Abstract
A range of severe human diseases called ciliopathies is caused by the dysfunction of primary cilia. Primary cilia are cytoplasmic protrusions consisting of the basal body (BB), the axoneme, and the transition zone (TZ). The BB is a modified mother centriole from which the axoneme, the microtubule-based ciliary scaffold, is formed. At the proximal end of the axoneme, the TZ functions as the ciliary gate governing ciliary protein entry and exit. Since ciliopathies often develop due to mutations in genes encoding proteins that localize to the TZ, the understanding of the mechanisms underlying TZ function is of eminent importance. Here, we show that the ciliopathy protein Rpgrip1l governs ciliary gating by ensuring the proper amount of Cep290 at the vertebrate TZ. Further, we identified the flavonoid eupatilin as a potential agent to tackle ciliopathies caused by mutations in RPGRIP1L as it rescues ciliary gating in the absence of Rpgrip1l.
INTRODUCTION
The spatiotemporal regulation of cellular processes such as proliferation, apoptosis, migration, specification, and differentiation depends on the cell’s ability to transduce signals from the environment into the cell’s interior. In nearly all mammalian cells, the primary cilium is dedicated to signal reception and transduction. Consequently, dysfunctional primary cilia result in severe often deadly human diseases, collectively called ciliopathies (Reiter and Leroux, 2017). The current treatment of ciliopathies is restricted to symptomatic therapies and a curative medication against ciliopathies is missing (McIntyre et al., 2013). In many cases, ciliopathies are caused by mutations in genes encoding transition zone (TZ) proteins (Hildebrandt et al., 2011; Czarnecki and Shah, 2012). As the TZ functions as the ciliary gatekeeper governing ciliary protein import and export (Betleja and Cole, 2010; Craige et al., 2010; Omran, 2010; Benzing and Schermer, 2011; Czarnecki and Shah, 2012; Garcia-Gonzalo and Reiter, 2012; Reiter et al., 2012; Garcia-Gonzalo and Reiter, 2017; Jensen and Leroux, 2017), a defective TZ can affect the proper formation of cilia and alter the transduction of signaling pathways (Reiter and Skarnes, 2006; Vierkotten et al., 2007; Chih et al., 2011; Garcia-Gonzalo et al., 2011; Williams et al., 2011; Warburton-Pitt et al., 2012; Jensen et al., 2015; Yee et al., 2015; Li et al., 2016; Shi et al., 2017; Weng et al., 2018; Lewis et al., 2019). Thus, the investigation of the molecular mechanisms underlying TZ function is an important prerequisite for the development of curative ciliopathy therapies.
In this study, we shed light on the role of Rpgrip1l in regulating the ciliary gating function of the TZ. Our previous investigations revealed that mutations in RPGRIP1L cause deadly ciliopathies (Delous et al., 2007), that Rpgrip1l localizes to the vertebrate TZ (Gerhardt et al., 2015), and that it is a decisive factor in vertebrate TZ assembly (Wiegering et al., 2018a). Our current work demonstrates that Rpgrip1l deficiency results in an altered ciliary protein composition and that Rpgrip1l governs ciliary gating by ensuring the proper amount of Cep290 at the vertebrate TZ. Further, we revealed that the flavonoid eupatilin rescues ciliary gating in the absence of Rpgrip1l. Consequently, eupatilin might represent a potential agent for the development of therapies against ciliopathies caused by mutations in RPGRIP1L.
RESULTS
Rpgrip1l and Cep290 but not nephrocystin (Nphp)1, Nphp4, and Inversin (Invs) function as ciliary gatekeepers in mouse embryonic fibroblasts (MEFs)
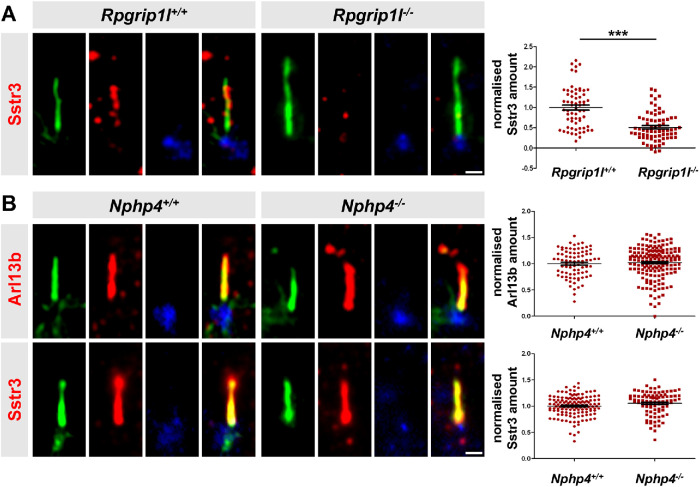
Among the proteins allowed to cross the TZ are receptors and mediators of signaling pathways essential for proper development. Examples for such proteins are ADP ribosylation factor-like GTPase 13B (Arl13b), somatostatin receptor 3 (Sstr3), smoothened (Smo), polycystin 2, or adenylate cyclase 3 (Ac3) which are often used as indicators to evaluate whether the gate function of the TZ is impaired (Hu et al., 2010; Chih et al., 2011; Garcia-Gonzalo et al., 2011; Roberson et al., 2015; Li et al., 2016; Shi et al., 2017; Takao et al., 2017; Ye et al., 2018). In a former study, we showed that Rpgrip1l deficiency leads to a reduction of the ciliary Arl13b amount in all analyzed mouse cells in vitro and in vivo (in MEFs, in mouse embryonic kidneys, and in mouse limb buds; Wiegering et al., 2018a). However, we could not detect an alteration of the ciliary Smo amount in Rpgrip1l–/– MEFs (Gerhardt et al., 2015), raising the question whether the effect of Rpgrip1l is Arl13b-specific or whether it functions as a more general ciliary gatekeeper at the vertebrate TZ. To answer this question, we analyzed the ciliary Sstr3 amount. It has been known for a long time that Sstr3 localizes to cilia of neuronal cells (Händel et al., 1999). More recently, it was shown that Sstr3 is also present in cilia of retinal pigment epithelial (RPE-1) cells (Klinger et al., 2014). We were now able to visualize endogenous Sstr3 in cilia of MEFs. Importantly, the amount of Sstr3 was decreased in Rpgrip1l–/– MEFs (Figure 1A) demonstrating that Rpgrip1l exerts a TZ gatekeeper function.
FIGURE 1:
In mouse cells, ciliary gating is disturbed by the loss of Rpgrip1l but not by the loss of Nphp4. (A) Immunofluorescence on MEFs obtained from WT (n = 5) and Rpgrip1l–/– (n = 5) embryos. At least 10 cilia per embryo were used for quantifications (Σ(WT) = 62 cilia, Σ(Rpgrip1l–/–) = 72 cilia). (B) Immunofluorescence on MEFs obtained from WT (Arl13b: n = 4; Sstr3: n = 3) and Nphp4–/– (Arl13b: n = 4; Sstr3: n = 3) embryos. At least 20 cilia per embryo were used for quantifications (Arl13b: Σ(WT) = 83 cilia, Σ(Nphp4–/–) = 171 cilia; Sstr3: Σ(WT) = 134 cilia, Σ(Nphp4–/–) = 92 cilia). (A, B) The ciliary axoneme is stained in green by acetylated α-tubulin, the BB is stained in blue by γ-tubulin. The scale bars represent a length of 0.5 µm. Data are shown as mean ± SEM. Asterisks denote statistical significance according unpaired t tests with Welch´s correction (***P < 0.001) (A: t (25) = 6.54, P < 0.0001; B: Arl13b: t (46) = 0.7483, P < 0.4581; Sstr3: t (80) = 1.807, P < 0.0745).
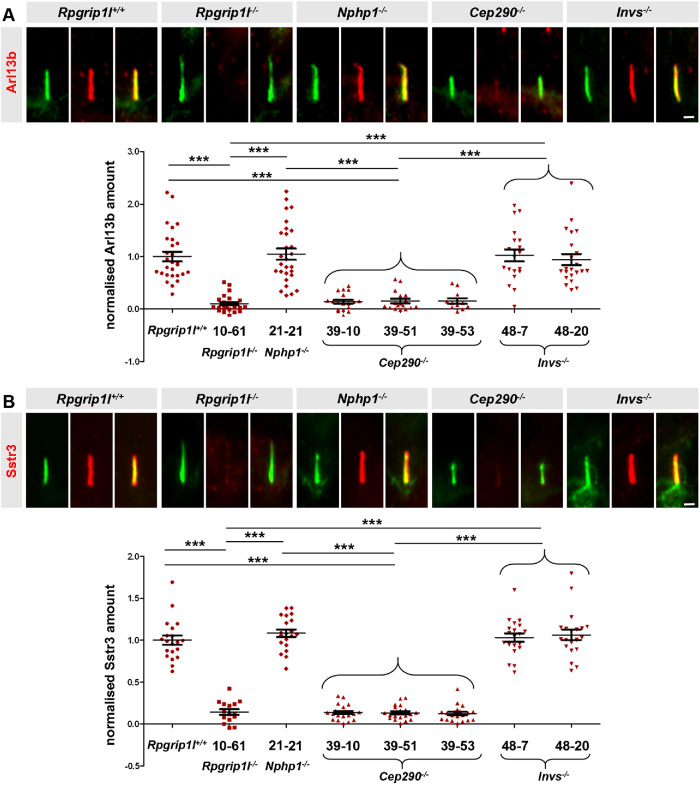
We next aimed to investigate how Rpgrip1l implements this function. Several proteins function as ciliary gatekeepers and it is an important task to unveil possible relationships between these gatekeeper proteins in order to understand the mechanisms that govern ciliary protein composition. In this context, centrosome and spindle pole-associated protein 1 (Cspp1) was an interesting object of investigation because its truncation in humans results in a reduced ciliary amount of Arl13b and Ac3 (Tuz et al., 2014) and because it interacts with Rpgrip1l (Patzke et al., 2010; Gerhardt et al., 2015). Previously, we and others reported that Rpgrip1l ensures the proper amount of many proteins at the base of vertebrate cilia (Shi et al., 2017; Wiegering et al., 2018a), suggesting that it serves as a vital scaffold protein. Thus, we quantified the amount of Cspp1 at the ciliary base of Rpgrip1l–/– MEFs but we could not detect any alteration (Supplemental Figure S1A), indicating that Rpgrip1l exerts its ciliary gatekeeper function independently of Cspp1. Recently, a potential link between Rpgrip1l and the septin proteins was discussed (Patnaik et al., 2018). Septins are known ciliary gatekeepers in vertebrates localizing at the proximal end of the axoneme in MEFs (Hu et al., 2010). We measured the ciliary amount of two different septins, Septin 2 (Sept2) and Septin 7 (Sept7), in Rpgrip1l–/– MEFs. Neither Sept2 nor Sept7 was altered by the loss of Rpgrip1l (Supplemental Figure S1, B and C). We then turned to the TZ proteins Nphp4, Nphp1, centrosomal protein 290 (Cep290) and Invs whose amount at the vertebrate TZ is decreased in the absence of Rpgrip1l (Wiegering et al., 2018a). Previous studies in the invertebrates Chlamydomonas reinhardtii and/or Caenorhabditis elegans demonstrated that ciliary gating is regulated by the TZ proteins Nphp4, Nphp1, Cep290, and Invs (Craige et al., 2010; Williams et al., 2011; Warburton-Pitt et al., 2012; Awata et al., 2014; Li et al., 2016), raising the possibility that Rpgrip1l might govern ciliary gating by controlling the amount of one of these proteins at the TZ. However, it is unknown whether these proteins function as ciliary gatekeepers in vertebrates and hence we quantified the ciliary amount of Arl13b and Sstr3 in Nphp4–/–, Nphp1–/–, Cep290–/–, and Invs–/– mouse cells. The ciliary amount of both Arl13b and Sstr3 was unaffected in Nphp4–/– MEFs (Figure 1B). To analyze TZ assembly, we had previously inactivated Nphp1, Cep290, and Invs in NIH3T3 cells (immortalized MEFs; Wiegering et al., 2018a). In the current study, we used these cells to investigate a possible involvement of these proteins in ciliary gating. To be able to perform comparative analyses in this context, we also inactivated Rpgrip1l in NIH3T3 cells (Supplemental Figure S2). As observed in MEFs (Gerhardt et al., 2015), ciliary length was increased in Rpgrip1l–/– NIH3T3 cells (Supplemental Figure S2C). Moreover, the ciliary amount of Arl13b and Sstr3 was decreased in Rpgrip1l–/– NIH3T3 cells (Figure 2, A and B). In contrast to these cells, Nphp1–/– and Invs–/– NIH3T3 cells did not show an altered ciliary amount of Arl13b and Sstr3 (Figure 2, A and B) indicating that the decreased amount of Nphp1 and Invs in Rpgrip1l–/– MEFs is not the reason for the reduced ciliary Arl13b and Sstr3 amount. Importantly, Cep290 deficiency causes a decrease of the ciliary Arl13b and Sstr3 amount (Figure 2, A and B) making it conceivable that the reduced amount of Cep290 might provoke the diminished amount of Arl13b and Sstr3 in the absence of Rpgrip1l.
