Abstract
Autophagy is a cellular degradation system widely conserved among eukaryotes. During autophagy, cytoplasmic materials fated for degradation are compartmentalized in double membrane–bound organelles called autophagosomes. After fusing with the vacuole, their inner membrane–bound structures are released into the vacuolar lumen to become autophagic bodies and eventually degraded by vacuolar hydrolases. Atg15 is a lipase that is essential for disintegration of autophagic body membranes and has a transmembrane domain at the N-terminus and a lipase domain at the C-terminus. However, the roles of the two domains in vivo are not well understood. In this study, we found that the N-terminal domain alone can travel to the vacuole via the multivesicular body pathway, and that targeting of the C-terminal lipase domain to the vacuole is required for degradation of autophagic bodies. Moreover, we found that the C-terminal domain could disintegrate autophagic bodies when it was transported to the vacuole via the Pho8 pathway instead of the multivesicular body pathway. Finally, we identified H435 as one of the residues composing the putative catalytic triad and W466 as an important residue for degradation of autophagic bodies. This study may provide a clue to how the C-terminal lipase domain recognizes autophagic bodies to degrade them.
INTRODUCTION
Macroautophagy (hereafter called “autophagy”) is a bulk degradation system widely conserved among eukaryotes from yeast to mammals (Mizushima et al., 2011). Autophagy enables cells to survive severe environmental conditions, such as nutrient limitation, by recycling cellular building blocks including amino acids and nucleic acids (Kuma et al., 2004; Onodera and Ohsumi, 2005; Huang et al., 2015).
In Saccharomyces cerevisiae, upon induction of autophagy by starvation or rapamycin treatment, the scaffold for autophagosome formation is organized on the vacuolar membrane (Kawamata et al., 2008; Fujioka et al., 2020). Subsequently, downstream Atg proteins, including Atg8, are recruited to the scaffold, where they generate an isolation membrane. Following expansion of the isolation membrane, a double membrane–bound organelle called an autophagosome is formed by closure of the open region of the isolation membrane. After the outer membrane of the autophagosome fuses with the vacuolar membrane, a single membrane–bound structure derived from the inner membrane of an autophagosome, called an autophagic body, is released into the vacuolar lumen and disintegrated. Membranes of autophagic bodies must be disintegrated to allow vacuolar hydrolases to access their cargoes for degradation.
On the other hand, a specific type of autophagic pathway is active under growing conditions in the yeast Saccharomyces cerevisiae. This pathway, the cytoplasm-to-vacuole targeting (Cvt) pathway, selectively encloses the vacuolar hydrolase aminopeptidase I (Ape1). Ape1 is synthesized in precursor form (prApe1) and assembles into the Ape1 complex in the cytoplasm (Shintani et al., 2002; Yamasaki et al., 2020). The Ape1 complex is transported to the vacuole by the Cvt pathway under nutrient-rich conditions, but by autophagy under starvation conditions (Baba et al., 1997). The Cvt vesicle, a double-membrane compartment, is responsible for Ape1 transport via the Cvt pathway (Baba et al., 1997). The outer membranes of Cvt vesicles fuse with the vacuolar membrane, and then Cvt bodies are released into the vacuole. After disintegration of the limiting membranes of Cvt bodies, the propeptide in prApe1 is cleaved by vacuolar proteinases; consequently, prApe1 is converted to mature Ape1 (mApe1; Klionsky et al., 1992). The decrease in the molecular weight of prApe1, detected by immunoblot analysis, can be used as an indicator of degradation of Cvt bodies and autophagic bodies.
Pep4 and Prb1, two vacuolar proteinases of Saccharomyces cerevisiae (Ammerer et al., 1986; Woolford et al., 1986; Moehle et al., 1987), are required for disintegration of Cvt bodies/autophagic bodies (Takeshige et al., 1992). The function of Pep4 in the vacuole is to activate other vacuolar hydrolases, such as Pho8, Prb1, and Prc1, and degrade cellular components (Ammerer et al., 1986). Although Pep4 and Prb1 are unlikely to have lipase activity, Cvt bodies/autophagic bodies remain intact in pep4Δ and prb1Δ cells. A longstanding question in the field is how Pep4 and Prb1 contribute to disintegration of the membranes of Cvt bodies/autophagic bodies.
Atg15/Aut5/Cvt17, a glycosylated integral membrane protein that is essential for disintegration of Cvt bodies/autophagic bodies, is the only vacuolar lipase in Saccharomyces cerevisiae (Epple et al., 2001; Teter et al., 2001). Although the molecular machinery of autophagosome formation is conserved across eukaryotes, orthologues of Atg15 are only found in fungi (Epple et al., 2001; Teter et al., 2001). Atg15 has a transmembrane domain (TMD) at its N-terminus and a lipase domain at the C-terminus (Epple et al., 2001). The S332 residue in the C-terminal domain is thought to be the active center for Atg15 lipase activity (Epple et al., 2001; Teter et al., 2001; Ramya and Rajasekharan, 2016). Localization analysis using fixed cells suggests that Atg15 is localized to the endoplasmic reticulum (ER) and transported to the vacuole via the multivesicular body (MVB) pathway (Epple et al., 2001), but its extreme N-terminal cytoplasmic region (residues 2–12) is not required for the localization (Epple et al., 2003). However, it remains unknown whether the N-terminal TMD plays a role in the Atg15 activity and whether the C-terminal lipase domain actually functions in the vacuolar lumen. In this study, we sought to answer these questions by analyzing the N- and C-terminal domains of Atg15 separately.
RESULTS
Expansion of autophagic membranes in atg15Δ cells is normal during autophagy
The primary role of Atg15 is to degrade autophagic bodies by its lipase activity (Epple et al., 2001; Teter et al., 2001). An electron microscopic analysis shows that autophagic bodies accumulate in the vacuoles of atg15Δ cells (Epple et al., 2001), indicating that autophagosomes are formed. However, we still do not know whether the size of autophagosomes is normal in atg15Δ cells, due to lack of quantitative analysis. Thus, we first examined whether the defect in degradation of autophagic bodies in atg15Δ cells directly or indirectly affected the size of autophagosomes by quantitative analysis. Because conventional assays of autophagic activity, that is, the Ape1 maturation assay, alkaline phosphatase assay, and GFP-Atg8 cleavage assay, require degradation of autophagic bodies (Klionsky et al., 1992; Noda et al., 1995; Shintani and Klionsky, 2004), previously there was no suitable method for measuring the activity of expansion of autophagic membranes (AM) in mutants defective in degradation of autophagic bodies.
Recently, we developed a system for measuring the activity of AM expansion by measuring AM length: the quantitative autophagy-related structure analysis system (Qautas; Kawaoka et al., 2017). Qautas is a high-throughput and comprehensive system for morphological analysis of AMs labeled with Atg8. In Qautas, morphological parameters of Atg8-labeled structures are extracted from fluorescence microscopic images, and then the structures are classified into dot-shaped or elongated structures by machine learning based on their morphological parameters. By focusing on the elongated structures, the AM expansion activity of each strain, which correlates with the activity of autophagosome formation, can be estimated. In this study, we used mNeonGreen-Atg8 in cells overexpressing prApe1 to visualize AMs (Suzuki et al., 2013; Hirata et al., 2017). We used the value “perimeter/2,” one of the parameters indicating the size of a structure, to estimate AM expansion activity (Kawaoka et al., 2017). Hereafter, we refer to this quantity as “AM length.”
Atg2 is essential for autophagosome formation (Shintani et al., 2001; Wang et al., 2001). In prApe1-overexpressing atg2Δ cells, Atg8 remains a dot close to the vacuole during autophagy, indicating that Atg8 can be targeted to the scaffold, but AM expansion is abrogated (Suzuki et al., 2013; Hirata et al., 2017).
During autophagy, we visualized AMs as cup-shaped structures in wild-type cells, but as dots in atg2Δ cells (Figure 1A). In atg15Δ cells, cup-shaped AMs were also detected (Figure 1A). Quantitative analysis revealed that the AM length of atg2Δ cells (median 0.64 μm) was significantly less than that of wild-type cells (median 0.82 μm; Figure 1B). In contrast, the AM length of atg15Δ cells (median 0.82 μm) was comparable to that in wild-type cells (Figure 1B). These results indicated that AM expansion is normal in atg15Δ cells. Moreover, we detected no significant differences in AM length between atg15Δ cells and wild-type cells cultured under nitrogen starvation conditions for 6 h (Supplemental Figure S1). This result suggests that a shortage of degradative products due to a defect in degradation of autophagic bodies in atg15Δ cells does not have a large impact on AM expansion activity.
