sigma-Aldrich. Mouse anti-DYKDDDDK (Flag)-tag mAb (#018-22386) was from Fujifilm WAKO (Osaka, Japan).
Rat anti-tricellulin mAb and rat anti–claudin-2 mAb were house-made. The DNA fragment encoding the N-terminal region of human tricellulin containing aa 1–153 (Supplemental Figure S9) was amplified from the cDNA library of Caco-2 cells by PCR using PrimeSTAR DNA Polymerase (Takara Bio, Shiga, Japan) and specific primers with BamHI and EcoRI sites. The fragment was cloned into pGEX-4T3 (GE Healthcare, USA), and the DNA sequence was verified (Macrogen Japan, Kyoto, Japan). The recombinant GST-tricellulin NT fusion protein was produced in Escherichia coli strain BL21 (Takara Bio) and purified with glutathione beads (Takara Bio) according to the manufacturer’s instruction. A peptide (NH2-CRPGQPPKVKSEFNS-COOH) corresponding to the cytoplasmic terminus of human claudin-2 was synthesized by Eurofins Genomics (Tokyo, Japan) and conjugated with Keyhole limpet hemocyanin (#77605; Thermo Fisher Scientific). Then, the proteins were dialyzed against phosphate-buffered saline (PBS) and mixed with Freund’s complete adjuvant (BD Biosciences) to make an emulsion. The emulsion was subcutaneously injected into the footpads of 8-wk-old Wistar rats. The experiments conducted using animals strictly adhered to the compliance standards of Japanese Guidelines for Proper Conduct of Animals Experiments. The protocols of the animal experiments (#29098, #30112, and #2019023) were reviewed by the Fukushima Medical University’s Animal Care and Use Committee and were approved by the president of the University. After 2 wk, the rats were killed by cervical dislocation under deep anesthesia using sevoflurane (Fujifilm Wako). The iliac lymph nodes were dissected out aseptically and minced, and the lymphocytes were isolated by filtration with a nylon cell strainer (#352350 Corning, USA). The lymphocytes were fused with mouse myeloma cell line SP2 using PEG4000 (#1.09727.0100; Merck Millipore, Darmstadt, Germany), and hybridoma cells were selected using GIT medium (Fujifilm Wako) containing hypoxanthine–aminopterin–thymidine (HAT) (Life Technologies, USA) supplemented with 10% BM-Condimed H1 (Sigma-Aldrich) for 7–10 d. The initial screening was conducted with a solid-phase enzyme-linked immunosorbent assay (ELISA) using a GST-tricellulin NT- or claudin-2 peptide–coated 96-well microplate (#655101; Greiner bio-one, Kremsmünster, Austria). GST alone and PBS were used as negative controls, respectively. Positive clones were expanded and further tested for immunofluorescence staining, and one clone with high specificity and a good titer was selected for each immunogen (clone 1E2 for tricellulin; clone 2D7 for claudin-2).
For the secondary antibodies used in the immunofluorescence staining, Alexa Fluor 488–conjugated donkey anti-mouse immunoglobulin G (IgG) pAb (#715-545-150), Alexa Fluor 488–conjugated donkey anti-rabbit IgG pAb (#711-545-152), Alexa Fluor 488–conjugated donkey anti-rat IgG pAb (#712-545-153), Cy3-conjugated donkey anti-mouse IgG pAb (#715-165-151), Cy3-conjugated donkey anti-rabbit IgG pAb (#711-165-152), and Cy3-conjugated donkey anti-rat IgG pAb (#712-165-153) were purchased from Jackson ImmunoResearch Laboratories, USA). For immunoblotting, HRP-linked sheep anti-mouse IgG pAb (#NA931V; GE Healthcare), HRP-linked goat anti-rabbit IgG pAb (#7074P; Cell Signaling Technology) and HRP-linked goat anti-rat IgG pAb (#NA935V; GE Healthcare) were used.
Generation of the Cldn2-KO, Tric-KO, Ocln-KO, and Tric/ Ocln-dKO cell lines
Cldn2-KO cells (designated “Ctrl” cells in this paper) were generated by CRISPR/Cas9-mediated genome editing from MDCK II cells, and Tric-KO, Ocln-KO, and Tric/Ocln-dKO cells were generated from parental “Ctrl” Cldn-2-KO cells. The DNA oligos encoding single-guide RNAs (sgRNAs) (Supplemental Figures S1 and S3) were synthesized (Macrogen Japan), and the annealed DNA oligos were cloned into pSpCas9 (BB)-2A-Puro (PX459) plasmids (Addgene, USA; plasmid #62988) (Ran, Hsu, et al., 2013) at the BbsI cleavage sites. Next, 7 × 104 to 1 × 105 cells were transiently transfected with a pair of plasmids encoding gRNAs (CRISPR-1 and CRISPR-2, 1 µg each) by PEI-max (Polysciences, USA) in a 12-well plate overnight. The transfected cells were selected with 3 µg/ml puromycin (Sigma-Aldrich) for 1 d. The cells were then sparsely seeded onto 10-cm dishes, and the cell clones were obtained by scraping off the cell colonies. After establishment of cell clones, we confirmed that the cells were no longer resistant to puromycin.