FIGURE 2:
Loss of Cep290 but not of Nphp1 or Invs impairs ciliary gating in mouse cells. (A, B) Immunofluorescence on NIH3T3 cells. The ciliary axoneme is stained in green by acetylated α-tubulin. The scale bars represent a length of 0.5 µm. At least 20 cilia per clone were used for quantification. Data are shown as mean ± SEM. Asterisks denote statistical significance according to one-way ANOVA and Tukey HSD tests (***P < 0.001) (A: F (7,160) = 26.09, P < 0.0001; B: F (7,147) = 133.6, P < 0.0001).
Restoration of the Cep290 amount at the TZ of Rpgrip1l –/– NIH3T3 and RPGRIP1L –/– HEK293 cells rescues the ciliary Arl13b and Sstr3 amount
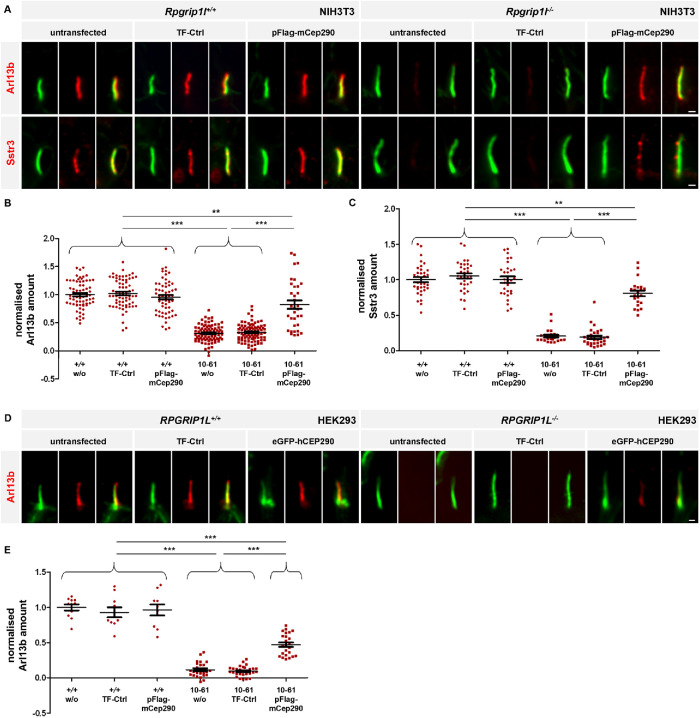
To test this hypothesis, we enhanced the amount of Cep290 at the TZ of Rpgrip1l–/– NIH3T3 cells by transfecting a plasmid that encodes a Flag-mCep290 fusion protein. In addition, to test for the conservation of the functional relationship between these two TZ proteins in humans, we transfected a plasmid that encodes a GFP-hCEP290 fusion protein into RPGRIP1L–/– HEK293 cells. In a previous study, we revealed a reduced amount of Cep290 at the TZ of Rpgrip1l–/– MEFs and RPGRIP1L–/– HEK293 cells (Wiegering et al., 2018a). In line with this, a reduction of Cep290 at the ciliary TZ of Rpgrip1l–/– NIH3T3 cells was observed in the present study (Supplemental Figure S4, E and F). In NIH3T3 and HEK293 cells, the transfected Cep290 fusion protein was located at the TZ (Supplemental Figure S4, A and H). In contrast to the Flag-mCep290 fusion protein in NIH3T3 cells and the GFP-hCEP290 fusion protein in HEK293 cells, we could not detect a transfected Myc-mNphp1 fusion protein at the TZ of Rpgrip1l–/– NIH3T3 or RPGRIP1L–/– HEK293 cells (Supplemental Figure S3). Most likely, Rpgrip1l functions as a TZ scaffold for Nphp1 but not for Cep290.
The transfection of the plasmid encoding the Flag-mCep290 fusion protein into Rpgrip1l–/– NIH3T3 cells as well as the transfection of the plasmid encoding the GFP-hCEP290 fusion protein into RPGRIP1L–/– HEK293 cells at least partially restored the amount of Cep290 at the TZ (Supplemental Figure S4, E, F, K, and L). Importantly, the rescued amount of Cep290 restored the ciliary amount of Arl13b and Sstr3 in Rpgrip1l–/– NIH3T3 (Figure 3, A–C) and the ciliary amount of Arl13b in RPGRIP1L–/– HEK293 cells (Figure 3, D and E). HEK293 cells lack Sstr3, preventing the analysis of its localization in the absence of RPGRIP1L and its rescue by CEP290 overexpression (War and Kumar, 2012). Our results indicate that Rpgrip1l controls ciliary gating via ensuring the proper amount of Cep290 at the TZ, both in mouse and human cells.
FIGURE 3:
Rescue of the Cep290 amount at the TZ of Rpgrip1l–/– NIH3T3 and RPGRIP1L–/– HEK293 cells restores the ciliary Arl13b and Sstr3 amount. (A–C) Rpgrip1l+/+ and Rpgrip1l–/– NIH3T3 cells were either untransfected or transfected with TF-Ctrl or pFlag-mCep290. (A) Immunofluorescence on Rpgrip1l+/+ and Rpgrip1l–/– (clone 10-61) NIH3T3 cells. The ciliary axoneme is stained in green by acetylated α-tubulin and Arl13b or Sstr3 is stained in red. The scale bars represent a length of 0.5 µm. (B, C) Normalized ciliary amount of Arl13b (B) and Sstr3 (C). At least 30 cilia per clone were used for quantification. Data are shown as mean ± SEM. Asterisks denote statistical significance according to one-way ANOVA and Tukey HSD tests (**P < 0.01; ***P < 0.001) (B: F (5, 400) = 146.8, P < 0.0001; C: F (5, 166) = 128.6, P < 0.0001). (D, E) RPGRIP1L+/+ and RPGRIP1L–/– HEK293 cells were either untransfected or transfected with eGFP-hCEP290 or TF-Ctrl. (D) Immunofluorescence on RPGRIP1L+/+ and RPGRIP1L–/– (clone 1–7) HEK293 cells. The ciliary axoneme is stained in green by acetylated α-tubulin and Arl13b is stained in red. The scale bars represent a length of 0.5 µm. (E) Normalized ciliary amount of Arl13b. At least 10 cilia (RPGRIP1L+/+HEK293 cells) or 20 cilia (RPGRIP1L–/– HEK293 cells) per clone were used for quantification. Data are shown as mean ± SEM. Asterisks denote statistical significance according to one-way ANOVA and Tukey HSD tests (***P < 0.001) (F (5, 104) = 142, P < 0.0001).
Cilia of NIH3T3 and HEK293 cells are elongated by the loss of Rpgrip1l (Wiegering et al., 2018a) (Supplemental Figure S4, C and J). Interestingly, the increased cilia length in the absence of Rpgrip1l was not rescued by the expression of the Flag-mCep290 fusion protein in Rpgrip1l–/– NIH3T3 cells or the GFP-hCEP290 fusion protein in RPGRIP1L–/– HEK293 cells (Supplemental Figure S4, C and J). Thus, contrary to its ciliary gating function, the role of Rpgrip1l in controlling ciliary length is not mediated by Cep290.
To verify the functionality of the Cep290 fusion protein, we transfected the plasmid encoding Flag-mCep290 into Cep290–/– NIH3T3 cells. The transfected Cep290 fusion protein was located at the TZ in Cep290–/– NIH3T3 cells (Supplemental Figure S4A) and the amount of Cep290 at the TZ was restored (Supplemental Figure S4, E and G). Moreover, the decreased cilia length observed in the absence of Cep290 (Wiegering et al., 2018a) was rescued by the expression of the Flag-mCep290 fusion protein (Supplemental Figure S4D), demonstrating the functionality of the transfected protein in NIH3T3 cells.
Eupatilin treatment rescues ciliary gating in Rpgrip1l-negative MEFs
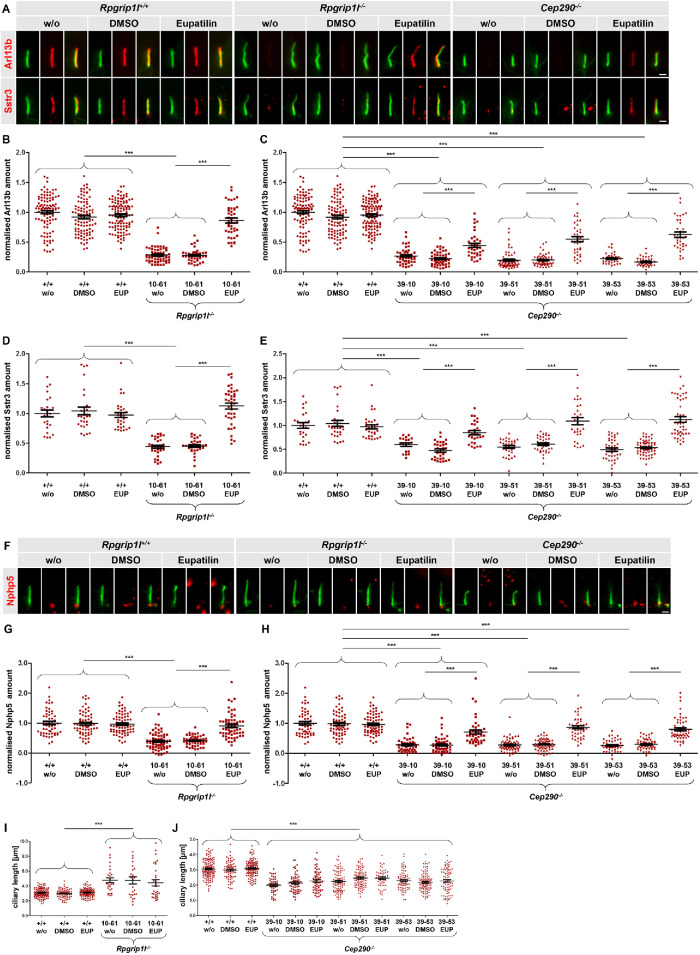
A recent report showed that eupatilin rescues ciliary gating in CEP290–/– human cells by replacing the function of CEP290 in TZ recruitment of Nphp 5 (NPHP5; alias IQCB1) (Kim et al., 2018). Since we showed above that Rpgrip1l function on ciliary gating was mediated by the control of Cep290 TZ amounts, we hypothesized that eupatilin would also rescue the ciliary gating defect in Rpgrip1l-deficient cells. Indeed, the treatment of Rpgrip1l–/– NIH3T3 cells with eupatilin restored the ciliary amount of both Arl13b and Sstr3 (Figure 4, A, B, and D), confirming our hypothesis. The enhanced cilia length in Rpgrip1l–/– NIH3T3 cells was not rescued by eupatilin (Figure 4I). We also verified that eupatilin treatment of Cep290–/–NIH3T3 cells restored the reduced amount of Arl13b and Sstr3 (Figure 4, A, C, and E), but not ciliary length alteration (Figure 4J), in these cells. Furthermore, we analyzed the TZ amount of Nphp5 and found that it was significantly reduced in both Rpgrip1l–/– and Cep290–/– NIH3T3 cells (Figure 4, F–H). Eupatilin treatment rescued the amount of Nphp5 completely in Rpgrip1l–/– NIH3T3 cells (Figure 4G) and more partially in Cep290–/– NIH3T3 cells (Figure 4H). Thus, we conclude that Rpgrip1l functions in ciliary gating upstream of Nphp5, via ensuring the proper amount of Cep290 at the TZ.
