Abstract
During Caenorhabditis elegans larval development, an inductive signal mediated by the LET-23 EGFR (epidermal growth factor receptor), specifies three of six vulva precursor cells (VPCs) to adopt vulval cell fates. An evolutionarily conserved complex consisting of PDZ domain-containing scaffold proteins LIN-2 (CASK), LIN-7 (Lin7 or Veli), and LIN-10 (APBA1 or Mint1) (LIN-2/7/10) mediates basolateral LET-23 EGFR localization in the VPCs to permit signal transmission and development of the vulva. We recently found that the LIN-2/7/10 complex likely forms at Golgi ministacks; however, the mechanism through which the complex targets the receptor to the basolateral membrane remains unknown. Here we found that overexpression of LIN-10 or LIN-7 can compensate for loss of their complex components by promoting LET-23 EGFR signaling through previously unknown complex-independent and receptor-dependent pathways. In particular, LIN-10 can independently promote basolateral LET-23 EGFR localization, and its complex-independent function uniquely requires its PDZ domains that also regulate its localization to Golgi. These studies point to a novel complex-independent function for LIN-7 and LIN-10 that broadens our understanding of how this complex regulates targeted sorting of membrane proteins.
INTRODUCTION
The hermaphroditic nematode Caenorhabditis elegans has served as an invaluable model in the discovery of the epidermal growth factor receptor (EGFR)/Ras/ERK signaling cascade and in our general understanding of how cellular signaling and development are regulated. Development of the vulva, required for egg laying and mating, is initiated by activation of the sole EGFR homologue, LET-23, in a set of progenitor epithelial cells called the vulva precursor cells (VPCs). The spatial organization of the signaling pathway within the VPCs is a critical dimension of signaling regulation: LET-23 EGFR must be present on the basolateral membrane of the VPCs to engage with the EGF-like ligand LIN-3, released from the overlying gonad, and to initiate vulval development (Aroian et al., 1990; Hill and Sternberg, 1992; Simske et al., 1996).
All six VPCs, named P3.p–P8.p, express LET-23 EGFR and can be induced into vulval cell fates; however, only the cell closest to the source of the ligand (P6.p) receives sufficient inductive signal to activate the downstream LET-60 Ras/MPK-1 ERK signaling cascade to specify the primary (1°) vulval cell fate (Sternberg and Horvitz, 1986; Aroian et al., 1990; Han et al., 1990). P6.p in turn signals to adjacent P5.p and P7.p cells through LIN-12 Notch signaling to induce them to adopt a secondary (2°) cell fate (Figure 1A), aided in part by graded inductive signaling through an alternate Ras/Ral GTPase pathway downstream of LET-23 EGFR (Greenwald et al., 1983; Zand et al., 2011). The 1° and 2° cells undergo cell divisions and morphogenic changes characteristic of their respective cell fates to produce a functioning vulva. The remaining three uninduced VPCs fuse with the surrounding hypodermis after one round of cell division (Sternberg and Horvitz, 1986).
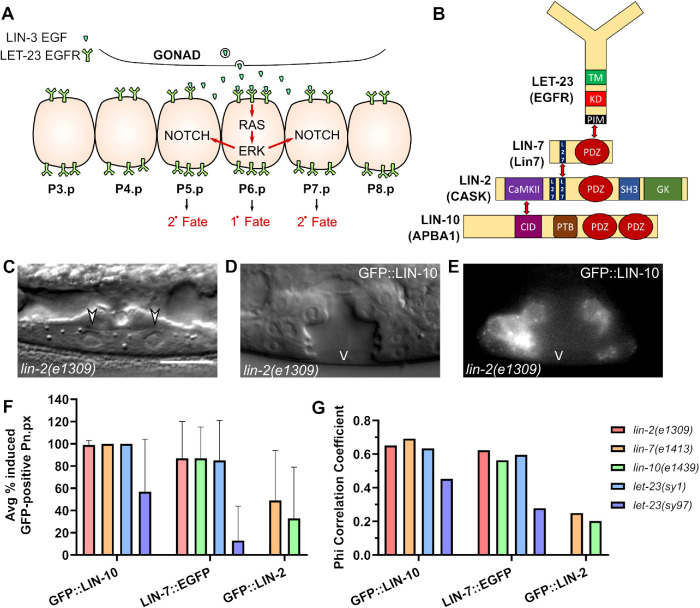
FIGURE 1:
LIN-10 and LIN-7 promote VPC cell fate induction independently of their complex components. (A) Model of vulval cell fate induction in the VPCs. The VPC closest to the source of the LIN-3 EGF-like ligand, P6.p, receives sufficient LET-23 EGFR activation on the basolateral membrane to activate the Ras/ERK signaling cascade, which induces the primary (1°) cell fate and initiates lateral signaling to adjacent P5.p and P7.p through Notch to induce the secondary (2°) vulval cell fate in those cells. (B) Known interactions and domains between LET-23 EGFR and the LIN-2/7/10 complex in C. elegans. The PDZ (PSD95/Dlg1/ZO-1) domain of LIN-7 interacts with the C-terminal PDZ-interaction motif (PIM) of LET-23 EGFR. LIN-7 and LIN-2 interact through shared L27 (LIN-2/LIN-7) domains. The CaMKII (calcium/calmodulin kinase II) domain of LIN-2 interacts with the CID (CASK-interacting domain) of LIN-10. TM: Transmembrane domain. KD: Kinase domain. SH3: Src homology domain 3. GK: Guanylate kinase domain. PTB: Phosphotyrosine-binding domain. (C) DIC image of a vulvaless lin-2(e1309) mutant with arrows marking uninduced P6.p daughter cells. Scale bar: 10 μm. (D, E) DIC and epifluorescence images of an induced vulva (the lumen marked with “V”) of a lin-2 mutant overexpressing GFP::LIN-10a (vhEx37). Scale as in C. (F) Average percentage of cells (Pn.px lineages) expressing the indicated extrachromosomal arrays that are induced into vulval cell fate lineages. Error bars are SD. Legend as in G. Sample sizes for corresponding genotypes in Tables 1 and 2. (G) Phi correlation of extrachromosomal array expression and vulval cell fate induction for the indicated transgenes and mutant backgrounds. Sample sizes for corresponding genotypes in Tables 1 and 2.
Basolateral localization of LET-23 EGFR, which is indispensable for signaling activation, is mediated by a protein complex consisting of three PDZ (PSD95/Dlg1/ZO-1) protein interaction domain-containing scaffold proteins: LIN-2, LIN-7, and LIN-10 (Figure 1B). Named for their involvement in the vulval cell fate lineage, these three proteins are essential for vulval development: loss of any individual complex component, or loss of the interaction between the C-terminal PDZ interaction motif of LET-23 EGFR to the PDZ domain of LIN-7, results in exclusive apical receptor localization and no vulval development (Ferguson and Horvitz, 1985; Simske et al., 1996; Kaech et al., 1998). This developmental defect prevents the worms from laying eggs, producing a striking “bag of worms” phenotype as the eggs accumulate and hatch inside the vulvaless (Vul) mother. Mammalian Lin7A-C, also known as Veli1-3 or Mals1-3, also interact with the C-terminal cytoplasmic tail of EGFR paralogues ErbB2 and ErbB4 to regulate their basolateral localization (Shelly et al., 2003); however, involvement of the other complex components in receptor localization has not been determined.
The LIN-2/LIN-7/LIN-10 (LIN-2/7/10) complex has been well-defined biochemically: LIN-7 and LIN-2 interact through N-terminal L27 domains on each protein, and the calcium/calmodulin kinase II (CaMKII) domain of LIN-2 interacts with the CASK-interacting domain (CID) of LIN-10 (Figure 1B) (Borg et al., 1998; Butz et al., 1998; Kaech et al., 1998). Still, little is known about how the complex regulates polarized LET-23 EGFR localization. LIN-2 also has a second L27 domain, which has been shown to interact with EPS-8 to prevent the internalization of LET-23 EGFR (Stetak et al., 2006). Similar to other membrane-associated guanylate kinase (MAGUK) scaffold proteins, LIN-2 has a type II PDZ domain (defined by specificity for consensus interaction motifs [ Songyang et al., 1997]), a Src homology 3 (SH3) domain, and a C-terminal guanylate kinase domain, though these domains have not been shown to be important for vulval development. LIN-10 has a phosphotyrosine-binding domain (PTB) and two tandem PDZ domains on its C-terminus with unique consensus interaction motifs that most closely resemble type III PDZ domains (Long et al., 2005; Swistowski et al., 2009). The PTB domain, but not the PDZ domains, was previously found to be involved in vulval development through an unknown mechanism (Whitfield et al., 1999).
