Abstract
Nucleoli are dynamic nuclear condensates in eukaryotic cells that originate through ribosome biogenesis at loci that harbor the ribosomal DNA. These loci are known as nucleolar organizer regions (NORs), and there are 10 in a human diploid genome. While there are 10 NORs, however, the number of nucleoli observed in cells is variable. Furthermore, changes in number are associated with disease, with increased numbers and size common in aggressive cancers. In the near-diploid human breast epithelial cell line, MCF10A, the most frequently observed number of nucleoli is two to three per cell. Here, to identify novel regulators of ribosome biogenesis we used high-throughput quantitative imaging of MCF10A cells to identify proteins that, when depleted, increase the percentage of nuclei with ≥5 nucleoli. Unexpectedly, this unique screening approach led to identification of proteins associated with the cell cycle. Functional analysis on a subset of hits further revealed not only proteins required for progression through the S and G2/M phase, but also proteins required explicitly for the regulation of RNA polymerase I transcription and protein synthesis. Thus, results from this screen for increased nucleolar number highlight the significance of the nucleolus in human cell cycle regulation, linking RNA polymerase I transcription to cell cycle progression.
INTRODUCTION
In eukaryotic organisms, the biogenesis of ribosomes initiates in membraneless, phase-separated, nuclear condensates known as nucleoli. In mammalian cells, the nucleoli form around tandemly repeated ribosomal DNA (rDNA) loci upon initiation of transcription by RNA polymerase I (RNAPI) (Hernandez-Verdun, 2011; Grob et al., 2014; Bersaglieri and Santoro, 2019; Potapova and Gerton, 2019). These loci, called nucleolar organizer regions (NORs), are transcribed as precursor RNAs (47S) that comprise three of the four mature rRNA species (18S, 5.8S, and 28S), along with external and internal transcribed spacer sequences that must be removed to assemble a functional ribosome. As a result, in coordination with RNAPI, trans-acting factors are recruited to process, modify, and assemble the pre-rRNA (47S) with the ∼80 ribosomal proteins (r-proteins) and the 5S ribonucleoprotein (RNP) to produce the mature small (40S) and large (60S) subunits of the translationally competent ribosome (Henras et al., 2015; Aubert et al., 2018; Bassler and Hurt, 2019).
Mammalian nucleoli are highly dynamic organelles. In the human genome there are 10 nucleolar organizing regions localized on the short arms of the five acrocentric chromosomes (chr) (chr13–15, 21, and 22; Henderson et al., 1972; Floutsakou et al., 2013). Yet high-throughput resolution of nucleolar number by fluorescence microscopy in diverse cell lines reveals few cells with 10 nucleoli per nucleus, with many averaging as few as three nucleoli per nucleus (Farley et al., 2015). The nucleolar number, however, is not static. We have previously reported a reduction in the number of nucleoli upon depletion of proteins required for ribosome biogenesis (Freed et al., 2012; Farley-Barnes et al., 2018). It is also well-known to cancer pathologists that increased nucleolar number and size are correlated with increased proliferation and poor prognosis (Derenzini et al., 2009). Dynamic remodeling of nucleolar structure, however, is not restricted to changes in number. The presence of various stressors causes large changes to the nucleolar proteome (Boisvert et al., 2010; Moore et al., 2011) and suppression of ribosome biogenesis (Grummt, 2013). This has been studied most extensively in the nucleolar response to DNA damage (Kruhlak et al., 2007; Larsen et al., 2014), and most notably upon treatment with actinomycin D, where RNAPI transcription is silenced and nucleolar caps are formed at the nucleolar periphery (Reynolds et al., 1964; Floutsakou et al., 2013). Taken together, these data support our understanding that the nucleolus is a highly responsive organelle that integrates signals from a vast network of cellular processes.
Because ribosome biogenesis is an essential and highly conserved process among all living organisms, mechanistic details on the process and its regulation in eukaryotes have been most widely studied in the budding yeast, Saccharomyces cerevisiae (Woolford and Baserga, 2013; Bohnsack and Bohnsack, 2019). However, advances in technologies and the growing evidence linking nucleolar dysfunction to several human congenital diseases (Narla and Ebert, 2010; Warren, 2018; Farley-Barnes et al., 2019), cancer (Ruggero, 2012; Penzo et al., 2019; Sulima et al., 2019; Bursac et al., 2021), viral infections (Jarboui et al., 2012; Rawlinson et al., 2018), and aging (Hetman and Pietrzak, 2012; Tiku and Antebi, 2018), has led several laboratories to mount screening campaigns to explore the complexities of ribosome biogenesis in higher eukaryotes. To date, each screen has utilized different methodologies to reach this endpoint. In Drosophila melanogaster cells, a genome-wide screen for proteins that impact nucleolar size were explored (Neumuller et al., 2013), whereas in human cell lines, screens for nucleolar disruption (Stamatopoulou et al., 2018), pre-rRNA processing defects (Tafforeau et al., 2013), and nucleolar-retained fluorescently tagged r-proteins were carried out (Wild et al., 2010; Badertscher et al., 2015), with the latter being performed at a genome-wide scale.
We have also performed a genome-wide small interfering RNA (siRNA) screen in MCF10A human breast epithelial cells to identify proteins required to maintain the typical two to three nucleoli per nucleus. MCF10A cells are a near-diploid non–cancer-derived cell line. Karyotype analysis revealed only minor rearrangements of chr 3, 6, and 9, as well as one extra copy of both chr 6 and 18 (Soule et al., 1990). Therefore, the chromosomes harboring the nucleolar organizing regions were not impacted in this cell line, leading to the selection of the MCF10A line for our high-throughput screen. In our initial study, we reported more than 100 proteins that caused an increase in the percentage of cells where the nucleolar number was reduced from 2–3 to 1 per nucleus using wide-field, multicolor, fluorescence microscopy (Farley-Barnes et al., 2018). Of these hits, many had no previously reported functional roles in ribosome biogenesis, and further investigation on a subset revealed varied deficits in ribosome biogenesis upon depletion, ranging from RNAPI transcriptional silencing, aberrant pre-rRNA processing, and decreased protein synthesis. Furthermore, these data supported the link between nucleolar number and nucleolar function.
In parallel, our screening campaign also revealed proteins that when depleted caused an increase in the percentage of cells with ≥5 nucleoli. Here, we report on these results, which reveal a wholly distinct and unexpected subset of proteins not typically associated with ribosome biogenesis. Instead, these hits are associated with the cell cycle and specifically with progression through the S and G2/M phase. Furthermore, in a functional analysis on a subset of hits, we found that most proteins were required explicitly for the transcriptional regulation of RNAPI, uncovering unique regulators of nucleolar number and strengthening the support for a link between nucleolar number and function.
RESULTS
Genome-wide siRNA screen for increased nucleolar number reveals 113 hits
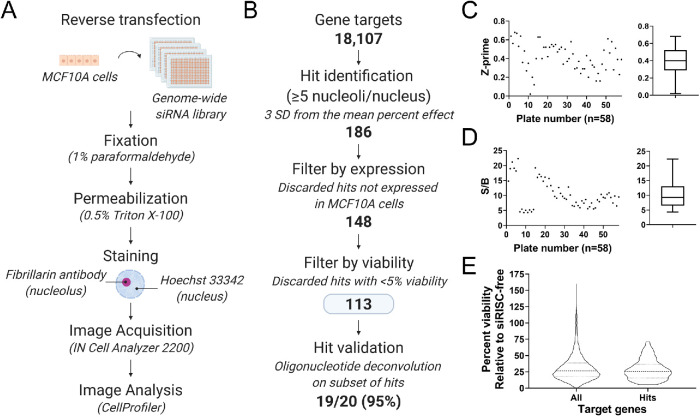
Here we report the results from a phenotypic, genome-wide siRNA screen for increased nucleolar number in the human breast epithelial cell line, MCF10A (Figure 1A). We previously reported that hits from this screen that caused a reduction in nucleolar number from two to three per nucleus to one per nucleus impacted ribosome biogenesis (Farley-Barnes et al., 2018). In the current study, we asked whether an observed increase in nucleolar number can also identify proteins required for ribosome biogenesis. To answer this question, we analyzed the images from the previously reported screen to derive a hit list of proteins that when depleted yielded an increase in the percentage of nuclei with ≥5 nucleoli. Consistent with our previous reporting, 18,107 gene targets were screened with pools of four siRNAs against each target (Figure 1B). The raw nucleolar number per nucleus data was quantified for each gene target using a pipeline in CellProfiler (Carpenter et al., 2006; McQuin et al., 2018). These data were then normalized to the 16 negative and 16 positive control wells that were run on each plate, which allowed for the comparison of siRNA treatments across all 58 screening plates. In this analysis, siRISC-free was the negative control and set to a 0 percent effect (PE), and siKIF11 was the positive control and set to a 100 PE. siRISC-free, the negative control, is a modified siRNA that cannot be loaded into the RNA-induced silencing complex (RISC) and therefore cannot modulate mRNA levels. siKIF11, the positive control, targets a kinesin motor protein required for spindle assembly during mitosis (Blangy et al., 1995) and is a central driver in cancer pathogenesis (Venere et al., 2015; Pei et al., 2017, 2019). Depletion of KIF11 was initially included as a positive transfection control (Weil et al., 2002; Zanin et al., 2013; von Stechow et al., 2015); however, due to the reproducibility with which KIF11 depletion led to an increase in the percentage of nuclei with ≥5 nucleoli and the favorable and reproducible Z-prime values between the two controls, siKIF11 was selected as the screen positive control. The mean normalized percent effect (NPE) of nuclei with ≥5 nucleoli was then determined for each gene target, and hits were identified.
FIGURE 1:
High-content, genome-wide siRNA screen in human MCF10A cells revealed 113 hits that increase the percentage of nuclei with ≥5 nucleoli. (A) Screen workflow. MCF10A cells were reverse transfected into 384-well plates containing the siGENOME SMARTpool siRNA genome library (Horizon Discovery). After 72 h, cells were fixed, permeabilized, and stained with an antibody to the nucleolar protein fibrillarin and Hoechst dye to stain the nucleus. Cell images were collected on an IN Cell Analyzer 2200 wide-field, multicolor, fluorescence microscope and nucleolar number quantified using a pipeline in CellProfiler (Carpenter et al., 2006; McQuin et al., 2018). (B) Screen analysis workflow. A total of 18,107 genes were screened, and hits were identified based on a cutoff of ≥3 SD from the mean percent effect (PE) normalized to the positive (siKIF11, 100 PE) and negative (siRISC-free, 0 PE) controls. Viability relative to siRISC-free negative control was quantified based on Hoechst-stained nuclei, and hits were then discarded if not expressed in MCF10A cells and if viability was <5%. One hundred thirteen high-confidence hits remained, and of those we validated a subset (n = 19/20; 95%) by oligonucleotide deconvolution. (C) Z-prime statistic by plate (left) and as a minimum to maximum box-and-whiskers plot (right) indicated a strong, screenable phenotype with an average Z-prime of 0.41 and a Z-prime on all plates of >0. (D) Signal-to-background (S/B) ratio by plate (left) and as a minimum to maximum box-and-whiskers plot (right) indicated a strong S/B with an average S/B of 10.29. (E) Violin plot of the percent viability of all target genes and the 113 hits relative to siRISC-free. A and B were created with BioRender.com.
One hundred eighty-six (186) hits were identified based on a stringent cutoff of three SDs from the mean NPE of all targets (NPE = 25.0–87.4; Figure 1B), yielding an overall hit rate of 1%. We then filtered the list to determine a high-confidence set of hits, discarding ones not expressed in MCF10A cells based on a transcriptome analysis (n = 3; FPKM = 0; GEO Accession #GSE154764) and ones with a viability of <5% relative to the siRISC-free treatment. This analysis left us with 113 high-confidence hits, including our positive control KIF11, that when depleted caused an increase in the percentage of nuclei with ≥5 nucleoli (NPE = 25.0–68.75; Supplemental Table S1). Validation of the screen was performed using oligonucleotide deconvolution of 20 selected high-confidence hits in triplicate, where each siRNA in the pool was tested individually in the screening assay (Sigoillot and King, 2011; Z-prime = 0.61; Table 1). Hits were subjectively selected to be representative of the data set based on bioinformatic analyses described below, including mitotic and DNA repair factors, nucleolar and nonnucleolar proteins, and proteins with putative RNA-binding domains. For 19 hits (19/20), at least 2/4 individual siRNAs in the pool caused an increase in nucleolar number (NPE ≥ 15.0).