Screening of KO cell clones
Genomic DNA was extracted from the cell clones by incubating cell suspension with 200 µg/ml proteinase K (Promega, USA) for 20 min at 55°C followed by inactivation of proteinase K by heating at 95°C for 5 min. The DNA fragments of the Cldn2, Tric, and Ocln loci were amplified by PCR using GoTaq DNA polymerase (Promega) and the specific primers (Supplemental Table S2 and Supplemental Figure S3). For screening of the Tric/Ocln-dKO clones, the Primer A/Primer C and Primer A/Primer E were used for the detection of wild-type (WT) and KO alleles, respectively. PCR products were subcloned into a cloning vector using the TA-cloning method (pGEM-T-easy; Promega), and the DNA sequences were analyzed (Macrogen Japan). At least six independent subclones encompassing PCR products were analyzed for each cell clone to ensure that the sequences of both alleles were examined.
Establishment of stable cell clones expressing Flag-tricellulin, Flag-occludin, or GFP-nls
A cDNA library of MDCK II cells were synthesized using the PrimeScript II 1st strand cDNA Synthesis Kit (Takara Bio). cDNAs encoding dog Tric and dog Ocln were amplified using PrimeSTAR GXL DNA Polymerase and were cloned into the pCAG vector (Niwa et al., 1991) with N-terminal 1xFlag (DYKDDDDK) and 2xStrep II (WSHPQFEK) tags (previously described [ Higashi et al., 2013]) with a C-terminal vector-derived sequence (EFTPQVQAAYQKVVAGVANALAHKYH). The vector encoding nuclear GFP (GFP-nls) was constructed by cloning the DNA fragments encoding IRES, EGFP, and the 3x nuclear localization signal (DPKKKRKVRS) into the pCAG vector. To establish the Tric/Ocln-dKO cell clones stably expressing exogenous Tric or Ocln and the Ctrl cell clone expressing GFP-nls, the cells were transfected with the vectors using PEI-max and were selected with 200 µg/ml G418 (Sigma-Aldrich) for 9 d. The cells were then screened by immunostaining and fluorescence microscopy.
Immunofluorescence microscopy and image acquisition
For fluorescence microscopy, cells cultured on coverslips or tissue sections were fixed with 99.9% methanol (Fujifilm Wako) for 15 min at –20°C or 1% formaldehyde (Fujifilm Wako) for 30 min at room temperature (RT), permeabilized with 0.2% Triton X-100 in PBS for 5 min at RT, and washed with PBS three times. After blocking with 2% bovine serum albumin (BSA) in PBS, the coverslips were incubated with primary antibodies diluted in Signal Booster solution F (Beacle, Kyoto, Japan) for 2 h at RT or overnight at 4°C. After incubation with secondary antibodies and three washes with PBS, cells were embedded with FLUORO-GEL II with DAPI (Electron Microscopy Sciences, USA). The samples were observed with a laser scanning confocal microscope (FV1000; Olympus, Tokyo, Japan) with a 60× oil-immersion objective lens (UPlanSApo 60×; Olympus) at laser wavelengths of 405, 488, and 559 nm or a fluorescence microscope (BX61; Olympus) with a 40× objective lens (UPlanSApo 40×; Olympus) equipped with a mercury lamp, dichroic filter sets (NIBA, WIG, and WU), and a cooled charge-coupled device (CCD) camera (DP71; Olympus). Images were acquired with fluoview ver. 4.2b (Olympus) or cellSens ver. 1.14 (Olympus) and processed with Photoshop (Adobe, USA). Fluorescence intensity along the cell–cell junctions was quantified with ImageJ software by subtracting the background signal at the cytoplasm. Graphs were generated with Excel (Microsoft).
Immunoblotting
Cells were washed with PBS twice, lysed in CelLytic MT (Sigma-Aldrich) supplemented with cOmplete Mini protease inhibitor cocktail (Sigma-Aldrich) and PhosSTOP phosphatase inhibitor cocktail (Sigma-Aldrich), and diluted in the SDS sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.005% bromophenol blue [BPB]). Sample protein concentration was determined by BCA Protein Assay (Thermo Fisher Scientific), and equal amounts of protein were loaded. Proteins were separated by SDS–PAGE using 7.5% or 5–20% gradient gels (Fujifilm WAKO, Osaka, Japan) and transferred to the polyvinylidene fluoride (PVDF) membrane (Immobilon, Merck). The membrane was blocked with 5% nonfat dried milk in Tris-buffered saline (TBS) with 0.1% Tween-20 (TBS-T) for 30 min at RT and incubated with a primary antibody diluted in Signal Booster solution A (Beacle) overnight at 4°C. After three washes with TBS-T, the membrane was incubated with HRP-conjugated secondary antibody in Signal Booster solution B (Beacle) for 1 h at RT. After three washes with TBS-T, the membranes were incubated with ECL prime (GE Healthcare) and developed using LAS4000 (GE Healthcare). The membranes were reblotted with β-actin as a loading control.