In addition to vulval cell fate induction, the LIN-2/7/10 complex has been found to regulate infection sensitivity in the hypodermis through an interaction between the PDZ domain of LIN-2 and the insulin receptor DAF-2 in C. elegans (Sem et al., 2012). In mammals, the homologous CASK (LIN-2)/Lin7/APBA1 (LIN-10) complex has been well-studied for its role in regulating synaptic localization of the NMDA receptor subunit NR2B (Jo et al., 1999; Setou et al., 2000) and the cellular adhesion protein neurexin (Butz et al., 1998; Fairless et al., 2008). Lin7 is often implicated in polarized membrane sorting of internalized target proteins (Perego et al., 1999; Straight et al., 2001; Shelly et al., 2003; Alewine et al., 2007) and, along with CASK, typically localizes to the basolateral membrane in epithelial cells (Cohen et al., 1998; Straight et al., 2000; Stetak et al., 2006). On the other hand, Golgi-associated APBA1, also known as Mint1 or X11α, can regulate protein trafficking by interacting with the neuron-specific kinesin KIF17 (Setou et al., 2000). APBA1 can also regulate secretion by interacting with the SNARE-interacting protein Munc18 (Okamoto and Sudhof, 1997; Biederer and Sudhof, 2000; Schutz et al., 2005). However, this interaction is likely not conserved in C. elegans due to the absence of a Munc18-interacting domain in LIN-10. Furthermore, the C. elegans homologues of KIF17 and Munc18 (OSM-3 and UNC-18, respectively) are restricted to neurons (Gengyo-Ando et al., 1993; Tabish et al., 1995); therefore, they are unlikely to be involved in LIN-2/7/10 complex function in epithelial cells.
Recently, we found that LIN-10 colocalizes with LIN-2, LIN-7, and LET-23 EGFR on cytosolic punctae and that LIN-10 and LIN-7 localize to the Golgi, consistent with a model in which the complex helps target the receptor to the basolateral membrane of the VPCs. LIN-10 is strongly punctate while LIN-2 and LIN-7 are largely cytosolic, but are recruited to punctae by LIN-10. We also found that LIN-2 colocalized and interacted strongly with LIN-7 and relatively weakly with LIN-10 in vivo, further highlighting unique features of LIN-10 in the complex (Gauthier and Rocheleau, 2021).
Here, we report that overexpression of LIN-10 or LIN-7 can compensate for loss of their fellow complex components and restore vulval development in Vul mutants, pointing to a novel complex-independent function for both proteins. This effect is independent of the LET-23 EGFR C-terminal PDZ interaction motif, but is LET-23 EGFR-dependent. We also found a differential requirement between the PTB and PDZ domains of LIN-10 in mediating its function and localization, suggesting that LIN-10 can work through a secondary pathway to promote LET-23 EGFR signaling and vulval development. These studies reveal novel contributions of LIN-10 and LIN-7 in regulating polarized protein sorting in epithelial cells.
RESULTS
LIN-10 and LIN-7 can promote VPC induction in a complex-independent manner
Loss of either lin-2, lin-7, or lin-10 alone has previously been shown to inhibit vulval cell fate induction due to the mislocalization of LET-23 EGFR, likely leading to a loss of signaling. Recently, we tested whether loss of these genes would also disrupt localization of their complex components (Gauthier and Rocheleau, 2021). In passing, we noticed an unusually high proportion of egg laying–competent lin-2(e1309) mutant worms expressing a green fluorescent protein (GFP)-tagged LIN-10 transgene, which contrasted with lin-2 mutants lacking the transgene that are normally Vul and egg laying–defective. This led us to suspect that there may be some compensatory mechanisms among members of the LIN-2/7/10 complex.
To test this, we scored vulval development in mutant L4 worms in which VPC lineages expressed extrachromosomal LIN-2, LIN-7, or LIN-10 transgenes that rescue their respective mutants (Gauthier and Rocheleau, 2021). These transgenes are inherently overexpressed due to expression of their respective endogenous genes and are expressed mosaically among cells. We found that overexpression of GFP::LIN-10a (vhEx37) and mCherry::LIN-10a (vhEx63) strongly rescued VPC induction in lin-2 and lin-7(e1413) null mutants without inducing defects in vulval development when overexpressed in wild-type worms (Figure 1, C–F; Table 1, rows 1, 4, 5, and 8). Overexpression of LIN-7a::EGFP (vhEx87) was also able to partially rescue vulval development in lin-2 and lin-10(e1439) mutants (Table 1, rows 3 and 11), whereas overexpression of GFP::LIN-2a (vhEx58) failed to rescue lin-7 or lin-10 (Table 1, rows 7 and 10).
TABLE 1:
LIN-10 and LIN-7, but not LIN-2, can promote vulval cell fate induction independently of their complex components.
| Genotype | %Vul | %Muv | Avg. # of VPCs induced | n | |
|---|---|---|---|---|---|
| Wild type (N2) | 0 | 0 | 3.00 | Many | |
| 1 | GFP::LIN-10a | 0 | 0 | 3.00 | 17 |
| 2 | lin-2(e1309)† | 95 | 0 | 0.35 | 42 |
| 3 | lin-2(e1309); LIN-7a::EGFP | 70 | 0 | 1.83**** | 27 |
| 4 | lin-2(e1309); GFP::LIN-10a | 16**** | 8 | 2.85**** | 37 |
| 5 | lin-2(e1309); mCh::LIN-10a‡ | 78 | 0 | 1.30**** | 27 |
| 6 | lin-7(e1413) | 78 | 2 | 1.15 | 51 |
| 7 | lin-7(e1413); GFP::LIN-2a | 79 | 0 | 1.42 | 33 |
| 8 | lin-7(e1413); GFP::LIN-10a | 23**** | 13 | 2.80**** | 30 |
| 9 | lin-10(e1439) | 87 | 0 | 0.79 | 39 |
| 10 | lin-10(e1439); GFP::LIN-2a | 86 | 0 | 0.90 | 21 |
| 11 | lin-10(e1439); LIN-7a::EGFP | 55** | 0 | 2.28**** | 29 |
** p < 0.01; **** p < 0.0001; Fisher’s exact test (%Muv and %Vul); one-way ANOVA with multiple comparisons (Avg. # of VPCs induced). Rows 3–5 were compared with row 2; rows 7 and 8 were compared with row 6; rows 10 and 11 were compared with row 9. †Control data were pooled with lin-2(e1309); vhEx58(plin-31::gfp::lin-2a) as published in Gauthier and Rocheleau (2021). ‡Worms selected for scoring based on expression of coinjection marker (Pttx-3::GFP) because mCherry fluorescence is undetectable in VPCs with an epifluorescence microscope.
Virtually every Pn.px lineage with a detectable GFP::LIN-10 transgene was induced to adopt vulval cell fates in lin-2 and lin-7 mutants, suggesting a strong association between LIN-10 overexpression and cell fate induction, and suggesting that LIN-10 is working cell autonomously (Figure 1F). This is supported by relatively strong Phi correlation coefficients of 0.65 and 0.69 in lin-2 and lin-7 mutants, respectively (Figure 1G). Correlation was similar for LIN-7 overexpression, with coefficients of 0.62 and 0.56 in lin-2 and lin-10 mutants, and with roughly 80–87% of LIN-7–overexpressing Pn.px lineages induced (Figure 1, F and G). Similar results were seen with a second transgene (Supplemental Table 1). On the other hand, LIN-2 overexpression had minimal correlation with VPC cell fate induction (Figure 1, F and G), further indicating that induction is specific to LIN-10 and LIN-7 overexpression, and not an artifact of GFP or transgene expression.
LIN-10 and LIN-7 overexpression rescue let-23(sy1), but not signaling-defective let-23(sy97)
LIN-7 interacts with LET-23 EGFR via its C-terminal type I PDZ domain. LIN-10 has two PDZ domains that have previously been found to recognize distinct consensus sequences (Swistowski et al., 2009). Nevertheless, there is some overlap in interacting networks between different PDZ domains. The second PDZ domain of mammalian LIN-10 homologue APBA1 typically recognizes X–Φ–V/L/I motifs (where Φ is a hydrophobic residue), although it can also interact with polar residues at the –1 position (Swistowski et al., 2009). Therefore, the PDZ-interaction motif of LET-23 EGFR (–TCL) is a potential match for the second PDZ domain of LIN-10.
To check whether LIN-10 overexpression might independently promote signaling by interacting with LET-23 EGFR via its own PDZ domains, we tested for rescue of the sy1 allele of let-23 that has an early stop codon resulting in a truncation of its C-terminal PDZ interaction motif (Aroian et al., 1994; Kaech et al., 1998). This allele is otherwise functional for let-23 signaling events and only causes abnormalities in vulval development (Aroian et al., 1994), where interaction with LIN-7 is necessary for receptor localization and function. LIN-10 overexpression strongly rescues a let-23(sy1) mutant (Table 2), suggesting that LIN-10 does not require an interaction between the receptor and LIN-7 to promote vulval development, nor is it likely that the PDZ domains of LIN-10 interact with the PDZ interaction motif of LET-23 EGFR. LIN-10 expression was once again strongly correlated to cell fate induction (Figure 1, F and G). Unexpectedly, LIN-7 overexpression also partially rescued the let-23(sy1) allele (Table 2; Figure 1, F and G), suggesting that LIN-7 can also promote vulval induction independently of its interaction with the LET-23 EGFR PDZ interaction motif.