TABLE 1:
High-confidence screen hits validated by oligonucleotide deconvolution.
| Protein name | HGNC symbol | Aliases | Validated (Y/N) | Nucleolar (Y/N) | Description |
|---|---|---|---|---|---|
| ATP Binding Cassette Subfamily E Member 1 | ABCE1 | RNASEL1, RNASELI, RNS4I | Y (−01, −02, −04, −17) | Y | Inhibits endoribonuclease activity through inhibition of RNase L. Also a ribosome recycling factor. |
| ATPase family AAA domain containing 5 | ATAD5 | C17orf41, ELG1, FRAG1 | Y (−01, −02, −03, −04) | N | DNA replication factor C–like complex subunit. |
| Cellular communication network factor 4 | CCN4 | Wisp1 | Y (−01, −02, −03, −17) | N | Wnt1-inducible signaling pathway protein. |
| Cell division cycle associated 8 | CDCA8 | Borealin, BOR, DasraB, Nbl1p | Y (−01, −02, −03, −04) | Y | Mitotic chromosomal passenger complex member. |
| Cytosolic Iron-Sulfur Assembly Component 2B | CIAO2B | FAM96B | Y (−01, −03, −04, −18) | N | Mediates incorporation of Fe/S proteins. Component of mitotic spindle–associated MMXD complex. |
| Dynein Cytoplasmic 1 Heavy Chain 1 | DYNC1H1 | DNECL, DNCL, DNCH1 | Y (−01, −02, −03, −04) | Y | Microtubule-activated molecular motor. Mitotic spindle assembly and metaphase plate congression factor. |
| ENY2 transcription and export complex 2 subunit | ENY2 | Sus1 | Y (−01, −02, −03, −04) | N | Transcriptional coactivator through association with the SAGA complex and others. |
| Family with sequence similarity 98 member A | FAM98A | Y (−01, −02, −03, −04) | N | Regulator of arginine methyltransferase, PRMT1, and contains a putative RNA-binding domain. | |
| H1 histone family member X | H1-10 | H1FX | Y (−01, −02, −03, −04) | Y | H1 linker histone. |
| Inner centromere protein | INCENP | Y (−01, −02, −03, −04) | Y | Mitotic chromosomal passenger complex member. | |
| Inka box actin regulator 1 | INKA1 | FAM212A, C3orf54 | Y (−01, −02, −03, −04) | N | PAK4 (P21-activated kinase) inhibitor. |
| Kinectin 1 | KTN1 | Y (−17, −18, −19, −20) | N | Binds kinesins and elongation factor-delta in endoplasmic reticulum. | |
| LUC7-like | LUC7L | Luc7 | Y (−01, −02, −03, −04) | N | Putative RNA-binding protein similar to yeast Luc7p subunit of the U1 snRNP splicing complex. |
| Midasin AAA ATPase 1 | MDN1 | Rea1 | Y (−03, −17, −18, −19) | Y | Large ribosomal subunit maturation factor. |
| Rac GTPase Activating Protein 1 | RACGAP1 | Y (−01, −02, −03, −04) | N | Mitotic centralspindlin complex member. | |
| Replication factor C subunit 1 | RFC1 | Y (−01, −02, −03, −04) | Y | DNA replication factor C complex subunit. | |
| Serine/threonine kinase 24 | STK24 | MST-3 | Y (−05, −21, −22, −23) | Y | GCK-3 family kinase involved in MAPK signaling. |
| TPX2 microtubule nucleation factor | TPX2 | Y (−01, −02, −03, −04) | Y | Mitotic spindle assembly factor and activator of Aurora A kinase signaling. | |
| WD repeat containing antisense to TP53 | WRAP53 | WDR79, Tcab1 | Y (−19, −20, −21, −22) | N | Telomerase holoenzyme member. |
| x-ray repair cross-complementing protein 5 | XRCC5 | Ku80 | N (−01, −02, −03, −04) | Y | Nonhomologous end joining (NHEJ) DNA repair factor. |
Twenty hits were selected for validation by oligonucleotide deconvolution.The four individual siRNAs tested are listed in parenthesis in the “Validated” column, and those that did not validate (NPE < 15.0) are in red. The number following the hyphen is the last two digits of the Horizon Discovery product number associated with the individual siRNA. The 14 hits selected for further analysis have the HGNC symbol in bold.
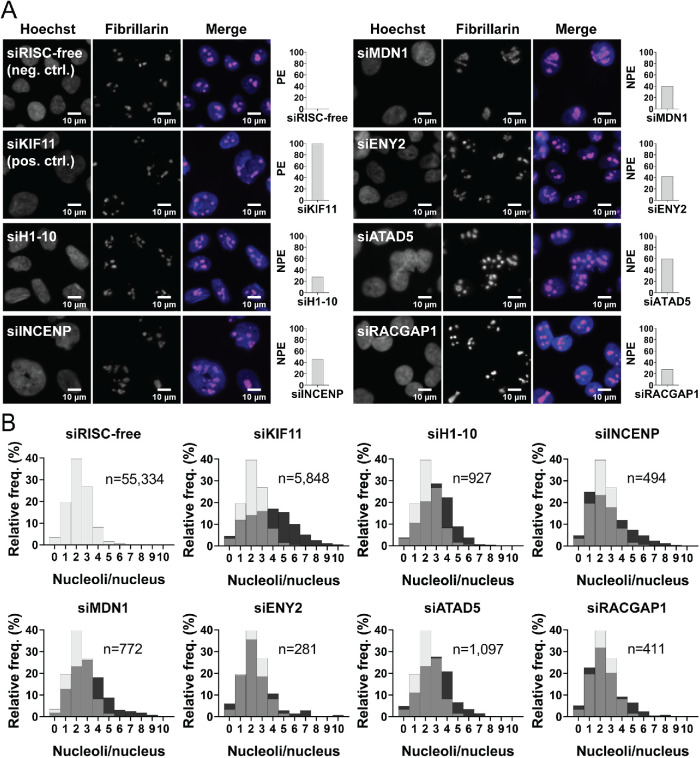
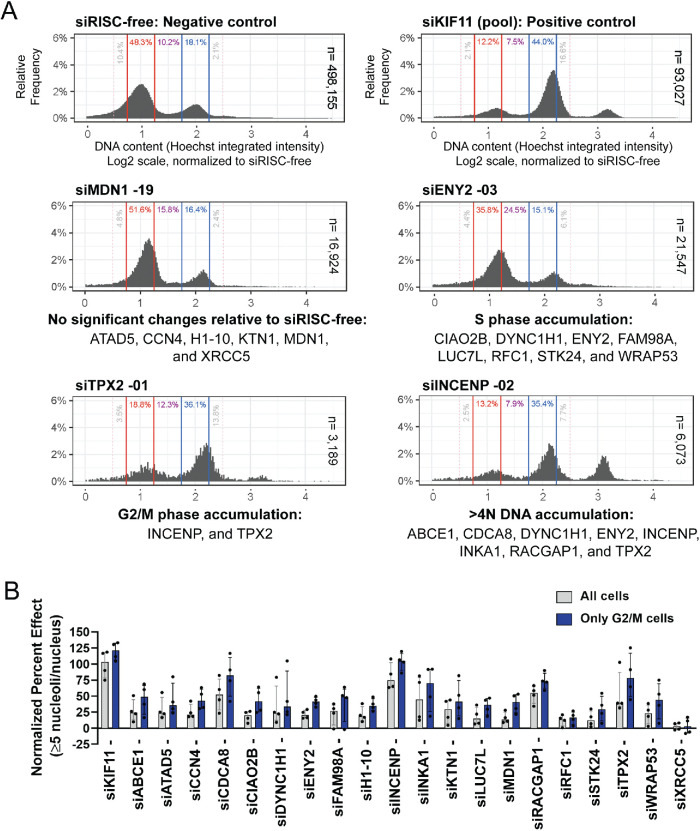
Screen performance was monitored using the Z-prime statistic and signal-to-background. The average Z-prime for the screen was 0.41 (range, 0.02–0.68; Figure 1C), suggesting a strong, screenable phenotype with good separation between the controls. The ≥5 nucleoli per nucleus signal-to-background between controls was also strong, with an average of 10.29 (range, 4.32–22.33; Figure 1D). Additionally, we monitored viability using cell numbers relative to siRISC-free. Viability was highly variable, both overall, ranging from 0.4 to 160.2%, and among the high-confidence hits, ranging from 5.2 to 71.3% (Figure 1E). A Pearson r correlation analysis reveals that there is no linear correlation between the ≥5 nucleoli per nucleus NPE and percent viability (r2 = 0.08; Supplemental Table S1). A representative subset of images and NPE from the screen are shown (Figure 2A), including the frequency distribution of nucleoli per nucleus based on the single cell data, which shows a flattening and rightward shift in the distribution from 2–3 nucleoli per nucleus to ≥5 (Figure 2B).
FIGURE 2:
Representative hits showing an increase in nuclei with ≥5 nucleoli. (A) Representative images of hits from the screen and the normalized percent effects (NPE). Shown are a selection of nuclei (100 × 100 μM) stained with Hoechst 33342 (blue) and an antibody to the nucleolar protein fibrillarin (72B9 [ Reimer et al., 1987]; pink) from the negative control (siRISC-free, 0 PE), positive control (siKIF11, PE), and representative screen hits (siH1-10, siINCENP, siMDN1, siENY2, siATAD5, and siRACGAP1) enlarged threefold using bicubic interpolation from a single field of view (left) and a bar graph of the NPE (right). (B) Histograms of the relative frequency of nucleoli per nucleus and the number of nuclei quantified are shown for the controls and representative hits in A. Relative to siRISC-free (gray bars), among the hits (and KIF11; black bars) there is a clear decrease in nuclei with 2–3 nucleoli and an increase in nuclei with ≥5 nucleoli (gray bars = overlap between siRISC-free and hit). Histograms for siRISC-free and siKIF11 are representative and were generated from the images collected from a single screening plate (Plate 1; 16 wells; 48 fields of view). Histograms for each hit were made from the images collected from their respective well and plate in the screen (three fields of view). The x-axis was limited to 10 nucleoli per nucleus to aid in visualization; protein depletion conditions with nuclei with >10 nucleoli include siRISC-free (n = 3), siKIF11 (n = 36), siH1-10 (n = 1), siINCENP (n = 2), siMDN1 (n = 3), siENY2 (n = 4), and siRACGAP1 (n = 2).
Bioinformatic analysis of hits reveals proteins required for cell cycle processes
We performed a range of bioinformatic analyses on the 113 high-confidence hits to determine their conservation status, how they partition into biological processes, and whether they localize to the nucleolus. First, we identified the proportion of hits that are conserved to S. cerevisiae through manual curation of available database tools and the literature. Using e!Ensembl’s BioMart data mining tool (Kinsella et al., 2011), yeast orthologues were identified for 22/113 hits. Using the Saccharomyces Genome Database’s YeastMine tool (Balakrishnan et al., 2012), yeast orthologues were identified for 26/113 hits. Manual curation of these data in combination with a review of the literature revealed that 39/113 hits have orthologues in S. cerevisiae, suggesting that neither tool on its own provides a complete picture. For a subset of hits, there also exists a lack of consensus regarding the specific orthologue and thus multiple gene names are listed (Supplemental Table S1). Thus, these data suggest that while one-third of hits may be conserved regulators of nucleolar form, the remaining majority are regulators of nucleolar number that are not present in yeast.
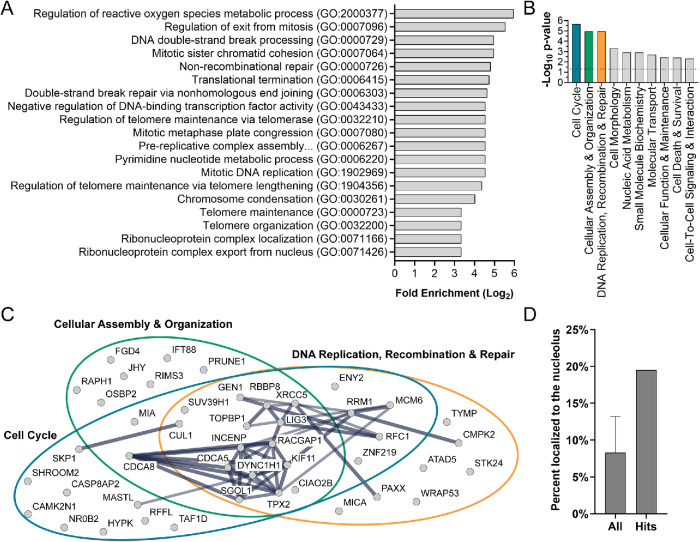
Second, we performed a Gene Ontology (GO) overrepresentation analysis using PANTHER (Mi et al., 2017). This analysis revealed a significant enrichment (p < 0.05, binomial test, Bonferroni correction) of 19 largely overlapping GO-Slim categories associated with biological process (log2 fold enrichment > 3.33; Figure 3A). The categories with the largest log2 fold enrichment include regulation of the reactive oxygen species metabolic process (GO:2000377), regulation of exit from mitosis (GO:0007096), DNA double-strand break processing (GO:0000729), mitotic sister chromatid cohesion (GO:0007064), and nonrecombinational repair (GO:0000726). These data were echoed in our core analysis in Ingenuity Pathway Analysis (IPA; Qiagen). Results revealed a significant association (–log10 p > 1.3, equivalent to p < 0.05) of the hits with 23 molecular and cellular functions (Supplemental Table S2). The top functions based on the highest –log10p value are shown and include Cell Cycle, Cellular Assembly and Organization, and DNA Replication, Recombination, and Repair (Figure 3B). We analyzed the hits included in these three top IPA categories as STRINGdb high-confidence interaction networks (Szklarczyk et al., 2019; Figure 3C), which revealed a large degree of overlap among the categories and interconnectedness among the hits. Interestingly, while multiple hits are known to be associated with ribosome biogenesis (ABCE1 [ Pisarev et al., 2010; Young et al., 2015], MDN1 [ Galani et al., 2004; Bassler et al., 2010], SUV39H1 [ Murayama et al., 2008], and TAF1D [ Gorski et al., 2007]), ribosome biogenesis-associated categories were not revealed in either the GO or the IPA analysis. Thus, these analyses suggest that this screen uncovered a unique subset of proteins associated with the cell cycle, and specifically S and G2/M phase, that are required for maintaining the typical number of nucleoli in MCF10A cells.