FIGURE 1:
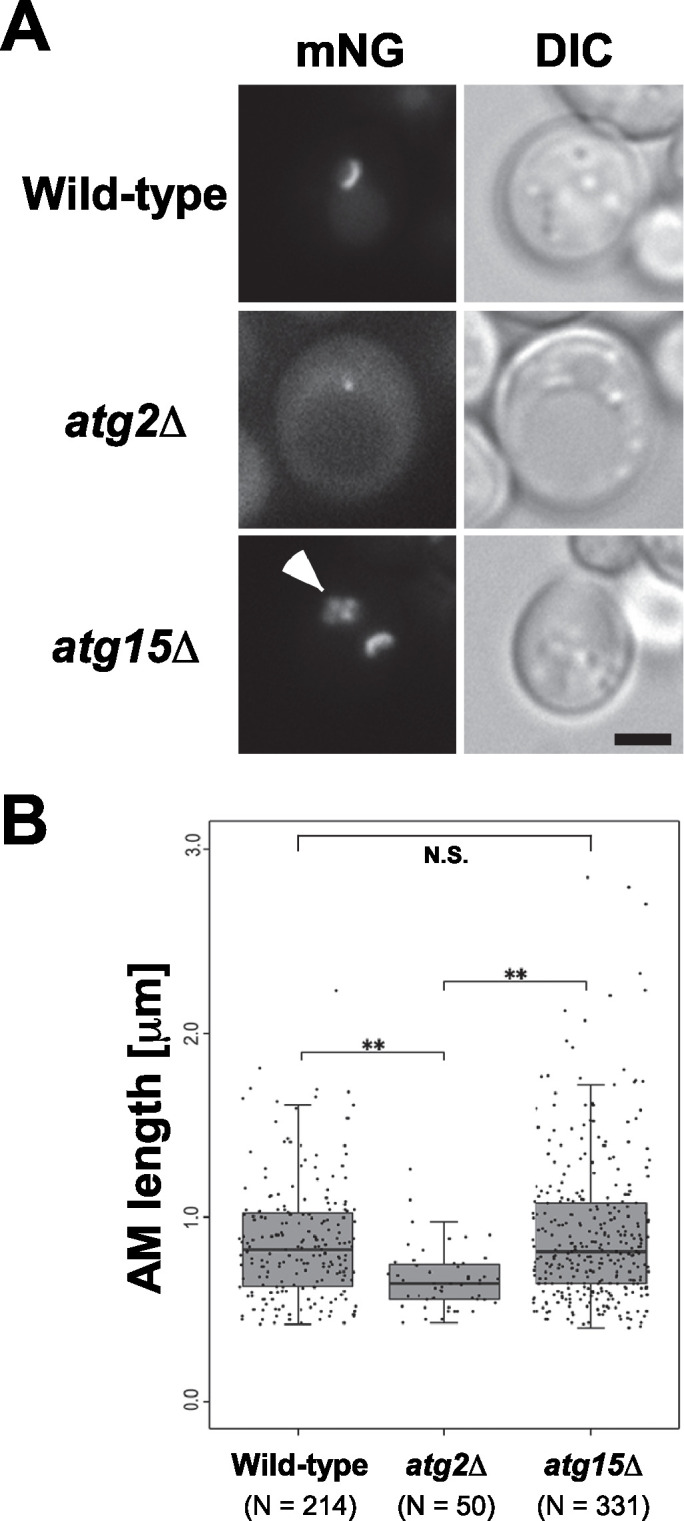
Expansion of autophagic membranes in atg15Δ cells is normal during autophagy. Cells carrying the pRS314[mNeonGreen-Atg8] and pYEX-BX[prApe1] plasmids were grown in SDCA medium containing CuSO4 and were treated with rapamycin for 1.5 h before observation. (A) Fluorescence microscopic images. Arrowhead indicates autophagic bodies inside the vacuole. Scale bar represents 2 μm. (B) Length of autophagic membranes (AM length) calculated with Qautas. N.S. indicates not significant. **P < 0.01 (Mann–Whitney U test with Bonferroni correction). Wild-type (SEY6210), atg2Δ (STY757), and atg15Δ (GYS1298) cells were used as host strains.
Atg15 is delivered to the vacuole via the multivesicular body pathway
Atg15 is a glycosylated transmembrane protein required for disintegration of autophagic bodies (Epple et al., 2001; Teter et al., 2001). Immunofluorescence microscopy has shown that Atg15 is transported to the vacuole via the MVB pathway (Epple et al., 2001, 2003). To visualize the dynamics of Atg15 in living cells, we constructed a strain that expressed C-terminally 2 × GFP-tagged Atg15 (hereafter Atg15-GFP) instead of endogenous Atg15, and found that Atg15-GFP labeled the vacuolar lumen efficiently with a weak ER staining in wild-type cells (Figure 2). To analyze the transport pathway of Atg15-GFP, we disrupted the VPS4 gene, which is required for the MVB pathway. Vps4 is an AAA-ATPase required for the MVB pathway, and in the absence of Vps4, an aberrant MVB termed the class E compartment accumulates (Babst et al., 1997). The lipophilic dye FM 4-64 labels vacuolar membranes and class E compartments in living cells (Vida and Emr, 1995). We stained Atg15-GFP cells with FM 4-64, which exclusively labeled vacuolar membranes in wild-type cells (Figure 2). In vps4Δ cells, Atg15-GFP accumulated in the class E compartment, which was also labeled with FM 4-64 (Figure 2). This result strongly supports the idea that Atg15 is transported to the vacuole via the MVB pathway. Pep4 is an aspartyl protease that plays a central role in maturation of vacuolar hydrolases (Ammerer et al., 1986; Woolford et al., 1986; Epple et al., 2001). In pep4Δ cells, Atg15-GFP was visualized as small dots moving around inside the vacuole (Supplemental Video S1). This result is well consistent with a previous report that Atg15-localizing vesicles derived from the MVB pathway accumulate in the vacuolar lumen in pep4Δ cells (Epple et al., 2001).
FIGURE 2:
Atg15 is delivered to the vacuole via the multivesicular body pathway. Cells expressing C-terminally GFP-fused Atg15 were grown in SDCA medium, labeled with FM 4-64, and subsequently, treated with rapamycin for 2 h. Wild-type (GYS1162), vps4Δ (GYS1166), and pep4Δ (GYS1168) cells were used. Scale bar represents 5 μm.
Video S1.
Cells expressing C-terminally GFP-fused Atg15 were grown in SDCA medium to mid-log phase and treated with rapamycin for 2 h before observation. All videos are played at 0.8 times speed. (A) Wild-type cells. (B) pep4Δ cells.
Video S1.
Cells expressing C-terminally GFP-fused Atg15 were grown in SDCA medium to mid-log phase and treated with rapamycin for 2 h before observation. All videos are played at 0.8 times speed. (A) Wild-type cells. (B) pep4Δ cells.
We next examined the expression of Atg15-GFP in these strains by immunoblot analysis. Full-length Atg15-GFP was detected at almost the same level in all strains (Figure 3A). In contrast, the amount of GFP moiety derived from Atg15-GFP differed among the strains (Figure 3A). In wild-type cells, free GFP was detected during nutrient growth and its level increased during autophagy (Figure 3A). The level of GFP in vps4Δ cells was reduced, but exhibited a pattern similar to that in wild-type cells (Figure 3A). Little free GFP was detected in pep4Δ cells (Figure 3A), suggesting that cleavage of Atg15-GFP occurred inside the vacuole dependent on Pep4. Ypt7 is a Rab GTPase required for homotypic fusion of vacuoles and heterotypic fusion of vesicles with the vacuole (Wada et al., 1992; Wichmann et al., 1992). Thus, protein sorting to the vacuole, including autophagic pathways, is retarded in ypt7Δ cells (Wichmann et al., 1992; Kirisako et al., 1999). The fact that free GFP decreased in ypt7Δ cells (Figure 3A) also supports the assumption that free GFP is cleaved off of Atg15-GFP by vacuolar hydrolases.
FIGURE 3:
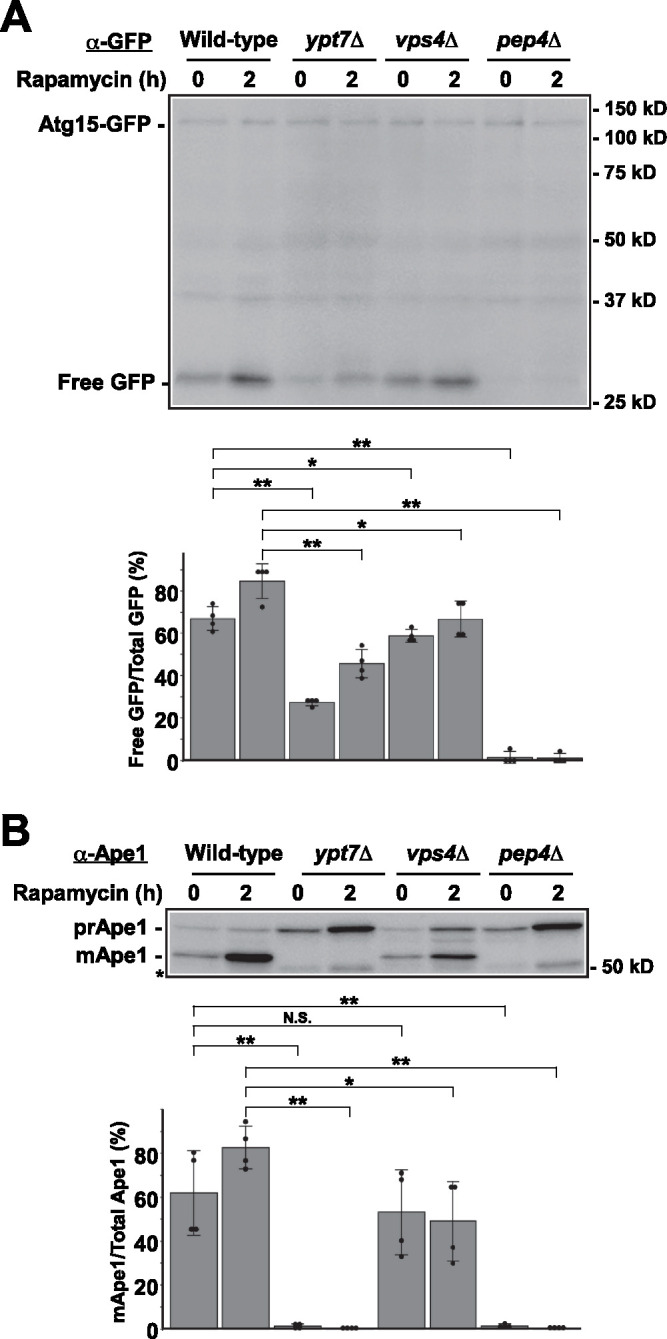
Analysis of Atg15-GFP strains by immunoblot. Cells were grown in SDCA medium and treated with rapamycin. After incubation for the indicated periods, cells were collected and cell lysates were prepared by the alkaline lysis method. Cell lysates equivalent to 0.15 OD600 units were subjected to immunoblot analysis with (A) anti-GFP antibodies or (B) an anti-Ape1 antiserum. The intensity of the bands was quantified for each panel. Asterisk indicates nonspecific bands. Wild-type (GYS1162), ypt7Δ(GYS1164), vps4Δ(GYS1166), and pep4Δ(GYS1168) cells were used in these experiments. N.S. indicates not significant. *P < 0.05, **P < 0.01 (N = 4, two-tailed Student’s t test).