TABLE 2:
LIN-10 and LIN-7 overexpression rescues a PDZ interaction-deficient let-23(sy1) receptor mutant, but not signaling-defective let-23(sy97).
| Genotype | % Vul | % Muv | Avg. # of VPCs induced | n | |
|---|---|---|---|---|---|
| 1 | let-23(sy1) | 92 | 0 | 0.40 | 36 |
| 2 | let-23(sy1); GFP::LIN-10 | 30**** | 10 | 2.72**** | 30 |
| 3 | let-23(sy1); LIN-7::EGFP | 52*** | 0 | 2.08**** | 25 |
| 4 | let-23(sy97) | 95 | 0 | 0.48 | 43 |
| 5 | let-23(sy97); GFP::LIN-10 | 96 | 0 | 0.72 | 25 |
| 6 | let-23(sy97); LIN-7::EGFP | 100 | 0 | 0.25 | 22 |
*** p < 0.001; **** p < 0.0001; Fisher’s exact test (%Muv and %Vul); one-way ANOVA with multiple comparisons (Avg # VPCs induced). Rows 2 and 3 were compared with row 1; rows 5 and 6 were compared with row 4.
To test whether LIN-10 and LIN-7 overexpression might be activating signaling downstream of the receptor, we tested for rescue of a signaling-defective let-23(sy97) mutant that is unable to activate the downstream LET-60 Ras/MPK-1 MAPK pathway. This allele has other phenotypes associated with loss of LET-60 Ras activation, such as a rod-like lethal phenotype in L1 worms (Aroian et al., 1994). Although LIN-10 overexpression moderately correlates with VPC induction in let-23(sy97) (Figure 1, F and G), both LIN-10 and LIN-7 overexpression fails to rescue the let-23(sy97) allele (Table 2), suggesting that both proteins require a functioning receptor and do not promote VPC induction by activating the signaling cascade downstream of LET-23 EGFR.
LIN-10 independently promotes basolateral LET-23 EGFR localization
The association between LIN-10 and LET-23 EGFR has previously been shown to be indirect via LIN-2 and LIN-7 (Kaech et al., 1998); therefore, it is particularly surprising to find that its overexpression can promote vulval cell fate induction independently of LIN-2 and LIN-7. This suggests that LIN-10 may be working through an alternate, complex-independent pathway to promote LET-23 EGFR signaling activation.
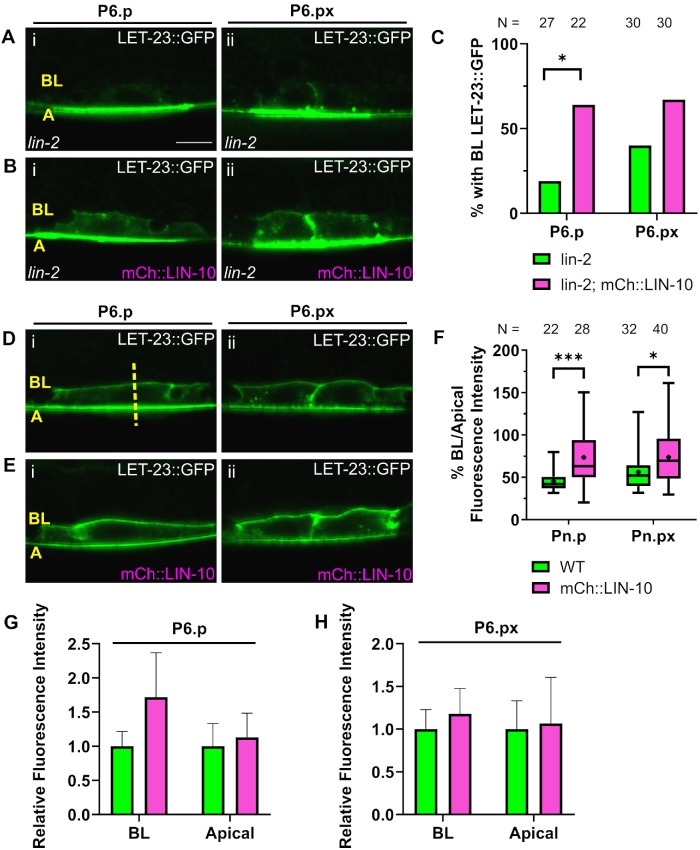
To better understand how LIN-10 overexpression can independently promote signaling, we next tested the effect of extrachromosomal mCh::LIN-10a expression (vhEx63) on LET-23::GFP (zhIs035) localization (Haag et al., 2014). We found that overexpression of LIN-10 was able to restore some basolateral receptor localization in a lin-2 mutant (Figure 2, A–C). Our lab has previously reported a similarly modest restoration of basolateral LET-23 EGFR localization in a lin-2 mutant by loss of negative regulators, such as the ARF guanine exchange factor AGEF-1 or the AP-1 clathrin adaptor µ-subunit UNC-101 (Skorobogata et al., 2014). In a wild-type background, LIN-10 overexpression also increased the basal/apical ratio of LET-23::GFP localization (Figure 2, D–F). An increase in fluorescence intensity on the basal membrane can be detected in one-cell P6.p, but not in two-cell P6.px (Figure 2, G and H). This suggests that LIN-10 independently regulates LET-23 EGFR signaling by promoting basolateral receptor localization.
FIGURE 2:
LIN-10 overexpression promotes basolateral targeting of LET-23::GFP. (A) LET-23::GFP (zhIs035) is localized on apical membranes of lin-2(e1309) mutant P6.p (i) and P6.px (ii) cells. (B) Overexpression of mCh::LIN-10 (vhEx63) restores basolateral LET-23::GFP in a lin-2 mutant. (C) Quantification of VPCs with basolateral LET-23::GFP localization in A and B. * p < 0.05 Fisher’s exact test. (D) Polarized LET-23::GFP localization in wild-type P6.p (i) and P6.px (ii) cells. (E) LET-23::GFP localization in VPCs expressing extrachromosomal mCh::LIN-10. (F) Quantification of basolateral/apical peak fluorescence intensity of images represented in D and E. *p < 0.05, ***p < 0.001 Two-tailed Student’s t test. (G, H) Comparing relative fluorescence intensities of basolateral and apical membranes in P6.p (G) and P6.px (H) with or without LIN-10 overexpression for the set of images represented in D and E. Legend and sample sizes as in F. Intensities normalized to fluorescence intensity of wild-type worms (no mCh::LIN-10 expressed) for basolateral and apical membranes. Error bars are SD. BL = Basolateral. A = Apical. WT = Wild type. Scale bar: 5 μm. Images are all at the same scale.
LIN-10 C-terminal domains mediate punctate localization
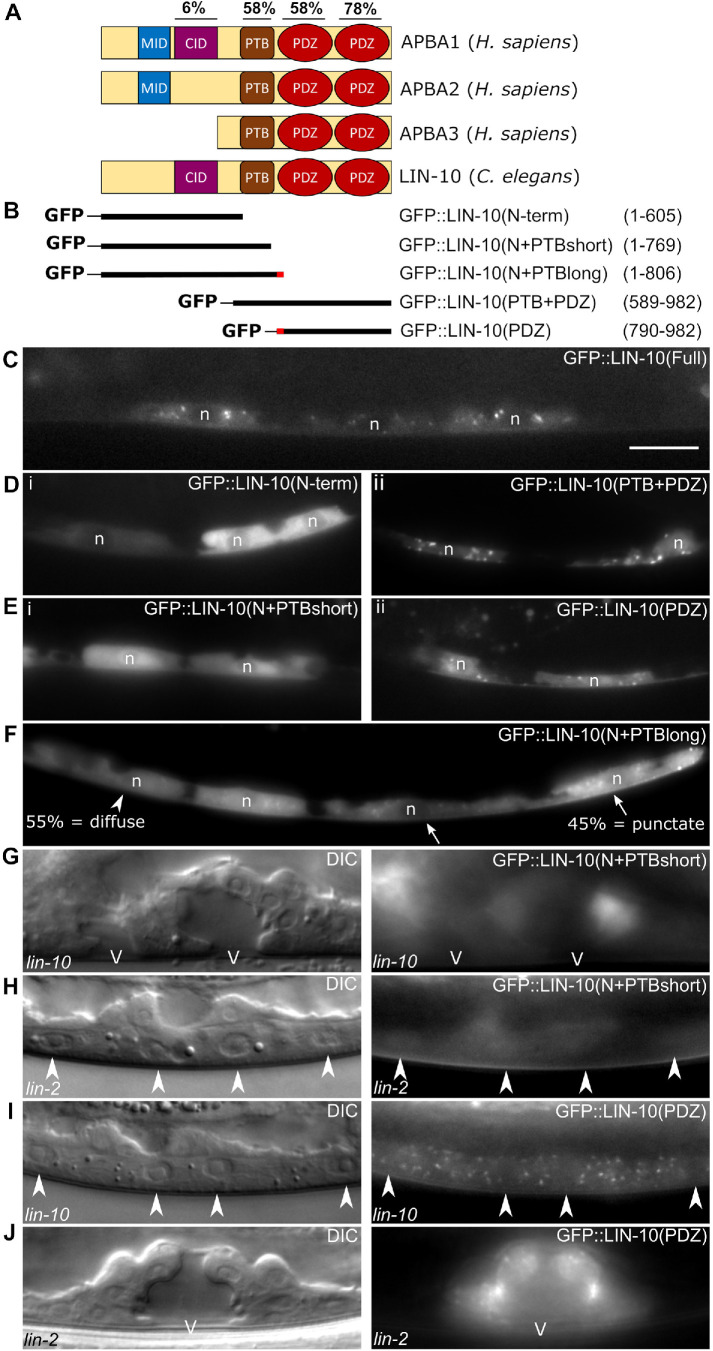
LIN-10 localizes to Golgi ministacks and recycling endosomes in neurons and intestinal epithelia (Rongo et al., 1998; Glodowski, Wright, Martinowich, et al., 2005; Zhang et al., 2012) and shows strong punctate localization that colocalizes with Golgi markers in the VPCs (Gauthier and Rocheleau, 2021). To better understand the function of LIN-10, we performed a structure-function analysis to identify the protein domains that regulate its localization and function (Figure 3, A and B). The N-terminal half of LIN-10 has no characterized domains other than the LIN-2–interacting CID, shared only with mammalian APBA1. Overall, the N-terminus of LIN-10 shares low sequence homology with APBA1 and APBA2, which in turn share low sequence homology in their N-terminal domains with each other, despite both sharing MIDs (Figure 3A and Glodowski, Wright, Martinowich. et al., 2005). Despite divergent sequences, the function of the CID is conserved between LIN-10 and APBA1, as C. elegans LIN-10 can interact with mammalian CASK (Borg et al., 1998). On the other hand, the C-terminal half, containing the PTB and tandem PDZ domains, has a much higher sequence identity among LIN-10 and APBA1-3 paralogues (Figure 3A and Glodowski, Wright, Martinowich, et al., 2005).