FIGURE 3:
Bioinformatic analysis on the 113 hits reveals a unique set of proteins required for maintaining normal nucleolar number. (A) Gene Ontology (GO) overrepresentation analysis using PANTHER is shown as a bar graph of the top enriched GO-Slim categories associated with biological process (Log2 fold enrichment >3.33; binomial test, Bonferroni correction, p < 0.05). Processes associated with mitosis (e.g., regulation of exit from mitosis [GO:0007096] and mitotic sister chromatid cohesion [GO:0007064]) and DNA replication and repair (e.g., DNA double-strand break processing [GO:0000729] and nonrecombinational repair [GO:0000726]) are common among the top enriched categories, but processes associated with ribosome biogenesis are strikingly absent. (B) Molecular and Cellular Function analysis in Ingenuity Pathway Analysis (IPA; Qiagen) is shown as a bar graph of the top 10 molecular and cellular functions associated with the 113 hits using the Fisher’s exact test scoring method in IPA (p < 0.05). Top associated functions include Cell Cycle (n = 31), Cellular Assembly and Organization (n = 24), and DNA Replication, Recombination, and Repair (n = 27). Colored bars are associated with the colored circles in C. All significantly associated categories and genes are listed in Supplemental Table S2. (C) Interaction networks of the hits in the top three categories in B are shown as STRING high-confidence (≥0.700 interaction score) interaction networks and reveal a large degree of overlap among the categories and interconnectedness among the hits. The heavier weighted lines represent the highest degree of confidence (≥0.900 interaction score). (D) Nucleolar proteins are enriched among the 113 hits. The percent of proteins in the human proteome (left) and of the hits (right) that localize to the nucleolus are shown as a bar graph. Nineteen and one-half percent (19.5%) of hits localize to the nucleolus, whereas the total number of nucleolar proteins in the human proteome ranges from 4.4 to 13.8%. These estimates were based on three published data sets (Leung et al., 2006; Ahmad et al., 2009; Jarboui et al., 2011; Thul et al., 2017) and a total number of proteins equal to 19,670 based on Thul et al. (2017). In our calculation based on NOPdb, we used 2717 proteins as the number of nucleolar proteins based on the last available data set accessed on 01/22/2009. All = mean ± SD.
Finally, we questioned whether the hits were enriched in nucleolar proteins. To broadly analyze all our screen hits, we identified proteins that localize to the nucleolus based on presence in just one of three available published data sets (Table 1; Supplemental Table S1). These data sets include the nucleolar proteome database (NOPdb; Leung et al., 2006; Ahmad et al., 2009), a T-cell nucleolar proteome (Jarboui et al., 2011), and the Human Protein Atlas Cell Atlas organelle proteome (nucleoli and nucleoli fibrillar centers; Thul et al., 2017). Based on this analysis, ∼20% of hits localize to the nucleolus (n= 22), which is enriched when compared with the ∼4–14% of nucleolar proteins in the entire human proteome based on calculations using these data sets (Figure 3D; Table 1). These data suggest that despite the lack of association with ribosome biogenesis categories in analysis of biological function, the hits are enriched for nucleolar proteins. We have thus discovered a unique subset of proteins required for the regulation of nucleolar number that may harbor nucleolar functions.
Comparison of hits to other screens for nucleolar form and function reveals a unique subset of proteins
We further compared our 113 high-confidence hits to other published screens for regulators of nucleolar form and function (Wild et al., 2010; Neumuller et al., 2013; Tafforeau et al., 2013; Badertscher et al., 2015; Farley-Barnes et al., 2018). Intriguingly, few of our hits overlapped with hits from other screens, with overlap ranging from two to six hits (<2%; Table 2). Notably, in screens for regulators of nucleolar size and/or fragmentation in S. cerevisiae and D. melanogaster (Neumuller et al., 2013), our positive control and hit, KIF11/Cin8/Klp61f, was identified in both data sets. In screens focused on identifying human ribosome biogenesis factors (Wild et al., 2010; Tafforeau et al., 2013; Badertscher et al., 2015), ABCE1, MDN1, DYNC1H1, CDCA8, SUV39H1, and TOPBP1 all overlapped with hits from our screen. Furthermore, our hits were completely nonoverlapping with the hits from our screen that, when depleted, caused a decrease in the number of nucleoli per nucleus (Farley-Barnes et al., 2018). Thus, while our screen validated hits revealed by other methods of screening for regulators of nucleolar form and function, these data support our unique screening approach in MCF10A cells and its ability to uncover a distinctive subset of proteins.
TABLE 2:
Comparison of hits to other screens for nucleolar form and function reveals a unique subset of proteins.
| Neumuller et al., S. cerevisiae (4/388) | Neumuller et al. D. melanogaster (6/757) | Wild et al. HeLa cells(2/153) | Badertscher et al. HeLa cells(2/300) | Tafforeau et al. HeLa cells(4/286) |
|---|---|---|---|---|
| KIF11/Cin8 | IFT88/nompB | ABCE1 | ABCE1 | CDCA8 |
| PMM2/Sec53 | INCENP | MDN1 | DYNC1H1 | MDN1 |
| SKP1/Skp1 | KIF11/Klp61f | SUV39H1 | ||
| YIPF7/Yip1 | LIG3 | TOPBP1 | ||
| MAN1A1/alpha-Man-I | ||||
| RAP2C/Rap2I |
The 113 high-confidence hits were compared with the hit lists from screens in human cell lines (Wild et al., 2010; Tafforeau et al., 2013; Badertscher et al., 2015), S. cerevisiae (Neumuller et al., 2013), and D. melanogaster (Neumuller et al., 2013), as indicated. Indicated in parentheses is the number of overlapping hits compared with the total number of hits identified by the screening approach. Gene names of the overlapping hits are listed.
Nuclear area is significantly larger in nuclei with ≥5 nucleoli
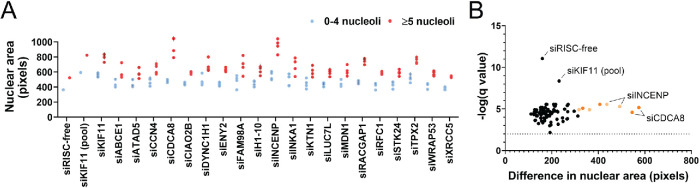
Observations of images from the screen, like those shown in Figure 2A, suggest that the nuclei of screen hits with ≥5 nucleoli may be larger than the nuclei in the siRISC-free control. To test whether nuclei are larger, using the images collected for the subset of hits analyzed in our screen validation, we used CellProfiler to classify nuclei by nucleolar number (0–4 vs. ≥5) and quantify the nuclear area of the Hoechst stain. Our analysis revealed that the nuclear area of nuclei with ≥5 nucleoli is significantly larger than that of nuclei with 0–4 nucleoli (n = 3 or 6; q < 0.01; Figure 4, A and B; Supplemental Table S3). Interestingly, however, this result is observed not only when screen hits are depleted, but also in the negative control cells. There is some variability in the nuclear size increase among some hits; notably, depletion of CDCA8 and INCENP resulted in a ≥2-fold increase in the nuclear area of nuclei with ≥5 nucleoli compared with siRISC-free and a majority of the screen hits (Figure 4B). These proteins are known mitotic inhibitors, and thus it is suggested that this increase may be driven by a failure in cell division. As a result, and in addition to the bioinformatic analyses revealing significant association of screen hits with the cell cycle, these data suggest that cell cycle profiling is warranted to address whether failed cell cycle progression, specifically in mitosis, is a unifying feature of cells treated with these siRNAs.
FIGURE 4:
Nuclear area is significantly greater in nuclei with ≥5 nucleoli. (A) Nuclear area is greater in nuclei with ≥5 nucleoli, including in the siRISC-free treatment. Nuclear area was quantified in pixels using analysis of the Hoechst-stained images collected for screen validation by oligonucleotide deconvolution. Three replicates were analyzed for each screen hit depletion, and six replicates were analyzed in this analysis for the controls, siRISC-free and siKIF11 (pool). Blue dots = nuclei with 0–4 nucleoli. Red dots = nuclei with ≥5 nucleoli. Each dot represents the mean ± SD of an individual siRNA (SD = black vertical line). For each blue dot there is a corresponding red dot (Supplemental Table S3). (B) Volcano plot of the statistical analysis of the data in A reveals that in all depletion conditions, including siRISC-free, nuclei with ≥5 nucleoli are significantly larger that nuclei with 0–4 nucleoli. Unpaired t tests were performed, and significance was determined based on a false discovery rate approach using the two-stage step-up method of Benjamini et al. (2006) (n = 3 or 6; q < 0.01/-log q-value > 2; Supplemental Table S3). The x-axis represents the difference in nuclear area between nuclei with 0–4 nucleoli and ≥5 nucleoli. The purple dots = individual siRNAs with the greatest difference between the two categories. Light orange dots = siINCENP individual siRNAs. Dark orange dots = siCDCA8 individual siRNAs.
Cell cycle analysis reveals proteins required for progression through S and G2/M phase
To evaluate whether failed cell cycle progression upon depletion of screen hits is a unifying theme, we used high-content image analysis of the Hoechst-stained nuclei as previously reported (Chan et al., 2013; Roukos et al., 2013, 2015; Gomes et al., 2018). Using the images collected for screen validation by oligonucleotide deconvolution (Table 1), the integrated intensity of the Hoechst stain for each nucleus was quantified and log2 values were plotted as histograms for each of the four individual siRNAs from the 20 hits. Cell cycle phases were normalized to the siRISC-free 2N and 4N peaks as described in Chan et al. (2013). We concluded cell cycle accumulation conservatively when depletion of at least two of four individual siRNAs resulted in a significant ≥2-fold decrease or increase in the percent of nuclei in a phase relative to siRISC-free (q < 0.01). As expected, depletion of KIF11, a mitotic kinesin, resulted in an accumulation of cells in G2/M phase (Figure 5A; Supplemental Table S4). However, depletion of only 2/20 hits caused an accumulation of cells in G2/M (INCENP and TPX2), with an additional 6/20 hits yielding a significant increase in >4N DNA content (ABCE1, CDCA8, DYNC1H1, ENY2, INKA1, and RACGAP1; Figure 5A; Supplemental Table S4). Mitosis-associated factors, CDCA8, INCENP, and RACGAP1, yielded the greatest accumulation of nuclei with >4N DNA content (>10% nuclei), while the increase among the other hits was more modest (<10% nuclei).