Next, we examined autophagic activity of Atg15-GFP cells by monitoring maturation of Ape1. We found that prApe1 was efficiently converted to mApe1 in wild-type cells during vegetative growth and autophagy (Figure 3B), indicating that Atg15-GFP was capable of disintegrating autophagic bodies efficiently. In ypt7Δ and pep4Δ cells, mApe1 was barely detectable, as previously reported (Klionsky et al., 1992; Kirisako et al., 1999; Figure 3B). Ape1 maturation in vps4Δ cells was normal during vegetative growth but partially defective during autophagy (Figure 3B), probably because a small amount of Atg15-GFP was still transported to the vacuolar lumen even in the absence of Vps4 (Figure 3A). These results suggest that Atg15 functions in degradation of autophagic bodies inside the vacuole after it is transported into the organelle, not outside the vacuole during transport.
The N-terminal transmembrane domain of Atg15 is a signal sequence for the multivascular body pathway
A previous analysis showed that deletion of most of the N-terminal cytosolic domain of Atg15 affects neither Atg15 transport to the vacuole nor Atg15 activity (Epple et al., 2003). This result suggests that the N-terminal cytosolic domain plays a minor role, if any, in the activity of Atg15. Thus, we decided to examine the role of the N-terminal domain, including the transmembrane domain of Atg15 (hereafter Atg15TMD), in the activity of Atg15. For this purpose, we fused GFP to the Atg15TMD, 2–36 amino acid residues of Atg15, and examined its localization (Figure 4A). GFP-Atg15TMD was transported to the vacuole in wild-type and atg15Δ cells, but its transport was blocked in vps4Δ and vps4Δatg15Δ cells (Figure 4B). Quantification of the GFP-labeled vacuolar lumen in these images revealed that the transport of GFP-Atg15TMD to the vacuolar lumen was significantly impaired in the presence of Atg15 (Figure 4C), suggesting that endogenous Atg15 competes with GFP-Atg15TMD for transport to the vacuole. FM 4-64 staining of these cells showed that GFP-Atg15TMD accumulated in the class E compartment in vps4Δ cells (Figure 4B). These results showed clearly that Atg15TMD is transported to the vacuole via the MVB pathway as Atg15 is. We conclude that the N-terminal region (amino acids 2–36) of Atg15 is sufficient for Atg15 delivery to the vacuole via the MVB pathway.
FIGURE 4:
The N-terminal transmembrane domain of Atg15 is a signal sequence for the MVB pathway. (A) Schematic diagram of the GFP-Atg15TMD construct. (B) Cells carrying the pRS316[GFP-Atg15TMD] plasmid were grown in SDCA medium, labeled with FM 4-64, and observed by fluorescence microscopy. Scale bar represents 5 μm. (C) Quantification of cells with a GFP-labeled vacuolar lumen using the images described in (B). N.S. indicates not significant. **P < 0.01 (two-tailed Student’s t test). Wild-type (SEY6210), atg15Δ (GYS1298), vps4Δ (MBY3), and atg15Δvps4Δ (GYS1336) cells were used.
Next, we investigated whether the Atg15TMD is important for the activity of Atg15. In addition to GFP-Atg15TMD, mentioned above, we constructed N-terminally GFP-fused full-length Atg15 (GFP-Atg15FL) and N-terminally GFP-fused Atg15 lacking the TMD (GFP-Atg15ΔTMD; Figure 5A). GFP-Atg15FL and GFP-Atg15TMD were localized to the ER and vacuolar lumen under growing conditions (Figure 5B), and their vacuolar lumen staining became more remarkable during autophagy (Figure 5B). By contrast, GFP-Atg15ΔTMD exhibited a cytoplasmic pattern with slight staining of the nucleus (Figure 5B), suggesting that vacuolar localization of Atg15 was defective in the absence of the Atg15TMD. Emergence of free GFP by immunoblot analysis is one of the indicators to monitor transport of GFP-fused proteins to the vacuolar lumen. Free GFP was detected in GFP-Atg15FL and GFP-Atg15TMD cells but not in GFP-Atg15ΔTMD cells (Figure 5C), supporting that Atg15TMD is important for its transport to the vacuolar lumen. Maturation of Ape1 was not observed in GFP-Atg15TMD cells, and was severely defective in GFP-Atg15ΔTMD cells (Figure 5, C and D). However, a significant amount of mApe1 was still detected in GFP-Atg15ΔTMD cells (Figure 5, C and D). This result implies a second MVB targeting site on Atg15 lacking TMD or alternative pathways of Atg15ΔTMD to the vacuolar lumen. These results suggest that the C-terminal domain of Atg15 is critical for its activity, and that transport of the C-terminal domain to the vacuolar lumen is important.
FIGURE 5:
The N-terminal transmembrane domain of Atg15 is important for the activity of Atg15. (A) Schematic diagram of GFP-Atg15 constructs. (B) atg15Δ cells carrying the pRS316[GFP-Atg15FL], pRS316[GFP-Atg15TMD], and pRS316[GFP-Atg15ΔTMD] plasmids were grown in SDCA medium and treated with rapamycin for 2 h before fluorescence microscopy. Scale bar represents 5 μm. (C) In addition to the plasmids listed in (A), atg15Δ cells carrying the pRS316 empty vector (EV) were used as a negative control. Cells were grown in SDCA medium to mid-log phase and collected. Cell lysates equivalent to 0.15 OD600 units prepared by the alkaline lysis method were subjected to immunoblot analysis with anti-GFP antibodies and anti-Ape1 antiserum. (D) Maturation of Ape1 was quantified based on the immunoblot images shown in (C). N.S. indicates not significant. **P < 0.01 (N = 3, two-tailed Student’s t test). atg15Δ (GYS1298) cells were used as the host strain.
Sorting of Atg15 via the AP-3 pathway maintains its hydrolytic activity
The C-terminal lipase domain is important for the function of Atg15 (Epple et al., 2001; Teter et al., 2001), but it is not known whether this domain is sufficient for Atg15 activity. To investigate this issue, we elected to alter the transport pathway of Atg15 by replacing its original N-terminal domain with the N-terminal domain of Pho8, which is transported to the vacuole via the Pho8 pathway (Cowles et al., 1997b). Because the 52 N-terminal residues of Pho8 serve as the sorting signal sequence of the Pho8 pathway (Klionsky and Emr, 1990; Cowles et al., 1997b), we fused GFP to the N-terminus of Pho8TMD (GFP-Pho8TMD). Furthermore, we fused the C-terminal region of Atg15 (amino acid residues 37–520, including the lipase domain) to generate GFP-Pho8TMD-Atg15C (Figure 6A).
FIGURE 6:
The C-terminal lipase domain is sufficient for enzymatic activity of Atg15. (A) Schematic diagram of the GFP-Pho8TMD constructs. (B) Wild-type and atg15Δ cells carrying pRS316 (EV), pRS316[GFP-Pho8TMD], and pRS316[GFP-Pho8TMD-Atg15C] plasmids were grown in SDCA medium to mid-log phase and collected. Cell lysates equivalent to 0.15 OD600 units prepared by the alkaline lysis method were subjected to immunoblot analysis with anti-GFP antibodies and anti-Ape1 antiserum. Wild-type (SEY6210) and atg15Δ (GYS1298) cells were used. (C) Maturation of Ape1 was quantified with immunoblot images shown in B. N.S. indicates not significant. **P < 0.01 (N = 3, two-tailed Student’s t test). (D) mRFP-prApe1 expressing wild-type and mRFP-prApe1 expressing atg15Δ cells carrying indicated plasmids were grown in SDCA medium and treated with rapamycin for 2 h before fluorescence microscopy. Percentages of cells possessing autophagic bodies labeled with mRFP-Ape1 are shown. ± represent standard deviations. N.S. indicates not significant. **P < 0.01 (N = 3, two-tailed Student’s t test). Wild-type (GYS638) and atg15Δ (GYS1300) cells were used. Scale bar represents 2 μm. (E) Activity of nonselective autophagy was quantified by the ALP assay. Cells carrying the indicated plasmids were grown in SDCA medium (+N), transferred to nitrogen starvation medium, and incubated for 4 h (–N). Wild-type (KVY55) and atg15Δ (GYS1349) cells were used as host strains. N.S. indicates not significant. **P < 0.01 (N = 3, two-tailed Student’s t test).
As expected, GFP-Pho8TMD-Atg15C expressed in atg15Δ cells was transported to the vacuolar membrane (Supplemental Figure S2A). Next, we investigated whether GFP-Pho8TMD-Atg15C bypassed the MVB pathway using fab1Δ, vps4Δ, vps27Δ, and vps28Δ, which were previously reported to be defective in the MVB pathway. In vps4Δ, vps27Δ, and vps28Δ cells, Atg15 accumulates to the class E compartment (Figure 2; Babst et al., 1997; Epple et al., 2001). We examined the localization of GFP-Pho8TMD-Atg15C in these cells. Fluorescence of GFP was detected normally at the vacuolar membrane, whereas the class E compartments were observed by FM 4-64 staining (Supplemental Figure S2A), suggesting that GFP-Pho8TMD-Atg15C bypasses the MVB pathway.
Transport of Pho8 requires the AP-3 adaptor complex, and in the absence of its subunits Apl5, Apl6, Apm3, and Aps3, Pho8 transport to the vacuole is delayed but still occurs (Cowles et al., 1997a). Moreover, the vacuolar localization of Pho8 is severely abrogated by simultaneous deletion of VPS4 in addition to the AP-3 subunits (Cowles et al., 1997a). Thus, we disrupted the VPS4 gene in AP-3 adaptor complex mutants to generate apl5Δvps4Δ, apl6Δvps4Δ, apm3Δvps4Δ, and aps3Δvps4Δ cells and then examined the localization of GFP-Pho8TMD-Atg15C. In these mutants, GFP-Pho8TMD-Atg15C was localized to fragmented vacuolar membranes (Supplemental Figure S2B). In homotypic vacuole fusion mutants, vesicles targeted to the vacuole do not fuse with the vacuole, but instead disperse throughout the cytoplasm (Conibear and Stevens, 1998). In ypt7Δ, vam6Δ, vam7Δ, and vps41Δ cells, GFP-Pho8TMD-Atg15C was detected as dots (Supplemental Figure S2C).