FIGURE 3:
C-terminal domains regulate punctate localization and function of LIN-10. (A) Protein domains of human APBA1-3 and C. elegans LIN-10. Percent identities compare amino acid sequences of CASK interaction domain (CID), phosphotyrosine-binding domain (PTB), and PDZ1/2 domains of LIN-10a to those of APBA1. (B) Truncations of LIN-10a generated to compare localization and function. Red region of N+PTBlong and PDZ indicates 16 amino acids overlapping between these two truncations. (C–F) Localization of full-length GFP::LIN-10 (vhEx37) (C) and the N-term (vhEx52) (Di), PTB+PDZ (vhEx55) (Dii), N+PTBshort (vhEx74) (Ei), PDZ (vhEx66) (Eii), and N+PTBlong (vhEx72) (F) truncated LIN-10 fragments as shown in B. n in F = 29 cells imaged (from 19 worms). Arrowhead: VPC with diffuse cytosolic localization. Arrows: VPCs with punctate localization. (G, H) Expression of the N+PTBshort construct rescues lin-10(e1439) (G) but not lin-2(e1309) (H). A pseudovulva, and a partially induced vulva can be seen in G. (I, J) Expression of the PDZ domains fails to rescue a lin-10(e1439) mutant (I) but does rescue a lin-2(e1309) mutant (J). Arrowheads in H and I are nuclei of uninduced primary (two in the center) and secondary (on left and right edges of image) Pn.px cells. n: Nucleus. V: Vulval lumen. Scale bar: 10 μm. Images are all at the same scale.
We cloned the CID-containing N-terminal half (LIN-10[N-term], aa 1–605) and the PTB and PDZ-containing C-terminal half (LIN-10[PTB+PDZ], aa 589–982) of LIN-10 to identify which region of LIN-10 regulates its localization and function (Figure 3B). Whereas full-length LIN-10 localizes to cytosolic punctae in all VPCs (Figure 3C), we found that the LIN-10(N-term) fragment was diffusely expressed in the cytosol (Figure 3Di). In contrast, the C-terminal LIN-10(PTB+PDZ) showed robust localization to punctae (Figure 3Dii), indicating that these domains mediate localization to the Golgi. Inclusion of the PTB domain in the N-terminal fragment, LIN-10(N+PTBshort) (aa 1–769), was not sufficient to localize to punctae (Figure 3Ei), whereas omitting the PTB domain from the C-terminal fragment, as in LIN-10(PDZ) (aa 790–982), did not prevent punctate localization (Figure 3Eii), suggesting that the PDZ domains mediate this localization pattern. Interestingly, extending the N-terminal half beyond the PTB domain into the flexible linker region between the PTB and the first PDZ domain, as in LIN-10(N+PTBlong) (aa 1–806), recovered some punctate localization, even in a lin-10 mutant (Figure 3F). This LIN-10(N+PTBlong) fragment overlaps with the LIN-10(PDZ) domain fragment by 16 amino acids covering a small segment of the linker and PDZ1 domain (Figure 3B), suggesting that there may be a regulatory region in this segment that in part determines punctate localization with the PDZ domains.
Mammalian APBA1 has been shown to have intramolecular interactions in its PDZ1 domain (Long et al., 2005), and C. elegans LIN-10 has been found to self-interact in a yeast two-hybrid screen (Walhout et al., 2000). To verify that the localization pattern of the PDZ domains was not due to oligomerization with endogenous full-length LIN-10, we looked at its localization in a lin-10 mutant and still found that the PDZ domains were punctate (Figure 3I). Therefore, these results demonstrate that the C-terminal PDZ domains, and likely a portion of the linker region, mediate Golgi localization of LIN-10.
The C-terminal PDZ domains mediate the complex-independent function of LIN-10, whereas the PTB domain is required for overall LIN-10 function
To further characterize the complex-independent function of LIN-10, we tested which domains of LIN-10 were necessary for the rescue of lin-2 and lin-10. We found that the C-terminal PDZ domains that mediated punctate localization of LIN-10 were necessary and sufficient to rescue VPC induction in a lin-2 mutant (Table 3, rows 8, 9, and 11; Figure 3, H and J). However, expression of these domains alone was not sufficient to rescue lin-10 (Table 3, row 6; Figure 3I). On the other hand, while truncation of the PDZ domains in the LIN-10(N+PTBshort) fragment failed to rescue a lin-2 mutant, this fragment did rescue lin-10 (Table 3, rows 3 and 9; Figure 3, G and H). We found that the presence of the PTB domain, whether on the N-terminal cytosolic half (N+PTBshort or N+PTBlong) or added to the C-terminal punctate domains (PTB+PDZ), was specifically required for rescue of a lin-10 mutant. However, lin-10 rescue by the LIN-10(PTB+PDZ) fragment was mild compared with the N-terminal fragments, consistent with the CID-mediated interaction with LIN-2 being important for LIN-10 complex–dependent function (Table 3, rows 3–5). Furthermore, LIN-10(N+PTBlong) rescue is stronger than N+PTBshort, suggesting that Golgi localization is functionally important (Table 3, rows 3 and 4). Interestingly, LIN-10(PDZ) rescues the lin-2 Vul phenotype slightly better than LIN-10(PTB+PDZ), suggesting that the PTB domain might interfere with the PDZ-mediated lin-2 rescue (Table 3, rows 10 and 11). These results suggest that the PTB and PDZ domains are differentially required for LIN-10 function: general LIN-10 function requires the PTB domain, and is less dependent on the subcellular localization, whereas the LIN-2–independent function of LIN-10 is likely restricted to Golgi subcellular compartments and is mediated by the C-terminal PDZ domains.
TABLE 3:
Differential requirements of the LIN-10 PTB and PDZ domains for rescue of lin-2 and lin-10 mutants.
| Genotype | %Vul | %Muv | Avg # of VPCs induced | n | |
|---|---|---|---|---|---|
| 1 | lin-10(e1439) | 90 | 0 | 0.86 | 49 |
| 2 | lin-10(e1439); GFP::LIN-10(N-term) | 91 | 3 | 1.27 | 35 |
| 3 | lin-10(e1439); GFP::LIN-10(N+PTBshort) | 52*** | 11* | 2.39**** | 27 |
| 4 | lin-10(e1439); GFP::LIN-10(N+PTBlong) | 4**** | 4 | 3.00****,† | 24 |
| 5 | lin-10(e1439); GFP::LIN-10(PTB+PDZ) | 77 | 3 | 1.85**** | 30 |
| 6 | lin-10(e1439); GFP::LIN-10(PDZ) | 95 | 0 | 0.73 | 22 |
| 7 | lin-2(e1309) | 88 | 0 | 0.64 | 60 |
| 8 | lin-2(e1309); GFP::LIN-10(N-term) | 81 | 0 | 1.08 | 31 |
| 9 | lin-2(e1309); GFP::LIN-10(N+PTBshort) | 100 | 0 | 0.62 | 26 |
| 10 | lin-2(e1309); GFP::LIN-10(PTB+PDZ) | 74 | 0 | 1.73**** | 31 |
| 11 | lin-2(e1309); GFP::LIN-10(PDZ) | 54** | 4 | 2.25**** | 24 |
* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; Fisher’s exact test (%Muv and %Vul); one-way ANOVA with multiple comparisons (Avg # VPC induced). Rows 2–6 were compared with row 1. Rows 8–11 were compared with row 7. †p < 0.01 Student’s two-tailed t test comparing row 4 to row 3.