FIGURE 5:
Cell cycle analysis reveals that hits are required for progression through either S or G2/M phase. (A) Representative histograms of DNA content by quantification of Hoechst 33342 log2 integrated intensity. The log2 integrated intensities of nuclei in the negative, siRISC-free, control (sum of 48 replicates; n = 498,155 nuclei) were plotted and the G0/G1 peak set to 1.0 (red lines and text) and G2/M peak set to 2.0 (blue lines and text), and all other depletion conditions were then normalized to siRISC-free. Phases were assigned based on Chan et al. (2013), with G2/M phase including late G2 nuclei; G0/G1 = 0.75–1.25; S = 1.25–1.75; G2/M = 1.75–2.25 and 2.25–2.50; >4N = >2.50. Depletion of the positive control, siKIF11 (pool) resulted in the expected accumulation of cells in G2/M phase and a subset of cells with a >4N DNA content (sum of 48 replicates; n = 93,027 nuclei). Cell cycle profiling reveals that several hits are required for progression through either S or G2/M phase. Representative histograms for screen hits are shown as a sum of the three replicates, yet each replicate for every depletion condition was characterized individually to perform statistical testing (Supplemental Table S4). Significance was determined by unpaired t tests relative to siRISC-free and a false discovery rate approach using the two-stage step-up method of Benjamini et al. (2006) (n = 3 or 48; q < 0.01 = *; q < 0.001 = **; q < 0.0001 = ***; Supplemental Table S4). Cell cycle defects were concluded based on a conservative threshold of whether treatment with ≥2 of 4 individual siRNAs resulted in a ≥2-fold significant increase or decrease in the percent of nuclei in a phase relative to siRISC-free. Each hit is listed below one of four representative histograms for the statistically significant cell cycle defects identified. (B) The total of ≥5 nucleoliper nucleus NPE is greater when restricting the analysis of nucleolar number to cells in G2/M phase. We restricted our analysis of the ≥5 nucleoli per nucleus NPE for each of the 20 hits based on cell cycle phase. Only when considering nuclei in G2/M phase was there a statistical separation between siRISC-free and siKIF11 (Z-prime = 0.47). The NPE for each individual siRNA is depicted as dots. The bars show the median NPE + interquartile range for the four individual siRNAs for each hit, for all nuclei (gray) and nuclei in G2/M phase only (blue).
Interestingly, these cell cycle results are consistent with a post hoc analysis of the screen images that we performed to evaluate how our CellProfiler pipeline segmented atypical nuclei and nucleoli. We observed annular and semiannular nuclei among some hits (e.g., siINCENP), which are indicative of late mitotic defects, that were counted as both one and more than one nucleus potentially skewing estimates of nucleolar number (Supplemental Figure S1A; Verstraeten et al., 2011). We also observed “stretched” nucleoli, reminiscent of anaphase bridges and mitotic defects (e.g., siMDN1; Daniloski et al., 2019), that in some cases could lead to an overestimate of nucleolar number (Supplemental Figure S1B). Regardless, while defects in G2/M phase progression and cytokinesis failures were present among the hits tested, they were not observed in all cases.
Other aspects of the cell cycle were also affected to various degrees. In addition to hits that, when depleted, caused an accumulation of cells in G2/M phase, our analysis also revealed that depletion of 8/20 hits caused a significant accumulation of cells in S phase (q < 0.01; CIAO2B, DYNC1H1, ENY2, FAM98A, LUC7L, RFC1, STK24, and WRAP53; Figure 5A; Supplemental Table S4). Interestingly, two of these (DYNC1H1 and ENY2) were hits that also led to a significant increase in nuclei with >4N DNA content, suggesting that defects in S phase progression may also contribute to failures in cell division. Furthermore, four hits (CDCA8, INCENP, RACGAP1, and TPX2) resulted in a significant decrease in nuclei in G0/G1 phase and correlate with the hits that resulted in an accumulation of cells either in G2/M or with a >4N DNA content (q < 0.01). Finally, depletion of 6/20 hits (ATAD5, CCN4, H1-10, KTN1, MDN1, and XRCC5) showed no change in cell cycle distribution based on our designated threshold, although significant minor differences were observed that may be meaningful (Figure 5A; Supplemental Table S4). Finally, when considering whether an individual siRNA treatment that caused a significant cell cycle accumulation also resulted in an increase in the ≥5 nucleoli per nucleus NPE, there are instances where a change in cell cycle distribution is observed, but no concomitant increase in nucleolar number is observed (CIAO2B, DYNC1H1, INKA1, LUC7L, MDN1, RFC1, STK24, and XRCC5; Figure 5A; Supplemental Table S4). Taking the results together, our cell cycle analysis using DNA content suggests that despite an increase in the nuclear area of nuclei with ≥5 nucleoli, failures in G2/M phase progression and cytokinesis may in only some cases explain the increased numbers of nucleoli that we observe, and the contribution of other mechanisms may be in part responsible.
To further investigate the link between the cell cycle and ≥5 nucleoli per nucleus NPE, we asked whether the occurrence of the ≥5 nucleoli per nucleus phenotype correlated with an individual phase of the cell cycle. Interestingly, when we restricted our analysis of nucleolar number by cell cycle phase and calculated the ≥5 nucleoli per nucleus NPE for each of the 20 hits, we found that the median NPE of the four individual siRNAs is greater when considering cells in the G2/M phase of the cell cycle (19/20; Figure 5B; Supplemental Table S5). The one exception was XRCC5, which was also the only hit that did not pass our initial validation. In addition, only when considering cells in the G2/M phase is there an observable difference in PE between siRISC-free and siKIF11 (Z-prime = 0.47). The Z-prime statistics were negative for both cells in G0/G1 phase (Z-prime = –0.08) and S phase (Z-prime = –0.12), suggesting no significant distinction between the negative and positive controls. Furthermore, it has been reported that nuclear volume scales with cellular volume, which gradually increases through the cell cycle (Jorgensen et al., 2007; Neumann and Nurse, 2007; Maeshima et al., 2011; Cantwell and Nurse, 2019); therefore, these data are consistent with our observation that nuclei with ≥5 nucleoli are significantly larger (Figure 4, A and B). Taken together, while these data suggest that some hits are required for S and G2/M phase progression, in most cases it is likely the cells specifically in G2/M phase that are driving the increase in the percentage of nuclei with ≥5 nucleoli that we observe.
Small molecule inhibition of DNA replication and mitosis increases nucleolar number
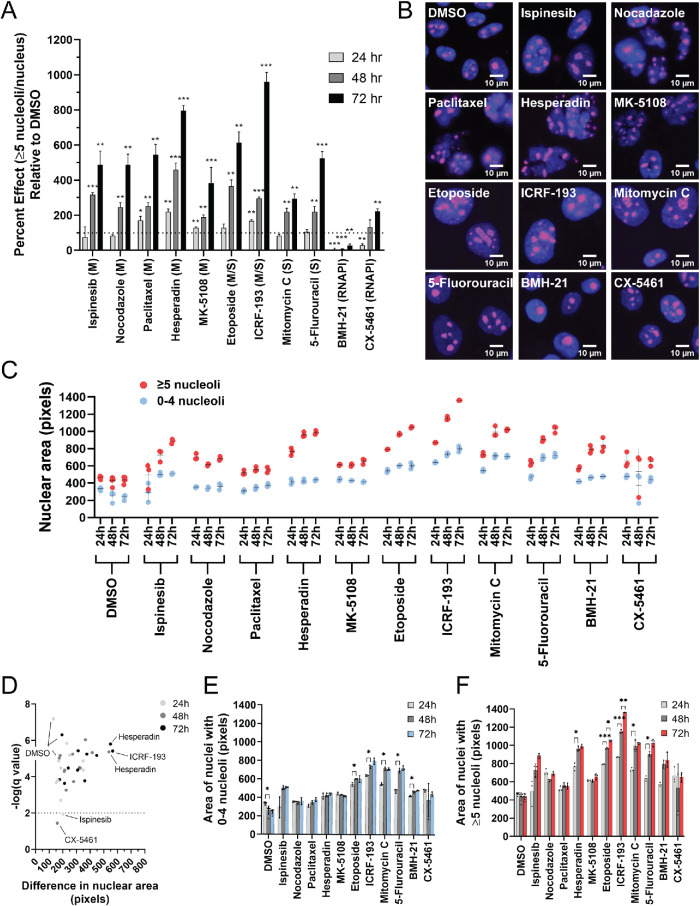
On the basis of the results from our cell cycle analysis that suggest that a proportion of hits are required for faithful progression through S and G2/M phase, we asked as validation whether inhibition of DNA replication and mitosis using small molecule drugs can cause an increase in the percentage of cells with ≥5 nucleoli per nucleus (Weiss et al., 2007). Inhibitors of mitosis tested included the KIF11 inhibitor, ispinesib, as well as nocodazole, paclitaxel, and the Aurora kinase inhibitors, hesperidin and MK-5108. We treated MCF10A cells at low doses of the respective drugs in triplicate for prolonged periods of time (24, 48, and 72 h) to best mimic the prolonged treatment with siRNAs from our screen. The ≥5 nucleoli per nucleus PE relative to dimethyl sulfoxide (DMSO) was then quantified (DMSO was set to 100 PE; Figure 6, A and B). Notably, treatment with ispinesib resulted in a significant increase in PE at 48 and 72 h, but not at 24 h, confirming the role for KIF11 in regulating nucleolar number (n = 3; adjusted p < 0.05). Microtubule-targeted inhibitors nocodazole and paclitaxel also resulted in significant increases to nucleolar number at 48 and 72 h, with paclitaxel additionally showing a modest increase at 24 h (n = 3; adjusted p < 0.05). Finally, we also tested selective Aurora A (MK-5108) and Aurora B (hesperadin) kinase inhibitors (de Groot et al., 2015), because hits INCENP, CDCA8, and TPX2 are associated with these kinases. Strikingly, both inhibitors also caused significant increases to the PE at all time points (n = 3; p < 0.05). These data support the conclusion that prolonged inhibition of mitosis is sufficient to yield an increase in the percentage of nuclei with ≥5 nucleoli.
FIGURE 6:
Inhibition of mitosis and DNA replication, but not RNAPI, increases the percentage of nuclei with ≥5 nucleoli. (A) Inhibition of mitosis and DNA replication, but not RNAPI, increases the percentage of nuclei with ≥5 nucleoli. The ≥5 nucleoli per nucleus PE was quantified relative to the negative control, DMSO (set to 100 PE), in MCF10A cells treated with a panel of small molecule inhibitors of the cell cycle for 24 h (light gray), 48 h (medium gray), and 72 h (dark gray) in triplicate (72–96 replicates of DMSO were performed). A dotted line is drawn at 100 PE. M = inhibitors of mitosis (ispinesib, nocodazole, paclitaxel, hesperadin, and MK-5108). S = inhibitors of DNA replication (mitomycin C and 5-fluorouracil). M/S = inhibitors of both mitosis and DNA replication (topoisomerase II inhibitors: etoposide and ICRF-193). RNAPI = inhibitors of RNAPI transcription (BMH-21 and CX-5461). Statistical significance was calculated by unpaired t tests with the Holm–Sidak method of correction for multiple comparisons (* = p < 0.05, ** = p < 0.01, *** = p < 0.001; n = 3). (B) Representative merged immunofluorescent images of the data collected for the analysis of nucleolar number at the 72 h time point in A. Shown are a selection of nuclei (100 × 100 μM) enlarged threefold using bicubic interpolation from a single field of view. Hoechst 33342 = nuclei (blue) and an antibody to the nucleolar protein fibrillarin (72B9; Reimer et al., 1987) = nucleoli (pink). (C) Nuclear area is greater in nuclei with ≥5 nucleoli. Nuclear area was quantified in pixels using CellProfiler analysis of the images collected for the nucleolar number analysis in A. Three replicates were analyzed for each drug treatment and time point, and six replicates were analyzed for the control, DMSO, treatment at each time point. Each dot represents a single replicate, and the mean ± SD is shown as a black line. Red dots = nuclei with ≥5 nucleoli. Blue dots = nuclei with 0–4 nucleoli. (D) Volcano plot of the statistical analysis of the data in C reveals that with all but two treatments (ispinesib [24 h] and CX-5461 [48 h]), including DMSO treatment, nuclei with ≥5 nucleoli are significantly larger than nuclei with 0–4 nucleoli. Unpaired t tests were performed, and significance was determined based on a false discovery rate approach using the two-stage step-up method of Benjamini et al. (2006) (n = 3 or 6; q < 0.01/-log q-value > 2). Light gray dots = 24 h treatments. Medium gray dots = 48 h treatments. Dark gray dots = 72 h treatments. The x-axis represents the difference in nuclear area between nuclei with 0–4 nucleoli and ≥5 nucleoli. The greatest difference in nuclear area between the nuclei with 0–4 nucleoli and ≥5 nucleoli are observed with hesperadin treatment (48 and 72 h) and ICRF-193 treatment (72 h). (E) Treatment with a subset of drugs, but not all, led to a significant increase in nuclear area from 24 to 48 h in nuclei with 0–4 nucleoli. Significance was determined by unpaired t tests and a false discovery rate approach using the two-stage step-up method of Benjamini et al. (2006) (n = 3 or 6; q < 0.01 = *). (F) Treatment with a subset of drugs, but not all, led to a significant increase in the nuclear area at subsequent time points of nuclei with ≥5 nucleoli. Significance was determined by unpaired t tests and a false discovery rate approach using the two-stage step-up method of Benjamini et al. (2006) (n = 3 or 6; q < 0.01 = *; q < 0.001 = **; q < 0.0001 = ***).