The C-terminal lipase domain is sufficient for enzymatic activity of Atg15
After GFP-Pho8TMD-Atg15C is transported to the vacuolar membrane, its Atg15C region should face the vacuolar lumen. Hence, we investigated whether GFP-Pho8TMD-Atg15C is functional. To this end, we first examined the activity of the Cvt pathway by monitoring maturation of Ape1. In atg15Δ cells expressing GFP-Pho8TMD, Ape1 transport was completely blocked, indicating that disintegration of Cvt bodies is blocked (Figure 6, B and C). On the other hand, atg15Δ cells expressing GFP-Pho8TMD-Atg15C exhibited normal Ape1 maturation (Figure 6, B and C). This result shows that GFP-Pho8TMD-Atg15C can function instead of Atg15 in the Cvt pathway.
When GFP-Pho8TMD-Atg15C was expressed in vps4Δatg15Δ cells, maturation of Ape1 was detected (Supplemental Figure S3A, lane 6 and S3B). This observation indicates that GFP-Pho8TMD-Atg15C can degrade Cvt bodies in cells defective in the MVB pathway. In apl5Δvps4Δatg15Δ cells, mApe1 was detected without expression of Atg15 (Supplemental Figure S3A, lane 7 and S3B). This suggests that, for unknown reasons, one or more proteases are accessible to prApe1 in these cells. We examined Ape1 transport in apl5Δvps4Δatg1Δ and apl5Δvps4Δatg2Δ cells but mApe1 was not detected in these cells (Supplemental Figure S3C). This result suggests that the completion of autophagosome formation is required for Ape1 maturation in apl5Δvps4Δ background cells. When GFP-Pho8TMD-Atg15C was expressed in apl5Δvps4Δatg15Δ cells, the ratio of mApe1 to total Ape1 increased (Supplemental Figure S3B), indicating that GFP-Pho8TMD-Atg15C is functional in these cells.
Next, we examined disintegration of autophagic bodies under a fluorescence microscope. Monomeric red fluorescent protein (mRFP) fused with the N-terminus of prApe1 (mRFP-prApe1) is selectively enclosed in autophagosomes and then transported to the vacuole (Suzuki et al., 2011). Therefore, this chimeric protein can be used as a marker for autophagic bodies. We assumed that mRFP-prApe1 would be detectable as dots inside the vacuole when autophagic bodies were intact, but diffused throughout the vacuolar lumen when autophagic bodies were disintegrated. As expected, mRFP-prApe1 was observed as dots inside the vacuole in atg15Δ cells expressing GFP-Pho8TMD (Figure 6D). By contrast, atg15Δ cells expressing GFP-Pho8TMD-Atg15C exhibited diffuse localization inside the vacuole (Figure 6D). It is worth mentioning that all these GFP-Pho8TMD proteins were correctly targeted to the vacuolar membrane (Figure 6D).
Subsequently, we estimated the activity of non-selective autophagy using the alkaline phosphatase assay. The activity of nonselective autophagy of GFP-Pho8TMD-Atg15C–expressing atg15Δ cells was comparable to that of wild-type cells (Figure 6E). By contrast, the activity of nonselective autophagy was significantly lower in apl5Δvps4Δatg15Δ cells expressing GFP-Pho8TMD-Atg15C than in atg15Δ cells expressing GFP-Pho8TMD-Atg15C (Supplemental Figure S3D).
These observations show that the C-terminal region of Atg15 transported to the vacuole via the Pho8 pathway is normally capable of disintegration of Cvt bodies/autophagic bodies. Moreover, it is unlikely that the transport of Atg15 via the MVB pathway is essential. Rather, the C-terminal region transported to the vacuole is critical for its activity. It is very plausible that the main role of the N-terminal Atg15TMD is to transport its C-terminal domain to the vacuole.
Residues S332, D387, and H435 are the putative catalytic triad
Previous studies have shown that S332 is essential for the activity of Atg15 (Epple et al., 2001; Teter et al., 2001). Moreover, Teter et al. suggest that several highly conserved residues constitute the putative catalytic triad: D387 or D421 and H435 (Supplemental Figure S4). We generated the D387A, D421A, and H435A mutants and examined their autophagic activities (Figure 7A). Degradation of Cvt bodies/autophagic bodies was normal in cells expressing D421A, but completely blocked in cells expressing D387A and H435A (Figure 7, B–E). From these results, we conclude that S332, D387, and H435 are plausible residues that constitute the catalytic triad of Atg15. Structural analysis will be necessary to prove whether these residues truly consist of the catalytic triad.
FIGURE 7:
Residues S332, D387, and H435 are the putative catalytic triad. (A) Schematic diagram of the catalytic mutants generated. (B) atg15Δ cells carrying pRS316[GFP-Pho8TMD-Atg15C], pRS316[GFP-Pho8TMD], or a series of mutant plasmids were grown in SDCA medium to midlog phase and collected. Cell lysates equivalent to 0.15 OD600 units, prepared by the alkaline lysis method, were subjected to immunoblot analysis with anti-GFP antibodies and anti-Ape1 antiserum. atg15Δ (GYS1298) cells were used. (C) Maturation of Ape1 was quantified from the immunoblot images shown in B. N.S. indicates not significant. **P < 0.01 (N = 3, two-tailed Student’s t test). (D) mRFP-prApe1 expressing atg15Δ cells carrying the indicated plasmids were grown in SDCA medium, treated with rapamycin for 2 h, and then subjected to fluorescence microscopy. Percentages of cells possessing autophagic bodies labeled with mRFP-Ape1 are shown. ± represent standard deviations. N.S. indicates not significant. **P < 0.01 (N = 3, two-tailed Student’s t test). atg15Δ (GYS1300) cells were used for this experiment. Scale bar represents 2 μm. (E) Activity of nonselective autophagy was quantified by the ALP assay. Cells carrying the indicated plasmids were grown in SDCA medium (+), transferred to nitrogen starvation medium, and incubated for 4 h (–). atg15Δ (GYS1349) cells were used. N.S. indicates not significant. **P < 0.01 (N = 3, two-tailed Student’s t test).
The C-terminal 467–520 residues are dispensable for Atg15 activity
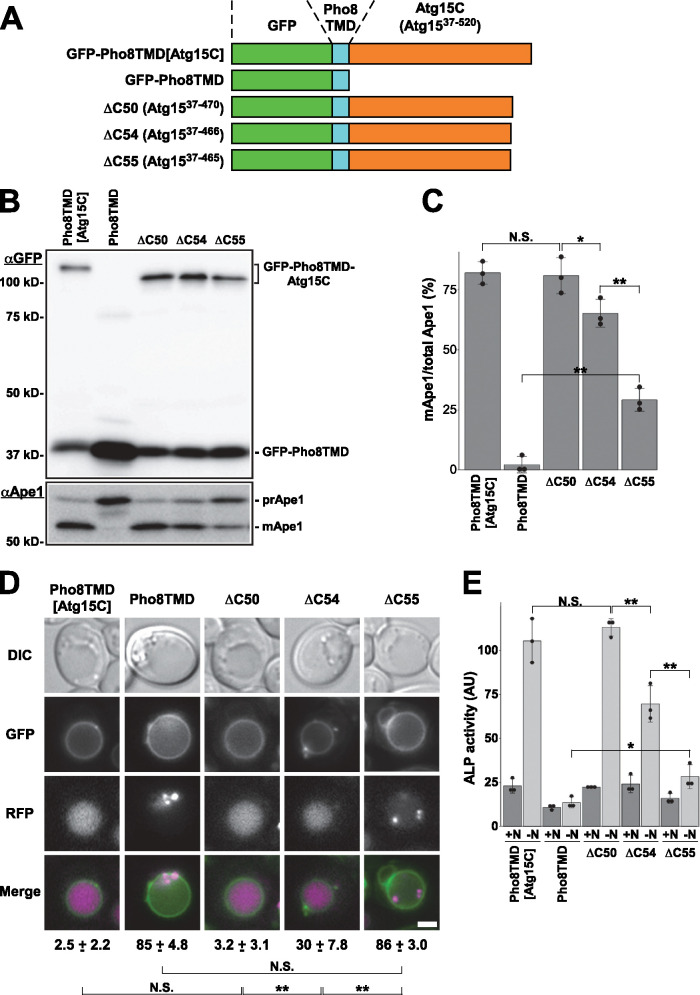
The results described above suggest that the C-terminal region of Atg15 (residues 37–520) is essential for its activity. As shown above, the GFP moiety is cleaved off from Atg15-GFP in a manner dependent on Pep4 (Figure 3A). Pep4 is a vacuolar endopeptidase that activates Prb1 and Pho8 by cleaving them (Ammerer et al., 1986). If Atg15 is activated by Pep4, a proper Atg15 truncation mutant may be active in pep4Δ cells. Alignment of Atg15 orthologues in various fungal species revealed that overall, the region between W164 and W466 is highly conserved (Supplemental Figure S4). To examine the necessity of the extreme C-terminus, we fused a series of truncated Atg15C mutants–Atg15ΔC50 (Atg1537–470), Atg15ΔC54 (Atg1537–466), and Atg15ΔC55 (Atg1537–465), which lack 50, 54, and 55 residues at the extreme C-terminus, respectively–with GFP-Pho8TMD, and then compared their activities (Figure 8A).
FIGURE 8:
The C-terminal 467–520 residues are dispensable for Atg15 activity. (A) Schematic diagram of the GFP-Pho8TMD-Atg15C constructs. (B) atg15Δ cells carrying pRS316[GFP-Pho8TMD-Atg15C], pRS316[GFP-Pho8TMD], and a series of mutant plasmids were grown in SDCA medium to midlog phase and collected. Cell lysates equivalent to 0.15 OD600 units, prepared by the alkaline lysis method, were subjected to immunoblot analysis with anti-GFP antibodies and anti-Ape1 antiserum. atg15Δ (GYS1298) cells were used. (C) Maturation of Ape1 is quantified with immunoblot images shown in B. N.S. indicates not significant. *P < 0.05, **P < 0.01 (N = 3, two-tailed Student’s t test). (D) mRFP-prApe1 expressing atg15Δ cells carrying indicated plasmids were grown in SDCA medium and treated with rapamycin for 2 h before fluorescence microscopy. Percentages of cells possessing autophagic bodies labeled with mRFP-Ape1 are shown. ± represent standard deviations. N.S. indicates not significant. **P < 0.01 (N = 3, two-tailed Student’s t test). atg15Δ (GYS1300) cells were used for this experiment. Scale bar represents 2 μm. (E) Activity of nonselective autophagy was quantified by the ALP assay. atg15Δ (GYS1349) cells carrying the indicated plasmids were grown in SDCA medium (+N), transferred to nitrogen starvation medium, and incubated for 4 h (–N). N.S. indicates not significant. *P < 0.05, **P < 0.01 (N = 3, two-tailed Student’s t test).