FIGURE 4:
Eupatilin treatment rescues ciliary gating in the absence of Rpgrip1l. (A, B) Immunofluorescence on Rpgrip1l+/+, Rpgrip1l–/– (clone 10-61) and Cep290–/– NIH3T3 cells. The ciliary axoneme is stained in green by acetylated α-tubulin. Arl13b or Sstr3 is stained in red. The scale bars represent a length of 1 µm. (B–E) Normalized ciliary amount of Arl13b (B, C) and Sstr3 (D, E). At least 20 (D, E) or 30 cilia (B, C) per clone were used for quantification. The same quantification of WT serves as comparison to Rpgri1l-negative and Cep290-negative cells, respectively (B and C; D and E). Data are shown as mean ± SEM. Asterisks denote statistical significance according to one-way ANOVA and Tukey HSD tests (***P < 0.001) (B: F (5, 447) = 117.1, P < 0.0001; C: F (11, 690) = 163.4, P < 0.0001; D: F (5, 183) = 47.48, P < 0.0001; E: F (11, 393) = 35.27, P < 0.0001). (F) Immunofluorescence on Rpgrip1l+/+, Rpgrip1l–/– (clone 10-61) and Cep290–/– (clones 39-10, 39-51, 39-53) NIH3T3 cells. The ciliary axoneme is stained in green by acetylated α-tubulin. Nphp5 is stained in red. The scale bars represent a length of 1 µm. (G, H) Normalized ciliary amount of Nphp5. At least 40 cilia per clone were used for quantification. The same quantification of WT serves as comparison to Rpgrip1l-negative and Cep290-negative cells. Data are shown as mean ± SEM. Asterisks denote statistical significance according to one-way ANOVA and Tukey HSD tests (***P < 0.001) (G: F (5, 354) = 44.34, P < 0.0001; H: F (11, 638) = 64.13, P < 0.0001). (I, J) Ciliary length measurements. At least 30 cilia (I) or 50 cilia (J) per clone were used for quantification. The same quantification of the WT serves as comparison to Rpgrip1l-negative and Cep290-negative cells. Data are shown as mean ± SEM. Asterisks denote statistical significance according to one-way ANOVA and Tukey HSD tests (***P < 0.001) (I: F (5, 402) = 28.71, P < 0.0001; J: F (11, 986) = 36,73, P < 0.0001).
DISCUSSION
Primary cilia mediate numerous signaling pathways thereby ensuring proper development and homeostasis. In this context, the intraciliary concentration of proteins involved in these signaling pathways is of enormous importance. Consequently, ciliary import and export and hence ciliary protein composition has to be tightly controlled. This control is implemented by the TZ. Since mutations in genes encoding TZ proteins result in ciliopathies (Hildebrandt et al., 2011; Czarnecki and Shah, 2012; Reiter and Leroux, 2017), current cilia research aims to uncover mechanisms underlying TZ assembly and function. However, little is known about these mechanisms in vertebrates. Recently, we described Rpgrip1l as a decisive factor in vertebrate TZ assembly (Wiegering et al., 2018a). In this context, Rpgrip1l deficiency leads to a reduced amount of Cep290, Nphp1, Nphp4, and Invs at the TZ (Wiegering et al., 2018a). Rpgrip1l, Cep290, Nphp1, Nphp4, and Invs were previously shown to govern ciliary gating in C. reinhardtii and/or C. elegans (Craige et al., 2010; Williams et al., 2011; Warburton-Pitt et al., 2012; Awata et al., 2014; Li et al., 2016; Lin et al., 2018). However, loss of Nphp1, Nphp4, and Invs did not alter the ciliary amount of Arl13b or Sstr3 in MEFs and NIH3T3 cells (Figures 1B and 2), indicating that they are not involved in gating these proteins in vertebrate primary cilia. Remarkably, several reports point to cell type-specific functions of some TZ proteins (Garcia-Gonzalo et al., 2011; Rachel et al., 2015; Lambacher et al., 2016; Wiegering et al., 2018a; Lewis et al., 2019) making a potential regulation of ciliary gating by Nphp1, Nphp4, and Invs in other vertebrate cell types conceivable. Rpgrip1l–/– and Cep290–/– mice have a much more severe phenotype than Nphp1–/–, Nphp4–/– and Invs–/– mice (Mochizuki et al., 1998; Morgan et al., 1998; Chang et al., 2006; Delous et al., 2007; McEwen et al., 2007; Vierkotten et al., 2007; Jiang et al., 2008; Jiang et al., 2009; Louie et al., 2010; Besse et al., 2011; Lancaster et al., 2011; Won et al., 2011; Gerhardt et al., 2013; Hynes et al., 2014; Chen et al., 2015; Laclef et al., 2015; Li et al., 2015; Rachel et al., 2015; Wiegering et al., 2018a; Andreu-Cervera et al., 2019; Choi et al., 2019). Moreover, mutations in RPGRIP1L and CEP290 result in more severe human ciliopathies than mutations in NPHP1, NPHP4, or INVS (Zaghloul and Katsanis, 2010; Szymanska and Johnson, 2012; Madhivanan and Aguilar, 2014; Mitchison and Valente, 2017). On the one hand, these differences might reflect that Rpgrip1l and Cep290 function as ciliary gatekeepers in vertebrates while Nphp1, Nphp4, and Invs do not. On the other hand, these differences might be based on the fact that Rpgrip1l and Cep290 exert additional functions in the cytoplasm, for example, the regulation of protein degradation systems (Gerhardt et al., 2015; Struchtrup et al., 2018) or the organization of the cytoplasmic microtubule network (Kim et al., 2008).
Rpgrip1l does not function as scaffold for Cep290 and Cspp1 at the TZ
Formerly, we demonstrated that Rpgrip1l deficiency does not affect the overall cellular amount of Cep290 but its proper amount at the vertebrate TZ (Wiegering et al., 2018a). There is a perennial debate about the function(s) of Rpgrip1l. Does it predominantly serve as a structural TZ anchor or scaffold protein and interacts with other proteins, thereby ensuring their localization and proper amount at the TZ, or does it control the TZ localization and amount of proteins by exerting additional functions, for example, regulating protein degradation systems, functioning as a TZ assembly factor, establishing a ciliary zone of exclusion that excludes signal transduction proteins, etc. (Coene et al., 2011; Williams et al., 2011; Gerhardt et al., 2015; Jensen et al., 2015; Assis et al., 2017; Shi et al., 2017; Struchtrup et al., 2018; Wiegering et al., 2018a; Wiegering et al., 2018b)? In this study, the Flag-mCep290 and the GFP-hCEP290 fusion proteins were able to localize to the TZ in the absence of Rpgrip1l (Supplemental Figure S4, A and H), indicating that Rpgrip1l does not function as a structural scaffold for the TZ presence of Cep290. In line with this assumption, it was not shown yet that Rpgrip1l interacts with Cep290. To stress this point, we also transfected a plasmid encoding a Myc-mNphp1 fusion protein into Rpgrip1l–/– NIH3T3 and RPGRIP1L–/– HEK293 cells (Supplemental Figure S3). It was reported before that Nphp1 interacts with Rpgrip1l (Sang et al., 2011). Since Myc-mNphp1 was not present at the TZ in the absence of Rpgrip1l (Supplemental Figure S3, B and D), we suggest that Rpgrip1l functions as a structural anchor for Nphp1 but not for Cep290. The mechanism by which Rpgrip1l regulates the TZ amount of Cep290 is thus not understood, and it will be an exciting future challenge to address this question.
For example, it would be conceivable that Rpgrip1l regulates the amount of Cep290 via interaction with centriolar satellites. It is predicted that Cep290 is part of a satellite subnetwork consisting of pericentriolar material 1 (PCM1), SSX family member 2 interacting protein (SSX2IP), orofaciaodigital syndrome protein 1 (OFD1, centriole and centriolar satellite protein), synaptic vesicle glycoprotein 2B (SV2B; also known as KIAA0735), and CEP290 (Gupta et al., 2015), and that Rpgrip1l is a potential interaction partner of this satellite subnetwork by interacting at least with PCM1 and SSX2IP (Gupta et al., 2015). PCM1 is a major component of centriolar satellites involved in the recruitment of Ceps such as centrin and ninein as well as the organization of a cytoplasmic microtubule network (Kubo et al., 1999; Dammermann and Merdes, 2002; Kubo and Tsukita, 2003). Several studies have shown physical and functional interactions between PCM1 and CEP290 (Chang et al., 2006; Kim et al., 2008; Gupta et al., 2015). However, the loss of PCM1 decreases the localization of CEP290 at centriolar satellites but does not affect the centrosomal/basal body (BB) accumulation of CEP290 (Kim et al., 2008; Stowe et al., 2012; Odabasi et al., 2019). In contrast to that, loss of SSX2IP leads to a reduced amount of CEP290 at the TZ (Klinger et al., 2014). SSX2IP is known to be involved in microtubule anchoring, centrosome maturation, and ciliogenesis as well as being an important effector protein in the FOXJ1 regulatory network (Bärenz et al., 2013; Hori et al., 2014; Hori et al., 2015; Mukherjee et al., 2019). On ciliogenesis, SSX2IP accumulates around the BB (Klinger et al., 2014) and loss of SSX2IP decreases ciliary length in RPE-1 cells (Hori et al., 2014; Klinger et al., 2014). Taken together, Rpgrip1l could regulate the TZ amount of Cep290 by directly regulating SSX2IP, which in turn regulates Cep290.