ARF-1 colocalizes with LIN-10 and could function in a common pathway
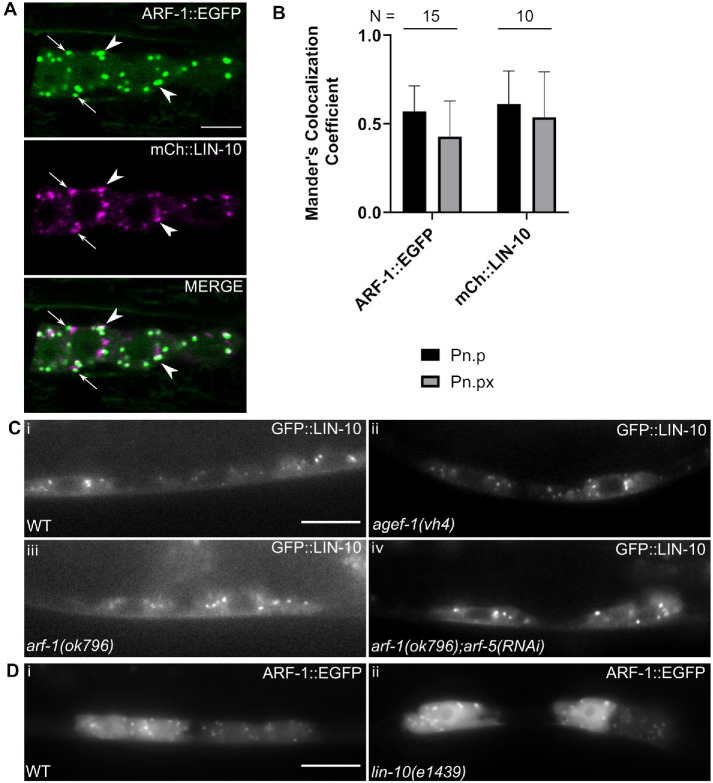
The finding that the C-terminal PDZ domains of LIN-10 mediate its complex-independent function suggests that LIN-10 has additional binding partners with which it promotes basolateral LET-23 EGFR targeting and signaling. The C-terminus (PDZ domains and part of the PTB domain) of all three mammalian APBA proteins mediates an interaction with GTP-bound Class I (Arf3) and Class II (Arf4) Arf GTPases in neurons to regulate amyloid precursor protein (APP) trafficking (Hill et al., 2003). APBA proteins were also found to localize to the Golgi in a brefeldin A (BFA)-sensitive manner, a compound that disrupts the Arf GEFs BIG1/2 and GBF1 (Donaldson et al., 1992a,b; Togawa et al., 1999; Niu et al., 2005), and are associated with clathrin-coated secretory vesicles (Hill et al., 2003; Shrivastava-Ranjan et al., 2008). Our lab has previously reported that the sole C. elegans Class I and Class II Arf GTPases (ARF-1 and ARF-5, respectively) work in opposition to the LIN-2/7/10 complex with the Arf GEF AGEF-1 (homologous to BIG1/2) and the AP-1 clathrin adaptor complex to negatively regulate LET-23 EGFR signaling and basolateral localization (Skorobogata et al., 2014), possibly by promoting the apical targeting of the receptor. LIN-10 overexpression phenocopies inhibition of the AGEF-1/ARF/AP-1 pathway with respect to rescue of VPC induction and increasing basolateral LET-23 EGFR localization. Therefore, the complex-independent function of LIN-10 might involve an interaction with ARF GTPases that could inhibit or compete with the AP-1–mediated pathway.
ARF-1::EGFP localizes to cytoplasmic foci when expressed in the VPCs (O Skorobogata and CER, unpublished results). We found that LIN-10 frequently colocalizes with ARF-1 cytoplasmic foci or can localize to adjacent foci at Pn.p and Pn.px stages (Figure 4, A and B). Unlike in mammals, LIN-10 does not require active ARF GTPases for its localization, as punctate localization is not disrupted in arf-1 loss-of-function mutants, arf-1; arf-5(RNAi)-treated mutants, or agef-1(vh4) hypomorphic mutants (Figure 4C). Similarly, loss of lin-10 does not disrupt recruitment of ARF-1 to cytosolic punctae (Figure 4D). If LIN-10 overexpression functions to inhibit ARF-1 and ARF-5 activity, then we would expect loss of arf-1 to not enhance the suppression of lin-2 Vul by LIN-10 overexpression. If LIN-10 overexpression functions in a parallel pathway, then loss of arf-1 might enhance the suppression of lin-2 Vul by LIN-10 overexpression. We found no change in vulval induction in arf-1; lin-2; GFP::LIN-10 compared with lin-2; GFP::LIN-10 (Table 4). The lack of enhanced rescue is most consistent with LIN-10 overexpression inhibiting the ARFs.
FIGURE 4:
LIN-10 colocalizes with ARF-1 but is not dependent on the ARFs for localization. (A) LIN-10 (vhEx63) frequently localizes with (arrowheads) or near (arrows) ARF-1 GTPase (vhEx5). Scale bar: 5 μm. (B) Average Mander’s correlation coefficients reveal moderate colocalization between ARF-1 and LIN-10 in C. elegans VPCs. Error bars are SD. (C) LIN-10 (vhEx37) localization to cytosolic punctae (i) is not altered in a hypomorphic agef-1(vh4) mutant (ii), a loss-of-function arf-1(ok796) mutant (iii), and an arf-1 mutant treated with arf-5(RNAi). (D) ARF-1 (vhEx7) localization to cytosolic punctae (i) is not altered in a loss-of-function lin-10(e1439) mutant (ii). Scale bars: 10 μm.
TABLE 4:
arf-1(ok796) is not additive with LIN-10 overexpression.
| Genotype | %Vul | %Muv | Avg # of VPCs induced | n | |
|---|---|---|---|---|---|
| 1 | lin-2(e1309); GFP::LIN-10a† | 16 | 8 | 2.85 | 37 |
| 2 | arf-1(ok796); lin-2(e1309) | 85 | 5 | 1.88**** | 20 |
| 3 | arf-1(ok796); lin-2(e1309); GFP::LIN-10a | 24 | 12 | 2.94 | 17 |
****p < 0.0001; Fisher’s exact test (%Muv and %Vul); one-way ANOVA with multiple comparisons (Avg # VPCs induced). Rows 2 and 3 were compared with row 1. †Data collected along with those in Table 1.
DISCUSSION
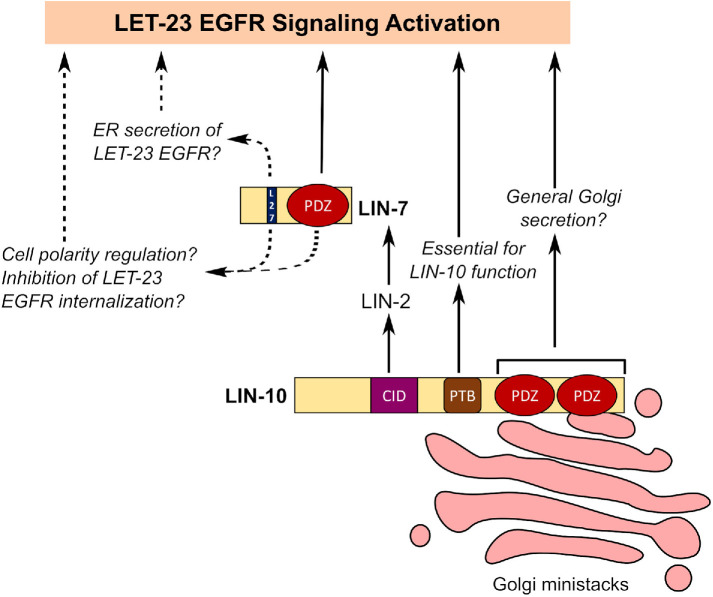
Here we identified a novel, complex-independent function for LIN-7 and LIN-10 in promoting LET-23 EGFR signaling and vulval cell fate induction. Overexpression of LIN-7 and LIN-10 rescued vulval development in lin-2/10 and lin-2/7 mutants, respectively, as well as in let-23(sy1) mutants deficient for an interaction with the LIN-7 PDZ domain. Focusing on LIN-10, the most removed from LET-23 EGFR, we found that it promoted basolateral receptor targeting even in the absence of its complex components. We found that the LIN-10 PTB domain, but not the PDZ domains, was required for the rescue of lin-10 mutants. Conversely, we found that the complex-independent function of LIN-10 required the C-terminal PDZ domains, which are largely responsible for its punctate localization, and not the PTB domain. Finally, LIN-10 colocalizes with ARF-1 in the VPCs, and the lack genetic enhancement between arf-1 mutant and LIN-10 overexpression is consistent with LIN-10 functioning to inhibit the ARF-1 and ARF-5 GTPases.
Previous research on LIN-2, LIN-7, and LIN-10 have led toward a model in which these proteins function in vivo as a complex, or at least as interacting components of a linear pathway (Kaech et al., 1998). Therefore, we were surprised to find that overexpression of either LIN-7 or LIN-10 can compensate for loss of the other components. This suggests that our assumptions about how the complex functions are incorrect, or that LIN-7 and LIN-10 can promote LET-23 EGFR signaling via complex-independent mechanisms. Overexpression of LIN-7 can rescue the Vul phenotypes of lin-2 and lin-10 mutants, but not as strongly as rescue of lin-7 mutants (Gauthier and Rocheleau, 2021), suggesting that compensation for loss of LIN-2 and LIN-10 is partial. Because LIN-7 interacts directly with LET-23 EGFR, it is perhaps not too surprising that increased levels of LIN-7 would rescue lin-2 or lin-10 mutants. Mammalian Lin7 is involved in recycling of internalized cargo, including the EGFR paralogue ErbB2, at the basolateral membrane, with no known involvement of mammalian LIN-2 or LIN-10 (CASK and APBA1-3, respectively) (Perego et al., 1999; Shelly et al., 2003); therefore, LIN-7 might similarly promote recycling of a small amount of LET-23 EGFR that makes it to the basolateral membrane in the absence of LIN-2 or LIN-10.
We recently found that LIN-7, but not LIN-2 or LIN-10, colocalizes with LET-23 EGFR on basolateral membranes of VPCs (Gauthier and Rocheleau, 2021), consistent with LIN-7 potentially having uncharacterized complex-independent roles in basolateral membrane traffic. Furthermore, LIN-7 basolateral localization was not dependent on the LET-23 EGFR PDZ interaction motif (Gauthier and Rocheleau, 2021). Similarly, we found that LIN-7 overexpression rescues a let-23(sy1) receptor mutant lacking a PDZ interaction motif, suggesting that LIN-7 can promote signaling independently of this interaction. Mammalian Lin7A was found to have a second point of interaction with the kinase domain of all four human EGFR paralogues through its extended N-terminus that helps transit the receptor from the endoplasmic reticulum to the Golgi (Shelly et al., 2003). Although C. elegans LIN-7 has only a portion of this N-terminal sequence (Butz et al., 1998; Shelly et al., 2003), a similar interaction could explain how LIN-7 overexpression may rescue let-23(sy1) by up-regulating LET-23 EGFR secretion (Figure 5). Alternatively, LIN-7 orthologues are also known to maintain epithelial cell polarity through L27 and PDZ domain–mediated interactions with the Crumbs complex and the cadherin-catenin complex at cell junctions (Perego et al., 2000; Straight et al., 2006). Thus, LIN-7 overexpression in the VPCs might have more global impacts on epithelial cell polarity that could indirectly shift the balance between apical and basolateral cargo sorting (Figure 5), thereby restoring some LET-23 EGFR signaling activation.
FIGURE 5:
Hypothetical models for complex-independent function of LIN-7 and LIN-10 in promoting LET-23 EGFR signaling. LIN-10 targets LET-23 EGFR to the basolateral membrane through LIN-2 and LIN-7 and requires its PTB domain for its function in the VPCs. However, overexpression of its PDZ domains can bypass the requirement for its complex to promote LET-23 EGFR signaling, possibly through general up-regulation of Golgi secretion. LIN-7 was also found to have a complex-independent function (dashed arrows). This may involve promoting the secretion of LET-23 EGFR from the ER (endoplasmic reticulum), inhibiting LET-23 EGFR internalization, or regulating cell polarity, similar to its mammalian orthologues.
LIN-2 is the only member of the complex that has been shown to have an additional binding partner in EPS-8 to regulate LET-23 EGFR trafficking (Stetak et al., 2006). While overexpression of LIN-2 strongly rescues a lin-2 mutant (Gauthier and Rocheleau, 2021), it could not bypass its complex components to activate signaling. The strong binding, colocalization, and similar expression dynamics between LIN-2 and LIN-7 (Gauthier and Rocheleau, 2021) may point to a requirement for some dependence on stoichiometry with LIN-7 for LIN-2 function.
The finding that LIN-10 overexpression can rescue lin-2 and lin-7 mutants and partly restore LET-23 EGFR localization to the basolateral membrane of the VPCs was particularly unexpected because its interaction with LET-23 EGFR is mediated by LIN-2 and LIN-7 (Kaech et al., 1998). One intriguing possibility was that the PDZ domains of LIN-10 might interact directly with the PDZ interaction motif of LET-23 EGFR. Consistent with previous protein interaction assays that did not find a direct interaction between LIN-10 and LET-23 EGFR (Kaech et al., 1998), we found that LIN-10 overexpression was able to rescue the let-23(sy1) mutation lacking the PDZ interaction motif, suggesting that LIN-10 was not promoting LET-23 localization via a direct interaction with its PDZ domains.
The mammalian homologues of LIN-10 (APBA1-3) have well-established roles in regulating secretion and recycling. APBA2/3 lack the CASK/LIN-2–interacting CID, indicating that APBA proteins do not rely on CASK and mLin7 for their function. In C. elegans neurons, LIN-10 regulates the trafficking and recycling of the glutamate receptor GLR-1 independent of LIN-2 and LIN-7, and without a direct interaction to GLR-1 (Rongo et al., 1998). In general, LIN-10 has relatively weak colocalization and in vivo association with LIN-2 and LIN-7 (Gauthier and Rocheleau, 2021). Throughout vulva development, LIN-2 and LIN-7 show dynamic changes in expression and localization not shared by LIN-10 (Gauthier and Rocheleau, 2021). These are consistent with our findings here that LIN-10 can function independently of the complex.
We recently demonstrated that LIN-10 recruits LIN-2 and LIN-7 to the Golgi, where it also colocalizes with LET-23 EGFR (Gauthier and Rocheleau, 2021). The LIN-10 C-terminal PDZ fragment that shows the strongest localization to these subcellular compartments, despite being involved only in the complex-independent function of LIN-10. The N+PTBshort fragment, despite not localizing to punctae, is able to rescue a lin-10 mutant, suggesting that LIN-10 function may not be entirely dependent on its endomembrane localization. Still, extending this N-terminal fragment into the linker region confers an even stronger rescue while also partially restoring some punctate localization, indicating that the Golgi is likely the primary site of LIN-10 function both with and without LIN-2 and LIN-7.
We found differential requirements for the PTB and PDZ domains for rescue of lin-10 and lin-2 mutants. A previous study by Glodowski, Wright, Martinowich, et al. (2005) queried which domains of LIN-10 were required for GLR-1 localization in neurons and vulva induction by testing for rescue of associated lin-10 phenotypes. They found that the PTB domain is essential for lin-10 rescue in both tissues, whereas the PDZ domains were essential for rescue of GLR-1 localization in neurons, but not for vulval development (Glodowski, Wright, Martinowich, et al., 2005). Our results support their findings that the PTB domain is required for rescue of the lin-10 Vul phenotype (Figure 5) and that the PDZ domains are not. We found that the PTB domain, whether part of the LIN-10 N-terminal constructs containing the CID or part of the LIN-10 C-terminal fragment containing the PDZ domains, rescues VPC induction in lin-10 mutants. However, only the N-terminal fragments significantly suppressed the frequency of Vul worms in lin-10 mutants, consistent with the N-terminal CID-mediated interaction with LIN-2 being important for LIN-10 function. How the PTB domain promotes LIN-10 function in VPC induction remains unclear.
Interestingly, the PDZ domains alone were necessary and sufficient for the rescue of lin-2 mutants, but not for lin-10. The PDZ domains did not require endogenous LIN-10 for their localization to punctae, suggesting that they may be similarly independent of endogenous LIN-10 for the rescue of lin-2. Testing for the rescue of a lin-10; lin-2 double mutant might help tease apart the requirement for endogenous LIN-10; however, this experiment would be difficult to interpret because the PDZ domains do not rescue lin-10.
The strong punctate localization and rescue of lin-2 by the C-terminal PDZ domain suggests that LIN-10 engages with additional interactors in the VPCs through its PDZ domains to regulate LET-23 EGFR trafficking independently of LIN-2 and LIN-7. These interactions may support general secretion from the Golgi (Figure 5), as has been hypothesized for the PDZ domains of APBA2 (Saito et al., 2011). Alternatively, the increase in basolateral LET-23 EGFR caused by LIN-10 overexpression may indicate that LIN-10 helps to redirect internalized LET-23 EGFR away from degradation pathways and instead toward recycling through a Golgi-associated pathway. The class I and II ARF-1 and ARF-5 GTPases are strong candidate effectors for the PDZ-mediated complex-independent function of LIN-10. Mammalian class I and II Arf GTPases interact with and might recruit APBA proteins to the Golgi to regulate secretion of APP, and APBA proteins are hypothesized to serve as clathrin adaptor proteins (Hill et al., 2003). If the interaction between APBA proteins and Arf GTPases is conserved in C. elegans, LIN-10 might compete with AP-1 for binding to ARF-1 and ARF-5 to promote basolateral targeting of LET-23 EGFR. Alternatively, an interaction between LIN-10 and the ARFs might suppress the normally inhibitory function of ARF-1 without directly promoting basolateral targeting. The relatively strong colocalization between LIN-10 and ARF-1 supports such a model, despite the AGEF-1 and the ARFs not being required for LIN-10 localization. The effects of LIN-10 overexpression and arf-1 mutations on vulval development were not additive, consistent with these two proteins working in the same pathway; however, more studies are required to validate this hypothesis.
Thus, we have identified a complex-independent role for LIN-10 in promoting LET-23 EGFR trafficking to the basolateral membrane. This function is likely secondary to its involvement in the LIN-2/7/10 complex, as normal expression levels of LIN-10 cannot compensate for loss of lin-2 or lin-7. Our results reveal a potential mechanism through which the PTB domain directs the general requirement of LIN-10 for VPC induction, and the PDZ domains promote both the Golgi localization of LIN-10 and basolateral localization of LET-23 EGFR. This may represent novel recycling or secretory pathways that facilitate polarized signaling in epithelial cells.
MATERIALS AND METHODS
Strains and maintenance
All strains were maintained at 20°C on nematode growth media seeded with HB101 Escherichia coli (Brenner, 1974). A complete strain list can be found in Supplemental Table 2. The arf-1 and arf-5 genes were previously named arf-1.2 and arf-3, and the names were recently changed to be in better accordance with their mammalian homologues (wormbase.org).
Vulval cell fate induction scoring
VPC induction scores were analyzed at the L4 larval stage by counting the induced vulval cells derived from each VPC. Wild-type worms have a VPC induction score of 3 because only three of six VPCs (P5.p–P7.p) are induced. Worms were classified as Vul if they had a VPC induction score of less than 3, or multivulva (Muv) if they had an induction score of greater than 3 (Gauthier and Rocheleau, 2017). Worms with detectable fluorescence signal in their vulval cells (driven by the lin-31 promoter) and in neurons in their head (driven by the ttx-3 promoter of the coinjection marker) were compared with siblings taken from the same plate with no fluorescence signal in both their vulval cells and neurons as negative controls. Because the mCherry::LIN-10 transgene was not bright enough to be detected by epifluorescence, worms were selected based solely on expression of the coinjection marker without knowledge of their expression in the VPCs, which may account for the lower level of rescue than that seen with GFP::LIN-10.
LIN-10 subdomain cloning
Subdomains for the LIN-10a isoform were mapped according to the domains previously identified for the LIN-10b isoform (Glodowski, Wright, Martinowich, et al., 2005). Primers for cloning are listed in Supplemental Table 3. Subdomains were cloned into a plin-31::gfp::lin-10a expression vector by replacing the full-length lin-10a open reading frame (Gauthier and Rocheleau, 2021). All constructs were verified by sequencing. Amino acid sequence identity between LIN-10a domains and mammalian APBA1 was calculated according to sequence identities identified by BLAST (National Center for Biotechnology Information, National Institutes of Health) and Clustal Omega (European Molecular Biology Laboratory, European Bioinformatics Institute). The total number of similar amino acids was divided by the total number of amino acids in the respective LIN-10 protein domain to calculate sequence identity.
Microscopy and image analysis
Worms were immersed in 10 mM levamisole on a 2% agarose pad on a glass slide before imaging and scoring on an Axio A1 Imager epifluorescence microscope (Zeiss). Images were analyzed using FIJI (ImageJ). Confocal (colorized) images were taken using an LSM780 laser-scanning confocal microscope. Mander’s correlation coefficients for ARF-1::EGFP and mCherry::LIN-10 were measured using Zen 2012 software (Zeiss) and quantified by tracing the cells of interest, yielding a software-generated scatterplot of green and red fluorescence intensity values per pixel. Cross-hairs along the X- and Y-axes divide the graph into four quadrants and are used to omit background fluorescence, such that only pixels in the upper right-hand quadrant are calculated for colocalization. The cross-hairs were adjusted to omit most of the cytosolic signal in order to capture colocalization at punctae.
Correlation analysis for extrachromosomal array expression and VPC induction
In addition to scoring VPC cell fate induction, we kept track of which specific Pn.px lineages expressed the extrachromosomal arrays (determined by visible GFP fluorescence on a Zeiss Axio A1 imager at 100×) in developing vulvas of early to mid-L4 larvae. For each worm, we counted the number of GFP-expressing Pn.px lineages that were induced into vulval cell fates (either the primary or secondary cell fates) or that were uninduced (did not continue dividing).
To assess the correlation between extrachromosomal array expression and VPC cell fate induction, Pn.px lineages of all worms per genotype were categorized into four groups: those that expressed GFP and that were induced to assume a vulval cell fate (A), those with no visible GFP that were induced (B), those with GFP expression that were not induced (C), and those with no visible GFP that were not induced (D). Using these groups, the Phi (Φ) correlation coefficient between GFP expression and VPC induction was calculated for each worm using the formula below, and the average correlation coefficient for each genotype was used for analysis:
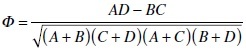 |
Analyzing LET-23 EGFR polarized distribution
Polarized membrane distribution of LET-23 EGFR was analyzed by drawing a 20-pixel-wide line across P6.p cells (as in Figure 2Di) for each worm and measuring the averaged peak fluorescence intensity on the basal and apical membrane using FIJI. For worms at the two-cell stage, one line was drawn across each P6.px cell. To limit bias, lines were drawn across differential interference contrast (DIC) images over the center of the nucleus and transposed to the fluorescence image.
To assess restoration of basolateral LET-23 EGFR localization in lin-2 mutants, we similarly drew a 20-pixel-wide line across the basal membrane of P6.p and P6.px cells. Cells with a peak in fluorescence intensity across the basal membrane that was at least twice as bright as the cytosol were considered to have basolateral localization of LET-23 EGFR.
RNA interference (RNAi) was performed by feeding as previously described (Kamath et al., 2001). All RNAi experiments were performed at 20°C. The RNAi clone for arf-5 (IV-4E13) was obtained from the Ahringer RNAi library (Wellcome CRC Institute, University of Cambridge, UK).
Statistical analysis
Average VPC induction scores and average peak basal/apical fluorescence intensities were compared using Student’s t test or one-way analysis of variance (ANOVA) with Dunnett’s correction for multiple comparisons. Proportions of Vul and Muv phenotypes and proportion of P6.p and P6.px cells with basolateral LET-23 EGFR localization were compared using Fisher’s exact test. Statistical analyses were performed using GraphPad Prism 8.0.
Supplementary Material
Acknowledgments
We thank Jung Hwa Seo, Min Fu, and ShiBo Feng for technical assistance; Richard Roy for sharing his RNAi library; and WormBase for strain information. Confocal microscopy was conducted in the Molecular Imaging Platform of the Research Institute–McGill University Health Centre. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by a National Institutes of Health Office of Research Infrastructure Program grant (P40 OD010440). This work was funded by an Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (RGPIN-2018-05673) to C.E.R. K.D.G. was funded by a Canada Graduate Scholarship Master’s Award and Post-Graduate Scholarship Doctoral fellowships from NSERC and Master’s and Doctoral Awards from Fonds de recherche du Québec–Santé.
Abbreviations used:
- CID
CASK-interacting domain
- Muv
multivulva
- VPC
vulva precursor cell
- Vul
vulvaless.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-07-0490) on January 21, 2021.
REFERENCES
Boldface type denotes co–first authors.