We also tested inhibitors of DNA replication, including mitomycin C and 5-fluorouracil. Notably, 5-fluorouracil inhibits the synthesis of thymidine by thymidylate synthase; therefore we tested it because the thymidine salvage pathway protein thymidine phosphorylase, TYMP, was a hit in this screen. Furthermore, we also tested topoisomerase inhibitors, etoposide and ICRF-193, known for roles in both DNA replication and mitosis, and particularly in the resolution of the rDNA in anaphase (Daniloski et al., 2019). Treatments with these drugs all resulted in the significant increase in the ≥5 nucleoli per nucleus PE at 48 and 72 h, with ICRF-193 treatment also resulting in a significant increase in PE at 24 h (n = 3; adjusted p < 0.05; Figure 6, A and B). Prolonged inhibition of DNA replication was also sufficient to cause an increase in the percentage of nuclei with ≥5 nucleoli.
Finally, we tested RNAPI transcription inhibitors, actinomycin D, BMH-21, and CX-5461, to test the extent to which inhibitors of ribosome biogenesis contribute to increased nucleolar numbers. Intriguingly, prolonged treatment with BMH-21 did not cause an increase in the ≥5 nucleoli per nucleus PE, and at most time points PE was significantly decreased (n = 3; adjusted p < 0.05; Figure 6, A and B). CX-5461 treatment also resulted in a significant decrease in PE at 24 h and no effect at 48 h, but did show a modest increase in PE at 72 h (n = 3; adjusted p < 0.05), which may be due in part to its reported effect on topoisomerase II (Bruno et al., 2020). Prolonged treatment with actinomycin D resulted in low viability, and therefore we were unable to confidently quantify changes in nucleolar number. These data suggest that inhibition of ribosome biogenesis on its own with these drugs is not likely sufficient to cause an increase in nucleolar number.
Furthermore, among the 11 drug treatments analyzed, except for BMH-21, there was a trend toward an increase in the ≥5 nucleoli per nucleus PE from 24 to 48 to 72 h, respectively, suggesting that progression through the cell cycle may be an important aspect of the increased numbers of nucleoli that we observe (Figure 6A). We therefore asked whether nuclear area was also increased with these drug treatments, and as we observed with depletion of our screen hits (Figure 4, A and B), in all conditions the nuclei of cells with ≥5 nucleoli are significantly larger, even in the DMSO control condition (n = 3 or 6; q < 0.01; Figure 6, C and D). Again, however, we do observe some variability in the size increase (e.g., ICRF-193 [72 h], hesperadin [48 h], and hesperadin [72 h]; Figure 6D). However, in only a subset of conditions does nuclear area significantly increase over time and thus does not strictly correlate with the ≥5 nucleoli per nucleus PE. The increase that we do observe, however, was in both nuclei with zero to four nucleoli (etoposide, ICRF-193, mitomycin C, and BMH-21; Figure 6E), as well as in nuclei with ≥5 nucleoli (hesperadin, etoposide, ICRF-193, mitomycin C, and 5-fluorouracil); Figure 6F). Inhibition of mitosis and DNA replication with these select inhibitors is therefore sufficient to yield an increase in the percentage of cells with ≥5 nucleoli; yet nuclear area does not necessarily increase with an increased ≥5 nucleoli per nucleus PE.
Hits are required for RNA polymerase I transcription and protein synthesis
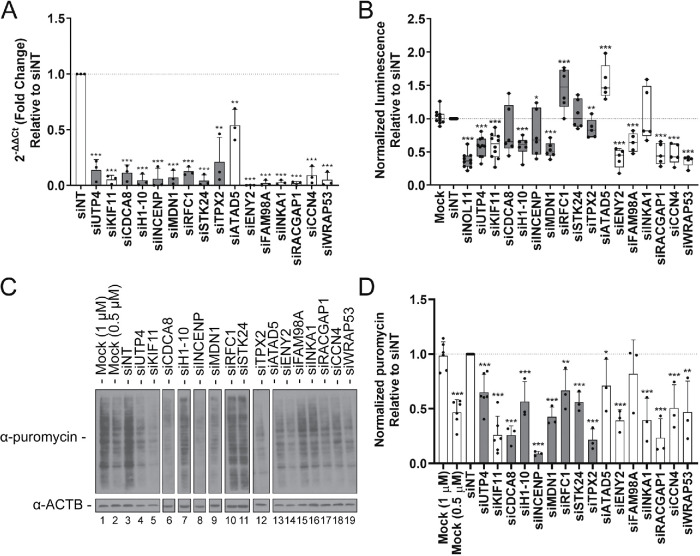
Finally, as we have been previously successful in identifying novel regulators of ribosome biogenesis revealed by a decrease in nucleolar number (Farley-Barnes et al., 2018), we wanted to ask whether increased nucleolar number also uncovered proteins required for the nucleolar function of ribosome biogenesis. To test whether an increase in the percentage of cells with ≥5 nucleoli revealed proteins required for ribosome biogenesis, we utilized a panel of assays to test the screen hits for roles in RNAPI transcription, pre-rRNA processing, and global protein synthesis. Cancer pathologists have known that increased nucleolar number and area are associated with increased nucleolar activity and poor prognosis in cancer patients (Derenzini et al., 2009). Thus, we hypothesized that the ≥5 nucleoli per nucleus phenotype may reflect increased RNAPI transcription. To test this hypothesis, we used a dual-luciferase reporter assay where firefly luciferase expression is driven by the human rDNA promoter (Ghoshal et al., 2004). We then depleted MCF10A cells of 14 validated hits using the siRNA pools from the screen (Table 1, bold; Figure 7A), in addition to the screen positive control, KIF11 and known ribosome biogenesis factor, UTP4. Firefly luminescence was normalized to Renilla luminescence and plotted relative to siNT (Figure 7B). Strikingly, depletion of 11/14 hits significantly affected RNAPI transcription (p < 0.05). Of these, however, depletion of only two caused significant increases in transcription (RFC1 and ATAD5), whereas the other nine caused significant decreases in transcription (H1-10, INCENP, MDN1, TPX2, ENY2, FAM98A, RACGAP1, CCN4, and WRAP53). Based on the current available data regarding protein localization, of these hits, INCENP, H1-10, MDN1, TPX2, and RFC1 localize to the nucleolus, suggesting that this regulation may be direct at the rDNA locus. ENY2, FAM98A, RACGAP1, CCN4, WRAP53, and ATAD5 have not been reported in the nucleolus, and thus their regulation may be indirect. Furthermore, depletion of KIF11 and known RNAPI transcription cofactors, UTP4 and NOL11, also decreased RNAPI transcription (Freed et al., 2012), whereas mock treatments revealed no effect. These data suggest that the screen was successful in identifying novel regulators of RNAPI transcription.
FIGURE 7:
Functional assessment of hits reveals proteins required for RNA polymerase I transcription and global protein synthesis. (A) qPCR analysis confirms depletion of a subset of validated nucleolar (n = 7; gray) and nonnucleolar (n = 7; white) screen hits in MCF10A cells. After depletion using pools of siRNAs targeting the indicated genes, respectively, or nontargeting siRNA control (siNT), the mRNA levels were quantified relative to β-actin mRNA expression. Relative expression values were calculated using the comparative CT method. Statistical significance for three biological replicates, each with three technical replicates, was performed using a two-tailed, unpaired t test. All comparisons are relative to siNT (p < 0.05 = *, p < 0.01 = **, p < 0.001 = ***; n = 3). Data are shown as a bar graph (mean ± SD) and with each replicate shown as a dot. (B) Depletion of the selected hits reveals 11/14 significantly decrease or increase RNAPI transcription. RNAPI transcription was assayed using a dual-luciferase reporter system, with firefly luciferase gene expression controlled by the human rDNA promoter (–410 to +314; Ghoshal et al., 2004). Data were normalized to Renilla luciferase expression controlled by the constitutive CMV promoter. Statistical significance for fivr or six replicates relative to siNT was calculated by two-tailed, unpaired t tests (* = p < 0.05, ** = p < 0.01, *** = p < 0.001; n = 5 or 6). Data are shown as minimum to maximum box-and-whiskers plots, and with each replicate represented as a dot. Gray = nucleolar proteins; white = nonnucleolar proteins. (C) Protein synthesis was significantly decreased in 13/14 of the hit depletion conditions. Shown are representative Western blots from the total protein harvested from hit-depleted MCF10A cells treated with 1 μM puromycin for 1 h. Protein was quantified by Bradford assay and run on a 10% SDS–PAGE gel followed by Western blots using an antibody to puromycin to test for puromycin incorporation into the nascent peptides. β-Actin (ACTB) was used as a loading control. Mock (0.5 μM) cells were treated with half the concentration of puromycin. (D) Quantification of results in C from three replicates. ImageJ was used to quantify the differences in puromycin signal intensity, normalized to the β-actin signal intensity. Statistical significance for the three replicates relative to siNT was calculated by two-tailed, unpaired t tests (* = p < 0.05, ** = p < 0.01, *** = p < 0.001; n = 3). Data are shown as a bar graph (mean ± SD), and with each replicate represented as a dot. Gray = nucleolar proteins; white = nonnucleolar proteins.
Because some factors required for pre-rRNA transcription are also known to affect pre-rRNA processing (Gallagher et al., 2004; Freed et al., 2012; Calo et al., 2015; Farley-Barnes et al., 2018), we tested by Northern blot whether depletion of these hits affects steady-state levels of the pre-rRNA intermediates or precursor-product relationships. Using oligonucleotide probes that detect different intermediates in the known pre-rRNA processing pathways (Supplemental Figure S2A), Northern blots were performed in MCF10A cells depleted of this same subset of hits (Supplemental Figure S2, B–E). Intermediates were then quantified by phosphorimager, and the Ratio Analysis of Multiple Precursors (RAMP; Wang et al., 2014) profiles were plotted relative to the siNT control (Supplemental Figures S3 and S4). As expected, the mock treatment had no effect on the ratios of pre-rRNA intermediates, whereas the positive control, UTP4 depletion (Freed et al., 2012), caused a significant increase in the 30S+1 pre-rRNA precursor and a decrease in the 21S product. Overall, while depletion of these hits led to some ratios from individual probes being statistically significant, only depletion of MDN1 caused more than a twofold change in the ratios of the intermediates, consistent with its known role in large subunit maturation. Taken together, these data suggest that this screen for increased nucleolar numbers did not likely uncover novel factors required for processing the pre-rRNA.
From these Northern blots, we also evaluated whether overall levels of pre-rRNA intermediates were affected by depletion of the selected proteins. Individual pre-rRNA intermediates were thus quantified relative to the RNA component of the signal recognition particle, 7SL, which was used as a loading control (Supplemental Figure S5). Mock-treated cells had little impact on the levels of steady-state intermediates, whereas UTP4-depleted cells showed a significant increase in the primary transcript plus (43S-47S; PTP) and 30S+1 and a decrease in the 30S and 21S pre-rRNAs as expected. While overall these data suggest a general trend toward a decrease in the steady-state pre-rRNA levels consistent with the RNAPI transcription results, only a few hits resulted in statistically significant decreases in a given intermediate, and of those, most differences were modest (e.g., siCDCA8, siSTK24, siENY2, siINKA1, and siKIF11).
Finally, we asked whether protein synthesis is impacted by depletion of these 14 hits, with the rationale that if the nucleolar function of making ribosomes is impacted, then ribosomal function in translation will also be affected. To test this, we used a puromycin-labeling assay followed by Western blot (Kelleher et al., 2013; Figure 7, C and D). Results revealed that 13/14 hits and both UTP4 and KIF11 caused significant decreases in global protein synthesis (p < 0.05). Additionally, mock-treated control cells showed no impact on protein synthesis, and mock-treated cells treated with half the concentration of puromycin (0.5 μM) showed a decrease in the signal by half. Interestingly, while depletion of ATAD5 and RFC1 caused an increase in RNAPI transcription, protein synthesis was decreased, suggesting that the failure of the increased transcription to yield increased translation. Conversely, depletion of INKA1, STK24, and CDCA8 resulted in a significant decrease in protein synthesis, but they were not identified as transcription or processing factors, suggesting that they may be involved in some other aspect of ribosome biogenesis. Finally, siRNA depletion of FAM98A did not show an effect on protein synthesis but did reveal a significant decrease in RNAPI transcription by nearly half, suggesting a compensatory response or limitations in the sensitivity of this assay. This screen was therefore successful in identifying proteins required for global protein synthesis.