It was shown before that the proper amount of Rpgrip1l at the TZ depends on Cspp1 and that Rpgrip1l and Cspp1 are directly interacting (Patzke et al., 2010). Moreover, mutations in CSPP1 disturb ciliary protein composition (e.g., reduced ciliary Arl13b amount; Tuz et al., 2014) and cause Joubert syndrome and Meckel syndrome (Shaheen et al., 2014). Interestingly, Rpgrip1l was not required for the ciliary localization of Cspp1 (Supplemental Figure S1A). Based on these facts, we propose that Cspp1 is at the top of the “ciliary gating hierarchy,” and it would be an interesting future task to monitor if gating defects in Cspp1-negative cells are indeed mediated by a decreased TZ amount of Rpgrip1l.
Rpgrip1l controls ciliary gating via Cep290
Here we show that Rpgrip1l controls ciliary gating via ensuring the proper amount of Cep290 at the TZ (Figure 3). The role of Cep290 in ciliary gating is addressed by several studies in which a reduced amount of ciliary membrane proteins like Arl13b and Ac3 in the absence of Cep290 has been shown (Craige et al., 2010; Li et al., 2016; Shimada et al., 2017; Kilander et al., 2018; Kim et al., 2018; Molinari et al., 2019). How Cep290 implements this function is not yet clearly understood, but it was shown that Cep290 governs ciliary protein composition by interacting with Nphp5 (Barbelanne et al., 2015a; Li et al., 2016; Shimada et al., 2017; Kim et al., 2018). Cep290 binds Nphp5 thereby covering the calmodulin binding of Nphp5 and promoting the recruitment of Nphp5 to the TZ (Kim et al., 2018). In the absence of Cep290, the Nphp5 amount at the TZ is reduced, and this amount is restored by eupatilin treatment, which inhibits calmodulin binding to Nphp5 (Kim et al., 2018). In our study, we observed a similar rescue of ciliary amounts of Arl13b and Sstr3, and of Nphp5 TZ amounts, by eupatilin treatment in Rpgrip1l-deficient cells (Figure 4). Together with the rescue of the Arl13b and Sstr3 amount by transfection of tagged Cep290 (Figure 3), these data underpin our assertion that Rpgrip1l exerts is gatekeeper function via Cep290 and Nphp5 (Figure 5). Interestingly, the phenotype of Nphp5-negative mice is not as striking as the phenotype of Cep290-mutant or Rpgrip1l-mutant mice, raising the question whether ciliary gating can really be regulated by Nphp5. Nonetheless, it was shown that the phenotype of Nphp5-mutant mice shows similarities to the phenotype of Cep290-mutant mice and that patients with mutations in NPHP5 can develop similar ciliopathy syndromes than patients with mutations in CEP290 (Otto et al., 2005; Chang et al., 2006; Helou et al., 2007; Stone et al., 2011; Ronquillo et al., 2016). It was also shown that the interaction of Cep290 and Nphp5 is required for ciliogenesis (Barbelanne et al., 2013) and that Nphp5 as well as Cep290 regulates components of the BBSome (octameric protein complex consisting of Bardet-Biedl syndrome) (Barbelanne et al., 2015a). In addition, Nphp5 interacts with components of the exocyst complex, a protein complex involved in exocytosis and thereby ciliogenesis (Zuo et al., 2009; Sang et al., 2011). Together with the fact we have previously discussed in this paper, that Cep290 and Rpgrip1l exert additional function that may lead to more severe phenotypes in these mutants, it is conceivable that the gating defect in Rpgrip1l- and Cep290-negative cells is actually mediated by Nphp5.
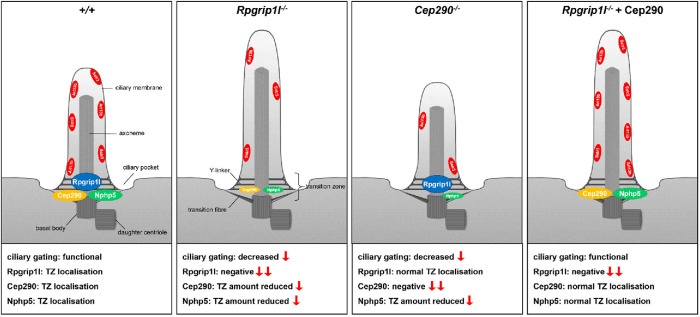
FIGURE 5:
Graphical abstract of ciliary gating defects and their rescue in Rpgrip1l- and Cep290-negative NIH3T3 cells. Rpgrip1l controls ciliary gating via ensuring the proper amount of Cep290 at the TZ, which in turn regulates the ciliary amount of Nphp5. In the absence of Rpgrip1l, the ciliary amount of Cep290 and Nphp5 is reduced. This leads to ciliary gating defects indicated by lower ciliary Arl13b and Sstr3 levels. In Cep290-negative cells, the ciliary amount of Rpgrip1l is unaltered, whereas the ciliary amount of Nphp5 is reduced. This leads to down-regulation of ciliary gating in Cep290–/– cells. Restoration of the Cep290 amount in Rpgrip1l–/– cells via transfection of full-length Cep290 rescues ciliary gating defects. We propose that the rescue of ciliary gating in transfected Rpgrip1l–/–cells is mediated by a restored ciliary amount of Nphp5.
Taken together, our results show that eupatilin may serve as a potential agent for the treatment of ciliopathies caused by mutations in RPGRIP1L and CEP290. We assume that the rescue of the ciliary gating defect in patients with variants of RPGRIP1L and CEP290 would bring enormous benefits, even though it may only represent one part of the complex disease pattern. To further validate this treatment strategy, we propose to test the eupatilin treatment in in vivo models by treating Rpgrip1l–/– and Cep290–/– mouse embryos in utero or ex vivo.
Ciliary length control and ciliary gating is mediated by different mechanisms
Strikingly, the ciliary length alterations in Rpgrip1l-deficient cells were not rescued by the eupatilin treatment (Figure 4I) or by the transfection of Cep290 fusion proteins (Supplemental Figure S4, C and J), indicating that ciliary length alterations and ciliary gating defects are caused by different mechanisms. Ciliogenesis and the associated regulation of ciliary length involve complex regulatory mechanisms that have been intensively studied but are not yet well understood. Many studies have investigated mechanisms involved in timing of cilium formation, cilium maintenance, and cilium disassembly, highlighting the importance of the control of the cell cycle and the conversion of centrioles into BBs, vesicle and membrane trafficking, IFT machinery, ciliary gating, as well as actin-based regulation mechanisms (Ishikawa and Marshall, 2011; Avasthi and Marshall, 2012; Keeling et al., 2016; Wang and Dynlacht, 2018; Copeland, 2020; Kumar and Reiter, 2020).
Taking into consideration that Rpgrip1l-mutant and Cep290-mutant NIH3T3 cells feature opposite ciliary length alterations (Figures 4, I and J and Supplemental Figure S4, C and D), which probably cannot be explained by the corresponding defect in ciliary gating, different mechanisms have to be affected in the respective mutant. It has been shown that Cep290 regulates ciliary localization of BBS4 via interaction with PCM-1, thereby regulating BBSome integrity and ciliary trafficking (Stowe et al., 2012; Klinger et al., 2014; Kobayashi et al., 2014; Barbelanne et al., 2015a). In this context, Cep290 is involved in the recruitment of the small GTPase Rab8 to the cilium. Rab8 regulates vesicle trafficking and has been shown to collaborate with the BBS protein complex involved in ciliary membrane formation (Nachury et al., 2007; Yoshimura et al., 2007; Kim et al., 2008; Tsang et al., 2008). Taken together, the reduction of ciliary length in Cep290–/– NIH3T3 cells could result from a failed recruitment of BBSome components and Rab8 followed by a misregulated vesicle trafficking and disrupted ciliary membrane elongation. In this regard, the reduction of Cep290 at the TZ in Rpgrip1l–/– NIH3T3 cells by around 50% does not seem strong enough to disrupt Cep290-dependent vesicle trafficking and ciliary membrane elongation in this mutant.
Instead it has been shown that Rpgrip1l interacts with Myosin Va (Assis et al., 2017), a motor protein required for preciliary vesicle transportation to the mother centriole during ciliogenesis (Wu et al., 2018). The loss of Myosin Va leads to decreased ciliogenesis in RPE-1 and murine inner medullary collecting duct (IMCD3) cells and it is assumed that an increased Myosin Va amount at the ciliary base leads to elongated cilia (Assis et al., 2017; Kohli et al., 2017; Copeland, 2020). As we discussed in a previous study (Wiegering et al., 2018b), mammalian Myosin Va is closely related to Myosin V in Drosophila melanogaster (Bonafé and Sellers, 1998). Myosin V is a substrate of proteasomal degradation (Pocha et al., 2011), making it very likely that Myosin Va is likewise degraded by the proteasome. Since Rpgrip1l regulates the proteasome specifically at the ciliary base (Gerhardt et al., 2015), Rpgrip1l could regulate the degradation of Myosin Va via the ciliary proteasome. A loss of Rpgrip1l would lead to an increased amount of Myosin Va at the ciliary base, resulting in an increased vesicle transport during ciliogenesis and an increased ciliary length. Remarkably, we previously showed that Rpgrip1l deficiency leads to a reduced autophagic activity and that the treatment of Rpgrip1l–/– MEFs with autophagy activators rescues cilia length (Struchtrup et al., 2018). For this reason, Rpgrip1l might regulate cilia length by controlling autophagy. Investigating the complex mechanisms of ciliogenesis will be a major task for the future of cilia research.
The role of TZ proteins in ciliary gating
Interestingly, Garcia-Gonzalo et al. revealed that the loss of the TZ gatekeeper protein Tmem67 diminishes the ciliary amount of Arl13b and Ac3 but the ciliary amount of Smo remains normal (Garcia-Gonzalo et al., 2011), demonstrating the existence of a specificity between the gatekeeper proteins and the proteins which are allowed to cross the TZ. In Rpgrip1l–/– MEFs, the ciliary amount of Smo is also unaltered (Gerhardt et al., 2015). The lack of Cep290 and Nphp5 in RPE-1 cells results in a reduced ciliary amount of Smo (Barbelanne et al., 2015b). Since the amount of Cep290 is reduced in Rpgrip1l–/– MEFs (Wiegering et al., 2018a), the expectation would be that the amount of Smo was decreased in these MEFs. However, these findings have been made in different cell types making it possible that the ciliary gating function of these proteins might be cell type specific. The analysis of this hypothesis is an exciting subject of future studies which would shed further light on the ciliary gating function of the TZ.
Many ciliopathies can be attributed to mutations in genes encoding TZ proteins (Hildebrandt et al., 2011; Czarnecki and Shah, 2012). For this reason, the assembly and function of the TZ is a hot topic in biomedical research. A lot of proteins participate in TZ assembly and/or function as ciliary gatekeepers at the TZ (Craige et al., 2010; Chih et al., 2011; Garcia-Gonzalo et al., 2011; Huang et al., 2011; Sang et al., 2011; Williams et al., 2011; Aubusson-Fleury et al., 2012; Cevik et al., 2013; Wang et al., 2013; Awata et al., 2014; Basiri et al., 2014; Klinger et al., 2014; Tuz et al., 2014; Bachmann-Gagescu et al., 2015; Barbelanne et al., 2015b; Damerla et al., 2015; Roberson et al., 2015; Yee et al., 2015; Lambacher et al., 2016; Li et al., 2016; Pratt et al., 2016; Slaats et al., 2016; Vieillard et al., 2016; Wei et al., 2016; Dyson et al., 2017; Lu et al., 2017; Schou et al., 2017; Shi et al., 2017; Takao et al., 2017; Jensen et al., 2018; Scheidel and Blacque, 2018; Wiegering et al., 2018a; Jack et al., 2019; Lapart et al., 2019; Lewis et al., 2019). However, the relationships between these proteins and hence the mechanisms underlying ciliary gating at the TZ remain largely elusive. Recently, we showed that Rpgrip1l represents a central factor in vertebrate TZ assembly (Wiegering et al., 2018a). Our current study reveals that Rpgrip1l also regulates ciliary gating by ensuring the proper amount of Cep290 at the vertebrate TZ. Combining our results with previous findings, we suggest a protein hierarchy regulating ciliary gating in which Cspp1, Rpgrip1l, Cep290, and Nphp5 are involved. Our work is an important piece of a puzzle depicting this fundamental ciliary process. The completion of this puzzle will be one of the most important tasks of cilia research in the next few years.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
See Table 1 for key resources.