Transmission electron microscopy of ultrathin sections
Cells (2 × 105 cells/ml) were seeded onto polycarbonate Transwells with a 0.4-µm pore size (#3401; Corning, USA) and cultured for 6 d. The cells were washed with 0.1 M phosphate buffer (PB) and prefixed with 2% glutaraldehyde in 0.1 M PB at 4°C for overnight. The filters were excised with scalpels and washed with 0.1 M cacodylate buffer (pH 7.4) three times. Next, the filters were postfixed with 0.1% OsO4 in 0.1 M cacodylate buffer for 30 min at 4°C and washed with water three times. The filters were subsequently stained en bloc with 0.5% aqueous uranyl acetate for 30 min at RT. After three washes with water, the filters were dehydrated with a graded ethanol series (65%/75%/85%, for 10 min at each concentration), further dehydrated in 95% and 99.5% ethanol for 15 min each, and transferred to 100% ethanol for two 15 min incubations. After incubating in propylene oxide for 1 min, the filters were incubated in a 1:1 mixture of propylene oxide/Quetol 812 resin (Nissin EM) overnight, and after three transfers through Quetol 812, the resin was polymerized at 60°C for >48 h. Semithin sections (0.5 mm) were cut and stained with toluidine blue to examine the sample preparation. Ultrathin sections (50–70 nm) were cut and mounted on 200-mesh copper grids. The sections were stained with 0.5% aqueous uranyl acetate in the dark for 3 min at RT, washed with water, further stained with Sato’s lead solution for 3 min at RT, washed with water again, and then allowed to dry. The samples were observed with a JEM1010 transmission EM (JEOL, Tokyo, Japan) at 100 kV accelerating voltage. Images were captured with a Veleta CCD camera using iTEM software (Olympus Soft Imaging Solutions). Cell–cell junctions at the most apical cell contacts were observed.
Transmission electron microscopy of freeze-fracture replicas
Freeze-fracture replicas were produced using a method described previously (Tarusawa et al., 2009) with some modifications. The prefixed cells on the filters with a 0.4-µm pore size (#353090; Corning) were prepared in the same way as the samples for ultrathin sections. After three washes with 0.1 M PB, the samples were cryoprotected with 30% glycerol in 0.1 M PB at 4°C overnight and then rapidly frozen in between two copper carriers by using a high-pressure freezing machine (HPM010; BAL-TEC, Balzers, Liechtenstein). The cells were then fractured by separation of the two carriers at –120°C and replicated by platinum (45° unidirectional from horizontal level, 2 nm thick) and carbon (20 nm thick) in a freeze-fracture replica machine (BAF060; BAL-TEC). The replicated materials were transferred to a solution containing kitchen bleach (50%) and incubated with shaking until the cell debris was removed from the replicas. The replicas were washed twice with distilled water and picked up onto grids coated with pioloform (Agar Scientific, Stansted, Essex, UK) for TEM observation. All freeze-fracture images were presented apical-side up in the figures.
Transepithelial electric resistance measurement
Cells (1 × 105 cells/ml) were seeded onto polycarbonate Transwells with 0.4-µm pore size (#3401; Corning) and cultured for 8 d. The alternating-current (AC) electric resistance between the apical and basal compartments was measured using a volt-ohm-meter (Millicell ERS-2; EMD Millipore, USA). All measurements were subtracted by a blank measurement of a Transwell filter without cells and then multiplied by the culture area of the Transwell filter to calculate the unit area resistance.
Tracer flux measurement
After TER measurement, the basal medium was replaced with phenol red-free DMEM (Life Technologies) supplemented with 10% FBS. The apical medium was replaced by the medium with 1 mg/ml FITC-dextran of molecular mass of 3–5 kDa (FD4; Sigma-Aldrich), 10 kDa (FD10S; Sigma-Aldrich), or 250 kDa (FD250S; Sigma-Aldrich). The cells were incubated in a 5% CO2 incubator at 37°C for 2 h, and the basal medium was collected. Fluorescence intensity of the medium was measured by microplate reader equipment (VARIOSKAN LUX; Thermo Fisher Scientific) in a black-bottomed 96-well plate (Corning). Standard curves were determined by measuring the fluorescence intensities of a serial dilution series of the FITC-dextran. Blank measurement of the medium without FITC-dextran was subtracted from the sample values. The apparent permeability coefficient (Papp) was calculated using the following equation:
 |
where dQ is the amount of tracer transported to the acceptor (basal) compartment during the time period dt, A is the area of the filter, and C is the initial concentration of the donor (apical) compartment.
Statistical analysis
The results are presented as mean ± SD unless otherwise indicated. Statistical significance of differences was evaluated by the Welch’s t test in Supplemental Figures S1E, S2, F and I, and S5, D and E, using Excel software; by the Welch’s t test with Bonferroni’s correction in Figures 3D and 6D and Supplemental Figure S4B using Excel software; by the Poisson’s exact test in Figures 3C and 6C and Supplemental Figure S2D using Excel software; by the weighted Welch’s t test in Figure 2E using R software; and by the Tukey–Kramer’s test in Figures 4 and 6F using R software, and a p < 0.05 was considered to be statistically significant.