Immunoblot analysis revealed that these chimeric proteins were expressed efficiently (Figure 8B). We examined the degradation of Cvt bodies by maturation of Ape1. Ape1 maturation was normal in Atg15ΔC50 cells and slightly defective in Atg15ΔC54 cells, whereas maturation of Ape1 was severely affected in Atg15ΔC55 cells (Figure 8, B and C). Next, we treated the cells with rapamycin to induce autophagy and then monitored disintegration of autophagic bodies as reflected by diffusion of mRFP-prApe1 in the vacuole. All of the GFP-Pho8TMD fusion proteins were correctly targeted to the vacuolar membranes (Figure 8D). Atg15ΔC50-expressing atg15Δ cells possessed uniformly mRFP-labeled vacuoles, and a small population of Atg15ΔC54 expressing atg15Δ cells contained autophagic bodies labeled with mRFP-prApe1, whereas mRFP-prApe1 in most Atg15ΔC55 cells were visualized as dots inside the vacuole (Figure 8D). The alkaline phosphatase assay also revealed that degradation of autophagic bodies was slightly defective in Atg15ΔC54 cells, but severely defective in Atg15ΔC55 cells (Figure 8E). Based on these observations, we concluded that the C-terminal 467–520 residues of Atg15 were dispensable for disintegration of Cvt bodies/autophagic bodies. We examined whether these truncation mutants could bypass the activity of Pep4 with atg15Δpep4Δ cells expressing them by the Ape1 maturation assay, showing that no Ape1 maturation was detected in any truncation mutants (Supplemental Figure S5).
Residue W466 is important for degradation of autophagic bodies
The observations described above suggest that Atg15 lacking the 55th residue from the extreme C-terminus (W466) is severely defective in degradation of Cvt bodies/autophagic bodies (Figure 8). Moreover, W466 is a highly conserved residue at the extreme C-terminal region among Atg15 orthologs (Supplemental Figure S4). Hence, to examine the importance of the residue, we generated the W466A mutant and examined its autophagic activities. Mature Ape1 appeared in W466A cells under nutrient-rich conditions, but maturation was defective during autophagy (Figure 9, A and B). In addition, degradation of autophagic bodies and nonselective autophagy were severely affected in W466A cells (Figure 9, C and D). These results indicate that autophagic pathways were affected in W466A cells, especially under autophagy-inducing conditions.
FIGURE 9:
Autophagic pathways were affected in W466A cells, especially under autophagy-inducing conditions. (A) atg15Δ cells carrying pRS316[GFP-Pho8TMD-Atg15C], pRS316[GFP-Pho8TMD], and pRS316[GFP-Pho8TMD(W466A)] plasmids were grown to midlog phase in SDCA medium and treated with rapamycin for 2 h. atg15Δ (GYS1298) cell lysates equivalent to 0.15 OD600 units, prepared by the alkaline lysis method, were subjected to immunoblot analysis with anti-GFP antibodies and anti-Ape1 antiserum. (B) Maturation of Ape1 was quantified from the immunoblotting images shown in (A). **P < 0.01 (N = 3, two-tailed Student’s t test). (C) mRFP-prApe1–expressing atg15Δ (GYS1300) cells carrying the indicated plasmids were grown in SDCA medium, treated with rapamycin for 2 h, and then subjected to fluorescence microscopy. Percentages of cells possessing autophagic bodies labeled with mRFP-Ape1 are shown. ± represent standard deviations. **P < 0.01 (N = 3, two-tailed Student’s t test). Scale bar represents 2 μm. (D) Activity of nonselective autophagy was quantified by the ALP assay. atg15Δ (GYS1349) cells carrying the indicated plasmids were grown in SDCA medium (+N), transferred to nitrogen starvation medium, and incubated for 4 h (–N). **P < 0.01 (N = 4, two-tailed Student’s t test).
ATG11 and ATG17 are specifically required for the Cvt pathway and autophagy, respectively (Kamada et al., 2000; Kim et al., 2001). Without Atg17, the formation of autophagosomes is inhibited, but Ape1 is selectively delivered to the vacuole by Cvt vesicles during vegetative growth and by small autophagosomes during autophagy (Cheong et al., 2005; Kabeya et al., 2005). On the other hand, in the absence of Atg11, the formation of Cvt vesicles is inhibited, but Ape1 is nonselectively delivered to the vacuole by autophagosomes (Kim et al., 2001). In atg17Δ cells expressing wild-type Atg15C or W466A, mApe1 was detected during both vegetative growth and autophagy (Figure 10, A and B), suggesting that the Cvt bodies and small autophagic bodies are degraded in W466A mutant cells (Cheong et al., 2005; Kabeya et al., 2005). In atg11Δ cells expressing Atg15C, maturation of Ape1 was not detected under nutrient-rich or autophagy-inducing conditions (Figure 10, A and B). In addition, we quantified the activity of nonselective autophagy by the alkaline phosphatase (ALP) assay. Autophagic activity significantly decreased in W466A-expressing atg11Δ cells (Figure 10C). These results suggest that degradation of autophagic bodies is defective in W466A mutant cells. Taken together, these findings indicate that W466 is critical for degradation of autophagic bodies.
FIGURE 10:

Residue W466 is important for degradation of autophagic bodies. (A, B) Maturation of Ape1 was examined by immunoblot analysis and quantified. atg15Δ (GYS1298), atg11Δatg17Δatg15Δ (KYS103), atg11Δatg15Δ (KYS101), and atg17Δatg15Δ (KYS102) cells were used. N.S. indicates not significant. *P < 0.05, **P < 0.01 (N = 3, two-tailed Student’s t test). (C) Activity of nonselective autophagy was quantified by the ALP assay. atg15Δ (KYS200), atg2Δatg15Δ (KYS201), atg11Δatg15Δ (KYS202), and atg17Δatg15Δ (KYS203) cells carrying the indicated plasmids were grown in SDCA medium (+), transferred to nitrogen starvation medium, and incubated for 4 h (–). *P < 0.05, **P < 0.01 (N = 3, two-tailed Student’s t test).
Residues 50–466 are the essential region for Atg15 activity
Next, we constructed the N-terminal truncations Atg15ΔN42 (Atg1543–520), Atg15ΔN49 (Atg1550–520), Atg15ΔN63 (Atg1564–520), Atg15ΔN70 (Atg1571–520), and Atg15ΔN90 (Atg1591–520) fused with GFP-Pho8TMD. Fluorescence microscopy revealed that degradation of autophagic bodies was normal in Atg15ΔN42 and Atg15ΔN49 cells, but not in Atg15ΔN63, Atg15ΔN70, or Atg15ΔN90 cells (Supplemental Figure S6). Moreover, Atg15ΔN42 and Atg15ΔN49 were targeted normally to the vacuolar membrane, whereas Atg15ΔN63, Atg15ΔN70, and Atg15ΔN90 cells were defective in vacuolar membrane targeting (Supplemental Figure S6). These results show that the N-terminal residues 37–49 are dispensable for the activity of Atg15.
Above, we showed that Atg15ΔC54 and Atg15ΔN49 exhibited normal localization and autophagic activities, suggesting that residues 37–49 and 467–520 are dispensable for the activity of Atg15. To explore this further, we generated the Atg15ΔN49ΔC54 mutant (Figure 11A). GFP-Pho8TMD-Atg15ΔN49ΔC54 could be detected by immunoblot analysis (Figure 11B). In Atg15ΔC54 cells, mApe1 was detected, but was slightly defective in comparison with wild-type cells (Figure 11, B and C). Ape1 maturation was comparable between Atg15ΔC54 and Atg15ΔN49ΔC54 cells. This result suggests that residues 37–49 and 467–520 are not essential for the activity of Atg15. Moreover, disintegration of autophagic bodies appeared normal in Atg15ΔN49ΔC54 cells (Figure 11D). Fluorescence microscopy also showed that GFP-Pho8TMD-Atg15ΔN49ΔC54 was correctly targeted to the vacuolar membrane (Figure 11D). In addition, the alkaline phosphatase assay showed that nonselective autophagy was active in Atg15ΔN49ΔC54 cells, albeit to a lesser extent than in wild-type cells (Figure 11E). These results show that Atg15ΔN49ΔC54 can degrade autophagic bodies. Taken together, these findings indicate that residues 50–466 in Atg15 are essential for disintegration of Cvt bodies/autophagic bodies inside the vacuole. Finally, we examined whether Atg15ΔN49ΔC54 requires activation by Pep4. mApe1 did not appear in atg15Δpep4Δ cells expressing Atg15ΔN49ΔC54 (Supplemental Figure S7), showing that this truncation mutant cannot bypass activation by Pep4.
FIGURE 11:
Residues 50–466 are the essential region for Atg15 activity. (A) Schematic diagram of the GFP-Pho8TMD-Atg15C constructs. (B) atg15Δ cells carrying pRS316[GFP-Pho8TMD-Atg15C], pRS316[GFP-Pho8TMD], and the indicated mutant plasmids were grown in SDCA medium to mid-log phase and collected. Cell lysates equivalent to 0.15 OD600 units prepared by the alkaline lysis method were subjected to immunoblot analysis with anti-GFP antibodies and anti-Ape1 antiserum. atg15Δ (GYS1298) cells were used for this analysis. (C) Maturation of Ape1 was quantified from immunoblot images shown in B. N.S. indicates not significant. *P < 0.05, **P < 0.01 (N = 3, two-tailed Student’s t test). (D) mRFP-prApe1 expressing atg15Δ cells carrying indicated plasmids were grown in SDCA medium and treated with rapamycin for 2 h before fluorescence microscopy. Percentages of cells possessing autophagic bodies labeled with mRFP-Ape1 are shown. atg15Δ (GYS1300) cells were used. ± represent standard deviations. N.S. indicates not significant. **P < 0.01 (N = 3, two-tailed Student’s t test). Scale bar represents 2 μm. (E) Activity of nonselective autophagy was quantified by the ALP assay. atg15Δ (GYS1349) cells carrying the indicated plasmids were grown in SDCA medium (+), transferred to nitrogen starvation medium, and incubated for 4 h (–). **P < 0.01 (N = 3, two-tailed Student’s t test).