TABLE 1:
Key resources.
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Actin | Sigma-Aldrich | #A2066 |
| Rabbit polyclonal anti-Arl13b | Proteintech | #17711-1-AP |
| Mouse monoclonal anti-Arl13b | Antibodies Incorporated | #75-287 |
| Rabbit polyclonal anti-Cep290 | Abcam | #ab84870 |
| Rabbit polyclonal anti-Cspp1 | Proteintech | #11931-1-AP |
| Rabbit polyclonal anti-Flag | Sigma-Aldrich | #F7425 |
| Rabbit polyclonal anti-Gapdh | Abcam | #ab9485 |
| Rabbit polyclonal anti-GFP | Thermo Fisher Scientific | #A-6455 |
| Rabbit polyclonal anti-Nphp5/Iqcb1 | Proteintech | #15747-1-AP |
| Rabbit polyclonal anti-c-Myc | Santa Cruz Biotechnology | #sc-789 |
| Rabbit polyclonal anti-Sept2 | Proteintech | #11397-1-AP |
| Rabbit polyclonal anti-Sept7 | Proteintech | #13818-1-AP |
| Rabbit polyclonal anti-Sstr3 | Proteintech | #20696-1-AP |
| Rabbit polyclonal anti-Sstr3 | Pierce Biotechnology | #PA3-207 |
| Goat polyclonal anti-Sstr3 | Santa Cruz Biotechnology |
#sc-11617 |
| Mouse monoclonal anti-acetylated α-Tubulin | Santa Cruz Biotechnology | #sc-23950 |
| Mouse monoclonal anti-acetylated α-Tubulin | Sigma-Aldrich | #T-6739 |
| Goat polyclonal anti-γ-Tubulin | Santa Cruz Biotechnology. | #sc-7396 |
| Goat polyclonal anti-mouse IgG2b Alexa Fluor 488 | Thermo Fisher Scientific | #A21141 |
| Goat polyclonal anti-mouse IgG2a Alexa Fluor 488 | Thermo Fisher Scientific | #A21131 |
| Goat polyclonal anti-mouse IgG1 Alexa Fluor 488 | Thermo Fisher Scientific | #A21121 |
| Goat polyclonal anti-mouse IgG1 Alexa Fluor 568 | Thermo Fisher Scientific | #A21124 |
| Goat polyclonal anti-mouse IgG2b Alexa Fluor 594 | Thermo Fisher Scientific | #21145 |
| Goat polyclonal anti-mouse IgG2a Alexa Fluor 594 | Thermo FisherScientific | #A21135 |
| Donkey polyclonal anti-goat Dylight405 | Jackson ImmunoResearch | #AB_2340426 |
| Donkey polyclonal anti-mouse Alexa488 | Jackson ImmunoResearch | #AB_2340846 |
| Donkey polyclonal anit-rabbit Cy3 | Jackson ImmunoResearch via Dianova | # 111-165-045 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMSO | Sigma-Aldrich | #W387520 |
| Eupatilin | Sigma-Aldrich | #SML1689 |
| Experimental Models: Cell Lines | ||
| HEK293 | DSMZ | #ACC305 |
| NIH3T3 | DSMZ | #ACC59 |
| Oligonucleotides | ||
| Rpgrip1l-T3b-for:GAATGGCCACCAAGTTAATACGGCTAG | This study | N/A |
| Rpgrip1l-T3b-rev:CTTCAGGATCTGACAGAGAGCAAGCCTC | This study | N/A |
| T3 off-1 for:CTGTCAGGTTTCCCAGTGTGCAG | This study | N/A |
| T3 off-1 rev:CTCTCAGCTCCTTTTAGGTCTCCAG | This study | N/A |
| T3 off-2 for:ATCCAGCCAAACCCTGCCTGTTC | This study | N/A |
| T3 off-2 rev:GGTTTGTCTCTGTCCTGACATGTCAC | This study | N/A |
| T3 off-3 for:GTCTCCTTCAGACCCACTGAAGTG | This study | N/A |
| T3 off-3 rev:GTCCCAGGAAGCCAGGCTGTTG | This study | N/A |
| Recombinant DNA | ||
| pMyc-mNphp1 | Kindly provided by Sophie Saunier | N/A |
| pFlag-mCep290 | Addgene | #27381 |
| eGFP-hCEP290 | Kindly provided by Hemant Khanna | N/A |
| Software and Algorithms | ||
| ImageJ | National Institutes of Heath | https://imagej.nih.gov/ij |
| AxioVision Rel. 4.8 software package | Carl Zeiss AG | https://www.zeiss.de/mikroskopie |
| NIS-Elements | Nikon | https://www.microscope.healthcare.nikon.com |
| Adobe Photoshop CS2 | Adobe | https://www.adobe.com |
| GraphPad Prism | GraphPad | https://www.graphpad.com |
Cell lines
We used two different cell lines in this study. NIH3T3 cells (#ACC59) and HEK293 cells (#ACC35) were both purchased by the German Collection of Microorganisms and Cell Cultures GmbH (DSMZ). Cells were grown in DMEM supplemented with 10% fetal calf serum (FCS), 1/100 (vol/vol) L-glutamine (Life Technologies), 1/100 (vol/vol) sodium pyruvate (Life Technologies), 1/100 (vol/vol) nonessential amino acids (Life Technologies), and 1/100 (vol/vol) pen/strep (Life Technologies) at 37°C and 5% CO2. The following clones were used: Rpgrip1l–/– NIH3T3 cells (clone 10-61), Cep290–/– NIH3T3 cells (clones 39-10, 39-51, 39-53) (Wiegering et al., 2018a), Nphp1–/– NIH3T3 cells (clone 21-21) (Wiegering et al., 2018a), Invs–/– NIH3T3 cells (clones 48-7, 48-20) (Wiegering et al., 2018a), and RPGRIP1L–/– HEK293 cells (clone 1-7) (Wiegering et al., 2018a).
Primary cell culture
We isolated MEFs from single mouse embryos (male and female) at embryonic stage (E) 12.5 after standard procedures. MEFs were grown in DMEM supplemented with 10% FCS, 1/100 (vol/vol) L-glutamine (Life Technologies), 1/100 (vol/vol) sodium pyruvate (Life Technologies), 1/100 (vol/vol) nonessential amino acids (Life Technologies), and 1/100 (vol/vol) pen/strep (Life Technologies) at 37°C and 5% CO2. The following mutant mice were used: Rpgrip1l-mutant mice on a C3H-background (Vierkotten et al., 2007) and Nphp4-mutant mice on a C57BL/6J-background (Wiegering et al., 2018a).
Cell culture, transfection, and drug treatment
Ciliogenesis in confluent-grown MEFs and NIH3T3 cells was induced by serum starvation (0.5% FCS) for at least 24 h. For DNA transfection, Lipofectamin 3000 (Invitrogen) was used following the manufacturer’s guidelines. Appropriate empty vectors were used as transfection control (TF-Ctrl). NIH3T3 cells were treated with 20 μM eupatilin (#SML1689; Sigma-Aldrich) or DMSO as a solvent control for 24 h.
Antibodies and plasmids
Cells were immunolabeled with primary antibodies targeting Arl13b (#17711-1-AP; Proteintech and #75-287; Antibodies Incorporated), Cep290 (#ab84870; Abcam), Cspp1 (#11931-1-AP; Proteintech), Flag (#F7425; Sigma-Aldrich), Myc (#sc-789; Santa Cruz Biotechnology), Nphp5 (#15747-1-AP; Proteintech), Sept2 (#11397-1-AP; Proteintech), Sept7 (#13818-1-AP; Proteintech), Sstr3 (#20696-1-AP; Proteintech; #PA3-207; Pierce Biotechnology and #11617; Santa Cruz Biotechnology), acetylated α-tubulin (#sc-23950; Santa Cruz Biotechnology; #T-6793; Sigma-Aldrich), and γ-tubulin (#sc-7396; Santa Cruz Biotechnology). The generation of the polyclonal antibody against Rpgrip1l was described formerly (Vierkotten et al., 2007).
The following plasmids were used: pMyc-mNphp1 (kindly provided by Sophie Saunier), eGFP-hCEP290 (kindly provided by Hemant Khanna), and pFlag-mCep290 (#27381; Addgene). pMyc-mNphp1 encodes for the murine full-length Nphp1 protein fused to a Myc-tag (vector: CMV), GFP-hCEP290 encodes for the human full-length Cep290 protein fused to a GFP-tag (vector: CMV), and pFlag-mCep290 encodes for the murine full-length Cep290 protein fused to a Flag-tag (vector: CMV2).
CRISPR/Cas9-mediated gene inactivation
Inactivation of mouse Rpgrip1l in NIH3T3 cells was performed as previously described (Wiegering et al., 2018a). We choose a target site which is located in exon3 of the gene (Supplemental Figure S2). After inactivation and single-cell cloning, eight clones, which on RFLP analysis appeared to have lost the diagnostic EagI recognition sequence, were further analyzed. To establish the genotype, individual alleles were cloned and sequenced (Supplemental Figure S2).
Sequences of the target site and primer pairs used to amplify the targeted region are as follows:
Rpgrip1l –T3: CTCGAGTTAACACCGGCCGCCGG
Rpgrip1l-T3b-for: GAATGGCCACCAAGTTAATACGGCTAG
Rpgrip1l-T3b-rev: CTTCAGGATCTGACAGAGAGCAAGCCTC.
Off-target analyses
Off-target analyses were performed by RFLP analyses as previously described (Wiegering et al., 2018a). For the on-target Rpgrip1l-T3 there exist no off-targets carrying one, two, or three mismatches. From the remaining four-mismatch off-targets, we tested the top-three ranking sites (Hsu et al., 2013) (crispr.mit.edu/) on the DNAs from the same eight-set clones analyzed for targeting of the on-target. In summary, we did not detect any mutations (unpublished data).
Sequences of the off-target sites and primer pairs used for amplification are as follows:
T3 offtarget-1: CACGAGTCAGCACCGGCCACTGG
T3 off-1 for: CTGTCAGGTTTCCCAGTGTGCAG
T3 off-1 rev: CTCTCAGCTCCTTTTAGGTCTCCAG
T3 offtarget-2: CTCTACTGAACAACGGCCGCAGG
T3 off-2 for: ATCCAGCCAAACCCTGCCTGTTC
T3 off-2 rev: GGTTTGTCTCTGTCCTGACATGTCAC
T3 offtarget-3: ATCCAGTTGACACCGGCCTCTGG
T3 off-3 for: GTCTCCTTCAGACCCACTGAAGTG
T3 off-3 rev: GTCCCAGGAAGCCAGGCTGTTG.
The following restriction enzymes were used: BslI (T3 off-target-1), EagI (T3 off-target-2), and BslI (T3 off-target-1).