Mathematical model
To evaluate the contribution of the TJ-strand complexity on the barrier function of TJs, a simplified multistrand mathematical model for the TER and macromolecular permeability was generated. The basic scheme of the model was essentially based on a previously reported computational dynamic strand model (Tervonen et al., 2019).
The permeation pathways are divided into bicellular and tricellular pathways. The bicellular pathway model is based on the simplified tile-like two-dimensional structure, which consists of horizontal rows and vertical partitions mimicking the complicated anastomosing TJ-strand networks (Figure 5A). TJ strands undergo stochastic breaks (break probability pbreak = 0.05 [s–1µm–1]) and reseal (resealing probability pseal = 0.033 [s–1]). The resealing probability pseal was defined based on previously reported experimental data (Sasaki, Matsui, et al., 2003; Van Itallie et al., 2017).
For the simulation of macromolecule permeability, we created a diffusion model using small particles with 4 kDa (14 Å radius) and 10 kDa (23 Å radius) and simulated the diffusion of particles among the compartments. The temporal change of the number of particles in each compartment is described by the following equation (Tervonen et al., 2019):
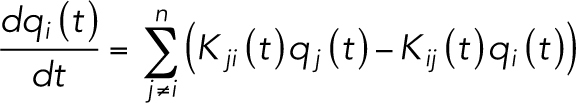 |
1 |
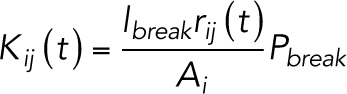 |
2 |
where Kji (t) [s–1] is the time-dependent rate coefficient of permeation of particles from compartment j to i, qi (t) is the number of particles in the compartment i at time t, lbreak is the size of the break in the strand (40 nm) (Sasaki, Matsui, et al., 2003; Van Itallie et al., 2017), rij (t) is a function describing the intact-broken state of the strand between compartments i and j (0: intact, 1:broken), Ai [m2] is the area of compartment i, and Pbreak [m/s] is the permeability coefficient of the strand break of indefinite length. The number of particles in the donor and acceptor compartments are referred to qdonor (t) and qacceptor (t), respectively. Initial parameters at time t = 0 are set to qdonor (0) = 6.022 × 1011 and qi (0) = qacceptor (0) = 0. Because the donor compartment is assumed to be connected to the abundant pool of particles, qdonor (t) is set to remain constant (dqdonor (t)/dt = 0). Additionally, because the acceptor region is connected to a large space, which serves as a sink, the backflow from the acceptor compartment into the upper small compartments was ignored. The Pbreak is proportional to the relative area of TJs per area of epithelium, so that the permeability reflects the complexity of cell boundary shape and size of the cells:
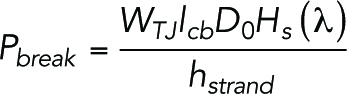 |
3 |
 |
4 |
where WTJ is the TJ width (8 nm) (Suzuki, Tani, et al., 2015), lcb is the cell boundary length per epithelium area (0.28 µm–1) analyzed from our experimental data, D0 [m2 s–1] is the diffusion coefficient of the permeating particle, hstrand is the strand height (6 nm) (Adson et al., 1994), and Hs (λ) is the slit hindrance factor that depends on the particle diameter dm [m] and WTJ (λ = dm/WTJ) (Dechadilok and Deen, 2006). The diffusion coefficient D0 is determined based on the diffusion coefficients of FITC-dextran with molecular weight Mw [Da] of 4–2000 kDa as reported previously (Gribbon and Hardingham, 1998):
| 5 |
Intact strands stochastically break with the probability pbreak [m–1s–1], and broken strands reseal with the probability pseal [s–1]. The break probability pbreak is proportional to the length of the sections between compartments so that the longer sections undergo more frequent breaks.
The initial state of the strands (intact or broken) is determined by the infinite state of Markov chain:
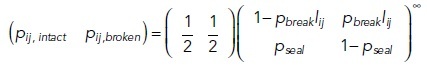 |
6 |
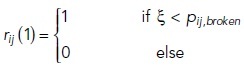 |
7 |
where pij,intact and pij,broken are the probabilities of the strand section between compartments i and j initially being in intact or broken states, respectively, lij [m] being the length of the strand section between those compartments and ξ being the pseudo-random number in the range of 0 to less than 1.
The strand state (rij (t)) is described by the following equation:
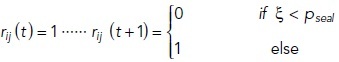 |
 |
8 |
The concentration for each compartment is set to
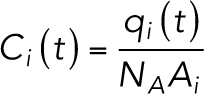 |
9 |
where ci (t) [M] is the concentration of particles in each compartment at time t and NA is the Avogadro’s constant (6.022 × 1023 mol–1). The volume is replaced with Ai [m2] because the model is two-dimensional. The differential equations described by Eqs. 1 and 2 is solved using Matlab’s (Release 2020a: The Math Works, Natick, MA) ode23 differential equation solver, which uses second- and third-order Runge–Kutta formulas. The simulation is run multiple times, and the linear phase of the average qacceptor curve is used to calculate the bTJ permeability coefficient:
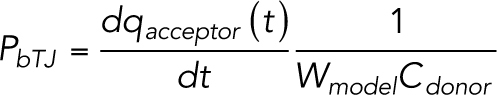 |
10 |
where Wmodel is the entire width of the model (5000 nm), which replaces the area in this two-dimensional model. A first-degree polynomial line is fitted to the linear phase of the average qacceptor to obtain the slope.