DISCUSSION
Disintegration of Cvt bodies/autophagic bodies before degradation of their enclosed cargo in the vacuole is important. However, little is known about the mechanism. Our findings in this study demonstrate that the N-terminal and C-terminal regions of Atg15 have distinct functions for the activity of Atg15; the N-terminal region acts as a targeting signal for the MVB pathway, and the C-terminal region is responsible for its enzymatic activity. Furthermore, we showed that residues 50–466, including the putative catalytic triad, are essential for Atg15 activity.
The N-terminal region of Atg15 acts as a signal sequence for the multivascular body pathway
A previous study using fixed cells showed that Atg15 is transported to the vacuole via the MVB pathway (Epple et al., 2003). We examined Atg15 transport in living cells by tagging Atg15 with GFP. Our observation also supports the idea that Atg15 is delivered to the vacuole via the MVB pathway (Figure 2). Moreover, this targeting largely relies on its N-terminal domain, including the TMD (residues 2–36), which is necessary and sufficient for transport of Atg15 to the vacuole (Figures 4 and 5). Even in the absence of the N-terminal domain, disintegration of Cvt bodies/autophagic bodies could be accomplished by replacing this N-terminal region with GFP-Pho8TMD (Figure 6), indicating that the Atg15 N-terminal domain largely acts as a signal sequence for the MVB pathway. Moreover, it is unlikely that Atg15 plays a role during transit via the MVB pathway.
Activity of Atg15 in mutants of the multivascular body pathway
Epple et al. (2003) showed that maturation of Ape1 and degradation of autophagic bodies is normal in class E mutants such as vps23Δ and vps28Δ, although the transport of Atg15 to the vacuolar lumen is inhibited in these cells. To explain this phenotype, they mentioned two possibilities: first, that Atg15 is mislocalized to the vacuolar membranes in these mutants, and second, that the small amount of Atg15 that reaches the vacuolar lumen is sufficient for degradation of autophagic bodies. To determine which possibility is more plausible, they attempted to decrease the amount of Atg15 that exited from the ER by fusing Atg15 to an HDEL sequence, which acts as an ER retention signal in yeast. The majority of Atg15-HDEL is found in the ER, and the only a small amount of Atg15 is detected in the vacuolar lumen (Epple et al., 2003). In Atg15-HDEL–expressing cells, the activity of the Cvt pathway decreased. Moreover, the maturation of Ape1 was almost completely abrogated in cells lacking Vps28. This inhibition was partially recovered by overexpression of Atg15-HDEL. These observations support their idea that the small amount of Atg15-HDEL that leaves the ER and reaches the vacuole is sufficient for degradation of Cvt bodies / autophagic bodies. However, there was a possibility that Atg15 functions at the MVB.
We examined the localization of Atg15-GFP in living vps4Δ cells and found that Atg15 was not localized to vacuolar membranes. Instead, a small amount of Atg15-GFP was delivered to the vacuolar lumen (Figure 2). Moreover, immunoblot analysis with anti-GFP antibodies showed that Atg15-GFP was degraded inside the vacuole in vps4Δ cells (Figure 3A). These observations support the possibility that the small amount of Atg15 delivered to the vacuolar lumen is sufficient for degradation of autophagic bodies. It is unlikely that Atg15 functions at the MVB, because autophagic activities were normal in atg15Δ cells expressing GFP-Pho8TMD-Atg15C, which bypasses the MVB pathway (Figure 6). Therefore, we conclude that Atg15 that reaches the vacuolar lumen participates in degradation of Cvt bodies / autophagic bodies in the vacuolar lumen.
The C-terminal region of Atg15 is responsible for its enzymatic activity
Atg15 is a short-lived protein, with a half-life of 45 to 70 min in wild-type cells (Epple et al., 2001; Teter et al., 2001). Because Atg15 is transported to the vacuole via the MVB pathway, it is reasonable that free GFP is generated dependent on Pep4, a key proteinase in the vacuole (Figure 3A). This observation also indicates that the C-terminal region of Atg15 is exposed to the vacuolar lumen. Thus, the GFP signals localized to the vacuolar lumen probably reflect the amounts of free GFP in wild-type and vps4Δ cells and the amount of Atg15-GFP in pep4Δ cells. Free GFP increased upon addition of rapamycin in wild-type and vps4Δ cells (Figure 3A). Consistent with this, the GFP signals in the vacuolar lumen were markedly elevated in these cells (Figure 2). In pep4Δ cells, free GFP does not emerge (Figure 3A), and GFP signals in the vacuolar lumen were unchanged by rapamycin treatment (Figure 2). In both cases, the amount of Atg15-GFP was maintained at constant levels in all cells examined (Figure 3A). Based on these results, we assume that there is a system that maintains the level of intact Atg15 by compensating for Atg15 degraded in the vacuole.
Previous studies showed that full-length Atg15 has phospholipase or triacylglycerol lipase activity (van Zutphen et al., 2014; Ramya and Rajasekharan, 2016). However, these studies did not take the intracellular localization of Atg15 into account. We estimated the enzymatic activity of Atg15 by monitoring conventional Ape1 maturation and newly developed mRFP-prApe1 diffusion in the vacuolar lumen (Figures 6 – 9 and 11). We replaced the N-terminal domain of Atg15 with Pho8TMD to generate GFP-Pho8TMD-Atg15C; this chimeric protein can bypass the MVB pathway and is transported to the vacuole via the Pho8 pathway (Supplemental Figure S2A). This protein is targeted to the vacuolar membrane by the Pho8TMD region, and its C-terminal Atg15C domain must be exposed to the vacuolar lumen. As a result, this chimeric protein can complement the defect in atg15Δ cells (Figure 6). These results show that the C-terminal region of Atg15 delivered into the vacuolar lumen is important for the activity of Atg15.
The mechanisms of Atg15 activation
In this study, we showed that transport of Atg15 through the MVB pathway is not important for disintegration of autophagic bodies, whereas the C-terminal domain of Atg15 inside the vacuole is essential. This fact prompted us to hypothesize that Atg15 is inactive before it reaches the vacuole and is activated once inside. Pep4 is an aspartyl endopeptidase that cleaves Prb1 and Pho8 to activate them (Ammerer et al., 1986). Similarly, Atg15 might be activated by the action of Pep4. If this assumption is true, a proper Atg15 truncation mutant should be active even in the absence of Pep4. We investigated whether Atg15 truncation could bypass the activity of Pep4 by expressing truncation mutants in atg15Δpep4Δ cells. However, even the minimal essential region of Atg15 (residues 50–466), which we determined in this study, could not disintegrate autophagic bodies in the absence of Pep4 (Supplemental Figure S7).
This result suggested two potential explanations for the cause of this defect. The first possibility is that the GFP-Pho8TMD moiety and the Atg15ΔN49ΔC54 moiety must be cleaved by Pep4. The GFP-Pho8TMD moiety derived from GFP-Pho8TMD-Atg15ΔN49ΔC54 could be detected by immunoblot analysis (Figure 11B), suggesting that cleavage of the GFP-Pho8TMD-Atg15ΔN49ΔC54 occurred inside the vacuole. By contrast, we have never detected the Atg15ΔN49ΔC54 moiety in atg15Δ cells by immunoblot analysis (Unpublished observations). A previous study showed that full-length Atg15 is a short-lived protein with a half-life less than 1 h (Epple et al., 2001; Teter et al., 2001). This might be one of the reasons that processed Atg15 could not be detected. The second possibility is that the role of Pep4 is not restricted to the processing of Atg15, and that Pep4 cooperates with Atg15 in the disintegration of autophagic bodies. Currently, we cannot exclude this possibility.
The substrate specificity of Atg15
Atg15 is responsible for disintegration of intravacuolar vesicles derived from the MVB pathway as well as those from the autophagic pathway (Epple et al., 2003). One fundamental question remains: Given that Atg15 must be active inside the vacuole, if it is capable of nonspecifically degrading biological membranes, why does it not disintegrate the vacuolar membrane? We can think of two possible answers to this conundrum. One is that the enzymatic activity of Atg15 is specific for intravacuolar vesicles rather than the vacuolar membrane. The substrate of Atg15 might be a specific lipid residing on the membrane of intravacuolar vesicles, or alternatively, Atg15 might recognize the positive curvature of intravacuolar vesicles. A second possibility is that the vacuolar membrane is actually degraded by Atg15 but is rapidly repaired by unknown mechanisms.
To answer this question, we need to establish an in vitro system to evaluate the activity of Atg15. Currently, we know that active Atg15 is present at least inside the vacuole. Accordingly, it should be straightforward to purify Atg15 from the vacuole. As a substrate, autophagic bodies are preferable. Isolation of subvacuolar vesicles, which contain prApe1, has already been achieved (Scott et al., 1997), and this method should be applicable to isolation of Cvt bodies/autophagic bodies. In this study, using N-terminally mRFP-fused prApe1, we monitored degradation of autophagic bodies in vivo (Figures 6 – 9 and 11). For the in vitro assay, mRFP-prApe1 could be used as a sensitive marker to monitor degradation of Cvt bodies / autophagic bodies.
In summary, we have shown that Atg15 has two distinct functional domains. The N-terminal transmembrane domain is a targeting sequence to the vacuole via the MVB pathway, whereas C-terminal lipase domain is responsible for its enzymatic activity. Establishment of in vitro monitoring system for the activity of Atg15 should help elucidate the mechanisms of disintegration of Cvt bodies/autophagic bodies.
MATERIALS AND METHODS
Strains, media, and growth conditions
The yeast strains used in this study are listed in Table 1. Cells were cultured in SDCA (0.17% Difco yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 0.5% Bacto casamino acids, and 2% glucose) or SD (0.17% Difco yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, and 2% glucose) medium supplemented with appropriate nutrients for plasmid selection. Autophagy was induced by addition of 200–400 ng/ml rapamycin or shifting cells to SD(-N) medium (0.17% Difco yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose). Amplification of plasmids was carried out using Escherichia coli cells grown in LB medium (1% Bacto tryptone, 0.5% Bacto yeast extract, 1% NaCl). When relevant, ampicillin was added to LB medium at a concentration of 60 µg/ml. To drive the Cu2+-inducible CUP1 promoter, cells were cultured for 1 day in medium containing 250 μM CuSO4 prior to experiments.