- Alewine C, Kim BY, Hegde V, Welling PA (2007). Lin-7 targets the Kir 2.3 channel on the basolateral membrane via a L27 domain interaction with CASK. Am J Physiol Cell Physiol 293, C1733–C1741. [DOI] [PubMed] [Google Scholar]
- Aroian RV, Koga M, Mendel JE, Ohshima Y, Sternberg PW (1990). The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature 348, 693–699. [DOI] [PubMed] [Google Scholar]
- Aroian RV, Lesa GM, Sternberg PW (1994). Mutations in the Caenorhabditis elegans let-23 EGFR-like gene define elements important for cell-type specificity and function. EMBO J 13, 360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Sudhof TC (2000). Mints as adaptors. Direct binding to neurexins and recruitment of munc18. J Biol Chem 275, 39803–39806. [DOI] [PubMed] [Google Scholar]
- Borg JP, Straight SW, Kaech SM, De Taddeo-Borg M, Kroon DE, Karnak D, Turner RS, Kim SK, Margolis B (1998). Identification of an evolutionarily conserved heterotrimeric protein complex involved in protein targeting. J Biol Chem 273, 31633–31636. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz S, Okamoto M, Sudhof TC (1998). A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell 94, 773–782. [DOI] [PubMed] [Google Scholar]
- Cohen AR, Woods DF, Marfatia SM, Walther Z, Chishti AH, Anderson JM (1998). Human CASK/LIN-2 binds syndecan-2 and protein 4.1 and localizes to the basolateral membrane of epithelial cells. J Cell Biol 142, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Cassel D, Kahn RA, Klausner RD (1992a). ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein beta-COP to Golgi membranes. Proc Natl Acad Sci USA 89, 6408–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Finazzi D, Klausner RD (1992b). Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature 360, 350–352. [DOI] [PubMed] [Google Scholar]
- Fairless R, Masius H, Rohlmann A, Heupel K, Ahmad M, Reissner C, Dresbach T, Missler M (2008). Polarized targeting of neurexins to synapses is regulated by their C-terminal sequences. J Neurosci 28, 12969–12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Horvitz HR (1985). Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110, 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier K, Rocheleau CE (2017). C. elegans vulva induction: an in vivo model to study epidermal growth factor receptor signaling and trafficking. Methods Mol Biol 1652, 43–61. [DOI] [PubMed] [Google Scholar]
- Gauthier KD, Rocheleau CE (2021). Golgi localization of the LIN-2/7/10 complex points to a role in basolateral secretion of LET-23 EGFR in the Caenorhabditis elegans vulval precursor cells. Development, 148, dev194167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K, Kamiya Y, Yamakawa A, Kodaira K, Nishiwaki K, Miwa J, Hori I, Hosono R (1993). The C. elegans unc-18 gene encodes a protein expressed in motor neurons. Neuron 11, 703–711. [DOI] [PubMed] [Google Scholar]
- Glodowski DR, Wright T, Martinowich K, Chang HC, Beach D, Rongo C (2005). Distinct LIN-10 domains are required for its neuronal function, its epithelial function, and its synaptic localization. Mol Biol Cell 16, 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, Horvitz HR (1983). The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34, 435–444. [DOI] [PubMed] [Google Scholar]
- Haag A, Gutierrez P, Buhler A, Walser M, Yang Q, Langouet M, Kradolfer D, Frohli E, Herrmann CJ, Hajnal A, et al. (2014). An in vivo EGF receptor localization screen in C. elegans identifies the Ezrin homolog ERM-1 as a temporal regulator of signaling. PLoS Genet 10, e1004341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Aroian RV, Sternberg PW (1990). The let-60 locus controls the switch between vulval and nonvulval cell fates in Caenorhabditis elegans. Genetics 126, 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Li Y, Bennett M, Mckay M, Zhu X, Shern J, Torre E, Lah JJ, Levey AI, Kahn RA (2003). Munc18 interacting proteins: ADP-ribosylation factor-dependent coat proteins that regulate the traffic of beta-Alzheimer’s precursor protein. J Biol Chem 278, 36032–36040. [DOI] [PubMed] [Google Scholar]
- Hill RJ, Sternberg PW (1992). The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature 358, 470–476. [DOI] [PubMed] [Google Scholar]
- Jo K, Derin R, Li M, Bredt DS (1999). Characterization of MALS/Velis-1, -2, and -3: a family of mammalian LIN-7 homologs enriched at brain synapses in association with the postsynaptic density-95/NMDA receptor postsynaptic complex. J Neurosci 19, 4189–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Whitfield CW, Kim SK (1998). The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell 94, 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2, RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JF, Feng W, Wang R, Chan LN, Ip FC, Xia J, Ip NY, Zhang M (2005). Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem. Nat Struct Mol Biol 12, 722–728. [DOI] [PubMed] [Google Scholar]
- Niu TK, Pfeifer AC, Lippincott-Schwartz J, Jackson CL (2005). Dynamics of GBF1, a brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol Biol Cell 16, 1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Sudhof TC (1997). Mints, Munc18-interacting proteins in synaptic vesicle exocytosis. J Biol Chem 272, 31459–31464. [DOI] [PubMed] [Google Scholar]
- Perego C, Vanoni C, Massari S, Longhi R, Pietrini G (2000). Mammalian LIN-7 PDZ proteins associate with beta-catenin at the cell-cell junctions of epithelia and neurons. EMBO J 19, 3978–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego C, Vanoni C, Villa A, Longhi R, Kaech SM, Frohli E, Hajnal A, Kim SK, Pietrini G (1999). PDZ-mediated interactions retain the epithelial GABA transporter on the basolateral surface of polarized epithelial cells. EMBO J 18, 2384–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongo C, Whitfield CW, Rodal A, Kim SK, Kaplan JM (1998). LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell 94, 751–759. [DOI] [PubMed] [Google Scholar]
- Saito Y, Akiyama M, Araki Y, Sumioka A, Shiono M, Taru H, Nakaya T, Yamamoto T, Suzuki T (2011). Intracellular trafficking of the amyloid beta-protein precursor (APP) regulated by novel function of X11-like. PLoS One 6, e22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz D, Zilly F, Lang T, Jahn R, Bruns D (2005). A dual function for Munc-18 in exocytosis of PC12 cells. Eur J Neurosci 21, 2419–2432. [DOI] [PubMed] [Google Scholar]
- Sem X, Kreisberg JF, Kawli T, Tan MW, Rhen M, Tan P (2012). Modulation of Caenorhabditis elegans infection sensitivity by the LIN-7 cell junction protein. Cell Microbiol 14, 1584–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setou M, Nakagawa T, Seog DH, Hirokawa N (2000). Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science 288, 1796–1802. [DOI] [PubMed] [Google Scholar]
- Shelly M, Mosesson Y, Citri A, Lavi S, Zwang Y, Melamed-Book N, Aroeti B, Yarden Y (2003). Polar expression of ErbB-2/HER2 in epithelia. Bimodal regulation by Lin-7. Dev Cell 5, 475–486. [DOI] [PubMed] [Google Scholar]
- Shrivastava-Ranjan P, Faundez V, Fang G, Rees H, Lah JJ, Levey AI, Kahn RA (2008). Mint3/X11gamma is an ADP-ribosylation factor-dependent adaptor that regulates the traffic of the Alzheimer’s precursor protein from the trans-Golgi network. Mol Biol Cell 19, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simske JS, Kaech SM, Harp SA, Kim SK (1996). LET-23 receptor localization by the cell junction protein LIN-7 during C. elegans vulval induction. Cell 85, 195–204. [DOI] [PubMed] [Google Scholar]
- Skorobogata O, Escobar-Restrepo JM, Rocheleau CE (2014). An AGEF-1/Arf GTPase/AP-1 ensemble antagonizes LET-23 EGFR basolateral localization and signaling during C. elegans vulva induction. PLoS Genet 10, e1004728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC (1997). Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275, 73–77. [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Horvitz HR (1986). Pattern formation during vulval development in C. elegans. Cell 44, 761–772. [DOI] [PubMed] [Google Scholar]
- Stetak A, Hoier EF, Croce A, Cassata G, Di Fiore PP, Hajnal A (2006). Cell fate-specific regulation of EGF receptor trafficking during Caenorhabditis elegans vulval development. EMBO J 25, 2347–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight SW, Chen L, Karnak D, Margolis B (2001). Interaction with mLin-7 alters the targeting of endocytosed transmembrane proteins in mammalian epithelial cells. Mol Biol Cell 12, 1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight SW, Karnak D, Borg JP, Kamberov E, Dare H, Margolis B, Wade JB (2000). mLin-7 is localized to the basolateral surface of renal epithelia via its NH(2) terminus. Am J Physiol Renal Physiol 278, F464–F475. [DOI] [PubMed] [Google Scholar]
- Straight SW, Pieczynski JN, Whiteman EL, Liu CJ, Margolis B (2006). Mammalian lin-7 stabilizes polarity protein complexes. J Biol Chem 281, 37738–37747. [DOI] [PubMed] [Google Scholar]
- Swistowski A, Zhang Q, Orcholski ME, Crippen D, Vitelli C, Kurakin A, Bredesen DE (2009). Novel mediators of amyloid precursor protein signaling. J Neurosci 29, 15703–15712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabish M, Siddiqui ZK, Nishikawa K, Siddiqui SS (1995). Exclusive expression of C. elegans osm-3 kinesin gene in chemosensory neurons open to the external environment. J Mol Biol 247, 377–389. [DOI] [PubMed] [Google Scholar]
- Togawa A, Morinaga N, Ogasawara M, Moss J, Vaughan M (1999). Purification and cloning of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. J Biol Chem 274, 12308–12315. [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M (2000). Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287, 116–122. [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Benard C, Barnes T, Hekimi S, Kim SK (1999). Basolateral localization of the Caenorhabditis elegans epidermal growth factor receptor in epithelial cells by the PDZ protein LIN-10. Mol Biol Cell 10, 2087–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zand TP, Reiner DJ, Der CJ (2011). Ras effector switching promotes divergent cell fates in C. elegans vulval patterning. Dev Cell 20, 84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Isack NR, Glodowski DR, Liu J, Chen CC, Xu XZ, Grant BD, Rongo C (2012). RAB-6.2 and the retromer regulate glutamate receptor recycling through a retrograde pathway. J Cell Biol 196, 85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.