The permeability model of the tTJ central tubes is assumed to be the static pores and is described by the following equation (Tervonen et al., 2019):
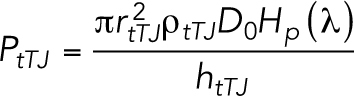 |
11 |
 |
12 |
where rtTJ is the radius of the tricellular pore (5 nm) (Staehelin, 1973), ρtTJ is the density of the tricellular junctions (0.014 µm–2) (Tervonen et al., 2019), Hp (λ) is the pore hindrance factor (λ = dm/2rtTJ), and htTJ is the tricellular pore height (1 µm) (Staehelin, 1973; Krug et al., 2009). The equation for Hp was derived from a previous report (Dechadilok and Deen, 2006).
The total epithelial TJ permeability is finally calculated based on the parallel connection between the two pathways as
| 13 |
The simulation time for the bicellular TJ model is 7200 [s], and the stochastic behavior is further averaged by running the simulations 512 times.
The partition density ρpd (Figure 5B) is determined based on the following equations:
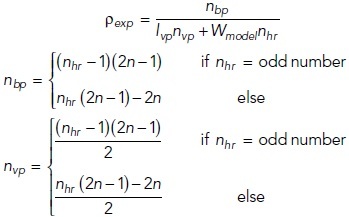 |
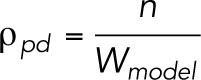 |
14 |
where ρexp is the frequency of the TJ-strand branching points determined experimentally [m–1] (Figures 3C and 6C), nbp is the total number of branching points, lvp is the length of the vertical partition (0.1 µm), nvp is the number of vertical partitions, nhr is the number of horizontal rows, n is the number of compartments in the first row.
In the TER model, the bicellular and tricellular pathways are calculated separately and connected in parallel. The bicellular TJ resistance model is constructed as a network of dynamic resistors based on the simplified tile-like two-dimensional structure (Figure 5A). The system is solved using a nodal analysis and Kirchhoff’s circuit laws. For each current loop i, the equation is described by the following equation (Tervonen et al., 2019):
 |
15 |
where Rij (t) [Ω] is the time-dependent resistance of the section of strand that is shared by current loops i and j, Ii [A] is the current in loop i, and V [V] is the voltage applied by the external source in the outer loop.
The strand dynamics are incorporated into the resistances. Ions can also pass through the intact strands, and Rij (t) depends on both the intact strand and break resistances:
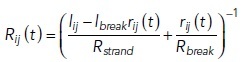 |
16 |
where lij, lbreak, and rij (t) have been described in the bTJ permeability model, Rbreak [Ω] is the resistance of a break region, and Rstrand is the intact strand resistance per strand length (45 GΩ·µm). Because we used Cldn-2 KO cells in this experiment, the Rstrand is set to higher than the previous mathematical model (Tervonen et al., 2019). The break resistance is calculated with the following equation:
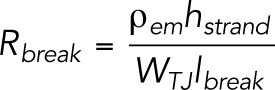 |
17 |
where ρem is the resistivity of the extracellular medium (0.537 Ω·m) (Krug et al., 2009). The bicellular TJ resistance model is simulated for a long time to obtain average results, although the TER measurement is basically instantaneous. The current flowing in the outer loop Iouter [A] is used to calculate the bicellular TJ resistance at each time point with Ohm’s law:
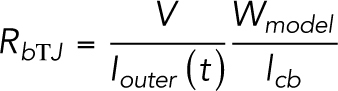 |
18 |
where the factor Wmodel/lcb scales the results for the whole epithelium. To solve the bicellular TJ model, the linear system defined by Eq. 16 is transformed into matrix form and solved using Matlab.
The tTJ tubes are modeled as static, and their resistance is calculated as
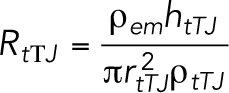 |
19 |
For each simulation time point of the bicellular TJ resistance model, the total TER is calculated based on the parallel connection as
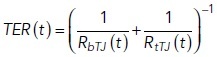 |
20 |
To obtain the average TER for the simulation, the time average is taken from the results. The simulation time is 106 s, and the simulations were run using Matlab.