Image acquisition
Image acquisition and data analysis were carried out at room temperature using a Zeiss Imager.A2 microscope, 100×, NA 1.46 oil immersion objective lens (Carl Zeiss AG), a monochrome charge-coupled device camera (AxioCam MRm, Carl Zeiss AG), and the AxioVision Rel. 4.8 software package (Carl Zeiss AG), or a TI-Eclipse Nikon inverted microscope, 100×, 1.49 oil immersion objective lens coupled with a 95B Prime 22 mm Photometrics sCMOS camera. A Nikon fluorescent lamp and a quadriband dichroic block were used to detect blue, green, and red fluorescence. The acquisition software NIS was used. Three single-plane images per cilium were obtained in an 8-bit grayscale modus respectively covering the specific spectrum of the used fluorochrome. Appropriate anti-mouse, anti-rabbit and anti-goat Alexa405, Cy3, Alexa594, and Alexa488 antibodies were used as fluorochromes.
Immunofluorescence
For immunofluorescence on MEFs, NIH3T3 cells, and HEK293 cells, cells were plated on coverslips until confluency. MEFs and NIH3T3 cells were serum-starved for at least 24 h. Cells were fixed with 4% paraformaldehyde (for staining with the antibodies to Cep290, Rpgrip1l, Flag, Myc, Sept7, and Arl13b) or methanol (for staining with the antibodies to Cspp1, Nphp5, Sept2, and Sstr3). Fixed cells were rinsed three times with phosphate-buffered saline (PBS), followed by a permeabilization step with PBS/0.5% Triton X-100 for 10 min. The samples were rinsed three times with PBS. Samples were incubated for at least 10 min at room temperature in PBST (PBS/0.1% Triton X-100) containing 10% donkey serum or 10% normal goat serum. Diluted primary antibodies (in blocking solution) were incubated overnight at 4°C. After three washing steps with PBST, incubation with fluorescent secondary antibody (diluted in blocking solution) was performed at room temperature for 1 h followed by several washing steps and subsequent embedding with Mowiol optionally containing DAPI.
Western blotting
Whole-cell lysates were obtained by lysis with radioimmunoprecipitation buffer (150 mM sodium chloride, 50 mM Tris-HCl, pH 7.4, 0.1% sodium deoxycholate, and 1 mM EDTA). Protein content was measured by the Bradford method, and samples were normalized. Total protein (20 mg ) was separated by SDS–PAGE on polyacrylamide gels (#456-1093; Bio-Rad Laboratories) and transferred to a PVDF membrane (#162-0176; Bio-Rad). The membrane was probed with antibodies against Flag (#F7425; Sigma-Aldrich), GFP (#A-6455; Thermo Fisher Scientific), and Myc (#sc-789; Santa Cruz Biotechnology). Anti-Gapdh (#ab9485; Abcam) antibody was used as loading control. Proteins were detected with secondary antibodies conjugated to horseradish peroxidase (RPN4201 and RPN4301) and the SuperSignal West Pico PLUS detection kit (#34580; Thermo Fisher Scientific). Visualization of protein bands was realized by GBox (SYNGENE).
Quantification and presentation
Ciliary protein staining and protein bands intensity were quantified using ImageJ (National Institutes of Health). Intensity measurement of proteins based on immunofluorescence staining was performed as described before (Garcia-Gonzalo et al., 2011; Garcia-Gonzalo et al., 2015; Gerhardt et al., 2015; Roberson et al., 2015; Yee et al., 2015; Struchtrup et al., 2018; Wiegering et al., 2018a). Triplets of 8-bit single-plane grayscale images were merged via ImageJ. The merged images were not further processed and the signal intensities were measured. The ciliary length has to be taken into account while quantifying the ciliary amount of Arl13b and Sstr3 in different genotypes. Therefore we used the area marked by acetylated α-tubulin as a reference and quantified the average pixel intensity of the Arl13 and Sstr3 staining. For all other ciliary protein intensities (Cep290, GFP, Flag, Cspp1, Myc, Nphp5, Rpgrip1l, Sept2, and Sept7), we selected the region labeled by γ-tubulin (for BB proteins) or the area in between the γ-tubulin staining and the proximal part of the acetylated α-tubulin staining and measured the total pixel intensity. To exclude unspecific staining from the measurements, we subtracted the mean value of the average pixel intensity (in the case of Arl13b and Sstr3) or of the total pixel intensity (all other ciliary proteins) of three neighboring regions free from specific staining.
Representative images were processed after quantification of ciliary protein staining was completed. The images were processed by means of background subtraction and contrast settings via Adobe Photoshop CS2.
Statistical analysis
Data are presented as mean ± SEM. Two-tailed t test with Welch´s correction was performed for all data in which two datasets were compared. Analysis of variance (ANOVA) and Tukey honest significance difference (HSD) tests were used for all data in which more than two datasets were compared. *P < 0.05 was considered to be statistically significant, **P < 0.01 was defined as statistically very significant, and ***P < 0.001 was noted as statistically high significant. Sample sizes are indicated in the figure legends and the power of statistical tests was verified via post-hoc power calculations.
All statistical data analysis and graph illustrations were performed by using GraphPad Prism (GraphPad Software) and the Post-hoc Power Calculator (https://clincalc.com/Stats/).
Data and code availability
This study did not generate datasets.
Supplementary Material
Acknowledgments
The authors thank Matias Zurbriggen and Leonie-Alexa Koch for their generous help to enable the continuation of the study. Moreover, we are grateful to Sophie Saunier for providing the Myc-mNphp1 construct. We thank the cell imaging facility of the IBPS (Institut de Biologie Paris-Seine FR3631, Sorbonne Université, CNRS, Paris, France) for their technical assistance. This work was funded by the Fondation ARC pour la Recherche sur le Cancer (Project ARC PJA 20171206591 to S.S.M.), the Fondation pour la Recherche Médicale (Equipe FRM EQU201903007943 to S.S.M.), and the German Research Foundation (DFG; Grant No. WI 5451/1-1 to A.W.).
Abbreviations used:
- Ac3
adenylate cyclase 3
- ANOVA
analysis of variance
- Arl13b
ADP ribosylation factor-like GTPase 13B
- BB
basal body
- BBS
Bardet-Biedl syndrome
- Cspp1
centrosome and spindle pole-associated protein 1
- Cep
centrosomal protein
- FCS
fetal calf serum
- HSD
honest significance difference
- Invs
Inversin
- MEF
mouse embryonic fibroblast
- Nphp
nephrocystin
- PBS
phosphate-buffered saline
- PCM1
pericentriolar material 1
- RPE
retinal pigment epithelial
- Sept
septin
- Smo
smoothened
- Sstr3
somatostatin receptor 3
- SSX2IP
SSX family member 2 interacting protein
- TF-Ctrl
transfection control
- TZ
transition zone.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-03-0190) on February 24, 2021.
REFERENCES
- Andreu-Cervera A, Anselme I, Karam A, Laclef C, Catala M, Schneider-Maunoury S (2019). The ciliopathy gene ftm/rpgrip1l controls mouse forebrain patterning via region-specific modulation of hedgehog/gli signaling. J Neurosci 39, 2398–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis L, Silva-Junior R, Dolce L, Alborghetti M, Honorato R, Nascimento A, Melo-Hanchuk T, Trindade D, Tonoli C, Santos C, et al. (2017). The molecular motor Myosin Va interacts with the cilia-centrosomal protein RPGRIP1L. Sci Rep 7, 43692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubusson-Fleury A, Lemullois M, de Loubresse N, Laligné C, Cohen J, Rosnet O, Jerka-Dziadosz M, Beisson J, Koll F (2012). The conserved centrosomal protein FOR20 is required for assembly of the transition zone and basal body docking at the cell surface. J Cell Sci 125, 4395–4404. [DOI] [PubMed] [Google Scholar]
- Avasthi P, Marshall W (2012). Stages of ciliogenesis and regulation of ciliary length. Differentiation 83, 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awata J, Takada S, Standley C, Lechtreck K, Bellvé K, Pazour G, Fogarty K, Witman G (2014). NPHP4 controls ciliary trafficking of membrane proteins and large soluble proteins at the transition zone. J Cell Sci 127, 4714–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann-Gagescu R, Dona M, Hetterschijt L, Tonnaer E, Peters T, de Vrieze E, Mans D, van Beersum S, Phelps I, Arts H, et al. (2015). The ciliopathy protein CC2D2A associates with NINL and functions in RAB8-MICAL3-regulated vesicle trafficking. PLoS Genet 11, e1005575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbelanne M, Hossain D, Chan D, Peränen J, Tsang W (2015a). Nephrocystin proteins NPHP5 and Cep290 regulate BBSome integrity, ciliary trafficking and cargo delivery. Hum Mol Genet 24, 2185–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbelanne M, Hossain D, Chan D, Peränen J, Tsang W (2015b). Nephrocystin proteins NPHP5 and Cep290 regulate BBSome integrity, ciliary trafficking and cargo delivery. Hum Mol Genet 24, 2185–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbelanne M, Song J, Ahmadzai M, Tsang W (2013). Pathogenic NPHP5 mutations impair protein interaction with Cep290, a prerequisite for ciliogenesis. Hum Mol Genet 22, 2482–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bärenz F, Inoue D, Yokoyama H, Tegha-Dunghu J, Freiss S, Draeger S, Mayilo D, Cado I, Merker S, Klinger M, et al. (2013). The centriolar satellite protein SSX2IP promotes centrosome maturation. J Cell Biol 202, 91–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basiri M, Ha A, Chadha A, Clark N, Polyanovsky A, Cook B, Avidor-Reiss T (2014). A migrating ciliary gate compartmentalizes the site of axoneme assembly in Drosophila spermatids. Curr Biol 24, 2622–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzing T, Schermer B (2011). Transition zone proteins and cilia dynamics. Nat Genet 43, 723–724. [DOI] [PubMed] [Google Scholar]
- Besse L, Neti M, Anselme I, Gerhardt C, Rüther U, Laclef C, Schneider-Maunoury S (2011). Primary cilia control telencephalic patterning and morphogenesis via Gli3 proteolytic processing. Development 138, 2079–2088. [DOI] [PubMed] [Google Scholar]
- Betleja E, Cole D (2010). Ciliary trafficking: CEP290 guards a gated community. Curr Biol 20, R928–R931. [DOI] [PubMed] [Google Scholar]
- Bonafé N, Sellers J (1998). Molecular characterization of myosin V from Drosophila melanogaster. J Muscle Res Cell Motil 19, 129–141. [DOI] [PubMed] [Google Scholar]
- Cevik S, Sanders A, Van Wijk E, Boldt K, Clarke L, van Reeuwijk J, Hori Y, Horn N, Hetterschijt L, Wdowicz A, et al. (2013). Active transport and diffusion barriers restrict Joubert Syndrome-associated ARL13B/ARL-13 to an Inv-like ciliary membrane subdomain. PLoS Genet 9, e1003977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Khanna H, Hawes N, Jimeno D, He S, Lillo C, Parapuram S, Cheng H, Scott A, Hurd R, et al. (2006). In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet 15, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Laclef C, Moncayo A, Snedecor E, Yang N, Li L, Takemaru K, Paus R, Schneider-Maunoury S, Clark R (2015). The ciliopathy gene Rpgrip1l is essential for hair follicle development. J Invest Dermatol 135, 701–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Liu P, Chinn Y, Chalouni C, Komuves L, Hass P, Sandoval W, Peterson A (2011). A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol 14, 61–72. [DOI] [PubMed] [Google Scholar]
- Choi Y, Laclef C, Yang N, Andreu-Cervera A, Lewis J, Mao X, Li L, Snedecor E, Takemaru K, Qin C, et al. (2019). RPGRIP1L is required for stabilizing epidermal keratinocyte adhesion through regulating desmoglein endocytosis. PLoS Genet 15, e1007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coene K, Mans D, Boldt K, Gloeckner C, van Reeuwijk J, Bolat E, Roosing S, Letteboer S, Peters T, Cremers F, et al. (2011). The ciliopathy-associated protein homologs RPGRIP1 and RPGRIP1L are linked to cilium integrity through interaction with Nek4 serine/threonine kinase. Hum Mol Genet 20, 3592–3605. [DOI] [PubMed] [Google Scholar]