TABLE 1:
Strains used in this study.
| Name | Genotype | Source | Figures |
|---|---|---|---|
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 lys2-801 | ( Robinson et al., 1988) | 1, 4B–C, 6B–C, S1 |
| STY757 | SEY6210; atg2Δ::LEU2 | ( Obara et al., 2008) | 1, S1 |
| GYS1298 | SEY6210; atg15Δ::hphNT1 | This study | 1, 4B–C, 5B–D, 6B–C, 7B–C, 8B–C, 9A–B, 10A–B, 11B–C, S1, S5, S7 |
| GYS1162 | SEY6210; atg15Δ::ATG15-2xGFP:kanMX6 | This study | 2, 3, SVideo1A |
| GYS1166 | GYS1162; vps4Δ::hphNT1 | This study | 2, 3 |
| GYS1168 | GYS1162; pep4Δ::hphNT1 | This study | 2, 3, SVideo1B |
| GYS1164 | GYS1162; ypt7Δ::LEU2 | This study | 3 |
| MBY3 | SEY6210; vps4Δ::TRP1 | ( Babst et al., 1997) | 4B–C |
| GYS1336 | SEY6210; vps4Δ::TRP1 atg15Δ::natNT2 | This study | 4B–C |
| BY4741 | MATahis3Δ1 met15Δ0 ura3Δ0 leu2Δ0 | ( Brachmann et al., 1998) | |
| Y5789 | BY4741; atg15Δ::kanMX4 | EUROSCARF | S2A, S3 |
| Y7080 | BY4741; fab1Δ::kanMX4 | EUROSCARF | S2A |
| Y5588 | BY4741; vps4Δ::kanMX4 | EUROSCARF | S2A |
| Y5381 | BY4741; vps27Δ::kanMX4 | EUROSCARF | S2A |
| Y2763 | BY4741; vps28Δ::kanMX4 | EUROSCARF | S2A |
| YOC5156 | BY4741; apl5Δ::kanMX4 vps4Δ::hphNT1 | This study | S2B |
| YOC5157 | BY4741; apl6Δ::kanMX4 vps4Δ::hphNT1 | This study | S2B |
| YOC5158 | BY4741; apm3Δ::kanMX4 vps4Δ::hphNT1 | This study | S2B |
| YOC5159 | BY4741; aps3Δ::kanMX4 vps4Δ::hphNT1 | This study | S2B |
| Y575 | BY4741; ypt7Δ::kanMX4 | EUROSCARF | S2C |
| Y3774 | BY4741; vam6Δ::kanMX4 | EUROSCARF | S2C |
| Y4578 | BY4741; vam7Δ::kanMX4 | EUROSCARF | S2C |
| Y4015 | BY4741; vps41Δ::kanMX4 | EUROSCARF | S2C |
| GYS638 | SEY6210; leu2Δ::mRFP-prAPE1:LEU2 | ( Suzuki et al., 2011) | 6D |
| GYS1300 | GYS638; atg15Δ::hphNT1 | This study | 6D, 7D, 8D, 9C, 11D |
| KVY55 | SEY6210; pho8Δ::pho8Δ60 | ( Ishihara et al., 2001) | 6E |
| GYS1349 | KVY55; atg15Δ::natNT2 | This study | 6E, 7E, 8E, 9D, 11E |
| KYS103 | SEY6210; atg11Δ::LEU2 atg17Δ::spHIS5 atg15Δ::natNT2 | This study | 10A–B |
| KYS101 | SEY6210; atg11Δ::LEU2 atg15Δ::natNT2 | This study | 10A–B |
| KYS102 | SEY6210; atg17Δ::spHIS5 atg15Δ::natNT2 | This study | 10A–B |
| KYS200 | SEY6210; pho8Δ::kanMX4-GPDp-pho8Δ60 atg15Δ::hphNT1 | This study | 10C |
| KYS201 | KYS200; atg2Δ::natNT2 | This study | 10C |
| KYS202 | KYS200; atg11Δ::natNT2 | This study | 10C |
| KYS203 | KYS200; atg17Δ::natNT2 | This study | 10C |
| GYS1348 | BY4741; vps4Δ::kanMX4 atg15Δ::natNT2 | This study | S3 |
| GYS1338 | BY4741; apl5Δ::kanMX4 vps4Δ::hphNT1 atg15Δ::natNT2 | This study | S3 |
| GYS1415 | BY4741; apl5Δ::kanMX4 vps4Δ::hphNT1 atg1Δ::LEU2 | This study | S3 |
| GYS1416 | BY4741; apl5Δ::kanMX4 vps4Δ::hphNT1 atg2Δ::LEU2 | This study | S3 |
| GYS1332 | BY4741; apl5Δ::kanMX4 vps4Δ::hphNT1 | This study | S3 |
| GYS1299 | SEY6210; atg15Δ::hphNT1 pep4Δ::LEU2 | This study | S5, S7 |
| YOC5121 | Y5789; leu2Δ::mRFP-prAPE1:LEU2 | This study | S6 |
Cells expressing C-terminally 2 × GFP-fused Atg15 were generated using a PCR-based gene-modification method using pFA6a-2 × YEGFP-kanMX as a template. The ATG15, PEP4, and VPS4 genes were disrupted by DNA fragments amplified by polymerase chain reaction (PCR) from plasmid pFA6a-hphNT1 or pFA6a-natNT2 (Janke et al., 2004). For YPT7 disruption, genomic DNA obtained from KVY4 was used as a template for amplification of the DNA fragment (Kihara et al., 2001).
Plasmids
The plasmids used in this study are listed in Table 2. For construction of pRS316[GFP-Pho8TMD], a PCR overlap-extension strategy was used to generate the GFP-Pho8TMD cassette. ADH1 promoter–GFP with a KpnI site at the 5′ end of the ADH1 promoter and a homologous region of the Pho8TMD at the 3′ end of YEGFP (Yeast Enhanced GFP) was generated by PCR using pYM-N9 as a template (Janke et al., 2004). The Pho8TMD fragment with a homologous region of GFP at the 5′ end and the ADH1 terminator at the 3′ end was generated by PCR using YGPM21o18, which contains the PHO8 gene as a template (Yeast Genomic Tiling Collection, Dharmacon). The ADH1 fragment with a homologous region of the Pho8TMD at the 5′ end and a HindIII site at the 3′ end was generated by PCR using pYM-N9 as a template. These fragments were fused with overlap extension PCR. The GFP-Pho8TMD cassette was cloned into the pRS316 plasmid after digestion with KpnI and HindIII to generate pRS316[GFP-Pho8TMD].
TABLE 2:
Plasmids used in this study.
| Name | Properties | Source | Figures |
|---|---|---|---|
| pFA6a-2 × YEGFP-kanMX | PCR template for generation of C-terminally 2 × GFP-fused Atg15 | Laboratory stock | |
| pYM-N9 | PCR template for amplification of the ADH1 promoter-YEGFP fragment | ( Janke et al., 2004) | |
| pFA6a-hphNT1 | PCR template for gene deletion | ( Janke et al., 2004) | |
| pFA6a-natNT2 | PCR template for gene deletion | ( Janke et al., 2004) | |
| YGPM21o18 | PCR template for amplification of the PHO8 fragment | Dharmacon | |
| YPGM23k01 | PCR template for amplification of the ATG15 fragment | Dharmacon | |
| pYEX-BX | 2μ plasmid for expression of a protein from the CUP1 promoter | Clontech | |
| pYEX-BX[prApe1] | 2μ plasmid for expression of prApe1 from the CUP1 promoter | ( Suzuki et al., 2013) | 1, S1 |
| pRS314 | Centromeric plasmid (TRP1 marker) | ( Sikorski and Hieter, 1989) | |
| pRS314[mNeonGreen-Atg8] | Centromeric plasmid for expression of mNeonGreen-Atg8 from the ATG8 promoter | ( Hirata et al., 2017) | 1, S1 |
| pRS316 | Centromeric plasmid (URA3 marker) | ( Sikorski and Hieter, 1989) | 5C–D, 6B–E, S3 |
| pRS316[GFP-Atg15TMD] | Centromeric plasmid for expression of YEGFP-Atg15(2–36) from the ADH1 promoter | This study | 4B–C, 5B–D |
| pRS316[GFP-Atg15FL] | Centromeric plasmid for expression of YEGFP-Atg15(2–520) from the ADH1 promoter | This study | 5B–D, S3 |
| pRS316[GFP-Atg15ΔTMD] | Centromeric plasmid for expression of YEGFP-Atg15(37–520) from the ADH1 promoter | This study | 5B–D |
| pRS316[GFP-Pho8TMD-Atg15C] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(37–520) from the ADH1 promoter | This study | S2, 6B–E, 7B–E, 8B–E, 9, 10, 11B–E, S2, S3, S5, S6, S7 |
| pRS316[GFP-Pho8TMD] | Centromeric plasmid for expression of YEGFP-Pho8TMD from the ADH1 promoter | This study | 6B–E, 7B–E, 8B–E, 9, 10, 11B–E, S5, S6, S7 |
| pRS316[GFP-Pho8TMD-Atg15C(S332A)] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(S332A) from the ADH1 promoter | This study | 7B–E |
| pRS316[GFP-Pho8TMD-Atg15C(D387A)] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(D387A) from the ADH1 promoter | This study | 7B–E |
| pRS316[GFP-Pho8TMD-Atg15C(D421A)] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(D421A) from the ADH1 promoter | This study | 7B–E |
| pRS316[GFP-Pho8TMD-Atg15C(H435A)] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(H435A) from the ADH1 promoter | This study | 7B–E |
| pRS316[GFP-Pho8TMD-Atg15ΔC50] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(37–470) from the ADH1 promoter | This study | 8B–E, S5 |
| pRS316[GFP-Pho8TMD-Atg15ΔC54] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(37–466) from the ADH1 promoter | This study | 8B–E, 11B–E, S5, S7 |
| pRS316[GFP-Pho8TMD-Atg15ΔC55] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(37–465) from the ADH1 promoter | This study | 8B–E, S5 |
| pRS316[GFP-Pho8TMD-Atg15C(W466A)] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(W466A) from the ADH1 promoter | This study | 9, 10 |
| pRS316[GFP-Pho8TMD-Atg15ΔN49] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(50–520) from the ADH1 promoter | This study | 11B–E, S6, S7 |
| pRS316[GFP-Pho8TMD-Atg15ΔN49ΔC54] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(50–466) from the ADH1 promoter | This study | 11B–E, S7 |
| pRS423 | 2μ plasmid (HIS3 marker) | ( Sikorski and Hieter, 1989) | |
| pCK3 | pRS423 background plasmid for overexpression of pho8Δ60 | This study | S3 |
| pRS316[GFP-Pho8TMD-Atg15ΔN42] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(43–520) from the ADH1 promoter | This study | S6 |
| pRS316[GFP-Pho8TMD-Atg15ΔN63] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(64–520) from the ADH1 promoter | This study | S6 |
| pRS316[GFP-Pho8TMD-Atg15ΔN70] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(71–520) from the ADH1 promoter | This study | S6 |
| pRS316[GFP-Pho8TMD-Atg15ΔN90] | Centromeric plasmid for expression of YEGFP-Pho8TMD-Atg15(91–520) from the ADH1 promoter | This study | S6 |
For construction of truncation mutants of Atg15C, each Atg15 fragment amplified by PCR with BamHI sites on both ends was cloned into BglII-digested pRS316[GFP-Pho8TMD]. Point mutants of Atg15C were generated by site-directed mutagenesis.
pRS316[GFP-Atg15TMD], pRS316[GFP-Atg15FL], and pRS316[GFP-Atg15ΔTMD] were generated by assembling pRS316[GFP] fragment amplified from pRS316[GFP-Pho8TMD] and Atg15 fragments amplified from YPGM23k01, which contains the ATG15 gene, using the NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs).
Fluorescence microscopy
Cells were cultured in SDCA medium to midlog phase, and autophagy was induced when necessary. Cells were harvested and subjected to fluorescence microscopy on an IX83 inverted system microscope (Olympus) equipped with a UPlanSApo100×/1.40 Oil (Olympus) and a CoolSNAP HQ CCD camera (Nippon Roper). A U-FGFP and U-FRFP filter sets (Olympus) were used for visualization of GFP/mNeonGreen and mRFP/FM 4-64, respectively. Images were acquired using the MetaVue imaging software (Molecular Devices). For determination of AM length, the fluorescence images of mNeonGreen-Atg8 were analyzed using Qautas (Kawaoka et al., 2017). FM 4-64 staining was performed as previously described (Suzuki et al., 2002). Data analysis was performed using R (https://www.r-project.org). Boxplots were drawn using the default settings of R.
Immunoblotting analysis
Cell lysates were prepared by the alkaline lysis method (Horvath and Riezman, 1994) and subjected to SDS–PAGE. Proteins were transferred to polyvinylidene fluoride membranes (Immobilon-P, Millipore) utilizing a Trans-Blot Turbo Transfer System (Bio-Rad). Following transfer, membranes were blocked with 2% skim milk in Tris-buffered saline containing 0.05% Tween 20 (TBST) for 30 min at room temperature. Membranes were then incubated for 60 min at room temperature with anti-GFP antibodies (1:10000; JL-8, Clontech) or an anti-Ape1 antiserum (1:10000) (Suzuki et al., 2002). Subsequently, membranes were washed once with TBST and treated with horseradish peroxidase–labeled anti-rabbit or -mouse secondary antibodies (Promega) at a dilution of 1:5000 for 30 min, followed by three washes in TBST. Chemiluminescent signals generated using ECL Prime (GE Healthcare) or ImmunoStar LD (Wako) were detected on an IR-LAS 1000 imaging system (FUJIFILM).
Alkaline phosphatase assay
Nonselective autophagic activity was estimated using the ALP assay (Noda et al., 1995). Briefly, the proform of the phosphatase (Pho8Δ60), expressed from the genome or a plasmid (pCK3), is transported to the vacuole via bulk autophagy and processed to its active form. After cell lysates were prepared by glass bead disruption, ALP activity was measured using α-naphtyl phosphate (Sigma) as a substrate.
Supplementary Material
Acknowledgments
We thank Emi Kato and Minako Miki for technical assistance. This work was supported by a grant from the Naito Foundation (to KS) and by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (18J13429 to EH, 19J11061 to TK, and 16H06280, 18H04853, and 20H05313 to Kuninori S).
Abbreviations used:
- ALP
alkaline phosphatase
- AM
autophagic membrane
- Atg proteins
autophagy-related proteins
- Cvt
cytoplasm-to-vacuole targeting
- DIC
differential interference contrast
- ER
endoplasmic reticulum
- EV
empty vector
- GFP
green fluorescent protein
- mApe1
mature aminopeptidase I
- mNG
mNeonGreen
- mRFP
monomeric red fluorescent protein
- MVB pathway
multivesicular body pathway
- PAS
preautophagosomal structure
- prApe1
precursor aminopeptidase I
- Qautas
quantitative autophagy-related structure analysis system
- SDCA medium
synthetic dextrose medium containing casamino acids
- TBST
Tris-buffered saline containing Tween 20
- TMD
transmembrane domain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-07-0500) on February 24, 2021.
REFERENCES
- Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH (1986). PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol 6, 2490–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y (1997). Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol 139, 1687–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD (1997). Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J 16, 1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang CW, Klionsky DJ (2005). Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell 16, 3438–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH (1998). Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim Biophys Acta 1404, 211–230. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD (1997a). The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell 91, 109–118. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Snyder WB, Burd CG, Emr SD (1997b). Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J 16, 2769–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple UD, Eskelinen EL, Thumm M (2003). Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J Biol Chem 278, 7810–7821. [DOI] [PubMed] [Google Scholar]
- Epple UD, Suriapranata I, Eskelinen EL, Thumm M (2001). Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J Bacteriol 183, 5942–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Alam JM, Noshiro D, Mouri K, Ando T, Okada Y, May AI, Knorr RL, Suzuki K, Ohsumi Y, Noda NN (2020). Phase separation organizes the site of autophagosome formation. Nature 578, 301–305. [DOI] [PubMed] [Google Scholar]
- Hirata E, Ohya Y, Suzuki K (2017). Atg4 plays an important role in efficient expansion of autophagic isolation membranes by cleaving lipidated Atg8 in Saccharomyces cerevisiae. PLoS One 12, e0181047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A, Riezman H (1994). Rapid protein extraction from Saccharomyces cerevisiae. Yeast 10, 1305–1310. [DOI] [PubMed] [Google Scholar]
- Huang H, Kawamata T, Horie T, Tsugawa H, Nakayama Y, Ohsumi Y, Fukusaki E (2015). Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J 34, 154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y (2001). Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell 12, 3690–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M (2004). A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y (2005). Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell 16, 2544–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y (2000). Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150, 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y (2008). Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell 19, 2039–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka T, Ohnuki S, Ohya Y, Suzuki K (2017). Morphometric analysis of autophagy-related structures in Saccharomyces cerevisiae. Autophagy 13, 2104–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 152, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Klionsky DJ (2001). Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol 153, 381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 147, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS (1992). Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol 119, 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD (1990). A new class of lysosomal/vacuolar protein sorting signals. J Biol Chem 265, 5349–5352. [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N (2004). The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y (2011). The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27, 107–132. [DOI] [PubMed] [Google Scholar]
- Moehle CM, Tizard R, Lemmon SK, Smart J, Jones EW (1987). Protease B of the lysosomelike vacuole of the yeast Saccharomyces cerevisiae is homologous to the subtilisin family of serine proteases. Mol Cell Biol 7, 4390–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Matsuura A, Wada Y, Ohsumi Y (1995). Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun 210, 126–132. [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y (2008). The Atg18–Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem 283, 23972–23980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y (2005). Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem 280, 31582–31586. [DOI] [PubMed] [Google Scholar]
- Ramya V, Rajasekharan R (2016). ATG15 encodes a phospholipase and is transcriptionally regulated by YAP1 in Saccharomyces cerevisiae. FEBS Lett 590, 3155–3167. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD (1988). Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol 8, 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ (1997). Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol 138, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang WP, Stromhaug PE, Klionsky DJ (2002). Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell 3, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ (2004). Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem 279, 29889–29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Suzuki K, Kamada Y, Noda T, Ohsumi Y (2001). Apg2p functions in autophagosome formation on the perivacuolar structure. J Biol Chem 276, 30452–30460. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y (2013). Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci 126, 2534–2544. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kamada Y, Ohsumi Y (2002). Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev Cell 3, 815–824. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Morimoto M, Kondo C, Ohsumi Y (2011). Selective autophagy regulates insertional mutagenesis by the Ty1 retrotransposon in Saccharomyces cerevisiae. Dev Cell 21, 358–365. [DOI] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y (1992). Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 119, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter SA, Eggerton KP, Scott SV, Kim J, Fischer AM, Klionsky DJ (2001). Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J Biol Chem 276, 2083–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer HF, Wolinski H, Veenhuis M, van der Klei IJ, Kohlwein SD (2014). Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell 25, 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Ohsumi Y, Anraku Y (1992). Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J Biol Chem 267, 18665–18670. [PubMed] [Google Scholar]
- Wang CW, Kim J, Huang WP, Abeliovich H, Stromhaug PE, Dunn WA, Klionsky DJ (2001). Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J Biol Chem 276, 30442–30451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann H, Hengst L, Gallwitz D (1992). Endocytosis in yeast: evidence for the involvement of a small GTP-binding protein (Ypt7p). Cell 71, 1131–1142. [DOI] [PubMed] [Google Scholar]
- Woolford CA, Daniels LB, Park FJ, Jones EW, Van Arsdell JN, Innis MA (1986). The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacuolar hydrolases. Mol Cell Biol 6, 2500–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki A, Alam JM, Noshiro D, Hirata E, Fujioka Y, Suzuki K, Ohsumi Y, Noda NN (2020). Liquidity is a critical determinant for selective autophagy of protein condensates. Mol Cell 77, 1–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.