Considering that we identified defects in cell cycle progression for several of the screen hits analyzed, it may be possible that these defects could lead to RNAPI transcription and translation defects through TP53 (p53) repression of RNAPI (Beckerman and Prives, 2010). On the contrary, it is also possible that defects in RNAPI transcription and translation could lead to cell cycle arrest through both p53-mediated and p53-independent mechanisms (Rubbi and Milner, 2003; James et al., 2014). As a result, we also evaluated levels of p53 by Western blot and found that depletion of only 2/14 screen hits caused a significant increase in p53 (RFC1 and RACGAP1; Supplemental Figure S6). In conclusion, screening for increased nucleolar number was successful in identifying novel regulators of ribosome biogenesis.
DISCUSSION
A genome-wide siRNA screen in human MCF10A breast epithelial cells revealed 113 hits that caused an increase in the percentage of nuclei with ≥5 nucleoli. Our analysis revealed few proteins that are traditionally associated with the nucleolar function of ribosome biogenesis, yet the hits were enriched in proteins that localize to the nucleolus (∼20%). We further revealed that the hits were largely associated with the cell cycle and that a majority are not conserved to S. cerevisiae. In addition, cell cycle analysis revealed that depletion of several hits caused the accumulation of cells in S and G2/M phase, and follow-up using biochemical analyses revealed that depletion of the majority of hits tested led to aberrant RNAPI transcription (11/14) and decreased protein synthesis (13/14). Pre-rRNA processing defects, however, were largely absent among the subset of hits tested (1/14), except for the depletion of the known LSU maturation factor, MDN1. While in some cases an increase in the number of nucleoli could be explained by failed cytokinesis and a duplicate number of NORs per nucleus, only 8/20 hits led to an increase in the percentage of nuclei with a >4N DNA content. We thus conclude that our results revealed a unique subset of proteins required for the transcriptional regulation of RNAPI that, when depleted, are concomitant with increased numbers of nucleoli. In addition, we have found a strong interdependence between faithful cell cycle progression and the regulation of ribosome biogenesis.
The 113 hits are a unique subset of proteins required for the regulation of nucleolar number and function. In comparison to our prior screening results for decreased nucleolar number (Farley-Barnes et al., 2018), there was no overlap among the hits or among the overrepresented GO categories (fold enrichment ≥5, p < 0.05), suggesting that the mechanisms producing these two phenotypes may likewise be distinct. Furthermore, the prior data set generated for decreased nucleolar number revealed that, of the hits tested, a majority were associated with defects in pre-rRNA processing (16/20; Farley-Barnes et al., 2018). Proteins required for RNAPI transcription were also uncovered (7/20); however, only one of these proteins was not concomitant with a pre-rRNA processing defect. This is in stark contrast to this data set (≥5 nucleoli per nucleus) that revealed proteins associated primarily with RNAPI transcription and only one, MDN1, associated with pre-rRNA processing. Interestingly, depletion of MDN1 also caused a decrease in RNAPI transcription, suggesting a role for this protein that has not previously been reported. Furthermore, when compared with other screens for regulators of nucleolar function in eukaryotes, the overlap among the hits was minimal. However, differences among the model systems employed or experimental readouts evaluated may largely explain the lack of overlap. For instance, the screens for human ribosome biogenesis factors were all performed in the aneuploid HeLa cervical cancer cell line (Wild et al., 2010; Tafforeau et al., 2013; Badertscher et al., 2015), whereas this study was performed in a near-diploid non–cancer-derived cell line, MCF10A. Each screen also utilized different experimental methodologies and readouts to establish ribosome biogenesis factors, where it is conceivable that the proteins required for the regulation of nucleolar number may be different from proteins that regulate ribosomal subunit export. Despite minimal overlap, however, comparisons of the 113 hits to the screens for changes in nucleolar size in S. cerevisiae and D. melanogaster both reveal links between the nucleolus and mitosis (Neumuller et al., 2013), suggesting that they may be evolutionarily conserved. These results are significant because they broaden our understanding of the regulation of nucleolar function by revealing proteins that are distinct from those revealed by other screens.
The hits revealed here add to the evidence supporting a significant cross-talk between the nucleolus and mitosis. Our screen identified proteins required for faithful progression through the G2/M phase that also function in the regulation of RNAPI transcription. Included among these are proteins required for mitotic spindle assembly, including KIF11 and TPX2, as well as the Aurora B kinase–associated protein, INCENP (Uehara et al., 2013). Furthermore, in addition to the aforementioned hits, sister chromatid cohesion proteins (CDCA5 and SGOL1) and regulators of cell cycle progression (CUL1, SKP1, and MASTL Nakayama and Nakayama, 2006) were identified as hits that caused an increase in nucleolar number when depleted. There has long been an appreciation for the role of the nucleolus in cell cycle regulation, including in the nucleolar sequestration of proteins required for cell cycle progression (Visintin and Amon, 2000; Boisvert et al., 2007). It is also well-known that the nucleolus undergoes dynamic remodeling as a consequence of the cell cycle, exemplified by cyclin-dependent kinase 1 (CDK1)-mediated silencing of RNAPI and the disassembly and reformation of nucleoli in mitosis (Hernandez-Verdun, 2011). It is thus interesting to speculate that these proteins may be required for the reinitiation of RNAPI transcription upon mitotic exit that, if disrupted, would lead to failure in the reformation and coalescence of nascent nucleoli. Furthermore, a novel complex of ribosome biogenesis factors was recently implicated in the regulation of mitotic entry, chromatid cohesion, and spindle assembly through AURKB (Fujimura et al., 2020), strengthening the support for a cross-talk between ribosome biogenesis factors, nucleolar proteins, and mitosis. Thus, while it is possible that our screen hits function in ribosome biogenesis outside of mitosis, as reported previously for KIF11 in translation (Bartoli et al., 2011), the enrichment of mitotic factors among the hits strongly supports the conclusion that the role for these proteins in mitosis also includes the regulation of RNAPI transcription.
Our results also support the link between the nucleolus and faithful DNA replication. In addition to proteins required for G2/M phase progression, our screen also identified hits required for progression through S phase, and specifically with the cellular processes of DNA replication, recombination, and repair. Of the proteins tested, a subset was also required for RNAPI transcription. Depletion of the proliferating cell nuclear antigen (PCNA)-associated proteins, RFC1 and ATAD5, for example, resulted in a significant increase in RNAPI transcription, suggesting a role for these proteins in rDNA transcriptional silencing. Interestingly, RFC1 has previously been reported to regulate rDNA transcriptional silencing in S. cerevisiae (Smith et al., 1999) and in plants (Liu et al., 2010), and ATAD5 in yeast has previously been associated with increased genome instability and recombination events (Ben-Aroya et al., 2003; Sikdar et al., 2009), as well as defects in sister chromatid cohesion (Parnas et al., 2009). The rDNA loci are the most highly transcribed loci in eukaryotic genomes, and conflict between the transcription and replication machinery can lead to genome instability at replication forks (Lindstrom et al., 2018). Furthermore, replication stress, particularly at fragile site loci like the rDNA, has been associated with defects in mitosis including increased DNA bridges in anaphase, chromosome breakage, and cancer (Chan et al., 2009; Stults et al., 2009; Franchitto, 2013; Warmerdam and Wolthuis, 2019). We also observed increases in the percent of nuclei with a >4N DNA content among hits that caused S phase accumulation that is consistent with this observation. As a result, mechanisms have evolved to ensure replication fidelity, including the recent evidence for transient silencing of RNAPI in response to DNA damage (Kruhlak et al., 2007; Ciccia et al., 2014; Larsen et al., 2014; Larsen and Stucki, 2016). The identification of DNA replication–associated proteins that also regulate RNAPI transcription is not a first (Ogawa and Baserga, 2017), and our results further support this important link between the nucleolus, genome integrity, and the fidelity of cellular growth and proliferation.
Drug screening for the ≥5 nucleoli per nucleus phenotype also supports the association of the cell cycle and increased nucleolar number. We found that prolonged inhibition of mitosis and DNA replication with several inhibitors led to significant increases in the percentage of nuclei with ≥5 nucleoli, including the KIF11 inhibitor, ispinesib. Additionally, inhibition of topoisomerases known to aid in the resolution of DNA both in anaphase (Daniloski et al., 2019; Gemble et al., 2020) and during replication (Vesela et al., 2017) further supports our conclusions regarding the interdependence between faithful cell cycle progression and nucleolar form and function. Even more intriguing is that prolonged inhibition of RNAPI transcription by BMH-21 did not result in an increase in the percentage of nuclei with ≥5 nucleoli, suggesting that RNAPI inhibition alone is not sufficient to cause increased nucleolar number. Taken together, these data validate our results from our evaluation of the screen hits that faithful cell cycle progression plays an important role in maintaining typical numbers of nucleoli.
To conclude, we have identified a unique subset of proteins required for maintaining normal numbers of nucleoli in cells that are also involved in regulating RNAPI transcription and protein synthesis. While ribosome biogenesis is the primary function associated with nucleoli, it is clear that the nucleolus is central to the regulation of multiple cellular processes. Screening in a human cell line for an increased percentage of nuclei with ≥5 nucleoli has validated this observation. Furthermore, the increase in nucleolar number (≥5 nucleoli per nucleus) that we observe primarily in G2/M phase upon depletion of screen hits may likely be a consequence of replication stress and failures in the resolution of the rDNA in mitosis. These data identified in this study thus advance our understanding of the processes and factors required to maintain normal nucleolar numbers through RNAPI transcriptional regulation and are significant for broadening our understanding of the regulation of ribosome biogenesis in higher eukaryotes that highlights the interdependence between the nucleolus and the mammalian cell cycle.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell lines
The human breast epithelial cell line, MCF 10A (American Type Culture Collection [ATCC]; CRL-10317), was 2D subcultured in DMEM/F-12 (Life Technologies; 1130-032) supplemented with 5% horse serum (Life Technologies; 16050), 10 μg/ml insulin (Sigma; I1882), 0.5 μg/ml hydrocortisone (Sigma; H0135), 100 ng/ml cholera toxin (Sigma; C8052), and 20 ng/ml epidermal growth factor (Peprotech; AF-100-15).
RNAi
For screen validation, nuclear area analysis, and cell cycle profiling, the individual siGENOME set of four siRNAs (Horizon Discovery) that comprised the pools for each hit was used. The last two digits of the Horizon Discovery/Dharmacon product number associated with the individual siRNAs are listed in Table 1. For biochemical assays on the subset of validated hits, the siGENOME SMARTpool siRNAs (Horizon Discovery) were used, except for with the siNT negative control, which used the ON-TARGETplus Non-targeting pool (D-001810-10-20). Unless otherwise noted, subconfluent cells (log phase) were transfected with siRNAs (20–30 nM, final concentration) using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific; 13778150) and incubated for 72 h before the experimental assays.
High-content genome-wide RNAi screen
The high-content genome-wide siRNA screen was performed as reported in Farley-Barnes et al. (2018) using the human siGENOME SMARTpool siRNA library that contained 18,107 pools of four siRNAs against each target (Horizon Discovery; Figure 1A). As described previously, cells were imaged on an IN Cell Analyzer 2200 (Cytiva), which is a wide-field, multicolor, fluorescence microscope. Three fields of view (20X; 665.63 μm x 665.63 μm) were acquired per well, and high-throughput image analysis was performed using CellProfiler (Carpenter et al., 2006; McQuin et al., 2018) to segment nucleoli based on fibrillarin staining (72B9; Reimer et al., 1987) and nuclei based on Hoechst 33342 staining. In this analysis, raw nucleolar number data were normalized to the 16 negative (siRISC-free; 0 PE) and 16 positive (siKIF11; 100 PE) control wells run on the same plate and averaged across the fields of view to yield a mean NPE.
Hits with ≥5 nucleoli per nucleus were identified based on a stringent cutoff of three SD from the mean NPE yielding 186 hits (Figure 1B). Hits were then filtered by expression, discarding hits not expressed in MCF10A cells based on a poly(A) transcriptome analysis by RNA sequencing performed at the Yale Center for Genome Analysis (West Haven, CT; GEO accession GSE154764), and analyzed using a pipeline developed in Partek Flow (Partek, St. Louis, MO). Sequencing reads were aligned to the human genome (hg19 assembly) using Bowtie 2 (v2.2.5) and quantified to the transcriptome (RefSeq 16 08 01 v2) using Cufflinks (v2.2.1) (n = 3; FPKM>0). Hits were also filtered by viability, discarding hits with <5% viability relative to the siRISC-free control, to limit hits called based on a small number of cells. This resulted in 113 high-confidence hits that, when depleted, caused an increase in nucleolar number to ≥5 per nucleus.
Screen performance was monitored by Z-prime factors (Figure 1C) and signal-to-background (S/B; Figure 1D), with an average Z-prime of 0.41 (range, 0.02–0.68) and an average S/B of 10.29 (range, 4.32–22.33).