- Copeland J (2020). Actin-based regulation of ciliogenesis - The long and the short of it. Semin Cell Dev Biol 102, 132–138. [DOI] [PubMed] [Google Scholar]
- Craige B, Tsao C, Diener D, Hou Y, Lechtreck K, Rosenbaum J, Witman G (2010). CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol 190, 927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki PG, Shah JV (2012). The ciliary transition zone: from morphology and molecules to medicine. Trends Cell Biol 22, 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerla R, Cui C, Gabriel G, Liu X, Craige B, Gibbs B, Francis R, Li Y, Chatterjee B, San Agustin J, et al. (2015). Novel Jbts17 mutant mouse model of Joubert syndrome with cilia transition zone defects and cerebellar and other ciliopathy related anomalies. Hum Mol Genet 24, 3994–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Merdes A (2002). Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol 159, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delous M, Baala L, Salomon R, Laclef C, Vierkotten J, Tory K, Golzio C, Lacoste T, Besse L, Ozilou C, et al. (2007). The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat Genet 39, 875–881. [DOI] [PubMed] [Google Scholar]
- Dyson J, Conduit S, Feeney S, Hakim S, DiTommaso T, Fulcher A, Sriratana A, Ramm G, Horan K, Gurung R, et al. (2017). INPP5E regulates phosphoinositide-dependent cilia transition zone function. J Cell Biol 216, 247–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F, Corbit K, Sirerol-Piquer M, Ramaswami G, Otto E, Noriega T, Seol A, Robinson J, Bennett C, Josifova D, et al. (2011). A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet 43, 776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F, Phua S, Roberson E, Garcia Gr, Abedin M, Schurmans S, Inoue T, Reiter J (2015). Phosphoinositides regulate ciliary protein trafficking to modulate hedgehog signaling. Dev Cell 34, 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F, Reiter J (2012). Scoring a backstage pass: Mechanisms of ciliogenesis and ciliary access. J Cell Biol 197, 697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F, Reiter J (2017). Open sesame: how transition fibers and the transition zone control ciliary composition. Cold Spring Harb Perspect Biol 9, pii: a028134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt C, Lier J, Burmühl S, Struchtrup A, Deutschmann K, Vetter M, Leu T, Reeg S, Grune T, Rüther U (2015). The transition zone protein Rpgrip1l regulates proteasomal activity at the primary cilium. J Cell Biol 210, 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt C, Lier J, Kuschel S, Rüther U (2013). The ciliary protein Ftm is required for ventricular wall and septal development. PLoS One 8, e57545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G, Coyaud E, Gonçalves J, Mojarad B, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach J, Cheung S, et al. (2015). A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell 163, 1484–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Händel M, Schulz S, Stanarius A, Schreff M, Erdtmann-Vourliotis M, Schmidt H, Wolf G, Höllt V (1999). Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience 89, 909–926. [DOI] [PubMed] [Google Scholar]
- Helou J, Otto E, Attanasio M, Allen S, Parisi M, Glass I, Utsch B, Hashmi S, Fazzi E, Omran H, et al. (2007). Mutation analysis of NPHP6/CEP290 in patients with Joubert syndrome and Senior-Løken syndrome. J Med Genet 44, 657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N (2011). Ciliopathies. N Engl J Med 364, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A, Ikebe C, Tada M, Toda T (2014). Msd1/SSX2IP-dependent microtubule anchorage ensures spindle orientation and primary cilia formation. EMBO Rep 15, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A, Peddie C, Collinson L, Toda T (2015). Centriolar satellite- and hMsd1/SSX2IP-dependent microtubule anchoring is critical for centriole assembly. Mol Biol Cell 26, 2005–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P, Scott D, Weinstein J, Ran F, Konermann S, Agarwala V, Li Y, Fine E, Wu X, Shalem O, et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Milenkovic L, Jin H, Scott M, Nachury M, Spiliotis E, Nelson W (2010). A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329, 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Szymanska K, Jensen V, Janecke A, Innes A, Davis E, Frosk P, Li C, Willer J, Chodirker B, et al. (2011). TMEM237 is mutated in individuals with a Joubert syndrome related disorder and expands the role of the TMEM family at the ciliary transition zone. Am J Hum Genet 89, 713–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes A, Giles R, Srivastava S, Eley L, Whitehead J, Danilenko M, Raman S, Slaats G, Colville J, Ajzenberg H, et al. (2014). Murine Joubert syndrome reveals Hedgehog signaling defects as a potential therapeutic target for nephronophthisis. Proc Natl Acad Sci USA 111, 9893–9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall W (2011). Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol 12, 222–234. [DOI] [PubMed] [Google Scholar]
- Jack B, Mueller D, Fee A, Tetlow A, Avasthi P. (2019). Partially redundant actin genes in chlamydomonas control transition zone organization and flagellum-directed traffic. Cell Rep 27, 2459–2467.e2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen V, Lambacher N, Li C, Mohan S, Williams C, Inglis P, Yoder B, Blacque O, Leroux M (2018). Role for intraflagellar transport in building a functional transition zone. EMBO Rep 19, pii: e45862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen V, Leroux M (2017). Gates for soluble and membrane proteins, two trafficking systems (IFT and LIFT), establish a dynamic ciliary signaling compartment. Curr Opin Cell Biol 47, 83–91. [DOI] [PubMed] [Google Scholar]
- Jensen V, Li C, Bowie R, Clarke L, Mohan S, Blacque O, Leroux M (2015). Formation of the transition zone by Mks5/Rpgrip1L establishes a ciliary zone of exclusion (CIZE) that compartmentalises ciliary signalling proteins and controls PIP2 ciliary abundance. EMBO J 34, 2537–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Chiou Y, Wang E, Chien Y, Ho H, Tsai F, Lin C, Tsai S, Li H (2009). Essential role of nephrocystin in photoreceptor intraflagellar transport in mouse. Hum Mol Genet 18, 1566–1577. [DOI] [PubMed] [Google Scholar]
- Jiang S, Chiou Y, Wang E, Lin H, Lee S, Lu H, Wang C, Tang M, Li H (2008). Targeted disruption of Nphp1 causes male infertility due to defects in the later steps of sperm morphogenesis in mice. Hum Mol Genet 17, 3368–3379. [DOI] [PubMed] [Google Scholar]
- Keeling J, Tsiokas L, Maskey D. (2016). Cellular mechanisms of ciliary length control. Cells. 5, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilander M, Wang C, Chang C, Nestor J, Herold K, Tsai J, Nestor M, Lin Y (2018). A rare human CEP290 variant disrupts the molecular integrity of the primary cilium and impairs Sonic Hedgehog machinery. Sci Rep 8, 17335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kim S, Jung Y, Jung E, Kwon H, Kim J (2018). Eupatilin Rescues Ciliary Transition Zone Defects to Ameliorate Ciliopathy-Related Phenotypes. J Clin Invest 128, 3642–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krishnaswami S, Gleeson J (2008). CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum Mol Genet 17, 3796–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger M, Wang W, Kuhns S, Bärenz F, Dräger-Meurer S, Pereira G, Gruss O (2014). The novel centriolar satellite protein SSX2IP targets Cep290 to the ciliary transition zone. Mol Biol Cell 25, 495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Kim S, Lin Y, Inoue T, Dynlacht B (2014). The CP110-interacting proteins Talpid3 and Cep290 play overlapping and distinct roles in cilia assembly. J Cell Biol 204, 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli P, Höhne M, Jüngst C, Bertsch S, Ebert L, Schauss A, Benzing T, Rinschen M, Schermer B (2017). The ciliary membrane-associated proteome reveals actin-binding proteins as key components of cilia. EMBO Rep 18, 1521–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N (1999). Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol 147, 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Tsukita S (2003). Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle-dependent assembly/disassembly. J Cell Sci 116, 919–928. [DOI] [PubMed] [Google Scholar]
- Kumar D, Reiter J (2020). How the centriole builds its cilium: of mothers, daughters, and the acquisition of appendages. Curr Opin Struct Biol 66, 41–48. [DOI] [PubMed] [Google Scholar]
- Laclef C, Anselme I, Besse L, Catala M, Palmyre A, Baas D, Paschaki M, Pedraza M, Métin C, Durand B, Schneider-Maunoury S (2015). The role of primary cilia in corpus callosum formation is mediated by production of the Gli3 repressor. Hum Mol Genet 24, 4997–5014. [DOI] [PubMed] [Google Scholar]
- Lambacher N, Bruel A, van Dam T, Szymańska K, Slaats G, Kuhns S, McManus G, Kennedy J, Gaff K, Wu K, et al. (2016). TMEM107 recruits ciliopathy proteins to subdomains of the ciliary transition zone and causes Joubert syndrome. Nat Cell Biol 18, 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M, Gopal D, Kim J, Saleem S, Silhavy J, Louie C, Thacker B, Williams Y, Zaki M, Gleeson J (2011). Defective Wnt-dependent cerebellar midline fusion in a mouse model of Joubert syndrome. Nat Med 17, 726–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapart J, Gottardo M, Cortier E, Duteyrat J, Augière C, Mangé A, Jerber J, Solassol J, Gopalakrishnan J, Thomas J, Durand B (2019). Dzip1 and Fam92 form a ciliary transition zone complex with cell type specific roles in Drosophila. Elife 8, e49307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis W, Bales K, Revell D, Croyle M, Engle S, Song C, Malarkey E, Uytingco C, Shan D, Antonellis P, et al. (2019). Mks6 mutations reveal tissue- and cell type-specific roles for the cilia transition zone. FASEB J 33, 1440–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Jensen V, Park K, Kennedy J, Garcia-Gonzalo F, Romani M, De Mori R, Bruel A, Gaillard D, Doray B, et al. (2016). MKS5 and CEP290 dependent assembly pathway of the ciliary transition zone. PLoS Biol 14, e1002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Klena N, Gabriel G, Liu X, Kim A, Lemke K, Chen Y, Chatterjee B, Devine W, Damerla R, et al. (2015). Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 521, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Guo S, Dutcher S (2018). RPGRIP1L helps to establish the ciliary gate for entry of proteins. J Cell Sci 131, pii: jcs220905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie C, Caridi G, Lopes V, Brancati F, Kispert A, Lancaster M, Schlossman A, Otto E, Leitges M, Gröne H, et al. (2010). AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat Genet 42, 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Galeano M, Ott E, Kaeslin G, Kausalya P, Kramer C, Ortiz-Brüchle N, Hilger N, Metzis V, Hiersche M, et al. (2017). Mutations in DZIP1L, which encodes a ciliary-transition-zone protein, cause autosomal recessive polycystic kidney disease. Nat Genet 49, 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhivanan K, Aguilar R (2014). Ciliopathies: the trafficking connection. Traffic 15, 1031–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen D, Koenekoop R, Khanna H, Jenkins P, Lopez I, Swaroop A, Martens J (2007). Hypomorphic CEP290/NPHP6 mutations result in anosmia caused by the selective loss of G proteins in cilia of olfactory sensory neurons. Proc Natl Acad Sci USA 104, 15917–15922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre J, Williams C, Martens J (2013). Smelling the roses and seeing the light: gene therapy for ciliopathies. Trends Biotechnol 31, 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison H, Valente E (2017). Motile and non-motile cilia in human pathology: from function to phenotypes. J Pathol 241, 294–309. [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Saijoh Y, Tsuchiya K, Shirayoshi Y, Takai S, Taya C, Yonekawa H, Yamada K, Nihei H, Nakatsuji N, et al. (1998). Cloning of inv, a gene that controls left/right asymmetry and kidney development. Nature 395, 177–181. [DOI] [PubMed] [Google Scholar]