Supplementary Material
Acknowledgments
We express our gratitude to Taichi Sugawara for sharing unpublished data, Ritsuko Arai for her assistance with the TEM experiments, the members of the Laboratory Animal Research Center of Fukushima Medical University for maintenance of animals, Kyoko Furuse, Ayumi Hozumi, Keiko Watari, and Takako Maegawa for their technical assistance, and the members of the Furuse lab and Chiba lab for their helpful discussion. This work was supported by the Cooperative Study Program (19-136) of the National Institute for Physiological Sciences and a Grant-in-Aid for Scientific Research on Innovative Areas—Platforms for Advanced Technologies and Research Resources “Advanced Bioimaging Support (ABiS).” We also express our gratitude to the Science English Editing Section of Fukushima Medical University for their proofreading of our manuscript. This work was funded by JSPS (Japanese Society for the Promotion of Science) KAKENHI (Grant Number 18K15085) to A.C.S. and was partly supported by JSPS KAKENHI (Grant Number 18K06223) and the Medical Research Fund from Takeda Science Foundation to T. H.
Abbreviations used:
- FFEM
freeze-fracture replica electron microscopy
- Papp
apparent permeability
- SJ
septate junction
- TAMP
TJ-Associated MARVEL Protein
- TEM
transmission electron microscopy
- TER
transepithelial electric resistance
- TJ
tight junction
- tTJ
tricellular tight junction.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-07-0464) on February 10, 2021.
REFERENCES
Boldface names denote co–first authors.
- Adson A, Raub TJ, Burton PS, Barsuhn CL, Hilgers AR, Audus KL, Ho NF (1994). Quantitative approaches to delineate paracellular diffusion in cultured epithelial cell monolayers. J Pharm Sci 83, 1529–1536. [DOI] [PubMed] [Google Scholar]
- Anderson JM, Van Itallie CM (2009). Physiology and function of the tight junction. Cold Spring Harb Perspect Biol 1, a002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalcroft JP, Bullivant S (1970). An interpretation of liver cell membrane and junction structure based on observation of freeze-fracture replicas of both sides of the fracture. J Cell Biol 47, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba H, Osanai M, Murata M, Kojima T, Sawada N (2008). Transmembrane proteins of tight junctions. Biochim Biophys Acta 1778, 588–600. [DOI] [PubMed] [Google Scholar]
- Claude P (1978). Morphological factors influencing transepithelial permeability: a model for the resistance of the zonula occludens. J Membr Biol 39, 219–232. [DOI] [PubMed] [Google Scholar]
- Claude P, Goodenough DA (1973). Fracture faces of zonulae occludentes from “tight” and “leaky” epithelia. J Cell Biol 58, 390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cording J, Berg J, Käding N, Bellmann C, Tscheik C, Westphal JK, Milatz S, Günzel D, Wolburg H, Piontek J, et al. (2013). In tight junctions, claudins regulate the interactions between occludin, tricellulin and marvelD3, which, inversely, modulate claudin oligomerization. J Cell Sci 126, 554–564. [DOI] [PubMed] [Google Scholar]
- Dechadilok P, Deen WM (2006). Hindrance factors for diffusion and convection in pores. Ind Eng Chem Res 45, 6953–6959. [Google Scholar]
- Farquhar MG, Palade GE (1963). Junctional complexes in various epithelia. J Cell Biol 17, 375–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S (1998a). Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol 141, 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Furuse K, Sasaki H, Tsukita S (2001). Conversion of zonulae occludentes from tight to leaky strand type by introducing claudin-2 into Madin-Darby canine kidney I cells. J Cell Biol 153, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S (1993). Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol 123, 1777–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Sasaki H, Fujimoto K, Tsukita S (1998b). A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol 143, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Tsukita S (2006). Claudins in occluding junctions of humans and flies. Trends Cell Biol 16, 181–188. [DOI] [PubMed] [Google Scholar]
- Gong Y, Renigunta V, Zhou Y, Sunq A, Wang J, Yang J, Renigunta A, Baker LA, Hou J (2015). Biochemical and biophysical analyses of tight junction permeability made of claudin-16 and claudin-19 dimerization. Mol Biol Cell 26, 4333–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A, Davies C, Southwood CM, Frolenkov G, Chrustowski M, Ng L, Yamauchi D, Marcus DC, Kachar B (2004). Deafness in claudin 11-null mice reveals the critical contribution of basal cell tight junctions to stria vascularis function. J Neurosci 24, 7051–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A, Southwood CM, Li JS, Pariali M, Riordan GP, Brodie SE, Danias J, Bronstein JM, Kachar B, Lazzarini RA (1999). CNS myelin and sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell 99, 649–659. [DOI] [PubMed] [Google Scholar]
- Gribbon P, Hardingham TE (1998). Macromolecular diffusion of biological polymers measured by confocal fluorescence recovery after photobleaching. Biophys J 75, 1032–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T, Chiba H (2020). Molecular organization, regulation and function of tricellular junctions. Biochim Biophys Acta 1862, 183143. [DOI] [PubMed] [Google Scholar]