Oligonucleotide deconvolution
We validated a subset of the high-confidence hits (19/20; Table 1) by oligonucleotide deconvolution, as is standard in high-throughput siRNA screens (Sigoillot and King, 2011). The individual siRNAs in the pools of four against the selected hits were tested in triplicate, six fields of view imaged for each replicate (20X) in the screening assay to quantify the NPE for the ≥5 nucleoli per nucleus phenotype (Z-prime = 0.61). Validated hits were determined by an NPE ≥ 15.0 in at least 2/4 siRNAs (Table 1; Supplemental Table S4). Of the validated hits, 14 were selected for biochemical analysis (Table 1, in bold).
Cell cycle analysis
We analyzed the images of the 20 hits collected for oligonucleotide deconvolution. Integrated intensity of the Hoechst 33342 DNA stain was measured per nucleus to determine cell cycle distribution. A histogram of the log2 integrated intensities for the negative control (siRISC-free; 48 replicates; six fields of view) was plotted and the G1 peak set to 1.0 and G2 peak set to 2.0. Each hit depletion condition (three replicates per siRNA; six fields of view) was then normalized to siRISC-free, including siKIF11 both as individual siRNAs (three replicates per siRNA; six fields of view) and as a pool (48 replicates; six fields of view). Cell cycle phases were defined as in Chan et al. (2013). G0/G1 phase nuclei were defined as normalized log2 integrated intensities of 0.75–1.25, S phase nuclei were defined as 1.25–1.75, and G2/M phase nuclei were defined as 1.75–2.50 (including late G2 nuclei), and nuclei with >4N DNA content were defined as >2.50.
Pharmacological inhibition of the cell cycle
MCF10A cells were treated with inhibitors of the cell cycle for 24, 48, and 72 h in triplicate and nine fields of view imaged per replicate (20X). Drugs were all dissolved in DMSO, and doses were selected based on reported EC50 values in cell culture conditions for each drug, with final DMSO concentration = 0.1%. The ≥5 nucleoli per nucleus PE were quantified by the same CellProfiler pipeline used in the initial screen and analyzed relative to the 0.1% DMSO treatment, which was set to 100 PE. Tested inhibitors of mitosis included ispinesib (0.082 μM; Cayman Chemical; 18014), nocodazole (0.741 μM; Cayman Chemical; 13857), paclitaxel (0.0274 μM; Sigma; T7402), hesperadin (0.082 μM; Cayman Chemical; 24199), and MK-5108 (0.247 μM; Cayman Chemical; 19167). Inhibitors of DNA replication included mitomycin C (0.741 μM; Cayman Chemical; 11435) and 5-fluorouracil (0.741 μM; Sigma; F6627). Topoisomerase inhibitors included etoposide (0.741 μM; Sigma; E1383) and ICRF-193 (6.67 μM; Sigma; I4659), and inhibitors of RNAPI transcription included actinomycin D (0.00914 μM; Sigma; A1410), BMH-21 (0.741 μM; Sigma; SML1183), and CX-5461 (0.741 μM; Cayman Chemical; 18392). Doses were selected based on doses used at prolonged time points as reported in the literature.
Quantitative PCR
Total RNA was extracted from siRNA-depleted cells using TRIzol reagent (Thermo Fisher Scientific; 15596018) as per the manufacturer’s instructions. After validating that A260/280 values were >1.80 and A260/230 values were >1.7, cDNA synthesis was performed using the iScript gDNA Clear cDNA Synthesis Kit (Bio-Rad; 172-5035) using 1 μg of total RNA and a mix of random hexamer and oligo dT primers. Previously published primers were used to test mRNA levels of ATAD5 (Bell et al., 2011) and RFC1 (Stielow et al., 2014). BioRad PrimePCR Assay gene-specific primers were used to test mRNA levels of the remaining hits (Bio-Rad, 10025636; UTP4, qHsaCID0021354; KIF11, qHsaCID0015908; CDCA8, qHsaCED0044566; H1-10, qHsaCED0019411; INCENP, qHsaCID0010103; MDN1, qHsaCID0006754; STK24, qHsaCID0012429; TPX2, qHsaCID0016024; ENY2, qHsaCED0003040; FAM98A, qHsaCID0010948; INKA, qHsaCED0020031; RACGAP1, qHsaCID0011308; CCN4, qHsaCED0036389; WRAP53, qHsaCID0006849). β-Actin primers were designed in our laboratory (intron-spanning; forward 5′-ATT GGC AAT GAG CGG TTC-3′ and reverse 5′-CGT GGA TGC CAC AGG ACT-3′). All quantitative PCRs (qPCRs) were completed using the iTaq Universal SYBR Green Supermix (Bio-Rad; 172-5121). To verify the amplification of a single PCR product, melt curves were generated for each sample. Three biological replicates, each with three technical replicates, were measured for each of the 14 tested hits as well as siUTP4, siKIF11, and the negative nontargeting control (siNT). Amplification of the β-actin mRNA was used as an internal control, and analysis was completed using the comparative CT method (ΔΔCT).
Dual-luciferase reporter assay
Following siRNA-depletion of hits for 48 h, cells were transfected with 1000 ng of pHrD-IRES-Luc (Ghoshal et al., 2004) and 0.1 ng of a plasmid that constitutively expresses Renilla luciferase (Freed et al., 2012) using Lipofectamine 3000 Reagent (Thermo Fisher Scientific; L3000015). After 72 h of siRNA-depletion and 24 h of incubation with the reporter plasmids, luminescence was detected using the Dual-Luciferase Reporter Assay System (Promega; E1910) and a GloMax 20/20 luminometer (Promega). In addition to incubation with the 1X passive lysis buffer for 15 min, MCF10A cells were scraped beforecollection for luminescence readings.
Northern blots
Total RNA was extracted from siRNA-depleted cells using TRIzol Reagent. To assay for changes in levels of pre-rRNA intermediates, 4 μg of total RNA was run on a 1% agarose/1.25% formaldehyde gel in a 1.5 M tricine/1.5 M triethanolamine buffer. RNA was transferred overnight to a Hybond XL nylon membrane (GE Healthcare; RPN 303S) by capillary transfer in 10X saline-sodium citrate (SSC) transfer buffer after a brief 15 min soak in a 0.5 M sodium hydroxide solution. Membranes were then exposed to UV (254 nm) to immobilize the RNA, incubated with denatured yeast tRNA for 1 h at 42°C, and hybridized overnight at 37°C with 5′ end radiolabeled oligonucleotide probes in a solution of 7.5X Denhardt’s solution, 5X sodium chloride-sodium phosphate-EDTA (SSPE) buffer, and 0.1% SDS as in Farley-Barnes et al. (2018).
Puromycin-labeling assay
Following siRNA-depletion of hits for 72 h, cells were treated as described in Farley-Barnes et al. (2018), with the exception that puromycin antibody (Kerafast; EQ0001) was used at a 1:500 dilution.
Western blots
Following siRNA-depletion of hits for 72 h, total protein was harvested by the same method as used in the puromycin-labeling assay described in Farley-Barnes et al. (2018). The protein concentration was quantified by Bradford assay, and 30 μg of total protein was run by SDS–PAGE on a 10% gel with a 5% stacking gel. Protein was transferred to a polyvinylidene difluoride membrane using a Trans-Blot Turbo Transfer System (Bio-Rad) and blocked for 1 h with 5% milk in phosphate-buffered saline (PBST) before being incubated overnight with horseradish peroxidase–-conjugated p53 antibody (Santa Cruz Biotechnology; sc-126) diluted in PBST (1:5000). Following imaging on a ChemiDoc Imaging System (Bio-Rad), blots were stripped and reprobed for β-actin as performed in the puromycin-labeling assay and quantified using ImageJ.
Statistical analyses
All statistical analyses were performed in GraphPad Prism 8.2.1 (GraphPad Software) using the tests described in the figure captionss.
Supplementary Material
Acknowledgments
We thank past and present members of the Baserga laboratory for their insights and optimism throughout this project and in the preparation of this article. We also thank Joan Steitz, Anthony Koleske, Megan King, and Lilian Kabeche for their influence on this project through many perceptive discussions. We acknowledge our funding sources including National Institutes of Health Training Grant T32GM007223 (to L.M.O. and S.J.B.), Ruth L. Kirschstein National Research Service Award (NRSA) Individual Predoctoral Fellowship F31AG058405 (to L.M.O.), a Yale Cancer Center Pilot Grant (to S.J.B.), and National Institute of General Medical Sciences Grant 1R35GM131687 (to S.J.B.).
Abbreviations used:
- NPE
normalized percent effect
- PE
percent effect
- rDNA
ribosomal DNA
- RNAPI
RNA polymerase I
- siRNA
small interfering RNA.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-10-0670) on March 10, 2021.
REFERENCES
- Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI (2009). NOPdb: Nucleolar Proteome Database–2008 update. Nucleic Acids Res 37, D181–D184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert M, O’Donohue MF, Lebaron S, Gleizes PE (2018). Pre-ribosomal RNA processing in human cells: from mechanisms to congenital diseases. Biomolecules 8, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badertscher L, Wild T, Montellese C, Alexander LT, Bammert L, Sarazova M, Stebler M, Csucs G, Mayer TU, Zamboni N, et al. (2015). Genome-wide RNAi screening identifies protein modules required for 40S subunit synthesis in human cells. Cell Rep 13, 2879–2891. [DOI] [PubMed] [Google Scholar]
- Balakrishnan R, Park J, Karra K, Hitz BC, Binkley G, Hong EL, Sullivan J, Micklem G, Cherry JM (2012). YeastMine–an integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database (Oxford) 2012, bar062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli KM, Jakovljevic J, Woolford JL Jr, Saunders WS (2011). Kinesin molecular motor Eg5 functions during polypeptide synthesis. Mol Biol Cell 22, 3420–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler J, Hurt E (2019). Eukaryotic ribosome assembly. Annu Rev Biochem 88, 281–306. [DOI] [PubMed] [Google Scholar]
- Bassler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, Hurt E (2010). The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol Cell 38, 712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerman R, Prives C (2010). Transcriptional regulation by p53. Cold Spring Harb Perspect Biol 2, a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DW, Sikdar N, Lee KY, Price JC, Chatterjee R, Park HD, Fox J, Ishiai M, Rudd ML, Pollock LM, et al. (2011). Predisposition to cancer caused by genetic and functional defects of mammalian Atad5. PLoS Genet 7, e1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S, Koren A, Liefshitz B, Steinlauf R, Kupiec M (2003). ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc Natl Acad Sci USA 100, 9906–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Krieger AM, Yekutieli D (2006). Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93, 491–507. [Google Scholar]
- Bersaglieri C, Santoro R (2019). Genome organization in and around the nucleolus. Cells 8, 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d’Herin P, Harper M, Kress M, Nigg EA (1995). Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83, 1159–1169. [DOI] [PubMed] [Google Scholar]
- Bohnsack KE, Bohnsack MT (2019). Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J 38, e100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Lam YW, Lamont D, Lamond AI (2010). A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics 9, 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007). The multifunctional nucleolus. Nat Rev Mol Cell Biol 8, 574–585. [DOI] [PubMed] [Google Scholar]
- Bruno PM, Lu M, Dennis KA, Inam H, Moore CJ, Sheehe J, Elledge SJ, Hemann MT, Pritchard JR (2020). The primary mechanism of cytotoxicity of the chemotherapeutic agent CX-5461 is topoisomerase II poisoning. Proc Natl Acad Sci USA 117, 4053–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursac S, Prodan Y, Pullen N, Bartek J, Volarevic S (2021). Dysregulated ribosome biogenesis reveals therapeutic liabilities in cancer. Trends Cancer 7, 57–76. [DOI] [PubMed] [Google Scholar]
- Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, Wysocka J (2015). RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantwell H, Nurse P (2019). Unravelling nuclear size control. Curr Genet 65, 1281–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GK, Kleinheinz TL, Peterson D, Moffat JG (2013). A simple high-content cell cycle assay reveals frequent discrepancies between cell number and ATP and MTS proliferation assays. PLoS One 8, e63583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID (2009). Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11, 753–760. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Huang JW, Izhar L, Sowa ME, Harper JW, Elledge SJ (2014). Treacher Collins syndrome TCOF1 protein cooperates with NBS1 in the DNA damage response. Proc Natl Acad Sci USA 111, 18631–18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z, Bisht KK, McStay B, Smith S (2019). Resolution of human ribosomal DNA occurs in anaphase, dependent on tankyrase 1, condensin II, and topoisomerase IIalpha. Genes Dev 33, 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot CO, Hsia JE, Anzola JV, Motamedi A, Yoon M, Wong YL, Jenkins D, Lee HJ, Martinez MB, Davis RL, et al. (2015). A cell biologist’s field guide to aurora kinase inhibitors. Front Oncol 5, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini M, Montanaro L, Trere D (2009). What the nucleolus says to a tumour pathologist. Histopathology 54, 753–762. [DOI] [PubMed] [Google Scholar]
- Farley-Barnes KI, McCann KL, Ogawa LM, Merkel J, Surovtseva YV, Baserga SJ (2018). Diverse regulators of human ribosome biogenesis discovered by changes in nucleolar number. Cell Rep 22, 1923–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley-Barnes KI, Ogawa LM, Baserga SJ (2019). Ribosomopathies: old concepts, new controversies. Trends Genet 35, 754–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley KI, Surovtseva Y, Merkel J, Baserga SJ (2015). Determinants of mammalian nucleolar architecture. Chromosoma 124, 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floutsakou I, Agrawal S, Nguyen TT, Seoighe C, Ganley AR, McStay B (2013). The shared genomic architecture of human nucleolar organizer regions. Genome Res 23, 2003–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto A (2013). Genome instability at common fragile sites: searching for the cause of their instability. Biomed Res Int 2013, 730714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EF, Prieto JL, McCann KL, McStay B, Baserga SJ (2012). NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet 8, e1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura A, Hayashi Y, Kato K, Kogure Y, Kameyama M, Shimamoto H, Daitoku H, Fukamizu A, Hirota T, Kimura K (2020). Identification of a novel nucleolar protein complex required for mitotic chromosome segregation through centromeric accumulation of Aurora B. Nucleic Acids Res 48, 6583–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani K, Nissan TA, Petfalski E, Tollervey D, Hurt E (2004). Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J Biol Chem 279, 55411–55418. [DOI] [PubMed] [Google Scholar]
- Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ (2004). RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev 18, 2506–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemble S, Buhagiar-Labarchede G, Onclercq-Delic R, Fontaine G, Lambert S, Amor-Gueret M (2020). Topoisomerase IIalpha prevents ultrafine anaphase bridges by two mechanisms. Open Biol 10, 190259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K, Majumder S, Datta J, Motiwala T, Bai S, Sharma SM, Frankel W, Jacob ST (2004). Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem 279, 6783–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes CJ, Harman MW, Centuori SM, Wolgemuth CW, Martinez JD (2018). Measuring DNA content in live cells by fluorescence microscopy. Cell Div 13, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JJ, Pathak S, Panov K, Kasciukovic T, Panova T, Russell J, Zomerdijk JC (2007). A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J 26, 1560–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob A, Colleran C, McStay B (2014). Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division. Genes Dev 28, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I (2013). The nucleolus-guardian of cellular homeostasis and genome integrity. Chromosoma 122, 487–497. [DOI] [PubMed] [Google Scholar]
- Henderson AS, Warburton D, Atwood KC (1972). Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci USA 69, 3394–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Plisson-Chastang C, O’Donohue MF, Chakraborty A, Gleizes PE (2015). An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6, 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D (2011). Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2, 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetman M, Pietrzak M (2012). Emerging roles of the neuronal nucleolus. Trends Neurosci 35, 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A, Wang Y, Raje H, Rosby R, DiMario P (2014). Nucleolar stress with and without p53. Nucleus 5, 402–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarboui MA, Bidoia C, Woods E, Roe B, Wynne K, Elia G, Hall WW, Gautier VW (2012). Nucleolar protein trafficking in response to HIV-1 Tat: rewiring the nucleolus. PLoS One 7, e48702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarboui MA, Wynne K, Elia G, Hall WW, Gautier VW (2011). Proteomic profiling of the human T-cell nucleolus. Mol Immunol 49, 441–452. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B (2007). The size of the nucleus increases as yeast cells grow. Mol Biol Cell 18, 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher AR, Kimball SR, Dennis MD, Schilder RJ, Jefferson LS (2013). The mTORC1 signaling repressors REDD1/2 are rapidly induced and activation of p70S6K1 by leucine is defective in skeletal muscle of an immobilized rat hindlimb. Am J Physiol Endocrinol Metab 304, E229–E236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. (2011). Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011, bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R (2007). The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 447, 730–734. [DOI] [PubMed] [Google Scholar]
- Larsen DH, Hari F, Clapperton JA, Gwerder M, Gutsche K, Altmeyer M, Jungmichel S, Toledo LI, Fink D, Rask MB, et al. (2014). The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat Cell Biol 16, 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen DH, Stucki M (2016). Nucleolar responses to DNA double-strand breaks. Nucleic Acids Res 44, 538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Trinkle-Mulcahy L, Lam YW, Andersen JS, Mann M, Lamond AI (2006). NOPdb: Nucleolar Proteome Database. Nucleic Acids Res 34, D218–D220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS, Jurada D, Bursac S, Orsolic I, Bartek J, Volarevic S (2018). Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 37, 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang J, Miki D, Xia R, Yu W, He J, Zheng Z, Zhu JK, Gong Z (2010). DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell 22, 2336–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Iino H, Hihara S, Imamoto N (2011). Nuclear size, nuclear pore number and cell cycle. Nucleus 2, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, et al. (2018). CellProfiler 3.0: next-generation image processing for biology. PLoS Biol 16, e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD (2017). PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45, D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore HM, Bai B, Boisvert FM, Latonen L, Rantanen V, Simpson JC, Pepperkok R, Lamond AI, Laiho M (2011). Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol Cell Proteomics 10, M111 009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, et al. (2008). Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133, 627–639. [DOI] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K (2006). Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 6, 369–381. [DOI] [PubMed] [Google Scholar]
- Narla A, Ebert BL (2010). Ribosomopathies: human disorders of ribosome dysfunction. Blood 115, 3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann FR, Nurse P (2007). Nuclear size control in fission yeast. J Cell Biol 179, 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Gross T, Samsonova AA, Vinayagam A, Buckner M, Founk K, Hu Y, Sharifpoor S, Rosebrock AP, Andrews B, et al. (2013). Conserved regulators of nucleolar size revealed by global phenotypic analyses. Sci Signal 6, ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa LM, Baserga SJ (2017). Crosstalk between the nucleolus and the DNA damage response. Mol Biosyst 13, 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnas O, Zipin-Roitman A, Mazor Y, Liefshitz B, Ben-Aroya S, Kupiec M (2009). The ELG1 clamp loader plays a role in sister chromatid cohesion. PLoS One 4, e5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei YY, Li GC, Ran J, Wan XH, Wei FX, Wang L (2019). Kinesin family member 11 enhances the self-renewal ability of breast cancer cells by participating in the Wnt/beta-catenin pathway. J Breast Cancer 22, 522–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei YY, Li GC, Ran J, Wei FX (2017). Kinesin family member 11 contributes to the progression and prognosis of human breast cancer. Oncol Lett 14, 6618–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzo M, Montanaro L, Trere D, Derenzini M (2019). The ribosome biogenesis-cancer connection. Cells 8, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV (2010). The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37, 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapova TA, Gerton JL (2019). Ribosomal DNA and the nucleolus in the context of genome organization. Chromosome Res 27, 109–127. [DOI] [PubMed] [Google Scholar]
- Rawlinson SM, Zhao T, Rozario AM, Rootes CL, McMillan PJ, Purcell AW, Woon A, Marsh GA, Lieu KG, Wang LF, et al. (2018). Viral regulation of host cell biology by hijacking of the nucleolar DNA-damage response. Nat Commun 9, 3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer G, Pollard KM, Penning CA, Ochs RL, Lischwe MA, Busch H, Tan EM (1987). Monoclonal autoantibody from a (New Zealand black x New Zealand white)F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum 30, 793–800. [DOI] [PubMed] [Google Scholar]
- Reynolds RC, Montgomery PO, Hughes B (1964). Nucleolar “caps” produced by actinomycin D. Cancer Res 24, 1269–1277. [PubMed] [Google Scholar]
- Roukos V, Pegoraro G, Voss TC, Misteli T (2015). Cell cycle staging of individual cells by fluorescence microscopy. Nat Protoc 10, 334–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T (2013). Spatial dynamics of chromosome translocations in living cells. Science 341, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi CP, Milner J (2003). Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22, 6068–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D (2012). Revisiting the nucleolus: from marker to dynamic integrator of cancer signaling. Sci Signal 5, pe38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigoillot FD, King RW (2011). Vigilance and validation: keys to success in RNAi screening. ACS Chem Biol 6, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikdar N, Banerjee S, Lee KY, Wincovitch S, Pak E, Nakanishi K, Jasin M, Dutra A, Myung K (2009). DNA damage responses by human ELG1 in S phase are important to maintain genomic integrity. Cell Cycle 8, 3199–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Caputo E, Boeke JD (1999). A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol 19, 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule HD, Maloney TM, Wolman SR, Peterson WD Jr, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC (1990). Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res 50, 6075–6086. [PubMed] [Google Scholar]
- Stamatopoulou V, Parisot P, De Vleeschouwer C, Lafontaine DLJ (2018). Use of the iNo score to discriminate normal from altered nucleolar morphology, with applications in basic cell biology and potential in human disease diagnostics. Nat Protoc 13, 2387–2406. [DOI] [PubMed] [Google Scholar]
- Stielow C, Stielow B, Finkernagel F, Scharfe M, Jarek M, Suske G (2014). SUMOylation of the polycomb group protein L3MBTL2 facilitates repression of its target genes. Nucleic Acids Res 42, 3044–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Williamson EP, Hourigan JS, Vargas HD, Arnold SM, Moscow JA, Pierce AJ (2009). Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res 69, 9096–9104. [DOI] [PubMed] [Google Scholar]
- Sulima SO, Kampen KR, De Keersmaecker K (2019). Cancer biogenesis in ribosomopathies. Cells 8, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47, D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL (2013). The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Mol Cell 51, 539–551. [DOI] [PubMed] [Google Scholar]
- Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, et al. (2017). A subcellular map of the human proteome. Science 356, eaal3321. [DOI] [PubMed] [Google Scholar]
- Tiku V, Antebi A (2018). Nucleolar function in lifespan regulation. Trends Cell Biol 28, 662–672. [DOI] [PubMed] [Google Scholar]
- Uehara R, Tsukada Y, Kamasaki T, Poser I, Yoda K, Gerlich DW, Goshima G (2013). Aurora B and Kif2A control microtubule length for assembly of a functional central spindle during anaphase. J Cell Biol 202, 623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venere M, Horbinski C, Crish JF, Jin X, Vasanji A, Major J, Burrows AC, Chang C, Prokop J, Wu Q, et al. (2015). The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Sci Transl Med 7, 304ra143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten VL, Peckham LA, Olive M, Capell BC, Collins FS, Nabel EG, Young SG, Fong LG, Lammerding J (2011). Protein farnesylation inhibitors cause donut-shaped cell nuclei attributable to a centrosome separation defect. Proc Natl Acad Sci USA 108, 4997–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesela E, Chroma K, Turi Z, Mistrik M (2017). Common chemical inductors of replication stress: focus on cell-based studies. Biomolecules 7, 19. [Google Scholar]
- Visintin R, Amon A (2000). The nucleolus: the magician’s hat for cell cycle tricks. Curr Opin Cell Biol 12, 372–377. [DOI] [PubMed] [Google Scholar]
- von Stechow L, Typas D, Carreras Puigvert J, Oort L, Siddappa R, Pines A, Vrieling H, van de Water B, Mullenders LH, Danen EH (2015). The E3 ubiquitin ligase ARIH1 protects against genotoxic stress by initiating a 4EHP-mediated mRNA translation arrest. Mol Cell Biol 35, 1254–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Anikin L, Pestov DG (2014). Two orthogonal cleavages separate subunit RNAs in mouse ribosome biogenesis. Nucleic Acids Res 42, 11180–11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmerdam DO, Wolthuis RMF (2019). Keeping ribosomal DNA intact: a repeating challenge. Chromosome Res 27, 57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren AJ (2018). Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome. Adv Biol Regul 67, 109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil D, Garcon L, Harper M, Dumenil D, Dautry F, Kress M (2002). Targeting the kinesin Eg5 to monitor siRNA transfection in mammalian cells. Biotechniques 33, 1244–1248. [DOI] [PubMed] [Google Scholar]
- Weiss WA, Taylor SS, Shokat KM (2007). Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat Chem Biol 3, 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E, Kutay U (2010). A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol 8, e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL Jr, Baserga SJ (2013). Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R (2015). Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3’UTRs in vivo. Cell 162, 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin E, Desai A, Poser I, Toyoda Y, Andree C, Moebius C, Bickle M, Conradt B, Piekny A, Oegema K (2013). A conserved RhoGAP limits M phase contractility and coordinates with microtubule asters to confine RhoA during cytokinesis. Dev Cell 26, 496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.