- Molinari E, Ramsbottom S, Srivastava S, Booth P, Alkanderi S, McLafferty S, Devlin L, White K, Gunay-Aygun M, Miles C, Sayer J (2019). Targeted exon skipping rescues ciliary protein composition defects in Joubert syndrome patient fibroblasts. Sci Rep 9, 10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D, Turnpenny L, Goodship J, Dai W, Majumder K, Matthews L, Gardner A, Schuster G, Vien L, Harrison W, et al. (1998). Inversin, a novel gene in the vertebrate left-right axis pathway, is partially deleted in the inv mouse. Nat Genet 20, 149–156. [DOI] [PubMed] [Google Scholar]
- Mukherjee I, Roy S, Chakrabarti S (2019). Identification of important effector proteins in the FOXJ1 transcriptional network associated with ciliogenesis and ciliary function. Front Genet 10, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M, Loktev A, Zhang Q, Westlake C, Peränen J, Merdes A, Slusarski D, Scheller R, Bazan J, Sheffield V, Jackson P. (2007). A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213. [DOI] [PubMed] [Google Scholar]
- Odabasi E, Gul S, Kavakli I, Firat-Karalar E (2019). Centriolar satellites are required for efficient ciliogenesis and ciliary content regulation. EMBO Rep. 20, e47723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omran H (2010). NPHP proteins: gatekeepers of the ciliary compartment. J Cell Biol 190, 715–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto E, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, Muerb U, O’Toole J, Helou J, Attanasio M, et al. (2005). Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat Genet 37, 282–288. [DOI] [PubMed] [Google Scholar]
- Patnaik S, Zhang X, Biswas L, Akhtar S, Zhou X, Kusuluri D, Reilly J, May-Simera H, Chalmers S, McCarron J, Shu X (2018). RPGR protein complex regulates proteasome activity and mediates store-operated calcium entry. Oncotarget 9, 23183–23197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzke S, Redick S, Warsame A, Murga-Zamalloa C, Khanna H, Doxsey S, Stokke T (2010). CSPP is a ciliary protein interacting with Nephrocystin 8 and required for cilia formation. Mol Biol Cell 21, 2555–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocha S, Shevchenko A, Knust E (2011). Crumbs regulates rhodopsin transport by interacting with and stabilizing myosin V. J Cell Biol 195, 827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt M, Titlow J, Davis I, Barker A, Dawe H, Raff J, Roque H (2016). Drosophila sensory cilia lacking MKS proteins exhibit striking defects in development but only subtle defects in adults. J Cell Sci 129, 3732–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachel R, Yamamoto E, Dewanjee M, May-Simera H, Sergeev Y, Hackett A, Pohida K, Munasinghe J, Gotoh N, Wickstead B, et al. (2015). CEP290 alleles in mice disrupt tissue-specific cilia biogenesis and recapitulate features of syndromic ciliopathies. Hum Mol Genet 24, 3775–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J, Blacque O, Leroux M (2012). The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep 13, 608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J, Leroux M (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J, Skarnes W (2006). Tectonic, a novel regulator of the Hedgehog pathway required for both activation and inhibition. Genes Dev 20, 22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson E, Dowdle W, Ozanturk A, Garcia-Gonzalo F, Li C, Halbritter J, Elkhartoufi N, Porath J, Cope H, Ashley-Koch A, et al. (2015). TMEM231, mutated in orofaciodigital and Meckel syndromes, organizes the ciliary transition zone. J Cell Biol 209, 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquillo C, Hanke-Gogokhia C, Revelo M, Frederick J, Jiang L, Baehr W (2016). Ciliopathy-associated IQCB1/NPHP5 protein is required for mouse photoreceptor outer segment formation. FASEB J 30, 3400–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L, Miller J, Corbit K, Giles R, Brauer M, Otto E, Baye L, Wen X, Scales S, Kwong M, et al. (2011). Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell 145, 513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidel N, Blacque O (2018). Intraflagellar transport complex A genes differentially regulate cilium formation and transition zone gating. Curr Biol 28, 3279–3287.e3272. [DOI] [PubMed] [Google Scholar]
- Schou K, Mogensen J, Morthorst S, Nielsen B, Aleliunaite A, Serra-Marques A, Fürstenberg N, Saunier S, Bizet A, Veland I, et al. (2017). KIF13B establishes a CAV1-enriched microdomain at the ciliary transition zone to promote Sonic hedgehog signalling. Nat Commun 8, 14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R, Shamseldin H, Loucks C, Seidahmed M, Ansari S, Ibrahim Khalil M, Al-Yacoub N, Davis E, Mola N, Szymanska K, et al. (2014). Mutations in CSPP1, encoding a core centrosomal protein, cause a range of ciliopathy phenotypes in humans. Am J Hum Genet 94, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Garcia Gr, Van De Weghe J, McGorty R, Pazour G, Doherty D, Huang B, Reiter J (2017). Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nat Cell Biol 19, 1178–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Lu Q, Insinna-Kettenhofen C, Nagashima K, English M, Semler E, Mahgerefteh J, Cideciyan A, Li T, Brooks B, et al. (2017). In Vitro modeling using ciliopathy-patient-derived cells reveals distinct cilia dysfunctions caused by CEP290 mutations. Cell Rep 20, 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaats G, Isabella C, Kroes H, Dempsey J, Gremmels H, Monroe G, Phelps I, Duran K, Adkins J, Kumar S, et al. (2016). MKS1 regulates ciliary INPP5E levels in Joubert syndrome. J Med Genet 53, 62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E, Cideciyan A, Aleman T, Scheetz T, umaroka A, Ehlinger M, Schwartz S, Fishman G, Traboulsi E, Lam B, et al. (2011). Variations in NPHP5 in patients with nonsyndromic leber congenital amaurosis and Senior-Loken Syndrome. Arch Ophthalmol 129, 81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe T, Wilkinson C, Iqbal A, Stearnsa T (2012). The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol Biol Cell 23, 3322–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struchtrup A, Wiegering A, Stork B, Rüther U, Gerhardt C (2018). The ciliary protein RPGRIP1L governs autophagy independently of its proteasome-regulating function at the ciliary base in mouse embryonic fibroblasts. Autophagy 14, 567–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanska K, Johnson C (2012). The transition zone: an essential functional compartment of cilia. Cilia 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao D, Wang L, Boss A, Verhey K (2017). Protein interaction analysis provides a map of the spatial and temporal organization of the ciliary gating zone. Curr Biol 27, 2296–2306.e2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W, Bossard C, Khanna H, Peränen J, Swaroop S, Malhotra V, Dynlacht B (2008). CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell 15, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuz K, Bachmann-Gagescu R, O’Day D, Hua K, Isabella C, Phelps I, Stolarski A, O’Roak B, Dempsey J, Lourenco C, et al. (2014). Mutations in CSPP1 cause primary cilia abnormalities and Joubert syndrome with or without Jeune asphyxiating thoracic dystrophy. Am J Hum Genet 94, 62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieillard J, Paschaki M, Duteyrat J, Augière C, Cortier E, Lapart J, Thomas J, Durand B (2016). Transition zone assembly and its contribution to axoneme formation in Drosophila male germ cells. J Cell Biol 214, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierkotten J, Dildrop R, Peters T, Wang B, Rüther U (2007). Ftm is a novel basal body protein of cilia involved in Shh signalling. Development 134, 2569–2577. [DOI] [PubMed] [Google Scholar]
- Wang L, Dynlacht B (2018). The regulation of cilium assembly and disassembly in development and disease. Development 145, dev151407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Tay H, Soni R, Perumal G, Goll M, Macaluso F, Asara J, Amack J, Tsou M (2013). CEP162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base. Nat Cell Biol 15, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- War S, Kumar U (2012). Coexpression of human somatostatin receptor-2 (SSTR2) and SSTR3 modulates antiproliferative signaling and apoptosis. J Mol Signal 7, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton-Pitt S, Jauregui A, Li C, Wang J, Leroux M, Barr M (2012). Ciliogenesis in Caenorhabditis elegans requires genetic interactions between ciliary middle segment localized NPHP-2 (inversin) and transition zone-associated proteins. J Cell Sci 125, 2592–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Zhang Y, Schouteden C, Zhang Y, Zhang Q, Dong J, Wonesch V, Ling K, Dammermann A, Hu J (2016). The hydrolethalus syndrome protein HYLS-1 regulates formation of the ciliary gate. Nat Commun 7, 12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng R, Yang T, Huang C, Chang C, Wang W, Liao J (2018). Super-Resolution Imaging Reveals TCTN2 Depletion-Induced IFT88 Lumen Leakage and Ciliary Weakening. Biophys J 115, 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegering A, Dildrop R, Kalfhues L, Spychala A, Kuschel S, Lier J, Zobel T, Dahmen S, Leu T, Struchtrup A, et al. (2018a). Cell type-specific regulation of ciliary transition zone assembly in vertebrates. EMBO J 37, pii: e97791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegering A, Rüther U, Gerhardt C (2018b). The ciliary protein Rpgrip1l in development and disease. Dev Biol 442, 60–68. [DOI] [PubMed] [Google Scholar]
- Williams C, Li C, Kida K, Inglis P, Mohan S, Semenec L, Bialas N, Stupay R, Chen N, Blacque O, et al. (2011). MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol 192, 1023–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won J, Marín de Evsikova C, Smith R, Hicks W, Edwards M, Longo-Guess C, Li T, Naggert J, Nishina P (2011). NPHP4 is necessary for normal photoreceptor ribbon synapse maintenance and outer segment formation, and for sperm development. Hum Mol Genet 20, 482–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Chen H, Tang T (2018). Myosin-Va is required for preciliary vesicle transportation to the mother centriole during ciliogenesis. Nat Cell Biol 20, 175–185. [DOI] [PubMed] [Google Scholar]
- Ye F, Nager A, Nachury M (2018). BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J Cell Biol 217, 1847–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee L, Garcia-Gonzalo F, Bowie R, Li C, Kennedy J, Ashrafi K, Blacque O, Leroux M, Reiter J (2015). Conserved genetic interactions between ciliopathy complexes cooperatively support ciliogenesis and ciliary signaling. PLoS Genet 11, e1005627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Egerer J, Fuchs E, Haas A, Barr F (2007). Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol 178, 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghloul N, Katsanis N (2010). Functional modules, mutational load and human genetic disease. Trends Genet 26, 168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Guo W, Lipschutz J (2009). The exocyst protein Sec10 is necessary for primary ciliogenesis and cystogenesis in vitro. Mol Biol Cell 20, 2522–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets.