Abstract
The fission yeast cells Schizosaccharomyces pombe divide at constant cell size regulated by environmental stimuli. An important pathway of cell size control involves the membrane-associated DYRK-family kinase Pom1, which forms decreasing concentration gradients from cell poles and inhibits mitotic inducers at midcell. Here, we identify the phosphatase 2C Ptc1 as negative regulator of Pom1. Ptc1 localizes to cell poles in a manner dependent on polarity and cell-wall integrity factors. We show that Ptc1 directly binds Pom1 and can dephosphorylate it in vitro but modulates Pom1 localization indirectly upon growth in low-glucose conditions by influencing microtubule stability. Thus, Ptc1 phosphatase plays both direct and indirect roles in the Pom1 cell size control pathway.
INTRODUCTION
Cells coordinate growth and division to maintain cell size over generations and to adapt it to environmental conditions. The rod-shaped fission yeast Schizosaccharomyces pombe is widely used to dissect the molecular mechanisms of cell size control. This organism grows in length by tip extension during interphase, stops growth, and divides by medial fission at a constant cell size influenced by nutrients. At the core of the decision to divide is the cyclin-dependent kinase CDK1, which drives the G2→M phase transition. CDK1 activity is directly inhibited by Wee1 kinase (Russell and Nurse, 1987; Gould and Nurse, 1989; Featherstone and Russell, 1991; Lundgren et al., 1991) and activated by Cdc25 phosphatase (Russell and Nurse, 1986; Gautier et al., 1991; Kumagai and Dunphy, 1991) and the balance between these activities controls division timing (Nurse, 1990; Martin, 2009; Wood and Nurse, 2015).
Over the past decade, it has become appreciated that Wee1 kinase integrates information from a cell geometry-sensing signaling cascade into the decision to divide. In this pathway, the DYRK-family Pom1 kinase forms plasma membrane–associated gradients with highest concentration at cell poles and serves to inhibit the SAD-family Cdr2 kinase at midcell (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). Mechanistically, Pom1 directly phosphorylates Cdr2 C-terminal tail (Bhatia et al., 2014; Deng et al., 2014), which prevents Cdr2 activation by the cytosolic Ssp1 CaMKK (Deng et al., 2014). At midcell, Cdr2 forms stable megadalton-large membrane-associated clusters, called nodes, which also contain a second SAD-family kinase Cdr1 that phosphorylates Wee1 to inhibit its activity (Young and Fantes, 1987; Breeding et al., 1998; Kanoh and Russell, 1998; Opalko et al., 2019). Wee1 phosphorylation likely occurs during its transient visits of the nodes (Allard et al., 2018).
The spatial organization of the Pom1-Cdr2 pathway is thought to help monitor a geometric feature of cell size, coordinating cell growth with mitotic entry during the vegetative cell cycle. Recent work proposed that this feature is cell surface area (Pan et al., 2014; Facchetti et al., 2019), consistent with the membrane-associated localization of all components of the pathway. As Pom1 levels at midcell diminish and Wee1 visits increase as cells grow, the pathway is well positioned to act as cell size sensor by linking Wee1 inhibition with cell dimensions for size homeostasis (Allard et al., 2018; Gerganova and Martin, 2018; Gerganova et al., 2019). However, as both pom1 and cdr2 mutants retain homeostatic capacity, this pathway may function in parallel to other homeostatic systems (Wood and Nurse, 2013; Facchetti et al., 2019). The pathway is also sensitive to changes in nutritional status. Specifically, Pom1 graded localization is altered when cells encounter a low-glucose environment (Kelkar and Martin, 2015; Allard et al., 2019).
Pom1 distribution at the plasma membrane is controlled by a cycle of autophosphorylation and localized dephosphorylation (Hachet et al., 2011). Dephosphorylation of a series of autophosphorylation sites in the Pom1 cortical-binding domain by the Tea4-associated PP1 Dis2 phosphatase promotes association of Pom1 at the plasma membrane (Gerganova et al., 2019). This dephosphorylation normally takes place at cell poles, where Tea4 and its binding partner Tea1 are deposited by microtubules (Martin and Arkowitz, 2014; Chiou et al., 2017). Tea1 and Tea4 are further tethered at the membrane through direct interaction of Tea1 with the prenylated Mod5 protein (Snaith and Sawin, 2003; Martin et al., 2005), which promotes the formation of a Tea1-Mod5 polymeric network (Bicho et al., 2010). At the plasma membrane, Pom1 autophosphorylates to promote its return to the cytosol. It also forms cortical clusters that support its graded distribution from cell poles to midcell (Allard et al., 2019; Gerganova et al., 2019). These clusters, together with autophosphorylation-mediated detachment, promote robustness of the gradient (Saunders et al., 2012; Hersch et al., 2015).
Apart from the Tea4-PP1 phosphatase that controls Pom1 membrane association, no phosphatase has been shown to regulate the Pom1-Cdr2-Cdr1-Wee1 kinase cascade. To uncover potential regulatory phosphatases of the Pom1-Cdr2 network, we conducted a minimal screen for phosphatase deletions with altered cell length at division. In this work, we identify the protein phosphatase 2C (PP2C) Ptc1 as a novel interactor of Pom1 and Mod5. PP2C phosphatases are usually monomeric metal-dependent serine/threonine-specific protein phosphatases involved in the regulation of cell growth and stress signaling (Shi, 2009). In fungi, PP2C phosphatases have been strongly implicated as negative regulators of all MAPK signaling cascades (Arino et al., 2011). In fission yeast, Ptc1 negatively regulates the stress-signaling MAPK Sty1 after heat and osmotic stress through both direct dephosphorylation and indirect downstream effects (Shiozaki et al., 1994; Shiozaki and Russell, 1995; Gaits et al., 1997; Nguyen and Shiozaki, 1999). Ptc1 also acts as negative regulator of the Pmk1 cell integrity MAPK pathway (Takada et al., 2007). Ptc1 localizes to cell poles in a manner partly dependent on the SH3 domain protein Skb5, a negative regulator of the cell integrity MAPK pathway (Stanger et al., 2012; Kanda et al., 2016). Ptc1 has also recently been involved in the dephosphorylation of AMPK to regulate TOR activity in response to nitrogen limitation (Davie et al., 2015).
Here, we report that Ptc1 controls cell size at division through the Pom1 pathway. We show that Ptc1 localization at cell tips depends on the polarity protein Mod5 in addition to Skb5. We also report that Ptc1 moderates the previously described Pom1 gradient redistribution in low-glucose conditions (Kelkar and Martin, 2015). Because Ptc1 directly binds Pom1 but exerts effects on Pom1 distribution by influencing microtubule stability and Tea4 deposition, Ptc1 likely plays both direct and indirect roles in modulating the Pom1 pathway.
RESULTS
Ptc1 affects cell size at division and localizes to the cell cortex
To search for phosphatases that may regulate the Pom1-Cdr2 network, we conducted a limited screen through phosphatase deletion strains, searching for those with altered cell length at division. Because fission yeast cells grow only during interphase, cell length at division serves as a proxy for cell cycle length. Several phosphatase deletions showed altered size at division (Supplemental Table S1). Here, we focus on the protein phosphatase 2C Ptc1 because its deletion and overexpression showed opposite effects on cell size at division (see below). As ptc1 is known to be important for heat-shock response (Shiozaki et al., 1994), we first investigated whether growth of ptc1∆ cells was affected at different temperatures. ptc1∆ cells behaved similarly to wild-type cells at both high and low temperatures, as assessed by colony size formation in spot assays (Supplemental Figure S1A). In this study, we did not investigate the effect of temperature and performed all experiments at 30°C, unless stated otherwise, to probe the role of Ptc1 in cell size regulation.
Ptc1 localization to the cell cortex depends on Skb5 and Mod5
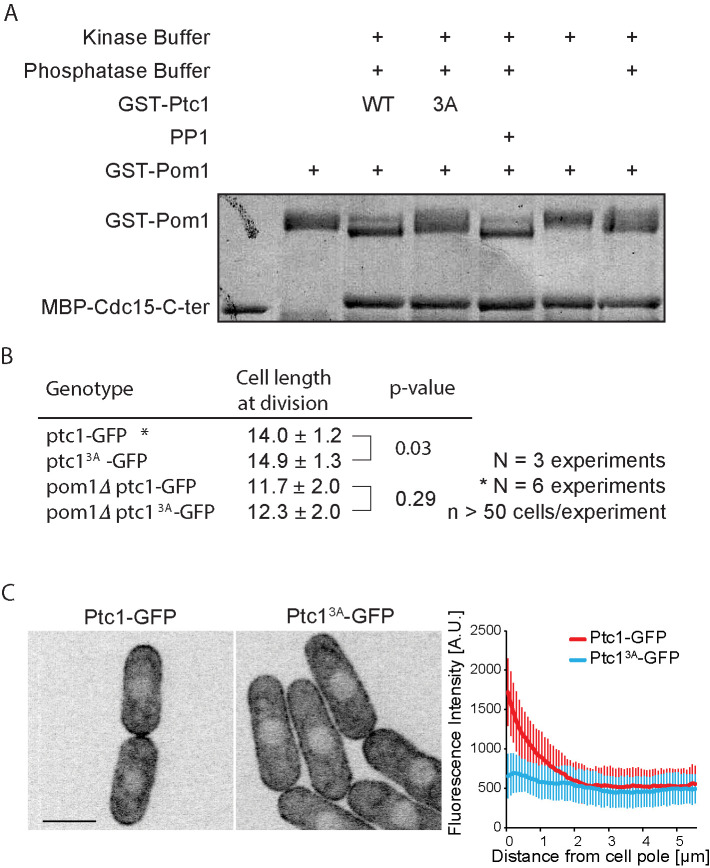
Previous work suggested that Ptc1 localizes to the cell poles as well as in the cytoplasm and cell nucleus (Kanda et al., 2016). We confirmed these observations by tagging of ptc1 at the endogenous locus with GFP, which yielded an apparently functional protein, as assessed by cell length measurements (Supplemental Table S1; also see Figure 3B later in the paper). Ptc1-GFP was indeed observed at the cell cortex and at the septum of dividing cells, as well as in the cytoplasm and weakly in the nucleus (Figure 1A and Supplemental Movie 1). Quantitative measurement of Ptc1-GFP cortical signal revealed higher intensities at the cell tips than at cell middle (Figure 1A, right panel), indicating a shallow gradient-like pattern.
FIGURE 3:
Ptc1 dephosphorylates Pom1 in vitro and its activity contributes to its localization. (A) In vitro phosphatase assay, using recombinant proteins. Autophosphorylated recombinant GST-Pom1 was used as substrate and incubated with recombinant GST-Ptc1 (WT or 3A mutant) or commercial PP1 as indicated. Autophosphorylated Pom1 migrates as a smear, which collapses to two fast-migrating bands upon dephosphorylation. Phosphatase-treated Pom1 was then used in a kinase assay to monitor Pom1 activity. Recombinant MBP-Cdc15C was used as Pom1 substrate, as this fragment is quantitatively phosphorylated by Pom1 leading to slower migration (Bhattacharjee et al., 2020). Note that the phosphorylation status of Pom1 does not alter its activity toward Cdc15C. (B) Cell lengths at division quantified from more than 160 cells in three independent experiments; statistical significance measured by Student’s t test against wild type or pom1∆. (C) Localization of Ptc1-GFP and Ptc13A-GFP (left) and quantification of cortical profiles (right). Scale bar: 5 µm.
FIGURE 1:
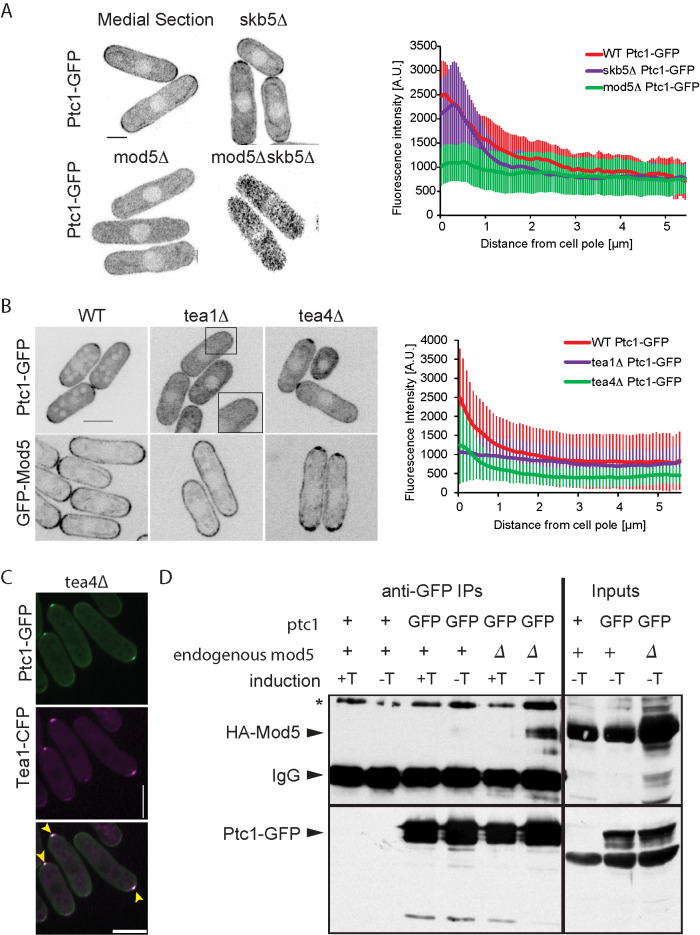
Ptc1 localizes to the cell cortex. (A) Localization of Ptc1-GFP in wild-type, skb5Δ, mod5Δ, and mod5Δskb5Δ fission yeast cells. Left, a medial section of Airyscan imaging acquired with identical settings. Contrasting was increased postacquisition on the mod5Δskb5Δ image. Right, Average cortical profile of Ptc1-GFP in wild-type cells (n = 20 cells). Error bars are standard deviations. (B) Ptc1-GFP and GFP-Mod5 localization in tea1Δ and tea4Δ. Left, a medial section of spinning-disk imaging acquired with identical settings. Right, average cortical profile of Ptc1-GFP in wild-type, tea1∆, and tea4∆ cells (n = 20 cells). Error bars are standard deviations. (C) Spinning-disk imaging of colocalization of Tea1-CFP and Ptc1-GFP (acquired in the YFP channel) in tea4∆ cells. Yellow arrowheads indicate colocalizing Tea1 and Ptc1 dots. False color image of Tea1-CFP. (D) Coimmunoprecipitation of HA-Mod5 and Ptc1-GFP from mod5∆ cells expressing HA-Mod5 on a plasmid under the inducible Pnmt1 promoter. HA-Mod5 induction was performed by growing cells in absence of thiamine (−T). Ptc1-GFP was immunoprecipitated using anti-GFP antibody and coimmunoprecipitation of Mod5 was revealed using anti-HA antibody. Note that coimmunoprecipitation of HA-Mod5 was more marked in absence of endogenous untagged Mod5. IgG serves as the immunoprecipitation control. Inputs are shown on the right. + symbols indicate untagged wild-type Ptc1 or Pom1. Images are sum projections of five confocal images acquired over time. Scale bars: 3 µm in A; 5 µm in B and C.
Movie S1.
Movie S1.
To uncover factors responsible for Ptc1 localization, we first investigated the possible role of Skb5, previously shown to bind Ptc1 (Stanger et al., 2012; Kanda et al., 2016). Skb5 is an SH3 domain protein, named after its Shk1 kinase binding function (Yang et al., 1999), which was shown to regulate cell integrity Pmk1 MAPK signaling by controlling the localization of the MAPKKK Mkh1 (Kanda et al., 2016). Skb5-GFP itself weakly localizes to cell tips (Supplemental Figure S2A; Kanda et al., 2016) when expressed from its endogenous locus. In skb5Δ cells the Ptc1 signal could still be detected at cell tips, but formed a narrower zone comprised of nonuniform dispersed individual clusters, rather than a smooth signal localization across the entire tip (Figure 1A). Due to this particular localization, the quantified Ptc1 cortical profile in this mutant (Figure 1A, right) does not start at a maximum. Thus, Skb5 is one, but not the only, contributor to Ptc1 localization to cell tips.
We investigated other possible determinants of Ptc1 cortical localization. Ptc1 localization was not affected by acute depolymerization of the actin or microtubule cytoskeleton (Supplemental Figure S2B). By contrast, the loss of polarity landmarks Tea1 (Mata and Nurse, 1997), Tea4 (Martin et al., 2005; Tatebe et al., 2005), or Mod5 (Snaith and Sawin, 2003) altered Ptc1 localization (Figure 1, A and B). Deletion of Tea3 (Arellano et al., 2002) or Pom1 had minor effects (Supplemental Figure S2C). Deletion of Bud6 (Glynn et al., 2001) produced no detectable change in Ptc1 distribution (Supplemental Figure S2C). Several observations pointed to Mod5 acting as a direct Ptc1 link to the membrane. First, deletion of mod5 had the strongest effect on Ptc1 localization (Figure 1A): in mod5∆ Ptc1-GFP localization ranged from completely cytosolic to a faint uniform cortical signal in some cells. Second, Ptc1-GFP was spread more evenly around the cell cortex in tea1∆ cells, in which Mod5 is also not restricted to cell poles (Figure 1B; Snaith and Sawin, 2003). Third, Ptc1-GFP formed a dot-like structure at the nongrowing end of tea4∆ cells, where Tea1 and Mod5 accumulate (Figure 1B; Martin et al., 2005). Indeed, in tea4∆ cells Ptc1-GFP colocalized with Tea1 (Figure 1C). Furthermore, the Ptc1-GFP dot was completely lost upon additional deletion of mod5 (Supplemental Figure S2C). Conversely, ptc1∆ did not affect the localization of Tea1, Tea4, or Mod5 (Supplemental Figure S2D). Thus, the prenylated protein Mod5 may serve as an anchor for Ptc1 at the cell cortex, similar to its role in anchoring Tea1 (Snaith and Sawin, 2003).
To test whether Mod5 associates with Ptc1, we performed coimmunoprecipitation experiments, using cells expressing HA-Mod5 from an inducible plasmid and Ptc1-GFP from the endogenous locus. Mod5 coimmunoprecipitated with Ptc1 from cell extracts (Figure 1D). We conclude that Mod5 forms a complex with Ptc1 and can dictate its binding at the cell cortex additionally to Skb5.
Finally, because Skb5 and Mod5 are two Ptc1 interactors that control its localization, we probed the relationship between these two proteins. In skb5Δ mod5Δ double mutant cells, Ptc1 was fully cytosolic in interphase cells (Figure 1A), suggesting that each interacting protein contributes to the residual cortical localization observed in single mutant cells. We note that in dividing cells Ptc1 retained the ability to localize at the septum, indicating the existence of at least a third interactor (Supplemental Figure S2C). Mod5 and Skb5 do not play a role in each other’s localization, as no major changes of distribution of GFP-Mod5 were observed in skb5Δ and vice versa (Supplemental Figure S2A). These data establish that Ptc1 localizes to cell poles through interaction with two independent proteins, Skb5 and Mod5.
Ptc1 regulates cell size through Pom1 kinase
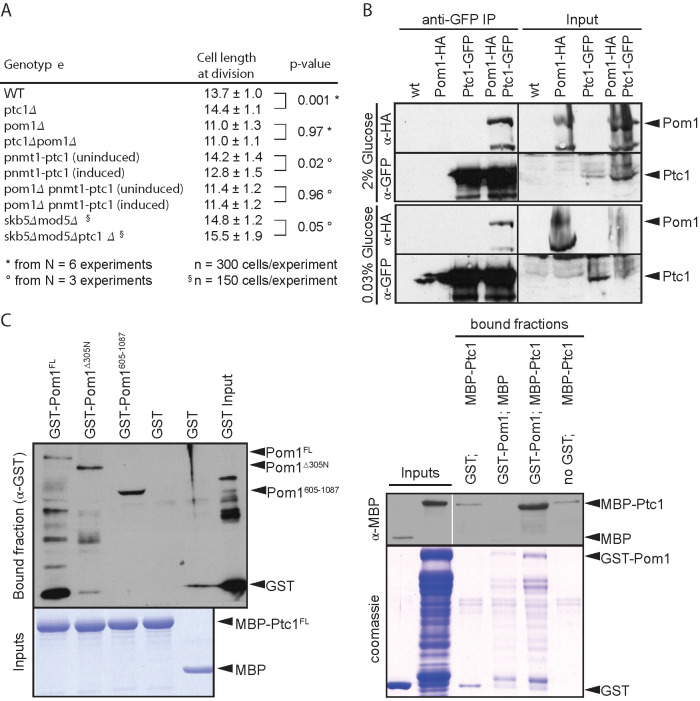
The slightly longer size at division of ptc1∆ cells and the cortical localization of Ptc1 prompted us to further probe its role in cell size regulation in relation with the cortical kinase Pom1. In exponentially growing ptc1∆ cells, calcofluor-staining revealed a slight yet significant increase of cell length at division (Figure 2A). Other phenotypes such as septum positioning and polarized growth were not significantly different from the wild type (Supplemental Figure S1B). Conversely, overexpression of ptc1 from the inducible pnmt1 promoter integrated at the endogenous locus led to a significant reduction in cell length at division (Figure 2A), consistent with previous data (Gaits et al., 1997). Interestingly, ptc1 deletion and overexpression had no effect on cell size at division in pom1∆ cells (Figure 2A). Therefore, pom1∆ is epistatic to ptc1∆, suggesting that Ptc1 regulates cell size at division through Pom1. These data establish the phosphatase Ptc1 as a novel regulator of the Pom1 pathway in fission yeast cells.
FIGURE 2:
Ptc1 localization to the plasma membrane is dependent on Skb5 and Mod5. (A) Cell lengths at division quantified from at least three independent experiments; statistical significance measured by Student’s t test across experiments against either wild-type or uninduced counterpart. The indicated variation is the SD across cells. (B) Coimmunoprecipitation of Ptc1-GFP and Pom1-HA. Protein extracts were made from cells grown in 2% glucose for 24 h and from cells shifted to 0.03% glucose for 2 h. Whole cell extract (2 mg) was used for immunoprecipitation assay. Inputs (100 µg) were loaded. Note that the levels of Pom1-HA detected in the inputs are variable across experiments. (C) Pom1 binds Ptc1 in vitro. Bacterially expressed proteins assayed by affinity columns. GST tagged Pom1FL and fragments Pom1Δaa305N and Pom1aa605-1087 tested for interaction with recombinant MBP-tagged Ptc1FL. Left, MBP-bound proteins were immobilized on amylose columns. Right, GST-bound proteins were bound to glutathione beads.
We also tested whether Ptc1 localization at cell poles was important for its role in regulating cell size at division. We found that mod5∆ skb5∆ double mutant cells, in which Ptc1 is cytosolic, were slightly longer than wild-type cells, suggesting that cytosolic Ptc1 may be less efficient at regulating the Pom1 pathway. However, additional deletion of ptc1 in these cells, led to even longer cells at division, indicating that Ptc1 still influences cell size from a cytosolic localization.
Consistent with the idea that Ptc1 regulates Pom1, we found that the two proteins form a complex both in vivo and in vitro. In vivo, Pom1-HA coimmunoprecipitated with Ptc1-GFP in several growth conditions (Figure 2B). In vitro, we found that recombinant GST-Pom1 directly binds recombinant MBP-Ptc1, whether MBP-Ptc1 was coupled to amylose resin (Figure 2C, left) or GST-Pom1 was coupled to glutathione beads (Figure 2C, right). This interaction was independent of Pom1 N-terminal membrane-binding region. Thus, Ptc1 phosphatase and Pom1 kinase bind directly to each other and Ptc1 modulates cell length at division in a manner dependent on Pom1.
Ptc1 activity is required for function and localization
We investigated the role of Ptc1’s phosphatase activity. To verify whether Ptc1 works as an active phosphatase in vitro, we purified GST-Ptc1 from bacterial cells and used it in an in vitro phosphatase assay with recombinant, autophosphorylated Pom1 as substrate. Recombinant Pom1 runs as a smear because of autophosphorylation and shows a clear shift in mobility upon dephosphorylation by the phosphatase PP1 (Hachet et al., 2011). Ptc1 induced a similar shift in Pom1 mobility (Figure 3A), suggesting that Ptc1 is active as a phosphatase in vitro. Through sequence alignment with Ptc1 homologue in Saccharomyces cerevisiae, we first identified four amino acids (aa; D109, G110, D275, and D314) within the phosphatase domain of fission yeast Ptc1 predicted to be involved in divalent metal ion binding and therefore important for catalytic activity. Mutation of one or several aa identified above, yielding Ptc11A (D275A) and Ptc13A (D109A/G110A/D314A) prevented the shift in mobility (Figure 3A and Supplemental Figure S3) confirming the phosphatase activity of Ptc1. We note that Pom1 activity toward its substrate Cdc15 (Bhattacharjee et al., 2020) was not dramatically affected by dephosphorylation, though we cannot exclude subtle changes. The Ptc13A catalytic-dead mutant was used further in the study.
To test the role of phosphatase activity in vivo, we introduced a ptc13A-GFP allele as a sole copy of ptc1 at the endogenous ptc1 locus. ptc13A-GFP cells phenocopied ptc1∆ mutants: these cells were slightly, but significantly, longer at division than wild-type cells, and pom1∆ was epistatic over this phenotype (Figure 3B). Interestingly, Ptc13A-GFP failed to be enriched at cell poles and instead spread all along the cortex (Figure 3C). Therefore, the catalytic activity of Ptc1 is critical for its function in cell size control as well as for its cell tip enrichment.
Localization and function of Ptc1 in glucose-limited conditions
Ptc1 is involved in the regulation of MAPK pathway in fission yeast under different kinds of environmental stresses (Shiozaki et al., 1994; Shiozaki and Russell, 1995; Gaits et al., 1997; Nguyen and Shiozaki, 1999; Takada et al., 2007). To further understand its role in cell size regulation, we tested whether Ptc1 itself is affected under stress, and whether it is involved in cell size regulation under these conditions. We first tested the localization of Ptc1 in cells subjected to osmostress (sorbitol and KCl), nutritional stress (lack of nitrogen and low-glucose conditions), and high temperature. Under all conditions except low glucose, Ptc1 remained well localized at the cortex (Supplemental Figure S4, A and B). In low glucose (0.08% glucose) the Ptc1 gradient flattened around the cell cortex (Figure 4A). Ptc1-GFP intensity measurements around the cell cortex indeed revealed a clear spread of signal from cell tips to the cell middle in 0.08% glucose growth conditions.
FIGURE 4:
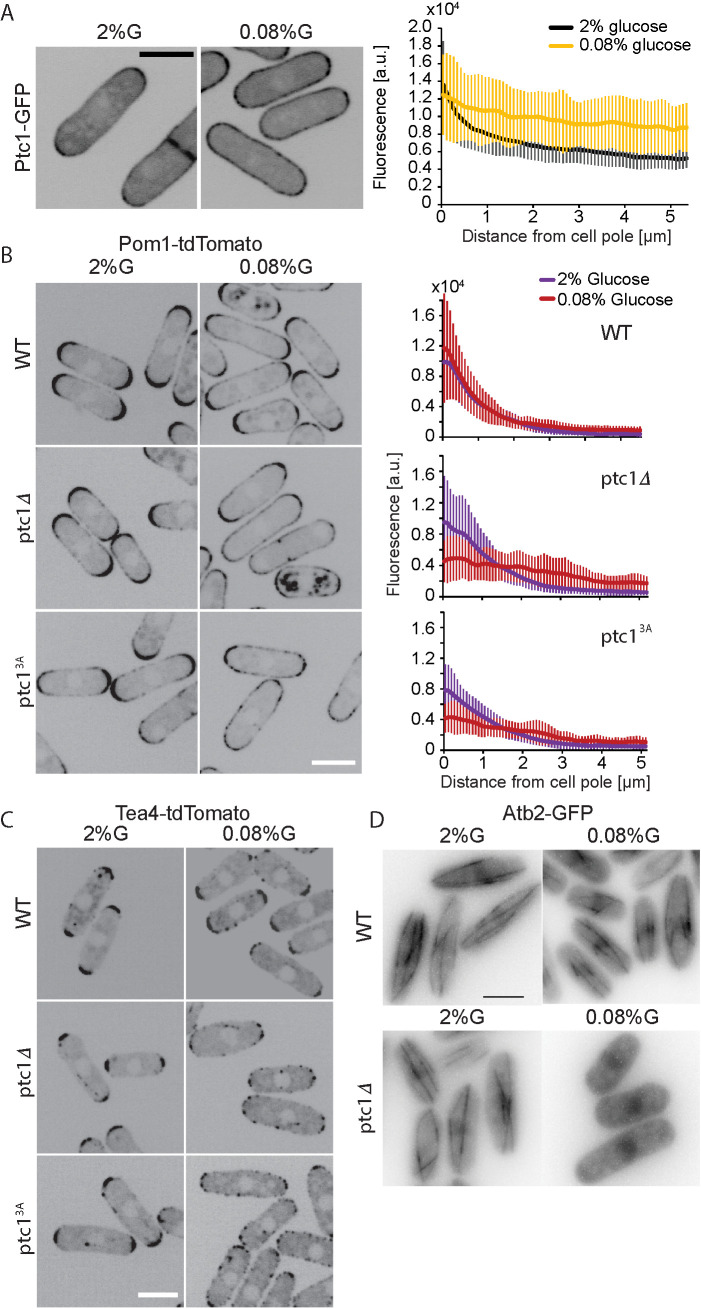
Ptc1 keeps Pom1 from spreading to the cell sides in limited glucose conditions. (A) Localization of Ptc1-GFP in EMM-ALU 2% glucose and 0.08% glucose. Graph on right shows average profiles of wild-type Ptc1-GFP from n = 20 cells. Individual profiles are shown in Supplemental Figure S4B. Error bars are standard deviations. (B) Localization of Pom1-tdTomato in wild type, ptc1∆, and ptc13A-GFP (GFP not shown). Graphs on the right show average fluorescence profiles of Pom1-tdTomato obtained from individual cells (n = 20). Error bars are standard deviations. (C) Localization of Tea4-tdTomato in wild-type, ptc1∆, and ptc13A-GFP backgrounds (GFP not shown). (D) Atb2-GFP signal in wild-type and ptc1∆ backgrounds. Widefield microscopy. The indicated time is the exact time point during the imaging interval. In all panels, cells were grown in 2% glucose (G) for 24 h and shifted to 0.08% glucose for 1 h before imaging. In A–C, images are sum projections of five confocal images acquired over time. In D, the images shown were taken 1 h 30 min (top) and 1 h 10 min (bottom) after transfer to 0.08% glucose. Scale bars: 5 µm.
Because of the epistasis of pom1∆ over ptc1∆ and because Pom1 localization is itself altered in low-glucose conditions, we tested whether loss or inactivation of Ptc1 affected Pom1 localization. Indeed, our previous work had shown that complete glucose starvation (≤0.03%) causes a uniform redistribution of Pom1 around the cell cortex, whereas limiting glucose conditions (0.08%) causes a much milder redistribution of Pom1 to the cell middle (Kelkar and Martin, 2015). While loss of Ptc1 or its activity had no major effect on Pom1 localization in 2% glucose (Figure 4B), it had a significant effect in 0.08% glucose: in ptc1∆ and ptc13A cells, Pom1 nearly completely lost its gradient shape and displayed in an almost flat profile around the cortex as compared with wild type (Figure 4B, right top panel). Thus, loss of Ptc1 function exacerbates the redistribution of Pom1 in low-glucose conditions.
Pom1 localization to the cell cortex depends on local dephosphorylation by the Tea4-Dis2 PP1 complex. In glucose-rich conditions, Tea4 is deposited by microtubules at cell poles, where it nucleates the formation of Pom1 concentration gradients (Hachet et al., 2011). In glucose-poor conditions, destabilization of microtubules (MTs) leads to Tea4 delivery at the cell sides, resulting in Pom1 relocalization around the cell periphery (Kelkar and Martin, 2015). We thus asked whether Ptc1 controls Tea4 deposition and/or microtubule organization. In ptc1∆ and ptc13A-GFP, Tea4 localized correctly at cell poles in 2% glucose. By contrast, in glucose-limited conditions (0.08%), Tea4 was strongly delocalized from the poles with Tea4 dots spread around the cortex (Figure 4C). We made similar observations regarding microtubules: in 2% glucose no major differences in microtubule dynamics between the wild type and the mutant were observed, but exposure of ptc1Δ cells to 0.08% glucose conditions destabilized microtubules. Indeed, we found that, 30 min to 1 h after transfer to 0.08% glucose, microtubules were depolymerized in ptc1∆ cells, but not in wild-type cells (Figure 4D). In summary, these data suggest that destabilization of MTs in ptc1Δ cells leads to aberrant Tea4 deposition at cell sides and subsequent loss of the Pom1 gradient shape.
DISCUSSION
In this work, we present an initial dissection of the mode of localization and action of the PP2C phosphatase Ptc1 in regulating cell size. In agreement with previous data (Gaits et al., 1997), we have found that Ptc1 promotes division at a shorter cell size, as shown by the extended length at division of cells carrying ptc1-deletion and -inactive alleles and the shorter length of cells overexpressing it. Our genetic data indicate that it does so through Pom1. What may be the mechanism?
A first simple scenario would be that Ptc1 serves to reverse Pom1 phosphorylation. This possibility is evoked from our discovery that Ptc1 forms complexes with Pom1 in vivo and directly binds and dephosphorylates it in vitro. In previous work, we showed that Pom1 gradients are patterned by a cycle of phosphorylation and dephosphorylation, where dephosphorylation of Pom1 by the Tea4-associated PP1 phosphatase within Pom1’s membrane-binding region promotes membrane attachment, and autophosphorylation reduces membrane binding (Hachet et al., 2011; Gerganova et al., 2019). If Ptc1 was acting to dephosphorylate these sites, one would expect that in ptc1∆ cells Pom1 exhibits higher phosphorylation and reduced membrane binding, as in tea4∆ cells. However, our observations are not consistent with this prediction: in glucose-rich conditions, Pom1 distribution was not significantly altered, and in glucose-limiting conditions Pom1 showed an opposite phenotype, decorating the entire cell cortex. We conclude that Ptc1 is highly unlikely to dephosphorylate Pom1 within the cortex-binding region. As Pom1 autophosphorylates on nearly 40 other serines and threonines (Hachet et al., 2011), one possibility is that Ptc1 reverses other phosphorylation events. The opposite cell size phenotypes of ptc1∆ and pom1∆ predicts that dephosphorylation by Ptc1 may serve to reduce Pom1 activity.
A second scenario is that Ptc1 controls Pom1 indirectly. Although Ptc1 likely does not modulate Pom1 localization through dephosphorylation of the Pom1 cortex-binding domain, Ptc1 clearly influences Pom1 distribution. Indeed, in glucose-limiting conditions, where Pom1 gradient distribution is less robust due to destabilization of microtubules, deletion of ptc1 caused a strong redistribution of Pom1 around the entire cell cortex. We hypothesize that this relocalization reflects an indirect function of Ptc1 on Pom1 distribution. Our previous work had shown that microtubule destabilization in glucose-limited conditions leads to deposition of Tea4 and consequent Pom1 redistribution at cell sides (Kelkar and Martin, 2015). The microtubule destabilization relies on PKA inhibiting CLASP-dependent microtubule rescue. We have now observed that ptc1∆ exacerbates the microtubule destabilization phenotype, leading to Pom1 redistribution. While additional work will be required to understand how Ptc1 exerts this function, it may, for instance, counteract PKA-dependent phosphorylation.
The observation that Ptc1 itself localizes at the plasma membrane suggests this may be its place of action to modulate the Pom1-Cdr2 geometry-sensing cell size pathway. Ptc1 is somewhat enriched at cell poles dependent on several factors. We found that Ptc1 recruitment to the cell cortex requires two proteins that Ptc1 associates with: the prenylated protein Mod5 and the SH3-domain protein Skb5 (this work and Kanda et al., 2016). Ptc1 may thus integrate information from the polarity/microtubule machinery through Mod5 and from the cell-wall/growth status through the cell integrity pathway-dependent Skb5 protein. Ptc1’s restriction to cell poles also depends on its own activity. These observations raise the question of the importance of Ptc1 localization for its cell size–modulatory function. We can speculate that the increased amounts of Ptc1 at cell poles may skew stabilization of microtubules toward cell poles, thus favoring the delivery of Tea4 at these locations. Similarly, they may help delay Pom1 autophosphorylation that promotes its detachment by lowering Pom1 activity at these locations. However, the finding that deletion of ptc1∆ led to cells of increased length also in a skb5∆ mod5∆ mutant background, in which Ptc1 is cytosolic, indicates that Ptc1 can exert its function in cell size regulation from the cytosol, similar to the CaMKK kinase Ssp1 acting on Cdr2 (Deng et al., 2014). The identification of Ptc1 as a first phosphatase regulating Pom1 adds to a growing number of regulatory factors modulating the Pom1-Cdr2 geometry-sensing cell size pathway, such as, for instance, the methyl transferase Skb1 (Deng and Moseley, 2013). These regulators, themselves important modulators of stress-signaling cascades, may help cells adapt their size to environmental stimuli.
MATERIALS AND METHODS
Yeast genetics and culture
Schizosaccharomyces pombe strains obtained from genetic crosses were selected by tetrad dissection and replica plating with appropriate supplements or drugs. All transformations were performed using the lithium acetate-DMSO (dimethyl sulfoxide) method. Tagged and deletion strains were constructed by using a PCR-based approach (Bähler et al., 1998) and confirmed by PCR. Ptc1 overexpressing strains were obtained by introducing Pnmt1 promoter upstream of ptc1 at its endogenous locus using a PCR-based approach. All strains were genotyped using PCR and are listed in Supplemental Table S2.
Cells were grown in standard Edinburgh minimal media (EMM) with appropriate supplements adenine, leucine, and uracil when required. For glucose limitation assays, cells were grown in 2% glucose to midlog phase, washed three times with EMM 0.08% glucose, and incubated in the same medium before imaging. For cell size experiments, cells were grown in either EMM 2% glucose or EMM 0.08% glucose for 24 h before imaging.
For expression of HA-Mod5, yeast cells were transformed with pREP41-HA-Mod5 plasmid and selected on EMM supplemented with adenine and uracil. Cells were grown in EMM adenine, uracil, and with or without thiamine (5 µg/ml) at 30°C for 24 h before pelleting down for Western blot or immunoprecipitation experiments.
For overexpression from nmt1 promoters, cells were grown for 24 h at 30°C in EMM supplemented with adenine, uracil, leucine, and with or without thiamine (15 µM).
For cell size measurement, calcofluor (Sigma) was added at a final concentration of 5 μg/ml from a 200X stock solution.
Plasmid construction
For GST-Ptc1, Ptc1 amplified from gDNA was cloned into pGEX-4T-1 GST vector between EcoR1 and Xho1 sites. Mutant GST-Ptc1 plasmids were generated by site-directed mutagenesis on GST-Ptc1 plasmid. Mod5 was cloned in a pREP41-HA (N) vector between Sal1 and BamH1 sites for the production of HA-Mod5. All plasmids constructed were confirmed by sequencing.
Microscopy
Images on live cells were acquired at room temperature, except stated otherwise, on Airyscan, a spinning-disk confocal microscope or on a DeltaVision epifluorescence microscope. AiryScan images in Figure 1A and Supplemental Figure S2A were acquired on a Zeiss LSM 980 system with Plan-Apochromat 63×/1.40 oil DIC objective and acquired by the ZEN Blue software (Zeiss). Imaging was set in superresolution mode with frame unidirectional maximum speed scanning. Laser power was kept at 0.4%, gain 850 V, with pixel time of 6.6 µs and frame acquisition time 3.89 s. All other settings were optimized as recommended by the ZEN Blue software. Confocal images were acquired using a Perkin Elmer Leica DMI4000B inverted microscope equipped with an HCX PL APO 100×/1.46 numerical aperture (NA) oil objective and a PerkinElmer Volocity Confocal system spinning-disk microscope including a Yokagawa CSU22 real-time confocal scanning head, a solid-state laser line and a cooled 14-bit frame transfer EMCCD C9100-50 camera, as described (Bendezu et al., 2013). Stacks of z-series confocal sections were acquired at 0.3-μm intervals with the Volocity software. For the rest of the images, five images were acquired over time. Widefield microscopy (Figure 4D) was performed using the conventional imaging mode of a DeltaVision OMX SR imaging system, equipped with a 60× 1.49 NA TIRF oil objective (oil 1.514) and a front illuminated sCMOS camera size 2560 × 2160 pixels (manufacturer PCO).
Image analysis
Fluorescence intensity quantifications shown in Figures 1, 3, 4, and Supplemental Figure S4 were performed on individual cells on a sum projection of spinning-disk confocal images (five consecutive medial plane images acquired over time). For measurement of fluorescence intensity, the cell was split into four quadrants and a 3-pixel-wide (on 7-pixel wide for Airyscan images) segmented line in ImageJ was used to trace the Ptc1 gradient by hand from the middle of the cell tip to the middle of the cell side, therefore producing four individual gradient profiles per cell. The fluorescence intensity measured was corrected for background fluorescence intensity (measured just outside the cell examined). Note that the reported fluorescence values are arbitrary and vary between experiments due to variation in acquisition settings on different days.
Cell length measurements were made on DeltaVision acquired calcofluor stained images. The line segment tool in ImageJ was used to measure the distance between cell ends of dividing cells.
Spot assays
Cultures were adjusted to the same OD600 for the highest concentration sample and subsequent 10-fold serial dilutions were made in a total of 200 μl. Diluted culture (5 μl) was spotted on YE plates and grown at respective temperatures.
Recombinant protein production
Expression of GST-Pom1, GST-Ptc1, and GST alone was induced in BL21 bacteria from the pGEX-4T-1–derived plasmids as described above. Expression of MBP-Ptc1 was induced from a pMAL-derived vector. Briefly, cells were grown overnight in 25 ml Lysogeny Broth (LB) with 100 µg/ml ampicillin at 37°C. LB–ampicillin (250 ml) was inoculated with 6.25 ml of the saturated culture, grown 3 h at 37°C. Protein expression was induced by the addition of 100 µM IPTG for 5 h at 18°C. Bacterial pellets were resuspended in 10 ml cold phosphate-buffered saline (PBS) and sonicated three times for 30 s each (50% amplitude). The sonicates were incubated with 1% Triton X-100 at 4°C, and centrifuged 15 min at 4°C at 10,000 × g. Soluble extract was incubated with 200 µl of glutathione sepharose beads (50% slurry) for 2 h at 4°C. Finally, beads were washed three times with cold PBS, and eluted in three steps in 100 µl elution buffer (15 mM reduced glutathione in 50 mM Tris-HCl, pH 8) elution buffer.
Immunoprecipitation and Western blotting
For Western blots and immunoprecipitation assays, whole cell extracts (WCE) were prepared in CXS buffer (50 mM HEPES, pH 7.0, 20 mM KCl, 1 mM MgCl, 2 mM EDTA, pH 7.5 and protease inhibitor cocktail, Roche) from cells grown in EMM with appropriate glucose concentrations and with or without thiamine (5µg/ml), using glass beads and a bead beater. Total protein concentration was measured using standard Bradford assay. 2 mg WCE was used for immunoprecipitation. Briefly, 50µl Dynabeads (Thermofisher) per sample was washed 3 times with cold 1X PBS and 2µg anti-GFP monoclonal antibody (Roche) per sample was conjugated with Dynabeads in 1 ml cold PBS at 4°C for 4 h. Antibody- conjugated beads were then washed 2 times with cold PBS and once with CXS buffer. In low-protein binding tubes, 2 mg WCE was combined with 50µl antibody-conjugated beads in 1 ml CXS buffer supplemented with 100 mM NaCl and 0.1% NP-40 and left at 4°C for overnight on a rotation wheel. Next day, any unbound antibody was removed by washing the WCE three times with CXS with 0.1% NP-40 and increasing amounts of NaCl (100 mM, 150 mM and 200 mM, respectively). Samples were boiled with 1X Laemmli buffer and 100µg of WCE were loaded together with the immunoprecipitates on SDS–PAGE gels. Bands were revealed using anti-GFP polyclonal antibody (SantaCruz) and mouse anti-HA monoclonal antibody (HA.11; Covance).
In vitro phosphatase assay
In vitro phosphatase assay was performed using purified GST-Pom1 and GST-Ptc1 wild-type and GST-Ptc1 mutant proteins. Briefly, 200 ng GST-Pom1 was incubated with 1µg GST-Ptc1, GST-Ptc1 mutants, or commercial PP1 (NEB #P0754S) in phosphatase buffer with 5X composition: 250 mM Tris-Cl pH7.5, 1 mM EDTA, 0,5% β-mercaptoethanol, 100 mM MgCl2 at 37°C for 1.5 h. The reaction was stopped using 1X Laemmli buffer. Samples were boiled at 95°C for 5 mins before loading on SDS–PAGE gel and stained with silver stain. The gel was run slowly at 80V for 3 h to enable Pom1 shift. Simultaneously, 1µg GST-Ptc1 and GST-Ptc1 mutants were loaded on another SDS–PAGE gel for Coomassie staining.
Supplementary Material
Acknowledgments
This work was supported by Swiss National Science Foundation research grants (31003A_138177, 310030B_176396 and CRSII3_160728) to SGM.
Abbreviations used:
- AMPK
AMP-activated protein kinase
- CaMKK
calcium/calmodulin-dependent protein kinase kinase
- CLASP
CLIP-170 associated protein
- DYRK-family
dual-specificity tyrosine-regulated kinases
- MAPK
mitogen-activated protein kinase
- PKA
protein kinase A
- PP1
protein phosphatase 1
- PP2C
protein phosphatase 2C
- SAD kinase
synapses of the amphid defective kinases
- SH3 domain
SRC homology 3 domain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-08-0508) on February 24, 2021.
REFERENCES
- Allard CAH, Opalko HE, Liu KW, Medoh U, Moseley JB (2018). Cell size-dependent regulation of Wee1 localization by Cdr2 cortical nodes. J Cell Biol 217, 1589–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard CAH, Opalko HE, Moseley JB (2019). Stable Pom1 clusters form a glucose-modulated concentration gradient that regulates mitotic entry. eLife 8, e46003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano M, Niccoli T, Nurse P (2002). Tea3p is a cell end marker activating polarized growth in Schizosaccharomyces pombe. Curr Biol 12, 751–756. [DOI] [PubMed] [Google Scholar]
- Arino J, Casamayor A, Gonzalez A (2011). Type 2C protein phosphatases in fungi. Eukaryot Cell 10, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Bendezu FO, Martin SG (2013). Cdc42 explores the cell periphery for mate selection in fission yeast. Curr Biol 23, 42–47. [DOI] [PubMed] [Google Scholar]
- Bhatia P, Hachet O, Hersch M, Rincon SA, Berthelot-Grosjean M, Dalessi S, Basterra L, Bergmann S, Paoletti A, Martin SG (2014). Distinct levels in Pom1 gradients limit Cdr2 activity and localization to time and position division. Cell Cycle 13, 538–552. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee R, Mangione MC, Wos M, Chen JS, Snider CE, Roberts-Galbraith RH, McDonald NA, Presti LL, Martin SG, Gould KL (2020). DYRK kinase Pom1 drives F-BAR protein Cdc15 from the membrane to promote medial division. Mol Biol Cell 31, 917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicho CC, Kelly DA, Snaith HA, Goryachev AB, Sawin KE (2010). A catalytic role for Mod5 in the formation of the Tea1 cell polarity landmark. Curr Biol 20, 1752–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeding CS, Hudson J, Balasubramanian MK, Hemmingsen SM, Young PG, Gould KL (1998). The cdr2+ gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomyces pombe. Mol Biol Cell 9, 3399–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou JG, Balasubramanian MK, Lew DJ (2017). Cell polarity in yeast. Annu Rev Cell Dev Biol 33, 77–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie E, Forte GM, Petersen J (2015). Nitrogen regulates AMPK to control TORC1 signaling. Curr Biol 25, 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Baldissard S, Kettenbach AN, Gerber SA, Moseley JB (2014). Dueling kinases regulate cell size at division through the SAD kinase Cdr2. Curr Biol 24, 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Moseley JB (2013). Compartmentalized nodes control mitotic entry signaling in fission yeast. Mol Biol Cell 24, 1872–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchetti G, Knapp B, Flor-Parra I, Chang F, Howard M (2019). Reprogramming Cdr2-dependent geometry-based cell size control in fission yeast. Curr Biol 29, 350–358.e354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone C, Russell P (1991). Fission yeast p107wee1 mitotic inhibitor is a tyrosine/serine kinase. Nature 349, 808–811. [DOI] [PubMed] [Google Scholar]
- Gaits F, Shiozaki K, Russell P (1997). Protein phosphatase 2C acts independently of stress-activated kinase cascade to regulate the stress response in fission yeast. J Biol Chem 272, 17873–17879. [DOI] [PubMed] [Google Scholar]
- Gautier J, Solomon MJ, Booher RN, Bazan JF, Kirschner MW (1991). cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell 67, 197–211. [DOI] [PubMed] [Google Scholar]
- Gerganova V, Floderer C, Archetti A, Michon L, Carlini L, Reichler T, Manley S, Martin SG (2019). Multi-phosphorylation reaction and clustering tune Pom1 gradient mid-cell levels according to cell size. eLife 8, e45983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerganova V, Martin SG (2018). Dynamic visits of cortical structures probe for cell size. J Cell Biol 217, 1559–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn JM, Lustig RJ, Berlin A, Chang F (2001). Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr Biol 11, 836–845. [DOI] [PubMed] [Google Scholar]
- Gould KL, Nurse P (1989). Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature 342, 39–45. [DOI] [PubMed] [Google Scholar]
- Hachet O, Berthelot-Grosjean M, Kokkoris K, Vincenzetti V, Moosbrugger J, Martin SG (2011). A phosphorylation cycle shapes gradients of the DYRK family kinase Pom1 at the plasma membrane. Cell 145, 1116–1128. [DOI] [PubMed] [Google Scholar]
- Hersch M, Hachet O, Dalessi S, Ullal P, Bhatia P, Bergmann S, Martin SG (2015). Pom1 gradient buffering through intermolecular auto-phosphorylation. Mol Syst Biol 11, 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda Y, Satoh R, Matsumoto S, Ikeda C, Inutsuka N, Hagihara K, Matzno S, Tsujimoto S, Kita A, Sugiura R (2016). Skb5, an SH3 adaptor protein, regulates Pmk1 MAPK signaling by controlling the intracellular localization of the MAPKKK Mkh1. J Cell Sci 129, 3189–3202. [DOI] [PubMed] [Google Scholar]
- Kanoh J, Russell P (1998). The protein kinase Cdr2, related to Nim1/Cdr1 mitotic inducer, regulates the onset of mitosis in fission yeast. Mol Biol Cell 9, 3321–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar M, Martin SG (2015). PKA antagonizes CLASP-dependent microtubule stabilization to re-localize Pom1 and buffer cell size upon glucose limitation. Nat Commun 6, 8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG (1991). The cdc25 protein controls tyrosine dephosphorylation of the cdc2 protein in a cell-free system. Cell 64, 903–914. [DOI] [PubMed] [Google Scholar]
- Lundgren K, Walworth N, Booher R, Dembski M, Kirschner M, Beach D (1991). mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell 64, 1111–1122. [DOI] [PubMed] [Google Scholar]
- Martin SG (2009). Geometric control of the cell cycle. Cell Cycle 8, 3643–3647. [DOI] [PubMed] [Google Scholar]
- Martin SG, Arkowitz RA (2014). Cell polarization in budding and fission yeasts. FEMS Microbiol Rev 38, 228–253. [DOI] [PubMed] [Google Scholar]
- Martin SG, Berthelot-Grosjean M (2009). Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature 459, 852–856. [DOI] [PubMed] [Google Scholar]
- Martin SG, McDonald WH, Yates JR 3rd, Chang F (2005). Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev Cell 8, 479–491. [DOI] [PubMed] [Google Scholar]
- Mata J, Nurse P (1997). tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89, 939–949. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Mayeux A, Paoletti A, Nurse P (2009). A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature 459, 857–860. [DOI] [PubMed] [Google Scholar]
- Nguyen AN, Shiozaki K (1999). Heat-shock-induced activation of stress MAP kinase is regulated by threonine- and tyrosine-specific phosphatases. Genes Dev 13, 1653–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P (1990). Universal control mechanism regulating onset of M-phase. Nature 344, 503–508. [DOI] [PubMed] [Google Scholar]
- Opalko HE, Nasa I, Kettenbach AN, Moseley JB (2019). A mechanism for how Cdr1/Nim1 kinase promotes mitotic entry by inhibiting Wee1. Mol Biol Cell 30, 3015–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan KZ, Saunders TE, Flor-Parra I, Howard M, Chang F (2014). Cortical regulation of cell size by a sizer cdr2p. eLife 3, e02040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P, Nurse P (1986). cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 45, 145–153. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P (1987). Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell 49, 559–567. [DOI] [PubMed] [Google Scholar]
- Saunders TE, Pan KZ, Angel A, Guan Y, Shah JV, Howard M, Chang F (2012). Noise reduction in the intracellular pom1p gradient by a dynamic clustering mechanism. Dev Cell 22, 558–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y (2009). Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484. [DOI] [PubMed] [Google Scholar]
- Shiozaki K, Akhavan-Niaki H, McGowan CH, Russell P (1994). Protein phosphatase 2C, encoded by ptc1+, is important in the heat shock response of Schizosaccharomyces pombe. Mol Cell Biol 14, 3742–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K, Russell P (1995). Counteractive roles of protein phosphatase 2C (PP2C) and a MAP kinase kinase homolog in the osmoregulation of fission yeast. EMBO J 14, 492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith HA, Sawin KE (2003). Fission yeast mod5p regulates polarized growth through anchoring of tea1p at cell tips. Nature 423, 647–651. [DOI] [PubMed] [Google Scholar]
- Stanger K, Gorelik M, Davidson AR (2012). Yeast adaptor protein, Nbp2p, is conserved regulator of fungal Ptc1p phosphatases and is involved in multiple signaling pathways. J Biol Chem 287, 22133–22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada H, Nishimura M, Asayama Y, Mannse Y, Ishiwata S, Kita A, Doi A, Nishida A, Kai N, Moriuchi S, et al. (2007). Atf1 is a target of the mitogen-activated protein kinase Pmk1 and regulates cell integrity in fission yeast. Mol Biol Cell 18, 4794–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Shimada K, Uzawa S, Morigasaki S, Shiozaki K (2005). Wsh3/Tea4 is a novel cell-end factor essential for bipolar distribution of Tea1 and protects cell polarity under environmental stress in S. pombe. Curr Biol 15, 1006–1015. [DOI] [PubMed] [Google Scholar]
- Wood E, Nurse P (2013). Pom1 and cell size homeostasis in fission yeast. Cell Cycle 12, 3228–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood E, Nurse P (2015). Sizing up to divide: mitotic cell-size control in fission yeast. Annu Rev Cell Dev Biol 31, 11–29. [DOI] [PubMed] [Google Scholar]
- Yang P, Pimental R, Lai H, Marcus S (1999). Direct activation of the fission yeast PAK Shk1 by the novel SH3 domain protein, Skb5. J Biol Chem 274, 36052–36057. [DOI] [PubMed] [Google Scholar]
- Young PG, Fantes PA (1987). Schizosaccharomyces pombe mutants affected in their division response to starvation. J Cell Sci 88 (Pt 3), 295–304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.