- Higashi T, Miller AL (2017). Tricellular junctions: how to build junctions at the TRICkiest points of epithelial cells. Mol Biol Cell 28, 2023–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T, Tokuda S, Kitajiri S, Masuda S, Nakamura H, Oda Y, Furuse M (2013). Analysis of the “angulin” proteins LSR, ILDR1 and ILDR2–tricellulin recruitment, epithelial barrier function and implication in deafness pathogenesis. J Cell Sci 126, 966–977. [DOI] [PubMed] [Google Scholar]
- Hou J, Renigunta A, Konrad M, Gomes AS, Schneeberger EE, Paul DL, Waldegger S, Goodenough DA (2008). Claudin-16 and claudin-19 interact and form a cation-selective tight junction complex. J Clin Invest 118, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S, Tsukita S (2005). Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol 171, 939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J, Sasaki H, Tsukita S, Furuse M, Tsukita S (2008). Loss of occludin affects tricellular localization of tricellulin. Mol Biol Cell 19, 4687–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto N, Higashi T, Furuse M (2014). Localization of angulin-1/LSR and tricellulin at tricellular contacts of brain and retinal endothelial cells in vivo. Cell Struct Funct 39, 1–8. [DOI] [PubMed] [Google Scholar]
- Kitajiri S, Miyamoto T, Mineharu A, Sonoda N, Furuse K, Hata M, Sasaki H, Mori Y, Kubota T, Ito J, et al. (2004). Compartmentalization established by claudin-11-based tight junctions in stria vascularis is required for hearing through generation of endocochlear potential. J Cell Sci 117, 5087–5096. [DOI] [PubMed] [Google Scholar]
- Kitajiri S-I, Katsuno T, Sasaki H, Ito J, Furuse M, Tsukita S (2014). Deafness in occludin-deficient mice with dislocation of tricellulin and progressive apoptosis of the hair cells. Biol Open 3, 759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug SM (2017). Contribution of the tricellular tight junction to paracellular permeability in leaky and tight epithelia. Ann NY Acad Sci 1397, 219–230. [DOI] [PubMed] [Google Scholar]
- Krug SM, Amasheh S, Richter JF, Milatz S, Günzel D, Westphal JK, Huber O, Schulzke JD, Fromm M (2009). Tricellulin forms a barrier to macromolecules in tricellular tight junctions without affecting ion permeability. Mol Biol Cell 20, 3713–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug SM, Günzel D, Conrad MP, Lee I-FM, Amasheh S, Fromm M, Yu ASL (2012). Charge-selective claudin channels. Ann NY Acad Sci 1257, 20–28. [DOI] [PubMed] [Google Scholar]
- Lynn KS, Peterson RJ, Koval M (2020). Ruffles and spikes: control of tight junction morphology and permeability by claudins. Biochim Biophys Acta 1862, 183339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madara JL, Dharmsathaphorn K (1985). Occluding junction structure-function relationships in a cultured epithelial monolayer. J Cell Biol 101, 2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Oda Y, Sasaki H, Ikenouchi J, Higashi T, Akashi M, Nishi E, Furuse M (2011). LSR defines cell corners for tricellular tight junction formation in epithelial cells. J Cell Sci 124, 548–555. [DOI] [PubMed] [Google Scholar]
- Morita K, Sasaki H, Fujimoto K, Furuse M, Tsukita S (1999). Claudin-11/OSP-based tight junctions of myelin sheaths in brain and Sertoli cells in testis. J Cell Biol 145, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroi S, Saitou M, Fujimoto K, Sakakibara A, Furuse M, Yoshida O, Tsukita S (1998). Occludin is concentrated at tight junctions of mouse/rat but not human/guinea pig Sertoli cells in testes. Am J Physiol 274, C1708–C1717. [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J (1991). Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199. [DOI] [PubMed] [Google Scholar]
- Noirot-Timothee C, Noirot C (1980). Septate and scalariform junctions in arthropods. Int Rev Cytol 63, 97–140. [DOI] [PubMed] [Google Scholar]
- Oda Y, Sugawara T, Fukata Y, Izumi Y, Otani T, Higashi T, Fukata M, Furuse M (2020). The extracellular domain of angulin-1 and palmitoylation of its cytoplasmic region are required for angulin-1 assembly at tricellular contacts. J Biol Chem 295, 4289–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani T, Nguyen TP, Tokuda S, Sugihara K, Sugawara T, Furuse K, Miura T, Ebnet K, Furuse M (2019). Claudins and JAM-A coordinately regulate tight junction formation and epithelial polarity. J Cell Biol 218, 3372–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontek J, Krug SM, Protze J, Krause G, Fromm M (2020). Molecular architecture and assembly of the tight junction backbone. Biochim Biophys Acta 1862, 183279. [DOI] [PubMed] [Google Scholar]
- Raleigh DR, Marchiando AM, Zhang Y, Shen L, Sasaki H, Wang Y, Long M, Turner JR (2010). Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol Biol Cell 21, 1200–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Ando-Akatsuka Y, Itoh M, Furuse M, Inazawa J, Fujimoto K, Tsukita S (1997). Mammalian occludin in epithelial cells: its expression and subcellular distribution. Eur J Cell Biol 73, 222–231. [PubMed] [Google Scholar]
- Saitou M, Fujimoto K, Doi Y, Itoh M, Fujimoto T, Furuse M, Takano H, Noda T, Tsukita S (1998). Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions. J Cell Biol 141, 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Furuse M, Sasaki H, Schulzke JD, Fromm M, Takano H, Noda T, Tsukita S (2000). Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol Biol Cell 11, 4131–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Matsui C, Furuse K, Mimori-Kiyosue Y, Furuse M, Tsukita S (2003). Dynamic behavior of paired claudin strands within apposing plasma membranes. Proc Natl Acad Sci USA 100, 3971–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger EE, Lynch RD (2004). The tight junction: a multifunctional complex. Am J Physiol Cell Physiol 286, C1213–C1228. [DOI] [PubMed] [Google Scholar]
- Schulzke JD, Gitter AH, Mankertz J, Spiegel S, Seidler U, Amasheh S, Saitou M, Tsukita S, Fromm M (2005). Epithelial transport and barrier function in occludin-deficient mice. Biochim Biophys Acta 1669, 34–42. [DOI] [PubMed] [Google Scholar]
- Shen L, Weber CR, Turner JR (2008). The tight junction protein complex undergoes rapid and continuous molecular remodeling at steady state. J Cell Biol 181, 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Weber CR, Raleigh DR, Yu D, Turner JR (2011). Tight junction pore and leak pathways: a dynamic duo. Annu Rev Physiol 73, 283–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA (1973). Further observations on the fine structure of freeze-cleaved tight junctions. J Cell Sci 13, 763–786. [DOI] [PubMed] [Google Scholar]
- Steed E, Rodrigues NTL, Balda MS, Matter K (2009). Identification of MarvelD3 as a tight junction-associated transmembrane protein of the occludin family. BMC Cell Biol 10, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Tani K, Tamura A, Tsukita S, Fujiyoshi Y (2015). Model for the architecture of claudin-based paracellular ion channels through tight junctions. J Mol Biol 427, 291–297. [DOI] [PubMed] [Google Scholar]
- Tarusawa E, Matsui K, Budisantoso T, Molnár E, Watanabe M, Matsui M, Fukazawa Y, Shigemoto R (2009). Input-specific intrasynaptic arrangements of ionotropic glutamate receptors and their impact on postsynaptic responses. J Neurosci 29, 12896–12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervonen A, Ihalainen TO, Nymark S, Hyttinen J (2019). Structural dynamics of tight junctions modulate the properties of the epithelial barrier. PLoS One 14, e0214876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda S, Furuse M (2015). Claudin-2 knockout by TALEN-mediated gene targeting in MDCK cells: claudin-2 independently determines the leaky property of tight junctions in MDCK cells. PLoS One 10, e0119869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Furuse M, Itoh M (2001). Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol 2, 285–293. [DOI] [PubMed] [Google Scholar]
- Turner JR, Buschmann MM, Romero-Calvo I, Sailer A, Shen L (2014). The role of molecular remodeling in differential regulation of tight junction permeability. Semin Cell Dev Biol 36, 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M, Tsukita S (2006). ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 126, 741–754. [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM (2014). Architecture of tight junctions and principles of molecular composition. Semin Cell Dev Biol 36, 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie CM, Fanning AS, Holmes J, Anderson JM (2010). Occludin is required for cytokine-induced regulation of tight junction barriers. J Cell Sci 123, 2844–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie CM, Lidman KF, Tietgens AJ, Anderson JM (2019). Newly synthesized claudins but not occludin are added to the basal side of the tight junction. Mol Biol Cell 30, 1406–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie CM, Tietgens AJ, Anderson JM (2017). Visualizing the dynamic coupling of claudin strands to the actin cytoskeleton through ZO-1. Mol Biol Cell 28, 524–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan S, Stephenson RE, Miller AL (2019). Multiscale dynamics of tight junction remodeling. J Cell Sci 132, jcs.229286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JB, Karnovsky MJ (1974). The structure of the zonula occludens. A single fibril model based on freeze-fracture. J Cell Biol 60, 168–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CJ, Rowland M, Warhurst G (2001). Functional modeling of tight junctions in intestinal cell monolayers using polyethylene glycol oligomers. Am J Physiol Cell Physiol 281, C388–C397. [DOI] [PubMed] [Google Scholar]
- Weber CR, Liang GH, Wang Y, Das S, Shen L, Yu ASL, Nelson DJ, Turner JR (2015). Claudin-2-dependent paracellular channels are dynamically gated. eLife 4, e09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ASL, Cheng MH, Angelow S, Günzel D, Kanzawa SA, Schneeberger EE, Fromm M, Coalson RD (2009). Molecular basis for cation selectivity in claudin-2-based paracellular pores: identification of an electrostatic interaction site. J Gen Physiol 133, 111–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ASL, McCarthy KM, Francis SA, McCormack JM, Lai J, Rogers RA, Lynch RD, Schneeberger EE (2005). Knockdown of occludin expression leads to diverse phenotypic alterations in epithelial cells. Am J Physiol Cell Physiol 288, C1231–C1241. [DOI] [PubMed] [Google Scholar]
- Zihni C, Mills C, Matter K, Balda MS (2016). Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol 17, 564–580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.